The crystal structure of a serine protease inhibitor from B. bauhinioides was determined at 1.4 Å resolution and compared with the structures of other Kunitz-type inhibitors, including the closely related dual-specificity inhibitor BbCI.
Keywords: β-trefoil, Kunitz inhibitor, kallikrein, crystal structure
Abstract
A serine protease inhibitor from Bauhinia bauhinioides (BbKI) belongs to the Kunitz family of plant inhibitors, which are common in plant seeds. BbKI does not contain any disulfides, unlike most other members of this family. It is a potent inhibitor of plasma kallikrein, in addition to other serine proteases, and thus exhibits antithrombotic activity. A high-resolution crystal structure of recombinantly expressed BbKI was determined (at 1.4 Å resolution) and was compared with the structures of other members of the family. Modeling of a complex of BbKI with plasma kallikrein indicates that changes in the local structure of the reactive loop that includes the specificity-determining Arg64 are necessary in order to explain the tight binding. An R64A mutant of BbKI was found to be a weaker inhibitor of plasma kallikrein, but was much more potent against plasmin, suggesting that this mutant may be useful for preventing the breakup of fibrin and maintaining clot stability, thus preventing excessive bleeding.
1. Introduction
The seeds of legumes are recognized as excellent sources of proteins of both nutritional and therapeutic value. The most extensively studied proteins from these plants are inhibitors, which interact with different classes of proteolytic enzymes. Protease inhibitors exhibit different degrees of specificity, thus contributing to the elucidation of the biochemical processes involved in coagulation, inflammation or the formation or suppression of tumors. In some specific cases, such inhibitors can also be developed as potential therapeutic agents (Birk, 2003 ▸; Oliva et al., 2011 ▸; Oliva & Sampaio, 2008 ▸; Sałaga et al., 2013 ▸).
The Kunitz family of plant protease inhibitors was named after the discoverer of the soybean trypsin inhibitor (STI), who isolated and crystallized it (Kunitz, 1947 ▸). Inhibitors of the Kunitz type are single-chain proteins of molecular weight ranging from 18 to 21 kDa and containing 160–180 amino-acid residues, or they may also be domains of larger proteins. These inhibitors may contain one or two disulfides, but may even entirely lack cysteine residues (Oliva et al., 2010 ▸; Oliva & Sampaio, 2008 ▸).
Plant-derived Kunitz-type inhibitors have been studied very extensively. Their inhibitory properties have been determined for a number of blood-clotting enzymes such as plasma kallikrein, factor XIIa, factor Xa and thrombin, for enzymes involved in digestive processes, such as trypsin and chymotrypsin, and for enzymes involved in inflammatory processes, such as elastase (Batista et al., 1996 ▸; Odei-Addo et al., 2014 ▸; Oliva & Sampaio, 2008 ▸; Souza-Pinto et al., 1996 ▸; Vadivel et al., 2014 ▸).
Two Kunitz-type inhibitors, BbCI and BbKI, have been isolated from the seeds of Bauhinia bauhinioides. These 18 kDa proteins show high similarity in their primary structures, being much more similar to each other than to other Kunitz-type inhibitors from plants. One of the distinct features of these two inhibitors is a lack of disulfides, with BbCI lacking cysteines entirely, whereas BbKI has only one such residue. Although the primary structure of BbKI is 84% identical to that of BbCI, the specificities of these two inhibitors are quite distinct (De Oliveira et al., 2001 ▸; Oliva et al., 2001 ▸). BbCI has the ability to inhibit members of two different classes of proteases: serine proteases, including human neutrophil elastase and porcine pancreatic elastase, as well as the cysteine proteases cathepsin L and cruzain/cruzipain. BbKI is not active on cysteine proteases but inhibits bovine trypsin, human plasma kallikrein and plasmin, whereas BbCI does not interfere with the activity of any enzymes involved in blood coagulation cascades (De Oliveira et al., 2001 ▸; Neuhof et al., 2003 ▸). BbKI is currently the only known inhibitor isolated from plants which inhibits tissue kallikrein in addition to plasma kallikrein (Oliva et al., 2001 ▸, 2010 ▸).
Because of the highly restricted specificity of blood coagulation enzymes, it is easy to understand why so few plant inhibitors are able to block their activities (Birk, 2003 ▸; Oliva & Sampaio, 2008 ▸). As is the case for digestive enzymes such as trypsin and chymotrypsin, the enzymes of blood coagulation are serine proteases. However, unlike trypsin, the proteases of the coagulation cascade have acquired a high degree of specificity during the course of evolution, since they cleave only a limited number of peptide bonds involving basic amino-acid residues (Krishnaswamy, 2005 ▸).
BbKI (or its recombinant form rBbKI) has become an attractive molecule for studying pathological models of the circulatory system, since this protein acts on enzymes involved in coagulation, fibrinolysis and inflammation. One of the targets of BbKI is human plasma kallikrein, an enzyme that participates in the processes of blood coagulation, platelet aggregation and muscle contraction (Botos & Wlodawer, 2007 ▸; Pampalakis & Sotiropoulou, 2007 ▸; Turk, 2006 ▸). Indeed, rBbKI is effective in inhibiting the viability of tumor cell lines (Nakahata et al., 2011 ▸). Other important aspects of the inhibitor have been reported by Brito et al. (2014 ▸), who found that BbKI may prolong the formation of blood clots in vitro and that it exhibits antithrombotic activity in venous and arterial thrombosis in vivo models.
The structures of a number of Kunitz-type inhibitors have been determined to date (Renko et al., 2012 ▸). These proteins belong to a very large family of proteins with a common fold defined as the β-trefoil fold. The family members contain a very similar core with pseudo-threefold symmetry, whereas they differ quite significantly in the structure of the loops connecting the central β-strands. Their functional properties are largely defined by the structure of their variable parts. Here, we describe the high-resolution crystal structure of BbKI and present some biochemical properties of this potent inhibitor of plasma kallikrein.
2. Materials and methods
2.1. Preparation and purification of rBbKI and its complex with bovine trypsin
The overexpression of N-terminally His-tagged rBbKI in Escherichia coli BL21(DE3) cells and its subsequent purification were carried out using previously described methods (Araújo et al., 2005 ▸) with some modifications. Briefly, cells containing the target gene cloned into the expression vector pET-28a (Novagen) were grown in Luria–Bertani medium supplemented with 30 µg ml−1 kanamycin (Invitrogen) at 37°C to an optical density OD600 nm of 0.5, followed by the induction of fusion-protein expression with isopropyl β-d-1-thiogalactopyranoside (Invitrogen). IPTG was added to a final concentration of 1 mM and the culture was grown for an additional 3 h. Subsequently, the cells were harvested by centrifugation (4000g, 20 min, 4°C; Hitachi himac CR21-GIII). The pellets were then resuspended in 10 ml lysis buffer (0.05 M Tris–HCl pH 8.0, 0.15 M NaCl). The bacteria were then disrupted by sonication (Unique, Campinas, Brazil), lysed in 0.5% Triton X-114 (Sigma–Aldrich) (12 cycles of ultrasound for 30 s at 40 W) and isolated by centrifugation (4000g, 20 min, 4°C). Lipopolysaccharide found on the wall of the bacteria was removed by the addition of chloroform [3:1(v:v)] at 4°C to the fractions containing the protein, leading to phase separation. The upper phase was removed by centrifugation. The fusion protein was then purified by Ni–NTA affinity chromatography and eluted with an imidazole gradient (100–250 mM) in the same sonication buffer. Fractions containing the fusion protein were combined and dialyzed against 0.05 M Tris–HCl pH 8.0, 0.15 M NaCl buffer in order to remove the imidazole and to exchange the buffer for subsequent enzyme digestion with 1 U thrombin (GE Healthcare) per milligram of fusion protein for 4 h at 18°C. As a consequence of the introduced thrombin cleavage site in the expression vector, the rBbKI contained three non-native amino acids at the N-terminus (Ser-Gly-His). The His tag was then separated from the rBbKI protein by size-exclusion chromatography (Superdex 75 HR 10/30 column; GE Healthcare) in 0.05 M Tris–HCl pH 8.0, 0.15 M NaCl.
For preparation of the complex of rBbKI with trypsin, equimolar amounts of rBbKI and bovine pancreatic trypsin (Sigma) were mixed and incubated overnight at room temperature in 50 mM Tris pH 8.0, 100 mM NaCl. The mixture was then applied onto a Sephacryl S-100 HR column (GE Healthcare) pre-equilibrated with the same buffer. Fractions of 2 ml in volume were collected at a flow rate of 0.2 ml min−1. The identity of the samples pooled from different peaks was verified by SDS–PAGE. The fractions corresponding to the rBbKI–trypsin complex were selected and concentrated to around 13 mg ml−1. The freshly prepared complex sample was immediately used for crystallization trials.
2.2. Protein crystallization and X-ray data collection and processing
rBbKI was concentrated to 6.3 mg ml−1 and crystallized using the conditions reported previously, namely 8% PEG 4000, 0.1 M sodium acetate pH 4.6 (Navarro et al., 2005 ▸). Crystals appeared in two weeks at 20°C and grew to their final size in about a month. Crystals of the putative complex of rBbKI and bovine trypsin were obtained with several conditions from Crystal Screen HT (Hampton Research) at 20°C. Diffraction-quality crystals, although very small, were obtained from conditions C6 (0.2 M ammonium sulfate, 30% PEG 8000) and E2 (0.5 M sodium chloride, 0.01 M magnesium chloride hexahydrate, 0.01 M hexadecyltrimethylammonium bromide).
Diffraction data were collected on the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID at the Advanced Photon Source, Argonne National Laboratory. Single crystals were transferred to a cryoprotectant solution (mother liquor with an extra 25% glycerol) for approximately 2 min and were then flash-cooled at 100 K in a stream of liquid nitrogen. Good-quality diffraction of the crystals of rBbKI was observed to a resolution of 1.4 Å; data extending beyond this limit were excluded from processing. Diffraction data were indexed, integrated and scaled with HKL-2000 (Otwinowski & Minor, 1997 ▸). The crystals belonged to space group P212121, with unit-cell parameters a = 46.7, b = 59.5, c = 64.0 Å. The estimated Matthews coefficient is 2.13 Å3 Da−1, corresponding to 42% solvent content with one rBbKI molecule in the asymmetric unit. Data-processing statistics are shown in Table 1 ▸. Diffraction data extending to a resolution of 2.1 Å for the putative rBbKI–trypsin complex were collected using a protocol similar to that used for the crystals of uncomplexed rBbKI. The crystals belonged to space group P43, with unit-cell parameters a = b = 45.4, c = 153.1 Å, and were very highly twinned.
Table 1. Data collection and structure refinement.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P212121 |
| Molecules in asymmetric unit | 1 |
| Unit-cell parameters () | a = 46.7, b = 59.5, c = 64.0 |
| Resolution () | 50.01.4 (1.421.40) |
| R merge † (%) | 4.8 (62.8) |
| No. of reflections (measured/unique) | 171809/34445 |
| I/(I) | 32.8 (2.34) |
| Completeness (%) | 96.1 (93.3) |
| Multiplicity | 5.0 (4.8) |
| Refinement | |
| Resolution () | 43.571.40 |
| No. of reflections (refinement/R free) | 32686/1726 |
| R/R free ‡ | 0.123/0.167 |
| No. of atoms | |
| Protein | 1314 |
| Ligands | |
| Water | 213 |
| R.m.s. deviations from ideality | |
| Bond lengths () | 0.014 |
| Bond angles () | 1.78 |
| PDB code | 4zot |
R
merge = 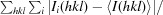
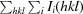 , where I
i(hkl) is the observed intensity of the ith measurement of reflection hkl and I(hkl) is the average intensity of that reflection obtained from multiple observations.
, where I
i(hkl) is the observed intensity of the ith measurement of reflection hkl and I(hkl) is the average intensity of that reflection obtained from multiple observations.
R = 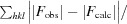
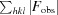 , where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is defined as in Brnger (1992 ▸).
, where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is defined as in Brnger (1992 ▸).
2.3. Structure determination and refinement
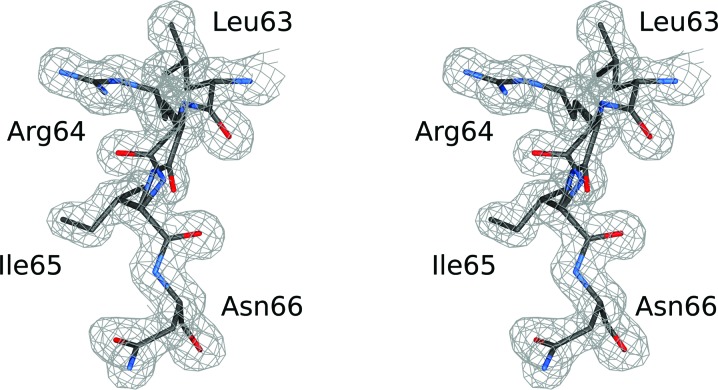
The crystals of rBbKI reported here were isomorphous to those used previously for structure determination and refinement at 1.87 Å resolution (PDB entry 2go2; Navarro et al., 2005 ▸). Since the definition of the unit cell of the previously deposited coordinates was nonstandard, Phaser (McCoy et al., 2007 ▸) was used to locate the rBbKI molecule in the cell with redefined axes. Further refinement was performed with REFMAC5 (Murshudov et al., 2011 ▸) within the HKL-3000 package (Minor et al., 2006 ▸), using all data between 20 and 1.4 Å resolution, after setting aside a randomly selected 5% of reflections (1726 in total) for the calculation of R free. Manual corrections were applied using Coot (Emsley & Cowtan, 2004 ▸). Anisotropic individual temperature factors were added in the final stages of refinement, resulting in a model characterized by an R factor of 12.3% and an R free of 16.7% (Table 1 ▸). The resulting maps were of high quality (Fig. 1 ▸).
Figure 1.
A stereoimage showing a fragment of the final model covered by the 2F o − F c electron-density map, contoured at 1σ, in the part of the reactive loop that includes the specificity-determining Arg64. Two orientations of the main chain of this residue are clearly visible.
An attempt to determine the structure of the putative complex of rBbKI with trypsin was performed with Phaser (McCoy et al., 2007 ▸) using the coordinates extracted from PDB entries 2go2 (rBbKI) and 2bza (bovine trypsin; Ota et al., 1999 ▸) as starting models. However, preliminary refinement of the molecular-replacement solution revealed that only two inhibitor molecules were present in the asymmetric unit of the crystals, regardless of whether the crystallization medium contained a high salt concentration or did not include added salt. For this reason, these data were not evaluated any further.
2.4. BbKI mutagenesis
The gene sequence encoding the reactive-site P1 residue Arg64 was mutated to alanine (BbKI-R64A) through site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Germany). The vector used was pET28aBbKI and the two primers, used at 100 pmol each, were BbKI-R64A_FORWARD, 5′-GGA AAC GAA GCA GAG CCA GCG GCA GTT GTT TTG GAT CCT CAC C-3′, and BbKI-R64A_ REVERSE, 5′-GGT GAG GAT CCA AAA CAA CTG CCG CTG GCT CTG CTT CGT TTC C-3′.
2.5. Enzymatic activity and inhibition assays
The inhibitory activity of rBbKI and rBbKI-R64A against a variety of proteolytic enzymes was investigated using previously described methods (Araújo et al., 2005 ▸). The inhibitor and protease were incubated at 37°C with one of the following proteases and their respective substrates: cathepsin L (18 nM) and cruzain (4.0 nM) activated with 100 mM sodium phosphate buffer pH 6.3 containing 10 mM EDTA, 400 mM NaCl and 2 mM dithiothreitol, substrate 0.3 mM Z-Phe-Arg-MCA; trypsin (7.0 nM in 0.05 M Tris–HCl pH 8.0, 0.02% CaCl2), substrate 1.0 mM BAPA; chymotrypsin (10 nM in 0.1 M Tris–HCl pH 8.0, 0.02% CaCl2), substrate 2.0 mM Suc-Phe-pNan; HuPK (human plasma kallikrein; 4.0 nM in 0.05 M Tris–HCl pH 8.0), substrate 0.4 mM H-d-Pro-Phe-Arg-pNA; PPE (porcine pancreatic elastase; 24 nM in 0.05 M Tris–HCl pH 8.0, 0.5 M NaCl), substrate 1.0 mM MeO-Suc-Ala-Ala-Pro-Val-pNA; HNE (human neutrophil elastase; 25 nM in 0.05 M Tris–HCl pH 7.0, 0.5 M NaCl), substrate 1.0 mM MeO-Suc-Ala-Ala-Pro-Val-pNA; and plasmin (3.5 nM in 0.1 M Tris–HCl pH 7.4 containing 0.2 M NaCl), substrate 1.0 mM H-d-Val-Leu-Lys-pNan. K i,app values were determined using the GraFit 3.01 program (Morrison, 1982 ▸) by adjusting the experimental points to the equation for tight binding using nonlinear regression.
As an example of an assay, 20 µl 0.042 µM HuPK was pre-incubated at 37°C with increasing concentrations of rBbKI or rBbKI-R64A in 50 mM Tris–HCl pH 8.0 containing 0.5 M NaCl pH 8.0. After 10 min, 20 µl 5 mM H-d-Pro-Phe-Arg-pNA substrate was added in a final volume of 250 µl. The reaction was followed for 30 min and stopped by the addition of 20 µl 30%(v/v) acetic acid. Substrate hydrolysis was followed by the absorbance at 405 nm. Apparent K i values were determined by adjusting the experimental points to the equation for a slow tight-binding mechanism (Morrison, 1982 ▸) using a nonlinear fitting adjusted by the GraFit 3.01 program.
3. Results and discussion
3.1. A comparison with the previously determined medium-resolution structure
rBbKI has previously been crystallized and its structure has been solved and refined at 1.87 Å resolution (Navarro et al., 2005 ▸). However, although the resulting coordinates were deposited in the Protein Data Bank (PDB entry 2go2), only a cursory mention of this structure is found in a publication describing the structure of the related cruzipain inhibitor BbCI (Hansen et al., 2007 ▸). The protein utilized in the current work was prepared from a recombinant source and was purified in the same manner as described previously (Araújo et al., 2005 ▸; Navarro et al., 2005 ▸). However, the crystals obtained in this work yielded diffraction data extending to the much higher resolution of 1.4 Å, particularly since the data were now collected on a synchrotron beamline rather than on a rotating-anode X-ray generator as in the past. This increase in resolution resulted in almost 2.4 times more structure factors (32 686 versus 13 850) that were now available for refinement.
Both the current and the previously determined structures of rBbKI are complete between Ser1 and Thr163, although the electron density in the current structure is weak and not completely unambiguous near His26 and in the stretch Gly37-Asn38-Glu39. The side chains of His28 and Asp143 were not modeled, and the side chain of Arg141 was modeled only partially owing to ambiguous electron density. Ser1, although present in the gene sequence of rBbKI, must be considered as an expression artefact, since it corresponds to the C-terminal residue of the signal peptide. Some electron density is also present for the preceding His0, although this residue, which is also an expression artefact, was not modeled. Two C-terminal residues present in the expressed protein, Asp164 and Glu165, were disordered both in the original structure and in the coordinates described here, and thus were not included in the models. Superposition of the previous and the current sets of coordinates of rBbKI yielded an r.m.s.d. of 0.144 Å for all 163 Cα atoms found in the two models, although two Ramachandran outliers present in the older structure have been corrected. Thus, the tracing of the main chain is identical in the two structures within the estimated coordinate error, and the orientations of almost all of the side chains are also very similar. One major exception is Phe157, which is unambiguously present in a single orientation in the lower resolution structure but has two different orientations in the structure described here. Other residues with double orientations of parts of the side chains in the high-resolution structure are Ser13, His26, Leu47, Ile68, Thr103, Asp104, Ser107, Ile123, Glu128, Thr132, Cys155 and Arg160. The main chain in the vicinity of Arg64, which resides in the reactive loop and is responsible for the specificity of this class of inhibitors for trypsin-like enzymes (Bhattacharjee et al., 2014 ▸), is found in two distinct but similar conformations, whereas the conformation of the side chain is virtually identical. Unlike in the previous structure, however, the side chain of Arg64 shows only one orientation starting at Cβ, whereas previously two similar but distinct orientations of the side chain were modeled.
The positions of water molecules were determined independently in the two studies, but nevertheless are very similar. The number of water molecules in the previously determined structure is 159, whereas 213 water molecules were identified in the present study. These numbers are within the expected ranges for the structures determined at different resolutions. For 128 water molecules located within 1.4 Å of each other in the two structures the r.m.s.d. is 0.248 Å, indicating very good reproducibility of the placement of water molecules which are, unlike the polypeptide chain, not subject to geometrical restraints other than van der Waals interactions (hydrogen-bonded interactions are not directly restrained).
3.2. rBbKI is a typical β-trefoil protein, but lacks disulfide bridges
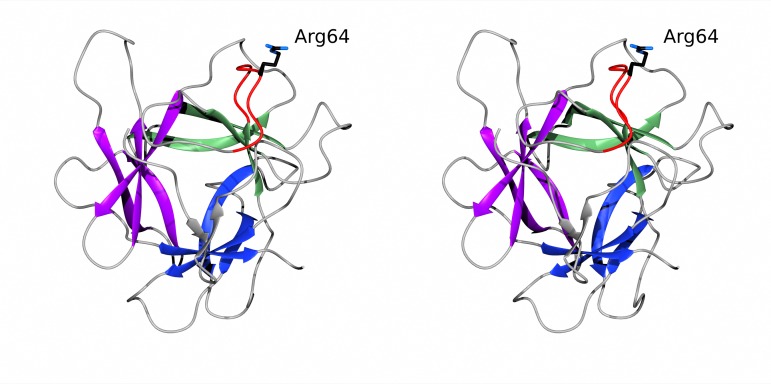
The fold of rBbKI is defined as a β-trefoil, a very common polypeptide framework found not only in protease inhibitors but also in a variety of lectins, cytokines, actin cross-linking proteins and DNA-binding proteins (Renko et al., 2012 ▸). This class of proteins contains a structurally conserved core (although the conservation of the sequence may be very low) and variable loops (Murzin et al., 1992 ▸). The core of the BbKI molecule is composed of six β-strands forming a barrel (18–23, 57–60, 73–78, 112–117, 120–125 and 157–161) and three hairpins (30–34 and 44–48, 87–93 and 96–103, and 134–141 and 144–149) exhibiting pseudo-threefold symmetry (Fig. 2 ▸).
Figure 2.
A stereoimage of BbKI viewed along the pseudo-threefold axis. The three β-sheets forming the core of a β-trefoil are colored blue, purple and green. The reactive loop responsible for the inhibitory properties of the inhibitor is red, and the specificity-determining Arg64 is shown in stick representation.
It has been suggested that the β-trefoil fold originated from two successive gene-duplication events of an ancestral protein (Mukhopadhyay, 2000 ▸). Evolutionary events and selective pressures led to many differences in the resulting proteins, generating a sequential hypervariability in functional regions (Mukhopadhyay, 2000 ▸). For example, plant Kunitz-type inhibitors such as soybean trypsin inhibitor (STI; Sweet et al., 1974 ▸) and trypsin inhibitors from Erythrina caffra (ETI; Onesti et al., 1991 ▸) and Enterolobium contortisiliquum (EcTI; Zhou et al., 2013 ▸), as well as the bifunctional inhibitor/lectin from Crataeva tapia (Ferreira et al., 2013 ▸), show the same β-trefoil fold as interleukin-1β and interleukin-1α (Graves et al., 1990 ▸; Priestle et al., 1988 ▸) and fibroblast growth factors (Zhu et al., 1991 ▸). Although these proteins have very similar tertiary structures, their sequences exhibit no significant similarities (Murzin et al., 1992 ▸). In fact, during the course of evolution, the three-dimensional structure of proteins is more conserved than their primary structure. Observations of the β-trefoil protein structure show that this type of conformation is very well preserved; changes in some specific regions give the immense variety of functions. Thus, Kunitz-type inhibitors impair the functionality of proteases, interleukins mediate the immune response and plant toxins act in defense against predatory organisms, but all of these functions are involved in recognition.
3.3. A comparison with the cysteine-less inhibitor BbCI
The structure of only one other disulfide-free protease β-trefoil inhibitor, rBbCI, has previously been published. Although the amino-acid sequence of rBbCI is very similar to that of rBbKI, with 82% identity, the specificities of the two inhibitors are very different. Whereas rBbKI inhibits trypsin-like serine proteases, in particular plasma kallikrein and plasmin, BbCI is primarily specific for cruzipain, a cysteine protease isolated from Trypanosoma cruzi. However, the specificity of BbCI is not limited to cysteine proteases, since it can also inhibit serine proteases such as porcine pancreatic elastase, human neutrophil elastase and cathepsin L, although with lower effect (Table 2 ▸).
Table 2. Inhibitory properties of rBbKI and its R64A mutant, compared with rBbCI.
K i,app values are shown in nM. n.i. indicates a lack of measurable inhibition.
| rBbKI | rBbKI-R64A | rBbCI | |
|---|---|---|---|
| Human plasma kallikrein | 2.0 | 98 | n.i |
| Bovine trypsin | 20 | 25 | n.i. |
| Bovine chymotrypsin | n.i. | n.i. | n.i. |
| Human plasmin | 33 | 2.6 | n.i. |
| Cathepsin L | n.i. | n.i. | 9.0 |
| Cruzain | n.i. | n.i. | 0.3 |
| Bovine pancreatic elastase | n.i. | n.i. | 47 |
| Human neutrophil elastase | n.i. | n.i. | 1.7 |
The published structure of BbCI (Hansen et al., 2007 ▸), determined at 1.7 Å resolution and deposited in the PDB as entry 2gzb, contains two molecules in the asymmetric unit. Since these two molecules are virtually identical (r.m.s.d. of 0.21 Å for all 164 Cα atoms), we used molecule A for comparison with the structure of rBbKI. Superposition of these two proteins using the SSM module of Coot results in an r.m.s.d. of only 1.15 Å for 159 Cα atoms. However, several loops exhibit very significant differences between these two closely related proteins (Fig. 3 ▸). These loops include residues 24–29 (position of Gly27 shifted by 5.9 Å), loop 78–87 (position of Ser82 shifted by as much as 9.5 Å), loop 138–146 (position of Asn142 shifted by 3.3 Å) and loop 150–154 (position of Cα of Gly152 shifted by 2.9 Å). The loop 24–29 is not involved in forming crystal contacts in any of the structures; thus, the differences in the loop are likely to be intrinsic properties of this structural element. Loop 78–87 makes no crystal contacts in rBbKI structure, but it forms a hydrogen bond between the O atom of Ser80 and the symmetry-related NH2 atom of Arg57 in the BbCI structure. Glycines 79 and 86 of rBbKI correspond to larger residues in BbCI, Val and Glu, respectively, and these significant differences may also be responsible for the variations in the structures. Loop 138–146 makes no crystal contacts in the rBbKI structure, but has a salt bridge between Arg140 and the symmetry-related Glu152 in the BbCI structure. Differences in loop 138–146 are present even though the two sequences are identical. The carbonyl of Gly152 in loop 150–154 makes a hydrogen bond to the NE2 atom of a symmetry-related His51 and a salt bridge to a symmetry-related guanidine group of Arg52 in the rBbKI structure and a salt bridge with a symmetry-related guanidine group of Arg140 in the BbCI structure. Therefore, the differences in loops 78–87, 138–146 and 150–154 could be the result of complex interplay between intrinsic properties of these structural elements and crystallographic contacts.
Figure 3.
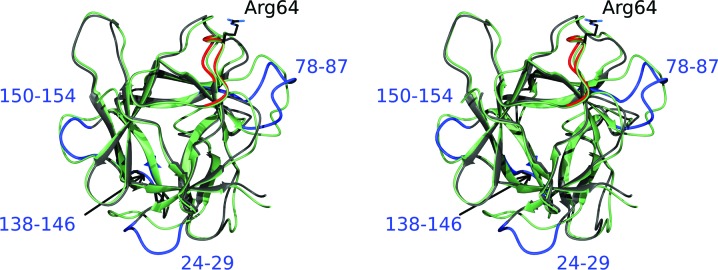
A stereoimage showing the superposition of the main chains of BbKI (grey) and BbCI (green) in an orientation similar to that in Fig. 2 ▸. The reactive loop of BbKI responsible for the inhibitory properties is shown in red; the specificity-determining Arg64 in BbKI is shown in stick representation. The four loops in BbKI that differ most from BbCI (see text for details) are shown in blue and labeled.
The sole cysteine residue present in rBbKI (Cys155) has two conformations and is located in a highly hydrophobic flexible environment consisting of the twofold disordered residues His26, Leu47 and Phe157, as well as Leu138 and Leu147 which are confidently modeled with one conformation each (Fig. 4 ▸). When compared with the equivalent areas in other related inhibitors, it can be seen that Cys155 in rBbKI is replaced by Phe in rBbCI and by Leu in EcTI. It can thus be concluded that the role of Cys155 is simply as another hydrophobic residue that has resulted from a conservative substitution of a hydrophobic residue for another residue.
Figure 4.
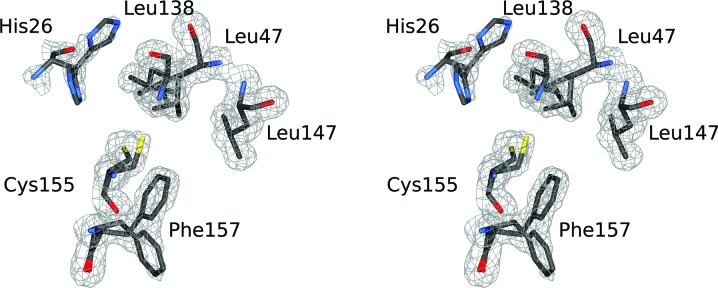
A stereoimage showing the hydrophobic environment of the sole cysteine in BbKI (Cys155).
The differences in the structures discussed above make only a secondary, if any, contribution to the specificity of the two inhibitors, since the most important structural difference is the replacement of Arg64 in rBbKI by an alanine in rBbCI. The side chain of this residue fills the S1 pocket of the trypsin-like proteases, which contain an aspartic acid at the bottom of the pocket, and thus provides the primary source of their specificity.
3.4. Modeling of the interactions between rBbKI and kallikrein
A model of the complex between plasma kallikrein and rBbKI was built on the basis of an experimentally determined structure of EcTI complexed with bovine trypsin (PDB entry 4j2y; Zhou et al., 2013 ▸). The coordinates of human plasma kallikrein refined at 1.4 Å resolution (PDB entry 2any; Tang et al., 2005 ▸) were superimposed on those of trypsin (r.m.s.d. of 1.15 Å for 214 Cα pairs), whereas rBbKI was separately superimposed on EcTI (r.m.s.d. of 1.26 Å for 146 Cα pairs). In the resulting model of the complex the specificity-determining residue in the reactive loop of rBbKI, Arg64, points into the middle of the β-sheet, between residues 196 and 213 of kallikrein, making impossibly short contacts with the main chain. It is very likely that the reactive loop will need to reposition itself by relatively small rotations involving the main-chain torsion angles of residues 63 and 65, as is also the case for EcTI, where the position of the side chain of Arg64 is quite different in the complex and in the free inhibitor (Zhou et al., 2013 ▸). A corresponding modification of the conformation of the reactive loop in rBbKI could be accomplished without deviation from the allowed Ramachandran angles and would lead to the creation of an ion pair between Arg64 of rBbKI and Asp189 of kallikrein.
A secondary contribution to the specific binding of rBbKI to kallikrein may be provided by the observed interactions between the N-terminus of rBbKI and its loop 69–71 and the 57–60 stretch of kallikrein, as well as residues 10–17 and 81–83 of rBbKI with several residues of kallikrein, including Lys192, Phe143, Glu146 and Lys147. Some adjustment of the relative positions of the residues involved in these interactions is however necessary. Lys106 of rBbKI could form an ion pair with Glu217 of kallikrein, also contributing to the strong binding of these two proteins. However, the details of the interactions between the enzyme and its specific inhibitor can only be fully evaluated by experimental determination of the structure of the complex.
3.5. Inhibitory properties of wild-type and mutated rBbKI
The inhibitory properties of the wild type and the R64A mutant of rBbKI were tested against several proteolytic enzymes and compared with the properties of the closely related BbCI. The purpose of the mutation was to investigate whether the removal of the P1 Arg and its replacement by the alanine that is found in BbCI would make the properties of the two inhibitors more similar. The values of the apparent K i (K i,app) are summarized in Table 2 ▸. As expected, the inhibition constant of the mutant for human plasma kallikrein was much higher than that of the wild-type protein, although rBbKI-R64A is still a comparatively good inhibitor of this enzyme, whereas rBbCI does not inhibit it at all. However, the R64A mutant became a potent inhibitor of plasmin and, although it lost its potency against kallikrein, it still remained a relatively good inhibitor of the latter enzyme. Thus, this mutant may prevent the breakup of fibrin and maintain clot stability, preventing excessive bleeding, in a manner similar to the mode of action of aprotinin (Henry et al., 2009 ▸), another proteinaceous inhibitor of serine proteases which acts on both plasmin and kallikrein. More surprising was the almost unchanged inhibition of bovine trypsin by rBbKI-R64A despite the lack of the specificity-determining basic P1 residue. Trypsin was not inhibited by rBbCI, and the cysteine proteases cathepsin L and cruzain, which are strongly inhibited by rBbCI, were not inhibited by the rBbKI mutant. These results show that despite the overall 84% sequence identity between BbCI and BbKI, removal of the basic P1 side chain does not switch the specificity of the latter inhibitor to make it more similar to the former. However, identification of the other residues that determine the specificity of these inhibitors may require determination of the structures of their complexes with the target proteases. Studies of the structure of the complex between rBbKI and the catalytic domain of human plasma kallikrein (Tang et al., 2005 ▸) are currently under way.
Supplementary Material
PDB reference: BbKI, 4zot
Acknowledgments
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT) located at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and in part by CAPES, CNPq and FAPESP 2009/53766-5.
References
- Araújo, A. P. U., Hansen, D., Vieira, D. F., Oliveira, C., Santana, L. A., Beltramini, L. M., Sampaio, C. A. M., Sampaio, M. U. & Oliva, M. L. V. (2005). Biol. Chem. 386, 561–568. [DOI] [PubMed]
- Batista, I. F. C., Oliva, M. L. V., Araujo, M. S., Sampaio, M. U., Richardson, M., Fritz, H. & Sampaio, C. A. M. (1996). Phytochemistry, 41, 1017–1022. [DOI] [PubMed]
- Bhattacharjee, N., Banerjee, S. & Dutta, S. K. (2014). Protein Expr. Purif. 96, 26–31. [DOI] [PubMed]
- Birk, Y. (2003). Plant Protease Inhibitors. Berlin: Springer Verlag.
- Botos, I. & Wlodawer, A. (2007). Curr. Opin. Struct. Biol. 17, 683–690. [DOI] [PMC free article] [PubMed]
- Brito, M. V., de Oliveira, C., Salu, B. R., Andrade, S. A., Malloy, P. M. D., Sato, A. C., Vicente, C. P., Sampaio, M. U., Maffei, F. H. A. & Oliva, M. L. V. (2014). Thromb. Res. 133, 945–951. [DOI] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Ferreira, R. da S., Zhou, D., Ferreira, J. G., Silva, M. C. C., Silva-Lucca, R. A., Mentele, R., Paredes-Gamero, E. J., Bertolin, T. C., dos Santos Correia, M. T., Paiva, P. M. G., Gustchina, A., Wlodawer, A. & Oliva, M. L. (2013). PLoS One, 8, e64426. [DOI] [PMC free article] [PubMed]
- Graves, B. J., Hatada, M. H., Hendrickson, W. A., Miller, J. K., Madison, V. S. & Satow, Y. (1990). Biochemistry, 29, 2679–2684. [DOI] [PubMed]
- Hansen, D., Macedo-Ribeiro, S., Veríssimo, P., Yoo Im, S., Sampaio, M. U. & Oliva, M. L. V. (2007). Biochem. Biophys. Res. Commun. 360, 735–740. [DOI] [PubMed]
- Henry, D., Carless, P., Fergusson, D. & Laupacis, A. (2009). Can. Med. Assoc. J. 180, 183–193. [DOI] [PMC free article] [PubMed]
- Krishnaswamy, S. (2005). J. Thromb. Haemost. 3, 54–67. [DOI] [PubMed]
- Kunitz, M. (1947). J. Gen. Physiol. 30, 311–320. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Morrison, J. F. (1982). Trends Biochem. Sci. 7, 102–105.
- Mukhopadhyay, D. (2000). J. Mol. Evol. 50, 214–223. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Murzin, A. G., Lesk, A. M. & Chothia, C. (1992). J. Mol. Biol. 223, 531–543. [DOI] [PubMed]
- Nakahata, A. M., Mayer, B., Ries, C., de Paula, C. A. A., Karow, M., Neth, P., Sampaio, M. U., Jochum, M. & Oliva, M. L. V. (2011). Biol. Chem. 392, 327–336. [DOI] [PubMed]
- Navarro, M. V. A. S., Vierira, D. F., Nagem, R. A. P., de Araújo, A. P. U., Oliva, M. L. V. & Garratt, R. C. (2005). Acta Cryst. F61, 910–913. [DOI] [PMC free article] [PubMed]
- Neuhof, C., Oliva, M. L. V., Maybauer, D., Maybauer, M., de Oliveira, C., Sampaio, M. U., Sampaio, C. A. M. & Neuhof, H. (2003). Biol. Chem. 384, 939–944. [DOI] [PubMed]
- Odei-Addo, F., Frost, C., Smith, N., Ogawa, T., Muramoto, K., Oliva, M. L. V., Gráf, L. & Naude, R. (2014). J. Enzyme Inhib. Med. Chem. 29, 633–638. [DOI] [PubMed]
- Oliva, M. L. V., Ferreira, R. da S., Ferreira, J. G., de Paula, C. A. A., Salas, C. E. & Sampaio, M. U. (2011). Curr. Protein Pept. Sci. 12, 348–357. [DOI] [PubMed]
- Oliva, M. L. V., Mendes, C. R., Santomauro-Vaz, E. M., Juliano, M. A., Mentele, R., Auerswald, E. A., Sampaio, M. U. & Sampaio, C. A. M. (2001). Curr. Med. Chem. 8, 977–984. [DOI] [PubMed]
- Oliva, M. L. V. & Sampaio, U. M. (2008). Biol. Chem. 389, 1007–1013. [DOI] [PubMed]
- Oliva, M. L. V., Silva, M. C. C., Sallai, R. C., Brito, M. V. & Sampaio, M. U. (2010). Biochimie, 92, 1667–1673. [DOI] [PubMed]
- Oliveira, C. de, Santana, L. A., Carmona, A. K., Cezari, M. H., Sampaio, M. U., Sampaio, C. A. M. & Oliva, M. L. V. (2001). Biol. Chem. 382, 847–852. [DOI] [PubMed]
- Onesti, S., Brick, P. & Blow, D. M. (1991). J. Mol. Biol. 217, 153–176. [DOI] [PubMed]
- Ota, N., Stroupe, C., Ferreira-da-Silva, J. M. S., Shah, S. A., Mares-Guia, M. & Brunger, A. T. (1999). Proteins, 37, 641–653. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pampalakis, G. & Sotiropoulou, G. (2007). Biochim. Biophys. Acta, 1776, 22–31. [DOI] [PubMed]
- Priestle, J. P., Schär, H.-P. & Grütter, M. G. (1988). EMBO J. 7, 339–343. [DOI] [PMC free article] [PubMed]
- Renko, M., Sabotič, J. & Turk, D. (2012). Biol. Chem. 393, 1043–1054. [DOI] [PubMed]
- Sałaga, M., Sobczak, M. & Fichna, J. (2013). Drug Discov. Today, 18, 708–715. [DOI] [PubMed]
- Souza-Pinto, J. C., Oliva, M. L. V., Sampaio, C. A. M., Damas, J., Auerswald, E. A., Limãos, E., Fritz, H. & Sampaio, M. U. (1996). Immunopharmacology, 33, 330–332. [DOI] [PubMed]
- Sweet, E. M., Wright, H. T., Janin, J., Chothia, C. H. & Blow, D. M. (1974). Biochemistry, 13, 4212–4228. [DOI] [PubMed]
- Tang, J., Yu, C. L., Williams, S. R., Springman, E., Jeffery, D., Sprengeler, P. A., Estevez, A., Sampang, J., Shrader, W., Spencer, J., Young, W., McGrath, M. & Katz, B. A. (2005). J. Biol. Chem. 280, 41077–41089. [DOI] [PubMed]
- Turk, B. (2006). Nature Rev. Drug Discov. 5, 785–799. [DOI] [PubMed]
- Vadivel, K., Ponnuraj, S. M., Kumar, Y., Zaiss, A. K., Bunce, M. W., Camire, R. M., Wu, L., Evseenko, D., Herschman, H. R., Bajaj, M. S. & Bajaj, S. P. (2014). J. Biol. Chem. 289, 31647–31661. [DOI] [PMC free article] [PubMed]
- Zhou, D., Lobo, Y. A., Batista, I. F., Marques-Porto, R., Gustchina, A., Oliva, M. L. & Wlodawer, A. (2013). PLoS One, 8, e62252. [DOI] [PMC free article] [PubMed]
- Zhu, X., Komiya, A., Chirino, A., Faham, S., Fox, G. M., Arakawa, T., Hsu, B. T. & Rees, D. C. (1991). Science, 251, 90–93. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: BbKI, 4zot