Invertase activity is affected by a protein complex to modulate accumulation of reducing sugars in cold-stored potato tubers.
Abstract
Slowing down cold-induced sweetening (CIS) of potato (Solanum tuberosum) tubers is of economic importance for the potato industry to ensure high-quality products. The conversion of sucrose to reducing sugars by the acid invertase StvacINV1 is thought to be critical for CIS. Identification of the specific StvacINV1 inhibitor StInvInh2B and the α- and β-subunits of the interacting protein SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE from the wild potato species Solanum berthaultii (SbSnRK1) has led to speculation that invertase activity may be regulated via a posttranslational mechanism that remains to be elucidated. Using bimolecular fluorescence complementation assays, this study confirmed the protein complex by pairwise interactions. In vitro kinase assays and protein phosphorylation analysis revealed that phosphorylation of SbSnRK1α is causal for StvacINV1 activity and that its active form blocks the inhibition of StInvInh2B by SbSnRK1β, whereas its inactive form restores the function of SbSnRK1β that prevents StInvInh2B from repressing StvacINV1. Overexpression of SbSnRK1α in CIS-sensitive potato confirmed that SbSnRK1α has significant effects on acid invertase-associated sucrose degradation. A higher level of SbSnRK1α expression was accompanied by elevated SbSnRK1α phosphorylation, reduced acid invertase activity, a higher sucrose-hexose ratio, and improved chip color. Our results lend new insights into a subtle regulatory mode of invertase activity and provide a novel approach for potato CIS improvement.
Suc is essential for growth and development in higher plants. Invertase (EC 3.2.1.26; β-fructofuranosidase) is an enzyme that mediates the hydrolytic cleavage of Suc into hexose monomers to meet the plant’s physiological requirements for carbohydrate transport, sugar signaling, and the stress response (Roitsch and González, 2004). A great concern in potato (Solanum tuberosum) tubers is the accumulation of reducing sugars (RS) when tubers are stored at low temperature (normally less than 10°C), a process known as cold-induced sweetening (CIS). CIS enhances the Maillard reaction, which results in undesired browning of potato chips and acrylamide formation in processed products (Tareke et al., 2002). The conversion of Suc into Glc and Fru has been reported to be one of the main pathways involved in potato CIS, and variation in the invertase activity of cold-stored tubers has been elucidated as a key factor determining the CIS-resistance level of potato (Cheng et al., 2004; Liu et al., 2011).
The acid invertases, which localize to the cell wall or vacuole and cleave Suc most efficiently between pH 4.5 and 5, are more relevant to potato CIS than neutral/alkaline invertases localized to the cytoplasm that cleave Suc most efficiently between pH 6.8 and 8 (Zrenner et al., 1996; Marian et al., 2005). Potato has at least six acid invertase genes, four cell wall invertase genes, and two vacuolar invertase genes (Liu et al., 2011). Among three acid invertase genes detected in potato tubers, the vacuolar invertase gene StvacINV1 was found to have the highest expression level and to be strongly induced by low temperature (Liu et al., 2011). Suppression of acid invertase activity by silencing StvacINV1 in potato resulted in a strong reduction of RS accumulation in cold-stored tubers, indicating that StvacINV1 activity is a major cause of CIS (Bhaskar et al., 2010; Liu et al., 2011; Wu et al., 2011). However, the transcript abundance of StvacINV1 does not always correlate with RS content in cold-stored tubers (Matsuura-Endo et al., 2004; Liu et al., 2011). These findings have led to the hypothesis of posttranslational regulation of StvacINV1 activity (Liu et al., 2011).
Since the early 1960s, invertase inhibitors have been postulated to modulate invertase activity (Schwimmer et al., 1961; Pressey, 1967). With the molecular characterization of two tobacco (Nicotiana tabacum) invertase inhibitor genes in the 1990s, the regulatory role of invertase inhibitors has been supported (Greiner et al., 1998, 1999). Further research has grouped the invertase inhibitors into two families, the pectin methylesterase-related proteins (PMEI-RPs), which share conserved domains with inhibitors of pectin methylesterase but have different functions (Hothorn et al., 2004), and the Kunitz-type proteinase inhibitors (KPIs), which contain a Kunitz domain and are able to inhibit the activity of soluble tuber invertase (Glaczinski et al., 2002). Overexpression of the representative for each type of inhibitor in transgenic potato plants revealed that they play similar roles but with varied levels in regulating potato CIS. Overexpression of a tobacco PMEI-RP gene, NtInvInh2, strongly reduced RS accumulation in potato tubers (Greiner et al., 1999). Similarly but much more weakly, the expression of a potato KPI gene, StInh, showed less effect on acid invertase activity and RS accumulation than NtInvInh2 (Liu et al., 2013a), implying that the PMEI-RPs may be more efficient than KPIs for repressing the acid invertase. There are two potato PMEI-RP invertase inhibitor genes, StInvInh2A and StInvInh2B, identified so far (Liu et al., 2010). They are isoforms that play similar roles of specifically inhibiting StvacINV1 activity in potato; the StvacINV1 activity is strongly regulated by alterations of these inhibitors’ expression in cold-stored tubers of the transgenic potatoes (Liu et al., 2013b).
However, this negative relationship between invertase activity and StInvInh2 expression seems to be regulated by environmental factors, as we found no detectable variations of invertase activity in the potato tubers stored at 20°C (Liu et al., 2013a, 2013b). Other reports also propose that the inhibitor expression may be regulated by both environmental and developmental signals and to be genotype dependent (Johnson and Ryan, 1990; Turrà et al., 2009). These results strongly suggest that the invertase activity could be modulated by a more complicated mechanism rather than only by a direct inhibition of its inhibitor.
SnRK1, a family of SUCROSE NONFERMENTING1 (SNF1)-related protein kinases, has been shown to be involved in Suc regulation and sugar signaling (Halford et al., 2011; O’Hara et al., 2013). In plants, SnRK1 is a heterotrimeric enzyme similar to yeast (Saccharomyces cerevisiae) SNF1, which consists of a catalytic α-subunit (Ser/Thr kinase subunit), a regulatory β-subunit that is required for kinase function and confers substrate specificity, and an activating γ-subunit (Davies et al., 1994; Mitchelhill et al., 1994; Jiang and Carlson, 1997; Bouly et al., 1999). In yeast, an SNF1 knockout mutant is unable to induce invertase expression on Suc-containing medium (Celenza and Carlson, 1986), and potato SnRK1 is able to complement this mutation (Alderson et al., 1991). More relevantly, overexpression of SnRK1 is able to decrease the Glc content of potato tubers (McKibbin et al., 2006). Using the yeast two-hybrid (Y2H) system, we previously showed that StvacINV1 directly interacts with the potato SnRK1 α-subunit while StInvInh2B binds to the potato SnRK1 β-subunit (Lin et al., 2013). These results imply that SnRK1 may interact with StvacINV1 and StInvInh2B and play an important role in regulating invertase activity.
Little information is available for how SnRK1 acts on invertases or invertase inhibitors. The knowledge we have to date is that SnRK1 can transfer phosphate groups to target proteins, thereby altering their function (Manning et al., 2002). Several target proteins have been described, such as 3-hydroxy-3-methylglutaryl-CoA reductase (Ball et al., 1995; Barker et al., 1996), nitrate reductase (Douglas et al., 1997; Sugden et al., 1999b), sucrose phosphate synthase (SPS; Sugden et al., 1999b), trehalose phosphate synthase5 (Harthill et al., 2006), 6-phosphofructo-2-kinase/Fru-2,6-bisphosphatase (Kulma et al., 2004), the barley (Hordeum vulgare) heat shock protein BHSP17, and the transcription factor FUSCA3 (Tsai and Gazzarrini, 2012), but no evidence has yet been obtained showing that SnRK1 phosphorylates invertases or invertase inhibitors.
Here, based on the cloning and functional assessment of the wild potato (Solanum berthaultii) SnRK1 gene SbSnRK1 and confirmation of the interactions between the two subunits of SbSnRK1, StInvInh2B and StvacINV1, we report the subtle regulatory mode of potato invertase activity by the invertase-regulation protein complex (IRPC) composed of StvacINV1, StInvInh2B, and SbSnRK1. The inhibition of StvacINV1 by StInvInh2B is blocked by SbSnRK1β and is restored by the phosphorylated form of SbSnRK1α. Inactivated SbSnRK1α is thus critical to maintain invertase activity for promoting potato CIS.
RESULTS
Cloning of the SbSnRK1α and SbSnRK1β Genes
By means of Y2H screening, we previously captured two clones from potato tubers encoding the deduced C-terminal parts of the α- and β-subunits of SnRK1. The α-subunit was found to bind to StvacINV1, while the β-subunit bound to StInvInh2B (Lin et al., 2013). Based on the complete sequences of StubSNF1 (α-subunit of potato SnRK1) and StubGal83 (β-subunit of potato SnRK1), full-length coding sequences of these two subunits were amplified from S. berthaultii complementary DNA (cDNA) using the primer pairs SbSnRK1α-F/R and SbSnRK1β-F/R (Supplemental Table S1). The sequence of the former shares 97% identity with StubSNF1 and is designated SbSnRK1α, and that of the latter shares 98% identity with StubGal83 and is designated SbSnRK1β. Similar sequences were also amplified from the potato ‘E-Potato 3’ (E3) and are respectively designated StSnRK1α and StSnRK1β. The deduced SbSnRK1α protein contains 514 amino acids, which is identical to the deduced StSnRK1α protein (Supplemental Fig. S1, A and C), including a conserved kinase domain in the N terminus, an internal ubiquitin-associated domain, an autoinhibitory sequence domain, and a β-subunit interaction domain that is considered responsible for β-subunit binding and the formation of the SnRK1 complex (Ghillebert et al., 2011; Supplemental Fig. S1B). Phylogenetic analysis revealed that SbSnRK1α belongs to the same clade as potato StubSNF1 and Capsicum annuum SnRK1 (Supplemental Fig. S1C). The deduced protein sequence of SbSnRK1β has 289 amino acids (Supplemental Fig. S1D) harboring an N-terminal glycogen-binding domain and a C-terminal domain of association with the SNF1 complex (Supplemental Fig. S1E). The protein is clustered in the same clade as potato and tomato (Solanum lycopersicum) Gal83s (Supplemental Fig. S1F).
A Protein Complex of SbSnRK1, StvacINV1, and StInvInh2B Is Confirmed in Tobacco Cells
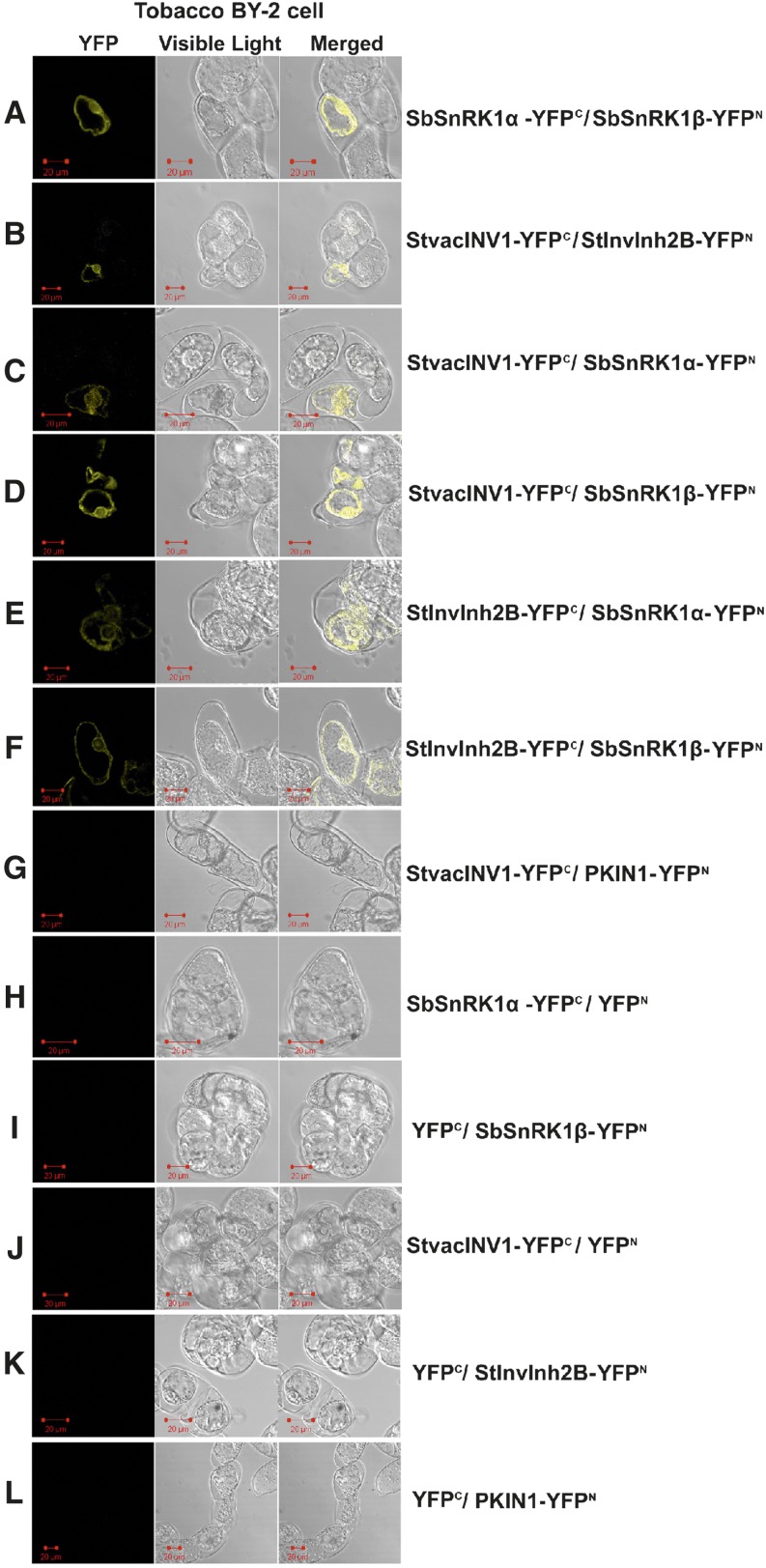
To confirm in-plant protein-protein interactions between the target proteins previously identified, bimolecular fluorescence complementation (BiFC) was conducted in the tobacco ‘Bright Yellow 2’ (BY-2) cell line. For this assay, individual full-length genes encoding StvacINV1, StInvInh2B, SbSnRK1α, and SbSnRK1β were fused to fragments encoding the N- or C-terminal part of the yellow fluorescent protein (YFP). Protein interactions were monitored by detecting YFP fluorescence of the coexpressed candidate proteins fused either to the C- or N-terminal part of YFP. The results verified the interactions between SbSnRK1α and SbSnRK1β, the two subunits of the potato SnRK1 protein (Fig. 1A), and between StvacINV1 and StInvInh2B, the specific counterparts in potato tubers (Fig. 1B). Notably, both StvacINV1 and StInvInh2B interacted with each of the two SbSnRK1 subunits (Fig. 1, C–F), demonstrating that these proteins are likely to bind together to exert their biological function. The specificity of the BiFC assay was tested by including negative controls. PKIN1, the α-subunit of one of the two potato SnRK1s (Man et al., 1997) that was not identified to interact with StvacINV1 in our previous Y2H assay (Lin et al., 2013), did not show a detectable florescence signal when coexpressed with StvacINV1 (Fig. 1G). In addition, no florescence signals were observed when the selected genes were expressed alone (Fig. 1, H–L). These results confirm in plant cells the protein complex composed of StvacINV1, StInvInh2B, and SbSnRK1.
Figure 1.
BiFC visualization of the target interaction partners in cv BY-2 tobacco cells under 514-nm excitation. The counterpart proteins were tested in a pairwise fashion by fusing the full-length genes separately to each of the N- and C-terminal fragments of YFP. The transformed cells were incubated at 23°C for 20 h in the dark and then partially plasmolyzed. Six pairwise combinations, any two proteins of StvacINV1, StInvInh2B, SbSnRK1α, and SbSnRK1β, are presented. No fluorescence was observed when any gene was coexpressed with empty vector or when PKIN1 (an α-subunit of one of the two potato SnRK1s identified) was coexpressed with StvacINV1. The test was conducted with three biological replicates. Bars = 20 μm.
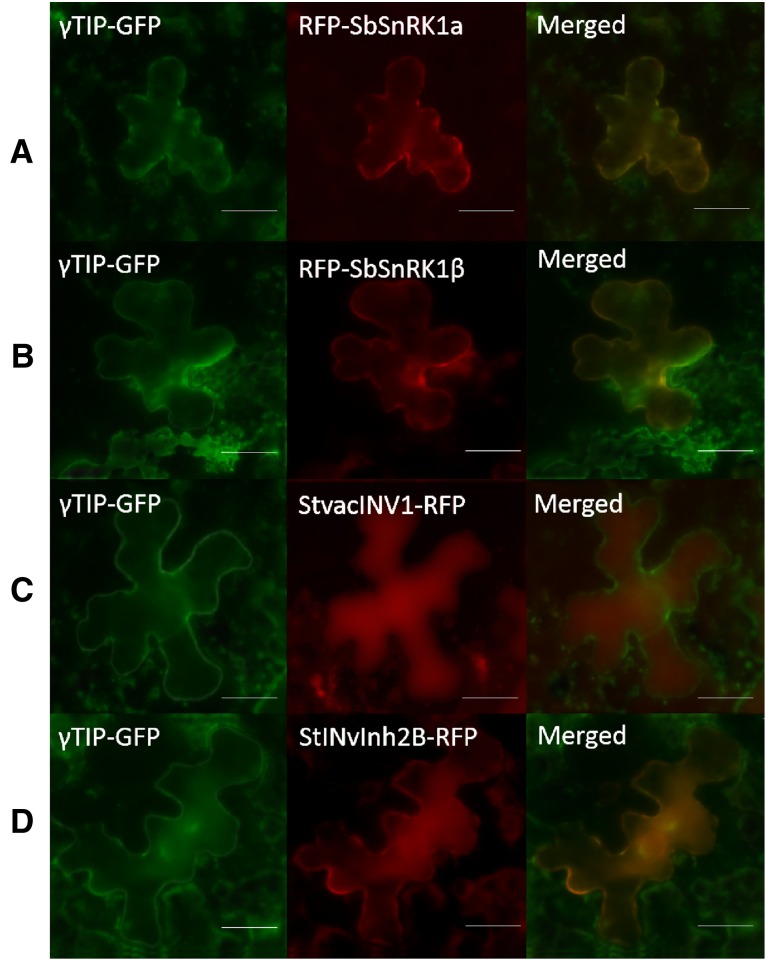
This protein complex was reinforced by subcellular localization of each component in tobacco leaf cells. Full-length StvacINV1, StInvInh2B, SbSnRK1α, and SbSnRK1β genes were separately fused to red fluorescent protein (RFP). The subcellular location was indicated by coexpression of γ-tonoplast intrinsic protein from Arabidopsis (Arabidopsis thaliana; γTIP) that was fused to the N terminus of GFP (Nelson et al., 2007). The results showed that SbSnRK1α and SbSnRK1β were localized to the vacuolar membrane, the same as γTIP (Fig. 2, A and B), while StvacINV1 was in the vacuole (Fig. 2C) and the florescence signals of StInvInh2B were detected in both the membrane and inside of the vacuole (Fig. 2D). These observations suggested that SbSnRK1 could be a specific form with vacuolar membrane localization to confer physiological functions by forming a protein complex with StvacINV1 and StInvInh2B at the membrane or inside of the vacuole in plant cells.
Figure 2.
Subcellular localization of target proteins in Nicotiana benthamiana. RFP-SbSnRK1α, RFP-SbSnRK1β, StvacINV1-RFP, and StInvInh2B-RFP were coexpressed with the vacuole membrane marker γTIP-GFP in vacuoles of N. benthamiana leaves as indicated. Bars = 20 μm.
The Inhibitory Function of StInvInh2B on StvacINV1 Can Be Blocked by SbSnRK1β
Although binding among StvacINV1, StInvInh2B, and SbSnRK1 could be established, their roles in the complex still remain elusive. To gain insight into their potential relationships, StvacINV1, StInvInh2B, SbSnRK1α, and SbSnRK1β proteins were separately expressed in vitro and purified (Fig. 3). Subsequently, StvacINV1 was mixed with all possible combinations of StInvInh2B, SbSnRK1α, and SbSnRK1β, and StvacINV1 activity was measured (Table I). The results showed that StvacINV1 activity remained unaltered if the protein was mixed with SbSnRK1α or SbSnRK1β (Table I, nos. 2–4), indicating that SbSnRK1α and SbSnRK1β do not directly impact StvacINV1 activity either individually or collectively. In contrast, StInvInh2B reduced StvacINV1 activity to 42.2% of the original (Table I, no. 5). This reduction was maintained when SbSnRK1α was added (Table I, no. 6), suggesting that SbSnRK1α may not directly affect StInvInh2B. However, adding SbSnRK1β to StvacINV1 + StInvInh2B or to StvacINV1 + StInvInh2B + SbSnRK1α, the activity of StvacINV1 was not affected by StInvInh2B (Table I, nos. 7 and 8), revealing that the inhibitory function of StInvInh2B on StvacINV1 is blocked specifically by SbSnRK1β.
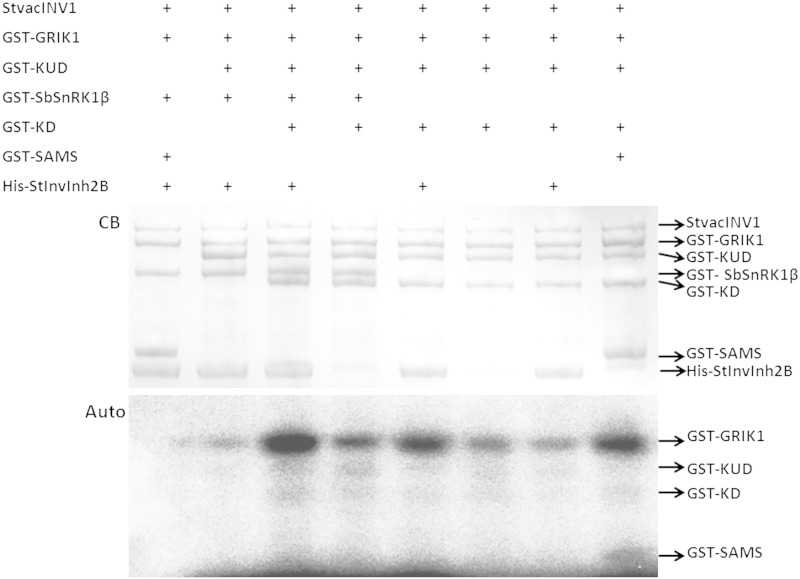
Figure 3.
In vitro phosphorylation of the kinase and its potential substrates. Auto, Autoradiograph; CB, Coomassie Blue staining; KD, kinase domain of SbSnRK1α (amino acids 1–271); KUD, kinase and ubiquitin-associated domains of SbSnRK1α (amino acids 1–329). The results show that no protein in the StvacINV1-StInvInh2B-SbSnRK1 complex could be phosphorylated by SbSnRK1.
Table I. Effects of combinations of SbSnRK1 subunits and StInvInh2 on StvacINV1 activity.
| No. | Protein Mixturea | StvacINV1 Residual Activityb | Significancec |
|---|---|---|---|
| 1 | StvacINV1 | 1.000 ± 0.024 | a |
| 2 | StvacINV1 + SbSnRK1α | 1.057 ± 0.134 | a |
| 3 | StvacINV1 + SbSnRK1β | 1.001 ± 0.112 | a |
| 4 | StvacINV1 + SbSnRK1α + SbSnRK1β | 0.986 ± 0.109 | a |
| 5 | StvacINV1 + StInvInh2B | 0.422 ± 0.101 | b |
| 6 | StvacINV1 + StInvInh2B + SbSnRK1α | 0.430 ± 0.047 | b |
| 7 | StvacINV1 + StInvInh2B + SbSnRK1β | 0.949 ± 0.033 | a |
| 8 | StvacINV1 + StInvInh2B + SbSnRK1α + SbSnRK1β | 0.938 ± 0.091 | a |
The target proteins were mixed in 0.01 m phosphate-buffered saline (PBS) buffer where SbSnRK1α was not phosphorylated. bEach activity value is the mean ± se of five biological replicates, which is calculated by the StvacINV1 activity with or without other component protein(s)/the StvacINV1 activity measured when it presented alone. cDifferent letter presents significance at P = 0.05 by Student’s t test.
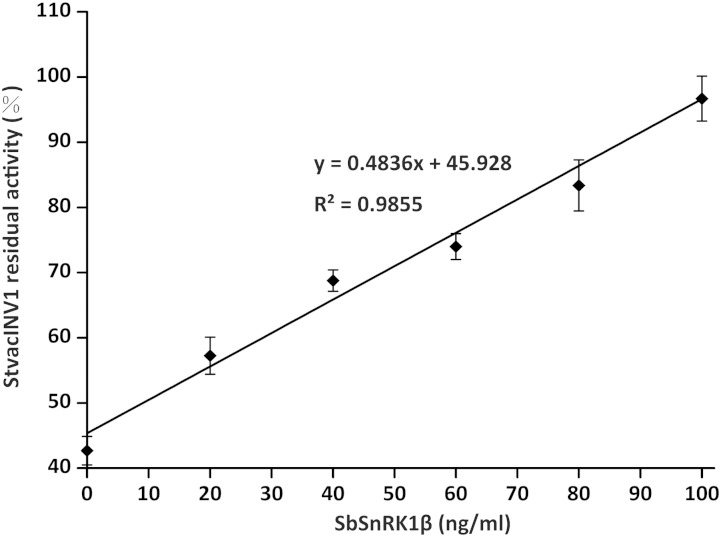
A dosage effect of SbSnRK1β on the function of StInvInh2B was further demonstrated in vitro (Fig. 4). In the presence of sufficient recombinant StInvInh2B protein, the residual activity of recombinant StvacINV1 increased from approximately 42% to 100% as the concentration of SbSnRK1β increased from 0 to 100 ng mL−1. A very significant linear relationship was established between the SbSnRK1β concentration and the residual StvacINV1 activity (r2 = 0.9855), reinforcing that SbSnRK1β is the subunit of SbSnRK1 protein that interacts directly with StInvInh2B and suppresses its inhibition of StvacINV1.
Figure 4.
The inhibitory function of StInvInh2B can be blocked by SbSnRK1β. Dose-dependent effects of the recombinant SbSnRK1β protein on the activity of recombinant StvacINV1 protein (107 ng mL−1) mixed with recombinant StInvInh2B protein (80 ng mL−1) are shown. Residual invertase activity was measured at pH 4.6 and 37°C after a 1-h preincubation of the protein mixtures.
Phosphorylation of SbSnRK1α Restores StInvInh2B Function by Counteracting SbSnRK1β
As a putative kinase, the function of SbSnRK1 could be achieved by phosphorylating its substrates or by some unknown mechanism. To elucidate whether SbSnRK1α could phosphorylate its counterparts in the protein complex, a phosphorylation assay in the presence of SbSnRK1α and γ-32P-labeled ATP was carried out. Because the substrates were hardly phosphorylated by the full-length SbSnRK1α (data not shown), the assay used two fragments of SbSnRK1α, the KD region and the KUD region. While SAMS (HMRSAMSGLHLVKRR), a specific and sensitive substrate peptide for SnRK1 (Man et al., 1997), was phosphorylated by both KD and KUD, no phosphorylation of SbSnRK1β, StvacINV1, or StInvInh2B was detected (Fig. 3). These results demonstrate that SbSnRK1α is unable to phosphorylate StvacINV1, StInvInh2B, or SbSnRK1β.
This finding was confirmed by protein phosphorylation analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS) with protein mixtures containing Geminivirus Replication-Interacting Kinase1 (GRIK1) from Arabidopsis, a kinase reported to be upstream of the SnRK1 α-subunit (Shen and Hanley-Bowdoin, 2006), and all known components of the protein complex (SbSnRK1β, StvacINV1, StInvInh2B, and full-length SbSnRK1α or the KD or KUD fragment). Although Ser and Thr phosphorylation sites were identified in the phosphorylated sequences of SbSnRK1α (Thr-175 and Ser-176) and GRIK1 (Thr-104, Ser-261, and Thr-290), no phosphorylation sites were found in StvacINV1 or StInvInh2B (Supplemental Table S2). Together, our results indicate that SbSnRK1β, StvacINV1, and StInvInh2B are not phosphorylation substrates of SbSnRK1α.
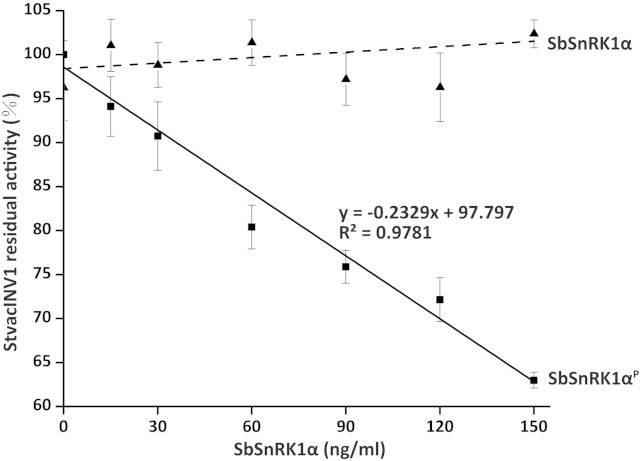
Based on the findings above, we hypothesized that the phosphorylation status of SbSnRK1α may be essential for a diametrical function in the protein complex. As shown in Figure 3, radioactive signals were detected from the two SbSnRK1α fragments, suggesting that they were phosphorylated by GRIK1. Further investigations were carried out to clarify whether phosphorylated SbSnRK1α has impacts on StvacINV1 activity and by which mechanism it may exert its function. StvacINV1 was incubated in all possible combinations with StInvInh2B, SbSnRK1α, and the KD and KUD fragments of SbSnRK1α and SbSnRK1β in the presence or absence of GRIK1. The results showed that, in the absence of StInvInh2B, none of the above proteins could affect the activity of StvacINV1, whether or not GRIK1 was added, implying that phosphorylated SbSnRK1α alone, or in the presence of SbSnRK1β, has no influence on the StvacINV1 activity (Table II). The results further demonstrated that the inhibitory effect of StInvInh2B on StvacINV1 activity that was blocked by SbSnRK1β could be restored by adding phosphorylated SbSnRK1α or its phosphorylated KD or KUD fragment (Table II). This finding was confirmed by the results shown in Figure 5. In protein mixtures with recombinant StvacINV1, StInvInh2B, and SbSnRK1β, a linear decrease in StvacINV1 activity was observed with increasing amounts of phosphorylated SbSnRK1α (r2 = 0.9781), whereas no obvious changes occurred when adding nonphosphorylated SbSnRK1α.
Table II. StvacINV1 residual activity in different protein mixtures.
| No. | Protein Mixturea | StvacINV1 Residual Activityb | Significancec |
|---|---|---|---|
| 1 | StvacINV1 | 1.000 ± 0.069 | a |
| 2 | StvacINV1 + GRIK1 | 1.005 ± 0.077 | a |
| 3 | StvacINV1 + SbSnRK1α | 1.016 ± 0.132 | a |
| 4 | StvacINV1 + SbSnRK1α + GRIK1 | 1.000 ± 0.125 | a |
| 5 | StvacINV1 + KD | 1.032 ± 0.091 | a |
| 6 | StvacINV1 + KD + GRIK1 | 0.988 ± 0.101 | a |
| 7 | StvacINV1 + KUD | 0.907 ± 0.080 | a |
| 8 | StvacINV1 + KUD + GRIK1 | 0.954 ± 0.096 | a |
| 9 | StvacINV1 + SbSnRK1β | 1.039 ± 0.104 | a |
| 10 | StvacINV1 + SbSnRK1β + SbSnRK1α | 1.090 ± 0.122 | a |
| 11 | StvacINV1 + SbSnRK1β + SbSnRK1α + GRIK1 | 0.948 ± 0.090 | a |
| 12 | StvacINV1 + SbSnRK1β + KD | 1.005 ± 0.024 | a |
| 13 | StvacINV1 + SbSnRK1β + KD + GRIK1 | 0.999 ± 0.051 | a |
| 14 | StvacINV1 + SbSnRK1β + KUD | 1.000 ± 0.091 | a |
| 15 | StvacINV1 + SbSnRK1β + KUD + GRIK1 | 0.979 ± 0.062 | a |
| 16 | StvacINV1 + StInvInh2B | 0.412 ± 0.011 | b |
| 17 | StvacINV1 + StInvInh2B + SbSnRK1α | 0.427 ± 0.035 | b |
| 18 | StvacINV1 + StInvInh2B + SbSnRK1α + GRIK1 | 0.498 ± 0.046 | b |
| 19 | StvacINV1 + StInvInh2B + KD | 0.422 ± 0.059 | b |
| 20 | StvacINV1 + StInvInh2B + KD + GRIK1 | 0.453 ± 0.042 | b |
| 21 | StvacINV1 + StInvInh2B + KUD | 0.407 ± 0.095 | b |
| 22 | StvacINV1 + StInvInh2B + KUD + GRIK1 | 0.461 ± 0.050 | b |
| 23 | StvacINV1 + StInvInh2B + SbSnRK1β | 1.027 ± 0.060 | a |
| 24 | StvacINV1 + StInvInh2B + SbSnRK1β + SbSnRK1α | 0.930 ± 0.045 | a |
| 25 | StvacINV1 + StInvInh2B + SbSnRK1β + SbSnRK1α + GRIK1 | 0.423 ± 0.073 | b |
| 26 | StvacINV1 + StInvInh2B + SbSnRK1β + KD | 0.967 ± 0.035 | a |
| 27 | StvacINV1 + StInvInh2B + SbSnRK1β + KD + GRIK1 | 0.470 ± 0.068 | b |
| 28 | StvacINV1 + StInvInh2B + SbSnRK1β + KUD | 0.923 ± 0.061 | a |
| 29 | StvacINV1 + StInvInh2B + SbSnRK1β + KUD + GRIK1 | 0.449 ± 0.050 | b |
The target proteins were mixed in kinase reaction buffer that supplied ATP for kinase activation. KD and KUD are different regions of SbSnRK1α. bEach activity value is the mean ± se of five biological replicates, which is calculated by the StvacINV1 activity with or without other component protein(s)/the StvacINV1 activity measured when it presented alone. cDifferent letters indicate significance at P = 0.05 by Student’s t test.
Figure 5.
Phosphorylation of SbSnRK1α restored the inhibition of StInvInh2B in the complex. The effects of the recombinant SbSnRK1α protein (the solid line shows phosphorylated SbSnRK1α and the dashed line indicates the unphosphorylated protein) on the activity of recombinant StvacINV1 protein (107 ng mL−1) mixed with recombinant StInvInh2B protein (60 ng mL−1) and recombinant SbSnRK1β protein (100 ng mL−1) are shown. Residual invertase activity was measured at pH 4.6 and 37°C after a 1-h preincubation of the protein mixtures. P, The SbSnRK1α protein is phosphorylated by the upstream kinase GRIK1 in kinase reaction buffer.
The amino acid Thr at position 175 was reported to be a phosphorylation site in Arabidopsis SNF1-related protein kinase catalytic subunit alpha KIN10 (Sugden et al., 1999a). The same site was detected in SbSnRK1α in our research here (Supplemental Table S2). In addition, the amino acid Ser at position 176 was also detected as another phosphorylation site (Supplemental Table S2). To identify phosphorylation sites of SbSnRK1α, Thr-175 and Ser-176 were mutated to Ala separately and simultaneously, and the derived mutants were denoted SbSnRK1αT175A, SbSnRK1αS176A, and SbSnRK1αT175A/S176A, respectively. The mutant proteins were expressed in Escherichia coli as glutathione S-transferase (GST) fusion proteins and used in the StvacINV1 enzyme activity assay. The results indicated that neither SbSnRK1αT175A nor SbSnRK1αT175A/S176A was able to restrict the function of SbSnRK1β when GRIK1 was present (Supplemental Table S3, nos. 8 and 10) compared with wild-type SbSnRK1α (Supplemental Table S3, no. 4). Although SbSnRK1αS176A partially repressed the function of SbSnRK1β (Supplemental Table S3, no. 7), the invertase activity was significantly lower than that of SbSnRK1αT175A, demonstrating that Thr-175 is a main phosphorylation site of SbSnRK1α and reinforcing the idea that phosphorylation of SbSnRK1α is a prerequisite for the protein complex that blocks the function of SbSnRK1β, which in turn impacts invertase activity.
SbSnRK1α Phosphorylation Plays Critical Roles in Potato CIS
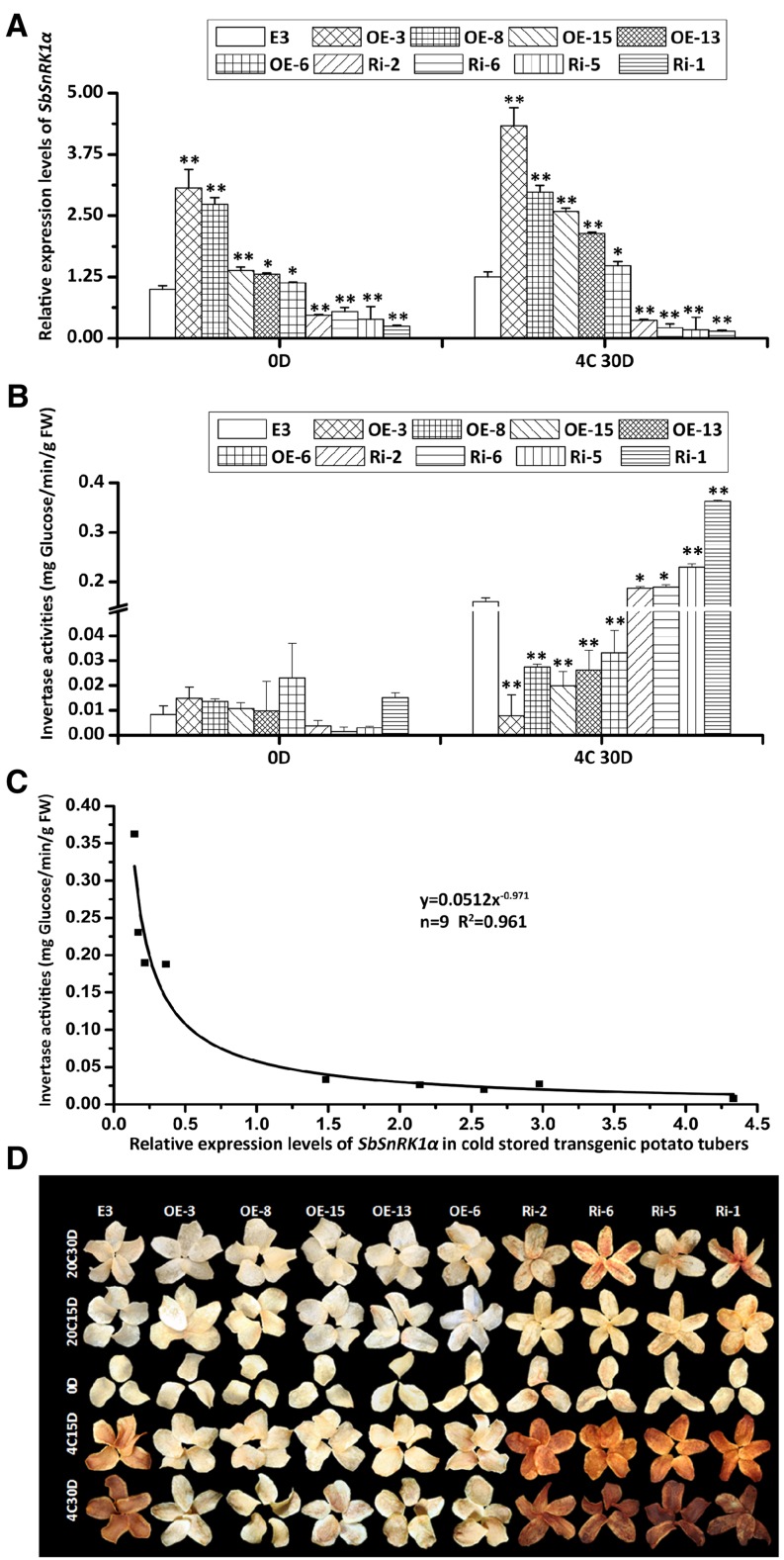
To test whether SbSnRK1α plays a significant role in the plant and to study its impact on potato CIS, the SbSnRK1α gene was transformed into the CIS-sensitive potato cv E3 by Agrobacterium tumefaciens-mediated overexpression and RNA interference. In total, five overexpression lines and four RNA interference lines were obtained. The SbSnRK1α transcripts of the overexpression tubers were 1.13- to 3.07-fold higher than wild-type tubers before storage, while those of the RNA interference tubers were 46% to 75% lower (Fig. 6A, 0D). These differences were increased when the tubers were stored at 4°C for 30 d (Fig. 6A, 4C30D). After cold storage, the invertase activity in all overexpression tubers was reduced by 83% to 95%, while it was increased by 30% to 100% in the RNA interference tubers (Fig. 6B). SbSnRK1α transcript levels in cold-stored transgenic tubers showed a negative correlation with invertase activity that can be represented by y = 0.0512x−0.971 (r2 = 0.9601), where y is invertase activity (ng Glc min−1 g−1 fresh weight) and x is the relative expression level of the SbSnRK1α gene (Fig. 6C). Accordingly, chip color of the overexpression tubers was obviously lighter, while that of the RNA interference tubers was darker than untransformed E3 after storage at 4°C for 15 or 30 d (Fig. 6D). These results suggest that the transcriptional elevation of SbSnRK1α markedly improved chip color by suppressing invertase activity.
Figure 6.
Relative expression levels, chip colors, and invertase activities of wild-type cv E3 and different SbSnRK1α transgenic lines. A, Quantitative real-time (RT)-PCR analysis revealed that SbSnRK1α was overexpressed in the tubers of overexpression lines (OE) and suppressed in the tubers of RNA interference lines (Ri). The expression level in wild-type cv E3 was taken as 1 for calculating the fold change in SbSnRK1α expression of each transgenic line. The transgenic tubers were compared with wild-type cv E3 separately for 4°C and 20°C storage. B, Invertase activity of potato tubers. FW, Fresh weight. C, Correlation between SbSnRK1α transcript level and invertase activity. Data from transgenic tubers stored at 4°C for 30 d were used. *, P < 0.05; **, P < 0.01 by Student’s t test. D, Color of potato chips from tubers stored at 4°C or 20°C for 0 d (0D), 15 d (4C15D and 20C15D), and 30 d (4C30D and 20C30D).
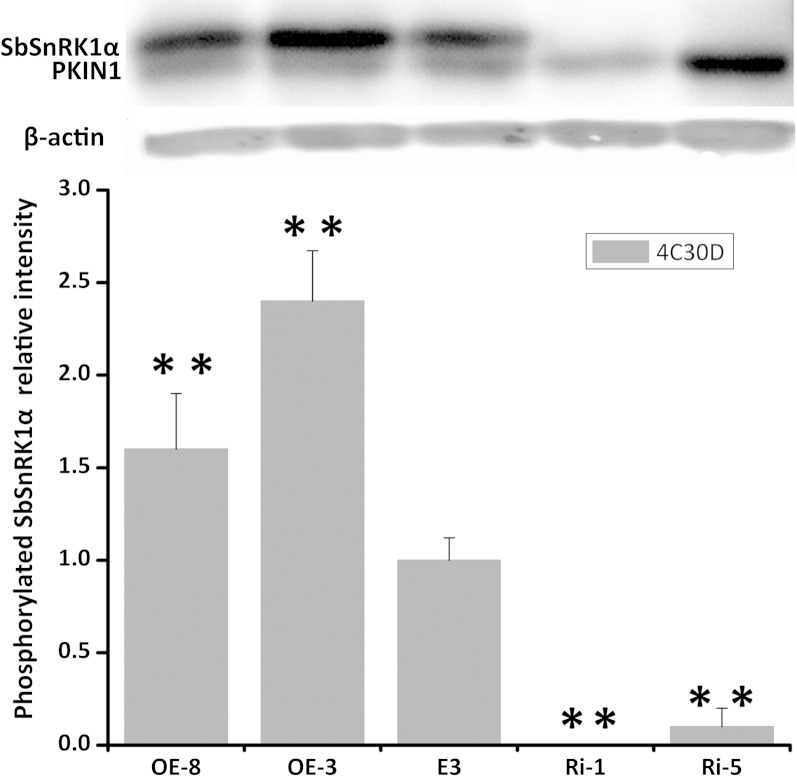
Based on transcript abundance, four extreme transgenic lines, two overexpression lines (OE-3 and OE-8) and two RNA interference lines (Ri-1 and Ri-5), were selected to analyze the level of SbSnRK1α phosphorylation. All of the transgenic lines showed normal plant morphology and tuber development under greenhouse conditions (Supplemental Fig. S2). Semiquantifying phosphorylation signals in western-blot analysis with phospho-specific SnRK1 antibodies revealed that the SbSnRK1α phosphorylation level was higher by 1.6- to 2.4-fold in cold-stored tubers of the two overexpression lines compared with cv E3. This phosphorylation could hardly be detected in the RNA interference tubers, whereas the phosphorylation of PKIN1 (the α-subunit of another potato SnRK1 protein) was observed in all of the tubers and was even very abundant in one of the two RNA interference lines (Fig. 7). The results indicate that the phosphorylation intensity of SbSnRK1α is likely proportional to its transcript abundance, and it could be causal for alteration of the invertase activity in potato tubers.
Figure 7.
SbSnRK1 and PKIN1 phosphorylation in potato tubers stored at 4°C for 30 d detected by western-blot analysis with a phospho-specific antibody. Each density was normalized to the corresponding β-actin density and averaged from three replicates. The density of cv E3 was taken as 1 for the comparisons. **, P < 0.01 by Student’s t test.
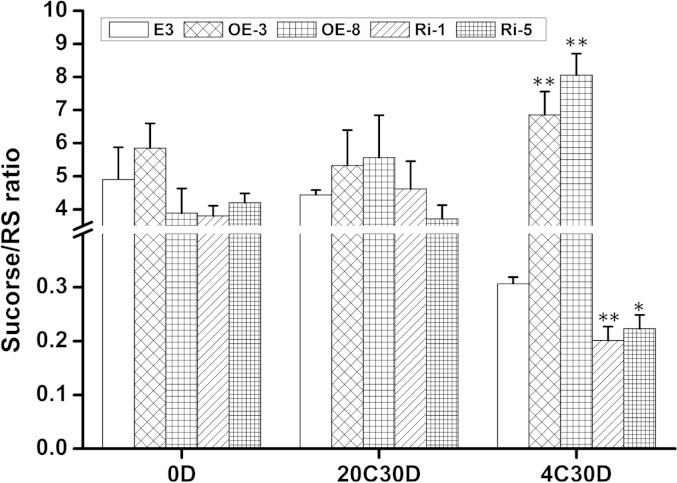
To elucidate if other carbohydrate pathways are influenced by SbSnRK1α, the tubers from the transgenic and wild-type plants were sampled for analysis of the transcripts and enzyme activities of ADP-glucose pyrophosphorylase (AGPase), sucrose synthase (SS), and SPS. Moreover, steady-state levels of 22 metabolites of different carbohydrate pathways were monitored, including Suc, reducing sugars, UDP-Glc, d-Fru-6-P, and Suc-6(F)-P (Supplementary Table S4). No large differences were detected in either the transcript abundance or the activity of AGPase, SS, or SPS (Supplemental Fig. S3), indicating that SbSnRK1α does not influence the activity of these enzymes in potato tubers. Metabolite analysis revealed that reducing sugars were lower in cold-stored overexpression tubers and higher in RNA interference tubers compared with wild-type cv E3 (Supplemental Table S5), which is in accordance with the invertase activity levels illustrated in Figure 6B. When stored at 4°C, a significantly higher Suc-RS ratio was observed in the overexpression tubers, whereas this ratio was remarkably lower in the RNA interference tubers when compared with the wild type (Fig. 8). There were no notable differences or regular trends observed in other metabolites analyzed when comparing the transgenic lines with the control cv E3 (Supplemental Table S4), demonstrating that SbSnRK1α is primarily associated with acid invertase-catalyzed Suc degradation in cold-stored potato tubers.
Figure 8.
Suc-RS ratios of wild-type cv E3 and SbSnRK1α transgenic tubers. The data were derived from the Suc and RS contents in Supplemental Table S5. *, P < 0.05; **, P < 0.01 by Student’s t test.
DISCUSSION
A Protein Complex of StvacINV1-StInvInh2B-SbSnRK1 Is Identified in Potato Tubers
Despite vital roles of StvacINV1 in Suc metabolism and stress responses, the precise regulatory mechanism of StvacINV1 activity is still elusive. We previously hypothesized that an StvacINV1-StInvInh2B-SbSnRK1 complex may regulate the activity of StvacINV1, because StInvInh2B is a counterpart of StvacINV1 (Liu et al., 2013b) and the two subunits of SbSnRK1 interact separately with StvacINV1 and StInvInh2B (Lin et al., 2013). In this study, an in vivo BiFC assay in tobacco cells confirmed all possible pairwise interactions between SbSnRK1α, SbSnRK1β, StvacINV1, and StInvInh2B (Fig. 1). The subcellular location of the counterparts suggests that the protein complex is most likely formed in the vacuole membrane and inside the vacuole (Fig. 2), where Suc is converted into reducing sugars by vacuolar invertase in potato tubers. Notably, SbSnRK1 could be another specific form of plant SnRK1s, with vacuolar membrane location where it can exert its function, in addition to the cytosolic type of SnRK1 for overall energy homeostasis (Williams et al., 2014). To our knowledge, this is the first report providing evidence for the existence of the StvacINV1-StInvInh2B-SbSnRK1 complex in planta.
StvacINV1 Activity Is Subtly Regulated by StInvInh2B and SbSnRK1
The mode of action of SbSnRK1 seems to be different from that of other reported SnRK1 proteins. As is well known, Snf1 in yeast, AMP-activated protein kinase in mammals, and SnRK1 in plants possess kinase cascades for the phosphorylation of their substrates. Once activated, broad ranges of downstream substrates are phosphorylated that affect cellular stress responses (Baena-González and Sheen, 2008; Carling et al., 2012). However, in our research here, both the kinase assay including the autoinhibitory sequence domain truncation that prevents SbSnRK1α from phosphorylating substrates and protein phosphorylation analysis provide evidence that, although the kinase subunit SbSnRK1α interacts with StvacINV1 and StInvInh2B (Fig. 1), neither is a phosphorylation substrate of SbSnRK1α (Fig. 3; Supplemental Tables S2 and S3). Instead, the phosphorylation status of SbSnRK1α fine-tunes StvacINV1 activity in coordination with SbSnRK1β and StInvInh2B.
Each counterpart of a protein complex has a unique function in an interrelated, interactive, and mutually checking manner. Our results reveal that StvacINV1 activity is directly inhibited by StInvInh2B (Table I), which confirms our previous finding that these two proteins specifically interact in potato tubers (Liu et al., 2013b). Our findings are also in accordance with other reports that overexpression of an invertase inhibitor in potato tubers reduces acid invertase activity (McKenzie et al., 2013), suggesting posttranslational regulation of invertase activity (Liu et al., 2013a). In fact, invertase inhibitors coexist with invertases widely in plant cells (Liu et al., 2010, 2011); therefore, it is worth investigating how invertase activity can be released in the competition with its inhibitor for developmental and stress responses.
Our research substantially elucidates that SbSnRK1β restrains StInvInh2B to promote StvacINV1 (Table I). Ingeniously, the phosphorylation of SbSnRK1α deactivates SbSnRK1β to restore the inhibition of StvacINV1 by StInvInh2B (Table II). The linear relationships established in the protein competition assays by plotting the residual activity of StvacINV1 against the levels of SbSnRK1β (Fig. 4) and phosphorylated SbSnRK1α (Fig. 5) prove that the inhibitory factors in these pairwise races are superior to their opponents in the protein complex. Our results further reveal that Thr-175 of SbSnRK1α is a dominant site for phosphorylation by the upstream kinase, although mutation of the Ser at position 176 also partially restores the function of StInvInh2B (Supplemental Table S3). This finding is in agreement with a report that the corresponding Thr residue in the SnRK1 activation loop (Thr-175) is the only residue phosphorylated by the GRIKs (Shen et al., 2009).
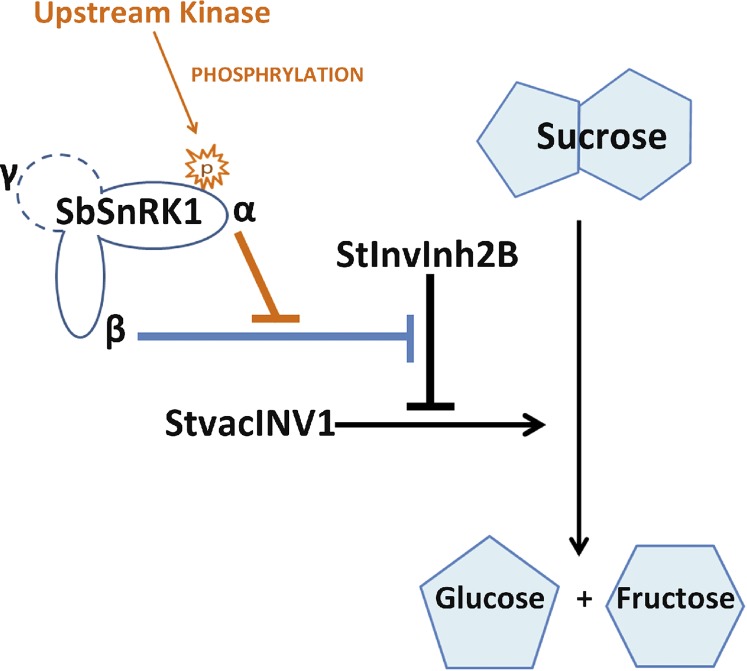
The evidence gained so far indicates a subtle mode of StvacINV1 activity regulation by a complex consisting of StvacINV1, StInvInh2B, SbSnRK1α, and SbSnRK1β, which can be denoted as the IRPC (Fig. 9). Without phosphorylation, SbSnRK1 acts as a negative regulator of the invertase inhibitor StInvInh2B. Once phosphorylated, SbSnRK1 switches to become a positive regulator of StInvInh2B. A similar conclusion was drawn that SnRK1 might have a role as a positive or negative regulator of starch synthesis depending on the levels of different metabolites and tissue type (O’Hara et al., 2013). The paradox created by these two apparently incompatible roles of SnRK1 might be resolved by differential effects of Suc compared with Glc and other hexoses (Halford and Hey, 2009). Intimate cooperation between the α-, β-, and γ-subunits is considered essential for this heterotrimeric enzyme’s function (Ghillebert et al., 2011). This type of subunit/protein behavior in complex assembly was recently reported to be important for biological function (Marsh et al., 2013). Our results clearly demonstrate that, within the IRPC, StvacINV1 activity is finely controlled by the phosphorylation status of SbSnRK1α and its subsequent impacts on SbSnRK1β for the roles of StInvInh2B.
Figure 9.
Proposed mode of Suc degradation regulated by the IRPC composed of StvacINV1, StInvInh2B, and the two subunits of SbSnRK1 (SbSnRK1α and SbSnRK1β). Absent SbSnRK1α phosphorylation, the inhibitory function of StInvInh2B is blocked by SbSnRK1β (blue line) and StvacINV1 is able to catalyze Suc cleavage. Once phosphorylated by an upstream kinase, SbSnRK1α restores StInvInh2B function by counteracting SbSnRK1β (orange lines) and the Suc cleavage catalyzed by StvacINV1 is prevented. The dashed circle represents a potential SbSnRK1 γ-subunit that has not been identified in potato.
SbSnRK1α Plays Vital Roles in Potato CIS through Regulating Invertase Activity
SnRK1 in plants is considered to be a global regulator of metabolism, controlling energy-conserving processes and the remobilization of alternative energy sources, including Suc, starch, cell wall compounds, amino acids, and lipids (Ghillebert et al., 2011). As reviewed by Halford and Hey (2009), SnRK1 functions to control metabolism at multiple levels by phosphorylating and inactivating many enzymes, such as 3-hydroxy-3-methylglutaryl-CoA reductase, SPS, nitrate reductase, trehalose-phosphate synthase, and 6-phosphofructo-2-kinase/Fru-2,6-bisphosphatase. In potato, antisense expression of PKIN1 was found to decrease the expression of SS in tubers and eliminate the Suc inducibility of SS transcripts in leaves (Purcell et al., 1998). It was also observed that starch synthesis is higher in PKIN1-overexpressing potato tubers (McKibbin et al., 2006). Although SnRK1 is thought to be associated with plant responses to environmental stresses, little is known about how SnRK1 impacts sugar levels in potato tubers exposed to low temperature, an efficiency measure taken to prevent serious economic loss in stored industrial raw materials and in which the SnRK1 metabolic pathway is implicated.
Here, the results suggested that, in cold-stored potato tubers, enhancing the expression of SbSnRK1α may promote a higher SbSnRK1α phosphorylation level, which results in an obvious decline in the accumulation of reducing sugars (Fig. 7; Supplemental Table S5). This reduction was related to a high Suc-RS ratio (Fig. 8) in concert with lower invertase activity (Fig. 6B). Our results further indicated that SbSnRK1α was involved neither in the starch synthesis and Suc pathways associated with SS and SPS nor in other carbon fluxes (Supplemental Fig. S3; Supplemental Table S4). These findings strongly suggest that, unlike the PKIN1 form that controls SS and AGPase gene expression and therefore starch synthesis, SbSnRK1 plays critical roles in potato CIS, mainly through altering invertase activity to modulate Suc cleavage.
As a component of a heterotrimeric kinase, the γ-subunit of SNF1/AMPK/SnRK1 is considered to be a regulatory subunit that responds to energy balance (Polge and Thomas, 2007). In plants with different β-subunits, γ-subunit isoforms were reported to exhibit similar expression patterns during development and upon stress (Bouly et al., 1999; Buitink et al., 2004). However, the potato SnRK1 γ-subunit has not been characterized. We also failed to identify its interaction with either StvacINV1 or StInvInh2B in the Y2H system (Lin et al., 2013). Indeed, two tandem transcripts putatively encoding SnRK1 γ-subunit proteins (PGSC0003DMT400039410 and PGSC0003DMT400039411) can be aligned in the Potato Genome Sequencing Consortium database (http://potato.plantbiology.msu.edu/cgi-bin/annotation_report.cgi). Therefore, its potential roles in the heterotrimeric kinase SbSnRK1, particularly in the IRPC for invertase regulation, deserve further investigation.
CONCLUSION
This research provides insight into a mechanism of subtle regulation of invertase activity in cold-stored potato tubers by the IRPC. The phosphorylation status of SbSnRK1α is a determinant of invertase activity. Activated SbSnRK1α suppresses SbSnRK1β, thereby releasing the StvacINV1 inhibitor StInvInh2B, by which potato CIS resistance can be significantly improved.
MATERIALS AND METHODS
Gene Cloning and Vector Development
The primers used for subcloning and plasmid construction are given in Supplemental Table S1. The plasmids used in this study are listed in Supplemental Table S6.
Based on complete nucleotide sequences of potato (Solanum tuberosum) StubSNF1 and StubGal83, full-length SbSnRK1α and SbSnRK1β were amplified from CIS-resistant wild potato species Solanum berthaultii cDNA with specific primer pairs (Supplemental Table S1).
To identify specific interaction in the BiFC assay, PKIN1 (a member of the potato SnRK1 α-subunit gene family) was cloned from S. berthaultii cDNA with the primers listed in Supplemental Table S1.
For the construction of SbSnRK1α mutant expression vectors, site-directed mutagenesis was performed to alter the SbSnRK1α Thr-175 triplet ACA to the Ala-coding triplet GCA, the SbSnRK1α Ser-176 triplet AGT to the Ala-coding triplet GCT, and both Thr-175 and Ser-176 to Ala to obtain the mutants SbSnRK1αT175A, SbSnRK1αS176A, and SbSnRK1αT175A/S176A, respectively. The single-base mutations were generated by overlapping PCR (Tao et al., 2002) using the complementary primer pairs listed in Supplemental Table S1.
For SnRK1 kinase analysis, GRIK1 (Arabidopsis [Arabidopsis thaliana] calcium/calmodulin-dependent protein kinase [NM_001203084]; Shen and Hanley-Bowdoin, 2006) was cloned from Arabidopsis leaf cDNA and subcloned into an expression vector to produce the upstream kinase of SbSnRK1α using the primer pair listed in Supplemental Table S1. At the same time, a SAMS peptide (HMRSAMSGLHLVKRR, a specific and sensitive substrate peptide for SnRK1; Man et al., 1997) was employed as a specific substrate for SbSnRK1α.
BiFC
To confirm the protein complex in vivo, pairwise interactions between StvacINV1, StInvInh2B, SbSnRK1α, and SbSnRK1β were tested in tobacco (Nicotiana tabacum ‘Bright Yellow 2’) cells using the BiFC assay (Walter et al., 2004). For vector construction, full-length counterpart genes were cloned and inserted into the BamHI/KpnI restriction site of the BiFC vectors pUC-SPYNE/pSPYNE-35S and pUC-SPYCE/pSPYCE-35S. Each fusion construct was verified by sequencing. The pairwise vectors were then transformed into cv BY-2 cells by particle bombardment as described (Xu et al., 2007). The fluorescence detection and recording were carried out as described (Lin et al., 2013).
Subcellular Localization
To analyze protein subcellular localization in vivo, StvacINV1, StInvInh2B, SbSnRK1α, and SbSnRK1β were fused with RFP, and the vacuole membrane marker γTIP was fused with GFP. For vector construction, the open reading frame of SbSnRK1α and SbSnRK1β with termination codon and γTIP without termination codon were amplified with specific primers (Supplemental Table S1), PCR products were purified and recombined into pDONR221 (Invitrogen) to generate entry clones via attB × attP reactions using Gateway technology (Invitrogen), N-terminal RFP fusions RFP-SbSnRK1α and SbSnRK1β were made by recombining the entry clones using LR Clonase (Invitrogen) with pK7WGR2, and the C-terminal GFP fusion γTIP-GFP was with pK7FWG2. To generate C-terminal RFP fusions of StvacINV1 and StInvInh2B-RFP, full-length StvacINV1 and StInvInh2B without termination codon were amplified with specific primers (Supplemental Table S1), cloned into pJCV55 obtained by restriction with BglII using Exnase II (Vazyme), and then transformed into the Agrobacterium tumefaciens strain GV3101 containing each construct. The A. tumefaciens harboring RFP-SbSnRK1α, RFP-SbSnRK1β, StvacINV1-RFP, or StInvInh2B-RFP together with A. tumefaciens harboring γTIP-GFP were infiltrated into the leaves of six-leaf-stage Nicotiana benthamiana plants. Two days after incubation, fluorescence was analyzed by Axio Observer A1 (Zeiss) according to its instructions.
Protein Expression and Purification
To analyze protein function in vitro, all of the recombinant proteins fused with a GST tag were expressed in the Escherichia coli prokaryotic expression system. Eight target proteins, GRIK1, SbSnRK1β, SbSnRK1α, SbSnRK1α partial regions KD (amino acids 1–271) and KUD (amino acids 1–329), and the three SbSnRK1α mutants, SbSnRK1αT175A, SbSnRK1αS176A, and SbSnRK1αT175A/S176A, were employed. For fusion expression vector construction, the coding sequences of the target genes were individually inserted into the BamHI-XhoI sites of the prokaryotic expression vector pGEX-6p-1. The SAMS-pGEX plasmid (Shen et al., 2011) was kindly provided by Zhou Xueping.
Recombinant proteins were produced in E. coli BL21 (DE3) induced with 0.01 mm isopropyl β-d-thiogalactoside for 4 h at 16°C. Bacterial cells collected from 2 L of medium were disrupted by sonication. The GST-fused proteins were purified using GST-binding Glutathione Sepharose 4B resin (GE Healthcare) according to the manufacturer’s protocol. The StvacINV1 and His-StInvInh2B recombinant proteins were produced previously (Liu et al., 2013b).
StvacINV1 Enzyme Activity Analyses in Vitro
To clarify the effects of SbSnRK1 on StvacINV1 and StInvInh2B, the residual activity of StvacINV1 was measured after mixing with different proteins. Based on the kinase characteristics of SbSnRK1α, two buffers were used after protein concentration homogenization. The first was 0.01× PBS buffer (135 mm NaCl, 2.7 mm KCl, 1.5 mm KH2PO4, and 8 mm K2HPO4, pH 7.5), which does not supply energy for kinase activation. StvacINV1 protein was mixed with other recombinant proteins in this buffer and incubated under 30°C for 1 h. Then, 5× invertase reaction buffer (30 mm NaAc, pH 4.7, and 30 mm Suc) was added before incubation at 37°C for 1 h. The invertase activity was measured in mg reducing sugar min−1 mg−1 invertase as described (Liu et al., 2010). After adding 3,5-dinitro salicylic acid solution in equal volume to the sample solution, the mixture was boiled at 95°C for 5 min and the absorbance rate was determined at 540-nm wavelength.
The second was a kinase reaction buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm ATP, and 1 mm dithiothreitol) that supplied ATP to the kinase activation. With this buffer, all of the steps were similar to those with the first buffer.
The effect of SbSnRK1β on StInvInh2B was monitored in the first 0.01 m PBS buffer. The protein solution samples were mixed by adding into the buffer StvacINV1 (107 ng mL−1), recombinant StInvInh2B (80 ng mL−1), and grades of recombinant SbSnRK1β (0–100 ng mL−1). Similarly, the effect of SbSnRK1α phosphorylation on StInvInh2B was monitored in the second kinase reaction buffer. The protein solution samples were mixed by adding into the buffer StvacINV1 (107 ng mL−1), recombinant StInvInh2B (60 ng mL−1), recombinant SbSnRK1β (100 ng mL−1), recombinant GRIK1 (200 ng mL−1), and grades of recombinant SbSnRK1α (0–150 ng mL−1). The residual activity of StvacINV1 in these two experiments was measured as mentioned above.
SnRK1 Kinase Assay Involving γ-32P-Labeled ATP
To assay whether SbSnRK1α could phosphorylate StvacINV1 and StInvInh2B in vitro, a kinase assay involving γ-32P-labeled ATP was performed as described (Shen et al., 2011), with minor modifications. Purified proteins including recombinant GRIK along with the KD and KUD fragments of SbSnRK1α were coincubated with other recombinant proteins in kinase reaction buffer (mentioned above) in a total volume of 20 μL. The SnRK1 substrate GST-SAMS was used as a positive control. Reactions were initiated by the addition of 5 μCi of [γ-32P]ATP and transferred to 30°C for 1 h. Loading buffer (6× SDS, 4 μL) was added to terminate the reaction. After boiling at 95°C for 5 min, proteins were separated on a 10% SDS-PAGE gel followed by staining with Coomassie Brilliant Blue R-250 (Takara). Radioactive signals were visualized through autoradiography.
Protein Phosphorylation Analysis Using LC-MS/MS
To reconfirm the SnRK1 kinase assay results and identify the phosphorylation sites of the target proteins, protein phosphorylation analysis by LC-MS/MS was carried out as reported (Zhou et al., 2001) at Shanghai Applied Protein Technology. For this assay, three protein mixtures were used. The first mixture was composed of recombinant GRIK1 and all proteins (SbSnRK1α, SbSnRK1β, StvacINV1, and StInvInh2B) of the IRPC, and the other two contained the same proteins but SbSnRK1α was replaced by recombinant KD or KUD.
Protein digestion was performed according to the filter-assisted sample preparation procedure described by Wiśniewski et al. (2009). Then, electrospray ionization-LC-MS/MS analysis was performed on a Q Exactive mass spectrometer that was coupled to an Easy nLC (Proxeon Biosystems [now part of Thermo Fisher Scientific]). For sequence database searching and data analysis, tandem mass spectra were searched using the MASCOT engine (Matrix Science; version 2.2) against the Uniprot potato database (2,826 sequences; download January 14, 2012) as described (Olsen et al., 2006; Cox and Mann, 2008). For protein identification, the following options were used: peptide mass tolerance, 20 ppm; tandem mass spectrometry tolerance, 0.1 D; enzyme, trypsin; missed cleavage, 2; fixed modification, carbamidomethyl (Cys); variable modification, oxidation (Met); phosphorylation (Ser, Thr, and Tyr); and decoy database pattern, reverse. All reported data were based on 99% confidence for protein identification as determined by a false discovery rate of 1% or less.
Plant Transformation
To construct the class I patatin promoter::SbSnRK1α overexpression vectors, the open reading frame of SbSnRK1α obtained by restriction with BamHI and SacI was subcloned in the sense orientation into the binary vector pBI121 containing the class I patatin promoter (Zhu et al., 2008). To construct the 35S::SbSnRK1α RNA interference vector, a 250-bp fragment starting from 781 bp downstream of the start codon was amplified from the SbSnRK1α cDNA with specific primers (Supplemental Table S1). It is expected that the deduced construct would bring about posttranscriptional gene silencing of only SbSnRK1α, because this fragment is specific for SbSnRK1α and not PKIN1. The amplified PCR products were purified using the QIA Quick PCR Purification Kit (Qiagen), gel verified, and cloned into the pENTR/D cloning vector (Invitrogen). The fragment was subcloned into the pHellsGate8 vector by a recombination method (Helliwell et al., 2002). Sequences in the recombinant pHellsGate8-SbSnRK1α plasmid were confirmed by restriction digestion (XhoI and XbaI), and the inserts were sequenced to ensure that the SbSnRK1α sequences recombined in the correct sense and antisense orientations. Both the overexpression and RNA interference constructs were introduced into Agrobacterium tumefaciens strain LBA4404 and transformed into the commercial CIS-sensitive potato cv E3 by A. tumefaciens-mediated transformation as described previously (Si et al., 2003). The regenerated plants were designated as OE and Ri lines to represent overexpression and RNA interference silencing of SbSnRK1α, respectively.
Plant Growth and Tuber Treatments
Transgenic and wild-type plants, 36 of each, were grown in pots of 24-cm diameter in a greenhouse at 18°C to 25°C at the National Centre for Vegetable Improvement (Central China), Huazhong Agricultural University (Wuhan, China). When the leaves had senesced naturally, the tubers were harvested. Mature tubers of similar size (approximately 5–7 cm in diameter) were taken from each transgenic line and the corresponding wild-type control. They were kept in the dark at 20°C for 10 d for skin set and then divided into two sets; one set was stored at 4°C, and the other was stored at 20°C to serve for comparison. Six tubers were sampled from each transgenic and control line of the two sets after 0, 15, and 30 d of storage. Each sampled tuber was cut into two equal parts. One part was used for chipping, and the other was frozen in liquid nitrogen and stored at −80°C for molecular and biochemical analyses.
Quantitative RT-PCR, Enzyme Activities, Sugar Content, Western Blotting, Chipping, and Metabolite Analysis
The frozen samples were ground to a fine powder in liquid nitrogen for RNA, enzyme, sugar content, SbSnRK1 phosphorylation, and metabolite analyses. Quantitative RT-PCR, invertase activity, sugar content, and chipping were assayed as described previously (Liu et al., 2013b). For quantitative RT-PCR, the potato elongation factor1α gene was used as a reference gene (Nicot et al., 2005). The relative quantification expression levels were calculated by the 2-ΔΔCq (ΔΔ Cycle threshold or ddCt algorithm) method as described by Bio-Rad, and significance was tested by Student’s t test.
For SbSnRK1 phosphorylation, frozen samples were homogenized and incubated in lysis buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.5, 1 mm EDTA, 0.05% [v/v] Triton X-100, 1:500 protease inhibitor cocktail [Sigma; P9599], and 1% [v/v] phosphatase inhibitor cocktail 3 [Sigma; P0044]) for 5 min. After centrifugation at 18,000g at 4°C for 15 min, the protein concentration was quantified via the manufacturer’s protocol. Equal amounts of lysate (300 μg) were separated on a 10% (w/v) SDS-PAGE gel and then blotted onto a polyvinylidene difluoride membrane (0.45 μm; Millipore) that was analyzed with the phospho-AMPKα (Thr-172) polyclonal antibody (Cell Signaling Technology), which recognizes the highly conserved sequence of phosphorylated SbSnRK1 or PKIN1.
Assays of AGPase, SS, and SPS were performed according to Li et al. (2002). The metabolites were measured as reported previously (Horst et al., 2010; Debast et al., 2011).
Statistical Analyses
Biological triplicates were used for each measurement, except that five replicates were used for enzyme activity analysis, and the data are presented as means ± sd. Regression analysis of the data were conducted using the Microsoft Excel program (Microsoft Office 2003). The significance between treatments was tested by Student’s t test with the software SPSS20.0 for Windows (SPSS).
Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: SbSnRK1α (KC184124), SbSnRK1β (KF366002), StubSNF1 (AF143743), StubGal83 (AJ012215), PKIN (X95996.1), StInvInh2B (GU321342), StvacINV1 (AY341425), GRIK1 (NM_001203084), StSnRK1α (KR069088), StSnRK1β (KR069089), ef1α (AB061263), Capsicum annuum SnRK1 (HQ176416.1), Nicotiana attenuata SnRK1 (AY462018.1), tobacco NPK5 (BAA05649.1), Glycine max SnRK1 (XP_003552545.1), Pisum sativum SnRK1 (CAI96819), Arabidopsis AKIN11 (CAA67671.1), Zea mays AKIN10 (EU961757.1), tomato Gal83 (NP_001266255.1), N. attenuata GAL83 (AAS19201.1), Theobroma cacao AMP-activated protein kinase β-2-subunit protein isoform 1 (XP_007016710.1), P. sativum SNF1-related protein kinase regulatory β-subunit 1 (CAI96820.1), Arabidopsis lyrata AMP-activated protein kinase (XP_002871972.1), Arabidopsis SNF1-related protein kinase regulatory subunit β-1 (NP_197615.1), and Arabidopsis AKIN-γ (CAB64718.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence analysis of SbSnRK1α and SbSnRK1β.
Supplemental Figure S2. Plant and tuber morphology of SbSnRK1α transgenic potato grown in a greenhouse.
Supplemental Figure S3. Transcripts abundance and activities of SS, SPS, and AGPase.
Supplemental Table S1. Primers used in this research.
Supplemental Table S2. Protein phosphorylation analysis.
Supplemental Table S3. StvacINV1 residual activity in different protein mixtures.
Supplemental Table S4. Metabolite contents of potato tubers.
Supplemental Table S5. Reducing sugars and Suc contents in wild-type and transgenic potato tubers.
Supplemental Table S6. Plasmids used in this research.
Supplementary Material
Acknowledgments
We thank Luo Jie (Huazhong Agriculture University) for critical reading and discussion of the article, Dr. Wu Xuna (Max-Planck-Institute of Molecular Plant Physiology) for assistance in protein phosphorylation analysis, Dr. Zhang Youjun (Max-Planck-Institute of Molecular Plant Physiology) for useful discussion on protein interaction analysis, and Jörg Hofmann, David Pscheidt, and Alfred Schmiedle (Friedrich-Alexander-University Erlangen-Nuernberg), who performed metabolome analysis.
Glossary
- RS
reducing sugars
- CIS
cold-induced sweetening
- PMEI-RP
pectin methylesterase-related protein
- KPI
Kunitz-type proteinase inhibitor
- Y2H
yeast two-hybrid
- IRPC
invertase-regulation protein complex
- cDNA
complementary DNA
- E3
E-Potato 3
- BiFC
bimolecular fluorescence complementation
- BY-2
Bright Yellow 2
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- PBS
phosphate-buffered saline
- RT
real-time
Footnotes
This work was supported by the National Science Foundation of China (grant nos. 30800754 and 31201258) and the Earmarked Fund for Modern Agro-industry Technology Research System of China (grant no. CARS–10–P06).
References
- Alderson A, Sabelli PA, Dickinson JR, Cole D, Richardson M, Kreis M, Shewry PR, Halford NG (1991) Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc Natl Acad Sci USA 88: 8602–8605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball KL, Barker J, Halford NG, Hardie DG (1995) Immunological evidence that HMG-CoA reductase kinase-A is the cauliflower homologue of the RKIN1 subfamily of plant protein kinases. FEBS Lett 377: 189–192 [DOI] [PubMed] [Google Scholar]
- Barker JH, Slocombe SP, Ball KL, Hardie DG, Shewry PR, Halford NG (1996) Evidence that barley 3-hydroxy-3-methylglutaryl-coenzyme A reductase kinase is a member of the sucrose nonfermenting-1-related protein kinase family. Plant Physiol 112: 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PB, Wu L, Busse JS, Whitty BR, Hamernik AJ, Jansky SH, Buell CR, Bethke PC, Jiang J (2010) Suppression of the vacuolar invertase gene prevents cold-induced sweetening in potato. Plant Physiol 154: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly JP, Gissot L, Lessard P, Kreis M, Thomas M (1999) Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINalpha1, an SNF1-like protein kinase. Plant J 18: 541–550 [DOI] [PubMed] [Google Scholar]
- Buitink J, Thomas M, Gissot L, Leprince O (2004) Starvation, osmotic stress and desiccation tolerance lead to expression of different genes of the regulatory beta and gamma subunits of the SnRK1 complex in germinating seeds of Medicago truncatula. Plant Cell Environ 27: 55–67 [Google Scholar]
- Carling D, Thornton C, Woods A, Sanders MJ (2012) AMP-activated protein kinase: new regulation, new roles? Biochem J 445: 11–27 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1986) A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233: 1175–1180 [DOI] [PubMed] [Google Scholar]
- Cheng SH, Su Z, Xie CH, Liu J (2004) Effects of variation in activities of starch-sugar metabolic enzymes on reducing sugar accumulation and processing quality of potato tubers. Agric Sci Sin 3: 1904–1910 [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Davies SP, Hawley SA, Woods A, Carling D, Haystead TA, Hardie DG (1994) Purification of the AMP-activated protein kinase on ATP-gamma-Sepharose and analysis of its subunit structure. Eur J Biochem 223: 351–357 [DOI] [PubMed] [Google Scholar]
- Debast S, Nunes-Nesi A, Hajirezaei MR, Hofmann J, Sonnewald U, Fernie AR, Börnke F (2011) Altering trehalose-6-phosphate content in transgenic potato tubers affects tuber growth and alters responsiveness to hormones during sprouting. Plant Physiol 156: 1754–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Pigaglio E, Ferrer A, Halfords NG, MacKintosh C (1997) Three spinach leaf nitrate reductase-3-hydroxy-3-methylglutaryl-CoA reductase kinases that are regulated by reversible phosphorylation and/or Ca2+ ions. Biochem J 325: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, Rolland F, Winderickx J (2011) The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J 278: 3978–3990 [DOI] [PubMed] [Google Scholar]
- Glaczinski H, Heibges A, Salamini R, Gebhardt C (2002) Members of the Kunitz-type protease inhibitor gene family of potato inhibit soluble tuber invertase in vitro. Potato Res 45: 163–176 [Google Scholar]
- Greiner S, Krausgrill S, Rausch T (1998) Cloning of a tobacco apoplasmic invertase inhibitor: proof of function of the recombinant protein and expression analysis during plant development. Plant Physiol 116: 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Rausch T, Sonnewald U, Herbers K (1999) Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat Biotechnol 17: 708–711 [DOI] [PubMed] [Google Scholar]
- Halford NG, Curtis TY, Muttucumaru N, Postles J, Mottram DS (2011) Sugars in crop plants. Ann Appl Biol 158: 1–25 [Google Scholar]
- Halford NG, Hey SJ (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Harthill JE, Meek SEM, Morrice N, Peggie MW, Borch J, Wong BHC, Mackintosh C (2006) Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J 47: 211–223 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Horst RJ, Doehlemann G, Wahl R, Hofmann J, Schmiedl A, Kahmann R, Kämper J, Sonnewald U, Voll LM (2010) Ustilago maydis infection strongly alters organic nitrogen allocation in maize and stimulates productivity of systemic source leaves. Plant Physiol 152: 293–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, D’Angelo I, Márquez JA, Greiner S, Scheffzek K (2004) The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension. J Mol Biol 335: 987–995 [DOI] [PubMed] [Google Scholar]
- Jiang R, Carlson M (1997) The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol 17: 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Ryan CA (1990) Wound-inducible potato inhibitor II genes: enhancement of expression by sucrose. Plant Mol Biol 14: 527–536 [DOI] [PubMed] [Google Scholar]
- Kulma A, Villadsen D, Campbell DG, Meek SEM, Harthill JE, Nielsen TH, MacKintosh C (2004) Phosphorylation and 14-3-3 binding of Arabidopsis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Plant J 37: 654–667 [DOI] [PubMed] [Google Scholar]
- Li CR, Gan LJ, Xia K, Zhou X, Hew CS (2002) Responses of carboxylating enzymes, sucrose metabolizing enzymes and plant hormones in a tropical epiphytic CAM orchid to CO2 enrichment. Plant Cell Environ 25: 369–377 [Google Scholar]
- Lin Y, Liu J, Liu X, Ou Y, Li M, Zhang H, Song B, Xie C (2013) Interaction proteins of invertase and invertase inhibitor in cold-stored potato tubers suggested a protein complex underlying post-translational regulation of invertase. Plant Physiol Biochem 73: 237–244 [DOI] [PubMed] [Google Scholar]
- Liu X, Cheng SH, Liu J, Ou YB, Song BT, Zhang C, Lin Y, Li XQ, Xie CH (2013a) The potato protease inhibitor gene, St-Inh, plays roles in the cold-induced sweetening of potato tubers by modulating invertase activity. Postharvest Biol Technol 86: 265–271 [Google Scholar]
- Liu X, Lin Y, Liu J, Song B, Ou Y, Zhang H, Li M, Xie C (2013b) StInvInh2 as an inhibitor of StvacINV1 regulates the cold-induced sweetening of potato tubers by specifically capping vacuolar invertase activity. Plant Biotechnol J 11: 640–647 [DOI] [PubMed] [Google Scholar]
- Liu X, Song B, Zhang H, Li XQ, Xie C, Liu J (2010) Cloning and molecular characterization of putative invertase inhibitor genes and their possible contributions to cold-induced sweetening of potato tubers. Mol Genet Genomics 284: 147–159 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang C, Ou Y, Lin Y, Song B, Xie C, Liu J, Li XQ (2011) Systematic analysis of potato acid invertase genes reveals that a cold-responsive member, StvacINV1, regulates cold-induced sweetening of tubers. Mol Genet Genomics 286: 109–118 [DOI] [PubMed] [Google Scholar]
- Man AL, Purcell PC, Hannappel U, Halford NG (1997) Potato SNF1-related protein kinase: molecular cloning, expression analysis and peptide kinase activity measurements. Plant Mol Biol 34: 31–43 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Marian JM, Joseph RS, Irene MS, Sanjay KG, Rebecca RL, John ADA (2005) Investigations on the role of acid invertase and UDP-glucose pyrophosphorylase in potato clones with varying resistance to cold-induced sweetening. Am J Potato Res 82: 231–239 [Google Scholar]
- Marsh JA, Hernández H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA (2013) Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell 153: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Endo C, Kobayashi A, Noda T, Takigawa S, Yamauchi H, Mori M (2004) Changes in sugar content and activity of vacuolar acid invertase during low-temperature storage of potato tubers from six Japanese cultivars. J Plant Res 117: 131–137 [DOI] [PubMed] [Google Scholar]
- McKenzie MJ, Chen RK, Harris JC, Ashworth MJ, Brummell DA (2013) Post-translational regulation of acid invertase activity by vacuolar invertase inhibitor affects resistance to cold-induced sweetening of potato tubers. Plant Cell Environ 36: 176–185 [DOI] [PubMed] [Google Scholar]
- McKibbin RS, Muttucumaru N, Paul MJ, Powers SJ, Burrell MM, Coates S, Purcell PC, Tiessen A, Geigenberger P, Halford NG (2006) Production of high-starch, low-glucose potatoes through over-expression of the metabolic regulator SnRK1. Plant Biotechnol J 4: 409–418 [DOI] [PubMed] [Google Scholar]
- Mitchelhill KI, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters LA, Kemp BE (1994) Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem 269: 2361–2364 [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914 [DOI] [PubMed] [Google Scholar]
- O’Hara LE, Paul MJ, Wingler A (2013) How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Mol Plant 6: 261–274 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Pressey R. (1967) Invertase inhibitor from potatoes: purification, characterization, and reactivity with plant invertases. Plant Physiol 42: 1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG (1998) Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J 14: 195–202 [Google Scholar]
- Roitsch T, González MC (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci 9: 606–613 [DOI] [PubMed] [Google Scholar]
- Schwimmer S, Makower RU, Rorem ES (1961) Invertase & invertase inhibitor in potato. Plant Physiol 36: 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Liu Z, Song F, Xie Q, Hanley-Bowdoin L, Zhou X (2011) Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiol 157: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Hanley-Bowdoin L (2006) Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol 142: 1642–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley-Bowdoin L (2009) Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol 150: 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si HJ, Xie CH, Liu J (2003) An efficient protocol for Agrobacterium-mediated transformation with microtuber and the introduction of an antisense class I patatin gene into potato. Acta Agron Sin 29: 801–805 [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG (1999a) Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J 19: 433–439 [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG (1999b) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao XR, Zhou XP, Li GX, Yu JL (2002) The pathogenicity on legumes of Cucumber mosaic virus was determined by 243 nucleotides on 2a polymerase gene of viral RNA2. Chin Sci Bull 47: 748–750 [Google Scholar]
- Tareke E, Rydberg P, Karlsson P, Eriksson S, Törnqvist M (2002) Analysis of acrylamide, a carcinogen formed in heated foodstuffs. J Agric Food Chem 50: 4998–5006 [DOI] [PubMed] [Google Scholar]
- Tsai AYL, Gazzarrini S (2012) AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J 69: 809–821 [DOI] [PubMed] [Google Scholar]
- Turrà D, Bellin D, Lorito M, Gebhardt C (2009) Genotype-dependent expression of specific members of potato protease inhibitor gene families in different tissues and in response to wounding and nematode infection. J Plant Physiol 166: 762–774 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Williams SP, Rangarajan P, Donahue JL, Hess JE, Gillaspy GE (2014) Regulation of Sucrose non-Fermenting Related Kinase 1 genes in Arabidopsis thaliana. Front Plant Sci 5: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Wu L, Bhaskar PB, Busse JS, Zhang RF, Bethke PC, Jiang JM (2011) Developing cold-chipping potato varieties by silencing the vacuolar invertase gene. Crop Sci 51: 981–990 [Google Scholar]
- Xu X, Soutto M, Xie Q, Servick S, Subramanian C, von Arnim AG, Johnson CH (2007) Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc Natl Acad Sci USA 104: 10264–10269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Watts JD, Aebersold R (2001) A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol 19: 375–378 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Song B, Zhang C, Ou Y, Xie C, Liu J (2008) Construction and functional characteristics of tuber-specific and cold-inducible chimeric promoters in potato. Plant Cell Rep 27: 47–55 [DOI] [PubMed] [Google Scholar]
- Zrenner R, Schüler K, Sonnewald U (1996) Soluble acid invertase determines the hexose-to-sucrose ratio in cold-stored potato tubers. Planta 198: 246–252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.