Epigenetic analysis of chlorophyll synthase expression demonstrates its effects on chlorophyll and tocopherol synthesis.
Abstract
Chlorophyll synthase catalyzes the final step in chlorophyll biosynthesis: the esterification of chlorophyllide with either geranylgeranyl diphosphate or phytyl diphosphate (PDP). Recent studies have pointed to the involvement of chlorophyll-linked reduction of geranylgeranyl by geranylgeranyl reductase as a major pathway for the synthesis of the PDP precursor of tocopherols. This indirect pathway of PDP synthesis suggests a key role of chlorophyll synthase in tocopherol production to generate the geranylgeranyl-chlorophyll substrate for geranylgeranyl reductase. In this study, contributions of chlorophyll synthase to tocopherol formation in Arabidopsis (Arabidopsis thaliana) were explored by disrupting and altering expression of the corresponding gene CHLOROPHYLL SYNTHASE (CHLSYN; At3g51820). Leaves from the homozygous chlysyn1-1 null mutant were nearly devoid of tocopherols, whereas seeds contained only approximately 25% of wild-type tocopherol levels. Leaves of RNA interference lines with partial suppression of CHLSYN displayed marked reductions in chlorophyll but up to a 2-fold increase in tocopherol concentrations. Cauliflower mosaic virus35S-mediated overexpression of CHLSYN unexpectedly caused a cosuppression phenotype at high frequencies accompanied by strongly reduced chlorophyll content and increased tocopherol levels. This phenotype and the associated detection of CHLSYN-derived small interfering RNAs were reversed with CHLSYN overexpression in rna-directed rna polymerase6 (rdr6), which is defective in RNA-dependent RNA polymerase6, a key enzyme in sense transgene-induced small interfering RNA production. CHLSYN overexpression in rdr6 had little effect on chlorophyll content but resulted in up to a 30% reduction in tocopherol levels in leaves. These findings show that altered CHLSYN expression impacts tocopherol levels and also, show a strong epigenetic surveillance of CHLSYN to control chlorophyll and tocopherol synthesis.
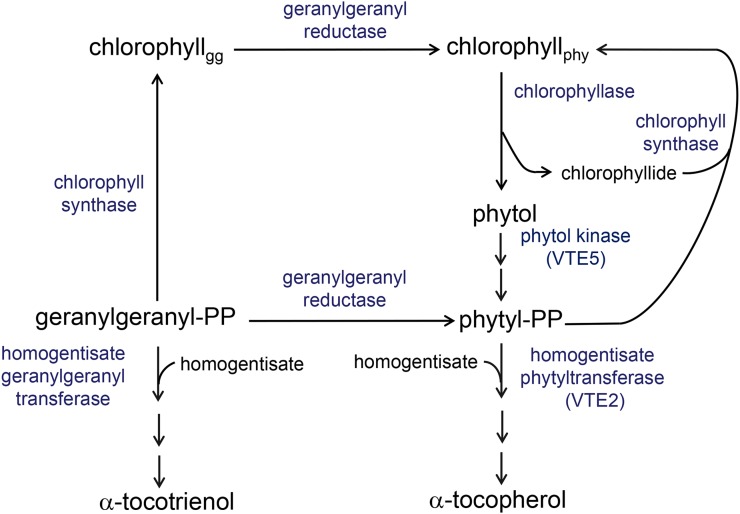
The tocopherol form of vitamin E is synthesized in plastids by condensation of the saturated C20 isoprenoid phytyl diphosphate (PDP) with homogentisate through the activity of homogentisate phytyltransferase (HPT; Soll et al., 1980; Collakova and DellaPenna, 2003). PDP is the product of the reduction of geranylgeranyl by the enzyme geranylgeranyl reductase (GGR; Keller et al., 1998). In vitro assays of Arabidopsis (Arabidopsis thaliana) GGR have shown that this enzyme can directly convert geranylgeranyl diphosphate (GGDP) to PDP or use geranylgeranyl-chlorophyll as a substrate (Keller et al., 1998). To be of physiological significance, the latter pathway would require the prenylation of chlorophyllide with geranylgeranyl from GGDP by chlorophyll synthase activity and the release of phytol formed by GGR activity with geranylgeranyl-chlorophyll (Fig. 1). The released phytol would then require two phosphorylation steps to generate the PDP substrate for tocopherol synthesis. The existence and quantitative significance of this indirect pathway for tocopherol production in Arabidopsis are supported by the identification of the VITAMIN E PATHWAY GENE5 (VTE5)-encoded phytol kinase that catalyzes the first phosphorylation step of phytol released from chlorophyll (Valentin et al., 2006). The Arabidopsis vte5-1 mutant accumulates only 35% of wild-type levels of tocopherols in leaves and 20% of wild-type levels of tocopherols in seeds (Valentin et al., 2006). Similarly, disruption of the Synechocystis sp. PCC 6803 homolog of VTE5 (encoded by open reading frame slr1652) results in a 50% reduction of tocopherol levels. The Arabidopsis and Synechocystis sp. PCC 6803 mutants also accumulate free phytol (Valentin et al., 2006).
Figure 1.
Pathways for de novo PDP synthesis and tocopherol biosynthesis in photosynthetic organisms. Tocopherol biosynthesis is initiated by the prenylation of homogentisate with PDP catalyzed by HPT. PDP is synthesized by the reduction of geranylgeranyl either in the diphosphate form or as the hydrophobic sidechain of chlorophyll by the activity of GGR, and recent evidence has pointed to that at least 80% of PDP synthesis proceeds through the reduction of geranylgeranyl bound to chlorophyll in Arabidopsis (Ischebeck et al., 2006; Valentin et al., 2006). This pathway for PDP synthesis requires an array of enzymes in addition to GGR, including chlorophyll synthase to esterify geranylgeranyl to chlorophyll, chlorophyllase to release phytol from chlorophyll, and two phytol kinases, including VTE5, to convert released phytol to PDP. The esterification of chlorophyllide with GGDP catalyzed by chlorophyll synthase is the primary reaction for chlorophyll biosynthesis, and esterification of chlorophyllide with PDP is a secondary reaction for chlorophyll synthesis catalyzed by chlorophyll synthase using PDP derived from chlorophyll degradation. An alternative fate for GGDP is condensation with homogentisate for biosynthesis of the tocotrienol form of vitamin E (Cahoon et al., 2003; Yang et al., 2011). chlorophyllgg, Chlorophyll linked to a geranylgeranyl moiety; chlorophyllphy, chlorophyll linked to a phytyl moiety.
Tocopherol synthesis has been a major target for biotechnological improvement of the antioxidant content and nutritive value of plants (Karunanandaa et al., 2005; Raclaru et al., 2006). Significant progress has been made in these efforts by enhancing expression of genes for enzymes, such as HPT (Soll et al., 1980; Collakova and DellaPenna, 2003). However, the indirect pathway of PDP formation likely represents a bottleneck for achieving maximal tocopherol production, particularly in green seeds, such as Arabidopsis, soybean (Glycine max), and rapeseed (Brassica napus; Karunanandaa et al., 2005; Raclaru et al., 2006; Seo et al., 2011; Yang et al., 2011; Zhang et al., 2013). For example, HPT overexpression in these seeds, even with the combined enhancement of homogentisate synthesis, has failed to achieve high levels of tocopherol enhancement (Savidge et al., 2002; Karunanandaa et al., 2005). This can be attributed, in part, to the complexity of PDP production through the chlorophyll-linked pathway. As such, a more in-depth understanding of the steps in this pathway is likely to provide important insights to guide tocopherol biofortification efforts.
Central to the indirect pathway of PDP synthesis for tocopherol production is the activity of chlorophyll synthase to generate the geranylgeranyl-chlorophyll substrate for GGR. Although chlorophyll synthase has been extensively studied for its role in chlorophyll synthesis, the importance of this enzyme for tocopherol synthesis has not been previously examined. Chlorophyll synthases from a variety of photosynthetic organisms have been shown to use not only PDP as a substrate for prenylation of chlorophylls a and b but also, GGDP as a substrate (Oster and Rüdiger, 1997; Oster et al., 1997; Wu et al., 2007). In fact, the enzyme encoded by the single chlorophyll synthase gene in Arabidopsis (CHLOROPHYLL SYNTHASE [CHLSYN]; At3g51820) displays higher in vitro activity with GGDP than with PDP for the esterification of chlorophyllide to generate chlorophyll (Oster and Rüdiger, 1997). The Synechocystis sp. PCC 6803 chlorophyll synthase, however, has higher activity with PDP than with GGDP (Oster et al., 1997).
Previous studies that have targeted chlorophyll synthase for genetic manipulation in planta have only focused on chlorophyll synthesis and not on tocopherol synthesis. For example, a missense mutation in chlorophyll synthase in rice (Oryza sativa) was shown to reduce enzymatic activity and chlorophyll content in leaves as well (Wu et al., 2007). In addition, antisense suppression of the tobacco (Nicotiana tabacum) chlorophyll synthase gene resulted in coordinate reduction of transcript levels for genes encoding enzymes involved in synthesis of the chlorophyllide porphyrin ring and chlorophyll binding proteins (Shalygo et al., 2009).
In this report, we explore the in planta contributions of chlorophyll synthase to tocopherol synthesis by examination of a transfer DNA (T-DNA) mutant for CHLSYN and by up- or down-regulation of this gene. Our findings provide additional support for the quantitative importance of the indirect biosynthetic pathway for the tocopherol precursor PDP in Arabidopsis leaves and seeds. They also show that altered chlorophyll synthase activity inversely impacts tocopherol levels in Arabidopsis leaves. Furthermore, it was discovered that CHLSYN is subject to strong epigenetic surveillance that affects both chlorophyll and tocopherol biosynthesis, because CHLSYN overexpression induces the production of small interfering RNAs (siRNAs) at a very high frequency.
RESULTS
Leaves and Seeds of an Arabidopsis Chlorophyll Synthase Mutant Are Deficient in Tocopherol
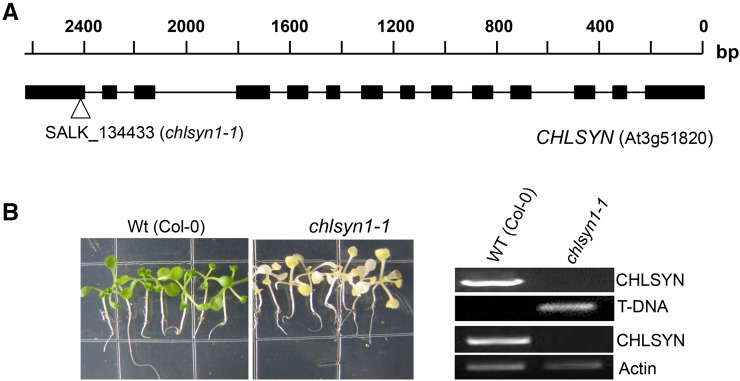
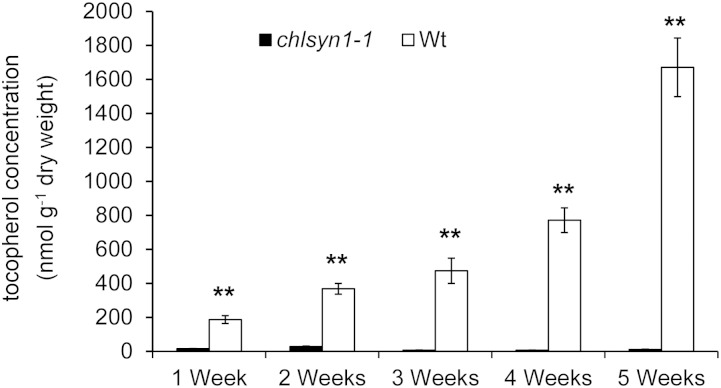
A T-DNA insertion mutant for chlorophyll synthase (SALK_134433; designated chlsyn1-1) was characterized to assess the role of this enzyme in tocopherol biosynthesis. The T-DNA insertion of chlsyn1-1 was confirmed to be present in the last exon of the Arabidopsis CHLSYN gene (Fig. 2A). The heterozygous T-DNA insertion line, which was confirmed by PCR analysis, displayed a segregation ratio of 3:1 for green seeds:yellow seeds in siliques. Although seed abortion was not detected in siliques, no plants homozygous for the T-DNA insertion were obtained from soil-sown seeds from self-pollinated plants genotyped as heterozygous chlsyn1-1 plants. However, albino plants genotyped as homozygous chlsyn1-1 plants were recovered by germination of these seeds on media containing 2% (w/v) Suc at the expected segregation ratio of 1:3. These plants lacked detectable CHLSYN transcript as assessed by reverse transcription (RT)-PCR analysis of leaf RNA (Fig. 2B). Homozygous chlsyn1-1 plants retained viability with vigorous growth on Suc-containing media for >5 weeks, although they were deficient in chlorophyll. Analysis of leaves of plants at weekly intervals from 1 to 5 weeks of growth revealed only low tocopherol concentrations in homozygous chlsyn1-1 plants relative to wild-type plants (Fig. 3; Supplemental Fig. S1). At 2 weeks of growth, homozygous chlsyn1-1 plants had 10% of wild-type tocopherol concentration, which was the highest ratio during the 5-week growth period. Leaves of homozygous chlsyn1-1 plants at this growth stage had 50% of wild-type total fatty acid concentration (Supplemental Fig. S2), indicating that plastids are still metabolically active, despite chlorophyll deficiency. These results suggest a strong link between chlorophyll synthase activity and vitamin E production in Arabidopsis.
Figure 2.
Gene structure and characterization of the mutant for CHLSYN. A, Gene structure of CHLSYN and T-DNA insertion site in SALK134433 (chlsyn1-1). Black bars represent exons, and lines indicate introns. The white triangle indicates the T-DNA insertion site. B, Plants shown are wild-type (WT) and null mutant chlsyn1-1, which were genetically and transcriptionally confirmed by PCR and RT-PCR, respectively. For RT-PCR analysis, the UBC gene (At5g25760) was used as an internal control.
Figure 3.
Tocopherol accumulation is reduced in leaves of the null mutant of chlsyn1-1. Measurements of tocopherol concentrations were conducted weekly with leaves from 1- to 5-week-old plants. Data represent means ± se of three independent biological replicates. Wt, Wild type; **, P < 0.01 (Student’s t test).
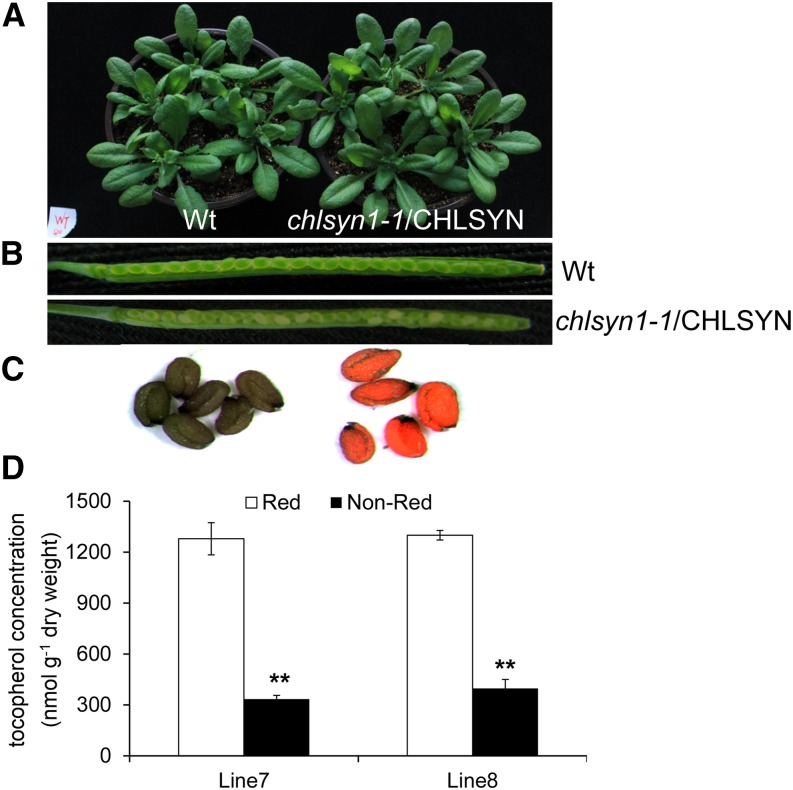
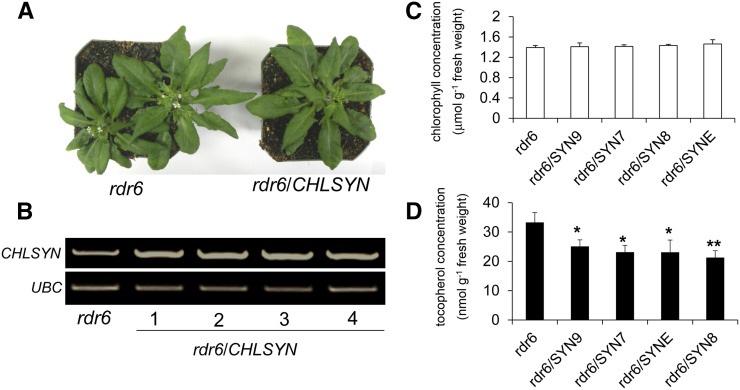
To gain additional evidence in support of this conclusion, genetic complementation studies of chlsyn1-1 were conducted. For these studies, the Arabidopsis rna-directed rna polymerase6 (rdr6) mutant was transformed with a CHLSYN complementary DNA (cDNA) under control of a Cauliflower mosaic virus35S (CaMV35S) promoter and linked to a DsRed fluorescent protein selection marker. This background was used to mitigate the high frequency of CHLSYN silencing observed with overexpression of this gene in Arabidopsis Columbia-0 (Col-0), which is described in more detail below. Transgenic lines were then crossed into heterozygous chlsyn1-1 plants. After selection and genetic confirmation, DsRed-positive F3 plants homozygous for the chlsyn1-1 allele displayed growth and pigmentation identical to those of wild-type plants (Fig. 4A), indicating that At3g51820 is responsible in vivo for chlorophyll synthesis, which was expected. Plants homozygous for the chlsyn1-1 allele but segregating for the transgene (Fig. 4, B and C) provided a tool for additional examination of the involvement of chlorophyll synthase in tocopherol synthesis. DsRed-positive seeds from the two lines corresponding to chlsyn1-1 complementation had tocopherol content comparable with that in wild-type seeds. However, non-DsRed seeds from the two lines corresponding to a lack of chylsyn1-1 complementation had tocopherol contents at 20% and 26% of wild-type seed content (Fig. 4D). These reductions in tocopherol content are nearly the same as those previously reported in the vte5 mutant (Valentin et al., 2006) and provide additional support for a central role of chlorophyll synthase in tocopherol biosynthesis.
Figure 4.
Tocopherol accumulation is reduced in mature seeds of chlsyn1-1. Seedlings and seed morphologies (A and B) of chlsyn1-1 plants with genetic complementation of AtCHLSYN. Also shown are green seeds lacking DsRed fluorescence corresponding to noncomplemented seeds (C, left), seeds with DsRed fluorescence corresponding to complemented seeds (C, right), and comparison of tocopherol concentrations in seeds with chlsyn1-1 complementation (D; red) and noncomplemented chlsyn1-1 (D; nonred) seeds. Data represent means ± se of three independent biological replicates. Wt, Wild type; **, P < 0.01 (Student’s t test).
RNA Interference-Mediated Silencing of CHLSYN Leads to Enhancement of Tocopherol Content in Arabidopsis Leaves
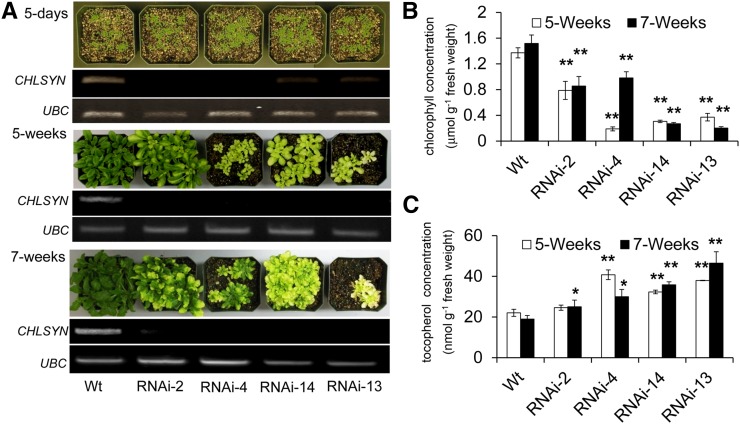
As shown in the model presented in Figure 1, chlorophyll synthase is not only required for phytol synthesis by GGR, but also, it catalyzes a biosynthetic reaction for chlorophyll formation that competes for PDP with HPT, the initial tocopherol biosynthetic enzyme. As such, it is not clear how alteration of CHLSYN would affect tocopherol biosynthesis. To examine this question, RNA interference (RNAi) suppression of CHLSYN was initially conducted. As expected for a gene encoding a central enzyme in chlorophyll synthesis, suppression of CHLSYN resulted in extensive chlorosis in transformants (Fig. 5, A and B). This phenotype, although not visually observed in 5-d-old seedlings, was strongly apparent in 5-week-old plants. In lines RNAi-13 and RNAi-14 with the most extreme chlorotic phenotype, chlorophyll levels were reduced in leaves at 5 and 7 weeks of growth by up to 65% relative to those in leaves of wild-type plants of the same age. Reductions in chlorophyll concentrations in leaves of transformants were accompanied by a general inverse correlation in tocopherol concentrations (Fig. 5C). In lines RNAi-13 and RNAi-14, for example, tocopherol concentrations were 1.5- to 2-fold higher in leaves of 5- and 7-week-old plants relative than those in wild-type plants of the same age.
Figure 5.
RNAi suppression of CHLSYN results in reduced chlorophyll accumulation but enhanced tocopherol concentrations. A, Plants shown are Col-0 plants and Col-0 plants transformed with the pH8CHLSYN RNAi construct at 5 d, 5 weeks, and 7 weeks grown under a 14 h of light/10 h of dark cycle. Semiquantitative RT-PCR analysis of the CHLSYN expression level in leaves from corresponding plants. The wild type (Wt) and RNAi-2, RNAi-4, RNAi-13, and RNAi-14 are representative of plants of Col-0 and independent transgenic events of Col-0 plants transformed with the pH8CHLSYN RNAi construct, respectively. The gene encoding UBC (At5g25670) was used as an internal control in RT-PCR analyses. Chlorophyll (B) and tocopherol (C) concentrations of leaves from Col-0 plants and Col-0 plants transformed with the pH8CHLSYN RNAi construct from sampling of 5- and 7-week-old plants. Data represent means ± se of three independent biological replicates. *, P < 0.05; **, P < 0.01 (Student’s t test).
Overexpression of Arabidopsis Chlorophyll Synthase Triggers CHLSYN Sense Suppression
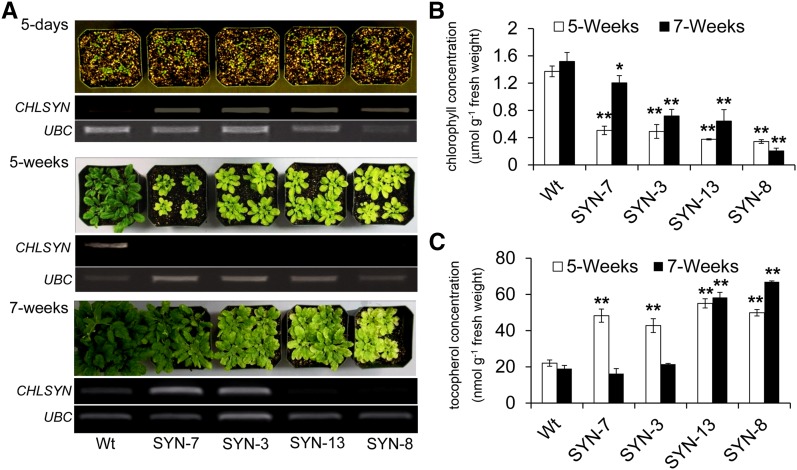
To complement the studies above, experiments were also conducted to examine the effects of overexpression of CHLSYN on tocopherol concentrations in Arabidopsis leaves. For these experiments, a CHLSYN cDNA was expressed under control of the constitutive CaMV35S promoter. Unexpectedly, approximately 90% of the transgenic plants obtained displayed varying degrees of chlorosis in leaves after 2 weeks of growth. As determined by RT-PCR, CHLSYN transcript was detected in leaves of 5-d-old seedling in four lines selected for more detailed characterization. However, no transcript was detectable in leaves of 5-week-old plants in any of these lines (Fig. 6A). Consistent with this, chlorophyll concentrations in leaves of these lines ranged from 25% to 30% of those in leaves of wild-type plants of the same age (Fig. 6B). However, CHLSYN suppression and leaf chlorosis were not stable over the entire lifecycle for two of these lines. For example, CHLSYN transcript was detectable in leaves of lines Wt/SYN3 and Wt/SYN7 in 7-week-old plants, and sharp increases in leaf chlorophyll concentration were measured in Wt/SYN7 between 5 and 7 weeks of age.
Figure 6.
Overexpression of CHLSYN triggers sense suppression and alters chlorophyll and tocopherol concentrations in leaves. A, Plants shown are Col-0 and Col-0 expressing chlorophyll synthase under control of the CaMV35S promoter at 5 d, 5 weeks, and 7 weeks of age and maintained in a 14 h of light/10 h of dark cycle. Semiquantitative RT-PCR analysis of expression of chlorophyll synthase in leaves from these plants is shown in A, middle. The wild type (Wt) and Wt/SYN-3, Wt/SYN-7, Wt/SYN-8, and Wt/SYN-13 are representative of plants of Col-0 and four independent transgenic chlorophyll synthase overexpression lines, respectively. The gene encoding UBC (At5g25670) was used as an internal control in RT-PCR analysis. Shown are chlorophyll (B) and tocopherol (C) concentrations of leaves from Wt plants and plants engineered for chlorophyll synthase overexpression from sampling of 5- and 7-week-old plants. Data represent means ± se of three independent biological replicates. *, P < 0.05; **, P < 0.01 (Student’s t test).
The effects of chlorophyll reductions on tocopherol concentrations in leaves of lines engineered for CHLSYN overexpression were similar but larger than those detected in leaves of plants engineered for CHLSYN RNAi suppression (Fig. 6C). In leaves of lines Wt/Syn8 and Wt/Syn13, which displayed the largest reductions in leaf chlorophyll concentrations relative to those of leaves of wild-type plants, tocopherol concentrations were 2- to 3-fold higher than those in leaves of wild-type plants in both 5- and 7-week-old plants.
Overexpression of CHLSYN Decreases Tocopherol Content of Leaves
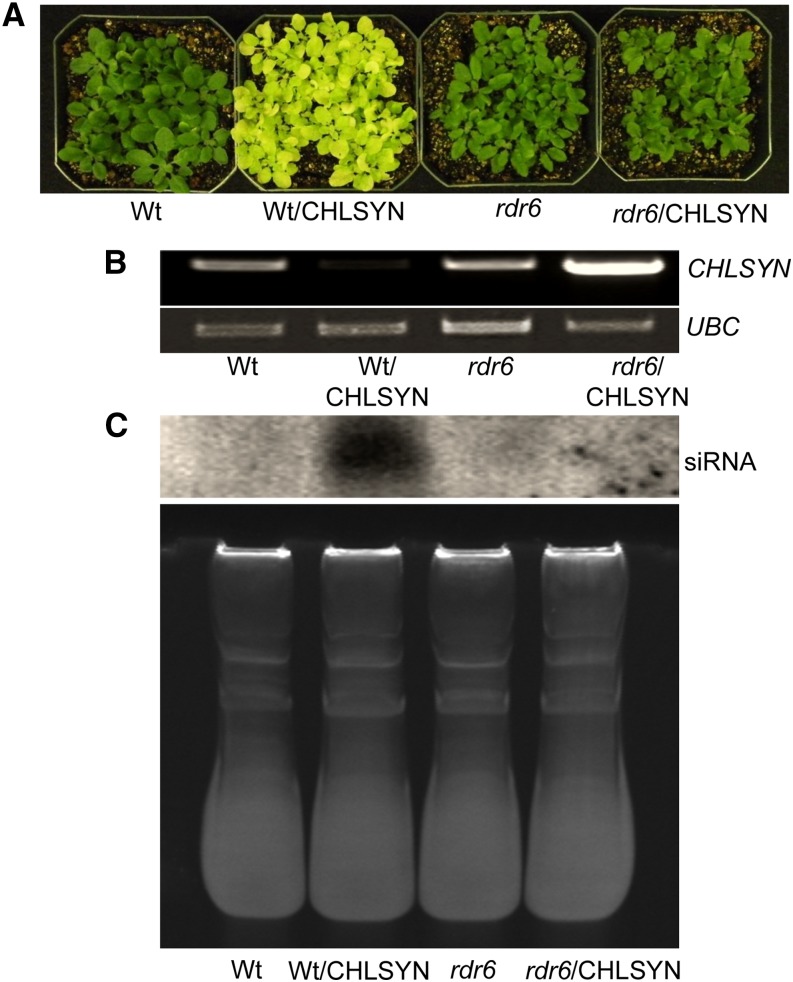
The observation of high rates of CHLSYN cosuppression triggered by overexpression of this gene was suggestive of sense transgene-induced posttranscriptional gene silencing similar to that reported for ARGONAUTE1 (Mallory and Vaucheret, 2009). This process was shown to be dependent primarily on RDR6, SUPPRESSOR OF GENE SILENCING3, and SILENCING DEFECTIVE5 (Mallory and Vaucheret, 2009). Based on this prior report, the CaMV35S-AtCHLSYN construct described above was introduced into the Arabidopsis rdr6 mutant that is defective in sense transgene-induced posttranscriptional gene silencing. In >30 independent lines obtained from this transformation, no leaf chlorosis phenotype was observed (Fig. 7A). Consistent with this, the CHLSYN transcript was elevated in four randomly selected transformants (Fig. 7B). Additional examination of RNA isolated from leaves of overexpression lines in the Col-0 and rdr6 backgrounds revealed high accumulation of CHLSYN-derived siRNAs in Col-0 that were not detectable in the rdr6 background (Fig. 8). Measurements conducted on extracts from leaves of four independent overexpression lines in the rdr6 background revealed no significant change in chlorophyll concentrations but approximately 30% reduction in tocopherol concentrations (Fig. 7, C and D) relative to leaves of nontransformed rdr6 plants. These results together with those from CHLSYN, RNAi, or sense suppression indicate a negative correlation between CHLSYN expression levels and tocopherol concentrations in Arabidopsis leaves.
Figure 7.
Overexpression of CHLSYN in rdr6 background results in modest reductions of leaf tocopherol concentrations. A, Plants shown are rdr6 and rdr6 transformed with CHLSYN under control of the CaMV35S promoter. B, Expression of CHLSYN in rdr6 plants and rdr6 plants from four independent events overexpressing CHLSYN is confirmed by RT-PCR (upper) and expression of an internal control gene (UBC; At5g25670; lower). Shown are measurements of chlorophyll (C) and tocopherol (D) concentrations of leaves from 3-week-old plants from rdr6 and four independent events of rdr6 transformed with CHLSYN. Data represent means ± se of three independent biological replicates. *, P < 0.05; **, P < 0.01 (Student’s t test).
Figure 8.
Sense suppression line accumulates CHLSYN-specific siRNAs. A, Plants shown from left to right are the wild type (Wt), sense suppression (Wt/CHLSYN), rdr6, and rdr6 transformed with CHLSYN (rdr6/CHLSYN) under control of the CaMV35S promoter. Also, RT-PCR analysis of CHLSYN transcript levels is shown in B. Shown in C, top is the accumulation of CHLSYN siRNA (21–22 nucleotides) in leaves from Wt and rdr6 plants nontransformed or engineered for CaMV35S-mediated overexpression of CHLSYN. C, bottom shows ethidium bromide staining of low-molecular weight RNA.
DISCUSSION
Previous findings from the vte5 mutant have highlighted the importance of metabolic flux of GGDP through chlorophyll for synthesis of the phytol sidechain of tocopherol by the activity of GGR in Arabidopsis (Valentin et al., 2006). Central to this pathway is chlorophyll synthase, which has the dual role of catalyzing the attachment of geranylgeranyl as a diphosphate substrate to chlorophyllide to initiate the synthesis of phytol and catalyzing the attachment of released phytol as a diphosphate substrate to chlorophyllide for synthesis of the major chlorophyll forms used in photosynthesis (Oster et al., 1997; Fig. 1). The studies presented here were conducted to examine the quantitative importance of chlorophyll synthase in tocopherol synthesis and explore its use as a target for altering tocopherol levels in leaves. A useful tool for our studies was a CHLSYN T-DNA insertion mutant chlsyn1-1. Although lacking chlorophyll, chlsyn1-1 seedlings were viable when grown on Suc and had 10% of wild-type tocopherol concentration. This mutant coupled with the use of a CHLSYN overexpression transgene linked to a DsRed fluorescence marker also provided a unique approach for investigating the quantitative significance of chlorophyll synthase in seed tocopherol synthesis. Plants homozygous for the chlsyn1-1 allele but segregating for the overexpression transgene were phenotypically similar to wild-type plants in appearance and leaf tocopherol concentrations. Seeds from these plants could be readily segregated based on the DsRed marker: genetically complemented chlsyn1-1 seeds were fluorescent, whereas seeds without the transgene and homozygous for the chlsyn1-1 allele were not fluorescent. The latter seeds, lacking a functional chlorophyll synthase, had approximately 20% of the tocopherol concentration of wild-type seeds, which is identical to the tocopherol reductions measured previously in vte5-1 seeds. In addition to providing useful information in our studies, a similar approach using DsRed as a screenable marker for complementation should have general use for examining seed phenotypes in mutants with compromised vegetative growth.
Because chlorophyll synthase has dual functions in phytol and chlorophyll synthesis (Oster et al., 1997; Ischebeck et al., 2006; Lin et al., 2014), the impact of suppressed or enhanced CHLSYN expression on tocopherol concentrations could not be easily predicted. Our findings indicate that suppression of CHLSYN expression increases tocopherol concentrations in leaves, whereas enhanced expression of this gene reduces tocopherol concentrations in leaves (Fig. 9). These observations can largely be attributed to the role of chlorophyll synthase in phytol attachment to chlorophyllide. Partial suppression of CHLSYN, which was achieved by RNAi or sense suppression, would likely reduce the competition between HPT (the initial step in tocopherol synthesis) and chlorophyll synthase for PDP. The result would be increased availability of PDP for tocopherol synthesis. The increases in tocopherol concentrations may indicate that some level of chlorophyll turnover occurs in leaves that can provide additional PDP for the tocopherol biosynthetic pathway in the CHLSYN suppression lines. Conversely, enhanced expression of CHLSYN may shunt additional PDP to chlorophyll synthesis at the expense of tocopherol production. Because chlorophyll concentrations are >100-fold higher than tocopherol concentrations in leaves, increased PDP flux into chlorophyll may have quantitative significance for tocopherol concentrations but little measurable effect on chlorophyll concentrations, which was observed in our studies.
Figure 9.
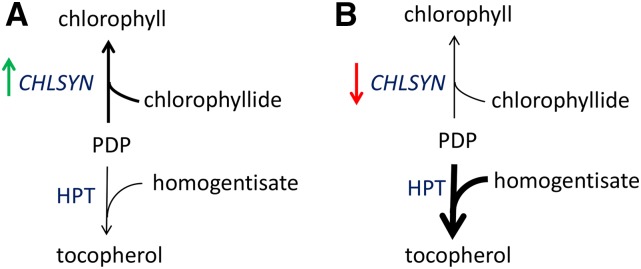
Model for the effects of altered chlorophyll synthase expression on tocopherol concentrations in Arabidopsis leaves. As indicated, HPT, which catalyzes the initial reaction in tocopherol biosynthesis, competes for the PDP substrate with chlorophyll synthase. Based on results from RNAi and cosuppression lines, reduced expression of CHLSYN increases PDP concentrations for HPT, leading to enhanced tocopherol production in Arabidopsis leaves (A). Conversely, based on results from overexpression lines, increased expression of CHLSYN decreases PDP concentrations for HPT, leading to reduced tocopherol production in Arabidopsis leaves (B).
An unexpected finding was the high incidence of CHLSYN suppression in response to the introduction of the CaMV35S-AtCHLSYN overexpression transgene in Arabidopsis Col-0. This effect was overcome by the use of the Col-0 rdr6 mutant as the overexpression background. Because RDR6 is required for generation of transgene-induced siRNAs and because CHLSYN overexpression in Col-0 resulted in formation of CHLSYN-specific siRNAs at very high frequency, the observed effect is clearly epigenetic based and suggests that chlorophyll biosynthesis is under the stringent surveillance of epigenetic machinery in Arabidopsis. Notably, the easily visualized chlorotic phenotype associated with CHLSYN overexpression may provide a useful tool for studies of siRNA-mediated gene silencing.
Our findings with chlsyn1-1 together with previous findings from vte5-1 (Ischebeck et al., 2006; Valentin et al., 2006) indicate that flux of geranylgeranyl through chlorophyll is the primary route of phytyl formation for tocopherol synthesis. However, in both mutants, seeds retain approximately 20% of wild-type tocopherol levels, and in leaves of the vte5-1 mutant, 35% of wild-type tocopherol levels are detected (Valentin et al., 2006). These findings are consistent with lower but significant contributions of direct conversion of GGDP to PDP for tocopherol synthesis. It is likely that this pathway is also a major contributor to tocopherol production in CHLSYN suppression lines. Like Arabidopsis, seeds from many other species, including commercially important oilseeds, such as rapeseed and soybean, retain chlorophyll throughout most of their development. As such, it is expected that the chlorophyll route of phytol synthesis is quantitatively important for tocopherol production in seeds of these species. However, many nongreen oilseeds are also enriched in tocopherol. Castor bean (Ricinus communis) seeds, for example, have reported tocopherol concentrations of approximately 600 µg g−1 seed weight (Velasco et al., 2005), which is 200 to 300 µg g−1 higher than tocopherol concentrations typically found in rapeseed and soybean seeds (Karunanandaa et al., 2005). Because castor bean and other nongreen seeds lack chlorophyll, it is possible that GGR in these seeds has evolved for more efficient direct conversion of GGDP to PDP. Tocopherol biosynthesis in these seeds also does not compete with chlorophyll biosynthesis for PDP, similar to the metabolic scenario in leaves of Arabidopsis CHLSYN suppression lines. Overall, more complete understanding of phytol formation in nongreen seeds may provide clues for enhancing tocopherol concentrations in green seeds through metabolic routes that bypass chlorophyll. This is important for tocopherol biofortification efforts in green seeds, which to date, have been relatively ineffective by approaches such as HPT overexpression (Savidge et al., 2002; Karunanandaa et al., 2005), likely because of insufficient pools of PDP.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type and mutant Arabidopsis (Arabidopsis thaliana) lines used for these studies were of the Col-0 ecotype. SALK_134433, a chlsyn1-1 heterozygous T-DNA insertion line, was obtained from the Arabidopsis Biological Resource Center (Alonso et al., 2003). Seeds of SALK_134433 were surface sterilized, grown on one-half-strength Murashige and Skoog medium agar plates supplemented with 2% (w/v) Suc and 100 mg L−1 of ampicillin, and grown under a 16-h-light (100 μmol m−2 s−1)/8-h-dark cycle at 22°C. Plants were genotyped by two PCR reactions using a gene-specific primer pair (P1, 5′-AAACTGCAAAATGGATATGCG-3′ and P2, 5′-AACAAAAACATTCTGAAAGCAGAG-3′) to determine homozygosity and primer P2 and the T-DNA left border primer PLB (5′-TGGTTCACGTAGTGGGCCATCG-3′) to test for the presence of the T-DNA. Cosuppression lines and RNAi-mediated gene silencing lines grown on soil were maintained at 22°C and 50% humidity under conditions with a 14-h-light (100 μmol m−2 s−1)/10-h-dark cycle. Unless indicated, plants were grown on soil under a long-day regime at 22°C and 50% humidity with a 16-h-light (100 μmol m−2 s−1)/8-h-dark cycle.
Vector Construction
All PCR amplifications were conducted with Phusion Polymerase (New England Biolabs), and products were verified by sequencing. Arabidopsis CHLSYN was expressed under control of the CaMV35S promoter in wild-type Arabidopsis and mutant rdr6. This construct was prepared by PCR amplification of the Arabidopsis flower cDNA library with forward and reverse oligonucleotides that contain added restriction enzyme sites (underlined): 5′-TATACAATTGGTTCTATGACTTCGATTCTCAAC-3′ and 5′-TATTCTCGAGTCAGTGTTGCGATGCTAATGCC-3′. The PCR reaction product was digested with MfeI/XhoI and ligated into EcoRI/XhoI sites of the binary vector pBin35SRed to produce pCaMV35S-CHLSYN with the insert flanked 5′ by the CaMV35S promoter and 3′ by the 3′ untranslated region of the nopaline synthase gene. This vector contains a DsRed fluorescent protein marker for selection of transgenic seeds.
An AtCHLSYN RNAi suppression construct was generated using the pHellsgate8 RNAi Gateway Vector System (Helliwell and Waterhouse, 2003). A 349-bp fragment was amplified from the AtCHLSYN 3′ untranslated region using forward and reverse oligonucleotides: 5′-CACCACTTTCTCAAGGACCCTGTCAAATACG-3′ and 5′-CAGAACTAACATAAACCCCAATCCTCG-3′, and the product was TOPO cloned into pENTR-TOPO clone vector (Invitrogen) to yield pENT-CHLSYN. The resulting plasmid was then moved into the destination vector pHellsgate8 by LR recombination reaction according to the manufacturer's protocol (Invitrogen) to generate the final RNAi plasmid, pH8CHLSYN.
Arabidopsis Transformation and Selection
Binary vectors pCaMV35S-CHLSYN and pH8CHLSYN were introduced into Agrobacterium tumefaciens C58 by electroporation. Arabidopsis (Col-0) plants or rdr6 mutants were transformed with the recombinant Agrobacterium tumefaciens-containing vector of pCaMV35S-CHLSYN, and Col-0 plants were transformed with recombinant A. tumefaciens-harboring pH8CHLSYN by use of the floral dip method as described (Clough and Bent, 1998). Transgenic seeds transformed with pCaMV35S-CHLSYN were identified by DsRed fluorescence as described previously (Jach et al., 2001; Pidkowich et al., 2007), and plants transformed with pH8CHLSYN were selected by resistance to Basta.
HPLC Analysis of Tocochromanols
For measuring tocopherol content of leaves from different types of plants, the procedure was used as described by Yang et al. (2011) with modification for vitamin E extraction: the freshly harvested or dried leaves were homogenized in 2 mL of 9:1 (v/v) methanol:dichloromethane containing 0.01% (w/v) butylatedhydroxytoluene. After 20 min of incubation and centrifugation, the supernatant was then recovered for HPLC analysis as described by Yang et al. (2011); 5,7-dimethyltocol (Matreya) was added to each sample as an internal standard before extraction. Sample components were identified by mobility relative to standards and quantified relative to the internal standard.
Chlorophyll Measurement
The method for spectrophotometric determination of chlorophyll concentrations was as described (Strain et al., 1971). For extraction of chlorophyll, freshly harvested leaf tissue was weighed and homogenized in liquid nitrogen, and subsequently, it was extracted in 3 volumes of 80% (v/v) acetone containing 1 μm KOH. After centrifugation for 2 min at 16,000g, supernatants were combined and used for spectrophotometric analysis.
Gene Expression Analysis
Total RNA was isolated from Arabidopsis leaves using the RNeasy Plant Kit (Qiagen) following the manufacturer's protocol. Total RNA (1 μg) was first treated with DNase (Roche), and then it was reverse transcribed for synthesizing the first-strand cDNA using SuperScript III Reverse Transcriptase (Life Technologies) and oligo(dT) primer according to the manufacturer's instructions. The resulting first-strand cDNA template of each sample was adjusted to the same concentration according to the PCR signal generated from the internal standard housekeeping gene Ubiquitin-Conjugating Enzyme (UBC; At5g25670; Czechowski et al., 2005). For semiquantitative RT-PCR analysis, a 2-μL aliquot of each adjusted first-strand cDNA was used as the template to amplify AtCHLSYN or the gene for the UBC (At5g25760) in a 20-μL reaction. The primers used for the amplification of AtCHLSYN-derived cDNA were 5′-CTCAACACTGTCTCCACCATCC-3′ and 5′-GCAAAATTTCCAACCCATCC-3′. The primers used for the amplification of the UBC-derived cDNA were 5′-ATGCAGGCATCAAGAGCGCGACTGT-3′ and 5′-CACCGCCTTCGTAAGGAGTCTCCGA-3′.
Small RNA Northern Blot
RNA isolation and hybridization were performed as described by Park et al. (2002). The cDNA template for synE siRNA detection was amplified using primer sets forward: 5′- ATGACTTCGATTCTCAAC-3′ and reverse: 5′-TCAGTGTTGCGATGCTAATGCC-3′. P32-labeled probes were further prepared by random prime labeling using the Ready-to-Go DNA Labeling Beads Kit (GE Healthcare).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Morphologies of Col-0 and chlsyn1-1.
Supplemental Figure S2. Total fatty acid accumulation in leaves from Col-0 and chlsyn1-1 plants.
Supplementary Material
Acknowledgments
We thank Liesheng Zheng (College of Plant Science and Technology, Huazhong Agricultural University) for providing assistance for a portion of the tocopherol content measurements and Tara Nazarenus (University of Nebraska, Lincoln) for photography of DsRed-expressing seeds.
Glossary
- cDNA
complementary DNA
- Col-0
Columbia-0
- GGDP
geranylgeranyl diphosphate
- GGR
geranylgeranyl reductase
- PDP
phytyl diphosphate
- RNAi
RNA interference
- RT
reverse transcription
- siRNA
small interfering RNA
- T-DNA
transfer DNA
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31370286 to C.Z.), the Program for New Century Excellent Talents in University (grant no. NCET–13–0813 to C.Z.), the International Cooperation Project of the Ministry of Science and Technology (grant no. 2014DFA32210 to Y.Z.), the Fundamental Research Funds for the Central Universities (grant no. 2012ZYTS047 to C.Z.), the U.S. Department of Agriculture National Institute of Food and Agriculture (grant no. 2014–04261 to E.B.C.), and the Huazhong Agricultural University Scientific and Technological Self-Innovation Foundation (program no. 2015RC010 to E.B.C.).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ (2003) Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotechnol 21: 1082–1087 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D (2003) Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol 131: 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C, Waterhouse P (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289–295 [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Zbierzak AM, Kanwischer M, Dörmann P (2006) A salvage pathway for phytol metabolism in Arabidopsis. J Biol Chem 281: 2470–2477 [DOI] [PubMed] [Google Scholar]
- Jach G, Binot E, Frings S, Luxa K, Schell J (2001) Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. Plant J 28: 483–491 [DOI] [PubMed] [Google Scholar]
- Karunanandaa B, Qi Q, Hao M, Baszis SR, Jensen PK, Wong YHH, Jiang J, Venkatramesh M, Gruys KJ, Moshiri F, et al. (2005) Metabolically engineered oilseed crops with enhanced seed tocopherol. Metab Eng 7: 384–400 [DOI] [PubMed] [Google Scholar]
- Keller Y, Bouvier F, d’Harlingue A, Camara B (1998) Metabolic compartmentation of plastid prenyllipid biosynthesis: evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur J Biochem 251: 413–417 [DOI] [PubMed] [Google Scholar]
- Lin YP, Lee TY, Tanaka A, Charng YY (2014) Analysis of an Arabidopsis heat-sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J 80: 14–26 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2009) ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep 10: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster U, Bauer CE, Rüdiger W (1997) Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J Biol Chem 272: 9671–9676 [DOI] [PubMed] [Google Scholar]
- Oster U, Rüdiger W (1997) The G4 gene of Arabidopsis thaliana encodes a chlorophyll synthase of etiolated plants. Bot Acta 110: 420–423 [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidkowich MS, Nguyen HT, Heilmann I, Ischebeck T, Shanklin J (2007) Modulating seed β-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc Natl Acad Sci USA 104: 4742–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raclaru M, Gruber J, Kumar R, Sadre R, Hs W, Zarhloul M, Friedt W, Frentzen M, Weier D (2006) Increase of the tocochromanol content in transgenic Brassica napus seeds by overexpression of key enzymes involved in prenylquinone biosynthesis. Mol Breed 18: 93–107 [Google Scholar]
- Savidge B, Weiss JD, Wong YH, Lassner MW, Mitsky TA, Shewmaker CK, Post-Beittenmiller D, Valentin HE (2002) Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol 129: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YS, Kim SJ, Harn CH, Kim WT (2011) Ectopic expression of apple fruit homogentisate phytyltransferase gene (MdHPT1) increases tocopherol in transgenic tomato (Solanum lycopersicum cv. Micro-Tom) leaves and fruits. Phytochemistry 72: 321–329 [DOI] [PubMed] [Google Scholar]
- Shalygo N, Czarnecki O, Peter E, Grimm B (2009) Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Mol Biol 71: 425–436 [DOI] [PubMed] [Google Scholar]
- Soll J, Kemmerling M, Schultz G (1980) Tocopherol and plastoquinone synthesis in spinach chloroplasts subfractions. Arch Biochem Biophys 204: 544–550 [DOI] [PubMed] [Google Scholar]
- Strain HH, Cope BT, Svec WA (1971) Analytical procedures for the isolation, identification, estimation, and investigation of the chlorophylls. In Anthony San P, ed, Methods in Enzymology, Vol 23 Academic Press, New York, pp 452–476 [Google Scholar]
- Valentin HE, Lincoln K, Moshiri F, Jensen PK, Qi Q, Venkatesh TV, Karunanandaa B, Baszis SR, Norris SR, Savidge B, et al. (2006) The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco L, Rojas-Barros P, Fernandez-Martinez JA (2005) Fatty acid and tocopherol accumulation in the seeds of a high oleic acid castor mutant. Ind Crops Prod 22: 201–206 [Google Scholar]
- Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, et al. (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145: 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Cahoon RE, Hunter SC, Zhang C, Han J, Borgschulte T, Cahoon EB (2011) Vitamin E biosynthesis: functional characterization of the monocot homogentisate geranylgeranyl transferase. Plant J 65: 206–217 [DOI] [PubMed] [Google Scholar]
- Zhang C, Cahoon RE, Hunter SC, Chen M, Han J, Cahoon EB (2013) Genetic and biochemical basis for alternative routes of tocotrienol biosynthesis for enhanced vitamin E antioxidant production. Plant J 73: 628–639 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.