The soybean cyst nematode induces genomewide differential DNA methylation that impacts a large number of structural genes and biological functions.
Abstract
The soybean cyst nematode (SCN; Heterodera glycines) induces the formation of a multinucleated feeding site, or syncytium, whose etiology includes massive gene expression changes. Nevertheless, the genetic networks underlying gene expression control in the syncytium are poorly understood. DNA methylation is a critical epigenetic mark that plays a key role in regulating gene expression. To determine the extent to which DNA methylation is altered in soybean (Glycine max) roots during the susceptible interaction with SCN, we generated whole-genome cytosine methylation maps at single-nucleotide resolution. The methylome analysis revealed that SCN induces hypomethylation to a much higher extent than hypermethylation. We identified 2,465 differentially hypermethylated regions and 4,692 hypomethylated regions in the infected roots compared with the noninfected control. In addition, 703 and 1,346 unique genes were identified as overlapping with hyper- or hypomethylated regions, respectively. The differential methylation in genes apparently occurs independently of gene size and GC content but exhibits strong preference for recently duplicated paralogs. Furthermore, a set of 278 genes was identified as specifically syncytium differentially methylated genes. Of these, we found genes associated with epigenetic regulation, phytohormone signaling, cell wall architecture, signal transduction, and ubiquitination. This study provides, to our knowledge, new evidence that differential methylation is part of the regulatory mechanisms controlling gene expression changes in the nematode-induced syncytium.
The soybean cyst nematode (SCN; Heterodera glycines) is a sedentary parasite of soybean (Glycine max) roots and is considered to be the most serious pathogen problem in soybean production worldwide (Koenning and Wrather, 2010). SCN undergoes a first molt inside the egg to develop from a first-stage (J1) to a second-stage juvenile (J2) before hatching. Hatched, preparasitic J2s then penetrate into soybean roots by breaching epidermal cell walls with their protrusible stylet, a hollow oral spear, aided by cell wall-digesting enzymes and other effector proteins (Hewezi and Baum, 2013). The parasitic J2s migrate to the vicinity of the vascular tissues, where the nematodes become sedentary and begin feeding. To maintain the sedentary lifestyle, SCN induces the formation of a multinucleated feeding site in roots, the syncytium, whose etiology includes considerable cell-to-cell fusion, in addition to dramatic cytoplasmic and nuclear modifications (Williamson and Hussey, 1996). Molecular and genetic studies in the last few years have provided new insights into the molecular mechanisms associated with SCN parasitism. Global gene expression profiling using microarrays has been extensively studied during plant-nematode interactions. Initially, these studies used the whole soybean root system to quantify transcriptional changes associated with the compatible interaction (Khan et al., 2004; Alkharouf et al., 2006). More comprehensive gene expression studies were conducted on syncytial cells using laser capture microdissection (LCM) coupled with microarray analysis (Ithal et al., 2007; Klink et al., 2007, 2009, 2010; Kandoth et al., 2011). These cell-specific gene expression analyses yielded lists of thousands of syncytial differentially expressed genes and provided new information about the underlying molecular events occurring during syncytium formation and function. However, the genetic networks underlying gene expression regulation in nematode-infected roots and particularly in the nematode-induced feeding sites are poorly understood. Because epigenetic modifications function in concert with genetic mechanisms to regulate gene expression in normal cells and are often dysregulated in infected cells, these modifications may contribute significantly to the regulation of the transcriptional activity of syncytial cells.
Epigenetics include biochemical modifications of DNA and associated proteins that regulate gene expression and chromosome structure and function, without changing DNA nucleotide sequences. DNA methylation, the most common epigenetic modification, is the addition or removal of a methyl group (CH3), mostly where cytosine bases occur repeatedly. In plants, DNA methylation occurs in symmetric (CpG and CHG) and asymmetric (CHH) contexts, where H refers to any nucleotide but G. The CpG and CHG patterns are symmetric across the two DNA strands, which are believed to be important for the maintenance of methylation at these sites following DNA replication. DNA cytosine methylation controls gene expression networks and hence plays essential roles in different aspects of plant growth, development, and response to biotic stress (Alvarez et al., 2010; Zhang et al., 2010; He et al., 2011; Dowen et al., 2012). In plants, de novo DNA methylation in all sequence contexts is mainly mediated through the activity of DOMAINS REARRANGED METHYLTRANSFERASEs (DRMs), homologs of the mammalian DNA methyltransferase3 enzymes (Cao and Jacobsen, 2002; Cao et al., 2003). The DRMs are guided to specific DNA sequences through the RNA-directed DNA methylation pathway, which involves the generation of approximately 23- to 24-nucleotide small interfering RNAs (siRNAs) by RNA-dependent RNA polymerase2 (RDR2) and DICER-Like3 (DCL3; Henderson and Jacobsen, 2007). These siRNAs are loaded into a complex containing the argonaute (AGO) proteins AGO4 and AGO6 to guide DRMs to target loci by a currently unknown mechanism (Matzke et al., 2009).
While DNA methylation has been initially reported to control various developmental processes in plants (He et al., 2011), recent studies revealed that this silencing pathway plays a key role in modulating plant defense responses during biotrophic interactions. Compelling evidence of dynamic changes in DNA methylation in response to infection by the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pst) has been recently reported (Dowen et al., 2012). Using deep sequencing of bisulfite-treated DNA, Dowen et al. (2012) found that differentially methylated regions were preferentially associated with genes involved in defense response and that hypomethylation in differentially methylated regions was frequently accompanied by activation of the proximal genes, specifically those with defense response function. Similarly, another recent study indicated that DNA demethylation restricted the multiplication and vascular propagation of the Pst and, consequently, some immune response genes were repressed by DNA methylation (Yu et al., 2013).
The role of DNA methylation in regulating the plant immune system was further supported by the finding that mutant lines entirely defective in maintenance of CpG methylation or non-CpG methylation were resistance to Pst infection (Dowen et al., 2012). Likewise, mutants partially impaired in CpG methylation (decrease in DNA methylation1) or non-CpG methylation (RNA-dependent RNA polymerase mutants rdr1, rdr2, and rdr6 and DICER-like triple mutant dcl2/dcl3/dcl4) showed modest increases in Pst resistance (Dowen et al., 2012). These data are consistent with our previously published results in which we found that several Arabidopsis (Arabidopsis thaliana) dcl and rdr mutants were more resistant to the beet cyst nematode (Heterodera schachtii) compared with the wild type (Hewezi et al., 2008). Chemical demethylation of the silenced resistance Xa21-like gene, Xa21G, in rice (Oryza sativa) reestablished its resistance function against Xanthomonas oryzae (Akimoto et al., 2007). Similarly, induced DNA hypomethylation at the Nucleotide-Binding Site-Leucine-Rich Repeat gene clusters by the Tobacco mosaic virus was associated with increased genomic rearrangements at these genomic loci (Boyko et al., 2007). The expression difference between the resistant alleles of the Medicago truncatula REP1 (for Resistance to Erysiphe pisi race1) gene, which confers resistance against the powdery mildew disease caused by the biotrophic fungus Erysiphe pisi, was found to be correlated with the methylation status of the promoter regions (Yang et al., 2013). In soybean, differential hypermethylation patterns at the genomic regions that contain multiple copies of the SCN resistance gene Resistance to Heterodera glycines1 have been recently identified (Cook et al., 2014). Collectively, these results indicate that DNA methylation plays a crucial role in regulating the immune system in response to infection by plant pathogens, including cyst nematodes.
In this study, we used the Methyl-Seq method to generate high-coverage genomewide DNA methylation maps encompassing the methylation status of single cytosine bases throughout the genome of soybean roots during the susceptible interactions with SCN. A high number of differentially methylated regions and overlapping protein-coding genes were identified. In addition, we found a significant portion of the differentially methylated genes was among genes reported to change expression in the soybean syncytium, pointing to a unique role of SCN-induced DNA methylation in regulating gene expression changes during parasitism.
RESULTS
Genomewide DNA Methylation Profiling of Soybean Roots during the Susceptible Interaction with SCN
To profile the DNA methylation patterns at single-nucleotide resolution during the susceptible interaction with SCN, we construction six whole-genome bisulfite-treated DNA libraries. In these experiments, we inoculated soybean ‘Williams 82’ with SCN (H. glycines-type 0 or race 3) and root tissues were collected at 5 d post inoculation from both infected and noninfected soybean roots from three independent experiments. Two libraries (infected and control) from each experiment for a total of six libraries were generated and sequenced using the Illumina HiSeq platform. The sequencing data were grouped into six different files, and a total number of 175 million 100-bp reads for SCN-infected samples and 182 million reads for noninfected control were obtained. Because of the paleopolyploid nature of the soybean genome, a significant portion of these reads were mapped to more than one genomic location, and thus, these reads were discarded. After quality filtering, a total of about 60 million reads for each of the SCN-infected and noninfected samples were of high quality and uniquely mapped to the soybean genome (Wm82.a2.v1; Table I; Schmutz et al., 2010). These high-quality sequence reads represented about 10× genome coverage, a depth greater than previously reported in Arabidopsis and soybean (Dowen et al., 2012; Schmitz et al., 2013). Bisulfite conversion efficiency was higher than 99%, as determined using the nonmethylated λ phage genome. The percentage of methylated cytosines in CpG, CHG, and CHH contexts was very similar across the three biological replicates for the SCN infected and the noninfected control (Supplemental Table S1). The SCN-infected samples had an average of 78.6%, 56.4%, and 5.0% methylation in overall cytosines occurring in CpG, CHG, or CHH contexts, respectively (Table I). Similarly, the control samples had an average of 81%, 59.3%, and 5.5% methylation in overall cytosines occurring in CpG, CHG, or CHH contexts, respectively (Table I). These data indicated that overall average methylation levels were very similar between the SCN-infected and control samples.
Table I. Summary of cytosine methylation in control and SCN-infected samples.
| Context | Control | SCN Infected |
|---|---|---|
| Total no. of high-quality reads analyzed | 91,560,375 | 87,972,505 |
| No. of reads with unique hits | 59,900,029 | 59,875,941 |
| Mapping efficiency percentage | 65.40 | 68.06 |
| Bisulfite conversion efficiency percentage | 99.6 | 99.6 |
| Total no. of C’s analyzed | 1,874,012,046 | 1,792,959,026 |
| Percentage of C’s in CpG context that are methylated | 81.00 | 78.60 |
| Percentage of C’s in CHG context that are methylated | 59.30 | 56.40 |
| Percentage of C’s in CHH context that are methylated | 5.50 | 5 |
Identification of Differentially Methylated Regions and Associated Genes
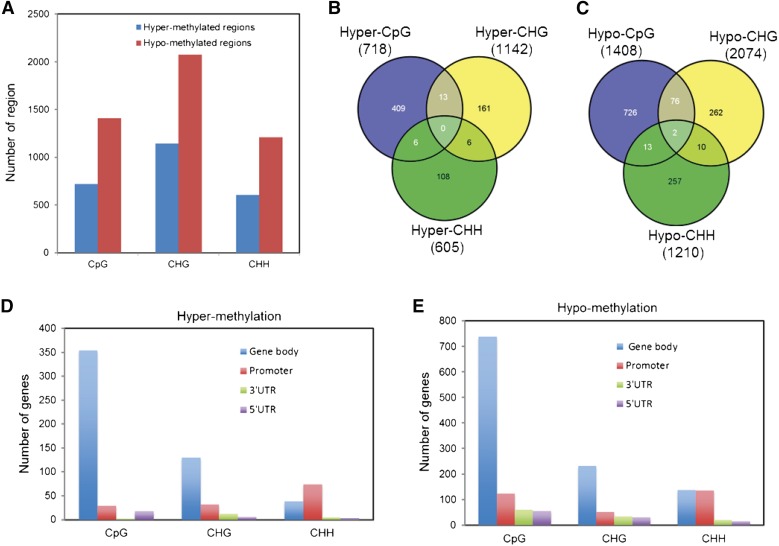
Because DNA methylation tends to cluster in specific regions of the genome (Zhang et al., 2006; Vaughn et al., 2007; Becker et al., 2011) and differences in single cytosines may not be of functional importance, a 200-bp nonoverlapping sliding window approach was used across the soybean genome to identify differentially methylated regions and overlapping genes. Differentially hyper- and hypomethylated regions between the SCN infected and the control in CpG, CHG, and CHH contexts were identified using P value < 0.01 and a percentage methylation difference larger than 25%. Interestingly, in CpG context, 718 hypermethylated regions and 1,408 hypomethylated regions were identified in the infected roots compared with the noninfected control (Fig. 1A). Similarly, in CHG context, 1,142 hypermethylated regions and 2,074 hypomethylated regions were identified (Fig. 1A), whereas, in CHH context, 605 hypermethylated regions and 1,210 hypomethylated regions were identified (Fig. 1A). These results indicate that SCN induces hypomethylation to much higher extent than hypermethylation.
Figure 1.
Characterization of differentially methylated regions in soybean roots in response to SCN infection. A, Number of differentially hyper- and hypomethylated regions between SCN-infected and noninfected roots in CpG, CHG, and CHH contexts. B and C, Number of protein-coding genes overlapping with differentially hypermethylated regions (B) or with hypomethylated regions (C) in CpG, CHG, and CHH contexts. The total number of differentially methylated regions in each context is shown in parentheses. D and E, Distribution of differentially hypermethylated regions (D) and hypomethylated regions (E) to various annotated features of protein-coding genes. UTR, Untranslated region.
We assigned each differentially methylated region to proximal protein-coding genes based on the genomic coordinates. We found 60% (428 genes), 16% (180 genes), and 20% (120 genes) of the hypermethylated regions in CpG, CHG, and CHH contexts, respectively, overlapped with protein-coding genes (Fig. 1B; Supplemental Tables S2–S4). Similarly, 58% (817 genes), 17% (350 genes), and 23% (282 genes) of the hypomethylated regions in CpG, CHG, and CHH contexts, respectively, overlapped with protein-coding genes (Fig. 1C; Supplemental Tables S5–S7). As a result, a total number of 703 and 1,346 unique genes were identified as hyper- and hypomethylated, respectively (Supplemental Tables S8 and S9). We next examined whether various methylation contexts occur in individual genes. Interestingly, we identified 25 genes that were hypermethylated in more than one context. Six of these genes were found to be hypermethylated in both CHH and CHG contexts (χ2 = 34.862 and P = 1.30E-07), six genes in CHH and CpG (χ2 = 9.275 and P = 0.026), and 13 genes in CpG and CHG contexts (χ2 = 37.786 and P = 3.138E-08; Fig. 2B; Supplemental Table S8). Also, we identified 12 genes that were hypomethylated in both CHH and CHG contexts (χ2 = 20.059 and P = 0.00017), 15 genes in CHH and CpG (χ2 = 5.392 and P = 0.145), and 78 genes in CpG and CHG contexts (χ2 = 457.025 and P = 9.797E-99; Fig. 2C; Supplemental Table S9).
Figure 2.
Genomewide mapping of differentially methylated regions and associated genes. A, A circle diagram showing chromosomal locations of the differentially hyper- and hypomethylated regions in CpG, CHG, and CHH contexts and overlapping genes into the 20 soybean chromosomes (Chr 1–Chr 20). B and C, Distribution of the differentially hypermethylated regions (B) and hypomethylated regions (C) in the 20 soybean chromosomes.
To determine if certain genic regions are associated with a specific methylation context, we examined the distribution of differentially hyper- and hypomethylated genes in various annotated features of genic regions, including promoter regions, 1,000 bp upstream of the transcription start site, exons, and 5′ and 3′ untranslated regions. The hyper- and hypomethylation in CpG and CHG contexts occur predominantly in the gene body and to much less extent in the flanking regions, whereas CHH hypermethylation was mainly located in the promoter regions (Fig. 1, D and E; Supplemental Tables S8 and S9). A set of 45 genes showed both hyper- and hypomethylation in various genic regions (Supplemental Tables S8 and S9).
To examine whether specific genomic regions are preferentially targeted for differential methylation during SCN infection, we mapped the differentially methylated regions to the 20 soybean chromosomes. The genomic distribution of the differentially methylated regions revealed that CpG and CHH regions were most enriched near the ends of the chromosomes (subtelomeric regions) and to a lesser extent in nontelomeric and centromeric regions (Fig. 2A). By contrast, CHG regions were mainly localized in nontelomeric regions (Fig. 2A), consistent with the preference of CHG methylation of targeting transposon-rich regions (Lister et al., 2008), which are located in the chromosome centers (Schmutz et al., 2010). The numbers of differentially hyper- and hypomethylated regions in CpG, CHG, and CHH were found to be significantly correlated with the chromosome size (P < 0.05). Thus, the differentially methylated regions seem to be equally distributed across the 20 chromosomes, taking into consideration chromosome size (Fig. 2, B and C).
It has been suggested that DNA methylation, particularly gene body methylation, exhibits a bias toward long genes versus short genes (Takuno and Gaut, 2013). Thus, we tested whether a correlation between DNA methylation/demethylation and gene size occurs. The methylation levels were plotted against the size of differentially hyper- and hypomethylated genes. No bias toward long genes was observed in any sequence context (Supplemental Fig. S1, A–C), suggesting that methylation/demethylation occurs independently of gene size. Similarly, we asked whether GC content in the differentially methylated regions (200 bp) correlates with the methylation levels. The differential methylation levels in various sequence contexts were plotted against the GC content of the differentially hyper- and hypomethylated regions. A constant profile of methylation levels versus various levels of GC content was observed (Supplemental Fig. S1, D and E), indicating that differential methylation occurs independent of GC content.
Recently Duplicated Soybean Genes Exhibit Strong Preference for Differential Methylation in Response to SCN Infection
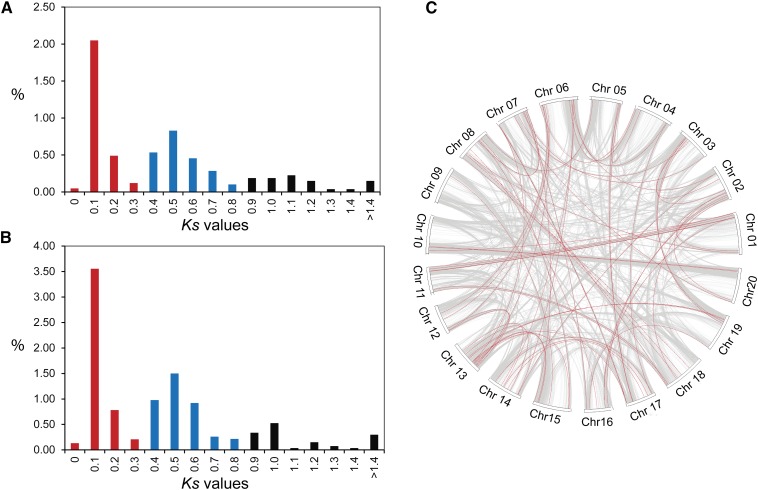
It has been suggested that the soybean genome experienced two whole genomewide duplication (WGD) events approximately 59 and 13 million years ago (Myr; Schmutz et al., 2010). We examined if differential methylation targets specifically genes contributed by a specific WGD event. We scanned the soybean genome for duplicate blocks that contain the differentially hyper- and hypomethylated genes and identified 673 collinearity events among the 703 hypermethylated genes and 1,163 events among the 1,346 hypomethylated genes. When we calculated the synonymous distance (Ks) values of these events, we found that recently duplicated genes contributed by the 13-Myr WGD (Ks = 0.06–0.39) are preferentially targeted for differential methylation compared with those genes contributed by the 59-Myr WGD (Ks = 0.40–0.80). Specifically, 2.71% (453 genes) of the paralogs contributed by the 13-Myr WGD were hypermethylated (χ2 = 18.228 and P = 3.946E-04) versus 2.2% (194 genes) of the paralogs contributed by the 59-Myr WGD (χ2 = 1.831 and P = 0.608; Fig. 3A). Likewise, 4.68% (783 genes) of the paralogs contributed by the 13-Myr WGD were hypomethylated (χ2 = 32.194 and P = 4.763E-07) versus 3.87% (341 genes) of the paralogs contributed by the 59-Myr WGD (χ2 = 2.025 and P = 0.567; Fig. 3B). We further asked whether homologous genes are targeted similarly for differential methylation. Careful examination of the collinear genomic regions among the differentially hyper- and hypomethylated genes resulted in the identification of 69 duplicated regions. These 69 duplicated regions contained 133 differentially methylated genes (Fig. 3C). Homologs among these 133 genes are connected by red lines in Figure 3C.
Figure 3.
Recently duplicated soybean genes are preferentially targeted for differential methylation in response to SCN infection. A, Percentages of hypermethylated genes with various Ks values relative to the total number of paralogs that were contributed by the 13-Myr WGD (red bars; Ks = 0.06–0.39) or by the 59-Myr WGD (blue bars; Ks = 0.40–0.80). B, Percentages of the hypomethylated genes with various Ks values relative to the total number of paralogs that were contributed by the 13- or 59-Myr WGD events. The Ks values for all differentially methylated genes were calculated using MCScanX program. C, Homologous genes were targeted correspondingly for differential methylation. Shown are 69 collinearity events (red lines) among 133 differentially methylated genes. Gray lines represent whole-genome duplicated genes contributed by the 13- and 59-Myr WGD events. Chr, Chromosome.
Differential Methylation Impacts Gene Expression Levels in SCN-Infected Roots
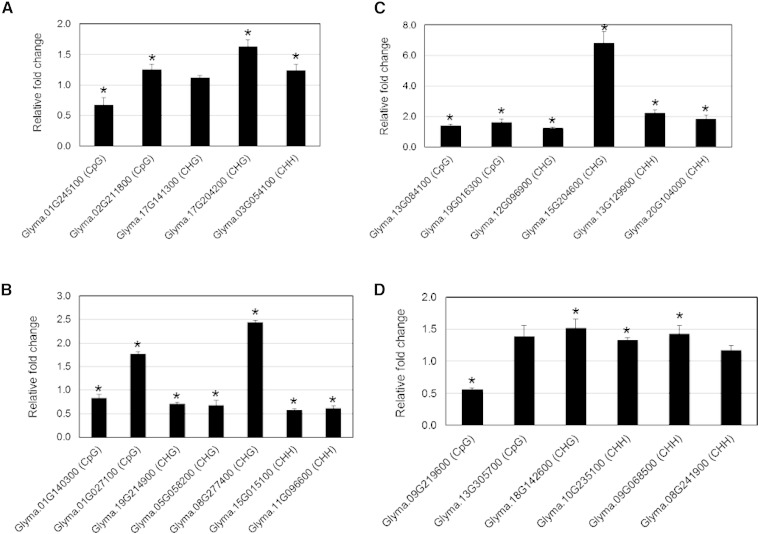
Recent studies have indicated that the effects of DNA methylation on gene expression levels differ depending on the genic regions and methylation context (Gent et al., 2013; Schmitz et al., 2013; Yang et al., 2015). Thus, we assayed the impact of DNA methylation in gene body and promoter regions on gene expression levels. RNA was isolated from the same SCN-infected and control samples and used in quantitative PCR (qPCR) assays. A set of 24 genes (eight for each sequence context) was randomly selected to represent hyper- and hypomethylation in gene body and promoter regions. Twenty-one of these 24 genes showed significant changes in mRNA expression levels in response to SCN infection relative to noninfected control (Fig. 4, A–D). qPCR analysis also revealed an association between gene body hypermethylation and both increased and decreased gene expression levels (Fig. 4A). By contrast, hypomethylation in gene body showed a general trend of increased gene expression levels (Fig. 4C). Also, increased CpG, CHG, and CHH methylation in the promoter regions showed positive or negative impacts on expression levels (Fig. 4B), whereas demethylation showed a trend of increased gene expression levels (Fig. 4D). These data suggest that differential methylation associated with SCN infection may have heterogeneous effects on gene expression levels.
Figure 4.
qPCR quantification of the impact of DNA methylation in gene body and promoter regions on gene expression levels. A and B, Hypermethylation in gene body (A) or promoter regions (B) in various sequence contexts showed heterogeneous effects on gene expression levels. C and D, Hypomethylation in gene body (C) or promoter regions (D) in various sequence contexts was mainly associated with increased gene expression levels. The relative fold-change values were calculated using the 2−ΔΔCT method and represent changes of the expression levels in SCN-infected root tissues relative to noninfected controls. Data are averages of three biologically independent experiments ± se. Mean values significantly different from 1.0 (no change) are indicated by an asterisk as determined by Student’s t tests (P < 0.05). Soybean ubiquitin (Glyma.20G141600) was used as an internal control to normalize gene expression levels.
Differentially Methylated Genes of SCN-Infected Roots Are Associated with Various Molecular Functions
We classified the differentially methylated region-associated genes into different groups by molecular function using the Gene Ontology (GO) categorization from the AgriGO database (Du et al., 2010) and conducted an enrichment analysis using Fisher’s exact test (P < 0.05). Molecular function groups corresponding to binding activity, transferase activity, catalytic activity, and hydrolase activity are significantly enriched among the differentially methylated genes (Supplemental Tables S10 and S11). While it seems likely that methylation and demethylation impact various molecular function categories to a similar degree, we observed a significant difference in a number of molecular function groups between the differentially hyper- and hypomethylated genes, specifically those associated with nucleic acid binding, RNA binding, RNA polymerase activity, hydrolase activity, microtubule and cytoskeletal protein binding, and kinase activity (Supplemental Tables S10 and S11).
Identification of Syncytium Genes That Are under Methylation Control
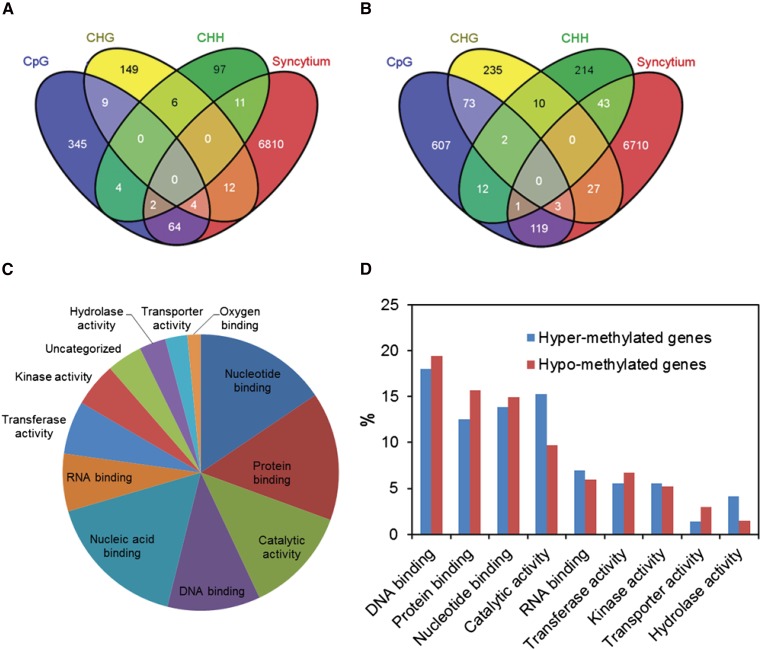
To identify syncytial genes that are under methylation control, we compared the differentially hyper- and hypomethylated genes identified by us with our reference list of genes that change the expression in the syncytium induced by SCN (6,903 genes; Ithal et al., 2007; Klink et al., 2009, 2010; Kandoth et al., 2011). We found 70, 16, and 13 genes of the differentially hypermethylated genes in CpG, CHG, and CHH contexts, respectively, that overlapped with syncytial differentially expressed genes (Fig. 5A). Similarly, 123, 30, and 44 genes of the differentially hypomethylated genes in CpG, CHG, and CHH contexts, respectively, were found to be overlapped with syncytial differentially expressed genes (Fig. 5B). After eliminating duplicated genes that were differentially methylated in more than one context, we identified 93 of the differentially hypermethylated genes (Supplemental Table S12) and 193 of the differentially hypomethylated genes (Supplemental Table S13) as overlapping with the 6,903 syncytium-regulated genes. Eight genes showed both hyper-and hypomethylation in different genic regions, resulting in 278 unique syncytium differentially methylated genes. These 278 genes were classified using GO terms into various molecular function categories, including binding activity, catalytic activity, transferase activity, kinase activity, and hydrolase activity (Fig. 5C). These categories were distributed to a similar extent in hyper- and hypomethylated genes overlapping with syncytium (Fig. 5D). Enrichment analysis revealed that genes involved in structural molecule activity and cofactor binding were significantly enriched among the 93 differentially hypermethylated genes. Genes involved in sequence-specific DNA binding, hydrolase activity, and peptidase activity were significantly enriched among the 193 differentially hypomethylated genes.
Figure 5.
Identification of syncytium genes that are under methylation control. A and B, Venn diagrams showing overlap between syncytium differentially expressed genes and those identified as hypermethylated genes (A) or hypomethylated genes (B) in various sequence contexts. Ninety-three of the differentially hypermethylated genes and 193 of the differentially hypomethylated genes were identified as overlapping with the 6,903 syncytium differentially expressed genes. C, GO categorization of the molecular function of 278 unique syncytium differentially methylated genes. Only categories represented by at least three genes were included. D, Comparison of GO categorization of the molecular function between the 93 hypermethylated and the 193 hypomethylated genes overlapping with syncytium differentially expressed genes.
DISCUSSION
We examined the impacts of SCN infection on the genomewide DNA methylation patterns in soybean roots and identified both differentially methylated regions and the associated genes in CpG, CHH, and CHG contexts. Our analysis provided several, to our knowledge, new insights into DNA methylation changes associated with SCN parasitism of soybean roots. We found that SCN induces hypomethylation to a much higher degree compared with hypermethylation in all sequence contexts. This may explain the increased number of up-regulated SCN-responsive genes relative to down-regulated genes reported in various global gene expression studies (Ithal et al., 2007; Klink et al., 2009; Kandoth et al., 2011). A high proportion of demethylated cytosines has also been reported to occur in response to bacterial infection (Dowen et al., 2012). When the differentially methylated regions were assigned to overlapping protein-coding genes, we found that methylation/demethylation occurs predominantly in the gene body in CpG and CHG contexts (Fig. 1, E and D), similar to the methylation patterns of Arabidopsis plants exposed to various biotic stresses (Dowen et al., 2012). The enrichment of non-CpG methylation in gene body is in contrast to recent findings in which gene body methylation was limited to CpG context (Cokus et al., 2008; Lister et al., 2008; Greaves et al., 2012; Takuno and Gaut, 2013). This controversy is presumably due to the fact that these studies used unchallenged plants. Whether differential gene body methylation in non-CpG contexts is a specific signature of plant response to biotic stresses needs to be investigated further.
It has been demonstrated that variable methylation over the gene body is a general feature of plant and animal genomes (Cokus et al., 2008; Lister et al., 2008; Ball et al., 2009; Laurent et al., 2010). While the suite of functions of this methylation is not fully understood, it has been suggested that it might regulate alternative splicing efficiency and prevent aberrant transcription of long genes (Zilberman et al., 2007; Luco et al., 2010; Maunakea et al., 2010; Regulski et al., 2013). Gene body methylation/demethylation, specifically in CpG and CHH contexts, does not seem to severely impact gene expression in response to SCN infection. A moderate up- and down-regulation of differentially methylated genes was observed in our qPCR assays (Fig. 4, A and C), suggesting that gene body methylation may fine-tune the transcriptional activity of targeted genes (Zilberman et al., 2007). The heterogeneous effect of body methylation on gene expression could be linked to the extent of methylation in the target genes. Moderate methylation tends to enhance gene expression, whereas low or high methylation tends to inhibit gene expression (Wang et al., 2013). The impact of gene body methylation on gene expression may also depend on the genic regions. Strong associations between DNA methylation in the first exon and gene down-regulation have been reported in plants and animals (Brenet et al., 2011; Yang et al., 2015). First exon methylation was found to block transcript initiation and cause proximal polymerase pausing in human cells (Okitsu and Hsieh, 2007), and a similar mode of function has been suggested to occur in plants (Zilberman et al., 2007).
In addition to gene body methylation, we found the promoters of 447 genes were differentially methylated in CpG (153 genes), CHG (84 genes), and CHH (210 genes) contexts. Differential methylation, particularly in the promoter regions, may impact the accessibility of regulatory factors to their cis-elements, thereby regulating the transcriptional activity of SCN-responsive genes. Although DNA methylation in the promoter regions is often associated with decreased transcriptional activity, we observed a positive association between DNA methylation in the promoter and the expression levels of two genes in our qPCR analysis. Hypermethylation in the promoter region of these two genes may overlap with the binding sites of transcriptional repressor factors, where inefficient binding associated with increased DNA methylation may result in increased gene transcription. Similarly, CHH methylation in the promoter regions was found to positively impact gene expression in maize (Zea mays; Gent et al., 2013).
Interestingly, a small portion of the differentially methylated genes (126/2,049) were found to be methylated in more than one sequence context simultaneously. This suggests that differential methylation in various contexts mostly occurs independently of each other, which is consistent with the fact that DNA methylation in CpG, CHG, and CHH are mediated by three different enzymes, including DNA METHYLTRANSFERASE1, the plant-specific CHROMOMETHYLASE3, and DRMs, respectively (Lindroth et al., 2001; Cao and Jacobsen, 2002; Kankel et al., 2003). Although the biological significance of simultaneous methylation in various sequence contexts remains an important question, a coordinated function of these enzymes may occur and could involve other epigenetic regulating factors and various forms of RNA polymerase activity.
SCN dramatically alters the transcription machinery of infected host cells (Klink et al., 2009, 2010; Kandoth et al., 2011), a process that may generate aberrant transcripts. These aberrant transcripts are processed by the RNA-induced silencing complex into siRNAs, which can induce DNA methylation through the RNA-directed DNA methylation pathway (Chan et al., 2005; Henderson and Jacobsen, 2007). The finding that 24-nucleotide small RNA reads are the most abundant class among other small RNA populations in nematode-infected root samples (Hewezi et al., 2008; Li et al., 2012) supports this hypothesis. SCN infection seems to trigger systemic hyper- and hypomethylation in root cells that are not in direct contact with the nematodes. This is consistent with our finding that several of the differentially methylated genes, identified using the whole root system, change the expression in response to SCN infection, but only 4% (278/6,903) of these genes were among the syncytium differentially expressed genes. The mechanism through which SCN induces methylation changes in a systemic manner is unknown, but this process may involve active gene expression changes of main epigenetic regulating genes and SCN-responsive siRNAs. Several epigenetic regulating genes were identified as differentially expressed in the SCN syncytium (Ithal et al., 2007; Klink et al., 2010; Kandoth et al., 2011). In addition, SCN induces differential accumulation of siRNAs (Li et al., 2012), and hence siRNA may function in a non-cell-autonomous fashion similar to the mobile epigenetically activated siRNAs in pollen (Slotkin et al., 2009) to shape the epigenome of nonsyncytial cells.
One striking finding of our analysis is that differential methylation was significantly enriched in the recently duplicated genes compared with early duplicated genes. In agreement with this finding, it has been recently reported that differential methylation in DNA occurs in a recently duplicated region of the genome (Schmitz et al., 2013; Keller and Yi, 2014). This pattern of methylation differences may contribute to gene dosage rebalance, thereby providing cell-type-specific regulation. Targeting recently duplicated genes for differential methylation by SCN could be due to the plausible association of the recently duplicated genes with disease and stress responses over the ancient copies, which are assumed to be involved in more basic and less stress-related cellular processes (Kondrashov, 2012).
Our finding that only 4% of the syncytium differentially expressed genes is differentially methylated suggests that regulation of gene expression in syncytium is heavily influenced by the well-known transcription factor-based regulatory mechanisms. Remarkably, we found several genes involved in DNA methylation and epigenetic regulation among the syncytium differentially methylated genes, including homologs of Arabidopsis O-acetyltransferases, S-adenosyl-l-Met-dependent methyltransferase protein, dicer-like4, DEFECTIVE IN MERISTEM SILENCING2, Suppressor of variegation 3-9 homolog9, chromatin remodeling complex subunit B, chromatin remodeling factor17, and a plant homeodomain finger family protein. This indicates that methylation changes in the syncytium are tightly controlled and a feedback loop may exist between various epigenetic components that regulates gene expression changes in the syncytium. A feedback regulatory loop between various epigenetic components has been recently suggested (Du et al., 2012).
Differential methylation induced by SCN in the syncytium seems to impact genes associated with various molecular functions. Suppression of defense responses is one of the main characteristics of the compatible interaction between SCN and soybean (Ithal et al., 2007; Hosseini and Matthews, 2014). Though a high proportion of defense-related genes was found to be down-regulated in the syncytium, only a few of these genes were differentially methylated. This is in contrast to the finding that differential methylation induced by the bacterial pathogen Pst in Arabidopsis is preferentially associated with genes involved in defense responses (Dowen et al., 2012; Yu et al., 2013).
Differential methylation may also contribute to the regulation of phytohormone-mediated soybean susceptibility to SCN. Various genes involved in auxin signaling were among the differentially methylated genes in the syncytium, including an auxin receptor homolog, auxin response factor8, and indole-3-acetic acid inducible9. Auxin plays crucial roles in the initiation and formation of the syncytium (Grunewald et al., 2009; Gheysen and Mitchum, 2011; Hewezi et al., 2014), and regulation of auxin signaling by DNA methylation was recently reported in Arabidopsis (Li et al., 2011). In addition, a homolog to Arabidopsis histone deacetylase1, which is involved in jasmonic acid and ethylene-dependent pathogen resistance, was differentially methylated (Zhou et al., 2005; Choi et al., 2012). Another homolog to the Arabidopsis ethylene insensitive3 (EIN3) family protein was also among the syncytium hypomethylated genes. EIN3 is a transcription factor that mediates downstream transcriptional cascades for ethylene responses (Chao et al., 1997; Binder et al., 2007), and various Arabidopsis EIN mutants showed reduced susceptibility to the beet cyst nematode (Wubben et al., 2001; Wubben et al., 2004).
The plant cytoskeleton plays a fundamental role in mediating changes in the morphology and structure of the syncytium (de Almeida Engler and Favery, 2011), and DNA demethylation may contribute to the regulation of this process. Genes involved in the dynamics of the cytoskeleton, including β tubulin, microtubule-associated protein, and Never in Mitosis A-related Ser/Thr kinase, were identified as differentially methylated in the syncytium. Similarly, cell wall biogenesis is extensively altered during syncytium initiation and expansion (Golinowski et al., 1996; Wieczorek et al., 2008; Siddique et al., 2009; Tucker et al., 2011). Several genes coding for enzymes with functions related to cell wall architecture and remodeling showed differential methylation accompanied with gene expression changes in the syncytium. This included, for example, cellulose synthase, pectate lyases, galacturonosyltransferase, and β galactosidase.
Signal transduction and regulatory genes, such as those encoding transcription factors and protein kinases, represent a significant portion of syncytium differentially methylated genes. Transcription factors belonging to Cys2-His2 zinc finger, basic region/Leu zipper motif, MYB, WRKY, calmodulin-binding transcription activator, B-Box Zinc Finger, Cys-rich polycomb-like protein, and homeobox-plant homeodomain family proteins were differentially methylated in the syncytium. Members of some of these gene families have been shown to be involved in defense-signaling pathways (Eulgem and Somssich, 2007; Reddy et al., 2011; Ambawat et al., 2013). In addition, a high number of protein kinases were highly represented among the syncytium differentially methylated genes, including components of mitogen-activated protein kinase signaling cascades. Thus, DNA methylation may be of particular importance in defining signal specificity associated with SCN parasitism. An important reprogramming of plant primary metabolism also occurs throughout syncytium formation and nematode feeding (Hofmann et al., 2010). Several genes involved in primary metabolism pathways, specifically those related to carbohydrate degradation and glycolysis, were differentially methylated. The potential transcriptional regulation of metabolic processes by DNA methylation may indicate that metabolite levels in the syncytium are tightly controlled.
Recent experimental evidence indicated that various plant pathogens have evolved the ability to manipulate the host ubiquitin/proteasome system (UPS) to cause disease (Dielen et al., 2010). UPS seems to play a vital role in syncytium formation, as many genes with UPS-related functions were found to be significantly up-regulated in the developing syncytia during a compatible interaction between SCN and soybean (Ithal et al., 2007; Klink et al., 2007). Demethylation of many of these genes, including, for example, proteasome component domain protein, MATH-BTB (for meprin and TRAF homology-broad complex, tramtrack, bric-a-brac) domain protein, and ubiquitin protein ligase, may be the cause of their up-regulation in the syncytium. The finding that cyst nematodes secrete effector proteins that may function in dysregulation of host UPS (Tytgat et al., 2004; Chronis et al., 2013) may point to a role of differential methylation as an additional mechanism of control to regulate particular cellular processes targeted by nematode effectors to promote parasitism.
In conclusion, our genomewide DNA methylation approach provides unprecedented insights into DNA methylation changes during the compatible interaction of SCN with soybean that impact a large number of protein-coding genes, locally in the syncytium and systemically in noninfected root cells, and their eventual transcriptional regulation.
MATERIALS AND METHODS
Plant and Nematode Material
Soybean (Glycine max) ‘Williams 82,’ an SCN (Heterodera glycines)-susceptible cultivar, was used in this study. SCN race 3 (H. glycines-type 0) was maintained in greenhouse culture on cv Williams 82 using standard procedures (Niblack et al., 2009).
Nematode Inoculation and Tissue Collection
Soybean seeds were soaked in running tap water for 30 min and surface sterilized in 10% (v/v) bleach solution for 10 min, followed by thorough rinsing in running tap water. Seeds were germinated for 3 d in rolled germination paper (rag dolls) at 26°C in the dark. Freshly hatched second-stage juveniles (J2) were extensively washed and suspended in 0.1% (w/v) sterile agarose at a concentration of 400 J2 per 100 μL. Healthy 3-d-old seedlings were then placed on moist blue blotter paper in the petri dish (150 × 15 mm) and inoculated with about 2,000 J2 distributed equally around the whole roots. Control samples were mock inoculated with 500 μL of 0.1% (w/v) agarose. SCN-inoculated and control roots were covered with sterile blue blotter paper soaked with MES-buffered sterile water (pH 6.5). The petri dishes were then covered with the lids and incubated in a plant growth chamber at 26°C in 16 h of light (75 µmol m–2 s–1)/8 h of dark conditions. The infection process was examined in 20% of the inoculated samples using acid fuschin stain as previously described by Daykin and Hussey (1985). The inoculation experiment was repeated three times for each of the two root samples (infected and control), for a total of six biologically independent samples that were collected at 5 d post infection. Each sample contained at least six seedlings.
Construction of Methyl-Seq Libraries
Whole-genome Methyl-Seq libraries were prepared using the Illumina TruSeq Library Prep Kit with minor modifications. Briefly, DNA was extracted from root tissues of control and inoculated seedling using DNeasy Plant Mini Kit (Qiagen). Approximately, 1.5 μg of genomic DNA plus unmethylated λ DNA was sheared using the Bioruptor (Diagenode) as per manufacturer instructions. Sheared genomic DNA was spiked with 1% to 2% (v/v) of fragmented, unmethylated λ DNA (Promega). Fragment size distribution of 250 to 400 bp for each sample was verified by a run on an Agilent Bioanalyzer using a 1,000 DNA chip (Agilent Technologies). Cytosine-methylated adapters provided by Illumina were ligated to the blunt ends of fragmented DNA. The ligated DNA was treated with sodium bisulfite using the MethylCode Bisulfite Conversion Kit (Invitrogen), and ligated fragments (400–500 bp) were selected on a Pippin prep (Sage Sciences). Post size selection, samples were enriched by 10 cycles of PCR reaction using 2.5 units of uracil-insensitive PfuTurboCxHotstart polymerase (Agilent), 5 μL of 10× PfuTurbo buffer, 0.4 μL of 100 nm deoxynucleoside triphosphate, and 5 μL of Illumina Truseq oligo mix. The PCR reactions were run using the following program: 95°C for 2 min and 10 cycles of 98°C for 15 s, 60°C for 30 s, and 72°C for 4 min, followed by a final elongation step of 10 min at 72°C. The amplification products were cleaned using Agencourt AMPure XP beads (Beckman Coulter). Purified PCR products were enriched further for five cycles of PCR amplification using Illumina’s protocol for library enrichment. Finally, Methyl-Seq libraries were validated on an Agilent Bioanalyzer using a 1,000 DNA chip (Agilent Technologies) and quantified using Illumina sequencing primers in a qPCR reaction before sequencing. Sequencing was performed at Oak Ridge National Laboratory using the Illumina HiSeq platform.
Data Analysis
MethylC-Seq reads for each biological replicate were filtered and trimmed using Trimmomatic with a phred quality threshold of 33 (Bolger et al., 2014). All sequencing reads were individually mapped using Bismark (Krueger and Andrews, 2011) to the soybean genome reference (Wm82.a2.v1). The alignments were done using the default software options for paired-end read alignment. The SAM alignment files generated by Bismark were used to identify differentially methylated cytosine bases or regions using the methylKit package in R with logistic regression (Akalin et al., 2012). Cytosines were called if the genomic region was covered by at least 10 reads, and three methylation call files for CpG, CHG, and CHH contexts were generated for both infected and control samples. Differentially methylated regions were identified using a nonoverlapping window of 200 bp. Hypo/hypermethylated regions covered by at least 10 reads with percentage methylation difference larger than 25% were identified using q-value < 0.01. Fisher’s exact test was used to calculate P values, and P values were adjusted to q-values using the SLIM method (Wang et al., 2011). The location and function of genes in the soybean genome version Wm82.a2.v1 were downloaded from Phytozome (Goodstein et al., 2012). Differentially methylated regions were mapped to various annotated features of genic regions, including protein-coding genes, and their subfeatures, including promoter regions, exons, and 5′ and 3′ untranslated regions, using a custom R script and the Bioconductor package rtracklayers (Lawrence et al., 2009). The promoter region was defined as 1,000 bases upstream of the first base of the mRNA annotated region. A positive overlap was recorded if the 200-bp region overlapped the feature or subfeature by 1 bp or more. GO enrichment analysis was performed using the AgriGO database (Du et al., 2010), and statistical significance was determined using Fisher’s exact test and a P value cutoff less than 0.05. Enrichment analyses of the differentially methylated genes were conducted using the latest annotated version of the soybean genome (Wm82.a2.v1) as background genes. Enrichment analyses of the differentially hyper- and hypomethylated genes that overlap with the syncytium differentially expressed genes were conducted using the total number of differentially methylated genes (2,004 genes) as background genes. χ2 tests of independence were used to evaluate the significance of overlaps between various gene lists as previously described by Hewezi et al. (2012).
RNA Isolation and qPCR Quantifications
Total RNA was extracted from frozen ground root tissues using the method previously described by Verwoerd et al. (1989). Total RNA was treated with DNase using DNase I kit (New England Biolabs). Fifty nanograms of DNase-treated RNA was used for complementary DNA synthesis and PCR amplification using the Verso One-Step qPCR SYBR Kit (Fisher Scientific) according to the manufacturer’s protocol. The reactions were run on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) using the following program: 50°C for 15 min, 95°C for 15 min, and 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 20 s. A dissociation curve was created after PCR amplification to detect nonspecific products using the following program: 95°C for 15 s, 50°C for 15 s, and a slow ramp from 50°C to 95°C. Three biologically independent samples of the SCN infected and noninfected control, each with four technical replicates, were used in qPCR analysis. The soybean ubiquitin (Glyma.20G141600), a constitutively expressed gene, was used as an internal control to normalize gene expression levels. Quantification of gene expression changes in SCN-infected samples relative to noninfected control were performed as previously described by Hewezi et al. (2015). The primer sequences used in qPCR analysis are provided in Supplemental Table S14.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Differential methylation levels are independent of gene size and GC content.
Supplemental Table S1. Summary of cytosine methylation of the Methyl-Seq libraries prepared from SCN-infected and control samples.
Supplemental Table S2. List of 428 hypermethylated genes in CpG context.
Supplemental Table S3. List of 180 hypermethylated genes in CHG context.
Supplemental Table S4. List of 120 hypermethylated genes in CHH context.
Supplemental Table S5. List of 817 hypomethylated genes in CpG context.
Supplemental Table S6. List of 350 hypomethylated genes in CHG context.
Supplemental Table S7. List of 282 hypomethylated genes in CHH context.
Supplemental Table S8. List of 703 unique hypermethylated genes in all sequence contexts.
Supplemental Table S9. List of 1,346 unique hypomethylated genes in all sequence contexts.
Supplemental Table S10. GO enrichment analysis of the differentially hypermethylated genes.
Supplemental Table S11. GO enrichment analysis of the differentially hypomethylated genes.
Supplemental Table S12. List of 93 syncytium differentially hypermethylated genes.
Supplemental Table S13. List of 193 syncytium differentially hypomethylated genes.
Supplemental Table S14. Primer sequences used in qPCR assays.
Supplementary Material
Glossary
- SCN
soybean cyst nematode
- Pst
Pseudomonas syringae pv tomato DC3000
- WGD
whole genomewide duplication
- Ks
synonymous distance
- qPCR
quantitative PCR
- GO
Gene Ontology
- UPS
ubiquitin/proteasome system
Footnotes
This work was supported by a grant from the Tennessee Soybean Promotion Board and by Hewezi Laboratory startup funds from the University of Tennessee, Institute of Agriculture.
Articles can be viewed without a subscription.
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE (2012) methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto K, Katakami H, Kim HJ, Ogawa E, Sano CM, Wada Y, Sano H (2007) Epigenetic inheritance in rice plants. Ann Bot (Lond) 100: 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224: 838–852 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Nota F, Cambiagno DA (2010) Epigenetic control of plant immunity. Mol Plant Pathol 11: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19: 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM (2009) Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD (2007) The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I (2007) Transgenerational changes in the genome stability and methylation in pathogen-infected plants: virus-induced plant genome instability. Nucleic Acids Res 35: 1714–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM (2011) DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 6: e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA (Suppl 4) 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Choi SM, Song HR, Han SK, Han M, Kim CY, Park J, Lee YH, Jeon JS, Noh YS, Noh B (2012) HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J 71: 135–146 [DOI] [PubMed] [Google Scholar]
- Chronis D, Chen S, Lu S, Hewezi T, Carpenter SC, Loria R, Baum TJ, Wang X (2013) A ubiquitin carboxyl extension protein secreted from a plant-parasitic nematode Globodera rostochiensis is cleaved in planta to promote plant parasitism. Plant J 74: 185–196 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Bayless AM, Wang K, Guo X, Song Q, Jiang J, Bent AF (2014) Distinct copy number, coding sequence, and locus methylation patterns underlie Rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiol 165: 630–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daykin M, Hussey R (1985) Staining and histopathological techniques in nematology. In Barker KR, Carter CC, Sasser JN, eds, An Advanced treatise on Meloidogyne, Vol 2 North Carolina State University Graphics, Raleigh, NC, 39–48. [Google Scholar]
- de Almeida Engler J, Favery B (2011) The plant cytoskeleton remodelling in nematode induced feeding sites. In Genomics and Molecular Genetics of Plant-Nematode Interactions. Springer Science + Business Media, Dordrecht, The Netherlands, pp 369–393 [Google Scholar]
- Dielen AS, Badaoui S, Candresse T, German-Retana S (2010) The ubiquitin/26S proteasome system in plant-pathogen interactions: a never-ending hide-and-seek game. Mol Plant Pathol 11: 293–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109: E2183–E2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK (2013) CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG (2011) How nematodes manipulate plant development pathways for infection. Curr Opin Plant Biol 14: 415–421 [DOI] [PubMed] [Google Scholar]
- Golinowski W, Grundler F, Sobczak M (1996) Changes in the structure of Arabidopsis thaliana during female development of the plant-parasitic nematode Heterodera schachtii. Protoplasma 194: 103–116 [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES (2012) Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA 109: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Cannoot B, Friml J, Gheysen G (2009) Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog 5: e1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Elling AA, Deng XW (2011) The epigenome and plant development. Annu Rev Plant Biol 62: 411–435 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE (2007) Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Baum TJ (2013) Manipulation of plant cells by cyst and root-knot nematode effectors. Mol Plant Microbe Interact 26: 9–16 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Howe P, Maier TR, Baum TJ (2008) Arabidopsis small RNAs and their targets during cyst nematode parasitism. Mol Plant Microbe Interact 21: 1622–1634 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Juvale PS, Piya S, Maier TR, Rambani A, Rice JH, Mitchum MG, Davis EL, Hussey RS, Baum TJ (2015) The cyst nematode effector protein 10A07 targets and recruits host posttranslational machinery to mediate its nuclear trafficking and to promote parasitism in Arabidopsis. Plant Cell 27: 891–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ (2012) The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol 159: 321–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Piya S, Richard G, Rice JH (2014) Spatial and temporal expression patterns of auxin response transcription factors in the syncytium induced by the beet cyst nematode Heterodera schachtii in Arabidopsis. Mol Plant Pathol 15: 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J, El Ashry AelN, Anwar S, Erban A, Kopka J, Grundler F (2010) Metabolic profiling reveals local and systemic responses of host plants to nematode parasitism. Plant J 62: 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini P, Matthews BF (2014) Regulatory interplay between soybean root and soybean cyst nematode during a resistant and susceptible reaction. BMC Plant Biol 14: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, Mitchum MG (2007) Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol Plant Microbe Interact 20: 510–525 [DOI] [PubMed] [Google Scholar]
- Kandoth PK, Ithal N, Recknor J, Maier T, Nettleton D, Baum TJ, Mitchum MG (2011) The soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress- and defense-related genes in degenerating feeding cells. Plant Physiol 155: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller TE, Yi SV (2014) DNA methylation and evolution of duplicate genes. Proc Natl Acad Sci USA 111: 5932–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R, Alkharouf N, Beard H, Macdonald M, Chouikha I, Meyer S, Grefenstette J, Knap H, Matthews B (2004) Microarray analysis of gene expression in soybean roots susceptible to the soybean cyst nematode two days post invasion. J Nematol 36: 241–248 [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Hosseini P, Matsye P, Alkharouf NW, Matthews BF (2009) A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol Biol 71: 525–567 [DOI] [PubMed] [Google Scholar]
- Klink VP, Hosseini P, Matsye PD, Alkharouf NW, Matthews BF (2010) Syncytium gene expression in Glycine max ([PI 88788]) roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines. Plant Physiol Biochem 48: 176–193 [DOI] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF (2007) Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 226: 1389–1409 [DOI] [PubMed] [Google Scholar]
- Koenning SR, Wrather JA (November 22, 2010) Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009. Plant Health Prog http://dx.doi.org/10.1094/PHP-2010-1122-01-RS [Google Scholar]
- Kondrashov FA. (2012) Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279: 5048–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. (2010) Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Gentleman R, Carey V (2009) rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25: 1841–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS (2011) DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet 7: e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, Zhang S, Liu D, Duan Y, Dong W (2012) Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS One 7: e39650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T (2010) Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. (2010) Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblack T, Tylka GL, Arelli P, Bond J, Diers B, Donald P, Faghihi J, Ferris VR, Gallo K, Heinz RD, et al. (2009) A standard greenhouse method for assessing soybean cyst nematode resistance in soybean: sCE08 (standardized cyst evaluation 2008). Plant Health Prog http://dx.doi.org/10.1094/PHP-2009-0513-01-RV [Google Scholar]
- Okitsu CY, Hsieh CL (2007) DNA methylation dictates histone H3K4 methylation. Mol Cell Biol 27: 2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Ali GS, Celesnik H, Day IS (2011) Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MT, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, He Y, Valdés-López O, Khan SM, Joshi T, Urich MA, Nery JR, Diers B, Xu D, Stacey G, et al. (2013) Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res 23: 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Siddique S, Endres S, Atkins JM, Szakasits D, Wieczorek K, Hofmann J, Blaukopf C, Urwin PE, Tenhaken R, Grundler FM, et al. (2009) Myo-inositol oxygenase genes are involved in the development of syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytol 184: 457–472 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS (2013) Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci USA 110: 1797–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ML, Murphy CA, Yang R (2011) Gene expression profiling and shared promoter motif for cell wall-modifying proteins expressed in soybean cyst nematode-infected roots. Plant Physiol 156: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat T, Vanholme B, De Meutter J, Claeys M, Couvreur M, Vanhoutte I, Gheysen G, Van Criekinge W, Borgonie G, Coomans A, et al. (2004) A new class of ubiquitin extension proteins secreted by the dorsal pharyngeal gland in plant parasitic cyst nematodes. Mol Plant Microbe Interact 17: 846–852 [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzić M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, et al. (2007) Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HQ, Tuominen LK, Tsai CJ (2011) SLIM: a sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics 27: 225–231 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang X, Lee TH, Mansoor S, Paterson AH (2013) Gene body methylation shows distinct patterns associated with different gene origins and duplication modes and has a heterogeneous relationship with gene expression in Oryza sativa (rice). New Phytol 198: 274–283 [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Hofmann J, Blöchl A, Szakasits D, Bohlmann H, Grundler FM (2008) Arabidopsis endo-1,4-β-glucanases are involved in the formation of root syncytia induced by Heterodera schachtii. Plant J 53: 336–351 [DOI] [PubMed] [Google Scholar]
- Williamson VM, Hussey RS (1996) Nematode pathogenesis and resistance in plants. Plant Cell 8: 1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubben MJE II, Rodermel SR, Baum TJ (2004) Mutation of a UDP-glucose-4-epimerase alters nematode susceptibility and ethylene responses in Arabidopsis roots. Plant J 40: 712–724 [DOI] [PubMed] [Google Scholar]
- Wubben MJE II, Su H, Rodermel SR, Baum TJ (2001) Susceptibility to the sugar beet cyst nematode is modulated by ethylene signal transduction in Arabidopsis thaliana. Mol Plant Microbe Interact 14: 1206–1212 [DOI] [PubMed] [Google Scholar]
- Yang H, Chang F, You C, Cui J, Zhu G, Wang L, Zheng Y, Qi J, Ma H (2015) Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis. Plant J 81: 268–281 [DOI] [PubMed] [Google Scholar]
- Yang S, Tang F, Caixeta ET, Zhu H (2013) Epigenetic regulation of a powdery mildew resistance gene in Medicago truncatula. Mol Plant 6: 2000–2003 [DOI] [PubMed] [Google Scholar]
- Yu A, Lepère G, Jay F, Wang J, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, et al. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA 110: 2389–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kimatu JN, Xu K, Liu B (2010) DNA cytosine methylation in plant development. J Genet Genomics 37: 1–12 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhou C, Zhang L, Duan J, Miki B, Wu K (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.