Two major chromoplasts are region-specifically formed in citrus and converted from amyloplast precursors.
Abstract
Globular and crystalloid chromoplasts were observed to be region specifically formed in sweet orange (Citrus sinensis) flesh and converted from amyloplasts during fruit maturation, which was associated with the composition of specific carotenoids and the expression of carotenogenic genes. Subsequent isobaric tag for relative and absolute quantitation (iTRAQ)-based quantitative proteomic analyses of purified plastids from the flesh during chromoplast differentiation and senescence identified 1,386 putative plastid-localized proteins, 1,016 of which were quantified by spectral counting. The iTRAQ values reflecting the expression abundance of three identified proteins were validated by immunoblotting. Based on iTRAQ data, chromoplastogenesis appeared to be associated with three major protein expression patterns: (1) marked decrease in abundance of the proteins participating in the translation machinery through ribosome assembly; (2) increase in abundance of the proteins involved in terpenoid biosynthesis (including carotenoids), stress responses (redox, ascorbate, and glutathione), and development; and (3) maintenance of the proteins for signaling and DNA and RNA. Interestingly, a strong increase in abundance of several plastoglobule-localized proteins coincided with the formation of plastoglobules in the chromoplast. The proteomic data also showed that stable functioning of protein import, suppression of ribosome assembly, and accumulation of chromoplast proteases are correlated with the amyloplast-to-chromoplast transition; thus, these processes may play a collective role in chromoplast biogenesis and differentiation. By contrast, the chromoplast senescence process was inferred to be associated with significant increases in stress response and energy supply. In conclusion, this comprehensive proteomic study identified many potentially new plastid-localized proteins and provides insights into the potential developmental and molecular mechanisms underlying chromoplast biogenesis, differentiation, and senescence in sweet orange flesh.
Chromoplasts are special organelles with superior ability to synthesize and store massive amounts of carotenoids, bringing vivid red, orange, and yellow colors to many flowers, fruits, and vegetables (Li and Yuan, 2013). Chromoplasts exhibit various morphologies, such as crystalline, globular, tubular, and membranous structures (Egea et al., 2010). The relationship between the architecture and carotenoid composition has been well stated in diverse pepper (Capsicum annuum) and tomato (Solanum lycopersicum) fruits (Kilcrease et al., 2013; Nogueira et al., 2013). Crystalline bodies have been observed in carrot (Daucus carota; Frey-Wyssling and Schwegler, 1965) and tomato (Harris and Spurr, 1969), which predominantly consist of β-carotene and lycopene, respectively. Globular and/or tubular-globular chromoplasts, in which numerous lipid droplets (also named plastoglobules), which act as passive storage compartments for triglycerides, sterol ester, and some pigments, are accumulated, were described for yellow fruits from kiwi (Actinidia deliciosa), papaya (Carica papaya), and mango (Mangifera indica), which contain lutein, β-cryptoxanthin, and β-carotene as the major pigments, respectively (Vasquez-Caicedo et al., 2006; Montefiori et al., 2009; Schweiggert et al., 2011). Carotenoid composition has been reported to be regulated by the expression of carotenogenic genes in the flesh of various citrus fruits differing in their internal colors (Fanciullino et al., 2006, 2008). Chromoplasts are frequently derived from fully developed chloroplasts, as seen during fruit ripening from green to red or yellow fruits in tomato and pepper (Egea et al., 2010). In some cases, chromoplasts also arise from nonphotosynthetic plastids, such as colorless proplastids, leucoplasts, or amyloplasts (Knoth et al., 1986; Schweiggert et al., 2011). To date, most studies on chromoplast differentiation have been focused on the synthesis of carotenoids by combining biochemical and molecular analyses (Cazzonelli and Pogson, 2010; Egea et al., 2010; Bian et al., 2011; Li and Yuan, 2013), and little is known about the molecular mechanisms underlying chromoplast biogenesis (Li and Yuan, 2013).
Recently, proteomics has become an efficient tool to study the protein composition of subcellular organelles such as chromoplasts and their dynamic changes during the development of a particular plant organ/tissue. The majority of chromoplast-related studies are concerned with the functions of these organelles in various crops, such as pepper, tomato, watermelon (Citrulis lanatus), carrot, cauliflower (Brassica oleracea), and papaya (Siddique et al., 2006; Wang et al., 2013). However, only a few of such studies addressed the mechanisms underlying plastid differentiation, such as the transition from proplastid to chloroplast in maize (Zea mays; Majeran et al., 2010), from etioplast to chloroplast in pea (Pisum sativum; Kanervo et al., 2008) and rice (Oryza sativa; Kleffmann et al., 2007), and from chloroplast to chromoplast in tomato (Barsan et al., 2012). In tomato, chromoplastogenesis appears to be associated with major metabolic shifts, including a strong decrease in abundance of the proteins involved in light reaction and an increase in terpenoid biosynthesis and stress-response proteins (Barsan et al., 2012). These changes in proteins are in agreement with the structural changes occurring in tomato during fruit ripening, which is characterized by the loss of chlorophyll and the synthesis of colored compounds. Chromoplast differentiation from nonphotosynthetic plastids occurs frequently in a number of plant tissues, such as watermelon flesh and carrot root (Kim et al., 2010; Wang et al., 2013). However, to the best of our knowledge, no large-scale proteomic study for understanding this developmental process has been reported.
Citrus is one of the most economically important fruit crops in the world. Different from the model fruit tomato, which represents climacteric fruits, citrus shows nonclimacteric characteristics during fruit maturation. Additionally, citrus fruits exhibit a unique anatomical fruit structure consisting of two major sections, the pericarp and the edible flesh. Considerable progress has been made in the understanding of chromoplast differentiation in the pericarp of citrus fruits (Eilati et al., 1969; Iglesias et al., 2007), which is a process similar to that of tomato and pepper (Egea et al., 2010). However, little is known about the molecular basis of chromoplast differentiation in the edible flesh, even though there is increasing evidence suggesting an essential role of carotenoid synthesis in inducing chromoplast differentiation (Egea et al., 2010; Bian et al., 2011; Li and Yuan, 2013). Recently, we successfully isolated and purified intact chromoplasts containing a large number of plastoglobules from the flesh of sweet orange (Citrus sinensis) fruits at the maturation stage (Zeng et al., 2011). The same method has also been used successfully to isolate plastids from sweet orange flesh in earlier maturation stages (Zeng et al., 2014), thus making comparative and quantitative proteomic analyses of plastid differentiation possible. In this study, we investigated how ultrastructural changes of plastids/chromoplasts during sweet orange fruit maturation might be associated with changes in the composition of carotenoids and the expression of carotenogenic genes in red and yellow flesh of the fruits. Furthermore, we employed the isobaric tag for relative and absolute quantitation (iTRAQ)-based technology to investigate how protein compositional changes might be correlated with metabolic and structural changes in the plastids of sweet orange flesh during their transformation from amyloplasts to chromoplasts.
RESULTS
Structural Diversity of Chromoplasts in Sweet Orange Flesh
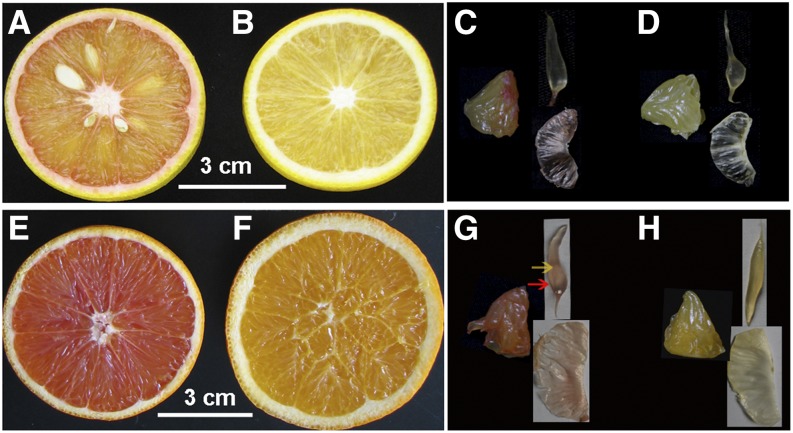
Citrus fruits are rich in carotenoids, and most of them have yellow flesh, but occasionally, red-fleshed fruits are produced due to spontaneous mutations. In this study, mutant cv Hong Anliu (Fig. 1A) and its wild-type fruit cv Anliu (Fig. 1B) as well as mutant cv Cara Cara (Fig. 1E) and its wild-type fruit cv Newhall Navel (Fig. 1F) at maturation were used as materials for observing chromoplast types. Based on the citrus anatomy, the edible flesh mainly consists of segment membrane and sac (Fig. 1, C, D, G, and H). In this study, we further separated the sac into two independent parts: one is sac membrane and the other is juice (Fig. 1G).
Figure 1.
Materials used in this experiment and their diagrammatic cross-section through a citrus fruit. A and C, Red-fleshed sweet orange ‘Hong Anliu’. B and D, Yellow-fleshed sweet orange ‘Anliu’. E and G, Red-fleshed sweet orange ‘Cara Cara’. F and H, Yellow-fleshed sweet orange ‘Newhall Navel’. C, D, G, and H show the small part of the pulp (left) and its corresponding sac (top right) and segment membrane (bottom right). The sac was gently separated into sac membrane and juice (yellow and red arrows denote juice and sac membrane, respectively).
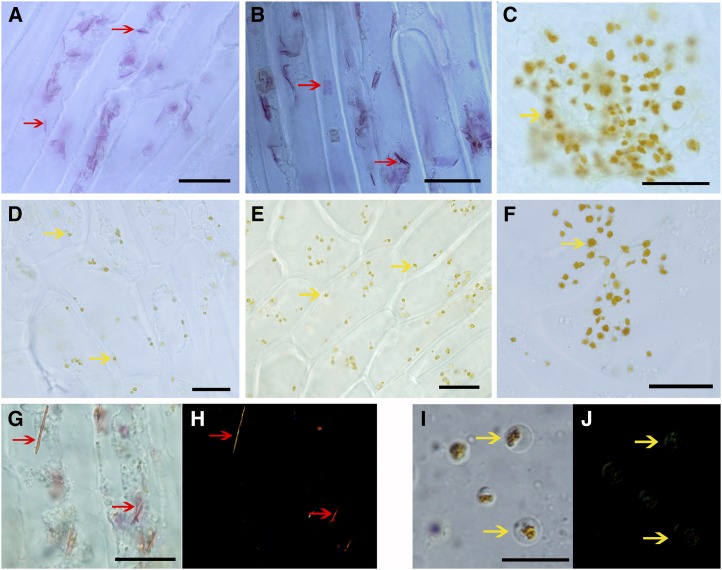
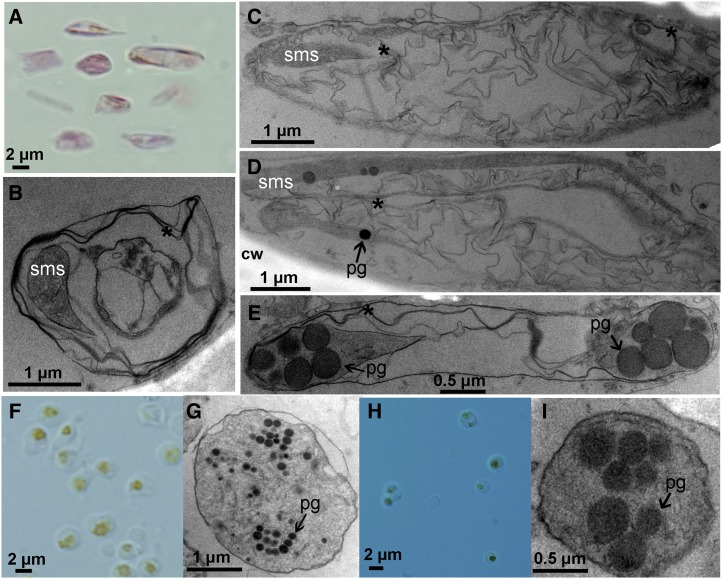
Crystalloid and globular chromoplast types were observed in the flesh of sweet orange. Light microscopy and transmission electron microscopy (TEM) were used to examine the formation of carotenoid sequestering structures in sweet orange. Visible red carotenoid sequestering structures were found in the membrane of sac and segment from cv Hong Anliu (Fig. 2, A and B) and cv Cara Cara (Supplemental Fig. S1, A and B), and polarization microscopy confirmed their crystal nature (Fig. 2, G and H; Supplemental Fig. S1, G and H). According to a protocol for isolating plastids (Zeng et al., 2011), abundant carotenoid crystals of 2 to 10 μm were detected in the isolation buffer (Fig. 3A). Their TEM images showed structural diversity in cv Hong Anliu (Fig. 3, B–E) and cv Cara Cara (Supplemental Fig. S2, A–D).
Figure 2.
Structural diversity of chromoplasts from the flesh of cv Hong Anliu and cv Anliu. A to F, Light microscopic inspection of the sac membrane, segment membrane, and juice separated from cv Hong Anliu (A–C) and cv Anliu (D–F) under normal light field microscopy. G and H, Carotenoid crystals within chromoplasts confirmed by polarization microscopy in sac membrane and segment membrane of cv Hong Anliu. I and J, Polarization micrographs of globular chromoplasts in cv Anliu flesh and juice of cv Hong Anliu. Red and yellow arrows indicate crystalloid chromoplasts and globular chromoplasts, respectively. Bars = 10 μm.
Figure 3.
Ultrastructures of chromoplasts from the flesh of cv Hong Anliu and cv Anliu. A to E, Pooled crystalloid chromoplasts from segment membrane and sac membrane of cv Hong Anliu (A), which showed several different ultrastructures, including those in B to E. F to I, Globular chromoplasts containing numerous plastoglobules (pg) were observed from the juice of cv Hong Anliu (F and G) and the flesh of cv Anliu (H and I). cw, Cell wall; sms, special membrane structure. Asterisks indicate crystal remnants.
Globe-shaped chromoplasts with yellow-orange color were frequently observed in the juice of cv Hong Anliu (Fig. 2C) and cv Cara Cara (Supplemental Fig. S1C) as well in the edible flesh of cv Anliu (Fig. 2, D–F) and cv Newhall Navel (Supplemental Fig. S1, D–F). TEM analysis showed that the globular chromoplasts isolated from different sweet orange tissues, including the juice of cv Hong Anliu (Fig. 3, F and G) and cv Cara Cara (Supplemental Fig. S2E) and the flesh of cv Anliu (Fig. 3, H and I) and cv Newhall Navel (Supplemental Fig. S2F), were found to contain plastoglobules varying in number, size, and electron density. Taken together, the above results suggest that the red color of the mutants is attributable to the membranes of sacs and segments rather than to the juice.
Relationship between the Architecture of Globular and Crystalloid Chromoplasts and Their Carotenoid Content and Composition
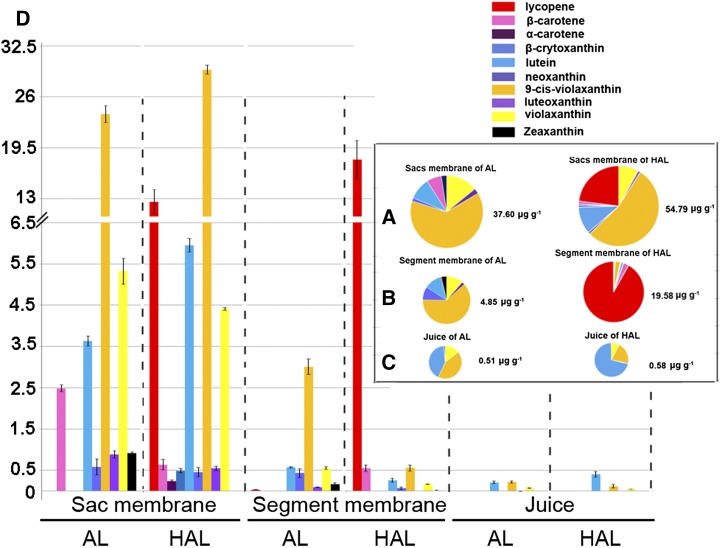
To determine whether the differences in chromoplast ultrastructure were associated with the alteration of pigment storage, we measured the content and composition of carotenoids in the membranes of sac and segment as well as in the juice fractions of cv Hong Anliu and cv Anliu. With regard to the sac membranes, we found that the total amount of carotenoids of cv Hong Anliu was about 1.5-fold higher than that of cv Anliu, and the levels of most carotenoids in both cultivars were similar, except for lycopene and β-carotene, which were relatively higher in cv Hong Anliu and cv Anliu, respectively (Fig. 4A). Interestingly, we observed remarkable differences of carotenoid content and composition in the segment membranes. The total amount of carotenoids in cv Hong Anliu was 4-fold higher than that of cv Anliu. In addition, the composition of carotenoids was also very different between these two cultivars, with lycopene accounting for 91.8% and violaxanthin for 3.6% of the total carotenoids in cv Hong Anliu (Fig. 4B, right) and with violaxanthin accounting for 73.4% (3.56 μg g−1 fresh weight; Fig. 4D) and lycopene being almost absent in the total carotenoids in cv Anliu (Fig. 4B, left). However, both cultivars had similar carotenoid profiles in their juice fractions, in which violaxanthin and lutein were dominant and lycopene was absent (Fig. 4C), suggesting that the chromoplasts from the juice may be structurally similar between these two cultivars.
Figure 4.
Chromoplast formation in relation to carotenoid content and composition. Pie charts illustrate the carotenoid compositions (fresh weight) in the sac membrane (A), segment membrane (B), and juice (C) of cv Hong Anliu (HAL) and cv Anliu (AL) at the maturation (Ma) stage. D, Detailed values (μg g−1) for each sample.
Ultrastructural Differentiation of Crystalloid and Globular Chromoplasts during Fruit Maturation
To understand the origin of chromoplasts, the sac membrane and juice of cv Hong Anliu fruit were examined throughout the fruit maturation period until typical crystalloid and globular chromoplasts were finally formed (Fig. 3, A and F). Amyloplasts with large starch granules were frequently observed in the sac membrane at the immature green (IMG) stage (for definition, see “Materials and Methods”), but plastoglobules were invisible (Fig. 5A). Subsequently, these starch granules gradually disappeared at the mature green (MG) stage, which coincided with the formation of plastids containing a small number of plastoglobules (Fig. 5B). These observations suggest that the chromoplasts might be transformed from amyloplasts. At the breaker (Br) stage, there was a dramatic increase in the number of plastoglobules, with the appearance of a few slightly undulated tuber bodies in the plastids (Fig. 5C). As fruit maturation proceeded, elongated (8–10 μm in length) or irregular crystalloid bodies were formed, which were often surrounded by a gray/whitish membrane structure (Fig. 5D) that disappeared before the formation of a crystalloid chromoplast filled with a few plastoglobules and some undulated membranes. The undulated membranes represented the remnants of membrane-coated carotenoid crystals, which were extracted during the fixation process of samples for TEM (Fig. 5E). It seems that the large accumulation of the crystalloid bodies might contribute to the change of the plastid shape from round to oval (Fig. 5, C–E).
Figure 5.
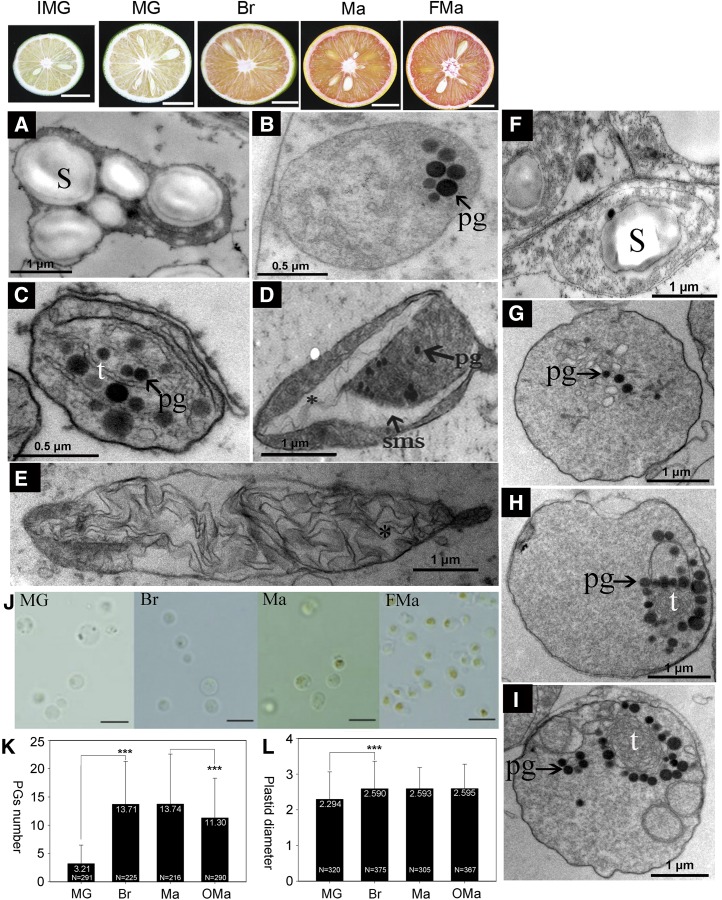
Ultrastructural changes during chromoplast differentiation at different maturation stages of cv Hong Anliu. A to E and F to I show micrographs of sac membrane and juice during fruit maturation (IMG, MG, Br, and full maturation [FMa] stages), respectively. A, Amyloplasts with starch granules at IMG stage. B, Plastids with limited number of plastoglobules (pg) at MG stage. C, Chromoplasts with numerous globular and tubular structures at Br stage. D and E, Crystalloid chromoplasts with crystal remnants at Ma and FMa stages. F, Amyloplasts with some starch granules at IMG stage. G to I, The formation of globular and tubular chromoplasts with an increase in the number of plastoglobules from MG to FMa stages. S, Starch granules; sms, special membrane structures; T, tubular elements. Asterisks indicate crystal remnants. J, Light micrographs of purified plastids, which show that the pigment-enriched areas gradually increased during chromoplast development. Bars = 5 μm. K, The number of plastoglobules increased significantly from MG to Br stages, then decreased slightly from Ma to overmaturation (OMa) stages. L, The size of plastids increased rapidly from MG to Br stages and then remained unchanged from Br to OMa stages. Data are expressed as means ± sd and were evaluated by Student’s t test using Microsoft Excel (***, P < 0.001); numbers in the bars represent the number of plastids for calculation using ImageJ software.
To investigate the pattern of plastid differentiation in the juice of sweet orange, we conducted cytological analyses using purified plastids from the flesh of cv Hong Anliu as described previously (Zeng et al., 2014). Based on our observations, plastids in the juice of cv Hong Anliu also appeared to be differentiated from amyloplasts (Fig. 5F), and the typical globular chromoplasts were strongly accumulated in the juice of cv Hong Anliu from MG stage to FMa stage (Fig. 5, G–I). Once the plastids appeared, the pigment accumulation areas within plastids gradually increased from IMG stage to Ma stage (Fig. 5J), which was largely correlated with a gradual increase of the average number of plastoglobules per plastid from 3.21 at the MG stage to approximately 13.7 at the Br stage and the FMa stage (Fig. 5K). However, the number of plastoglobules showed a slight decline to 11.3 at the OMa stage. Meanwhile, the size of the plastids increased from 2.29 μm in diameter at the MG stage to 2.59 μm at the Br stage and then stayed unchanged until the OMa stage (Fig. 5L). It is worth noting that the outer monolayer of plastoglobules appeared to be separated from the internal main body (Supplemental Fig. S3) at the OMa stage. We also noted that the chromoplasts from the juice maintained a round shape throughout the fruit maturation period and that typical globular chromoplasts often contained concentrically arranged tubular lamellae in the shape of a hollow structure (Fig. 5, H and I). More interestingly, crystal remnants frequently found in the chromoplasts of the sac membrane were not observed in the chromoplasts of the juice, suggesting that plastid differentiation in these two types of tissues may follow different patterns. To test if chloroplasts were present in the flesh of cv Hong Anliu during fruit maturation, we performed an immunoblot assay for two thylakoid-targeted proteins involved in photosynthesis (Supplemental Fig. S4). No signal for either protein was detected from the total proteins of flesh throughout the entire fruit maturation process, including the earlier maturation stage (30 d after flowering). As a positive control, both proteins were detected in the total protein extracts of the cv Hong Anliu leaves. These results suggest that the chromoplasts in the flesh were probably not directly derived from chloroplasts. Thus, it can be speculated that citrus flesh chromoplasts may be differentiated from amyloplasts, which is similar to the results of studies on the origin of chromoplasts in saffron (Crocus sativus) flowers, tobacco (Nicotiana tabacum) floral nectaries, and peach palm (Bactris gasipaes) fruit (Grilli Caiola and Canini, 2004; Horner et al., 2007; Hempel et al., 2014).
Relationship between the Architecture of Globular and Crystalloid Chromoplasts and Their Carotenogenic Transcript Levels
Carotenoid accumulation is a net result of different activities related to carotenoid metabolism, such as biosynthesis, turnover, and sequestration of carotenoids (Cazzonelli and Pogson, 2010). Hence, it is conceivable that carotenoid accumulation during the development of globular and crystalloid chromoplasts is an end result of the coordinated expression of genes involved in carotenoid biosynthesis, degradation, and storage. To test this assumption, the transcript levels of the 16 carotenoid genes were selected to be compared between the segment membranes of cv Hong Anliu and cv Anliu, where typical crystalloid and globular chromoplasts were formed during fruit maturation (Fig. 6). These genes included DXS and DXR involved in the plastidic 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway, eight genes in carotenoid biosynthesis (PSY, PDS, ZDS, CRTISO, LCYb2, LCYe, HYD, and ZEP), two in carotenoid storage (FBN1 and FBN4), and four in carotenoid degradation (NCED2, NCED3, CCD4a, and CCD4b; Supplemental Table S1).
Figure 6.
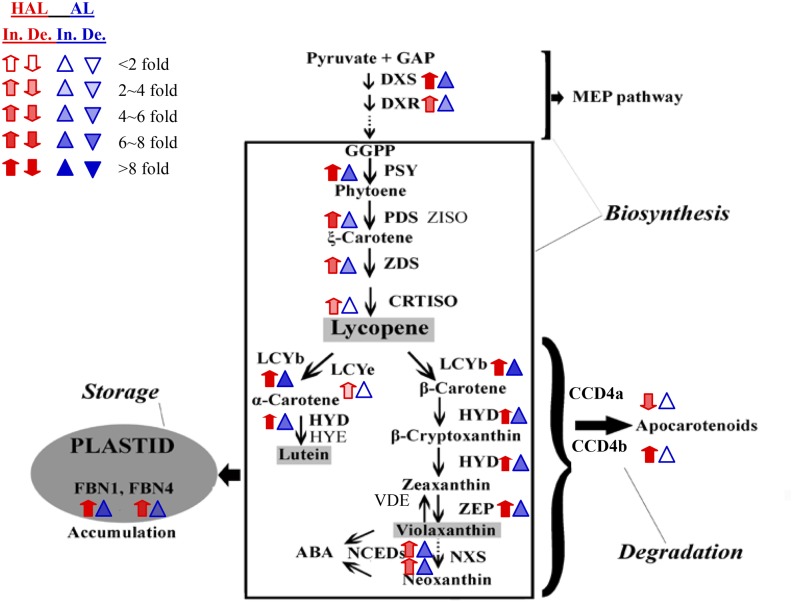
Comparative transcriptional analysis of carotenoid metabolic genes among FMa stage and IMG stage. The major carotenoids accumulated in chromoplasts are shaded in gray in a square box. The transcript levels are represented by segment membrane of cv Anliu (AL; blue arrowheads) and cv Hong Anliu (HAL; red arrows), and variation in the transcript levels is indicated by the different shades of the arrowheads and arrows. De., Decrease; In., increase. Three experimental replicates were used to indicate up-regulation (up arrow) or down-regulation (down arrow). The expression levels of ACTIN were used to normalize the expression for each sample. For detailed information, see Supplemental Figure S5. DXR, 1-Deoxy-d-xylulose 5-phosphate reductoisomerase; PSY, phytoene synthase; ZISO, ζ-carotene isomerase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRTISO, carotene isomerase; LCYb2, lycopene β-cyclase2; LCYe, lycopene ε-cyclase; HYD, flavedo-specific β-carotene hydroxylase; HYE, α-carotene hydroxylase; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; VDE, violaxanthin deepoxidase; NXS, neoxanthin synthase; FBN, fibrillin; CCD, carotenoid cleavage dioxygenease; ABA, abscisic acid.
As shown in Figure 6, most of the 16 genes showed a gradual increase in their transcript levels during the formation of globular chromoplasts in cv Anliu, except for CRTISO, LCYe, CCD4a, and CCD4b, whose expression was relatively stable (less than 2-fold change). The increased expression of most carotenoid biosynthesis genes and the steady expression level of carotenoid degradation genes (CCD4a and CCD4b) well explained the enhanced carotenoid accumulation. In contrast, nearly all the tested genes showed enhanced expression during the formation of crystalloid chromoplasts in cv Hong Anliu, except for CCD4a and CCD4b. The expression levels of CCD4a and CCD4b showed an approximately 3-fold decrease and a strong increase (11.2-fold change), respectively (Fig. 6), indicating that they might have different functions in the formation of two distinct chromoplast types.
Experimental Design, iTRAQ Analysis, and Identification of Plastid-Localized Proteins
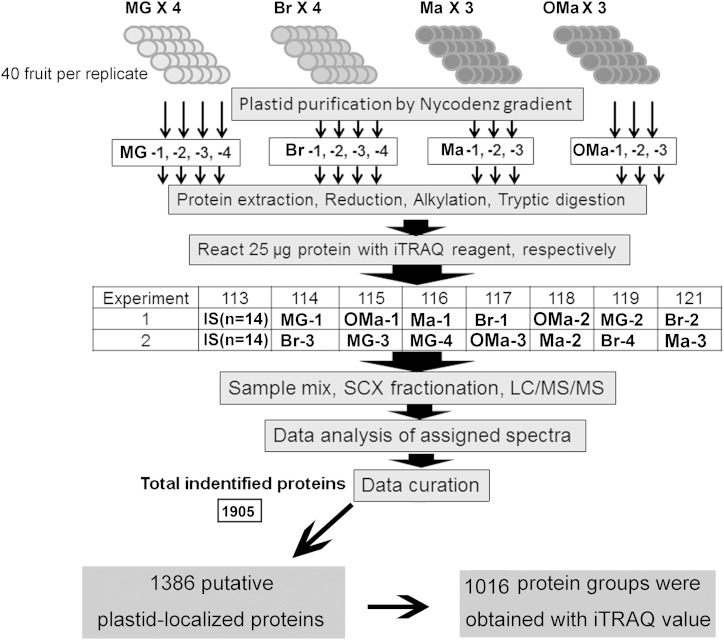
Levels of gene transcription may not always be correlated with the expression of the proteins of interest. To understand global protein changes during chromoplast differentiation, we conducted an iTRAQ-based quantitative proteomic analysis of plastids purified from the flesh of cv Hong Anliu at four key color stages (MG, Br, Ma, and OMa). iTRAQ tag labeling of all samples in each stage is summarized in Figure 7. To minimize system errors arising from variations of the iTRAQ-based quantification system, a protein mixture obtained by pooling an equal amount of all tested samples was labeled with 113 tags as internal standard. Based on the chosen criteria for peptide and protein identification (see “Materials and Methods”), a total of 9,236 unique peptides were identified (Supplemental Table S2). These peptides were matched to 1,905 unique protein groups in three independent biological samples, and 1,596 of the 1,905 unique protein groups were found to be mapped with at least two unique peptides (Supplemental Table S3), with the corresponding false discovery rate of each individual analysis lower than 1% (Sheng et al., 2012). The proteins detected are well distributed (i.e. approximately 10%–20% of the total proteins were identified in each molecular mass group) within a range of 10 to 60 kD (Supplemental Fig. S6A). In addition, a good peptide coverage was achieved for most of the identified proteins (Supplemental Fig. S6B). When the cutoff value was set as 50% variation, the coverage levels for the three biological replicates at four chromoplast differentiation stages varied from 98.99% to 99.38% (Supplemental Fig. S7A), and the coverage levels for the technological replicates at MG and Br stages were 99.29% and 99.30%, respectively (Supplemental Fig. S7B), indicating that there is a good biological and technological reproducibility of this iTRAQ analysis.
Figure 7.
Schematic of the experimental design for iTRAQ proteomic analysis. Plastids were purified from 40 fruits at each maturation stage for each independent replicate (for details, see “Materials and Methods”). A total of three biological replicates plus two technological replicates at MG and Br stages (MG 3+1, Br 3+1, Ma 3, and OMa 3) were performed. A total of 1,905 unique proteins were identified (Supplemental Table S3A), of which 1,386 proteins were predicted to be plastid localized, and 1,016 of which were further annotated with iTRAQ values as shown in Supplemental Table S3B. IS, Internal standard; SCX, strong cation-exchange.
The raw data obtained from the analysis of the replicates of the citrus plastid proteins were refined by comparing the set of proteins with five plastidial databases (AT-CHLORO, Plprot, PPDB, SUBA, and Uniprot) and three predictors for subcellular localization (TargetP, Predotar, and WoLF PSORT). Only proteins present in at least two plastidial databases or predicted to be plastid localized by one of the predictors were retained for further analysis. Additionally, manual curation was performed when sequence information was available in the literature. An inventory of 1,386 proteins (including 20 proteins encoded by the plastid genome; Supplemental Table S4) was constructed, and 1,016 of them were annotated with iTRAQ values (Supplemental Table S5). The majority of these annotated proteins (992 = 72.7%) were predicted to be plastid localized by at least one predictor. Specifically, WoLF PSORT, TargetP, and Predotar predicted 63.7% (870 proteins), 49.3% (673 proteins), and 39.5% (540 proteins), respectively, of the 1,386 annotated proteins as being plastid localized, with a considerable degree of overlapping prediction (Supplemental Fig. S8). For example, the percentage of proteins predicted to be plastid localized by all three programs was as high as 33.2% (453 proteins), suggesting that these proteins are likely plastid-targeted proteins. BLAST searches against the plastid protein databases suggest that 187 of the 1,386 proteins may be novel plastid-localized proteins (Supplemental Table S4).
Validation of Differentially Expressed Proteins by Western Blot
To test if the iTRAQ values of the predicted plastid-targeted proteins generally reflect the actual protein abundance, we selected three proteins that are known to have distinct plastid localizations and that show slight but clear development stage fluctuations for the iTRAQ values to determine protein abundance by immunoblot analysis. As shown in Figure 8, the abundance of the protein binding to the chloroplast membrane (TIC40, a component of the translocon complexes at the inner envelope membrane) showed a slight decreasing tendency during chromoplast differentiation, while plastoglobule-localized FBN1 and chloroplast stroma-localized RbcL displayed a slight increase from the MG stage to the Br stage, followed by a decrease at the Br to OMa stage. These results were in good agreement with the corresponding iTRAQ values, suggesting that the data from our proteomic analysis are of high quality and may be useful for exploring how changes in the abundance of plastid-localized proteins are associated with chromoplast differentiation at the whole-proteome scale.
Figure 8.
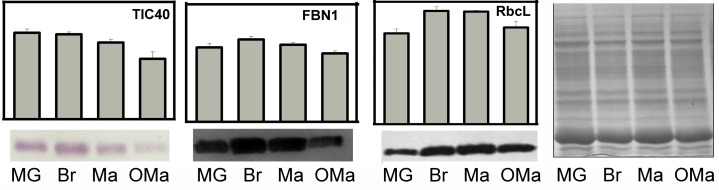
Comparison of protein abundance determined by proteomic analysis and immunoblotting. The abundance of proteins determined by the iTRAQ analysis is expressed as the value of individual/internal standard. A total of 30 μg of proteins extracted from purified plastids at the indicated stages was separated using SDS-PAGE, blotted, and subjected to immunoassays using specific antibodies that recognize the following three proteins: TIC40, a component of the translocon complexes at the inner envelope membrane; FBN1, targeted to the plastoglobules in plastids; and RbcL, targeted to the plastid stroma.
Patterns of Dynamic Changes in Protein Abundance during the Amyloplast-to-Chromoplast Transition
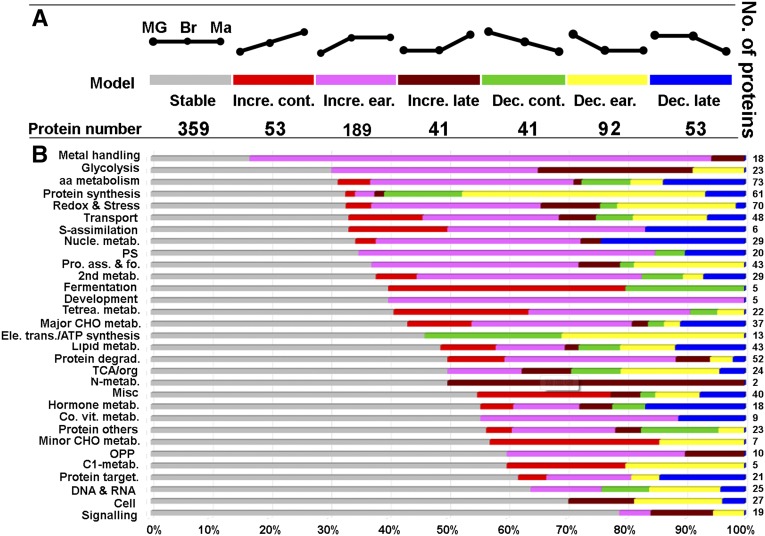
To test the idea mentioned above, we subjected the iTRAQ data of the samples prepared from fruit flesh of cv Hong Anliu at three key color stages (i.e. MG, Br, and Ma) to statistical analysis to infer if there is a significant change (P ≤ 0.05) of protein abundance for any target protein. The dynamic changes in protein abundance from the MG stage to the Ma stage could be classified into seven patterns: stable, continuous increase, early increase, late increase, continuous decrease, early decrease, and late decrease (Fig. 9A). Based on this classification, it was found that the proteins whose abundance remained constant (359) outnumbered those whose abundance underwent an increase (53 continuous + 189 early + 41 late = 283) or a decrease (41 continuous + 92 early + 53 late = 186). Notably, among those proteins with a change in abundance, 189 (approximately 22.8% of all 828 proteins examined) showed an early increase and 92 (approximately 11.1%) showed an early decrease, indicating that important changes in protein abundance occurred in the early MG to Br stage (Fig. 9A).
Figure 9.
Abundance model of plastid-localized proteins during chromoplast differentiation. A, Abundance model of plastid-localized proteins being classified into seven categories during fruit maturation (MG-Br-Ma). These seven categories are stable, early increase, late increase, continuous increase, early decrease, late decrease, and continuous decrease. Excluding the not assigned class and 91 proteins that lack a consistent and logical pattern of abundance, we established an abundance model for 828 proteins. B, Patterns of protein abundance summarized according to functional classes. Note that the number (right) and percentage (bottom) of proteins in the color-coded MapMan functional classes are shown in A. aa, Amino acid; CHO, Carbohydrate; PS, photosynthesis; OPP, oxidative pentose phosphate pathway; Pro. ass. & fo., protein assembly and folding; Co. vit. metab., cofactor/vitamin metabolism.
To get a clue of what specific cellular processes are stable or changing during chromoplast differentiation, we further studied the stability or changes of proteins in different functional classes. Interestingly, we found that 15 of 19 (78.9%) proteins presumably involved in the signaling functional class remained stable in abundance. In contrast, only three of 18 (16.7%) proteins predicted to be involved in metal handling remained constant. The percentage of proteins that showed no significant changes in abundance in the remaining 29 functional classes ranged from 70.4% to 30% (Fig. 9B). Conceivably, these proteins with constant abundance may be important for maintaining basal metabolic functions of the organelle during differentiation.
Proteins falling into the protein synthesis functional class showed the most remarkable decrease in abundance, with 41% of proteins having an early decrease, 13.1% having a continuous decrease, and 6.6% having a late decrease (Fig. 9B). The majority of the proteins in the electron transport/ATP synthesis functional class also showed decreases in abundance, with 30.8% having an early decrease and 23.1% having a continuous decrease. A decreasing pattern of proteins involved in protein and ATP synthesis during chromoplast differentiation can be anticipated, considering the lower metabolic activity of maturing organelles in general. Compared with the decreasing pattern, the increasing pattern is more popular among more functional classes during chromoplast differentiation. For example, 44.2% of proteins implicated in protein degradation showed an abundance increase during the chromoplast differentiation, with 28.8% having an early increase, 9.6% having a continuous increase, and 5.8% having a late increase. More strikingly, 83.3% of the proteins involved in metal handling showed either an early increase (77.8%) or a late increase (5.6%), suggesting that metal handling is implicated in chromoplast differentiation. Also notable is that 50% of the proteins involved in secondary metabolism and sulfur-metabolism showed increases in abundance mostly in the early stage, which is consistent with the anticipated increase in the synthesis of some important secondary metabolites, such as carotenoids, vitamin E, and terpene in chromoplasts at the Br stage. Additionally, 43.9% of redox- and stress-related proteins showed increases in abundance, possibly reflecting that more stresses were imposed on fruits, which in turn strengthened the stress responses during chromoplast differentiation.
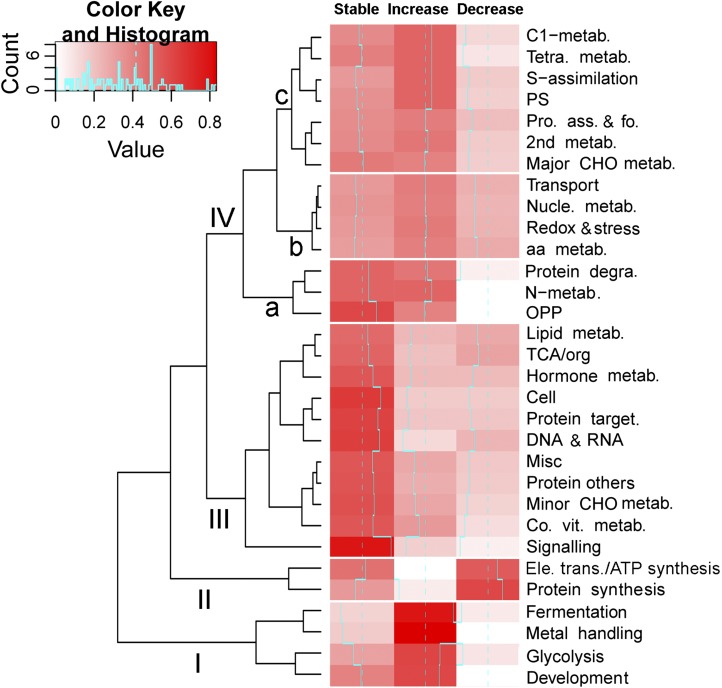
To further infer possible regulatory mechanisms underlying the changes of protein abundance, we constructed a heat map that represents the hierarchical clustering of the proteins into each functional class according to their abundance patterns. The results indicate that the four main clusters were distinguishable (Fig. 10). Among them, cluster III is enriched for stable proteins, including those involved in lipid metabolism, TCA/organic acid, hormone metabolism, cell, protein targeting, DNA and RNA, miscellaneous, protein others, minor carbohydrate metabolism, cofactor and vitamin metabolism, and signaling. Cluster II is the only cluster consisting of proteins with a strong decrease in abundance, including protein synthesis and electron transport/ATP synthesis, while cluster I is enriched for proteins with increased abundance that are involved in fermentation, metal handling, glycolysis, and development. Two subclusters of cluster IV (a and c) contain stable proteins or proteins with increased abundance and a few proteins with decreased abundance. Subcluster IVb contains proteins without a major bias in abundance patterns, and these proteins are involved in transport, nucleotide metabolism, redox and stress, and amino acid metabolism.
Figure 10.
Heat map showing the proportion of proteins of each functional class in each of the three abundance (stable, increase, and decrease) patterns during chromoplast differentiation. The magnitude of the percentage is represented by the color scale (top left) going from low (white) to high (red). aa, Amino acid; CHO, carbohydrate; PS, photosynthesis; OPP, oxidative pentose phosphate pathway; Pro. ass. & fo., protein assembly and folding; Co. vit. metab., cofactor/vitamin metabolism.
Functional Categories of Differentially Expressed Proteins between the OMa and Ma Stages
On-tree storage is an effective method to delay the senescence process and has been widely used in the citrus industry. During this process, a variety of physiological parameters are changed, such as the decrease of firmness and titratable acidity and the increase of color index and maturity index (Sánchez et al., 2013). However, the mechanisms underlying the regulation of chromoplast fate remains unclear. To screen for differentially expressed proteins during chromoplast senescence, the corresponding iTRAQ values of the samples from the OMa and Ma stages were averaged and compared. Since iTRAQ quantification might underestimate the amount of real fold change between the labeled samples (Karp et al., 2010), a protein with a difference of 1.4-fold or greater and P ≤ 0.05 after false discovery rate control was regarded as being differentially expressed between the OMa and Ma stages (Benjamini and Hochberg, 1995; Supplemental Table S6). Based on these criteria, the expression of 76 proteins was significantly higher in the OMa stage than in the Ma stage, while the expression of 28 proteins was the opposite. To gain insight into the biological significance of such differential expression, the 104 proteins were categorized into functional classes by MapMan. The 76 up-regulated proteins mainly fell into five categories: stress and redox, TCA/organic acid, transport, cell, and electron transport/ATP synthesis. With regard to the stress and redox class, 11 stress-related proteins had higher expression in the OMa stage than in the Ma stage, such as GLUTATHIONE PEROXIDASE6 proteins (Cs5g31640.1), early-responsive dehydration proteins (Cs2g08320.1), and MEMBRANE-ASSOCIATED PROGESTERONE-BINDING PROTEIN3 (Cs9g17130.1). The significant increase of these proteins involved in the stress response in this study is indicative of a high level of redox activity in citrus chromoplasts and suggestive of an important role of these proteins in sustaining the plant defense reaction during chromoplast senescence. Additionally, almost all the proteins categorized in the transport and electron transport/ATP synthesis classes showed dramatic increases in protein abundance at the OMa stage. Among them, there were up to 14 ATP synthases and ATP/ADP carriers, which were involved in energy supply (Supplemental Table S6). These proteins underwent dramatic and wide-ranging increases in protein abundance, suggesting that the ATP synthesis machinery was maintained at a good level during chromoplast senescence. The active supply of ATP might reflect the demand for energy to ensure normal biological processes in the course of chromoplast senescence. It is well known that ATP import or synthesis in nongreen plastids is required for the synthesis of several metabolites, such as amino acids and carbohydrates (Neuhaus and Emes, 2000). Consistently, several proteins for amino acid metabolism were significantly increased in abundance, and proteins involved in TCA/organic acid for the supply of energy to the plastids were also identified with a significant up-regulation. By contrast, some of the 28 down-regulated proteins are involved in major carbohydrate metabolism, hormone metabolism, protein degradation, and protein synthesis. Conceivably, the differential expression of so many plastid-localized proteins may directly or indirectly contribute to chromoplast senescence.
DISCUSSION
Two Types of Putative Amyloplast-to-Chromoplast Transition in Citrus Flesh
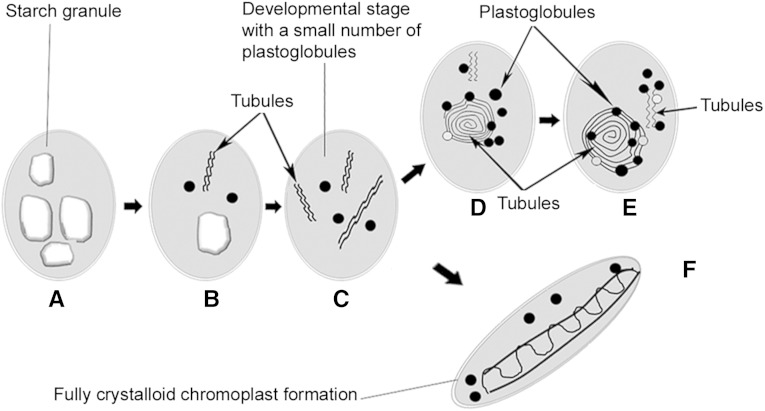
Unlike the flavedo of citrus fruits, where chromoplasts are converted from chloroplasts (Eilati et al., 1975), chromoplasts in the flesh of citrus fruits seem to be differentiated from amyloplasts. There are two putative different amyloplast-to-chromoplast patterns for citrus pulp, each with its own unique characteristics. Figure 11 depicts a tentative model for the sequential development of citrus flesh plastids. The starch granules were degraded rapidly and transformed into plastids with a few plastoglobules (Fig. 11, A and B). In Arabidopsis (Arabidopsis thaliana), the globules were documented to be swelled and separated from thylakoidal membranes (Ben-Shaul and Naftali, 1969; Austin et al., 2006). In our study, the mechanism underlying globular formation remains unknown, as the association of these plastoglobules with membranes or other related structural elements was not directly observed. However, we cannot rule out the possibility that the degradation of starch granules provides energy or metabolic precursors for the subsequent formation of globular elements, as plastoglobules were generally found around the degraded starch granules in the subsequent stage (Fig. 11D). Since key biosynthetic enzymes are targeted to plastoglobules (Ytterberg et al., 2006; Schweiggert et al., 2011; Davidi et al., 2015), carotenoid biosynthesis is supposed to be initiated in these globules. Additionally, the undulating lines in Figure 11C can also been interpreted as early carotenoid depositions in tomato (Harris and Spurr, 1969). In parallel with carotenoid accumulation, tubular structures apparently developed independently of the globular structure, resulting in the formation of tubular-globular chromoplasts (Fig. 11, D and E).
Figure 11.
Schematic diagram of chromoplast differentiation in the flesh of citrus fruits. A to E The globular chromoplast formation in yellow tissues. A to C and F, The crystalloid chromoplast development in red tissues. A, Amyloplasts. B and C, Starch granule degradation and the formation of plastids with globular and tubular structures. D and E, Typical globular and tubular chromoplasts. F, Chromoplasts with crystal remnants.
The crystal remnants were accumulated in the chromoplasts of red tissues, but the additional formation of comparably large crystalloids was apparently induced by the concomitant biosynthesis of lipid-soluble lycopene (Fig. 11F; Schweiggert et al., 2011). There are two hypotheses regarding the biogenesis of lycopene-rich structures: that the structures might (1) be associated with the thylakoid system in the transformation of tomato (Ben-Shaul and Naftali 1969) and Aglaonema commutatum (Knoth, 1981), where lycopene is tightly assembled into photosynthesis structures, or (2) derive from plastid membrane (Schweiggert et al., 2011). Since such crystal remnants were frequently found to be close to the plastid envelope (Fig. 11F) and thylakoid structures appeared to be absent within citrus flesh, the lycopenic membranes in red-fleshed cv Hong Anliu might be derived from the plastid envelop or other unknown structures specific to citrus flesh.
Complex Regulation of Carotenoid Metabolism during Globular Chromoplast Differentiation
Chromoplast development is accompanied by massive synthesis and the accumulation of carotenoids, and it is often regulated at the transcriptional level (Cazzonelli and Pogson, 2010), whereas the regulatory mechanism at the protein level is still poorly understood. In this study, we found that carotenoid metabolism is coordinately regulated by both transcription and protein levels (Figs. 6 and 12A). For example, four members of the MEP pathway, namely DXR (Cs5g05440.1), 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (Cs5g03050.1), hydroxymethylbutenyl 4-diphosphate synthase (Cs8g16700.2), and hydroxymethylbutenyl 4-diphosphate reductase (Cs8g07020.1), showed early or continuous increases (P < 0.05) in abundance during chromoplast differentiation, coinciding with the accumulation of isoprenoids and carotenoids. Additionally, the synthesis of violaxanthin and lutein needs to be facilitated by HYD catalysis (Cazzonelli and Pogson, 2010). In comparison with the significant increase of the HYD transcriptional level during chromoplast differentiation, which may explain the large accumulation of violaxanthin and lutein in globular chromoplasts, we only identified a limited number of enzymes participating in downstream carotenoid synthesis, for instance HYD (Cs9g19270.2) and NCED5 (Cs2g03270.1). Therefore, we presume that HYD mainly plays a key role in violaxanthin biosynthesis at the transcriptional level. Consistently, the expression of HYD promotes violaxanthin biosynthesis in Arabidopsis seeds (Yu et al., 2007) and citrus callus (Cao et al., 2012). The relatively higher abundance of upstream pathway proteins led to the biosynthesis of lycopene, while the downstream enzyme proteins were present at low abundance or not detected, which may be important for gauging the metabolic flux into the carotenoid synthesis pathway. Consistently, the carotenoid accumulation areas were formed as maturation proceeded (Fig. 5J).
Figure 12.
Abundance of proteins (y axis) involved in carotenoid accumulation, plastoglobule formation, ribosome assembly, as well as protein degradation and processing during amyloplast-to-chromoplast differentiation and chromoplast senescence. A, Proteins involved in the MEP pathway and carotenoid biosynthesis. B, Proteins involved in plastoglobule formation. C, Sum of all proteins involved in protein initiation, nucleus-encoded 40S subunit and 60S subunit as well as plastid-encoded 30S subunit and 50S subunit. D, Proteins involved in protein degradation and processing. Individual values are given in Supplemental Table S3B. Protein abundance is represented by the average of the ratios of iTRAQ values to internal standard ± sd. The full names of the proteins are indicated in the text and Supplemental Table S3B. HDR, Hydroxymethylbutenyl 4-diphosphate reductase; HDS, hydroxymethylbutenyl 4-diphosphate synthase (Cs8g16700.2); ISPF, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (Cs5g03050.1).
Plastoglobule Composition and Formation within Citrus Flesh Chromoplasts
A rapid increase in the number of plastoglobules is the most representative feature during chromoplastogenesis in citrus flesh. Therefore, we performed BLASTP of the citrus plastid proteome against the list of proteins of the plastoglobules established by Lundquist et al. (2012). Among the 30 plastoglobule-localized proteins in Arabidopsis, 24 were identified to be citrus homologs, 16 of which were quantified (Supplemental Table S7). Among the 16 quantified proteins, 11 had continuously/early increased expression (Fig. 12B), including ACTIVITY OF BC1 COMPLEX KINASE1 (ABC1K1), ABC1K3, ABC1K7, and ABC1K9, FBN2/4, ESTERASE1 (Cs2g14090.1, Cs2g14110.1, and Cs2g14120.1), FLAVIN REDUCTASE-RELATED2 (Cs2g03600.1), and an unknown protein (Cs5g28130.1).
Among the ABC1 kinase subfamily, six proteins (ABC1K1, ABC1K3, ABC1K5, ABC1K6, ABC1K7, and ABC1K9) predicted to localize in plastoglobules constitute the core components of the plastoglobule proteome in chromoplasts of citrus flesh (Supplemental Fig. S9), which is similar to the case of Arabidopsis chloroplasts (Lundquist et al., 2012). The Atabc1k7 mutant developed larger plastoglobules and lipid composition of the chloroplast membrane when compared with wide-type Arabidopsis (Manara et al., 2014, 2015). Additionally, ABC1K1/3, as a complex, binds with phosphorylated tocopherol cyclase to affect plastoquinone metabolism (Lundquist et al., 2013; Martinis et al., 2013, 2014; Manara et al., 2014). Their double mutation also leads to abnormal accumulation of plastoglobules in Arabidopsis. The increase of ABC1K1, ABC1K3, and ABC1K7 in abundance during chromoplastogenesis suggests that they might positively regulate plastoglobule development. A previous study revealed that ABC1K9 might be coexpressed with FBNs and MEP enzymes (Lundquist et al., 2012), suggesting that the increase of ABC1K9 in abundance might play an essential role in plastoglobule development and carotenoid metabolism. In this study, we observed an early or continuous increase in the abundance of ABC1K1, ABC1K3, ABC1K7, and ABC1K9 during the amyloplast-to-chromoplast transition, which confirms the speculation that these proteins may have distinct modulating functions in plastogobule development in citrus.
The FBN family is required for leaf or fruit development to regulate the early generation or extension of plastoglobules (Lundquist et al., 2012). In this study, the FBN family was the most abundant in chromoplast plastoglobules, accounting for 68.5% of the total identified peptides (Supplemental Fig. S9), which is similar to the case in Arabidopsis chloroplasts (Lundquist et al., 2012). It is not surprising to find an increasing abundance of FBN2/4 in our study during chromoplast differentiation, confirming its role in the structural development of plastoglobules (Singh and McNellis, 2011). FBN1 has been well established in several studies with a positive regulatory role in plastoglobule development (Singh and McNellis, 2011). In our proteomic analysis, it is noteworthy that FBN1 only showed a slight increase (P < 0.05) in abundance from MG to Br stages, followed by a slight decrease from Br to OMa stages (P < 0.05). This change tendency was confirmed by immunoblot analysis of FBN1 (Fig. 8), which is also in line with the variation in the number of plastoglobules from Br to OMa stages. However, the fold change in protein abundance was much lower than the rapid increase in the number of plastoglobules from MG to Br stages. To explain this discrepancy, we conducted another series of immunoblot analyses by including two earlier stages of fruit development (Supplemental Fig. S10), which confirmed the tendency of a slight increase in abundance from MG to Br stages (Fig. 8). More importantly, the abundance of FBN1 was enhanced rapidly from 90 to 120 d after flowering (DAF). These data indicate that FBN1 expression is sensitive at earlier stages, when the average number of plastoglobules per plastid increased from 0 to about 3.2, and the subsequent slowdown of the increase tendency might be due to the dull response of proteins to developmental conditions or environmental stresses on the basis of the preexisting plastoglobules or regulation via posttranslational modification, such as protein phosphorylation (Zeng et al., 2014). In this study, we selected the MG stage as the first stage for plastid differentiation analysis for two reasons: (1) the fruit at the MG stage gains the capacity to ripen, and its plastid ultrastructure shows an apparent difference from that in the subsequent maturation stages; and (2) the accumulation of high-density starches in amyloplasts at an earlier stage makes it difficult to use the same density gradient centrifugation to purify plastids. Taken together, our data concerning these plastoglobule-localized proteins suggest that they have important and exclusive functions in regulating the differentiation of chromoplasts in the flesh of citrus fruits.
Stability in Protein Import, Loss of Ribosome Assembly, and Buildup of Chromoplast Proteases
The translocation of proteins into plastids is of importance for chromoplast biogenesis (Egea et al., 2010). Here, we identified nearly all the elements of the protein import machinery and classified them into protein targeting, which showed a constant abundance during the amyloplast-to-chromoplast transition (Supplemental Table S5). Among these elements, translocon at the outer envelope membrane of chloroplasts159 (TOC159), which is tightly bound to the transit peptides, serves as a receptor for chloroplast-destined preproteins (Aronsson and Jarvis, 2008) and is thought to interact with CASEIN KINASE2 to affect chromoplast biogenesis (Agne et al., 2010; Zeng et al., 2014). TOC75 is the main pore of the TOC complex deeply embedded within the outer membrane, forming a translocation channel (Jarvis, 2008). Additionally, TIC110/40, major components of the TIC complex, play a critical role in protein import. These four proteins remained unchanged in abundance, among which TIC40 was confirmed by immunoblot analysis. The constant protein abundance of the TOC/TIC complex suggests the continuous import of nucleus-encoded proteins during chromoplast differentiation.
Consistent with a previous finding in the chloroplast-to-chromoplast transition in tomato (Barsan et al., 2012), the translation machinery of the citrus chromoplast underwent a great loss of efficiency with a strong decrease in abundance of ribosomal proteins of both the chloroplast-encoded small 30S subunit and the large 50S subunit complexes as well as those of the nucleus-encoded small 40S subunit and the large 60S subunit during chromoplast differentiation (Fig. 12C). This finding is also in agreement with the gradual down-regulation of plastid translation during tomato chromoplast differentiation observed by Kahlau and Bock (2008). These results indicate that there probably exists a conservative property of translation machinery in chromoplast biogenesis, which arises not only from nonphotosynthetic plastids like amyloplasts but also from photosynthetic chloroplasts.
Plastids undergo drastic changes morphologically and physiologically at different developmental stages and under different environmental conditions (Egea et al., 2010). Keeping a balance between protein biosynthesis and degradation/processing is crucial for accomplishing structurally dynamic changes of plastids and maintaining their homeostasis (Sakamoto, 2006). A set of caseinolytic peptidase (Clp) proteases increased in abundance during chromoplast differentiation, including five continuous increase, 15 early increase, and three late increase proteins, while only three proteins showed the decreasing pattern (Fig. 12D; Supplemental Table S5). These Clp proteases formed a chaperone complex to drive the import of proteins into the plastids, cleave the transit peptide, and recycle plastid components. In Arabidopsis, knocking out most members of the Clp protease family results in reduced plant growth with pale-green or variegated leaf phenotype, indicating the crucial role of the Clp complex in chloroplast development (Meinke et al., 2008; Liu et al., 2010; Olinares et al., 2011). Previous studies have revealed that the Clp complex, one of the prominent complexes, increases in abundance during the transition from proplastids or etioplasts into chloroplasts (Kanervo et al., 2008; Majeran et al., 2010). Our study shows that the proteins involved in protein degradation are among the proteins of the highest abundance during the amyloplast-to-chromoplast transition, while an apparent increasing tendency in protein abundance of the Clp complex suggests that the machinery of protein degradation or processing during chromoplast differentiation might have distinct modulating functions that are conserved during the biogenesis of both chloroplasts and chromoplasts.
The plastid proteome of citrus flesh also consists of a number of membrane-bound ATP-dependent zinc-metalloproteases (FtsH1, Cs3g14380; FtsH2, Cs2g15180; FtsH5, Cs3g27030; FtsH7, Cs8g19900; FtsH11, Cs9g18420; FtsH12, orange1.1t03252; FtsHi1, Cs5g02060; FtsHi2, Cs1g15540; and FtsHi3, Cs7g14690; Supplemental Table S5). Several FtsH proteins, such as FtsH1, FtsH2, and FtsH5, are key components of the thylakoid, which has been well demonstrated by previous studies (Zaltsman et al., 2005; Sakamoto, 2006; Kato and Sakamoto, 2010). Since citrus flesh lacks chloroplasts and cannot perform photosynthesis throughout the entire maturation stage, it is reasonable to speculate that these proteins are localized in other subcellular structures within plastids rather than thylakoids. Therefore, the identification of these proteins in nonphotosynthetic plastids may imply a basic role other than photosynthesis. Additionally, another type of ATP-independent metalloprotease present in the citrus flesh plastid proteome, such as stromal presequence proteases (PreP2; Cs8g05720.1) and M24 peptidase (Cs4g13510.1), has been described as necessary for plastid development (Cederholm et al., 2009; Kmiec and Glaser, 2012). The continuous increase of PreP2 and M24 peptidase probably contributed to chromoplast differentiation by means of the removal and degradation of transit peptides within the plastids during the amyloplast-to-chromoplast transition. In summary, our data suggest that chromoplast differentiation in citrus flesh is correlated not only with the repression of protein synthesis and the promotion of protein degradation/processing but also with the stable maintenance of protein import via the TOC/TIC complex. These dynamic changes are associated with the remodeling of protein systems and may have a crucial role in governing chromoplast biogenesis and differentiation.
CONCLUSION
In this study, we examined two tissue-specific chromoplast types converted from amyloplasts, which are associated with the composition of carotenoids in sweet orange. Subsequently, we used iTRAQ-based high-throughput proteomics to construct an inventory of the proteins present at different fruit maturation stages in sweet orange during plastid differentiation, which expands the knowledge of the plant plastid proteome available so far. Additionally, our results highlight several aspects of the regulation of global reorganization during chromoplast differentiation. The change pattern in protein abundance reported here is partially in agreement with the chromoplast differentiation in tomato (Barsan et al., 2012) but shows great differences from the proplastid/etioplast-to-chloroplast transition in maize/rice (Kleffmann et al., 2007; Majeran et al., 2010). The overall findings from this study provide useful information about chromoplast biogenesis in sweet orange flesh and facilitate a better understanding of its chromoplast differentiation and senescence.
MATERIALS AND METHODS
Plant Materials
Fruits of sweet orange (Citrus sinensis ‘Anliu’) and its red-fleshed mutant cv Hong Anliu as well as cv Newhall Navel and its red-fleshed mutant cv Cara Cara were harvested at the National Centre of Citrus Breeding located at Huazhong Agricultural University, Wuhan, China. Fruits of cv Hong Anliu and cv Anliu were collected at six maturation stages after careful selection: (1) IMG (90 DAF); (2) MG (140 DAF), at which fruit nearly reaches full size with green peel and without pigment accumulation in the flesh; (3) Br (170 DAF), at which fruit is characterized by a change in color from green to a bit of orange in the peel and a bit of red in the flesh; (4) Ma (200 DAF), at which fruit has a red color in the whole flesh and is harvested 30 d after the Br stage; (5) FMa (230 DAF); and (6) OMa (260 DAF), at which fruit is overmature and stored on the tree under a low temperature in winter. Fruits at each stage were harvested from three different trees, and 10 representative fruits were harvested from each tree; thus, a total of 30 fruits were obtained for each genotype. The edible citrus segment was then gently separated into segment membrane, sac membrane, and juice, followed by freezing in liquid nitrogen for subsequent extraction of carotenoids and RNA. Fruits of cv Hong Anliu at MG, Br, Ma, and OMa stages were sampled from three selected trees on each harvest day, with 40 representative fruits from each tree; thus, a total of 120 fruits per stage were harvested and divided into three aliquots for immediate chromoplast purification as described previously (Zeng et al., 2014). Plastids of flesh were prepared in three biological replicates, followed by immediate freezing in liquid nitrogen and storage at −80°C until use.
Microscopy Observation and Analysis
The segment membrane and sac membrane were carefully cut into small slices using a frozen sectioning technique (CM1900; Leica). A drop of juice was gently placed onto microscope slides. These fresh tissues were immediately observed with a microscope (BX61; Olympus) equipped with a DP70 camera to compare the structural characteristics of chromoplasts.
An aliquot of the same segment membrane and sac membrane was also used for TEM analysis. The plastids were enriched from the juice using gradient centrifugation as described by Zeng et al. (2014). Samples for TEM analysis were fixed, stained, and observed as described previously (Zeng et al., 2011). The diameter of the plastids was measured by (major axis + minor axis)/2 using ImageJ software (http://rsbweb.nih.gov/ij/).
Carotenoid Profile Analysis
Carotenoid extraction and analysis using reverse-phase HPLC were conducted as described previously (Cao et al., 2012). Approximately 4 g of citrus fruit fresh weight was extracted separately from the segment membrane, sac membrane, and juice from cv Anliu and cv Hong Anliu at the FMa stage. The carotenoids were identified by their characteristic absorption spectra and typical retention times based on the literature and standards from CaroNature Co. (Bern, Switzerland). The output data were obtained from three experimental replicates.
Quantitative Analysis of Gene Expression
RNA was extracted from segment membranes and juice of cv Anliu and cv Hong Anliu fruits as described by Liu et al. (2007). RNA quality and quantity measurement, first-strand complementary DNA synthesis, and real-time reverse transcription (RT)-PCR were performed according to Cao et al. (2012). The primer pairs used in this experiment came from Cao et al. (2012) or were designed using Primer Express 3 software (Applied Biosystems), both of which are shown in Supplemental Table S1. ACTIN was used as an endogenous control to normalize the expression in different samples. At least three replicates were conducted in real-time RT-PCR analysis.
Protein Extraction and Western Blot
Polyclonal antibodies were diluted as appropriate against RbcL (1:3,000), FBN1 (1:2,000), two thylakoid membrane marker proteins (PsbA|D1 protein of PSII, C terminal, 1:2,000; and Lhcb2|LHCII type II chlorophyll a/b-binding protein, 1:2,000), and chloroplast inner envelope membrane TIC40 (1:2,000). RbcL, FBN1, PsbA, and Lhcb2 were obtained from Agrisera, and TIC40 was provided by Paul Jarvis’s laboratory (University of Oxford). A total of 30 μg of protein was separated by 12% (w/v) SDS-PAGE, and detection by antibodies were performed as described by Zeng et al. (2011) or Ling et al.(2012). At least two independent replicates were conducted for each western-blot analysis.
Protein Preparation, Digestion, and iTRAQ Labeling
The pooled plastids were treated with acetone, and the precipitated proteins were resuspended by adding approximately 500 μL of STD buffer (4% [w/v] SDS, 1 mm dithiothreitol [DTT], and 150 mm Tris-HCl, pH 8, containing cocktail for proteases [EDTA free; Roche]). After 5 min of incubation in boiling water, the homogenate was sonicated on ice. The crude extract was then incubated in boiling water again and clarified by centrifugation at 14,000g for 40 min at 25°C before the supernatant was collected. The protein concentration was determined using a bicinchoninic acid protein assay kit (Beyotime).
Protein digestion was performed based on the Filter Assisted Sample Preparation method as described previously (Wiśniewski et al., 2009), and the resulting peptide mixtures were labeled with the eight-plex iTRAQ reagent by following the manufacturer’s instructions (Applied Biosystems). A total of 80 μg of protein from each sample was pooled as an internal standard. Briefly, 250 μg of protein for each sample (including the internal standard) was incorporated into 30 μL of 4% (w/v) SDS, 150 mm Tris-HCl, pH 8, and 100 mm DTT. UA buffer (8 m urea and 150 mm Tris-HCl, pH 8) was used to remove DTT, detergent, and other low-molecular-mass components by repeated ultrafiltration (Microcon units; 30 kD). Next, the samples were incubated in darkness for 30 min by adding 100 μL of 50 mm iodoacetamide into UA buffer to block the reduced Cys residues. The filters were washed with 100 μL of UA buffer three times followed by two washes with 100 μL of DS buffer (50 mm triethylammoniumbicarbonate, pH 8.5). Finally, the protein suspension was digested overnight at 37°C with 6 μg of trypsin (Promega) dissolved in 40 μL of DS buffer, and the resulting peptides were collected as a filtrate. The peptide content was estimated by UV light spectral density at 280 nm using an extinction coefficient of 1.1 of 0.1% (g L−1) solution that was calculated on the basis of the frequency of Trp and Tyr in vertebrate proteins.
Two iTRAQ reagent eight-plex kits (Applied Biosystems) were used to label the peptide samples (each 25 μg) of plastids isolated at each developmental stage according to the manufacturer’s specifications (Fig. 7). Three biological replicates were iTRAQ labeled for MG, Br, Ma, and OMa stages, as well as one technological replicate for MG and Br stages. Protein samples of each stage were equally mixed as internal standard and then labeled with iTRAQ 113 tag. MG-3_E2_115 and Br-1_E1_117 were technological replicates for MG-4_E2_116 and Br-2_E1_121, respectively.
Sample Fractionation Using Strong Cation-Exchange Chromatography
Prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, peptides were purified from excess labeling reagent by strong cation-exchange chromatography using the AKTA Purifier 100 (GE Healthcare). Peptides were dried in a vacuum concentrator and then dissolved in 2 mL of buffer A (10 mm KH2PO4 in 25% [v/v] acetonitrile [ACN], pH 3) to reconstitute and acidify the dried peptide mixtures, which were subsequently loaded onto a PolySULFOETHYL 4.6- × 100-mm column (5 µm, 200 Å; PolyLC) at a flow rate of 1 mL min−1. A suitable gradient elution (0%–10% for 32 min, 10%–20% for 10 min, 20%–45% for 15 min, and 45%–100% for 13 min) was applied to separate the peptides at a flow rate of 1 mL min−1 with buffer B (10 mm KH2PO4 and 500 mm KCl in 25% [v/v] ACN; pH 3). Eluted peptides were collected and desalted by an offline fraction collector. Finally, a total of 10 sample pools including 30 collected fractions were desalted on C18 cartridges (Empore SPE Cartridges [standard density], bed i.d. 7 mm, volume 3 mL; Sigma). Each final fraction was concentrated by a vacuum concentrator and reconstituted in 40 µL of 0.1% (v/v) trifluoroacetic acid. All samples were stored at −80°C for LC-MS/MS analysis.
LC-MS/MS Analysis by Q Exactive
A nanoflow HPLC instrument (Easy nLC; ProxeonBiosystems, now Thermo Fisher Scientific) was coupled online to a Q Exactive mass spectrometer. The peptide mixture (5 μg) was loaded onto a C18 reverse-phase column (Thermo Scientific Easy Column; 10 cm long, 75 μm i.d., 3 μm resin) in buffer A (0.1% [v/v] formic acid) and separated with a linear gradient of buffer B (0.1% [v/v] formic acid and 80% [v/v] ACN) at a flow rate of 250 nL min−1 controlled by IntelliFlow technology over 140 min.
Mass spectrometry data were acquired using a data-dependent top 10 method dynamically choosing the most abundant precursor ions from the survey scan (mass-to-charge ratio [m/z] 300–1,800) for higher energy collisional dissociation fragmentation. Determination of the target value was based on predictive automatic gain control. The dynamic exclusion duration was 60 s. Survey scans were acquired at a resolution of 70,000 at m/z 200, and the resolution for higher energy collisional dissociation spectra was set to 17,500 at m/z 200. Normalized collision energy was 27 eV, and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
Tandem mass spectrometry spectra were searched using the MASCOT engine (Matrix Science; version 2.2) embedded into Proteome Discoverer 1.3 (Thermo Electron) against the nonredundant Protein Index C. sinensis protein database (Huazhong Agricultural University; http://citrus.hzau.edu.cn) and the proteome sequences predicted from the chloroplastic genome (accession no. NC_008334). Proteins were identified with the following parameters: peptide mass tolerance of 20 ppm, fragment mass tolerance of 0.1 D, trypsin enzyme with the number of missed cleavages up to 2, carbamidomethyl Cys residues as fixed modification, and oxidation of Met residues as variable modification. Proteomics Tools (version 3.1.6) was used to further process the MASCOT search results of each strong cation-exchange elution. The BuildSummary program was used to assemble protein identifications according to a target decoy in shotgun proteomics. All reported data were based on 99% confidence intervals for protein identification as determined by false discovery rate of 1% or less. False discovery rate = N (decoy) × 2/(N (decoy) + N (target)), which is a widespread normalization process and considered a standard way to preprocess data in proteomics (Zeng et al., 2014; Zheng et al., 2014).
Search against Existing Databases, Functional Classification, Targeting Predictions, and Curation
Identified proteins were functionally classified according to MapMan (http://mapman.gabipd.org/web/guest/home) using annotations retrieved from databases and manual curation based on PPDB (http://ppdb.tc.cornell.edu). Homologies of the detected proteins were searched against five plastidial databases: (1) two databases specific to plastids (Plprot, http://www.plprot.ethz.ch/; and AT_CHLORO, http://www.grenoble.prabi.fr/at_chloro/); and (2) three databases comprising plastidial subsets (SUBA, http://suba.plantenergy.uwa.edu.au/; PPDB, http://ppdb.tc.cornell.edu/; and Uniprot, http://www.uniprot.org/). Predictions of subcellular localization were carried out using TargetP (http://www.cbs.dtu.dk/services/TargetP/), Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html), and WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html). Those proteins were tentatively considered to be plastid proteins if they met at least one of the following two criteria: (1) the protein candidates were predicted to localize in plastids by at least one of the three Web programs (Wolf PSORT, TargetP, and Predotar); and (2) the protein candidates were present in at least two of the five databases mentioned above (Plprot, AT_CHLORO, SUBA, PPDB, and Uniprot).
Differential Abundance Analysis
The final ratios of proteins were normalized by the median average protein ratio of an equal mix of different labeled samples. Only proteins detected in all runs (every biological replicate) were included in the data set. Data are expressed as means ± sd and were evaluated by Student’s t test using Microsoft Excel (TTEST function [two tailed]). The trends of changes in abundance of proteins between stages were calculated with a 5% significance level and referred to as 0 for no change, −1 for decrease, and +1 for increase.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Structural diversity of chromoplasts from the flesh of cv Cara Cara and cv Newhall Navel.
Supplemental Figure S2. Ultrastructures of chromoplasts from the flesh of cv Cara Cara and cv Newhall Navel.
Supplemental Figure S3. The outer monolayer of plastoglobules appeared to be separated from the internal body at OMa stage.
Supplemental Figure S4. Immunoblotting analysis reveals the absence of chloroplasts within citrus flesh during fruit maturity.
Supplemental Figure S5. Expression of carotenoid metabolic genes during the formation of chromoplasts at different maturation stages.
Supplemental Figure S6. Distribution of peptide molecular weight and of peptide coverage.
Supplemental Figure S7. Analysis of reproducibility of iTRAQ biological replicates and technological replicates of each development stage.
Supplemental Figure S8. Venn diagram of the number of proteins of the citrus plastid proteome predicted to be plastid localized by three predictors and five plastidial databases.
Supplemental Figure S9. Relative mass contributions of the 30 plastoglobule core proteins to the total core plastoglobule proteome for Arabidopsis chloroplasts and sweet orange flesh chromoplasts.
Supplemental Figure S10. Abundance of FBN1 during fruit maturation.
Supplemental Table S1. Primers used for real-time RT-PCR.
Supplemental Table S2. Identified peptides with iTRAQ values in this experiment.
Supplemental Table S3. iTRAQ data of proteomics analysis during chromoplast differentiation and a list of quantitative proteins.
Supplemental Table S4. Characteristics of the 1,386 citrus pulp plastid protein candidates identified by iTRAQ.
Supplemental Table S5. Set of 1,016 plastid proteins that have been quantified during chromoplast differentiation and senescence.
Supplemental Table S6. Set of 104 differentially expressed proteins between OMa and Ma stages.
Supplemental Table S7. The core proteome of plastoglobules in citrus chromoplasts.
Supplementary Material
Acknowledgments
We thank the Guangxi Citrus Research Institute for providing citrus materials and Shanghai Applied Protein Technology for technology support and helpful advice; Paul Jarvis for the generous gift of antibody (TIC40); Zuoxiong Liu and Hanchang Zhu for reading and editing the article; and Jianbo Cao (Huazhong Agricultural University, China) for technical assistance.
Glossary
- iTRAQ
isobaric tag for relative and absolute quantitation
- TEM
transmission electron microscopy
- IMG
immature green
- MG
mature green
- Br
breaker
- FMa
full maturation
- Ma
maturation
- OMa
overmaturation
- MEP
plastidic 2-C-methyl-d-erythritol 4-phosphate
- DAF
days after flowering
- Clp
caseinolytic peptidase
- DTT
dithiothreitol
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- ACN
acetonitrile
- m/z
mass-to-charge ratio
Footnotes
This work was supported by the National 973 Project of China (grant no. 2011CB100601) and the National Natural Science Foundation of China (grant nos. 31330066 and 31221062).
Articles can be viewed without a subscription.
References
- Agne B, Andrès C, Montandon C, Christ B, Ertan A, Jung F, Infanger S, Bischof S, Baginsky S, Kessler F (2010) The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol 153: 1016–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Jarvis P (2008) The chloroplast protein import apparatus, its components, and their roles. In AS Sandelius, H Aronsson, eds, The Chloroplast: Interactions with the Environment. Plant Cell Monographs, Vol 13. Springer-Verlag, Heidelberg, Germany, pp 89–123
- Austin JR II, Frost E, Vidi PA, Kessler F, Staehelin LA (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsan C, Zouine M, Maza E, Bian W, Egea I, Rossignol M, Bouyssie D, Pichereaux C, Purgatto E, Bouzayen M, et al. (2012) Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol 160: 708–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Ben-Shaul Y, Naftali Y (1969) The development and ultrastructure of lycopene bodies in chromoplasts of Lycopersicum esculentum. Protoplasma 67: 333–344 [Google Scholar]
- Bian W, Barsan C, Egea I, Purgatto E, Chervin C, Zouine M, Latché A, Bouzayen M, Pech JC (2011) Metabolic and molecular events occurring during chromoplast biogenesis. J Bot 2011: 289859 [Google Scholar]
- Cao H, Zhang J, Xu J, Ye J, Yun Z, Xu Q, Xu J, Deng X (2012) Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J Exp Bot 63: 4403–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Cederholm SN, Bäckman HG, Pesaresi P, Leister D, Glaser E (2009) Deletion of an organellar peptidasome PreP affects early development in Arabidopsis thaliana. Plant Mol Biol 71: 497–508 [DOI] [PubMed] [Google Scholar]
- Davidi L, Levin Y, Ben-Dor S, Pick U (2015) Proteome analysis of cytoplasmatic and plastidic β-carotene lipid droplets in Dunaliella bardawil. Plant Physiol 167: 60–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian W, Purgatto E, Latché A, Chervin C, Bouzayen M, Pech JC (2010) Chromoplast differentiation: current status and perspectives. Plant Cell Physiol 51: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Eilati SK, Budowskj P, Monselise SP (1975) Carotenoid changes in the ‘Shamouti’ orange peel during chloroplast-chromoplast transformation on and off the tree. J Exp Bot 26: 624–632 [Google Scholar]
- Eilati SK, Monselise SP, Budowski P (1969) Seasonal development of external color and carotenoid content in the peel of ripening ‘Shamouti’ oranges. J Am Soc Hortic Sci 94: 346–348 [Google Scholar]
- Fanciullino AL, Cerćos M, Dhique-Mayer, Froelicher Y, Talón M, Ollitrault P, Morillon R (2008) Changes in carotenoid content and biosynthetic gene expression in juice sacs of four orange varieties (Citrus sinensis) differing in flesh fruit color. J Agric Food Chem 56: 3628–3638 [DOI] [PubMed] [Google Scholar]
- Fanciullino AL, Dhuique-Mayer C, Luro F, Casanova J, Morillon R, Ollitrault P (2006) Carotenoid diversity in cultivated citrus is highly influenced by genetic factors. J Agric Food Chem 54: 4397–4406 [DOI] [PubMed] [Google Scholar]
- Frey-Wyssling A, Schwegler F (1965) Ultrastructure of the chromoplasts in the carrot root. J Ultrastruct Res 13: 543–559 [DOI] [PubMed] [Google Scholar]
- Grilli Caiola M, Canini A (2004) Ultrastructure of chromoplasts and other plastids in Crocus sativus L. (Iridaceae). Plant Biosyst 138: 43–52 [Google Scholar]
- Harris WM, Spurr AR (1969) Chromoplasts of tomato fruits. II. The red tomato. Am J Bot 56: 380–389 [Google Scholar]
- Hempel J, Amrehn E, Quesada S, Esquivel P, Jiménez VM, Heller A, Carle R, Schweiggert RM (2014) Lipid-dissolved γ-carotene, β-carotene, and lycopene in globular chromoplasts of peach palm (Bactris gasipaes Kunth) fruits. Planta 240: 1037–1050 [DOI] [PubMed] [Google Scholar]
- Horner HT, Healy RA, Ren G, Fritz D, Klyne A, Seames C, Thornburg RW (2007) Amyloplast to chromoplast conversion in developing ornamental tobacco floral nectaries provides sugar for nectar and antioxidants for protection. Am J Bot 94: 12–24 [DOI] [PubMed] [Google Scholar]
- Iglesias DJ, Cercós M, Colmenero-Flores JM, Naranjo MA, Ríos G, Carrera E, Ruiz-Rivero O, Lliso I, Morillon R, Tadeo FR (2007) Physiology of citrus fruiting. Braz J Plant Physiol 19: 333–362 [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Kahlau S, Bock R (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanervo E, Singh M, Suorsa M, Paakkarinen V, Aro E, Battchikova N, Aro EM (2008) Expression of protein complexes and individual proteins upon transition of etioplasts to chloroplasts in pea (Pisum sativum). Plant Cell Physiol 49: 396–410 [DOI] [PubMed] [Google Scholar]
- Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics 9: 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2010) New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Kilcrease J, Collins AM, Richins RD, Timlin JA, O’Connell MA (2013) Multiple microscopic approaches demonstrate linkage between chromoplast architecture and carotenoid composition in diverse Capsicum annuum fruit. Plant J 76: 1074–1083 [DOI] [PubMed] [Google Scholar]
- Kim JE, Rensing KH, Douglas CJ, Cheng KM (2010) Chromoplasts ultrastructure and estimated carotene content in root secondary phloem of different carrot varieties. Planta 231: 549–558 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, von Zychlinski A, Russenberger D, Hirsch-Hoffmann M, Gehrig P, Gruissem W, Baginsky S (2007) Proteome dynamics during plastid differentiation in rice. Plant Physiol 143: 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec B, Glaser E (2012) A novel mitochondrial and chloroplast peptidasome, PreP. Physiol Plant 145: 180–186 [DOI] [PubMed] [Google Scholar]
- Knoth R. (1981) Ultrastructure of lycopene containing chromoplasts in fruits of Aglaonema commutatum Schott (Araceae). Protoplasma 106: 249–259 [Google Scholar]
- Knoth R, Hansmann P, Sitte P (1986) Chromoplasts of Palisota barteri, and the molecular structure of chromoplast tubules. Planta 168: 167–174 [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H (2013) Chromoplast biogenesis and carotenoid accumulation. Arch Biochem Biophys 539: 102–109 [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659 [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu J, Liu Y, Zhao X, Deng X, Guo L, Gu J (2007) A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J Exp Bot 58: 4161–4171 [DOI] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S (2010) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J Integr Plant Biol 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol 158: 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Giacomelli L, Friso G, Appel M, McQuinn RP, Krasnoff SB, Rowland E, Ponnala L, Sun Q, et al. (2013) Loss of plastoglobule kinases ABC1K1 and ABC1K3 causes conditional degreening, modified prenyl-lipids, and recruitment of the jasmonic acid pathway. Plant Cell 25: 1818–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel E, Zhang C, Asakura Y, Bhuiyan NH, Sun Q, et al. (2010) Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manara A, DalCorso G, Guzzo F, Furini A (2015) Loss of the atypical kinases ABC1K7 and ABC1K8 changes the lipid composition of the chloroplast membrane. Plant Cell Physiol 56: 1193–1204 [DOI] [PubMed] [Google Scholar]
- Manara A, DalCorso G, Leister D, Jahns P, Baldan B, Furini A (2014) AtSIA1 and AtOSA1: two Abc1 proteins involved in oxidative stress responses and iron distribution within chloroplasts. New Phytol 201: 452–465 [DOI] [PubMed] [Google Scholar]
- Martinis J, Glauser G, Valimareanu S, Kessler F (2013) A chloroplast ABC1-like kinase regulates vitamin E metabolism in Arabidopsis. Plant Physiol 162: 652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis J, Glauser G, Valimareanu S, Stettler M, Zeeman SC, Yamamoto H, Shikanai T, Kessler F (2014) ABC1K1/PGR6 kinase: a regulatory link between photosynthetic activity and chloroplast metabolism. Plant J 77: 269–283 [DOI] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Montefiori M, McGhie TK, Hallett IC, Costa G (2009) Changes in pigments and plastid ultrastructure during ripening of green-fleshed and yellow-fleshed kiwifruit. Sci Hortic (Amsterdam) 119: 377–387 [Google Scholar]
- Neuhaus HE, Emes MJ (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Nogueira M, Mora L, Enfissi EM, Bramley PM, Fraser PD (2013) Subchromoplast sequestration of carotenoids affects regulatory mechanisms in tomato lines expressing different carotenoid gene combinations. Plant Cell 25: 4560–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PD, Kim J, van Wijk KJ (2011) The Clp protease system: a central component of the chloroplast protease network. Biochim Biophys Acta 1807: 999–1011 [DOI] [PubMed] [Google Scholar]
- Sánchez MT, La Haba D, Pérez-Marín D (2013) Internal and external quality assessment of mandarins on-tree and at harvest using a portable NIR spectrophotometer. Comput Electron Agric 92: 66–74 [Google Scholar]
- Sakamoto W. (2006) Protein degradation machineries in plastids. Annu Rev Plant Biol 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Schweiggert RM, Steingass CB, Heller A, Esquivel P, Carle R (2011) Characterization of chromoplasts and carotenoids of red- and yellow-fleshed papaya (Carica papaya L.). Planta 234: 1031–1044 [DOI] [PubMed] [Google Scholar]
- Sheng Q, Dai J, Wu Y, Tang H, Zeng R (2012) BuildSummary: using a group-based approach to improve the sensitivity of peptide/protein identification in shotgun proteomics. J Proteome Res 11: 1494–1502 [DOI] [PubMed] [Google Scholar]
- Siddique MA, Grossmann J, Gruissem W, Baginsky S (2006) Proteome analysis of bell pepper (Capsicum annuum L.) chromoplasts. Plant Cell Physiol 47: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Singh DK, McNellis TW (2011) Fibrillin protein function: the tip of the iceberg? Trends Plant Sci 16: 432–441 [DOI] [PubMed] [Google Scholar]
- Vasquez-Caicedo AL, Heller A, Neidhart S, Carle R (2006) Chromoplast morphology and β-carotene accumulation during postharvest ripening of mango cv. ‘Tommy Atkins’. J Agric Food Chem 54: 5769–5776 [DOI] [PubMed] [Google Scholar]
- Wang YQ, Yang Y, Fei Z, Yuan H, Fish T, Thannhauser TW, Mazourek M, Kochian LV, Wang X, Li L (2013) Proteomic analysis of chromoplasts from six crop species reveals insights into chromoplast function and development. J Exp Bot 64: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts: a surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Lydiate DJ, Schäfer UA, Hannoufa A (2007) Characterization of a β-carotene hydroxylase of Adonis aestivalis and its expression in Arabidopsis thaliana. Planta 226: 181–192 [DOI] [PubMed] [Google Scholar]
- Zaltsman A, Ori N, Adam Z (2005) Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Pan Z, Ding Y, Zhu A, Cao H, Xu Q, Deng X (2011) A proteomic analysis of the chromoplasts isolated from sweet orange fruits [Citrus sinensis (L.) Osbeck]. J Exp Bot 62: 5297–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Pan Z, Wang L, Ding Y, Xu Q, Xiao S, Deng X (2014) Phosphoproteomic analysis of chromoplasts from sweet orange during fruit ripening. Physiol Plant 150: 252–270 [DOI] [PubMed] [Google Scholar]
- Zheng BB, Fang YN, Pan ZY, Sun L, Deng XX, Grosser JW, Guo WW (2014) iTRAQ-based quantitative proteomics analysis revealed alterations of carbohydrate metabolism pathways and mitochondrial proteins in a male sterile cybrid pummelo. J Proteome Res 13: 2998–3015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.