Roquin mediates mRNA degradation by recognizing the constitutive-decay element. Crystal structures of the ROQ domain of Roquin-2 were determined in ligand-free and RNA-bound forms, in which Roquin-2 recognized RNA forming a stem-loop structure.
Keywords: Roquin, mRNA degradation, winged-helix motif, stem-loop structure
Abstract
Roquin mediates mRNA degradation by recognizing the constitutive-decay element (CDE) in the 3′ untranslated region of the target gene followed by recruitment of the deadenylation machinery. Deficiency or dysfunction of Roquin has been associated with autoimmunity and inflammation. To establish the structural basis for the recognition of CDE RNA by Roquin, the crystal structure of the ROQ domain of human Roquin-2 was determined in ligand-free and CDE-derived RNA-bound forms. The ROQ domain of Roquin-2 folded into a winged-helix structure in which the wing region showed structural flexibility and acted as a lid for RNA binding. The CDE RNA, forming a stem-loop structure, bound to the positively charged surface of the ROQ domain and was mainly recognized via direct interactions with the phosphate backbone in the 5′ half of the stem-loop and its triloop and via indirect water-mediated interactions. Structural comparison with Roquin-1 revealed conserved features of the RNA-binding mode. Therefore, it is suggested that the Roquin proteins function redundantly in mRNA degradation.
1. Introduction
The intracellular level of post-transcriptional mRNA is critical to cell function and is regulated by various mechanisms. The constitutive-decay element (CDE) in the 3′ untranslated region (3′ UTR) of mRNA is involved in the degradation of mRNA (Stoecklin et al., 2003 ▸). The CDE is highly conserved in vertebrates, and many mRNAs containing CDE sequences encode proteins crucial for the regulation of development, transcription, inflammation and immunity, such as the inducible T-cell co-stimulator (ICOS), tumour necrosis factor α and Nfkbid, a member of the IκB family of NF-κB inhibitors (Leppek et al., 2013 ▸). Recent studies have demonstrated that Roquin (Roquin-1 and Roquin-2), a member of the RING-type ubiquitin ligase family, recognizes the CDE stem-loop structure from target mRNA and recruits the deadenylation and decapping machinery, leading to mRNA decay (Leppek et al., 2013 ▸; Glasmacher et al., 2010 ▸). Thus, Roquin has a central role in regulating the mRNA level and is involved in various pathological conditions such as autoimmune diseases (Vinuesa et al., 2005 ▸). The mRNAs of Roquin themselves also contain CDE sequences, which function as part of the feedback mechanism (Leppek et al., 2013 ▸).
Human Roquin-2 (residues 1–1191) contains a RING finger, a ROQ domain and a CCCH-type zinc finger in the N-terminal region and proline-rich and coiled-coil regions in the C-terminal region (Fig. 1 ▸ a). RING-finger domains are conserved in many E3 ubiquitin-ligases (Joazeiro & Weissman, 2000 ▸). Accordingly, Roquin-2 promotes the ubiquitylation and degradation of apoptosis signal-regulating kinase 1 and is involved in the regulation of stress responses (Maruyama et al., 2014 ▸). The function of the C-terminal region of Roquin-2 is poorly defined, but truncation of the C-terminal proline-rich and coiled-coil regions decreases the Roquin-mediated repression of ICOS (Glasmacher et al., 2010 ▸).
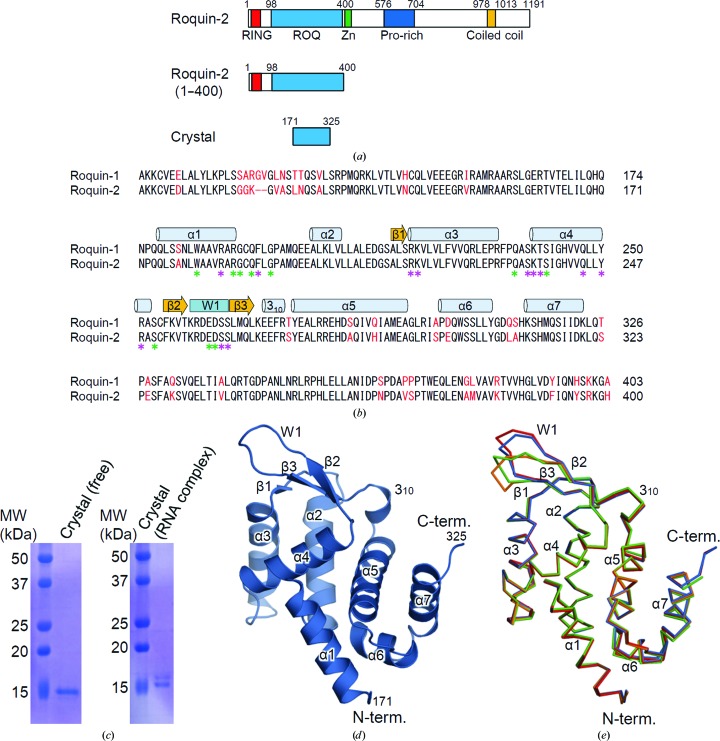
Figure 1.
Domain organization of human Roquin-2 and the structure of the ROQ domain of ligand-free Roquin-2. (a) Schematic representation of the domain structures of human Roquin-2. (b) Sequence alignment of the ROQ domain of human Roquin-1 and Roquin-2. Secondary-structure elements and the wing region are indicated above the alignment. The unconserved residues are highlighted in red. The residues forming the hydrogen bonds and other interactions with RNA are indicated by purple and green asterisks, respectively. (c) SDS–PAGE analyses of the dissolved crystals in the ligand-free form (left) and the RNA-complexed form using the 30 kDa fragment (right). (d) Structure of the ligand-free form of the ROQ domain. The N- and C-termini and the structural elements are labelled. (e) Superposition of the four molecules of the ROQ domain of Roquin-2 in the asymmetric unit. Molecules A, B, C and D are shown in blue, red, green and orange, respectively.
Roquin-1 and Roquin-2 are expressed ubiquitously and have high sequence similarity, with 88% sequence identity in the ROQ domains (Vogel et al., 2013 ▸; Fig. 1 ▸ b). T cells from mice that are deficient in both Roquin-1 and Roquin-2 show higher ICOS expression than T cells from mice deficient in either Roquin-1 or Roquin-2, suggesting that these proteins function redundantly in mRNA degradation (Vogel et al., 2013 ▸).
Recently, several groups have reported crystal structures of Roquin-1, revealing the overall structure of the ROQ domain of Roquin-1 and the recognition mode of CDE RNA by Roquin-1 (Srivastava et al., 2015 ▸; Tan et al., 2014 ▸; Schuetz et al., 2014 ▸; Schlundt et al., 2014 ▸). However, the structural details of Roquin-2 remain unknown. Here, we report the crystal structure of the human Roquin-2 ROQ domain and its complex with Roquin CDE RNA.
2. Materials and methods
2.1. Macromolecule production
The gene encoding human Roquin-2 (residues 1–400) was inserted into the expression vector pGEX6P-1 (GE Healthcare) between the BamHI and EcoRI sites. Escherichia coli BL21(DE3)pLysS cells were transformed with the vector and cultured at 37°C to a suitable cell density (OD600 of 0.6–0.7); protein expression was then induced by addition of 0.2 mM IPTG and cultivation was continued for 16 h at 18°C. Before induction, 10 µM zinc acetate was added to the culture. The cells were collected by centrifugation and lysed by sonication in a buffer consisting of 20 mM Tris–HCl pH 8.0, 500 mM NaCl, 1 mM DTT, 1 mM MgCl2, 0.2 mM zinc acetate. The proteins were purified from the cleared lysate by Glutathione Sepharose 4B (GE Healthcare) followed by GST-tag cleavage by PreScission protease. Further purification was performed using a Superdex 200 (GE Healthcare) gel-filtration column and a Glutathione Sepharose 4B column (GE Healthcare). The proteins were concentrated to about 36 mg ml−1 in a buffer consisting of 10 mM Tris–HCl pH 8.0, 500 mM NaCl.
Before crystallization, purified Roquin-2 (1–400) was digested with a 1:400(w:w) ratio of trypsin for 30 min at room temperature. Limited proteolysis of Roquin-2 yielded two polypeptide fragments of 15 and 30 kDa, which were further purified by HiTrap SP (GE Healthcare) and concentrated to about 15 mg ml−1.
2.2. Crystallization
The concentration of Roquin-2 (1–400) was adjusted to 10 mg ml−1 in a buffer consisting of 10 mM Tris–HCl pH 8.0, 150 mM NaCl. Crystals of the ligand-free form of Roquin-2 were grown by the sitting-drop vapour-diffusion method at 20°C by mixing the protein solution with an equal volume of reservoir solution (15% PEG 6000, 100 mM HEPES pH 7.5, 100 mM KCl).
The Roquin-2–RNA complex was prepared using the 30 kDa fragment derived from trypsin-digested Roquin-2 (1–400) with 17 nt Roquin CDE RNA (5′-UAACUUCUGUGAAGUUG-3′) at a molar ratio of 1:1.5. The concentration of Roquin-2–RNA was adjusted to 7.5 mg ml−1 in a buffer consisting of 10 mM Tris–HCl pH 8.0, 5 mM sodium acetate pH 5.0, 160 mM NaCl. Crystals of the Roquin-2–RNA complex were grown at 20°C by the sitting-drop vapour-diffusion method by mixing the protein solution with an equal volume of reservoir solution (20% PEG 8000, 100 mM Tris–HCl pH 8.0, 200 mM LiCl).
2.3. Data collection and processing
X-ray diffraction data for both ligand-free and RNA-complexed Roquin-2 were collected (λ = 1.0000 Å) on beamline BL41XU at SPring-8, Hyogo, Japan under cryogenic conditions at 100 K. Crystals were equilibrated in a cryoprotectant solution consisting of reservoir solution supplemented with 25% ethylene glycol (ligand-free form) or 25% glycerol (RNA complex) prior to flash-cooling. X-ray diffraction data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▸; Table 1 ▸).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the outer shell.
| Ligand-free | RNA complex | |
|---|---|---|
| Data collection | ||
| Diffraction source | BL41XU, SPring-8 | BL41XU, SPring-8 |
| Wavelength () | 1.0000 | 1.0000 |
| Temperature (K) | 100 | 100 |
| Detector | Pilatus 6M | Pilatus 6M |
| Crystal-to-detector distance (mm) | 380 | 250 |
| Rotation range per image () | 0.4 | 0.2 |
| Total rotation range () | 360 | 180 |
| Exposure time per image (s) | 0.2 | 0.2 |
| Space group | P212121 | C2221 |
| a, b, c () | 60.8, 60.9, 169.5 | 59.2, 90.0, 84.6 |
| , , () | 90, 90, 90 | 90, 90, 90 |
| Mosaicity () | 0.450.85 | 0.190.31 |
| Resolution range () | 50.02.70 (2.752.70) | 50.01.60 (1.631.60) |
| Total No. of reflections | 220208 | 192964 |
| No. of unique reflections | 17819 | 29045 |
| Completeness (%) | 99.1 (94.9) | 95.9 (87.7) |
| Multiplicity | 12.3 (8.5) | 6.6 (5.4) |
| I/(I) | 14.4 (2.1) | 24.9 (3.1) |
| R meas † | 0.176 (0.823) | 0.053 (0.334) |
| CC1/2 | 0.979 (0.907) | 0.989 (0.964) |
| Overall B factor from Wilson plot (2) | 26.8 | 16.4 |
| Refinement | ||
| Resolution range () | 50.02.7 | 49.51.6 |
| Cutoff | None | None |
| No. of reflections, working set | 16909 | 27572 |
| No. of reflections, test set | 910 | 1473 |
| Final R cryst (%) | 22.4 | 15.5 |
| Final R free (%) | 26.0 | 17.6 |
| Cruickshank DPI | 0.076 | |
| No. of non-H atoms | ||
| Protein | 4748 | 1243 |
| RNA | 357 | |
| Glycerol | 24 | |
| Water | 41 | 186 |
| Total | 4789 | 1810 |
| R.m.s. deviations | ||
| Bonds () | 0.010 | 0.007 |
| Angles () | 1.43 | 1.26 |
| Average B factors (2) | ||
| Protein | 29.8 | 22.2 |
| RNA | 19.4 | |
| Glycerol | 37.0 | |
| Water | 35.6 | 32.4 |
| Ramachandran plot | ||
| Most favoured (%) | 98.8 | 99.3 |
| Allowed (%) | 1.2 | 0.7 |
| Outliers (%) | 0.0 | 0.0 |
| Rotamer outliers (%) | 1.0 | 0.0 |
R
meas = 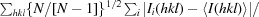
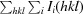 calculated for all data.
calculated for all data.
2.4. Structure solution and refinement
The crystal structure of the Roquin-2–RNA complex was solved by molecular replacement using MOLREP (Vagin & Teplyakov, 2010 ▸) using the ROQ domain of the mouse Roquin-1 structure (PDB entry 4qi0; Schlundt et al., 2014 ▸) as a search model. The crystal structure of the ligand-free form of Roquin-2 was solved by molecular replacement using MOLREP with the refined model of the Roquin-2–RNA complex. The models were subjected to the iterative cycles of manual model building using Coot (Emsley & Cowtan, 2004 ▸) and restrained refinement using REFMAC (Murshudov et al., 2011 ▸) (Table 1 ▸). The qualities of the refined models were evaluated with MolProbity (Chen et al., 2010 ▸). The structural figures were prepared with PyMOL (DeLano, 2002 ▸). The coordinates and structure-factor data for the ligand-free form and the RNA complex have been deposited in the PDB as PDB entries 4zlc and 4zld, respectively.
3. Results
3.1. Structure determination of the ROQ domain of human Roquin-2
Although we prepared recombinant human Roquin-2 (residues 1–400) consisting of the RING-finger and ROQ domains for crystallization (Figs. 1 ▸ a and 1 ▸ b), we obtained crystals of a 15 kDa fragment from the purified protein, which was possibly cleaved off during crystallization (Fig. 1 ▸ c). We determined its structure at 2.7 Å resolution in the ligand-free form (Fig. 1 ▸ d, Table 1 ▸). The asymmetric unit of the crystals contained four molecules of Roquin-2 (molecule A, residues 171–325; molecule B, residues 173–325; molecule C, residues 177–323; molecule D, residues 171–321) (Fig. 1 ▸ e), all of which corresponded to the 15 kDa fragment. We hereafter refer to the region encompassing residues 171–325 as the ROQ domain.
Despite extensive trials to obtain RNA-bound forms of crystals using Roquin-2 (residues 1–400), we failed to obtain crystals suitable for structural analyses. Hence, we conducted limited proteolysis of the purified protein using trypsin, yielding 30 and 15 kDa fragments. We screened for crystallization conditions using both fragments in a mixture with a 17 nt ssRNA derived from the CDE in the human Roquin 3′ UTR. We obtained crystals using both fragments and we found that both crystals contained the 15 kDa fragment (Fig. 1 ▸ c). It is possible that the 30 kDa fragment was degraded into the 15 kDa fragment during crystallization, suggesting that the 15 kDa fragment represents the minimal RNA-binding region. We have succeeded in determining the crystal structure of Roquin-2 in complex with Roquin CDE RNA at 1.6 Å resolution (Fig. 2 ▸ a, Table 1 ▸). The refined model contained one complex of the ROQ domain (residues 171–321) and RNA (17 nt) with 1:1 stoichiometry. The electron density of the 17 nt RNA was clearly visible (Fig. 2 ▸ b).
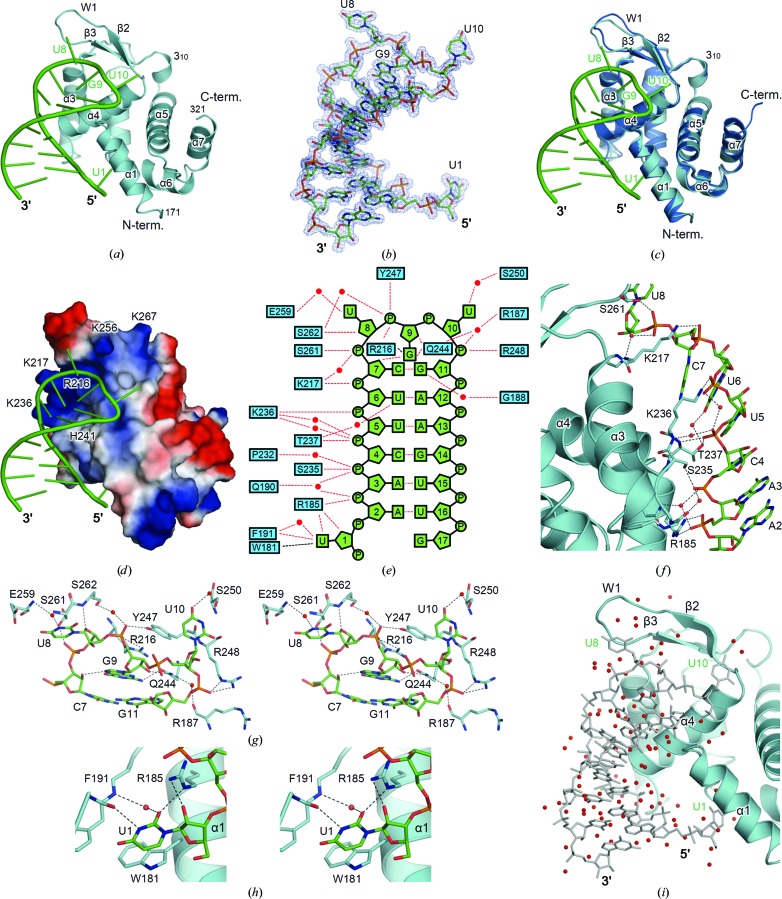
Figure 2.
Structure of the RNA-bound form of Roquin-2. (a) Structure of the RNA-bound form of the ROQ domain of Roquin-2. Roquin-2 and RNA are shown in cyan and green, respectively. (b) The 2F o − F c difference electron-density map of RNA. The map is contoured at the 1.5σ level with a blue mesh. (c) Superposition of the ligand-free and the RNA-bound forms of the ROQ domain of Roquin-2. The ligand-free and the RNA-bound forms are shown in blue and cyan, respectively. The bound RNA in the complex is shown in green. (d) Electrostatic surface potential of the ROQ domain of Roquin-2. The residues forming the positively charged groove are labelled. Positive and negative electrostatic potentials are shown in blue and red, respectively. (e) Schematic summary of the interactions between Roquin-2 and RNA. Hydrogen bonds and other interactions are indicated by red and black dashed lines, respectively. (f) Detailed view of the interactions between the 5′ half of the stem of RNA and Roquin-2. (g) Detailed stereoview of the interactions between the triloop region of RNA and Roquin-2. (h) Detailed stereoview of the interactions between U1 of the RNA and Roquin-2. (i) Water-mediated interaction of Roquin-2 and RNA. In (f)–(i), residues involved in the recognition of RNA are shown as stick structures and labelled. Red spheres represent water molecules. Hydrogen bonds are indicated by dashed lines.
3.2. Crystal structure of the ligand-free form of the ROQ domain
The structure of the ROQ domain of Roquin-2 has a compact globular shape with dimensions of approximately 25 × 45 × 35 Å composed of seven α-helices (α1–α7), one 310-helix and a β-sheet of three strands (β1–β3) (Fig. 1 ▸ d). The ROQ domain contained a winged-helix (WH) motif consisting of three α-helices (α2–α4) and three β-strands (β1–β3). The region connecting the WH motif to the rest of the domain (α5–α7) formed a 310-helix. The four molecules in the asymmetric unit were essentially the same, with root-mean-square deviations (r.m.s.d.) ranging from 0.4 to 0.7 Å (Fig. 1 ▸ e). Large structural deviations among the four molecules in the asymmetric unit were observed in the wing region of the WH motif (W1), suggesting that this region is intrinsically flexible in the absence of RNA (Fig. 1 ▸ e).
3.3. Crystal structure of the RNA-bound form of the ROQ domain
The structure of the ROQ domain in complex with RNA was very similar to those of the ligand-free forms, with an r.m.s.d. ranging from 0.5 to 0.6 Å (Fig. 2 ▸ c). The conformation of W1 was stabilized upon RNA binding. The surface groove formed by α3, α4 and W1 of the WH motif was highly positively charged because of a large patch of basic residues (Arg216, Lys217, Lys236, His241, Lys256 and Lys267), suitable for interacting with negatively charged RNA (Fig. 2 ▸ d). Accordingly, the CDE RNA, which formed a stem-loop structure, bound to the positively charged groove (Figs. 2 ▸ a and 2 ▸ d). The stem-loop consisted of the stem region of six Watson–Crick base pairs A2–U16, A3–U15, C4–G14, U5–A13, U6–A12 and C7–G11, the triloop region U8-G9-U10 and the flanking region of the stem (U1 and G17) (Fig. 2 ▸ e).
The interactions between the stem of RNA and Roquin-2 were mainly mediated by the backbone phosphate groups of RNA (Figs. 2 ▸ e and 2 ▸ f). Almost all of the phosphate groups of the 5′ half of the stem made specific interactions with Roquin-2: P3 with Arg185, P4 with Ser235, P5 with Thr237, P6 with Lys236, P7 with Lys217 and P8 with Ser261. The triloop region U8-G9-U10 mainly interacted with the W1 and α4 regions of the WH motif (Figs. 2 ▸ a and 2 ▸ c). The bases of the first and third positions of the triloop (U8 and U10) flipped out of the duplex, while that of the second position (G9) flipped in (Fig. 2 ▸ g). The base of G9 made intimate contacts with Roquin-2 and the other part of the RNA, thus acting as an important residue in the triloop region for specific binding. The base of G9 was sandwiched between the last base pair of the stem (C7–G11) and the side chain of Arg216, which simultaneously made hydrogen bonds between N7 and O2′ of C7 and between N2 and Gln244. On the other hand, the bases of U8 and U10 were in loose contact with Roquin-2. N3 of U8 and O4 of U10 interact with Glu259 and Ser250 via a water molecule, respectively. The backbone atoms of the triloop region made several hydrogen bonds to Roquin-2: from O4′ of U8 to Ser262, from P9 to Arg216 and Tyr247, from O2′ of G9 to Gln244 and from P11 to Arg248. The flexible W1 region in the ligand-free form acted as a lid for the triloop binding site (Fig. 2 ▸ c). Owing to the flexibility of the W1 region, the ROQ domain would accommodate a broader range of target RNA molecules.
In addition to the interactions in the triloop and stem regions, the 5′ flanking residue (U1) made specific interactions with Roquin-2 (Figs. 2 ▸ e and 2 ▸ h). The base of U1 stacked onto the side chain of Trp181 and made a hydrogen bond to Phe191. The O2′ in the backbone ribose moiety made a hydrogen bond to Arg185.
It should be noted that our structure determined at high resolution made it possible to identify numerous water molecules. Water molecules filled the space between RNA and Roquin-2, and some water molecules bridged the two molecules (Fig. 2 ▸ i). Although no direct interactions were observed between the backbone phosphate groups of the 3′ half of the stem and Roquin-2, these water-mediated indirect interactions contributed to RNA recognition.
4. Discussion
In this study, we establish the structural basis for the recognition of CDE RNA by human Roquin-2. We first described the structure of Roquin-2, and the high quality of the electron-density map of the RNA-bound form of Roquin-2 enabled us to elucidate the interactions between Roquin-2 and RNA. Recently, several groups have reported crystal structures of Roquin-1 (Srivastava et al., 2015 ▸; Tan et al., 2014 ▸; Schuetz et al., 2014 ▸; Schlundt et al., 2014 ▸). Schlundt and coworkers reported the structure of the ROQ domain of mouse Roquin-1 in complex with the 23 nt Tnf CDE RNA (PDB entry 4qi2; Schlundt et al., 2014 ▸). The ROQ domain of mouse Roquin-1 was almost the same region as our crystallization construct, suggesting that the ROQ domain was the stably folded domain. Tan and coworkers reported the structure of human Roquin-1 comprised of the ROQ domain and an additional helical domain in complex with the 19 nt Hmgxb3 CDE RNA (PDB entry 4qil; Tan et al., 2014 ▸). The Tnf CDE RNA, Hmgxb3 CDE RNA and Roquin CDE RNAs differ in the stem and its flanking regions, while they have the UGU triloop in common (Fig. 3 ▸). Despite the differences in the crystallization constructs and RNA sequences used in the structural analyses, the structures of the Roquin proteins and the modes of RNA recognition are essentially the same in the three Roquin structures in complex with RNA (Fig. 3 ▸).
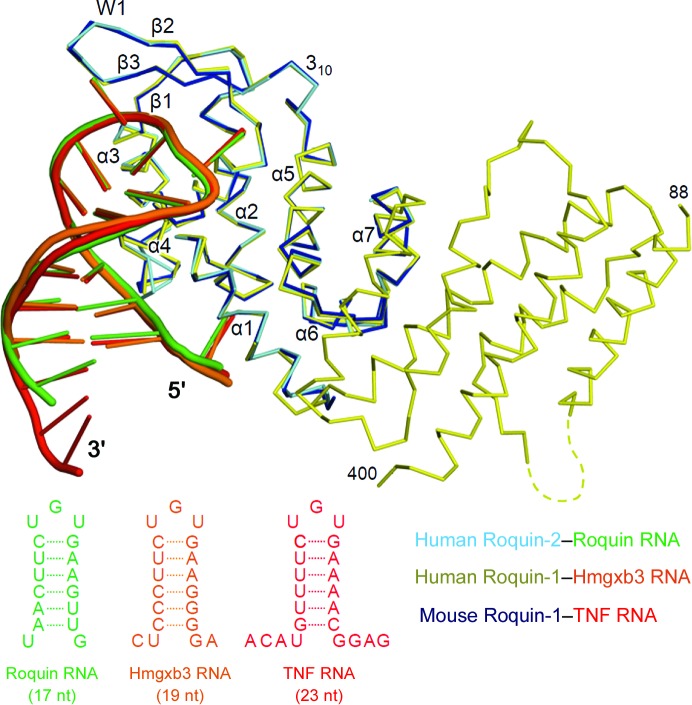
Figure 3.
Comparison of the RNA-binding modes of Roquin-1 and Roquin-2. Superposition of the RNA-bound forms of the Roquin proteins. Roquin CDE-bound human Roquin-2 (this study), Hmgxb3 CDE-bound human Roquin-1 (Tan et al., 2014 ▸) and TNF CDE-bound mouse Roquin-1 (Schlundt et al., 2014 ▸) are shown in cyan, yellow and blue, respectively. The CDE RNA of Roquin, Hmgxb3 and TNF are coloured in green, orange and red, respectively, and are shown schematically.
The structures of the ROQ domains of Roquin-1 and Roquin-2 in complex with RNA were very similar, with r.m.s.d. ranging from 0.4 to 0.7 Å. The Roquin–RNA complexes have three key features of the RNA recognition in common: the backbone of the 5′ half of the stem and the triloop, and the flanking region of the stem (Fig. 3 ▸). Of note, the residues involved in recognition of CDE RNA are perfectly conserved between Roquin-1 (human and mouse) and Roquin-2 (human). Moreover, our high-resolution structure revealed that many water-mediated interactions also contributed to RNA recognition. In fact, several phosphate groups and bases are indirectly recognized by Roquin, suggesting that RNA recognition is achieved by direct and indirect interactions.
The structural analyses of Roquin-2 demonstrate that the RNA-recognition mechanism is highly conserved among Roquin proteins and that the ROQ domain is essential for recognition, suggesting that Roquin-2 functions redundantly in Roquin-mediated mRNA degradation.
Supplementary Material
PDB reference: ROQ domain of Roquin-2, ligand-free, 4zlc
PDB reference: RNA complex, 4zld
Acknowledgments
We acknowledge Dr Yusuke Sekine and Professor Hidenori Ichijo for providing the cDNA of human Roquin-2. We also thank the beamline staff members at the Photon Factory and SPring-8 for their assistance with data collection. This work was supported by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science and Technology (UO and TS), the Takeda Science Foundation (UO and TS) and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (UO).
References
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Glasmacher, E., Hoefig, K. P., Vogel, K. U., Rath, N., Du, L., Wolf, C., Kremmer, E., Wang, X. & Heissmeyer, V. (2010). Nature Immunol. 11, 725–733. [DOI] [PubMed]
- Joazeiro, C. A. P. & Weissman, A. M. (2000). Cell, 102, 549–552. [DOI] [PubMed]
- Leppek, K., Schott, J., Reitter, S., Poetz, F., Hammond, M. C. & Stoecklin, G. (2013). Cell, 153, 869–881. [DOI] [PubMed]
- Maruyama, T., Araki, T., Kawarazaki, Y., Naguro, I., Heynen, S., Aza-Blanc, P., Ronai, Z., Matsuzawa, A. & Ichijo, H. (2014). Sci. Signal. 7, ra8. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, A. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Schlundt, A., Heinz, G. A., Janowski, R., Geerlof, A., Stehle, R., Heissmeyer, V., Niessing, D. & Sattler, M. (2014). Nature Struct. Mol. Biol. 21, 671–678. [DOI] [PubMed]
- Schuetz, A., Murakawa, Y., Rosenbaum, E., Landthaler, M. & Heinemann, U. (2014). Nature Commun. 5, 5701. [DOI] [PubMed]
- Srivastava, M. et al. (2015). Nature Commun. 6, 6253. [DOI] [PMC free article] [PubMed]
- Stoecklin, G., Lu, M., Rattenbacher, B. & Moroni, C. (2003). Mol. Cell. Biol. 23, 3506–3515. [DOI] [PMC free article] [PubMed]
- Tan, D., Zhou, M., Kiledjian, M. & Tong, L. (2014). Nature Struct. Mol. Biol., 21, 679–685. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vinuesa, C. G., Cook, M. C., Angelucci, C., Athanasopoulos, V., Rui, L., Hill, K. M., Yu, D., Domaschenz, H., Whittle, B., Lambe, T., Roberts, I. S., Copley, R. R., Bell, J. I., Cornall, R. J. & Goodnow, C. C. (2005). Nature (London), 435, 452–458. [DOI] [PubMed]
- Vogel, K. U. et al. (2013). Immunity, 38, 655–668. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ROQ domain of Roquin-2, ligand-free, 4zlc
PDB reference: RNA complex, 4zld