A MADS box gene that affects ethylene signaling also regulates floral organ senescence and abscission.
Abstract
In this study of Arabidopsis (Arabidopsis thaliana), we investigated the relationship between FOREVER YOUNG FLOWER (FYF) and Ethylene Response DNA-binding Factors (EDFs) and functionally analyzed a key FYF target, an Ethylene-Responsive Factor (ERF), that controls flower senescence/abscission. Ectopic expression of EDF1/2/3/4 caused promotion of flower senescence/abscission and the activation of the senescence-associated genes. The presence of a repressor domain in EDFs and the enhancement of the promotion of senescence/abscission in EDF1/2/3/4+SRDX (converting EDFs to strong repressors by fusion with the ERF-associated amphiphilic repression motif repression domain SRDX) transgenic plants suggested that EDFs act as repressors. The significant reduction of β-glucuronidase (GUS) expression by 35S:FYF in EDF1/2/3/4:GUS plants indicates that EDF1/2/3/4 functions downstream of FYF in regulating flower senescence/abscission. In this study, we also characterized an ERF gene, FOREVER YOUNG FLOWER UP-REGULATING FACTOR1 (FUF1), which is up-regulated by FYF during flower development. Ectopic expression of FUF1 caused similar delayed flower senescence/abscission as seen in 35S:FYF plants. This phenotype was correlated with deficient abscission zone formation, ethylene insensitivity, and down-regulation of EDF1/2/3/4 and abscission-associated genes in 35S:FUF1 flowers. In contrast, significant promotion of flower senescence/abscission and up-regulation of EDF1/2/3/4 were observed in 35S:FUF1+SRDX transgenic dominant-negative plants, in which FUF1 is converted to a potent repressor by fusion to an SRDX-suppressing motif. Thus, FUF1 acts as an activator in suppressing EDF1/2/3/4 function and senescence/abscission of the flowers. Our results reveal that FYF regulates flower senescence/abscission by negatively regulating EDF1/2/3/4, which is the downstream gene in the ethylene response, by activating FUF1 in Arabidopsis.
Senescence and abscission of floral organs usually occur after pollination. However, these two processes are not perfectly coupled during flower development, suggesting that both overlapping and independent regulations exist (Rogers, 2013). The processes of flower senescence and abscission, which are triggered by pollination, are primarily controlled by two hormones: ethylene and auxin (Roberts et al., 2002; Rogers, 2013). In Arabidopsis (Arabidopsis thaliana), ethylene accelerates flower senescence/abscission, and mutations in the genes involved in the ethylene signaling pathway, such as ethylene receptor1 (etr1) and ethylene insensitive2 (ein2), cause delayed senescence and abscission (Patterson and Bleecker, 2004; Graham et al., 2012). By contrast, a mutation in the CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) gene causes a constitutive ethylene response and early senescence and abscission of the flowers (Kieber et al., 1993; Huang et al., 2003). Auxin is thought to inhibit senescence and abscission (Roberts et al., 2002; Basu et al., 2013).
Although the key genes involved in the Arabidopsis ethylene signaling pathway have been identified (Chen et al., 2005; Wang et al., 2013), the putative functions of the candidate downstream genes induced by this pathway, such as the Ethylene Response Factor (ERF)/APETALA2 (AP2) transcription factors, which include ERFs and the Ethylene Response DNA-binding Factors (EDFs; Alonso et al., 2003; Stepanova and Alonso, 2005; Castillejo and Pelaz, 2008; Xu et al., 2011; Schaller, 2012), still remain to be investigated. A possible role for EDF as a downstream gene in the ethylene response was supported by several recent reports (Woo et al., 2010; Chen et al., 2011; Lumba et al., 2012; Chang et al., 2013). It has been shown that EDF1/2 was down-regulated in plants that ectopically expressed FOREVER YOUNG FLOWER (FYF), which caused the delay of flower senescence and abscission and ethylene insensitivity (Chen et al., 2011). EDF genes have also been reported to be regulated by FUSCA3 (FUS3), which regulates the vegetative-phase transitions (Lumba et al., 2012). EDF2/4 expression was up-regulated, and the protein stability of a key transcriptional regulator of ethylene signaling, EIN3, was increased in germinating fus3 mutants (Lumba et al., 2012). The increased levels of the EDF transcripts in the fus3 mutants were partially suppressed by treatment with an ethylene signaling inhibitor: silver ions (AgNO3; Lumba et al., 2012). Interestingly, EDF4/RELATED TO ABI3/VP1 1 (RAV1) has been shown to be involved in regulating leaf senescence (Woo et al., 2010). Furthermore, it has been reported that EIN3 could bind to the promoters of EDF1/2/3/4 (Chang et al., 2013). Although these results suggest that the EDF genes may be regulated by FYF, FUS3, and EIN3 in distinct developmental processes (Chen et al., 2011; Lumba et al., 2012; Schaller, 2012; Chang et al., 2013), another functional analysis of the EDF genes is required to determine if they play a role in regulating senescence and abscission or vegetative-phase transitions in the downstream steps of the ethylene response.
In delaying flower senescence and abscission, FYF not only suppresses EDF expression but also, negatively regulates some abscission-associated genes, including BLADE-ON-PETIOLE2 (BOP2) and INFLORESCENCE DEFICIENT IN ABSCISSION (IDA; Chen et al., 2011). BOP1/2 is necessary for the formation of the floral abscission zone (AZ; Hepworth et al., 2005; Norberg et al., 2005; McKim et al., 2008; Wu et al., 2012), whereas IDA proteins interact with the receptor-like kinases HAESA (HAE) and HAESA-LIKE2 (HSL2) to control the abscission initiation of floral organs (Cho et al., 2008; Stenvik et al., 2008; Shi et al., 2011; Liljegren, 2012). The functions of BOP1/2 and IDA are thought to be independent of enzymes, such as polygalacturonase, that may be involved in AZ cell wall degradation (Roberts et al., 2002; Gonzalez-Carranza et al., 2007a, 2012). Thus, FYF inhibits flower senescence and abscission through both ethylene-dependent and -independent pathways (Chen et al., 2011; Rogers, 2013).
To further validate the relation between the EDFs and FYF in the ethylene signaling pathway, a functional analysis of the EDF genes was performed in this study. We showed that EDF1/2/3/4 functioned as a repressor to cause the senescence and abscission in flower organs by suppressing a mechanism that suppresses senescence and abscission in the ethylene signaling pathway. To further delineate the mechanisms regulated by FYF, we identified and investigated an ERF gene (termed FOREVER YOUNG FLOWER UP-REGULATING FACTOR1 [FUF1]; At1g71450) in this study. We showed that ectopically expressing FUF1 caused ethylene insensitivity and delayed the senescence/abscission of the floral organs by suppressing the EDFs, BOP1/2, and IDA/HAE/HSL2/HAWAIIAN SKIRT (HWS). The promotion of flower senescence/abscission in FUF1+SRDX plants, which are transgenic dominant-negative mutants, in which FUF1 is converted to a potent repressor by fusion to an SRDX-suppressing motif (Chen et al., 2011), further supported the idea that FUF1 acts as an activator in promoting a mechanism that suppresses senescence/abscission. Our findings further extend the knowledge regarding the pathway by which FYF suppresses EDF1/2/3/4, the downstream gene in the ethylene signaling pathway, by activating the ERF gene FUF1 in Arabidopsis.
RESULTS
The Detection of EDF Expression during Flower Development
The EDF1/2/3/4 proteins contain ERF/AP2 and B3 DNA-binding domains and belong to the RAV subfamily of ERF/AP2-related proteins (Supplemental Fig. S1; Sakuma et al., 2002; Nakano et al., 2006). To investigate the expression patterns of the EDF1/2/3/4 genes, EDF1/2/3/4:GUS plants were obtained. In contrast with FYF:GUS flowers, in which strong GUS staining was detected in the sepals and petals of young flower buds and weaker staining decreased in young and mature flowers (Chen et al., 2011; Supplemental Fig. S2), the GUS activity of the EDF1:GUS flowers was not detected during the early flower development stages (before stage 8), and their expression increased from stage 9 to the late developmental stages (Supplemental Fig. S3A). Similar GUS staining pattern was observed in plants carrying EDF2:GUS (Supplemental Fig. S3B), EDF3:GUS (Supplemental Fig. S3C), and EDF4:GUS (Supplemental Fig. S3D), respectively. When the expression of EDF1/2/3/4 in flowers in different developmental stages was further analyzed, significantly higher expression of EDF1/2/3/4 was observed in late flower development (after stage 12) than in early development stages (before stage 9; Supplemental Fig. S3, E–H). These results indicated that the temporal expression patterns of EDF1/2/3/4 were the opposite of those patterns of FYF during flower development.
The Ectopic Expression of EDFs Promotes Flower Senescence and Abscission in Transgenic Arabidopsis Plants
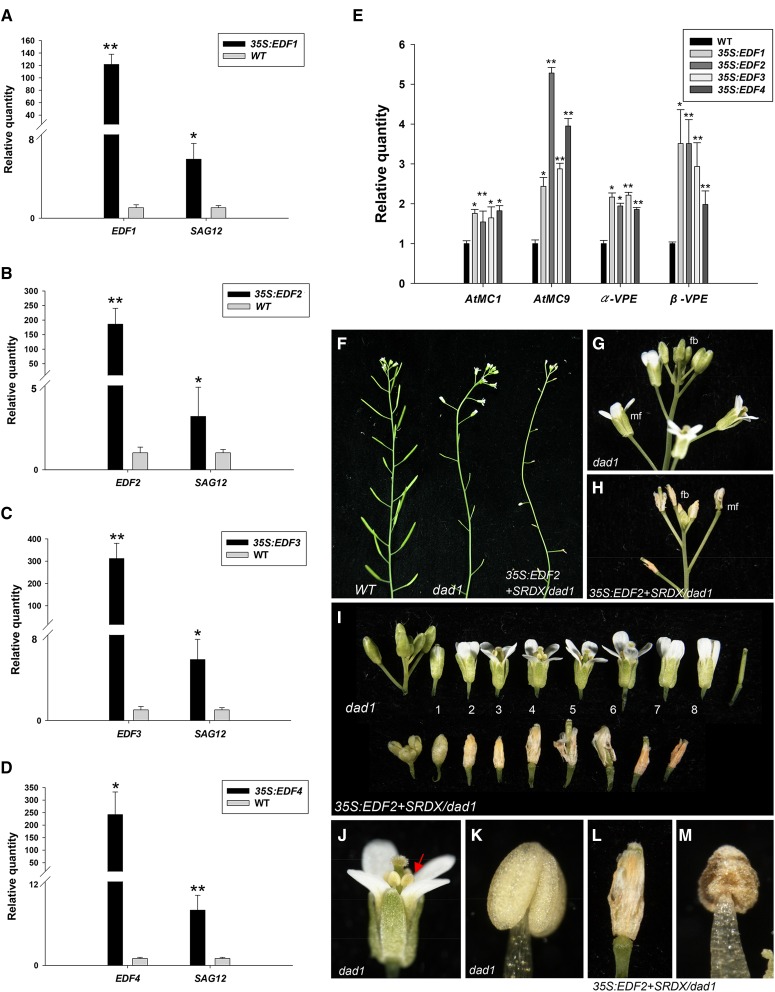
To investigate the functions of the EDF genes, the 35S:EDF1/2/3/4 was transformed into Arabidopsis. Of 140 kanamycin-resistant plants analyzed for each construct, 47, 59, 59, and 43 plants displayed flower senescence and abscission promotion for 35S:EDF1, 35S:EDF2, 35S:EDF3, and 35S:EDF4, respectively (Supplemental Table S1). The perianths of wild-type flowers abscised at approximately position 5 (Fig. 1, A and B), whereas the perianth organs of the 35S:EDF1/2/3/4 flowers senesced and abscised at positions 3 and 4 (Fig. 1, A, C, and D). The position was counted starting from the first flower with visible white petals at the top of the inflorescence (Bleecker and Patterson, 1997), and flower organ senescence and abscission were measured as described previously (Chen et al., 2011). Although the 35S:EDF1/2/3/4 perianth organs were completely senescent and abscised from the base of the flower after positions 3 and 4, these organs remained hanging on the siliques (Fig. 1A), even after these organs had been abscised from the flowers for 2 weeks (Fig. 1, A, C, and D). This phenotype resembled that observed in the ctr1 mutants (Fig. 1F), which have been shown to cause a constitutive ethylene response.
Figure 1.
The ectopic expression of the EDFs and the EDF+SRDX constructs promotes flower senescence and abscission. A, The flowers along the inflorescence of the wild-type (WT; row 1), 35S:EDF1 (row 2), 35S:EDF2 (row 3), 35S:EDF3 (row 4), and 35S:EDF4 (row 5) plants. The numbers indicate the positions of the flowers. B to D, The perianth organs of the wild-type flowers (B) senesced and abscised after position 5 (arrow), whereas the perianth organs were completely senescent and abscised after position 3 (arrows) in 35S:EDF2 (C) and 35S:EDF4 (D) flowers. E, A schematic representation of the EDF+SRDX constructs. The red box indicates the conserved repressor sequence (RLFGV), and the green box indicates the SRDX domain (LDLDLELRLGFA). NLS, Nucleus localization signal. F, The flowers along the inflorescences of ctr1 (row 1), 35S:EDF1+SRDX (row 2), 35S:EDF2+SRDX (row 3), 35S:EDF3+SRDX (row 4), and 35S:EDF4+SRDX (row 5) plants. G and H, The inflorescence stems of 35S:EDF1+SRDX (G) and 35S:EDF4+SRDX (H) plants. The perianth organs were completely senescent and abscised after position 2 (arrows) flowers.
35S:EDFs+SRDX Transgenic Arabidopsis Exhibits Enhanced Promotion of Flower Senescence and Abscission
When the sequences of EDF1/2/3/4 proteins were analyzed, a conserved RLFGV sequence, similar to that of the ERF/AP2 repressors (Ikeda and Ohme-Takagi, 2009), was identified in the C-terminal region (Fig. 1E; Supplemental Fig. S1). The presence of this sequence indicated that the EDF1/2/3/4 genes might encode transcriptional repressors. To examine this assumption, a conserved SRDX-suppressing motif that contains a 12-amino acid repressor sequence (LDLDLELRLGFA; Chen et al., 2011) was fused to EDF1/2/3/4 (EDF1/2/3/4+SRDX; Fig. 1E). We expected that the overexpression of EDF1/2/3/4+SRDX would enhance the repression, resulting in a more severe phenotype than that observed in the 35S:EDF1/2/3/4 plants.
Of the 180 kanamycin-resistant plants analyzed for each construct, 52, 60, 68, and 59 plants showed the same abnormal phenotype for 35S:EDF1+SRDX, 35S:EDF2+SRDX, 35S:EDF3+SRDX, and 35S:EDF4+SRDX, respectively (Supplemental Table S2). Interestingly, senescence and abscission of the perianths of the 35S:EDF1/2/3/4+SRDX plants (Fig. 1F) were enhanced compared with plants carrying 35S:EDF1/2/3/4 (Fig. 1A). The perianths of the 35S:EDF1/2/3/4+SRDX flowers senesced and abscised at positions 2 and 3 (Fig. 1, F–H), which were earlier than in 35S:EDF1/2/3/4 flowers (position 3 and 4) or wild-type flowers (position 5; Fig. 1A). The completely senescent perianth organs that abscised from the base of the flowers after position 2 remained hanging on the 35S:EDF1/2/3/4+SRDX siliques (Fig. 1, F–H) for several weeks, such as in the ctr1 mutants (Fig. 1F). These results support the hypothesis that EDF1/2/3/4 plays a repressor role in regulating flower senescence and abscission.
Senescence-Associated Gene12 and Programmed Cell Death-Associated Gene Expression Levels Are Up-Regulated in Plants Carrying 35S:EDFs and 35S:EDFs+SRDX
Because flower senescence and abscission were promoted in 35S:EDFs+SRDX transgenic Arabidopsis, thus, we analyzed the expression of senescence marker gene senescence-associated gene12 (SAG12; Noh and Amasino, 1999) and the marker genes for programmed cell death (PCD), such as vacuolar processing enzymes (VPEs; with caspase-1 activity) and Metacaspase (MCs) genes (Rojo et al., 2004; Wagstaff et al., 2009; Coll et al., 2010) in 35S:EDFs and 35S:EDFs+SRDX plants. The result indicated that the expressions of SAG12, AtMC1, AtMC9, α-VPE, and β-VPE were significantly up-regulated in 35S:EDFs (Fig. 2, A–E) and 35S:EDF1/2/3/4+SRDX plants (Supplemental Fig. S4, A–E). Thus, 35S:EDFs promoted the senescence and abscission of the flower organs by activating senescence- and PCD-associated genes.
Figure 2.
The detection of senescence-associated gene expression in 35S:EDF plants and a phenotypic analysis of flower senescence and abscission in dad1 plants ectopically expressing the EDF2+SRDX gene. A to D, The expression of the SAG12 gene in 35S:EDF1 (A), 35S:EDF2 (B), 35S:EDF3 (C), and 35S:EDF4 (D) plants. E, The expression of AtMC1, AtMC9, α-VPE, and β-VPE genes in 35S:EDF plants. The mRNA levels were determined by real-time quantitative PCR. Total RNA was isolated from the floral buds and the open flowers at positions 1 to 3 from one wild-type (WT) Col-0 plant and one 35S:EDFs plant. The transcript levels of these genes were determined using two to three replicates and normalized using UBQ10. The expression of each gene is given relative to that of the wild-type plant, which was set at one. The asterisks in A to E indicate a significant difference from the wild-type value. *, P ≤ 0.05; **, P ≤ 0.01. F, The inflorescences of wild-type (left), dad1 (center), and 35S:EDF2+SRDX/dad1 (right) plants. No silique elongation was observed in dad1 and 35S:EDF2+SRDX/dad1 plants. G and H, Close-up of the flower buds (fbs) and mature flowers (mfs) for dad1 (G) and 35S:EDF2+SRDX/dad1 (H) plants from F. I, The flowers along an inflorescence of the dad1 (upper) and 35S:EDF2+SRDX/dad1 (lower) plants. The perianth organs did not senesce and abscised after position 8 for dad1 mutants. The numbers indicate the positions of the flowers. The perianth organs of the flowers in 35S:EDF2+SRDX/dad1 plants were completely senescent and abscised at positions 2 and 3. J and K, Close-up view of a mature flower at position 4 (J) and an indehiscent anther (K) for the dad1 plant from I. An arrow indicates the indehiscent anthers in J. L and M, Close-up view of a mature flower at position 4 (L) and an indehiscent anther (M) of the 35S:EDF2+SRDX/dad1 plant from I. Expression statistical analysis was measured by Student’s t test (each n = 3).
35S:EDF2+SRDX Promotes Flower Senescence and Abscission Independent of the Male Sterility Phenotype in defective in anther dehiscence1 Mutants
The ectopic expression of EDF1/2/3/4 clearly affects flower organ senescence and abscission and seems to render plants sterile, because delayed or no silique elongation was also observed in 35S:EDFs and 35S:EDFs+SRDX transgenic plants (Fig. 1, A and F). If the ectopic expression of EDF1/2/3/4 does affect fertility, then this expression might have a consequence on the timing of floral organ senescence and abscission. To exclude this possibility, we generated transgenic plants containing 35S:EDF2+SRDX in male sterility mutant defective in anther dehiscence1 (dad1), which has defects in anther dehiscence (Fig. 2, F, J, and K; Ishiguro et al., 2001). The senescence/abscission of the dad1 flowers was slightly delayed compared with the senescence/abscission of wild-type flowers (Fig. 2, F, G, and I). The result indicated that, in addition to producing a male sterility phenotype (Fig. 2, F, L, and M), a significant promotion of senescence and abscission was observed in 35S:EDF2+SRDX/dad1 flowers (Fig. 2, H, I, and L), similar to that observed for 35S:EDF2+SRDX transgenic plants (Fig. 1F). This result strongly indicated that the 35S:EDF-induced early senescence/abscission of the flower organs was not caused by the effect of the male sterility.
35S:EDFs+SRDX Constructs Promote Flower Senescence and Abscission in the etr1 and ein2 Mutants
To further confirm the relation between EDF1/2/3/4 and the ethylene signaling pathway, the 35S:EDF1/2/3/4+SRDX constructs were separately transformed into the Arabidopsis ethylene-insensitive mutants etr1 and ein2, in which floral senescence and abscission are delayed to approximately position 8 (Supplemental Figs. S5A and S6A; Bleecker et al., 1988; Ecker, 1995; Patterson and Bleecker, 2004). The phenotypes of 35S:EDF1/2/3/4+SRDX/etr1-1 plants resembled the phenotypes of the 35S:EDF1/2/3/4+SRDX plants. These plants displayed earlier senescence/abscission of the perianth organs (positions before 3 and 4) than the senescence/abscission of the etr1-1 mutants (position 8; Supplemental Fig. S5, A–C; Supplemental Table S3). The promotion of floral senescence and abscission in 35S:EDF1/2/3/4+SRDX/etr1-1 correlated with the expression of EDF1/2/3/4+SRDX (Supplemental Fig. S5, D–G). Similarly, earlier senescence/abscission of the perianth organs was observed (positions before 3–5) for 35S:EDF1/2/3/4+SRDX/ein2-1 than for the ein2-1 mutants (position 8; Supplemental Fig. S6, A–C; Supplemental Table S4). The promotion of floral senescence/abscission for 35S:EDF1/2/3/4+SRDX/ein2-1 also correlated with the expression of EDF1/2/3/4+SRDX (Supplemental Fig. S6, D–G). This promotion further indicated that EDF1/2/3/4+SRDX functions downstream of ETR1 and EIN2 in regulating flower senescence/abscission.
35S:EDFs+SRDX Is Epistatic to 35S:FYF in Regulating Flower Senescence and Abscission
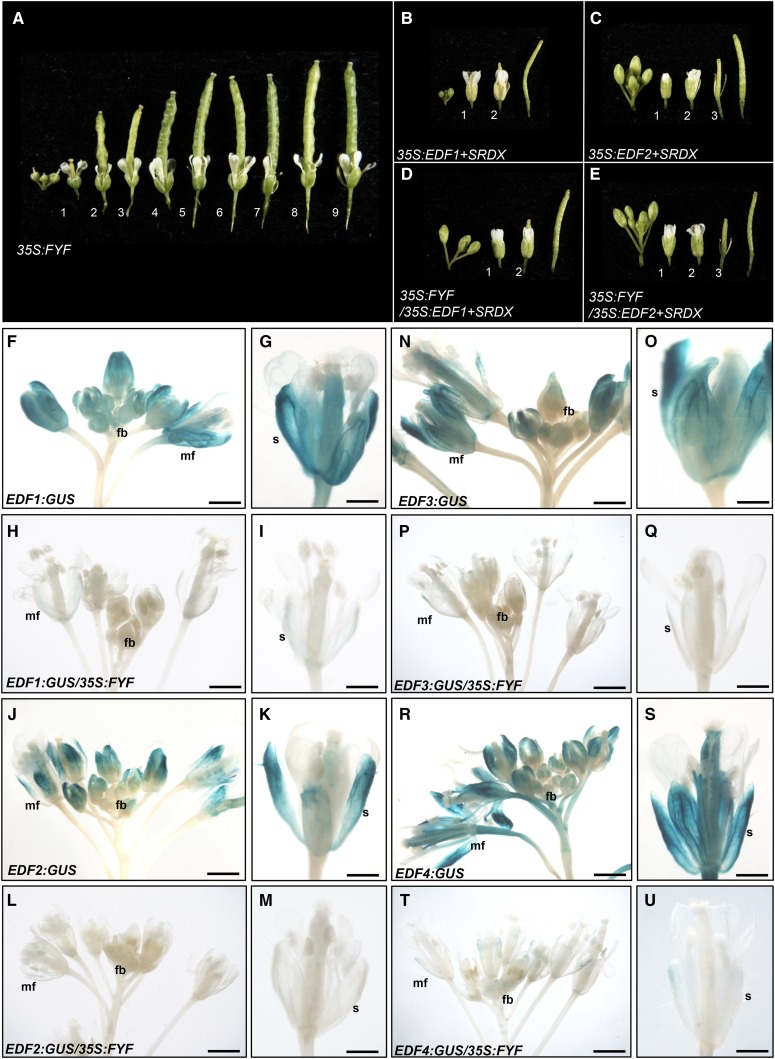
To confirm the relationship between EDF1/2/3/4 and FYF, transgenic plants containing both 35S:EDF1/2+SRDX and 35S:FYF were generated by genetic crossing. In the 35S:FYF Arabidopsis, floral senescence/abscission was delayed until after position 9 (Fig. 3A; Chen et al., 2011). The phenotype of 35S:FYF/35S:EDF1/2+SRDX resembled that of the 35S:EDF1/2+SRDX plants (Fig. 3, B and C); they displayed early senescence/abscission of the perianth organs (positions before 3; Fig. 3, D and E), which was earlier than that in wild-type flowers (position 5; Fig. 1A). This promotion indicated that 35S:EDF1/2+SRDX is epistatic to 35S:FYF in regulating flower senescence/abscission in the ethylene-insensitive response.
Figure 3.
A phenotypic analysis of flower senescence and abscission in 35S:FYF plants ectopically expressing the EDF1,2+SRDX genes and GUS staining patterns in EDFs:GUS plants ectopically expressing 35S:FYF. A, The perianth organs did not senesce and abscised after position 9 of the 35S:FYF inflorescence. B to E, Inflorescence stems of 35S:EDF1+SRDX (B), 35S:EDF2+SRDX (C), 35S:FYF/35S:EDF1+SRDX (D), and 35S:FYF/35S:EDF2+SRDX (E) plants. The perianth organs of the flowers in these four plants were completely senescent and abscised at positions 2 and 3. F to U, GUS staining was weak in the flower buds (fbs) and stronger in the mature flowers (mfs) of EDF1:GUS (F and G), EDF2:GUS (J and K), EDF3:GUS (N and O), and EDF4:GUS (R and S) Arabidopsis. GUS staining was undetectable in the fbs and mature mfs of EDF1:GUS/35S:FYF (H and I), EDF2:GUS/35S:FYF (L and M), EDF3:GUS/35S:FYF (P and Q), and EDF4:GUS/35S:FYF (T and U) Arabidopsis. G, I, K, M, O, Q, S, and U are stage 13 mature flowers from F, H, J, L, N, P, R, and T, respectively. s, Sepal. Bars = 2 mm (F, H, J, L, N, P, R, and T) and 1 mm (G, I, K, M, O, Q, S, and U).
35S:FYF Significantly Suppressed the GUS Activity in EDFs:GUS Plants
To clearly show that the FYF negatively regulated the expression of the EDFs during flower development, transgenic plants containing both EDF1/2/3/4:GUS and 35S:FYF were generated. The result clearly indicated that a significant reduction in the expression of GUS was observed in EDF1/2/3/4:GUS/35S:FYF plants. In contrast with EDF1:GUS flowers, in which strong GUS staining was detected in the sepals and petals of mature flowers (Fig. 3, F and G), GUS activity was almost undetectable in the EDF1:GUS/35S:FYF flower buds and mature flowers (Fig. 3, H and I). Similarly, compared with the high GUS activity in the mature flowers of EDF2:GUS (Fig. 3, J and K), EDF3:GUS (Fig. 3, N and O), and EDF4:GUS (Fig. 3, R and S), GUS staining was undetected in the floral buds and mature flowers of plants carrying EDF2:GUS/35S:FYF (Fig. 3, L and M), EDF3:GUS/35S:FYF (Fig. 3, P and Q), and EDF4:GUS/35S:FYF (Fig. 3, T and U). This result strongly indicated that EDF1/2/3/4 was negatively regulated by FYF during flower development.
The Identification of FYF-Regulated Genes by a Microarray Analysis
To further identify the downstream genes regulated by FYF, FYF was fused to the glucocorticoid receptor (GR; 35S:FYF+GR) and transformed into Arabidopsis. The 35S:FYF+GR plants grew normally and were indistinguishable from wild-type plants without dexamethasone (DEX) treatment (Supplemental Fig. S7, A and B). Upon DEX treatment, the 35S:FYF+GR plants flowered early (Supplemental Fig. S7A) and exhibited delayed flower senescence and abscission (Supplemental Fig. S7, C and D), similar to the 35S:FYF plants (Chen et al., 2011). This result indicated that this inducible strategy can be successfully used to study the genes regulated by FYF.
Thus, we performed a microarray analysis using the RNA extracted from the flowers of DEX-treated (3 h) and untreated 35S:FYF+GR Arabidopsis; 107 up-regulated (2-fold or greater) and 10 down-regulated (0.5-fold or less) genes with P values less than 0.05 were identified in the DEX-treated plants (Supplemental Tables S5 and S6). Among 107 up-regulated genes, five ERFs—AtERF19 (At1g22810), AtERF21 (At1g71450), AtERF95 (At3g23220), AtERF96 (At5g43410), and AtERF98 (At3g23230)—were identified (Supplemental Fig. S8, A and B; Supplemental Table S5). Because EDF1-4 was not detected in this array experiment, this indicated that these EDFs are likely the indirect targets for FYF gene.
The Analysis of FUF1 Expression
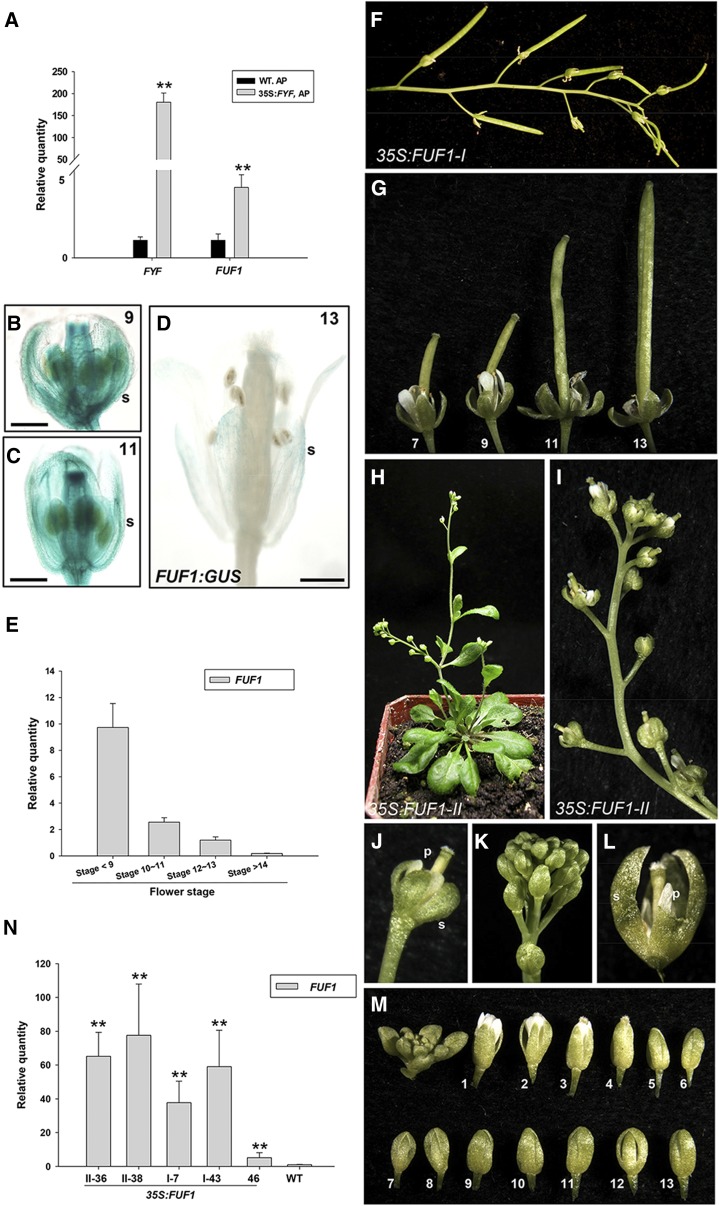
The ERF gene AtERF21 (At1g71450), termed FUF1, is up-regulated by FYF (Fig. 4A; Supplemental Table S5) and was further characterized. Additional analysis indicated that a CArG box was found in the promoter (position −240) of the FUF1 gene. The FUF1 protein contains an ERF/AP2 domain, which is conserved among the ERF/AP2-related proteins (Supplemental Fig. S8A; Sakuma et al., 2002; Nakano et al., 2006). To investigate the expression pattern of the FUF1 gene, an FUF1:GUS construct was generated and transformed into Arabidopsis. Similar to the GUS staining observed in FYF:GUS flowers (Supplemental Fig. S2), strong GUS staining was detected in sepals and petals of the flower buds and young flowers (Fig. 4, B and C) and significantly decreased in mature flowers after stage 13 of FUF1:GUS plants (Fig. 4D). When FUF1 expression was further analyzed in flowers at different developmental stages, significantly higher FUF1 expression was observed during early flower development (before stage 9) than during late developmental stages (after stage 12; Fig. 4E). These results indicate that the spatial and temporal expression pattern of FUF1 is similar to that for FYF during flower development.
Figure 4.
The ectopic expression of FUF1 delays flower senescence and abscission. A, The expression levels of FYF and FUF1 in flowers after stage 12 and pollination (AP) in wild-type (WT) and 35S:FYF plants as determined by real-time quantitative RT-PCR. **, Significant difference from the wild-type value (P ≤ 0.01). B and C, GUS staining was strong in the flower buds at stages 9 (B) and 11 (C) of FUF1:GUS Arabidopsis. s, Sepal. Bars = 0.5 mm (B), 1 mm (C). D, The GUS staining decreased in the stage 13 mature flowers of FUF1:GUS Arabidopsis. Bar = 1 mm. E, The detection of FUF1 expression in wild-type flowers at four different developmental stages (<9, 10–11, 12–13, and >14). The mRNA levels were determined by real-time quantitative PCR. F and G, An inflorescence (F) and flowers along the inflorescence (G) of a 35S:FUF1 type I plant. The perianth organs of the 35S:FUF1 flowers did not senesce and remained attached to the bases of the siliques after position 13. H and I, A 40-d-old 35S:FUF1 type II plant (H) and a close-up image of a type II inflorescence (I). The perianth organs of this 35S:FUF1 type II plant did not senesce and remained attached to the bases of the siliques in positions greater than 13. J, A close-up image of a 35S:FUF1 type II flower from I that contained short s and petal (p) and that did not senesce or abscise. K, A close-up image of the flowers in a 35S:FUF1 type II inflorescence. These flowers failed to open and did not senesce for a long period. L, Close-up image of a type II 35S:FUF1 flower from K that was manually opened. M, The flowers along an inflorescence of a 35S:FUF1 type II plant failed to open, and the perianth organs did not senesce or abscise even after position 13. N, The detection of FUF1 expression in 35S:FUF1 plants. The mRNA levels were determined by real-time quantitative PCR. Total RNA was isolated from one wild-type Col-0 plant, two type II plants (36 and 38), two type I plants (7 and 43), and one 35S:FUF1 plant (46) with a wild-type-like phenotype. Expression statistical analysis was measured by Student’s t test (each n = 3). **, Significant difference from the wild-type value (P ≤ 0.01).
The Ectopic Expression of FUF1 Delays Flower Senescence and Abscission in Transgenic Arabidopsis Plants
To investigate the function of FUF1, 35S:FUF1 was transformed into Arabidopsis; 80 of 200 35S:FUF1 plants showed delayed flower senescence/abscission. These plants were divided into two classes based on the degree of phenotypic alteration.
In type I plants, the delay of senescence/abscission of the perianth (Fig. 4, F and G) was very similar to that observed in 35S:FYF flowers (Fig. 3A; Chen et al., 2011). The perianth organs of these 35S:FUF1 transgenic flowers did not senesce and remained attached at the bases of the siliques for longer than position 13 (Fig. 4, F and G). Furthermore, the elongation of the siliques and the maturation of the seeds in these type I plants (Fig. 4, F and G) were normal, such as in 35S:FYF plants. In type II plants, the delay of senescence/abscission of the perianths (Fig. 4, H–M) was enhanced compared with type I and 35S:FYF flowers. These type II flowers produced short sepals and petals (Fig. 4, I and J), failed to open (Fig. 4, K–M) during almost the entire flower development, and did not senesce for longer than position 13 (Fig. 4, I and M). When these flowers were opened manually, we observed that the petals and stamens had failed to elongate (Fig. 4L). Interestingly, the enhancement of the delay in senescence/abscission of the perianths and the formation of the short sepals and petals in these type II flowers were similar to the phenotypes observed in the severe 35S:FYF+SRDX flowers (Chen et al., 2011). As shown in Figure 4N, relatively higher FUF1 expression was observed in type II plants compared with type I plants.
The Formation of AZ Is Deficient in 35S:FUF1 Flowers
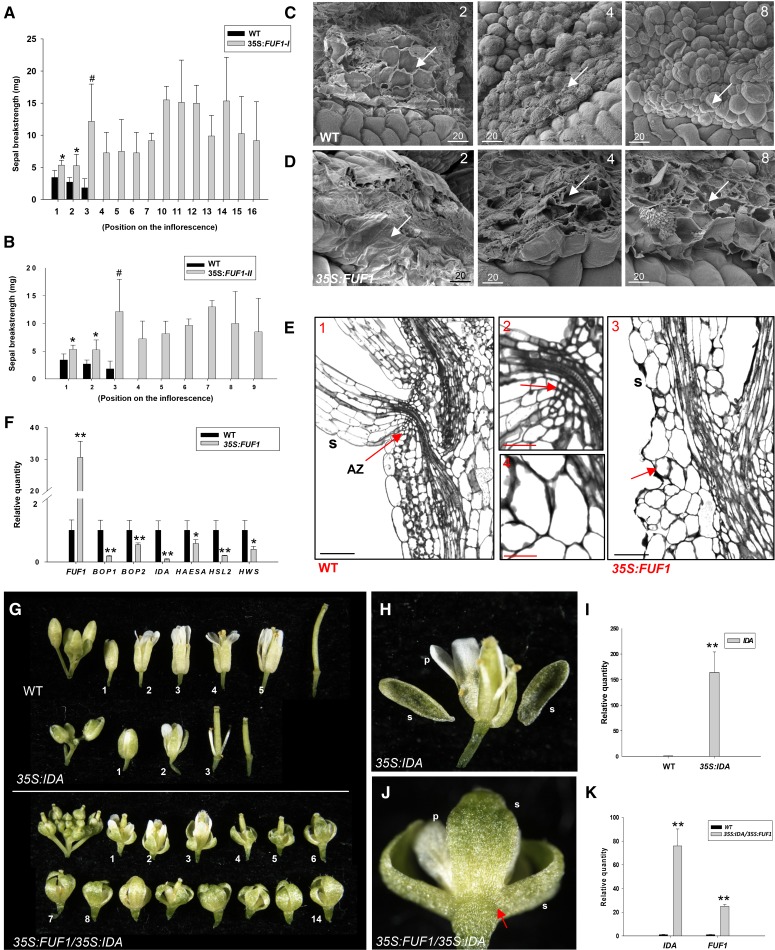
A break-strength meter was used to quantitatively measure the force required to remove the sepals from the plant as described previously (Chen et al., 2011). As shown in Figure 5A, we can only analyze the break strength of wild-type sepals at position 3. In contrast, the break strength of type I 35S:FUF1 sepals could be detected in the flowers at position 18 (Fig. 5A). In type II 35S:FUF1 plants, the sepals also could be detected in all nine flowers in the inflorescence (Fig. 5B).
Figure 5.
The detection sepal break strength, the morphology of the AZs, and the gene expression for 35S:FUF1 plants as well as a phenotypic analysis of flower senescence and abscission in 35S:FUF1 plants ectopically expressing the IDA gene. A and B, Sepal break strength measured from positions 1 to 3 for wild-type (WT), positions 1 to 16 for 35S:FUF1 type I (A), and positions 1 to 9 for 35S:FUF1 type II (B) plants along the inflorescence. C and D, SEM image of Arabidopsis sepal AZs (arrows) in wild-type (C) and 35S:FUF1 (D) flowers at positions 2, 4, and 8. Bars = 20 µm. E, Thin sections of embedded flowers at position 4 for wild-type flower (E, 1 and 2) and 35S:FUF1 (E, 3 and 4). E, 2 and 4 are close-up images of E, 1 and 3, respectively. Arrows indicate the positions of the AZ of the sepals (s). The small AZ cells were not formed in the 35S:FUF1 s. Bars = 25 μm (E, 1), 10 μm (E, 2), 50 μm (E, 3), and 20 μm (E, 4). F, The expression levels for FUF1, BOP1, BOP2, IDA, HAESA, HSL2, and HWS in wild-type and 35S:FUF1 flowers using real-time quantitative RT-PCR. G, The flowers along an inflorescence of the wild-type, 35S:IDA, and 35S:FUF1/35S:IDA plants. The perianth organs of the flowers in 35S:IDA plants were abscised at positions 2 and 3. The perianth organs did not senesce and abscised after position 14 for 35S:FUF1/35S:IDA. The numbers indicate the positions of the flowers. H, Close-up view of a flower at position 2 for the 35S:IDA plant from G. The s and petals (p) were abscised and easily dropped by a gentle touch. I, The expression levels for IDA in wild-type and 35S:IDA flowers using real-time quantitative RT-PCR. J, Close-up view of a flower at position 14 for the 35S:FUF1/35S:IDA plant from G. The s and p were not abscised and remained firmly attached to the base of the flowers (arrow). K, The expression levels for IDA and FUF1 in wild-type and 35S:FUF1/35S:IDA flowers using real-time quantitative RT-PCR. Expression statistical analysis was measured by Student’s t test (each n = 3). Symbols in A, B, F, I, and K indicate a significant difference from the wild type value. *, P ≤ 0.05; **, P ≤ 0.01; #, 0.1 ≤ P ≤ 0.05.
Scanning electron microscopy (SEM) was used to further examine whether a normal sepal AZ developed in the 35S:FUF1 flowers. Upon sepal removal at position 2 in wild-type and 35S:FUF1 flowers, broken cells were observed at the fracture planes of the sepal AZs (Fig. 5C, 2 and 5D, 2). At position 4, wild-type AZ cells began to display an initial rounded appearance without any obviously ruptured cells (Fig. 5C, 4). At position 8, wild-type AZ cells were fully round (Fig. 5C, 8). In contrast, the broken cells at the fracture plane could still be observed in the 35S:FUF1 flowers after the sepal removal at positions 4 and 8 (Fig. 5D, 4 and 8). This pattern indicated that the timing of cell separation was significantly delayed in the AZ of the 35S:FUF1 flowers.
To further examine the deficiency of abscission in 35S:FUF1, thin sections of embedded flowers were generated and analyzed. In the wild-type flower at position 4, small differentiated cells were clearly formed in the AZ of the sepals (Fig. 5E, 1 and 5E, 2). In contrast, the cells were not differentiated, and the AZ was not formed in the 35S:FUF1 sepal (Fig. 5E, 3 and 5E, 4). This result indicated that the deficiency of abscission in 35S:FUF1 was clearly caused by a deficiency or significant delay in floral AZ formation.
BOP1/2, IDA, HAE, HSL2, and HWS Expression Levels Are Down-Regulated in Plants Carrying 35S:FUF1
BOP1 and BOP2 are required for the formation of the floral AZ (McKim et al., 2008). The expression levels of BOP1/2 in wild-type and 35S:FUF1 flowers were analyzed. The results indicated that BOP1/2 was significantly down-regulated in the 35S:FUF1 flowers compared with the wild-type flowers (Fig. 5F). Thus, FUF1 likely functions upstream of BOP1/2 to negatively regulate AZ formation and flower abscission.
IDA has been proposed to interact with the receptor-like kinases HAESA and HSL2 to regulate flower organ abscission (Cho et al., 2008; Stenvik et al., 2008). A mutation in HWS also displays defects in organ abscission (Gonzalez-Carranza et al., 2007b). We thus analyzed the expression levels of IDA/HAESA/HSL2/HWS in wild-type and 35S:FUF1 flowers. These four genes were all significantly down-regulated in the 35S:FUF1 flowers compared with the wild-type flowers (Fig. 5F). Thus, FUF1 likely negatively regulates IDA/HAESA/HSL2/HWS to control flower abscission.
35S:FUF1 Delays Flower Senescence and Abscission in Plants Carrying 35S:IDA
To further explore the relation between FUF1 and IDA, the 35S:FUF1 construct was transformed into plants carrying the 35S:IDA construct. The abscission of flower organs was clearly promoted at positions 2 and 3 in the 35S:IDA transgenic plant (Fig. 5, G and H). This early abscission correlated with high levels of IDA gene expression in the transgenic plant (Fig. 5I). In contrast, 35S:IDA/35S:FUF1 double transgenic plants, which highly expressed both IDA and FUF1 (Fig. 5K), displayed a significant delay in flower organ abscission (Fig. 5, G and J), similar to that observed in 35S:FUF1 transgenic plants. This result indicated that the failure of AZ formation in 35S:FUF1 flowers prevented the abscission of the flower organs, even with the ectopic expression downstream of abscission-associated genes, such as IDA.
35S:FUF1 Arabidopsis Plants Are Insensitive to Ethylene Treatment
In the presence of air, the leaves remained green (Fig. 6, C and D, left), and the perianth organs remained attached to the flowers until position 5 in wild-type plants (Fig. 6A, upper) but did not senesce or abscise in the 35S:FUF1 transgenic plants (Fig. 6B, upper). When ethylene was added to wild-type plants, the leaves senesced (Fig. 6C, right), and the perianth organs abscised early (at position 2; Fig. 6A, lower). In contrast, in the presence of ethylene, the leaves of the 35S:FUF1 transgenic plants remained green (Fig. 6D, right; Supplemental Fig. S9), and the perianth organs were turgid and remained on the flowers (Fig. 6B, lower). These results indicated that the delay of senescence and abscission of the flower organs in the 35S:FUF1 Arabidopsis was unaffected by the ethylene treatment.
Figure 6.
An analysis of the effect of ethylene on 35S:FUF1 Arabidopsis and a phenotypic analysis of flower senescence and abscission in etr1-1, ein2-1, ctr1-1, and 35S:EDFs+SRDX plants ectopically expressing the FUF1 gene as well as in FUF1+SRDX plants. A and B, A comparison of wild-type (WT; A) and 35S:FUF1 (B) flowers exposed to either air (upper) or ethylene (lower). C and D, A rosette leaf from a wild-type plant (C, right) was senescent; however, a leaf from a 35S:FUF1 plant (D, right) remained green after ethylene treatment. E, The inflorescences of wild-type (left), etr1-1 (center), and 35S:FUF1/etr1-1 (right) plants. The floral organs (arrows) remained on the flowers and siliques of the etr1-1 and 35S:FUF1/etr1-1 plants until approximately positions 7 and 9, respectively. F, Close-up view of the flower at position 9 for the 35S:FUF1/etr1-1 plant from E. p, Petal; s, sepal. G, The inflorescences of wild-type (left), ein2-1 (center), and 35S:FUF1/ein2-1 (right) plants. The floral organs (arrows) remained on the flowers and siliques of the ein2-1 and 35S:FUF1/ein2-1 plants until approximately positions 8 and 18, respectively. H, Close-up view of the flower at position 18 for the 35S:FUF1/ein2-1 plant from G. I, Adult ctr1-1 (left), 35S:FUF1/ctr1-1 (center), and wild-type (right) plants. J, An inflorescence of a ctr1-1 mutant. The floral organs senesced (arrow) at an early stage. K, An inflorescence of a 35S:FUF1/ctr1-1 plant. The floral organs were green and remained on the flowers. L, The flowers along inflorescences of ctr1-1 (upper) and 35S:FUF1/ctr1-1 (lower) plants. The perianths of the 35S:FUF1/ctr1-1 flowers were green and displayed no signs of senescence or abscission at the late stages. M, The expression levels of FUF1, EDF1, EDF2, EDF3, and EDF4 in wild-type and 35S:FUF1 flowers using real-time quantitative RT-PCR. N, The flowers along the inflorescences of plants carrying 35S:FUF1 (type II plant; N, 1), 35S:FUF1/35S:EDF1+SRDX (N, 2), 35S:FUF1/35S:EDF2+SRDX (N, 3), 35S:FUF1/35S:EDF3+SRDX (N, 4), 35S:FUF1/35S:EDF4+SRDX (N, 5), 35S:FUF1+SRDX (N, 6), and FUF1:FUF1+SRDX (N, 7). O, The expression levels of FUF1, EDF1, EDF2, EDF3, and EDF4 in wild-type, 35S:FUF1+SRDX, and FUF1:FUF1+SRDX flowers using real-time quantitative RT-PCR. Expression statistical analysis was measured by Student’s t test (each n = 3). The asterisks in M and O indicate a significant difference from the wild-type value. *, P ≤ 0.05; **, P ≤ 0.01; #, 0.1 ≤ P ≤ 0.05.
35S:FUF1 Enhances the Delay of Flower Senescence and Abscission in etr1, ein2, and ctr1 Mutants
To further confirm the relation between FUF1 and the ethylene signaling pathway, the 35S:FUF1 construct was transformed into the Arabidopsis mutants etr1, ein2, and ctr1. The phenotypes of the 35S:FUF1/etr1-1 and 35S:FUF1/ein2-1 plants resembled the phenotype of the 35S:FUF1 plants; these plants displayed delayed senescence/abscission of the perianth organs at positions greater than 9 for 35S:FUF1/etr1-1 (Fig. 6, E and F) or greater than 18 for 35S:FUF1/ein2-1 (Fig. 6, G and H). This was later than that in the untransformed etr1-1 and ein2-1 mutants, which abscised at positions 7 and 8, respectively (Fig. 6, E and G, center). This delay indicated that 35S:FUF1 functions downstream of ETR1 and EIN2 in the ethylene-insensitive response.
Mutations in the CTR1 gene caused a constitutive ethylene response and early senescence/abscission of the flowers (Fig. 6, J and L, upper; Kieber et al., 1993; Huang et al., 2003). The size of the 35S:FUF1/ctr1-1 plants (Fig. 6I, center) was also reduced and resembled that of the untransformed ctr1-1 mutants (Fig. 6I, left). However, the senescence/abscission of the perianth organs was delayed in these plants (Fig. 6, K and L, lower), which was observed for the 35S:FUF1 plants. This delay indicates that 35S:FUF1 functions downstream of CTR1 and was epistatic to ctr1 mutants in regulating flower senescence/abscission.
FUF1 Inhibits EDF Genes in the Ethylene Response
To further investigate the involvement of the FUF1 gene in ethylene signaling, the expression levels of EDFs were analyzed in 35S:FUF1 plants. EDF1, EDF3, and EDF4 were clearly down-regulated in the 35S:FUF1 plants (Fig. 6M). These results indicated that the delayed flower senescence/abscission of the 35S:FUF1 plants was because of the FUF1-mediated inhibition of the downstream genes in the ethylene response.
To further confirm the relationship between FUF1 and the EDFs, transgenic plants containing both 35S:EDF1/2/3/4+SRDX and 35S:FUF1 were generated by genetic crossing. Floral senescence/abscission was delayed to later than position 10 (Fig. 6N, 1) in 35S:FUF1 plants. By contrast, 35S:FUF1/35S:EDF1/2/3/4+SRDX plants displayed early senescence/abscission of the perianth organs at positions less than 3 (Fig. 6N, 2–6N, 5), which is the same phenotype as the 35S:EDF1/2/3/4+SRDX plants (Fig. 1F). This phenotype confirmed that EDF1/2/3/4+SRDX functions downstream of FUF1 in the ethylene response.
Flower Senescence and Abscission Are Significantly Promoted, and EDFs Are Up-Regulated in the FUF1+SRDX Transgenic Dominant-Negative Plants
To determine if any phenotypes were produced in an FUF1 loss-of-function mutant, an FUF1 transfer DNA insertion line, SALK_136922 (containing an insertion in the 5′-untranslated region [UTR]), was analyzed. The flower organ senescence/abscission occurred similarly in the SALK_136922 flowers compared with the wild-type flowers (Supplemental Fig. S10). This indicated a possible functional redundancy for FUF1 and other unknown genes. To examine this assumption, a strategy to generate transgenic dominant loss-of-function mutant plants was performed by fusing the conserved SRDX-suppressing motif (Chen et al., 2011) to FUF1 (FUF1+SRDX). If FUF1 functions as an activator, we expect that 35S:FUF1+SRDX should convert FUF1 into a repressor and cause a dominant-negative effect, resulting in the opposite phenotype to that observed in the 35S:FUF1 plants.
In total, 41 of 100 independent 35S:FUF1+SRDX kanamycin-resistant Arabidopsis plants exhibited an abnormal phenotype of significantly early flower senescence/abscission. Unlike the perianths of the 35S:FUF1 flowers, which began to senesce and abscise in a turgid state at positions later than 10 (Fig. 6N, 1), early senescence/abscission was observed in the 35S:FUF1+SRDX plants at positions before 3 (Fig. 6N, 6).
We further expressed FUF1+SRDX under its own promoter (FUF1:FUF1+SRDX) through the transactivation system (Hsu, 2012). The resulting plants showed similar early senescence/abscission of the perianths (Fig. 6N, 7). These results indicated that the FUF1 gene encodes a transcriptional activator; 35S:FUF1+SRDX and FUF1:FUF1+SRDX result in the opposite phenotype of that observed in the 35S:FUF1 plants. In contrast to the gene expression detected in the 35S:FUF1 plants, the expression levels of EDF1/2/3/4 were clearly up-regulated in the 35S:FUF1+SRDX and FUF1:FUF1+SRDX plants (Fig. 6O).
Figure 7.
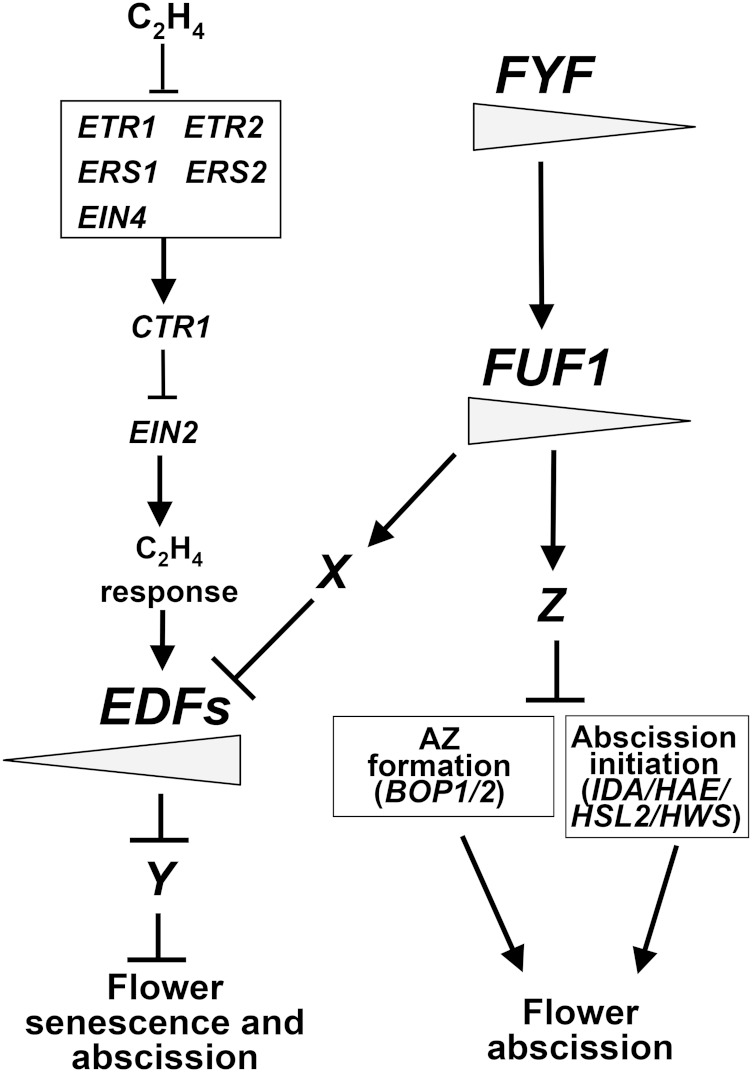
A model for the function of FYF and FUF1 genes in regulating flower senescence and abscission. In wild-type flowers, FYF negatively regulates the ethylene signaling pathway by activating (→) the expression of FUF1. In this pathway, FUF1 activates (→) an X gene, which negatively regulates (line with bar) the EDF1/2/3/4 downstream genes of the ethylene signaling pathway. EDF1/2/3/4 promotes the senescence/abscission by suppressing (line with bar) a Y gene that functions to suppress (line with bar) the senescence/abscission. The suppression of EDF1/2/3/4 before pollination results in the repression of senescence/abscission. This pathway also regulates abscission by activating (→) a Z gene that suppresses (line with bar) the expression of BOP1 and BOP2, which regulate the formation of the AZ and the expression of genes, such as IDA/HAESA/HSL2/HWS, that regulate the initiation of abscission. The ectopic expression of FYF or FUF1 suppressed EDF1/2/3/4, BOP1/2, and IDA/HAESA/HSL2/HWS and caused the inhibition of senescence/abscission during all stages of flower development. The gradient of FYF and FUF1 activity is illustrated as the gradual reduction in the size of the gray bar during flower maturation. In contrast, the gradient of EDF1/2/3/4 activity is illustrated as the gradual increase in the size of the gray bar during flower maturation.
DISCUSSION
EDFs Function as Downstream Genes in the Ethylene Response in Controlling Flower Senescence and Abscission
Our previous results suggested that EDFs may function as downstream genes in the ethylene response and may be suppressed by FYF in the control of flower senescence/abscission (Chen et al., 2011). In this study, we provide evidence to support this assumption through a functional analysis of the EDF1/2/3/4 genes.
First, weak EDF1/2/3/4 expression was detected in the young flower buds and increased significantly in mature flowers. This pattern was opposite of that for FYF (Chen et al., 2011) and reveals opposite roles for EDFs and FYF. Second, 35S:EDFs and 35S:EDFs+SRDX promoted flower senescence/abscission in transgenic Arabidopsis plants by activating SAG12 and the PCD-associated genes AtMCs and VPEs. This result indicated that EDF1/2/3/4 functions as a repressor to promote senescence/abscission in contrast to FYF. Third, 35S:EDFs+SRDX is epistatic to 35S:FYF. Fourth, GUS activity in EDFs:GUS plants was significantly suppressed by 35S:FYF. These results support the conclusion that EDF1/2/3/4 functions downstream of FYF and that the expression before pollination is suppressed by FYF. This suppression prevents the flowers from senescing and abscising. The decrease in FYF expression after pollination activates EDF1/2/3/4 and the ethylene signaling pathway, resulting in the senescence/abscission of the flowers.
The involvement of EDF1/2/3/4 in the ethylene signaling pathway was further supported by the analysis of the etr1 and ein2 mutants that ectopically expressed EDFs+SRDX. These plants displayed early senescence/abscission of the perianth organs, which was observed in 35S:EDFs+SRDX plants, suggesting that 35S:EDFs+SRDX is epistatic to both etr1 and ein2 mutants in the ethylene-insensitive response. This assumption was further supported by the result that EIN3 binds to the promoters of all four EDFs (Chang et al., 2013). Thus, the hypotheses that EDFs are downstream genes in the ethylene response and suppressed by FYF in controlling flower senescence and abscission were confirmed. Interestingly, although EDFs caused very similar effect on the promotion of the flower senescence/abscission, it seems that EDF2 has a relatively stronger effect than EDF1, EDF3, and EDF4 (Supplemental Tables S1–S4). This result may reflect the small functional difference among the genes with redundant activity.
FUF1 Acts as an Activator Downstream of FYF in Controlling Flower Organ Senescence and Abscission
FUF1, which up-regulated by FYF, was hypothesized to be related to the regulation of senescence/abscission, and this assumption was supported by three lines of evidence. The first line of evidence originates from the significant delay in floral organ senescence/abscission observed in the 35S:FUF1 plants, similar to the delay observed in the 35S:FYF plants. The second line of evidence originates from the expression pattern of the FUF1 gene during flower development. Similar to the GUS expression pattern in FYF:GUS Arabidopsis, strong GUS activity was detected in the young flower buds; however, this staining significantly decreased after anthesis in FUF1:GUS plants. The third line of evidence originates from the significant promotion of flower organ senescence/abscission in the 35S:FUF1+SRDX and FUF1:FUF1+SRDX dominant-negative plants in contrast to the phenotype of the 35S:FUF1 plants. These results indicated that FUF1 functions as a transcriptional activator to prevent the senescence/abscission of flowers during the early stages of floral development. This inhibition extends to late flower development in the 35S:FUF1 plants and results in the delay of flower senescence and abscission. In contrast, this inhibition was reversed in the FUF1+SRDX plants, which exhibited early senescence/abscission.
FUF1 Controls Senescence and Abscission through Inhibiting the EDFs, Which Are Downstream Genes in the Ethylene Response
Similar to the plants carrying 35S:FYF, the floral senescence/abscission of the 35S:FUF1 plants was also insensitive to ethylene treatment, further supporting the idea that FUF1 acts downstream of FYF to control floral senescence/abscission by suppressing the ethylene response.
This hypothesis was supported by the analysis of the etr1, ein2, and ctr1 mutants ectopically expressing FUF1. The results indicated that, similar to that in plants carrying 35S:FUF1, enhanced delay of the senescence/abscission of the perianth organs was observed in the 35S:FUF1/etr1-1 and 35S:FUF1/ein2-1 plants, suggesting that 35S:FUF1 is epistatic to both etr1 and ein2 mutants in the ethylene-insensitive response. Furthermore, the senescence/abscission of the perianth organs was significantly delayed in the constitutive ethylene response mutant ctr1-1 ectopically expressing FUF1 (35S:FUF1/ctr1-1). This result suggests that 35S:FUF1 repressed the genes that function downstream of CTR1 in controlling the constitutive ethylene response and regulating flower senescence/abscission. This assumption was supported by the observed down-regulation of EDFs in 35S:FUF1 plants. Conversely, the expression level of EDF1/2/3/4 was up-regulated in the 35S:FUF1+SRDX and FUF1:FUF1+SRDX dominant-negative plants, in which senescence/abscission was greatly promoted. In addition, 35S:EDFs+SRDX is epistatic to 35S:FUF1. These results support the conclusions that EDF1/2/3/4 functions downstream of FUF1 and that the expression before pollination is suppressed by FUF1.
The hypothesis that FUF1 functions downstream of FYF to regulate the ethylene response during plant growth and development was further supported by two additional results. First, neither 35S:FUF1 nor 35S:FYF rescued the reduction in plant size in the ctr1 mutant. This result suggests that FYF and FUF1 are involved in the ethylene response pathway that controls flower senescence and abscission but not the other ethylene response pathways that control plant growth and development, including plant size (Chen et al., 2011). Second, as observed for 35S:FYF, the senescence of the 35S:FUF1 leaves was insensitive to ethylene treatment, supporting the notion that the FYF/FUF1 pathway may also control leaf senescence in a similar manner to its control of flower senescence by repressing the ethylene response.
FUF1 Controls Abscission through the Down-Regulation of BOP1, BOP2, IDA, HAESA, HSL2, and HWS
In addition to regulating ethylene-responsive genes, a clear down-regulation of BOP1/2 expression was observed in the 35S:FUF1 flowers. Because the BOP1/2 genes function in the control of floral AZ formation (McKim et al., 2008), BOP1/2 suppression by FUF1 may block abscission. This hypothesis was supported by the lack or significant delay of the AZ formation in the 35S:FUF1 flowers, which was determined by SEM examination of a thin section of the 35S:FUF1 sepals. Thus, FUF1 likely suppresses the abscission of the floral organs primarily through the down-regulation of BOP1/2 and the repression of the AZ formation. In addition to BOP1/2, the expression of the abscission-associated genes IDA/HAESA/HSL2/HWS, which regulate the abscission initiation of the floral organs, was also down-regulated in the 35S:FUF1 flowers. FUF1 may be directly responsible for the suppression of IDA/HAESA/HSL2/HWS expression. Alternatively, if the ectopic expression of FUF1 caused deficient AZ formation, then this result might have a consequence on the down-regulation of IDA/HAESA/HSL2/HWS expression. To distinguish these two possibilities, the 35S:FUF1/35S:IDA double transgenic plants were generated, and the phenotype was analyzed. The result indicated that 35S:FUF1/35S:IDA plants showed the similar 35S:FUF1 phenotype, and thus, it supports the latter assumption. This conclusion is further supported by a previous study that showed that 35S:IDA phenotype was also suppressed by bop1/bop2 double mutants (McKim et al., 2008). It has been shown that 35S:IDA bop1/bop2 plants clearly lack floral organ abscission similar to bop1/bop2 and our 35S:FUF1/35S:IDA plants. Thus, the failure of AZ formation because of the suppression of BOP1/2 expression in 35S:FUF1 flowers prevented the abscission of the flower organs, even with the ectopic expression downstream of abscission-associated genes, such as IDA.
A model depicting the possible roles of the FYF and FUF1 genes in regulating genes associated with senescence, abscission, and ethylene signaling is illustrated in Figure 7. Thus, this report presents data that extend the knowledge of the genes in a pathway regulated by the FYF gene, which controls flower senescence and abscission.
MATERIALS AND METHODS
Plant Materials and Plant Transformation
The etr1-1 (CS237) Arabidopsis (Arabidopsis thaliana) seeds were obtained from Long-Chi Wang (Biotechnology Center, National Chung Hsing University). The ein2-1 (CS3071) and ctr1-1 (CS8057) mutant Arabidopsis seeds were obtained from the Arabidopsis Biological Resource Center. Seeds for Arabidopsis were germinated and grown as described previously (Chang et al., 2010). Arabidopsis seeds were sterilized and placed on agar plates containing one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) at 4°C for 2 d. Before being transplanted into soil, the seedlings were grown in growth chambers with the light intensity at 150 μE m−2 s−1 under long-day conditions (16-h-light/8-h-dark cycle) at 22°C for 10 d. A floral dip method as described elsewhere (Clough and Bent, 1998) was used to introduce constructs in the Agrobacterium tumefaciens strain GV3101 into Arabidopsis plants. Transformants that survived in medium containing kanamycin (50 µg mL−1) were further verified by PCR and reverse transcription (RT)-PCR analyses. More than 10 transgenic plants with the similar altered phenotype for each construct were phenotype analyzed to confirm the function of the transgene and avoid the effect that the 35S promoter may switch on the downstream gene in different tissues and with different timing to the native promoter in transgenic plants. To generate 35S:FUF1/ctr1-1 Arabidopsis, homozygous 35S:FUF1 plants were crossed with the ctr1-1 mutant in the Columbia-0 (Col-0) background, and F1 plants resistant to kanamycin were selected to generate the F2 generation. One-quarter of the F2 plants that were resistant to kanamycin were 35S:FUF1/ctr1-1 and selected for additional analysis.
Cloning of the Complementary DNAs for EDF1/2/3/4 from Arabidopsis
For 35S:EDF1/2/3/4 constructs, the complementary DNAs (cDNAs) for EDF1 (At1g25560), EDF2 (At1g68840), EDF3 (At3g25730), and EDF4 (At1g13260) were obtained by PCR amplification and sequence confirmed. The primers contained the XbaI and KpnI recognition sites for EDF1/2/4 and the BamHI and KpnI recognition sites for EDF3 to facilitate the cloning of the cDNAs. The XbaI-KpnI or BamHI-KpnI fragment containing the cDNA was cloned into the binary vector pEpyon-32K (Chen et al., 2011) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and used for plant transformation. Sequences for the primers are listed in Supplemental Table S7.
Cloning of the cDNA for FUF1 from Arabidopsis
For the 35S:FUF1 construct, the cDNA for FUF1 (At1g71450) was obtained by PCR amplification and sequence confirmed. The primers contained the XbaI and KpnI recognition sites to facilitate the cloning of the cDNA. The XbaI-KpnI fragment containing the cDNA was cloned into the binary vector pEpyon-12K (Chen et al., 2011) under the control of the CaMV 35S promoter and used for plant transformation. Sequences for the primers are listed in Supplemental Table S7.
Cloning of the Promoter DNA Fragment from Arabidopsis
For the EDF1/2/3/4:GUS constructs, the promoter regions that included the 5′-UTR for EDF1 (2.12 kb), EDF2 (2.07 kb), EDF3 (2.03 kb), and EDF4 (2.07 kb) were obtained by PCR amplification and sequence confirmed from the genomic DNA followed by cloning into the pGEM-T easy vector (Promega). These EDF1/2/3/4 promoter fragments were then subcloned into the linker region before the GUS coding region in the binary vector pEpyon01k (Peng et al., 2013). For the FUF1:GUS construct, the FUF1 promoter that included the 5′-UTR (2.46 kb) was obtained by PCR amplification from the genomic DNA; this was then cloned into the pGEM-T easy vector. The FUF1 promoter fragment was then subcloned into the linker region before the GUS coding region in the binary vector pEpyon01k. The sequences of the primers are listed in Supplemental Table S7.
Construction of the 35S:EDF1/2/3/4+SRDX and 35S:FUF1+SRDX Constructs
For the 35S:EDF1/2/3/4+SRDX construct, the cDNAs for EDF1/2/3/4 were obtained by PCR amplification and cloned into the pEpyon-3aK plasmid upstream of the SRDX (LDLDLELRLGFA*) sequence under the control of the CaMV 35S promoter as described previously (Chen et al., 2011). For the 35S:FUF1+SRDX construct, the cDNA for FUF1 was obtained by PCR amplification and cloned into the pEpyon-3aK in the same manner as 35S:EDF1/2/3/4+SRDX. The sequences of the primers are listed in Supplemental Table S7.
Generation of the FUF1:FUF1+SRDX Construct
The transactivation system was used to generate the FUF1:FUF1+SRDX construct. The FUF1+SRDX fragment was obtained by PCR amplification and cloned into the effector line vector pEpyon-7aK downstream of the LexA operator that was used as the upstream activating sequence (Hsu, 2012). The FUF1 promoter (2.46 kb) was cloned into the activator line vector pBroly-KXV upstream of the chimeric transcription factor LexA-VP16, which recognizes the LexA operator DNA sequence. These constructs (pEpyon-7aK and pBroly-KXV) were transformed into Arabidopsis plants separately. The transactivation line (FUF1:FUF1+SRDX) was obtained by crossing the effector line and the activator line.
GUS Staining and Microscopy
Histochemical staining was performed under standard methods described previously (Jefferson et al., 1987; Chou et al., 2001). SEM was performed according to the methods by Tzeng and Yang (2001), Hsu and Yang (2002), and Chang et al. (2010).
Sepal Break-Strength Analysis
For sepal break-strength analysis, a single flower sepal was clipped with a small clamp that was attached to a Brookfield CT3-4500 Break-Strength Meter (Brookfield Engineering Laboratories, Inc.). The sepal break strength was the measurement of the force necessary to remove the sepal from the flower, and it was measured using the CT3-4500 Break-Strength Meter. The flower position was counted starting from the first flower with visible white petals at the top of the inflorescence.
Ethylene Responses
For ethylene responses, mature wild-type and transgenic Arabidopsis plants were sealed in plastic chambers and gassed with air or air containing 10 μL L−1 ethylene for 3 d in a 16-h-light/8-h-dark cycle as described previously (Chen and Bleecker, 1995; Chen et al., 2011).
RNA Isolation and Real-Time PCR Analysis
Total RNA isolated from Arabidopsis plants used for RT-PCR analysis is described below. All of the equipment was sterilized two times and baked at 180°C for 12 h to prevent Rnase contamination. Plant tissues were ground to a fine power under liquid nitrogen and mixed with 1 mL of TRIzol Reagent (Invitrogen) to homogenize and lyse samples. A volume of 200 μL of chloroform was added into the lysate, incubated at room temperature for 5 min, and then, centrifuged at 13,000g for 10 min at 4°C. The supernatant was transferred to a new 1.5-mL tube, and an equal volume of isopropanol was added to precipitate RNA for 20 min at 4°C. After centrifugation at 13,000g for 10 min at 4°C, the supernatant was discarded, and the RNA pellet was suspended by 75% (v/v) ethanol and centrifuged at 7,500g for 5 min at 4°C. The RNA pellet was vacuum dried for 5 min to remove residual trace of ethanol and dissolved in 20 μL of diethyl pyrocarbonate-treated water.
For real-time quantitative RT-PCR, the reaction was performed on an MJ Opticon System (MJ Research) using SYBER Green Real-Time PCR Master Mix (TOYOBO Co., LTD.). The amplification condition was as described previously (Chen et al., 2011). Sequences for the primers used for real-time quantitative RT-PCR for FYF (At5g62165), FUF1 (At1g71450), IDA (At1g68765), HAESA (At4g28490), HSL2 (At5G65710), HWS (At3G61590), BOP1 (At3g57130), BOP2 (At2g41370), EDF1 (At1g25560), EDF2 (At1g68840), EDF3 (At3g25730), EDF4 (At1g13260), SAG12 (At5G45890), AtMC1 (At1G02170), AtMC9 (At5G04200), α-VPE (At2G25940), and β-VPE (At1G62710) are listed in Supplemental Table S7. The housekeeping gene UBIQUITIN10 (UBQ10) was used as normalization control with the following primers: RT-UBQ10-1 and RT-UBQ10-2. All experiments were repeated at least two times as biological replicates for reproducibility. Data were analyzed using Gene Expression Macro software (version 1.1; Bio-Rad).
Construction of the FYF+GR Construct and Microarray Analysis
For the 35S:FYF+GR construct, the cDNA for FYF was obtained by PCR amplification and sequence confirmed. The primers contained the BamHI recognition site to facilitate the cloning of the cDNA. The BamHI fragment containing the cDNA was cloned into the PBIGR plasmid upstream of the GR sequence under the control of the CaMV 35S promoter and used for plant transformation. Sequences for the primers are listed in Supplemental Table S7. For microarray analysis, inflorescence from 35S:FYF+GR Arabidopsis was cut and placed into the water containing 5 μm DEX for 6 h before the isolation of RNA. Total RNA was isolated from all the flower buds before opening of DEX-treated and -untreated 35S:FYF+GR Arabidopsis by TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol; 0.5 g of total RNA derived from paired RNA samples from DEX-treated and -untreated 35S:FYF+GR Arabidopsis was amplified by a Low RNA Input Fluor Linear Amp Kit (Agilent Technologies) and labeled with Cy3 or Cy5 (CyDye; PerkinElmer) during the in vitro transcription process. CyDye-labeled complementary RNA was fragmented to an average size of about 50 to 100 nucleotides by incubation with fragmentation buffer at 60°C for 30 min. Correspondingly fragmented labeled complementary RNA is then pooled and hybridized to Agilent Arabidopsis 3 Oligo 4x44K Microarray (Agilent Technologies) at 60°C for 17 h. After washing and drying by nitrogen gun blowing, microarrays are scanned with an Agilent Microarray Scanner (Agilent Technologies) at 535 nm for Cy3 and 625 nm for Cy5. Scanned images are analyzed by Feature Extraction 9.5.3 Software (Agilent Technologies), an image analysis and normalization software that is used to quantify signal and background intensity for each feature; it substantially normalized the data by rank-consistency-filtering locally weighted scatterplot smoothing method. The microarray experiments were done three times, and the data analysis described above was done by Welgene Biotech.
Microarray data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSM1700867.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A protein sequence comparison of EDF1/2/3/4, which is in the RAV family of the AP2/ERF-relative proteins.
Supplemental Figure S2. GUS staining patterns in FYF:GUS Arabidopsis plants.
Supplemental Figure S3. GUS staining patterns in EDFs:GUS Arabidopsis plants and the detection of EDFs expression in flowers at different developmental stages.
Supplemental Figure S4. The detection of senescence-associated gene expression in 35S:EDFs+SRDX plants.
Supplemental Figure S5. A phenotypic analysis of flower senescence and abscission in etr1-1 mutants ectopically expressing the EDFs+SRDX constructs.
Supplemental Figure S6. A phenotypic analysis of flower senescence and abscission in ein2-1 mutants ectopically expressing the EDFs+SRDX constructs.
Supplemental Figure S7. The ectopic expression of FYF+GR delayed flower senescence and abscission after DEX treatment.
Supplemental Figure S8. A protein sequence comparison of FUF1 (AtERF21) and four other ERF proteins (AtERF19, AtERF95, AtERF96, and AtERF98) with transcripts that are up-regulated by FYF.
Supplemental Figure S9. An analysis of the effect of ethylene on leaves of 35S:FUF1 Arabidopsis.
Supplemental Figure S10. Analysis of FUF1 transfer DNA insertion line SALK_136922.
Supplemental Table S1. Position of flower that showed senescence/abscission in the 35S:EDFs Arabidopsis.
Supplemental Table S2. Position of flower that showed senescence/abscission in the 35S:EDFs+SRDX Arabidopsis.
Supplemental Table S3. Position of flower that showed senescence/abscission in the 35S:EDFs+SRDX/etr1-1 Arabidopsis.
Supplemental Table S4. Position of flower that showed senescence/abscission in the 35S:EDFs+SRDX/ein2-1 Arabidopsis.
Supplemental Table S5. Arabidopsis genes up-regulated in the 35S:FYF+GR Arabidopsis.
Supplemental Table S6. Arabidopsis genes down-regulated in the 35S:FYF+GR Arabidopsis.
Supplemental Table S7. Oligo nucleotide sequence of primers used in gene cloning and PCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Long-Chi Wang (Biotechnology Center, National Chung Hsing University [NCHU]) for helpful discussion of the results and Dr. Ching-Chang Shiesh (Department of Horticulture, NCHU) for help in the ethylene treatment experiment.
Glossary
- AZ
abscission zone
- CaMV
Cauliflower mosaic virus
- cDNA
complementary DNA
- Col-0
Columbia-0
- DEX
dexamethasone
- PCD
programmed cell death
- RT
reverse transcription
- SEM
scanning electron microscopy
- UTR
untranslated region
Footnotes
This work was supported by the Ministry of Science and Technology (grant no. NSC101–2321–B–005–008 to C.-H.Y.) and the Ministry of Education, Taiwan, Republic of China (Aiming for the Top University Plan).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Basu MM, González-Carranza ZH, Azam-Ali S, Tang S, Shahid AA, Roberts JA (2013) The manipulation of auxin in the abscission zone cells of Arabidopsis flowers reveals that indoleacetic acid signaling is a prerequisite for organ shedding. Plant Physiol 162: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Kao NH, Li JY, Hsu WH, Liang YL, Wu JW, Yang CH (2010) Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol 152: 837–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MK, Hsu WH, Lee PF, Thiruvengadam M, Chen HI, Yang CH (2011) The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J 68: 168–185 [DOI] [PubMed] [Google Scholar]
- Chen QG, Bleecker AB (1995) Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol 108: 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller GE (2005) Ethylene signal transduction. Ann Bot (Lond) 95: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Haung MD, Yang CH (2001) EMF genes interact with late-flowering genes in regulating floral initiation genes during shoot development in Arabidopsis thaliana. Plant Cell Physiol 42: 499–507 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P (2010) Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Ecker JR. (1995) The ethylene signal transduction pathway in plants. Science 268: 667–675 [DOI] [PubMed] [Google Scholar]
- González-Carranza ZH, Elliott KA, Roberts JA (2007a) Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot 58: 3719–3730 [DOI] [PubMed] [Google Scholar]
- González-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA (2007b) Hawaiian skirt: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol 144: 1370–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Shahid AA, Zhang L, Liu Y, Ninsuwan U, Roberts JA (2012) A novel approach to dissect the abscission process in Arabidopsis. Plant Physiol 160: 1342–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LE, Schippers JHM, Dijkwel PP, Wagstaff C (2012) Ethylene and senescence processes. In MT McManus, ed, The Plant Hormone Ethylene: Annual Plant Reviews, Vol 44. Wiley-Blackwell, Oxford, pp 305–341 [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW (2005) BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HF, Yang CH (2002) An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol 43: 1198–1209 [DOI] [PubMed] [Google Scholar]
- Hsu WH (2012) Functional analysis of genes regulating cell division and gametophyte development in Arabidopsis. PhD dissertation. National Chung Hsing University, Taichung, Taiwan, China [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ohme-Takagi M (2009) A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 3: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ. (2012) Organ abscission: exit strategies require signals and moving traffic. Curr Opin Plant Biol 15: 670–676 [DOI] [PubMed] [Google Scholar]
- Lumba S, Tsuchiya Y, Delmas F, Hezky J, Provart NJ, Shi Lu Q, McCourt P, Gazzarrini S (2012) The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biol 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim SM, Stenvik GE, Butenko MA, Kristiansen W, Cho SK, Hepworth SR, Aalen RB, Haughn GW (2008) The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (1999) Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Norberg M, Holmlund M, Nilsson O (2005) The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132: 2203–2213 [DOI] [PubMed] [Google Scholar]
- Patterson SE, Bleecker AB (2004) Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol 134: 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YJ, Shih CF, Yang JY, Tan CM, Hsu WH, Huang YP, Liao PC, Yang CH (2013) A RING-type E3 ligase controls anther dehiscence by activating the jasmonate biosynthetic pathway gene DEFECTIVE IN ANTHER DEHISCENCE1 in Arabidopsis. Plant J 74: 310–327 [DOI] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53: 131–158 [DOI] [PubMed] [Google Scholar]
- Rogers HJ. (2013) From models to ornamentals: how is flower senescence regulated? Plant Mol Biol 82: 563–574 [DOI] [PubMed] [Google Scholar]
- Rojo E, Martín R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sánchez-Serrano JJ, Baker B, et al. (2004) VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Schaller GE. (2012) Ethylene and the regulation of plant development. BMC Biol 10: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA (2011) Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM (2005) Arabidopsis ethylene signaling pathway. Sci STKE 2005: cm4. [DOI] [PubMed] [Google Scholar]
- Tzeng TY, Yang CH (2001) A MADS box gene from lily (Lilium Longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol 42: 1156–1168 [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Yang TJ, Stead AD, Buchanan-Wollaston V, Roberts JA (2009) A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J 57: 690–705 [DOI] [PubMed] [Google Scholar]
- Wang F, Cui X, Sun Y, Dong CH (2013) Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep 32: 1099–1109 [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, Kim J, Lee U, Song IJ, Kim JH, Lee HY, Nam HG, Lim PO (2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot 61: 3947–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XM, Yu Y, Han LB, Li CL, Wang HY, Zhong NQ, Yao Y, Xia GX (2012) The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol 159: 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Chen M, Li LC, Ma YZ (2011) Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol 53: 570–585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.