Efficient repair of DNA damage in plants depends on chromatin remodeling factors, histone chaperones, and histone modifiers.
Abstract
The integrity of DNA molecules is constantly challenged. All organisms have developed mechanisms to detect and repair multiple types of DNA lesions. The basic principles of DNA damage repair (DDR) in prokaryotes and unicellular and multicellular eukaryotes are similar, but the association of DNA with nucleosomes in eukaryotic chromatin requires mechanisms that allow access of repair enzymes to the lesions. This is achieved by chromatin-remodeling factors, and their necessity for efficient DDR has recently been demonstrated for several organisms and repair pathways. Plants share many features of chromatin organization and DNA repair with fungi and animals, but they differ in other, important details, which are both interesting and relevant for our understanding of genome stability and genetic diversity. In this Update, we compare the knowledge of the role of chromatin and chromatin-modifying factors during DDR in plants with equivalent systems in yeast and humans. We emphasize plant-specific elements and discuss possible implications.
A DNA molecule in a eukaryotic chromosome has a diameter of 2 nm but a length in the range of centimeters. Being 107 times longer than it is wide would make it highly sensitive to breakage if it were not organized in compact and dynamic chromatin, with nucleosomes as basic units followed by multiple higher order levels of organization. Within this packaging, replication and transcription constitute an endogenous mechanical strain, while exposure of cells to physical or chemical hazard creates a wide range of DNA damage induced by external factors, including photoproducts, pyrimidine dimers, interstrand cross-linking, and single and double-strand breaks (DSBs). All organisms possess mechanisms that repair DNA molecules. DNA damage repair (DDR) systems for the various types of damage are overlapping as well as complementary, and their prevalence depends on the organism, the stage of the cell cycle, the site and the amount of damage, and the availability of intact templates. A coarse categorization distinguishes nucleotide excision repair, base excision repair, nonhomologous end joining (NHEJ), and homologous recombination (HR). Many schematic models for these DDR pathways describe the action and interaction of several repair components, but they are usually misleading in one aspect: DNA strands are illustrated as straight lines, neglecting the emerging role of chromatin for the location and the fate of DNA lesions. More realistically, we should envisage DNA lesions in the chromatin context like the leakage or burst of an in-ground pipe. Recognizing the defect, localizing it, excavating the broken parts, cleaning the ends, reconnecting them, and restoring the original state are mirrored in the subsequent steps of DDR: signaling, labeling, accessing, resecting, religating, and reassembling. Chromatin is expected to especially affect the access, resection, and restoration of the original arrangement, and several factors that can dissolve higher order structures, slide, evict, or exchange nucleosomes, or restore chromatin have been implicated in DDR (for review, see Lukas et al., 2011; Czaja et al., 2012; Dinant et al., 2012; Euskirchen et al., 2012; Lans et al., 2012; Soria et al., 2012; Altmeyer and Lukas, 2013; Dion and Gasser, 2013; Gospodinov and Herceg, 2013a, 2013b; Ohsawa et al., 2013; Papamichos-Chronakis and Peterson, 2013; Peterson and Almouzni, 2013; Price and D’Andrea, 2013; Stanley et al., 2013; House et al., 2014; Jeggo and Downs, 2014; Swygert and Peterson, 2014; Polo, 2015). The repair process in plants has been the focus of several reviews (Roth et al., 2012; Donà et al., 2013; Knoll et al., 2014a; Missirian et al., 2014). In this Update, we summarize recent literature describing the connection of repair and chromatin in plants, adding to earlier reviews (Balestrazzi et al., 2011; Zhu et al., 2011; Roy, 2014). Although plants are surprisingly much less investigated in this context when compared with other multicellular organisms, they are potentially rewarding for further research, as some repair- and chromatin-related genes have extended gene families, and several complete loss-of-function mutations that are lethal in animals are viable in plants. Due to length restriction, we will not cover replication-associated DNA repair, the role of chromatin for long-range interactions, or the recently emerging role of small RNA in DDR; instead, we focus on defects during interphase, local chromatin configuration, and DNA-associated proteins.
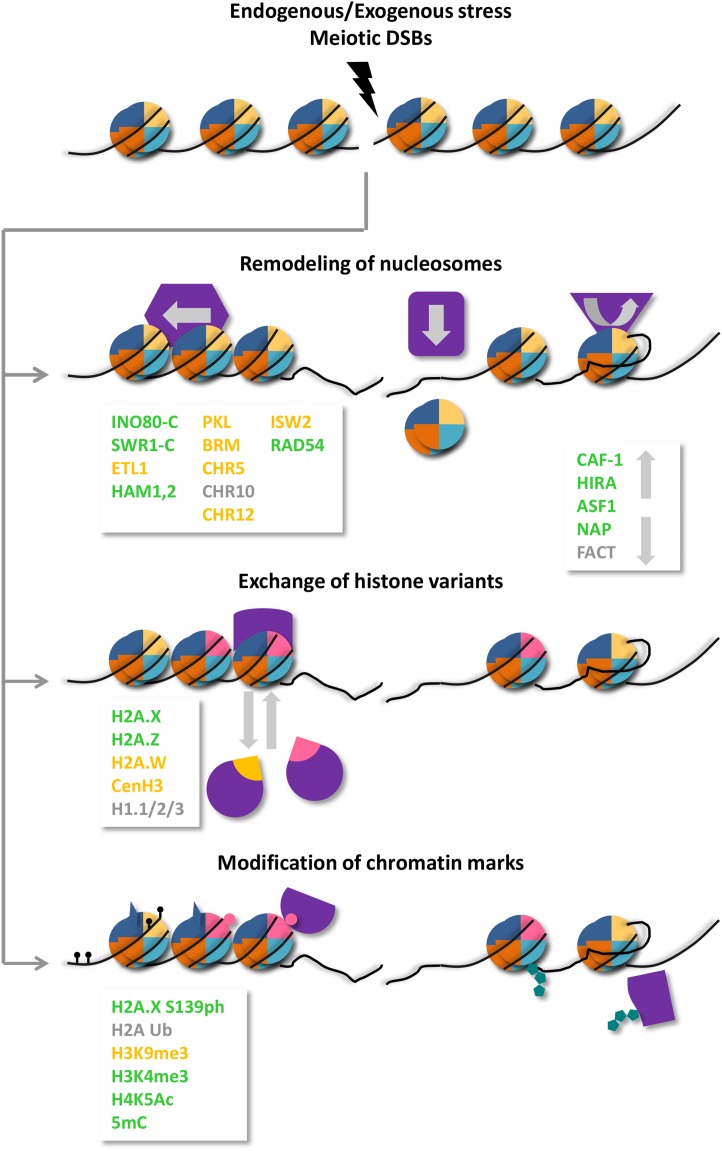
Chromatin consists mainly of nucleosomes, spherical octamers formed by two molecules of each of the four histone types H2A, H2B, H3, and H4 with 1.7 turns of DNA wrapped around their surface and sealed by H1 linker histones. Firm interaction between histones and DNA together with regular nucleosome spacing results in stable and periodic structures, but histone octamers can also become less tightly bound, change their position along the DNA, or be completely dismantled. The strength of the association can be modulated in three ways: (1) by energy-consuming remodeling processes that shift or remove nucleosomes; (2) by exchanging core histones with histone variants; and (3) by introducing posttranslational modifications (PTMs) in the histone subunits (Fig. 1). In the following three paragraphs, we review the role of these chromatin changes during DNA repair in plants.
Figure 1.
Schematic representation of chromatin changes connected with DNA damage. DDR is associated with chromatin at three different levels: nucleosomes can be shifted, evicted, or exchanged; their composition can be varied; and their subunits can be covalently modified. Arabidopsis proteins or protein complexes participating in these processes are listed, those with a known repair-related role in green, those with a reported chromatin connection in yellow; factors in gray have only sequence similarity to yeast or human genes necessary for DDR.
CHROMATIN-REMODELING COMPLEXES CONNECTED WITH DDR
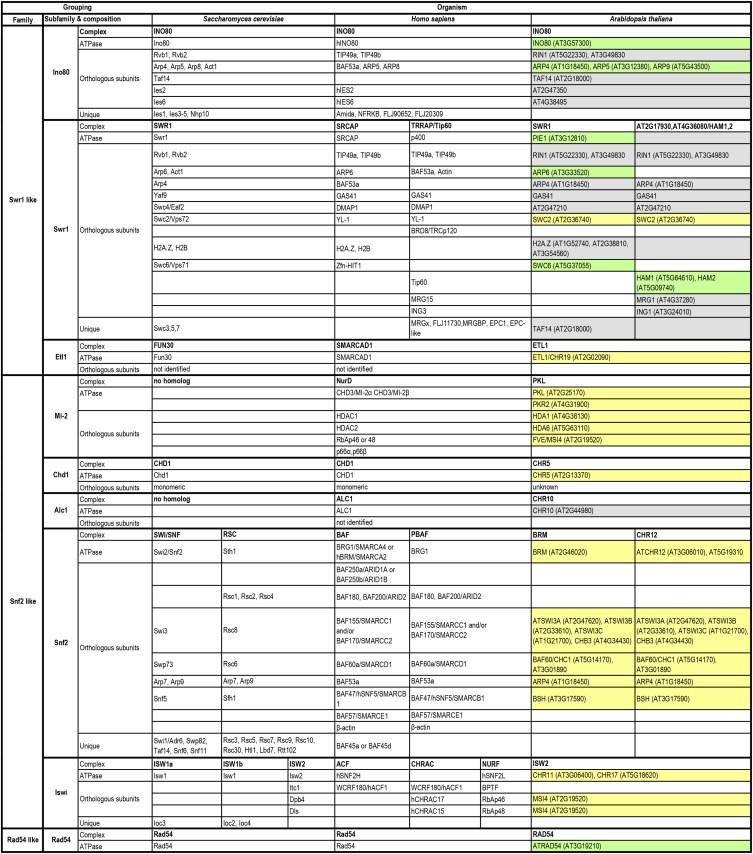
Chromatin remodeling complexes (remodelers) use ATP hydrolysis to slide, evict, or unwrap nucleosomes and can alter histone composition (for review, see Clapier and Cairns, 2009). They are grouped into different classes with unique features and specificity (Fig. 2) and are able to recognize a plethora of chromatin marks, either through diverse protein domains and/or through interaction with a large set of different subunits (Hopfner et al., 2012). This complexity reflects the multiple functions of remodelers in DNA-related processes, and it is not surprising that mutants lacking functional chromatin remodelers often exhibit increased sensitivity to DNA damage. However, only a few complexes have been linked unambiguously and directly to DDR. DNA repair requires nucleosome remodeling from the early steps through the final repair (Seeber et al., 2013; Tsabar and Haber, 2013). Recruitment of remodelers to the damage site changes the nucleosome occupancy of the surrounding area, ensuring accessibility for the repair factors. Moreover, as shown in the yeast Saccharomyces cerevisiae, chromatin remodeling is essential for modulating DNA strand resection, resolving intermediate structures during HR, and orchestrating the mobility of chromatin to enhance HR efficiency (for review, see Gospodinov and Herceg, 2013a; Seeber et al., 2013). Although unique features distinguish yeast and mammalian chromatin-remodeling complexes, the mechanisms underlying nucleosome alteration during DNA repair are highly conserved throughout evolution (Jasin and Rothstein, 2013; Seeber et al., 2013; Tsabar and Haber, 2013). Most of the repair-related chromatin-remodeling complexes described in yeast and mammals have orthologs in plants, and early characterization of several mutants indicated similar roles in repair (Shaked et al., 2006). However, the individual components were most often identified through their role in transcriptional regulation rather than in repair, as their loss of function caused striking deviations from normal development. Compared with the knowledge of yeast and mammalian complexes, there is relatively little known about the role of chromatin remodeling in DNA repair in plants. Unique structural and functional features of plant chromatin-remodeling complexes justify and highlight the need for more detailed studies (Knizewski et al., 2008). Therefore, we extend a recent comparison of the repair functions of chromatin remodelers in yeast and mammals (Seeber et al., 2013) to include current insights into their role in plant DDR (Fig. 2). We list the full gene names only for the most important components in our context; for the expansion of all abbreviated gene and protein complex names, see Supplemental Text S1.
Figure 2.
Overview of DNA damage-related chromatin-remodeling complexes. The data for the budding yeast and human complexes from Seeber et al. (2013) were complemented by orthologous complexes, subunits, or genes in Arabidopsis. Acronyms are followed by Arabidopsis Genome Initiative gene identity codes in parentheses; genes without acronyms have only their Arabidopsis Genome Initiative code. Green background, experimental evidence for a role in plant DDR; yellow background, connection with chromatin; gray background, only sequence similarity to yeast or human genes necessary for DDR.
The SWR1 (Swi2/Snf2-RELATED1) and INO80 (INOSITOL-REQUIRING MUTANT80) complexes are members of the family of SWR1-like chromatin remodelers. They share a conserved role in genome-wide regulation of transcription via installation and removal of the H2A.Z variant at transcriptional start sites. H2A.Z deposition affects genome stability, as SWR-C activity is coordinated with replicative polymerases to reduce mutation rates (Van et al., 2015). Both complexes also impact DDR by remodeling the nucleosomal neighborhood surrounding DSBs in yeast and mammals (for review, see Gerhold and Gasser, 2014). SWR1-C is also required during the early steps of mammalian HR to prevent the propagation of γH2A.X, the phosphorylated histone variant that labels the site of damage, and at later steps for loading RAD51 onto single-stranded DNA to promote successful repair (Courilleau et al., 2012). Conversely, loss of SWR1-C in yeast does not affect resection, which suggests a role in NHEJ rather than HR. Both complexes are also involved in larger scale mobility: the yeast SWR1-C and INO80-C affect the relocation of unrepaired DSBs to anchor sites at the nuclear envelope in a cell cycle-dependent manner (Horigome et al., 2014). Unrepaired DSBs in mammalian cells are also relocated to the nuclear envelope to prevent erroneous recombination and to activate alternative repair pathways that can overcome persistent damage (Lemaître and Soutoglou, 2015). The specificity of INO80-C and SWR1-C function is provided by different subunits that mediate the complexes’ interaction with DNA and proteins (Yen et al., 2013).
Also in plants, SWR1-C mediates the incorporation of the H2A.Z histone variant, besides being the most well-studied remodeling complex in the repair context. In Arabidopsis (Arabidopsis thaliana), the complex has a major role in transcriptional regulation, as SWR1-C mutants exhibit a characteristic early-flowering phenotype due to the dysregulation of several developmental genes (Noh and Amasino, 2003; Choi et al., 2005; Deal et al., 2005; Lázaro et al., 2008; March-Díaz and Reyes, 2009). A role of Arabidopsis SWR1-C in DNA repair has only recently been documented. Loss of one of several SWR1-C subunits, namely ACTIN-RELATED PROTEIN6 (ARP6), PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1, and SWR1 COMPONENT6, results in increased DNA damage sensitivity. This phenotype is aggravated by the simultaneous impairment of NHEJ, while SWR1-C and HR mutants are epistatic, suggesting a role of SWR1-C in HR specifically. The involvement in somatic HR is supported by reduced recombination frequencies between incomplete but overlapping parts of a marker gene in the AtSWR1-C mutant background. Furthermore, reduced fertility of the mutants also indicates defects in meiosis (Rosa et al., 2013). The proximate cause, however, is difficult to separate from the role of ARP6 for temporal and spatial regulation of meiotic genes (Qin et al., 2014).
The INO80 complex in Arabidopsis is also involved in developmental regulation as well as in DNA repair (Kandasamy et al., 2009; Zhang et al., 2015). Loss of INO80, the ATP-dependent helicase of the complex, results in impairment of HR under standard growth conditions, but, in contrast to the SWR1 mutants, HR is increased after induced DNA damage (Fritsch et al., 2004; Zhang et al., 2015). Transfer DNA integration, which relies upon illegitimate repair pathways such as NHEJ, is not affected (Fritsch et al., 2004). Loss of INO80 increases sensitivity to genotoxic treatments (Zhang et al., 2015), and also Arabidopsis mutants lacking the INO80 subunit ARP5, which is necessary for H2A.Z removal in yeast (Yen et al., 2013), are strongly affected by DNA-damaging agents (Kandasamy et al., 2009). Other plant INO80 subunits have not been challenged for their role in DDR.
Yeast and mammalian components of a third SWR1-like remodeler subfamily with a role in strand resection at DSBs, FUN30 and SMARCAD1, respectively, share the ETL1 complex like the Arabidopsis equivalent. Its ATPase CHR19 was identified as an interactor of SUVR2 that is involved in transcriptional silencing (Han et al., 2014), but a link to DDR has yet to be described.
Snf2-like complexes are the second large group of remodelers; however, their connection to DNA repair is less well established. The human NurD complex is not tethered to, but rather removed from, sites of lesions (Goodarzi et al., 2011), a response that may allow access for other complexes. Several subunits have equivalents in Arabidopsis, with prominent chromatin-interacting (PKL, PKR2, and FVE/MULTICOPY SUPPRESSOR OF IRA1 4 [MSI4]) and chromatin-modifying (HDA1 and HDA6) components. While central roles in developmental regulation (determination of cell identity, flowering time regulation, and several pleiotropic effects) are well documented (Ogas et al., 1999; Tian and Chen, 2001; Ausín et al., 2004; Kim et al., 2004; Aichinger et al., 2009; Hu et al., 2014), no repair-related function has been described. Interaction of the mammalian CHD1 and ALC1 complexes is positively correlated with DNA damage (for review, see Seeber et al., 2013), and their ATPase subunits are encoded as CHR5 and CHR10 in Arabidopsis. Again, only a developmentally relevant role is known (Shen et al., 2015). The same holds true for orthologs of the Snf2 complex subunits. They form two complexes regulating plant development, differing in their ATPase subunits BRM and CHR12. While the mammalian equivalents have a clear role in genome stability, especially in the context of cancer genesis (Wilson and Roberts, 2011), their involvement in DNA repair in plants remains to be investigated; the same holds true for all other subunits of the complexes. Among these subunits, BAF60 has a potential role in repair, as it is involved in the formation of DNA loops at the FLC locus controlling flowering time (Jégu et al., 2014). Such structural arrangements could also be important during repair processes, especially during intrachromosomal recombination. The only SNF2 family member investigated outside Arabidopsis is the ATPase ALT1 in rice (Oryza sativa), which regulates alkaline tolerance through the modulation of genes involved in reactive oxygen species metabolism and DNA repair. In spite of this rather indirect involvement in DDR, it is interesting that the same nucleosome remodeler acts in both ROS and DNA damage response, strengthening the link between these two stresses (Guo et al., 2014).
The ISWI family of remodelers has a prominent role in several pathways of DNA repair in mammals (for review, see Aydin et al., 2014). The human ATPase SMARCA5/SNF2H is targeted by PARP1 upon activation of DDR. CHR11 and CHR17 are the Arabidopsis orthologs, for which a function in plant development and chromatin remodeling is documented (Li et al., 2012b, 2014), although a direct link with DNA repair needs to be investigated. MSI4 of Arabidopsis is related to subunits of the CHRAC complex that is involved in DDR in Drosophila melanogaster and mammals (Lan et al., 2010; Mathew et al., 2014) and is a potential target of the repair-related kinases ATM/ATR (see below).
RAD54, another Snf2-related chromatin remodeler with a prominent role in HR, is so well conserved that the yeast and Arabidopsis genes can reciprocally complement mutants with regard to their DNA damage sensitivity (Klutstein et al., 2008), and overexpression of the yeast gene enhances gene targeting in Arabidopsis (Shaked et al., 2005). More recently, RAD54 was specified to participate mainly in the synthesis-dependent strand-annealing mechanism of DSB repair by HR (Roth et al., 2012).
Chromatin organization also affects DNA repair beyond the nucleosome level. Genes encoding STRUCTURAL MAINTENANCE OF CHROMOSOMES (SMC) complexes in Arabidopsis, namely AtRAD18, AtRAD21.1, and AtRAD21.3, are transcriptionally activated in response to DNA damage to increase the pool of cohesins in the nucleus. When mutated, the number of DSBs increases prior to and after DNA damage, and repair is severely delayed. Additional loss of KU80, a component of NHEJ, exacerbates this repair deficiency, suggesting a role of SMCs during early phases of DSB repair, likely for initiating de novo cohesion between sister chromatids to facilitate HR (Kozak et al., 2009; da Costa-Nunes et al., 2014). AtMMS21, interacting with SMC5, participates in DNA damage reduction in the root stem cell niche to preserve genome integrity (Xu et al., 2013).
RECQ helicases take part in the resolution of Holliday junctions that arise during HR or branch migration during replication. They have multiple functions, reflected in their representation by seven genes in Arabidopsis. AtRECQ4A counteracts recombination during the DNA damage response, while AtRECQ4B is involved in HR, unique among RECQ helicases (Knoll and Puchta, 2011). The AtRTEL1 helicase disrupts the D-loop structures arising from HR recombination intermediates or stalled replication forks to prevent inappropriate recombination between chromosome ends (Recker et al., 2014).
MORC2 ATPases contribute to the DNA damage response in human cells. Their nucleosome-remodeling activity at damage sites is triggered by PAK1-dependent phosphorylation, resulting in a subsequent decondensation of chromatin that provides accessibility to the lesion (Li et al., 2012a). Although MORC orthologs are present in Arabidopsis, their only described role is to establish heterochromatin regions (Lorković, 2012; Moissiard et al., 2012).
While not necessarily a remodeler, the REPLICATION PROTEIN A (RPA) complex is associated with DDR in several groups of organisms. Arabidopsis and rice have multiple genes for the three subunit types, and mutants of the RPA1 types show different DNA sensitivity phenotypes, indicating specialization and redundancy at the same time (Chang et al., 2009; Aklilu et al., 2014). RPA2 mutants are affected in crossover formation in rice (Li et al., 2013). The fact that RPA2 can be phosphorylated by kinases involved in both cell cycle and DNA damage signaling, as well as its epigenetic control function in gene silencing (Elmayan et al., 2005; Kapoor et al., 2005), make it a very interesting central component of the regulation and integration of several cellular processes. The same could be true for two other replication-associated proteins, the catalytic subunit of DNA polymerase-α and DNA replication factor C1, which upon mutation result in DNA damage sensitivity (Liu et al., 2010a, 2010b).
HISTONES AND HISTONE CHAPERONES CONNECTED WITH DDR
The basic nucleosome composition of 4 × 2 subunits and the sequences of the most abundant canonical histones are highly conserved across all organisms, but several more diverse histone variants (and histone modifications; see next paragraph) can modulate the nucleosome interaction with DNA and other proteins. Prior to their integration into chromatin, histones associate with chaperone complexes that prevent uncontrolled assembly or association and deliver them to the DNA. The mobility of nucleosomes along DNA, and transport of their building blocks to and from DNA, are also connected with the repair of DNA lesions (for review, see Ransom et al., 2010; Biterge and Schneider, 2014).
Among all histones, the H2A family is most diverse and expanded in plants (for review, see Kawashima et al., 2015), and several variants are demonstrated or assumed to be connected to DDR. H2A.X is a variant of the H2A subunit conserved between yeast, mammals, and plants. It is discussed below due to its phosphorylation during DNA damage signaling. Another H2A variant, H2A.Z, was already mentioned in connection with the SWR1 complex, but its involvement in loading repair proteins was so far shown only for mammalian cells (Xu et al., 2012b). It is not yet clear whether the role of SWR1 in DSB repair via HR is coupled to its role in H2A.Z incorporation (Rosa et al., 2013). MacroH2A variants, associated with repressive chromatin in mammals, are incorporated at DSBs in a PARP1-dependent manner (Xu et al., 2012a). MacroH2A is not present in plants, but it shares a conserved SPKK motif with another, plant-specific, variant, H2A.W, that is associated with heterochromatin in Arabidopsis (Yelagandula et al., 2014). Whether H2A.W is connected with repair is not yet known, but modification by PARP1 is also repair relevant in Arabidopsis (Jia et al., 2013). Histone H3 has two major variants in plants, H3.1 and H3.3, which are associated with different transcriptional activities. Although H3 chaperones have been shown to have a role in regulating DDR efficiency (see below), it is difficult to determine whether this role is mediated by the chaperone or by H3 directly. The special features of the H3 variant at centromeric repeats, Cse4 in budding yeast, CENP-A in mammals, and CenH3 in plants, likely necessitate an adapted repair process for lesions in these chromosomal regions. Again, the complex interaction of diverse centromeric proteins and their role in DNA repair and recombination (for review, see Osman and Whitby, 2013) make it difficult to dissect the involvement of individual components in all systems, including plants.
Besides the histone subunits within the nucleosomes, the presence, absence, and modification of linker histones are additional factors expected to influence DNA repair processes. A specific variant of H1 was connected with DSB repair via HR in chicken cells (Hashimoto et al., 2007). Arabidopsis has three H1 genes, and the role of H1 in chromatin dynamics is well documented (for review, see Over and Michaels, 2014); however, its role during DDR remains to be specified.
Although histones are by far the most abundant chromatin proteins, a group of other mostly chromatin-associated proteins should not be neglected: the HIGH MOBILITY GROUP (HMG) proteins, which are characterized by a common motif, the HMG box. Their multiplicity and variation along with partially contradictory experimental evidence impede a precise description of their involvement in DNA repair in mammalian cells (for review, see Stros, 2010). They also represent an interesting and diverse gene family in plants and, among many other functions, participate in recombination (Grasser et al., 2007; Antosch et al., 2012). Their mechanistic contribution to DDR deserves further investigation.
Besides the proteins described above, which are more or less tightly associated with DNA at all times, there are several complexes serving as chaperones that transport these proteins to and from the DNA. CHROMATIN ASSEMBLY FACTOR1 (CAF-1) is an H3/H4 chaperone with a major role in nucleosome assembly during replication. An active role for nucleosome dissociation prior to repair has not been documented, but the expression of its components is induced by genotoxic stress. The complex might serve as an acceptor for disassembled subunits, and it is clearly necessary for reassembly after repair in a manner similar to that during replication (for review, see Ransom et al., 2010). The CAF-1 complex is conserved in plants, and its impairment causes multiple phenotypes, including DDR defects. Mutants in the FAS1 gene, encoding the largest CAF-1 subunit, display increased rates of HR (Kirik et al., 2006) and transfer DNA integration (Endo et al., 2006), elevated expression levels of ATM-dependent repair genes (Hisanaga et al., 2013), shorter telomeres, and a reduced number of ribosomal DNA copies (Muchová et al., 2014). Some of these phenotypes were also found in fas2 mutants, devoid of the middle-sized CAF-1 subunit. MSI-1, the small subunit, is a member of several other complexes as well; therefore, the severe mutant phenotypes it displays are difficult to study. The interaction of CAF-1 with RecQ helicases in human cells (for review, see Ransom et al., 2010) could also be relevant in plants, as several RecQ family members in Arabidopsis and rice have been connected to genotoxic stress resistance and DDR repair pathway choice (Knoll and Puchta, 2011; Kwon et al., 2012, 2013; Knoll et al., 2014b; Recker et al., 2014; Schröpfer et al., 2014). HISTONE REGULATOR A (HIRA), another H3/H4 chaperone, acts mainly independent of replication and was recently shown to deposit nucleosome subunits at sites of DNA repair after UV-C light-induced damage in mammalian cells (Adam et al., 2013). All known HIRA subunits are conserved in plants (Nie et al., 2014), and the complex is necessary for regular nucleosome loading in Arabidopsis (Duc et al., 2015). Its link with plant DNA repair needs to be established. Another histone chaperone termed ANTI-SILENCING FUNCTION1 (ASF1), which works upstream of both major H3/H4 chaperones, is also conserved in plants and represented by two partially redundant variants in Arabidopsis that are involved in replication and cell cycle control (Zhu et al., 2011). ASF1 promotes nucleosome reassembly after UV light irradiation (Lario et al., 2013) and interacts with the histone acetyltransferases HAM1/2, which are also connected with efficient repair (Campi et al., 2012).
The H2A/H2B chaperone complex NUCLEOSOME ASSEMBLY PROTEIN (NAP) has also been identified in Arabidopsis and is important for HR (Gao et al., 2012; Zhou et al., 2015), as are the NAP-related proteins NRP1 and NRP2 (Zhu et al., 2006). The hyporecombinogenic phenotype of NAP mutants, in contrast to increased HR in CAF-1 mutants, points to important mechanistic differences between H3/H4 and H2A/H2B delivery during DNA repair processes. FACILITATES CHROMATIN TRANSCRIPTION is a histone chaperone that also mainly associates with H2A/H2B dimers. From the two human subunits, only SPT16 (but not the HMG box-containing SSRP1) is required for the efficient repair of UV light damage (Dinant et al., 2013; Oliveira et al., 2014). Both subunits have orthologs in Arabidopsis, and these are important for regular development (Lolas et al., 2010) and epigenetic regulation (Ikeda et al., 2011), but a link to DNA damage is not described.
CHROMATIN MODIFIERS CONNECTED WITH DDR
DNA can become covalently modified, most prominently in eukaryotes by the methylation of cytosine residues. Additionally, multiple amino acid residues of all the histone subunits can undergo PTMs, leading to steric or electrostatic changes with impacts on nucleosome/DNA association or on the recruitment of other proteins. The presence or absence of Ser and Thr phosphorylation, Lys acetylation, Lys and Arg methylation, Lys ubiquitylation, biotinylation and sumoylation, as well as poly-ADP-ribosylation at Arg and Glu residues offer a plethora of combinations. PTMs usually define larger chromatin regions associated with different functions, in first approximation between transcriptionally active and open euchromatin and less active and more condensed heterochromatin. Dynamic regulation of histone PTMs is tuned by specific writer and eraser enzymes with antagonistic functions, like histone acetyltransferases and histone deacetylases (HDACs) or histone methyltransferases and histone demethylases (Berr et al., 2011). Selected PTMs have been described in connection with DNA damage (for review, see Méndez-Acuña et al., 2010).
Phosphorylation of Ser-139 in the C-terminal portion of the H2A.X histone variant is strongly correlated to DDR, as it is one of the earliest events occurring after DSB induction. Phosphatidylinositol-3-kinases, such as ATM, ATR, and DNA-PKc, control the formation and propagation of phosphorylated γH2A.X next to the DSB site, which is essential for DNA damage signaling and repair factor recruitment. Swr1 and Ino80 chromatin-remodeling complexes (discussed above) interact with γH2A.X through the shared subunit Arp4 (Downs et al., 2004). Similar to yeast and animals, phosphorylation of H2A.X in Arabidopsis occurs early after DSB formation and is a trigger for both HR and NHEJ pathways (Charbonnel et al., 2011). It also interacts with the transcriptional activator E2F, likely in a cell cycle-dependent manner (Lang et al., 2012). H2A and H2A.X in mammalian cells are also targets for monoubiquitylation in the context of DDR. This modification is made at DSBs by PRC1, a ubiquitin ligase and component of the POLYCOMB REPRESSIVE COMPLEX. H2A ubiquitylation contributes to transcriptional repression in the proximity of the lesion and prompts the activation of the RNF8-RNF168-ubiquitin pathway, a key event in the DNA repair cascade (Vissers et al., 2012). Whether PRC1 has a similar role in plants remains to be investigated.
In mammals, ATM is further involved in fine-tuning of the chromatin state at DSBs, as it becomes activated by Tip60, an acetyltransferase that, in turn, is recruited by larger domains of H3K9me3 that are transiently formed around DSBs. The latter involves a complex containing the methyltransferase SUV39h1 and a self-reinforcing loop, due to the recruitment of the heterochromatin-binding proteins HP1 and kap-1, to mark larger domains (Ayrapetov et al., 2014). How these transient sites are distinguished from other regions with more permanent heterochromatin is unclear. The Arabidopsis TIP60 orthologs, HAM1 and HAM2, regulate developmental gene expression through the acetylation of H4K5 (Xiao et al., 2013), but their function is also connected to genotoxic stress. The ham1 and ham2 mutants accumulate persistent cyclobutane pyrimidine dimers after UV-B light treatment, resulting in increased UV-B light sensitivity (Campi et al., 2012). Recently, MSI4, a component of a histone deacetylase complex, was identified as a target of ATM/ATR kinases in response to DNA damage (Roitinger et al., 2015).
The histone deacetylase SIRTUIN6 (SIRT6) plays a central role in DNA repair in human cells. SIRT6 has a dual function during early events after DSB formation: enzymatically, it reduces the level of H3K56Ac, and as a scaffold protein, it recruits SNF2H, an example of how PTMs can interact with chromatin remodelers and control access to DSBs (Toiber et al., 2013). Histone acetylation in the context of DDR in plants is both varied and complex. For example, x-ray irradiation of Arabidopsis causes H3 hyperacetylation and H4 hypoacetylation, partially dependent on ATM (Drury et al., 2012). In contrast, γ-irradiation of wheat (Triticum aestivum) seedlings results in H3 hypoacetylation and H4 hyperacetylation (Raut and Sainis, 2012). Both studies demonstrate the need for further, detailed studies with higher resolution of modification specificity, location, kinetics, dependence on and interaction with other DDR pathways, as well as taking into account evolutionary conservation or species-specific adaptation. Histone methylation or demethylation, which has prominent roles in developmental regulation, has not been directly connected with DNA repair, as the suggested role for the Polycomb group protein CURLY LEAF in somatic recombination may be exerted by the up-regulation of DNA repair genes (Chen et al., 2014).
Methylation of the DNA itself can also have a strong effect on the ability to repair lesions. It has been known for a long time that Arabidopsis mutants that are directly or indirectly affected in DNA methylation display a DNA damage-sensitive phenotype (Gong et al., 2002; Elmayan et al., 2005; Shaked et al., 2006; Yao et al., 2012). However, the relation is complex, as it depends on both the type of damage and the type of repair pathway. Repair of UV-B light-induced damage in repressor of silencing1 (ros1), mutated in a gene for a glycosylase that specifically removes methylated cytosines from DNA, is impaired only in the dark, as the negative effect is compensated in light-grown ros1 mutants by the up-regulation of photorepair-specific genes (Qüesta et al., 2013). Interestingly, the demethylation process results in DNA lesions, as it requires base excision repair to restore the DNA sequence (Lee et al., 2014). The decreased in DNA methylation1 (ddm1) mutant, without an SNF2 chromatin-remodeling factor that is crucial for establishing a correct methylation pattern, is generally affected in the repair of UV light-induced damage (Qüesta et al., 2013). Whether or how these effects are linked to the methylation per se is not yet clear, as the repair function of LSH1, the mammalian protein closest to DDM1, is independent from its role in methylation but rather connected to the formation of γH2A.X foci (Burrage et al., 2012). One possibility is that DDM1 and LSH could modulate the accessibility of the lesion, as discussed previously for the other chromatin-remodeling factors.
The many different substrates of CK2, a Ser/Thr kinase, include several chromatin components (Stemmer et al., 2002; Krohn et al., 2003). CK2 is involved in DDR in Arabidopsis, as a dominant-negative mutant causes increased sensitivity to DNA-damaging agents and decreased HR efficiency (Moreno-Romero et al., 2012). The mutant plants have additional phenotypes similar to mutants of the histone chaperones NRP1 and NRP2, where chromatin decondensation leads to the overexpression of silenced loci upon genotoxic treatment. Therefore, it is likely that phosphorylation by CK2 controls genetic and epigenetic stability in some still to be investigated way. Although not yet shown to have chromatin modification activity, BRUSHY, a protein that is responsible for genetic stability and epigenetic maintenance in specific parts of the genome, shares the CK2 mutant phenotype of increased sensitivity to DNA damage combined with a release of transcriptional silencing (Takeda et al., 2004; Ohno et al., 2011).
THE ROLE OF CHROMATIN DURING PROGRAMMED DSB REPAIR DURING MEIOSIS
In addition to random DNA lesions resulting from adverse conditions, sexual propagation and gamete formation are preceded by numerous programmed DSBs during meiosis. These occur to promote HR between homologous chromosomes. The induction of DSBs is a finely tuned process controlled by different proteins, and chromatin organization is an important parameter for the location and fate of meiotic DSBs. After the synthesis of sister chromatids in the S phase prior to the first meiotic division, chromosomes are arranged in a particular structure consisting of chromatin loops that are anchored in a proteinaceous matrix termed the chromosome axis. SPO11, highly conserved among eukaryotes, catalyzes DSBs on DNA regions exposed by this loop configuration. In yeast, low-density nucleosome regions, such as promoters, are targeted more frequently by SPO11. In mammals, DSBs occur especially in regions marked by histones carrying Set1-dependent H3K4me3, as the chromatin reader PRDM9 recognizes this modification and recruits SPO11, thereby determining the localization of DSBs and recombinational hot spots (for review, see Borde and de Massy, 2013). In Arabidopsis, very few of the approximately 200 initial DSBs created by SPO11 per cell are finally converted into crossover events. Their location seems to be determined by a mix of genetic (sequence-determined) factors and chromatin parameters (for review, see Choi and Henderson, 2015). Genome-wide analysis of crossover events in Arabidopsis revealed that they are concentrated in regions enriched for CTT-repeat DNA motifs and epigenetic marks, including low nucleosome density, high H2A.Z and H3K4me3, and low DNA methylation (Choi et al., 2013; Wijnker et al., 2013). The role of H2A.Z is plausible due to its role in the HR repair pathway (Choi et al., 2013). Unexpectedly, however, higher transcription in heterochromatic regions in hypomethylated mutants did not result in an overall higher recombination rate but instead shifted the position of crossovers and the interference between them (Colomé-Tatché et al., 2012; Melamed-Bessudo and Levy, 2012; Mirouze et al., 2012; Yelina et al., 2012). In barley (Hordeum vulgare), the borders between euchromatin and heterochromatin appear as preferential recombination sites (Higgins et al., 2012). A recent study in maize (Zea mays) revealed that DNA sequence diversity, distance from telomeres, DNA methylation, GC content, and repeat content can predict the location of crossover events with high confidence (Rodgers-Melnick et al., 2015). Therefore, it is likely that recombinational hot spots are determined by many different parameters in a complex interplay between synergistic and competing factors.
CONCLUSION AND FUTURE CHALLENGES
The configuration of chromatin at the site of a DNA lesion and the factors that shape this configuration are important for the efficiency, and likely also for the outcome, of the DDR process (Aymard et al., 2014; Burgess et al., 2014). Reciprocally, chromatin organization might influence the frequency and type of damage along the genome. Besides the role of chromatin in a narrow sense, the involvement of other epigenetic regulators is emerging, such as the participation of coding, noncoding, and small RNAs (Wei et al., 2012; Chen et al., 2013; Keskin et al., 2014). Therefore, genetic and epigenetic (in)stability should be perceived as tightly interconnected in all organisms, including plants. It will remain difficult to distinguish whether apparently heritable epigenetic changes occurring after DNA damage (Mueller-Xing et al., 2014) are indeed independent from the accompanying genetic modifications, but the idea of stress-induced long-term plant adaption without genome sequence modification is widely discussed (for review, see Becker and Weigel, 2012; Pecinka and Mittelsten Scheid, 2012). It should be kept in mind that, especially for plants outside of well-controlled laboratory conditions, in natural habitats DNA-damaging conditions like UV light irradiation are usually connected with high light intensity, high temperature, or oxidizing conditions. These additional stress factors can directly or indirectly modify chromatin configurations and make the interplay between genetic and epigenetic effects even more complex (Turner and Caspari, 2014). Besides the role that chromatin features play in the repair of randomly occurring DNA damage, the emerging potential of genome-editing procedures (Cantos et al., 2014; Baltes and Voytas, 2015) might be enhanced, if the organization and accessibility of the target DNA in its chromatin context are considered. The combined application of tools to create targeted lesions together with sequence-specific chromatin remodelers or modifiers might further improve the precision and efficiency of gene-replacement attempts.
Supplemental Data
The following supplemental materials are available.
Supplemental Text S1. Expansions of all abbreviated gene and protein complex names and other abbreviations.
Supplementary Material
Acknowledgments
Due to the nature of an Update, we had to omit many other relevant publications, and we apologize to authors who might not find their work directly cited here. We thank J. Matthew Watson for careful editing.
Glossary
- DDR
DNA damage repair
- NHEJ
nonhomologous end joining
- HR
homologous recombination
- PTMs
posttranslational modifications
- DSB
double-strand break
Footnotes
This work was supported by the Austrian Academy of Sciences, the Austrian Science Fund (grant nos. FWF I1107, FWF I1477, and FWF W1238), and the Vienna Science and Technology Fund (grant no. LS13–057).
References
- Adam S, Polo SE, Almouzni G (2013) Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155: 94–106 [DOI] [PubMed] [Google Scholar]
- Aichinger E, Villar CBR, Farrona S, Reyes JC, Hennig L, Köhler C (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aklilu BB, Soderquist RS, Culligan KM (2014) Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res 42: 3104–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Lukas J (2013) To spread or not to spread: chromatin modifications in response to DNA damage. Curr Opin Genet Dev 23: 156–165 [DOI] [PubMed] [Google Scholar]
- Antosch M, Mortensen SA, Grasser KD (2012) Plant proteins containing high mobility group box DNA-binding domains modulate different nuclear processes. Plant Physiol 159: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Aydin OZ, Vermeulen W, Lans H (2014) ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 13: 3016–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, et al. (2014) Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21: 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD (2014) DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA 111: 9169–9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrazzi A, Confalonieri M, Macovei A, Donà M, Carbonera D (2011) Genotoxic stress and DNA repair in plants: emerging functions and tools for improving crop productivity. Plant Cell Rep 30: 287–295 [DOI] [PubMed] [Google Scholar]
- Baltes NJ, Voytas DF (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol 33: 120–131 [DOI] [PubMed] [Google Scholar]
- Becker C, Weigel D (2012) Epigenetic variation: origin and transgenerational inheritance. Curr Opin Plant Biol 15: 562–567 [DOI] [PubMed] [Google Scholar]
- Berr A, Shafiq S, Shen WH (2011) Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta 1809: 567–576 [DOI] [PubMed] [Google Scholar]
- Biterge B, Schneider R (2014) Histone variants: key players of chromatin. Cell Tissue Res 356: 457–466 [DOI] [PubMed] [Google Scholar]
- Borde V, de Massy B (2013) Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev 23: 147–155 [DOI] [PubMed] [Google Scholar]
- Burgess RC, Burman B, Kruhlak MJ, Misteli T (2014) Activation of DNA damage response signaling by condensed chromatin. Cell Rep 9: 1703–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrage J, Termanis A, Geissner A, Myant K, Gordon K, Stancheva I (2012) The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J Cell Sci 125: 5524–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campi M, D’Andrea L, Emiliani J, Casati P (2012) Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol 158: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantos C, Francisco P, Trijatmiko KR, Slamet-Loedin I, Chadha-Mohanty PK (2014) Identification of “safe harbor” loci in indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front Plant Sci 5: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Gong L, Yuan W, Li X, Chen G, Li X, Zhang Q, Wu C (2009) Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol 151: 2162–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel C, Allain E, Gallego ME, White CI (2011) Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair (Amst) 10: 611–619 [DOI] [PubMed] [Google Scholar]
- Chen H, Kobayashi K, Miyao A, Hirochika H, Yamaoka N, Nishiguchi M (2013) Both OsRecQ1 and OsRDR1 are required for the production of small RNA in response to DNA-damage in rice. PLoS ONE 8: e55252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhou WB, Wang YX, Dong AW, Yu Y (2014) Polycomb-group histone methyltransferase CLF is required for proper somatic recombination in Arabidopsis. J Integr Plant Biol 56: 550–558 [DOI] [PubMed] [Google Scholar]
- Choi K, Henderson IR (2015) Meiotic recombination hotspots: a comparative view. Plant J 83: 52–61 [DOI] [PubMed] [Google Scholar]
- Choi K, Kim S, Kim SY, Kim M, Hyun Y, Lee H, Choe S, Kim SG, Michaels S, Lee I (2005) SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17: 2647–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FCH, et al. (2013) Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Colomé-Tatché M, Cortijo S, Wardenaar R, Morgado L, Lahouze B, Sarazin A, Etcheverry M, Martin A, Feng S, Duvernois-Berthet E, et al. (2012) Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci USA 109: 16240–16245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, Boudsocq F, Trouche D, Canitrot Y (2012) The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. J Cell Biol 199: 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja W, Mao P, Smerdon MJ (2012) The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int J Mol Sci 13: 11954–11973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa-Nunes JA, Capitão C, Kozak J, Costa-Nunes P, Ducasa GM, Pontes O, Angelis KJ (2014) The AtRAD21.1 and AtRAD21.3 Arabidopsis cohesins play a synergistic role in somatic DNA double strand break damage repair. BMC Plant Biol 14: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Kandasamy MK, McKinney EC, Meagher RB (2005) The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, et al. (2013) Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell 51: 469–479 [DOI] [PubMed] [Google Scholar]
- Dinant C, Bartek J, Bekker-Jensen S (2012) Histone displacement during nucleotide excision repair. Int J Mol Sci 13: 13322–13337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Gasser SM (2013) Chromatin movement in the maintenance of genome stability. Cell 152: 1355–1364 [DOI] [PubMed] [Google Scholar]
- Donà M, Macovei A, Faè M, Carbonera D, Balestrazzi A (2013) Plant hormone signaling and modulation of DNA repair under stressful conditions. Plant Cell Rep 32: 1043–1052 [DOI] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J (2004) Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 16: 979–990 [DOI] [PubMed] [Google Scholar]
- Drury GE, Dowle AA, Ashford DA, Waterworth WM, Thomas J, West CE (2012) Dynamics of plant histone modifications in response to DNA damage. Biochem J 445: 393–401 [DOI] [PubMed] [Google Scholar]
- Duc C, Benoit M, Le Goff S, Simon L, Poulet A, Cotterell S, Tatout C, Probst AV (2015) The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J 81: 707–722 [DOI] [PubMed] [Google Scholar]
- Elmayan T, Proux F, Vaucheret H (2005) Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr Biol 15: 1919–1925 [DOI] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, et al. (2006) Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G, Auerbach RK, Snyder M (2012) SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem 287: 30897–30905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B (2004) The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol Cell 16: 479–485 [DOI] [PubMed] [Google Scholar]
- Gao J, Zhu Y, Zhou W, Molinier J, Dong A, Shen WH (2012) NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell 24: 1437–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold CB, Gasser SM (2014) INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol 24: 619–631 [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18: 831–839 [DOI] [PubMed] [Google Scholar]
- Gospodinov A, Herceg Z (2013a) Chromatin structure in double strand break repair. DNA Repair (Amst) 12: 800–810 [DOI] [PubMed] [Google Scholar]
- Gospodinov A, Herceg Z (2013b) Shaping chromatin for repair. Mutat Res 752: 45–60 [DOI] [PubMed] [Google Scholar]
- Grasser KD, Launholt D, Grasser M (2007) High mobility group proteins of the plant HMGB family: dynamic chromatin modulators. Biochim Biophys Acta 1769: 346–357 [DOI] [PubMed] [Google Scholar]
- Guo M, Wang R, Wang J, Hua K, Wang Y, Liu X, Yao S (2014) ALT1, a Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance through enhanced defense against oxidative stress in rice. PLoS ONE 9: e112515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YF, Dou K, Ma ZY, Zhang SW, Huang HW, Li L, Cai T, Chen S, Zhu JK, He XJ (2014) SUVR2 is involved in transcriptional gene silencing by associating with SNF2-related chromatin-remodeling proteins in Arabidopsis. Cell Res 24: 1445–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Sonoda E, Takami Y, Kimura H, Nakayama T, Tachibana M, Takeda S, Shinkai Y (2007) Histone H1 variant, H1R is involved in DNA damage response. DNA Repair (Amst) 6: 1584–1595 [DOI] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin FCH (2012) Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24: 4096–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisanaga T, Ferjani A, Horiguchi G, Ishikawa N, Fujikura U, Kubo M, Demura T, Fukuda H, Ishida T, Sugimoto K, et al. (2013) The ATM-dependent DNA damage response acts as an upstream trigger for compensation in the fas1 mutation during Arabidopsis leaf development. Plant Physiol 162: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Gerhold CB, Lakomek K, Wollmann P (2012) Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr Opin Struct Biol 22: 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Dion V, Harata M, Gasser SM (2014) SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell 55: 626–639 [DOI] [PubMed] [Google Scholar]
- House NCM, Koch MR, Freudenreich CH (2014) Chromatin modifications and DNA repair: beyond double-strand breaks. Front Genet 5: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lai Y, Zhu D (2014) Transcription regulation by CHD proteins to control plant development. Front Plant Sci 5: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, Higashiyama T, Kakutani T, Kinoshita T (2011) HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell 21: 589–596 [DOI] [PubMed] [Google Scholar]
- Jasin M, Rothstein R (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Downs JA (2014) Roles of chromatin remodellers in DNA double strand break repair. Exp Cell Res 329: 69–77 [DOI] [PubMed] [Google Scholar]
- Jégu T, Latrasse D, Delarue M, Hirt H, Domenichini S, Ariel F, Crespi M, Bergounioux C, Raynaud C, Benhamed M (2014) The BAF60 subunit of the SWI/SNF chromatin-remodeling complex directly controls the formation of a gene loop at FLOWERING LOCUS C in Arabidopsis. Plant Cell 26: 538–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, den Dulk-Ras A, Shen H, Hooykaas PJJ, de Pater S (2013) Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol Biol 82: 339–351 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Deal RB, Smith AP, Meagher RB (2009) Arabidopsis actin-related protein ARP5 in multicellular development and DNA repair. Dev Biol 335: 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK (2005) Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr Biol 15: 1912–1918 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Lorković ZJ, Nishihama R, Ishizaki K, Axelsson E, Yelagandula R, Kohchi T, Berger F (2015) Diversification of histone H2A variants during plant evolution. Trends Plant Sci pii: S1360–S1385 [DOI] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F (2014) Transcript-RNA-templated DNA recombination and repair. Nature 515: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Kim HJ, Lee MH, Moon J, Lee I, et al. (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Kirik A, Pecinka A, Wendeler E, Reiss B (2006) The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18: 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutstein M, Shaked H, Sherman A, Avivi-Ragolsky N, Shema E, Zenvirth D, Levy AA, Simchen G (2008) Functional conservation of the yeast and Arabidopsis RAD54-like genes. Genetics 178: 2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knizewski L, Ginalski K, Jerzmanowski A (2008) Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci 13: 557–565 [DOI] [PubMed] [Google Scholar]
- Knoll A, Fauser F, Puchta H (2014a) DNA recombination in somatic plant cells: mechanisms and evolutionary consequences. Chromosome Res 22: 191–201 [DOI] [PubMed] [Google Scholar]
- Knoll A, Puchta H (2011) The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J Exp Bot 62: 1565–1579 [DOI] [PubMed] [Google Scholar]
- Knoll A, Schröpfer S, Puchta H (2014b) The RTR complex as caretaker of genome stability and its unique meiotic function in plants. Front Plant Sci 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak J, West CE, White C, da Costa-Nunes JA, Angelis KJ (2009) Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst) 8: 413–419 [DOI] [PubMed] [Google Scholar]
- Krohn NM, Stemmer C, Fojan P, Grimm R, Grasser KD (2003) Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J Biol Chem 278: 12710–12715 [DOI] [PubMed] [Google Scholar]
- Kwon YI, Abe K, Endo M, Osakabe K, Ohtsuki N, Nishizawa-Yokoi A, Tagiri A, Saika H, Toki S (2013) DNA replication arrest leads to enhanced homologous recombination and cell death in meristems of rice OsRecQl4 mutants. BMC Plant Biol 13: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YI, Abe K, Osakabe K, Endo M, Nishizawa-Yokoi A, Saika H, Shimada H, Toki S (2012) Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol 53: 2142–2152 [DOI] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, et al. (2010) The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 40: 976–987 [DOI] [PubMed] [Google Scholar]
- Lang J, Smetana O, Sanchez-Calderon L, Lincker F, Genestier J, Schmit AC, Houlné G, Chabouté ME (2012) Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol 194: 353–363 [DOI] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Vermeulen W (2012) ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lario LD, Ramirez-Parra E, Gutierrez C, Spampinato CP, Casati P (2013) ANTI-SILENCING FUNCTION1 proteins are involved in ultraviolet-induced DNA damage repair and are cell cycle regulated by E2F transcription factors in Arabidopsis. Plant Physiol 162: 1164–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro A, Gómez-Zambrano A, López-González L, Piñeiro M, Jarillo JA (2008) Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J Exp Bot 59: 653–666 [DOI] [PubMed] [Google Scholar]
- Lee J, Jang H, Shin H, Choi WL, Mok YG, Huh JH (2014) AP endonucleases process 5-methylcytosine excision intermediates during active DNA demethylation in Arabidopsis. Nucleic Acids Res 42: 11408–11418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C, Soutoglou E (2015) DSB (im)mobility and DNA repair compartmentalization in mammalian cells. J Mol Biol 427: 652–658 [DOI] [PubMed] [Google Scholar]
- Li DQ, Nair SS, Ohshiro K, Kumar A, Nair VS, Pakala SB, Reddy SDN, Gajula RP, Eswaran J, Aravind L, et al. (2012a) MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep 2: 1657–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liu S, Wang J, He J, Huang H, Zhang Y, Xu L (2014) ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J 78: 706–714 [DOI] [PubMed] [Google Scholar]
- Li G, Zhang J, Li J, Yang Z, Huang H, Xu L (2012b) Imitation Switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis. Plant J 72: 261–270 [DOI] [PubMed] [Google Scholar]
- Li X, Chang Y, Xin X, Zhu C, Li X, Higgins JD, Wu C (2013) Replication protein A2c coupled with replication protein A1c regulates crossover formation during meiosis in rice. Plant Cell 25: 3885–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ren X, Yin H, Wang Y, Xia R, Wang Y, Gong Z (2010a) Mutation in the catalytic subunit of DNA polymerase alpha influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J 61: 36–45 [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang J, Miki D, Xia R, Yu W, He J, Zheng Z, Zhu JK, Gong Z (2010b) DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22: 2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD (2010) The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J 61: 686–697 [DOI] [PubMed] [Google Scholar]
- Lorković ZJ. (2012) MORC proteins and epigenetic regulation. Plant Signal Behav 7: 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J (2011) More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13: 1161–1169 [DOI] [PubMed] [Google Scholar]
- March-Díaz R, Reyes JC (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2: 565–577 [DOI] [PubMed] [Google Scholar]
- Mathew V, Pauleau AL, Steffen N, Bergner A, Becker PB, Erhardt S (2014) The histone-fold protein CHRAC14 influences chromatin composition in response to DNA damage. Cell Rep 7: 321–330 [DOI] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Levy AA (2012) Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci USA 109: E981–E988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Acuña L, Di Tomaso MV, Palitti F, Martínez-López W (2010) Histone post-translational modifications in DNA damage response. Cytogenet Genome Res 128: 28–36 [DOI] [PubMed] [Google Scholar]
- Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J (2012) Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci USA 109: 5880–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missirian V, Conklin PA, Culligan KM, Huefner ND, Britt AB (2014) High atomic weight, high-energy radiation (HZE) induces transcriptional responses shared with conventional stresses in addition to a core “DSB” response specific to clastogenic treatments. Front Plant Sci 5: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, et al. (2012) MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336: 1448–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Romero J, Armengot L, Marquès-Bueno MM, Britt A, Martínez MC (2012) CK2-defective Arabidopsis plants exhibit enhanced double-strand break repair rates and reduced survival after exposure to ionizing radiation. Plant J 71: 627–638 [DOI] [PubMed] [Google Scholar]
- Muchová V, Amiard S, Mozgová I, Dvořáčková M, Gallego ME, White C, Fajkus J (2015) Homology-dependent repair is involved in 45S rDNA loss in plant CAF-1 mutants. Plant J 81: 198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Xing R, Xing Q, Goodrich J (2014) Footprints of the sun: memory of UV and light stress in plants. Front Plant Sci 5: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Wang H, Li J, Holec S, Berger F (2014) The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol Open 3: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y, Narangajavana J, Yamamoto A, Hattori T, Kagaya Y, Paszkowski J, Gruissem W, Hennig L, Takeda S (2011) Ectopic gene expression and organogenesis in Arabidopsis mutants missing BRU1 required for genome maintenance. Genetics 189: 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Seol JH, Tyler JK (2013) At the intersection of non-coding transcription, DNA repair, chromatin structure, and cellular senescence. Front Genet 4: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DV, Kato A, Nakamura K, Ikura T, Okada M, Kobayashi J, Yanagihara H, Saito Y, Tauchi H, Komatsu K (2014) Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J Cell Sci 127: 763–772 [DOI] [PubMed] [Google Scholar]
- Osman F, Whitby MC (2013) Emerging roles for centromere-associated proteins in DNA repair and genetic recombination. Biochem Soc Trans 41: 1726–1730 [DOI] [PubMed] [Google Scholar]
- Over RS, Michaels SD (2014) Open and closed: the roles of linker histones in plants and animals. Mol Plant 7: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL (2013) Chromatin and the genome integrity network. Nat Rev Genet 14: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Mittelsten Scheid O (2012) Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol 53: 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Almouzni G (2013) Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harb Perspect Biol 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE. (2015) Reshaping chromatin after DNA damage: the choreography of histone proteins. J Mol Biol 427: 626–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD (2013) Chromatin remodeling at DNA double-strand breaks. Cell 152: 1344–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhao L, Skaggs MI, Andreuzza S, Tsukamoto T, Panoli A, Wallace KN, Smith S, Siddiqi I, Yang Z, et al. (2014) ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell 26: 1612–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qüesta JI, Fina JP, Casati P (2013) DDM1 and ROS1 have a role in UV-B induced- and oxidative DNA damage in A. thaliana. Front Plant Sci 4: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK (2010) Chaperoning histones during DNA replication and repair. Cell 140: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raut VV, Sainis JK (2012) 60Co-γ radiation induces differential acetylation and phosphorylation of histones H3 and H4 in wheat. Plant Biol (Stuttg) 14: 110–117 [DOI] [PubMed] [Google Scholar]
- Recker J, Knoll A, Puchta H (2014) The Arabidopsis thaliana homolog of the helicase RTEL1 plays multiple roles in preserving genome stability. Plant Cell 26: 4889–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Bradbury PJ, Elshire RJ, Glaubitz JC, Acharya CB, Mitchell SE, Li C, Li Y, Buckler ES (2015) Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc Natl Acad Sci USA 112: 3823–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitinger E, Hofer M, Köcher T, Pichler P, Novatchkova M, Yang J, Schlögelhofer P, Mechtler K (2015) Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics 14: 556–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Von Harder M, Cigliano RA, Schlögelhofer P, Mittelsten Scheid O (2013) The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25: 1990–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth N, Klimesch J, Dukowic-Schulze S, Pacher M, Mannuss A, Puchta H (2012) The requirement for recombination factors differs considerably between different pathways of homologous double-strand break repair in somatic plant cells. Plant J 72: 781–790 [DOI] [PubMed] [Google Scholar]
- Roy S. (2014) Maintenance of genome stability in plants: repairing DNA double strand breaks and chromatin structure stability. Front Plant Sci 5: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröpfer S, Kobbe D, Hartung F, Knoll A, Puchta H (2014) Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res 42: 1684–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Hauer M, Gasser SM (2013) Nucleosome remodelers in double-strand break repair. Curr Opin Genet Dev 23: 174–184 [DOI] [PubMed] [Google Scholar]
- Shaked H, Avivi-Ragolsky N, Levy AA (2006) Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173: 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Melamed-Bessudo C, Levy AA (2005) High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci USA 102: 12265–12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Devic M, Lepiniec L, Zhou D (2015) Chromodomain, helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol J 13: 811–820 [DOI] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G (2012) Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46: 722–734 [DOI] [PubMed] [Google Scholar]
- Stanley FKT, Moore S, Goodarzi AA (2013) CHD chromatin remodelling enzymes and the DNA damage response. Mutat Res 750: 31–44 [DOI] [PubMed] [Google Scholar]
- Stemmer C, Schwander A, Bauw G, Fojan P, Grasser KD (2002) Protein kinase CK2 differentially phosphorylates maize chromosomal high mobility group B (HMGB) proteins modulating their stability and DNA interactions. J Biol Chem 277: 1092–1098 [DOI] [PubMed] [Google Scholar]
- Stros M. (2010) HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 1799: 101–113 [DOI] [PubMed] [Google Scholar]
- Swygert SG, Peterson CL (2014) Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim Biophys Acta 1839: 728–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, et al. (2004) BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev 18: 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chen ZJ (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA 98: 200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. (2013) SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 51: 454–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsabar M, Haber JE (2013) Chromatin modifications and chromatin remodeling during DNA repair in budding yeast. Curr Opin Genet Dev 23: 166–173 [DOI] [PubMed] [Google Scholar]
- Turner T, Caspari T (2014) When heat casts a spell on the DNA damage checkpoints. Open Biol 4: 140008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van C, Williams JS, Kunkel TA, Peterson CL (2015) Deposition of histone H2A.Z by the SWR-C remodeling enzyme prevents genome instability. DNA Repair (Amst) 25: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers JHA, van Lohuizen M, Citterio E (2012) The emerging role of Polycomb repressors in the response to DNA damage. J Cell Sci 125: 3939–3948 [DOI] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y (2012) A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112 [DOI] [PubMed] [Google Scholar]
- Wijnker E, Velikkakam James G, Ding J, Becker F, Klasen JR, Rawat V, Rowan BA, de Jong DF, de Snoo CB, Zapata L, et al. (2013) The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2: e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CWM (2011) SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492 [DOI] [PubMed] [Google Scholar]
- Xiao J, Zhang H, Xing L, Xu S, Liu H, Chong K, Xu Y (2013) Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J Plant Physiol 170: 444–451 [DOI] [PubMed] [Google Scholar]
- Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD (2012a) The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett 586: 3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Yuan D, Liu M, Li C, Liu Y, Zhang S, Yao N, Yang C (2013) AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol 161: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD (2012b) Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell 48: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Bilichak A, Golubov A, Kovalchuk I (2012) ddm1 plants are sensitive to methyl methane sulfonate and NaCl stresses and are deficient in DNA repair. Plant Cell Rep 31: 1549–1561 [DOI] [PubMed] [Google Scholar]
- Yelagandula R, Stroud H, Holec S, Zhou K, Feng S, Zhong X, Muthurajan UM, Nie X, Kawashima T, Groth M, et al. (2014) The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Choi K, Chelysheva L, Macaulay M, de Snoo B, Wijnker E, Miller N, Drouaud J, Grelon M, Copenhaver GP, et al. (2012) Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet 8: e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF (2013) SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154: 1246–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cao L, Rong L, An Z, Zhou W, Ma J, Shen WH, Zhu Y, Dong A (2015) The chromatin-remodeling factor AtINO80 plays crucial roles in genome stability maintenance and in plant development. Plant J 82: 655–668 [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu Y, Dong A, Shen WH (2015) Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development. Plant J 83: 78–95 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Dong A, Meyer D, Pichon O, Renou JP, Cao K, Shen WH (2006) Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18: 2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dong A, Shen WH (2013) Histone variants and chromatin assembly in plant abiotic stress responses. Biochim Biophys Acta 1819: 343–348 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Weng M, Yang Y, Zhang C, Li Z, Shen WH, Dong A (2011) Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J 66: 443–455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.