Reference methylomes provide insights into the evolutionary role of DNA methylation in paleopolyploid genomes.
Abstract
Soybean (Glycine max) and common bean (Phaseolus vulgaris) share a paleopolyploidy (whole-genome duplication [WGD]) event, approximately 56.5 million years ago, followed by a genus Glycine-specific polyploidy, approximately 10 million years ago. Cytosine methylation is an epigenetic mark that plays an important role in the regulation of genes and transposable elements (TEs); however, the role of DNA methylation in the fate/evolution of genes following polyploidy and speciation has not been fully explored. Whole-genome bisulfite sequencing was used to produce nucleotide resolution methylomes for soybean and common bean. We found that, in soybean, CG body-methylated genes were abundant in WGD genes, which were, on average, more highly expressed than single-copy genes and had slower evolutionary rates than unmethylated genes, suggesting that WGD genes evolve more slowly than single-copy genes. CG body-methylated genes were also enriched in shared single-copy genes (single copy in both species) that may be responsible for the broad and high expression patterns of this class of genes. In addition, diverged methylation patterns in non-CG contexts between paralogs were due mostly to TEs in or near genes, suggesting a role for TEs and non-CG methylation in regulating gene expression post polyploidy. Reference methylomes for both soybean and common bean were constructed, providing resources for investigating epigenetic variation in legume crops. Also, the analysis of methylation patterns of duplicated and single-copy genes has provided insights into the functional consequences of polyploidy and epigenetic regulation in plant genomes.
In plant genomes, cytosine methylation is a meiotically and mitotically heritable, but reversible, DNA modification affecting chromatin structure and transcription without changing the DNA sequence. DNA methylation in plants exists in three sequence contexts, CG, CHG, and CHH (where H = A, C, or T), and is regulated by context-specific DNA methyltransferases and the RNA-directed DNA methylation (RdDM) pathway guided by 24-nucleotide small interfering RNAs (Law and Jacobsen, 2010; Stroud et al., 2013). The role and patterns of DNA methylation differ depending on the genomic features being targeted. Methylation of repetitive sequences, such as transposable elements (TEs), generally occurs in all three contexts, suppressing the transcription and proliferation of TEs to prevent the deleterious effects of TE insertions (Lisch, 2009). On the other hand, methylation of genes occurs primarily at CG sites in the transcribed region, referred to as CG gene-body methylation, and is associated with gene transcription (Zilberman et al., 2007; Coleman-Derr and Zilberman, 2012).
All angiosperms are paleopolyploids, having undergone at least two rounds of polyploidy in their past (Bowers et al., 2003; Paterson et al., 2010; Jiao et al., 2011; Vanneste et al., 2014). Polyploidization (or whole-genome duplication [WGD]) increases the genome size as well as the number of genes and is often followed by diploidization, or fractionation, thereby returning to a diploid state (Wolfe, 2001; Freeling, 2009). Although a majority of duplicated genes are lost through nonfunctionalization or pseudogenization (Lynch and Conery, 2000), many can be retained through balancing gene dosage (Papp et al., 2003; Birchler et al., 2005; Innan and Kondrashov, 2010) and/or functional divergence (Ohno, 1970; Force et al., 1999; Zhang and Cohn, 2008). While biased retention of duplicated genes can occur (Seoighe and Gehring, 2004; Freeling, 2009; Jiang et al., 2013), generally one of the paralogs is selectively lost, resulting in single-copy genes due to both neutral and selective forces. An interesting subset is shared single-copy genes, those that are repeatedly restored to singleton status after independent WGDs between lineages (Paterson et al., 2006; Duarte et al., 2010; De Smet et al., 2013). Both duplicated (paralogous) and single-copy genes are found in plant genomes, yet few studies have directly compared the differences in DNA methylation patterns and potential epigenetic regulation. Many previous studies have focused on immediate postpolyploidy changes in methylation by comparing natural and/or artificial polyploids with their parental diploid species (Madlung and Wendel, 2013). However, Thomas et al. (2006), by analysis of homologous regions in Arabidopsis (Arabidopsis thaliana), proposed that differential epigenetic marking of homologs might contribute to fractionation bias. Moreover, a recent study in soybean (Glycine max) revealed differential targeting of non-CG methylation between paralogs, indicating a possible role in the regulation of gene expression (Schmitz et al., 2013a). More comprehensive analyses are needed to clarify the underlying elements responsible for differential methylation among duplicate and single-copy genes and to determine how epigenetic repatterning after polyploidization, and following diploidization, influences the evolutionary fate of duplicated genes.
Soybean and common bean (Phaseolus vulgaris) are the most important legume sources of protein for human nutrition. These two species share a WGD event approximately 56.5 million years ago (MYA), prior to their divergence approximately 19.2 MYA (Lavin et al., 2005). After speciation, soybean underwent another lineage-specific WGD event approximately 10 MYA (Schmutz et al., 2010), resulting in a distinctive chromosome number for the genus Glycine (mostly 2n = 40) as compared with other members of the tribe Phaseoleae (mostly 2n = 22; Hadley and Hymowitz, 1973; Lackey, 1980). Due to a relatively recent WGD and lack of immediate diploidization (Kim et al., 2009), the soybean genome remains duplicated, with nearly 75% of the genes present in multiple copies (Schmutz et al., 2010), more than most diploid plant genomes (De Smet et al., 2013). Thus, the soybean genome provides a model to study the evolutionary aspects of epigenetic marks on retained duplicate and single-copy genes in comparison with related species, such as common bean, that lack the recent WGD event. Comparisons of DNA methylation between related species should help to elucidate the evolutionary role and impact of DNA methylation during and after polyploidy.
Here, we produced single-nucleotide resolution methylomes for both soybean and common bean using whole-genome bisulfite sequencing. Comparing the levels and distribution of DNA methylation at the whole-genome scale allowed us to gain a better understanding of the epigenetic landscape of these paleopolyploid genomes as well as insights into the association between methylation and gene expression. Of note, we found that diverged methylation patterns in non-CG contexts were due mostly to TEs in or near genes, suggesting a role for TE insertions and non-CG methylation in regulating gene expression post polyploidy.
RESULTS
DNA Methylomes of Soybean and Common Bean
Genome-wide DNA methylation profiling at the single-nucleotide resolution using whole-genome bisulfite sequencing was done for soybean (cv Williams 82) and common bean (cv G19833), both sources of reference genomes. With two biological replicates in both species, three different tissues (leaves, root hairs, and stripped roots) were used for soybean and leaf tissue for common bean. In total, eight whole-genome bisulfite sequencing (MethylC-seq) libraries were sequenced, resulting in approximately 141 to 348 million quality-filtered 101-bp paired-end reads per library that mapped uniquely to the reference genomes (Supplemental Table S1). Methylated cytosines were determined using a binomial test as described previously (Lister et al., 2009), with conversion efficiency of the sodium bisulfite reaction ranging from 99.35% to 99.83%, as estimated from the fraction of sequenced cytosines from spiked-in unmethylated λ-DNA.
Methylation levels in the three contexts (CG, CHG, and CHH) were compared between biological replicates and tissues, considering only cytosine sites supported by more than 20 sequence reads (Supplemental Figs. S1 and S2). Methylation levels of cytosines between biological replicates at symmetric sites (CG and CHG) were strongly correlated (r = 0.962–0.992), whereas those at asymmetric CHH sites were less correlated (r = 0.786–0.794), indicating that asymmetric sites were less stable than CG and CHG. However, both symmetric (r = 0.99–0.997) and asymmetric (r = 0.912–0.978) sites showed stronger correlation when the methylation levels of genes (not single-base positions) were compared (Supplemental Fig. S3). This suggests that methylation levels of individual cytosine sites can be variable but the sum of sites within a gene were maintained at constant levels between replicates. Between tissues, using merged replicates with 20- to 60-fold strand coverage (Supplemental Table S1), similar patterns were observed (Supplemental Fig. S2), with strong correlations at symmetric sites (r = 0.961–0.995) and lower, yet still correlated, levels at asymmetric CHH sites (r = 0.696–0.826).
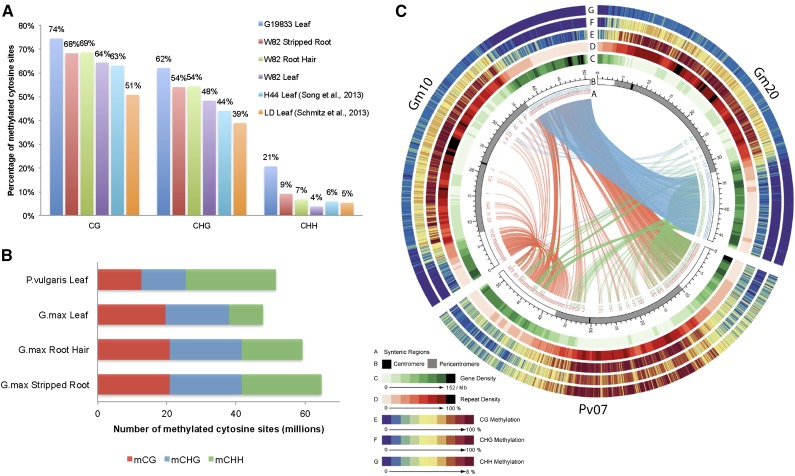
The overall percentage of methylated cytosines was compared between tissues and species (Fig. 1A) and revealed differences between soybean tissues, where root had more methylated cytosines (68%–69% CG, 54% CHG, and 7%–9% CHH) than leaf (64% CG, 48% CHG, and 4% CHH), varying from a previous analysis where leaf had more methylated cytosines than root (Song et al., 2013). However, differences with the previous studies (Schmitz et al., 2013a; Song et al., 2013) are likely due to the lack of biological replication, lower read depth, and coverage of cytosines as compared with this study (97%–99% versus 70%–80%) and possible genotype-specific methylation differences (cv Williams 82 versus cv Heinong 44 and cv LD00-2817P). Interestingly, common bean leaf had more methylated cytosine sites (74% CG, 62% CHG, and 21% CHH) than any soybean tissue. All these values are higher than methylation levels reported for Arabidopsis and rice (Oryza sativa; Greaves et al., 2012; Li et al., 2012). The soybean methylomes contain almost equal numbers of methylated CG (20,392,250–21,206,724) and methylated CHG (19,621,254–21,331,475) sites while differing in the number of methylated CHH sites (stripped root, 23,173,937; root hair, 17,461,816; leaf, 9,608,780; Fig. 1B). The common bean methylome had similar numbers of methylated CG (12,601,058) and methylated CHG (13,086,897) sites, much less than methylated CHH sites (26,019,193; Fig. 1B).
Figure 1.
Comparison between soybean and common bean methylomes. A, Percentage of methylated cytosine sites between soybean (cv Williams 82 [W82], cv Heinong 44 [H44], and cv LD00-2817P [LD]; this and previous studies) and common bean (cv G19833). B, Comparison of the number of methylated cytosine sites between soybean and common bean. C, Circos plot of two soybean chromosomes (Gm10 and Gm20) and one common bean chromosome (Pv07).
The chromosome-wide distribution of CG, CHG, and CHH methylation was highly correlated throughout the chromosomes and enriched in transposon-rich and gene-poor pericentromeric regions in both soybean and common bean (Fig. 1C; Supplemental Figs. S4 and S5; Supplemental Table S2). These results are consistent with distributions in Arabidopsis and rice (Cokus et al., 2008; Lister et al., 2008; Li et al., 2012) but in contrast with maize (Zea mays) and Brachypodium distachyon, where CHH methylation did not correlate with either CG or CHG methylation (Gent et al., 2013; Takuno and Gaut, 2013). However, CHG and CHH methylation levels were higher in common bean than in soybean, while those in the CG context were similar (Fig. 1C). In addition to methylome sequencing, immunodetection of 5-methylcytosine on meiotic pachytene chromosomes in both species showed that bright 5-methylcytosine signals coincided with heterochromatic regions at pericentromeres (Supplemental Fig. S6), consistent with the sequence-based analysis (Fig. 1C; Supplemental Figs. S4 and S5). In common bean, bright 5-methylcytosine signals were also detected at heterochromatic knobs on chromosomal termini (Supplemental Fig. S6, arrows). These knobs have been reported to consist of a satellite repeat, khipu (David et al., 2009), which we found to be highly methylated throughout the genome (Supplemental Table S3).
CG gene-body methylation, which is found in both plants and animals (Feng et al., 2010; Zemach et al., 2010), and hypermethylation of TEs in all three sequence contexts were also found in soybean and common bean (Supplemental Fig. S7). In soybean, CG gene-body methylation was similar between tissues but higher than in common bean. Both soybean and common bean had higher methylated CG levels than either Arabidopsis or Populus trichocarpa but lower than rice (Feng et al., 2010). Higher levels of methylation were found in flanking regions of common bean genes as compared with soybean, consistent with the overall higher methylation in common bean (Fig. 1A). CHG and CHH methylation levels of TEs in soybean leaf were lower than in soybean roots and common bean leaf, whereas CG methylation levels were similar (Supplemental Fig. S7). Similar trends in methylation levels were found chromosome wide (Fig. 1C).
Significantly Overexpressed Paralogs Are More CG Gene-Body Methylated
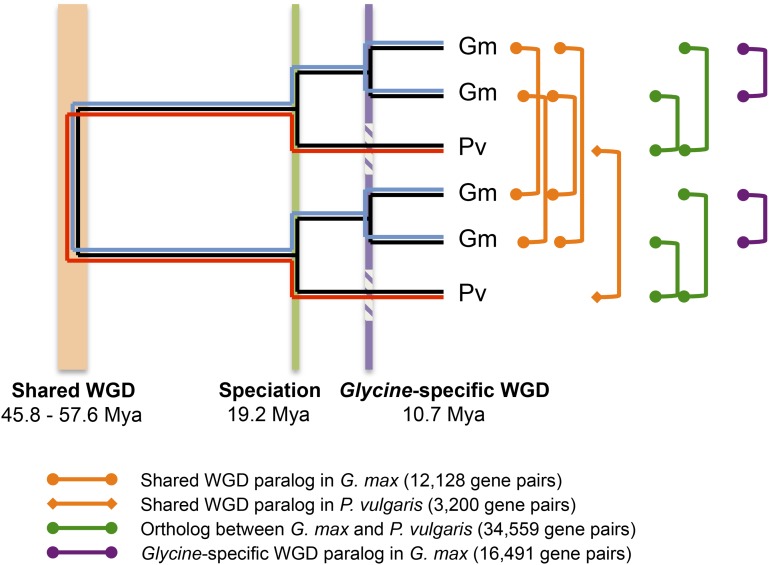
Soybean and common bean share a WGD (polyploidy) event approximately 56.5 MYA, followed by a WGD specific to the genus Glycine (Schmutz et al., 2010; Fig. 2). Due to the independent WGD and the relatively slow process of diploidization in soybean (Kim et al., 2009), nearly three-quarters (74.9%) of its genes are present in more than one copy (Table I). This is in contrast to common bean, where 42.5% of the genes are duplicated. Thus, common bean has nearly 3 times more single-copy genes (24.1%) than soybean (9.6%).
Figure 2.
Estimation of WGD and speciation events in soybean and common bean (adapted from Schmutz et al., 2014) and the paralogous and orthologous gene pairs used in this study.
Table I. Soybean and common bean genes by type.
–, Not applicable.
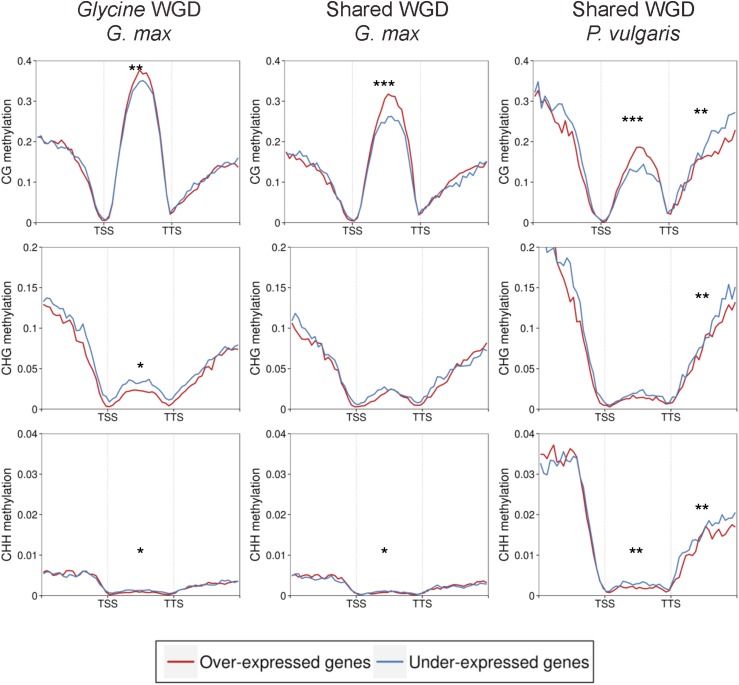
After a polyploidy event, the functional divergence of duplicated genes can result in subfunctionalization, neofunctionalization, or nonfunctionalization (Lynch and Conery, 2000). Our previous study indicated that nearly half of paralogs in soybean were differentially expressed and thus had undergone expression subfunctionalization/neofunctionalization (Roulin et al., 2013). In plants, it is known that CG gene-body methylation is positively correlated and CHG and CHH gene-body methylation is negatively associated with gene expression (Schmitz et al., 2013a; Seymour et al., 2014). In this study, similar associations between methylation and expression were also observed in soybean and common bean (Supplemental Fig. S8). To estimate the impact of methylation on the differential expression of duplicated genes, paralogs from the two WGDs with significant differences in expression were identified (Supplemental Table S4) and methylcytosine densities between overexpressed and underexpressed genes within a paralogous gene pair were compared (Fig. 3; Supplemental Fig. S9). Among 12,128 and 3,200 paralogous gene pairs derived from the shared WGD event in soybean and common bean, respectively, 45% to 51% and 71% were differentially expressed and 40% to 42% (6,868–7,157/16,941) were differentially expressed among the genus Glycine-specific WGD gene pairs in soybean. Within the gene body, significantly overexpressed genes showed higher levels of CG methylation and lower levels of non-CG methylation than the underexpressed paralogs, similar to the methylation and expression patterns for all genes (Supplemental Fig. S8). This was true for both the shared and the genus Glycine-specific WGDs regardless of species or tissue, except for CHG methylation in all tissues and CHH methylation in soybean root hair for shared WGD-derived gene pairs. No significant differences in methylation between overexpressed and underexpressed gene pairs were found in 5′ upstream regions, while some differences were found in 3′ downstream regions (Fig. 3).
Figure 3.
Average distribution of DNA methylation of overexpressed and underexpressed genes within paralogous gene pairs. Flanking regions are 1 kb. TSS, Transcription start site; TTS, transcription termination site. The methylation level of each cytosine context was compared using the Wilcoxon rank-sum test: ***, P < 0.001; **, P < 0.01; and *, P < 0.05.
Methylation and Expression Patterns of Duplicated and Single-Copy Genes
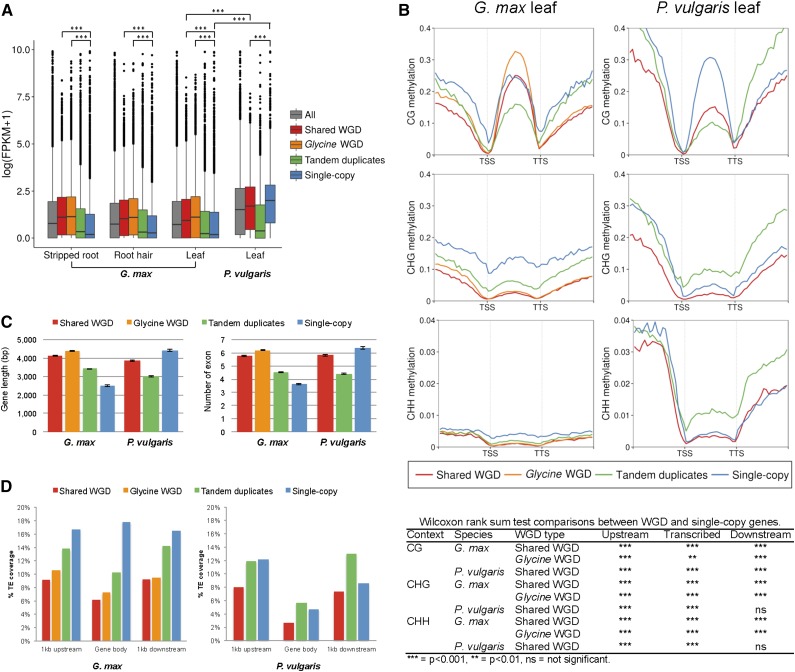
Soybean and common bean are close relatives that diverged approximately 19.2 MYA (Lavin et al., 2005) but differ in genome structure due to a genus Glycine-specific WGD and subsequent diploidization (Schmutz et al., 2014). The distinct evolutionary histories and genome structures between the two species may have contributed to differences in gene expression levels. To test this hypothesis, whole-transcriptome data of soybean and common bean were quantified and compared (Fig. 4A). In soybean, WGD genes were, on average, more highly expressed (P < 2.2 × 10−16; Wilcoxon rank-sum test, a permutation test that uses the sum of the ranks as a test statistic with random sampling from each of the two populations) than single-copy genes or tandemly duplicated genes, regardless of tissue. In common bean leaf, WGD genes were also more highly expressed than tandemly duplicated genes, but common bean leaf contrasted with soybean in that single-copy genes were more highly expressed. Interestingly, only 17.4% of soybean single-copy genes had orthologs in common bean, whereas 76.3% of single-copy genes in common bean had orthologs in soybean (Supplemental Table S5). Thus, the elevated expression of single-copy genes in common bean as compared with soybean may be due to soybean-specific genes not found in common bean, likely due to fractionation after species divergence.
Figure 4.
Comparison between different gene categories: duplicates from shared WGD, genus Glycine-specific WGD, and tandem and single-copy genes. A, Box plot of gene expression levels in common bean leaf tissue and three soybean tissues. Expression level was compared using the Wilcoxon rank-sum test: ***, P < 0.001. FPKM, Fragments per kilobase of exon per million fragments mapped. B, Graphs at top show the average distribution of DNA methylation in each cytosine context. Flanking regions are 1 kb. TSS, Transcription start site; TTS, transcription termination site. The table at bottom shows a pairwise comparison of methylation levels between WGD genes and single-copy genes using the Wilcoxon rank-sum test: ***, P < 0.001; **, P < 0.01; and ns, not significant. C, Comparison of gene length and exon number between gene categories in soybean and common bean. D, Percentage total TE in genic regions in gene categories in soybean and common bean.
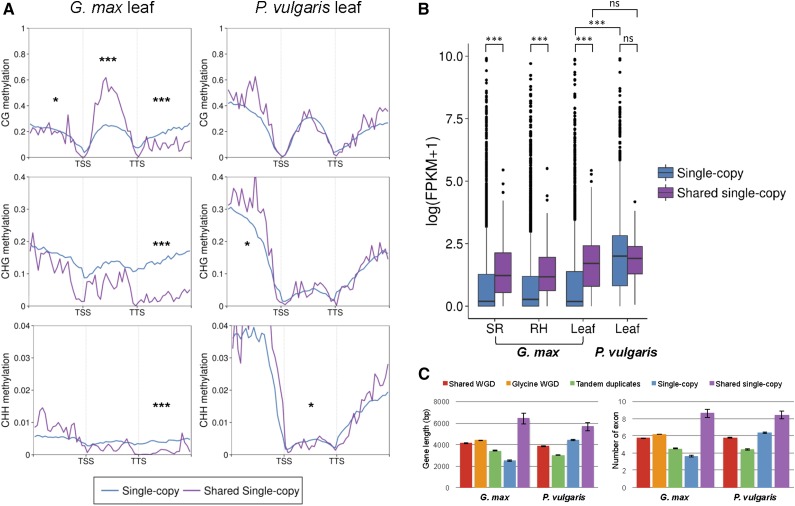
To better understand the role that DNA methylation played in genome evolution in soybean and common bean, we compared the methylation and expression profiles of single-copy and duplicated genes (Fig. 4, A and B; Supplemental Fig. S10). For comparison of CG gene-body methylation, genes that were also significantly enriched for either CHG or CHH methylation (PCHG < 0.05 or PCHH < 0.05) were considered heterochromatic marks or potential targets of RdDM pathways (hereafter referred to as C-methylated genes; Supplemental Table S6) and therefore excluded. In soybean, CG gene-body methylation was highest in WGD genes and lowest in tandemly duplicated genes, similar to gene expression levels. These results contrast with CHG and CHH gene-body methylation, where the highest levels were in single-copy genes, which had the lowest average expression levels. In common bean, a similar trend between CG gene-body methylation and gene expression patterns was observed, except that tandemly duplicated genes had high levels of non-CG methylation and low levels of gene expression. Therefore, as expected, within the gene body, CG methylation was positively associated with gene expression and non-CG methylation was negatively associated with gene expression.
The methylation levels of regions flanking the genes were also analyzed (Fig. 4B; Supplemental Fig. S10). In both species, the methylation trends in all three contexts in 1-kb flanking regions were identical to levels of non-CG methylation within gene-body regions. The exception was 5′ upstream regions of single-copy genes in common bean, where we observed highest levels of methylation in all three contexts.
CG Body-Methylated Genes
To further investigate the role of CG gene-body methylation among various types of genes, CG body-methylated genes were identified using a probabilistic approach (Takuno and Gaut, 2012). After removing C-methylated genes (PCHG < 0.05 or PCHH < 0.05), the probability distribution of CG methylation (PCG) was bimodal for both soybean and common bean (Supplemental Fig. S11), consistent with distributions in other plant species (Takuno and Gaut, 2012, 2013; Wang et al., 2013). In total, 10,007 (18.6% of total genes), 9,456 (17.5%), 9,445 (17.5%), and 2,992 (11%) body-methylated genes (PCG < 0.05) were identified in soybean leaf, root hair, stripped root, and common bean leaf, respectively (Supplemental Table S6). In both species, tandemly duplicated genes (10.1%–10.5% in soybean and 4.5% in common bean) were underrepresented as body-methylated genes. In soybean, WGD genes (17.1%–22.8%) were more likely to be body methylated than either tandemly duplicated genes (10.1%–10.5%) or single-copy genes (11.8%–12.4%). In common bean, in contrast, single-copy genes (18.2%) were more likely to be body methylated than duplicated genes (4.5%–9.5%), similar to CG gene-body methylation (Fig. 4B). The average CG methylation of body-methylated genes was similar between different gene categories (e.g. duplicated versus single copy; Supplemental Fig. S12), even though the absolute number of body-methylated genes varied by gene category.
CG body-methylated genes have been reported to be longer and to contain more exons than unmethylated genes (Takuno and Gaut, 2012, 2013). To test this hypothesis in soybean and common bean, the length of genes and the number of exons were calculated and compared (Fig. 4C). In soybean, duplicated genes generated by the genus Glycine-specific WGD, which were highly expressed and more CG body methylated, were longer (4,379 bp) and contained more exons (6.18 exons per gene) than other types of genes. Single-copy genes, for example, which were not highly expressed, had less CG body methylation and were the shortest (2,499.6 bp) and contained the fewest exons (3.63 exons per a gene). In common bean, however, single-copy genes were longest (4,427 bp), contained the most exons (6.36 exons per gene), and had the most CG body methylation. Therefore, gene length and number of exons are positively associated with gene expression and CG gene-body methylation in both species and may be a generally consistent phenomenon across plants.
CG body-methylated genes have been reported to evolve more slowly than unmethylated genes (Takuno and Gaut, 2012, 2013), even though increased mutation rates would be expected in CG body-methylated genes due to deamination of 5-methylcytosine leading to C→T transitions (Bird, 1980). Nonsynonymous (Ka) and synonymous (Ks) substitution rates were calculated for orthologs (Supplemental Table S7). In both species, the average Ka and Ks values were significantly lower (P < 2.2 × 10−16; Wilcoxon rank-sum test) in body-methylated genes than unmethylated genes, indicating lower evolutionary rates for body-methylated genes. In soybean, WGD-derived genes, which contained the highest portion of body-methylated genes, had the lowest Ka and Ks values. Similarly in common bean, single-copy genes had the highest portion of body-methylated genes while also having the lowest Ks values.
Transposons and Non-CG Methylation
An insertion of a TE in or near a gene may affect the methylation of the gene as well as its expression. The positions of TEs in the genome were defined using RepeatMasker with soybean (Du et al., 2010a) and common bean (Schmutz et al., 2014) TE databases. The percentage coverage of a gene and its flanking regions by TEs was calculated (Fig. 4D). In both species, duplicated genes generated by WGD had less TE coverage in both the gene body and flanking regions. In soybean, single-copy genes, which had the lowest expression levels (Fig. 4A), had more TE coverage as compared with duplicated genes, following non-CG, especially CHG, methylation levels (Fig. 4B). A similar trend between percentage TE coverage and non-CG methylation was also found in and/or near common bean genes (Fig. 4B), showing a positive correlation between the presence of TEs and non-CG methylation levels. Since TEs of different lengths have been shown to be methylated by different pathways (Zemach et al., 2013; Stroud et al., 2014), we compared long (greater than 4 kb) and short (less than 500 bp) TEs, and although the percentage coverage by TE category varied (more short than long), non-CG methylation trends were the same for both long and short TEs (Supplemental Fig. S13).
Methylation Patterns between Paralogous and Orthologous Genes
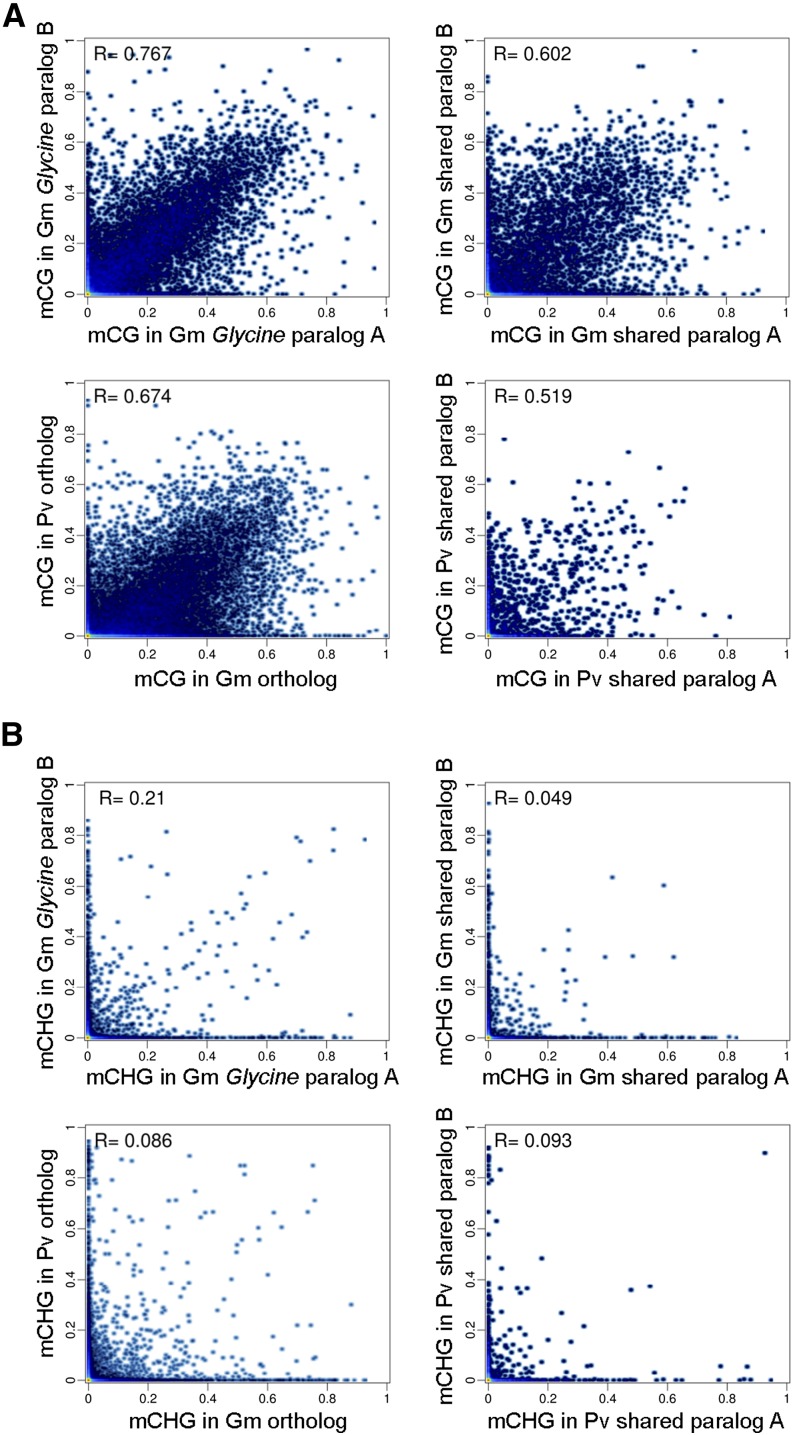
To gain insight into evolutionary changes in DNA methylation patterns after polyploidy and speciation, the methylation status of paralogous and orthologous genes from both species were compared (Fig. 5; Supplemental Fig. S14). A total of 34,559 orthologous gene pairs were identified between soybean and common bean, and 12,128, 16,491, and 3,200 paralogous gene pairs were determined as shared WGD pairs in soybean, genus Glycine-specific WGD pairs in soybean, and shared WGD pairs in common bean, respectively (Fig. 2). For a clear comparison of CG gene-body methylation, gene pairs that contain C-methylated genes (PCHG < 0.05 or PCHH < 0.05) were excluded. The scatterplot revealed that levels of CG methylation were highly correlated between orthologs (r = 0.674) as well as paralogs (r = 0.519–0.767; Fig. 5A; Supplemental Table S8), consistent with previous reports (Schmitz et al., 2013a; Takuno and Gaut, 2013). CG methylation was more conserved (P = 2.4 × 10−139 and P = 2.5 × 10−96, Fisher’s z transformation) between genus Glycine-specific WGD pairs (r = 0.767) and shared WGD pairs (r = 0.602 and 0.519), showing fewer changes in methylation between recently duplicated gene pairs. A shift in distribution was observed between soybean and common bean orthologs (Fig. 5A), where soybean had higher levels of CG gene-body methylation (also seen in Supplemental Fig. S7).
Figure 5.
Pairwise comparisons of methylation levels between paralogous and orthologous gene pairs in CG (A) and CHG (B) contexts. R = Pearson correlation coefficient.
In contrast, non-CG methylation was not well correlated between orthologs (r = 0.035–0.086) or paralogs (r = 0.001–0.210; Fig. 5B; Supplemental Fig. S14; Supplemental Table S8). Genes that were enriched in non-CG methylation (PCHG < 0.05 or PCHH < 0.05) were, on average, physically closer to TEs than unmethylated genes (PCG > 0.95; Supplemental Fig. S15), consistent with our results showing a positive correlation between the presence of TEs and non-CG methylation levels. Therefore, diverged patterns of non-CG methylation are most likely due to the presence of TEs in or near genes.
Most genes, 34,205 (63.4% of total) in soybean and 19,054 (70.4% of total) in common bean, were unmethylated (Supplemental Table S6), and as a result, 59% of the orthologous and 60% to 69% of paralogous gene pairs were unmethylated (PCG > 0.95; Supplemental Table S9). This indicates that unmethylated genes were likely maintained as unmethylated following polyploidy and/or speciation. Moreover, only a small portion (0.6%–2.1%) of the gene pairs showed significant enrichment in non-CG methylation for both gene copies, while there was a higher proportion (3.8%–15.7%) where both genes were body methylated. This indicates a higher level of conservation of CG methylation than non-CG methylation.
Between the two species, 3,256 orthologous gene pairs were identified with gene-body methylation that was either conserved following speciation or occurred independently in both lineages (Supplemental Table S9). Gene Ontology (GO) analysis using AgriGO (Du et al., 2010b) revealed that these conserved body-methylated genes were significantly enriched for several categories, such as binding (P = 1.1 × 10−43), helicase activity (P = 4.8 × 10−28), membrane coat (P = 2.8 × 10−13), coated membrane (P = 2.8 × 10−13), and cellular localization (P = 4.7 × 10−15; Supplemental Figs. S16–S18), similar to a previous report (Takuno and Gaut, 2013). A total of 419 orthologous gene pairs were identified as conserved C-methylated genes between the two species with no significantly enriched GO categories.
CG body-methylated genes are generally functionally important (Takuno and Gaut, 2012, 2013) and conserved between different species (Fig. 5; Supplemental Figs. S16–S18); however, some genes have lost or changed methylation states (e.g. from body methylated to unmethylated). Among 26,273 soybean genes that had unmethylated orthologs in common bean, 3,648 (13.9%) were body methylated, while in common bean, only 557 of 14,951 (3.7%) genes that had unmethylated orthologs in soybean were body methylated. Similarly, 724 of 4,575 (15.8%) soybean genes that had body-methylated orthologs in common bean were unmethylated, while 2,758 of 5,397 (51.1%) common bean genes that had body-methylated orthologs in soybean were unmethylated. This indicates that either more unmethylated genes became body methylated in soybean or, the converse, that more body-methylated genes became unmethylated in common bean. This is consistent with our results that soybean genes generally have higher levels of CG gene-body methylation than common bean (Fig. 5; Supplemental Fig. S7). Moreover, body-methylated genes in soybean with unmethylated orthologs in common bean were significantly enriched (P < 2.2 × 10−16; two-sample test of proportion using the prop.test function in R) for genus Glycine-specific WGD genes (3,243/3,648) as compared with the total body-methylated gene set (7,695/10,007).
Methylation and Expression Patterns of Shared Single-Copy Genes
Although most angiosperms have undergone WGD events, a subset of genes, even across species, return to single-copy status (i.e. one of the duplicate copies is lost). For example, 959 genes were identified as shared single-copy genes between Arabidopsis, P. trichocarpa, Vitis vinifera, and rice (Duarte et al., 2010), even though these species have experienced independent species-specific WGD events. To investigate methylation patterns of shared single-copy genes between soybean and common bean, soybean/common bean genes orthologous to the 959 genes that represent shared single-copy genes for angiosperms (Duarte et al., 2010) were identified (Supplemental Table S10). Among the 959 genes, 952 and 948 genes had homology in soybean and common bean, respectively. In common bean, 809 of 948 genes were single copy, while in soybean, only 196 of 952 genes were single copy, the difference due to the more recent genus Glycine-specific WGD event. A total of 144 genes were identified as shared single-copy genes between the six species, including soybean and common bean. Methylation and expression levels of the 144 shared single-copy genes were compared with other single-copy genes in each species (Fig. 6; Supplemental Fig. S19).
Figure 6.
Comparison of shared single-copy and species-specific single-copy genes. A, Average distribution of DNA methylation in each cytosine context. Flanking regions are 1 kb. TSS, Transcription start site; TTS, transcription termination site. The methylation level of each cytosine context was compared using the Wilcoxon rank-sum test: ***, P < 0.001; and *, P < 0.05. B, Box plot of gene expression levels in soybean (leaf, stripped root [SR], and root hair [RH]) and common bean leaf tissue. Expression level was compared using the Wilcoxon rank-sum test: ***, P < 0.001; and ns, not significant. FPKM, Fragments per kilobase of exon per million fragments mapped. C, Comparison of gene length and exon number between species-specific single-copy and shared single-copy genes from soybean and common bean.
In common bean, no significant differences (Wilcoxon rank-sum test) in expression and methylation were found between single-copy genes (n = 6,525) and shared single-copy genes (n = 144). In soybean, however, shared single-copy genes (n = 144) were more highly expressed (P = 3.1 × 10−15, P = 7.4 × 10−16, and P = 2.2 × 10−16; Wilcoxon rank-sum test) and had higher levels of CG body methylation (P = 8.5 × 10−5, P = 5.2 × 10−5, and P = 3.9 × 10−5; Wilcoxon rank-sum test) than did single-copy genes (n = 5,179), regardless of tissue. This was consistent with our results showing that a higher percentage of shared single-copy genes in soybean (41.7%; 60/144) were CG body-methylated genes (PCG < 0.05) than single-copy genes (12.4%; 644/5,179), while a similar portion of common bean genes (18.2% for single-copy genes and 17.4% for shared single-copy genes) were CG body methylated (Supplemental Table S6). Moreover, lower evolutionary rates (Ka and Ks) and higher gene length and exon numbers were found in shared single-copy genes for both species than other single-copy genes (Fig. 6C; Supplemental Table S7), also characteristics of CG body-methylated genes. Among the 144 shared single-copy genes, the soybean copies were more CG body-methylated genes and had fewer unmethylated genes than common bean, consistent with overall higher levels of CG gene body methylation in soybean (Fig. 5; Supplemental Fig. S7). Interestingly, non-CG methylation was enriched in 5′ upstream regions of shared single-copy genes in both species, but not in 3′ regions, corresponding to higher TE coverage in 5′ flanking regions (12.6% in both soybean and common bean) than in 3′ flanking regions (7.4% in soybean and 9.7% in common bean).
DISCUSSION
With the availability of reference genome sequences and advances in high-throughput sequencing technologies, recent studies have uncovered genome-wide patterns of cytosine methylation for several plant species, such as Arabidopsis, B. distachyon, rice, maize, tomato (Solanum lycopersicum), and wild cabbage (Brassica oleracea; Lister et al., 2008; Chodavarapu et al., 2012; Eichten et al., 2013; Takuno and Gaut, 2013; Zhong et al., 2013; Parkin et al., 2014). There have been two reports on methylation patterns in soybean (Schmitz et al., 2013a; Song et al., 2013), but with limited coverage (70%–80% of cytosines) and not for the reference genome, cv Williams 82. Here, we produced high-coverage methylomes for the reference genomes of both soybean and common bean, covering nearly every cytosine (97%–99%) in their respective genomes. Also, we focused on the contribution of methylation to the fate/evolution of genes following speciation and polyploidy.
TEs Shape Plant Methylomes
A large fraction of most plant genomes is composed of TEs, roughly proportional to genome size variation (Tenaillon et al., 2010; Vitte et al., 2014). Comparisons of the DNA methylation content of several plant species with different TE percentages have shown that the global methylation level clearly follows the TE density of a genome (Mirouze and Vitte, 2014). For example, a genome with a large portion of TEs, such as maize, is more methylated overall than Arabidopsis, which has a lower TE density. The association between TE density and global methylation was seen in this study, as overall methylation levels of soybean and common bean were higher than Arabidopsis and rice, with lower TE densities, and lower than maize and tomato, with higher TE densities (Fig. 1). However, global methylation levels as well as TE methylation levels in common bean were higher than in soybean (Figs. 1 and 2), even though the common bean genome has a lower percentage of TEs than soybean (Schmutz et al., 2014). This might be due to the recent burst of TEs in common bean (Schmutz et al., 2014), as the age of TEs was found to negatively correlate with DNA methylation levels (Vonholdt et al., 2012). Moreover, differences in methylation levels were largely found in non-CG contexts (Figs. 1 and 2), suggesting that RdDM may be more active in the common bean genome, again likely due to the recent TE activity.
CG Body Methylation Was High in Transcriptionally Active Single-Copy Genes
TEs and other heterochromatic sequences are methylated in all three contexts, whereas gene body methylation occurs primarily at CG sites (Diez et al., 2014). Unlike Arabidopsis, numerous genes in both soybean and common bean were found to contain densely methylated TE insertions that could complicate analysis. Therefore, genes enriched also in non-CG methylation were considered to be RdDM target loci or heterochromatic marks and were excluded from the analysis of CG gene-body methylation. Following this, we found a positive correlation between gene expression and CG gene-body methylation and a strong correlation between percentage TE coverage and non-CG methylation in or near a gene, which correlated with repressed gene expression (Figs. 3 and 4; Supplemental Fig. S8).
Expression profiles of duplicated and single-copy genes differed between soybean and common bean, and gene-body DNA methylation was correlated (Fig. 4). As shown in Arabidopsis (Zilberman et al., 2007; Schmitz et al., 2013b), single-copy genes in common bean were more transcriptionally active and more CG gene-body methylated; in contrast, however, single-copy genes in soybean had lower transcription levels and increased DNA methylation in all three contexts due mostly to TEs. However, WGD-derived paralogous genes in soybean were more actively expressed and enriched in CG body methylation.
Although duplicated and single-copy genes in soybean and common bean showed dissimilar expression patterns, shared single-copy genes in both species were similar in that they were transcriptionally active and enriched for CG body methylation. In a previous study, shared single-copy genes were found to be involved in essential housekeeping functions, expressed more highly and broadly than species-specific single-copy genes, and had higher sequence conservation, suggesting selection pressure to retain such genes as singletons (De Smet et al., 2013). Therefore, it was suggested that shared single-copy genes in soybean are true single-copy genes, while remaining single-copy genes are likely derived from a duplicated gene pair that lost one copy and the remaining copies are often located in hypermethylated pericentromeric regions (Du et al., 2012).
Methylation May Have Contributed to Biased Retention of Genes after WGD
In soybean, it is possible that CG gene-body methylation may have played a role in biased retention of WGD-derived genes following the genus Glycine-specific WGD event. Previous research examined the relationships between gene features and retention probability following WGD in six plant genomes, including soybean (Jiang et al., 2013). They concluded that retained WGD genes have either low evolutionary rates (Ka) and high and broad expression patterns (type I), high structural complexity, including longer gene length, greater number of introns, and gene isoforms (type II), or high GC/GC3 content (type III). Since CG body-methylated genes are biased for lower evolutionary rates (Ka), longer lengths (higher exon number), and lower GC content (Takuno and Gaut, 2012, 2013), the conservation of CG gene-body methylation in WGD-derived genes may have contributed to the retention of type I and II genes that were selected for dosage balance and subfunctionalization, respectively.
In addition, WGD genes with high expression patterns had higher levels of CG gene-body methylation (Fig. 3) and, therefore, may be retained as duplicates, as high gene expression has long been shown to be a potential marker for the retention of gene duplicates (Seoighe and Wolfe, 1999; Aury et al., 2006; Yang and Gaut, 2011). In rice, for example, older WGD-generated duplicates (higher Ks) tended to have higher body methylation, suggesting that the long-term retention of WGD genes was correlated with increased body methylation (Wang et al., 2013). In addition, lower evolutionary rates within CG body-methylated genes could contribute to the slow diploidization process in soybean, as a greater portion of CG body-methylated genes were found in soybean than in common bean. Also, most (more than 90%) of the soybean CG body-methylated genes with unmethylated orthologs in common bean were derived from the genus Glycine-specific WGD event. Taken together, this indicates that soybean acquired more CG gene-body methylation prior to the genus Glycine-specific WGD event, leading to the retention of more duplicated genes and the slow diploidization/fractionation process.
The role of methylation in the regulation of alternative splicing (AS) needs further exploration, as the characteristics of AS genes (significantly longer introns, more exons, higher expression levels, and lower GC content than non-AS genes; Shen et al., 2014) are similar to those of CG body-methylated genes, from this study and previous reports (Takuno and Gaut, 2012, 2013). Since retained duplicated genes are more prone to AS (type II; Jiang et al., 2013), CG body methylation could play a role in retention if there is an explicit link with AS. Further analysis of paralogous or orthologous gene pairs that have simultaneously lost both AS and CG body methylation could help to elucidate the role of gene-body methylation in transcript splicing and its role in the evolution of duplicated genes.
Differential Methylation among Duplicated Genes Can Serve as a Backup System in Plants
We and others have observed differential non-CG methylation among paralogous and orthologous gene pairs (Schmitz et al., 2013a). The majority of protein-coding genes in both soybean and common bean were unmethylated, and most paralogs and orthologs were also unmethylated. In contrast, non-CG methylation was not correlated between paralogs and orthologs, but this can be explained by the increase of non-CG methylation due to TE insertions in or near genes, resulting in asymmetry between paralog and ortholog pairs. A TE can introduce gene-body hypermethylation not only by insertion but through spreading of methylation to adjacent regions, as shown in Arabidopsis and maize (Ahmed et al., 2011; Eichten et al., 2012). This repressive mark may epigenetically silence duplicate copies in a stress-inducible and/or tissue-specific manner, resulting in an increase in the rate of gene fixation and a decrease in pseudogenization under purifying selection (Rodin and Riggs, 2003; Branciamore et al., 2014). Therefore, epigenetic variation, including DNA methylation, may provide a buffering system for duplicated genes to avoid pseudogenization, thereby retaining a large number of WGD-derived genes, as found in soybean. Calculating the insertion times of TEs responsible for the hypermethylation of genes may help to determine whether these insertions were contemporaneous to the WGD events, perhaps due to genomic shock. Also, highly methylated genes could be a path to pseudogenization followed by gene loss, but they may also serve as backup copies that may become demethylated and, under certain conditions, transcriptionally active (Pecinka et al., 2013). Collectively, genetic and epigenetic variation triggered by TEs may play an important role in the functional divergence of duplicated genes in soybean and common bean.
CONCLUSION
In this study, we produced high-coverage methylomes for the reference genomes of both soybean and common bean, the most important legume sources of protein for human nutrition. These reference methylomes are valuable resources for the analysis of epigenetic variation within and between species and provide baseline information for further population analyses. We found evidence that DNA methylation likely plays an important role post- and pre-WGD in how duplicated genes are retained and expressed and in potential buffering. TEs play an important role in this process, either establishing the methylation of genes spreading or by direct insertion into a gene, often responsible for asymmetric methylation of paralogs and orthologs. These data provide insights into the functional consequences of polyploidy in plant genomes and specifically into the evolutionary trajectories of these sister species that diverged approximately 20 MYA.
MATERIALS AND METHODS
Plant Materials and DNA Extraction
Soybean (Glycine max ‘Williams 82’) and common bean (Phaseolus vulgaris ‘G19833’) plants were grown in a greenhouse at the University of Georgia, and 18-d-old trifoliate leaves were harvested. Soybean root tissues were isolated at the University of Oklahoma as described (Wan et al., 2005; Qiao and Libault, 2013). Briefly, seeds were surface sterilized and were sown on a 3:1 mixture of vermiculite and perlite. Four-day-old seedlings were transferred in the ultrasound aeroponic system treated with nitrogen-free B&D liquid medium (Broughton and Dilworth, 1971). Root hair cells and stripped roots were isolated from 7-d-old seedlings. For each soybean and common bean tissue, two independent biological replicates were performed. Tissues were frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen) following the manufacturer’s recommendations.
MethylC-seq Library Preparation
Genomic DNA (3 μg) was spiked with 15 ng of unmethylated cl857 Sam7 λ-DNA (Promega). The DNA was sheared to an average of 350 bp with the Covaris S2 device using the following parameters: duty = 10%, intensity = 4, cycles/burst = 200, and time = 80 s. Sheared DNA was size selected for 200 to 500 bp with Agencourt AMPure XP beads (Beckman Coulter) using the double solid-phase reversible immobilization method as described (Lennon et al., 2010) with the following modifications: 0.65× volume beads was added for the first reaction and 0.15× volume beads was added for the second reaction. Size-selected DNA was blunted with deoxyribonucleotide triphosphate mix (no dCTP) and T4 DNA polymerase (New England Biolabs) for 20 min at 12°C, then 5′ phosphorylated with T4 polynucleotide kinase (New England Biolabs) for 15 min at 37°C. End-repaired DNA was A tailed with dATP and Klenow fragment (3′ to 5′ exonuclease-minus; New England Biolabs) for 30 min at 37°C. Paired-end Illumina adapters containing 5′-methylcytosine were annealed prior to ligation. A-tailed DNA was ligated to preannealed adapters with DNA Quick Ligase (New England Biolabs) for 25 min at room temperature. Adapter-ligated DNA was bisulfite treated twice using the EpiTect Bisulfite Kit (Qiagen) following the manufacturer’s protocol for DNA isolated from formalin-fixed, paraffin-embedded tissue samples, then enriched by PCR with ExTaq DNA polymerase (Takara) using the following thermocycling conditions: 1 min at 98°C followed by 10 cycles of 30 s at 95°C and 3 min at 62°C. After each step of the library preparation, the reaction products were purified with 1.8× volume AMPure beads, except the ligation product, which was purified twice with 1× volume beads, and the PCR product, which was purified with 0.8× volume beads.
Sequencing and Methylation Analysis
MethylC-seq libraries were paired-end sequenced for 101 cycles using an Illumina HiSeq 2000 at HudsonAlpha Institute. Image analysis, base calling, and quality calibration were performed using the standard Illumina pipeline. Only high-quality reads were used for data analysis after filtering low-quality reads and reads containing primer/adaptor sequences using NGS QC Toolkit version 2.3 (Patel and Jain, 2012) with default parameters. Quality filtered reads were aligned to either cv Williams 82 genome version 1.1 (Schmutz et al., 2010) or cv G19833 genome version 1.0 (Schmutz et al., 2014) using Bismark version 0.7.7 (Krueger and Andrews, 2011), and only uniquely mapped reads were retained. Reads that mapped to the same position in the genome were consolidated into a single read to remove potential clonal bias from PCR amplification. Mean DNA methylation levels for each position in the sequence reads were calculated to identify biases in methylation levels introduced during the end-repairing step of library preparation. The first six bases of read 1 and the last seven bases of read 2 were found to contain incorrect methylation calls and were excluded from further analysis. Methylated cytosines were identified using the binomial distribution as described (Lister et al., 2009). The error rates, which are composed of bisulfite nonconversion and sequencing errors, were estimated from the percentage of cytosine bases sequenced at reference cytosine positions in the unmethylated λ-genome (Supplemental Table S1).
RNA Sequencing Analysis
The published RNA sequencing data of soybean (Libault et al., 2010) and common bean (Schmutz et al., 2014) were used to determine the abundance of transcripts. High-quality reads were obtained after filtering low-quality reads and reads containing primer/adaptor sequences using NGS QC Toolkit version 2.3 (Patel and Jain, 2012) with default parameters. Quality filtered reads were aligned to either cv Williams 82 version 1.1 (Schmutz et al., 2010) or cv G19833 version 1.0 (Schmutz et al., 2014) transcriptome using TopHat2 version 2.0.7 (Kim et al., 2013). Uniquely mapped reads were provided as input to Cufflinks version 2.0.2 for transcript assembly and quantification (Trapnell et al., 2010). Transcript abundance is given as fragments per kilobase of exon per million fragments mapped. For each tissue sample, differentially expressed genes (overexpressed and underexpressed genes) between paralogs were determined using the exact conditional test (Gu et al., 2008) as described (Roulin et al., 2013).
Identification of Paralogous and Orthologous Genes between Soybean and Common Bean
Orthologous genes between soybean and common bean and paralogous genes within both genomes were identified using synteny and Ks rates correlated to the two WGD events in soybean (Schmutz et al., 2014). The gene states and synteny block order were predicted with DAGchainer (Haas et al., 2004) based on peptides, corresponding coding sequence, and genome coordinates and requiring at least four homologous genes to construct a synteny block. The Ks for gene pairs was calculated, and the mean values per synteny block were used to infer the time of duplication. Tandem duplicated genes were identified through BLAST between whole chromosomes for each genome in windows of 100 kb (Schmutz et al., 2014). Single-copy genes were detected by self-BLASTP. Any gene with BLAST hits other than itself with greater than 30% identity and 70% coverage was considered a multiple-copy gene and excluded from the single-copy genes.
Analysis of TEs
TE sequence libraries of soybean and common bean were downloaded from SoyBase (http://soybase.org/soytedb/) and Phytozome (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Pvulgaris/related_files/pvTEdatabase-V3.txt), respectively. In order to reduce the redundancy of highly repeated TE families, the UCLUST algorithm, which is part of the USEARCH software (http://www.drive5.com/usearch/manual/uclust_algo.html), was used. The nonredundant TE libraries of soybean and common bean were used to mask their respective genomes using RepeatMasker (http://www.repeatmasker.org/) with default parameters.
Immunodetection of 5-Methylcytosine
Flower buds of soybean ‘Williams 82’ and common bean ‘G19833’ were used to obtain pachytene chromosome spreads. Flower buds were harvested and fixed in 3:1 ethanol and glacial acetic acid for at least 24 h at room temperature and then stored at 4°C. After a 20-min rinse of flower buds in deionized water, anthers of appropriate size were dissected from the flower buds and digested with an enzyme mixture (0.3% [w/v] cellulase [MP Biomedicals], 0.3% [w/v] pectolyase [MP Biomedicals], and 0.3% [w/v] cytohelicase [Sigma-Aldrich]) in citric buffer (10 mm sodium citrate and 10 mm sodium EDTA, pH 5.5) at 37°C for 2 h. After rinsing digested anthers with deionized water, the anthers were macerated on glass slides in 20 µL of 60% (v/v) acetic acid at 50°C with fine forceps. Subsequently, 3:1 ethanol and glacial acetic acid was added to the slide, and the slide was dried.
Postfixation of the slides was performed according to a published protocol (Lysak et al., 2006). Briefly, the slides were fixed in 4% (w/v) formaldehyde in 1× phosphate-buffered saline for 10 min at room temperature, washed twice in 1× phosphate-buffered saline for 5 min each, and dehydrated in a 70%, 90%, and 100% (v/v) ethanol series. The immunodetection was performed according to Zhang et al. (2008) using mouse anti-5-methylcytosine (1:500; Eurogentec) detected with Alexa Flour 568 goat anti-mouse IgG (Life Technologies). The pachytene chromosomes were counterstained with the adenine-thymine-specific fluorochrome 4′,6-diamidino-2-phenylindole. Images were taken with the Zeiss Axio Imager M2 microscope, equipped with AxioCam MRm, controlled by Axio Vision 40 version 4.8.2.0. Adobe Photoshop CS5 (Adobe Systems) was used to produce publication images.
Whole-genome bisulfite sequencing data generated for this work have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA264602.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Scatterplot of cytosine methylation between biological replicates.
Supplemental Figure S2. Scatterplot of cytosine methylation between soybean tissues.
Supplemental Figure S3. Scatterplot of the weighted methylation level of protein-coding genes between biological replicates.
Supplemental Figure S4. Chromosome-wide distribution of methylation in soybean and correlation with annotated repeats in sliding 500-kb windows.
Supplemental Figure S5. Chromosome-wide distribution of methylation in common bean and correlation with annotated repeats in sliding 500-kb windows.
Supplemental Figure S6. Immunodetection of 5-methylcytosine on meiotic pachytene chromosomes of soybean (A) and common bean (B).
Supplemental Figure S7. Average distribution of DNA methylation over genes and TE categories in soybean and common bean.
Supplemental Figure S8. Relationship between DNA methylation and gene expression in soybean (A) and common bean (B).
Supplemental Figure S9. Average distribution of DNA methylation of overexpressed and underexpressed genes within paralogous gene pairs.
Supplemental Figure S10. Average distribution of DNA methylation from soybean root over different gene categories.
Supplemental Figure S11. Frequency distribution of PCG after removing C-methylated genes (PCHG < 0.05 or PCHH < 0.05) in soybean (A) and common bean (B).
Supplemental Figure S12. Average distribution of DNA methylation in CG body-methylated genes.
Supplemental Figure S13. Percentage total TE in genic regions in gene categories in soybean and common bean.
Supplemental Figure S14. Pairwise comparisons of CHH methylation levels between paralogous and orthologous gene pairs.
Supplemental Figure S15. Distance from the nearest TE for CG body-methylated (CGBM), unmethylated (CGUM), and C-methylated (CHHM) genes.
Supplemental Figure S16. GO categories (molecular function) enriched for conserved CG body-methylated genes.
Supplemental Figure S17. GO categories (cellular component) enriched for conserved CG body-methylated genes.
Supplemental Figure S18. GO categories (biological process) enriched for conserved CG body-methylated genes.
Supplemental Figure S19. Average distribution of DNA methylation from soybean root in shared single-copy and species-specific single-copy genes.
Supplemental Table S1. Summary of MethylC-seq libraries used in this study.
Supplemental Table S2. Correlation between methylation levels, gene density, and TE coverage in nonoverlapping 100-kb sliding windows.
Supplemental Table S3. Methylation levels of satellite repeat khipu throughout the common bean genome.
Supplemental Table S4. Number of significantly differentially expressed paralogs by species and tissue.
Supplemental Table S5. Number of orthologs between soybean and common bean by gene categories.
Supplemental Table S6. CG body-methylated genes and C-methylated genes from different categories.
Supplemental Table S7. Comparison of sequence evolutionary rates between genes from different gene categories.
Supplemental Table S8. Correlation of methylation levels between paralogous and orthologous gene pairs.
Supplemental Table S9. Summary of the methylation of gene pairs.
Supplemental Table S10. Number of soybean and common bean genes that were homologous to 959 shared single-copy genes (Duarte et al., 2010).
Supplementary Material
Acknowledgments
We thank Dr. Robert J. Schmitz for critical comments on the article.
Glossary
- RdDM
RNA-directed DNA methylation
- TE
transposable element
- WGD
whole-genome duplication
- MYA
million years ago
- Ka
nonsynonymous substitution
- Ks
synonymous substitution
- GO
Gene Ontology
- AS
alternative splicing
Footnotes
This work was supported by the United Soybean Board and the National Science Foundation (grant no. MCB 1339194).
Articles can be viewed without a subscription.
References
- Ahmed I, Sarazin A, Bowler C, Colot V, Quesneville H (2011) Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res 39: 6919–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, et al. (2006) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21: 219–226 [DOI] [PubMed] [Google Scholar]
- Bird AP. (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Branciamore S, Rodin AS, Riggs AD, Rodin SN (2014) Enhanced evolution by stochastically variable modification of epigenetic marks in the early embryo. Proc Natl Acad Sci USA 111: 6353–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, Jia Y, Wang GL, Meyers BC, Jacobsen SE, Pellegrini M (2012) Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci USA 109: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D (2012) Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P, Chen NW, Pedrosa-Harand A, Thareau V, Sevignac M, Cannon SB, Debouck D, Langin T, Geffroy V (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151: 1048–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Adams KL, Vandepoele K, Van Montagu MC, Maere S, Van de Peer Y (2013) Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci USA 110: 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez CM, Roessler K, Gaut BS (2014) Epigenetics and plant genome evolution. Curr Opin Plant Biol 18: 1–8 [DOI] [PubMed] [Google Scholar]
- Du J, Grant D, Tian Z, Nelson RT, Zhu L, Shoemaker RC, Ma J (2010a) SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tian Z, Sui Y, Zhao M, Song Q, Cannon SB, Cregan P, Ma J (2012) Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010b) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, Pires JC, Leebens-Mack J, dePamphilis CW (2010) Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Briskine R, Song J, Li Q, Swanson-Wagner R, Hermanson PJ, Waters AJ, Starr E, West PT, Tiffin P, et al. (2013) Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Ellis NA, Makarevitch I, Yeh CT, Gent JI, Guo L, McGinnis KM, Zhang X, Schnable PS, Vaughn MW, et al. (2012) Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK (2013) CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES (2012) Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA 109: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K, Ng HK, Tang ML, Schucany WR (2008) Testing the ratio of two Poisson rates. Biom J 50: 283–298 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20: 3643–3646 [DOI] [PubMed] [Google Scholar]
- Hadley HH, Hymowitz T (1973) Speciation and cytogenetics. In Caldwell BE, ed, Soybeans: Improvement, Production, and Uses. American Society of Agronomy, Madison, WI, pp 96–116 [Google Scholar]
- Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Jiang WK, Liu YL, Xia EH, Gao LZ (2013) Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol 161: 1844–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KD, Shin JH, Van K, Kim DH, Lee SH (2009) Dynamic rearrangements determine genome organization and useful traits in soybean. Plant Physiol 151: 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey JA. (1980) Chromosome numbers in the Phaseoleae (Fabaceae: Faboideae) and their relation to taxonomy. Am J Bot 67: 595–602 [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon NJ, Lintner RE, Anderson S, Alvarez P, Barry A, Brockman W, Daza R, Erlich RL, Giannoukos G, Green L, et al. (2010) A scalable, fully automated process for construction of sequence-ready barcoded libraries for 454. Genome Biol 11: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, et al. (2012) Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Lisch D. (2009) Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60: 43–66 [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Lysak M, Fransz P, Schubert I (2006) Cytogenetic analyses of Arabidopsis. Methods Mol Biol 323: 173–186 [DOI] [PubMed] [Google Scholar]
- Madlung A, Wendel JF (2013) Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet Genome Res 140: 270–285 [DOI] [PubMed] [Google Scholar]
- Mirouze M, Vitte C (2014) Transposable elements, a treasure trove to decipher epigenetic variation: insights from Arabidopsis and crop epigenomes. J Exp Bot 65: 2801–2812 [DOI] [PubMed] [Google Scholar]
- Ohno S. (1970) Evolution by Gene Duplication. Springer-Verlag, Berlin [Google Scholar]
- Papp B, Pál C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Parkin IA, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE, Town CD, Nixon J, Krishnakumar V, Bidwell SL, et al. (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC (2006) Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet 22: 597–602 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Freeling M, Tang H, Wang X (2010) Insights from the comparison of plant genome sequences. Annu Rev Plant Biol 61: 349–372 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Abdelsamad A, Vu GT (2013) Hidden genetic nature of epigenetic natural variation in plants. Trends Plant Sci 18: 625–632 [DOI] [PubMed] [Google Scholar]
- Qiao Z, Libault M (2013) Unleashing the potential of the root hair cell as a single plant cell type model in root systems biology. Front Plant Sci 4: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin SN, Riggs AD (2003) Epigenetic silencing may aid evolution by gene duplication. J Mol Evol 56: 718–729 [DOI] [PubMed] [Google Scholar]
- Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA (2013) The fate of duplicated genes in a polyploid plant genome. Plant J 73: 143–153 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, He Y, Valdés-López O, Khan SM, Joshi T, Urich MA, Nery JR, Diers B, Xu D, Stacey G, et al. (2013a) Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res 23: 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. (2013b) Patterns of population epigenomic diversity. Nature 495: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, et al. (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20: 461–464 [DOI] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH (1999) Yeast genome evolution in the post-genome era. Curr Opin Microbiol 2: 548–554 [DOI] [PubMed] [Google Scholar]
- Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D (2014) Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet 10: e1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhou Z, Wang Z, Li W, Fang C, Wu M, Ma Y, Liu T, Kong LA, Peng DL, et al. (2014) Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26: 996–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Lu X, Li QT, Chen H, Hu XY, Ma B, Zhang WK, Chen SY, Zhang JS (2013) Genome-wide analysis of DNA methylation in soybean. Mol Plant 6: 1961–1974 [DOI] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS (2012) Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol 29: 219–227 [DOI] [PubMed] [Google Scholar]
- Takuno S, Gaut BS (2013) Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci USA 110: 1797–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS (2010) A triptych of the evolution of plant transposable elements. Trends Plant Sci 15: 471–478 [DOI] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y (2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res 24: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Fustier MA, Alix K, Tenaillon MI (2014) The bright side of transposons in crop evolution. Brief Funct Genomics 13: 276–295 [DOI] [PubMed] [Google Scholar]
- Vonholdt BM, Takuno S, Gaut BS (2012) Recent retrotransposon insertions are methylated and phylogenetically clustered in japonica rice (Oryza sativa spp. japonica). Mol Biol Evol 29: 3193–3203 [DOI] [PubMed] [Google Scholar]
- Wan J, Torres M, Ganapathy A, Thelen J, DaGue BB, Mooney B, Xu D, Stacey G (2005) Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol Plant Microbe Interact 18: 458–467 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang X, Lee TH, Mansoor S, Paterson AH (2013) Gene body methylation shows distinct patterns associated with different gene origins and duplication modes and has a heterogeneous relationship with gene expression in Oryza sativa (rice). New Phytol 198: 274–283 [DOI] [PubMed] [Google Scholar]
- Wolfe KH. (2001) Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet 2: 333–341 [DOI] [PubMed] [Google Scholar]
- Yang L, Gaut BS (2011) Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol 28: 2359–2369 [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zhang G, Cohn MJ (2008) Genome duplication and the origin of the vertebrate skeleton. Curr Opin Genet Dev 18: 387–393 [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang X, Yu Q, Ming R, Jiang J (2008) DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res 18: 1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.