Biosynthesis of tricin requires specific hydroxylation catalyzed by a phylogenetically distinct hydroxylase.
Abstract
Flavones are ubiquitously accumulated in land plants, but their biosynthesis in monocots remained largely elusive until recent years. Recently, we demonstrated that the rice (Oryza sativa) cytochrome P450 enzymes CYP93G1 and CYP93G2 channel flavanones en route to flavone O-linked conjugates and C-glycosides, respectively. In tricin, the 3′,5′-dimethoxyflavone nucleus is formed before O-linked conjugations. Previously, flavonoid 3′,5′-hydroxylases belonging to the CYP75A subfamily were believed to generate tricetin from apigenin for 3′,5′-O-methylation to form tricin. However, we report here that CYP75B4 a unique flavonoid B-ring hydroxylase indispensable for tricin formation in rice. A CYP75B4 knockout mutant is tricin deficient, with unusual accumulation of chrysoeriol (a 3′-methoxylated flavone). CYP75B4 functions as a bona fide flavonoid 3′-hydroxylase by restoring the accumulation of 3′-hydroxylated flavonoids in Arabidopsis (Arabidopsis thaliana) transparent testa7 mutants and catalyzing in vitro 3′-hydroxylation of different flavonoids. In addition, overexpression of both CYP75B4 and CYP93G1 (a flavone synthase II) in Arabidopsis resulted in tricin accumulation. Specific 5′-hydroxylation of chrysoeriol to selgin by CYP75B4 was further demonstrated in vitro. The reaction steps leading to tricin biosynthesis are then reconstructed as naringenin → apigenin → luteolin → chrysoeriol → selgin → tricin. Hence, chrysoeriol, instead of tricetin, is an intermediate in tricin biosynthesis. CYP75B4 homologous sequences are highly conserved in Poaceae, and they are phylogenetically distinct from the canonical CYP75B flavonoid 3′-hydroxylase sequences. Recruitment of chrysoeriol-specific 5′-hydroxylase activity by an ancestral CYP75B sequence may represent a key event leading to the prevalence of tricin-derived metabolites in grasses and other monocots today.
Flavonoids are secondary metabolites widespread in land plants, but their composition is often unique in different plant lineages. Tricin (a 3′,5′-dimethoxyflavone) is a flavonoid typically distributed in sedges, palms, and grasses, including cereal crops such as rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), maize (Zea mays), and sorghum (Sorghum bicolor; Zhou and Ibrahim, 2010). Commonly present as O-linked conjugates, tricin and its derivatives serve as allelochemicals (Kong et al., 2010) and insect deterrents (Ling et al., 2007). Very recently, tricin was demonstrated to be a flavonoid monomer in monocot lignin, representing the first nonmonolignol monomer involved in lignification (Lan et al., 2015). In addition, tricin is widely recognized as a promising multifunctional nutraceutical because of its antioxidant (Duarte-Almeida et al., 2007), anticancer (Cai et al., 2007), anti-inflammatory (Shalini et al., 2012), and cardiovascular-protective (Chang et al., 2010) properties. The lipophilicity nature of methoxylated flavones is believed to enhance their access to target cells and tissues (Walle, 2007). In cereal, tricin accumulates mainly in vegetative tissues (leaves and stem; Zhou and Ibrahim, 2010), grain bran, and husk (Moheb et al., 2013). Elucidation of the metabolic steps for tricin biosynthesis will facilitate efforts to genetically engineer crop plants for tricin fortification and help understand how this unique flavonoid pathway is evolved in grasses or monocot.
Flavone biosynthesis in monocots was largely elusive until recent years. Chalcone synthase is a universal enzyme in land plants catalyzing the committed step for flavonoid production (Shih et al., 2008). The resulting chalcones are isomerized to flavanones, the common precursors for all major flavonoid classes. In dicots, two enzymatically distinct flavone synthases (FNSs), FNSI and FNSII, have been described for the conversion of flavanones to flavones (Martens and Mithöfer, 2005). FNSIs are soluble Fe2+/oxoglutarate-dependent dioxygenases restricted to Apiaceae, while FNSIIs are cytochrome P450 enzymes (CYP93B subfamily) identified in other flavone-accumulating dicots. Recently, we established CYP93G1 and CYP93G2 as branch point enzymes channeling flavanones to the formation of tricin O-linked conjugates (glycosides and flavanolignans) and flavone (apigenin, luteolin, and chrysoeriol) C-glycosides, respectively, in rice (Du et al., 2010; Lam et al., 2014). CYP93G1 functions as an FNSII, which generates flavone aglycones for different O-linked modifications (Lam et al., 2014). On the other hand, CYP93G2 is a flavanone 2-hydroxlase, which produces 2-hydroxyflavanones for immediate C-glycosylation, followed by the formation of the flavone nucleus (Du et al., 2010). Our previous work essentially confirmed the early speculation that separate biosynthetic routes (Wallace et al., 1969; Wallace and Grisebach, 1973) are responsible for these two predominant types of flavone-derived metabolites present in a variety of monocot species.
Tricin occurs naturally as O-glycosides or O-linked flavanolignans in grasses (Zhou and Ibrahim, 2010; Lam et al., 2014; Yang et al., 2014). In flavonoid biosynthesis, O-linked modifications generally occur as terminal steps after the final aglycone structures are formed (Winkel-Shirley, 2001). As a preferred substrate for CYP93G1 in planta, naringenin is desaturated to form apigenin (Lam et al., 2014), which requires further modifications to generate the tricin aglycone (Fig. 1). Tricetin (a 3′,5′-dihydroxylated flavone) has been proposed to be a precursor for tricin production (Cummins et al., 2006; Zhou and Ibrahim, 2010). In fact, sequential 3′,5′-O-methylation of tricetin to selgin (3′-methoxylated) and then tricin was demonstrated in vitro using recombinant O-methyltransferases (OMTs) isolated from rice (Kim et al., 2006; Lee et al., 2008) or wheat (Zhou et al., 2006, 2009). Accordingly, apigenin needs to be first converted to tricetin by a flavonoid 3′,5′-hydroxylase (F3′5′H) in a pathway leading to tricin formation. The vast majority of characterized F3′5′Hs are CYP enzymes belonging to the CYP75A subfamily (Seitz et al., 2006). Interestingly, several F3′5′Hs in Asteraceae fall into the same subfamily (CYP75B) with canonical flavonoid 3′-hydroxylases (F3′Hs), suggesting the recruitment of F3′5′H from F3′H members within this plant family (Seitz et al., 2006). F3′5′H activities enable plants to synthesize delphinidin-based anthocyanins, which confer violet/blue color in flowers and fruits, potentially a selective advantage in angiosperms for attraction of pollinators and seed dispersers (Harborne, 2014). A number of dicot F3′5′H genes have been isolated for engineering flower colors in plants without violet/blue pigmentation, such as roses (Rosa hybrida; Katsumoto et al., 2007), chrysanthemums (Chrysanthemum morifolium; Brugliera et al., 2013; Noda et al., 2013), and carnations (Dianthus caryophyllus; Tanaka and Brugliera, 2013). On the other hand, suppression of F3′5′H expression in torenia (Torenia hybrida; Suzuki et al., 2000) and cyclamen (Cyclamen persicum; Boase et al., 2010) was shown to shift the petal colors from violet/blue to red.
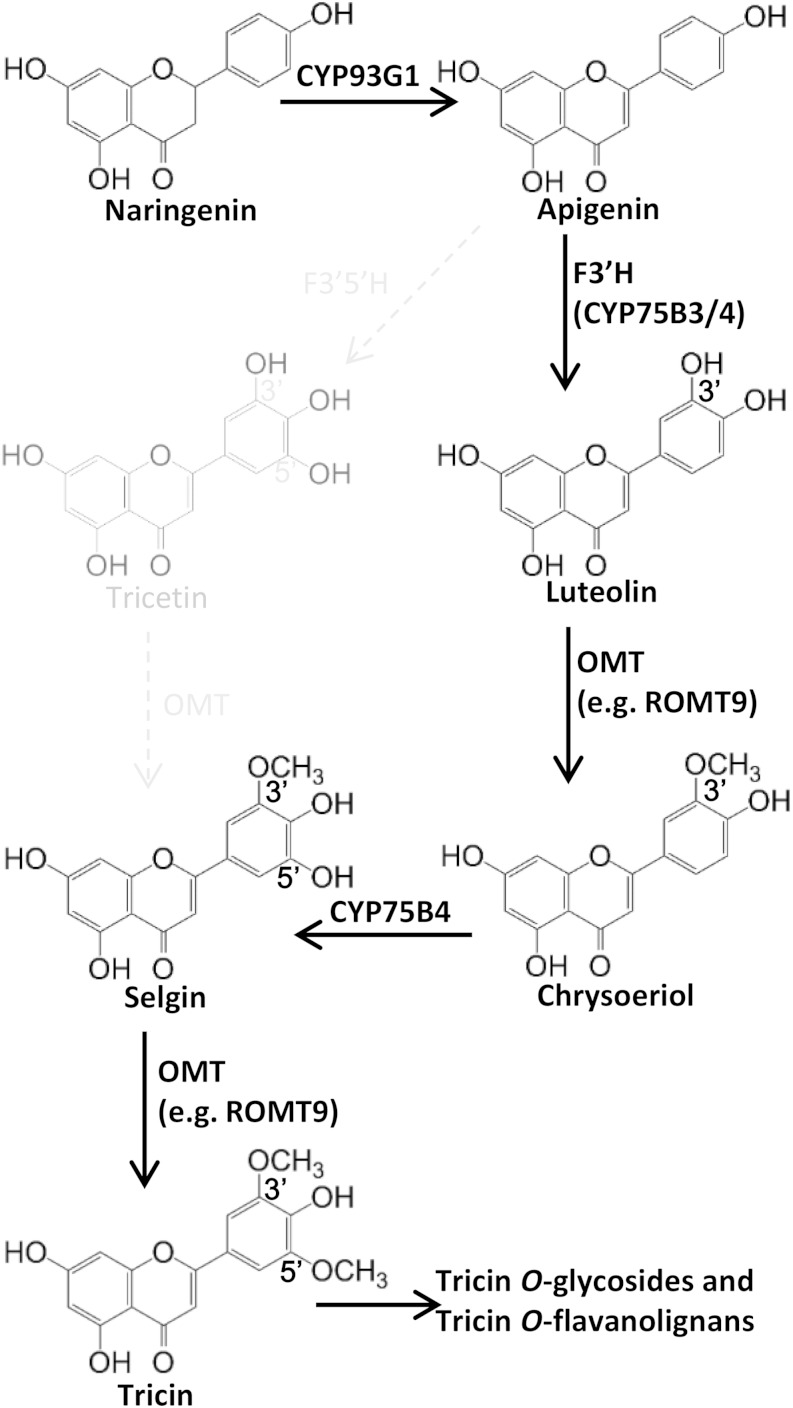
Figure 1.
Tricin biosynthesis pathway. Tricin occurs exclusively as O-linked conjugates in grasses, and the flavone nucleus is formed before O-linked modifications. Naringenin is desaturated to apigenin by CYP93G1, which functions as an FNSII. Previously, tricetin was proposed to be an intermediate in the pathway (illustrated in light color and dotted arrows). In this study, chrysoeriol is established as an intermediate during the biotransformation of apigenin to tricin. Hence, apigenin is converted to luteolin by an F3′H (CYP75B3 or CYP75B4) and then to chrysoeriol by an OMT (e.g. ROMT9). CYP75B4 functions as a chrysoeriol 5′-hydroxylase, which generates selgin, the immediate precursor of tricin.
The metabolic roles of F3′5′H in flavone biosynthesis, however, are not well understood. Recombinant CYP75A F3′5′H enzymes isolated from petunia (Petunia hybrida), Catharanthus roseus (Kaltenbach et al., 1999), and tomato (Solanum lycopersicum; Olsen et al., 2010) showed broad substrate specificity in vitro, producing 3′,5′-hydroxylated flavonoids, including flavones. Recently, tricin engineering in the flavonoid-deficient rice endosperm was achieved by overexpressing a cascade of genes encoding Viola spp. F3′5′H and soybean (Glycine max) FNSII as well as rice OMT, chalcone synthase, and Phe ammonia lyase (Ogo et al., 2013). However, endogenous F3′5′H genes in grasses essential for the widespread occurrence of tricin and its derivatives in vegetative tissues remain unidentified. In this study, we have filled the remaining gap in the tricin biosynthesis pathway through the functional characterizations of CYP75B4, which belongs to the same subfamily as the canonical F3′H enzymes. Rice CYP75B4 mutants are deficient in tricin production, while transgenic Arabidopsis (Arabidopsis thaliana) plants coexpressing CYP75B4 and CYP93G1 accumulate 5′-modified flavones. Further analysis revealed that CYP75B4 is a bona fide F3′H accepting different substrates. However, it also showed 5′-hydroxylase activity that was restricted to chrysoeriol (a 3′-methoxylated flavone). Hence, tricin biosynthesis in rice involves chrysoeriol, instead of tricetin, as an intermediate. As CYP75B4 is highly conserved in Poaceae, the 5′-hydroxylation of chrysoeriol to selgin is likely to represent a reaction specifically recruited for the production of tricin and its derivatives during the speciation of grasses or monocots.
RESULTS
Characterization of CYP75A11 in Yeast and Arabidopsis
Rice contains a single CYP75A member (CYP75A11), which is encoded by Os03g25150, presenting an apparent target for investigation of F3′5′H activities in relation to tricin biosynthesis. CYP75A11 expression was detected in rice seedlings from which the full-length complementary DNA (cDNA) was isolated and cloned into a yeast (Saccharomyces cerevisiae) expression vector. CYP75A11-expressing microsomes were assayed against different flavanones, flavones, dihydroflavonols, and flavonols. However, no F3′H or F3′5′H activities were detected for all the substrates tested (Supplemental Table S1). A well-characterized F3′5′H (CYP75A1) isolated from petunia petals was included as a positive control. In planta functions of CYP75A11 were then investigated by overexpression in an Arabidopsis transparent testa7 (tt7) mutant, which is deficient in F3′H activities. Consistent with the enzyme assay data, no complementation of seed coat and pigment phenotypes was observed (Supplemental Fig. S1A). Further metabolite analysis revealed the lack of 3′-hydroxylated and 3′,5′-hydroxylated flavonols in the CYP75A11 transgenic tt7 plants (Supplemental Fig. S1, B and C). These results indicated that CYP75A11 did not show F3′H or F3′5′H activities under our expression and assay conditions.
Flavone Profiling of Rice CYP75B4 Mutant Seedlings
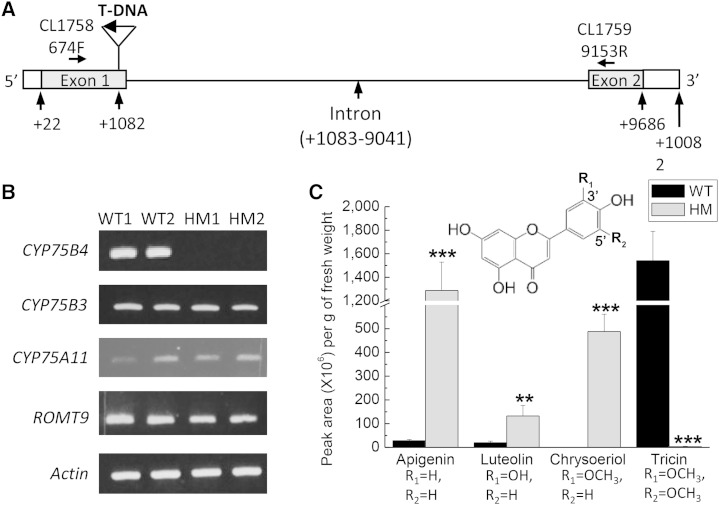
As an ongoing effort in our laboratory, we have been screening rice mutants for unique metabolite profiles to unravel new enzymatic functions involved in flavonoid biosynthesis. We report here a rice transfer DNA (T-DNA) insertion mutant for Os10g16974, which encodes CYP75B4, a close homolog of the rice F3′H (CYP75B3) encoded by Os10g17260 (Shih et al., 2008), with 68% sequence identity at amino acid level. The insertion occurs at the end of the first exon (Fig. 2A), and reverse transcription (RT)-PCR analysis confirmed the absence of an intact CYP75B4 transcript in the homologous insertion (HM) lines (Fig. 2B). On the other hand, both CYP75B3 and CYP75A11 were expressed normally in the HM plants (Fig. 2B), and no mutation was detected in their corresponding cDNA sequences (data not shown). Metabolites were then extracted from 3-week-old seedlings of wild-type and HM sibling lines derived from a hemizygous insertion parent. Liquid chromatography (LC)-mass spectrometry (MS) flavone profiling revealed the complete depletion of tricin derivatives in HM seedlings, while they were accumulated abundantly in wild-type seedlings (Fig. 2C). By contrast, HM seedlings accumulate substantially higher amounts of apigenin (45-fold) and luteolin (7-fold) than wild-type seedlings. Furthermore, chrysoeriol was detected in HM but not wild-type seedlings. Collectively, the CYP75B4 insertion mutant produces 3′-hydroxylated and methoxylated flavones (luteolin and chrysoeriol, respectively) but not 5′-modified flavones, strongly suggesting a role of this unexplored cytochrome P450 enzyme in 5′-hydroxylation of the flavonoid B-ring.
Figure 2.
Analysis of the rice CYP75B4 T-DNA insertion mutant. A, Gene structure of CYP75B4 (Os10g16974). The T-DNA is inserted in the first exon of the gene in the mutant. B, RT-PCR gene expression analysis of HM and wild-type (WT) seedlings. In the HM samples, the CYP75B4 RT-PCR product was absent when primers CL1758 and CL1759 were used. The expression of CYP75B3, CYP75A11, and ROMT9 was not affected. C, Relative quantities of apigenin, luteolin, chrysoeriol, and tricin detected in acid-hydrolyzed HM and wild-type extracts. Error bars represent sd (n = 5; **, P < 0.01; and ***, P < 0.001 by Student’s t test).
In addition to F3′5′H, the formation of tricin requires 3′- and 5′-OMT activities. The rice OMT, ROMT9 (Os08g06100), was previously demonstrated to be a flavonoid B-ring-specific OMT in enzyme activity assays (Kim et al., 2006). Here, a rice T-DNA insertion mutant for ROMT9 was also analyzed as described above (Supplemental Fig. S2). The HM lines showed 46% reduction in tricin accumulation compared with wild-type sibling lines, while luteolin and selgin were increased by 6 and 9.5 times, respectively. These data indicated that ROMT9 is a major OMT required for tricin biosynthesis in rice.
Transgenic Analysis of CYP75B4 in Arabidopsis Flavonoid Mutants
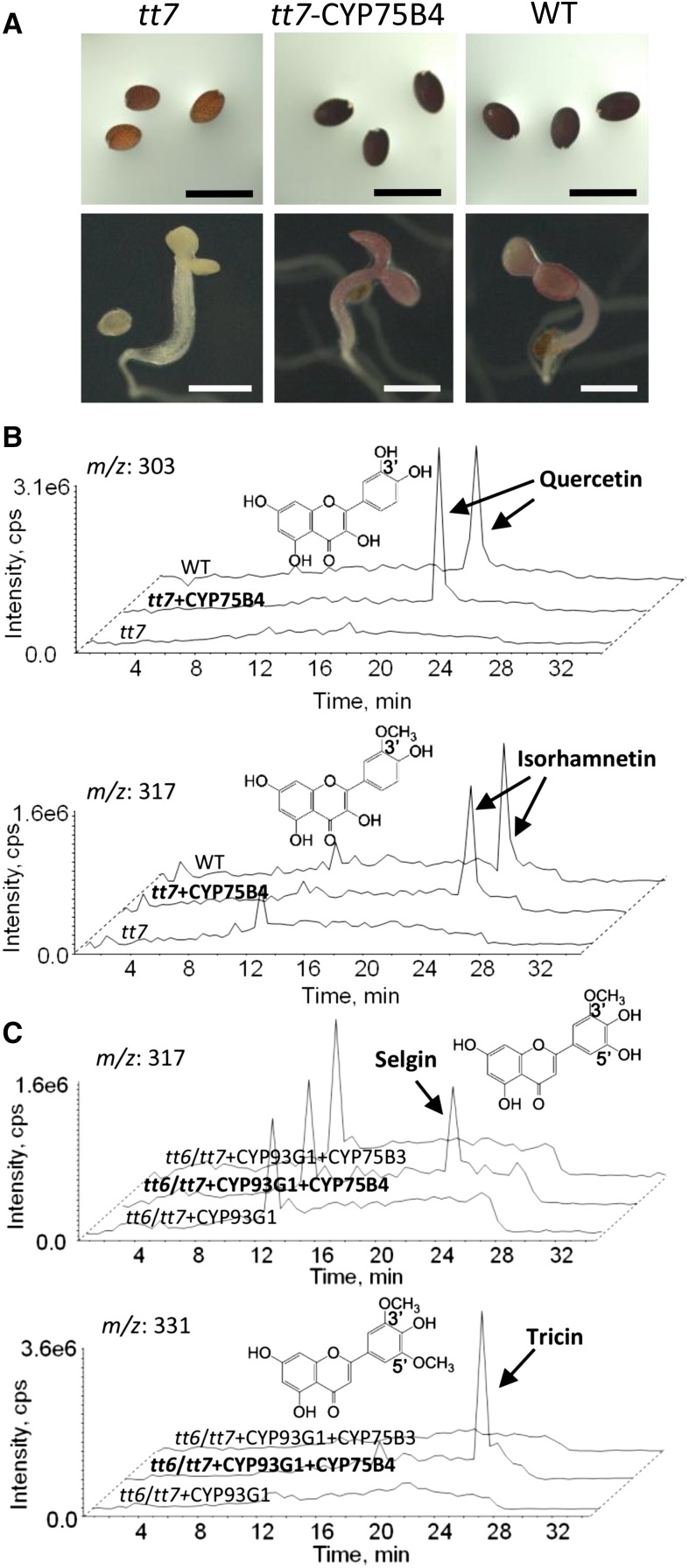
In planta functions of CYP75B4 were first evaluated by complementation analysis in an Arabidopsis tt7 mutant as described above. Transgenic expression of CYP75B4 in the mutant restored its brown seed coat and purple pigmentation under nitrogen stress (Fig. 3A). LC-tandem mass spectrometry (MS/MS) metabolite analysis further revealed the accumulation of 3′-modified flavonols (quercetin and isorhamnetin) in CYP75B4-overexpressing tt7 plants (Fig. 3B). Formation of isorhamnetin is likely to be resulted from endogenous OMT activities in Arabidopsis. We also searched for the presence of 3′,5′-modified flavonols (myricetin, laricitrin, and syringetin), but none of them were detected in the transgenic plant samples (Supplemental Fig. S3C). Hence, CYP75B4 is only able to introduce 3′-hydroxylation, but not 5′-hydroxylation, to the endogenous flavonoids in Arabidopsis.
Figure 3.
Transgenic analysis of CYP75B4 in the Arabidopsis mutants. A, CYP75B4 expression restored the ability of tt7 plants to accumulate proanthocyanin in seed coats and purple anthocyanin when grown on a medium devoid of nitrogen. B, CYP75B4 expression restored the accumulation of 3′-modified flavonols in tt7 plants. C, CYP75B4 plus CYP93G1 expression resulted in the accumulation of 5′-modified flavones in the tt6/tt7 double mutant. LC-MS chromatograms are representatives of at least three independent experiments. Metabolites in acid-hydrolyzed extracts were identified by retention time and MS/MS spectra in comparison with authentic standards. Selgin identification is described in Figure 4 and Supplemental Figure S4. WT, Wild type; cps, counts per second. Bar = 1 mm.
To further determine whether CYP75B4 is required for 5′-modification of flavones, it was coexpressed with CYP93G1 (a rice FNSII gene) in an Arabidopsis tt6/tt7 double mutant. Arabidopsis does not synthesize flavones naturally, and the tt6 mutation resulted in flavanone 3-hydroxylase deficiency, allowing higher flux of flavanones toward flavone formation in the presence of CYP93G1. Previously, CYP93G1-expressing tt6 plants were shown to produce apigenin, luteolin, and chrysoeriol (Lam et al., 2014). In addition to these flavones, selgin (5′-hydroxylated) and tricin (5′-methoxylated) were both detected in tt6/tt7 plants overexpressing CYP93G1 plus CYP75B4 (Fig. 3C). These 5′-modified flavones were not produced when CYP93G1 was coexpressed with CYP75B3 in the tt6/tt7 double mutant (only apigenin, luteolin, and chrysoeriol were detected; data not shown). Apparently, 5′-hydroxylation by CYP75B4 is highly specific to flavones (generated by CYP93G1) in the transgenic plants. On the other hand, endogenous OMT activities in Arabidopsis are likely to be responsible for the O-methylation reactions in the formation of selgin and tricin.
CYP75B4 in Vitro Enzyme Activity Assays
CYP75B4 was heterologously expressed in yeast cells, and microsomes were prepared for enzyme activity assays. Using flavonoids as substrates, CYP75B4 was found to catalyze their conversion to different 3′-hydroxylated products, i.e. apigenin to luteolin, naringenin to eriodictyol, and kaempferol to quercetin (Supplemental Table S1). However, prolonged incubation of these flavonoids with CYP75B4 did not result in the formation of 3′,5′-hydroxylated products. By contrast, both 3′- and 3′,5′-hydroxylated products were detected when CYP75A1 (petunia F3′5′H) was used in those reactions. The substrate range was extended to 3′-hydroxylated flavonoids in the CYP75B4 enzyme assays, but again no 3′,5′-hydroxylated products were identified (Supplemental Table S1).
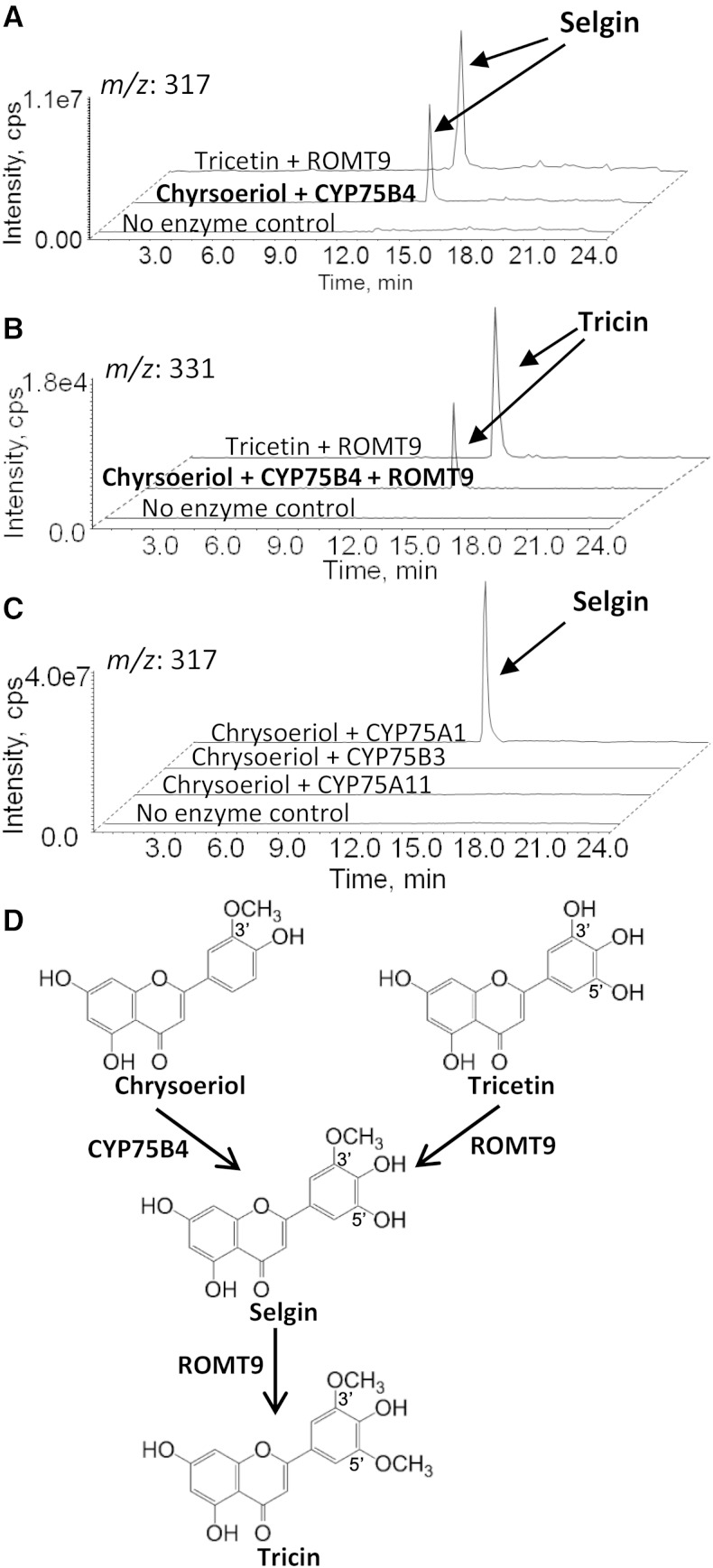
Interestingly, when CYP75B4-expressing microsomes were incubated with chrysoeriol (a 3′-methoxylated flavone), a major [M+H]+ ion (mass-to-charge ratio [m/z] 317) consistent with a selgin (5′-hydroxylated) ion was detected by LC-MS analysis (Fig. 4A). Because selgin is not available commercially, a purified recombinant rice OMT (ROMT9) was used to convert tricetin to selgin and tricin (Kim et al., 2006). The chrysoeriol plus CYP75B4 reaction product showed consistent retention time (Fig. 4A) and MS/MS spectrum (Supplemental Fig. S4) with those obtained for selgin generated in the tricetin plus ROMT9 reaction. Furthermore, incubation of the chrysoeriol plus CYP75B4 reaction product with ROMT9 resulted in the formation of tricin (Fig. 4B), further confirming its identity as selgin. Such product was also detected when CYP75A1-expressing microsomes (but not CYP75B3 or CYP75A11) were used in the chrysoeriol assay (Fig. 4C). CYP75B4 was further tested against other 3′-methoxylated flavonoids, including homoeriodictyol and isorhamnetin, but no 5′-hydroxylated products were detected (Supplemental Table S1). By contrast, these substrates were accepted by CYP75A1 for 5′-hydroxylation. Taken together, it is evident that the 5′-hydroxylation activity of CYP75B4 is highly specific to chrysoeriol.
Figure 4.
LC-MS analysis of in vitro enzyme assays. A, A major peak at m/z 317 was detected in the CYP75B4 reaction containing chrysoeriol as a substrate. The same peak was identified when tricetin was incubated with ROMT9, and the expected product was selgin. MS/MS spectra for the two products were identical (Supplemental Fig. S4). B, Addition of ROMT9 to the chrysoeriol plus CYP75B4 reaction further converted selgin to tricin. The same product was also detected in the tricetin plus ROMT9 reaction after prolonged incubation. C, No selgin was detected when chrysoeriol was incubated with CYP75A11 or CYP75B3. CYP75A1 (petunia F3′5′H) was used as a positive enzyme control. D, Enzymatic reaction steps for A and B. LC-MS chromatograms are representatives of at least three independent experiments. cps, Counts per second.
Phylogenetic Analysis of CYP75A and CYP75B Sequences
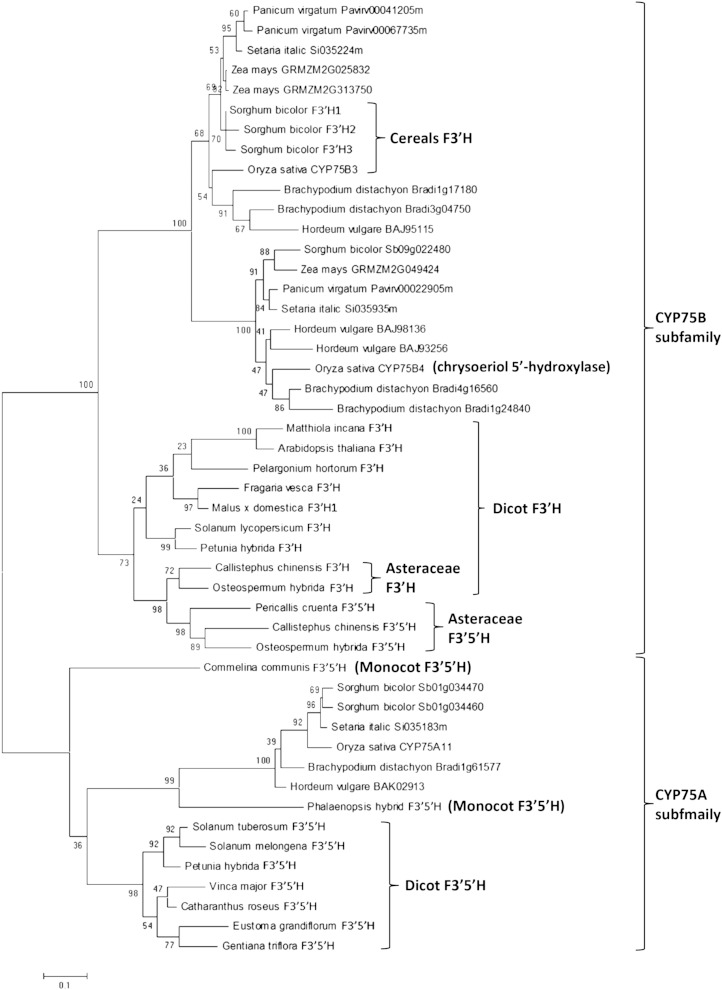
A phylogenetic analysis of F3′H and F3′5′H sequences from taxonomically diverse plant species was inferred using the maximum-likelihood method (Fig. 5). The phylogeny tree clearly shows two distinct clusters of CYP75A and CYP75B sequences. Each cluster contains separate clades of dicot and monocot members, implicating the functional divergence of F3′H and F3′5′H from an ancestral angiosperm CYP75 sequence. The monocot CYP75A members include CYP75A11 from rice (this study) as well as functional F3′5′Hs from a Phalaenopsis spp. orchid (Su and Hsu, 2003; Qi et al., 2013) and Commelina communis (Yuki et al., 2007). The C. communis F3′5′H was found to be primitive to all the other CYP75A members, suggesting that it may undergo independent evolution. In the CYP75B subfamily, a number of sequences from Poaceae, including several cereals, forage, and energy crops, are highly conserved with CYP75B4. These sequences are closely related to, but phylogenetically distinct from, CYP75B3 (a canonical F3′H) and its homologs from different grass species. These results strongly indicate the acquisition of chrysoeriol 5′-hydroxylase activities by CYP75B4 and related sequences in the monocot lineages after their divergence from the dicot lineages. An apparently similar scenario is found among the Asteraceae CYP75B sequences in which the F3′5′H members were clustering away from their sister F3′H members. Hence, distinct evolutionary events giving rise to 5′-hydroxylation in flavonoids appear to have occurred in Poaceae and Asteraceae independently.
Figure 5.
Phylogenetic analysis of CYP75A and CYP75B sequences. The unrooted tree was constructed by maximum-likelihood method using MEGA6. Bootstrapping with 1,000 replications was performed. Canonical F3′5′H and F3′H enzymes belong to CYP75A and CYP75B subfamilies, respectively. Functionally characterized F3′H and F3′5′H are annotated. Acquisition of 5′-hydroxylase activity by CYP75B members occurred independently in Poaceae and Asteraceae. Bar = 0.1 substitution per site.
DISCUSSION
Over the years, strong interests in engineering blue pigmentation in flowering plants have driven a large number of investigations on F3′5′Hs in diverse species. The majority of known F3′5′Hs are members of the CYP75A subfamily. Phylogenetic analysis implicated that the divergence of CYP75A and CYP75B (F3′H) members predated the speciation of angiosperms (Seitz et al., 2006). However, genome sequences of several dicot plants, such as Arabidopsis, carnation, and a few Rosaceae species, do not harbor any CYP75A homologous sequences, consistent with their absence of 5′-modified flavonoids (Tanaka and Brugliera, 2014). Hence, some plant lineages may have lost the CYP75A genes and the encoded F3′5′H enzyme activities during the course of their evolution (Tanaka and Brugliera, 2014). Interestingly, such loss was apparently compensated in some Asteraceae species (e.g. Pericallis cruenta, Osteospermum hybrida, and Callistephus chinensis) by recruitment of F3′5′H genes within the CYP75B subfamily, restoring their ability to synthesize delphinidin-type anthocyanin (Seitz et al., 2006). In monocots, the only CYP75A members reported to be functional F3′5′Hs were isolated from Phalaenopsis spp. (Su and Hsu, 2003; Qi et al., 2013) and C. communis (Yuki et al., 2007), presumably responsible for the violet/blue pigmentation in their petals. Other monocot families, such as Amaryllidaceae and Iridaceae, also produce violet/blue flowers. Moreover, some wheat grains were reported to accumulate delphinidin- and malvidin-based anthocyanins (Ficco et al., 2014). Whether CYP75A enzymes are involved in 3′,5′-hydroxylation of flavonoids in these monocot species remains to be elucidated.
Biosynthesis of tricin was generally considered to involve the intermediacy of tricetin (Cummins et al., 2006; Zhou and Ibrahim, 2010). The proposal was in accordance with the sequential O-methylation of tricetin by cereal OMTs to selgin and tricin demonstrated in vitro (Kim et al., 2006; Zhou et al., 2006, 2009; Lee et al., 2008). Recently, we described the accumulation of naringenin, but not eriodictyol or dihydrotricetin, when FNSII functions were blocked in rice CYP93G1 mutant seedlings, suggesting that the B-ring modifications take place after apigenin is formed from naringenin (Lam et al., 2014). In this study, our results indicated that CYP75A11 is unlikely to be involved in 5′-hydroxylation for tricin biosynthesis in rice. Apparently, the prevalent occurrence of tricin and its derivatives demanded an independent origin of F3′5′H activities in the grass family, or at least in rice. In fact, we established that CYP75B4 is indispensable for tricin biosynthesis by metabolite profiling of a rice T-DNA insertion mutant (Fig. 2). Transgenic analysis in Arabidopsis further suggested that CYP75B4 introduced 5′-hydroxylation in flavones in planta (Fig. 3). However, CYP75B4-expressing yeast microsomes converted apigenin to luteolin without further hydroxylation to produce tricetin. Similar 3′-hydroxylation activities were detected when other substrates of other flavonoid classes were assayed. Intriguingly, CYP75B4 catalyzed the specific 5′-hydroxylation of chrysoeriol to selgin (Fig. 4). This unique catalytic activity is consistent with the unusual accumulation of chrysoeriol in the CYP75B4 mutant seedlings.
Collectively, we have now completed the elucidation of the metabolic steps in tricin biosynthesis (Fig. 1). As opposed to the earlier proposals (Cummins et al., 2006; Zhou and Ibrahim, 2010), chrysoeriol, instead of tricetin, is an intermediate during the biotransformation of apigenin to tricin. Naringenin is first desaturated to apigenin by CYP93G1, which functions as an FNSII. Afterward, CYP75B3 or CYP75B4 (both are functional F3′Hs) catalyzes the 3′-hydroxylation to generate luteolin. Several wheat and rice OMTs, including TaOMT1, TaOMT2, ROMT9, ROMT15, and ROMT17, are able to modify luteolin to produce chrysoeriol (Kim et al., 2006; Zhou et al., 2006, 2009; Lee et al., 2008). CYP75B4 then participates as a 5′-hydroxylase in the newly explored step that converts chrysoeriol to selgin. Finally, the 5′-hydroxyl group in selgin can be O-methylated by ROMT9 or other OMTs to produce tricin (Kim et al., 2006; Zhou et al., 2006, 2009; Lee et al., 2008), followed by different O-linked modifications. The involvement of ROMT9 in tricin biosynthesis is supported by metabolite profiling of a T-DNA mutant, which showed reduced levels of tricin with simultaneous overaccumulation of luteolin and selgin (Supplemental Fig. S2).
Several studies have demonstrated the incorporation of tricin into cell wall lignin in a few monocot species (del Río et al., 2012; Rencoret et al., 2013; You et al., 2013). Recently, tricin was suggested to serve as a nucleation or polymer initiation site during lignification (Lan et al., 2015). Therefore, it is plausible that CYP75B4 mutation affects the formation and structures of lignin in rice. Because our CYP75B4 knockout mutant grows to maturity normally without observable phenotypes, it is not expected to have structural defects resulting from lignin deficiency. Given that lignin polymerization in many angiosperm species lacking tricin is naturally initiated by dimerization of conventional monolignols (Ralph et al., 2008), tricin depletion in our mutant is likely to shift the polymer starting moiety toward monolignol dimers and give rise to lignin polymers without tricin pendant units. In fact, it was suggested that lignin chains in maize also could be initiated by dimerization of acylated monolignols (Lan et al., 2015). Nevertheless, the existence of an independent biosynthetic pathway that channels tricin specifically for lignin formation in rice could not be excluded.
The acquisition of unique catalytic activity by a grass CYP75B member represents a recent evolutionary event distinct from the Asteraceae F3′5′H CYP75B sequences and the more ancient CYP75A enzymes. CYP75B4 remains a bona fide F3′H and plays redundant metabolic roles with CYP75B3. Hence, CYP75B4 mutants were still able to synthesize the 3′-modified flavones luteolin and chrysoeriol (Fig. 2). However, the specific 5′-hydroxylation of chrysoeriol by CYP75B4 restricts the 5′-modification to flavone aglycones in rice. By contrast, the CYP75B-derived F3′5′Hs in Asteraceae catalyzed sequential 3′- and 5′-hydroxylation of a range of flavonoid substrates (Seitz et al., 2006). The flavonoid B-ring was proposed to rotate after 3′-hydroxylation to allow 5′-hydroxylation to occur at the same active site in these F3′5′H enzymes (Seitz et al., 2007). However, the mechanism for the dual catalytic activities of CYP75B4 is potentially very different. After conversion from apigenin, luteolin does not undergo further hydroxylation and is instead released for O-methylation. The resulting chrysoeriol may dock the same or a different active site in CYP75B4 for the specific 5′-hydroxylation. Interestingly, the typical Thr-to-Ser or Thr-to-Ala substitution in the C-terminal region of F3′5′Hs versus F3′Hs (Seitz et al., 2007) is replaced by a Thr-to-Leu substitution in CYP75B4 and related sequences in grasses (Supplemental Fig. S5). Further biochemical characterizations will be necessary to generate mechanistic insights on this unique CYP flavonoid hydroxylase in the grass family.
Recruitment of lineage-specific metabolic pathways has resulted in the structural diversity of flavonoids among land plants. Previously, we established CYP93G1 and CYP93G2 as branch point enzymes channeling flavanones en route to flavone O-linked conjugates and C-glycosides, respectively (Du et al., 2010; Lam et al., 2014). They were believed to be derived from an ancestral CYP93G sequence following functional diversification (Lam et al., 2014). This study further revealed CYP75B4 as an evolution extension from F3′H with acquired chrysoeriol 5′-hydroxylase activity specific for the tricin biosynthesis pathway. These three unique P450 enzymes represent new molecular tools for metabolic engineering of crops that do not produce the health-beneficial flavones naturally. For example, coexpression of CYP93G1 plus CYP75B4 is sufficient to drive tricin biosynthesis in plants with endogenous flavanone production and OMT activities (Fig. 3). Interestingly, the absence of regular F3′5′H activities in rice is apparently due to the nonfunctional CYP75A11 rather than a gene loss event proposed for some dicot genomes. Finally, phylogenetic analyses revealed clusters of highly conserved homologs of CYP75A11, CYP75B3, and CYP75B4 (Fig. 5), as well as CYP93G1 (Lam et al., 2014) and CYP93G2 (Du et al., 2010), from the available grass genomes. Hence, our work also provides a framework for future molecular and evolutionary investigation of the biosynthetic pathways of different flavone-derived metabolites that are prevalent in many monocot species today.
MATERIALS AND METHODS
Rice T-DNA Insertion Mutants
Rice (Oryza sativa ssp. japonica) T-DNA insertion mutants of CYP75B4 (accession no. RMD-04Z11JZ82; cv Zhonghua 11) and ROMT9 (accession no. PFG_2B-50240; cv Hwayoung) were identified at the RiceGE database (http://signal.salk.edu/cgi-bin/RiceGE). CYP75B4 mutant seeds were acquired from the National Center of Plant Gene Research at Huazhong Agricultural University (Zhang et al., 2006). ROMT9 mutant seeds were acquired from the Crop Biotech Institute at Kyung Hee University. Seeds were incubated in 1% (v/v) nitric acid at room temperature in darkness for 20 to 24 h, washed three times in distilled water, and then incubated in darkness at 37°C for 2 d. Germinated seeds were transferred to rice growth medium (Yoshida et al., 1976) and incubated in a plant growth chamber (12 h of light at 28°C and 12 h of dark at 22°C). Genotypes of individual plants were determined by genomic PCR. Three-week-old seedlings were harvested for RNA and metabolite extraction.
Generation of Transgenic Arabidopsis Plants
Arabidopsis (Arabidopsis thaliana) tt6 mutants (SALK_113904), tt7 mutants (SALK_124157 and SALK 039417), and atomt1 mutants (SALK_020611) were obtained from the Arabidopsis Biological Resource Center. Double tt6/tt7 mutants were generated by crossing, and F2 homozygous lines were identified by genomic PCR. Full-length cDNA clones for CYP75B4 (AK070442) and ROMT9 (AK061859) were acquired from the National Institute of Agrobiological Sciences, and CYP75A11 cDNA was prepared from rice seedlings. The respective coding sequence was placed under the control of the Cauliflower mosaic virus 35S promoter and the nopaline 3′-terminator in the pCAMBIA1300 binary vector for overexpression in plants. The CYP93G1 binary vector is available from our previous work (Lam et al., 2014). Different expression constructs were transformed into the appropriate Arabidopsis mutants by the Agrobacterium tumefaciens strain GV3101 using floral dipping (Clough and Bent, 1998). Seeds harvested from transformed plants were germinated on Murashige and Skoog (Sigma; Murashige and Skoog, 1962) agar with 3% (w/v) Suc and 25 µg mL–1 hygromycin. Hygromycin-resistant individuals were transferred into soil and kept in growth chambers at 23°C (16 h of light and 8 h of dark). Successful transformants were confirmed by genomic PCR and RT-PCR. CYP93G1 transgenic plants were crossed with CYP75B4 transgenic plants (both T2 generation and tt6 background). Double transgenic (CYP93G1 plus CYP75B4) lines were identified in the F1 generation by genomic PCR.
For metabolite analysis, the transgenic seeds were surface sterilized and germinated on Murashige and Skoog agar without nitrogen source for 10 d at 23°C (16 h of light and 8 h of dark) to induce anthocyanin biosynthesis (Du et al., 2010). To detect proanthocyanidins (condensed tannin), seeds were incubated in a staining reagent containing 2% (w/v) dimethylaminocinnaldehyde and 3 m HCl-50% (w/v) methanol for 1 week (Shih et al., 2008). Seeds were washed three times by 70% (v/v) ethanol before observation.
RNA Extraction and RT-PCR Gene Expression Analysis
RNA samples were prepared from 3-week-old rice seedlings using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. First strand cDNA was synthesized from DNase I (Invitrogen)-treated RNA samples by a reverse transcriptase (M-MLV RT RNase [H–] Point Mutant; Promega) using an oligo(dT) primer. Expression of different genes was then analyzed by PCR using the cDNA templates and gene-specific primers. The PCR program was set as follows: preincubation (94°C for 10 min); 30 cycles of denaturation (94°C for 30 s), annealing (55°C for 30 s), and elongation (72°C for 1 min); and finial extension (72°C for 10 min).
Plant Metabolite Extraction
Rice seedlings (3 weeks old; 200 µg) and Arabidopsis seedlings (10 d old on N-depleted medium; 600 µg) were frozen by liquid nitrogen, ground into fine powder, and resuspended in HPLC-grade methanol (600 µL) for metabolite extraction. To release flavonoid aglycones from O-linked conjugates, acid hydrolysis was performed by incubation with an equal volume of 2 n HCl at 90°C for 1 h. The final preparations were analyzed by LC-MS/MS as described below.
CYP Protein Expression in Yeast and Enzyme Assays
The coding regions of CYP75A11, CYP75B3, CYP75B4, and CYP75A1 were cloned individually into the pYES2.1/V5-His-TOPO vector (Invitrogen). CYP75A1 cDNA was isolated from violet petals of petunia (Petunia hybrida; available in a local flower market) by RT-PCR. The different expression constructs were transformed into the yeast (Saccharomyces cerevisiae) strain INVSc1 (Invitrogen) according to the manufacturer’s instruction. Expression of the different P450 genes in transformed yeast cells was confirmed by RT-PCR. Yeast culture, induction of protein expression, and preparation of microsomes were performed as described previously (Du et al., 2010). Protein concentrations in the microsome preparations were measured by the Bradford method (Bio-Rad; Bradford., 1976). Enzyme assays were conducted by incubating 500 µg of microsomal protein in 100 mm potassium phosphate buffer (pH 7.0) containing 5 mm NADPH, 2 mm l-glutathione, and 100 µm flavonoid substrate at 30°C for 1 h. The reaction products were extracted twice in ethyl acetate, dried under vacuum, and resuspended in HPLC-grade methanol for LC-MS/MS analysis.
ROMT9 Protein Expression in Bacteria and Enzyme Assays
The coding region of ROMT9 amplified from a full-length cDNA clone (AK061859) was cloned into the pET28a vector (Novagen) and transformed into the Escherichia coli strain BL21. Bacterial culture, induction of protein expression, and purification of the His6-tag ROMT9 protein were performed according to the manufacturer’s instructions. For the OMT assay, 5 µg of the purified protein was incubated in 10 mm Tris-HCl (pH 7.5) buffer containing 2 mm dithiothreitol, 40 µm S-adenosyl-l-Met, and 100 µm flavonoid substrate at 37°C for 1 h. Reaction products were processed as described above for LC-MS/MS analysis.
LC-MS Analysis
Enzyme reaction products and plant extracts were filtered and separated on a Nucleosil 100-5 C18 column (5 μm, 150 × 2 mm; Agilent Technologies) connected to the HP1100 series HPLC system (Agilent Technologies). Separation of enzyme reaction products was performed using a solvent system of 0.5% (v/v) formic acid/water (A) and 0.5% (v/v) formic acid/methanol (B) with a linear gradient of 15% to 90% B over 15 min. Separation of acid-hydrolyzed plant extracts was carried out with a linear gradient of 10% to 60% B over 18 min. In both cases, the flow rate was maintained at 0.2 mL min–1 and the elution was analyzed by an AP3200-QTRAP mass spectrometer (AB SCIEX) operating in positive ionization mode. Enhanced product ion scan and MS/MS spectra were obtained and acquired as described previously (Lo et al., 2007; Shih et al., 2008). Compounds were identified by comparing their retention times and MS/MS spectra with those obtained for authentic standards available commercially.
Primers for Molecular Biology Experiments
Primers for genotyping of mutants and transgenic plants, molecular cloning, and RT-PCR analyses are listed in Supplemental Table S2.
Phylogenetic Analysis
Multiple sequence alignment was performed by ClustalW2. The unrooted phylogenetic tree was constructed by maximum-likelihood method using MEGA6. Bootstrapping with 1,000 replications was performed. GenBank accession numbers of the proteins are listed in Supplemental Table S3.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers AK070442 (CYP75B4).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transgenic analysis of CYP75A11 in Arabidopsis tt7 mutants.
Supplemental Figure S2. Analysis of the rice ROMT9 T-DNA insertion mutant.
Supplemental Figure S3. LC-MS analysis of 3′,5′-modified flavonols in CYP75B4-expressing Arabidopsis tt7 mutants.
Supplemental Figure S4. Identification of selgin ions (m/z 317) from different sources in this study.
Supplemental Figure S5. Multiple sequence alignment of CYP75A and CYP75B sequences (C-terminal region).
Supplemental Table S1. LC-MS analysis of CYP75A11, CYP75B4, and CYP75A1 enzyme assays using different flavonoids substrates.
Supplemental Table S2. Primers used for cloning, genotyping, and RT-PCR experiments.
Supplemental Table S3. GenBank accession numbers of proteins used for constructing the phylogenetic tree.
Supplementary Material
Acknowledgments
We thank Dr. Yuki Tobimatsu (Kyoto University) for helpful comments on the discussion of tricin and lignification.
Glossary
- OMT
O-methyltransferase
- F3′5′H
flavonoid 3′,5′-hydroxylase
- F3′H
flavonoid 3′-hydroxylase
- cDNA
complementary DNA
- T-DNA
transfer DNA
- RT
reverse transcription
- HM
homologous insertion
- LC
liquid chromatography
- MS
mass spectrometry
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region, China (grant no. HKU7736/11M).
References
- Boase MR, Lewis DH, Davies KM, Marshall GB, Patel D, Schwinn KE, Deroles SC (2010) Isolation and antisense suppression of flavonoid 3′,5′-hydroxylase modifies flower pigments and colour in cyclamen. BMC Plant Biol 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brugliera F, Tao GQ, Tems U, Kalc G, Mouradova E, Price K, Stevenson K, Nakamura N, Stacey I, Katsumoto Y, et al. (2013) Violet/blue chrysanthemums. Metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors. Plant Cell Physiol 54: 1696–1710 [DOI] [PubMed] [Google Scholar]
- Cai H, Boocock DJ, Steward WP, Gescher AJ (2007) Tissue distribution in mice and metabolism in murine and human liver of apigenin and tricin, flavones with putative cancer chemopreventive properties. Cancer Chemother Pharmacol 60: 257–266 [DOI] [PubMed] [Google Scholar]
- Chang CL, Wang GJ, Zhang LJ, Tsai WJ, Chen RY, Wu YC, Kuo YH (2010) Cardiovascular protective flavonolignans and flavonoids from Calamus quiquesetinervius. Phytochemistry 71: 271–279 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cummins I, Brazier-Hicks M, Stobiecki M, Fraǹski R, Edwards R (2006) Selective disruption of wheat secondary metabolism by herbicide safeners. Phytochemistry 67: 1722–1730 [DOI] [PubMed] [Google Scholar]
- del Río JC, Rencoret J, Prinsen P, Martínez ÁT, Ralph J, Gutiérrez A (2012) Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J Agric Food Chem 60: 5922–5935 [DOI] [PubMed] [Google Scholar]
- Du Y, Chu H, Chu IK, Lo C (2010) CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol 154: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte-Almeida JM, Negri G, Salatino A, de Carvalho JE, Lajolo FM (2007) Antiproliferative and antioxidant activities of a tricin acylated glycoside from sugarcane (Saccharum officinarum) juice. Phytochemistry 68: 1165–1171 [DOI] [PubMed] [Google Scholar]
- Ficco DBM, De Simone V, Colecchia SA, Pecorella I, Platani C, Nigro F, Finocchiaro F, Papa R, De Vita P (2014) Genetic variability in anthocyanin composition and nutritional properties of blue, purple, and red bread (Triticum aestivum L.) and durum (Triticum turgidum L. ssp. turgidum convar. durum) wheats. J Agric Food Chem 62: 8686–8695 [DOI] [PubMed] [Google Scholar]
- Harborne JB. (2014) Biochemistry of plant pollination. In Introduction to Ecological Biochemistry. Academic Press, London, pp 36–70 [Google Scholar]
- Kaltenbach M, Schröder G, Schmelzer E, Lutz V, Schröder J (1999) Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J 19: 183–193 [DOI] [PubMed] [Google Scholar]
- Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A, et al. (2007) Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 48: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67: 387–394 [DOI] [PubMed] [Google Scholar]
- Kong CH, Xu XH, Zhang M, Zhang SZ (2010) Allelochemical tricin in rice hull and its aurone isomer against rice seedling rot disease. Pest Manag Sci 66: 1018–1024 [DOI] [PubMed] [Google Scholar]
- Lam PY, Zhu FY, Chan WL, Liu H, Lo C (2014) Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol 165: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J (2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim BG, Chong Y, Lim Y, Ahn JH (2008) Cation dependent O-methyltransferases from rice. Planta 227: 641–647 [DOI] [PubMed] [Google Scholar]
- Ling B, Dong H, Zhang M, Xi D, Wang J (2007) Potential resistance of tricin in rice against brown planthopper. Nilaparvata lugens (Stål). Acta Ecol Sin 27: 1300–1306 [Google Scholar]
- Lo C, Le Blanc JC, Yu CK, Sze KH, Ng DC, Chu IK (2007) Detection, characterization, and quantification of resveratrol glycosides in transgenic Arabidopsis over-expressing a sorghum stilbene synthase gene by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 21: 4101–4108 [DOI] [PubMed] [Google Scholar]
- Martens S, Mithöfer A (2005) Flavones and flavone synthases. Phytochemistry 66: 2399–2407 [DOI] [PubMed] [Google Scholar]
- Moheb A, Grondin M, Ibrahim RK, Roy R, Sarhan F (2013) Winter wheat hull (husk) is a valuable source for tricin, a potential selective cytotoxic agent. Food Chem 138: 931–937 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Noda N, Aida R, Kishimoto S, Ishiguro K, Fukuchi-Mizutani M, Tanaka Y, Ohmiya A (2013) Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol 54: 1684–1695 [DOI] [PubMed] [Google Scholar]
- Ogo Y, Ozawa K, Ishimaru T, Murayama T, Takaiwa F (2013) Transgenic rice seed synthesizing diverse flavonoids at high levels: a new platform for flavonoid production with associated health benefits. Plant Biotechnol J 11: 734–746 [DOI] [PubMed] [Google Scholar]
- Olsen KM, Hehn A, Jugdé H, Slimestad R, Larbat R, Bourgaud F, Lillo C (2010) Identification and characterisation of CYP75A31, a new flavonoid 3′,5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol 10: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Lou Q, Quan Y, Liu Y, Wang Y (2013) Flower-specific expression of the Phalaenopsis flavonoid 3′,5′-hydroxylase modifies flower color pigmentation in Petunia and Lilium. Plant Cell Tiss Org 115: 263–273 [Google Scholar]
- Ralph J, Brunow G, Harris PJ, Dixon RA, Schatz PF, Boerjan W (2008) Lignification: are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? In Daayf F, El Hadrami A, Adam L, Balance GM, eds, Recent Advances in Polyphenol Research, Vol 1 Wiley-Blackwell Publishing, Oxford, pp 36–66 [Google Scholar]
- Rencoret J, Ralph J, Marques G, Gutiérrez A, Martínez Á, del Río JC (2013) Structural characterization of lignin isolated from coconut (Cocos nucifera) coir fibers. J Agric Food Chem 61: 2434–2445 [DOI] [PubMed] [Google Scholar]
- Seitz C, Ameres S, Forkmann G (2007) Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett 581: 3429–3434 [DOI] [PubMed] [Google Scholar]
- Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G (2006) Cloning, functional identification, and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol Biol 61: 365–381 [DOI] [PubMed] [Google Scholar]
- Shalini V, Bhaskar S, Kumar KS, Mohanlal S, Jayalekshmy A, Helen A (2012) Molecular mechanisms of anti-inflammatory action of the flavonoid tricin from Njavara rice (Oryza sativa L.) in human peripheral blood mononuclear cells: possible role in the inflammatory signaling. Int Immunopharmacol 14: 32–38 [DOI] [PubMed] [Google Scholar]
- Shih CH, Chu H, Tang LK, Sakamoto W, Maekawa M, Chu IK, Wang M, Lo C (2008) Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Su V, Hsu BD (2003) Cloning and expression of a putative cytochrome P450 gene that influences the colour of Phalaenopsis flowers. Biotechnol Lett 25: 1933–1939 [DOI] [PubMed] [Google Scholar]
- Suzuki KI, Xue HM, Tanaka Y, Fukui Y, Fukuchi-Mizutani M, Murakami Y, Katsumoto Y, Tsuda S, Kusumi T (2000) Flower color modifications of Torenia hybrida by cosuppression of anthocyanin biosynthesis genes. Mol Breed 6: 239–246 [Google Scholar]
- Tanaka Y, Brugliera F (2013) Flower colour and cytochromes P450. Philos Trans R Soc Lond B Biol Sci 368: 20120432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Brugliera F (2014) Metabolic engineering of flower color pathways using cytochromes P450. In Fifty Years of Cytochrome P450 Research. Springer, Tokyo, pp 207–229 [Google Scholar]
- Wallace JW, Grisebach H (1973) The in vivo incorporation of a flavanone into C-glycosylflavones. Biochim Biophys Acta 304: 837–841 [DOI] [PubMed] [Google Scholar]
- Wallace JW, Mabry TJ, Alston RE (1969) On the biogenesis of flavone O-glycosides and C-glycosides in the Lemnaceae. Phytochemistry 8: 93–99 [Google Scholar]
- Walle T. (2007) Methoxylated flavones, a superior cancer chemopreventive flavonoid subclass? Semin Cancer Biol 17: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nakabayashi R, Okazaki Y, Mori T, Takamatsu S, Kitanaka S, Kikuchi J, Saito K (2014) Toward better annotation in plant metabolomics: isolation and structure elucidation of 36 specialized metabolites from Oryza sativa (rice) by using MS/MS and NMR analyses. Metabolomics 10: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno D, Cock J, Gomez K (1976) Routine procedure for growing rice plants in culture solution. In Laboratory Manual for Physiological Studies of Rice. International Rice Research Institute, Los Banos, Philippines, pp 61–66 [Google Scholar]
- You TT, Mao JZ, Yuan TQ, Wen JL, Xu F (2013) Structural elucidation of the lignins from stems and foliage of Arundo donax Linn. J Agric Food Chem 61: 5361–5370 [DOI] [PubMed] [Google Scholar]
- Yuki S, Araki S, Suzuki T, inventors. March 15, 2007. Flavonoid-3′,5′-hydroxylase gene of Commedina Communis. U.S. Patent No. US8440879 B2
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S (2006) RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34: D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Gold ND, Martin VJJ, Wollenweber E, Ibrahim RK (2006) Sequential O-methylation of tricetin by a single gene product in wheat. Biochim Biophys Acta 1760: 1115–1124 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Ibrahim RK (2010) Tricin. A potential multifunctional nutraceutical. Phytochem Rev 9: 413–424 [Google Scholar]
- Zhou JM, Seo YW, Ibrahim RK (2009) Biochemical characterization of a putative wheat caffeic acid O-methyltransferase. Plant Physiol Biochem 47: 322–326 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.