Duplication and sequence divergence of a gene family for histone demethylases likely contributed to the enhanced, chromatin-based regulation in angiosperms and vertebrates.
Abstract
Histone modifications, such as methylation and demethylation, are crucial mechanisms altering chromatin structure and gene expression. Recent biochemical and molecular studies have uncovered a group of histone demethylases called Jumonji C (JmjC) domain proteins. However, their evolutionary history and patterns have not been examined systematically. Here, we report extensive analyses of eukaryotic JmjC genes and define 14 subfamilies, including the Lysine-Specific Demethylase3 (KDM3), KDM5, JMJD6, Putative-Lysine-Specific Demethylase11 (PKDM11), and PKDM13 subfamilies, shared by plants, animals, and fungi. Other subfamilies are detected in plants and animals but not in fungi (PKDM12) or in animals and fungi but not in plants (KDM2 and KDM4). PKDM7, PKDM8, and PKDM9 are plant-specific groups, whereas Jumonji, AT-Rich Interactive Domain2, KDM6, and PKDM10 are animal specific. In addition to known domains, most subfamilies have characteristic conserved amino acid motifs. Whole-genome duplication (WGD) was likely an important mechanism for JmjC duplications, with four pairs from an angiosperm-wide WGD and others from subsequent WGDs. Vertebrates also experienced JmjC duplications associated with the vertebrate ancestral WGDs, with additional mammalian paralogs from tandem duplication and possible transposition. The sequences of paralogs have diverged in both known functional domains and other regions, showing evidence of selection pressure. The increases of JmjC copy number and the divergences in sequence and expression might have contributed to the divergent functions of JmjC genes, allowing the angiosperms and vertebrates to adapt to a great number of ecological niches and contributing to their evolutionary successes.
Chromatin-based regulation is an important mechanism of modulating eukaryotic chromatin structure and gene expression by altering DNA and histone modifications rather than changing DNA sequences. The eukaryotic chromatin contains a group of highly conserved proteins called histones, including the core histones H2A, H2B, H3, and H4, as well as the linker histone H1. The core histones, with two copies each, form an eight-subunit complex, which is wrapped by 146 bp of DNA to form the nucleosome, the basic unit of chromatin. In each nucleosome, the hydrophobic C-terminal regions of the eight subunits occupy the interior, whereas the hydrophilic N-terminal regions (the tails) extend outward (Luger et al., 1997). The covalent histone modifications occur on the tails of the core histone proteins and encode epigenetic information that can be passed through mitosis, sometimes even meiosis, and alter chromatin structure and modulate genomic functions. Specifically, histone modifications include methylation, acetylation, phosphorylation, ubiquitination, and sumoylation. Chromatin structure can be regulated by three classes of proteins: DNA methylase and demethylases, chromatin remodelers that regulate nucleosome positioning, and enzymes for histone modifications.
Among various histone modifications, the role of methylation varies in different species (Feng et al., 2010; Liu et al., 2010) but is relatively stable and suited for the transmission of epigenetic information (Strahl and Allis, 2000), sometimes even transgenerationally, as supported by a recent study in Arabidopsis (Arabidopsis thaliana) showing that the vernalized state can be partially transgenerationally inherited due to a defect in the reduction of histone H3 lysine-27 trimethylation (H3K27me3) levels at FLOWERING LOCUS C (FLC; Crevillén et al., 2014). Histone methylation occurs on Arg and Lys residues and is involved in a wide range of biological processes, including gene expression, chromatin structure, dosage compensation, and epigenetic memory (Martin and Zhang, 2005). A Lys residue can be monomethylated, dimethylated, or trimethylated; however, an Arg residue can only be monomethylated or dimethylated. Different histone modifications regulate distinct functional outcomes within an epigenetic marking system: methylation at histone H3 Lys-4 (H3K4), H3K36, and H3K79 is correlated with higher gene expression, whereas methylation at H3K9, H3K27, and H4K20 is associated with lower gene expression, hence often termed activating or repressing marks, respectively, although the causal relationship between histone modification and transcriptional activity is still elusive (Martin and Zhang, 2005; Henikoff and Shilatifard, 2011; Dong and Weng, 2013).
Histone methylation has been regarded as an irreversible modification for a long time, because of the stable nature of the carbon-nitrogen bond; this idea is also supported by the similar half-lives of histone fractions and methyl Lys and methyl Arg marks (Byvoet et al., 1972). In 2004, the discovery of the first histone demethylase, known as LYSINE-SPECIFIC DEMETHYLASE1 (LSD1), provided experimental evidence for enzymatic demethylation (Shi et al., 2004). The single-copy LSD1 mediates oxidative demethylation on monomethylated or dimethylated H3K4 and/or H3K9 but not trimethylated H3K4 (Shi et al., 2004). Subsequently, a second and larger class of demethylases containing a Jumonji C (JmjC) domain was identified (Tsukada et al., 2006). Unlike LSD1, the JmjC proteins do not require a protonated nitrogen and can also reverse the trimethylated histone Lys state (Shi and Whetstine, 2007). Five amino acid residues within predicted cofactor-binding sites in the JmjC domain are conserved and important for enzymatic activity (Klose et al., 2006). Among these, three residues bind to the Fe(II) cofactor and two other residues are required for α-ketoglutarate (αKG) binding.
Several plant JmjC genes are known to have important functions in regulating development and environmental responses (Liu et al., 2010; Chen et al., 2011; Kooistra and Helin, 2012). For example, POLYCOMB REPRESSIVE COMPLEX2, a conserved and key transcriptional regulator in animals and plants, has been demonstrated to have H3K27me3 affinity (Cao et al., 2002). In Arabidopsis, Relative of Early Flowering6 (REF6)/AtPKDM9A and Early Flowering6 (ELF6)/AtPKDM9B are closely related JmjC paralogs and have H3K27me3 demethylase activities (Lu et al., 2011a; Crevillén et al., 2014) but have different roles in the regulation of flowering time (Noh et al., 2004). The elf6 mutant flowers early and exhibits reduced expression of the flowering repressor FLC (Noh et al., 2004), due to an elevated level of H3K27me3 at the FLC locus (Crevillén et al., 2014), but the ref6 mutant shows an FLC-dependent late-flowering phenotype (Noh et al., 2004). In addition, a recent study showed that loss of H3K27me3 was observed after salt priming in Arabidopsis seedlings (Sani et al., 2013), suggesting that some JmjC genes may be induced upon salt stress. To date, many members of the JmjC gene family have not been characterized genetically or biochemically; nevertheless, sequence and evolutionary analyses can help predict their functions in histone demethylation. However, the expression and functional divergence of JmjC duplicates have not been studied extensively, especially in response to abiotic stresses, such as abscisic acid (ABA), salt, and drought. In addition, members of the PKDM7 subfamily exhibited distinct expression patterns correlated with their different enzymatic activities (Lu et al., 2008), but the underlying sequence divergence and dynamic evolution are still unknown.
An early study of the JmjC gene family using sequences from Arabidopsis, rice (Oryza sativa), and human (Homo sapiens) defined 12 subfamilies according to phylogenetic relationships, with support from the presence of other domains (Zhou and Ma, 2008). These domains play potential regulatory roles in the demethylation process, such as the recognition of methylated histone marks (e.g. Plant homeodomain and Tudor domain), protein-protein interaction (e.g. F-box), and DNA binding (e.g. Cys2His2 zinc finger; Zhou and Ma, 2008). However, the previous reports have focused on a single species or a few species (Lu et al., 2008; Zhou and Ma, 2008); thus, there has not been a systematic study of the JmjC family in plants or animals. Recent progress in genome sequencing provides an opportunity to gain additional insights into the evolution of the JmjC family during the histories of angiosperms and vertebrates.
Histone methylation is mainly mediated by SET {for Suppressor of variegation [Su(var)3-9], Enhancer of zeste [E(z)] and Trithorax [Trx]} domain protein methyltransferase, and genome duplication has resulted in an increase of SET copy numbers. In Arabidopsis, five duplicated gene pairs are retained after recent genome duplication events, and 19 pairs are retained in poplar (Populus trichocarpa; Lei et al., 2012). It would also be helpful to learn whether the JmjC gene family has a similar evolutionary pattern to that of the SET genes. The evolutionary patterns of these two families could inform the balanced influence of demethylation and methylation on species evolution and divergence. In this study, we have characterized the evolution of JmjC genes in major eukaryotic lineages, including land plants, using phylogenetic and domain analyses, and we define 14 monophyletic subfamilies: Jumonji, AT-Rich Interactive Domain2, JMJD6, KDM2, KDM3, KDM4, KDM5, KDM6, PKDM7, PKDM8, PKDM9, PKDM10, PKDM11, PKDM12, and PKDM13. We show that some families underwent distinct gene duplication during evolution in angiosperms, especially in the ancestor of extant angiosperms.
RESULTS
Identification of JmjC Genes in Plants, Animals, and Fungi
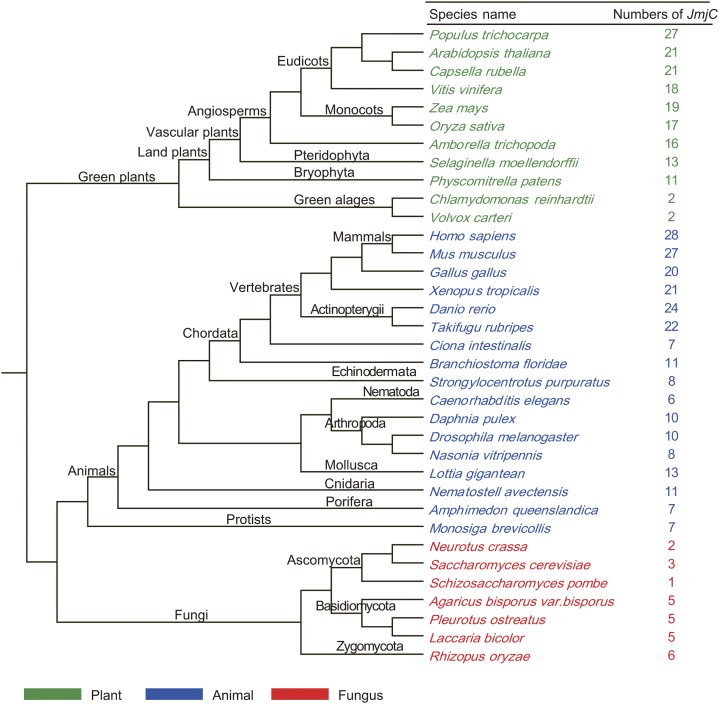
The complete set of JmjC genes was identified from a comprehensive data set that contains selected plants, animals, and fungi based on HMMER software. In all, 434 sequences, each containing a JmjC domain, were retrieved from 35 different organisms: 11 plants, 16 metazoans, seven fungi, and Monosiga brevicollis (a unicellular choanoflagellate, a protist related to animals; Fig. 1; Supplemental Tables S1 and S2). The identified JmjC proteins range in size from 266 to 2,740 amino acids. Among major lineages of plants, JmjC genes are present in algae, Bryophyta, Pteridophyta, and angiosperms. The copy number of JmjC genes varies considerably among plants, ranging from two in the green algae Chlamydomonas reinhardtii and Volvox carteri to 17 in rice (monocot) and 21 in Arabidopsis (eudicot), with the highest number of 27 in poplar (eudicot). JmjC genes are also widespread in animals, from simple invertebrates, such as the sponge Amphimedon queenslandica, to mammals, such as human, with the gene copy number ranging from six to 28, as well as in the unicellular M. brevicollis. Further investigation reveals that the copy number variation in animals is mainly due to gene duplications in vertebrates; also, the distribution of subgroup PKDM12 in different organisms is complex. In fungi, there are fewer than six genes for each species, such as one in Schizosaccharomyces pombe, three in Saccharomyces cerevisiae, and five in Agaricus bisporus var bisporus; altogether, 27 JmjC gene sequences were retrieved from seven fungi.
Figure 1.
Phylogenetic relationships of species that were used in this study. The total number of JmjC proteins found in the genome of each species is indicated. The cladogram is based on the current view of plant and eukaryotic phylogeny (Gough et al., 2001).
In order to standardize gene names and be consistent with the literature, we adopted a common nomenclature system based on the names of human genes according to the chromatin-modifying enzyme activities of animal members and Arabidopsis genes designated in previous studies as well as our phylogenetic analysis (Allis et al., 2007; Zhou and Ma, 2008). First, for human and Arabidopsis genes with known functions or previous research, the published gene names were retained and used as a reference. Second, the orthologs of these genes from plants, animals, and fungi were named following established references. Ultimately, recent paralogs were distinguished with an uppercase letter after the number, using the same letter for orthologs between organisms whenever possible.
Phylogenetic Classification of JmjC Genes into 14 Subfamilies
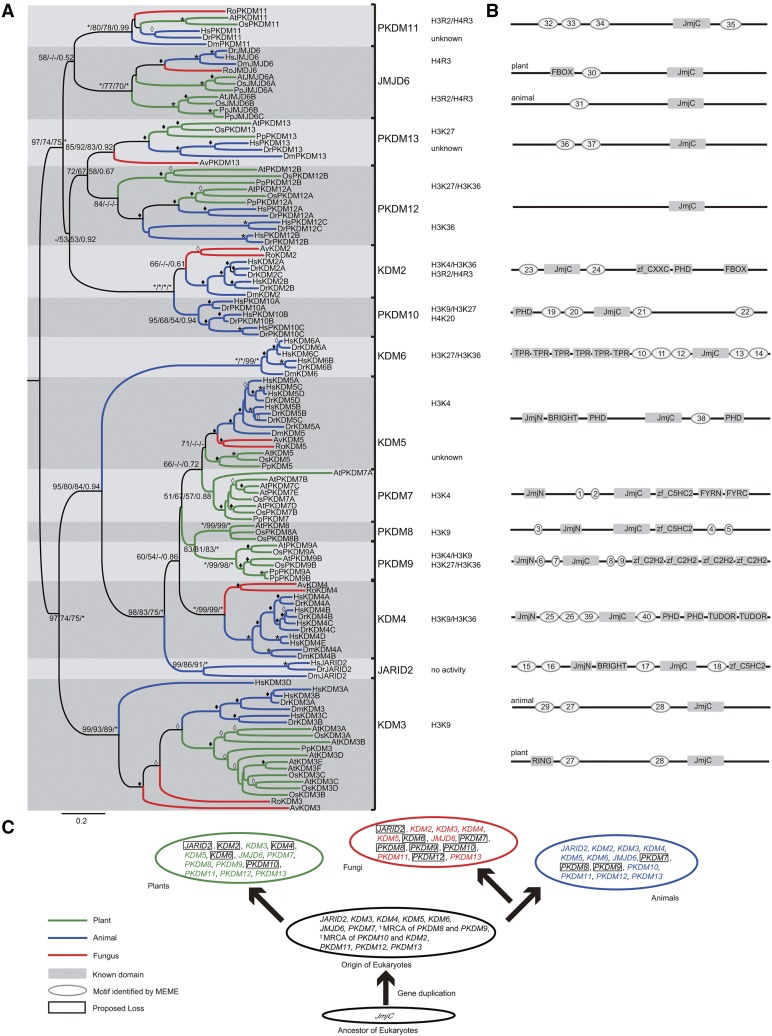
To explore the evolutionary relationships of eukaryotic JmjC genes, we conducted phylogenetic analyses with an alignment of the conserved JmjC domain from representative species using neighbor-joining (NJ), maximum likelihood (ML), and Bayesian methods. ML and Bayes analyses showed that proteins from different species cluster together in clades with high support values, with support from NJ analysis for most results. According to the results from phylogenetic and motif analyses, the eukaryotic JmjC genes can be divided into 14 subfamilies, designated as JARID2, JMJD6, KDM2, KDM3, KDM4, KDM5, KDM6, PKDM7, PKDM8, PKDM9, PKDM10, PKDM11, PKDM12, and PKDM13 (Fig. 2). Among these subfamilies, KDM3, KDM5, JMJD6, PKDM11, and PKDM13 each contain members from plants, animals, and fungi. On the other hand, PKDM12 lacks fungal members, and KDM2 and KDM4 lack plant members. In addition, PKDM7, PKDM8, and PKDM9 are plant-specific groups, and JARID2, KDM6, and PKDM10 are animal specific.
Figure 2.
Phylogeny of representative JmjC genes from plants, animals, and fungi. A, Evolutionary relationship between JmjC genes from eight representative species (Chordata: Hs, Homo sapiens; Dr, Danio rerio. Arthropoda: Dm, Drosophila melanogaster. Basidiomycota: Av, A. bisporus var bisporus. Zygomycota: Ro, Rhizopus oryzae. Angiosperms: At, Arabidopsis; Os, rice. Bryophyta: Pp, Physcomitrella patens). Tree topology constructed using MEGA is shown here. For major nodes, NJ and ML (ML bootstrap [BS] values were generated by PhyML (version 3.0) and RAxML (version 7.0), respectively), BS values above 50%, followed by Bayesian posterior probability (PP) values are shown. For minor nodes, empty diamonds represent BS values of 50% to 69% and solid diamonds mark BS values of 70% to 99% from NJ analysis. For major and minor nodes, asterisks indicate BS/PP values of 100%/1. Substrate specificities for residues of histone tails are indicated for all subfamilies with demonstrated demethylase activity. B, Highly conserved non-JmjC amino acid motifs in each JmjC subfamily. An idealized representation of a typical member of each JmjC subfamily is shown, with known domains drawn as shaded rectangles and other conserved motifs drawn as shaded ovals. The diagrams are not drawn to scale. The sequences of each motif in gene family members are given in Supplemental Table S3. C, Model of the evolutionary history of the JmjC gene family. In different kingdoms, the JmjC gene experienced several specific duplication events associated with the species divergence.
The fact that KDM3, KDM5, JMJD6, PKDM11, and PKDM13 subfamilies have members from major multicellular groups of eukaryotes, including plants, animals, and fungi, suggests that these clades originated from five respective ancestral genes in the most recent common ancestor (MRCA) of the three kingdoms. According to our phylogenetic analyses, JMJD6, PKDM11, and PKDM13 cluster together with strong support, but some of the internal relationships of these clades are not clear. The KDM2 and KDM4 subfamilies both contain animal and fungal JmjC genes, indicating that the two clades are derived from ancestral genes that were present before the separation of animals and fungi. The KDM4 clade is well supported by all phylogenetic methods, but the classification of KDM2 also relied on the protein domain structures. The PKDM7, PKDM8, and PKDM9 subfamilies are plant specific. According to the tree topology, the PKDM8 subfamily forms a sister group to PKDM9, and together they are from an ancestral gene in the MRCA of plants, animals, and fungi, because they are sister to a large clade with genes from all three kingdoms. Both PKDM7 and PKDM9 subfamilies contain genes from major lineages of land plants, including Bryophyta, Pteridophyta, and angiosperms, revealing that they are retained in land plants, whereas PKDM8 genes are conserved from Pteridophyta to angiosperms, suggesting a likely loss of this subfamily in nonvascular plants. The JARID2, KDM6, and PKDM10 subfamilies are composed of only animal genes. However, the sisterhood of JARID2 and KDM6 to other clades containing genes from all three kingdoms suggests the origins of these subfamilies in the MRCA of three kingdoms; similarly, the ancestral gene of PKDM10 was probably already present in the MRCA of animals and fungi. The early origins of JARID2 and PKDM10 were further supported by the identification of their homologs in M. brevicollis.
In summary, eukaryotic JmjC genes form 14 subfamilies. These subfamilies can be grouped into four categories: five shared by all three major eukaryotic kingdoms; one shared by animals and plants; two found in both animals and fungi, with the counterparts lost from plants; and three plant specific and three animal specific. It is likely that there were at least 12 ancestral JmjC genes present in the MRCA of three major eukaryotic kingdoms, with subsequent duplications and losses in specific kingdoms (Fig. 2C). Additionally, within most subfamilies, except for JARID2, PKDM11, and PKDM13, there are two or more gene copies in plants or animals, suggesting further functional divergence of JmjC genes.
Different Domain Architecture and Conserved Non-JmjC Motifs in Subfamilies
According to the phylogenetic tree shown in Figure 2A, 14 monophyletic subfamilies were identified in the JmjC gene family. These 14 subfamilies represent 12 different domain architectures, as three subfamilies (PKDM11, PKDM12, and PKDM13) possess only the JmjC domain. Among these, the plant-specific PKDM7, PKDM8, and PKDM9 subfamilies have similar domain architectures, but PKDM7 proteins possess extra FYRN and FYRC domains. To determine whether motifs outside the JmjC and other known domains were conserved between members of the same subfamily, we searched for motifs in our data set of JmjC proteins. We found 40 motifs with lengths greater than 10 amino acids that are specific within subfamilies (Supplemental Table S3; Supplemental Fig. S1). The subfamilies exhibited various combinations of motifs; in addition, for subfamilies KDM3 and JMJD6, different conserved motifs were found in plant and animal proteins (Fig. 2B). None of the 40 conserved motifs corresponded to known domains in the Pfam database. The highly conserved motifs shared by members of subfamilies of the JmjC domain proteins further support the classification presented here. The conservation of these additional domains in the respective subfamilies implies that they play important roles in the functions of these subfamilies.
Multiple Gene Duplication Events Identified in Angiosperms and Retention in Ancestral Angiosperms
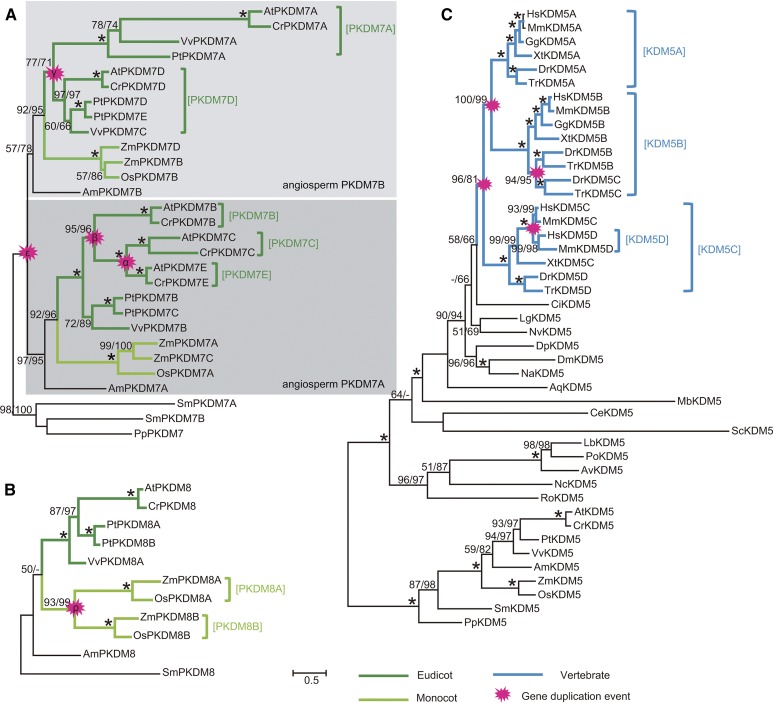
To investigate the evolutionary history of the JmjC gene family, we analyzed the phylogeny of each subfamily (Supplemental Figs. S2–S12). In the PKDM7 subfamily, AmPKDM7A from Amborella trichopoda (sister to other angiosperms) and AmPKDM7B are in two sister groups, each of which contains genes from both monocots and eudicots (Fig. 3). The Bryophyta and Pteridophyta PKDM7 genes constitute a clade separate from sister groups containing the A. trichopoda genes. Therefore, it is most likely that AmPKDM7A and AmPKDM7B were derived from gene duplication before the divergence of extant angiosperms but after the separation of angiosperms from other land plants. Additionally, in the PKDM7 subfamily, each of the PKDM7B, PKDM7C, and PKDM7E genes forms a monophyletic group that includes genes from Brassicaceae species, whereas the homologous genes in P. trichocarpa and Vitis vinifera form a separate basal clade. This topology is supported by multiple analyses and suggests that the duplication events likely occurred in the ancestor of the Brassicaceae. Similarly, in the PKDM8 subfamily, grass PKDM8A and PKDM8B genes are sister groups likely generated by duplication in the ancestor of grasses (Fig. 3B).
Figure 3.
Phylogenies of the PKDM7, PKDM8, and KDM5 subfamilies. Phylogenetic trees of the PKDM7 (A) and PKDM8 (B) genes from plants and KDM5 from major eukaryotes (C) constructed by ML using the Jones, Taylor, and Thorton model for amino acid substitution and γ-parameters are shown. For each clade, PhyML (version 3.0) and RAxML (version 7.0) percentage bootstrap values no less than 50% are presented. Asterisks indicate both bootstrap values of 100%.
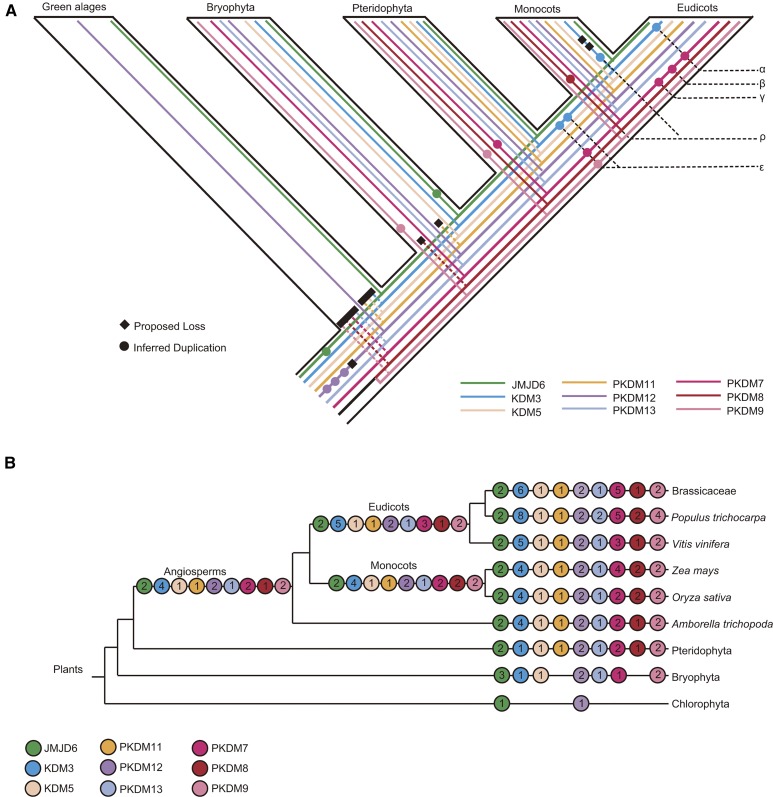
Whole-genome duplications (WGDs) are thought to be common in angiosperms and are also associated with the origin of vertebrate animals (Panopoulou et al., 2003; Jiao et al., 2011). To further ascertain whether these JmjC genes are caused by genome duplication events, we examined genomic regions containing duplicated genes in different species. Most members of these duplicated gene pairs are located in syntenic genomic regions, indicating that these JmjC gene copies are likely generated by chromosome segmental duplications or WGDs, such α and β in Brassicaceae, γ in early core eudicots, ρ and σ in grasses, and ε in early angiosperms (Jaillon et al., 2007; Barker et al., 2009; Tang et al., 2010; Jiao et al., 2011). Specifically, in the PKDM7 clade, the duplication of PKDM7C and PKDM7E in Brassicaceae is likely the result of α, and the combined clade of PKDM7C and PKDM7E form a sister to PKDM7B, likely due to β (Fig. 3A; Supplemental Fig. S13). The duplication in core eudicots that generated PKDM7A and PKDM7D corresponds to γ ( Supplemental Fig. S13C). A more ancient event, ε, is also detected in our results, generating the aforementioned pair of AmPKDM7A and AmPKDM7B belonging to the two angiosperm sister groups in the PKDM7 subfamily. In the PKDM8 clade, the occurrence of the PKDM8A and PKDM8B is consistent with the ρ duplication within Poaceae (Fig. 3B; Supplemental Fig. S13D; Paterson et al., 2004). Additionally, we identified four duplicated gene pairs in the ancestral angiosperm (Fig. 4).
Figure 4.
Duplication events in the plant JmjC family inferred from amino acid sequence analyses. A, Schematic depiction of the inferred history of gene duplication and loss within the green plants. Inferred duplications are marked with colored circles, and proposed losses are indicated with black diamonds. B, Schematic representation of the complement of JmjC proteins in major plant groups. A numbered circle represents the copy number of each JmjC subfamily. The numbers in the left-most circles represent the minimum inferred JmjC protein complement in the last common ancestor of each major plant group.
Multiple Gene Duplication Events Identified in Vertebrate Evolution and Lineage-Specific Losses
To better understand the evolutionary relationships of lineages within animals, we carried out similar analyses in subfamilies containing animal members. The KDM5 subfamily has three vertebrate groups named KDM5A, KDM5B, and KDM5C, but invertebrates, plants, and fungi have a single copy (Fig. 3C). KDM5A and KDM5B genes formed two well-supported sister groups that coincide with one of two WGDs (1R and 2R) before the divergence of vertebrates (Dehal and Boore, 2005). The combined clade of vertebrate KDM5A and KDM5B is sister to KDM5C, probably due to the earlier of the two WGDs in early vertebrates (Dehal and Boore, 2005). Further support for the origin of these groups from the WGDs is provided by the finding that HsKDM5A, HsKDM5B, and HsKDM5C are located in syntenic regions of three different chromosomes (Supplemental Fig. S14). In the KDM5B clade, a high degree of conservation of synteny was found in the two chromosome segments DrKDM5B and DrKDM5C (Supplemental Fig. S14B). These two genes were members of two fish-specific sister groups in the phylogenetic tree (Fig. 3C). The duplication generating DrKDM5B and DrKDM5C corresponds to the WGD (3R WGD) that is specific to a fish lineage (Kasahara, 2007).
In addition, phylogenetic analyses were performed using full-length sequences of JmjC proteins in other orthologous subfamilies. Most orthologous groups exhibited duplication events that correspond to WGDs, and 15 duplication events were shared by at least two animal species, in the phylogenetic trees for JMJD6, KDM2, KDM3, KDM4, KDM6, PKDM10, and PKDM12 (Supplemental Table S4; Supplemental Figs. S3–S9, S11, and S15). In addition to duplications produced by the 2R and 3R WGDs, a clade including only human and/or mouse genes was discovered in KDM3, KDM4, KDM5, and KDM6 groups, but the mechanisms of such mammal-specific duplication are unclear. Moreover, JARID2 contains animal-specific single-copy genes, and PKDM11 and PKDM13 subfamily members are single-copy orthologs not only in animals but also in plants and fungi (Supplemental Figs. S2, S10, and S12). Additionally, some of the duplicates were lost in some lineages. In KDM3, four human paralogs were located in a syntenic region, namely KDM3A, KDM3B, KDM3C, and KDM3D (Table I), but KDM3D was lost from birds and fishes. A phylogeny tree supports the origin of the four paralogs as results of the vertebrate WGDs 1R and 2R (Supplemental Fig. S5).
Table I. Number of human paralogous genes identified from the Synteny Database using KDM3 loci as seed genes.
The first number represents the number of paralogous genes obtained with a window of 100 genes, and the second number represents the number of paralogous genes obtained with a window of 200 genes.
| Gene | HsKDM3A |
HsKDM3B |
HsKDM3C |
HsKDM3D |
|---|---|---|---|---|
| Hsa2 | Hsa5 | Hsa10 | Hsa8 | |
| HsKDM3A | ||||
| HsKDM3B | 6/52 | |||
| HsKDM3C | 39/97 | 14/76 | ||
| HsKDM3D | 13/70 | 32/32 | 59/63 |
The KDM5 and KDM6 subfamilies are unusual in that they possess paralogs located on X and Y chromosomes, respectively. Our phylogenetic analyses indicate that KDM5D and KDM6C each has a sister group in mammals. Synteny analysis shows that these two JmjC genes are located on the Y chromosome and their respective paralogs are located on the X chromosome. A previous study showed that KDM5D arose about 154 million years ago in the common ancestor of marsupial and placental mammals, and KDM6C differentiated before the placental mammal radiation about 116 million years ago (Cortez et al., 2014). The human sex chromosome evolved from autosomes about 240 to 320 million years ago, which is more ancient than the occurrence of KDM5D and KDM6C (Lahn and Page, 1999; Bellott et al., 2010), suggesting that these paralogs could be the result of transposition from one sex chromosome to the other. On the other hand, the origins of KDM4D and KDM4E are not clear. KDM4D was found in mammals but not in other classes of vertebrates, and the latter is only found in primates. HsKDM4D and HsKDM4E are located close together on the same chromosome, indicating that the two genes were likely generated by a tandem duplication (Table II). KDM4D is only found in mammals and is nested in a clade of vertebrate KDM4B genes (Supplemental Fig. S6); the weak support of the clade with the KDM4D and fish KDM4B genes allows the possibility that the KDM4D genes originated in early vertebrates but were lost in birds and fishes. KDM4D and KDM4E both possess an N terminus but lack the C-terminal sequence that is conserved in KDM4B. About 20 introns were identified in HsKDM4A, HsKDM4B, and HsKDM4C, but only two in HsKDM4D and none in HsKDM4E, suggesting that the HsKDM4B to HsKDM4D genes derived from HsKDM4B by retroposition. It is possible that KDM4A, KDM4B, KDM4C, and KDM4D resulted from the vertebrate WGDs, if weakly supported relationships are not considered.
Table II. Number of human paralogous genes identified from the Synteny Database using KDM4 loci as seed genes.
The first number represents the number of paralogous genes obtained with a window of 100 genes, and the second number represents the number of paralogous genes obtained with a window of 200 genes.
| Gene | HsKDM4A |
HsKDM4B |
HsKDM4C |
HsKDM4D |
|---|---|---|---|---|
| Hsa1 | Hsa19 | Hsa9 | Hsa11 | |
| HsKDM4A | ||||
| HsKDM4B | 93/203 | |||
| HsKDM4C | 84/171 | 45/87 | ||
| HsKDM4D | 2/10 | 5/6 | 3/3 |
Expression and Functional Divergence of JmjC Duplicates
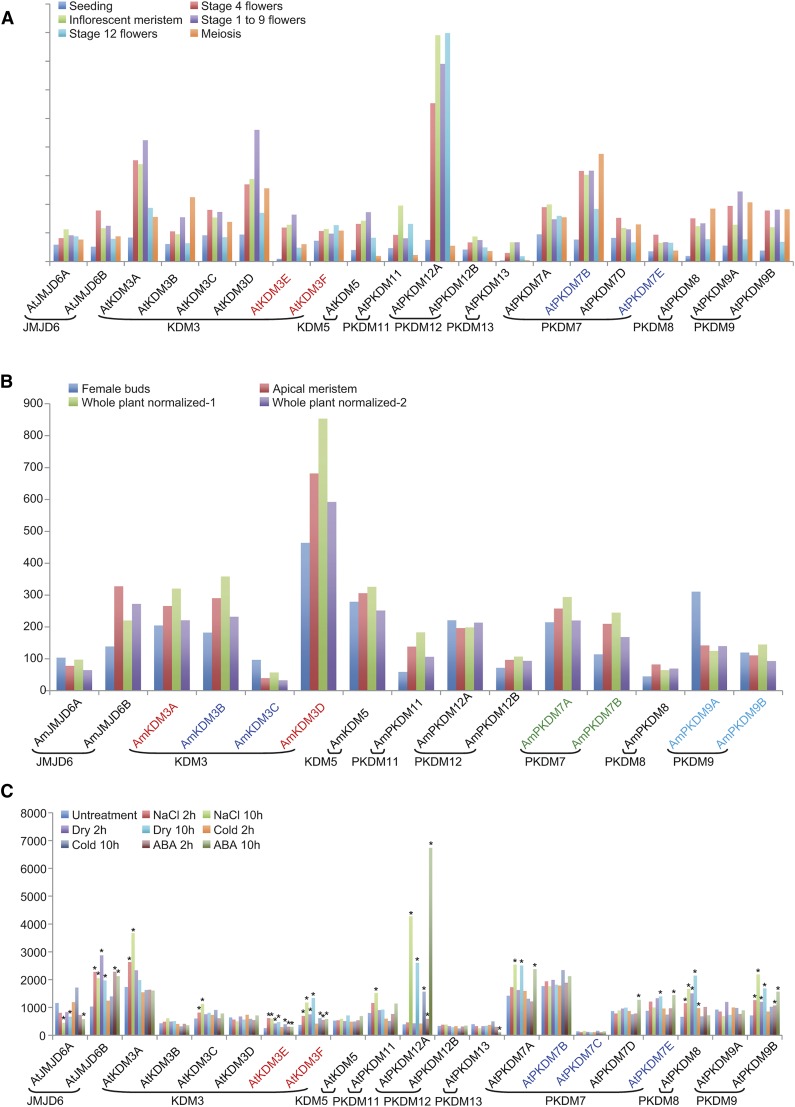
To obtain clues about possible functional divergence, we examined RNA-sequencing (RNA-seq) data of Arabidopsis and A. trichopoda and found that most JmjC genes were expressed in the developing flower and different tissues, except AtPKDM7C and AmPKDM12B (Fig. 5; Supplemental Table S5). More interestingly, AtKDM3D and AmKDM3D are both expressed more highly than other genes in the same subfamily. Transcriptomic comparison of Arabidopsis and A. trichopoda JmjC genes indicated that most were expressed at similar levels in different tissues, suggesting functional conservation in angiosperms. In addition, public tiling array data of Arabidopsis under drought, cold, high-salinity, and ABA treatments were examined for JmjC gene expression (Fig. 5C; Supplemental Table S6). Some JmjC genes showed differential expression upon cold treatment, including AtKDM3E, AtKDM3F, AtPKDM12A, and AtPKDM8, and many were affected by other treatments. The number of differentially expressed genes in a 10-h treatment was greater than that found after a 2-h treatment. These results suggest that plant JmjC genes play potential roles in stress responses.
Figure 5.
Expression of Arabidopsis and A. trichopoda JmjC genes. A, Expression of Arabidopsis JmjC genes in different stages or tissues. The x axis indicates different representative genes, and the y axis represents reads per kilobase of mRNA length per million mapped reads values. Genes resulting from recent duplications are shown in the same color. B, Expression of A. trichopoda JmjC genes in different tissues. C, Expression of Arabidopsis JmjC genes under drought, cold, high-salinity, and ABA treatment. Asterisks indicate differential expression under stress conditions compared with normal conditions.
Gene duplication increases the quantity of genetic material and improves the chance of functional innovations during evolution. To obtain clues about functional divergence, we examined some of the recent duplicates for expression changes in different tissues and upon abiotic stresses. In Arabidopsis, a trio of paralogs are retained after the Brassicaceae WGDs, AtPKDM7C, AtPKDM7E, and AtPKDM7B. Among them, AtPKDM7E was differentially expressed under drought, but not AtPKDM7B and AtPKDM7C (Fig. 5C; Supplemental Table S6). Also, two duplicates are retained after the core eudicot WGDs, AtPKDM7A and AtPKDM7D. AtPKDM7A was induced by drought and NaCl treatments, but AtPKDM7D was not (Fig. 5C; Supplemental Table S6). In A. trichopoda, four pairs of duplicates, AmKDM3A and AmKDM3D, AmKDM3B and AmKDM3C, AmPKDM7A and AmPKDM7B, and AmPKDM9A and AmPKDM9B, are retained from the WGD in the ancestral angiosperm, but only the AmPKDM7A and AmPKDM7B pair exhibited a minor difference in expression. AtPKDM9B, the Arabidopsis ortholog of AmPKDM9B, is expressed differentially under drought, NaCl, and ABA treatments, but AtPKDM9A, the Arabidopsis ortholog of AmPKDM9A, is not significantly induced. In conclusion, duplicated genes in several pairs differ in the induction of expression upon abiotic stresses and/or in different tissues, suggesting functional divergence between these paralogs.
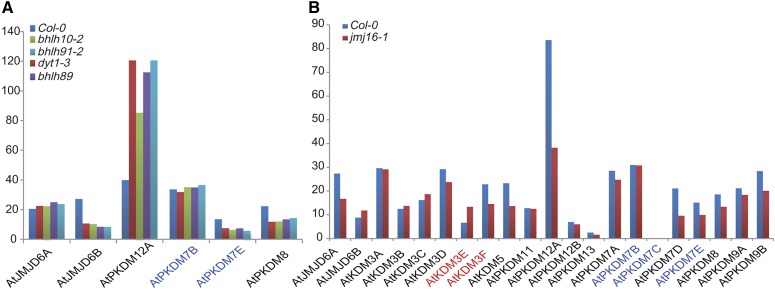
To further investigate functional divergence among JmjC genes, we examined transcriptomic data sets of the wild type and several anther mutants, including dysfunctional tapetum1 (dyt1), basic helix-loop-helix10 (bhlh10), bhlh89, and bhlh91 (E. Zhu, C. You, J. Cui, F. Chang, and H. Ma, unpublished data), as a case study of possible regulation of JmjC genes by such regulatory genes. The expression of most JmjC genes was not altered in these mutants, except for AtJMJD6B, AtPKDM12A, AtPKDM7E, and AtPKDM8, with 2-fold or greater changes (Fig. 6; Supplemental Table S7). DYT1 encodes a bHLH-type transcription factor and is important for normal tapetum development (Zhang et al., 2006; Feng et al., 2012), as are the other bHLH transcription factors; thus, the expression change of several JmjC genes in these mutants suggests a potential role in tapetum development. Additionally, we also compared JmjC gene expression between the wild type and the pkdm7d/jmj16 mutant (E. Zhu, C. You, J. Cui, F. Chang, and H. Ma, unpublished data) and found some JmjC genes with distinct expression patterns (Fig. 6; Supplemental Table S7), suggesting functional divergence among these genes. Specifically, AtPKDM12A was dramatically down-regulated in pkdm7d//jmj16, indicating that its transcription may be regulated by PKDM7D directly or indirectly. Together, JmjC gene expression from public and our own RNA-seq data strongly supports possible functional divergence.
Figure 6.
Expression of JmjC genes in Arabidopsis wild-type Columbia-0 (Col-0) and mutants by RNA-seq. A, Two-fold change in the expression of JmjC genes and their paralogs compared with dyt1 (bhlh22), bhlh10, bhlh89, and bhlh91 mutants and the wild type. B, Expression of JmjC genes in the pkdm7d/jmj16 mutants. The x axis represents different genes, and the y axis indicates reads per kilobase of mRNA length per million mapped reads values. Genes resulting from recent duplications are shown in the same color.
Sequence Analysis for Functional Diversification of JmjC Duplicates
In addition to the expression divergence of JmjC genes, functional diversification (FD) of JmjC paralogs could also occur in coding regions. To estimate FD between two paralogs, we used the DIVERGE3.0 program with amino acid sequence data: FD I was based on evolutionary rate (Gu, 1999), and FD II was based on differences in biochemical properties of amino acids (Gu, 2006; Table III). We found that the angiosperm-wide duplicates, PKDM7A and PKDM7B, have FD I with the highest significance (P = 1.9E-16). In addition, two other pairs, Brassicaceae PKDM7B versus PKDM7C (P = 3.1E-08) and grass PKDM8A versus PKDM8B (P = 8.9E-13), also have highly significant FD I.
Table III. Analysis of FD by DIVERGE.
| FD | WGD | Subfamilies | Coefficient θ ± se (P) | No.a |
|---|---|---|---|---|
| Type I | α/β | Brassicaceae PKDM7C versus PKDM7E | 0.225383 ± 0.101966 (2.23E-02) | 0 |
| Brassicaceae PKDM7B versus PKDM7C | 0.529252 ± 0.101985 (3.09E-08) | 16 | ||
| Brassicaceae PKDM7B versus PKDM7E | 0.360678 ± 0.117604 (1.45E-03) | 1 | ||
| α/β | Brassicaceae KDM3E versus KDM3F | 0.101601 ± 0.074635 | 0 | |
| γ | Eudicot PKDM7A versus PKDM7D | 0.008360 ± 0.152924 | 0 | |
| ρ | Grass PKDM8A versus PKDM8B | 0.547235 ± 0.085772 (8.92E-13) | 29 | |
| Grass KDM3C versus KDM3D | 0.333263 ± 0.083005 (1.22E-05) | 11 | ||
| ε | Angiosperm PKDM7A versus PKDM7B | 0.401872 ± 0.059802 (1.86E-16) | 51 | |
| Angiosperm PKDM9A versus PKDM9B | 0.128948 ± 0.038607 (3.21E-04) | 22 | ||
| Angiosperm KDM3A versus KDM3D | 0.349834 ± 0.149399 (1.37E-02) | 4 | ||
| Angiosperm KDM3B versus KDM3C | 0.343037 ± 0.089236 (1.04E-05) | 32 | ||
| Angiosperm KDM3AD versus KDM3BC | 0.115225 ± 0.063442 (4.91E-02) | 4 | ||
| Type II | α/β | Brassicaceae PKDM7C versus PKDM7E | −0.055129 ± 0.045874 | |
| Brassicaceae PKDM7B versus PKDM7C | 0.027313 ± 0.049140 | |||
| Brassicaceae PKDM7B versus PKDM7E | 0.139158 ± 0.032327 (1.67E-05) | |||
| α/β | Brassicaceae KDM3E versus KDM3F | −0.008820 ± 0.035777 | ||
| γ | Eudicot PKDM7A versus PKDM7D | −0.167973 ± 0.084460 (4.67E-02) | ||
| ρ | Grass PKDM8A versus PKDM8B | 0.151855 ± 0.040068 (1.51E-04) | ||
| Grass KDM3C versus KDM3D | −0.004414 ± 0.061494 | |||
| ε | Angiosperm PKDM7A versus PKDM7B | 0.049528 ± 0.100748 | ||
| Angiosperm PKDM9A versus PKDM9B | −0.015586 ± 0.068176 | |||
| Angiosperm KDM3A versus KDM3D | −0.064805 ± 0.133296 | |||
| Angiosperm KDM3B versus KDM3C | 0.035761 ± 0.132655 | |||
| Angiosperm KDM3AD versus KDM3BC | −0.181663 ± 0.213708 |
Number of critical amino acid sites detected as related to FD.
Using the same program, amino acid sites likely contributing to FD I could be identified. Between the angiosperm PKDM7A and PKDM7B following the ε WGD, 51 putative sites responsible for FD I were identified (Table III; Supplemental Table S8). In addition, 32 sites were found for the angiosperm pair KDM3B and KDM3C, also generated at the time of the ε WGD. The amino acid residues at these sites were highly conserved between plants within one paralogous group but diverse among the other group of paralogs. Some of the differences between paralogs involved functionally distinct amino acids, such as that at position 268, which is occupied by the hydrophobic Phe in PKDM7A proteins but by polar hydrophilic Ser in PKDM7B. Similarly, position 671 has a polar hydrophilic Arg in PKDM7B but hydrophobic amino acids (e.g. Ile, Trp, and Leu) in PKDM7A proteins (Supplemental Fig. S16). Next, we examined the amino acid sites identified using FD analysis for known functions and found that many such FD I-associated amino acids are located in the JmjC or zinc finger domain, suggesting that functional specialization could have occurred in enzymatic and/or binding activities (Supplemental Table S8). At the same time, other sites showing FD I have no defined role; further experiments are needed to test the possible functional differences at these sites.
DISCUSSION
WGDs Contributed to JmjC Gene Expansion Especially Near the Origin of Angiosperms and Vertebrates
WGDs are found widely, including in both plants and animals, such as those associated with the origins of angiosperms (ε), core eudicots (γ), and Brassicaceae (α and β), and those in the ancestral vertebrates (2R) and supported by synteny, age estimates of gene duplications, and gene family phylogeny (Jaillon et al., 2007; Kasahara, 2007; Barker et al., 2009; Jiao et al., 2011; Amborella Genome Project, 2013). We have identified four pairs of JmjC paralogs likely due to the angiosperm ancestral ε WGD (Amborella Genome Project, 2013); these pairs of paralogous groups are also supported by genes from A. trichopoda, the sister of all other extant angiosperms (Fig. 4A). These results suggest that duplicate JmjC genes from ancient duplications were associated with the origin and early evolution of angiosperms.
Additionally, synteny analysis indicated that most JmjC duplicates in vertebrates were formed by WGD, with additional fish-specific contribution from the 3R WGD. Globally, most duplicates generated by the 2R WGD have been lost (Dehal and Boore, 2005); thus, the relatively high rate of retention of KDM3 duplicates in vertebrates (Supplemental Fig. S5) suggests that this group might be important for chromatin-based regulation in vertebrates. Previous studies proposed that more than 90% of the increases in Arabidopsis regulatory genes were due to WGD during the last 150 million years (Maere et al., 2005). Consistently, our results suggest that WGDs could be the main mechanism for the expansion of the JmjC gene family in angiosperms and vertebrates.
This pattern of evolution is also very similar to that of the SET domain family of histone methylases (Lei et al., 2012). The JmjC and SET genes both control the methylation status of histones and are important regulators of chromatin structure; our study indicates that these regulators share evolutionary patterns and mechanisms with transcription factor genes and likely contribute to the evolution of regulatory networks. The expansion of genes for histone modification following WGDs near the origins of angiosperms and vertebrate animals and their major subgroups (core eudicots, Brassicaceae, and fish) suggests that epigenetic modulation of gene expression played an important role in the evolutionary successes of the dominating groups in both plants and animals.
Sequence Divergence of Specific JmjC Proteins Suggests Changes in Enzyme Activity and Biological Functions
The JmjC domain contains five conserved amino acid residues within cofactor-binding sites that are important for enzymatic activity. Among the five conserved residues, His, Glu, and His bind to the Fe(II) cofactor and Phe and Lys are required for binding to αKG. According to the phylogeny, AtPKDM7C, JMJ14/AtPKDM7B, JMJ16/AtPKDM7D, JMJ18/AtPKDM7E, and JMJ19/AtPKDM7A belong to an H3K4-specific demethylase group (Table IV); however, AtPKDM7A lacks the conserved residues for αKG binding, suggesting that enzymatic activity is lost in this protein (Lu et al., 2008). Similarly, the other proteins encoded by core eudicot members of the PKDM7A clade in the PKDM7 subfamily, CrPKDM7A, VvPKDM7A, and PtPKDM7A, also lack the same conserved residues; in contrast, other angiosperm proteins of the PKDM7 subfamily have all five conserved amino acids within the cofactor-binding sites (Supplemental Fig. S17), suggesting that the ancestral core eudicot PKDM7A gene had lost the conserved residues. Specifically, the two clades (PKDM7A and PKDM7D) are due to the γ WGD in early core eudicots, and PKDM7D proteins still have the conserved residues, suggesting sequence and functional divergence following the γ WGD but before the divergence of extant core eudicots. We also found that members of the KDM3A clade, including AmKDM3A, lack the conserved residues for Fe(II) and αKG binding; their paralogs and other homologs still retain the conserved amino acids, showing that the lost and possible functional change occurred before the divergence of A. trichopoda from other angiosperms. The paralogs KDM3A and KDM3D (Supplemental Fig. S5) were due to the duplication in early angiosperms, indicating that KDM3A had lost the conserved residues after gene duplication. Moreover, these genes are differentially expressed in various tissues and in response to environmental signals, indicating that they might have lost the ancestral functions but gained new functions after gene duplication.
Table IV. Functionally characterized JmjC genes from Arabidopsis.
| Gene Name | Locus Tag | Other Names | Substrates | Biological Function | Reference |
|---|---|---|---|---|---|
| Subfamily JMJD6 | |||||
| AtJMJD6A | AT5G06550 | JMJ22 | H4R3me2a | Seed germination | Cho et al. (2012) |
| AtJMJD6B | AT1G78280 | JMJ21 | |||
| Subfamily KDM3 | |||||
| AtKDM3A | AT1G09060 | JMJ24 | No activity | ||
| AtKDM3B | AT4G21430 | JMJ28 | No activity | ||
| AtKDM3C | AT3G07610 | JMJ25/IBM1 | H3K9me1/2 | RNA-directed DNA methylation | Saze et al. (2008); Fan et al. (2012) |
| AtKDM3D | AT4G00990 | JMJ27 | |||
| AtKDM3E | AT1G11950 | JMJ26 | |||
| AtKDM3F | AT1G62310 | JMJ29 | |||
| Subfamily KDM5 | |||||
| AtKDM5 | AT1G63490 | JMJ17 | |||
| Subfamily PKDM7 | |||||
| AtPKDM7A | AT2G38950 | JMJ19 | No activity | ||
| AtPKDM7B | AT4G20400 | JMJ14 | H3K4me1/2/3 | Flowering time, repression of the floral transition, maintenance of DNA methylation | Deleris et al. (2010); Lu et al. (2010); Yang et al. (2010) |
| AtPKDM7C | AT2G34880 | JMJ15/MEE27 | H3K4me2/3 | Salt tolerance, flowering time, female gametophyte development, early embryo and endosperm formation | Pagnussat et al. (2005); Day et al. (2008); Yang et al. (2012b); Shen et al. (2014) |
| AtPKDM7D | AT1G08620 | JMJ16 | |||
| AtPKDM7E | AT1G30810 | JMJ18 | H3K4me2/3 | Flowering time | Yang et al. (2012a) |
| Subfamily PKDM8 | |||||
| AtPKDM8 | AT5G46910 | JMJ13 | |||
| Subfamily PKDM9 | |||||
| AtPKDM9A | AT3G48430 | JMJ12/REF6 | H3K9me3, H3K27me2/3, H3K36me2/3 | Flowering time, repression of FLC | Noh et al. (2004); Yu et al. (2008); Ko et al. (2010); Lu et al. (2011a) |
| AtPKDM9B | AT5G04240 | JMJ11/ELF6 | H3K4me1/2/3, H3K9me3, H3K27me3 | Flowering time, photoperiod pathway, transgenerational inheritance of vernalized state, repression of flowering time | Noh et al. (2004); Yu et al. (2008); Jeong et al. (2009); Crevillén et al. (2014) |
| Subfamily PKDM11 | |||||
| AtPKDM11 | AT5G63080 | JMJ20 | H3R2me2, H4R3me2 | Seed germination | Cho et al. (2012) |
| Subfamily PKDM12 | |||||
| AtPKDM12A | AT3G20810 | JMJ30/JMJD5 | H3K27me3, H3K36me2/3 | Circadian system, flowering time | Jones et al. (2010); Jones and Harmer (2011); Lu et al. (2011b); Gan et al. (2014); Yan et al. (2014) |
| AtPKDM12B | AT5G19840 | JMJ31 | |||
| Subfamily PKDM13 | |||||
| AtPKDM13 | AT3G45880 | JMJ32 | H3K27me3 | Flowering time | Gan et al. (2014) |
These terms are defined as follows: H, histone; R, Arg; K, Lys; and me1/2/3, monomethylation, dimethylation, or trimethylation.
Increase in the Complexity of the JmjC-Mediated Histone Demethylation for Angiosperm and Vertebrate Evolution
The SET genes have experienced many duplication events, particularly in the Suv, Absent, small, or homeotic discs1, Trx, and E(z) subfamilies, which are responsible for catalyzing specific Lys methylation at H3K9, H4K36, H3K4, and H3K27 (Zhang and Ma, 2012). Our results showed that the JmjC genes share a similar evolutionary pattern, with expansion in many well-supported large clades, including lineage-specific subfamilies KDM3 to KDM6 and PKDM7 to PKDM9 (Fig. 2A). The JmjC gene family also has similar copy numbers to the SET family, suggesting that the two gene families might have coevolved, which is an important question for further investigation. In addition, the Arabidopsis JMJD6A and PKDM11 proteins are histone Arg demethylases (Table III), and their genes exist as single-copy genes belonging to highly conserved subfamilies (Supplemental Figs. S3 and S10). Similarly, histone Arg methylation is catalyzed by a small group of protein Arg methyltransferases, including PRMT5, PRMT10, and PRMT4 (Ahmad and Cao, 2012). The phylogenetic tree showed that members of these three subfamilies are also highly conserved and mostly single copy, except for two copies of PRMT4 (PRMT4A and PRMT4B) due to the lineage-specific expansion in Brassicaceae (Supplemental Fig. S18). Together, the highly stable single/low copies of these genes suggest that Arg methylases and demethylases are more conserved than those for Lys modification. In addition to duplication patterns, the JmjC and SET proteins also share some domains, such as Plant homeodomain, Tudor domain, and Really Interesting Gene, suggesting that they might have similar interactive partners.
It is worth noting that there are also some important differences between the SET and JmjC families. Among the Arabidopsis SET genes, the SETD and SMYD genes encode proteins with an insertion of 100 to 300 residues in the middle of the SET domain; however, the JmjC family does not have members with an insertion. Also, the human SET family has only 20 members in four subfamilies, due to fewer retained copies after the 2R WGD, compared with 28 human JmjC genes. Also, as mentioned before, some JmjC duplicates were generated by tandem duplication and possibly transpositions, but such mechanisms have not been detected for SET genes.
In angiosperms, ancient duplication events created at least four duplicate gene pairs, contributing to the expansion of JmjC genes. Among these, two pairs belong to the KDM3 subfamily for H3K9 demethylation related to heterochromatin. Histone H3 lysine-9 dimethylation (H3K9me2) is highly concentrated at the centromeric heterochromatin, and this modification is a prerequisite for DNA methylation and gene silencing in Arabidopsis (Jackson et al., 2004; Kavi and Birchler, 2009). In Arabidopsis, 15 SET domain-containing proteins are thought to mediate H3K9me2 deposition, but only six JmjC domain-containing proteins are related to H3K9 demethylation. During angiosperm evolution with additional WGDs, several duplicates in the SET family have been retained, but most of the recent JmjC duplicates have been lost. Our results also suggest that H3K9me2 modification and genome duplication are likely correlated. For instance, an elevated H3K9me2 level repressed redundant gene expression after WGD.
During the evolution of angiosperms with increasing species diversity, it is likely that greater complexity of the histone modification system, due to the expansion of both JmjC and SET genes, provided crucial functions for chromatin-based regulation. The rapidly advancing reverse genetic technologies, including the clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease system, will allow functional tests of the hypotheses raised here on the divergence between paralogs. In particular, the biochemical similarity of closely related JmjC family members can be examined experimentally, whereas functional differences of more distantly related members can be similarly tested. These studies in the near future will likely further improve the understanding of the function and evolution of histone modification enzymes and their impact on development and physiology.
MATERIALS AND METHODS
Data Retrieval
To establish an initial data set with as many JmjC genes as possible, we used several resources. The sequenced genomes and predicted proteomes of plants and animals were downloaded from Phytozome (version 9.0; http://www.phytozome.net/) and Ensembl (release 66; http://www.ensembl.org), respectively. The Joint Genome Institute (http://genome.jgi.doe.gov/) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) databases were additional sources for fungal and partial animal sequences with genome annotation. The sequences for Amborella trichopoda and Monosiga brevicollis were retrieved from the Amborella Genome Database (http://www.amborella.org/) and the Joint Genome Institute, respectively. In proteome data sets, if two or more proteins are annotated for the same gene from alternative splicing, we selected the longest form. The numbers of genes in each JmjC subfamily in different organisms can be found in Supplemental Table S1.
Homolog Searches
Three steps were carried out to identity all recognizable JmjC family members. First, regardless of the origin of proteome data, the HMMER program (version 3.0; Eddy, 1998) with the hidden Markov model was employed to retrieve all eukaryotic JmjC homologs. The hidden Markov model profile of the JmjC domain (PF02373 in the Pfam database) was downloaded and used as a query to find homologous sequences in proteome data sets (Finn et al., 2014). Second, to search for potential JmjC genes from unannotated genomic regions, the sequences acquired in the first step were employed as queries to search genomic sequence data sets using a software called Phoenix (Protein Homolog Extraction), which is based on TBLASTN and GeneWise (http://www.ebi.ac.uk/Tools/psa/genewise/; Altschul et al., 1990; Goujon et al., 2010). Additionally, the sequences from the first and second steps were verified using the PFAM database (http://pfam.xfam.org/search), CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) available from the National Center for Biotechnology Information, and the SMART database (http://smart.embl-heidelberg.de/), with a threshold e-value of less than 1e-10 (Marchler-Bauer et al., 2011; Letunic et al., 2012; Finn et al., 2014). The genes verified by all three steps were then included in our study.
Sequence Alignment
Preliminary multiple sequence alignment was performed using MUSLCE (version 3.8.425) with default parameters (Edgar, 2004). The alignment was then used to generate a preliminary ML tree using FastTree (version 2.1.7; Price et al., 2009). According to the tree topology, the JmjC family was divided into several subgroups. A second round of multiple sequence alignment was carried out for sequences in each subgroup and then adjusted manually in Jalview (version 2.8; Waterhouse et al., 2009). Subsequently, these alignments were combined together using the profile alignment function of MUSCLE.
Phylogenetic Analysis
Systematic phylogenetic analysis of the JmjC family was performed using NJ, ML, and Bayesian methods. For NJ analysis, we used MEGA (version 5.0) with the pairwise deletion option and Poisson correction model (Tamura et al., 2011). The reliability of internal branches was evaluated with a bootstrap test of 1,000 replicates. In this study, ProtTest (version 2.4) was used for model selection, crucial for ML and Bayesian analysis (Abascal et al., 2005). PhyML (version 3.0) and RAxML (version 7.0) were employed to construct ML trees with the Whelan and Goldman amino acid substitution model, γ-distribution, and 100 nonparametric bootstrap replicates (Guindon and Gascuel, 2003; Stamatakis, 2006). Bayesian trees were constructed using MrBayes (version 3.2.1), with the fixed Whelan and Goldman model, four Markov chains, and an average sd of 0.01 (Ronquist and Huelsenbeck, 2003).
Motif and Synteny Analyses
All verified JmjC genes were used to search against the PFAM, CDD, and SMART databases to uncover other known domains or motifs apart from the JmjC domain. Additionally, to discover novel conserved patterns in the amino acid sequences of JmjC proteins, all sequences were analyzed using the software MEME (version 4.9.0; Bailey and Elkan, 1994). The length of a motif was set between 10 and 120 amino acids, and the number of motifs was limited to no more than 10. All sequences within each subfamily were analyzed separately by MEME to identify conserved motifs within the group. Every motif was screened against the PFAM database, and the majority of these motifs are not recorded in the database. In addition, genome synteny between duplicate gene pairs was examined by phylogenetic analysis using the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/; last accessed in February 2015; Tang et al., 2008) and the Synteny Database (http://syntenydb.uoregon.edu/synteny_db/; last accessed in February 2015; Catchen et al., 2009) for plants and animals, respectively.
Expression Analysis
RNA-seq data were of the same sources and treatment as our previous work, including the following Arabidopsis (Arabidopsis thaliana) tissues: seedling, stage 4 flower, inflorescence meristem, stage 1 to 9 flowers, stage 12 flowers, and meiosis (Zhang and Ma, 2012; Zhang et al., 2015). RNA-seq data sets of the following A. trichopoda tissues (ftp://amborella-project.huck.psu.edu/Public/Amborella/transcriptome/) were mapped to the A. trichopoda genome sequences with Periodic Seed Mapping (https://code.google.com/p/perm/): apical meristem, female buds, whole-plant normalized-1, and whole-plant normalized-2. Only the uniquely mapped sequence reads were used further. The gene expression levels were measured by reads per kilobase of mRNA length per million mapped reads. Tiling array data of Arabidopsis JmjC homologs under drought, cold, high-salinity, and ABA treatment (Matsui et al., 2008) were also included in our analysis.
Functional Divergence Analysis
The analysis of FD between JmjC genes of paralogous clades was performed using DIVERGE version 3.0 software (Gu et al., 2013). DIVERGE used the model free estimation calculation of the θ and se type I and II coefficients of FD, based on the occurrence of altered selective constraints or radical shifts of physiochemical properties, respectively (Gu, 1999, 2006). The program also estimates the posterior probabilities of amino acid sites to be responsible for FD. A value of 0.6 was chosen as a cutoff to measure the degree of FD at the amino acid level.
Accession numbers and gene identifiers for sequences used in this study are provided in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Logos representing motifs conserved within each subfamily.
Supplemental Figure S2. The ML tree of the JARID2 subfamily generated by RAxML.
Supplemental Figure S3. The ML tree of the JMJD6 subfamily generated by RAxML.
Supplemental Figure S4. The ML tree of the KDM2 subfamily generated by RAxML.
Supplemental Figure S5. The ML tree of the KDM3 subfamily generated by RAxML.
Supplemental Figure S6. The ML tree of the KDM4 subfamily generated by RAxML.
Supplemental Figure S7. The ML tree of the KDM6 subfamily generated by RAxML.
Supplemental Figure S8. The ML tree of the PKDM9 subfamily generated by RAxML.
Supplemental Figure S9. The ML tree of the PKDM10 subfamily generated by RAxML.
Supplemental Figure S10. The ML tree of the PKDM11 subfamily generated by RAxML.
Supplemental Figure S11. The ML tree of the PKDM12 subfamily generated by RAxML.
Supplemental Figure S12. The ML tree of the PKDM13 subfamily generated by RAxML.
Supplemental Figure S13. An illustration of plants with the syntenic regions containing representative duplicated gene pairs from recent polyploidy events.
Supplemental Figure S14. An illustration of animals with duplicated gene pairs generated by recent polyploidy events.
Supplemental Figure S15. Duplication events in the JmjC family inferred from amino acid sequence analyses.
Supplemental Figure S16. Multiple sequence alignments of angiosperm PKDM7A and PKDM7B.
Supplemental Figure S17. An ML tree and multiple sequence alignments of PKDM7A and PKDM7D in core eudicots.
Supplemental Figure S18. An ML tree of PRMT5, PRMT10, and PRMT4 in green plants.
Supplemental Table S1. The number of different JmjC subfamily members in representative species.
Supplemental Table S2. A list of all JmjC genes included in this study.
Supplemental Table S3. The sequences of conserved motifs within individual subfamilies.
Supplemental Table S4. Number of human paralogous genes identified from Synteny Database at Uoregon site using KDM2 and PKDM10, KDM5, KDM6 loci as seed genes and Ciona intestinalis as an out outgroup.
Supplemental Table S5. Expression profile of JmjC genes at different stages of flower development in Arabidopsis and Amborella.
Supplemental Table S6. Tiling array profile of Arabidopsis JmjC genes under drought, cold, high-salinity, and ABA treatments.
Supplemental Table S7. Expression of JmjC genes in the wild type and the dyt1 (bhlh22), bhlh10, bhlh89, bhlh91, and pkdm7d jmj16 mutants by RNA-seq.
Supplemental Table S8. A list of sites with Functional Divergence type I in plant JmjC genes.
Supplementary Material
Acknowledgments
We thank Yaqiong Wang, Liping Zeng, Haifeng Wang, Qi Li, and Fei Chen for comments on the article and helpful discussion.
Glossary
- H3K27me3
histone H3 lysine-27 trimethylation
- αKG
α-ketoglutarate
- NJ
neighbor-joining
- ML
maximum likelihood
- MRCA
most recent common ancestor
- WGD
whole-genome duplication
- RNA-seq
RNA-sequencing
- FD
functional diversification
- H3K9me2
histone H3 lysine-9 dimethylation
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 91131007), the Ministry of Science and Technology of China (grant no. 2011CB944603), and the Program for Young Excellent Talents in Tongji University.
Articles can be viewed without a subscription.
References
- Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Ahmad A, Cao X (2012) Plant PRMTs broaden the scope of arginine methylation. J Genet Genomics 39: 195–208 [DOI] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. (2007) New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME (2009) Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol 1: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, et al. (2010) Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byvoet P, Shepherd GR, Hardin JM, Noland BJ (1972) The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys 148: 558–567 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Catchen JM, Conery JS, Postlethwait JH (2009) Automated identification of conserved synteny after whole-genome duplication. Genome Res 19: 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hu Y, Zhou DX (2011) Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim Biophys Acta 1809: 421–426 [DOI] [PubMed] [Google Scholar]
- Cho JN, Ryu JY, Jeong YM, Park J, Song JJ, Amasino RM, Noh B, Noh YS (2012) Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell 22: 736–748 [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H (2014) Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–493 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C (2014) Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Herridge RP, Ambrose BA, Macknight RC (2008) Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol 148: 1964–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Greenberg MV, Ausin I, Law RW, Moissiard G, Schubert D, Jacobsen SE (2010) Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep 11: 950–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Weng Z (2013) The correlation between histone modifications and gene expression. Epigenomics 5: 113–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Dai Y, Wang X, Wang Z, He H, Yang H, Cao Y, Deng XW, Ma L (2012) IBM1, a JmjC domain-containing histone demethylase, is involved in the regulation of RNA-directed DNA methylation through the epigenetic control of RDR2 and DCL3 expression in Arabidopsis. Nucleic Acids Res 40: 8905–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Lu D, Ma X, Peng Y, Sun Y, Ning G, Ma H (2012) Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J 72: 612–624 [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science 330: 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan ES, Xu YF, Wong JY, Goh JG, Sun B, Wee WY, Huang JB, Ito T (2014) Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun 5: 5098. [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313: 903–919 [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. (1999) Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol 16: 1664–1674 [DOI] [PubMed] [Google Scholar]
- Gu X. (2006) A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol 23: 1937–1945 [DOI] [PubMed] [Google Scholar]
- Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee Z, Zeng Y (2013) An update of DIVERGE software for functional divergence analysis of protein family. Mol Biol Evol 30: 1713–1719 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A (2011) Histone modification: cause or cog? Trends Genet 27: 389–396 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE (2004) Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112: 308–315 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jeong JH, Song HR, Ko JH, Jeong YM, Kwon YE, Seol JH, Amasino RM, Noh B, Noh YS (2009) Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 4: e8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL (2010) Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA 107: 21623–21628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Harmer S (2011) JMJD5 functions in concert with TOC1 in the Arabidopsis circadian system. Plant Signal Behav 6: 445–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M. (2007) The 2R hypothesis: an update. Curr Opin Immunol 19: 547–552 [DOI] [PubMed] [Google Scholar]
- Kavi HH, Birchler JA (2009) Interaction of RNA polymerase II and the small RNA machinery affects heterochromatic silencing in Drosophila. Epigenetics Chromatin 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, Amasino RM, Noh B, Noh YS (2010) Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J 29: 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K (2012) Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13: 297–311 [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC (1999) Four evolutionary strata on the human X chromosome. Science 286: 964–967 [DOI] [PubMed] [Google Scholar]
- Lei L, Zhou SL, Ma H, Zhang LS (2012) Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol Biol 12: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X (2011a) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Liu C, Cao X (2010) JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 20: 387–390 [DOI] [PubMed] [Google Scholar]
- Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X (2008) Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50: 886–896 [DOI] [PubMed] [Google Scholar]
- Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, Tobin EM (2011b) The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol 155: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Panopoulou G, Hennig S, Groth D, Krause A, Poustka AJ, Herwig R, Vingron M, Lehrach H (2003) New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res 13: 1056–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sani E, Herzyk P, Perrella G, Colot V, Amtmann A (2013) Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 14: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T (2008) Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319: 462–465 [DOI] [PubMed] [Google Scholar]
- Shen Y, Conde e Silva N, Audonnet L, Servet C, Wei W, Zhou DX (2014) Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front Plant Sci 5: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR (2007) Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 25: 1–14 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH (2010) Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA 107: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Shen L, Chen Y, Bao S, Thong Z, Yu H (2014) A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev Cell 30: 437–448 [DOI] [PubMed] [Google Scholar]
- Yang H, Han Z, Cao Y, Fan D, Li H, Mo H, Feng Y, Liu L, Wang Z, Yue Y, et al. (2012a) A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet 8: e1002664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Mo H, Fan D, Cao Y, Cui S, Ma L (2012b) Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep 31: 1297–1308 [DOI] [PubMed] [Google Scholar]
- Yang W, Jiang D, Jiang J, He Y (2010) A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 62: 663–673 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y (2008) Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma H (2012) Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol 195: 248–263 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang L, Yang Y, Cui J, Chang F, Wang Y, Ma H (2015) Analysis of Arabidopsis floral transcriptome: detection of new florally expressed genes and expansion of Brassicaceae-specific gene families. Front Plant Sci 5: 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhou X, Ma H (2008) Evolutionary history of histone demethylase families: distinct evolutionary patterns suggest functional divergence. BMC Evol Biol 8: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.