Reticulon proteins involved in membrane curvature are targeted to the developing cell plate and label desmotubules in primary plasmodesmata.
Abstract
Primary plasmodesmata (PD) arise at cytokinesis when the new cell plate forms. During this process, fine strands of endoplasmic reticulum (ER) are laid down between enlarging Golgi-derived vesicles to form nascent PD, each pore containing a desmotubule, a membranous rod derived from the cortical ER. Little is known about the forces that model the ER during cell plate formation. Here, we show that members of the reticulon (RTNLB) family of ER-tubulating proteins in Arabidopsis (Arabidopsis thaliana) may play a role in the formation of the desmotubule. RTNLB3 and RTNLB6, two RTNLBs present in the PD proteome, are recruited to the cell plate at late telophase, when primary PD are formed, and remain associated with primary PD in the mature cell wall. Both RTNLBs showed significant colocalization at PD with the viral movement protein of Tobacco mosaic virus, while superresolution imaging (three-dimensional structured illumination microscopy) of primary PD revealed the central desmotubule to be labeled by RTNLB6. Fluorescence recovery after photobleaching studies showed that these RTNLBs are mobile at the edge of the developing cell plate, where new wall materials are being delivered, but significantly less mobile at its center, where PD are forming. A truncated RTNLB3, unable to constrict the ER, was not recruited to the cell plate at cytokinesis. We discuss the potential roles of RTNLBs in desmotubule formation.
Plasmodesmata (PD), the small pores that connect higher plant cells, are complex structures of about 50 nm in diameter. Each PD pore is lined by the plasma membrane and contains an axial endoplasmic reticulum (ER)-derived structure known as the desmotubule (Overall and Blackman, 1996; Maule, 2008; Tilsner et al., 2011). The desmotubule is an enigmatic structure whose function has not been fully elucidated. The small spiraling space between the desmotubule and the plasma membrane, known as the cytoplasmic sleeve, is almost certainly a conduit for the movement of small molecules (Oparka et al., 1999). Some reports, however, suggest that the desmotubule may also function in cell-to-cell trafficking, providing an ER-derived pathway between cells along which macromolecules may diffuse (Cantrill et al., 1999). The desmotubule is one of the most tightly constricted membrane structures found in nature (Tilsner et al., 2011), but the forces that generate its intense curvature are not understood. In most PD, the desmotubule is a tightly furled tube of about 15 nm in diameter in which the membranes of the ER are in close contact along its length. The desmotubule may balloon out in the region of the middle lamella into a central cavity, but at the neck regions of the PD pore it is tightly constricted (Robinson-Beers and Evert, 1991; Ding et al., 1992; Glockmann and Kollmann, 1996; Overall and Blackman, 1996; Ehlers and Kollmann, 2001). Studies of PD using GFP targeted to the ER lumen (e.g. GFP-HDEL) have shown that GFP is excluded from the desmotubule due to the constriction of ER membranes in this structure (Oparka et al., 1999; Crawford and Zambryski, 2000; Martens et al., 2006; Guenoune-Gelbart et al., 2008). Therefore, lumenal GFP is unable to move between plant cells unless the membranes of the desmotubule become relaxed in some way. On the other hand, dyes and some proteins inserted into the ER membrane can apparently move through the desmotubule, either along the membrane or through the lumen, at least under some conditions (Grabski et al., 1993; Cantrill et al., 1999; Martens et al., 2006; Guenoune-Gelbart et al., 2008).
Recently, a number of proteins have been described in mammalian, yeast, and plant systems that induce extreme membrane curvature. Among these are the RETICULONS (RTNs), integral membrane proteins that induce curvature of the ER to form tubules (Voeltz et al., 2006; Hu et al., 2008; Tolley et al., 2008, 2010; Sparkes et al., 2010). In animals, RTNs have been shown to be involved in a wide array of endomembrane-related processes, including intracellular transport and vesicle formation, and as RTNs can also influence axonal growth, they may have roles in neurodegenerative disorders such as Alzheimer’s disease (Yang and Strittmatter, 2007). Arabidopsis (Arabidopsis thaliana) has 21 RTN homologs, known as RTNLBs (Nziengui et al., 2007; Sparkes et al., 2010), considerably more than in yeast or mammals, but most have not been examined. RTNLBs contain two unusually long hydrophobic helices that form reentrant loops (Voeltz et al., 2006; Hu et al., 2008; Sparkes et al., 2010; Tolley et al., 2010). These are thought to induce membrane curvature by the molecular wedge principle (Hu et al., 2008; Shibata et al., 2009). When RTNLBs are overexpressed transiently in cells expressing GFP-HDEL, the ER becomes tightly constricted and GFP-HDEL is excluded from the lumen of the constricted ER tubules (Tolley et al., 2008, 2010), a situation similar to that which occurs in desmotubules (Oparka et al., 1999; Crawford and Zambryski, 2000; Martens et al., 2006). In vitro studies with isolated membranes have shown that the degree of tubulation is proportional to the number and spacing of RTNLB proteins in the membrane (Hu et al., 2008). For example, to constrict the ER membrane into a structure of 15 nm, the diameter of a desmotubule, would require RTNLBs to be inserted every 2 nm or less along the desmotubule axis (Hu et al., 2008), potentially making the desmotubule an extremely protein-rich structure (Tilney et al., 1991). Interestingly, a number of RTNLB proteins appear in the recently described PD proteome (Fernandez-Calvino et al., 2011), suggesting that RTNLBs are good candidates for proteins that model the cortical ER into desmotubules.
Primary PD form at cytokinesis during the assembly of the cell plate (Hawes et al., 1981; Hepler, 1982). Of the numerous studies devoted to the structure of the cell plate, very few have examined the behavior of the ER during cytokinesis. During mitosis, elements of the ER are located in the spindle apparatus, separated from the cytoplasm (Hepler, 1980). Just prior to cytokinesis, there is a relative paucity of ER in the region destined to become the cell plate (Hepler, 1980; Hawes et al., 1981). The studies of Hawes et al. (1981) and Hepler (1982), exploiting heavy-metal impregnation of the ER, showed that during the formation of the new cell plate, strands of cortical ER are inserted across the developing wall, between the Golgi-derived vesicles that deposit wall materials. These ER strands become increasingly thinner during formation of the desmotubule, eventually excluding heavy metal stains from the ER lumen (Hepler, 1982). The center of the desmotubule often appears electron opaque in transmission electron microscopy images and has been referred to as the central rod (Overall and Blackman, 1996). This structure may consist of proteins that extend from the inner ER leaflets or may correspond to head groups of the membrane lipids themselves. In the fully formed primary PD, the desmotubule remains continuous with the cortical ER that runs close to the new cell wall (Hawes et al., 1981; Hepler, 1982; Oparka et al., 1994).
Here, we show that two of the RTNLBs present in the PD proteome, RTNLB3 and RTNLB6, become localized to the cell plate during the formation of primary PD. These RTNLBs remain associated with the desmotubule in fully formed PD and are immobile, as evidenced by fluorescence recovery after photobleaching (FRAP) studies. A truncated version of RTNLB3, in which the second hydrophobic region was deleted (Sparkes et al., 2010), was not recruited to the cell plate at cytokinesis. We suggest that RTNLBs play an important role in the formation of primary PD and discuss mechanisms by which these proteins may model the ER into desmotubules.
RESULTS
RTNLBs Appear at Cell Plates during Cytokinesis
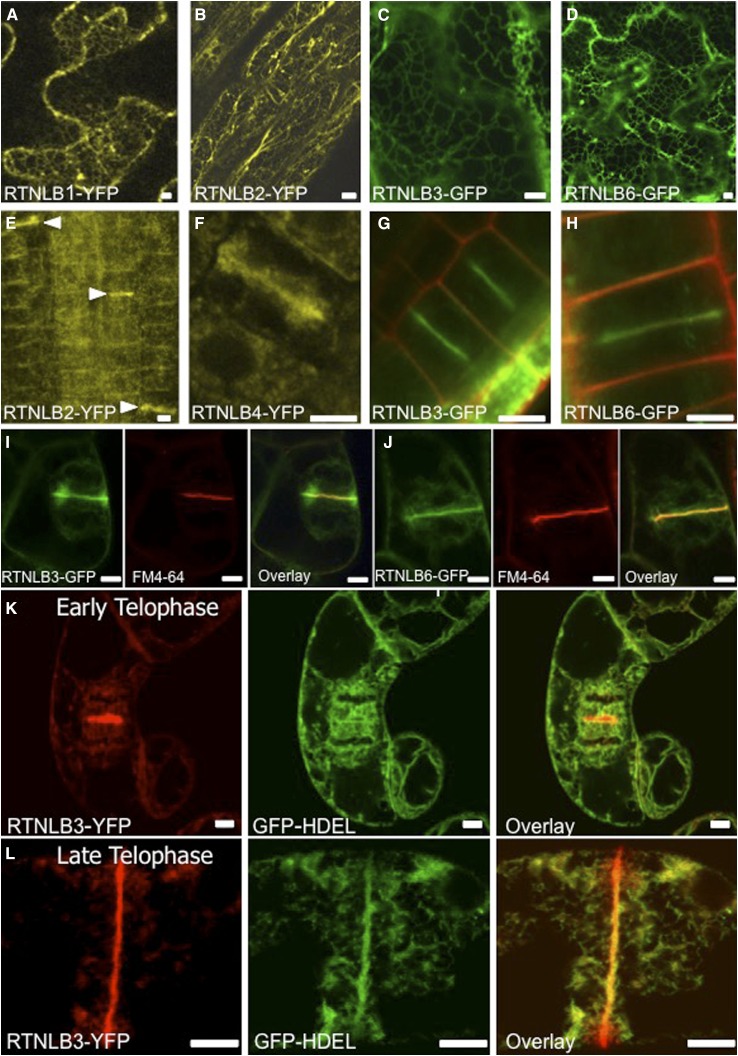
Of the 21 RTNLB homologs in the Arabidopsis database, we generated transgenic lines for RTNLB1 to RTNLB4 and RTNLB6, either as RTNLB-YFP (for yellow fluorescent protein) or RTNLB-GFP fusions under the control of Cauliflower mosaic virus 35S promoters. In all transgenic lines, RTNLBs labeled the ER in interphase cells (Fig. 1, A–D). We noticed for some RTNLB lines that fluorescence became localized to the cell plate in dividing root cells. This localization was recorded for RTNLB2, RTNLB3, RTNLB4, and RTNLB6 (Fig. 1, E–H). In particular, strong cell plate labeling was observed for RTNLB3 and RTNLB6 (Fig. 1, G and H), two RTNLBs present in the PD proteome (Fernandez-Calvino et al., 2011).
Figure 1.
RTN localization in Arabidopsis and tobacco ‘BY2’ cells. A to D, RTNLBs label the ER but not the nuclear envelope in Arabidopsis leaf epidermis. A, RTNLB1-YFP. B, RTNLB2-YFP. C, RTNLB3-GFP. D, RTNLB6-GFP. E to H, RTNLBs are recruited to the developing cell plate in Arabidopsis root cells. E, RTNLB2-YFP. F, RTNLB4-YFP. G, RTNLB3-GFP. H, RTNLB6-GFP. I and J, RTNLB3-GFP (I) and RTNLB6-GFP (J) are also recruited to the cell plate in cv BY2 cells. FM4-64 (red) strongly labels the cell plate. Overlaid images show that the RTNLB-GFP signal is more diffuse at the edges of the developing cell plate. K and L, RTNLB is recruited specifically from the cortical ER to the developing cell plate. During early telophase, RTNLB3-YFP and GFP-HDEL label both the cortical ER and the cell plate (K), but by late telophase, RTNLB3 becomes redistributed into the cell plate (L). Bars = 5 μm.
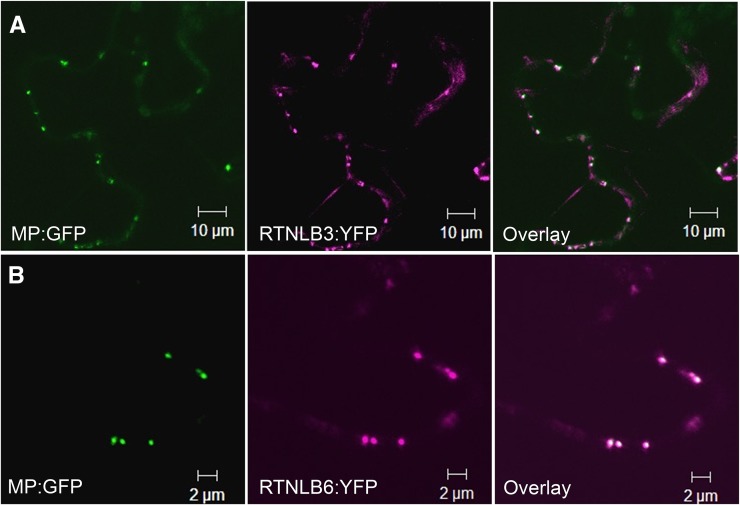
To examine RTNLB distribution during cell division, we expressed Arabidopsis RTNLB3 and RTNLB6 in tobacco (Nicotiana tabacum) ‘Bright Yellow 2’ (BY2) cells and followed their distribution in mitotically dividing cells. We used FM4-64 as a marker for the developing cell plate (Bolte et al., 2004) and found that both RTNLBs showed strong colocalization with FM4-64-labeled cell plate membranes (Fig. 1, I and J). We found a strong signal associated with the newly formed wall at the center and a more diffuse signal associated with the trailing edges of the cell plate (Fig. 1, I and J). Next, we expressed the lumenal ER marker GFP-HDEL together with RTNLB3-YFP. In cv BY2 cells at interphase, as in Arabidopsis, GFP-HDEL was restricted to pockets in the ER by the RTNLB3-YFP-mediated constriction of the ER lumen (Supplemental Fig. S1A). However, both RTNLB3-YFP and GFP-HDEL labeled the cortical ER uniformly throughout cell division (Fig. 1, K and L). However, RTNLB3 became redistributed from the cortical ER into the developing cell plate during mid and late telophase, features not seen with GFP-HDEL (Fig. 1L). We next agroinfiltrated constructs of RTNLB3-YFP and RTNLB6-YFP into Nicotiana benthamiana plants transgenically expressing the movement protein of Tobacco mosaic virus fused to GFP (MP-GFP; Oparka et al., 1999) that is known to locate to PD. In mature epidermal cells, we saw a significant colocalization of these RTNLB fusions with MP-GFP (Fig. 2).
Figure 2.
Colocalization of MP-GFP (green) with RTNLB3-YFP (A) and RTNLB6-YFP (B; magenta). PD showing colocalization signals appear white.
Superresolution Imaging of Primary PD
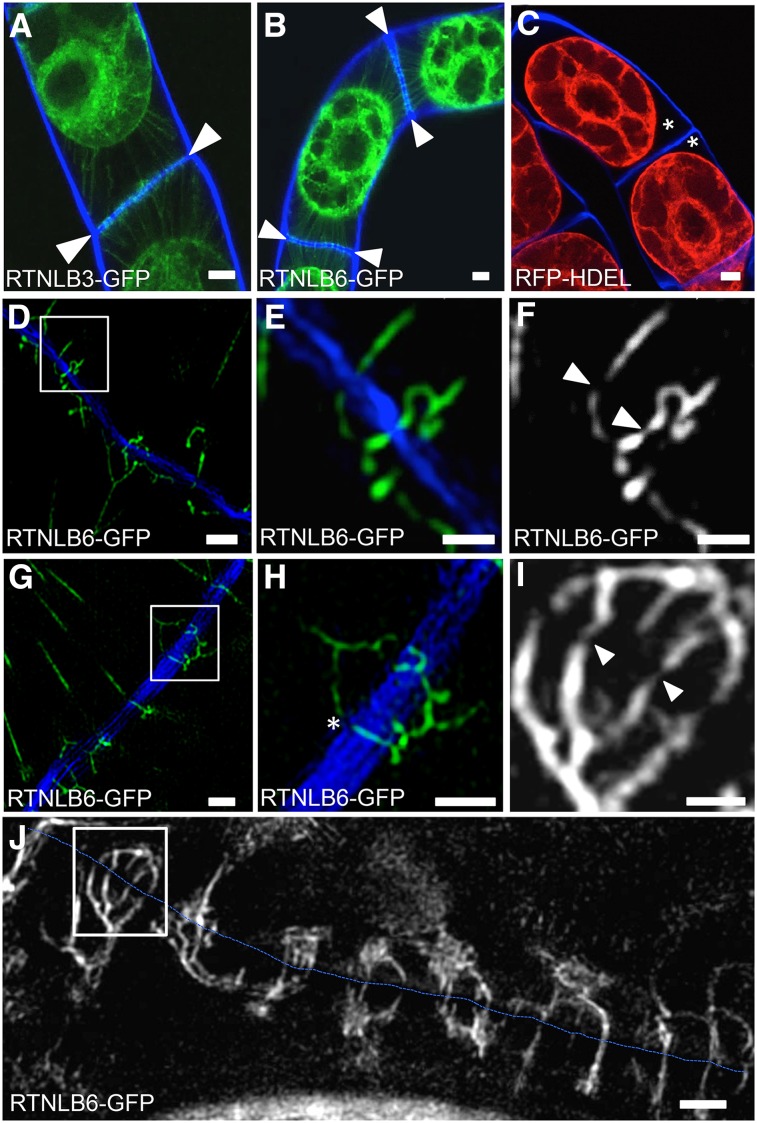
To confirm that RTNLBs were associated with desmotubules, we used three-dimensional structured illumination microscopy (3D-SIM; Fitzgibbon et al., 2010) to examine primary PD in the walls between cv BY2 cells. To image the ER associated with PD more clearly, we plasmolyzed adjacent cells expressing RTNLB3 or RTNLB6 and also imaged plasmolyzed cells expressing red fluorescent protein (RFP)-HDEL for comparison. By confocal microscopy, we observed a strong RTNLB signal associated with punctae at the end walls of plasmolyzed cells (Fig. 3, A and B). Interestingly, Hechtian strands connecting the retracting protoplasts with the end walls showed strong RTNLB labeling, indicating that a constricted tubule of ER runs through each Hechtian strand (Fig. 3, A and B). In contrast, we did not find RFP-HDEL within Hechtian strands or associated with PD (Fig. 3C), confirming previous reports that GFP targeted to the ER lumen is excluded from the center of the desmotubule, where the ER membranes are in close contact (Oparka et al., 1999; Crawford and Zambryski, 2000; Martens et al., 2006; Guenoune-Gelbart et al., 2008). RFP-HDEL was also excluded from the Hechtian strands (Fig. 3C), indicating that, during plasmolysis, RFP-HDEL is squeezed into the contracting protoplast.
Figure 3.
RTNLBs label the Hechtian strands of plasmolyzed cv BY2 cells and show continuous labeling through PD. A and B, Confocal images of RTNLB3-GFP (A) and RTNLB6-GFP (B) labeling of the Hechtian strands (green) in a plasmolyzed cv BY2 cell. Calcofluor (blue) was used to visualize the cell wall between plasmolyzed cells. GFP-labeled punctae were observed along the adjoining cell wall (arrowheads), indicating the positions of PD. C, RFP-HDEL does not reveal the Hechtian strands or PD, leaving the space between the plasma membrane and cell wall unlabeled (asterisks). D to J, 3D-SIM images of plasmolyzed cv BY2 cells expressing RTNLB6-GFP. D to F, RTNLB6-GFP-labeled ER in Hechtian strands is severely constricted as it passes through PD. E and F show closeups of the boxed area in D; arrowheads indicate areas of highly constricted ER, forming the desmotubule at the core of PD. G and H, The cortical ER shows abrupt changes in direction toward the entrance to a PD (asterisk). I, Single-channel closeup of the boxed region in J, showing the extreme constriction of the labeled ER. J, Section of the cell wall between cells (blue dashed line) showing that the RTNLB6-GFP-labeled ER is continuous across the cell wall. Bars = 5 μm (A–C), 1 μm (D–I), and 0.5 μm (F).
With 3D-SIM, we were able to resolve simple PD and found that desmotubules labeled with RTNLB6-GFP could be traced across individual cell walls (Fig. 3, D–H). Often, we observed the cortical ER undergoing abrupt changes in direction toward the entrance of a PD pore (Fig. 3, G and H). Figure 3J shows the cortical ER associated with a single postdivision wall. Note that multiple ER strands extend from the adjoining cells and converge at the entrances of PD, where they can be traced across the wall. An enlargement of a region of cortical ER (Fig. 3I) shows that desmotubules are labeled with RTNLB6. A movie depicting the three-dimensional arrangement of ER associated with PD is shown in Supplemental Video S1. The optical sections shown in Supplemental Figure S2 were taken 150 nm apart in the axial dimension and show that a single RTNLB6-labeled desmotubule can be tracked as it crosses the cell wall.
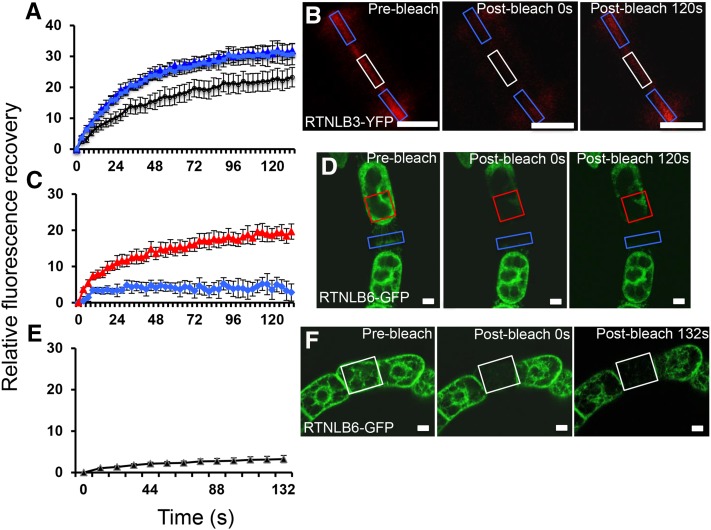
FRAP Reveals Reduced RTNLB Mobility during Cell Plate Formation
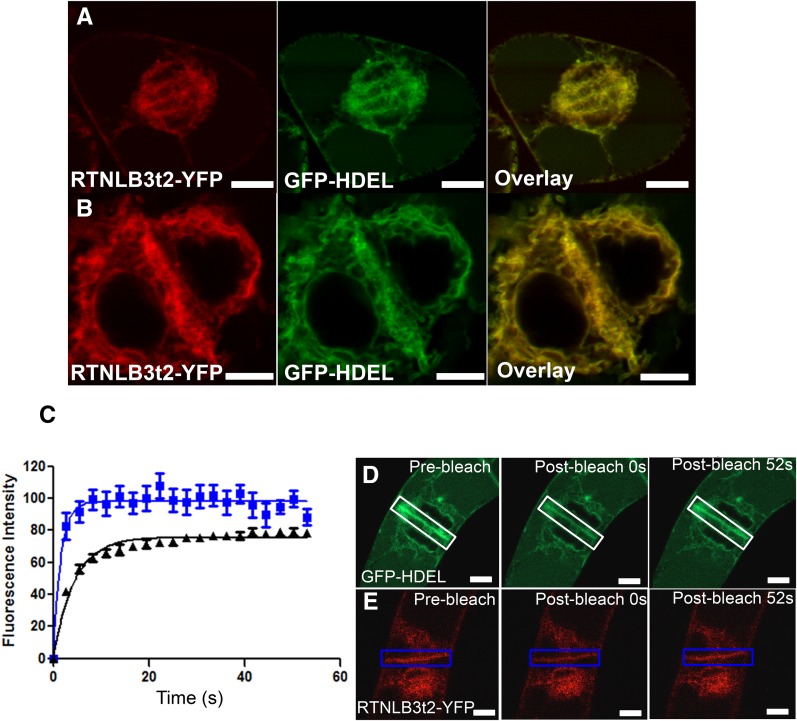
We used FRAP to study the mobility of RTNLBs in the developing cell plate. We compared the leading edge of the cell plate, where the RTNLB signal was diffuse (Fig. 4B), with the middle of the cell plate, where RTNLB distribution was tightly confined (and where PD formation had presumably begun; Hawes et al., 1981; Hepler, 1982). We found a higher rate of fluorescence recovery at the edge of the cell plate compared with the center, indicative of reduced RTNLB mobility during the formation of primary PD (Fig. 4A). At the completion of cytokinesis, we found that a strong RTNLB signal remained on the new cell wall (Fig. 1L). We then plasmolyzed cv BY2 cells expressing RTNLB3 or RTNLB6 and performed FRAP on the ER associated with the primary PD in the cell wall. There was no fluorescence recovery in this location, although ER in the center of the cell showed a characteristic fluorescence recovery (Fig. 4D). As cv BY2 cells often grow in linear chains, we also photobleached the ER of entire cells and monitored the bleached cell for fluorescence recovery. In this scenario, the only source of fluorescence is from neighboring cells, across the intervening PD (Grabski et al., 1993). We failed to detect a return of fluorescence into the bleached cell, indicating that, even though RTNLBs are present in the desmotubule, this structure does not form a conduit for RTNLB mobility between cells (Fig. 4, E and F).
Figure 4.
FRAP reveals that RTNLB mobility is reduced in areas of PD development during cell plate formation. A, FRAP comparing the leading edge of the developing cell plate (light and dark blue) with the center of the cell plate (black diamonds) in cv BY2 cells expressing RTNLB3-YFP. B, Representative prebleach and postbleach images. Boxes indicate the bleached regions. C, RTNLB6-GFP on the ER associated with the primary PD at the cell wall in plasmolyzed cv BY2 cells is immobile (blue diamonds), whereas within the protoplast, fluorescence recovers (red triangles), indicating RTNLB6-GFP mobility. D, Representative prebleach and postbleach images of RTNLB6-GFP-labeled plasmolyzed cv BY2 cells. Boxes indicate the bleached regions. E, Whole cells bleached within a chain show that RTNLBs are not mobile between cells. F, Representative prebleach and postbleach images of RTNLB6-GFP-labeled cv BY2 cells. Boxes indicate the bleached cell. Data are averages of at least nine separate experiments, and error bars indicate se. Bars = 5 μm.
An RTNLB Truncation Is Not Recruited to the Developing Cell Plate
Next, we utilized cv BY2 lines stably expressing a truncation of RTNLB3, comprising the first two transmembrane domains and cytosolic loop but lacking the C-terminal region predicted to form two transmembrane domains (RTNLB3t2; Sparkes et al., 2010). The ER in these cells was less tubular than in cells expressing the full-length protein (Supplemental Fig. S1B). This truncated protein was not strongly recruited to the cell plate at cytokinesis and remained associated with the cytoplasmic ER (Fig. 5, A and B). We then performed FRAP on cells expressing RTNLB3t2. RTNLB3t2-YFP fluorescence in the cell plate of these cells recovered much more quickly than in those expressing the full-length RTNLB-YFP proteins (compare Fig. 5C with Fig. 4, A and B) and also at faster rates than lumenal GFP-HDEL (Fig. 5C).
Figure 5.
A truncated version of RTNLB3 (RTNLB3t2) is not recruited to the cell plate. A and B, RTNLB3t2-YFP coexpressed with GFP-HDEL. The overlaid images show that the RTNLB3t2 has even distribution across the ER, identical to GFP-HDEL, with no specific recruitment to the cell plate. C, FRAP of the cell plate in cells expressing RTNLB3t2-YFP, showing that fluorescence recovers rapidly in the cell plate compared with GFP-HDEL and intact RTNLB3 (D; compare Fig. 4A). D and E, Representative prebleach (D) and postbleach (E) images. Bars = 5 μm.
DISCUSSION
The desmotubule, the intercellular strand of cortical ER that runs through PD, is an extremely constricted membrane tubule whose function remains unclear. Proteins of the RTN family are candidates for shaping the desmotubule as they constrict ER membranes and appear in the Arabidopsis PD proteome (Fernandez-Calvino et al., 2011). Here, we present data suggesting that RTNs are indeed involved in modeling the cortical ER into desmotubules during the formation of primary PD. Of the seven Arabidopsis RTNLBs analyzed so far, none was localized exclusively to PD, although both RTNLB3 and RTNLB6 were significantly enriched at PD. The remainder were associated generally with the cortical ER (Nziengui et al., 2007; Tolley et al., 2008, 2010; Sparkes et al., 2010; this study). However, RTNLB2 to RTNLB4 and RTNLB6 were recruited to the developing cell plate at cytokinesis, where substantial ER modifications occur during PD formation. Of these cell plate-localized RTNs, we focused on RTNLB3 and RTNLB6, both of which are present in the PD proteome (Fernandez-Calvino et al., 2011).
Our 3D-SIM data show that RTNLB6 remains associated with the highly constricted ER in the primary PD of mature cell walls and in Hechtian strands of plasmolyzed cells. Both of these ER regions allow diffusion of the membrane dye 3,3′-dihexyloxacarbocyanine iodide (Grabski et al., 1993; Oparka et al., 1994) but exclude lumenal GFP-HDEL (Oparka et al., 1999; Crawford and Zambryski, 2000; Martens et al., 2006; Guenoune-Gelbart et al., 2008). Tolley et al. (2008, 2010) showed that overexpression of RTNLB13 constricted the cortical ER to an extent that lumenal GFP-HDEL was excluded, being forced into lumenal pockets distributed along the tubules. Likewise, the ER membrane marker GFP-calnexin demonstrated that tubular ER could be constricted to fine threads (Sparkes et al., 2010). Thus, there seems to be no a priori reason why RTNLBs could not form a structure similar to the desmotubule. However, since the RTNLBs are distributed over the entire cortical ER tubules, their presence alone cannot be sufficient to produce the strong constriction of the desmotubule.
The degree of membrane constriction produced by RTNs is dependent on their concentration (Hu et al., 2008). Our experiments provide no information on whether RTNLBs are enriched within PD. However, the ability of RTNLBs to constrict ER tubules depends on their oligomerization, which in turn confers a loss of RTNLB mobility (Shibata et al., 2009; Tolley et al., 2010). Significantly, we observed reduced RTNLB3 and RTNLB6 mobility at the cell plate, and also in PD, as evidenced by FRAP analysis. Although RTNLB3 and RTNLB6 labeling was continuous between cells, no intercellular transport was detected, unlike that shown for the membrane dye 3,3′-dihexyloxacarbocyanine iodide (Grabski et al., 1993) and the ER transmembrane proteins calnexin and Arabidopsis Ca2+-ATP synthase (Guenoune-Gelbart et al., 2008). Truncated RTNLB3 (RTNLB3t2), in which the C-terminal proximal hydrophobic domain was deleted, a mutation that prevents oligomerization and membrane constriction (Sparkes et al., 2010; Tolley et al., 2010), compromised recruitment to the cell plate and caused increased mobility on the plate ER. The mobility of RTNLB3t2 was in fact greater than free lumenal GFP, indicating that the hydrophobic domain of RTNLB3 is likely required for the formation of low-mobility oligomers (Tolley et al., 2010). Collectively, our data suggest that RTNLB3 and RTNLB6, and potentially other RTNLBs, oligomerize preferentially on those ER strands that traverse the cell plate during the formation of desmotubules. The positional cues that trigger this process remain unknown. RTNLB1 to RTNLB7 form a cluster of closely related isoforms (Nziengui et al., 2007; Tolley et al., 2008), but their primary sequences contain no obvious clues to why they should localize to the developing cell plate. It is likely that interactions with other proteins present at the cell plate and in PD may play a role in this context.
The desmotubule within PD often dilates in the middle lamella region of the wall, creating a central cavity whose function is unknown. In vitro, the linear spacing of RTNs determines the degree of dilation between the RTN insertion points, and the greater the distance between the constricting proteins the larger the membrane bulge (Hu et al., 2008). Absence or removal of RTNLBs from the central cavity region of PD could provide a facile means of controlling ER dimensions within PD and may explain the dilation of the desmotubule in the central cavity. In the case of secondary PD, which form across already formed cell walls (Faulkner et al., 2008), RTNLBs may also play a role in ER modeling, as the cortical ER strands on either side of the wall must meet and fuse within secondary PD (Faulkner et al., 2008; Zhang and Hu, 2013).
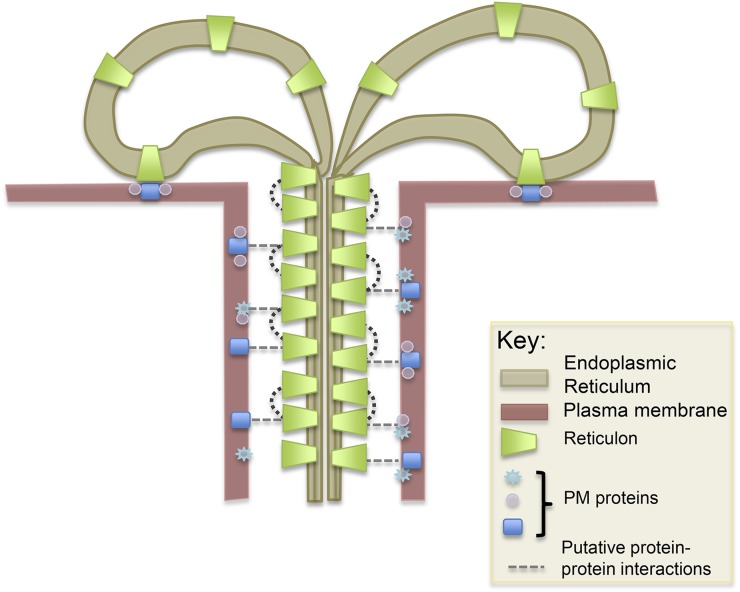
Using the electron microscope, globular proteins have been found along the length of the desmotubule, associated with the outer ER leaflet (Hepler, 1982; Ding et al., 1992; Ehlers and Kollmann, 2001). These have been suggested to be cytoskeletal elements (Overall and Blackman, 1996), but it is equally possible that they are proteins such as RTNLBs, responsible for maintaining the constriction of the desmotubule membranes and also linking the desmotubule to the PM (Tilsner et al., 2011). Several published electron micrographs show spoke-like extensions between these proteins and the plasma membrane (Ding et al., 1992; Schulz, 1995; Overall and Blackman, 1996; Ehlers and Kollmann, 2001). These links with the plasma membrane, as well as other desmotubule-localized proteins, could contribute to a localized enrichment and immobilization of RTNLBs. The extensive protein scaffold of the desmotubule is likely to impart considerable rigidity to this structure, and links to the plasma membrane would provide a means of exerting control of the PD size-exclusion limit (Tilsner et al., 2011). A schematic showing the putative distribution of RTNLB proteins on the desmotubule, and their links with the PM, is shown in Figure 6. We are currently searching for interactors of RTNLBs that may fulfill the above functions.
Figure 6.
Model showing the insertion of RTNLB proteins into the membranes of the desmotubule. Putative protein-protein interactions between RTNLBs and proteins resident in the plasma membrane are depicted as broken lines.
Many of the viral movement proteins that facilitate virus transport through PD interact with the cortical ER or are integral ER membrane proteins (Vilar et al., 2002; Krishnamurthy et al., 2003; Martínez-Gil et al., 2009; Peiró et al., 2014). The transmembrane movement proteins of Potato virus X accumulate in RTNLB-rich regions of curved cortical ER in Saccharomyces cerevisiae (Wu et al., 2011) and also in the desmotubules of N. benthamiana PD (Tilsner et al., 2013). Viral movement proteins may disrupt the desmotubule scaffold by perturbing protein-lipid or protein-protein interactions (Tilsner et al., 2011). The links between viral proteins and desmotubule proteins will be an interesting area for future research.
It remains to be shown how many of the RTNLB proteins in the Arabidopsis database are involved in membrane curvature alone, and RTNLBs may have additional functions at PD. A screen for proteins that interact with the FLAGELLIN SENSITIVE2 (FLS2) receptor identified RTNLB1 and RTNLB2 as interacting proteins, and these RTNLBs appear to regulate the transport of newly synthesized FLS2 from the ER to the plasma membrane (Lee et al., 2011). FLS2 is also enriched at PD (Faulkner et al., 2013). It will be interesting to determine whether other RTNLB members play roles in either intracellular or intercellular signaling.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) seeds were sterilized with 10% (v/v) bleach, rinsed once in 70% (v/v) ethanol, and then rinsed four times in sterile distilled, deionized water. Unless stated otherwise, seeds were plated in petri dishes on Murashige and Skoog medium with 1% (w/v) Suc, solidified with 1.2% (w/v) phytoagar, and grown in 16-h photoperiods with 200 μE m−2 s−1 at 18°C to 22°C.
Molecular Biology and Cloning
The generation of both the RTNLB-YFP and RTNLB3 truncation constructs has been described previously (Sparkes et al., 2010). RTNLB3 and RTNLB6 were amplified by PCR using clones from the Arabidopsis Biological Resource Center, recombined into Gateway vector pDONR201, and then both were recombined into binary vector pGWB405, creating C-terminal GFP fusions (Nakagawa et al., 2007). All binary vectors were then transformed into Agrobacterium tumefaciens GV3101 and then into Arabidopsis (Columbia-0) using the floral dip method (Clough and Bent, 1998).
Tobacco ‘BY2’ Cell Culture and Transformation
Tobacco (Nicotiana tabacum) ‘BY2’ cell lines (Nagata et al., 1992) were cultured aseptically in liquid medium containing 4.3 g L−1 Murashige and Skoog Basal Medium (Sigma; M5519), 200 μg mL−1 2,4-dichlorophenoxyacetic acid, and 30 g L−1 Suc. Cells were grown in the dark at 28°C on an orbital shaker and subcultured to fresh medium weekly. RTNLB3-GFP and RTNLB6-GFP transgenic lines were generated by A. tumefaciens-mediated transformation with the binary vectors described above. A total of 40 μL of 20 mm acetosyringone was added to 40 mL of 3-d-old cv BY2 cell culture, and cells were pipetted up and down 20 times to induce minor cell damage. The cells were then mixed with 100 μL of the appropriate A. tumefaciens overnight culture and incubated at 28°C in the dark for 3 d. Cells were then washed three times in sterile medium, resuspended in 5 mL, and plated on medium solidified with 0.75% (w/v) phytoagar, 50 μg mL−1 cefotaxime, and 50 μg mL−1 kanamycin. Plates were incubated for 3 to 4 weeks. Resultant calli were then subcultured onto fresh selective medium to confirm antibiotic resistance before expression was assessed by fluorescence microscopy and positive cell lines transferred and maintained in liquid culture. Cells were imaged at day 4 post subculture for cell plate studies and plasmolysis and days 4 to 6 for FRAP.
Confocal Imaging and Cell Staining
Seedlings and cv BY2 cells were imaged live on slides using either an SP2 confocal laser-scanning microscope (Leica Microsystems) with 40× and 63× water-immersion lenses (HCX PLAPO CS; Leica Microsystems) or a Zeiss LSM510 Meta confocal laser-scanning microscope with 40× and 63× oil-immersion objectives. For cell plate images, 3-d-old Arabidopsis seedlings were partially synchronized by transferring to medium containing 2 mm hydroxyurea for 15 to 17 h before imaging the roots. Arabidopsis roots were stained with 10 μg mL−1 propidium iodide, where indicated. The cv BY2 cells were stained with 8.5 μg mL−1 Calcafluor White for 1 h or 2 μm FM4-64 (Synaptored) for 10 min, and then medium was removed and replaced with fresh medium before imaging or plasmolysis.
Plasmolysis Experiments
The cv BY2 cells were plasmolyzed in a 0.45 m solution of mannitol in liquid cv BY2 medium for 20 min and then mounted on slides in the osmoticum to maintain plasmolysis throughout imaging.
FRAP
FRAP analyses were conducted either on a Zeiss LSM510 Meta confocal laser-scanning microscope or a Leica SP2 confocal laser-scanning microscope under the 63× objective using rapid switching between low-intensity imaging and high-intensity bleach mode. Prebleach and postbleach images were collected using 40% intensity of the 488-nm laser. Bleaching of the selected region, either an entire cell, a cell wall, or square regions of interest on the middle or edges of cell plates, was achieved with five to 10 scans at 100% intensity of the 488-nm laser. Subsequent recovery was monitored for up to 5 min, dependent on the experiment. Relative levels of fluorescence were normalized to the first postbleach reading. Data are means of at least nine replicates with error bars indicating the se.
3D-SIM
3D-SIM was fully described by Fitzgibbon et al. (2010) Briefly, the 3D-SIM images were obtained using a Deltavision OMX Blaze microscope (GE Healthcare) equipped with 405-, 488-, and 593-nm solid-state lasers and a UPlanSApochromat 100× 1.4 numerical aperture oil immersion objective (Olympus). To maintain the delicate ER structure, cv BY2 cells were imaged live. Exposure times were typically between 100 and 200 ms, and the power of each laser was adjusted to achieve optimal intensities of between 1,000 and 3,000 counts in a raw image of 15-bit dynamic range of the Edge sCMOS camera (PCO). As photobleaching of the GFP was an issue in the live cells, exposure times and laser power were adjusted to minimal levels. Unprocessed image stacks were composed of 15 images per z section (five phase-shifted images for each of three interference pattern angles). Superresolution three-dimensional image stacks were reconstructed with SoftWorx 6.0 (GE) using channel-specific optical transfer functions and Wiener filter setting of 0.002 (0.005 for the 4′,6-diamidino-2-phenylindole channel). Images from the different color channels, recorded on separate cameras, were registered with the SoftWorx 6.0 alignment tool (GE).
Sequence data for genes in this article can be found in the GenBank/EMBL databases using the following accession numbers: RTNLB1, At4g23630; RTNLB2, At4g11220; RTNLB3, At1g64090; RTNLB4, At5g41600; and RTNLB6, At3g61560.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RTNLB3 can constrict the ER in BY2 cells during interphase, whereas a truncated version cannot.
Supplemental Figure S2. RTNLB6-GFP labels desmotubules in BY2 cells.
Supplemental Video S1. 3D reconstruction of ER labeled with RTNLB6-GFP shows close association with PD in BY2 cells.
Supplementary Material
Acknowledgments
We thank Dr. Markus Posch (OMX facility, University of Dundee) for technical expertise and assistance.
Glossary
- PD
plasmodesmata
- ER
endoplasmic reticulum
- FRAP
fluorescence recovery after photobleaching
- BY2
Bright Yellow 2
- 3D-SIM
three-dimensional structured illumination microscopy
Footnotes
This work was supported by the British Biotechnology and Biological Sciences Research Council (grant no. BB/J004987/1 to K.O. and C.H.) and by the Leverhulme Trust (grant no. F/00 382/G to C.H.). Use of the OMX microscope was supported by a Medical Research Council Next Generational Optical Microscopy Award (grant no. MR/K015869/1).
References
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Cantrill LC, Overall RL, Goodwin PB (1999) Cell-to-cell communication via plant endomembranes. Cell Biol Int 23: 653–661 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC (2000) Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr Biol 10: 1032–1040 [DOI] [PubMed] [Google Scholar]
- Ding B, Turgeon R, Parthasarathy MV (1992) Substructure of freeze-substituted plasmodesmata. Protoplasma 169: 28–41 [Google Scholar]
- Ehlers K, Kollmann R (2001) Primary and secondary plasmodesmata: structure, origin, and functioning. Protoplasma 216: 1–30 [DOI] [PubMed] [Google Scholar]
- Faulkner C, Akman OE, Bell K, Jeffree C, Oparka K (2008) Peeking into pit fields: a multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell 20: 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A (2011) Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K (2010) Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol 153: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockmann C, Kollmann R (1996) Structure and development of cell connections in the phloem of Metasequoia glyptostroboides needles. I. Ultrastructural aspects of modified primary plasmodesmata in Strasburger cells. Protoplasma 193: 191–203 [Google Scholar]
- Grabski S, De Feijter AW, Schindler M (1993) Endoplasmic reticulum forms a dynamic continuum for lipid diffusion between contiguous soybean root cells. Plant Cell 5: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenoune-Gelbart D, Elbaum M, Sagi G, Levy A, Epel BL (2008) Tobacco mosaic virus (TMV) replicase and movement protein function synergistically in facilitating TMV spread by lateral diffusion in the plasmodesmal desmotubule of Nicotiana benthamiana. Mol Plant Microbe Interact 21: 335–345 [DOI] [PubMed] [Google Scholar]
- Hawes CR, Juniper BE, Horne JC (1981) Low and high voltage electron microscopy of mitosis and cytokinesis in maize roots. Planta 152: 397–407 [DOI] [PubMed] [Google Scholar]
- Hepler PK. (1980) Membranes in the mitotic apparatus of barley cells. J Cell Biol 86: 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK. (1982) Endoplasmic reticulum in the formation of the cell plate and plasmodesmata. Protoplasma 111: 121–133 [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA (2008) Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy K, Heppler M, Mitra R, Blancaflor E, Payton M, Nelson RS, Verchot-Lubicz J (2003) The Potato virus X TGBp3 protein associates with the ER network for virus cell-to-cell movement. Virology 309: 135–151 [DOI] [PubMed] [Google Scholar]
- Lee HY, Bowen CH, Popescu GV, Kang HG, Kato N, Ma S, Dinesh-Kumar S, Snyder M, Popescu SC (2011) Arabidopsis RTNLB1 and RTNLB2 Reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 23: 3374–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens HJ, Roberts AG, Oparka KJ, Schulz A (2006) Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiol 142: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Gil L, Sánchez-Navarro JA, Cruz A, Pallás V, Pérez-Gil J, Mingarro I (2009) Plant virus cell-to-cell movement is not dependent on the transmembrane disposition of its movement protein. J Virol 83: 5535–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ. (2008) Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol 11: 680–686 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the “He-La” cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nziengui H, Bouhidel K, Pillon D, Der C, Marty F, Schoefs B (2007) Reticulon-like proteins in Arabidopsis thaliana: structural organization and ER localization. FEBS Lett 581: 3356–3362 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Prior DAM, Crawford JW (1994) Behavior of plasma membrane, cortical ER and plasmodesmata during plasmolysis of onion epidermal cells. Plant Cell Environ 17: 163–171 [Google Scholar]
- Oparka KJ, Roberts AG, Boevink P, Santa Cruz S, Roberts I, Pradel KS, Imlau A, Kotlizky G, Sauer N, Epel B (1999) Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 743–754 [DOI] [PubMed] [Google Scholar]
- Overall RL, Blackman LM (1996) A model of the macro-molecular structure of plasmodesmata. Trends Plant Sci 1: 307–311 [Google Scholar]
- Peiró A, Martínez-Gil L, Tamborero S, Pallás V, Sánchez-Navarro JA, Mingarro I (2014) The tobacco mosaic virus movement protein associates with but does not integrate into biological membranes. J Virol 88: 3016–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Beers K, Evert RF (1991) Fine structure of plasmodesmata in mature leaves of sugarcane. Planta 184: 307–318 [DOI] [PubMed] [Google Scholar]
- Schulz A. (1995) Plasmodesmal widening accompanies the short-term increase in symplasmic phloem unloading in pea root-tips under osmotic stress. Protoplasma 188: 22–37 [Google Scholar]
- Shibata Y, Hu J, Kozlov MM, Rapoport TA (2009) Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol 25: 329–354 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Tolley N, Aller I, Svozil J, Osterrieder A, Botchway S, Mueller C, Frigerio L, Hawes C (2010) Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22: 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Cooke TJ, Connelly PS, Tilney MS (1991) The structure of plasmodesmata as revealed by plasmolysis, detergent extraction, and protease digestion. J Cell Biol 112: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Amari K, Torrance L (2011) Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma 248: 39–60 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN, Oparka KJ (2013) Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 201: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N, Sparkes I, Craddock CP, Eastmond PJ, Runions J, Hawes C, Frigerio L (2010) Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J 64: 411–418 [DOI] [PubMed] [Google Scholar]
- Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, Frigerio L (2008) Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9: 94–102 [DOI] [PubMed] [Google Scholar]
- Vilar M, Saurí A, Monné M, Marcos JF, von Heijne G, Pérez-Payá E, Mingarro I (2002) Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane. J Biol Chem 277: 23447–23452 [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Wu CH, Lee SC, Wang CW (2011) Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J Cell Biol 193: 521–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Strittmatter SM (2007) The reticulons: a family of proteins with diverse functions. Genome Biol 8: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Hu J (2013) Homotypic fusion of endoplasmic reticulum membranes in plant cells. Front Plant Sci 4: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.