Abstract
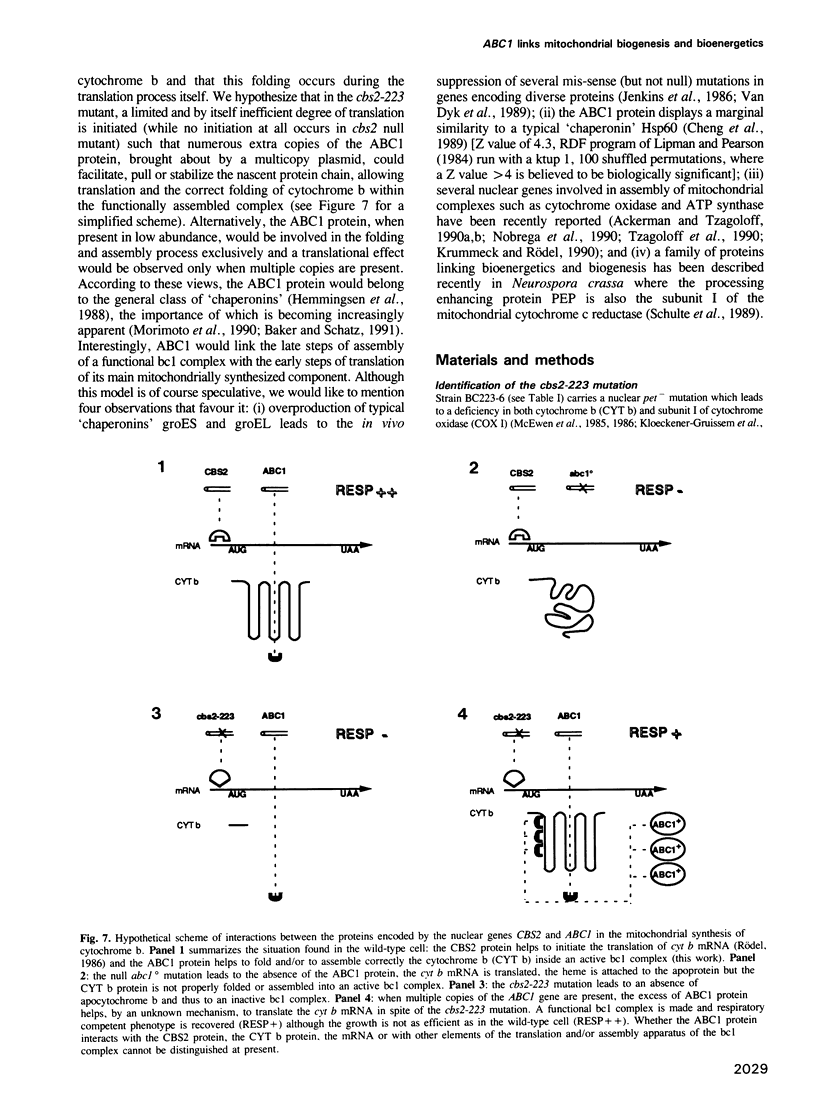
We have cloned and sequenced a novel yeast nuclear gene ABC1 which suppresses, in multicopy, the cytochrome b mRNA translation defect due to the nuclear mutation cbs2-223. Analysis of the ABC1 gene shows that it is weakly expressed, it could code for a protein of 501 amino acids which has a typical presequence of a protein imported into mitochondria and which does not display a strong similarity to any known protein. Inactivation of the ABC1 gene is not lethal to the cell but leads to a respiratory defect: no oxygen uptake and no growth on non-fermentable media. A total absence of NADH-cytochrome c oxidoreductase and succinate-cytochrome c oxidoreductase activities concomitant with the presence of specific dehydrogenases, suggests a block in the bc 1 segment of the respiratory chain. However, all the cytochromes are spectrally detectable. Cytochrome b is quite efficiently reduced while cytochromes c1 and c are not. The function of ABC1 in the suppression of a defect in apocytochrome b mRNA translation and in the activity of the bc1 complex suggests that the ABC1 protein would be a novel chaperonin involved both in biogenesis and bioenergetics of mitochondria.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman S. H., Tzagoloff A. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem. 1990 Jun 15;265(17):9952–9959. [PubMed] [Google Scholar]
- Ackerman S. H., Tzagoloff A. Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc Natl Acad Sci U S A. 1990 Jul;87(13):4986–4990. doi: 10.1073/pnas.87.13.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher E. B., Groudinsky O., Dujardin G., Altamura N., Kermorgant M., Slonimski P. P. Novel class of nuclear genes involved in both mRNA splicing and protein synthesis in Saccharomyces cerevisiae mitochondria. Mol Gen Genet. 1989 Feb;215(3):517–528. doi: 10.1007/BF00427051. [DOI] [PubMed] [Google Scholar]
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Baker K. P., Schatz G. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature. 1991 Jan 17;349(6306):205–208. doi: 10.1038/349205a0. [DOI] [PubMed] [Google Scholar]
- Banroques J., Delahodde A., Jacq C. A mitochondrial RNA maturase gene transferred to the yeast nucleus can control mitochondrial mRNA splicing. Cell. 1986 Sep 12;46(6):837–844. doi: 10.1016/0092-8674(86)90065-6. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bousquet I., Dujardin G., Poyton R. O., Slonimski P. P. Two group I mitochondrial introns in the cob-box and coxI genes require the same MRS1/PET157 nuclear gene product for splicing. Curr Genet. 1990 Aug;18(2):117–124. doi: 10.1007/BF00312599. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brivet-Chevillotte P., di Rago J. P. Electron-transfer restoration by vitamin K3 in a complex III-deficient mutant of S. cerevisiae and sequence of the corresponding cytochrome b mutation. FEBS Lett. 1989 Sep 11;255(1):5–9. doi: 10.1016/0014-5793(89)81050-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., Hallberg E. M., Hallberg R. L., Horwich A. L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989 Feb 16;337(6208):620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- Claisse M. L., Pajot P. F. Presence of cytochrome c1 in cytoplasmic "petite" mutants of Saccharomyces cerevisiae. Eur J Biochem. 1974 Nov 1;49(1):49–59. doi: 10.1111/j.1432-1033.1974.tb03810.x. [DOI] [PubMed] [Google Scholar]
- Claisse M. L., Péré-Aubert G. A., Clavilier L. P., Slonimski P. P. Méthode d'estimation de la concentration des cytochromes dans les cellules entières de levure. Eur J Biochem. 1970 Nov;16(3):430–438. doi: 10.1111/j.1432-1033.1970.tb01098.x. [DOI] [PubMed] [Google Scholar]
- Crivellone M. D., Wu M. A., Tzagoloff A. Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome c reductase complex. J Biol Chem. 1988 Oct 5;263(28):14323–14333. [PubMed] [Google Scholar]
- De La Salle H., Jacq C., Slonimski P. P. Critical sequences within mitochondrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell. 1982 Apr;28(4):721–732. doi: 10.1016/0092-8674(82)90051-4. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Koerner T. J., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4722–4731. [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Dujardin G., Pajot P., Groudinsky O., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol Gen Genet. 1980;179(3):469–482. doi: 10.1007/BF00271736. [DOI] [PubMed] [Google Scholar]
- Gallwitz D. Construction of a yeast actin gene intron deletion mutant that is defective in splicing and leads to the accumulation of precursor RNA in transformed yeast cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3493–3497. doi: 10.1073/pnas.79.11.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989 Jul 1;182(3):477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- Groudinsky O., Dujardin G., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. II. Genetic and biochemical analyses of suppressors which selectively alleviate the mitochondrial intron mutations. Mol Gen Genet. 1981;184(3):493–503. doi: 10.1007/BF00352529. [DOI] [PubMed] [Google Scholar]
- Hemmingsen S. M., Woolford C., van der Vies S. M., Tilly K., Dennis D. T., Georgopoulos C. P., Hendrix R. W., Ellis R. J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988 May 26;333(6171):330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Herbert C. J., Labouesse M., Dujardin G., Slonimski P. P. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 1988 Feb;7(2):473–483. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J., McGraw P., Tzagoloff A. A mutation in yeast mitochondrial DNA results in a precise excision of the terminal intron of the cytochrome b gene. J Biol Chem. 1985 Mar 25;260(6):3235–3238. [PubMed] [Google Scholar]
- Hurt E. C., Pesold-Hurt B., Suda K., Oppliger W., Schatz G. The first twelve amino acids (less than half of the pre-sequence) of an imported mitochondrial protein can direct mouse cytosolic dihydrofolate reductase into the yeast mitochondrial matrix. EMBO J. 1985 Aug;4(8):2061–2068. doi: 10.1002/j.1460-2075.1985.tb03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins A. J., March J. B., Oliver I. R., Masters M. A DNA fragment containing the groE genes can suppress mutations in the Escherichia coli dnaA gene. Mol Gen Genet. 1986 Mar;202(3):446–454. doi: 10.1007/BF00333275. [DOI] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B., McEwen J. E., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae: multiple trans-acting nuclear genes exert specific effects on expression of each of the cytochrome c oxidase subunits encoded on mitochondrial DNA. Curr Genet. 1987;12(5):311–322. doi: 10.1007/BF00405753. [DOI] [PubMed] [Google Scholar]
- Koll H., Schmidt C., Wiesenberger G., Schmelzer C. Three nuclear genes suppress a yeast mitochondrial splice defect when present in high copy number. Curr Genet. 1987;12(7):503–509. doi: 10.1007/BF00419559. [DOI] [PubMed] [Google Scholar]
- Kreike J., Schulze M., Ahne F., Lang B. F. A yeast nuclear gene, MRS1, involved in mitochondrial RNA splicing: nucleotide sequence and mutational analysis of two overlapping open reading frames on opposite strands. EMBO J. 1987 Jul;6(7):2123–2129. doi: 10.1002/j.1460-2075.1987.tb02479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummeck G., Rödel G. Yeast SCO1 protein is required for a post-translational step in the accumulation of mitochondrial cytochrome c oxidase subunits I and II. Curr Genet. 1990 Jul;18(1):13–15. doi: 10.1007/BF00321109. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Dujardin G., Slonimski P. P. The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell. 1985 May;41(1):133–143. doi: 10.1016/0092-8674(85)90068-6. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Slonimski P. P. Construction of novel cytochrome b genes in yeast mitochondria by subtraction or addition of introns. EMBO J. 1983;2(2):269–276. doi: 10.1002/j.1460-2075.1983.tb01416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Slonimski P. P. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2286–2290. doi: 10.1073/pnas.86.7.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Ljungdahl P. O., Pennoyer J. D., Robertson D. E., Trumpower B. L. Purification of highly active cytochrome bc1 complexes from phylogenetically diverse species by a single chromatographic procedure. Biochim Biophys Acta. 1987 May 6;891(3):227–241. doi: 10.1016/0005-2728(87)90218-0. [DOI] [PubMed] [Google Scholar]
- McDonald-Gibson R. G., Thorn M. B. Reversible activation of succinate dehydrogenase. Biochem J. 1969 Oct;114(4):775–784. doi: 10.1042/bj1140775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J. E., Cameron V. L., Poyton R. O. Rapid method for isolation and screening of cytochrome c oxidase-deficient mutants of Saccharomyces cerevisiae. J Bacteriol. 1985 Mar;161(3):831–835. doi: 10.1128/jb.161.3.831-835.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J. E., Ko C., Kloeckner-Gruissem B., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J Biol Chem. 1986 Sep 5;261(25):11872–11879. [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis U., Rödel G. Identification of CBS2 as a mitochondrial protein in Saccharomyces cerevisiae. Mol Gen Genet. 1990 Sep;223(3):394–400. doi: 10.1007/BF00264445. [DOI] [PubMed] [Google Scholar]
- Michaelis U., Schlapp T., Rödel G. Yeast nuclear gene CBS2, required for translational activation of cytochrome b, encodes a basic protein of 45 kDa. Mol Gen Genet. 1988 Oct;214(2):263–270. doi: 10.1007/BF00337720. [DOI] [PubMed] [Google Scholar]
- Muroff I., Tzagoloff A. CBP7 codes for a co-factor required in conjunction with a mitochondrial maturase for splicing of its cognate intervening sequence. EMBO J. 1990 Sep;9(9):2765–2773. doi: 10.1002/j.1460-2075.1990.tb07464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M. P., Nobrega F. G., Tzagoloff A. COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. J Biol Chem. 1990 Aug 25;265(24):14220–14226. [PubMed] [Google Scholar]
- Phillips J. D., Schmitt M. E., Brown T. A., Beckmann J. D., Trumpower B. L. Isolation and characterization of QCR9, a nuclear gene encoding the 7.3-kDa subunit 9 of the Saccharomyces cerevisiae ubiquinol-cytochrome c oxidoreductase complex. An intron-containing gene with a conserved sequence occurring in the intron of COX4. J Biol Chem. 1990 Dec 5;265(34):20813–20821. [PubMed] [Google Scholar]
- Roise D., Horvath S. J., Tomich J. M., Richards J. H., Schatz G. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986 Jun;5(6):1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Jensen R., Zoller M. J., Burke J., Errede B., Smith M., Herskowitz I. Structure of the Saccharomyces cerevisiae HO gene and analysis of its upstream regulatory region. Mol Cell Biol. 1986 Dec;6(12):4281–4294. doi: 10.1128/mcb.6.12.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel G., Fox T. D. The yeast nuclear gene CBS1 is required for translation of mitochondrial mRNAs bearing the cob 5' untranslated leader. Mol Gen Genet. 1987 Jan;206(1):45–50. doi: 10.1007/BF00326534. [DOI] [PubMed] [Google Scholar]
- Rödel G., Michaelis U., Forsbach V., Kreike J., Kaudewitz F. Molecular cloning of the yeast nuclear genes CBS1 and CBS2. Curr Genet. 1986;11(1):47–53. doi: 10.1007/BF00389425. [DOI] [PubMed] [Google Scholar]
- Rödel G. Two yeast nuclear genes, CBS1 and CBS2, are required for translation of mitochondrial transcripts bearing the 5'-untranslated COB leader. Curr Genet. 1986;11(1):41–45. doi: 10.1007/BF00389424. [DOI] [PubMed] [Google Scholar]
- SHERMAN F., SLONIMSKI P. P. RESPIRATION-DEFICIENT MUTANTS OF YEAST. II. BIOCHEMISTRY. Biochim Biophys Acta. 1964 Jul 15;90:1–15. doi: 10.1016/0304-4165(64)90113-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlapp T., Rödel G. Transcription of two divergently transcribed yeast genes initiates at a common oligo(dA-dT) tract. Mol Gen Genet. 1990 Sep;223(3):438–442. doi: 10.1007/BF00264451. [DOI] [PubMed] [Google Scholar]
- Schoppink P. J., Hemrika W., Berden J. A. The effect of deletion of the genes encoding the 40 kDa subunit II or the 17 kDa subunit VI on the steady-state kinetics of yeast ubiquinol-cytochrome-c oxidoreductase. Biochim Biophys Acta. 1989 May 8;974(2):192–201. doi: 10.1016/s0005-2728(89)80372-x. [DOI] [PubMed] [Google Scholar]
- Schulte U., Arretz M., Schneider H., Tropschug M., Wachter E., Neupert W., Weiss H. A family of mitochondrial proteins involved in bioenergetICS and biogenesis. Nature. 1989 May 11;339(6220):147–149. doi: 10.1038/339147a0. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Boulet A., Simon M., Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Boulet A., Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989 Jan 5;337(6202):84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpower B. L. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990 Jun;54(2):101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Capitanio N., Nobrega M. P., Gatti D. Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 1990 Sep;9(9):2759–2764. doi: 10.1002/j.1460-2075.1990.tb07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Dieckmann C. L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990 Sep;54(3):211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyk T. K., Gatenby A. A., LaRossa R. A. Demonstration by genetic suppression of interaction of GroE products with many proteins. Nature. 1989 Nov 23;342(6248):451–453. doi: 10.1038/342451a0. [DOI] [PubMed] [Google Scholar]
- Wu M., Tzagoloff A. Identification and characterization of a new gene (CBP3) required for the expression of yeast coenzyme QH2-cytochrome c reductase. J Biol Chem. 1989 Jul 5;264(19):11122–11130. [PubMed] [Google Scholar]
- de Vries S., Marres C. A. The mitochondrial respiratory chain of yeast. Structure and biosynthesis and the role in cellular metabolism. Biochim Biophys Acta. 1987;895(3):205–239. doi: 10.1016/s0304-4173(87)80003-4. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986 Jun;5(6):1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]