In vivo formation of protein crystals and their isolation and structural analysis by novel highly brilliant XFEL and synchrotron-radiation sources is described.
Keywords: in vivo crystallization, serial femtosecond crystallography, X-ray free-electron laser, expression systems
Abstract
During the last decade, the number of three-dimensional structures solved by X-ray crystallography has increased dramatically. By 2014, it had crossed the landmark of 100 000 biomolecular structures deposited in the Protein Data Bank. This tremendous increase in successfully crystallized proteins is primarily owing to improvements in cloning strategies, the automation of the crystallization process and new innovative approaches to monitor crystallization. However, these improvements are mainly restricted to soluble proteins, while the crystallization and structural analysis of membrane proteins or proteins that undergo major post-translational modifications remains challenging. In addition, the need for relatively large crystals for conventional X-ray crystallography usually prevents the analysis of dynamic processes within cells. Thus, the advent of high-brilliance synchrotron and X-ray free-electron laser (XFEL) sources and the establishment of serial crystallography (SFX) have opened new avenues in structural analysis using crystals that were formerly unusable. The successful structure elucidation of cathepsin B, accomplished by the use of microcrystals obtained by in vivo crystallization in baculovirus-infected Sf9 insect cells, clearly proved that crystals grown intracellularly are very well suited for X-ray analysis. Here, methods by which in vivo crystals can be obtained, isolated and used for structural analysis by novel highly brilliant XFEL and synchrotron-radiation sources are summarized and discussed.
1. Introduction
It is very well known that protein crystals may appear under in vivo conditions in living cells, such as peroxidase in peroxisomes, insulin in secretory vesicles of pancreatic cells and storage proteins in plant seeds, amongst others (Doye & Poon, 2006 ▸). These proteins are produced in large quantities and usually appear within organelles, surrounded by membranes, that offer only limited space and volume. The formation of protein crystals seems to be the simplest way to achieve both a high yield and concentration of individual protein molecules. As a consequence of the restricted space within the organelles, they may occur as a result of the displacement of surrounding water. Thus, the phenomenon of protein crystallization in vivo appears, in principle, to be not very different from crystal formation by the application of in vitro methods and crystallization compartments in the laboratory. However, since in vivo-grown crystals are fairly small and possess only low diffraction capabilities with high sensitivity to radiation damage, until recently they could not be used for conventional X-ray crystallography. On the other hand, the use of in vivo-grown crystals for structural analysis is very appealing, as they represent the nascent protein formed under defined cultivation conditions and include all post-translational modifications that occurred before the crystal was formed. In this way, one may also gain insight into dynamic processes within living cells. The question is whether or not the number of different proteins that are able to form in vivo crystals is restricted to just a few proteins, or whether many different proteins can form crystals within living cells, provided that the appropriate conditions can be experimentally designed. In this regard, the baculovirus-based expression system using Sf9 insect cells offers a perfect starting point, as it possesses a very effective polyhedrin promoter to promote the overexpression of the protein of interest, which replaces the viral polyhedrin gene (Rohel et al., 1983 ▸; Smith et al., 1983 ▸; Kroemer et al., 2015 ▸).
In natural infections of plant cells by baculoviruses, polyhedrin is expressed in high amounts, since it forms a matrix in which the virions are embedded (Rohrmann, 1986 ▸). On expressing cathepsin B from Trypanosoma brucei (CatB) using this heterologous expression system, it was readily recognized that the protein was not only expressed in considerable amounts (as judged by Western blotting and mass spectrometry) but that it formed in vivo crystals which grew as long needles with dimensions of up to 1 × 1 × 30 µm. These were readily observed with a light microscope using phase contrast and a 200-fold to 400-fold final magnification (Koopmann et al., 2012 ▸) and proved to be stable in both neutral and basic buffer solutions, even in the presence of detergents, but dissolved immediately under acidic conditions. Based on these observations, insect cells were lysed at pH 8 in RIPA buffer and the crystals were isolated by differential centrifugation. As mentioned previously, it was not possible to obtain suitable diffraction data by applying conventional X-ray crystallography and synchrotron radiation at the time. Thus, these crystals were dissolved at pH 5 and recrystallized using conventional vapour-diffusion crystallization techniques in vitro, thereby producing crystals that were suitable for structural analysis and yielded a resolution of 2.55 Å (Koopmann et al., 2012 ▸). At this time, the world’s first hard X-ray FEL, the LCLS in Stanford, California, USA, started user operation. Preliminary experiments showed convincing diffraction of CatB in vivo crystals to 7.5 Å resolution, only restricted by the technical resolution limit (Koopmann et al., 2012 ▸). Inspired by this highly promising success, we subsequently employed the high-resolution data-collection capability of the LCLS Coherent X-ray Imaging (CXI) instrument that was commissioned in 2011 (Boutet & Williams, 2010 ▸). A suspension containing 2 × 109 crystals per millilitre was injected into the pulsed FEL beam using a liquid micro-jet. This high number of crystals was obtained after crushing crystals isolated from liquid cultures with a cell density of 2 × 106 virus-infected Sf9 cells per millilitre. As the crystals are visible in phase-contrast microscopy, the number of crystals was simply calculated using a Neubauer haemocytometer, showing that approximately 70% of the cells contained one crystal. Any crystal hit by the intense XFEL pulses produced a diffraction pattern within femtoseconds just before the crystal was destroyed (Neutze et al., 2000 ▸; Chapman et al., 2011 ▸; Barty et al., 2012 ▸). Since the structure of this protein obtained under identical conditions had been solved from recrystallized material previously, molecular replacement could be used for phasing and the structure was refined to a resolution of 2.1 Å (Redecke et al., 2013 ▸). Parallel to the development of XFELs, third-generation synchrotron sources have been significantly improved. Today, microfocus X-ray beamlines are in routine operation, allowing diffraction data collection from crystals with volumes of less than 1000 µm3 (Coulibaly et al., 2007 ▸; Rasmussen et al., 2007 ▸). Thus, the same in vivo-grown CatB crystals have been used for diffraction data collection on beamline P14 of PETRA III, DESY, Hamburg, Germany, which is the most brilliant synchrotron source available to date. Applying a modified serial crystallography approach, a complete data set was successfully collected to 3.0 Å resolution by performing high-precision diffractometry and shutterless data acquisition with a pixel-array detector (Gati et al., 2014 ▸). These examples demonstrate that in vivo crystals are highly suitable for structure analysis and, in contrast to conventionally grown crystals, offer the possibility to obtain further insights into dynamic processes such as sequential glycosylation steps within the cell. In this review, we summarize the technical basis as well as a set of essential details and further considerations to obtain reproducibly high-quality in vivo crystals from different proteins.
2. Protein-expression systems and examples of in vivo crystallization
Several different cell-free or cell-bound expression systems for the heterologous formation of proteins from mRNA have previously been described in the literature, of which four systems seem to be principally suitable for in vivo protein crystal formation: (i) bacteria (Sawaya et al., 2014 ▸); (ii) insect cells (Fan et al., 1996 ▸; Coulibaly et al., 2007 ▸; Koopmann et al., 2012 ▸; Schönherr et al., 2015 ▸); (iii) yeast (Vonck & van Bruggen, 1992 ▸); and (iv) mammalian cells (Hasegawa et al., 2011 ▸; Gallat et al., 2014 ▸). In any case, the protein of interest has to be cloned, after which the respective gene has to be integrated either into the host organism’s DNA or, in the form of mRNA, into an extrachromosomal vector of some sort. In this particular case, codon usage may be considered a major problem inherent to this step. Although the genetic code is (with a few exceptions) universal, some of the 61 mRNA codons are used more frequently than others. In general, the so-called major codons are used for highly expressed genes, while minor codons are found in genes with a low expression level. Codon usage differs markedly in different organisms (see codon-usage databases, NCBI GenBank) and problems such as inhibited or interrupted translation, frame shifting or misincorporation of certain amino acids are frequently observed if considerable differences occur between the organisms of the donor protein and the host translation system. With regard to the protein of interest, other problems include the rate of overexpression, intracellular localization and post-translational modifications. These problems should be considered when choosing a translation system (Table 1 ▸).
Table 1. Advantages and disadvantages of different expression systems.
| Expression system | Advantages | Disadvantages |
|---|---|---|
| Bacteria | (i) Many different expression vectors | (i) Different codon usage |
| (ii) Different bacterial strains for different purposes | (ii) No post-translational modifications | |
| (iii) Easy overexpression of the protein of interest | (iii) No organelles and thus no protein sorting | |
| (iv) High protein yield | (iv) Inclusion-body formation | |
| (v) Cost-effective systems available | ||
| Yeasts | (i) Easy cell handling and high cell yield | (i) Protein yield sometimes limited |
| (ii) Protein sorting and secretion | (ii) N- and O-glycosylation might be different from higher eukaryotic cells | |
| (iii) Post-translational modifications possible | ||
| (iv) Cells can be genetically manipulated | ||
| (v) Strong promoters available | ||
| Mammalian cell lines (HEK, CHO) | (i) Post-translational modifications | (i) Expensive system |
| (ii) Protein sorting | (ii) Slow cell growth, limited cell yield | |
| (iii) Secretory proteins can be isolated from culture media | (iii) Limited overexpression rates | |
| (iv) Genetic manipulations are difficult | ||
| (v) Transfection of cells may be difficult | ||
| Baculovirus-infected insect cells | (i) Post-translational modifications | (i) Expensive system |
| (ii) Protein sorting | (ii) Time-consuming cultivation system | |
| (iii) Very potent promotors are available | ||
| (iv) Effective expression system |
The most widely used expression system is still the bacterial system, usually represented by Escherichia coli. Its major advantages include (i) many different expression vectors are available, which often contain an N- or C-terminal tag for easy detection and purification; (ii) different E. coli strains for special applications are available; (iii) overexpression of the respective protein is usually easy to perform and produces high yields; and (iv) the system is well established and cost-effective. The disadvantages are (i) compared with higher eukaryotes the codon usage is rather different; (ii) post-translational modifications as known in eukaryotic proteins do not occur; (iii) since organelles are absent, correct intracellular localization of eukaryotic proteins will not occur; and (iv) many heterologously expressed proteins end up in inclusion bodies and refolding often fails to restore the native conformation.
Yeasts are lower eukaryotic cells and several genera are commercially available as protein-expression systems, including Saccharomyces, Pichia and Kluyveromyces. The advantages are (i) yeast cells are easy to handle and grow rapidly to very high densities, (ii) as it is a eukaryotic system, proteins can be directed towards intracellular organelles or alternatively (a major advantage) to be secreted into the medium by adding the respective signal peptides, (iii) proteins can be post-translationally modified, especially glycosylated, (iv) yeast cells can be genetically manipulated and (v) strong promoters are available, which render the expression of heterologous proteins very effective. One major disadvantage of this system is that N-glycosylation as well as O-glycosylation shows significant differences when compared with higher eukaryotes.
Some mammalian cell lines have been adopted for heterologous protein expression, especially HEK (human embryonic kidney) and CHO (Chinese hamster ovary) cells. The advantages are (i) even complex post-translational modifications occur according to higher eukaryotic cellular regulation processes; (ii) the organelle localization of proteins can be studied; and (iii) secretory proteins are often expressed in high quantities and can then be easily isolated from cultivation media. The disadvantages include (i) culture media are expensive; (ii) cell growth is slow and the cell yield is low; (iii) the level of overexpression for intracellular proteins is usually rather limited; (iv) genetic manipulation of these cells is usually more difficult; and (v) transfection of these cells with a suitable vector may become quite tedious.
The use of baculovirus-infected insect cells is in many cases an interesting alternative. Insect cells have reached a higher eukaryotic status compared with yeast and thus are most useful to study post-translational modifications and proper protein folding as well as the correct intracellular translocation of nascent proteins. Additionally, the use of the insect-cell specific baculovirus introduces a highly potent promoter to the shuttle vector, called a bacmid, thus rendering insect ovary cells a very effective expression system. The disadvantages of this system include higher costs, time-consuming cultivation and a more laborious handling to produce the respective bacmid.
The formation of in vivo crystals has been observed in bacteria (Sawaya et al., 2014 ▸; Schnepf et al., 1998 ▸; Evdokimov et al., 2014 ▸), mammalian cells (Hasegawa et al., 2011 ▸; Gallat et al., 2014 ▸; Tsutsui et al., 2015 ▸), yeast (Vonck & van Bruggen, 1992 ▸) and baculovirus-infected insect cells (Fan et al., 1996 ▸; Coulibaly et al., 2007 ▸, 2009 ▸; Koopmann et al., 2012 ▸; Redecke et al., 2013 ▸; Axford et al., 2014 ▸; Schönherr et al., 2015 ▸).
3. In vivo crystal formation
So far, the most prominent examples of in vivo crystal formation from heterologously expressed genes are related to baculovirus-infected insect cells. The major advantage here is the use of a bacmid, which contains the highly effective polyhedrin promotor controlling the expression of the gene of interest. Baculoviruses have a large, circular, double-stranded DNA genome that permits large DNA insertions and can be easily propagated and grown to high titre (Hitchman et al., 2009 ▸). Infection of insect cells with recombinant baculoviruses containing foreign genes under strong baculovirus promoters (polyhedrin, p10 or p6.9) has become a rapid and robust method for target protein production (Kroemer et al., 2015 ▸). As insect cells perform extensive post-translational modifications such as glycosylation (James et al., 1995 ▸), phosphorylation (Héricourt et al., 2000 ▸) and disulfide-bond formation (Hodder et al., 1996 ▸) among others, the baculovirus expression-vector system (BEVS) is especially valuable for the expression of many complex eukaryotic proteins whose proper folding and biological activity require post-translational modifications.
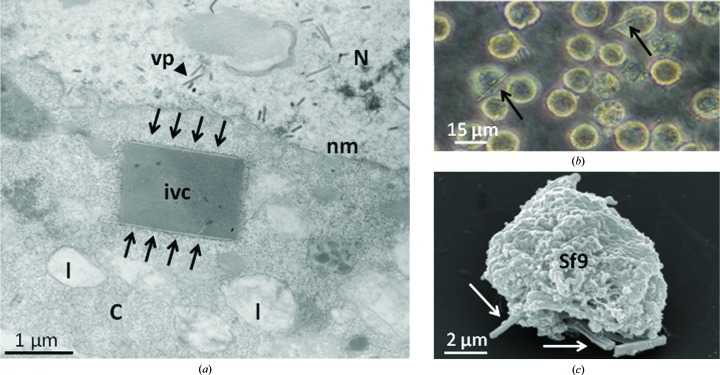
The question remains as to which parameters facilitate the appearance of in vivo crystals? Obviously, a high local concentration of the recombinant protein is required to render crystallization thermodynamically favourable. Intracellular accumulation of a respective protein within an organelle may be a prerequisite to obtain a sufficiently high local concentration. This is true for the heterologous expression of cathepsin B from T. brucei (Koopmann et al., 2012 ▸), for example. The gene for this enzyme encodes a signal peptide on its 5′ terminus that, on appearing outside the ribosome, is recognized by the signal recognition particle (SRP), which ensures the appearance of this protein within the lumen of the rough endoplasmic reticulum (rER) (Koopmann et al., 2012 ▸; Wild et al., 2004 ▸). This trypanosomal enzyme is not then transported via the Golgi apparatus to the lysosome, as would have been expected, but stays within the rER. Owing to the increasing concentration of this protein, the so-called unfolded protein response (UPR; Hetz, 2012 ▸), a stress response of the rER observed in higher eukaryotes as well as in yeasts, limits protein biosynthesis, but fails, likely owing to the insensitivity of the polyhedrin promoter (Koopmann et al., 2012 ▸). Under these conditions, it seems to be a logical consequence that a nucleation process then starts, eventually leading to crystallization of this protein. Accordingly, and by applying transmission electron microscopy, intracellular crystals have been observed, readily recognized by their lattice structures and surrounded by membranes decorated with ribosomes (Fig. 1 ▸). Once started, however, crystallization continues incessantly, forcing the cell to expand its plasma membrane until, eventually, the crystal perforates the membrane, leading to the death of the cell. This process was observed by both phase-contrast light microscopy and scanning electron microscopy (Fig. 1 ▸). As verified by mass spectrometry, Western blotting and enzyme-activity assays, these crystals were solely formed by CatB. Another example of in vivo crystal formation inside rER has been described for IgG in CHO cells (Hasegawa et al., 2011 ▸).
Figure 1.
In vivo crystals inside Sf9 cells. (a) Electron micrograph showing an in vivo crystal, surrounded by a membrane, within the cytoplasm. Arrows indicate ribosome-decorated membrane and an arrowhead indicates virus particles; C, cytoplasm; N, nucleus; nm, nuclear membrane; vp, virus particle; l, lysosome; ivc, in vivo crystal. (b) Light microscopic image showing Sf9 insect cells containing in vivo crystals. Arrows indicate in vivo crystals. (c) Scanning electron micrograph image of in vivo crystals protruding from Sf9 insect cells. Arrows indicate in vivo crystals.
A similar phenomenon was also shown for the heterologous expression of firefly luciferase in the same expression system (Schönherr et al., 2015 ▸). However, luciferase does not contain an rER signal peptide but a C-terminal SKL signal. Consequently, the protein was produced within the cytosol and finally translocated into peroxisomes. Co-localization studies using PEX3 and PEX25 verified that the respective in vivo crystals indeed occurred within peroxisomes. In this case, real-time life-imaging microscopy has shown not only the dynamics of crystal growth, but also the increase as well as the decrease of in vivo crystals (Schönherr et al., 2015 ▸). Another example of in vivo crystal formation concerns the trypanosomal inosine monophosphate dehydrogenase (IMPDH). When heterologously expressed in baculovirus-infected Sf9 cells, this enzyme also forms in vivo crystals, even though this is a cytosolic enzyme which lacks any signal peptide for organelle translocation. Despite this fact, intracellular IMPDH in vivo crystals detected by TEM were clearly surrounded by membranes. The most obvious explanation would be that this membrane could be of autophagosomal origin, as stress situations may induce autophagy in the respective cells. It has been observed that cell-toxic proteins are separated from the cytosol by inclusion into autophagosomes, which will eventually fuse with lysosomes for protein degradation (Tsutsui et al., 2015 ▸; Schönherr et al., 2015 ▸). Accordingly, crystal formation was inhibited by adding 3-methyladenine, a potent inhibitor of autophagy. These preliminary data (B. P. Sommer, unpublished work) also support the idea that the initial nucleation of in vivo crystals requires a high local concentration of the respective protein, which is most easily provided by localization within a cell organelle. However, there are three examples known that challenge this presumption: the formation of in vivo crystals of (i) the insecticidal Cry3A toxin in Bacillus thuringiensis (Sawaya et al., 2014 ▸), (ii) the insecticidal protein Cry1Ac from B. thuringiensis (Evdokimov et al., 2014 ▸) and (iii) μNS viral protein in baculovirus-infected Sf9 cells (Schönherr et al., 2015 ▸). The authors, however, reported the occurrence of sporulation-like processes in the former two cases, while the viral protein in the latter case is involved in viral production and initiated a nucleation process within the host cell anyway.
While it is not known how widely applicable in vivo crystallization may be, it is clear from the examples that we discuss that there is reason to be optimistic that it can be achieved in many cases. In addition, it is our expectation that on optimization of the procedures many initial failures will be converted to successes.
4. Factors that influence in vivo crystal formation
In natural infections of insect cells by nucleopolyhedroviruses, virions will be packed within a protein matrix of polyhedrin. This polyhedrin matrix is crystalline and is able to protect the virus against adverse environmental conditions, such as the surface of plant leaves. If these leaves are eaten by moths, the polyhedrin matrix is dissolved owing to the alkaline conditions within the digestive tract and the infection process starts (Vogel, 1986 ▸). Evidently, viral infection seems to induce a crystallization machinery within the insect cell, which was used to obtain chimeric protein crystals by linking the polyhedrin gene to another gene of interest (Coulibaly et al., 2007 ▸). However, in the cases of CatB, IMPDH, luciferase and others, the respective genes replaced the polyhedrin gene and crystallization did not appear within the nucleus (as in the case of polyhedrin) but elsewhere in the cell. In addition, in the case of luciferase, formation of in vivo crystals was observed even during transfection of the insect cell by the genetic information itself, although to a lesser extent (Schönherr et al., 2015 ▸). It can thus be ruled out that the baculovirus is inevitably needed for in vivo crystallization, while the polyhedrin promoter is undoubtedly most helpful to improve upon the overexpression of recombinant proteins.
The most commonly used cell lines for the baculovirus expression system are derived from Spodoptera frugiperda and are called Sf9 and Sf21. In addition, the so-called High-Five cell line from Trichoplusia ni has also been used successfully. In general, these cell lines have a generation time of between 24 and 48 h if grown at 27°C in culture media designed according to the composition of the haemolymphatic fluid of lepidopterans and hymenopterans (Wyatt et al., 1956 ▸). This requires a pH value between 6.2 and 6.5 and an osmotic concentration of about 350 mOs (Palomares et al., 2006 ▸). Insect cells can be grown in suspension culture, but also attach to a surface when cultured under static conditions. Although these monolayer cultures are often used for baculovirus transfection experiments and amplification of the virus stocks, in vivo crystals have been obtained in monolayers as well as in suspension cultures (R. Koopmann, doctoral thesis). Insect cells are grown to a desired cell density before they are transfected with a recombinant baculovirus containing the gene of interest, called a bacmid. Soon after transfection, the baculovirus takes control of the insect cell by inhibiting the transcription of cellular genes until the infected cell dies. In this way, P1 virus stock is obtained that can sequentially be further amplified, usually until a P3 virus stock is produced. Amplification of the baculovirus stock is one of the most important parts of this process, as low-quality stock results in very low productivity. There are two well known issues that affect the quality of a baculovirus stock. The first is genetic instability, where the gene of interest is lost during amplification of the stock by subsequent passaging (Kohlbrenner et al., 2005 ▸). This issue can be solved by carefully designing the vector to avoid an unstable construct. The second is the appearance of defective viruses, which owing to deletions comprise only 43% of the full genome length (Kool et al., 1991 ▸). The name ‘defective interfering particles’ (DIP) was proposed for these viruses by Huang & Baltimore (1970 ▸) to describe viruses that lack an essential part of the genome and thus require a helper virus in order to replicate (Holland, 1990 ▸). Kool et al. (1991 ▸) suggested two possible mechanisms for the emergence of DIP in serially propagated baculovirus preparations: firstly, mutant viruses with progressive deletions are generated over time; secondly, with continued passaging, a selection occurs for viruses with a genome of smaller size. These processes may lead to a more or less homogeneous population of DIPs, thus interfering with heterologous gene expression. For this reason, the multiplicity of infection (MOI) plays a key role in the propagation of baculovirus stocks. The MOI refers to the number of infectious particles that are added to a culture and is expressed as plaque-forming units per cell (pfu/cell). Therefore, to reduce the appearance of DIP, a very low MOI (≤0.1 pfu/cell) is recommended for the expansion of baculovirus stocks. In this way, the infection of a cell with more than one DIP becomes a rare event (Palomares et al., 2015 ▸). Yet even when caution is taken to reduce genetic instability and the appearance of DIP, the passage number of the baculovirus stock should be kept as low as possible, preferentially below six passages for heterologous protein expression (Palomares et al., 2015 ▸). In our hands, i.e. for in vivo crystal production, the passage number should not exceed 15. Determining the titre of baculovirus stocks by estimation of the concentration of active viral particles in a reproducible and precise way is a challenge. It can usually be performed through a plaque assay (King & Possee, 1992 ▸), quantitative real-time PCR (Hitchman et al., 2007 ▸) or, as we prefer, end-point dilution (Lynn, 1992 ▸; Hopkins & Esposito, 2009 ▸). Thus, following the construction of a recombinant baculovirus (validated by PCR and sequencing), production of the recombinant protein is started by adding P3 virus stock to a not yet confluent layer of the Sf9 cell line. The formation and the expression level of the respective protein is controlled by Western blotting using isolated infected cells and analysis of the excised SDS–PAGE gel band by means of mass spectrometry. At the same time, intracellular localization can be performed by light or electron microscopy using either fluorescent antibodies or immunogold-labelled antibodies, or (in the case of in vivo crystals) EM micrographs to watch for lattice structures. For most efficient in vivo crystallization of the respective protein we strongly recommend (i) using insect cells (cell density 1 × 106 cells ml−1, viability 80–90%) with a passage number below 15, (ii) growing cells to a confluence of about 70% and (iii) adding about 1‰(v/v) of P3 virus stock with an MOI of between 0.1 and 0.2 pfu ml−1.
5. Detection, isolation and handling of in vivo crystals
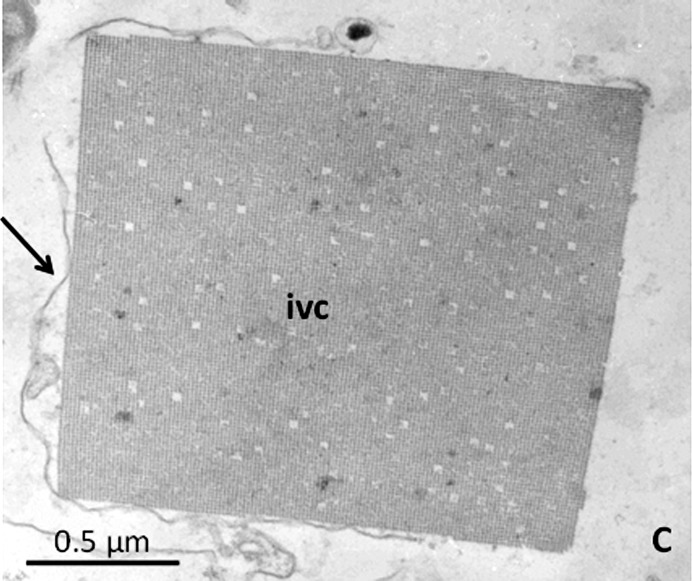
When the protein of interest is expressed, as verified for example by Western blotting, in vivo crystals may appear between 1 and 5 d after transfection with the virus P3 stock (Koopmann et al., 2012 ▸; Redecke et al., 2013 ▸) or within 2 d after direct transfection (Schönherr et al., 2015 ▸). During this time, the appearance of nanocrystals can be detected and monitored by electron microscopy. In the case of crystal growth, rectangular structures will appear within a specific cellular compartment/organelle of the transformed cells, usually membrane-surrounded, that show the typical lattice structure (Fig. 2 ▸). These structures can be fairly small and may not yet be observable by light microscopy. With time, however, these crystals will grow further and will finally be visible by phase-contrast microscopy of the insect cells. During this time, they can also be easily documented by light and especially scanning electron microscopy.
Figure 2.
Electron micrograph of an IMPDH in vivo crystal showing the typical lattice structure. The arrow indicates the membrane surrounding the in vivo crystal. C, cytoplasm; ivc, in vivo crystal.
To isolate CatB and IMPDH in vivo crystals, the medium is removed from the culture vessel and the cells are instead incubated with RIPA buffer [i.e. 25 mM Tris buffer pH 8.0 containing 0.5% Nonidet P-40 (NP-40; a nonionic, nondenaturing detergent), 0.25% sodium deoxycholate (an anionic detergent often used for the extraction of membrane proteins) and 0.05% sodium dodecyl sulfate (SDS; also an anionic tensid) in addition to 75 mM NaCl] for 10 min at room temperature (Koopmann et al., 2012 ▸; Redecke et al. 2013 ▸). However, not all intracellular crystals produced in insect cells remain stable after cell lysis, as has been observed for firefly luciferase (Schönherr et al., 2015 ▸). Here, cell lysis by hypotonic buffers supplemented with 0.1% Triton X-100 resulted in increased crystal stability. Further stabilization mechanisms for in vivo crystals outside the intact cell need to be investigated with regard to the respective protein. If in vivo crystals remain stable in solution, the subsequent isolation steps will depend significantly on the density of the crystals. CatB and IMPDH crystals are located in the supernatant following centrifugation at 3000g for 10 min. The supernatant containing the intact crystals is recovered, while cell debris is discarded with the pellet. The crystal-containing solution is then centrifuged at 17 000g for 10 min, followed by washing of the pellet with PBS or another appropriate buffer that confers sufficient crystal stability, which has to be individually tested. Soluble proteins from the lysed cells, together with remaining membranes and organelles, will remain in the supernatant and can thus be removed. The purity of the isolated crystals should be controlled by phase-contrast microscopy. If cell remnants, usually membranous structures, cannot be removed by differential centrifugation alone, crystals can be loaded onto a continuous or stepwise sucrose gradient ranging from 1 to 3 M. The flowthrough containing the crystals should be centrifuged again (3500g for 2 h). The remaining sucrose can be removed by dialysis. Finally, crystals can be concentrated by centrifugation, for example using Centricons with a 10 kDa pore size. This protocol has successfully been applied to the isolation of CatB and IMPDH in vivo crystals from Sf9 insect cells (Koopmann et al., 2012 ▸; Redecke et al., 2013 ▸). In any case, care has to be taken to ensure that the crystals are not destroyed during the purification procedure. Accordingly, the stability of the crystals within the appropriate pH range has to be closely checked. Additionally, no centrifugation parameters should be employed that may adversely affect crystal quality or integrity. Isolated crystals are produced which can be subjected to XFEL or high-brilliance synchrotron-radiation sources for diffraction data collection.
6. Diffraction data collection using in vivo crystals at XFEL and synchrotron-radiation sources
After the advent of microfocus beamlines (Riekel et al., 2005 ▸) at third-generation synchrotron light sources, in vivo-grown crystals could be used for crystallographic structure determination of biological macromolecules. At beamline X06SA of the Swiss Light Source (SLS), Coulibaly and coworkers used both recombinant and infectious silkworm cypovirus polyhedra in vivo crystals ranging in size from 5 to 12 µm for diffraction data collection (Coulibaly et al., 2007 ▸). Applying isomorphous replacement, they were able to solve and refine the structure of cypovirus polyhedra to 2.0 Å resolution. However, most in vivo crystals proved to be too small to be used in conventional diffraction data collection at synchrotron-radiation beamlines. The first hard X-ray free-electron laser (XFEL) LCLS (Emma et al., 2010 ▸) and the development of serial femtosecond crystallography (Chapman et al., 2011 ▸) paved the way for crystallography using submicrometre-sized crystals. Crystals of CatB grown in vivo were injected into the XFEL beam at the Coherent X-ray Imaging (CXI) instrument (Liang et al., 2015 ▸). The diffraction data collected to 2.1 Å resolution allowed the native structure of the protein to be solved and refined, including the pro-peptide and sugar moieties, which were absent from the structure obtained from the recrystallized protein (Redecke et al., 2013 ▸). Applying XFEL radiation to analyze cathepsin B provided the first structure and, in addition, revealed novel structural features. Subsequently, other groups implemented similar protocols and were able to obtain diffraction images from human neuraminidase crystals grown in CHO cells (Gallat et al., 2014 ▸) and from in vivo-grown CPV17 polyhedrin (Ginn et al., 2015 ▸). Inspired by the highly successful application of FEL radiation and new capabilities to process large data sets, cathepsin B in vivo crystals were also used to establish a serial synchrotron crystallography approach at the PETRA III synchrotron source located at DESY in Hamburg (Gati et al., 2014 ▸). Subsequently, a complete data set to 3.0 Å resolution was assembled by combining diffraction data from only 80 CatB crystals with an average volume of 9 µm3. The data allowed refinement of the structural model previously obtained using conventional X-ray crystallography, thus providing mutual validation for both serial crystallography approaches using FEL pulses and highly brilliant synchrotron radiation. In these cases, described by Koopmann and coworkers, in vivo-grown crystals were purified from cells by lysis followed by differential centrifugation (Koopmann et al., 2012 ▸). This has the distinct benefit that the diffraction of the crystals is not biased by residual cellular material, which will add diffuse scattering to the background. Conversely, not all in vivo-grown protein crystals improve in quality when subjected to this purification process. Crystal degradation can be avoided to some extent by optimizing the separation and purification procedures, although the separation of cell debris from crystalline material remains challenging and is clearly detrimental to optimized data-collection strategies. Recently, diffraction data have been collected from in vivo-grown protein crystals within cells at synchrotron microfocus beamlines (Axford et al., 2014 ▸) and at CXI/LCLS (Sawaya et al., 2014 ▸), thus omitting tedious purification steps. In both cases, the collected data were of sufficient quality to allow the refinement of the structural models. Axford and coworkers used CPV crystals (4–5 µm in diameter) in Sf9 cells and a standard mesh loop for data collection on beamline I24 at Diamond Light Source, UK under cryogenic conditions. Hence, they could refine the structure of CPV to 1.70 Å resolution. No significant differences in data-collection and refinement statistics were found when compared with those of the ex cellulo CPV crystals (Axford et al., 2014 ▸). Sawaya and coworkers used crystals of the natural insecticidal toxin Cry3A produced by B. thuringiensis israelensis strain 4Q7/pPFT3As during sporulation. They could refine the structure using the in cellulo diffraction data collected at CXI to 2.9 Å resolution (Sawaya et al., 2014 ▸). Very recently, Tsutsui and coworkers successfully recorded complete diffraction data from coral fluorescent protein to 2.9 Å resolution on beamline BL44XU at SPring-8, Japan from a single crystal inside the HEK cell in which the crystallization process occurred by mounting this cell at 100 K (Tsutsui et al., 2015 ▸). Thus, in cellulo diffraction data collection from in vivo-grown protein crystals might prove to be a highly suitable way to overcome stability problems during crystal isolation and purification. This has the potential to significantly increase the application of in vivo crystals as suitable targets for structural biology in the future.
Acknowledgments
These investigations were supported by the Röntgen-Angström-Cluster (project 05K12GU3), the German Federal Ministry of Education and Research (BMBF, projects 01KX0806 and 01KX0807) and the excellence cluster ‘The Hamburg Centre for Ultrafast Imaging-Structure, Dynamics, and Control of Matter at the Atomic Scale’ of the Deutsche Forschungsgemeinschaft (DFG). We are grateful to Christian Bosselmann for critically reading the manuscript.
References
- Axford, D., Ji, X., Stuart, D. I. & Sutton, G. (2014). In cellulo structure determination of a novel cypovirus polyhedrin. Acta Cryst. D70, 1435–1441. [DOI] [PMC free article] [PubMed]
- Barty, A. et al. (2012). Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nature Photonics, 6, 35–40. [DOI] [PMC free article] [PubMed]
- Boutet, S. & Williams, G. J. (2010). The Coherent X-ray Imaging (CXI) instrument at the LINAC Coherent Light Source (LCLS). New J. Phys. 12, 035024.
- Chapman, H. N. et al. (2011). Femtosecond X-ray protein nanocrystallography. Nature (London), 470, 73–77. [DOI] [PMC free article] [PubMed]
- Coulibaly, F., Chiu, E., Gutmann, S., Rajendran, C., Haebel, P. W., Ikeda, K., Mori, H., Ward, V. K., Schulze-Briese, C. & Metcalf, P. (2009). The atomic structure of baculovirus polyhedra reveals the independent emergence of infectious crystals in DNA and RNA viruses. Proc. Natl Acad. Sci. USA, 106, 22205–22210. [DOI] [PMC free article] [PubMed]
- Coulibaly, F., Chiu, E., Ikeda, K., Gutmann, S., Haebel, P. W., Schulze-Briese, C., Mori, H. & Metcalf, P. (2007). The molecular organization of cypovirus polyhedra. Nature (London), 446, 97–101. [DOI] [PubMed]
- Doye, J. P. K. & Poon, W. C. K. (2006). Protein crystallization in vivo. Curr. Opin. Colloid Interface Sci. 11, 40–46.
- Emma, P. et al. (2010). First lasing and operation of an ångstrom-wavelength free-electron laser. Nature Photonics, 4, 641–647.
- Evdokimov, A. G., Moshiri, F., Sturman, E. J., Rydel, T. J., Zheng, M., Seale, J. W. & Franklin, S. (2014). Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals. Protein Sci. 23, 1491–1497. [DOI] [PMC free article] [PubMed]
- Fan, G. Y., Maldonado, F., Zhang, Y., Kincaid, R., Ellisman, M. H. & Gastinel, L. N. (1996). In vivo calcineurin crystals formed using the baculovirus expression system. Microsc. Res. Tech. 34, 77–86. [DOI] [PubMed]
- Gallat, F. X. et al. (2014). In vivo crystallography at X-ray free-electron lasers: the next generation of structural biology? Philos. Trans. R. Soc. B Biol. Sci. 369, 20130497. [DOI] [PMC free article] [PubMed]
- Gati, C., Bourenkov, G., Klinge, M., Rehders, D., Stellato, F., Oberthür, D., Yefanov, O., Sommer, B. P., Mogk, S., Duszenko, M., Betzel, C., Schneider, T. R., Chapman, H. N. & Redecke, L. (2014). Serial crystallography on in vivo grown microcrystals using synchrotron radiation. IUCrJ, 1, 87–94. [DOI] [PMC free article] [PubMed]
- Ginn, H. M., Messerschmidt, M., Ji, X., Zhang, H., Axford, D., Gildea, R. J., Winter, G., Brewster, A. S., Hattne, J., Wagner, A., Grimes, J. M., Evans, G., Sauter, N. K., Sutton, G. & Stuart, D. I. (2015). Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data. Nature Commun. 6, 6435. [DOI] [PMC free article] [PubMed]
- Hasegawa, H., Wendling, J., He, F., Trilisky, E., Stevenson, R., Franey, H., Kinderman, F., Li, G., Piedmonte, D. M., Osslund, T., Shen, M. & Ketchem, R. R. (2011). In vivo crystallization of human IgG in the endoplasmic reticulum of engineered Chinese hamster ovary (CHO) cells. J. Biol. Chem. 286, 19917–19931. [DOI] [PMC free article] [PubMed]
- Hetz, C. (2012). The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature Rev. 13, 89–102. [DOI] [PubMed]
- Hitchman, R. B., Possee, R. D. & King, L. A. (2009). Baculovirus expression systems for recombinant protein production in insect cells. Recent Pat. Biotechnol. 3, 46–54. [DOI] [PubMed]
- Hitchman, R. B., Siaterli, E. A., Nixon, C. P. & King, L. A. (2007). Quantitative real-time PCR for rapid and accurate titration of recombinant baculovirus particles. Biotechnol. Bioeng. 96, 810–814. [DOI] [PubMed]
- Hodder, A. N., Crewther, P. E., Matthew, M. L., Reid, G. E., Moritz, R. L., Simpson, R. J. & Anders, R. F. (1996). The disulfide bond structure of plasmodium apical membrane antigen-1. J. Biol. Chem. 271, 29446–29452. [DOI] [PubMed]
- Holland, J. J. (1990). Defective viral genomes. Virology, edited by B. N. Fields & D. M. Knipe, pp. 151–165. New York: Raven.
- Hopkins, R. & Esposito, D. (2009). A rapid method for titrating baculovirus stocks using the Sf-9 Easy Titer cell line. Biotechniques, 47, 785–788. [DOI] [PubMed]
- Héricourt, F., Blanc, S., Redeker, V. & Jupin, I. (2000). Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus–insect cell system. Biochem. J. 349, 417–425. [DOI] [PMC free article] [PubMed]
- Huang, A. S. & Baltimore, D. (1970). Defective viral particles and viral disease processes. Nature (London), 226, 325–327. [DOI] [PubMed]
- James, D. C., Freedmann, R. B., Hoare, M., Ogonah, O. W., Rooney, B. C., Larinov, O. A., Dobrovolsky, V. N., Lagutin, O. V. & Jenkins, V. (1995). N-glycosylation of recombinant human interferon-gamma produced in different animal expression systems. Biotechnology, 13, 592–596. [DOI] [PubMed]
- King, L. A. & Possee, R. D. (1992). The Baculovirus Expression System: A Laboratory Guide. London: Chapman & Hall.
- Kohlbrenner, E., Aslanidi, G., Nash, K., Shklyaev, S., Campbell-Thompson, M., Byrne, B. J., Snyder, R. O., Muzyczka, N., Warrington, K. H. Jr & Zolotukhin, S. (2005). Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol. Ther. 12, 1217–1225. [DOI] [PMC free article] [PubMed]
- Kool, M., Voncken, J. W., Van Lier, F. L. J., Tramper, J. & Vlak, J. M. (1991). Detection and analysis of Autographa californica nuclear polyhedrosis virus mutants with defective interfering properties. Virology, 183, 739–746. [DOI] [PubMed]
- Koopmann, R. et al. (2012). In vivo protein crystallization opens new routes in structural biology. Nature Methods, 9, 259–262. [DOI] [PMC free article] [PubMed]
- Kroemer, J. A., Bonning, B. C. & Harrison, R. L. (2015). Expression, delivery and function of insecticidal proteins expressed by recombinant baculoviruses. Viruses, 7, 422–455. [DOI] [PMC free article] [PubMed]
- Liang, M. et al. (2015). The Coherent X-ray Imaging instrument at the Linac Coherent Light Source. J. Synchrotron Rad. 22, 514–519. [DOI] [PMC free article] [PubMed]
- Lynn, D. E. (1992). Improved efficiency in determining the titer of the Autographa californica baculovirus nonoccluded virus. Biotechniques, 13, 282–285. [PubMed]
- Neutze, R., Wouts, R., van der Spoel, D., Weckert, E. & Hajdu, J. (2000). Potential for biomolecular imaging with femtosecond X-ray pulses. Nature (London), 406, 752–757. [DOI] [PubMed]
- Palomares, L. A., Estrada-Mondaca, S. & Ramirez, O. T. (2006). Principles and applications of the insect cell–baculovirus expression vector system. Cell Culture Technology for Pharmaceutical and Cellular Applications, edited by S. Ozturk & W.-S. Hu, pp. 627–692. Boca Raton: CRC Press.
- Palomares, L. A., Realpe, M. & Ramirez, O. T. (2015). An overview of cell culture engineering for the insect cell–baculovirus expression vector system (BEVS). Animal Cell Culture, edited by M. Al-Rubeai, pp. 501–519. Heidelberg: Springer.
- Rasmussen, S. G. F., Choi, H.-J., Rosenbaum, D. M., Kobilka, T. S., Thian, F. S., Edwards, P. C., Burghammer, M., Ratnala, V. R., Sanishvili, R., Fischetti, R. F., Schertler, G. F., Weis, W. I. & Kobilka, B. K. (2007). Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature (London), 450, 383–387. [DOI] [PubMed]
- Redecke, L. et al. (2013). Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science, 339, 227–230. [DOI] [PMC free article] [PubMed]
- Riekel, C., Burghammer, M. & Schertler, G. (2005). Protein crystallography microdiffraction. Curr. Opin. Struct. Biol. 15, 556–562. [DOI] [PubMed]
- Rohel, D. Z., Cochran, M. A. & Faulkner, P. (1983). Characterization of two abundant mRNAs of Autographa californica nuclear polyhedrosis virus present late in infection. Virology, 124, 357–365. [DOI] [PubMed]
- Rohrmann, G. F. (1986). Polyhedrin structure. J. Gen. Virol. 67, 1499–1513. [DOI] [PubMed]
- Sawaya, M. R. et al. (2014). Protein crystal structure obtained at 2.9 Å resolution from injecting bacterial cells into an X-ray free-electron laser beam. Proc. Natl Acad. Sci. USA, 111, 12769–12774. [DOI] [PMC free article] [PubMed]
- Schnepf, E., Crickmore, N., Van Rie, J., Lereclus, D., Baum, J., Feitelson, J., Zeigler, D. R. & Dean, D. H. (1998). Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 775–806. [DOI] [PMC free article] [PubMed]
- Schönherr, R., Klinge, M., Rudolph, J. M., Fita, K., Rehders, D., Lubber, F., Schneegans, S., Majoul, I., Duszenko, M., Betzel, C., Brandaris-Nunez, J., Martines-Costas, J., Duden, R. & Redecke, L. (2015). In the press. [DOI] [PMC free article] [PubMed]
- Smith, G. E., Vlak, J. M. & Summers, M. D. (1983). Physical analysis of Autographa californica nuclear polyhedrosis virus transcripts for polyhedrin and 10,000-molecular-weight protein. J. Virol. 45, 215–225. [DOI] [PMC free article] [PubMed]
- Tsutsui, H., Jinno, Y., Shoda, K., Tomita, A., Matsuda, M., Yamashita, E., Katayama, H., Nakagawa, A. & Miyawaki, A. (2015). A diffraction-quality protein crystal processed as an autophagic cargo. Mol. Cell, 58, 186–193. [DOI] [PubMed]
- Vogel, S. (1986). Baculo-viren: Biologische Insektizide – Werkzeuge der Molekularbiologie. Chem. Unserer Zeit, 20, 77–83.
- Vonck, J. & van Bruggen, E. F. (1992). Architecture of peroxisomal alcohol oxidase crystals from the methylotrophic yeast Hansenula polymorpha as deduced by electron microscopy. J. Bacteriol. 174, 5391–5399. [DOI] [PMC free article] [PubMed]
- Wild, K., Halic, M., Sinning, I. & Beckmann, R. (2004). SRP meets the ribosome. Nature Struct. Mol. Biol. 11, 1049–1053. [DOI] [PubMed]
- Wyatt, G. R., Loughheed, T. C. & Wyatt, S. S. (1956). The chemistry of insect hemolymph; organic components of the hemolymph of the silkworm, Bombyx mori, and two other species. J. Gen. Physiol. 39, 853–868. [DOI] [PMC free article] [PubMed]