Recent work highlights the functional roles of alternative DNA-dependent RNA polymerases and unique non-coding sequences in mediating paramutation behaviors in maize.
Abstract
Paramutations represent locus-specific trans-homolog interactions affecting the heritable silencing properties of endogenous alleles. Although examples of paramutation are well studied in maize (Zea mays), the responsible mechanisms remain unclear. Genetic analyses indicate roles for plant-specific DNA-dependent RNA polymerases that generate small RNAs, and current working models hypothesize that these small RNAs direct heritable changes at sequences often acting as transcriptional enhancers. Several studies have defined specific sequences that mediate paramutation behaviors, and recent results identify a diversity of DNA-dependent RNA polymerase complexes operating in maize. Other reports ascribe broader roles for some of these complexes in normal genome function. This review highlights recent research to understand the molecular mechanisms of paramutation and examines evidence relevant to small RNA-based modes of transgenerational epigenetic inheritance.
Transgenerational epigenetic changes affecting gene regulation have been implicated in agricultural performance, human disease, and evolution (Groszmann et al., 2013; Jablonka, 2013; Heard and Martienssen, 2014). Paramutation represents one well-studied mechanism for generating and establishing such changes (Hollick, 2010). Alexander Brink first applied the term paramutation to describe directed, yet reversible, changes in maize (Zea mays) kernel pigment patterns resulting from trans-homolog interactions (THI) of certain red1 (r1) alleles (Brink, 1958). Mottled pigmentation conditioned by the R-r:standard (R-r) allele is invariably suppressed after exposure to R-stippled (R-st) in heterozygotes (Brink, 1956). The suppressed R-r state (denoted R-rʹ ) is meiotically heritable, induces a similar change to naive R-r (secondary paramutation), and gradually reverts back to an R-r reference state when maintained in hemizygous conditions (Styles and Brink, 1969). These behaviors classify R-r as an epiallele having variable silencing properties influenced by THI. This type of non-Mendelian inheritance has remained a great unsolved mystery of genetics research and is a topic of considerable interest.
Paramutation has a genetic definition intended for invariable occurrences of heritable, although potentially reversible, changes of an allele when heterozygous with another specific allele of the same gene (Brink, 1958). Used in the same sense originally assigned to mutation, paramutation represents both the process and outcome of these THI without reference to specific molecular features. Alleles inducing paramutation, such as R-st and R-rʹ, are termed paramutagenic. Alleles susceptible to paramutation, like R-r, are paramutable, and alleles neither paramutagenic nor paramutable are termed neutral. Some neutral alleles, similar to deficiencies, facilitate the reversion of an existing paramutation to the paramutable reference state, while others do not (Gross and Hollick, 2007). These behaviors can generate and maintain wide phenotypic variation from specific allelic combinations and/or transgenerational conditioning at a single locus.
The generality of paramutation in other eukaryotes remains unclear. Well-described examples occur among certain alleles of several other maize loci (Table I), including those, like r1, encoding transcription factors responsible for anthocyanin pigment biosynthesis: booster1 (b1; Coe, 1959), purple plant1 (pl1; Hollick et al., 1995), and pericarp color1 (p1; Sidorenko and Peterson, 2001). Another example occurs at the maize low phytic acid1 locus (Pilu et al., 2009). Paramutation-like behaviors have also been noted in other plants, fungi, and metazoans (for review, see Chandler and Stam, 2004), including more recent examples in Mus musculus (Rassoulzadegan et al., 2006; Wagner et al., 2008; Worch et al., 2008; Grandjean et al., 2009; Yuan et al., 2015), Drosophila melanogaster (Seong et al., 2011; de Vanssay et al., 2012), and Caenorhabditis elegans (Ashe et al., 2012; Shirayama et al., 2012; Seth et al., 2013; Wedeles et al., 2013; Sapetschnig et al., 2015). A common feature for many of these examples is the possible involvement of RNA-based mechanisms related to basal RNA interference (RNAi) machinery (for review, see Castel and Martienssen, 2013). Here, we present an RNAi-based working model for paramutation in maize and discuss experimental results addressing key aspects of the model.
Table I. Characteristics defining examples of paramutation in maize.
N/A, Not applicable.
| Locus | Allele | Paramutation Properties |
Proteins Required fora |
|||||
|---|---|---|---|---|---|---|---|---|
| Paramutable | Paramutagenic | Efficiency | Reversion | Secondary Paramutation | Somatic Repression | Induction of Paramutation | ||
| red1 (r1) | R-r:standardb | Yesb | Yes (R-r')b | 100%b | Yesb,c | Yesd,e | RPD1f | RPD1f |
| RDR2g | ||||||||
| RP(D/E)2ah | ||||||||
| [RMR1]i | ||||||||
| [RMR2]j | ||||||||
| R-stippledb | Nob | Yesb | 100%b | N/A | N/A | N/A | RPD1f | |
| RDR2g | ||||||||
| RP(D/E)2ah | ||||||||
| [RMR1]i | ||||||||
| [RMR2]j | ||||||||
| booster1 (b1) | B1-Intensek | Yesk,l | Yes (B')k,l | 100% when induced by B'k,l; less than100% when induced by most transgenesm,n | Not when induced by B'k,l,o; yes if initiated by transgenesm,n | Yes when induced by B'k,l; variable if initiated by transgenesm,n | RDR2g | RDR2g |
| RPD1f | RPD1f | |||||||
| RP(D/E)2ah,p | RP(D/E)2ah,p | |||||||
| [RMR1]i | Parent-of-origin effect | |||||||
| [RMR2]j | [RMR1]i | |||||||
| purple plant1 (pl1) | Pl1-Rhoadesq | Yesq | Yes (Pl')q | 100%q | Yesq,r,s | Yesq | RDR2g | RMR2j |
| RMR1i | RDR2g | |||||||
| RPD1f | RPD1f | |||||||
| RP(D/E)2ah,p | [RMR1]i | |||||||
| RMR2j | [RP(D/E)2a]h,p | |||||||
| pericarp color1 (p1) | P1-rrt,u | Yest,u | Yes (P1-rr')t,u | 0%–95%t,u,v,w | Yest | Yest,u,v | Putative ufo1 proteinw | RDR2v |
| [RDR2]u | ||||||||
| P1-pru | Nou | Yesu | Variableu | Yesu | Yesu | Putative ufo1 proteinu,w | Not tested | |
| low phytic acid1 (lpa1) | lpa1-241x | Nox | Yesx | <100%x | Yesx | Nox | [RDR2]x | Not tested |
Proteins shown not to be required for the indicated paramutation behaviors are bracketed. bBrink (1956). cStyles and Brink (1969). dBrink et al. (1960). eBrown and Brink (1960). fHollick et al. (2005). gDorweiler et al. (2000). hSidorenko et al. (2009). iHale et al. (2007). jBarbour et al. (2012). kCoe (1959). lCoe (1966). mArteaga-Vasquez et al. (2010). nBelele et al. (2013). oChandler et al. (2000). pStonaker et al. (2009). qHollick et al. (1995). rHollick and Chandler (1998). sGross and Hollick (2007). tSidorenko and Peterson (2001). uGoettel and Messing (2013). vSidorenko and Chandler (2008). wSekhon et al. (2012). xPilu et al. (2009).
A WORKING MODEL FOR PARAMUTATION IN MAIZE
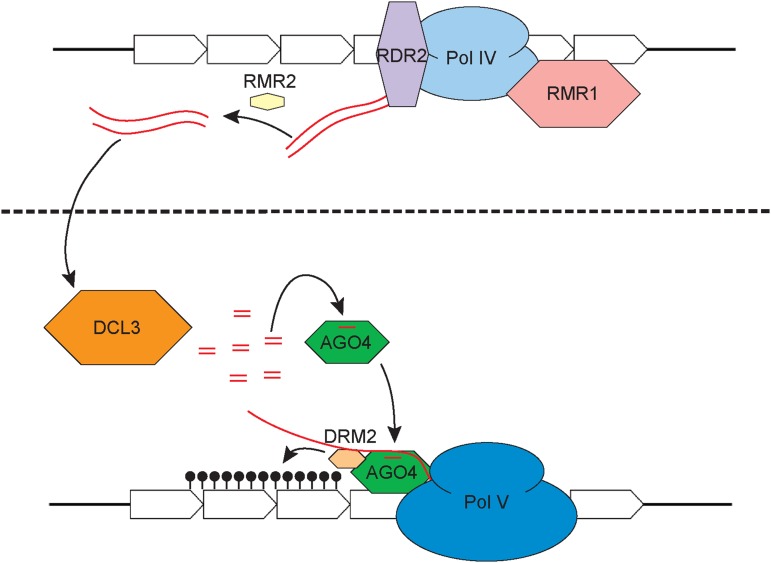
Forward genetic approaches have begun to identify molecules relevant to the paramutation mechanism. Recessive mutations of the required to maintain repression (rmr; Hollick and Chandler, 2001) and mediator of paramutation (mop; Dorweiler et al., 2000) loci identify functions required to maintain repressed pigmentation associated with paramutations (paramutant states) of pl1 and b1 alleles, respectively. A dominant mutation identifying the unstable factor for orange1 (ufo1) locus similarly affects b1 and p1 alleles (Sekhon et al., 2012). Among the estimated 14 rmr loci (J.B. Hollick, unpublished data), four encode orthologs of Arabidopsis (Arabidopsis thaliana) proteins required to generate 24-nucleotide RNAs, including catalytic subunits of a plant-specific DNA-dependent RNA polymerase IV (Pol IV; Erhard et al., 2009; Sidorenko et al., 2009; Stonaker et al., 2009), an RNA-dependent RNA polymerase (RDR2; Alleman et al., 2006), and a Rad54-like ATPase (RMR1; Hale et al., 2007). In addition to RMR2, a small (366-amino acid) pioneer molecule defining a plant-specific clade of novel proteins (Barbour et al., 2012), all these maize proteins are required for complete 24-nucleotide RNA biogenesis (Hale et al., 2007; Nobuta et al., 2008; Erhard et al., 2009; Stonaker et al., 2009; Barbour et al., 2012). In Arabidopsis, 24-nucleotide RNAs complexed with Argonaute-type proteins direct sequence-specific de novo cytosine methylation (5meC) through a process known as RNA-directed DNA methylation (RdDM; Matzke and Mosher, 2014; Matzke et al., 2015). Thus, current working models and hypotheses regarding the paramutation mechanisms in maize are largely based on a presumed RdDM framework (Fig. 1).
Figure 1.
An RdDM-type working model for paramutation. Maize components identified by mutations are presented above the dotted line, and presumed orthologs of select Arabidopsis RdDM proteins are presented below. In this schematic, Pol IV is transcribing a repetitive enhancer element (arrowed boxes), the nascent RNA is copied by RDR2, and the resulting double-stranded RNA (red lines) is cleaved by DCL3 to create 24-nucleotide RNAs. These small RNAs, in complex with argonaute4 (AGO4), facilitate the association with Pol V nascent RNAs and the recruitment of a de novo methyltransferase (domains-rearranged DNA methyltransferase2 [DRM2]) to accomplish site-directed cytosine methylation (black lollipops). All proteins are to relative scale based on amino acid content.
The general outline of an RdDM-type working model posits that important regulatory sequences of paramutagenic alleles are transcribed by a Pol IV:RDR2 complex (Haag et al., 2012; Li et al., 2015) to produce double-stranded RNAs. A dicer-like3 (DCL3) endoribonuclease cleaves these RNAs into 24-nucleotide fragments that are incorporated into an Argonaute protein and help recruit de novo DNA methyltransferases to identical or similar sequences in nascent transcripts produced from another DNA-dependent RNA polymerase (Pol V). In this model, 24-nucleotide RNAs represent a cache of epigenetic information that mediates THI with 24-nucleotide RNAs produced from a paramutagenic allele influencing 5meC patterns of a paramutable allele in trans. This model is supported by the requirement of the maize Pol IV largest subunit (rna polymerase d1 [RPD1]) and RDR2 for facilitating paramutation at multiple loci (for review, see Hollick, 2012); however, it fails to explain why other RMR components are either only involved in locus-specific behaviors or are only required to maintain repressed expression states in somatic lineages (Table I). These relationships indicate a complexity not reflected by the current Arabidopsis RdDM paradigm. Other observations inconsistent with a simplistic RdDM model are discussed below.
DIVERSE ROLES FOR MULTIPLE DNA-DEPENDENT RNA POLYMERASES
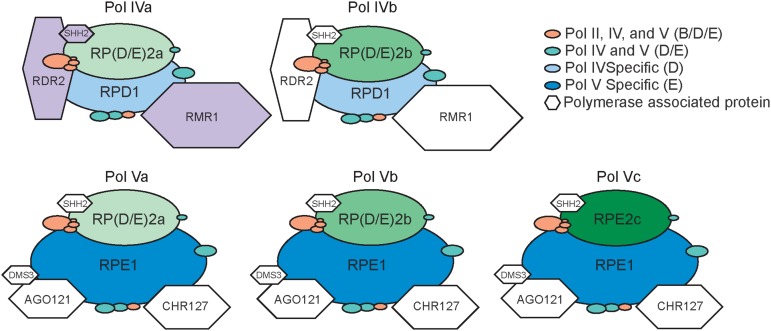
An RdDM-based model for paramutations occurring in maize likely involves multiple Pol IV and Pol V subtypes. Arabidopsis has only one functional second largest subunit shared by both Pol IV and Pol V, but maize and other grasses have three (Haag et al., 2014; Huang et al., 2015). Consistent with genetic data showing diverse functions of more than one Pol IV holoenzyme (Stonaker et al., 2009), recent proteomic profiles support the presence of at least two Pol IV (Pol IVa and Pol IVb) and three Pol V (Pol Va, Pol Vb, and Pol Vc) subtypes in maize callus (Fig. 2; Haag et al., 2014). Second largest subunits rna polymerase (d/e)2a [RP(D/E)2a] and RP(D/E)2b define Pol IV and Pol V subtypes a and b, respectively, and RPE2c specifies subtype Pol Vc. Both Pol IV and Pol V associate with distinct accessory proteins, although the specificities of these proteins for each polymerase subtype remain unresolved (Fig. 2; Haag et al., 2014). Both polymerase subunits and accessory proteins may specify unique functions of these distinct complexes at different loci, cell types, and/or in certain developmental phases. Such functional diversity might account for the different RMR requirements observed at specific paramutable alleles (Table I).
Figure 2.
Maize DNA-dependent RNA polymerase diversity. Polymerase subunit and associated protein compositions of Pol IV and Pol V subtypes are based on coimmunoprecipitation profiles using epitope-tagged versions of RPD1, RPE1, and RP(D/E)2a in maize callus and epitope-tagged versions of RP(D/E)2a and RDR2 in developing ears with 3- to 5-cm cobs (Haag et al., 2014). All proteins are represented by sizes reflecting primary amino acid contents. Polymerase subtypes (ovals) are distinguished by combinations of largest and second largest subunits. Subunits are color coded according to the legend at top right and arranged relative to Saccharomyces cerevisiae Pol II structural information (Cramer et al., 2000) and presumed Arabidopsis compositions (Ream et al., 2009). Polymerase-associated proteins (hexagons) are coded white or purple to indicate ambiguous or unambiguous assignments, respectively. Peptides specific for RMR1 and another RMR1-like protein (CHR167) coimmunoprecipitated with RPD1, and the unambiguous placement of RMR1 with at least one of possibly multiple Pol IVa complexes is inferred from genetic results (Hale et al., 2009; Stonaker et al., 2009). Peptides identifying two distinct SHH2-like proteins coimmunoprecipitated with RPE1, RP(D/E)2a, and RDR2, placing these unambiguously with the Pol IVa subtype and ambiguously with all other subtypes.
Recent results show that maize Pol IV also has more general effects on gene regulation, possibly unrelated to RdDM (Erhard et al., 2015). Loss of RPD1 function results in strong developmental defects, possibly explained by the ectopic expression of specific genes (Parkinson et al., 2007; Erhard et al., 2013, 2015). Yet, similar defects are not seen in rp(d/e)2a or other rmr mutant plants (Hale et al., 2007; Stonaker et al., 2009; Barbour et al., 2012), thus implicating Pol IVb in gene regulation. RDR2-dependent 24-nucleotide RNAs are found enriched at both genic transcription initiation sites and pretermination regions (Gent et al., 2013; Erhard et al., 2015), consistent with nascent transcript profiling showing that RPD1-containing complexes generally affect Pol II transcription at all gene boundaries (Erhard et al., 2015). These data support models in which Pol IV competes with Pol II for both genic and nongenic transcription (Hale et al., 2009; Stonaker et al., 2009; Erhard et al., 2015). Such models were proposed to account for the observations that polyadenylated RNAs (Pol II products) from certain transposable elements (TEs) accumulate in the absence of RPD1 but not in the absence of RdDM components [RMR1, RDR2, and RP(D/E)2a] required for 24-nucleotide RNA biogenesis (Hale et al., 2009; Stonaker et al., 2009). At least 183 alleles in the reference B73 genome (Schnable et al., 2009) are differentially transcribed in the absence of RPD1 (Erhard et al., 2015), and it is possible that some of these differences are responsible for the observed developmental defects.
Maize also has two Pol II subtypes defined by paralogous rna polymerase b2 subunits (Haag et al., 2014). In Arabidopsis, Pol II can provide scaffold transcripts for RdDM-type modifications (Zheng et al., 2009), so it is also possible that one or both of the maize Pol II subtypes play a role in the paramutation process. Run-on transcription results show that the sequences mediating paramutation at b1 are transcribed by Pol II (Arteaga-Vazquez et al., 2010), but it is unknown which subtypes are responsible.
Methylome profiles of rpd1, rp(d/e)2a, and rdr2 mutants show that maize Pol IV influences 5meC patterns (Li et al., 2014b), consistent with an RdDM mechanism. One or more of the maize Pol V complexes likely function in an RdDM-type capacity, as some RPE1-associated proteins are orthologous to those interacting with Arabidopsis Pol V, including DEFECTIVE IN MERISTEM SILENCING3, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION1 (CHR127), ARGONAUTE4 (AGO105 and/or AGO119), and AGO6-type (AGO121) proteins (Fig. 2; Haag et al., 2014). This evidence strongly indicates the existence and function of at least one maize RdDM pathway. The diversity of maize DNA-dependent RNA polymerases and AGO-type proteins (Singh et al., 2011) makes it likely that unique and/or expanded roles are assigned to various polymerase complexes. Paramutations may be dependent on one or more of these diversified functions.
GENOMIC FEATURES AFFECTING PARAMUTATION PROPERTIES
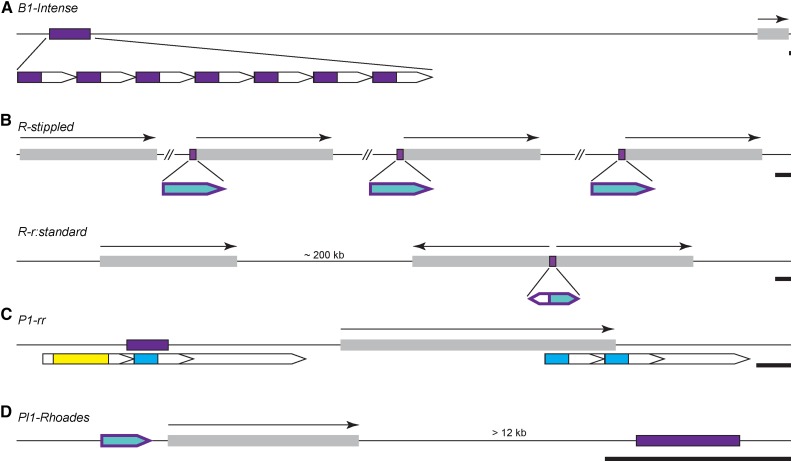
Paramutation only occurs among specific alleles having unique features responsible for their behaviors. In an RdDM-type working model (Fig. 1), these features would attract both Pol IV and Pol V complexes to mediate a 24-nucleotide RNA trans-acting signal. Previous studies functionally implicated transcriptional enhancers at both P1-rr and B1-Intense (B1-I; Sidorenko and Peterson, 2001; Stam et al., 2002), direct repeats at R-st, P1-rr, and B1-I (Kermicle et al., 1995; Sidorenko and Peterson, 2001; Stam et al., 2002), and promoter sequences at a subcomponent of the complex R-r haplotype (Fig. 3; Kermicle, 1996; Walker, 1998). All these features identified either genetically and/or by transgenesis have direct effects on the expression of the genes being assayed (Eggleston et al., 1995; Kermicle, 1996; Walker, 1998; Sidorenko and Peterson, 2001; Stam et al., 2002).
Figure 3.
Regulatory features implicated in paramutation behaviors. Schematic representations of alleles showing paramutation behaviors are drawn to scale (black bars = 1 kb) unless indicated otherwise. Gray boxes with overhead black arrows delimit transcribed genic regions. Pentagons identify repeated sequences. Purple features represent regions implicated as having regulatory importance to paramutation behaviors. A, The B1-I distal enhancer (purple box) is composed of seven 853-bp tandem repeat (TR) units (pentagons enlarged below) with important 413-bp subregions (purple; Stam et al., 2002; Belele et al., 2013). B, R-st has four tandem r gene duplications, with the centromere-distal three having promoter-proximate doppia sequences (purple boxes on the top line and purple-rimmed teal pentagons enlarged below; Matzke et al., 1996). The sizes and directions of these doppia sequences are unknown (Matzke et al., 1996). Hashed lines indicate distances of unknown length. R-r has three r gene-coding regions with a 226-bp doppia fragment (purple box on the top line and purple-rimmed teal pentagon enlarged below), a 161-bp region (purple-rimmed white pentagon), and a 15-nucleotide doppia terminus (purple tip) serving as a promoter for the inverted pair (Walker et al., 1995). C, The P1-rr coding region is flanked by a large duplication (large pentagons below the line) containing internal tandem duplications approximately 1.2 kb in length (smaller pentagons) each containing approximately 600 bp of unique sequence and a Mutator-like TE fragment (blue boxes; Goettel and Messing, 2013). The 5ʹ-most repeat contains a hAT TE (yellow box). The area highlighted by the purple box is sufficient to induce paramutation (Sidorenko and Peterson, 2001; Sidorenko and Chandler, 2008). D, Pl1-Rh has a promoter-proximate doppia fragment (purple-rimmed teal pentagon) required for kernel-specific expression but insufficient to facilitate paramutation. A genetically defined downstream transcriptional enhancer of unknown size (purple box) associated with paramutagenic function (Erhard et al., 2013) is highlighted.
TEs may also play a role in mediating paramutation behaviors (Walker, 1998; Erhard et al., 2013; Goettel and Messing, 2013), since TEs are a primary source of 24-nucleotide RNAs (Nobuta et al., 2008; Gent et al., 2014). Promoter sequences required for both kernel-specific expression and paramutation at R-r are mostly composed of one terminal end of a CACTA-type TE named doppia (Fig. 3B; Walker et al., 1995). At the paramutable Pl1-Rhoades (Pl1-Rh) allele, 5meC patterns at a similar promoter-proximal doppia fragment are dependent on RPD1, RDR2, and RMR1, indicating that Pol IV targets these features (Hale et al., 2007). In the absence of these Pol IV components, or RP(D/E)2a, Pl1-Rh is ectopically expressed in the aleurone layer of the endosperm (Erhard et al., 2013) in a mottled pattern reminiscent of Pl1-Blotched (Cocciolone and Cone, 1993), a related pl1 allele having an identical DNA sequence (including the doppia fragment) that does not participate in paramutation (Hollick et al., 2000; Gross and Hollick, 2007). Although these results indicate that the doppia sequences are insufficient for paramutation at Pl1-Rh, their potential necessity remains unknown. The doppia sequences, however, are very likely necessary for paramutation at R-r (Kermicle, 1996; Walker, 1998).
Because pigmentation from Pl1-Blotched is also enhanced in other rmr mutants, it is hypothesized that the doppia fragment serves as a Pol IV-regulated controlling element specifying aleurone expression (Erhard et al., 2013). By recurrent crossing of heterozygous (+/rpd1) and homozygous (rpd1/rpd1) mutant siblings, Pl1-Rh-specified aleurone expression continually intensifies to a solid-color phenotype (Erhard et al., 2013). This ectopic aleurone expression persists even in the presence of normal RPD1, indicating that heritable changes in gene regulation can be conditioned by this breeding scheme. Although the loss of RPD1 reduces the doppia fragment 5meC levels as expected, heritable increases in aleurone pigmentation seen in the presence of RPD1 are not associated with changes in 5meC patterns, leaving the nature of the meiotically heritable mark unknown. Because similar heritable conditioning is not seen in rdr2, rmr1, or rp(d/e)2a mutants (Erhard et al., 2013), it is inferred that some unique or complementary function of Pol IVb is required to define this transgenerational epigenetic variation. These results implicate the doppia region as a Pol IV-dependent regulatory element of pl1 potentially unrelated to the paramutation mechanism.
Similar to B1-I, Pl1-Rh also has a distal feature necessary for both high levels of expression and paramutation (Erhard et al., 2013). This 3ʹ-localized distal (greater than 12 kb) element (Fig. 3D) was genetically identified by a recombinant derivative of Pl1-Rh that coincidently lost its ability to undergo paramutation. The nature of these 3ʹ paramutation sequences is currently unknown, but paramutation-defective derivatives isolated from active Mutator TE lines and γ-irradiation coincidently lose high levels of pl1 expression (Gross, 2007; Gross and Hollick, 2007), indicating, as with B1-I, that enhancer function is related to paramutation behaviors. At B1-I, the 5ʹ distal (approximately 100 kb) enhancer (Fig. 3A) and two other intervening regions physically loop to the b1 gene promoter in frequencies correlated to the strength of b1 transcription (Louwers et al., 2009). The distal enhancers in both B1-I and Pl1-Rh represent independent examples in which paramutation affects long-distance regulatory interactions.
The B1-I enhancer is currently the best characterized feature necessary for paramutation, and recent transgenesis results indicate its sufficiency (Belele et al., 2013). Both paramutagenic strength and enhancer-promoter associations depend on the enhancer’s seven TRs of a unique 853-bp sequence (Fig. 3A; Stam et al., 2002; Louwers et al., 2009). Belele et al. (2013) found that either the entire unique sequence or just the 5ʹ half of this repeat unit (413 bp) recapitulates paramutation behaviors in an RDR2-dependent manner, but only in TR organizations. The remaining half of the repeat unit is insufficient, even in tandem arrays, to affect changes at endogenous B1-I. Not all TR transgenes containing the 413-bp subfragment were initially able to induce paramutation, but some acquired this ability following exposure to endogenous paramutagenic Bʹ. Thus, most of the TR transgenes could accomplish essentially the same behaviors as the endogenous 5ʹ heptarepeat enhancer (b1TR) but from various positions in the genome. The efficiency of these TR transgenes in inducing meiotically heritable Bʹ states, however, was often (eight of 11) less than 100% (Table I), indicating that other unknown aspects of the THI were not fully replicated. In opposition to the RdDM model, not all TR transgenes that produced small RNAs (sRNAs) could induce paramutation (Belele et al., 2013). This finding agrees with previous results showing no correlations between b1TR sRNAs and either induction or maintenance of B1-I paramutations (Arteaga-Vazquez et al., 2010).
NATURE OF THE MEIOTICALLY HERITABLE EPIGENETIC MARK
In maize, paramutation is genetically defined by a locus-specific behavior without the apparent contribution of cytoplasmic factors (Brink et al., 1960; Coe, 1966; Hollick et al., 1995; Hollick, 2012). In both D. melanogaster and C. elegans, meiotically heritable paramutation-like behaviors occur in germline lineages and require the cytoplasmic transmission of sRNAs (Ashe et al., 2012; de Vanssay et al., 2012; Shirayama et al., 2012; Seth et al., 2013; Wedeles et al., 2013; Sapetschnig et al., 2015). Analogous to the canonical Schizosaccharomyces pombe RNAi mechanism, C. elegans germline sRNAs utilize Argonaute proteins and Pol II scaffold transcripts to index self (germline-expressed genes) and nonself (e.g. TEs and foreign transgenes) sequences (for review, see Youngman and Claycomb, 2014). Perhaps the inheritance of paramutations occurring in plants similarly requires the germline transmission of sRNAs in addition to alleles poised to recruit Pol V. This scenario would account for the locus specificity of paramutation seen in plants as well as a requirement for extrachromosomal sRNAs. So far, however, paramutation-specific sRNAs have not been identified (Gross and Hollick, 2007; Arteaga-Vazquez et al., 2010; Belele et al., 2013).
Cytosine methylation is commonly cited as both a mitotically and meiotically heritable epigenetic mark in many organisms (Richards, 2006), although many species (e.g. S. pombe and D. melanogaster) with clear examples of meiotically heritable epigenetic information lack this methylation machinery. In Arabidopsis, Pol IV can be recruited to chromatin having generally repressive histone H3 modifications (unmethylated or monomethylated Lys-4 and dimethylated Lys-9 [H3K9me2] residues) via a tandem-Tudor domain found in the SAWADEE HOMEODOMAIN HOMOLOG1 (SHH1) protein (Law et al., 2013). Haag et al. (2014) found that at least one or more maize Pol IV and Pol V complexes contain related proteins (SHH2a/SHH2b; Fig. 2) that might provide similar recruitment functions. Arabidopsis Pol V is recruited directly to DNA via two redundant noncatalytic histone methyltransferases that bind 5meC (Johnson et al., 2014), and the H3K9 methyltransferase KRYPTONITE is similarly recruited to 5meC residues (Du et al., 2014). In combination with H3K9me2 binding of the DNA methyltransferases CHROMOMETHYLASE3 (CMT3) and CMT2, a self-reinforcing maintenance of non-CG-context 5meC and H3K9me2 is achieved. Thus, in Arabidopsis, non-CG 5meC patterns appear sufficient to initiate a feed-forward RdDM cycle. Maize apparently has no CMT2 ortholog, and recent methylome profiles show that Pol IV defines only a small fraction of the genome-wide 5meC patterns (Li et al., 2014b), unlike in Arabidopsis (Stroud et al., 2013).
Several studies have examined correlations between 5meC and paramutagenicity. The RdDM-based working model for paramutation predicts that 5meC patterns at important features should be altered in manners coincident with gene repression. The 1.2-kb enhancer of P1-rr is one such feature recently evaluated in detail (Fig. 3C; Sekhon et al., 2012; Goettel and Messing, 2013). From transgenesis results, these P1-rr sequences can induce paramutation at naive P1-rr (Sidorenko and Peterson, 2001; Sidorenko and Chandler, 2008); however, similar sequences found in the neutral P1-ww allele are insufficient to acquire paramutagenicity (Sidorenko and Chandler, 2008). The observation that higher 5meC levels at P1-rr enhancers are associated with paramutagenic reference states agrees with the RdDM model (Haring et al., 2010; Sekhon et al., 2012). These correlations were further delineated across the upstream 1.2-kb enhancer to short p1-specific repeated sequences (approximately 600 bp) flanked and/or interrupted by TE fragments (hAT [for hobo Activator Tam3] and Mutator-like elements) not present in the otherwise nearly identical P1-ww enhancer sequences (Goettel and Messing, 2013). Goettel and Messing (2013) hypothesize that transitive spreading of 5meC from these highly methylated TEs across the intervening sequence could be responsible for the paramutation behavior of these enhancer sequences at P1-rr.
Evidence that 5meC changes represent either paramutation signatures or triggers remains equivocal, particularly at B1-I (Haring et al., 2010; Belele et al., 2013). Belele et al. (2013) found that paramutagenic b1TR-based transgenes could have 5meC levels (measured with methylation-sensitive restriction digests) even lower than those seen at nonparamutagenic B1-I alleles, implying that extensive 5meC modifications are not an essential feature of paramutagenicity. The efficacy of transgene-induced paramutagenicity at endogenous b1TRs, however, was generally correlated with increased 5meC levels. Other correlations were noted during the induction of paramutation at B1-I (Haring et al., 2010) and more recently at the p1 enhancer sequences (Goettel and Messing, 2013). During the induction of B1-I paramutation, significant reductions in pigmentation precede major 5meC changes (Haring et al., 2010) at the b1TRs, although minor changes detected immediately following germination may be responsible for inhibiting b1TR enhancer activities. Maximum Bʹ-like 5meC levels are only acquired late in development (Haring et al., 2010), at a time consistent with genetic mosaic results showing that somatically heritable repression of B1-I is not established until after the 10-leaf stage (Coe, 1966). It appears as though some form of transrepression occurs during early development and is followed by progressive increases in 5meC. For paramutation induction at P1-rr, Goettel and Messing (2013) found no correlation between the repression of cob pigmentation and 5meC levels using bisulfite sequencing of leaf-derived DNA, leading them to speculate that 5meC changes lag behind the establishment of a repressed epigenetic expression state. These observations at both b1 and p1 are consistent with the idea that heritable changes, possibly related to extensive 5meC modifications, do not occur until very late in development, perhaps during meiosis (Coe, 1966).
In both maize and Arabidopsis, recent methylome comparisons between parents and hybrids identify dominant 5meC patterns suggestive of paramutation-like events (Eichten et al., 2013; Regulski et al., 2013; Greaves et al., 2014). Importantly, however, neither the locus specificity (showing that methylation functions genetically map to a specific allele or locus), functional relevance to gene regulation, nor subsequent trans-silencing properties of these events is known. Several studies have now catalogued thousands of differentially methylated regions among inbreds or ecotypes, and many of these 5meC patterns show strong transgenerational stability (Becker et al., 2011; Schmitz et al., 2011; Li et al., 2014a). Changes to these differentially methylated region 5meC patterns have been highlighted as potential examples of paramutation (Eichten et al., 2013; Regulski et al., 2013; Greaves et al., 2014) without considering other available mechanisms or explanations that may account for these changes. Further pedigree-based methylome studies combined with allele-specific transcriptome profiling should begin to distinguish these possible mechanisms.
Another possible meiotically heritable mark accounting for transgenerational inheritance of paramutagenic states may be histone based. There is currently little information regarding specific histone modifications at the b1 and p1 enhancer sequences (Haring et al., 2010; Sekhon et al., 2012), and no histone-modifying components have been reported among rmr and mop loci. Nonetheless, given the apparent integral role of H3K9me2 in maintaining 5meC patterns in Arabidopsis (Du et al., 2014), it is predicted that such modifications would similarly correlate with paramutable and paramutagenic states. Indeed, both b1 and p1 enhancers have elevated H3K9me2 levels coincident with the reduced gene expression associated with paramutagenic states, but Haring et al. (2010) showed that this was a tissue-restricted correlation, which was not expected from a persistent somatically inherited mark of paramutation. The dominant ufo1-1 mutation also coincidently affects both 5meC and H3K9me2 levels in pericarp tissues where gene expression is monitored (Sekhon et al., 2012). These initial results indicate that H3K9 modifications are likely to be correlated with gene function, but their role as potential meiotically heritable changes remains unclear.
Other histone modifications may be important, including changes at H3 Lys-27 (H3K27) residues (Haring et al., 2010). Recently, Crevillén et al. (2014) implicated reversible modifications of H3K27 in the transgenerational reprogramming necessary for vernalization in Arabidopsis. Intriguingly, Li et al. (2015) speculate that Arabidopsis Pol IV recruitment at specific regions may require H3K27 monomethylation marks, a feature potentially heritable by replication-dependent deposition of H3.1 variant histones (Jacob et al., 2014). These features remain to be evaluated for inducible, stable, and reversible paramutagenic states. Although the nature of the meiotically heritable epigenetic marks responsible for the transgenerational changes associated with paramutation remains unclear, there are now multiple paramutation-specific sequences and mutant stocks available for detailed molecular study.
CHALLENGES TO THE EMERGING RdDM-TYPE WORKING MODEL
Based on the design of genetic screens performed to date, all rmr and mop factors are required to maintain the somatic repression of pigmentation coincident with paramutagenic states. Only some of these factors, however, are necessary to maintain the meiotically heritable information specifying paramutagenic function. For instance, paramutagenic Pl1-Rh can revert back to a meiotically stable paramutable form in the absence of RMR1 (Hollick and Chandler, 2001; Hale et al., 2007; Barbour et al., 2012) but not in the absence of RMR2 (Barbour et al., 2012). Such reversion events occurring in specific mutant backgrounds can make it difficult to distinguish between functions required for establishing versus maintaining paramutant states (for review, see Hollick, 2012). In considering evidence relevant to an RdDM-based working model, it is important to distinguish between these various functions because they may reflect mechanistically distinct features. Accordingly, the following observations need to be accommodated by any paramutation working model.
Components Required Downstream of 24-Nucleotide Biogenesis Have Not Been Implicated in Examples of Paramutation
Despite nearly saturation-level screens for rmr factors (J.B. Hollick, unpublished data), all cloned loci encode proteins specific for the 24-nucleotide RNA biogenesis part of a presumed RdDM pathway. The maize genome encodes downstream RdDM orthologs, including de novo methyltransferases (Li et al., 2014b), yet none have been discovered that affect paramutation behaviors. It remains possible that one or more of the nine remaining uncharacterized rmr loci will identify such molecules. This part of the pathway may be essential for gametophyte function and/or proper embryonic development, and some components may have retained paralogs with functional redundancy. These possibilities are consistent with failed attempts to obtain maize double mutants for some of the presumed CMT3-like orthologs (Li et al., 2014b). Given the large number of Pol V-associated proteins required for Arabidopsis RdDM (Matzke et al., 2015) and the identification of maize orthologs by sequence similarity, it is surprising that none of these maize orthologs have been identified in the rmr and mop screens. It remains plausible that paramutation is dependent on strictly Pol IV-based functions that may or may not require sRNAs. It is also possible that these screens, designed to identify functions required to maintain the somatic repression associated with paramutant states, do not always detect functions related to RdDM initiation.
Paramutation-Specific RNAs Have Not Been Identified
The b1TR enhancer sRNAs are also produced, in seemingly similar quantities, from neutral b1 alleles (Arteaga-Vazquez et al., 2010) and even from nonparamutagenic b1TR-derived transgenes (Belele et al., 2013), indicating that the presence of these sRNA species alone is insufficient. However, the fact that inverted repeat transgenes producing Pol IV-independent 24-nucleotide b1TR RNAs can induce paramutation (Arteaga-Vazquez et al., 2010; Sloan et al., 2014) indicates direct actions of these 24-nucleotide species. Overexpression of a CXC domain-containing protein that binds to the insufficient portion of the b1TR (Belele et al., 2013) can also induce B1-I paramutation (Brzeska et al., 2010), indicating that non-sRNA triggers can suffice. Thus, (1) sRNAs are not causal; (2) the tissue(s), phase(s), or time point(s) in which a difference in sRNAs occurs has not been assayed; (3) important differences in sRNA levels have been too subtle to discern; or perhaps (4) there are other features, such as developmentally regulated or cell type-specific Pol V scaffold transcripts, required to interpret the sRNA signals.
Mutations of rpd1 and Other rmr Genes May Not Specifically Affect Paramutation
Because pigmentation from Pl1-Blotched is intensified in the absence of RPD1 (Erhard et al., 2013), this allele could have been used to identify rmr-type factors. Pl1-Blotched does not show paramutation behaviors, despite having the same doppia element and coding sequences as Pl1-Rh (Gross and Hollick, 2007), so it is possible that the genetic screens are not specific for the paramutation mechanism per se. Mutations affecting the general behavior of TEs may have either direct or indirect regulatory consequences on the regulation of specific alleles. Given that the maize genome composition is greater than 85% TE (Baucom et al., 2009; Schnable et al., 2009) and that many TEs flanking genes are likely sources or targets of RdDM (Gent et al., 2014), there is great potential for such heritable regulatory effects (McClintock, 1951).
Paramutation Can Occur in the Absence of Some sRNA Biogenesis Components
The absolute genetic requirement for rdr2 (Dorweiler et al., 2000) and rpd1 (Hollick et al., 2005) functions for the induction of r1, b1, and pl1 paramutation strongly indicates the role of 24-nucleotide RNAs, but paramutations can occur in other 24-nucleotide-deficient mutants. Paramutations at r1, b1, and pl1 all occur in the absence of RMR1, an ATPase associated with one or both Pol IV complexes (Haag et al., 2014), despite a major depletion of 24-nucleotide RNAs (Hale et al., 2007). Another RMR1-like protein, CHR167, also copurifies with RPD1 (Haag et al., 2014), so it remains possible that a Pol IV complex defined by CHR167 provides essential sRNAs. Loss of RMR2 also depletes 24-nucleotide RNAs yet has no effect on the induction of paramutation at r1 and is only partially required for induction at pl1 (Barbour et al., 2012). Neither RMR2 nor its two paralogs was found complexed with RPD1, RPE1, RP(D/E)2a, or RDR2 (Haag et al., 2014), but these paralogous proteins could provide semiredundant functions.
Paramutation at b1 Can Be Induced in the Absence of Pol IVa in an Apparent Parent-of-Origin Fashion
Paramutation could be induced in rp(d/e)2a mutant offspring when paramutagenic Bʹ was transmitted from rp(d/e)2a mutant mothers but not rp(d/e)2a mutant fathers (Stonaker et al., 2009). With recent proteomic data confirming the existence of Pol IVb subtypes, this finding may indicate that maternal-specific information is required for paramutation induction. One speculative hypothesis is that maternally provided Pol IVb-derived sRNAs are amplified only in female gametophytes transmitting paramutagenic templates. Developing cobs at the stage of egg sac development are a highly abundant source of 24-nucleotide RNAs (Hale et al., 2007; Nobuta et al., 2008; Erhard et al., 2009; Stonaker et al., 2009; Barbour et al., 2012), so perhaps these represent sRNAs sufficient to induce paramutation in the absence of Pol IVa. In Arabidopsis, there is compelling evidence of pollen-transmitted sRNA information (Slotkin et al., 2009; Calarco et al., 2012). Maize sperm cells, however, do not appear to transmit paramutagenic sRNAs by themselves (J.B. Hollick, unpublished data; for discussion, see Hollick, 2012), so paramutagenic alleles transmitted from male gametophytes may require Pol IVa in the newly formed zygote to produce the types of sRNAs that induce paramutation. Because some paramutation-like behaviors in metazoans also appear to involve the maternal transmission of sRNAs, similar ideas deserve further experimental attention in plants.
These apparent inconsistencies mentioned here no doubt highlight interesting aspects of paramutation biology yet to be discovered. Many unanswered questions remain to be addressed, but an important role for a basal RNAi mechanism appears increasingly certain.
CONCLUSION
The potential diversity of maize RdDM-type pathways combined with the non-RdDM functions of Pol IV represent a different landscape of epigenetic control relative to Arabidopsis (Li et al., 2014b; Erhard et al., 2015). Whether examples of paramutation remain unique to this landscape remains to be seen. Nonetheless, the Arabidopsis RdDM pathway provides a useful paradigm with which to model paramutation behaviors and generate meaningful hypotheses. It is widely expected that continued research on examples of paramutation in maize will broaden our understanding of sRNA biology while exposing increasing evidence of rapid regulatory evolution in the grasses.
Acknowledgments
We thank current members of the Hollick laboratory and an anonymous reviewer for helpful comments and opinions.
Glossary
- THI
trans-homolog interactions
- RNAi
RNA interference
- RdDM
RNA-directed DNA methylation
- 5meC
de novo cytosine methylation
- TE
transposable element
- sRNA
small RNA
- TR
tandem repeat
- H3K9me2
histone H3 dimethylated lysine-9
References
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL (2006) An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442: 295–298 [DOI] [PubMed] [Google Scholar]
- Arteaga-Vazquez M, Sidorenko L, Rabanal FA, Shrivistava R, Nobuta K, Green PJ, Meyers BC, Chandler VL (2010) RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc Natl Acad Sci USA 107: 12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. (2012) piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour JE, Liao IT, Stonaker JL, Lim JP, Lee CC, Parkinson SE, Kermicle J, Simon SA, Meyers BC, Williams-Carrier R, et al. (2012) required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize. Plant Cell 24: 1761–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Belele CL, Sidorenko L, Stam M, Bader R, Arteaga-Vazquez MA, Chandler VL (2013) Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing. PLoS Genet 9: e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink RA. (1956) A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41: 872–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink RA. (1958) Paramutation at the R locus in maize. Cold Spring Harb Symp Quant Biol 23: 379–391 [DOI] [PubMed] [Google Scholar]
- Brink RA, Brown DF, Kermicle J, Weyers WH (1960) Locus dependence of the paramutant R phenotype in maize. Genetics 45: 1297–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DF, Brink RA (1960) Paramutagenic action of paramutant Rr and Rg alleles in maize. Genetics 45: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska K, Brzeski J, Smith J, Chandler VL (2010) Transgenic expression of CBBP, a CXC domain protein, establishes paramutation in maize. Proc Natl Acad Sci USA 107: 5516–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MTA, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL, Eggleston WB, Dorweiler JE (2000) Paramutation in maize. Plant Mol Biol 43: 121–145 [DOI] [PubMed] [Google Scholar]
- Chandler VL, Stam M (2004) Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet 5: 532–544 [DOI] [PubMed] [Google Scholar]
- Cocciolone SM, Cone KC (1993) Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe EH., Jr (1959) A regular and continuing conversion-type phenomenon at the B locus in maize. Proc Natl Acad Sci USA 45: 828–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe EH., Jr (1966) The properties, origin, and mechanism of conversion-type inheritance at the B locus in maize. Genetics 53: 1035–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288: 640–649 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C (2014) Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bougé AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S (2012) Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490: 112–115 [DOI] [PubMed] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL (2000) mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12: 2101–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Groth M, Feng S, Hale CJ, Li S, Vashisht AA, Gallego-Bartolome J, Wohlschlegel JA, Patel DJ, et al. (2014) Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol Cell 55: 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston WB, Alleman M, Kermicle JL (1995) Molecular organization and germinal instability of R-stippled maize. Genetics 141: 347–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Briskine R, Song J, Li Q, Swanson-Wagner R, Hermanson PJ, Waters AJ, Starr E, West PT, Tiffin P, et al. (2013) Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF Jr, Parkinson SE, Gross SM, Barbour JE, Lim JP, Hollick JB (2013) Maize RNA polymerase IV defines trans-generational epigenetic variation. Plant Cell 25: 808–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF Jr, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB (2009) RNA polymerase IV functions in paramutation in Zea mays. Science 323: 1201–1205 [DOI] [PubMed] [Google Scholar]
- Erhard KF Jr, Talbot JE, Deans NC, McClish AE, Hollick JB (2015) Nascent transcription affected by RNA polymerase IV in Zea mays. Genetics 199: 1107–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK (2013) CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Madzima TF, Bader R, Kent MR, Zhang X, Stam M, McGinnis KM, Dawe RK (2014) Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goettel W, Messing J (2013) Paramutagenicity of a p1 epiallele in maize. Theor Appl Genet 126: 159–177 [DOI] [PubMed] [Google Scholar]
- Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M (2009) The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development 136: 3647–3655 [DOI] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES (2014) Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci USA 111: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SM (2007) Trans-sensing interactions and structural features of the maize Pl1-Rhoades allele. PhD thesis. University of California, Berkeley [Google Scholar]
- Gross SM, Hollick JB (2007) Multiple trans-sensing interactions affect meiotically heritable epigenetic states at the maize pl1 locus. Genetics 176: 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES (2013) The role of epigenetics in hybrid vigour. Trends Genet 29: 684–690 [DOI] [PubMed] [Google Scholar]
- Haag JR, Brower-Toland B, Krieger EK, Sidorenko L, Nicora CD, Norbeck AD, Irsigler A, LaRue H, Brzeski J, McGinnis K, et al. (2014) Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep 9: 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS (2012) In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell 48: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CJ, Erhard KF Jr, Lisch D, Hollick JB (2009) Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CJ, Stonaker JL, Gross SM, Hollick JB (2007) A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol 5: e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Bader R, Louwers M, Schwabe A, van Driel R, Stam M (2010) The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J 63: 366–378 [DOI] [PubMed] [Google Scholar]
- Heard E, Martienssen RA (2014) Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157: 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB. (2010) Paramutation and development. Annu Rev Cell Dev Biol 26: 557–579 [DOI] [PubMed] [Google Scholar]
- Hollick JB. (2012) Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr Opin Plant Biol 15: 536–543 [DOI] [PubMed] [Google Scholar]
- Hollick JB, Chandler VL (1998) Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics 150: 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB, Chandler VL (2001) Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics 157: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB, Kermicle JL, Parkinson SE (2005) Rmr6 maintains meiotic inheritance of paramutant states in Zea mays. Genetics 171: 725–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB, Patterson GI, Asmundsson IM, Chandler VL (2000) Paramutation alters regulatory control of the maize pl locus. Genetics 154: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB, Patterson GI, Coe EH Jr, Cone KC, Chandler VL (1995) Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Kendall T, Forsythe ES, Dorantes-Acosta A, Li S, Caballero-Pérez J, Chen X, Arteaga-Vázquez M, Beilstein MA, Mosher RA (2015) Ancient origin and recent innovations of RNA polymerase IV and V. Mol Biol Evol 32: 1788–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E. (2013) Epigenetic inheritance and plasticity: the responsive germline. Prog Biophys Mol Biol 111: 99–107 [DOI] [PubMed] [Google Scholar]
- Jacob Y, Bergamin E, Donoghue MTA, Mongeon V, LeBlanc C, Voigt P, Underwood CJ, Brunzelle JS, Michaels SD, Reinberg D, et al. (2014) Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343: 1249–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. (2014) SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507: 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL. (1996) Epigenetic silencing and activation of a maize r gene. In Russo VEA, Riggs AD, Martienssen RA, eds, Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 267–287 [Google Scholar]
- Kermicle JL, Eggleston WB, Alleman M (1995) Organization of paramutagenicity in R-stippled maize. Genetics 141: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AMS, Strahl BD, Patel DJ, Jacobsen SE (2013) Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Eichten SR, Hermanson PJ, Springer NM (2014a) Inheritance patterns and stability of DNA methylation variation in maize near-isogenic lines. Genetics 196: 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Eichten SR, Hermanson PJ, Zaunbrecher VM, Song J, Wendt J, Rosenbaum H, Madzima TF, Sloan AE, Huang J, et al. (2014b) Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Vandivier LE, Tu B, Gao L, Won SY, Li S, Zheng B, Gregory BD, Chen X (2015) Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 25: 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers M, Bader R, Haring M, van Driel R, de Laat W, Stam M (2009) Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 21: 832–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJM (2015) RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66: 243–267 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJM, Eggleston WB (1996) Paramutation and transgene silencing: a common response to invasive DNA? Trends Plant Sci 1: 382–388 [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1951) Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol 16: 13–47 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. (2008) Distinct size distribution of endogeneous siRNAs in maize: evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci USA 105: 14958–14963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson SE, Gross SM, Hollick JB (2007) Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev Biol 308: 462–473 [DOI] [PubMed] [Google Scholar]
- Pilu R, Panzeri D, Cassani E, Cerino Badone F, Landoni M, Nielsen E (2009) A paramutation phenomenon is involved in the genetics of maize low phytic acid1-241 (lpa1-241) trait. Heredity (Edinb) 102: 236–245 [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F (2006) RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441: 469–474 [DOI] [PubMed] [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolić L, Pikaard CS (2009) Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 33: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MTA, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. (2006) Inherited epigenetic variation: revisiting soft inheritance. Nat Rev Genet 7: 395–401 [DOI] [PubMed] [Google Scholar]
- Sapetschnig A, Sarkies P, Lehrbach NJ, Miska EA (2015) Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet 11: e1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Wang PH, Sidorenko L, Chandler VL, Chopra S (2012) Maize Unstable factor for orange1 is required for maintaining silencing associated with paramutation at the pericarp color1 and booster1 loci. PLoS Genet 8: e1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong KH, Li D, Shimizu H, Nakamura R, Ishii S (2011) Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145: 1049–1061 [DOI] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D Jr, Mello CC (2013) The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell 27: 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr, Mello CC (2012) piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L, Chandler V (2008) RNA-dependent RNA polymerase is required for enhancer-mediated transcriptional silencing associated with paramutation at the maize p1 gene. Genetics 180: 1983–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L, Dorweiler JE, Cigan AM, Arteaga-Vazquez M, Vyas M, Kermicle J, Jurcin D, Brzeski J, Cai Y, Chandler VL (2009) A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet 5: e1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko LV, Peterson T (2001) Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell 13: 319–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D (2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan AE, Sidorenko L, McGinnis KM (2014) Diverse gene-silencing mechanisms with distinct requirements for RNA polymerase subunits in Zea mays. Genetics 198: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, Belele C, Dorweiler JE, Chandler VL (2002) Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev 16: 1906–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonaker JL, Lim JP, Erhard KF Jr, Hollick JB (2009) Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet 5: e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles ED, Brink RA (1969) The metastable nature of paramuatable R alleles in maize. IV. Parallel enhancement of R action in heterozygotes with r and in hemizygotes. Genetics 61: 801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M (2008) RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell 14: 962–969 [DOI] [PubMed] [Google Scholar]
- Walker EL. (1998) Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148: 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Robbins TP, Bureau TE, Kermicle J, Dellaporta SL (1995) Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J 14: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, Claycomb JM (2013) Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev Cell 27: 664–671 [DOI] [PubMed] [Google Scholar]
- Worch S, Hansmann I, Schlote D (2008) Paramutation-like effects at the mouse scapinin (Phactr3) locus. J Mol Biol 377: 605–608 [DOI] [PubMed] [Google Scholar]
- Youngman EM, Claycomb JM (2014) From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front Genet 5: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Oliver D, Schuster A, Zheng H, Yan W (2015) Breeding scheme and maternal small RNAs affect the efficiency of transgenerational inheritance of a paramutation in mice. Sci Rep 5: 9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X (2009) Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]