Although DNA methylation within transcribed genes is commonly found in diverse animals and plants, recent advances highlight the remaining mystery regarding intragenic DNA methylation.
Abstract
DNA methylation within transcribed genes is commonly found in diverse animals and plants. Here, we provide an overview of recent advances and the remaining mystery regarding intragenic DNA methylation.
In plant genomes, DNA methylation is found not only in promoters but also within transcribed regions (Fig. 1; Table I). The characteristics of DNA methylation within transcribed regions differ from those in promoters. For promoters, high levels of DNA methylation are found only in silent genes and transposable elements (TEs). In contrast, actively transcribed genes do not have much DNA methylation around their promoter regions. However, in regions sufficiently downstream (approximately 500 bp) from the transcription start sites (TSSs), substantial amounts of DNA methylation are often found even in transcribed genes (Zhang et al., 2006; Zilberman et al., 2007; Fig. 1). DNA methylation within transcribed genes is commonly found in plants and animals. Furthermore, intragenic DNA methylation has unique features that are evolutionarily conserved among these organisms (Zemach et al., 2010), implicating one or more basic functions. However, the control and biological role of intragenic methylation still remain largely unknown. Here, we provide an overview of recent findings about intragenic DNA methylation and discuss remaining questions.
Figure 1.
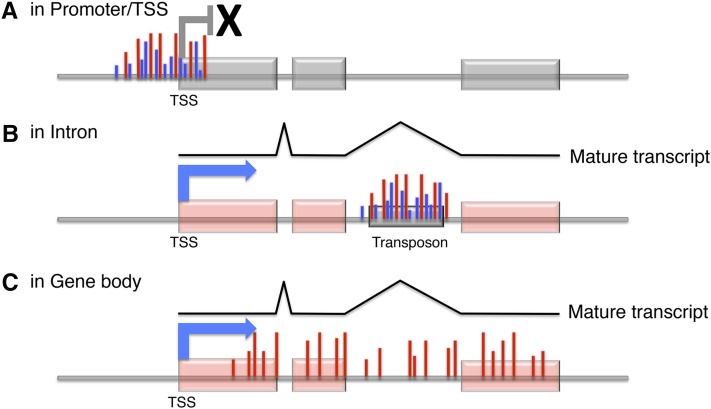
Schematic diagram of intragenic DNA methylation in Arabidopsis (Arabidopsis thaliana). A, DNA methylation at TSSs and/or in promoters. Intensive CpG methylation (red bars) and non-CpG methylation (blue bars) are found within the TSSs and promoters of silent genes and TEs. Their transcription is repressed by the methylation. B, DNA methylation in intron. TEs or repeats located in the introns of transcribed genes are methylated in both CpG and non-CpG contexts. When such DNA methylation is lost, immature transcripts are detected, probably because of the cryptic poly(A) addition signal within the repeats. C, DNA methylation in gene bodies. Only CpG methylation is found in the gene bodies of actively transcribed genes. Both exons and introns are methylated except for TSS-proximal regions.
Table I. DNA methylation in the promoter and transcribed region.
| Methylation in Promoter | Methylation in Transcribed Region |
||
|---|---|---|---|
| Intragenic Heterochromatin | Gene Body Methylation | ||
| Transcription | Silent | Any type | Housekeeping |
| Contexts | Both CpG and non-CpG | Both CpG and non-CpG | CpG |
| Exon/intron | Mainly in intron | Both exon and intron | |
We review two types of intragenic methylation, which differ in relation to transcription activity, exon/intron distribution, and the context of methylated cytosine (Table I). In plant genomes, cytosine can be methylated in both CpG and non-CpG contexts. Non-CpG methylation is associated with methylation of histone H3 Lys 9 (H3K9me), which is an epigenetic mark of silent chromatin conserved among eukaryotes (Johnson et al., 2007; Inagaki et al., 2010). Methylation is found in both CpG and non-CpG contexts in promoters of silent genes and TEs. The first type of intragenic methylation we discuss is found mainly within intron, and both CpG and non-CpG contexts can be methylated. Therefore, this type of DNA methylation can be understood as islands of silent chromatin found in introns of active genes (Fig. 1).
The other type of intragenic methylation, called gene body methylation, is found primarily in exons but also in introns (Chodavarapu et al., 2010), and almost always only CpG sites are methylated. Very interestingly, this type of intragenic methylation is found in constitutively transcribed genes. Inducible genes and developmentally regulated genes generally do not have CpG gene body methylation (Aceituno et al., 2008; Coleman-Derr and Zilberman 2012). In this review, we discuss these two types of intragenic DNA methylation separately, dealing first with intronic heterochromatin and subsequently with gene body methylation in constitutively transcribed genes.
CONTROL OF INTRAGENIC HETEROCHROMATIN
Substantial numbers of intragenic heterochromatin islands are found for both animal and plant genes, especially within the introns (van de Lagemaat et al., 2006; Sela et al., 2007; Nystedt et al., 2013; Seymour et al., 2014; West et al., 2014). Most of these examples of intronic heterochromatin reflect insertions of TEs. In the human genome, 60% of TEs are localized within introns that comprise only 24% of the genome; therefore, TEs are much enriched in introns. Similarly, most plant genomes have many TEs in introns. One exception is the genome of Arabidopsis, which contains relatively few intronic TEs. A recent analysis estimated that only 0.7% of annotated genes in the Arabidopsis genome contain intronic TEs (Le et al., 2015). However, genome organization of Arabidopsis is exceptional; most plant species have a large number of TEs within introns. A recent maize (Zea mays) epigenome study showed that approximately 10% of genes contain intronic TEs >1 kb in length (West et al., 2014). The introns of Arabidopsis lyrata, which diverged from Arabidopsis around 10 million years ago, contain many more TEs than those of Arabidopsis, reflecting the lineage-specific expansion/contraction of TE sequence within genic regions (Seymour et al., 2014). The genome of Norway spruce (Picea abies), the first genome available for gymnosperm, is 100 times larger than Arabidopsis, but the number of genes (approximately 28,000 genes) and exon sizes are similar (Nystedt et al., 2013). In contrast, the P. abies genome contains many genes harboring long introns, mainly reflecting insertion of long-terminal-repeat-type retrotransposons. These findings suggest that genomes are relatively tolerant of the presence of TEs, especially in intronic sequences, and intronic TEs are widespread in the plant genomes.
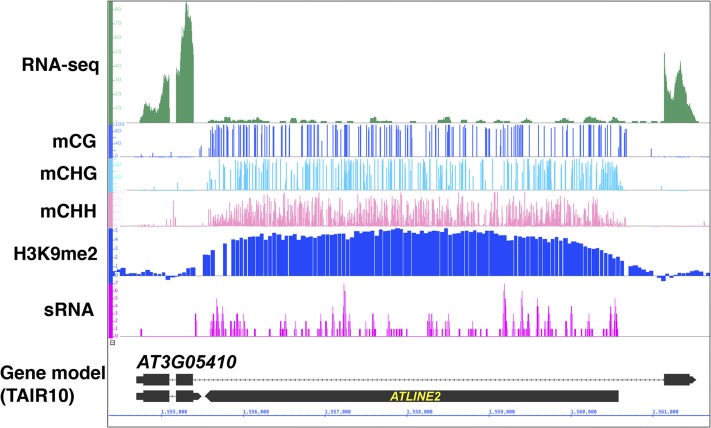
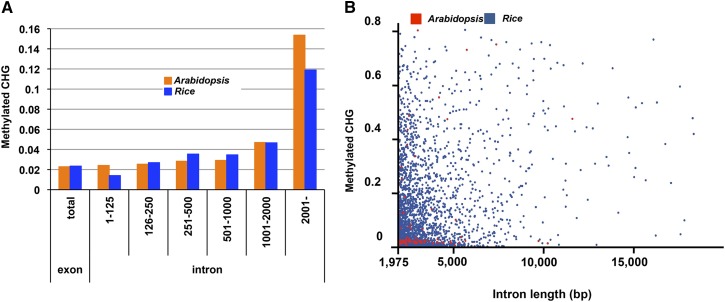
Many of these genes with intronic TEs are actively transcribed. Although the presence of TEs negatively correlates with transcription of nearby genes for a genome-wide trend (Hollister and Gaut, 2009; Wang et al., 2013b), a reasonably high level of expression is often found in the TE-bearing genes, comparable with that of genes without TE insertion (Nystedt et al., 2013; West et al., 2014). Importantly, TEs within introns often have repressive epigenetic marks, such as DNA methylation and H3K9me (Fig. 2). That is the case for intronic TEs in both maize and A. lyrata. Even in the exceptionally compact genome Arabidopsis, long introns often contain non-CpG methylation, mainly associated with TE insertions there (Saze et al., 2013; Le et al., 2015; Fig. 1). The level of non-CpG methylation in long introns in Arabidopsis is comparable with that in rice (Oryza sativa), although the number of such heterochromatic introns is much higher in rice (Fig. 3), as in other plant species. The intronic TEs are sometimes indistinguishable from copies in intergenic regions in terms of silent marks, such as non-CpG methylation, H3K9me, and small RNAs (Fig. 2).
Figure 2.
An intronic TE associated with heterochromatic epigenetic marks within the actively transcribed gene AT3G05410 in Arabidopsis. Top to bottom tracks: RNA-sequencing reads (green) in wild-type Columbia (Col; Saze et al., 2013), DNA methylation at CpG (blue), CHG (light blue), CHH sites (pink; H. Saze, unpublished) in wild-type Col, chromatin immunoprecipitation-Chip hybridization signals of H3K9me2 in wild-type Col (blue; Inagaki et al., 2010), small RNAs (magenta; Lister et al., 2008), and Arabidopsis gene model (The Arabidopsis Information Resource 10 [https://www.arabidopsis.org]). Non-long-terminal-repeat retrotransposon ATLINE2 is inserted in the antisense orientation relative to AT3G05410.
Figure 3.
Heterochromatic introns in the genomes of Arabidopsis and rice. The figure is adapted from Saze et al. (2013). A, Proportion of methylated cytosine at CHG sites compared among introns of different length. The value was derived from the sum of mapped cytosines in each class. Long introns tend to have more CHG methylation in both Arabidopsis and rice. B, The rice genome contains many more long heterochromatic introns than the Arabidopsis genome. Introns longer than the seventh intron of IBM1 (>1,975 bp) are plotted with the proportion of methylated cytosine in CHG sites, a hallmark of heterochromatin. Red and blue dots represent introns of Arabidopsis and rice, respectively.
Genes containing islands of heterochromatin within their introns are generally transcribed properly. An interesting question is what mechanisms allow plants to mask the deleterious effects of intronic TE sequences associated with repressive epigenetic marks. Indeed, recent studies identified factors in plants required for proper transcription of genes containing repressive epigenetic marks in intronic regions (Saze et al., 2013; Wang et al., 2013a; Coustham et al., 2014; Lei et al., 2014). One of the factors, ENHANCED DOWNY MILDEW2 (EDM2), was initially identified as a factor required for plant resistance to pathogen and was subsequently found to be required for proper transcription of a disease resistance gene, Resistance to Peronospora parasitica7 (RPP7), that contains a number of intronic TEs forming heterochromatic domains (Tsuchiya and Eulgem, 2013). EDM2 has Plant Homeodomain domains that recognize H3K9me and a putative RNA methyltransferase domain in its C-terminal part (Lei et al., 2014; Tsuchiya and Eulgem, 2014). Another factor, INCREASE IN BONSAI MATHYLATION2 (IBM2)/ANTI-SILENCING1 (ASI1)/SHOOT GROWTH1 (SG1), was identified as a gene responsible for DNA hypermethylation of gene bodies (Saze et al., 2013), an antisilencing effect for a transgene (Wang et al., 2013a), and pleiotropic developmental abnormalities (Coustham et al., 2014) in these Arabidopsis mutants. IBM2 contains the bromo-adjacent homology domain that likely binds to chromatin and an RNA recognition motif, although the direct target of IBM2 is still unclear. The mutant phenotypes are partly due to a transcription defect at the histone H3K9 demethylase gene IBM1, the seventh intron of which contains a repetitive sequence, which is similar to organellar genomes and modified with CpG and non-CpG DNA methylation (Rigal et al., 2012). The ibm1 mutation is known to cause a genome-wide genic DNA hypermethylation in non-CpG sites, accompanied by pleiotropic developmental defects (Saze et al., 2008). Indeed, the IBM1 transgene without the sequence of heterochromatic domain in the intron can rescue these ibm1-like phenotypes of ibm2. In edm2 and ibm2 mutants, in addition to the IBM1 gene, the full-length transcript of genes containing heterochromatic TE was reduced, and instead transcripts were prematurely terminated and polyadenylated within the associated TE sequences (Saze et al., 2013; Tsuchiya and Eulgem, 2013). Interestingly, RNA polymerase II elongation over the heterochromatic domains within introns is not affected in ibm2, suggesting that IBM2 is not required for passage of PolII; it more likely affects posttranscriptional processes, such as efficient splicing of heterochromatic introns and/or suppression of cryptic poly(A) signal sequences in the intronic repeats. Although EDM2 and IBM2 preferentially localize to the intronic heterochromatin, it is currently unclear how the maintenance of repressive heterochromatic states within the actively transcribed region is achieved.
More counterintuitively, maintenance of heterochromatin marks such as DNA methylation and H3K9me within introns can be important for proper expression of genes containing that, as loss of heterochromatic modifications in mutants such as decrease in DNA methylation1 and methyltransferase1 (met1) leads to a transcription defect of these genes (Le et al., 2015). For example, a reduction of DNA methylation of the repetitive element in the IBM1 gene in the met1 mutant results in a premature termination of the transcripts in the upstream of the repeat sequence (Rigal et al., 2012). In addition, a reduction of H3K9me at intronic TEs in the triple mutant of H3K9 methylase genes suvh4-suvh5-suvh6 affects transcription of the RPP7 gene (Tsuchiya and Eulgem, 2013). A relationship between non-CpG methylation in the intronic region and transcription also occurs in maize, where non-CpG methylation at exon-intron junctions inhibits alternative splicing (Regulski et al., 2013). In winter wheat (Triticum aestivum), cold treatment for induction of vernalization induces non-CpG methylation at TEs present in the intron of the VARNALIZATION-A1 gene, which is associated with the transcriptional activation of the gene (Khan et al., 2013). Thus, repressive epigenetic modifications on repeats and TEs in intronic regions might have a role in regulation of gene activation, beyond the silencing of TEs. Indeed, the intronic repeat in the IBM1 gene could function as a sensor for a fine-tuning mechanism for global changes in DNA methylation, where a reduction of DNA methylation at the repeat can reduce IBM1 transcripts, which eventually induce a genome-wide DNA hypermethylation (Rigal et al., 2012). The IBM1 intronic repeat emerged before the speciation of Arabidopsis, suggesting that an acquisition of feedback regulatory mechanisms for DNA methylation might be beneficial (Rigal et al., 2012; Saze et al., 2013). On the other hand, intronic TEs often show intraspecies insertion/deletion polymorphisms (Liu et al., 2004; Ziolkowski et al., 2009), suggesting that the modification of intronic TEs with repressive epigenetic marks might be a short-term adaptation mechanism to neutralize the deleterious effects of TE insertion.
GENE BODY METHYLATION
We have discussed intronic DNA methylation at both CpG and non-CpG sites, a signature of heterochromatin. The other type of intragenic DNA methylation is gene body methylation, which is found in euchromatic regions and is predominant in CpG sites (Lister et al., 2008; Feng et al., 2010; Zemach et al., 2010; Regulski et al., 2013). Gene body methylation of transcriptionally active genes is a common feature in diverse eukaryotes, suggesting a conserved function(s) (Feng et al., 2010; Zemach et al., 2010). Gene body methylation level positively correlates with gene expression level (Lister et al., 2008; Feng et al., 2010; Zemach et al., 2010). The connection of gene body methylation to transcription level can also be seen in the allelic pair of genes in X chromosomes of female mammals; genes on the active X chromosome have higher levels of gene body methylation than their counterparts on the inactive X chromosome, despite their nearly identical sequences (Hellman and Chess, 2007). Interestingly, the correlation between gene body methylation and transcription is more clearly observed in proliferating cells than in nonproliferating cells, suggesting that gene body methylation might be connected to cell proliferation in mammals (Aran et al., 2011). In plants, although TEs and silent genes have cytosine methylation in both CpG and non-CpG contexts, gene body methylation is found only in the CpG context (Table I). Gene body methylation is maintained by the maintenance methylase DNA METHYLTRANSFERASE1 (MET1) and its cofactor proteins in the VARIANT IN METHYLATION family (VIM1–VIM3) in Arabidopsis. Both classes of proteins are conserved in mammals; MET1 and VIMs are Arabidopsis orthologs of DNA Methyltransferase1 and Ubiquitin-Like with Plant Homeodomain and Ring Finger Domains1 in vertebrates, respectively (Finnegan and Kovac, 2000; Woo et al., 2008).
In regard to gene body methylation, two important questions remain unsolved: what is the biological role(s) of gene body methylation, and how is the gene body methylation pattern generated?
To address the second question, one approach is to identify mutants affecting gene body methylation. When DNA methylation is examined genome wide in 86 mutants of Arabidopsis known to have DNA methylation defects, none of the mutants other than met1 or vim1 vim2 vim3 triple mutants abolished gene body methylation (Stroud et al., 2013). Histone modifications and non-CpG methylation in heterochromatin are controlled by multiple pathways (Matzke and Mosher, 2014), but CpG methylation in gene bodies seems to be controlled by a simpler manner, by the maintenance methylation machinery. This simplicity may make sense, because CpG methylation in gene bodies tends to be maintained very stably and can even be inherited over generations (Vongs et al., 1993; Kakutani et al., 1999; Kankel et al., 2003; Saze et al., 2003). Nonetheless, it would be interesting to know how the pattern of gene body methylation is generated initially, before it is maintained transgenerationally.
Recently, several new results have been published with regard to the first question: the biological role of gene body methylation. Takuno and Gaut (2012, 2013) addressed the evolutional role of gene body methylation. Comparison of genomic bisulfite sequencing data with the phenotypic database showed that phenotypic effects could be seen in 55.7% of body-methylated genes, whereas such effects were only associated with 26.2% of the undermethylated genes, indicating that body-methylated genes are functionally more important. In addition, body-methylated genes show significantly lower nonsynonymous to synonymous substitution rates, which is consistent with the phenotypic results; amino acid sequences of functionally important genes should be conserved. Interestingly, both nonsynonymous and synonymous substitution rates are lower in body-methylated genes. This was especially surprising considering that methylcytosine is expected to be mutagenic and increase C/G-to-T/A mutation rates through spontaneous deamination (Bird, 1980; Pfeifer, 2006). The low mutation rate may be due to a low frequency of CpG sites, or due to low nucleosome occupancy, which may allow easy access of repair machinery (Meier and Thoma, 2005; Ataian and Krebs, 2006), although, according to the calculation by Takuno and Gaut (2012), each of these factors alone does not fully explain the low mutation rate of body-methylated genes.
Body-methylated genes also evolve slower in mammals and invertebrates. The comparative analysis of human DNA methylome data with human-macaque and human-mouse protein evolutionary rates revealed that gene body methylation is negatively correlated with protein evolutionary rate, whereas promoter methylation correlates positively (Chuang and Chiang, 2014). Similarly, genes with heavy body methylation are evolutionarily conserved and enriched for housekeeping functions in invertebrates (Sarda et al., 2012).
In mammals and other animals, accumulating evidence suggests a link between gene body methylation and alternative splicing. Depletion of DNA methylation by the inhibitor 5-aza-dC causes the failure of methyl-CpG-binding protein2 recruitment to the specific exons, resulting in exon skipping of the alternatively spliced sites (Maunakea et al., 2013). DNA methylation can also control alternative splicing by modulating binding of CCCTC-binding factor and RNA polymerase II pausing (Shukla et al., 2011). Interestingly, artificially established repressive marks driven by small interfering RNAs can cause an increase in DNA methylation and ectopic inclusion of noncanonical exons (Schor et al., 2013). A role of gene body methylation in alternative splicing is also observed during caste differentiation of social insects (Lyko et al., 2010; Bonasio et al., 2012; Terrapon et al., 2014). It would be interesting to know how certain methylated exons, but not other methylated exons nearby, can be specifically recognized by these factors.
The role of gene body methylation may be related to the interaction of the host with TEs. One possible role could be to shelter genes from TE insertion. Maize transposon Robertson’s Mutator (Mu) inserts preferentially within genes. Genome-wide mapping of insertion sites of Mu in the maize genome revealed that it preferentially inserts within unmethylated regions (Liu et al., 2009; Regulski et al., 2013). It would be interesting to examine if this can be applied to insertion sites of TEs other than Mu. Another possible role of gene body methylation could be a trigger to silence TEs but not genes, generating their differential methylation. If the host methylates the body of transcribed sequences, this may affect transcription of TEs more than that of genes. The small difference could be amplified by positive feedbacks of active and inactive states (see Inagaki and Kakutani, 2012 for a detailed discussion).
An interesting feature of gene body methylation is that it is found in housekeeping genes (Aceituno et al., 2008; Sarda et al., 2012). In other words, genes responding to environmental or developmental signals generally do not have body methylation. The link between gene body methylation and responsiveness has been suggested recently. In diverse eukaryotes including mammals, fish, and plants, DNA methylation anticorrelates with a histone variant H2A.Z (Zilberman et al., 2008; Conerly et al., 2010; Zemach et al., 2010; Valdés-Mora et al., 2012). H2A.Z is a histone variant evolutionarily conserved from yeast (Saccharomyces cerevisiae) to higher eukaryotes (Raisner and Madhani, 2006). It is predominantly localized near TSSs and is required for the poised state of transcription initiation, reducing nucleosome density and increasing DNA accessibility around TSSs (Fan et al., 2002; Guillemette et al., 2005; Tirosh and Barkai, 2008; Hu et al., 2013). These antagonistic epigenetic marks at TSSs may decide whether the gene is silenced (methylated) or kept poised for activation (H2A.Z). In Arabidopsis, the anticorrelation of H2A.Z against DNA methylation is also observed within gene bodies (Zilberman et al., 2008; Coleman-Derr and Zilberman, 2012). The genes containing H2A.Z over gene bodies are enriched in genes responsive to environmental or developmental stimuli (Coleman-Derr and Zilberman, 2012). Conversely, gene body methylation is found predominantly at constitutively expressed housekeeping genes and showed clear negative correlation against gene responsiveness (Aceituno et al., 2008). This global anticorrelation may imply that DNA methylation and H2A.Z exclude each other. Actually, the global loss of CpG methylation in Arabidopsis met1 causes an increase in H2A.Z occupancies in regions normally methylated in the wild type (Zilberman et al., 2008). On the other hand, disruption of the H2A.Z gene rarely alters the levels and patterns of gene body methylation, suggesting that H2A.Z does not exclude DNA methylation (Coleman-Derr and Zilberman, 2012). Therefore, gene body methylation might function upstream by preventing the incorporation of H2A.Z within gene bodies of housekeeping genes. It will be interesting to determine if the gene body methylation affects transcription stability/instability.
CONCLUSION AND PERSPECTIVE
Here, we discussed two topics: (1) mechanisms to handle intronic heterochromatin, and (2) gene body methylation of housekeeping genes.
Mechanisms to control intronic heterochromatin were discussed first. Although the Arabidopsis genome does not have much heterochromatin in introns, genomes of typical plant species have a large amount of intronic heterochromatin. The mechanism to mask the deleterious effect of heterochromatin in intron by EDM2 and IBM2/ASI1/SG1 should be important for evolution. An interesting possibility is that the intronic heterochromatin could also be a source for unique gene controls. For example, the intronic repeat in the IBM1 gene could function as a sensor for a fine-tuning mechanism for global changes in DNA methylation (Rigal et al., 2012). It might be interesting to search conserved intronic heterochromatin in various species with a potential regulatory role of gene expression through its epigenetic changes.
We also discussed control and biological role of gene body methylation. In plants, gene body methylation pattern, which is prevalently found in CpG sites, should be established transgenerationally. For the transgenerational establishment of gene body methylation, possible effects of histone variants and histone modifications as well as environmental and genetic variations could be an exciting research area for future exploration. A big question would be possible biological roles of gene body methylation. Despite conservation during evolution, the roles still remain mysterious.
Acknowledgments
We thank Eric Richards and Shohei Takuno for comments on the article.
Glossary
- TE
transposable element
- TSS
transcription start site
- Mu
Robertson’s Mutator
References
- Aceituno FF, Moseyko N, Rhee SY, Gutiérrez RA (2008) The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics 9: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Toperoff G, Rosenberg M, Hellman A (2011) Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet 20: 670–680 [DOI] [PubMed] [Google Scholar]
- Ataian Y, Krebs JE (2006) Five repair pathways in one context: chromatin modification during DNA repair. Biochem Cell Biol 84: 490–504 [DOI] [PubMed] [Google Scholar]
- Bird AP. (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. (2012) Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol 22: 1755–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TJ, Chiang TW (2014) Impacts of pretranscriptional DNA methylation, transcriptional transcription factor, and posttranscriptional microRNA regulations on protein evolutionary rate. Genome Biol Evol 6: 1530–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D (2012) Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S (2010) Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res 20: 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V, Vlad D, Deremetz A, Gy I, Cubillos FA, Kerdaffrec E, Loudet O, Bouché N (2014) SHOOT GROWTH1 maintains Arabidopsis epigenomes by regulating IBM1. PLoS One 9: e84687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ (2002) The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA (2000) Plant DNA methyltransferases. Plant Mol Biol 43: 189–201 [DOI] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L (2005) Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Chess A (2007) Gene body-specific methylation on the active X chromosome. Science 315: 1141–1143 [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS (2009) Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res 19: 1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K (2013) H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12: 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Kakutani T (2012) What triggers differential DNA methylation of genes and TEs: contribution of body methylation? Cold Spring Harb Symp Quant Biol 77: 155–160 [DOI] [PubMed] [Google Scholar]
- Inagaki S, Miura-Kamio A, Nakamura Y, Lu F, Cui X, Cao X, Kimura H, Saze H, Kakutani T (2010) Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. EMBO J 29: 3496–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE (2007) The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Munakata K, Richards EJ, Hirochika H (1999) Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Enjalbert J, Marsollier A-C, Rousselet A, Goldringer I, Vitte C (2013) Vernalization treatment induces site-specific DNA hypermethylation at the VERNALIZATION-A1 (VRN-A1) locus in hexaploid winter wheat. BMC Plant Biol 13: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TN, Miyazaki Y, Takuno S, Saze H (2015) Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acids Res 43: 3911–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, La H, Lu K, Wang P, Miki D, Ren Z, Duan CG, Wang X, Tang K, Zeng L, et al. (2014) Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc Natl Acad Sci USA 111: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He Y, Amasino R, Chen X (2004) siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev 18: 2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Ji T, Ying K, Wu H, Tang HM, Fu Y, Nettleton D, Schnable PS (2009) Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet 5: e1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R (2010) The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol 8: e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Chepelev I, Cui K, Zhao K (2013) Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res 23: 1256–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A, Thoma F (2005) RNA polymerase I transcription factors in active yeast rRNA gene promoters enhance UV damage formation and inhibit repair. Mol Cell Biol 25: 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y-C, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. (2006) Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol 301: 259–281 [DOI] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD (2006) Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev 16: 119–124 [DOI] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MTA, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal M, Kevei Z, Pélissier T, Mathieu O (2012) DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J 31: 2981–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV (2012) The evolution of invertebrate gene body methylation. Mol Biol Evol 29: 1907–1916 [DOI] [PubMed] [Google Scholar]
- Saze H, Kitayama J, Takashima K, Miura S, Harukawa Y, Ito T, Kakutani T (2013) Mechanism for full-length RNA processing of Arabidopsis genes containing intragenic heterochromatin. Nat Commun 4: 2301. [DOI] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T (2008) Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319: 462–465 [DOI] [PubMed] [Google Scholar]
- Schor IE, Fiszbein A, Petrillo E, Kornblihtt AR (2013) Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J 32: 2264–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G (2007) Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol 8: R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D (2014) Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet 10: e1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS (2012) Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol 29: 219–227 [DOI] [PubMed] [Google Scholar]
- Takuno S, Gaut BS (2013) Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci USA 110: 1797–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrapon N, Li C, Robertson HM, Ji L, Meng X, Booth W, Chen Z, Childers CP, Glastad KM, Gokhale K, et al. (2014) Molecular traces of alternative social organization in a termite genome. Nat Commun 5: 3636. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N (2008) Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Eulgem T (2013) An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc Natl Acad Sci USA 110: E3535–E3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Eulgem T (2014) The PHD-finger module of the Arabidopsis thaliana defense regulator EDM2 can recognize triply modified histone H3 peptides. Plant Signal Behav 9: e29202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ (2012) Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res 22: 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat LN, Medstrand P, Mager DL (2006) Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol 7: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wang X, Duan CG, Tang K, Wang B, Zhang H, Lei M, Lu K, Mangrauthia SK, Wang P, Zhu G. , et al. (2013a) RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc Natl Acad Sci USA 110: 15467–15472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM (2013b) Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet 9: e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West PT, Li Q, Ji L, Eichten SR, Song J, Vaughn MW, Schmitz RJ, Springer NM (2014) Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9: e105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer TA, Richards EJ (2008) Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet 4: e1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE. , et al. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S (2008) Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
- Ziolkowski PA, Koczyk G, Galganski L, Sadowski J (2009) Genome sequence comparison of Col and Ler lines reveals the dynamic nature of Arabidopsis chromosomes. Nucleic Acids Res 37: 3189–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]