Light-dependent phytochrome A-regulated transcription of floral repressors affects photoperiodic induction of flowering.
Abstract
Photoperiodism is a rhythmic change of sensitivity to light, which helps plants to adjust flowering time according to seasonal changes in daylength and to adapt to growing conditions at various latitudes. To reveal the molecular basis of photoperiodism in soybean (Glycine max), a facultative short-day plant, we analyzed the transcriptional profiles of the maturity gene E1 family and two FLOWERING LOCUS T (FT) orthologs (FT2a and FT5a). E1, a repressor for FT2a and FT5a, and its two homologs, E1-like-a (E1La) and E1Lb, exhibited two peaks of expression in long days. Using two different approaches (experiments with transition between light and dark phases and night-break experiments), we revealed that the E1 family genes were expressed only during light periods and that their induction after dawn in long days required a period of light before dusk the previous day. In the cultivar Toyomusume, which lacks the E1 gene, virus-induced silencing of E1La and E1Lb up-regulated the expression of FT2a and FT5a and led to early flowering. Therefore, E1, E1La, and E1Lb function similarly in flowering. Regulation of E1 and E1L expression by light was under the control of E3 and E4, which encode phytochrome A proteins. Our data suggest that phytochrome A-mediated transcriptional induction of E1 and its homologs by light plays a critical role in photoperiodic induction of flowering in soybean.
Daylength is an important environmental cue used by plants to help them cope with seasonal changes and adjust flowering time to adapt to growing conditions at various latitudes. Garner and Allard (1920) unambiguously demonstrated that cv Biloxi of soybean (Glycine max), a facultative short-day (SD) plant, and the SD cv Maryland Mammoth of tobacco (Nicotiana tabacum) flowered only when daylength decreased below a certain threshold, and they proposed the notion of photoperiodism as a causal factor for different responses of flowering to varying daylengths. Two different approaches were used to show that photoperiodism involves a rhythmic change of sensitivity to light (for review, see Thomas and Vince-Prue, 1997; Kobayashi and Weigel, 2007). One approach uses night-break (NB) experiments, in which a short photoperiod is combined with a dark period of various lengths (up to 64 h) and the dark period is interrupted by short light periods (night breaks). Another approach uses cycle-length experiments, in which the total cycle length is set by combining a constant photoperiod with various lengths of the dark period. The cv Biloxi exhibited different responses to light given at various times during a long night or at various cycle lengths in the two types of experiments; its flowering was promoted when light was given at cycle lengths of 24, 48, and 72 h or as night breaks at 24 and 48 h during a night of 64 h after an 8-h light phase, but it was inhibited when light was given at cycle lengths of 36 and 60 h or as night breaks at 12, 36, and 60 h (Bünning, 1960, 1979; Nanda and Hamner, 1962; Coulter and Hamner, 1964; Hamner and Takimoto, 1964). These findings led to the development of a model in which a 24-h day is divided into light-sensitive and dark-sensitive phases, the rhythmic alternation of which is prescribed by an endogenous circadian oscillator; the response of flowering is determined by coincidence or noncoincidence of light with the light-sensitive phase. In long-day (LD) plants, light during the dark-sensitive phase promotes flowering, whereas in SD plants, it inhibits flowering (Thomas and Vince-Prue, 1997).
Understanding of the molecular mechanisms involved in photoperiodic induction of flowering has advanced considerably since it was discovered that FLOWERING LOCUS T (FT) in Arabidopsis (Arabidopsis thaliana) and its ortholog, Heading date 3a (Hd3a) in rice (Oryza sativa), encode florigens, which function as leaf-derived mobile, long-distance signals promoting floral transition (Corbesier et al., 2007; Jaeger and Wigge, 2007; Tamaki et al., 2007; Notaguchi et al., 2008; for review, see Itoh and Izawa, 2013). The FT/Hd3a protein is conserved across a wide range of plant species and is involved in development, including floral induction (for review, see Pin and Nilsson, 2012; Tsuji et al., 2013). Soybean possesses at least 10 FT homologs, which are arranged as five pairs of linked genes in different homologous chromosomal regions (Kong et al., 2010). At least six of these genes promote early flowering when they are ectopically expressed in Arabidopsis (Kong et al., 2010; Thakare et al., 2011; Fan et al., 2014), whereas FT4 has an antagonistic effect (Zhai et al., 2014). Among these six genes, only FT2a and FT5a exhibit transcriptional patterns closely associated with changes in photoperiod (Kong et al., 2010). This photoperiodic control of FT2a and FT5a expression is conditioned by E3 and E4, which encode the phytochrome A (PHYA) proteins GmPHYA3 and GmPHYA2, respectively (Liu et al., 2008; Watanabe et al., 2009; Kong et al., 2010). Furthermore, overexpression of FT2a and FT5a promotes soybean flowering even under noninductive conditions (Sun et al., 2011; Nan et al., 2014). The maturity gene E1, which has the largest effect on flowering in soybean (McBlain et al., 1987; Upadhyay et al., 1994; Tsubokura et al., 2014), is a floral repressor encoding a possible transcription factor that contains a putative bipartite nuclear localization signal and a region related to the DNA-binding B3 domain; E1 overexpression strongly suppresses the expression of FT2a and FT5a and inhibits the floral induction of a photoperiod-insensitive cultivar (Xia et al., 2012). The E1 transcript is up-regulated under LD conditions but down-regulated under SD conditions: in the e3 and e4 genetic background, the up-regulation by long days was abolished, suggesting that the expression of E1 is under the photoperiodic control of E3 and E4 (Xia et al., 2012). Even in the e3 and e4 background, however, E1 still inhibits flowering, particularly under the far-red light-enriched LD condition with red:far-red light quantum ratios of less than 1 (Cober et al., 1996). The soybean genome has at least three functional PHYA genes, E3, E4, and GmPHYA1; the latter two genes are homologs and redundantly affect hypocotyl elongation under far-red light-enriched light (Liu et al., 2008). Thus, the three PHYA genes may be involved in the photoperiodic control of E1 expression, which, in turn, may regulate the photoperiodic control of FT2a and FT5a expression in soybean.
The three loci (E1, E3, and E4), together with E2, an ortholog of Arabidopsis GIGANTEA (GI; Watanabe et al., 2011), are major contributors to the variation in flowering time among soybean cultivars (Xu et al., 2013; Tsubokura et al., 2014). Photoperiod insensitivity in cultivars adapted to high-latitude environments has been independently and repeatedly generated through mutations at E1, E3, and E4 (Tsubokura et al., 2013; Xu et al., 2013). However, it remains to be determined what molecular mechanisms are involved in the responses of soybean to changes in photoperiod and to short periods of light given during the dark phase. We focused on the effect of light on the transcriptional pattern of the E1 family (E1, E1-like-a [E1La], and E1Lb) because of the strong inhibitory effect of E1 on the expression of FT2a and FT5a. Here, we report that soybean has a molecular mechanism for photoperiodic induction of flowering that appears to be distinct from those known in Arabidopsis and rice. Induction of E1 transcription mediated by PHYA (encoded by E3 and E4) by light may play a central role in the control of photoperiodic responses of flowering in soybean.
RESULTS
Structure of the E1L Genes
The soybean genome has two E1 homologs whose functions remain undetermined, Glyma04g24640 and Glyma18g22670 (Xia et al., 2012). In version a2.v1 of the cv Williams 82 genome sequence database, Glyma18g22670 is renamed as Glyma.04G143300.1 and placed 10,640 kb apart from Glyma04g24640 (renamed as Glyma.04G156400.1) in the pericentromeric region of chromosome 4, which is homologous to the chromosome 6 region where E1 is located (Schmutz et al., 2010). In this study, we designated Glyma04g24640 and Glyma18g22670 as E1La and E1Lb, respectively. RACE analysis of cv Toyomusume revealed that the full-length complementary DNAs (cDNAs) for both E1L genes had 77-bp 5′ untranslated regions (UTRs); E1La (800 bp) had a 144-bp 3′ UTR, whereas E1Lb (805 bp) had a 149-bp 3′ UTR (Supplemental Fig. S1A). Both genes were predicted to encode proteins of 192 amino acid residues, which is 19 amino acids longer than those predicted previously (Xia et al., 2012). The two proteins were 96% identical to each other and 91% identical to E1. A total of 11 amino acid substitutions and three gaps were found in the three proteins (Supplemental Fig. S1B). The sequences of E1La and E1Lb in cv Harosoy and cv Toyomusume were identical.
Induction of E1 and E1L Expression Is Regulated by Light
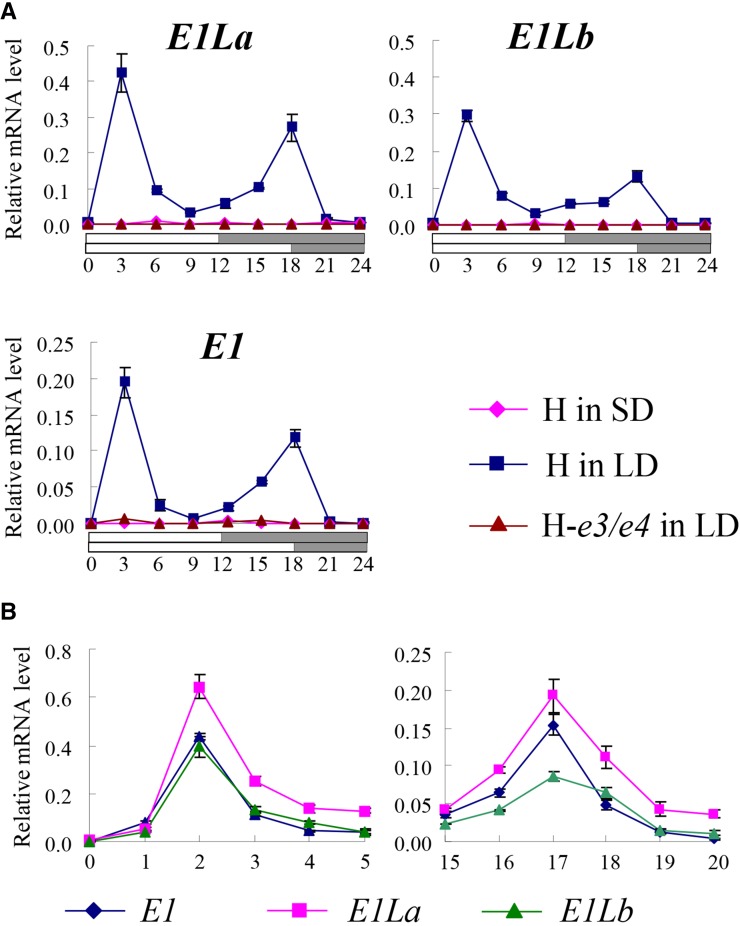
We compared the diurnal patterns of expression of E1, E1La, and E1Lb at 20 d after emergence (DAE) in photoperiod-sensitive cv Harosoy (e1-as/e1-as, e2/e2, E3/E3, and E4/E4) and its insensitive near-isogenic line (NIL) for e3 and e4 (H-e3/e4). In cv Harosoy grown under LD conditions, all three genes had two peaks of expression, at zeitgeber time 3 (ZT3) and ZT18 (Fig. 1A), in line with the observation of Xia et al. (2012). Expression was not observed in cv Harosoy grown under SD conditions and was markedly low in H-e3/e4 grown under LD conditions (Fig. 1A). The transcript levels changed sharply around each of the two peaks; therefore, we analyzed the expression of E1 and E1L with a higher temporal resolution (every 1 h around the peaks instead of 3 h). The transcript abundance of each gene increased sharply from ZT1 to a maximum at ZT2 (the first peak) and then decreased to approximately one-fifth of the peak level at ZT4; it reached the second peak at ZT17 and decreased to the baseline level at ZT19 to ZT20 during the dark phase (Fig. 1B).
Figure 1.
Diurnal expression of the E1 and E1L genes in the photoperiod-sensitive cv Harosoy (H) and its insensitive NIL for e3 and e4 (H-e3/e4). A, Diurnal expression under SD and LD conditions. The cv Harosoy exhibited two peaks of expression at ZT3 and ZT18 under LD conditions. The expression of E1 and E1L was not observed in cv Harosoy under SD conditions and in H-e3/e4 under LD conditions (Supplemental Fig. S2). B, Changes in transcript abundance in cv Harosoy around two peaks under LD conditions. Relative mRNA levels are expressed as the ratios to β-tubulin transcript levels. Average and se values for three replications are given for each data point.
The E1 and E1L transcript abundance around the two peaks when part of the light phase was replaced by dark is shown in Figure 2. Under LD conditions, cv Harosoy produced two peaks of expression, the second peak before dusk (19 DAE) and the first peak after dawn the next day (20 DAE), in line with the experiment shown in Figure 1, although the second peak was low. When the second half of the light phase (ZT12–ZT18) was changed to a dark period at 20 DAE, the second peak did not appear, and no expression after dawn was observed on the next day (LD19SD1; 21 DAE). This inhibition of expression by short days was also observed 2 d later (LD19SD3; 23 DAE). When the dark phase at ZT12 to ZT18 was changed back to the light phase, the second peak was restored to some degree (LD19SD3LD1; 23DAE). This dark-to-light transition induced expression at ZT2 (after dawn) on the next day (LD19SD3LD1; 24 DAE); the expression level was similar to that observed in plants of 20 DAE. Thus, light before dusk induces E1 and E1L expression not only before dusk but also after dawn the next day. Two days later (LD19SD3LD3; 25–26 DAE), the first peak remained at the same level but the second peak increased. Similar to the expression profiles in cv Harosoy, two peaks were induced under LD conditions in H-e3/e4, but the transcript abundance was markedly low, and no expression was observed when the second half of the light phase was replaced by dark (Supplemental Fig. S2).
Figure 2.
Effects of light on the induction of E1 and E1L expression. The cv Harosoy plants were grown under LD conditions until 19 DAE (LD19). The next day, the light phase from ZT12 to ZT18 was replaced with a dark period to shorten the daylength to 12 h (LD19SD1). Three days later (LD19SD3), this dark period was changed back to the light phase (LD19SD3LD1). Relative mRNA levels are expressed as the ratios to β-tubulin transcript levels. Average and se values for three replications are given for each data point.
Next, we analyzed the effect of light given during different time periods (Fig. 3A). The transition from light to dark during ZT0 to ZT6 and ZT6 to ZT12 did not affect expression after dawn the next day (Fig. 3B), which was up-regulated to the maximum levels at ZT2 or ZT3 as in plants of 20 DAE grown under LD conditions (LD19 in Fig. 2). The expression was also induced before dusk, although at low levels in both treatments. We also examined which periods between ZT12 and ZT18 influenced the induction of E1 and E1L expression after dawn the next day by interrupting the light phase by 2-h dark periods (Fig. 3A, treatments ZT12–ZT14, ZT14–ZT16, and ZT16–ZT18). All three treatments strongly suppressed the induction of expression after dawn (Fig. 3B). Taken together, these data suggest that the induction of E1 and both E1L genes depends on light given at the right time, not on the length of the photoperiod. These genes were expressed only during the light phase, and their expression after dawn required a period of light before dusk the previous day.
Figure 3.
Effects of dark periods at different times during the light phase of the LD conditions on E1 and E1L expression. A, Dark periods during the light phase. The cv Harosoy plants were grown under LD conditions until 19 DAE, and then dark periods were introduced during the light phase at different time intervals on 20 DAE. B, E1 and E1L expression around the second peak (ZT16–ZT20; left) on 20 DAE and around the first peak (ZT0–ZT5; right and bottom) on 21 DAE. Relative mRNA levels are expressed as the ratios to β-tubulin transcript levels. Average and se values for three replications are given for each data point.
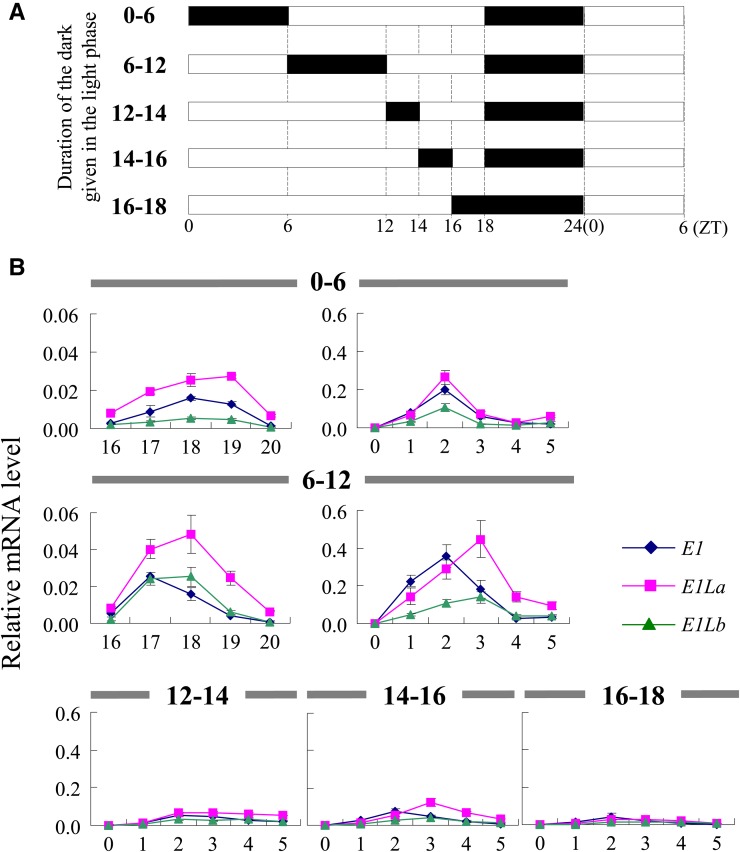
Timing of the Night Break Is Critical for E1 and E1L Expression
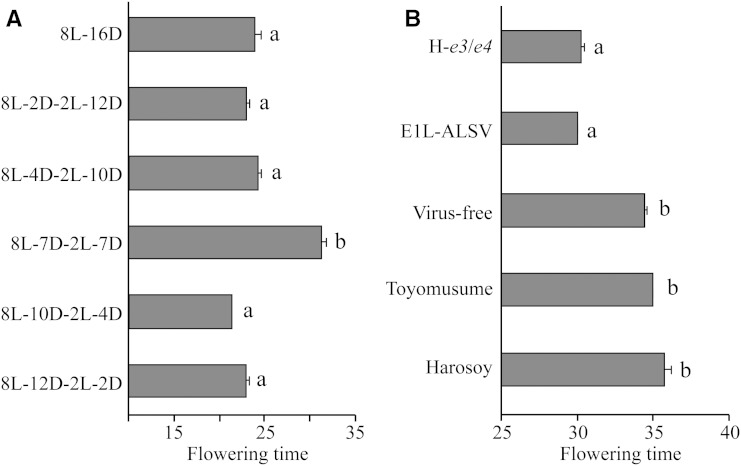
Soybean flowering exhibits a distinct and characteristic NB response (Bünning, 1960, 1979; Nanda and Hamner, 1962; Coulter and Hamner, 1964; Hamner and Takimoto, 1964). We analyzed the effect of NB treatment on transcription of the E1 and E1L genes and two FT genes using cv Harosoy, H-e3/e4, and a NIL for E1 (H-E1). The cv Harosoy has a recessive allele, e1-as, at the E1 locus; e1-as encodes a protein with a defect in nuclear localization (Xia et al., 2012). In the cv Harosoy background, the E1 allele delayed flowering by 20 d or more relative to the e1-as allele (Xia et al., 2012). Plants were grown under different NB treatments (2 h of light given at different time periods during the night after an 8-h light phase), and their flowering dates were recorded (Fig. 4). Without NB treatment, all lines initiated flowering at 23 to 25 DAE. The largest effect of NB treatment was observed when light was given at the midpoint of the dark period (8L-7D-2L-7D in Fig. 4). With this treatment, flowering was delayed in comparison with SD conditions by 7 d in cv Harosoy, whereas H-E1 did not produce any flower buds until 50 DAE (when the experiment was stopped). In contrast, the photoperiod-insensitive H-e3/e4 flowered at almost the same time as cv Harosoy grown under SD conditions. The effects of other NB regimes were very small in cv Harosoy (differences of less than 3 d), whereas H-E1 was still sensitive to NB treatment, particularly the 8L-4D-2L-10D treatment (Fig. 4). These results suggest that the NB response depends on the genotype at E1 and is under the control of PHYA encoded by E3 and E4 and that the timing of the night break is crucial for the inhibition of flowering in soybean, as in rice (Ishikawa et al., 2005, 2009; Itoh et al., 2010) and morning glory (Ipomoea nil; Hamner and Takimoto, 1964).
Figure 4.
Effects of the NB timing on flowering in cv Harosoy (H) and its NILs for E1 (H-E1) and for e3 and e4 (H-e3/e4). Plants were exposed to light for 2 h at various time intervals during the dark phase of 16 h from emergence to 50 DAE. The most marked delay in flowering was observed when light was given at the midpoint of the dark phase. H-E1 did not flower until the end of the experiment (50 DAE). Average and se values for eight plants are given for each data point.
We then analyzed the expression patterns of E1 and E1L under the three NB treatments (8L-4D-2L-10D, 8L-7D-2L-7D, and 8L-10D-2L-4D). E1 and E1L expression was induced by 8L-7D-2L-7D but not by the two other NB treatments (Fig. 5; Supplemental Fig. S3). There was no difference in the E1 and E1L expression patterns between cv Harosoy and H-E1. Unlike under the LD conditions (Figs. 1 and 2), the first peak of expression after dawn did not appear, and the expression increased before dusk and then reached a maximum at the end of the night break. In H-e3/e4, similar expression patterns of E1 and E1L were observed, but their transcript abundances were markedly low (Supplemental Fig. S4).
Figure 5.
Expression profiles of the E1 and FT genes in cv Harosoy and its NIL for E1 (H-E1) under NB treatments. The E1 transcript was up-regulated by NB treatment only when light was given at the midpoint of the dark phase of 16 h in cv Harosoy (left) and H-E1 (right) and reached a maximum at the end of the 2-h NB treatment, with another peak just before dusk. FT2a and FT5a were up-regulated under the 8L-4D-2L-10D and 8L-10D-2L-4D treatments to similar levels in cv Harosoy and H-E1, although the time-course patterns were slightly different between the two lines. Under 8L-7D-2L-7D, no FT2a expression was observed in both cv Harosoy and H-E1, whereas FT5a expression was observed in cv Harosoy but not in H-E1. Relative mRNA levels are expressed as the ratios to β-tubulin transcript levels. Average and se values for three replications are given for each data point.
The 8L-7D-2L-7D treatment inhibited FT2a expression in both cv Harosoy and H-E1, whereas FT5a expression was inhibited in H-E1 but was slightly induced (particularly during the night break) in cv Harosoy (Fig. 5). In H-e3/e4, both FT2a and FT5a were expressed (Supplemental Fig. S4). In two other treatments, the expression of both FT2a and FT5a was induced; a high level of FT2a transcription was observed after dawn (ZT3–ZT4) in both cv Harosoy and H-E1, whereas FT5a was expressed at a higher level in cv Harosoy than in H-E1.
RNA Interference-Mediated Suppression of E1L Promotes Flowering
To determine the function of E1L genes, we developed RNA interference (RNAi) plants by virus-induced gene silencing (VIGS) using the vector from Apple latent spherical virus (ALSV). In soybean, ALSV does not considerably affect vegetative and reproductive growth and plant morphogenesis, and it is transmitted to the next generation at a rate of 20% to 30%, which is particularly suitable for knocking down target gene expression at early seedling stages (Yamagishi and Yoshikawa, 2009, 2011). We used cv Toyomusume for viral infection, because it lacks the genomic region (of around 130 kb) containing the entire E1 gene region (the e1-nl allele at E1 locus; Xia et al., 2012) but possesses the same maturity genotype at the other three loci (e2/e2, E3/E3, and E4/E4) as cv Harosoy (Tsubokura et al., 2014). The fragment used for VIGS was a 207-bp E1La region, which is identical to the corresponding region of E1Lb except for one single-nucleotide polymorphism. The ALSV vector carrying the fragment was designated as E1L-ALSV.
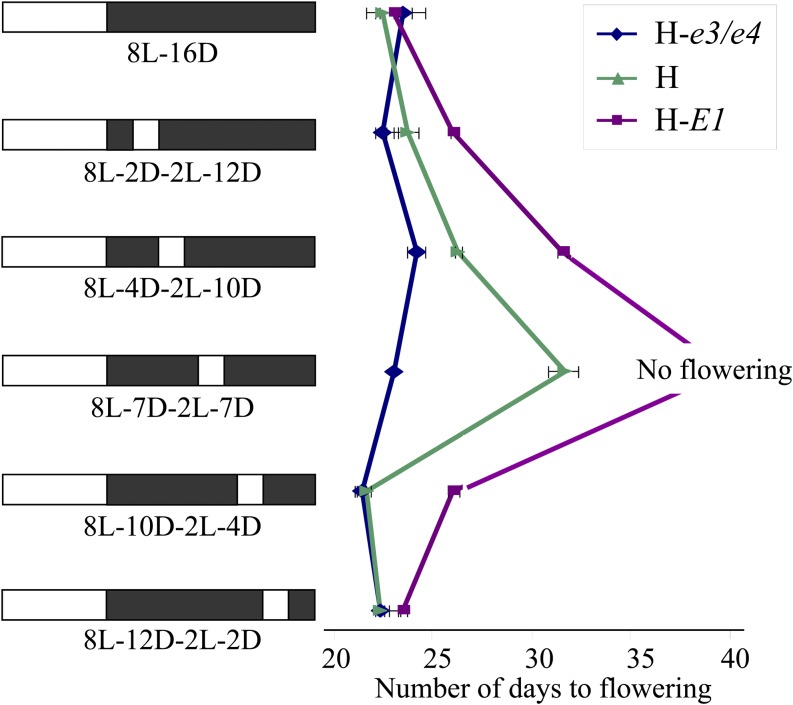
The E1L-ALSV-infected plants flowered earlier than noninfected plants and those infected with wild-type ALSV without the E1La fragment (WT-ALSV); there was no difference in flowering time between the latter two (Supplemental Fig. S5A). WT-ALSV infection did not affect the levels of any E1L transcripts (Supplemental Fig. S5B). We compared the effect of RNAi on flowering and gene expression in infected and virus-free progeny from three E1L-ALSV-infected plants. Infected progeny initiated flowering, on average, 2 weeks earlier and five nodes lower and matured earlier than virus-free progeny of the same plants (Fig. 6).
Figure 6.
Effects of E1L RNAi on flowering, maturation, and stem termination of cv Toyomusume. The maturity genotype at the E2, E3, and E4 loci of cv Toyomusume is identical to that of cv Harosoy (e2/e2, E3/E3, and E4/E4), but cv Toyomusume lacks the E1 gene (e1-nl/e1-nl). A and B, Flowering time (A) and the number of nodes at flowering (B) of plants carrying E1L-ALSV (+) and virus-free plants (−) among the progeny of three E1L-ALSV-infected plants (#1, #3, and #5). C, Plants carrying E1L-ALSV (+) showed earlier maturation than virus-free plants (−).
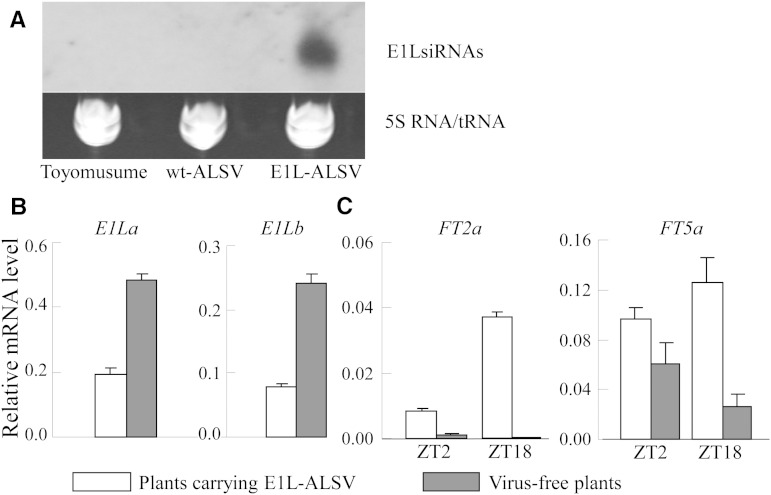
Infected plants produced small RNAs corresponding to the E1La gene (Fig. 7A). Not only E1La but also E1Lb was down-regulated in E1L-ALSV-infected plants in comparison with virus-free plants used as a control (Fig. 7B). In contrast, FT2a and FT5a were up-regulated in E1L-ALSV-infected plants in comparison with the control plants at ZT2 and ZT18 (Fig. 7C). These results demonstrate that one or both E1L genes inhibit flowering by suppressing FT2a and FT5a, similar to the E1 gene.
Figure 7.
Effects of E1L RNAi on FT2a and FT5a expression in cv Toyomusume grown under LD conditions. A, E1L RNAi produced small interfering RNAs (siRNAs) in plants carrying E1L-ALSV. wt-ALSV, Plants infected with wild-type ALSV without the E1La sequence. B, Knockdown of E1La and E1Lb expression at ZT2. C, Up-regulation of FT2a and FT5a expression at ZT2 and ZT18 by E1L RNAi. Transcript abundance was analyzed in fully expanded trifoliate leaves of 22 DAE. Relative mRNA levels are expressed as the ratios to β-tubulin transcript levels. Average and se values for three replications are given for each data point.
The NB Response Is Also Observed in the Absence of E1
The NB response of cv Harosoy and its NILs strongly suggests that E1 controls this response (Fig. 4). To determine the role of the E1L genes in the NB response, we analyzed the NB response of cv Toyomusume. This cultivar showed a significant NB response when light was given at the midpoint during the dark phase (treatment 8L-7D-2L-7D; Fig. 8A). The delay in flowering (10 d) was similar to that of cv Harosoy (Fig. 4), suggesting that a gene or genes other than E1 may also be involved in the NB response of soybean. Similar to the expression patterns of E1L in cv Harosoy and H-E1, the 8L-7D-2L-7D treatment induced E1L expression in cv Toyomusume (Supplemental Fig. S6). FT2a expression was inhibited, whereas FT5a expression was induced, at a higher level in cv Toyomusume than in cv Harosoy (Fig. 5).
Figure 8.
Effects of NB treatment and E1L RNAi on flowering time of cv Toyomusume. A, NB responses. Plants exhibited an NB response under the 8L-7D-2L-7D treatment (similar to cv Harosoy; Fig. 4). B, Effects of E1L RNAi under the 8L-7D-2L-7D treatment. Plants carrying E1L-ALSV among the progeny of E1L-ALSV-infected plants flowered earlier than the virus-free plants and noninfected cv Toyomusume plants and as early as the photoperiod-insensitive cv Harosoy NIL (H-e3/e4). The data for flowering time (number of days from emergence to flowering) are means ± se (n = 4). Different letters indicate statistically significant (P = 0.05) differences between treatments, as evaluated by the Tukey-Kramer method.
The RNAi plants initiated flowering at almost the same time as H-e3/e4 under the 8L-7D-2L-7D treatment (Fig. 8B). The difference in flowering time between the RNAi plants and controls (both virus-free progeny and noninfected cv Toyomusume) was smaller (approximately 5 d) but was statistically significant (F = 3.82, P = 0.023). Taken together, our data suggest that the E1L genes delay flowering by inhibiting the expression of FT2a and FT5a, but their effect appears to be smaller than the effect of E1, because flowering was strongly inhibited by the 8L-7D-2L-7D treatment in H-E1 (Fig. 4).
DISCUSSION
Alleles at the E1 Locus Differentially Regulate FT5a Expression
The E1 locus consists of multiple alleles, including the E1 allele, one hypomorphic allele (e1-as), two dysfunctional alleles (e1-nl and e1-fs), and two uncharacterized alleles (e1-re and e1-pe; Xia et al., 2012; Tsubokura et al., 2014). The allele e1-as, traditionally designated as e1, encodes a protein that is dysfunctional in its nuclear localization because of a point mutation in the putative nuclear localization signal (Xia et al., 2012). A comparison of flowering time under LD conditions among cultivars with different E1 alleles revealed that e1-as is a leaky allele and may retain partial E1 function (Xia et al., 2012). Although E1 overexpression strongly inhibited the expression of both FT2a and FT5a (Xia et al., 2012), the expression analyses of plants subjected to NB treatment in this study revealed that E1 and e1-as differently regulated the abundance of the FT5a transcript; FT5a expression was detected at a low level in cv Harosoy but was not detected in H-E1 (Fig. 5). Under the NB treatment that delayed flowering, FT5a expression was higher in cv Toyomusume (e1-nl; Supplemental Fig. S6) than in cv Harosoy (Fig. 5). E1 thus had the strongest inhibitory effect on FT5a expression, most likely followed by e1-as and e1-nl, in accordance with the allelic effects observed among the cultivars (Xia et al., 2012). No clear tendency for FT2a regulation by different alleles at the E1 locus was detected (Fig. 5), suggesting that one or more factors other than E1 is (are) involved in the control of FT2a expression. Watanabe et al. (2011) found that the E2 gene (a soybean ortholog of GI) mainly controls flowering time through the regulation of FT2a, not FT5a. Different expression profiles of FT2a and FT5a indicate that, in addition to the PHYA-mediated E1 pathway, another mechanism may control the two FT genes (Watanabe et al., 2012). Further studies are needed to better understand how E1 and other factors control the expression of FT2a and FT5a.
Two E1 Homologs Inhibit Flowering
The high structural similarity of E1, E1La, and E1Lb suggests their functional similarity; at the same time, a number of amino acid substitutions and insertions/deletions may indicate certain subfunctionalization between E1 and the two E1L genes (Xia et al., 2012). To determine the function of the E1L genes in the control of flowering, we developed cv Toyomusume plants with E1L expression down-regulated by RNAi. We took advantage of the transferability of ALSV into seeds to analyze the effect of E1L down-regulation on flowering and on the expression of FT2a and FT5a by comparing seedlings with or without the virus among the progeny of three E1L-ALSV-infected plants. Down-regulation of E1L expression up-regulated the expression of FT2a and FT5a and promoted flowering (Figs. 6 and 7). Under the NB conditions, the E1L-ALSV-infected plants initiated flowering almost at the same time as H-e3/e4. Thus, similar to E1, the two E1L genes appear to inhibit flowering by down-regulating the FT2a and FT5a genes. However, their effect on flowering may be weaker than that of E1 or their function(s) could be subfunctionalized from those of E1, because a complete E1 loss in cv Toyomusume (e1-nl), or partial lack of E1 function in cv Harosoy (e1-as), is not compensated for by the presence of functional E1L genes in these two cultivars.
E1 and E1L Genes Are Key Players in the NB Response of Soybean
The most intriguing finding of this study is that the induction and expression of E1 and E1Ls depended on the coincidence of their light-sensitive phase and the light signal; in particular, the induction of expression after dawn required a period of light before dusk the previous day (Figs. 2 and 3). The replacement of the light phase with a dark period during 6 h before dusk abolished the first peak of E1 and E1L expression, whereas other durations of light period did not have any inhibitory effect on E1 and E1L expression the next day (Figs. 2 and 3). The analysis of the NB response further supports the critical role of irradiation timing in the induction and expression of the E1 and E1L genes (Fig. 5). Only light given at the midpoint of an 8-h dark period induced E1 and E1L expression. Therefore, our data suggest that light given at the right time is needed for the induction of E1 and E1L expression, which in turn inhibits the expression of FT2a and FT5a. Different responses to NB treatments among cv Harosoy and its NILs for E1 and for e3 and e4 further suggest that the NB response depends on the genotype at E1 and is under the control of PHYA encoded by E3 and E4 (Fig. 4). Furthermore, despite lacking the E1 gene, cv Toyomusume responded to NB treatment as cv Harosoy did, but this response was abolished by down-regulation of E1L genes by VIGS (Fig. 8). Taken together, these data suggest that PHYA-mediated induction of E1 and E1L genes by light is crucial for the photoperiodic response of flowering in soybean. Therefore, the flowering response of soybean cv Biloxi in NB or cycle-length experiments that was observed in classical studies (Bünning, 1960, 1979; Nanda and Hamner, 1962; Coulter and Hamner, 1964; Hamner and Takimoto, 1964) could be accounted for, at least in part, by light-dependent regulation of the E1 family genes. However, H-e3/e4 had a markedly low E1 and E1L expression under LD and 8L-7D-2L-7D NB treatments, but the expression patterns were similar to those in cv Harosoy and H-E1. The data obtained in this study thus suggest that two PHYA genes, E3 and E4, may control the amplitude of expression levels of the E1 and E1L genes but not the circadian rhythm itself. The weak but detectable up-regulation of the E1 and E1L genes in the first peak under LD conditions (Supplemental Fig. S2) and during the night break (Supplemental Fig. S4) may be caused by GmPHYA1, which is a homolog of E4. Both GmPHYA1 and E4 redundantly control photomorphogenesis under light, with red:far-red light quantum ratios of less than 1 (Liu et al., 2008). Furthermore, in the e3/e3 e4/e4 genetic background, E1 delayed flowering under far-red light-enriched LD conditions, although e1-as did not, suggesting that GmPHYA1 controls floral induction under far-red light-enriched LD conditions. Further studies are needed to determine how PHYA mediates the induction of E1 and E1L expression and what mechanisms are involved in the generation of circadian rhythms.
The E1 family is distantly related to the genes encoding proteins with the plant-specific B3 domain (Xia et al., 2012). The B3 superfamily encompasses many gene families, which have diverse functions in plant growth and development. Among the genes with well-characterized functions, Arabidopsis TEMPRANILLO and rice LEAFY COTYLEDON2 and FUSCA3-Like1 genes delay flowering (Peng et al., 2007; Castillejo and Pelaz, 2008). However, there is no distinct homolog of the E1 family in the Arabidopsis and rice genomes (Xia et al., 2012). In Arabidopsis, CONSTANS (CO), a transcriptional activator of FT (Kobayashi et al., 1999; Samach et al., 2000; Suárez-López et al., 2001; Yanovsky and Kay, 2002), plays a key role in the regulation of photoperiodic flowering (for review, see Kobayashi and Weigel, 2007; Turck et al., 2008; Andrés and Coupland, 2012; Song et al., 2013). Transcriptional and posttranscriptional up-regulation of CO results in accumulation of the CO protein in the late afternoon under LD conditions, which in turn activates FT expression. The rice CO ortholog Hd1 acts as an activator of Hd3a expression under SD conditions but as a suppressor under LD conditions (Yano et al., 2000; Izawa et al., 2002; Kojima et al., 2002; Hayama et al., 2003; Ishikawa et al., 2005, 2011). This functional switch, which is absent in Arabidopsis, is controlled by phytochrome B (Ishikawa et al., 2011), in contrast to CO, which only acts as an activator under inductive LD conditions. In addition, two gating mechanisms that act on a floral promoter, the B-type response regulator Early heading date1 and a repressor, the CCT domain protein Grain number, plant height, and heading date7, fine-tune critical daylength recognition in rice (Itoh et al., 2010). Therefore, different systems may be involved in the control of photoperiodic responses in LD (Arabidopsis) and SD (rice) plant species (for review, see Andrés and Coupland, 2012) and even in different SD plants, such as rice (Itoh et al., 2010; Ishikawa et al., 2011; Itoh and Izawa, 2013) and soybean (this study).
Dominant alleles at E1 to E4 loci inhibit flowering under LD conditions (Bernard, 1971; Buzzell, 1971; Buzzell and Voldeng, 1980; Saindon et al., 1989; Cober et al., 1996; Abe et al., 2003). All four cultivars or lines used in this study had dysfunctional e2 alleles, but all except for the photoperiod-insensitive H-e3/e4 responded to NB treatment, suggesting that E2 is not involved in the light-dependent control of photoperiodism. Soybean possesses three orthologs of Arabidopsis GI, including E2 (Watanabe et al., 2011); therefore, alternatively, the remaining two GI genes might compensate for the loss of E2 function. Although E2 has the second largest effect (after E1) on flowering time among the four maturity genes (Tsubokura et al., 2014), it may be involved in the inhibition of flowering under LD conditions through a pathway different from the PHYA-E1 pathway. Soybean has 26 CO-like genes, of which GmCOL1a/GmCOL1b and GmCOL2a/GmCOL2b show the highest sequence similarity to Arabidopsis CO (Wu et al., 2014). These four CO homologs fully complemented the late-flowering phenotype of the Arabidopsis co-1 mutant, suggesting that they are potential inducers of flowering in soybean (Wu et al., 2014). However, their function in soybean remains undetermined. The functional roles of the GI-CO module in the regulation of FT2a and FT5a expression need to be studied to improve our understanding of the photoperiodic responses of flowering in soybean.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Soybean (Glycine max ‘Harosoy’; L58-266; e1-as/e1-as, e2/e2, E3/E3, and E4/E4) and its NILs for E1 (L68-694; H-E1) and for two dysfunctional phyA genes, e3 and e4 (OT89-5; H-e3/e4), and cv Toyomusume (e1-nl/e1-nl, e2/e2, E3/E3, and E4/E4) were used in this study. The cv Harosoy and cv Toyomusume are early-maturing and photoperiod-sensitive cultivars. Four germinated seeds (3 d after imbibition) of each line with tap roots of almost the same size were transplanted in Wagner pots (16-cm diameter) or plastic pots (12-cm diameter) and put in growth chambers at a constant air temperature of 23°C or 25°C and an average photosynthetic photon flux density of 200 μmol m–2 s–1 supplied only by fluorescent lights (for NB treatments) or 300 μmol m–2 s–1 supplied by fluorescent and incandescent lights. Daylength was 8 or 12 h for SD conditions and 18 h for LD conditions.
Transfer between Different Daylength Conditions
The cv Harosoy plants were grown under LD conditions until 19 DAE. In one set of experiments, the light phase was shortened to 12 h by turning off the light between ZT12 and ZT18. Three days later, the dark phase between ZT12 and ZT18 returned to the light phase. In another set of experiments, 6- or 2-h dark periods were introduced between ZT0 and ZT18. Pieces of young, fully developed trifoliate leaves were sampled every 1 h from ZT15 or ZT16 to ZT20 and from ZT0 to ZT5 the next day and bulked from four plants.
NB Treatments
The NB response of flowering was analyzed in cv Harosoy, H-E1, H-e3/e4, and cv Toyomusume. The SD (8 h of light) conditions were used as a control, and five different NB treatments were set, in which a night break of 2 h was given at 2, 4, 7, 10, or 12 h after dusk during the 16-h dark phase. Pieces of young fully developed trifoliate leaves were sampled every 1 h from ZT1 or ZT2 to ZT8 and 1 and 2 h after the start of NB treatment and bulked from four plants at 20 DAE for expression analyses. Flowering time (number of days from emergence to flowering) was recorded for each plant.
Quantitative Reverse Transcription-PCR Analysis
Sampled tissues were immediately frozen in liquid N2 and stored at −80°C. Total RNA was isolated from frozen tissues following the lithium chloride precipitation procedure (Napoli et al., 1990), except that we removed genomic DNA from the RNA fraction using DNase I (Takara Bio). Purification of mRNA and synthesis of cDNA were performed according to Dwiyanti et al. (2011). The PCR mixture contained 1 µL of cDNA, 5 µL of 1.2 µm primer premix (for primer sequences, see Supplemental Table S1), 10 µL of SYBR Premix ExTaq Perfect Real Time (Takara Bio), and water to a final volume of 20 µL. Quantitative PCR was performed by using the CFX96 Real-Time System (Bio-Rad Laboratories Japan). The PCR cycling conditions were 95°C for 3 min followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, 72°C for 20 s, and 78°C for 2 s. Fluorescence was quantified before and after the incubation at 78°C to monitor the formation of primer dimers. The mRNA for β-tubulin was used as a control. A reaction mixture without reverse transcriptase was also used as a control to confirm the absence of genomic DNA contamination. Amplification of a single DNA species was confirmed by melting-curve analysis of quantitative PCR and gel electrophoresis of the PCR products.
RACE and Sequencing
RNA was extracted from leaves of cv Harosoy and cv Toyomusume. 5′ and 3′ RACE was performed to obtain full-length cDNA sequences of cv Toyomusume E1La and E1Lb using the SMARTer RACE cDNA Amplification Kit (Clontech, Takara Bio). Primer sequences were designed based on the cv Williams 82 sequences (Glyma04g24640 and Glyma18g22670; Supplemental Table S1). The cv Harosoy coding regions of E1La and E1Lb were amplified by using the cDNA as a template, and the amplified products were cloned and sequenced. The primers used for the PCR amplification are listed in Supplemental Table S1.
RNAi-Mediated Silencing of the E1L Genes
The ALSV vector was used to down-regulate E1L expression. E1L-ALSV carrying a 207-bp fragment of the E1La gene was prepared as follows. An E1La cDNA fragment was amplified from a plasmid carrying the E1La gene by PCR using the primers E1La-220Xho+ (5′-TACATCTCGAGCCTTGGAAGATCAAGAAGACG-3′, corresponding to nucleotides 220–240 of E1La; the XhoI site is underlined) and E1La-426Bam- (5′-TACATGGATCCAGACCATCGCTTTAGAACGAG-3′, corresponding to positions 406–426 of E1La; the BamHI site is underlined). The amplified fragment was digested with XhoI and BamHI and ligated into pEALSR2L5R5GFP (Yamagishi and Yoshikawa, 2009) digested with the same enzymes. Two cDNAs comprising the viral genome, pEALSR1 and pEALSR2L5R5E1La, were then mechanically inoculated into Chenopodium quinoa (Li et al., 2004), and the resulting virus was designated E1L-ALSV. The wild-type ALSV without the target sequence was used as a control. Soybean seeds were sown on vermiculite soaked with water in a petri dish and incubated in a growth chamber (25°C) overnight. Imbibed seeds were used for viral inoculation using the Helios Gene Gun System (Bio-Rad) as described previously (Yamagishi and Yoshikawa, 2009). After particle bombardment, seeds were sown in soil in pots and placed into a growth chamber (25°C/20°C, 18-h photoperiod). The effects of RNAi on flowering time and gene expression were evaluated for plants carrying the virus and for virus-free plants selected from the progeny of three E1L-ALSV-infected plants.
Detection of Small Interfering RNAs
Small interfering RNA detection was carried out at ZT2 in fully expanded trifoliate leaves of 22-d-old E1L-ALSV-infected plants. Small interfering RNA was detected by northern-blot analysis according to a modified protocol of Goto et al. (2003), as described previously (Kasai et al., 2013). Small RNAs were isolated, separated by electrophoresis on a 15% polyacrylamide-Tris-borate-EDTA-urea gel, and transferred to a Hybond-N+ membrane. After baking and UV light cross-linking, the membrane was hybridized with a digoxigenin-labeled sense RNA probe (nucleotide positions 220–426 of E1La).
Sequence data from this study have been deposited in the GenBank/EMBL/DNA Data Bank of Japan databases under accession numbers LC003236 and LC003237 for the full-length cDNA sequences of E1La and E1Lb of cv Toyomusume, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. E1 and E1L structures.
Supplemental Figure S2. Effects of light on the induction of E1 and E1L expression in a cv Harosoy NIL for e3 and e4 (H-e3/e4).
Supplemental Figure S3. Expression profiles of the E1L genes in cv Harosoy and its NIL for E1 (H-E1) under NB treatments.
Supplemental Figure S4. Expression profiles of the E1 and E1L genes in a cv Harosoy NIL for e3 and e4 (H-e3/e4) under the 8L-7D-2L-7D NB treatment.
Supplemental Figure S5. Flowering time and E1L transcript abundance in cv Toyomusume plants infected with wild-type ALSV (wt-ALSV) and virus-free plants.
Supplemental Figure S6. Expression profiles of the E1L and FT genes in cv Toyomusume under NB treatments.
Supplemental Table S1. List of primers for quantitative reverse transcription-PCR, RACE, and sequencing analyses used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Elroy R. Cober (Eastern Cereal and Oilseed Research Centre, Agriculture and Agri-Food Canada) and Dr. Randall L. Nelson (Department of Crop Sciences, University of Illinois) for supplying seeds of the cv Harosoy isolines.
Glossary
- SD
short-day
- NB
night-break
- LD
long-day
- cDNA
complementary DNA
- UTR
untranslated region
- DAE
days after emergence
- NIL
near-isogenic line
- RNAi
RNA interference
- VIGS
virus-induced gene silencing
- ALSV
Apple latent spherical virus
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31430065 and 31171579 to B.L.) and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant no. 23880001 to J.A.).
Articles can be viewed without a subscription.
References
- Abe J, Xu DH, Miyano A, Komatsu K, Kanazawa A, Shimamoto Y (2003) Photoperiod-insensitive Japanese soybean landraces differ at two maturity loci. Crop Sci 43: 1300–1304 [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Bernard RL. (1971) Two genes for time of flowering in soybeans. Crop Sci 11: 242–244 [Google Scholar]
- Bünning E. (1960) Circadian rhythms and the time measurement in phototropism. Cold Spring Harb Symp Quant Biol 25: 249–256 [Google Scholar]
- Bünning E. (1979) Circadian rhythm, light, and photoperiodism: a re-evaluation. Bot Mag Tokyo 92: 89–103 [Google Scholar]
- Buzzell RI. (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can J Genet Cytol 13: 703–707 [Google Scholar]
- Buzzell RI, Voldeng HD (1980) Inheritance of insensitivity to long daylength. Soybean Genet Newsl 7: 26–29 [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Cober ER, Tanner JW, Voldeng HD (1996) Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci 36: 606–610 [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Coulter MW, Hamner KC (1964) Photoperiodic flowering response of Biloxi soybean in 72-hour cycles. Plant Physiol 39: 848–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwiyanti MS, Yamada T, Sato M, Abe J, Kitamura K (2011) Genetic variation of γ-tocopherol methyltransferase gene contributes to elevated α-tocopherol content in soybean seeds. BMC Plant Biol 11: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Hu R, Zhang X, Wang X, Zhang W, Zhang Q, Ma J, Fu YF (2014) Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biol 14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner WW, Allard HA (1920) Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res 18: 553–606 [Google Scholar]
- Goto K, Kanazawa A, Kusaba M, Masuta C (2003) A simple and rapid method to detect plant siRNAs using nonradioactive probes. Plant Mol Biol Rep 21: 51–58 [Google Scholar]
- Hamner KC, Takimoto A (1964) Circadian rhythms and plant photoperiodism. Am Nat 98: 295–322 [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K (2011) Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genomics 285: 461–470 [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Shinomura T, Takano M, Shimamoto K (2009) Phytochrome dependent quantitative control of Hd3a transcription is the basis of the night break effect in rice flowering. Genes Genet Syst 84: 179–184 [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, Takano M, Shimamoto K (2005) Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17: 3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Izawa T (2013) The coincidence of critical day length recognition for florigen gene expression and floral transition under long-day conditions in rice. Mol Plant 6: 635–649 [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Kasai M, Matsumura H, Yoshida K, Terauchi R, Taneda A, Kanazawa A (2013) Deep sequencing uncovers commonality in small RNA profiles between transgene-induced and naturally occurring RNA silencing of chalcone synthase-A gene in petunia. BMC Genomics 14: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D (2007) Move on up, it’s time for change: mobile signals controlling photoperiod-dependent flowering. Genes Dev 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Kong F, Liu B, Xia Z, Sato S, Kim BM, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, et al. (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154: 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Sasaki N, Isogai M, Yoshikawa N (2004) Stable expression of foreign proteins in herbaceous and apple plants using Apple latent spherical virus RNA2 vectors. Arch Virol 149: 1541–1558 [DOI] [PubMed] [Google Scholar]
- Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlain BA, Hesketh JD, Bernard RL (1987) Genetic effect on reproductive phenology in soybean isolines differing in maturity genes. Can J Plant Sci 67: 105–116 [Google Scholar]
- Nan H, Cao D, Zhang D, Li Y, Lu S, Tang L, Yuan X, Liu B, Kong F (2014) GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS ONE 9: e97669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda KK, Hamner KC (1962) Investigations on the effect of light break on the nature of the endogenous rhythm in the flowering response of Biloxi soybean (Glycine max, L. Merr.). Planta 58: 164–174 [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T (2008) Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol 49: 1645–1658 [DOI] [PubMed] [Google Scholar]
- Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL (2007) Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem Biophys Res Commun 360: 251–256 [DOI] [PubMed] [Google Scholar]
- Pin PA, Nilsson O (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ 35: 1742–1755 [DOI] [PubMed] [Google Scholar]
- Saindon G, Voldeng HD, Beversdorf WD, Buzzell RI (1989) Genetic control of long daylength response in soybean. Crop Sci 29: 1436–1439 [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sun H, Jia Z, Cao D, Jiang B, Wu C, Hou W, Liu Y, Fei Z, Zhao D, Han T (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 6: e29238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Thakare D, Kumudini S, Dinkins RD (2011) The alleles at the E1 locus impact the expression pattern of two soybean FT-like genes shown to induce flowering in Arabidopsis. Planta 234: 933–943 [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue B (1997) Photoperiodism in Plants, Ed 2 Academic Press, San Diego [Google Scholar]
- Tsubokura Y, Matsumura H, Xu M, Nakashima H, Liu B, Anai T, Kong F, Yuan X, Kanamori H, Katayose Y, et al. (2013) Genetic variation in soybean at the maturity locus E4 is involved in adaptation to long days at high latitudes. Agronomy 3: 117–134 [Google Scholar]
- Tsubokura Y, Watanabe S, Xia Z, Kanamori H, Yamagata H, Kaga A, Katayose Y, Abe J, Ishimoto M, Harada K (2014) Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann Bot (Lond) 113: 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K (2013) Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr Opin Plant Biol 16: 228–235 [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Upadhyay AP, Ellis RH, Summerfield RJ, Roberts EH, Qi A (1994) Characterization of photothermal flowering responses in maturity isolines of soyabean [Glycine max (L.) Merrill] cv. Clark. Ann Bot (Lond) 74: 87–96 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Harada K, Abe J (2012) Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed Sci 61: 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, et al. (2009) Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, et al. (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188: 395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Price BW, Haider W, Seufferheld G, Nelson R, Hanzawa Y (2014) Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS ONE 9: e85754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Watanabe S, Yamada T, Tsubokura Y, Nakashima H, Zhai H, Anai T, Sato S, Yamazaki T, Lü S, et al. (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci USA 109: E2155–E2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Xu Z, Liu B, Kong F, Tsubokura Y, Watanabe S, Xia Z, Harada K, Kanazawa A, Yamada T, et al. (2013) Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol 13: 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, Yoshikawa N (2009) Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with Apple latent spherical virus vectors. Plant Mol Biol 71: 15–24 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Yoshikawa N (2011) Virus-induced gene silencing of endogenous genes and promotion of flowering in soybean by Apple latent spherical virus-based vectors. In Sudaric A. ed, Soybean: Molecular Aspects of Breeding. InTech China, Shanghai, pp 43–56 [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Zhai H, Lü S, Liang S, Wu H, Zhang X, Liu B, Kong F, Yuan X, Li J, Xia Z (2014) GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 9: e89030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.