Florigen-encoding genes have different chromatin signatures associated with the floral transition in ancestral and modern maize.
Abstract
The activity of the maize (Zea mays) florigen gene ZEA CENTRORADIALIS8 (ZCN8) is associated with the floral transition in both day-neutral temperate maize and short-day (SD)-requiring tropical maize. We analyzed transcription and chromatin modifications at the ZCN8 locus and its nearly identical paralog ZCN7 during the floral transition. This analysis was performed with day-neutral maize (Zea mays ssp. mays), where flowering is promoted almost exclusively via the autonomous pathway through the activity of the regulatory gene indeterminate1 (id1), and tropical teosinte (Zea mays ssp. parviglumis) under floral inductive and noninductive photoperiods. Comparison of ZCN7/ZCN8 histone modification profiles in immature leaves of nonflowering id1 mutants and teosinte grown under floral inhibitory photoperiods reveals that both id1 floral inductive activity and SD-mediated induction result in histone modification patterns that are compatible with the formation of transcriptionally competent chromatin environments. Specific histone modifications are maintained during leaf development and may represent a chromatin signature that favors the production of processed ZCN7/ZCN8 messenger RNA in florigen-producing mature leaf. However, whereas id1 function promotes histone H3 hyperacetylation, SD induction is associated with increased histone H3 dimethylation and trimethylation at lysine-4. In addition, id1 and SD differently affect the production of ZCN7/ZCN8 antisense transcript. These observations suggest that distinct mechanisms distinguish florigen regulation in response to autonomous and photoperiod pathways. Finally, the identical expression and histone modification profiles of ZCN7 and ZCN8 in response to floral induction suggest that ZCN7 may represent a second maize florigen.
The transition from vegetative to reproductive development is triggered by a leaf-derived, mobile floral-promoting signal named florigen (Chailakhyan, 1937; Giakountis and Coupland, 2008). In Arabidopsis (Arabidopsis thaliana), the protein encoded by the FLOWERING LOCUS T (FT) gene has florigen activity (Corbesier et al., 2007; Jaeger and Wigge, 2007). All Arabidopsis floral regulatory pathways, including the autonomous, GA, photoperiod, and vernalization pathways, converge on the FT gene, indicating that FT is a key integrator of floral inductive signals (Turck et al., 2008). In the photoperiod pathway, FT transcription in the vasculature of mature leaves is regulated by the direct binding of a protein encoded by CONSTANS (CO) in response to long-day growing conditions (Samach et al., 2000; Tiwari et al., 2010). FT, a phosphatidylethanolamine-binding protein, then moves from leaves through the phloem to the shoot apical meristem (Corbesier et al., 2007; Jaeger and Wigge, 2007), where it interacts with a basic Leu zipper domain transcription factor encoded by FLOWERING LOCUS D (FD) to induce the expression of floral identity genes, such as APETALA1 (Abe et al., 2005; Wigge et al., 2005). Recently, it was shown that chromatin-related mechanisms play an important role in regulating FT expression (He, 2012; Gu et al., 2013; Bu et al., 2014; López-González et al., 2014; Wang et al., 2014).
FT orthologs have been discovered in diverse plant species, and many of these FT-related genes are regulated differently depending on the photoperiod sensitivity or other floral inductive requirements of the species (Andrés and Coupland, 2012). In maize (Zea mays), a large phosphatidylethanolamine-binding protein gene family named ZEA CENTRORADIALIS (ZCN) was identified and found to include putative FT orthologs (Danilevskaya et al., 2008). One of these genes, ZCN8, was shown to have the requisite characteristics for florigenic activity (Lazakis et al., 2011; Meng et al., 2011). Indeed, ZCN8 has high sequence similarity to FT and exhibits a leaf-exclusive expression pattern, and ZCN8 protein interacts with Delayed Flowering (DLF), the cognate maize ortholog of the Arabidopsis FD protein (Muszynski et al., 2006; Danilevskaya et al., 2008). Moreover, phloem-specific ZCN8 expression in Arabidopsis rescues the ft mutation (Lazakis et al., 2011), and in maize, ectopic ZCN8 expression causes earlier flowering in transgenic plants, while silencing by artificial microRNA results in delayed flowering (Meng et al., 2011). Quantitative trait locus (QTL) studies show that ZCN8 is closely associated with Vegetative to generative transition2, a major-effect QTL for flowering time variation (Bouchet et al., 2013). Finally, ZCN8 is up-regulated upon the floral transition in day-neutral temperate maize and photoperiod-sensitive tropical maize (Lazakis et al., 2011; Meng et al., 2011). The floral transition in maize temperate lines (e.g. the inbred line B73) occurs almost exclusively in response to endogenous signals by means of the autonomous pathway, and ZCN8 expression in mature leaf is activated by the indeterminate1 (id1) gene, which encodes a monocot-specific zinc finger transcriptional regulator (Colasanti et al., 1998; Colasanti and Coneva, 2009; Lazakis et al., 2011). Conversely, in tropical maize, including the progenitor of temperate maize, teosinte (Zea mays ssp. parviglumis), which has an obligate requirement for short-day (SD) photoperiods to induce flowering, ZCN8 is highly up-regulated in leaves under inductive photoperiods (Lazakis et al., 2011; Meng et al., 2011). Therefore, these extreme maize varieties present a unique opportunity to investigate the differences in florigen regulation between the autonomous and photoperiod pathways.
The nature of the upstream mechanisms that control ZCN8 expression in the leaf remains an open question. A potential maize ortholog of Arabidopsis CO, CONZ1, was identified (Miller et al., 2008), yet there is no indication that CONZ1 directly activates ZCN8 expression or that it regulates maize flowering. In addition, although id1 activates ZCN8, the temporal and spatial expression patterns of the id1 and ZCN8 genes do not overlap, because id1 gene activity is confined to immature developing leaves (Colasanti et al., 1998; Wong and Colasanti, 2007), whereas ZCN8 expression is detected only in mature leaves (Danilevskaya et al., 2008). Therefore, it is unlikely that ZCN8 is a direct target of the ID1 protein. One possibility is that ID1 protein operates by means of epigenetic mechanisms to promote the formation of a transcriptionally competent ZCN8 chromatin in immature leaf, which is maintained throughout leaf development to enable the production of florigenic signals in the mature leaf. The relevance of epigenetic mechanisms in maize flowering was demonstrated recently with the finding that the nucleosome remodeling factor complex component101 (nfc101) and nfc102 genes encode WD-repeat proteins that bind both id1 and ZCN8 sequences, thus regulating their expression and chromatin modifications (Mascheretti et al., 2013).
In this study, we provide evidence that chromatin modifications are involved in id1-mediated regulation of ZCN8 expression in temperate, day-neutral maize. Unexpectedly, a different histone modification pattern of ZCN8 chromatin during leaf development was detected in response to SD-dependent floral induction in teosinte. These findings highlight both conserved and divergent features of florigen regulation between autonomous and photoperiod-regulated pathways. Furthermore, analysis of ZCN7, a highly similar paralog of ZCN8 (Danilevskaya et al., 2008), shows that both genes have identical expression and chromatin profiles, suggesting that ZCN7 may encode another maize florigen.
RESULTS
Identification and Isolation of ZCN7 and ZCN8 Gene Sequences
A previous phylogenetic analysis of ZCN genes suggests that ZCN7 and ZCN8 represent paralogs arising from the tetraploid ancestry of maize (Danilevskaya et al., 2008). ZCN7 and ZCN8 genomic DNA and complementary DNA (cDNA) sequences are publicly available. However, the ZCN8 sequence was identified in the maize B73 inbred line (GenBank accession no. EU241899), while the ZCN7 sequence was identified in the Gaspé Flint early-flowering line. In addition, the ZCN8 cDNA corresponding to the processed mRNA was isolated from mature leaf of the B73 inbred line, whereas only the unspliced version of the ZCN7 cDNA was obtained in the Gaspé Flint line (Danilevskaya et al., 2008). In this study, we identified ZCN7 genomic DNA and cDNA sequences from the B73 inbred line (GenBank accession no. KP202720) and isolated a sequence representing the processed mRNA for the ZCN7 gene from mature leaf of this line (Supplemental Fig. S1; for details regarding specific ZCN7 and ZCN8 transcript analysis, see below).
For the purposes of analyzing expression in teosinte, we sequenced the teosinte ZCN7 and ZCN8 genes and cDNAs. The ZCN7 and ZCN8 genomic sequences were identified using a PCR-based method (Supplemental Text S1), and their respective coding sequences were subsequently identified by synthesizing cDNA from total RNA of teosinte mature leaf followed by reverse transcription (RT)-PCR with primers located close to the start and stop codons. Even though teosinte is considered a wild relative of maize, we detected no sequence heterogeneity in the ZCN genes isolated from teosinte. Sequence similarity between line B73 ZCN7 and ZCN8 genes and their orthologs in teosinte are reported in Supplemental Table S1.
Experimental Conditions for Analyzing ZCN7 and ZCN8 Regulation during Leaf Development and in Response to Floral Induction
The major aim of this study is to analyze ZCN7 and ZCN8 regulation when the plant is committed to flowering and in early and later stages of leaf development. This provides a framework for determining whether and how chromatin modification patterns might be established at florigen genes in response to floral inductive cues and maintained during leaf development. To this end, immature and mature leaf samples were obtained from id1 mutant segregating plants introgressed into the B73 background at the V6/V7 developmental stage (i.e. seedlings with the sixth or seventh leaf fully extended with collars and the eighth/ninth leaves visible; Fig. 1). This stage corresponds to the vegetative-to-reproductive phase transition in the B73 inbred line (Coneva et al., 2007; Lazakis et al., 2011). Moreover, the immature leaf tissue used for analysis represents where the id1 gene is expressed, while the ZCN8 processed mRNA is not yet produced (Colasanti et al., 1998; Wong and Colasanti, 2007; Meng et al., 2011). In contrast, mature leaf expresses ZCN8 mRNA but id1 transcript and ID1 protein are not detected. To characterize more precisely ZCN7 and ZCN8 mRNA localization during leaf development, we carried out quantitative reverse transcription (qRT)-PCR with primers conserved in the two paralogs and using RNA obtained following leaf blade dissection of pretransition (V4), transition (V6), and posttransition (V8) stage plants (Supplemental Fig. S2). This analysis showed that ZCN7/ZCN8 mRNA is most abundant in the distal portions of adult leaf blades and much less so in juvenile leaves at all three stages of development. ZCN7/ZCN8 expression was not detected in blade sheaths or in nonphotosynthetic regions of immature leaves. Interestingly, transcript levels continued to increase well after the floral transition (V8 leaves), suggesting a possible role for ZCN7/ZCN8 in reproductive development. On the basis of these observations, we decided to employ the leaf blades of V6-to-V7 transition stage plants for the analysis of ZCN7/ZCN8 transcripts and chromatin modifications (Supplemental Fig. S3). A parallel analysis of the same tissues was performed with teosinte plants harvested at the analogous developmental stage. In this case, the floral transition was induced by growing plants under SD conditions, while flowering was inhibited in a parallel set of plants by interrupting the long night with 1 h of light. We previously showed that this night break (NB) regimen effectively prevents flowering in teosinte without greatly altering the total amount of light received by nonflowering plants relative to those grown under SD conditions (Fig. 1; Lazakis et al., 2011; Coneva et al., 2012).
Figure 1.
Photographs of line B73 maize and teosinte plants induced and uninduced for flowering. A, Wild-type (wt) plant at V7 stage. The inset shows a meristem at the floral transition stage. B, Maize id1 mutant plant at V7 stage. The inset shows a meristem at the vegetative stage. C, Teosinte grown under NB regimen for 27 d and induced for 10 d under SD conditions. The inset shows an inflorescence meristem. D, Teosinte plant grown under noninductive NB conditions for 37 d. The inset shows an uninduced meristem. Teosinte plants exhibited extensive tillering, but only leaves from the main shoot were harvested for analysis. Bars = 0.25 mm for all inset photographs.
Analysis of Specific ZCN7 and ZCN8 Transcripts
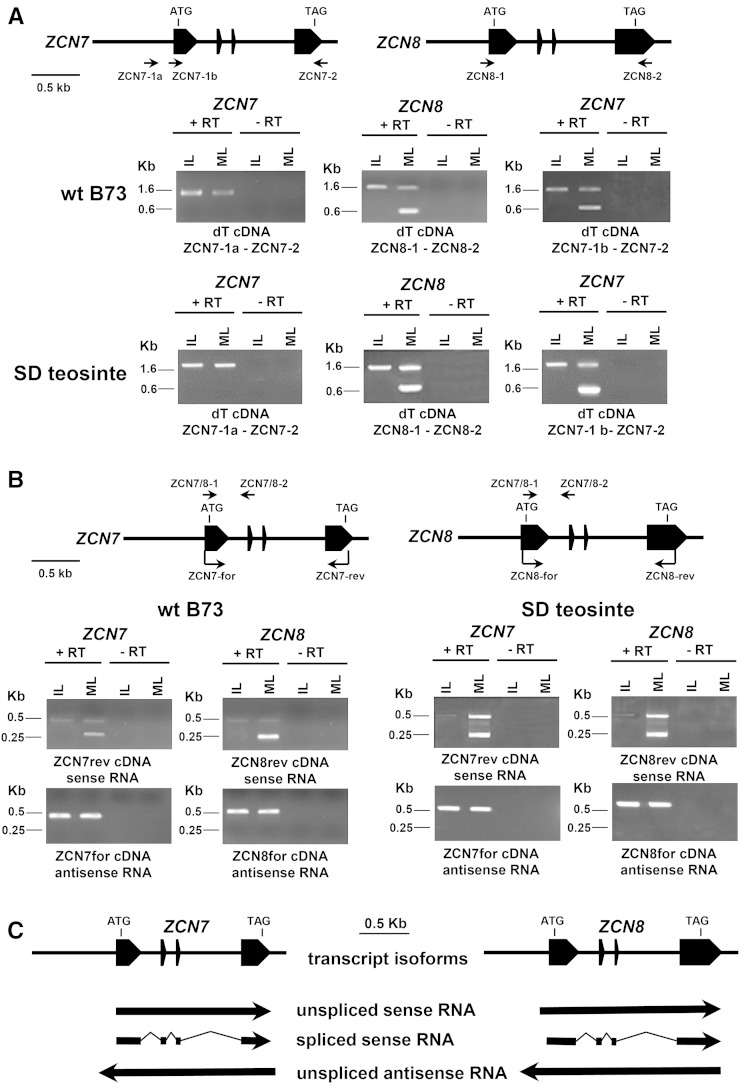
A previous analysis of ZCN7 and ZCN8 transcription using locus-specific primers located near the start and stop codons revealed that the ZCN8 locus gives rise to a mixture of spliced and unspliced ZCN8 transcripts in leaf blade of the maize B73 inbred line, while only an unspliced form was detected in immature leaf and other maize tissues (Danilevskaya et al., 2008). Conversely, only unspliced ZCN7 transcript was detected in all maize tissues analyzed in the maize Gaspé Flint line. Using primer combinations to the same regions, we confirmed these results in line B73 immature and mature leaf samples (Fig. 2A). However, when we used a ZCN7-specific forward primer located 75 bp downstream of that employed by Danilevskaya et al. (2008; Fig. 2A, primer ZCN7-1b instead of ZCN7-1a), the ZCN7 gene showed the same pattern of transcript accumulation reported for ZCN8, including the production of spliced mRNA in mature leaf (Fig. 2A). Similar results were obtained in mature and immature leaves of teosinte plants grown under floral inductive SD conditions. This indicates that, in mature leaves of the B73 inbred line and in teosinte, ZCN7 produces a processed mRNA that encodes a putative polypeptide. It is worth mentioning that the expression pattern we observed for ZCN7 and ZCN8 is supported by data from RNA sequencing studies (Supplemental Table S2; Sekhon et al., 2013), which can distinguish among highly similar ZCN paralogs, thus indicating that both ZCN7 and ZCN8 are detected predominantly in mature leaf blade.
Figure 2.
RT-PCR analysis of ZCN7 and ZCN8 transcripts in line B73 and teosinte plants. A, RT-PCR was performed with oligo(dT)-primed cDNA prepared with RNA extracted from mature (ML) and immature (IL) leaves and with (+) or without (−) the addition of reverse transcriptase (RT). Schemes of ZCN7 and ZCN8 genes and positions of primers used for RT-PCR are reported. The structures of both genes are conserved in line B73 and teosinte. B, Strand-specific RT-PCR carried out using forward (ZCN7-for and ZCN8-for) and reverse (ZCN7-rev and ZCN8-rev) primers for synthesizing cDNA produced by the antisense and sense RNA strands, respectively. Subsequent RT-PCR with ZCN7/8-1 and ZCN7/8-2 primers permits the detection of both spliced and unspliced RNAs. C, Diagram schematizing the three transcript isoforms produced by ZCN7 and ZCN8 genes. RNA length and position with respect to the gene structure are based on strand-specific RT-PCR results with line B73 plants.
A recent report used a combination of RNA blotting with strand-specific probes and RT-PCR with locus- and strand-specific primers to show that ZCN8 produces three transcript isoforms (Mascheretti et al., 2013). These isoforms correspond to (1) the processed sense mRNA, which accumulates only in mature leaf; (2) a very low amount of unspliced sense-strand premRNA present both in meristematic enriched regions and in mature leaf; and (3) an unspliced antisense RNA strand, which represents the prevalent form of unspliced transcript and is produced not only in leaves but also in various other tissues (Fig. 2C). This observation indicates that, to study ZCN8 transcription in detail, it is necessary to employ a method that can distinguish all three RNA isoforms. Accordingly, we performed strand-specific RT-PCR to differentiate the ZCN7 and ZCN8 transcript isoforms present in leaves of wild-type line B73 and SD-induced teosinte plants. Our results reveal that ZCN7 and ZCN8 produce the same three transcript isoforms in the B73 inbred line and that these isoforms are present also in teosinte plants (Fig. 2, B and C). We cloned and sequenced cDNAs derived from the ZCN7 and ZCN8 antisense RNAs of B73 and teosinte plants and found that they encode very short putative open reading frames with no homology to known proteins (Supplemental Fig. S4; Mascheretti et al., 2013). Therefore, these antisense transcripts may represent long noncoding RNAs (lncRNAs). Collectively, our results indicate that the highly similar ZCN7 and ZCN8 transcripts also exhibit an analogous expression profile in line B73 and teosinte.
Variation of ZCN7 and ZCN8 Transcript Isoform Levels under Floral Inductive versus Noninductive Conditions
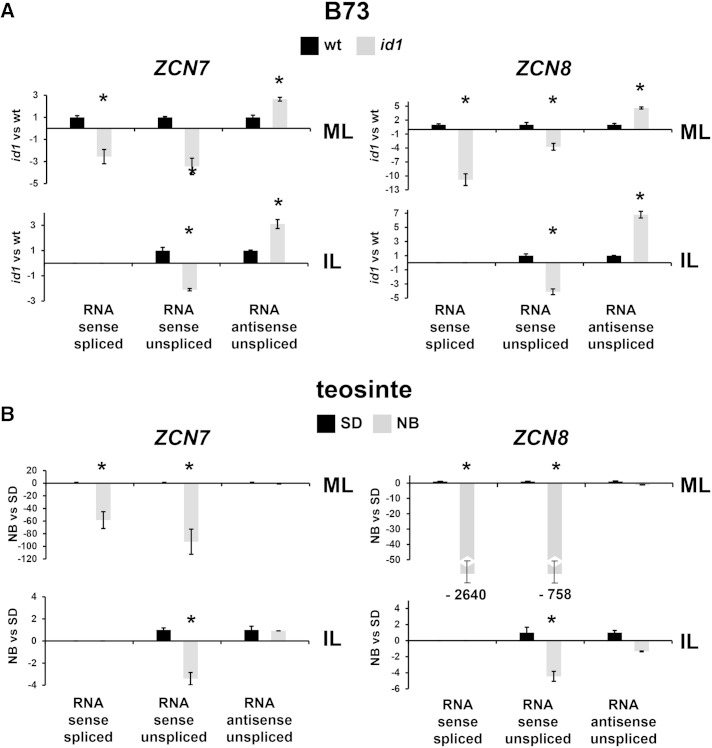
Strand-specific qRT-PCR was employed to analyze the abundance of ZCN7 and ZCN8 transcript isoforms under conditions that reduce florigen production, namely, the absence of id1 function in temperate maize and noninductive NB conditions in tropical teosinte plants. We found that processed sense mRNA levels of ZCN7 and ZCN8 decrease in mature leaves of the id1 mutant compared with wild-type mature leaves and that this occurs with a concomitant reduction of unspliced sense pre-mRNA and an increase of unspliced antisense strand in the same tissues (Fig. 3A). The same pattern was observed in B73 immature leaf, except that in these samples, spliced sense mRNA was not detected. In contrast, a different pattern of isoform transcript variation was observed in teosinte plants. Mature leaves from plants grown under noninductive NB conditions have less ZCN7 and ZCN8 processed and unspliced sense-oriented mRNA levels compared with plants grown under inductive SD conditions; however, antisense-strand levels were unaffected (Fig. 3B). No variation in antisense RNA accumulation was observed, and a decrease in unspliced pre-mRNA levels was also detected in teosinte immature leaf. It is worth noting that the magnitude of the decrease of ZCN7 and ZCN8 sense transcript levels is much higher in mature leaf of teosinte under noninductive NB conditions than in mature leaf of the id1 late-flowering mutant (Fig. 3). This effect may be correlated with the higher ZCN7 and ZCN8 transcript levels detected in mature leaves of teosinte under floral inductive SD conditions compared with wild-type line B73 (Supplemental Fig. S5; Lazakis et al., 2011). Although higher levels of ZCN7 and ZCN8 RNA are more pronounced for the sense pre-mRNA and processed mRNA, it is also evident for the antisense strand. Conversely, no differences in transcript isoform abundance were detected in immature leaves of line B73 and teosinte.
Figure 3.
Strand-specific qRT-PCR analysis of ZCN7 and ZCN8 transcript isoforms in B73 and teosinte plants. Real-time qRT-PCR quantification is shown for ZCN7 and ZCN8 sense and antisense transcripts from mature (ML) and immature (IL) leaves of wild-type (wt) and id1 mutant B73 plants (A) and of teosinte plants grown under inductive SD and inhibitory NB flowering conditions (B). Bar diagrams show mean values of transcript amounts for one biological replicate, normalized to glyceraldehyde-3-phosphate dehydrogenase2 (gapc2) sense mRNA. Wild-type and SD values are set to 1, and the amount and direction (increase or decrease) of change for each RNA isoform in id1 mutants versus the wild type (A) or in SD versus NB conditions (B) was calculated using the 2−ΔΔCt method as described by Rossi et al. (2007; for values less than 1, the negative value was obtained by applying the formula −1/2−ΔΔCt). Asterisks indicate statistically significant changes (P ≤ 0.01) achieved in separate analyses of the two biological replicates.
Overall, these results indicate that both ZCN7 and ZCN8 mRNAs are positively regulated by factors that induce flowering (i.e. id1 gene function in autonomously flowering maize and SD photoperiod-requiring teosinte). However, only functional id1 concomitantly affects the abundance of the antisense-orientated RNA strand of these genes, suggesting that the regulation of ZCN7/ZCN8 putative antisense lncRNAs is limited to the id1-regulated flowering pathway and not the photoperiod pathway.
Correlation of Histone Modifications in Line B73 ZCN7 and ZCN8 Chromatin with id1 Gene Activity
Histone modifications are marks of chromatin transcriptional status (Lauria and Rossi, 2011). Therefore, we analyzed the histone profiles of the ZCN7 and ZCN8 genes in temperate maize to understand whether and how histone modifications are correlated with the id1-mediated activation of these genes. Chromatin immunoprecipitation (ChIP) assays were performed to analyze histone modifications in different regions of the ZCN7 and ZCN8 genes (Fig. 4; Supplemental Fig. S6). We selected histone marks that are usually located within genes and that were previously analyzed for florigen genes of other plant species (Jiang et al., 2008; Adrian et al., 2010; He, 2012; Sun et al., 2012; Gu et al., 2013; Mascheretti et al., 2013; López-González et al., 2014). Four histone marks usually associated with active transcription were selected for chromatin analysis. These are histone H3 acetylated at lysine-9 and lysine-14 (H3ac), histone H3 dimethylated and trimethylated at lysine-4 (H3K4me2 and H3K4me3), and histone H3 dimethylated at lysine-36 (H3K36me2). In addition, we examined histone H3 trimethylation at lysine-27 (H3K27me3) as a representative repressive euchromatin mark. Finally, an antibody against the histone H3 C-terminal region that recognizes histone H3 independently of any posttranscriptional modifications was used as an estimate of nucleosome density, which is a useful metric for normalizing data generated in histone modification analyses (Rossi et al., 2007).
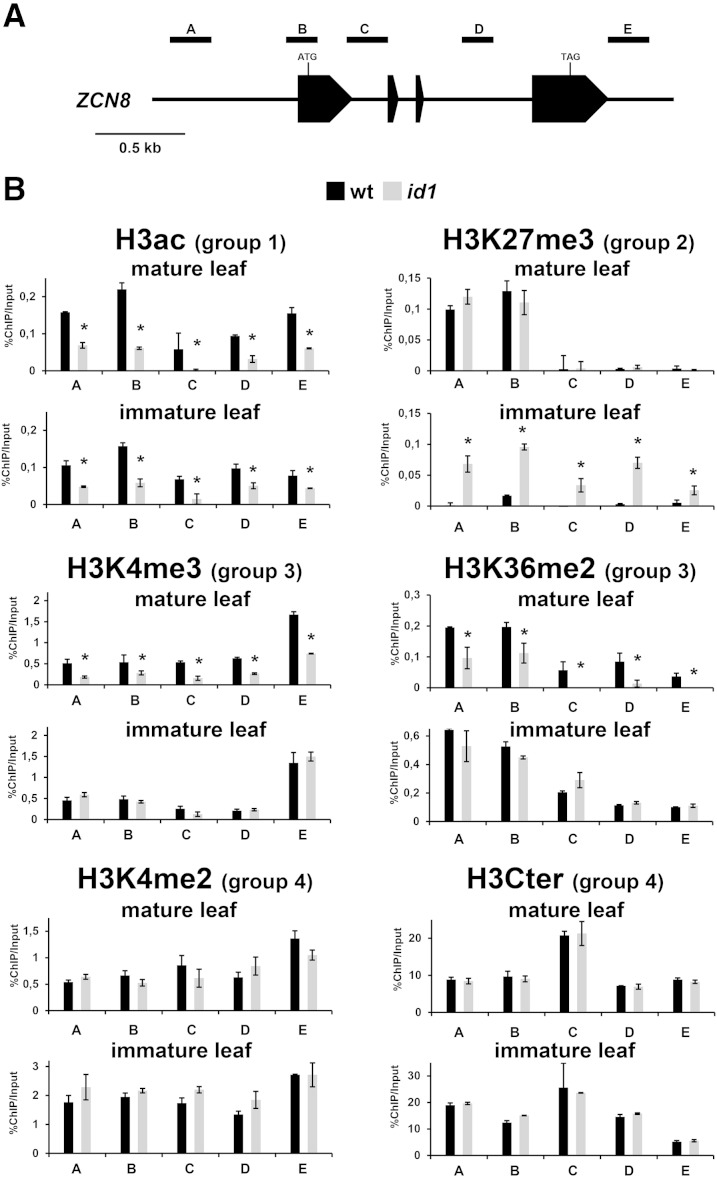
Figure 4.
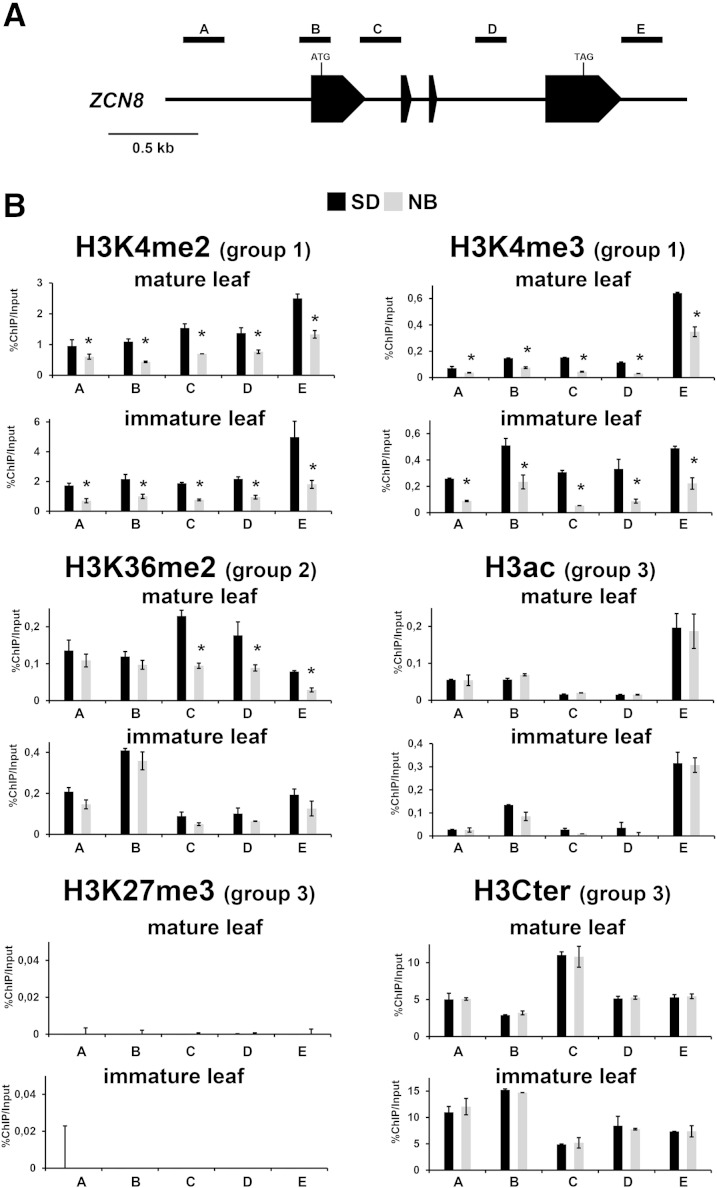
Analysis of ZCN8 histone modifications in line B73 wild-type (wt) and id1 mutant plants. A, Schematic depiction of the ZCN8 gene, with black boxes representing exons. The positions of the regions analyzed in ChIP assays are indicated. B, Bar diagrams represent real-time PCR quantification of ChIP DNA, reported as percentage of the chromatin input, from assays performed using the indicated antibodies. The data are average values from two independent ChIP assays and from three PCR repetitions for each ChIP assay and are reported by subtracting the background signal, measured by omitting antibody during the ChIP procedure. Asterisks indicate statistically significant changes (P ≤ 0.01) in the id1 mutant versus the wild type. The grouping of histone marks on the basis of how its variation associates with the activity of the id1 gene is indicated in parentheses. Similar results were obtained after correction for nucleosome occupancy measured as reported by Rossi et al. (2007). H3Cter, Histone H3 C-terminal region.
We observed that the pattern of histone modifications at ZCN7 and ZCN8 chromatin was very similar and that the histone marks analyzed can be divided into four groups based on their correlation with the activity of the id1 gene. These results are illustrated in Figure 4 and Supplemental Figure S6 and are summarized schematically in Figure 6. The first group includes H3ac, which exhibits significantly higher levels in both immature and mature leaves of wild-type plants relative to id1 mutant leaves in all genomic regions analyzed. Conversely, in the second group, the repressive H3K27me3 mark was detected at higher levels in immature leaves of the id1 mutant relative to the wild type, while in mature leaves, H3K27me3 was prevalent at the 5′ end regions of ZCN7 and ZCN8 independently of normal id1 function. The active marks H3K4me3 and H3K36me2, similar to H3ac, were higher in mature leaves of wild-type plants relative to id1 mutant mature leaves. However, unlike H3ac, these marks were present at similar levels in immature leaves of both wild-type and id1 mutant plants, thus defining a third group. Finally, the fourth group includes H3K4me2 and nucleosome occupancy, which were independent of id1 gene activity for both immature and mature leaf. Since nucleosome occupancy is not affected by id1, the statistical significance of the observed changes in histone modifications was not influenced by the correction for nucleosome density (Fig. 4; Supplemental Fig. S6).
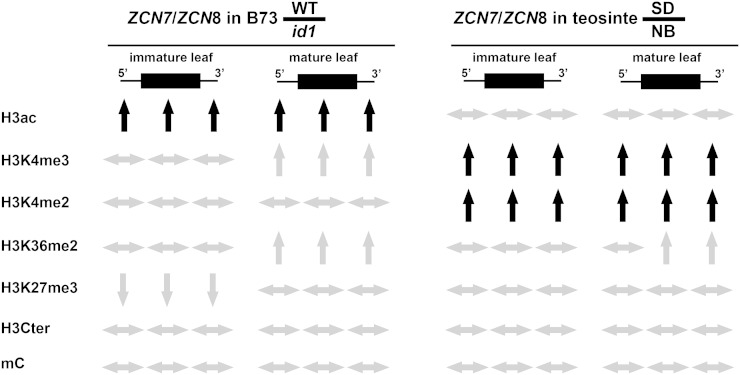
Figure 6.
Summary of ZCN7 and ZCN8 epigenetic pattern variations in line B73 and teosinte plants. The diagram is based on data reported in Figures 4 and 5 and in Supplemental Figures S6 and S7. The direction of each arrow indicates the direction of the change for the histone modification listed at left and for cytosine methylation (mC) measured by means of restriction with the MspJI enzyme. Variations of epigenetic mark levels that occur in immature leaf and that are maintained in mature leaf are highlighted by black arrows. WT, Wild-type.
This analysis also provides information about the features of chromatin modification at florigen genes in temperate maize. First, the changes detected are consistent for all analyzed genomic regions of ZCN7 and ZCN8. Second, the ZCN7 and ZCN8 loci in both immature and mature leaves of florally competent wild-type plants are characterized by the presence of histone marks associated with active chromatin, suggesting that an open chromatin structure is present through all stages of leaf development. Third, the localization of some histone marks within the ZCN7 and ZCN8 loci was different than expected. That is, H3K4me3 was enriched in the 3′ end, while it usually exhibits a peak in the 5′ end region, and H3K27me3 accumulated in the 5′ end region, while it usually is located in the gene body (Lauria and Rossi, 2011).
Correlation of Histone Modifications in Teosinte ZCN7 and ZCN8 Chromatin with SD-Induced Flowering
The same ChIP assays used to analyze histone modifications in line B73 plants were carried out with SD (induced) and NB (uninduced) teosinte plants (Figs. 5 and 6; Supplemental Fig. S7). As described above for line B73 with respect to id1 gene activity, the histone modification variation between SD and NB conditions allows the subdivision of the histone marks into different groups based on changes in response to SD floral induction. The first group includes H3K4me2 and H3K4me3, which were enriched in ZCN7 and ZCN8 in immature leaves of SD-grown plants and are maintained at high levels in mature leaf. The second group is typified by H3K36me2, which increased in response to SD conditions only in mature leaf. The third group includes H3ac, H3K27me3, and the nucleosome density assay, which displayed no variation between SD and NB conditions. It is worth noting that histone marks altered in response to SD floral induction in teosinte are different compared with those affected by id1 in the B73 inbred line (Fig. 6). In addition, H3K27me3 was not detected in ZCN7 and ZCN8 chromatin of teosinte plants. Although a direct comparison between photoperiodic and autonomous flowering may require consideration of the different growth conditions, these observations suggest that distinct chromatin modification mechanisms occur for regulating florigen expression in response to the autonomous and photoperiod pathways. Nevertheless, our analysis also reveals that some of the chromatin modification features reported for temperate maize are similarly found in teosinte. These include a similar histone modification pattern correlated with SD floral induction exhibited by the ZCN7 and ZCN8 teosinte paralogs, the absence of nucleosome occupancy variation between SD and NB conditions, the presence of histone marks linked to active chromatin already detectable in immature leaf, and the unexpected distribution pattern of H3K4me3.
Figure 5.
Analysis of ZCN8 histone modifications in teosinte SD and NB plants. A, Schematic depiction of the ZCN8 gene, with black boxes representing exons. The positions of the regions analyzed in ChIP assays are indicated. B, Bar diagrams represent real-time PCR quantification of ChIP DNA, reported as percentages of chromatin input, from assays using the indicated antibodies. The data are average values from two independent ChIP assays and from three PCR repetitions for each ChIP assay and are reported by subtracting the background signal, measured by omitting antibody during the ChIP procedure. Asterisks indicate statistically significant changes (P ≤ 0.01) in SD versus NB plants. The grouping of histone marks on the basis of how its variation associates with SD floral induction is indicated in parentheses. Similar results were obtained after correction for nucleosome occupancy measured as reported by Rossi et al. (2007). H3Cter, Histone H3 C-terminal region.
DNA Methylation at ZCN7 and ZCN8 in Line B73 and Teosinte Plants
DNA methylation is another epigenetic mark that can affect gene transcription (Lauria and Rossi, 2011). Therefore, we analyzed whether DNA methylation variation at the ZCN7 and ZCN8 loci is associated with id1 gene activity in temperate maize or SD induction in tropical teosinte. First, we estimated the global mC level in the same genomic regions previously analyzed by ChIP assays. This was done with the methylation-dependent restriction enzyme MspJI, which cleaves methylated DNA and can detect mC in all sequence contexts (CG, CHG, and CHH, where H = A, C, or T; Cohen-Karni et al., 2011). The results indicate that all regions analyzed are methylated, but mC levels in both immature and mature leaf were not different in id1 mutant tissue compared with wild-type line B73, nor did it vary in teosinte grown under SD and NB conditions (Supplemental Fig. S8). To further validate these results and to examine possible mC variation in a specific sequence context, three ZCN7 and ZCN8 genomic regions, representing the 5′ end, gene body, and 3′ end of the genes, were selected for bisulfite sequencing analysis with genomic DNA extracted from line B73 and teosinte immature leaves. We focused on this tissue to assess whether mC variation is already established at ZCN7 and ZCN8 loci in immature leaf and in response to floral inductive conditions. We also carried out bisulfite sequencing analysis of the 5′ end putative cis-regulatory region of ZCN8 in mature leaf. We detected no statistically significant variation of mC in any of the sequence contexts in the comparison of wild-type line B73 with the id1 mutant or the comparison of teosinte SD with NB plants (Supplemental Figs. S9–S12; Supplemental Table S3). These results suggest that mC is not a factor in ZCN7 and ZCN8 regulation with respect to floral induction as mediated by id1 function in temperate maize or by SD photoperiod in tropical teosinte. Nonetheless, our analysis indicates that ZCN7 and ZCN8 genes are methylated and that methylation occurs almost exclusively in the CG sequence context.
DISCUSSION
Specific Histone Modification Patterns in ZCN7 and ZCN8 Chromatin Are Associated with Competence to Flower in Response to id1 Gene Activity in Temperate Maize
In the development of modern temperate maize from its tropical progenitor, teosinte, ancient farmers selected variants with a flowering habit that is less influenced by photoperiod. The id1 gene is a key regulator of flowering in temperate maize, with loss of id1 function causing a severe delay in the floral transition (Colasanti and Muszynski, 2009). Thus, comparison of id1 mutants with normal-flowering plants provides an opportunity to study elements of the autonomous flowering pathway in maize. This study finds that id1-mediated regulation of ZCN7 and ZCN8 expression in temperate maize is associated, at least in part, with chromatin-related mechanisms (Fig. 7). An interesting observation from our study is that, in immature leaves of wild-type maize plants at the floral transition, despite not producing florigen, the ZCN7/ZCN8 loci are characterized by a histone modification pattern (i.e. the presence of H3ac, H3K4me2, H3K4me3, and H3K36me2) that is usually linked to active chromatin. In addition, the high CG methylation level that we detect at ZCN7 and ZCN8 is compatible with a transcriptionally competent or active status, as supported by an mC genome-wide distribution study in maize (Regulski et al., 2013). In agreement with these observations, low-level accumulation of ZCN7 and ZCN8 sense pre-mRNA and a higher level of the antisense RNA are detected in immature leaf. ChIP assays of id1 mutant immature leaves indicate that the majority of histone marks analyzed are not related to id1 function, suggesting that additional factors must be involved. For example, some of these factors could be chromatin-remodeling complexes containing the WD-repeat proteins NFC101/NFC102, which directly bind ZCN8 to negatively regulate its expression and H3K4me2 levels in meristem-enriched tissues (Mascheretti et al., 2013).
Figure 7.
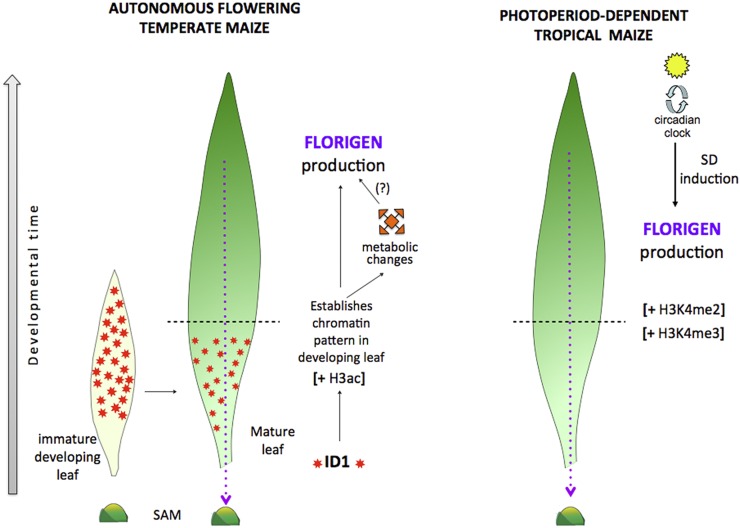
Model of florigen regulation in autonomous maize and photoperiod-induced teosinte. Autonomous maize (left) requires ID1 regulatory protein activity (orange stars) in developing leaves to establish chromatin modifications that allow the expression of florigen genes (ZCN7 and ZCN8). Thus, the id1 gene acts in immature leaves to establish a chromatin signature and prime the leaf for florigen synthesis as the leaf develops. Active chromatin is specified by H3ac. Once the distal portion of the leaf develops, another signal (unknown) activates florigen production in leaf vasculature, which then migrates to the shoot apical meristem (SAM) to activate flowering genes (purple dotted line). The autonomous signal may consist, partly, of changes in metabolic activity. Metabolic changes could also indirectly activate florigen production (?). In teosinte (right), floral induction is dependent on SD photoperiods and the circadian clock to activate florigen production. Similar to id1, the photoperiod pathway also establishes chromatin modifications in immature leaves, which enable florigen synthesis in mature leaves, but the pattern of histone modifications related to its activity is different from the one created by id1 in the autonomous pathway (i.e. open chromatin is specified by H3K4me2/H3K4me3). The horizontal dashed lines across the mature maize and teosinte leaves delineate the regions of the immature, developing leaf zone (bottom) from the mature leaf blade (top).
Nevertheless, we report that id1 function is required to specify high H3ac levels and to prevent H3K27me3 accumulation at ZCN7/ZCN8 chromatin in immature leaf. More importantly, id1 activity is essential to maintain histone H3 hyperacetylation in mature leaf, where the id1 gene is not expressed. This supports the involvement of epigenetic mechanisms, mediated by id1 activity, which facilitate the production of ZCN7/ZCN8 processed mRNAs in mature leaf, even though id1 mRNA and ID1 protein accumulation do not coincide with ZCN7/ZCN8 expression. Thus, in the proposed model (Fig. 7), id1 induces histone H3 hyperacetylation at florigen genes to establish a transcriptionally competent chromatin environment in early stages of leaf development. A previous study showed that chromatin modifications that are established at the Arabidopsis FLOWERING LOCUS C in actively dividing cells of developing tissues are maintained in later stages of development (Finnegan and Dennis, 2007). Accordingly, our findings also suggest that the chromatin status established at ZCN7/ZCN8 in developing maize leaves by the id1-mediated autonomous pathway is maintained throughout leaf development, until the formation of mature leaves, where it may facilitate the synthesis of ZCN7/ZCN8 processed mRNAs by means of still unknown factors. In this scenario, id1 acts as a gatekeeper that primes floral induction in day-neutral temperate maize. Previous transcriptome comparisons of flowering and nonflowering maize suggest that id1 also regulates genes involved in primary metabolism to establish a physiological state associated with readiness for flowering (Coneva et al., 2007, 2012), thus connecting florigen production with metabolic rate to facilitate the transition to reproductive growth.
Our study shows that id1 function is correlated with increased H3K4me3 and H3K36me2 levels at ZCN7/ZCN8 chromatin only in mature leaf, although the id1 gene is not expressed in these tissues. This could be explained by an indirect effect related to id1-dependent activation of ZCN7/ZCN8 sense mRNA levels in mature leaf. Indeed, it is widely documented that, in many cases, transcription drives the accumulation of histone marks associated with active chromatin and not vice versa (Henikoff and Shilatifard, 2011). In addition, our analysis provides no evidence for whether ID1 protein directly or indirectly regulates histone modifications at ZCN7 and ZCN8. However, ZCN7 and ZCN8 sequences have no apparent ID1 protein-binding sites (Kozaki et al., 2004). Moreover, the putative rice (Oryza sativa) id1 ortholog (Rice Indeterminate1 [RID1], OsId1, or Early heading date2 [Ehd2]) activates Hd3a florigen by promoting the expression of the transcription factor Ehd1 (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008). Therefore, it is possible that the ID1 protein does not interact directly with florigen gene regulatory elements.
The Photoperiod Floral Inductive Pathway Alters ZCN7 and ZCN8 Chromatin Modifications in Tropical Teosinte Differently Than the Autonomous Pathway in Line B73 Temperate Maize
Teosinte was selected for this analysis because it represents maize with an obligate requirement for SD photoperiods to induce flowering. Although it is difficult to make direct comparisons of photoperiod-induced versus autonomously regulated flowering because the plants were exposed to different growth conditions, examination of chromatin modifications of florigen genes could reveal some commonalities and differences. Whereas autonomous flowering is controlled by endogenous signals, photoperiod-induced flowering represents an immediate response to inductive signals. Therefore, comparison of the chromatin profiles associated with these different pathways could reveal underlying mechanisms implicit to these different floral induction strategies. Similar to id1-controlled flowering via the autonomous pathway in temperate maize, we found that SD-induced florigen activation in teosinte promotes the formation of a particular histone modification pattern at ZCN7/ZCN8 loci during leaf development (Fig. 7). However, unlike id1 regulation in line B73, SD floral induction in teosinte promotes increased H3K4me2 and H3K4me3 levels in both immature and mature leaves but does not affect histone H3 acetylation. This suggests that distinct mechanisms, perhaps reflecting the different ways that various histone modifications influence transcription (Bannister and Kouzarides, 2011), distinguish the id1- and SD-related regulation of ZCN7/ZCN8 expression. Another difference between the two florigen regulatory pathways is that the H3K27me3 repressive mark is present at the 5′ end of ZCN7/ZCN8 chromatin only in mature leaf of wild-type maize but is not detected in teosinte. This difference could have functional implications, given that a genome-wide analysis found limited H3K27me3 variation in the same tissues of different maize inbred lines (Makarevitch et al., 2013). In particular, the presence of H3K27me3 may be associated with overall lower levels of the ZCN7 and ZCN8 transcript isoforms detected in mature maize leaf compared with mature leaf of teosinte. Indeed, H3K27me3 in the 5′ regions attenuates the transcription of expressed mammalian genes (Young et al., 2011). This could also explain the unusual localization of H3K27me3 in the 5′ end region of florigen chromatin only (Lauria and Rossi, 2011). Alternatively, the unexpected localizations of H3K27me3 and H3K4me3 in the 5′ and 3′ end regions, respectively, could be associated with the transcription of ZCN7/ZCN8 antisense RNA. Indications from extensively characterized eukaryotic transcriptional regulatory models suggest that ZCN7/ZCN8 antisense lncRNAs may be involved in florigen regulation through various mechanisms, including chromatin modification (De Lucia and Dean, 2011). Although precise characterization of ZCN7 and ZCN8 antisense transcripts is beyond the scope of this study, we provide evidence that their regulation is different with respect to autonomous and photoperiod flowering pathways. Specifically, the antisense RNA level is inversely correlated with id1 gene activity in line B73 maize yet unaffected by photoperiod in teosinte. The significance of this difference is an intriguing feature in future studies to investigate the function of ZCN7/ZCN8 antisense RNAs.
ZCN7 May Be a Second Maize Florigen Gene
We found that the specific accumulation pattern of a spliced sense mRNA in mature leaf previously detected for ZCN8 (Danilevskaya et al., 2008) occurs also for its paralog ZCN7. The discrepancy between our findings and those reported by Danilevskaya et al. (2008) may be due to genotype-related differences of ZCN7 in producing transcript-splicing variants, since that early study analyzed ZCN7 expression in the Gaspé Flint line, while our study analyzed inbred line B73 and teosinte. Alternatively, the forward primer used to amplify ZCN7 cDNA in the previous study may not be within the ZCN7 processed sense mRNA, because using this primer we were also unable to detect the spliced variant in line B73 and teosinte. In addition to the ability of both paralogs to produce the processed mRNA, our results provide evidence that ZCN7 and ZCN8 exhibit a pattern of transcript isoform accumulation and histone modifications that are affected identically by the id1 gene in wild-type maize and SD photoperiod in teosinte. Collectively, these findings support a functional analogy between ZCN7 and ZCN8, implying that ZCN7 may encode a second maize florigen. Meng et al. (2011) reported that the ZCN7 sequence identified in the Gaspé Flint line does not exhibit some of the properties of a florigen. For example, they found that, unlike ZCN8, ZCN7 interacted only weakly with the maize FD ortholog DLF1. Furthermore, ectopic expression of ZCN7 cDNA in transgenic maize did not cause the minor yet significant acceleration of flowering displayed by ZCN8-expressing lines. Since the genomic ZCN7 sequence was used for transgenic plant production, it is possible that the absence of the early-flowering phenotype observed by Meng et al. (2011) could be due to the above-mentioned inability of the Gaspé Flint line ZCN7 sequence to produce processed mRNA. Alternatively, the function of ZCN7 and ZCN8 paralogs may not be fully redundant, with ZCN8 having higher florigenic activity. Nevertheless, here we show that, similar to ZCN8, ZCN7 is responsive to both id1-regulated and SD-mediated floral induction, an important florigen feature that was not tested for the ZCN7 gene (Meng et al., 2011). In addition, ZCN7 lies within a flowering time QTL, as does ZCN8 (Bouchet et al., 2013). Further extensive studies would be required to definitively demonstrate whether ZCN7 possesses florigenic activity similar to ZCN8.
Unique Features of Maize Florigen Regulation Compared with Other Plants
In this study, we describe features of florigen regulation that appear to be unique to maize. First, the production of florigen antisense transcripts in both temperate maize and teosinte has not been reported for other species. Second, we identified ZCN7 and ZCN8 unspliced sense pre-mRNAs that have not been detected for florigen genes in other species. These pre-mRNAs are particularly abundant in teosinte mature leaf tissue, and their presence suggests that posttranscriptional RNA-processing mechanisms may play a role in florigen regulation (Mascheretti et al., 2013). Third, the patterns of histone modifications at ZCN7/ZCN8 loci in maize and teosinte are somewhat different from those described for florigen genes from other species. For example, the presence of H3K27me3 in Arabidopsis FT chromatin is correlated with repression of this gene (Turck et al., 2007; Adrian et al., 2010; Wang et al., 2014), whereas in line B73 maize, H3K27me3 at ZCN7/ZCN8 is detected only in mature leaf, which is the tissue with maximum florigen production. This suggests that, unlike Arabidopsis, H3K27me3 deposition in maize mature leaf is not associated with the formation of fully repressive chromatin. Nevertheless, it is worth noting that, in immature leaves of the id1 mutant, H3K27me3 accumulates in all ZCN7/ZCN8 genomic regions. Since id1 is also involved in the control of the ZCN7/ZCN8 sense and antisense unspliced RNA isoform production at this developmental stage (i.e. id1 activity is correlated with the increase of sense and decrease of antisense RNA strands), these findings indicate a possible role for id1 in modulating the rate of sense/antisense production through chromatin modifications. Histone acetylation represents another difference between maize and Arabidopsis florigen genes. Indeed, in Arabidopsis, H3ac is correlated with the photoperiod regulation of FT (Adrian et al., 2010; Gu et al., 2013), but it is unaffected by photoperiod in teosinte and altered in the id1-controlled autonomous pathway in maize.
Overall, we find that maize employs unique mechanisms to regulate florigen production. We also show that distinct chromatin modification patterns characterize florigen genes in leaves of autonomously regulated maize plants compared with teosinte plants that rely on photoperiod induction. These findings reveal a key role for chromatin-related epigenetic mechanisms in controlling environmental adaptations to maize flowering and are thus important for elucidating the regulatory underpinnings of flowering.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Maize (Zea mays ssp. mays) seeds segregating the id1-m1 mutant allele backcrossed 10 times into the B73 inbred line background were planted in soil (50% Sunshine Mix and 50% Turface clay) in Conviron growth chambers under conditions of broad-spectrum light at 1,000 μmol m−2 s−1. The id1-m1 mutant allele and wild-type segregating plants (Colasanti et al., 1998) were identified by PCR genotyping as described previously (Wong and Colasanti, 2007). All maize plants were grown under long-day conditions (14 h of light/10 h of dark) with day temperatures of 25°C and night temperatures of 21°C. Conversely, teosinte (Zea mays ssp. parviglumis) plants were grown initially in a growth chamber under noninductive NB conditions of 9 h of light/7 h of dark/1 h of light/7 h of dark for 27 d (approximately eight visible leaves), after which one-half of the plants were transferred to an SD growth chamber at 10 h of light/14 h of dark. Prior to growing plants for this experiment, we determined that plants exposed to 1 h of daylight conditions in the middle of the long dark period (NB) were effectively inhibited in flowering in 100% of teosinte plants, as determined by examining shoot apex structure at various time points (Fig. 1; K. Turner and J. Colasanti, unpublished data).
Plants at the V6/V7 stage corresponding to the floral transition in the maize B73 inbred line or 10 d after growth of teosinte plants under NB conditions (Fig. 1) were used for isolation of the tissues employed in the analysis of ZCN7/ZCN8 transcripts and in ChIP assays. Mature and immature leaf tissues were sampled. Specifically, for maize B73 inbred line plants, tissues of mature leaves were obtained by isolating the distal 30 cm of blade tissue from leaf 8, while the immature leaf tissue from the same plant consisted of the central cylinder of nonphotosynthetic developing leaves from 2 to 12 cm above the shoot apex, sampled as reported by Colasanti et al. (1998). Details of the sampling strategy are illustrated in Supplemental Figure S3. For induced and uninduced teosinte plants, the mature leaf samples were obtained by harvesting the distal 30 cm of the youngest mature leaf with an exposed collar, usually leaf 8 or 9. Immature leaves, 2 to 12 cm above the shoot apex, were harvested as described for maize. For each biological replicate, a minimum of five plants were harvested. For ZCN7/ZCN8 mRNA expression analysis during the course of maize development (Supplemental Fig. S2), leaves from the B73 inbred line were harvested from V4, V6, and V8 plants at the midpoint of the daylight cycle. Whole leaf blade was used for small leaves (less than 10 cm), or leaves were dissected into halves, thirds, or quarters for larger leaves, as shown in Supplemental Figure S2.
cDNA Synthesis and RT-PCR
Total RNA extraction and oligo(dT)-primed and strand-specific cDNA synthesis were performed as described previously (Mascheretti et al., 2013). Sequences of locus-specific primers used for strand-specific RT are reported in Supplemental Table S4. Conditions for PCR amplification of maize line B73 and teosinte full-length ZCN7 and ZCN8 cDNAs were as described by Danilevskaya et al. (2008), with primers located close to the start and stop codons (Supplemental Table S5). Conditions for strand-specific RT-PCR amplification and subsequent cloning of line B73 and teosinte ZCN7/ZCN8 cDNAs produced by unspliced sense and antisense RNAs were as reported previously (Mascheretti et al., 2013; for the list of PCR primers, see Supplemental Table S5). Real-time qRT-PCR was performed and the amount of changes between samples was calculated using the 2−ΔΔCt method as described previously by Rossi et al. (2007). cDNA preparations from two biological replicates were made, and three replicates of qRT-PCR were performed for each cDNA preparation. To account for possible differences in cDNA synthesis and amplification efficiency, the data were normalized to the transcript amount of gapc2. Similar results were obtained when the data were normalized to the transcript amount of Elongation Factor-1α (EF-1α). ANOVA (P ≤ 0.01) was applied separately to each of the two biological replicates, and statistical significance was considered only when reported for both replicates.
ChIP Assay
Chromatin preparation was performed using frozen tissues according to the protocol described by Luo et al. (2013), which was adapted for maize and teosinte plants. ChIP assays were carried out as described by Locatelli et al. (2009) with minor modifications (for details regarding the protocol, see Supplemental Text S1). The following antibodies were used: 6 μg of α-H3ac (Millipore; 07-352), 10 μg of α-H3K4me2 (Millipore; 07-030), 4 μg of α-H3K4me3 (Active Motif; 39159), 10 μg of α-H3K36me2 (Millipore; 07-369), 12 μg of α-H3K27me3 (Millipore; 07-449), and 4 μg of α-H3Cter (for histone H3 C-terminal region; Abcam; ab1791). A no-antibody negative control was performed by adding no antibody during incubation. Two independent ChIP experiments were performed, and quantitative PCR and statistical analysis (ANOVA; P ≤ 0.01) were carried out as described by Rossi et al. (2007). The sequences of primers used for ChIP assays are reported in Supplemental Table S5.
DNA Methylation Analysis
Digestion with MspJI (New England Biolabs) was performed following the manufacturer’s instructions. Two independent MspJI treatments and three repetitions of quantitative PCR analysis were carried out, and statistical analysis was performed as described by Locatelli et al. (2009). Genomic DNA bisulfite treatment was performed with the EZ DNA Methylation kit (Zymo Research) following the manufacturer’s instructions. Primers for the mC analysis of the upper strand were designed using the Kismeth Web-based tool (http://katahdin.mssm.edu/kismeth/revpage.pl; Gruntman et al., 2008; Supplemental Table S5). One microliter of bisulfite-treated DNA was used for PCR under the following conditions: 1.5 mm MgCl2, 0.2 mm of each deoxyribonucleoside triphosphate, 0.4 μm of each primer, and 1.25 units of Taq Platinum (Life Technologies). The PCR program was as follows: 5 min at 95°C; 40 cycles of 30 s at 95°C, 1 min at 45°C, and 1 min at 72°C; 10 min at 72°C; and held at 4°C. Amplified fragments were cloned, and 10 independent clones were sequenced for each fragment.
The GenBank accession numbers of the genes cited in this study are as follows, with the gene model number according the line B73 maize sequencing project (http://www.maizegdb.org/) in parentheses: line B73 ZCN7, KP202720 (GRMZM2G141756); line B73 ZCN8, EU241899 (GRMZM2G179264); teosinte ZCN7, KP172200; teosinte ZCN8, KP172201; line B73 id1, (GRMZM22G011357); line B73 gapc2, U45855; line B73 EF-1a, U76259.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic depiction of ZCN7 and ZCN8 genes and transcripts of the maize B73 inbred line.
Supplemental Figure S2. Examination of ZCN7/ZCN8 in plants at pretransition (V4), transition (V6), and posttransition reproductive (V8) stages.
Supplemental Figure S3. Sampling strategy.
Supplemental Figure S4. Nucleotide sequence of the ZCN7 and ZCN8 antisense RNA strand.
Supplemental Figure S5. Comparison of ZCN7 and ZCN8 transcript isoforms levels in line B73 and teosinte plants under inductive floral conditions.
Supplemental Figure S6. Analysis of ZCN7 histone modifications in line B73 wild-type and id1 mutant plants.
Supplemental Figure S7. Analysis of ZCN7 histone modifications in teosinte SD and NB plants.
Supplemental Figure S8. Analysis of mC levels at ZCN7 and ZCN8 by MspJI restriction.
Supplemental Figure S9. Bisulfite sequencing analysis of mC at line B73 ZCN7.
Supplemental Figure S10. Bisulfite sequencing analysis of mC at line B73 ZCN8.
Supplemental Figure S11. Bisulfite sequencing analysis of mC at teosinte ZCN7.
Supplemental Figure S12. Bisulfite sequencing analysis of mC at teosinte ZCN8.
Supplemental Table S1. Sequence similarity of line B73 and teosinte ZCN7 and ZCN8 genes.
Supplemental Table S2. ZCN7 and ZCN8 transcript levels obtained from RNA sequencing experiments using different line B73 tissues.
Supplemental Table S3. Analysis of the mC profile of ZCN7 and ZCN8 by bisulfite sequencing.
Supplemental Table S4. List of primers used for line B73 and teosinte cDNA synthesis in strand-specific RT.
Supplemental Table S5. List of primers used in PCR.
Supplemental Text S1.Materials and Methods.
Supplementary Material
Acknowledgments
We thank Fabio Fornara, Massimiliano Lauria, Lewis Lukens, and Steven Rothstein for critical reading of the article, Raul Pirona for in silico reconstitution of the teosinte genomic sequences, and Michael Mucci and Tannis Slimmon for providing expert plant care at the Guelph Phytotron.
Glossary
- QTL
quantitative trait locus
- SD
short-day
- cDNA
complementary DNA
- RT
reverse transcription
- qRT
quantitative reverse transcription
- NB
night-break
- lncRNAs
long noncoding RNAs
- ChIP
chromatin immunoprecipitation
- H3ac
histone H3 acetylated at lysine-9 and lysine-14
- H3K4me2
histone H3 dimethylated at lysine-4
- H3K4me3
histone H3 trimethylated at lysine-4
- H3K36me2
histone H3 dimethylated at lysine-36
- H3K27me3
histone H3 trimethylation at lysine-27
- mC
cytosine methylation
Footnotes
This work was supported by the Italian Ministry of Education, University, and Research and the National Research Council of Italy for the Epigenomics Flagship Project, by the European Commission (AENEAS FP7 project no. KBBE–2009–226477), by the Research Agricultural Council in agreement with the University of Padova, Italy (Ph.D. fellowship to I.M.), by institutional grants from Italian Ministry of Agriculture, Food and Forestry Policies (to V.R.), and by the Natural Sciences and Engineering Research Council of Canada (Discovery grant to J.C.).
Articles can be viewed without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet S, Servin B, Bertin P, Madur D, Combes V, Dumas F, Brunel D, Laborde J, Charcosset A, Nicolas S (2013) Adaptation of maize to temperate climates: mid-density genome-wide association genetics and diversity patterns reveal key genomic regions, with a major contribution of the Vgt2 (ZCN8) locus. PLoS ONE 8: e71377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Z, Yu Y, Li Z, Liu Y, Jiang W, Huang Y, Dong AW (2014) Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet 10: e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chailakhyan MK. (1937) Concerning the hormonal nature of plant development processes. Dokl Akad Nauk SSSR 16: 227–230 [Google Scholar]
- Cohen-Karni D, Xu D, Apone L, Fomenkov A, Sun Z, Davis PJ, Kinney SR, Yamada-Mabuchi M, Xu SY, Davis T, et al. (2011) The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc Natl Acad Sci USA 108: 11040–11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Coneva V (2009) Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiol 149: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Muszynski MG (2009) The maize floral transition. In Hake SC, Bennetzen JL, eds, Handbook of Maize: Its Biology, Vol 1 Springer Science, New York, pp 41–55 [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Coneva V, Guevara D, Rothstein SJ, Colasanti J (2012) Transcript and metabolite signature of maize source leaves suggests a link between transitory starch to sucrose balance and the autonomous floral transition. J Exp Bot 63: 5079–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V, Zhu T, Colasanti J (2007) Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J Exp Bot 58: 3679–3693 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR (2008) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Dean C (2011) Long non-coding RNAs and chromatin regulation. Curr Opin Plant Biol 14: 168–173 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES (2007) Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol 17: 1978–1983 [DOI] [PubMed] [Google Scholar]
- Giakountis A, Coupland G (2008) Phloem transport of flowering signals. Curr Opin Plant Biol 11: 687–694 [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Wang Y, He Y (2013) Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol 11: e1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. (2012) Chromatin regulation of flowering. Trends Plant Sci 17: 556–562 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A (2011) Histone modification: cause or cog? Trends Genet 27: 389–396 [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria M, Rossi V (2011) Epigenetic control of gene regulation in plants. Biochim Biophys Acta 1809: 369–378 [DOI] [PubMed] [Google Scholar]
- Lazakis CM, Coneva V, Colasanti J (2011) ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J Exp Bot 62: 4833–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli S, Piatti P, Motto M, Rossi V (2009) Chromatin and DNA modifications in the Opaque2-mediated regulation of gene transcription during maize endosperm development. Plant Cell 21: 1410–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-González L, Mouriz A, Narro-Diego L, Bustos R, Martínez-Zapater JM, Jarillo JA, Piñeiro M (2014) Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell 26: 3922–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Sidote DJ, Zhang Y, Kerstetter RA, Michael TP, Lam E (2013) Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J 73: 77–90 [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Eichten SR, Briskine R, Waters AJ, Danilevskaya ON, Meeley RB, Myers CL, Vaughn MW, Springer NM (2013) Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascheretti I, Battaglia R, Mainieri D, Altana A, Lauria M, Rossi V (2013) The WD40-repeat proteins NFC101 and NFC102 regulate different aspects of maize development through chromatin modification. Plant Cell 25: 404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON (2011) The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23: 942–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TA, Muslin EH, Dorweiler JE (2008) A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227: 1377–1388 [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON (2006) delayed flowering1 Encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol 142: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Park SH, Xuan YH, et al. (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J 56: 1018–1029 [DOI] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MT, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V, Locatelli S, Varotto S, Donn G, Pirona R, Henderson DA, Hartings H, Motto M (2007) Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell 19: 1145–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM (2013) Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Fang J, Zhao T, Xu B, Zhang F, Liu L, Tang J, Zhang G, Deng X, Chen F, et al. (2012) The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24: 3235–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gu X, Yuan W, Schmitz RJ, He Y (2014) Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell 28: 727–736 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wong AY, Colasanti J (2007) Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J Exp Bot 58: 403–414 [DOI] [PubMed] [Google Scholar]
- Wu C, You C, Li C, Long T, Chen G, Byrne ME, Zhang Q (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA 105: 12915–12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME, Oshlack A, Majewski IJ (2011) ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res 39: 7415–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.