The genetic basis of dynamic salinity stress responses is elucidated using image-based phenomics and functional association analysis.
Abstract
Salinity affects a significant portion of arable land and is particularly detrimental for irrigated agriculture, which provides one-third of the global food supply. Rice (Oryza sativa), the most important food crop, is salt sensitive. The genetic resources for salt tolerance in rice germplasm exist but are underutilized due to the difficulty in capturing the dynamic nature of physiological responses to salt stress. The genetic basis of these physiological responses is predicted to be polygenic. In an effort to address this challenge, we generated temporal imaging data from 378 diverse rice genotypes across 14 d of 90 mm NaCl stress and developed a statistical model to assess the genetic architecture of dynamic salinity-induced growth responses in rice germplasm. A genomic region on chromosome 3 was strongly associated with the early growth response and was captured using visible range imaging. Fluorescence imaging identified four genomic regions linked to salinity-induced fluorescence responses. A region on chromosome 1 regulates both the fluorescence shift indicative of the longer term ionic stress and the early growth rate decline during salinity stress. We present, to our knowledge, a new approach to capture the dynamic plant responses to its environment and elucidate the genetic basis of these responses using a longitudinal genome-wide association model.
Nearly one-third of the 54 million ha of the highly saline soils in the world are located in South and Southeast Asia. Rice (Oryza sativa), which is the primary source of calories and protein for these two regions, is very sensitive to salinity stress, with even moderate salinity levels known to decrease yields by 50% (Zeng et al., 2002). Projected sea level rise due to climate change is expected to increase saltwater ingress in coastal rice-growing regions of South and Southeast Asia. Therefore, development of salt-tolerant rice cultivars is essential to maintain rice productivity in the salinity-affected regions globally.
Salt tolerance, defined as the ability to maintain growth and productivity in saline conditions, is a complex polygenic trait that may be influenced by distinct physiological mechanisms (Munns et al., 1982; Munns and Termaat, 1986; Cheeseman, 1988; Munns and Tester, 2008; Horie et al., 2012; for a comprehensive review of genes involved in salinity tolerance in rice, see Negrão et al., 2011) At the cellular level, plants respond to saline conditions in two phases, namely an osmotic (shoot ion independent) and an ionic stress phase, which can occur in an overlapping manner with varying intensity during the course of salinity stress (Munns and Termaat, 1986; Munns, 2002; Munns and James, 2003; Munns and Tester, 2008; Horie et al., 2012). During the osmotic stress phase, which occurs soon after the onset of salinity, the reduction in external osmotic potential disrupts water uptake and impedes cell expansion, which, at the whole plant level, leads to reduced growth rate (Matsuda and Riazi, 1981; Munns and Passioura, 1984; Rawson and Munns, 1984; Azaizeh and Steudle, 1991; Fricke and Peters, 2002; Fricke, 2004; Boursiac et al., 2005). As salinity stress persists over several days and weeks, sodium ions (Na+) accumulate to toxic levels, resulting in cell death and precocious leaf senescence (Lutts and Bouharmont, 1996; Munns, 2002; Munns and James, 2003; Ghanem et al., 2008). This is typically observed during the ionic phase of the salinity response (Munns, 2002; Munns and James, 2003; Munns and Tester, 2008). Plants possess distinct mechanisms to adapt to these osmotic and ionic stresses that are controlled by a suite of genes (Apse et al., 1999; Carvajal et al., 1999; Halfter et al., 2000; Ishitani et al., 2000; Shi et al., 2000; Zeng and Shannon, 2000; Rus et al., 2001; Berthomieu et al., 2003; Martínez-Ballesta et al., 2003; Boursiac et al., 2005, 2008; Ren et al., 2005; Huang et al., 2006; Davenport et al., 2007; Obata et al., 2007; Székely et al., 2008; Horie et al., 2011; Rivandi et al., 2011; Asano et al., 2012; Munns et al., 2012; Latz et al., 2013; Schmidt et al., 2013; Campo et al., 2014; Choi et al., 2014; Liu et al., 2014). The genetic basis of temporal adaptive responses to salinity stress remains to be explored in rice and other crops. This is primarily due to challenges in capturing the dynamic physiological responses to salinity for a large number of genotypes in a nondestructive manner. Manual phenotyping to detect incremental changes in growth rate during the osmotic stress and slight shifts in leaf color due to ionic stress is difficult to quantify for a large number of genotypes.
In rice, at least one major quantitative trait loci (QTL; saltol) for salinity tolerance has been characterized based on end point measurements of biomass, senescence/injury, and Na+ and K+ concentrations (Bonilla et al., 2002; Lin et al., 2004; Thomson et al., 2010). SHOOT K+ CONTENT1 (SKC1) is the causative gene underlying the saltol region. SKC1 encodes a Na+-selective high-affinity potassium transporter that regulates Na+/K+ homeostasis during salinity stress (Ren et al., 2005). High levels of Na+ displace cellular K+, an essential element for several enzymatic reactions and physiological processes (Gierth and Mäser, 2007). The ability to maintain cellular K+ levels during salinity through the action of Na+-selective potassium transporters or Na+/H+ antiporters is a well-characterized tolerance mechanism in cereals including rice (Ren et al., 2005; Sunarpi et al., 2005; Huang et al., 2006; Møller et al., 2009; Mian et al., 2011; Munns et al., 2012).
Numerous studies have utilized conventional linkage mapping to identify QTL for morphological and physiological responses to salinity in rice using discrete end point measurements (Bonilla et al., 2002; Lin et al., 2004; Ren et al., 2005; Negrão et al., 2011; Wang et al., 2012). However, the physiological adaptation to saline conditions is a complex continuous process that develops over time. While some accessions will exhibit similar end point phenotypic values, the genetic and physiological mechanisms giving rise to the similar phenotypes may be very different and the growth trajectories throughout the experiment may be distinct. Although single time point studies have yielded important information regarding the genetic basis of salinity tolerance, such approaches are too simple to reveal the genetic architecture of stress adaptation. With the advent of high-throughput image-based phenotyping platforms, it is now feasible to quantify dynamic responses during the stress treatment for a large number of genotypes (Berger et al., 2010; Golzarian et al., 2011; Chen et al., 2014; Honsdorf et al., 2014).
Image-based phenotyping has been combined with genome-wide association studies (GWAS) and linkage mapping to examine the genetic basis of complex developmental processes (Busemeyer et al., 2013; Moore et al., 2013; Topp et al., 2013; Slovak et al., 2014; Würschum et al., 2014; Yang et al., 2014; Bac-Molenaar et al., 2015). Moreover, the introduction of the time axis provides a better understanding of the physiological processes underlying complex stress and developmental responses compared with single end point measurements (Zhang et al., 2012; Moore et al., 2013; Brown et al., 2014; Chen et al., 2014; Slovak et al., 2014; Bac-Molenaar et al., 2015). However, to date, no studies have implemented an association mapping approach using image-derived phenotypes to address the genetic basis of dynamic stress responses in plants. Image-based phenotyping offers several advantages over conventional phenotyping: (1) quantitative measurements can be recorded over discrete time points to capture morphological and physiological responses in a nondestructive manner, and (2) the use of various types of spectral imaging address phenotypes that are not detectable to the human eye such as chlorophyll fluorescence and leaf water content. Integrating dynamic phenotypic data and association mapping has the potential to query genetic diversity across hundreds of accessions for complex traits and provides much higher resolution compared with conventional linkage mapping. Here, we explored the dynamic growth and chlorophyll responses to salinity of a diverse set of rice accessions using high-throughput visible and fluorescence imaging. To assess the genetic basis of plant growth in saline conditions, a logistic model was used to describe the temporal growth responses and was incorporated into the statistical framework necessary for association mapping. Coupled with temporal fluorescence imaging, we present, to our knowledge, new insights into the genetic architecture of osmotic and ionic responses during salinity stress in rice.
RESULTS
Capturing Osmotic and Ionic Components of Salinity Stress Using High-Throughput Imaging
To assess the ionic and osmotic components of salinity response in rice, a diversity panel consisting of 373 rice lines was exposed to 90 mm NaCl during the early tillering stage (Supplemental Figs. S1A and S2). Morphological and physiological responses were monitored over a period of 14 d after 90 mm NaCl treatment using nondestructive imaging (Supplemental Fig. S1B). Two types of imaging systems, red, green, blue (RGB)/visible spectrum and fluorescence (FLUO), were utilized to address osmotic and ionic components of salinity stress, respectively. RGB/visible imaging allows growth and other morphological parameters to be quantified in a nondestructive manner and has been used to monitor growth shortly (1–10 d) after the onset of salinity to address primarily the shoot ion-independent phase (osmotic phase) of salinity stress (Rajendran et al., 2009). The effects of salinity on chlorophyll and other fluorophores can be monitored by detecting changes in the color or intensity of pixels, thus providing important information regarding the ionic component of salinity stress (Berger et al., 2010).
We developed an open-source image processing software named Image Harvest to extract plant pixels from images and quantify fluorescence color ranges (“Materials and Methods;” Supplemental Fig. S1C). Image Harvest is publically available for download and is optimized for processing high-volume plant image data using parallel computing. The data set generated for the current study consisted of 142,671 RGB images and 95,118 fluorescence images, which were processed using LemnaGrid and Image Harvest, respectively. The entire data set is available on iPlant (http://mirrors.iplantcollaborative.org/browse/iplant/home/shared/walia_rice_salt). The resulting output consists of temporal data for 97 digital (pixel-based) traits, seven of which are used to describe plant morphology and growth traits, while the remaining 90 traits are used to describe fluorescence responses (Supplemental Table S1).
Assessing Salinity-Induced Growth Responses in the Rice Diversity Panel
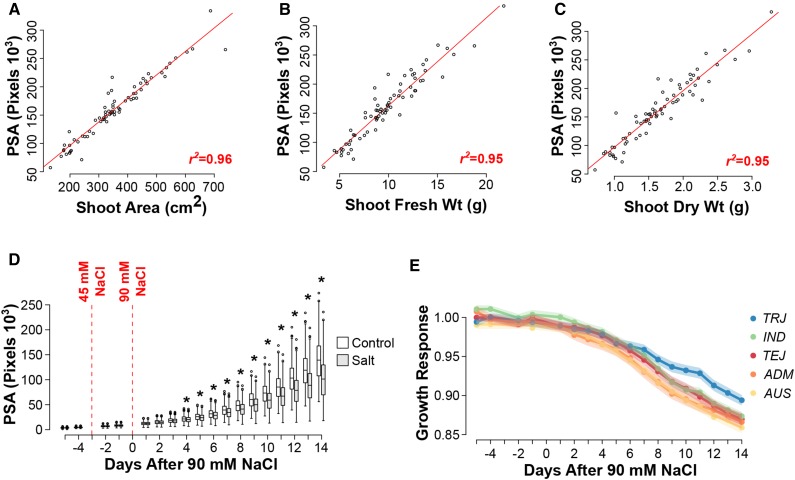
To determine how accurately the biomass-related, image-derived growth metrics represented actual biomass and total shoot area, plants from 72 accessions were harvested on the last day of the imaging experiment, fresh and dry mass was recorded, and total plant area was directly measured using a leaf area meter (LI-3100C; LI-COR). Correlation analysis was done using seven biomass-related metrics and three manual measurements. Of the seven biomass-related metrics derived from color images, projected shoot area (PSA), which is defined as the sum of all pixels from all three RGB images, showed the highest correlation with all manual biomass-related measurements (Supplemental Table S2). As expected, PSA showed the strongest positive correlation with total plant area as well (r2 = 0.96, P < 0.001, n = 72; Fig. 1A). Shoot fresh and dry weight showed a strong positive correlation with PSA, although at a slightly lower correlation compared with total plant area (r2 = 0.95, P < 0.001 for both shoot fresh and dry weight; Fig. 1, B and C, respectively). A significant difference in PSA was detected between treatments using a one-way blocked ANOVA (where accession is considered as a block) beginning at day 4 after 90 mm NaCl (P < 0.0028; Fig. 1D). These results indicate that PSA is an accurate and sensitive metric for assessing plant biomass accumulation in response to salinity.
Figure 1.
Salinity-induced growth responses in a rice diversity panel. A to C, Relationship between PSA and conventional biomass metrics. Pearson correlation analyses were performed between PSA and shoot area (A), shoot fresh mass (B), and shoot dry mass (C). D, Comparisons of PSA between treatments at each of the 18 d of imaging. Differences between treatments at each time point were determined using a one-way blocked ANOVA, where accession is considered as a block (P < 0.0027). E, Comparison of salinity-induced growth response models between each of the five subpopulations defined by Zhao et al. (2011). The salinity-induced growth response was modeled with a decreasing logistic curve, and pairwise comparisons were made between each subpopulation. Aromatic accessions were excluded due to low n. Mean growth responses for each subpopulation are denoted by solid lines, while the se for each subpopulation is indicated by shadows. TRJ, Tropical japonica; TEJ, temperate japonica; IND, indica; ADM, admix.
To determine whether there were any differences in the salinity response among the five subpopulations as classified by Zhao et al. (2011), we calculated the salinity-induced growth response as the ratio of PSA in salt-treated plants over control plants. Aromatic lines were excluded due to small number of accessions. For each subpopulation, the salinity-induced growth response was modeled across all time points with a decreasing logistic curve. Therefore, on day 1 of salt treatment, the growth response is 1, and it begins to decrease after the onset of salinity stress and eventually flattens out as vegetative growth declines and plants transition to reproductive phase. Pairwise comparisons of growth response models revealed significant differences between several subpopulations (Table I). Notably, the tropical japonica subpopulation showed a significantly lower growth reduction in response to salinity when compared with other subpopulations, suggesting that this varietal group may be an important source for osmotic stress tolerance response during early stages of salinity stress (Fig. 1E). The admix and aus subpopulations showed the most severe reduction in growth. Aus accessions displayed the earliest reduction in PSA, with a significant difference observed 4 d after 90 mm salt application (P < 6.18 × 10–5). While for the admix subpopulation, significant differences between treatments were observed beginning 7 d after the onset of 90 mm salinity (P < 1.31 × 10–6).
Table I. Comparison of salinity-induced growth response models between subpopulations.
The mean growth response was calculated for each subpopulation and fitted to a decreasing logistic curve. Aromatic accessions were omitted due to low n. Subpopulations with the same letter indicate no significant differences (P < 10–4). Asym, Horizontal asymptote; xmid, value of x at the inflection point of the curve; scal, rate coefficient; AIC, Akaike Information Criterion; BIC, Bayesian Information Criterion; TeJ, temperate japonica; TrJ, tropical japonica.
| Subpopulation | Asym | xmid | scal | AIC | BIC | logLik | P > 10−4 |
|---|---|---|---|---|---|---|---|
| Admix | 0.997 | 32.4 | −6.66 | −3,560.161 | −3,536.05 | 1,785.08 | A |
| Aus | 0.999 | 28.9 | −4.82 | −4,015.505 | −3,991.30 | 2,012.75 | A, B |
| Indica | 0.979 | 30.0 | −5.14 | −5,152.778 | −5,126.87 | 2,581.39 | A |
| TeJ | 0.982 | 28.7 | −4.21 | −6,093.456 | −6,066.51 | 3,051.73 | B |
| TrJ | 0.983 | 30.8 | −4.49 | −5,465.48 | −5,439.12 | 2,737.74 |
Assessing Salinity-Induced Chlorophyll Responses
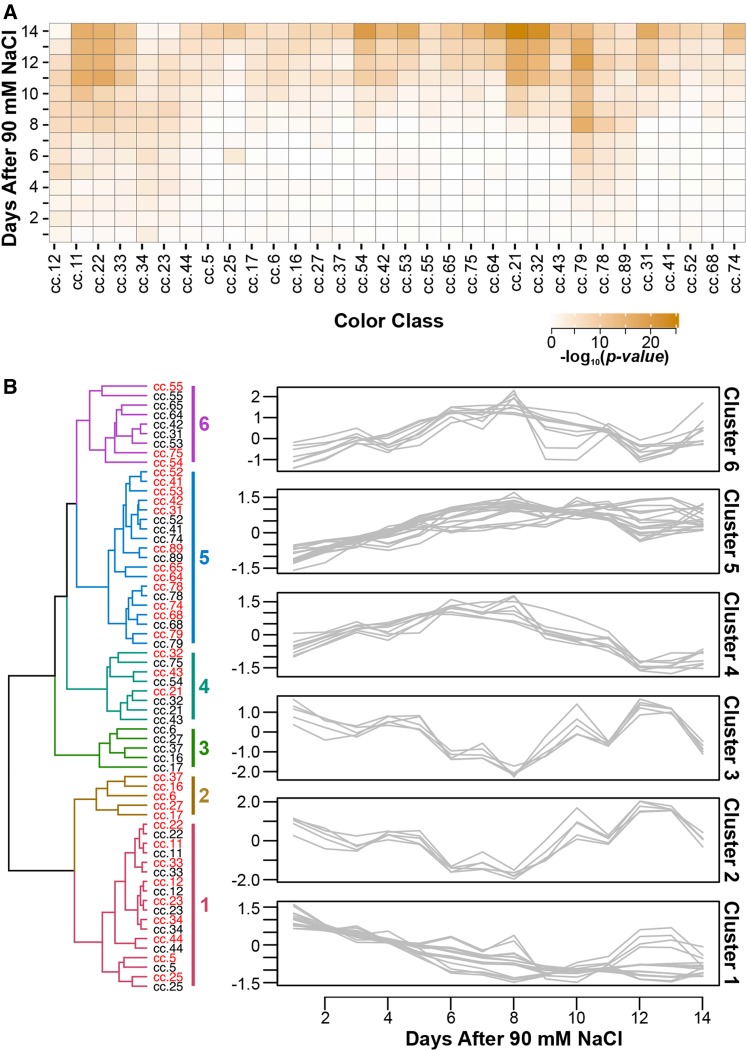
To assess the effects of salinity stress on leaf senescence, plants were imaged in a separate fluorescence-imaging chamber. Because the available functions are limited in LemnaGrid software, we developed an open-source processing software called Image Harvest to extract several spectral metrics from the 95,118 fluorescence images. Color ranges that may be indicative of salinity-induced chlorophyll responses were identified by utilizing an ad hoc image segmentation strategy that classified the range of colors present in all fluorescence images into 90 classes of color ranges. Based on our pixel classification strategy, we identified 32 color classes that showed significant differences between treatments after three or more days of salt stress across all 373 accessions (P < 0.00056; Fig. 2A). No difference between treatments was observed for any of the 32 color classes before the application of NaCl. Canonical correlation analysis between pixel classes within each time point showed strong correlations between color classes (Supplemental Data Sets S1 and S2). As senescence progresses, the color properties of the fluorescence signal emitted from stressed tissue will change over time. Because our ad hoc segmentation approach classifies pixels into discrete color classes, it is likely that over time, pixels that represent stressed tissue will change membership between classes with similar color properties. Therefore, to examine the relationship between color classes across time points, we performed hierarchal clustering analysis (HCA) using the mean temporal trend for each color class across all 373 accessions for each treatment. No clear distinction between treatments could be observed from hierarchal clustering. Rather, clustering seemed to be driven largely by the behavior of color classes over time (Fig. 2B). If fluorescence signals were influenced only by salinity stress, one would expect clusters to be formed based on treatment. These results suggest that developmental processes likely influence the fluorescence signals. However, the significant differences observed between treatments suggest that the onset of salinity may affect the timing or magnitude of these signals.
Figure 2.
The development of image-based fluorescence traits for monitoring chlorophyll responses to salinity. A, Salinity-responsive color classes were identified through comparisons between treatments at each time point via one-way ANOVA. Color classes were considered to be responsive to saline conditions if significant differences between treatments were observed in 3 or more days of 90 mm NaCl stress (P < 0.00056). B, Identification of color classes exhibiting similar trends over 14 d of 90 mm NaCl. HCA with complete linkage was performed using the mean value in each treatment for each color class. The six clusters are depicted to the right of the dendrogram. Labels in red indicate mean response in saline conditions, while those in black indicate control conditions. The right section summarizes the temporal trend captured by each cluster. The mean values for each color class were scaled and centered prior to clustering, so that the mean is 0 and variance is 1, and are represented on the y axis.
Significant differences between treatments were generally observed during the later time points, with the majority of color classes displaying significant differences after day 11 of 90 mm NaCl (Fig. 2A). However, color classes that displayed significant differences during the early stages of salinity stress tended to populate cluster 1. For instance, at 7 d after 90 mm NaCl, five of the seven color classes that displayed significant differences between treatments were members of cluster 1. This suggests that fluorescence responses represented by cluster 1 may be important digital markers for monitoring the early effects of salinity stress on chlorophyll responses in rice.
Development of Model for Association Analysis of Longitudinal Salinity-Induced Growth Responses
Because PSA was most strongly correlated with conventional biomass-related measurements and displayed significant differences between treatments during the early phase of salt treatment, we sought to identify genetic loci associated with the salinity-induced growth response (defined here as the square root-transformed ratio of the PSA of plants under salt stress over that of plants in control conditions) of 360 rice accessions for which phenotypic and genetic information was available (Famoso et al., 2011; Zhao et al., 2011). Briefly, for each accession and single nucleotide polymorphism (SNP), the growth response over 18 d is fitted with a decreasing logistic curve for each genotype. To perform statistical inference, the kinship matrix between lines is used to account for the population structure of the diversity panel and a first-order autoregressive covariance structure is used to account for the dependency between time points. For each SNP, a likelihood ratio test is carried out on whether the growth response curves are the same for the two genotypes. The model accounts for both genetic relationships between accessions due to rice subpopulation structure and the inherent nonindependent nature of daily observations.
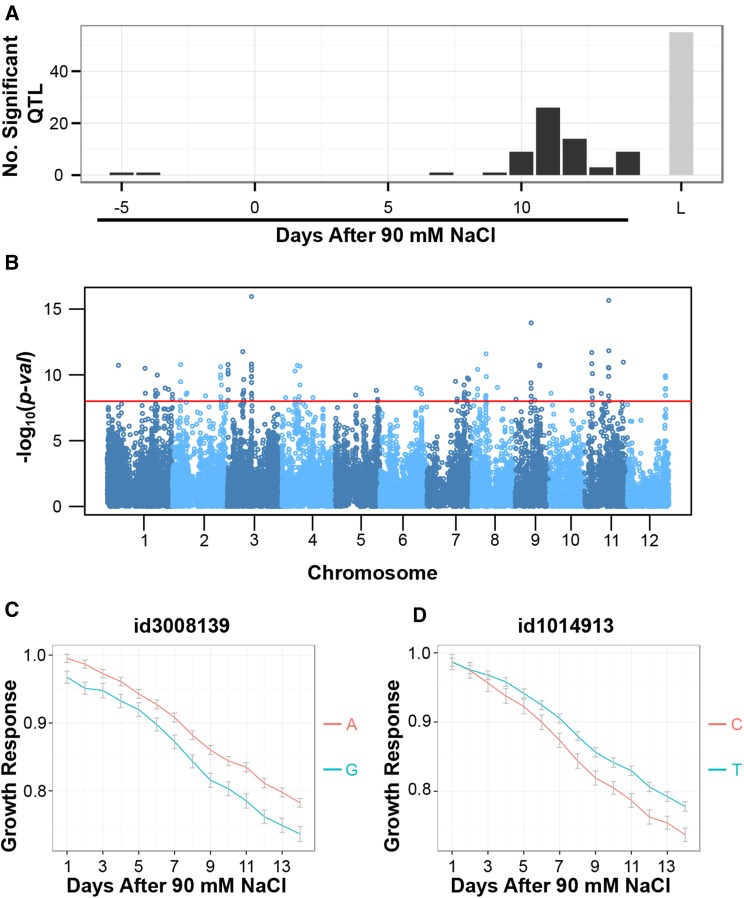
The longitudinal growth response model was compared to a conventional mixed model in which the response ratio at each individual time point was used as the phenotypic variable. While several peaks were identified with both methods, we observed a considerably higher number of significant QTL and considerable lower P values with the longitudinal model. For instance, a total of 115 highly significant SNPs, which corresponds to 55 QTL, were identified when the longitudinal model was used (P < 10–8; Fig. 3A; Supplemental Data Set S3). By contrast, the maximum number of significant QTL identified at only one time point was 26 when the segmented model was used (P < 10–4; Fig. 3A; Supplemental Data Set S4). Though the number of significant SNPs and the number of loci are not proportional, these results indicate that the collective analysis of multiple time points significantly improved the ability to identify loci associated with complex polygenic traits such as salinity-induced growth responses.
Figure 3.
Examining the genetic architecture of salinity-induced growth responses using conventional mixed-model and logistic growth response association analysis. A, Comparison of conventional mixed-model association mapping approach with logistic growth response association mapping approach. A conventional association mapping approach was performed at each time point using the salinity-induced growth response as a phenotypic measure. With the mixed-model approach, an SNP was determined to be significant if P < 10–4, while a threshold of P < 10–8 was used for the logistic growth response model. Significant SNPs within a 200-kb window were combined and considered as a single QTL. L, Logistic growth response model. B, Manhattan plot for the logistic growth response association analysis. The red horizontal line indicates a significance threshold of P < 10–8. C, Comparison of growth response trajectories between allelic groups for the significant association observed at approximately 16.3 Mb on chromosome 3. A indicates the major allele (frequency, 0.68), and G indicates the minor allele (frequency, 0.32). D, Growth trajectories of major and minor allele accessions for the signal observed at approximately 25 Mb on chromosome 1. T indicates the major allele (frequency, 0.79), and C indicates the minor allele (frequency, 0.21).
Genetic Architecture of Salinity-Induced Temporal Growth Response
Our genetic association analysis using the longitudinal model identified several highly significant peaks in close proximity to genes with putative roles in salt tolerance and or ion homeostasis in rice and other species based on published literature. A highly significant cluster of SNPs was located at approximately 16.3 Mb on chromosome 3 (id3008139; P < 1.11 × 10–16; Fig. 3B). This region was identified with both the longitudinal growth response and mixed-model genome-wide association (GWA) methods. The large peak on chromosome 3 is populated by 10 highly significant SNPs converging on an approximately 240-kb region and includes 89 gene models based on the Michigan State University Rice Annotation Project Release 7 (Supplemental Data Set S3). We found a gene within a 200-kb window surrounding the most significant SNP (id3008139) on chromosome 3 that encodes a putative Shaker family inward-rectifying potassium channel (POTASSIUM TRANSPORTER1; LOC_Os03g28120) gene (Supplemental Table S3). These K+ channels mediate cellular potassium uptake and have been shown to increase salinity tolerance via lowered Na+-to-K+ ratios in yeast (Saccharomyces cerevisiae) and rice (Obata et al., 2007). The accessions with the minor allele at this locus displayed a greater reduction in plant growth in response to saline conditions when compared with the major allele genotypes (Fig. 3C). The minor allele was represented at a greater proportion in the temperate japonica, aus, and admix subpopulations, with approximately 77%, 36%, and 46% of accessions retaining the allele, respectively, suggesting that salt sensitivity associated with this locus is present in multiple subpopulations.
On chromosome 1, a cluster of highly significant SNPs was detected around approximately 25 Mb (id1014913; P < 3.18 × 10–11). The minor allele at SNP id1014913 was significantly underrepresented in the japonica varietal groups, with no tropical japonica accessions and only a single temperate japonica variety possessing the minor allele, suggesting that this locus may be fixed between indica and japonica subspecies. In general, the accessions with the minor allele at id1014913 displayed a greater reduction in growth in response to salinity when compared with the major allele group, with a slight difference observed as early as 4 d after reaching the 90 mm NaCl and progressively greater difference during later time points (Fig. 3D). GWAS conducted with leaf Na+ content showed a minor peak at approximately 30 Mb, suggesting that this 5-Mb region on chromosome 1 may influence both growth as well as Na+ ion homeostasis (id1018154, 30.2 Mb, P < 8.44 × 10–5; Supplemental Fig. S3A). Varieties with the major allele at id1014913 had higher Na+ content and Na+-to-K+ ratio, as well as lower K+ content (P < 0.001; Supplemental Fig. S4, A and B). These results suggest that the mechanism underlying the more tolerant response observed in the major allelic group might be independent of ion homeostasis or accession from this allelic group may be able to tolerate higher cellular Na+ content without detrimental effects on growth.
Genetic Architecture of Salinity-Induced Chlorophyll Responses
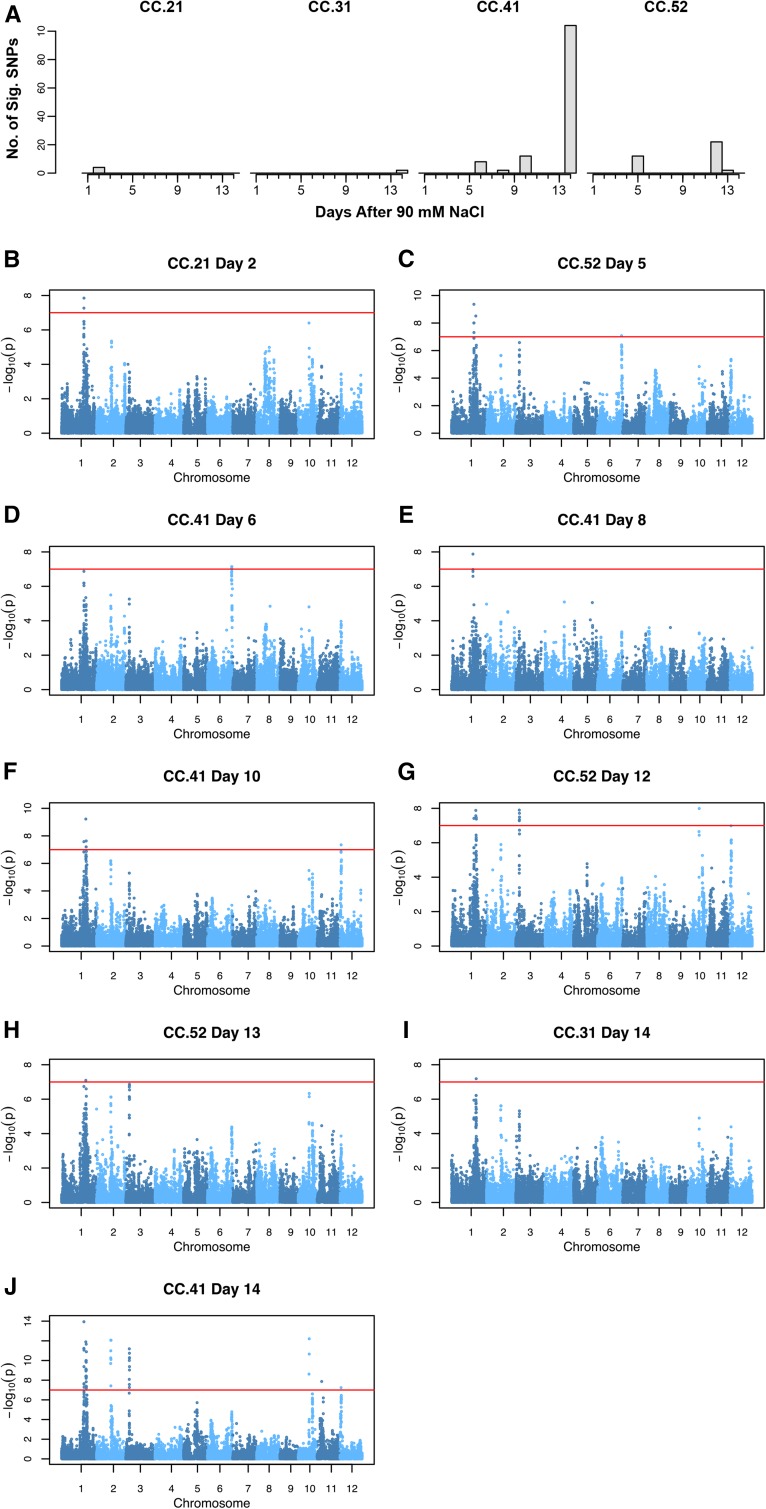
To identify loci associated with salinity-induced chlorophyll responses, association mapping was performed at each time point individually using a conventional mixed model for each of the 32 salinity-responsive color classes (those with P < 0.00056). For each color class and accession, we calculated the salinity response, which is defined as the percentage of total pixels from two side-view images accounted for by the color class in saline conditions minus the percentage of total pixels accounted by the class in control conditions. In total, four classes showed significant associations at one or more time points throughout the experiment (P < 1.0 × 10–7; Fig. 4A; Supplemental Figs. S5–S18; Supplemental Data Set S5). Highly significant signals were detected on chromosomes 1, 3, 6, and 10 for color classes 21, 31, 41, and 52 at multiple time points. HCA of color classes in control and saline conditions revealed strong similarities in the temporal behavior of color classes 31, 41, and 52 (Supplemental Figs. S19 and S20). Interestingly, HCA showed a clear separation between treatments for these color classes (Fig. 2B). The salinity response of classes 31, 41, and 52 were tightly grouped in cluster 5, while classes 41 and 52 displayed similar behavior in control conditions. These results indicate that these fluorescence-based metrics are measuring similar phenotypic responses, which may explain the overlapping genomic regions associated with these metrics.
Figure 4.
Examining the genetic architecture of salinity-induced fluorescence responses. A, Summary of the significant signals detected for color classes 21, 31, 41, and 52 at each time point after 90 mm NaCl. The x axis indicates the number of days after 90 mm NaCl application, while the y axis shows the number of SNPs with P < 10–7 that were detected with the conventional mixed-model GWA approach. B to J, Genome-wide P values for each fluorescent color class exhibiting significant genetic associations. The red horizontal line indicates a significance threshold of P < 10–7. The corresponding color class and day after 90 mm NaCl is given in the title above each plot.
The most significant association was observed on day 14 after 90 mm NaCl for class 41 (P < 1.14 × 10–14; id1015984). The corresponding SNP was located at approximately 27.6 Mb on chromosome 1. Notably, this region was also associated with several other metrics at later time points during the experiment. For instance, the earliest significant signal for class 21 localized to this same region on chromosome 1 and was detected only on day 2 after 90 mm NaCl. However, significant associations were not observed again in this region until day 5 after 90 mm NaCl (class 52; P < 9.91 × 10–9; id1015991).
DISCUSSION
Excess soil sodium reduces plant growth through osmotic and ionic effects. Shortly after exposure to excess Na+ (before Na+ accumulates in the cell), the osmotic potential outside the cell is reduced, which impairs cell expansion at the cellular level and disrupts plant-water relations, carbon assimilation, and transpiration at the plant level. In time, Na+ accumulates in the cytoplasm to toxic levels, resulting in cell death and senescence. The temporal nature of excess Na+ on plant growth adds an additional level of complexity and complicates the study of physiological mechanisms, conferring tolerance and the underlying genetic basis of these polygenic traits.
When quantifying these dynamic processes, the choice of a sampling time point for end point measurements is critical for detecting variance for the trait of interest. When evaluating large diversity panels for a given trait, determining the appropriate time point to sample is very difficult, because such germplasm collections are designed to capture a large portion of the genetic diversity in a species and should exhibit a wide range of phenotypic values. Therefore, sampling at only a single time point for the trait of interest may yield a significant underrepresentation of the variance for the trait in the population. The quantification of the trait across many time points, which, in some instances, may be laborious and unfeasible, eliminates many of the pitfalls associated with single end point measurements and adds another dimension that may help dissect and further elucidate the complex biological processes underlying the phenotype. However, with the advent of high-throughput image-based phenomics platforms, the quantification of morphological and physiological traits across multiple time points is expected to become routine for many laboratories.
Recent studies by Moore et al. (2013), Yang et al. (2014), and Würschum et al. (2014) utilized image-based phenomics to identify QTL involved with temporal developmental dynamics in Arabidopsis, rice, and triticale (× Triticosecale), respectively (Moore et al., 2013; Würschum et al., 2014; Yang et al., 2014). Genetic analysis conducted at discrete time points identified several time-specific QTL that partially influence the final phenotype but would only be detected at a specific time point. In this study, a similar univariate association analysis was performed at each time point for the 32 fluorescence color classes. The significant signals identified from fluorescence imaging displayed a high degree of temporal dynamics. For instance, a region localized to approximately 27.6 Mb on chromosome 1 displayed significant associations in both the early and later time points of the experiment, with the earliest significant signal in this region being detected on day 2 after 90 mm NaCl for class 21, but were not observed again in this region until day 5 (class 52; 9.91 × 10–9; id1015991). While on the final day of the experiment, significant associations were detected only for classes 41 and 31. The presence of these signals at discontinuous intervals across time points suggests that these loci may influence the processes that give rise to the fluorescence phenotype and would have been missed if fluorescence responses were measured at a single time point. The inclusion of multiple time points provides additional insight into the genetic architecture underlying dynamic fluorescence responses.
The presence of favorable alleles in close proximity provides an opportunity to utilize this region for breeding programs. Several significant associations were identified for growth response, chlorophyll health, and leaf Na+ content in a region spanning approximately 5 Mb on chromosome 1 (approximately 25–30 Mb). This overlap among multiple traits is supported by canonical correlation analysis of multiple traits derived from image-based and conventional phenotyping (Supplemental Data Set S2). Class 41 showed a strong negative correlation (r2 = –0.50 at day 12 after 90 mm salt application) with growth (PSA) reduction during salinity, indicating that lines exhibiting a more tolerant growth response also exhibited reduced senescence (Supplemental Data Set S2). No significant/strong correlations were observed for image-based traits and leaf ion content. Our results suggest that this 5-Mb region on chromosome 1 may influence both growth and senescence during salinity and may serve as an important source of genetic diversity for development of salt-tolerant rice varieties that are able to maintain growth and minimize senescence in saline conditions. Interestingly, several other studies have identified QTL associated with seedling survival, Na+ uptake and Na+-to-K+ ratio, chlorophyll content, and ion homeostasis spanning this region on chromosome 1 (Koyama et al., 2001; Lin et al., 2004; Thomson et al., 2010). These studies were conducted during the early vegetative growth stage of rice using a similar NaCl concentration as this study and support the conclusion that these QTL may regulate aspects of salinity tolerance during the early tillering stage in rice. While plants have distinct mechanisms to respond at the cellular level to the osmotic and ionic effects of salinity, salt tolerance, defined as the ability to maintain growth in saline conditions, may be the result of tolerant ionic or osmotic responses or a combination of both. However, dissecting the precise physiological response mechanisms (osmotic or ionic) underlying these loci requires further experimentation.
Functional data analysis allows complex longitudinal phenotypic data to be reduced to a single mathematic function with a limited set of parameters that fully capture and describe the dynamics of biological processes (Paine et al., 2012). This approach simplifies phenotypic comparisons between individuals across multiple time points and can be used to accurately predict future outcomes. Time series data sets derived from image-based phenotyping have been combined with functional data analysis to examine shoot and root growth in response to various environmental conditions (Walter et al., 2002; van der Weele et al., 2003; Chen et al., 2014; Poiré et al., 2014; Bac-Molenaar et al., 2015). Functional data analysis can be combined with conventional genetic analysis such as linkage mapping and GWAS to identify loci that may be regulating dynamic processes (Cui et al., 2006; Wu and Lin, 2006; He et al., 2010; Das et al., 2011; Bac-Molenaar et al., 2015).
In our experiments, the temporal and polygenic nature of plant salinity responses required the development of unique statistical approaches that combine information across time points to identify loci with minor effects that regulate the adaptation of growth to saline conditions. In this study, we have leveraged unique image processing software and longitudinal GWA methods to examine the genetic architecture of salinity-induced growth responses in rice. The salinity-induced growth response GWA model greatly increased the ability to detect SNPs with minor effects compared with the conventional mixed-model approach. A highly significant cluster of SNPs was located at approximately 16.3 Mb on chromosome 3 (id3008139; P = 1.11 × 10–16) and was identified with both the longitudinal growth response and mixed-model GWA methods. However, additional highly significant signals that impacted growth, senescence, and Na+ content were detected only with the longitudinal model. Similar power gains for detecting genetic associations from longitudinal phenotypes have been reported by Xu et al. (2014) and Wu and Lin (2006). However, to date, no studies have leveraged longitudinal genetic algorithms for large-scale association studies of image-based data from plants. Our results suggest that longitudinal phenotypes can provide an advantage over typical cross-sectional (i.e. end point) phenotypic data for studying the genetic architecture of complex abiotic stress responses in plants. This study highlights the potential of these new technologies and provides a framework for future studies to integrate next-generation phenotyping technology with association mapping to understand the genetic architecture of complex polygenic traits such as salinity tolerance.
MATERIALS AND METHODS
Greenhouse Conditions and Salt Treatment
Seeds from 373 genotypes from the rice (Oryza sativa) diversity panel were surface sterilized with fungicide, thiram, and germinated on moist paper towels in plastic boxes for 3 d (Famoso et al., 2011; Zhao et al., 2011). Three uniformly germinated seeds of each genotype were transplanted to pots (150-mm diameter × 200-mm height) filled with 2.6 kg of UC Mix and placed into square containers to allow for water to collect. Plants were thinned to one seedling per pot 6 d after transplanting (DAT). For the first 7 DAT, each pot was watered daily with approximately 150 mL from the top of the pot. Over the course of the three experiments, the greenhouse temperatures during the day averaged 28.8°C (±2.02°C, sd) and 26.0°C (±1.01°C, sd) at night. Relative humidity was maintained at 63.4% (±9.04%, sd) during the day and 69.7% (±1.73%, sd) at night (Rotation Atomizer Defensor ABS3, Condair).
Eight DAT, each pot was watered to a uniform weight so that approximately 600 mL of water was maintained in the soil. For the salt-stressed plants, 100 mL of NaCl solution (270 mm NaCl:9.9 mm CaClβ) was applied to the square dish, and small holes in the bottom of the pots allowed for the infiltration of salt into the soil through capillary action. Salt treatment was applied in two steps of 45 mm to reach a final concentration of 90 mm at 10 and 13 DAT (Supplemental Fig. S1A). Control plants received 100 mL of water on days 10 and 13 (Supplemental Fig. S2).
The experiment involved two Smarthouses that were used consecutively for three periods, the periods forming blocks. In each Smarthouse, 432 pots were situated in 24 lanes × 18 positions. A split-plot design was employed with two consecutive pots having the same genotype, but with the two different salt treatments randomly assigned to them. For each period, a blocked, partially replicated design was used to allocate the 378 genotypes to the 432 pairs of pots in the two Smarthouses and was generated using DiGGer, a package for the R statistical computing environment (Coombes, 2009; R Core Team, 2014). There were five check genotypes that were always included in a Smarthouse and 373 genotypes, of which 49 were duplicated in a period and the remaining 324 were unreplicated. The duplicated genotypes differed between periods so that 147 genotypes were duplicated in total. The entire data set of RGB and FLUO images can be accessed through the iPlant Collaborative (http://mirrors.iplantcollaborative.org/browse/iplant/home/shared/walia_rice_salt).
RGB/Visible Spectrum Image Acquisition and Processing
Plant imaging was initiated 2 d before the first salt application (8 DAT) to provide a baseline for the determination of growth rate. To assess plant growth and morphological traits, plants were imaged in an imaging chamber using a 5-megapixel visible/RGB camera (Basler Pilot piA2400-12gc; Supplemental Fig. S1C). Lighting conditions, plant positioning, and camera settings were fixed throughout the experiment. Plants were imaged for 14 d after the final salt application (until 27 DAT). For each plant, two side-view images, in which the pot was rotated 90°, and a single top-view image were acquired daily.
The 142,671 color (RGB) images were processed in LemnaGrid (LemnaTec) to remove nonplant pixels from images. RGB processing consisted of two main steps: (1) color classification for object extraction and (2) noise reduction (Supplemental Fig. S21). To extract plant pixels from the background, the colors of the image were assigned to object (plant) and background (nonplant) color classes (Supplemental Fig. S22). Briefly, a set of predefined colors was selected manually to represent the range of colors present in shoot tissues. For each image, pixels are assigned to color classes using the nearest neighbor method. The nearest neighbor method searches in a set of predefined colors to find the most similar (nearest neighbor), defined as the smallest Euclidean distance, for any given pixel. Pixels are then assigned membership to the most similar predefined colors. Nonplant pixels were further removed from the processed images using a series of erosion and dilation steps. The LemnaGrid devices used for processing RGB images and corresponding Image Harvest/OpenCV functions are listed in Supplemental Table S4. Seven traits were derived from RGB images and were used to describe plant growth (Supplemental Table S1). To determine the effects of salinity treatment on each trait, a one-way ANOVA was performed at each individual time point, where treatment was considered as a fixed effect and accession was considered as a random effect. Raw P values were adjusted using a Bonferroni correction of α = 0.05, which corresponds to a raw P value < 0.0028.
Fluorescence Image Acquisition and Processing
To assess the effects of salinity stress on chlorophyll content or leaf senescence, plants were imaged in a separate fluorescence-imaging chamber illuminated with a constant blue light (400–500 nm). Fluorescence images were captured using a 1.4-megapixel camera (Basler Scout scA1400-17gc) with a high-pass filter, which captures steady-state chlorophyll fluorescence from 500 to 700 nm. To prevent the detection of fluorescence signals from algae, only images captured from two side-view angles were used for further analysis (Supplemental Fig. S1C).
To process the 95,118 fluorescence images, we developed an open-source software called Image Harvest (https://git.unl.edu/aknecht2/ih). Image Harvest is written in Python and utilizes functions in OpenCV to extract plant pixels from LemnaTec images (Bradski, 2000). For the execution of the image processing pipelines on high-performance computing clusters, Image Harvest utilizes Pegasus, which translates a series of computational tasks into a Directed Acyclic Graph and utilizes HTCondor to execute the jobs in parallel on the Open Science Grid (Thain et al., 2005; Sfiligoi et al., 2009; Deelman et al., 2015). The processing workflow was developed using a set of 95,118 side-view images, and the quality of each processing step was determined manually. The workflow consists of three main steps: pot masking, background removal, and image cropping. The background removal steps are accomplished using the colorFilter function. Due to the bright plant and dark background, filtering pixels by intensity serves as the primary tool to distinguish between plant and nonplant pixels. Specifically, filtering based on the net intensity (r + g + b) and filtering by the difference between the red and green channels are the major operations. The first filtering keeps pixels from both the plant and pot, whereas the second threshold removes the green pixels of the pot, as well as orange pixels that may show up from debris fluorescing in the image. The image is then cropped based on contours (connected areas of pixels) in the image through the use of the contourCut function.
To identify color ranges that may be representative of senescent tissue, histograms representing the range of pixel values for red and green color channels present in all fluorescence images were generated and segmented into 90 color classes. All possible pairwise combinations of R and G classes were generated, resulting in a total of 90 final color classes that represented the full range of color values in each fluorescence image. For each image, pixels were allocated to each bin depending on the color value. The number of pixels for each bin were summed from both side-view images and expressed as a percentage of the plant area from the two side-view images. To identify color classes displaying differences between treatments across all 373 accessions, a one-way blocked ANOVA was performed at each individual time point, where treatment was considered as a fixed effect and accession as a block. Raw P values were adjusted using a Bonferroni correction of α = 0.05, which corresponds to a raw P value < 0.00056.
Determination of Sodium and Potassium Content
The newest expanding leaf (third leaf) was marked on each plant at the time of the first salt application. At the end of the experiment, this leaf was harvested, rinsed with milliQ water, and patted dry. The samples were placed into 50-mL conical tubes, and after being dried at 60°C overnight, the dry weight was determined for each leaf sample. Leaves were digested in 10 mL of 1% (w/w) nitric acid (70% [w/w] Nitric Acid; Chem-Supply NA001-500M, Gillman) at 70°C for 8 h. Samples were diluted in milliQ water at a 1:5 or 1:10 ratio, and Na+ and K+ content was determined against the appropriate Na+ and K+ standard (100:1,000–500:1,000 μm NaCl:KCl) using flame photometry (Model 420 Flame Photometer, Sherwood Scientific). Ion content was calculated per gram of dry mass as described by Munns et al. (2010).
Hierarchical Clustering Analysis of Fluorescence Color Classes
Hierarchical clustering analysis was used to identify and visualize similar temporal trends between fluorescence color classes. For each color class and treatment, the mean response across all 373 accessions was determined. Because the values differ by orders of magnitude between color classes, the raw mean values were transformed to Z-scores using the scale function in R (R Core Team, 2014). Clustering was done with the complete-linkage method using Dynamic Time Warping as a distance metric.
Canonical Correlation Analysis
Pearson correlation analysis was conducted between fluorescence and growth image-based metrics at each time point to examine the relationships between traits. At each time point, correlation analysis was done using the rcorr function with the Pearson option in the Hmisc package in R (Harrell, 2014; R Core Team, 2014).
Comparison of Salinity-Induced Growth Response Models between Subpopulations
To determine whether there were any differences in the salinity response among the five subpopulations (admix, aus, temperate japonica, tropical japonica, and indica), the mean salinity-induced growth response, defined here as the square root-transformed ratio of the PSA of plants under salt stress over that of plants under normal growth situations, was calculated for each subpopulation. Aromatic accessions were excluded from the analysis due to low n. The mean response for each subpopulation was fitted to a decreasing logistic function with a first-order autoregressive covariance structure over time using gnls and SSLogis functions in the nlme package in R (R Core Team, 2014; Pinheiro et al., 2015). For each pairwise comparison between subpopulations, two models were fit: one where the parameters of the model are considered to be different between subpopulations and one where the parameters are considered to be similar between subpopulations. An ANOVA was used to test the null hypothesis that the parameters are similar between subpopulations.
Longitudinal Salinity-Induced Growth Response Association Analysis
To identify the SNPs associated with characteristics of the growth pattern, a unique functional association-mapping model was applied (Supplemental Fig. S1C). The salt stress-induced growth response is measured as the square root-transformed ratio of the PSA of plants under salt stress over that of plants under normal growth situations. For each line and a specific SNP, the growth response over 18 d is fitted with a decreasing logistic curve for each genotype, with a first-order autoregressive covariance structure over time. For each SNP, a likelihood ratio test is carried out on whether the growth response curves are the same for the two genotypes. At each time point, on the other hand, the relatedness between lines is accounted for by a random line effect with the covariance matrix proportional to the kinship matrix estimated by the software package EMMA (Kang et al., 2008).
In summary, for each line, the growth response is modeled with a nonlinear mixed model. While at each time point, the model is the linear mixed model commonly used for plant association studies as in Kang et al. (2008) and others using the kinship matrix between lines to account for the population structure (Yu et al., 2006; Malosetti et al., 2007; Zhao et al., 2007, 2011; Huang et al., 2010; Famoso et al., 2011). For each line across time points, on the other hand, the model reduces to a nonlinear mixed model with a first-order autoregressive covariance structure. Taken together, this results in an extended mixed model with correlation structure both across time and also across lines. This unique approach is required to account for the two types of dependency structure to obtain valid inference results. Further details on the extended mixed model can be found in Supplemental Methods S1.
From the 44,000 SNPs on the array, we removed SNPs with missing values for more than 10% of the lines and those with minor allele frequency less than 10% (Zhao et al., 2011). This results in 26,258 SNPs for association analysis. For each SNP, a likelihood ratio test is carried out on whether the growth response curves are the same for the two genotypes. The P values from the analysis of three replicates are combined with Fisher’s method to obtain the combined P value, which is subsequently used to identify significant SNPs (Hartung et al., 2011). Significant SNPs within a 200-kb window were combined and considered as a single QTL. The 200-kb window was chosen based on the estimated linkage disequilibrium decay in this diversity panel (Zhao et al., 2011).
Conventional Mixed-Model Genome-wide Association Analysis
A conventional mixed-model genome-wide association analysis was used to identify genomic regions associated with leaf ion content (Na+, Na+:K+, and K+) and fluorescence imaging traits. The mixed linear model can be summarized as: y = Xβ + Cγ + Zu + e, where β and γ represent coefficient vectors for SNP effects and subpopulation principal components, respectively, which are fixed effects, u is a random effect that accounts for population structure and relatedness, Z represents the corresponding design matrices, and e is the random error term. The mixed model was implemented using EMMA in R using the same 26,258 SNPs that were used for longitudinal salinity-induced growth response association analysis (Kang et al., 2008).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic illustrating the experimental design and pipeline for the analysis of high-throughput phenotyping data.
Supplemental Figure S2. Schematic illustrating NaCl application.
Supplemental Figure S3. GWA analysis of leaf Na+, K+, and Na+:K+ content.
Supplemental Figure S4. Distribution of leaf ion content observed between allelic groups at id1018154 and id1014913.
Supplemental Figure S5. GWA analysis of fluorescence color classes, day 1.
Supplemental Figure S6. GWA analysis of fluorescence color classes, day 2.
Supplemental Figure S7. GWA analysis of fluorescence color classes, day 3.
Supplemental Figure S8. GWA analysis of fluorescence color classes, day 4.
Supplemental Figure S9. GWA analysis of fluorescence color classes, day 5.
Supplemental Figure S10. GWA analysis of fluorescence color classes, day 6.
Supplemental Figure S11. GWA analysis of fluorescence color classes, day 7.
Supplemental Figure S12. GWA analysis of fluorescence color classes, day 8.
Supplemental Figure S13. GWA analysis of fluorescence color classes, day 9.
Supplemental Figure S14. GWA analysis of fluorescence color classes, day 10.
Supplemental Figure S15. GWA analysis of fluorescence color classes, day 11.
Supplemental Figure S16. GWA analysis of fluorescence color classes, day 12.
Supplemental Figure S17. GWA analysis of fluorescence color classes, day 13.
Supplemental Figure S18. GWA analysis of fluorescence color classes, day 14.
Supplemental Figure S19. Trajectories of the 32 salinity-responsive fluorescence color classes.
Supplemental Figure S20. Mean temporal trends of the 32 salinity-responsive fluorescence color classes.
Supplemental Figure S21. LemnaGrid pipeline used to process RGB side- and top-view images.
Supplemental Figure S22. Color classes used to define foreground and background pixels from RGB side- and top-view images for the LemnaGrid nearest-neighbor foreground-background color separation method.
Supplemental Table S1. Seven digital traits used to describe plant growth responses.
Supplemental Table S2. Accuracy of digital traits for describing biomass and shoot area.
Supplemental Table S3. Candidate genes underlying significant SNPs.
Supplemental Table S4. LemnaGrid devices used for processing RGB images and corresponding Image Harvest/OpenCV functions.
Supplemental Data Set S1. Pearson correlation between PSA and 32 fluorescence color classes in control and saline conditions.
Supplemental Data Set S2. Pearson correlation of salt-induced responses for PSA and fluorescence color classes.
Supplemental Data Set S3. Genes located within 200 kb of significant SNPs associated with salinity-induced growth responses identified with longitudinal GWA analysis (P < 10–8).
Supplemental Data Set S4. Genes located within 200 kb of significant SNPs associated with salinity-induced growth responses identified with conventional mixed model GWA analysis (P < 10–4).
Supplemental Data Set S5. Genes located within 200 kb of significant SNPs associated with salinity-induced fluorescence responses identified with conventional mixed model GWA analysis (P < 10–7).
Supplemental Methods S1. Extended mixed model for association mapping with a growth response curve.
Supplementary Material
Acknowledgments
We thank Dr. Aaron Schmitz for multiplying seed stocks; Dr. Rachel Burton, Helli Meinecke, Dr. Alex Garcia, Richard Norrish, Dr. Guntur Tanjung, Evi Guidolin, Robin Hosking, Lidia Mischis, Nicki Bond, and Fiona Groskreutz for providing technical and organizational support; and Dr. David Swanson and Dr. Adam Caprez for providing support in the area of high-performance computing.
Glossary
- QTL
quantitative trait loci
- PSA
projected shoot area
- HCA
hierarchal clustering analysis
- DAT
days after transplanting
Footnotes
This work was supported by the National Science Foundation Plant Genome Research (grant no. 1238125 to D.W. and H.W.) and the National Cooperative Research Infrastructure Strategy.
Articles can be viewed without a subscription.
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Asano T, Hayashi N, Kobayashi M, Aoki N, Miyao A, Mitsuhara I, Ichikawa H, Komatsu S, Hirochika H, Kikuchi S, et al. (2012) A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J 69: 26–36 [DOI] [PubMed] [Google Scholar]
- Azaizeh H, Steudle E (1991) Effects of salinity on water transport of excised maize (Zea mays L.) roots. Plant Physiol 97: 1136–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac-Molenaar JA, Vreugdenhil D, Granier C, Keurentjes JJB (April 28, 2015) Genome-wide association mapping of growth dynamics detects time-specific and general quantitative trait loci. J Exp Bot http://dx.doi.org/10.1093/jxb/erv176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Parent B, Tester M (2010) High-throughput shoot imaging to study drought responses. J Exp Bot 61: 3519–3528 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla P, Mackell D, Deal K, Gregorio G (2002) RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp Agric Sci 85: 68–76 [Google Scholar]
- Boursiac Y, Boudet J, Postaire O, Luu DT, Tournaire-Roux C, Maurel C (2008) Stimulus-induced down-regulation of root water transport involves reactive oxygen species-activated cell signalling and plasma membrane intrinsic protein internalization. Plant J 56: 207–218 [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C (2005) Early effects of salinity on water transport in Arabidopsis roots: molecular and cellular features of aquaporin expression. Plant Physiol 139: 790–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradski G (2000) The OpenCV Library. Dr Dobb's J Softw Tools 25: 120–126
- Brown TB, Cheng R, Sirault XRR, Rungrat T, Murray KD, Trtilek M, Furbank RT, Badger M, Pogson BJ, Borevitz JO (2014) TraitCapture: genomic and environment modelling of plant phenomic data. Curr Opin Plant Biol 18: 73–79 [DOI] [PubMed] [Google Scholar]
- Busemeyer L, Ruckelshausen A, Möller K, Melchinger AE, Alheit KV, Maurer HP, Hahn V, Weissmann EA, Reif JC, Würschum T (2013) Precision phenotyping of biomass accumulation in triticale reveals temporal genetic patterns of regulation. Sci Rep 3: 2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B (2014) Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165: 688–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal M, Martínez V, Alcaraz CF (1999) Physiological function of water channels as affected by salinity in roots of paprika pepper. Physiol Plant 105: 95–101 [Google Scholar]
- Cheeseman JM. (1988) Mechanisms of salinity tolerance in plants. Plant Physiol 87: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26: 4636–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S (2014) Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA 111: 6497–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes N (2009) Digger design search tool in R. http://www.austatgen.org/software (February 25, 2015)
- Cui Y, Zhu J, Wu R (2006) Functional mapping for genetic control of programmed cell death. Physiol Genomics 25: 458–469 [DOI] [PubMed] [Google Scholar]
- Das K, Li J, Wang Z, Tong C, Fu G, Li Y, Xu M, Ahn K, Mauger D, Li R, et al. (2011) A dynamic model for genome-wide association studies. Hum Genet 129: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30: 497–507 [DOI] [PubMed] [Google Scholar]
- Deelman E, Vahi K, Juve G, Rynge M, Callaghan S, Maechling PJ, Mayani R, Chen W, da Silva RF, Livny M, et al (2015) Pegasus, a workflow management system for science automation. Futur Gener Comput Syst 46: 17–13
- Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W. (2004) Rapid and tissue-specific accumulation of solutes in the growth zone of barley leaves in response to salinity. Planta 219: 515–525 [DOI] [PubMed] [Google Scholar]
- Fricke W, Peters WS (2002) The biophysics of leaf growth in salt-stressed barley: a study at the cell level. Plant Physiol 129: 374–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem ME, Albacete A, Martínez-Andújar C, Acosta M, Romero-Aranda R, Dodd IC, Lutts S, Pérez-Alfocea F (2008) Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.). J Exp Bot 59: 3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth M, Mäser P (2007) Potassium transporters in plants: involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett 581: 2348–2356 [DOI] [PubMed] [Google Scholar]
- Golzarian MR, Frick RA, Rajendran K, Berger B, Roy S, Tester M, Lun DS (2011) Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung J, Knapp G, Sinha BK (2011) Statistical Meta-Analysis with Applications. John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- He Q, Berg A, Li Y, Vallejos CE, Wu R (2010) Mapping genes for plant structure, development and evolution: functional mapping meets ontology. Trends Genet 26: 39–46 [DOI] [PubMed] [Google Scholar]
- Honsdorf N, March TJ, Berger B, Tester M, Pillen K (2014) High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS One 9: e97047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Kaneko T, Sugimoto G, Sasano S, Panda SK, Shibasaka M, Katsuhara M (2011) Mechanisms of water transport mediated by PIP aquaporins and their regulation via phosphorylation events under salinity stress in barley roots. Plant Cell Physiol 52: 663–675 [DOI] [PubMed] [Google Scholar]
- Horie T, Karahara I, Katsuhara M (2012) Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice (N Y) 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R (2006) A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 142: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42: 961–967 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FEH Jr (2014) Hmisc: Harrell Miscellaneous. http://biostat.mc.vanderbilt.edu/Hmisc (November 20, 2013)
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama ML, Levesley A, Koebner RMD, Flowers TJ, Yeo AR (2001) Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 1: 406–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz A, Mehlmer N, Zapf S, Mueller TD, Wurzinger B, Pfister B, Csaszar E, Hedrich R, Teige M, Becker D (2013) Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol Plant 6: 1274–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108: 253–260 [DOI] [PubMed] [Google Scholar]
- Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, Chu C, Wang X (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84: 19–36 [DOI] [PubMed] [Google Scholar]
- Lutts S, Bouharmont JMKJ (1996) NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann Bot (Lond) 5: 389–398 [Google Scholar]
- Malosetti M, van der Linden CG, Vosman B, van Eeuwijk FA (2007) A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Ballesta MC, Aparicio F, Pallás V, Martínez V, Carvajal M (2003) Influence of saline stress on root hydraulic conductance and PIP expression in Arabidopsis. J Plant Physiol 160: 689–697 [DOI] [PubMed] [Google Scholar]
- Matsuda K, Riazi A (1981) Stress-induced osmotic adjustment in growing regions of barley leaves. Plant Physiol 68: 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian A, Oomen RJFJ, Isayenkov S, Sentenac H, Maathuis FJM, Véry AA (2011) Overexpression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. Plant J 68: 468–479 [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CR, Johnson LS, Kwak IY, Livny M, Broman KW, Spalding EP (2013) High-throughput computer vision introduces the time axis to a quantitative trait map of a plant growth response. Genetics 195: 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239–250 [DOI] [PubMed] [Google Scholar]
- Munns R, Greenway H, Delane R, Gibbs J (1982) Ion concentration and carbohydrate status of the elongating leaf tissue of Hordeum vulgare growing at high external NaCl II. Cause of growth reduction. J Exp Bot 33: 574–583 [Google Scholar]
- Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253: 201–218 [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, et al. (2012) Wheat grain yield on saline soils is improved by an ancestral Na⁺ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Munns R, Passioura J (1984) Effect of prolonged exposure to NaCl on the osmotic pressure of leaf xylem sap from intact, transpiring barley plants. Aust J Plant Physiol 11: 497 [Google Scholar]
- Munns R, Termaat A (1986) Whole-plant responses to salinity. Aust J Plant Physiol 13: 143 [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Munns R, Wallace PA, Teakle NL, Colmer TD (2010) Measuring soluble ion concentrations (Na+, K+, Cl−) in salt-treated plants. Methods Mol Biol 639: 371–382 [DOI] [PubMed] [Google Scholar]
- Negrão S, Courtois B, Ahmadi N, Abreu I, Saibo N, Oliveira MM (2011) Recent updates on salinity stress in rice: from physiological to molecular responses. CRC Crit Rev Plant Sci 30: 329–377 [Google Scholar]
- Obata T, Kitamoto HK, Nakamura A, Fukuda A, Tanaka Y (2007) Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiol 144: 1978–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine CET, Marthews TR, Vogt DR, Purves D, Rees M, Hector A, Turnbull LA (2012) How to fit nonlinear plant growth models and calculate growth rates: an update for ecologists. Methods Ecol Evol 3: 245–256 [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D; R Core Team (2015) {nlme}: Linear and Nonlinear Mixed Effects Models. R package version 3.1-121. http://CRAN.R-project.org/package=nlme (June 30, 2015)
- Poiré R, Chochois V, Sirault XRR, Vogel JP, Watt M, Furbank RT (2014) Digital imaging approaches for phenotyping whole plant nitrogen and phosphorus response in Brachypodium distachyon. J Integr Plant Biol 56: 781–796 [DOI] [PubMed] [Google Scholar]
- R Core Team (2014) R: A Language and Environment for Statistical Computing. http://www.r-project.org (July 8, 2015)
- Rajendran K, Tester M, Roy SJ (2009) Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ 32: 237–249 [DOI] [PubMed] [Google Scholar]
- Rawson HM, Munns R (1984) Leaf expansion in sunflower as influenced by salinity and short-term changes in carbon fixation. Plant Cell Environ 7: 207–213 [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rivandi J, Miyazaki J, Hrmova M, Pallotta M, Tester M, Collins NC (2011) A SOS3 homologue maps to HvNax4, a barley locus controlling an environmentally sensitive Na+ exclusion trait. J Exp Bot 62: 1201–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Mieulet D, Hubberten H, Obata T, Hoefgen R, Fernie AR, Fisahn J, Segundo BS, Guiderdoni E, Schippers JHM, et al. (2013) SALT-RESPONSIVE ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfiligoi I, Bradley DC, Holzman B, Mhashilkar P, Padhi S, Wurthwein F (2009) The pilot way to grid resources using glideinWMS. WRI World Congr 2: 428–432 [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovak R, Göschl C, Su X, Shimotani K, Shiina T, Busch W (2014) A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell 26: 2390–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi HT, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al. (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al. (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53: 11–28 [DOI] [PubMed] [Google Scholar]
- Thain D, Tannenbaum T, Livny M (2005) Distributed computing in practice: the Condor experience. Concurr Comput Pract Exp 17: 323–356 [Google Scholar]
- Thomson MJ, Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, et al. (2010) Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice (N Y) 3: 148–160 [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee CR, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110: E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weele CM, Jiang HS, Palaniappan KK, Ivanov VB, Palaniappan K, Baskin TI (2003) A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth: roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiol 132: 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A, Spies H, Terjung S, Küsters R, Kirchgessner N, Schurr U (2002) Spatio-temporal dynamics of expansion growth in roots: automatic quantification of diurnal course and temperature response by digital image sequence processing. J Exp Bot 53: 689–698 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen Z, Cheng J, Lai Y, Wang J, Bao Y, Huang J, Zhang H (2012) QTL analysis of Na+ and K+ concentrations in roots and shoots under different levels of NaCl stress in rice (Oryza sativa L.). PLoS One 7: e51202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Lin M (2006) Functional mapping: how to map and study the genetic architecture of dynamic complex traits. Nat Rev Genet 7: 229–237 [DOI] [PubMed] [Google Scholar]
- Würschum T, Liu W, Busemeyer L, Tucker MR, Reif JC, Weissmann EA, Hahn V, Ruckelshausen A, Maurer HP (2014) Mapping dynamic QTL for plant height in triticale. BMC Genet 15: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Shen X, Pan W; Alzheimer’s Disease Neuroimaging Initiative (2014) Longitudinal analysis is more powerful than cross-sectional analysis in detecting genetic association with neuroimaging phenotypes. PLoS One 9: e102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, Xie W, Lian X, et al. (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5: 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zeng L, Shannon M (2000) Salinity effects on seedling growth and yield components of rice. Crop Sci 40: 996–1003 [Google Scholar]
- Zeng L, Shannon MC, Grieve CM (2002) Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica 127: 235–245 [Google Scholar]
- Zhang X, Hause RJ, Borevitz JO (2012) Natural genetic variation for growth and development revealed by high-throughput phenotyping in Arabidopsis thaliana. G3 (Bethesda) 2: 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, et al. (2007) An Arabidopsis example of association mapping in structured samples. PLoS Genet 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, et al. (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.