DNA methylome dynamics show crucial roles for DNA methylation in seed development.
Abstract
Seed development is an important process of reproductive development and consists of embryo and endosperm development; both comprise several key processes. To determine and investigate the functions of the dynamic DNA methylome during seed development, we profiled the DNA methylation genome wide in a series of developmental stages of rice (Oryza sativa) embryo and endosperm by methylcytosine immunoprecipitation followed by Illumina sequencing. The results showed that embryo is hypermethylated predominantly around non-transposable element (TE) genes, short DNA-TEs, and short interspersed TEs compared with endosperm, and non-TE genes have the most diverse methylation status across seed development. In addition, lowly expressed genes are significantly enriched in hypermethylated genes, but not vice versa, confirming the crucial role of DNA methylation in suppressing gene transcription. Further analysis revealed the significantly decreased methylation at early developing stages (from 2 to 3 d after pollination), indicating a predominant role of demethylation during early endosperm development and that genes with a consistent negative correlation between DNA methylation change and expression change may be potentially directly regulated by DNA methylation. Interestingly, comparative analysis of the DNA methylation profiles revealed that both rice indica and japonica subspecies showed robust fluctuant profiles of DNA methylation levels in embryo and endosperm across seed development, with the highest methylation level at 6 d after pollination (2 d after pollination of endosperm in japonica as well), indicating that a complex and finely controlled methylation pattern is closely associated with seed development regulation. The systemic characterization of the dynamic DNA methylome in developing rice seeds will help us understand the effects and mechanism of epigenetic regulation in seed development.
DNA methylation is essential for the repression of transposable elements (TEs) and the regulation of gene expression. In plants, DNA methylation occurs in the cytosine residue within three different sequence contexts (CG, CHG, and CHH; Cokus et al., 2008), of which methylation at CHG and CHH is predominantly found in TEs, while CG methylation occurs in TEs and genes (Feng et al., 2010; Zemach et al., 2010). Recent studies demonstrated the crucial roles of DNA methylation in plant development, including gametogenesis, seed development, and response to stresses (Boyko and Kovalchuk, 2008; Chen and Zhou, 2013).
Seed development is an important and finely controlled complex process of reproductive development. Seed development involves several key processes and initiates with the zygote and fertilized central cell. Early embryo development mainly involves apical-basal polarity establishment, epidermis differentiation, and shoot/root meristem formation (Goldberg et al., 1994). Endosperm development mainly involves coenocytic, cellularization, differentiation, and maturation. The structure and developmental program of the rice (Oryza sativa) embryo is different from that of Arabidopsis (Arabidopsis thaliana). An ordered cell division is observed in Arabidopsis embryo at the early stage, while cell divisions are not regular even at the very early stage of rice embryo (Itoh et al., 2005). In addition, Arabidopsis endosperm is consumed by developing embryo and disappears when seed matures, while that of rice is reserved, reflecting the fundamental difference of embryo and endosperm development between rice and Arabidopsis. Studies have shown that many factors are involved in the regulation of embryo and endosperm development, and genetics studies and functional genomics studies have identified some key regulators of seed development. Recent high-throughput transcriptome analysis using microarray and RNA sequencing provided detailed transcriptional profiles of genes at different developmental stages of developing seeds (Xu et al., 2012; Xue et al., 2012; Gao et al., 2013), which help to illustrate the regulatory networks of seed development.
In addition to the genetic approaches, recent studies also indicated a crucial role of epigenetic modification in developing seeds (Ikeda, 2012; Sreenivasulu and Wobus, 2013). The central cell chromatin is less methylated than that of somatic cells (Jullien and Berger, 2010) and then reprogrammed in the offspring after double fertilization. The endosperm DNA of Arabidopsis and rice is globally hypomethylated (Gehring et al., 2009; Hsieh et al., 2009; Zemach et al., 2010). Analysis of the quantified methylation in rice embryo and endosperm at the milky stage by bisulfite sequencing indicated the globally reduced non-CG methylation and local CG hypomethylation in endosperm compared with that in embryo, resembling the phenomenon in Arabidopsis (Zemach et al., 2010). In addition, comparative analysis of transcriptome and methylome between two inbred parents, cv Nipponbare and indica varieties, and their hybrid offspring showed the variable epigenetic heritability and strong association between small interfering RNA differences and epimutation levels (Chodavarapu et al., 2012).
Like most angiosperms, endosperm development of both rice and Arabidopsis follows the nuclear-type development manner, which consists of two main phases: an initial syncytial phase (free nuclear divisions without cytokinesis) followed by a cellularized phase (cell walls form around the free nuclei; Brown et al., 1996; Hehenberger et al., 2012). Endosperm cellularization shares multiple basic components with somatic cytokinesis (Sørensen et al., 2002), and analysis of the endosperm defective1 (ede1; Pignocchi et al., 2009) and spätzle (Sørensen et al., 2002) mutants indicated that endosperm cellularization may also have some unique components. However, the key components and regulatory mechanism of endosperm cellularization are still largely unknown.
Integrative analysis of epigenetics, especially genome-wide DNA methylation and histone modification, and transcriptomics will help to elucidate the molecular mechanism underlying seed development and may contribute to yield improvement by molecular breeding. To further determine and investigate the functions of the dynamic DNA methylome during seed development, we profiled DNA methylation genome wide in a series of developmental stages of rice embryo and endosperm. The results showed that endosperm is predominantly hypomethylated around short DNA-TEs, short interspersed transposable elements (SINEs), and non-TE genes, and the methylation of non-TE genes showed higher variation during early seed development. There is an obvious global demethylation of non-TE genes and long terminal repeats (LTRs) during cellularization. Methylation of LTRs is most stable, which shows typical negative association with the expression of surrounding genes. The candidate non-TE genes regulated by DNA methylation are involved in multiple processes. Moreover, genes related to the cell cycle and hormone response, especially responses to auxin and abscisic acid (ABA) stimulus, are up-regulated during endosperm cellularization.
RESULTS
Global DNA Methylation Patterns of Developing Rice Seeds
To characterize the dynamic DNA methylation during rice seed development, methylcytosine immunoprecipitation followed by Illumina sequencing (MeDIP-seq) was performed using Zhonghua11 (ZH11) embryo at three developmental stages (3, 6, and 9 d after pollination [DAP]) and endosperm at four stages (2, 3, 6, and 9 DAP; Supplemental Fig. S1). These developmental stages correspond to the key stages/events during embryo and endosperm development. Embryonic shoot apical meristem differentiates after 3 DAP (late globular stage), and first leaf primordium emerges at 5 to 6 DAP. Seed organs enlarge and morphogenesis completes at 9 to 10 DAP (Itoh et al., 2005). In endosperm, following fertilization, free nuclear division without cytokinesis occurs at the initial syncytial phase and then cell wall forms at 3 DAP. The cellularization process completes at 6 DAP, followed by endoreduplication at 8 to 10 DAP (Agarwal et al., 2011).
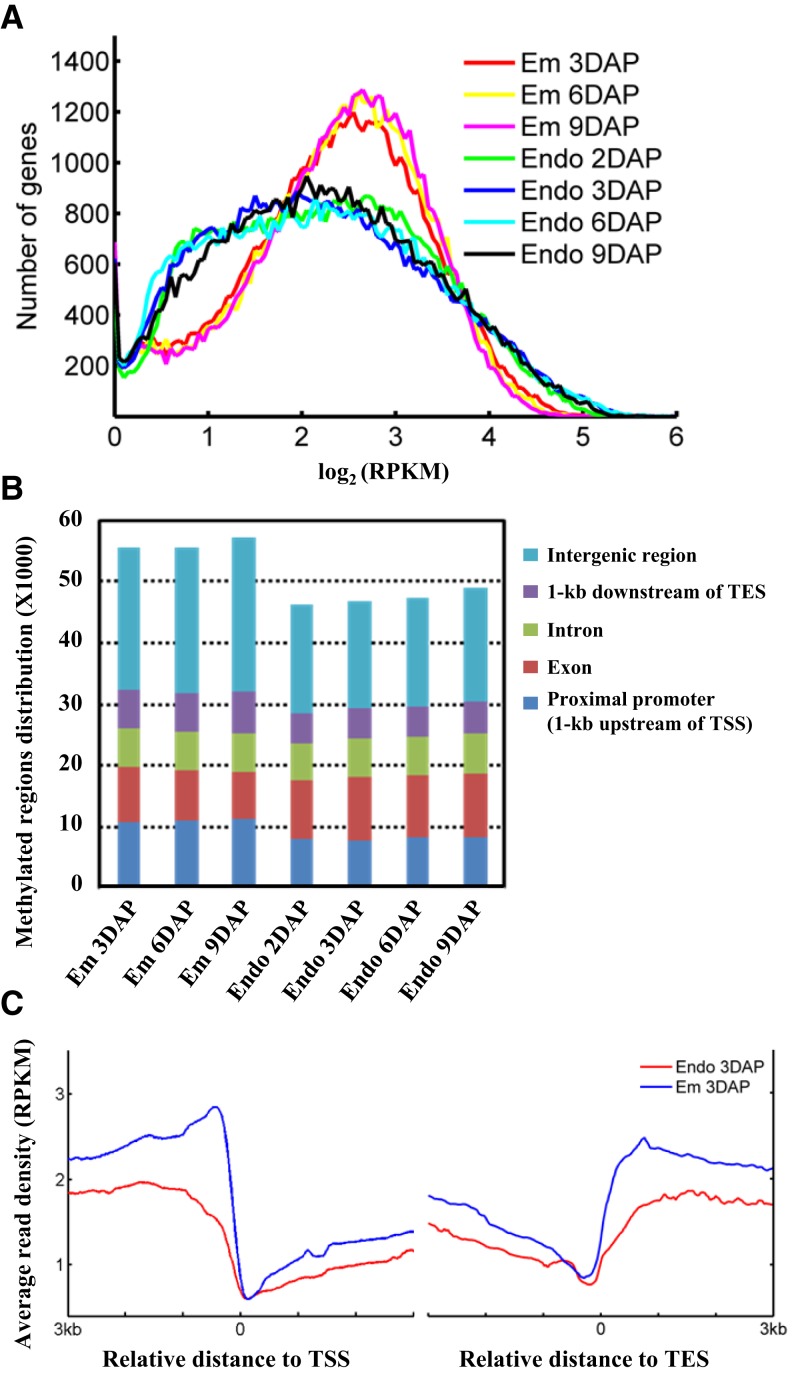
Genome-wide DNA methylation profiles were obtained, and calculation of the read intensity around all annotated genes revealed the higher level of DNA methylation in embryo compared with endosperm across all examined developmental stages (Fig. 1A; Supplemental Fig. S2), which is similar to previous studies from Arabidopsis torpedo-stage seeds (Gehring et al., 2009) or rice seeds at the milky stage (Zemach et al., 2010). Analysis of the methylation status at the promoter region of two randomly selected genes (LOC_Os01g59300 and LOC_Os04g18610) by bisulfite sequencing confirmed the results by MeDIP-seq (Supplemental Fig. S3). In addition, the differentially methylated genes identified by MeDIP-seq were compared with the data of bisulfite sequencing using rice embryo and endosperm at the milky stage, and the results showed that among 69 embryo-hypermethylated genes (Zemach et al., 2010), 51 genes showed similar methylation status (high methylation in embryo and significantly decreased methylation level in endosperm), further confirming the consistency and accuracy of the MeDIP-seq data.
Figure 1.
Global patterns of DNA methylation during rice seed development. A, Distribution of gene methylation density during rice endosperm and embryo development. The x axis represents the gene methylation level defined based on read density normalized by sequencing depth and gene length (reads per kilobase per million mapped reads [RPKM]) from 3 kb upstream of the transcriptional start site (TSS) to 3 kb downstream of the transcriptional end site (TES). Similar results were obtained for both promoter regions (Supplemental Fig. S2A) and the transcriptional termination region (1 kb downstream of the TES; Supplemental Fig. S2B). The Kolmogorov-Smirnov (K-S) test was performed and revealed a significant difference (P < 0.001) between embryo and endosperm across all developmental stages. B, Distribution of the methylated regions in relation to gene annotation. Genomic regions were divided into five categories: proximal promoter (1 kb upstream of the TSS), exon, intron, transcriptional termination region (1 kb downstream of the TES), and intergenic region. Student’s t test was performed and revealed a significant difference in the intergenic region between embryo and endosperm (P = 0.00156). C, Methylation profiles around the TSS and TES of non-TE genes for endosperm and embryo. The x axis represents the relative distance to the TSS (left) or the TES (right), and the y axis represents the average read density normalized by sequencing depth and window size of 20 bp (RPKM). Student’s t test was performed and revealed a significant difference in the methylation level around the TSS and TES between embryo and endosperm at 3 DAP (P < 0.001). The methylation pattern at 3 DAP is shown, and the other samples have the same pattern.
DNA methylation regions characterized by enriched MeDIP-seq reads were first detected to analyze the distribution of DNA methylation in relation to gene annotations. The genome is divided into consecutive 1-kb bins, and those overlapping with read-enriched regions are recorded. The results showed that embryo has more methylation regions (56,145 regions on average) compared with endosperm (47,400 regions on average; Supplemental Table S1). In addition, the number of methylated regions in intragenic regions is similar in endosperm and embryo, while more intergenic regions are methylated in embryo (Fig. 1B). Analysis of the read distribution around the TSS and the TES showed the high methylation level in embryo, which is more obvious in intergenic regions (Fig. 1C). The hypermethylation in the intergenic region of embryo possibly contributes to maintaining the high stability of the embryo genome.
Hypermethylation of TE Genes in Embryo and Hypomethylation of Small DNA-TEs in Endosperm
To compare the DNA methylation profile around TE genes and non-TE genes, patterns of read distribution around TSS and TES were plotted for 15,839 TE genes and 40,147 non-TE genes, respectively, annotated by Michigan State University Rice Genome Annotation Project version 7. Analysis showed that in both endosperm and embryo, TE genes are highly methylated compared with non-TE genes across all examined developmental stages. The gene body of TE genes is more highly methylated compared with intergenic regions, while non-TE genes showed lower methylation in the gene body than in intergenic regions (Supplemental Fig. S4A). Hypermethylation of TE genes is responsible for the suppression of active transposons.
Notably, non-TE genes in endosperm are significantly less methylated than those in embryo (Supplemental Fig. S4, B and C), and a total of 5,048 non-TE genes are consistently highly methylated in embryo compared with endosperm across the examined three developmental stages. Functional analysis of these highly methylated genes indicated that the majority of the enriched biological processes are closely associated with biosynthetic and metabolic processes (Supplemental Table S2). Lipids act as important components of the membrane, and genes with low methylation status in endosperm are enriched in steroid biosynthetic process (25 genes), lipid biosynthetic process (18 genes), and cell wall modification (12 genes), suggesting the involvement of lipid metabolism in endosperm cellularization. In addition, enrichment of amino acid biosynthetic process (Trp, Phe, and Val), endoplasmic reticulum-to-Golgi vesicle-mediated transport (eight genes), and extracellular polysaccharide biosynthetic process implies a relationship between the demethylation of endosperm and the synthesis/accumulation of storage compounds during seed development.
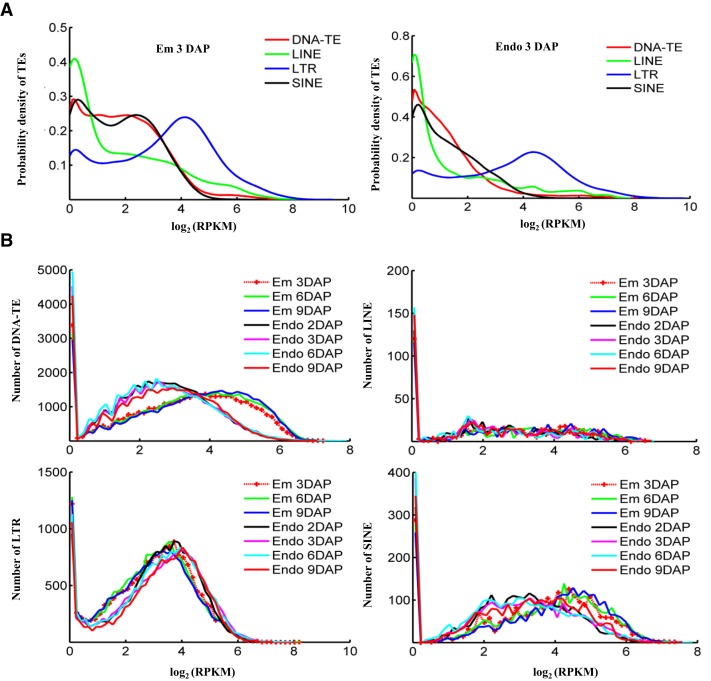
Four major types of TEs are identified by scanning the rice genome (i.e. 40,126 DNA-TEs whose transposition does not involve an RNA intermediate, and three classes of retrotransposons, including 17,267 LTRs, 2,609 SINEs, and 550 long interspersed transposable elements [LINEs]; Supplemental Table S3). Among these TEs, LTR and DNA-TE are most abundant and account for 12.11% and 5.6% of the rice genome. Analysis showed that LTR has correspondingly more methylation signals compared with other TEs (Fig. 2A) and that only DNA-TEs and SINEs present significant hypomethylation in endosperm, while other TEs did not show obvious differences in global methylation levels between endosperm and embryo (Fig. 2B), suggesting the distinct mechanism of demethylation in endosperm for different types of TEs. Previous studies showed that short TEs are predominantly demethylated in Arabidopsis endosperm (Ibarra et al., 2012), and consistently, short DNA-TEs and SINEs have significantly lower methylation levels across all examined developmental stages of rice endosperm (Fig. 3). Short DNA-TE accounts for 87.28% of all DNA-TEs, hypomethylation of which indicates that demethylation of DNA-TE is widespread in endosperm.
Figure 2.
Methylation patterns of TEs. A, Distribution of the methylation read density for DNA-TEs, LINEs, LTRs, and SINEs in embryo and endosperm. The methylation level of TE was defined based on average read density normalized by sequencing depth (RPKM). The read density distribution of embryo or endosperm at 3 DAP is shown, and the other samples have the same pattern. The K-S test was performed and revealed a significant difference between LTR and other TEs (P < 0.001). B, Read density distribution of TEs in all samples, with one plot for one type of TE. The K-S test was performed and revealed a significant difference in the numbers of DNA-TEs and SINEs between embryo and endosperm (P < 0.001).
Figure 3.
Methylation patterns of different types of TEs with different lengths. Read density distribution is shown for DNA-TEs (A), LINEs (B), LTRs (C), and SINEs (D) with length of less than 500 bp (SINEs included four additional subgroups with length of less than 100 bp, 100–200 bp, 200–300 bp, and 300–500 bp), 500 to 2,000 bp, and more than 2,000 bp across different developmental stages of rice embryo and endosperm. The K-S test was performed and revealed a significant difference in the numbers of short DNA-TEs (less than 500 bp) and SINEs (less than 500 bp) between the embryo and endosperm (P < 0.001).
Dynamic DNA Methylomes during Early Seed Development
Investigation of the dynamic methylation by comparing DNA methylation levels between consecutive developmental stages using MAnorm (Shao et al., 2012) showed that numbers of differentially methylated regions in embryo are only one-fourth to one-half of those in endosperm (Fig. 4A), indicating the relatively stable methylome of embryo. Of the differentially methylated regions during embryo development, approximately 24% occur in exon, while DNA methylation regions detected in embryo samples are only about 14% on average (Figs. 1B and 4B). Additionally, a slightly higher percentage of differential methylation regions in intron is also observed. A previous study in rice showed that DNA methylation in the gene body has a larger effect on transcription than methylation in the promoter region (Li et al., 2008). The high percentage of DNA methylation in exon suggests a more crucial role of DNA methylation in regulating gene transcription during embryo development.
Figure 4.
Dynamic DNA methylation of rice embryo and endosperm. A, Number of regions with differential DNA methylation. Increased or decreased DNA methylation between consecutive stages during embryo and endosperm development is shown. For the detection of differential methylation regions, see “Materials and Methods.” B, Distribution of differentially methylated regions in relation to gene annotation between consecutive developmental stages, which is compared with the average proportion of methylated region distribution for embryo and endosperm. Genomic regions were divided into five categories: proximal promoter (1 kb upstream of the TSS), exon, intron, transcriptional termination region (1 kb downstream of the TES), and intergenic region. Student’s t test was performed and revealed significantly more exonic regions for differentially methylated loci compared with all methylated loci in embryo (P = 0.0397), while no significant difference was detected in endosperm (P = 0.62). C, Cumulative distribution of the coefficient of variance in log2 scale for DNA-TEs, LINEs, LTRs, SINEs, and non-TE genes. The K-S test was performed and revealed a significant difference between non-TE genes and TEs, LTRs, and other TEs or non-TE genes (P < 0.001). D, DNA methylation level negatively correlates with the gene expression level in rice embryo and endosperm. The expression intensity in log2 scale of all examined genes was sorted in ascending order (left), and the methylation intensity around the TSS (middle; from 3 kb upstream of the TSS to 3 kb downstream of the TSS) and the TES (right; from 3 kb upstream of the TES to 3 kb downstream of the TES) of the corresponding genes was plotted. The methylation intensity around the TSS and TES is generated from the average read density normalized by sequencing depth and window size of 20 bp (RPKM). Each row in the heat map of expression represents a gene, and each column in the heat map of methylation intensity corresponds to the RPKM in each 20-bp bin around the TSS (middle) or the TES (right). The expression and methylation patterns of embryo or endosperm at 3 DAP are shown, and all other samples have the same pattern.
Further examination of the dynamic DNA methylation levels by calculating the variance coefficient showed that, in both embryo and endosperm, non-TE genes have the most significant changes, while LTR is the most stable element, with slight changes of DNA methylation across developmental stages (Fig. 4C).
In Arabidopsis, it has been assumed that demethylation of DNA-TEs in central cells could facilitate the production of 24-nucleotide small RNAs, which are transported to egg cells to suppress transposons by increasing the methylation in embryo (Hsieh et al., 2009; Ibarra et al., 2012). If this is the case, the hypomethylated TEs in the early stage of rice endosperm should result in higher methylation levels of TEs in embryo at later developmental stages. Indeed, statistical analysis revealed a significant enrichment of highly methylated regions of all TEs in embryo (3 DAP) compared with that in endosperm (3 DAP) or at late embryo (9 DAP) compared with that at early embryo (3 DAP; Supplemental Table S4).
DNA Hypermethylation Is Crucial for Suppressing Surrounding Genes during Seed Development
Furthermore, the DNA methylome was compared with the gene transcriptome of embryo and endosperm at different developmental stages (Xue et al., 2012) to assess the effects of DNA methylation on gene expression during seed development. Calculation of the average methylation read intensity showed that the extent of DNA methylation in the gene body is obviously negatively correlated with gene expression (Supplemental Fig. S5A).
Four gene sets, including genes stably hypomethylated or hypermethylated and genes with stably high or low expression across seed development, were extracted to investigate the long-term role of DNA methylation in the transcription/expression of surrounding genes. Consistent with the previous report in rice (He et al., 2010), Fisher’s exact test showed that, in both embryo and endosperm, hypermethylated genes are enriched in lowly expressed genes, but not vice versa (Supplemental Fig. S5B). Further analysis showed that hypermethylated genes account for approximately 20% of lowly expressed genes in embryo and 17% of lowly expressed genes in endosperm, while the hypermethylated genes are not enriched among the highly expressed genes (Supplemental Fig. S5C), indicating that DNA hypermethylation is responsible for gene expression suppression during seed development at different stages but that hypomethylation is not directly related to gene expression. Meanwhile, 1,847 TE genes were identified in the microarray data set, and comparison of the expression and methylation status showed that hypermethylation of TE genes is obviously correlated with low expression. Analysis by Fisher’s exact test showed that, in both embryo and endosperm, the hypermethylated TE genes are enriched in lowly expressed genes.
The DNA methylation level in the promoter region (1 kb upstream of the TSS) does not show obviously negative correlation as in other regions (Supplemental Fig. S5A). We hypothesized that this may due to the effect from some outliers; thus, we plotted the methylation level for all genes on a microarray ranked by expression intensity (from low to high). The read intensity gradually increased in gene bodies but not in promoter regions (Fig. 4D). It is possible that promoter regions are the major sites for transcriptional regulation by diverse mechanisms, while the methylation status in regions downstream of the TSS is responsible for maintenance of the expression level. Inspired by a similar pattern reported previously for miniature inverted-repeat TEs in rice (Zemach et al., 2010), we plotted the DNA methylation profile around the TSS and TES for different types of TEs. Analysis revealed that DNA-TEs showed negative correlation with the transcription of genes surrounding DNA-TEs in the gene body and the downstream region but not with the regions upstream of the TSS (Supplemental Fig. S5D). In contrast, methylation of LTRs is negatively correlated with the expression of genes surrounding LTRs around the whole intragenic region (Supplemental Fig. S5E). Given that the LTR is hypermethylated (Fig. 2A) and that hypermethylated genes are enriched in lowly expressed genes (Supplemental Fig. S5B), it is hypothesized that the LTR is closely associated with the stable repression of surrounding genes.
Genes Regulated by DNA Methylation Are Involved in Multiple Processes
Genes that are potentially negatively regulated by DNA methylation were identified by analyzing differential gene expression and methylation based on comparisons between consecutive stages. After calculating the Pearson correlation between the expression change and methylation change of each gene, 318 genes with negative association (Pearson correlation coefficient < −0.6 and P < 0.05) between expression and methylation change were recorded (Supplemental Table S5). The top enriched biological processes of these genes include small GTPase-mediated signal transduction (four genes) and oxidation reduction (15 genes; Table I; http://www.ricearray.org/index.shtml; Cao et al., 2012). Indeed, analysis of two randomly selected genes (LOC_Os12g37360, a small GTPase, and LOC_Os06g42000, a peroxiredoxin) confirmed the opposite methylation and expression across seed development (Supplemental Fig. S5F).
Table I. GO category enrichment of genes showing negative correlation between methylation change and expression change.
A total of 318 genes across five comparisons between consecutive developmental stages were analyzed (Pearson correlation coefficient < −0.6 and P < 0.05).
| GO Identifier | GO Name | Query Number | Hyper P Value | Fold Change (Log2) | Entries |
|---|---|---|---|---|---|
| GO:0006886 | Intracellular protein transport | 6 | 1.80E-03 | 4.55 | Os01g24060 |
| Os03g10370 | |||||
| Os03g27450 | |||||
| Os07g12170 | |||||
| Os07g48880 | |||||
| Os12g37360 | |||||
| GO:0007264 | Small GTPase-mediated signal transduction | 4 | 4.10E-03 | 5.95 | Os03g10370 |
| Os03g27450 | |||||
| Os07g12170 | |||||
| Os12g37360 | |||||
| GO:0055114 | Oxidation reduction | 15 | 4.50E-03 | 2.03 | Os01g22370 |
| Os02g08230 | |||||
| Os02g30714 | |||||
| Os02g47470 | |||||
| Os02g51880 | |||||
| Os03g45519 | |||||
| Os04g06790 | |||||
| Os04g20164 | |||||
| Os04g46960 | |||||
| Os06g42000 | |||||
| Os06g46680 | |||||
| Os06g46799 | |||||
| Os07g32570 | |||||
| Os10g30140 | |||||
| Os12g34874 | |||||
| GO:0045454 | Cell redox homeostasis | 3 | 3.94E-02 | 3.69 | Os02g51370 |
| Os06g42000 | |||||
| Os07g32570 | |||||
| GO:0015031 | Protein transport | 4 | 4.75E-02 | 2.65 | Os03g10370 |
| Os03g27450 | |||||
| Os07g12170 | |||||
| Os12g37360 |
Twelve transcription factors and eight transcription regulators (Table II) were annotated (rice transcription factor database PInTEDB; Pérez-Rodríguez et al., 2010) that belong to the ABA INSENSITIVE3 (ABI3)/VIVIPAROUS1(VP1), APETALA2 (AP2)-ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN (EREBP), MADS (for MINI CHROMOSOME MAINTENANCE1, AGAMOUS, DEFICIENS, and SERUM RESPONSE FACTOR), BASIC REGION/LEUCINE ZIPPER (bZIP), CCAAT-BINDING TRANSCRIPTION FACTOR (CCAAT), and AUXIN RESPONSE FACTOR (ARF) families. Some Arabidopsis homologs have been reported to be involved in seed development regulation. Arabidopsis FUSCA3 is a homolog of LOC_Os01g51610 (a B3 DNA-binding domain-containing protein) and regulates embryogenesis (Wang and Perry, 2013). In addition, several other regulators, including WD domain, protein kinase, zinc-finger, and C3HC4-type proteins involved in transcriptional regulation during seed maturation (Suzuki and McCarty, 2008; Sreenivasulu and Wobus, 2013), also showed negative associations between expression and DNA methylation.
Table II. Transcription factors and regulators showing negative correlation between methylation change and expression change.
A total of 318 genes across five comparisons between consecutive developmental stages were analyzed (Pearson correlation coefficient < −0.6 and P < 0.05).
| Type | Category | Annotation |
|---|---|---|
| Transcription factor | ||
| Os01g51610 | ABI3/VP1 | B3 DNA-binding domain-containing protein |
| Os02g26430 | WRKY | WRKY42 |
| Os02g39220 | FAR-RED-IMPAIRED RESPONSE1 | Transposon protein, putative, unclassified |
| Os04g36790 | TRIHELIX DNA-BINDING DOMAIN CONTAINING TRANSCRIPTION FACTOR | Transcription factor-like protein, putative |
| Os04g58010 | ABI3/VP1 | B3 DNA-binding domain-containing protein |
| Os06g07030 | AP2-EREBP | AP2 domain-containing protein |
| Os06g47150 | ARF | Auxin response factor18, putative |
| Os08g41960 | MADS | OsMADS37, MADS box family gene |
| Os09g28310 | bZIP | bZIP transcription factor, putative |
| Os11g24130 | PLANT AT-RICH SEQUENCE- and ZINC-BINDING PROTEIN | Zinc-binding protein, putative |
| Os11g35390 | MYELOBLASTOSIS | MYB family transcription factor, putative |
| Os12g25120 | CCAAT | Core histone H2A/H2B/H3/H4, putative |
| Transcription regulator | ||
| Os02g51880 | SWIRM (for SWI3, RSC8, and MOIRA) DOMAIN CONTAINING TRANSCRIPTION REGULATOR | Amine oxidase, putative |
| Os03g05680 | JUMONJI | Histone demethylase JARID1C, putative |
| Os06g07040 | AUXIN/INDOLE-3-ACETIC ACID (IAA) | OsIAA20 |
| Os07g39690 | GCN5-RELATED N-ACETYLTRANSFERASE | GCN5-related N-acetyltransferase, putative |
| Os09g36220 | Pseudo TYPE-B PHOSPHO-ACCEPTING RESPONSE REGULATOR (ARR-B) | Response regulator receiver domain-containing protein |
| Os11g40490 | BTB (for BRIC-À-BRAC, TRAMTRACK) DOMAIN CONTAINING TRANSCRIPTION REGULATOR (TRAF) | MBTB63-Bric-a-Brac, Tramtrack, Broad Complex BTB domain with Meprin and TRAF homology MATH domain |
Further analysis revealed that, among the embryo-preferentially expressed or endosperm-preferentially expressed genes, only a few are highly methylated (Table III), while most are lowly or median methylated, suggesting the possible role of DNA methylation in regulating the expression of seed (including both embryo and endosperm)-preferentially expressed genes. Gene Ontology (GO) analysis of the embryo-preferentially expressed genes with low DNA methylation (bottom 10%) revealed the involvement of them in protein/lipid/metal ion transport, glycolysis, transcription regulation, and multicellular organismal development, indicating that DNA methylation may be involved in embryo development through regulating the expression of embryo-preferential genes and related processes. The same phenomena were observed in endosperm (Supplemental Table S6).
Table III. Methylation levels of embryo-preferentially expressed and endosperm-preferentially expressed genes.
Genes with greater than 4-fold expression in embryo or endosperm compared with vegetative tissues were selected. High methylation indicates that the methylation level of a given gene is above top 10% based on read numbers from 1 kb upstream to 1 kb downstream of the gene body.
Starch is the main storage of rice seed, and its biosynthesis/metabolism is critical for yield and quality. Analysis showed that the methylation status of endosperm-specific starch biosynthesis-related genes, including ADP-GLUCOSE PYROPHOSPHORYLASE2b, GRANULE-BOUND STARCH SYNTHASE I, STARCH SYNTHASE IIa, and endosperm-preferentially expressed genes ADP-Glucosec Pyrophosphorylase Large Subunit1 (AGPL1), AGPL2, and ISOAMYLASE1 (Ohdan et al., 2005), were relatively low in endosperm at different stages (2, 3, 6, and 9 DAP; Supplemental Table S7), suggesting the crucial roles of DNA methylation in regulating starch synthesis. However, whether the constantly low DNA methylation level of these genes is directly responsible for the gradually increased starch synthesis during endosperm development would be an interesting issue for further study.
Involvement of DNA Methylation in Endosperm Cellularization
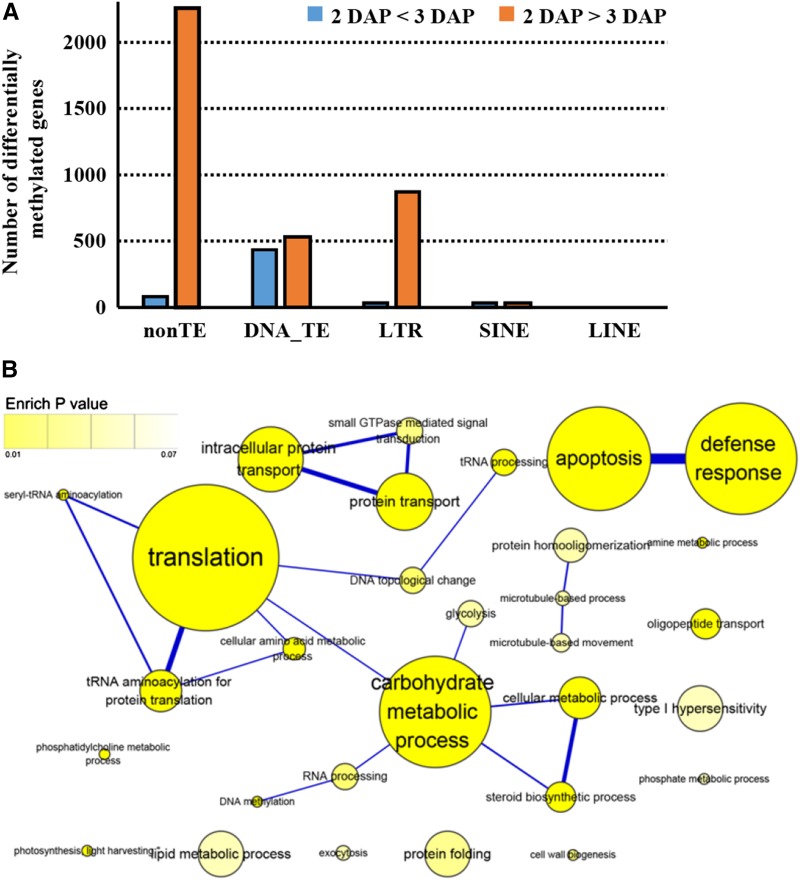
To investigate the role of DNA methylation during endosperm cellularization, global gene expression profiles between 2 DAP (free nuclei) and 3 DAP (cellularization starts) of endosperm were analyzed and compared with DNA methylation patterns. Analysis revealed 1,067 up-regulated and 1,303 down-regulated genes at 3 DAP, which are enriched in cell cycle and hormone responses, especially to auxin and ABA stimulus (Supplemental Fig. S6; Supplemental Table S8). Further analysis revealed the dramatically decreased global DNA methylation level from 2 to 3 DAP, and 12,246 regions had reduced, while only 4,766 regions had increased, methylation (Fig. 4A). A detailed examination of DNA methylation levels for different types of genomic features revealed that non-TE genes and LTRs undergo apparent demethylation from 2 to 3 DAP in endosperm, while other types of TEs show slight changes (Fig. 5A).
Figure 5.
DNA methylation status during early endosperm development. A, Numbers of differentially methylated elements at 2 and 3 DAP during early endosperm development. B, GO term enrichment network of highly methylated (log2 [ChIP-seq intensity ratio; M] M > 1 and P < 1e-5) genes at 2 DAP compared with 3 DAP in endosperm. Nodes represent enriched GO terms, and the color intensity is proportional to the significance of enrichment. The area of a node is proportional to the number of genes in the test set belonging to that GO category. The weight of the edges represents the number of genes shared by a pair of GO terms.
In addition, 1,233 non-TE genes that showed reduced levels of DNA methylation at 3 DAP are enriched in processes including translation/tRNA aminoacylation, protein transport, apoptosis, cell wall biogenesis, microtubule-based process, and DNA methylation (Fig. 5B; Supplemental Table S9). Overlap between differentially expressed genes and differentially methylated genes from 2 to 3 DAP found that 43 genes showed opposite patterns between expression change and DNA methylation alteration (Supplemental Fig. S7; Supplemental Table S10). Among them, LOC_Os01g60050, a microtubule organization protein, is a MAP215 family microtubule-associated protein that is required to establish interphase arrays of cortical microtubules in plant cells, and loss of function of Arabidopsis homolog MICROTUBULE ORGANIZATION1/GEMINI POLLEN1 results in defects in cytokinesis during pollen development (Twell et al., 2002). Interestingly, a key endosperm cellularization-related gene, EDE1, also encodes a microtubule-associated protein (Pignocchi et al., 2009). Considering that endosperm cellularization shares multiple basic components with somatic cytokinesis (Sørensen et al., 2002), it is speculated that LOC_Os01g60050 may play an important role during cellularization.
LOC_Os05g49440 is a DUF1264 domain-containing protein whose homolog in Arabidopsis is Tyr phosphorylated, and the phosphorylation status is modulated in response to ABA in seed (Ghelis et al., 2008). LOC_Os04g17660 (a Rhodanese-like domain-containing protein) and LOC_Os02g01170 (a HECT domain-containing protein) are involved in cell cycle control, and their homolog in Arabidopsis, known as KAKTUS, is involved in the regulation of the number of endoreduplication cycles in various organs (El Refy et al., 2003). LOC_Os02g29140 is an ankyrin repeat-containing protein possibly mediating protein-protein interactions among diverse groups of proteins, including those involved in signal transduction, cytoskeleton interactions, and regulation of the cell cycle; mutation of its homologs ANKYRIN REPEAT PROTEIN and EMBRYO-DEFECTIVE506 results in defective embryo development at the globular stage (Garcion et al., 2006).
TE Genes Showed High Variation of DNA Methylation between japonica and indica Varieties
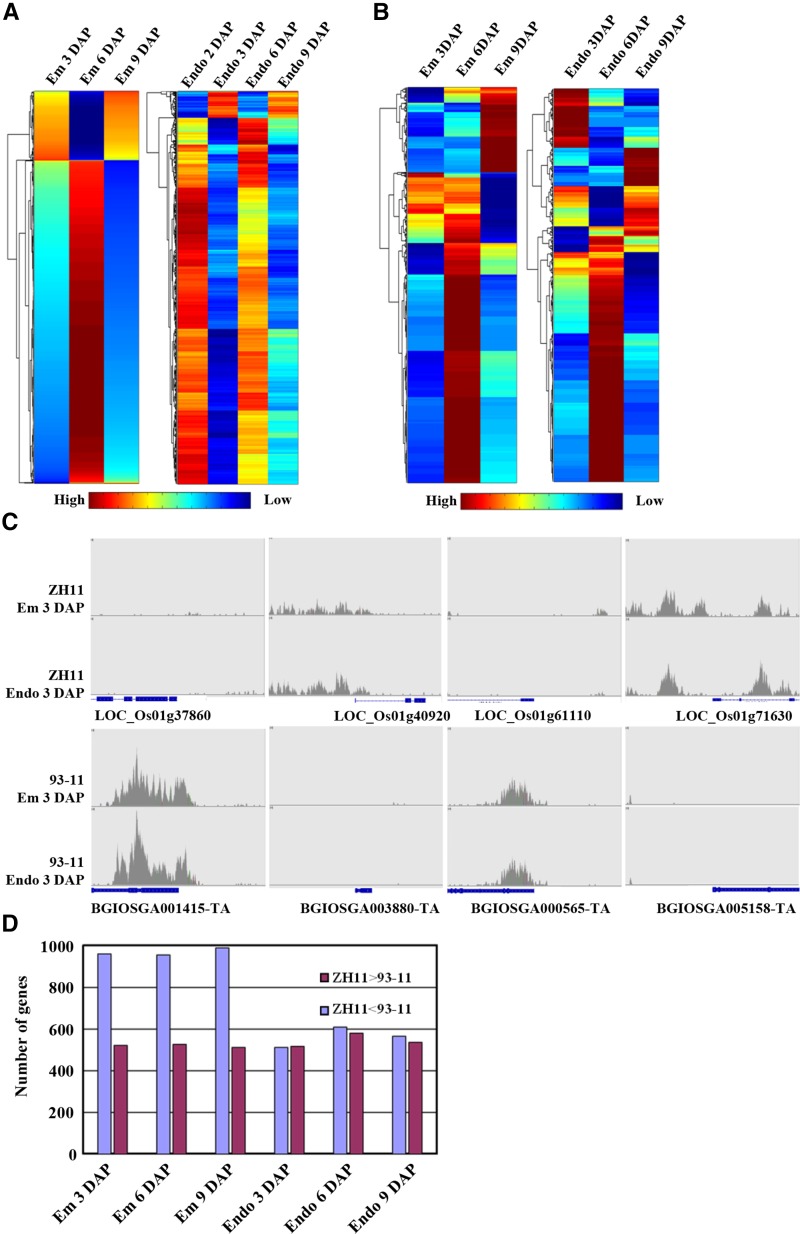
DNA methylation profiles of rice subspecies 93-11 (indica) in embryo and endosperm at 3, 6, and 9 DAP were also characterized by MeDIP-seq, and comparative analysis showed that both indica and japonica subspecies showed robust fluctuant profiles of DNA methylation levels in embryo and endosperm across development, with the highest methylation level at 6 DAP (Fig. 6, A and B). A total of 26,024 homologous genes in the collinear blocks of alignments were analyzed to investigate the methylation variation between ZH11 and 93-11, and the results showed that 2,166 genes in embryo and 1,681 genes in endosperm displayed differential methylation. Analysis of four randomly selected genes (LOC_Os01g37860, LOC_Os01g40920, LOC_Os01g61110, and LOC_Os01g71630) confirmed the differential methylation status between ZH11 and 93-11 (Fig. 6C). Among the differentially methylated genes, more genes of embryo had higher methylation levels in 93-11, while those of endosperm did not show this difference (Fig. 6D). GO enrichment of differentially methylated genes revealed the involvement of them in apoptosis, defense response, response to stress in both embryo and endosperm, and specifically enriched biological processes, including the activation of protein kinase C activity, endoplasmic reticulum-to-Golgi vesicle-mediated transport, tricarboxylic acid cycle, cell-matrix adhesion in embryo or xylan catabolic process, negative regulation of translation, translational elongation, and glycolysis in endosperm. In addition, interestingly, TE genes showed significant enrichment in differentially methylated genes (with enrichment P < 1e-21 and enrichment fold change > 2.3; Supplemental Table S11), indicating the possible large variation in epigenetic regulation of TE genes during seed development between indica and japonica subspecies.
Figure 6.
Methylation patterns of embryo and endosperm in ZH11 and 93-11. A and B, Heat map of read intensity for differentially methylated regions during endosperm and embryo development in ZH11 (A) and 93-11 (B). Each column represents a time point during embryo or endosperm development, and each row represents a differentially methylated region between all neighboring stages during embryo or endosperm development. The color scale in red (high), yellow (medium), and blue (low) represents the relative methylation level. C, Representative genes showing differential methylation between ZH11 and 93-11. D, Number of genes exhibiting differential DNA methylation between ZH11 and 93-11 at different developmental stages. The number of reads from 1 kb upstream of the TSS to 1 kb downstream of the TES was used for analysis.
DISCUSSION
Dynamic DNA Methylation of Rice Developing Seeds
Seed development is finely coordinated among embryo, endosperm, and seed coat development. Previous studies indicated that early seed development is regulated predominantly at the transcriptional level by a set of transcription factors (Le et al., 2010; Sreenivasulu et al., 2010; Sreenivasulu and Wobus, 2013), and genome-wide studies of the DNA methylome in Arabidopsis, rice, maize (Zea mays), and soybean (Glycine max; Hsieh et al., 2009; Zemach et al., 2010; Li et al., 2012; Eichten et al., 2013; Song et al., 2013) provided important information for studying the function of DNA methylation in seed development. However, most of these studies involved only one stage, and no study has been performed to characterize the dynamics of the DNA methylome and the association with seed development regulation.
By characterizing the dynamic DNA methylation profile genome wide across seed development, our studies showed that the DNA methylation level is more stable in embryo compared with endosperm and that DNA methylation of non-TE genes is more dynamic compared with TEs during both embryo and endosperm development. The relatively stable DNA methylation of TEs is responsible for the repression of transposon and the maintenance of genome stability. In addition, lowly expressed genes present high DNA methylation levels but not vice versa, indicating the major role of DNA methylation in maintaining the low expression of surrounding genes during rice seed development.
Endosperm at 2 DAP shows the highest methylation level compared with later stages, indicating a genome-wide demethylation in endosperm from the early stage, which is different from that of Arabidopsis, in which the central cell chromatin is less methylated than that of somatic cells (Jullien and Berger, 2010) and then reprogrammed in the offspring after double fertilization. In Arabidopsis, DEMETER (DME) is responsible and required for global CG hypomethylation during this process (Zemach et al., 2010); however, no DME orthologs of rice have been characterized yet. The mechanism of the differential endosperm demethylation between monocots and dicots needs further study (Chen and Zhou, 2013).
Endosperm cellularization begins at 2 DAP and involves many physiological processes, such as cell cycle and hormone responses and global high-DNA methylation at 2 DAP, and then demethylation results in the reactivation or up-regulation of genes and plays a direct role in regulating cellularization. In addition, there is an increase of DNA methylation at 6 DAP in both endosperm and embryo, which is observed in both indica (93-11) and japonica (ZH11) varieties. During early seed development, 6 DAP is a key time point, when the first leaf primordium emerges, the cellularization process completes, and filling starts. Based on the obviously negative correlation between hypermethylation and low expression (Supplemental Fig. S5B), it is reasonable that high methylation at 6 DAP in this stage may involve the temporal conversion of expression of many genes.
Although there is no direct evidence for how dynamic DNA methylation and differentially methylated genes regulate seed development, our results here may help to illustrate the molecular mechanism in gene expression regulation at distinct developmental stages via dynamic methylation. The exact functions of these differentially methylated genes during seed development will be interesting subjects for future studies.
Different Roles of DNA-TE and LTR during Seed Development
DNA demethylation in endosperm predominantly occurs in short DNA-TEs and SINEs. The DNA methylation levels of DNA-TE and LTR, the two most abundant types of TEs, showed distinct relationships with the expression of downstream genes (i.e. positive correlation for DNA-TE and negative correlation for LTR), while they both have similar methylation patterns in the gene body and the transcriptional termination region (1 kb downstream of the TES). It is supposed that LTR mainly contributes to maintain the suppression of surrounding genes during seed development based on the following data. First, LTR is the most abundant TE (12.11% of the genome) and displays a high level of DNA methylation; second, LTR shows consistently negative correlation with the expression of surrounding genes; and third, genes with high DNA methylation levels are significantly enriched in lowly expressed genes during seed development, but not vice versa.
Complex Regulation of Endosperm Cellularization
Endosperm cellularization that transits the syncytial phase to the cellularized phase is an important event during seed development in both rice and Arabidopsis, while the underlying mechanism is not well understood. There is a dramatically decreased global DNA methylation level from 2 to 3 DAP; however, hypermethylated genes are enriched in lowly expressed genes, but not vice versa, which suggested that only some of the demethylated genes from 2 to 3 DAP are involved in endosperm cellularization. A possible reason for the demethylation of other genes may be an early removal of methylation repression for the later stages.
The expression of many auxin and ABA response-related genes was significantly altered during cellularization, suggesting the crucial roles of hormones in this process. Auxin regulates seed development initiation (Zhou et al., 2013) and may control seed size through regulating endosperm cellularization. In maize, Z. mays PINFORMED1-mediated transport of auxin is involved in cellular differentiation in early endosperm development (Forestan et al., 2010). ABA is critical for seed maturation and dormancy, while little is known about its role at early stages. In barley (Hordeum vulgare), decreased ABA content causes abnormal endosperm development, including abnormal cellularization/differentiation of endosperm (Sreenivasulu et al., 2010). Recent studies showed that the ABA-deficient mutant aba2-1 presents delayed endosperm cellularization (Cheng et al., 2014), suggesting that ABA promotes cellularization, which is consistent with our finding revealing the up-regulation of ABA response-related genes during rice endosperm cellularization. The expression of a DUF1264 domain-containing protein (LOC_Os05g49440) during cellularization is regulated by methylation, and the phosphorylation state of its homolog in Arabidopsis is modulated by ABA (Ghelis et al., 2008), which suggests a possible role of LOC_Os05g49440 in cellularization in response to ABA. Functional studies of these hormone-responsive genes may provide more clues to how ABA and auxin regulate endosperm cellularization and, hence, seed development.
The cell cycle of endosperm cellularization is distinct from that of normal mitosis. Shortly after fertilization, the fertilized central cell goes through repeated rounds of mitosis without cell wall formation, resulting in syncytium formation by the free nucleus. Consistently, analysis showed that cell cycle-, cell division-, and mitosis-related GO terms were especially enriched in down-regulated gene sets of endosperm at 2 to 3 DAP, confirming a repression of the normal cell cycle during endosperm cellularization. Regulation of cell cycle-related genes by DNA methylation, including a Rhodanese-like domain-containing protein (LOC_Os04g17660), an HECT ubiquitin protein ligase family protein (LOC_Os02g01170), and an ankyrin repeat-containing protein (LOC_Os02g29140), suggests the crucial roles of DNA methylation in cell cycle regulation during endosperm cellularization. In addition, a large number of transport-related GO terms (lipid, iron ion, and carbohydrate transport) were enriched at 2 and 3 DAP, suggesting that they may serve as substrates or participate in signal transduction. Hexose content is increased in Arabidopsis mutants defective in endosperm cellularization (Hehenberger et al., 2012), indicating the important role of the transport or metabolism of hexose for cellularization. Due to the limited information on endosperm cellularization, especially in rice, exact roles for the transport remain to be studied.
Besides signal transduction and gene expression regulation, protein modification-related GO terms are also enriched in endosperm at 2 to 3 DAP, suggesting a complex regulation of endosperm cellularization. Until now, little was known about the transcriptional and posttranscriptional regulation of endosperm cellularization, and the only example is that the MADS box transcription factor AGL62, the direct target of FERTILIZATION-INDEPENDENT SEED POLYCOMB GROUP-REPRESSIVE COMPLEX2, acts as a suppressor of endosperm cellularization (Kang et al., 2008; Hehenberger et al., 2012). Functional analysis of the identified genes, especially the transcriptional factor and protein kinases regulated by DNA methylation, may help to identify other factors regulating endosperm cellularization and expand our understanding of the molecular mechanism of endosperm cellularization and early endosperm development.
Interestingly, DNA/RNA methylation is among the top enriched GO terms of genes with reduced DNA methylation during cellularization (Supplemental Table S9), suggesting a possible feedback for DNA methylation regulation during seed development.
Crucial Roles of DNA Methylation during Rice Seed Development
Alteration of DNA methylation around non-TE genes is more apparent as compared with TEs during rice seed development, and there is a consistent negative correlation between DNA methylation change and expression change (Supplemental Table S5). These genes potentially directly regulated by DNA methylation may regulate seed development by involvement in multiple biological processes (Table I). Several auxin-related proteins (auxin efflux carrier component, AUXIN/IAA20, and ARF18) and one ARR-B family member (LOC_Os09g36220, a response regulator receiver domain-containing protein) may regulate core cell cycle genes and cell fate determination in endosperm by interacting with cytokinin response factors. Proteolytic processes are thought to regulate seed size (Sreenivasulu and Wobus, 2013), and five F-box domain-containing proteins and one caseinolytic protease homolog are regulated by DNA methylation.
In addition, 25 genes reported to be involved in rice seed development regulation, especially in endosperm development, show different levels of methylation. Further comparison of the methylation read intensity and expression patterns revealed that nine genes, REPRESSOR OF SILENCING1, GRAIN WIDTH2, RICE SEED B-ZIPPER1, RICE CELL CYCLE SWITCH52A, the rice homolog of maize CRINKLY4, FERTILIZATION-INDEPENDENT ENDOSPERM1, NUCLEAR FACTOR Y TRANSCRIPTION FACTOR B1, FLOURY ENDOSPERM6, and SMALL KERNEL1, show negative correlation between gene expression and DNA methylation, suggesting that these genes may be regulated by DNA methylation during seed development. However, there is still no direct evidence confirming that the expression change of these genes is regulated by methylation, and further functional studies will help to reveal the cross talk of genetic and epigenetic regulation during embryo and endosperm development.
Low methylation of DNA-TE in endosperm contributes to the maintenance of embryo genome stability in Arabidopsis (Hsieh et al., 2009; Ibarra et al., 2012). Our results provide supporting evidence that not only DNA-TEs, but also hypomethylated retrotransposons, are enriched in early endosperm, indicating the similar regulation tendency of Arabidopsis and rice from the developmental aspect.
Significant correlation between consistently high methylation and surrounding consistently low expression was observed, while no obvious correlation was observed for consecutive developmental stages. It is possible that regulation by DNA methylation is relatively stable, and changing it does not result in the rapid expression change of surrounding genes.
CONCLUSION
We report the dynamic change of global DNA methylation during rice seed development and reveal that endosperm hypomethylation predominantly occurs in short DNA-TEs, SINEs, and non-TE genes. Our studies indicate the distinct roles of DNA-TE and LTR in regulating seed development and demethylation during early endosperm development. To our knowledge, this is the first systemic analysis characterizing the DNA methylome in developing seeds and will be of great help in studying the complex regulatory mechanism of, and to illustrate the effect of epigenetic regulation during, seed development.
MATERIALS AND METHODS
Plant Materials and DNA Isolation
Rice (Oryza sativa) varieties ZH11 (ssp. japonica) and 93-11 (ssp. indica) were used in this study. Rice endosperm was collected at 2, 3, 6, and 9 DAP, and embryo was collected at 3, 6, and 9 DAP. DNA was isolated using a cetyl-trimethyl-ammonium bromide method and treated with RNase, and then the MeDIP-seq assay were performed.
Read Mapping and Distribution Comparison for TEs and Non-TEs
A total of 12 million 2× 49-bp paired-end reads were generated for each sample, resulting in 13 MeDIP-seq samples, which were deposited in the Gene Expression Ominibus (GEO) with accession number GSE57706. BOWTIE2 was used for read mapping. All reads of samples from ZH11 were mapped to the japonica genome (version 7.0; http://rice.plantbiology.msu.edu/), and 93-11 samples were mapped to the indica genome (ftp://public.genomics.org.cn/BGI/rice/rise2/). Generally, more than 95% of the reads can be aligned to the reference genome with default settings (Supplemental Table S12). The duplicated reads were removed, and only alignments with MAPQ scores of 20 or greater were kept for further analysis. Syntenic regions between ZH11 and 93-11 were detected for comparison (Supplemental Fig. S8). Briefly, homologous regions of sequences shared by two genomes were obtained by pairwise alignment using BLASTN (version 2.2.25; Altschul et al., 1997), and alignments with E values < 1e-5 were retained. Synteny and collinearity detection were then applied to identify collinear blocks of alignments using MCScanX. Homologous genes in collinear blocks were used for further comparison of methylation levels between ZH11 and 93-11.
To compare the distribution patterns of DNA methylation between TE genes and non-TE genes in embryo and endosperm, 15,839 TE genes and 40,147 non-TE genes were obtained based on rice gene annotation from the MSU Web site (version 7.0; http://rice.plantbiology.msu.edu/). The average number of reads normalized by sequencing depth and bin length (RPKM) was then plotted around the TSS and TES, which cover most of the transcription information in the genome and are commonly chosen for analysis of DNA methylation distribution.
TE Characterization
The TEs were screened and annotated using the RepeatMasker program (version open-4.0.3; http://www.repeatmasker.org/). Annotation of TEs was obtained from Repbase (Repbase18.11.embl; http://www.girinst.org/server/archive/RepBase18.11/). Four types of TEs were identified: DNA-TEs, LINEs, LTRs, and SINEs (Supplemental Table S3). The divergence score of identified TE and non-TE genes overlapping with annotated genes (version 7.0; http://rice.plantbiology.msu.edu/) were compared, and a divergence score of 12 or less was chosen as the cutoff for TE identification. TEs of the same family located within 100 bp of one another were merged into a single TE annotation in order to prevent single TE insertions from being annotated as separate insertions.
Identification of Differentially Expressed Genes
Gene expression data were published previously (GEO accession nos. GSE11966 and GSE27856), except for the data set of endosperm at 2 DAP, which is deposited in the GEO with accession number GSE57615. Expression data sets of seven tested samples were divided into four quantiles ranked by gene expression intensities. Robust multiarray average was used for normalization, and differential expression was determined using the Bioconductor limma package (Smyth, 2005). Differentially expressed genes were defined with two criteria: differential expression of fold change > 2 and P < 0.05. The embryo-preferential or endosperm-preferential genes indicate genes that are predominantly expressed in embryo or in endosperm compared with the average expression level of vegetative tissues (fold change ≥ 4). The seed-preferential genes include both embryo-preferential and endosperm-preferential genes.
Identification of Differentially Methylated Regions
Enriched regions of DNA methylation were identified using MACS14 (Zhang et al., 2008). Each chromosome of the rice genome was divided into consecutive bins of 1 kb, and only the bins covered by peaks from any sample were selected for further comparison. Differentially methylated bins were detected using MAnorm (Shao et al., 2012), a program specifically designed for the identification of differential binding regions between chromatin immunoprecipitation sequencing samples. The bins with M > 0.5 (M indicates log2 [ChIP-seq intensity ratio]; corresponding to fold change close to 1.5) and P < 1e-5 were defined as differentially methylated regions.
Analysis of Dynamic DNA Methylomes
We collected all differentially methylated regions between consecutive developmental stages of embryo and endosperm. To investigate the dynamic changes of DNA methylation, the distribution of differential methylation regions (1-kb bin) in relation to different genomic features was compared. The cumulative distribution of the coefficient of variance of read density in the gene body was plotted to characterize the dynamic change between different types of TE and non-TE genes.
All raw data from this study have been submitted to the National Center for Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE57706, GSE11966, GSE27856, and GSE57615.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Different stages of ZH11 developing seeds.
Supplemental Figure S2. Global patterns of DNA methylation during rice seed development.
Supplemental Figure S3. Verification of methylation status of two randomly selected genes by bisulfite sequencing.
Supplemental Figure S4. Methylation patterns of TE genes and non-TE genes.
Supplemental Figure S5. DNA methylation negatively correlates with gene expression in rice embryo and endosperm at different stages.
Supplemental Figure S6. GO term enrichment network of up-regulated and down-regulated genes of endosperm at 2 to 3 DAP.
Supplemental Figure S7. Venn diagram showing the number of genes with increased methylation and decreased expression.
Supplemental Figure S8. Circosdiagram illustrating syntenicblocks between cv Nipponbare and 93-11.
Supplemental Table S1. Numbers of DNA methylated bins at different developmental stages of rice endosperm and embryo.
Supplemental Table S2. GO category enrichment of highly methylated genes in embryo compared to endosperm across all three examined developmental stages.
Supplemental Table S3. Number of TEs screened and annotated by RepeatMasker Program.
Supplemental Table S4. Enrichment of higher methylation regions in embryo at 3 DAP than in endosperm at 3 DAP and higher methylation regions in embryo at 9 DAP than in embryo at 3 DAP.
Supplemental Table S5. log2 fold change of methylation and expression for 318 genes with negative correlation between methylation change and expression change across 5 comparisons between each consecutive stage.
Supplemental Table S6. GO category enrichment of embryo- or endosperm-preferentially expressed genes with significantly low methylation.
Supplemental Table S7. Methylation status of starch-biosynthesis related genes.
Supplemental Table S8. GO category enrichment analysis of the differentially expressed genes of endosperm from 2 to 3 DAP.
Supplemental Table S9. GO category enrichment of highly methylated genes at 2 DAP compared with 3 DAP in endosperm.
Supplemental Table S10. Candidate methylation-regulated genes during endosperm cellularization.
Supplemental Table S11. Enrichment analysis of differentially methylated TE/non-TE genes in collinear blocks.
Supplemental Table S12. Reads statistics of Zhonghua11 and 93-11.
Supplementary Material
Glossary
- TE
transposable element
- SINE
short interspersed transposable element
- LTR
long terminal repeat
- MeDIP-seq
methylcytosine immunoprecipitation followed by Illumina sequencing
- ZH11
Zhonghua11
- DAP
days after pollination
- TSS
transcriptional start site
- TES
transcriptional end site
- LINE
long interspersed transposable element
- ABA
abscisic acid
- GO
Gene Ontology
- GEO
Gene Expression Ominibus
- RPKM
reads per kilobase per million mapped reads
- K-S
Kolmogorov-Smirnov
Footnotes
This work was supported by the State Key Project of Basic Research (grant no. 2012CB944804) and the Ministry of Science and Technology of China (grant no. 2012AA10A302).
Articles can be viewed without a subscription.
References
- Agarwal P, Kapoor S, Tyagi AK (2011) Transcription factors regulating the progression of monocot and dicot seed development. BioEssays 33: 189–202 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Kovalchuk I (2008) Epigenetic control of plant stress response. Environ Mol Mutagen 49: 61–72 [DOI] [PubMed] [Google Scholar]
- Brown R, Lemmon B, Olsen OA (1996) Development of the endosperm in rice (Oryza sativa L.): cellularization. J Plant Res 109: 301–313 [Google Scholar]
- Cao P, Jung KH, Choi D, Hwang D, Zhu J, Ronald PC (2012) The Rice Oligonucleotide Array Database: an atlas of rice gene expression. Rice (N Y) 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhou DX (2013) Rice epigenomics and epigenetics: challenges and opportunities. Curr Opin Plant Biol 16: 164–169 [DOI] [PubMed] [Google Scholar]
- Cheng ZJ, Zhao XY, Shao XX, Wang F, Zhou C, Liu YG, Zhang Y, Zhang XS (2014) Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26: 1053–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, Jia Y, Wang GL, Meyers BC, Jacobsen SE, Pellegrini M (2012) Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci USA 109: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Briskine R, Song J, Li Q, Swanson-Wagner R, Hermanson PJ, Waters AJE, Starr E, West PT, Tiffin P, et al. (2013) Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Refy A, Perazza D, Zekraoui L, Valay JG, Bechtold N, Brown S, Hülskamp M, Herzog M, Bonneville JM (2003) The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol Genet Genomics 270: 403–414 [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S (2010) ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol 152: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Xu H, Shen Y, Wang J (2013) Transcriptomic analysis of rice (Oryza sativa) endosperm using the RNA-Seq technique. Plant Mol Biol 81: 363–378 [DOI] [PubMed] [Google Scholar]
- Garcion C, Guilleminot J, Kroj T, Parcy F, Giraudat J, Devic M (2006) AKRP and EMB506 are two ankyrin repeat proteins essential for plastid differentiation and plant development in Arabidopsis. Plant J 48: 895–906 [DOI] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelis T, Bolbach G, Clodic G, Habricot Y, Miginiac E, Sotta B, Jeannette E (2008) Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol 148: 1668–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R (1994) Plant embryogenesis: zygote to seed. Science 266: 605–614 [DOI] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Köhler C (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039 [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y. (2012) Plant imprinted genes identified by genome-wide approaches and their regulatory mechanisms. Plant Cell Physiol 53: 809–816 [DOI] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Berger F (2010) DNA methylation reprogramming during plant sexual reproduction? Trends Genet 26: 394–399 [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, He K, Ma Y, Su N, He H, Stolc V, Tongprasit W, Jin W, Jiang J, et al. (2008) High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20: 259–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, et al. (2012) Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56: 3229–3244 [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38: D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Minns GE, Nesi N, Koumproglou R, Kitsios G, Benning C, Lloyd CW, Doonan JH, Hills MJ (2009) ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 21: 90–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ (2012) MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 13: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2005) Limma: linear models for microarray data. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds, Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Song QX, Lu X, Li QT, Chen H, Hu XY, Ma B, Zhang WK, Chen SY, Zhang JS (2013) Genome-wide analysis of DNA methylation in soybean. Mol Plant 6: 1961–1974 [DOI] [PubMed] [Google Scholar]
- Sørensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jürgens G, Somerville C, Lepiniec L, Berger F (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Borisjuk L, Junker BH, Mock HP, Rolletschek H, Seiffert U, Weschke W, Wobus U (2010) Barley grain development toward an integrative view. Int Rev Cell Mol Biol 281: 49–89 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Wobus U (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol 64: 189–217 [DOI] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Twell D, Park SK, Hawkins TJ, Schubert D, Schmidt R, Smertenko A, Hussey PJ (2002) MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat Cell Biol 4: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Gao Y, Wang J (2012) Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS ONE 7: e30646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue LJ, Zhang JJ, Xue HW (2012) Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PLoS ONE 7: e31081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SR, Yin LL, Xue HW (2013) Functional genomics based understanding of rice endosperm development. Curr Opin Plant Biol 16: 236–246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.