Abstract
Interleukin-33 (IL-33) is a novel member of the IL-1 family of cytokines that plays diverse roles in the regulation of immune responses. IL-33 exerts its effects through a heterodimeric receptor complex resulting in the production and release of pro-inflammatory cytokines. A detailed understanding of the signaling pathways activated by IL-33 is still unclear. To gain insights into the IL-33 mediated signaling mechanisms, we carried out a SILAC-based global quantitative phosphoproteomic analysis that resulted in the identification of 7,191 phosphorylation sites derived from 2,746 proteins. We observed alterations in the level of phosphorylation in 1,050 sites corresponding to 672 proteins upon IL-33 stimulation. We report, for the first time, phosphorylation of multiple protein kinases, including Mitogen-activated protein kinase-activated protein kinase 2 (Mapkapk2), Receptor (TNFRSF)-interacting serine-threonine kinase 1 (Ripk1) and NAD kinase (Nadk) that are induced by IL-33. In addition, we observed IL-33-induced phosphorylation of several protein phosphatases including Protein tyrosine phosphatase, Non-receptor type 12 (Ptpn12) and Inositol polyphosphate-5-phosphatase D (Inpp5d), which have not been reported previously. Network analysis revealed an enrichment of actin binding and cytoskeleton reorganization that could be important in macrophage activation induced by IL-33. Our study is the first quantitative analysis of IL-33-regulated phosphoproteome. Our findings significantly expand the understanding of IL-33 mediated signaling events and have the potential to provide novel therapeutic targets pertaining to immune related diseases such as asthma where dysregulation of IL-33 is observed.
Keywords: inflammation, immune disorders, immunoaffinity purification, RhoGTPases
1. Introduction
Interleukin-33 (IL-33) is a member of the interleukin-1 (IL-1) family of cytokines and has been associated primarily with the initiation and propagation of the T helper 2 (Th2) immune responses [1]. IL-33 contains an N-terminal nuclear localization signal (NLS), a helix-turn-helix (HTH) motif and a C-terminal region, which has structural homology to IL-1 cytokine family [2]. IL-33 mediates its effects through a heterodimeric receptor complex consisting of interleukin-1 receptor like 1 (IL1RL1) and an accessory receptor protein, IL-1RAcP. It is constitutively expressed in multiple cell types including endothelial cells, epithelial lining of the gut, lung, smooth muscle cells and adipocytes among others [1-4]. Similar to IL-1, IL-33 may be secreted through unconventional mechanisms or upon cellular damage and necrosis [5] and acts on a number of different cell types resulting in cell-type specific signaling. IL-33 is known to initiate a Th2 response independent of T-cell receptor triggering [1, 6]. In mast cells, IL-33 induces the secretion of chemokines and cytokines such as IL-6, IL-8, and IL-13 [7]. It activates dendritic cells which in turn promotes T cell proliferation and Th2 polarization [8], stimulates B1 cell proliferation [9] and recently has been shown to promote CD8+ T cell responses and thus direct Th1 responses as well [10, 11]. Apart from the role of IL-33 as a potent mediator in inflammatory responses, studies have also shown a cardioprotective role for IL-33 [12].
The exact mechanisms by which IL-33 exerts its effects are yet not fully established. It is believed that IL-33 acts in a manner similar to that of the IL-1 family of proteins [1]. IL-33 binds to the heterodimeric receptor complex, recruits adaptor proteins - MyD88 and the associated protein, IL-1R-associated kinase (IRAK), resulting in the activation of downstream mitogen-activated protein kinases (MAPK) and nuclear factor-kappa B (NF-κB) through TNF receptor-associated factor 6 (TRAF6). IL-33 is also known to increase the phosphorylation of ERK1/2, JNK1/2, AKT, JAK2 and SYK [1, 13] resulting in the production and release of pro-inflammatory cytokines. Although the involvement of IL-33 has been well studied in several diseases including asthma [14, 15], atopic dermatitis [16, 17], fibrosis [18-20], cancer and ulcerative colitis [21, 22], a detailed understanding of the pathways regulated by IL-33 still remains elusive. Therefore, characterization of the IL-33 mediated signaling mechanisms is essential to improve therapeutic modalities in these diseases.
To gain further insights into the IL-33 signaling network, we carried out quantitative phosphoproteomic analysis using SILAC to identify IL-33 mediated phosphorylation changes in a mouse macrophage cell line (RAW264.7). Macrophages are sentinels of the immune system and several immune-related disorders are associated with altered macrophage function [23]. The effects of IL-33 on human and mouse macrophages have been well documented. These cells are reported to constitutively express ST2L [24, 25] and amplify the expression of M2 markers resulting in an enhanced Th2 immune responses [25]. It is also known to increase LPS-induced production of TNF-α in macrophages. Therefore, we used macrophage cell line as a prototype to study IL-33 signaling. By employing metal-oxide affinity chromatography (TiO2 enrichment) and anti-phosphotyrosine antibody-based phosphopeptide enrichment, we identified 7,191 phosphorylation sites from 8,442 phosphopeptides. We report the identification of known downstream effectors including Erk1/2 and p38. In addition, our study led to the identification of several novel molecules that have not been reported previously to be regulated by IL-33. Our analysis also revealed enrichment of proteins involved in cytoskeleton reorganization and actin binding that have not been previously linked to IL-33 signaling suggesting a role for IL-33 signaling in macrophage activation. Thus, our study provides evidence of novel signaling modules regulated by IL-33.
2. Materials and methods
2.1 Reagents
DMEM with and without lysine and arginine, fetal bovine serum (FBS), L-glutamine, and antibiotics were purchased from Invitrogen (Carlsbad, CA). SILAC amino acids, 13C6-Lysine and 13C6-Arginine, were obtained from Cambridge Isotope Laboratories (Andover, MA). Recombinant IL-33 was purchased from R&D Systems (Minneapolis, MN). TPCK-treated trypsin was from Worthington Biochemical Corp. (Lakewood, NJ). Titansphere (TiO2, 5 μm beads) were from GL Sciences Inc. (Torrance, CA). 4G10 anti-phosphotyrosine (HRP conjugated) antibody was purchased from Millipore (Billerica, MA). Anti-phospho-p44/42 MAPK (Erk1/2) (T202/Y204), anti-phospho-MAPKAPK-2 (T334), anti-phospho-SAPK/JNK (T183/Y185), anti-phospho p38MAPK (T180/Y182), anti-p38MAPK, and anti- beta actin antibodies were all purchased from Cell Signaling Technology (Danvers, MA). For immunoaffinity purification of phosphopeptides, anti-phosphotyrosine rabbit monoclonal antibody (P-Tyr-1000) beads were obtained from Cell Signaling Technology (Danvers, MA). All other reagents used in this study were from Fisher Scientific (Pittsburgh, PA)
2.2 Cell culture and SILAC labeling
The murine macrophage cell line RAW264.7 was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin in a humidified incubator at 37 °C with 5.0% CO2. These cells were adapted to SILAC media as described earlier [26]. For SILAC labeling, the cells were maintained in DMEM without lysine and arginine supplemented with 5% FBS, 2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin, 50 mg/L arginine-12C6 monohydrochloride and 100 mg/L lysine-12C6 monohydrochloride (light) or 50 mg/L arginine-13C6 monohydrochloride and 100 mg/L lysine-13C6 monohydrochloride (heavy) (Cambridge Isotope Laboratories). Exponentially growing cells were washed six times with PBS and grown in serum free media for 12 h prior to stimulation with IL-33. Cells grown in 13C6-lysine/13C6-arginine-containing media were stimulated with IL-33 (100 ng/mL) (R&D Systems) for 10 min at 37 °C and cells grown in light medium were left unstimulated.
2.3 Cell lysis and protein digestion
The IL-33 stimulated and unstimulated RAW264.7 cells were washed with cold phosphate buffered saline and lysed in lysis buffer (20 mM HEPES pH8.0, 9 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate), sonicated and centrifuged at 16,000 × g at 15 °C for 20 min. The protein concentration was determined using BCA assay (Pierce, Waltham, MA). Equal amounts of protein (20 mg) were mixed, reduced using DTT at a final concentration of 5 mM at 60°C for 20 minutes and alkylated using 10mM iodoacetamide for 10 minutes at room temperature in the dark. The samples were then diluted such that urea was < 2M with 20 mM HEPES, pH 8.0 and subjected to digestion with TPCK treated trypsin (Worthington Biochemical Corp) overnight at room temperature. The peptide mixture was acidified using 1 % Triflouroacetic acid and desalted using C18 Sep-Pak cartridge (Waters, Cat#WAT051910). The extracted peptides were lyophilized and stored at -80°C until further analysis.
2.4 Immunoaffinity purification of tyrosine phosphopeptides
Immunoaffinity purification (IAP) of phosphopeptides was carried out as described previously [27]. Briefly, for each experiment, 30 mg of lyophilized peptide mixture was dissolved in 1.4 ml of IAP buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate, 50 mM NaCl) and the pH was adjusted to 7.2 using 1M Tris Base. Before IAP, the P-Tyr-1000 beads (Cell Signaling Technology) were washed with IAP buffer twice at 4 °C. The peptide mixture was then incubated with P-Tyr-1000 beads for 30 minutes with gentle rotation. The peptide bound beads were then washed three times with ice cold IAP buffer followed by two washes with ice cold water. Peptides were eluted twice from the beads with 0.15% TFA at room temperature and desalted using C18 StageTips as described earlier [28].
2.5 Basic pH reversed-phase liquid chromatography (bRPLC) and TiO2-based phosphopeptide enrichment
Peptides were fractionated by high pH reversed-phase liquid chromatography as described earlier [29]. Briefly, 10 mg of lyophilized peptides mixture was resuspended in 1 mL of bRPLC solvent A (10 mM TEABC pH 8.4, Sigma Aldrich) and fractionated by bRPLC chromatography on a XBridge C18, 5 μm, 250 × 4.6 mm column (Waters Corporation, Milford, MA) by employing an increasing gradient of bRPLC solvent B (10 mM TEABC in 90% acetonitrile, pH 8.4.) on an Agilent 1100 LC system with a flow rate of 1 mL/min. For each experiment, a total of 96 fractions were initially collected in 96- well plates with 0.1% formic acid added to each well. The fractions were then concatenated to 12 fractions in both the experiments and dried using speedvac. Each fraction was subjected to TiO2-based phosphopeptide enrichment. The TiO2 beads (Titansphere, GL Sciences Inc.) were incubated with DHB solution (80% ACN, 1% TFA, and 3% 2, 5-dihydroxybenzoic acid) for 1 h at room temperature. Each fraction was resuspended in 3% DHB solution and incubated with TiO2 beads at 1:1 ratio for 30 minutes at room temperature. Phosphopeptide-bound TiO2 beads were washed three times with DHB solution and twice with 40% ACN. Peptides were eluted three times with 40 μL of 2% ammonia solution into tubes containing 10 μL of 20% TFA on ice. The peptides were dried and resuspended in 30 μL of 0.1% TFA and desalted using C18 StageTips. The eluted peptides were subjected to LC-MS/MS analysis.
2.6 LC-MS/MS analysis of enriched peptides
The enriched phosphopeptides were analyzed on LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) interfaced with Easy-nLC II nanoflow liquid chromatography system (Thermo Scientific, Odense, Denmark). The peptide digests were reconstituted in 0.1% formic acid and loaded onto trap column (75 μm × 2 cm) packed in-house with Magic C18 AQ (Michrom Bioresources, Inc., Auburn, CA, USA). Peptides were resolved on an analytical column (75 μm × 20 cm) at a flow rate of 350 nL/min using a linear gradient of 10-35% solvent B (0.1% formic acid in 95% acetonitrile) over 80 minutes. The total run time including sample loading and column reconditioning was 120 min. Data dependent acquisition with full scans in 350-1700 m/z range was carried out using an Orbitrap mass analyzer at a mass resolution of 120,000 at 400 m/z. Fifteen most intense precursor ions from a survey scan were selected for MS/MS fragmented using HCD fragmentation with 32% normalized collision energy and detected at a mass resolution of 30,000 at 400 m/z. Dynamic exclusion was set for 30 seconds with a 10 ppm mass window. Internal calibration was carried out using lock mass option (m/z 445.1200025) from ambient air.
2.7 Data analysis
The mass spectrometry derived data were searched using MASCOT (Version 2.2.0) and SEQUEST search algorithms against a mouse RefSeq database (version 60 containing 27,798 entries with common contaminants) using Proteome Discoverer 1.4 (Version 1.4.0.288) (Thermo Fisher Scientific, Bremen, Germany). The search parameters for both algorithms included: carbamidomethylation of cysteine as a fixed modification; N-terminal acetylation, oxidation of methionine, phosphorylation at serine, threonine and tyrosine (+79.966 Da) and SILAC labeling (13C6) at lysine and arginine as variable modifications (6.02013 Da). MS/MS spectra were searched with a precursor mass tolerance of 10 ppm and fragment mass tolerance of 0.05 Da. Trypsin was specified as protease and a maximum of two missed cleavages were allowed. The data were searched against decoy database and the false discovery rate was set to 1% at the peptide level. The SILAC ratio for each phosphopeptide-spectrum match (phosphoPSM) was calculated by the quantitation node and the probability of phosphorylation for each Ser/Thr/Tyr site on each peptide was calculated by the phosphoRS 3.1 node in the Proteome Discoverer (version 1.4, Thermo Scientific). The SILAC ratios were normalized such that the log2 median ratio were 1. For further analysis, only those phosphopeptides with > 75% site localization probability were considered. The phosphorylation sites that were identified with >75% localization probability but were assigned to different site by the search algorithm were manually corrected based on the phosphoRS localization probability for a given residue. Peptides with ratios greater than 1.5-fold were considered as regulated and used for further bioinformatics analysis.
2.8 Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [30] (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000984.
2.9 Bioinformatics analysis
Molecular function of phosphoproteins was obtained from Panther Classification System [31]. KEGG [32] pathway mapping of IL-33-regulated phosphoproteins was performed using the DAVID bioinformatics functional annotation tool [33]. To analyze the predicted consensus phosphorylation motifs in the IL-33 regulated dataset, we used motif-x algorithm [34] to extract motifs. The parameters used were: (i) sequence window of 7 amino acids flanking each identified phosphorylation site. For phosphorylation sites that were localized at the region of the N- or C-termini, the surrounding sequence could not be extended and were excluded, (ii) the significance threshold was set to p < 0.02, (iii) the minimum occurrence of motifs was set to 10. All phosphorylation sites identified in this study (including phosphorylation sites that showed no change in response to IL-33) were used as background for this motif enrichment analysis.
2.10 Western Blot Analysis
Exponentially growing RAW264.7 cells were washed with PBS and cultured in serum free DMEM medium for 12 h. The cells were left untreated or stimulated with rIL-33 (100 ng/mL) at 37 °C for the time indicated in the figures. Cells were lysed in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate, and 1 mM sodium orthovanadate in the presence of protease inhibitors) followed by centrifugation. The protein lysates were resolved using SDS-PAGE and Western blotting was performed using phospho-specific antibodies followed by reprobing with antibodies against the corresponding proteins. β-actin was used as a loading control.
3. Results
To decipher the early signaling events occurring upon IL-33 stimulation, we studied the phosphorylation changes of Erk1/2 in a time dependent manner as it has been previously reported that IL-33 induces Erk phosphorylation in macrophages [24]. The cells were stimulated with 100 ng/mL of IL-33 for various times as indicated in Fig. 1A. We observed an increase in the phosphorylation levels of Erk1/2 upon IL-33 stimulation as early as 2 minutes with a peak signal at 10 minutes of stimulation (Fig. 1A). For further quantitative phosphoproteomic analysis, we focused on characterizing the molecular snapshot of changes at 10 min of IL-33 stimulation as a robust induction was observed at this time point. Additionally, to assess for changes in the tyrosine phosphorylation, we carried out an immunoblot analysis using phosphotyrosine antibody. We observed a very small increase in the level of tyrosine phosphorylation upon IL-33 stimulation (Fig. 1B) suggesting that tyrosine signaling by IL-33 was somewhat weak. The signaling mechanism of IL-33 has been presumed to be largely similar to IL-1, which implies mainly serine/threonine kinase-mediated signaling.
Figure 1. IL-33-induced phosphorylation.
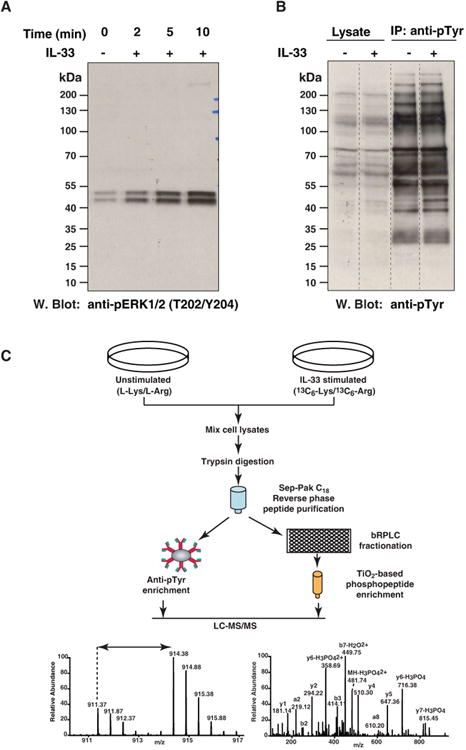
(A) Dose and time-dependent response to IL-33 stimulation. RAW264.7 cells were stimulated with 100 ng/ml IL-33 for the indicated times and cell lysates resolved by SDS-PAGE. Phosphorylation of Mapk1 and Mapk3 was probed by Western blotting with antibody that recognizes p-Mapk1 (T183/Y185) and p-Mapk3 (T203/Y205). The amount of total MAPK1 and MAPK3 were determined by reprobing the membrane with anti-MAPK antibody.
(B) Phosphotyrosine profile of IL-33 stimulated RAW264.7 cells was compared with that of unstimulated cells by Western blotting with an anti-phosphotyrosine antibody.
(C) Outline of the experimental strategy. RAW264.7 cells were cultured in “light” or “heavy” SILAC medium. The cells grown in heavy medium were stimulated with IL-33 for 10 min and the cells grown in light medium were left unstimulated. The samples were subjected to trypsin digestion and enriched for phosphopeptides using two approaches. One part of the peptide mixture was incubated with antiphosphotyrosine antibodies for enrichment of tyrosine-phosphorylated peptides. The other part was fractionated by bRPLC and phosphopeptides were enriched using TiO2 beads. The enriched phosphopeptides were analyzed by LC-MS/MS. The resulting high resolution mass spectra reveal IL-33-induced changes in the phosphorylation status on each site.
3.1 Quantitative phosphoproteomic analysis of IL-33 signaling
To identify IL-33 mediated phosphorylation changes, we carried out SILAC-based quantitative phosphoproteomic analysis. We performed two independent biological replicate experiments. In each replicate, the cells grown in ‘heavy’ SILAC media were stimulated for 10 minutes with IL-33 and the cells grown in ‘light’ media were left unstimulated. Equal amounts of lysates were pooled and subjected to in-solution trypsin digestion. The phosphopeptides were enriched by two different strategies - titanium dioxide (TiO2) chromatography [35] and immunoaffinity purification of phosphotyrosine containing peptides [27]. TiO2-based enrichment predominantly enriches pSerine/pThreonine containing peptides as they are more abundant; thus, to increase the identification of phosphotyrosine-containing peptides, we employed immunoaffinity purification. The enriched phosphopeptides were analyzed on LTQ-Orbitrap Elite mass spectrometer. The schematic workflow of SILAC-based IL-33 phosphoproteomics is shown in Fig. 1C.
A total of 26 LC-MS/MS runs from both the replicates were performed. The acquired data were processed and searched using MASCOT and SEQUEST search algorithms through Proteome Discoverer platform and stringently filtered for false discovery rate (FDR) of 1% at the peptide level. We identified 87,132 phosphopeptide-spectrum matches (Supporting Information Table S1) corresponding to a total of 10,265 phosphopeptides. A good correlation was observed between the replicate analysis for TiO2-based enrichment (Pearson correlation coefficient 0.76) and phosphotyrosine enrichment (Fig. 2A and Fig. 2B). Using the phosphoRS probability cutoff of 75% (hereafter referred to as class I phosphorylation sites), we identified 8,442 phosphopeptides that contained unambiguous localization of phosphorylation sites mapping to 2,746 proteins. Fig. 2C summarizes the results obtained from this analysis. We identified a total of 7,191 phosphorylation sites comprising of 5,237 serine, 861 threonine and 1,093 tyrosine sites. We identified 6,317 phosphopeptides with phosphorylation at serine residues and 1,038 phosphopeptides with threonine phosphorylation, the majority of which were obtained from TiO2-based enrichment method (Fig. 2D). We also identified 1,373 tyrosine-phosphorylated peptides mainly from anti-phosphotyrosine-based enrichment method. Analysis of the identified phosphopeptides revealed that most are singly phosphorylated (89%), and 10% of the phosphopeptides have two or more phosphorylation sites. Of these, ∼16% have not been previously reported (Supporting Information Table S2). We next analyzed the overlap of quantified class I phosphorylation sites among the biological replicates. Of the 7,191 phosphorylation sites identified, 6,806 were quantitated in at least one biological replicate.
Figure 2. Summary statistics of the analysis.
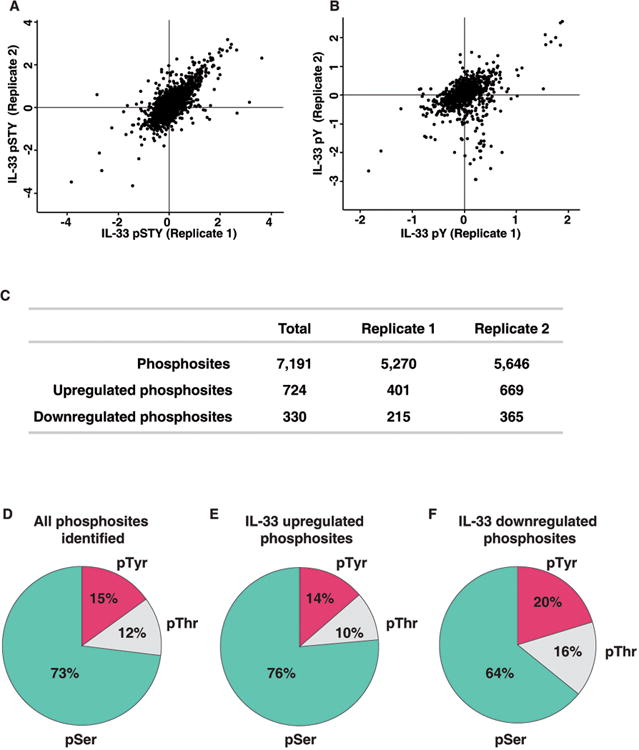
Correlation of the normalized SILAC ratio between replicate measurements of TiO2-based phosphopeptide enrichment (A) and anti-phosphotyrosine antibody enrichment method (B) (C) Summary of the number of phosphopeptides and class I phosphorylation sites that were identified and quantified in the two biological replicates.
(D) Distribution of pSerine, pThreonine and pTyrosine containing peptides.
(E) Distribution of pSerine, pThreonine and pTyrosine containing peptides in IL-33-upregulated phosphopeptides.
(F) Distribution of pSerine, pThreonine and pTyrosine containing peptides in IL-33-downregulated phosphopeptides.
Next, we looked at the phosphorylation sites that were differentially regulated upon IL-33 stimulation. Using a 1.5-fold cutoff for increased phosphorylation and a 0.67-fold cutoff for decreased phosphorylation following manual confirmation of the SILAC ratios, we identified 724 sites hyperphosphorylated and 330 hypophosphorylated upon IL-33 stimulation (Supporting Information Table S2, Fig. 2E and 2F). These hyperphosphorylated and hypophosphorylated sites corresponded to 488 proteins and 245 proteins, respectively. Based on our analysis, we observed that serine phosphorylation constitute a majority of the regulated sites (Fig. 2E and 2F).
3.2. Global view of IL-33 regulated phosphoproteome
To understand the role of the IL-33 regulated phosphoproteome, we carried out Gene Ontology (GO) analysis using PANTHER classification system. Our analysis revealed that most of the identified proteins were localized to the cytoskeleton followed by plasma membrane, nucleus and the cytosol. We next performed GO-based enrichment analyses to identify biological processes in which IL-33-regulated phosphorylation sites were significantly overrepresented. We considered those biological processes that exhibited greater than 1.5-fold enrichment for regulated phosphorylation sites in at least one of the two replicates. We observed that the majority of the IL-33 regulated phosphoproteome were involved in a broad spectrum of molecular functions (Supporting Information Fig. S1A, S1C). Majority of the proteins were found to be involved in catalytic activity, binding activity, and enzyme regulator activity. The classification based on the biological processes revealed a vast majority of the proteins regulated by IL-33 were involved in metabolic processes, followed by cellular process and biological regulation (Supporting Information Fig. S1B, S1D).
Additionally, our analysis led to identification of 44 protein kinases and 5 protein phosphatases that are regulated by IL-33 (Fig. 3, Table1, and Supporting Information Table S2). Of the protein kinases regulated by IL-33, we identified several members of the MAPK family. Some of these kinases have been previously implicated in IL-33 signaling. In addition, we also identified several adaptor proteins, members of the dedicator of cytokinesis protein family, Rho GTPases, guanine exchange nucleotide factors (GEFs) and several membrane bound receptors that have not been previously reported in IL-33 signaling (Fig. 3).
Figure 3. Concise map of IL-33 signaling.
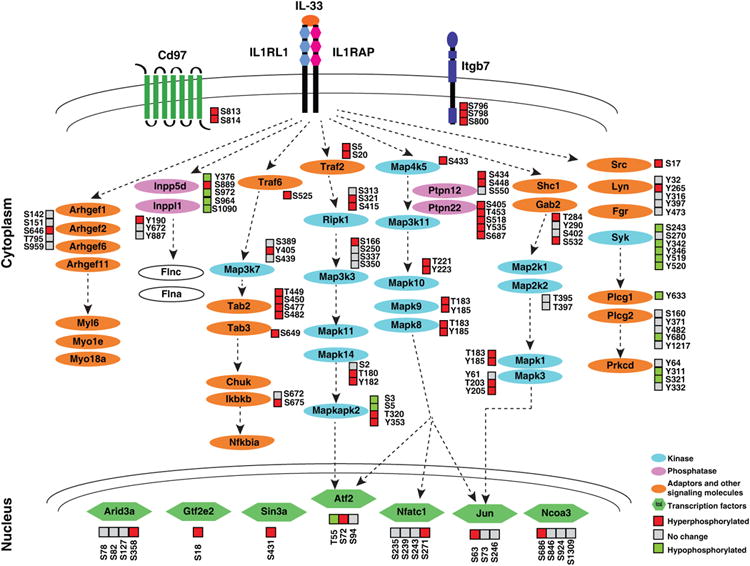
Overview of phosphoproteins regulated by IL-33 stimulation. The proteins identified from this study and previous studies have been combined to create a signaling map. The regulation of phosphorylation sites by IL-33 is indicated as increased (red), unchanged (gray) or decreased (green).
Table 1. A partial list of kinases/phosphatases modulated by IL-33 stimulation.
| Gene Symbol | Protein | Site | Phosphopeptide sequence | IL-33 stimulated/unstimulated |
|---|---|---|---|---|
| Mapk1 | Mitogen-activated protein kinase 1 (Erk2) | T183, Y185 | VADPDHDHTGFLtEyVATR | 3.6 |
| Mapk3 | Mitogen-activated protein kinase 3 (Erk1) | T203, Y205 | IADPEHDHTGFLtEyVATR | 2.8 |
| Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Y405 | IVATAAySKPK | 3.9 |
| Mapkapk2 | MAP kinase-activated protein kinase 2 | T320 | VPQtPLHTSR | 6.6 |
| Nadk | NAD kinase | S64 | sLHGPCPVTTFGPK | 2.4 |
| Ripk1 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 | S415 | RVsHDPFAQQR | 3.0 |
| Inpp5d | Inositol polyphosphate-5-phosphatase D | S889 | NLTsHDPMR | 4.9 |
| Psph | Phosphoserine phosphatase | S3 | MVsHSELR | 2.8 |
| Ppp1r12a | Protein phosphatase 1, regulatory (inhibitor) subunit 12A | S507 | LAsTSDIEEKENR | 2.0 |
| Ppp2r5d | Protein phosphatase 2, regulatory subunit B (B56), delta | S565 | RKsELPQDVYTIK | 2.2 |
To obtain information regarding the kinases that are responsible for these induced phosphorylation events, we used the motif-x algorithm to identify phosphorylation motifs in IL-33 mediated signaling. The hyperphosphorylated and hypophosphorylated peptides were compared to the background dataset derived from the whole phosphoproteome identified in our analysis. A sequence window of ±6 amino acids around the identified phosphorylated residue was considered. As shown in Fig. 4A, the motif-x algorithm identified 6 distinct phosphorylation motifs from the upregulated phosphopeptides. Our analysis revealed identification of motifs including four arginine-directed motifs (at -3 position) and two leucine-directed motifs (at -5 or +1 position).
Figure 4. Western blot validation and motif analysis.
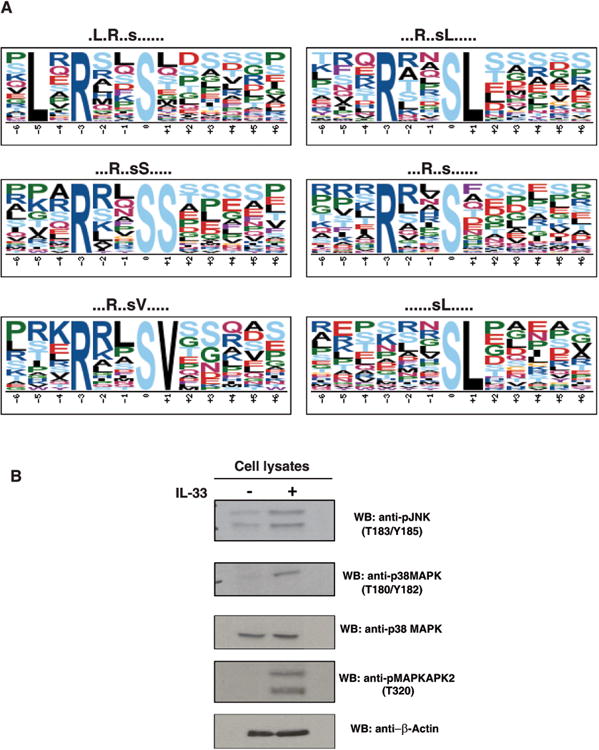
(A) IL-33-induced differentially regulated motifs. The motifs that were identified to be enriched in IL-33-induced differentially regulated phosphorylation sites dataset are depicted.
(B) Proteins identified to be differentially regulated by IL-33 from mass spectrometry data were validated by Western blot. RAW264.7 cell lysates were treated with anti-phospho antibodies for JNK (T183/Y185), p38MAPK (T180/Y182) and MAPKAPK2 (T320). Western blot confirmed the phosphorylation status of these proteins.
3.2 Recapitulation of known signaling molecules of the IL-33 pathway
It has been shown that upon IL-33 addition, many proteins including members of the interleukin-1 receptor associated kinase (IRAK) family- IRAK1 and IRAK4, adaptor proteins such as MyD88, TRAF6, members of the MAPK family and NF-κB [13, 36-39] are activated and initiate downstream signaling. Our quantitative mass spectrometry-based approach identified regulated phosphorylation of most of these proteins. We identified a four-fold increase in the phosphorylation of S525 on TRAF6. Phosphorylation of this site has not been reported previously; however, it is well known that TRAF6 is a key signal transducer in IL-33 mediated signal transduction [13, 37]. We observed an increased phosphorylation of T203/Y205 and T183/Y185 residues in Erk1 (3-fold) and Erk2 (3.6-fold), respectively (Supporting Information Fig. S2A), which is in agreement with our initial immunoblot analysis thus confirming that RAW264.7 cells indeed respond to IL-33. Consistent with previous studies, we also observed an increased phosphorylation of the conserved T183/Y185 residues in JNK (4-fold), the conserved T180/Y182 residues in p38 MAPK (3.3-fold) (Supporting Information Fig. S2B and S2C) and Map3k7 (Supporting Information Fig. S2D). Western blot analysis confirmed the phosphorylation dynamics of p38 Mapk, and Jnk (Fig. 4B). In addition, we also observed IL-33 mediated increased phosphorylation changes in c-Jun at S63, (2-fold) and Atf2 (3-fold) (Supporting Information Fig. S2E and S2F) that are known downstream effectors of p38Mapk signaling. Overall, our results suggest activation of the Erk1/2 and Jnk/c-Jun signaling pathways by IL-33 in murine macrophages, which is consistent with previous studies. Our analysis thus indicates the robustness of this approach in identifying downstream effectors of IL-33R.
3.3 Identification of novel signaling molecules regulated by IL-33
In addition to identifying several known molecules that are regulated by IL-33, we also identified proteins that are differentially regulated but have not been previously associated with IL-33 signaling. The majority of newly identified hyperphosphorylated kinases belong to the serine/threonine family of kinases including MKK3 (Map2k3), ERK3 (Mapk12) and MK2 (Mapkapk2). Among the kinases, we observed 3-fold higher phosphorylation of receptor-interacting protein (Ripk1) at S415 upon IL-33 stimulation (Fig. 5A). RIPK1 is a serine/threonine kinase that mediates cellular response to stress. It plays a key role in activation of the transcription factor NF-κB and is required for TNFα, TLR3, and DNA damage-induced NF-κB activation [40, 41]. Although phosphorylation of this site is known, its significance and role in cellular signaling is yet to be explored. We also identified several known substrates of p38 MAPK including MAP kinase-activated protein kinase 2 (Mapkapk2) also known as MK2. T320, which corresponds to T334 in humans, has been reported to be directly phosphorylated by p38 Mapk [42, 43]. In our data, we found Mapkapk2 to be hyperphosphorylated by 6-fold upon IL-33 stimulation (Fig. 5B) and the results were consistent with western blot analysis (Fig. 4B).
Figure 5. Novel molecules identified in IL-33 signaling pathway.
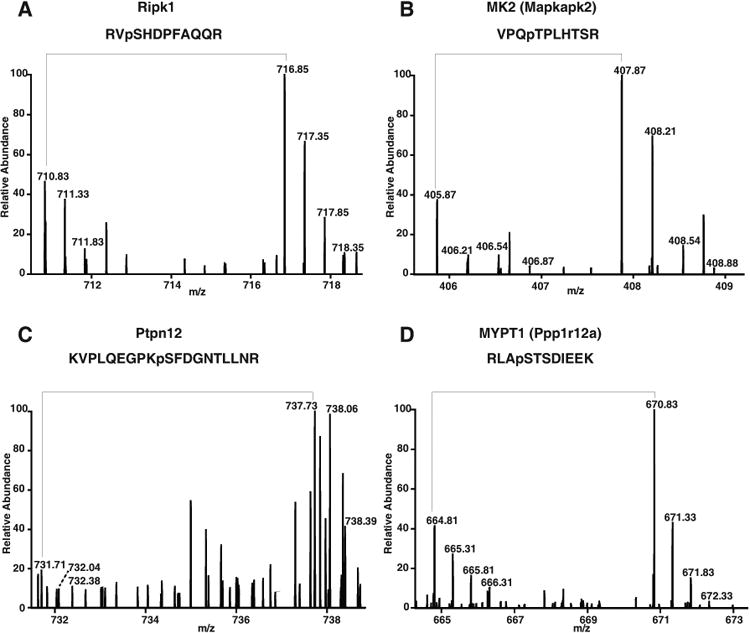
Representative MS spectra of novel phosphorylated kinases/phosphatases. A–D, phosphorylation of peptides on kinases (Ripk1 and Mapkapk2) and phosphatases (Ptpn12 and Ppp1r12a) was upregulated as evidenced by MS spectra showing the changes in the relative abundance of phosphopeptides.
Interestingly, from our analysis we observed serine and threonine residues in several tyrosine phosphatases including Ptpn12, Ptpn22, and Innp5d to be hyperphosphorylated upon IL-33 stimulation. Ptpn22, a non-receptor tyrosine phosphatase, was identified with 6 phosphorylation sites of which 2 sites- T453 and S558 were observed to be hyperphosphorylated 3-fold and 1.6-fold respectively upon IL-33 stimulation. In addition, we identified a 5-fold hyperphosphorylation of Ptpn12 at S448 (Fig. 5C). Protein phosphatase 1 regulatory subunit 12A (Ppp1r12a) also known as myosin phosphatase target subunit 1 (MYPT1 or MBS) is a 110 kDa target/regulatory subunit of the myosin phosphatase. We observed hyperphosphorylation of two sites - T694 and S507 on Mypt1 upon IL-33 stimulation (Fig. 5D). Phosphorylation of Mypt1 at T696 results in the inhibition of myosin phosphatase activity[44]. Ppp1r12c also known as MBS85 encodes a protein that in addition to Mypt1 regulates the catalytic activity of protein phosphatase 1 delta. We observed hyperphosphorylation of S403 and S411 residues of Ppp1r12c upon IL-33 stimulation. Our data thus suggests that, in addition to kinases, phosphatases also play an important role in IL-33-mediated signaling.
Functional analysis of the IL-33 regulated molecules by DAVID revealed activation of molecules involved in regulation of actin cytoskeleton (Fig. 6), MAPK signaling (Supporting Information Fig. S3), Toll like receptor (TLR) signaling and in adherens junction. Interestingly, we observed many molecules involved in the cdc42/Rho signaling pathway. Rho signaling is involved in many cellular processes including polarity [45], migration, gene regulation and actin cytoskeleton reorganization amongst others [46]. To date, there is no report of IL-33-induced Rho dependent signaling.
Figure 6. Proteins involved in the regulation of actin cytoskeleton.
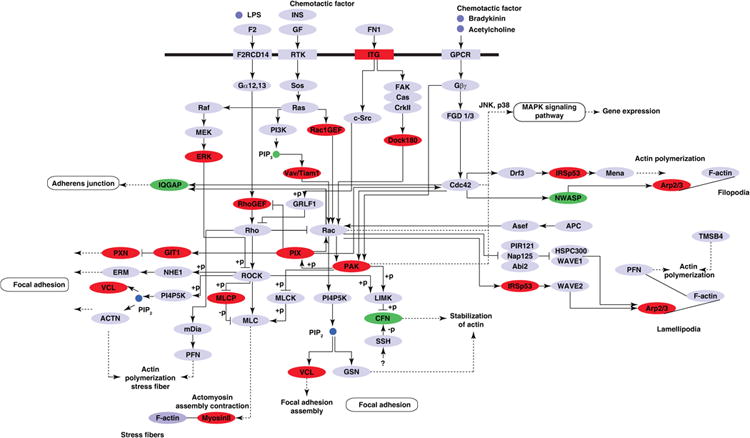
KEGG pathway analysis of proteins differentially phosphorylated by IL-33 using DAVID functional analysis tool indicated the enrichment of the Regulation of actin cytoskeleton pathway. Proteins identified to be regulated by IL-33 are represented in red (hyperphosphorylated) or green (hypophosphorylated).
4. Discussion
IL-33 signaling plays an important role in a number of biological processes. Dysregulated IL-33 signaling due to changes in the expression, secretion of IL-33 or downstream signaling, contributes to the development of a number of diseases, including asthma, arthritis and ulcerative colitis. Current understanding of the molecular mechanism by which IL-33 mediates signaling is limited. This study provides the first unbiased and quantitative investigation of the phosphoproteome and its dynamic changes in response to IL-33 stimulation using a mouse macrophage cell line. We employed a SILAC-based labeling approach coupled with TiO2-based phosphopeptide enrichment and anti-phosphotyrosine based phosphotyrosine enrichment coupled to high-accuracy mass spectrometry to reproducibly identify and quantify a large number of serine, threonine and tyrosine phosphorylation sites with high confidence.
Number of groups have used conventional approaches to study the IL-33 signal transduction pathway. ERK1/2 and NF-κB were the first molecules that were shown to be phosphorylated by IL-33 stimulation in both human and murine systems. Our proteomic data revealed the identification of several known signaling proteins especially proteins involved in MAPK signaling pathways [24, 39, 47, 48] and numerous novel proteins. In agreement with these reports, our proteomic data indicates that Erk1/2, Jnk1/2, and p38 were all inducibly phosphorylated by IL-33. Importantly, our proteomic analysis also led to the determination of the exact phosphorylation sites that were involved. In addition to MAPK signaling, IL-33 has been shown to mediate its activity through PI3K pathway [37] and JAK2 [49]. However, we did not identify effectors of the PI3K signaling module suggesting that either there is no activation of this pathway in macrophages upon IL-33 stimulation or the pathway may not be activated at the time point of analysis. Our findings are thus in agreement with previous studies showing cell- and gene-specific responses to IL-33 stimulation in the different cell types investigated.
Our study identified a number of kinases and phosphatases that are regulated upon IL-33 stimulation. The kinases predominantly belong to the serine/threonine family of kinases. Interestingly, we observed phosphorylation of serine/threonine residues on most tyrosine phosphatases. PTPN12 or PTP-PEST has been shown to be regulated by serine phosphorylation and is phosphorylated in vitro by both cyclic AMP-dependent protein kinase (PKA) and protein kinase C (PKC) at two major sites- S39 and S435 in humans [50]. Although phosphorylation of these sites have been reported previously, their role in regulation of the tyrosine phosphatase activity are not well characterized. A detailed understanding of the role of phosphorylation of serine/threonine residues of tyrosine phosphatases is essential to delineate the molecular mechanism of the activity of tyrosine phosphatases.
Network analysis revealed enrichment of proteins involved in the Rho-mediated signaling. Our analysis revealed protein products of several guanine exchange nucleotide factors (GEFs) including TIAM1, ARHGEF1, ARHGEF2, ARHGEF10 and ARHGEF 11 among others to be regulated by IL-33. We identified T-cell lymphoma invasion and metastasis 1 (TIAM1), a ubiquitous guanine nucleotide exchange factor (GEF) to be hyperphosphorylated at S231 (2- fold) upon IL-33 stimulation. TIAM1 has been shown to promote cell invasion, cell adhesion and also to involve in the regulation of axon formation at neuronal growth cones [51-53]. TIAM1 is reported to interact directly with spinophilin (PPP1R9B), a scaffold protein that plays a role in Rac-mediated actin reorganization [54]. Our data showed a 3-fold increase in the phosphorylation of Ppp1r9b at S100 suggesting IL-33 mediated regulation of cytoskeleton organization. Our data also revealed hyperphosphorylation of three sites (S182, S932 and S1352) on Dock7 upon IL-33 stimulation. DOCK7, an atypical guanine-nucleotide exchange factor has been recently reported to function as an essential downstream regulator of RAGE-mediated cellular migration through the formation of dendritic pseudopodia in cancer cells [55]. We identified vasodilator-stimulated phosphoprotein (Vasp) to be 2-fold hyperphosphorylated upon IL-33 stimulation at S317. VASP belongs to Ena/Vasp family which is involved in filopodia formation. PAK1 mediated phosphorylation of filamin A has been reported to be essential for PAK1-induced cytoskeletal reorganization [56]. PAK1 phosphorylates MYPT1 (PPP1R12A) at its inhibitory site, thus resulting in increased myosin light chain phosphorylation and contraction. We identified peptides from both Pak1 and Mypt1 to be differentially phosphorylated in response to IL-33 stimulation. The role of GTPases has been reported previously in the IL-1 signaling [57, 58]. Studies have shown that IL-1 caused increased GTP binding and hydrolysis in the membrane and also activated Rho and Rac [57]. In response to IL-1, dominant negative mutants of Rac and cdc42 completely blocked the activation of JNK and p38 MAPK signaling [59]. We have identified for the first time effectors of the Rho/Rac mediated pathways. This suggests that like IL-1, IL-33 may signal through this mechanism for the activation of p38 MAPKs.
Concluding Remarks
The data gathered from this study greatly expands the understanding of the IL-33 signaling pathway by the discovery of several known and novel IL-33-regulated phosphoproteins. As the next step of this study, a detailed analysis of the proteins identified in the phosphoproteome analysis is required. In addition, from our analysis as well as from earlier studies, it is clear that IL-33 is involved in cell specific signaling. Hence, comparison of signaling across different cell types will further broaden our understanding of IL-33 mediating signaling. Furthermore, temporal analysis of IL-33 mediated signaling will provide valuable information on the signaling modules activated and help us to understand the complex signaling networks mediated by IL-33.
Supplementary Material
Supporting Information Figure S1. Functional annotation of phosphoproteins. Molecular function and biological processes of phosphoproteins were obtained from Panther Classification System to calculate the distribution of molecular functions of phosphoproteins identified (A) Molecular function and (B) Biological process of IL-33-upregulated phosphoproteins. (C) Molecular function and (D) Biological process of IL-33-downregulated phosphoproteins.
Supporting Information Figure S2. Known molecules identified in IL-33 signaling pathway. Representative MS spectra of known phosphorylated proteins. A–D, phosphorylation status of Map kinases—Mapk1, Mapk8, Mapk14 and Map3k7, and transcription regulators—c-Jun (E) and Atf2 (F) were upregulated as evidenced by MS spectra showing the changes in the relative abundance of phosphopeptides.
Supporting Information Figure S3. MAPK signaling pathway. KEGG pathway analysis of proteins differentially phosphorylated by IL-33 using DAVID functional analysis identified the MAPK signaling pathway to be enriched. Proteins colored in red were found to be differentially phosphorylated in response to IL-33.
Supporting Information Table S1. A list of phosphoPSMs identified by MASCOT and SEQUEST.
Supporting Information Table S2. A list of phosphopeptides quantitated by SILAC.
Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics. SMP, RSN are recipients of Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), Government of India. SSM and YS are recipients of Senior Research Fellowship from the University Grants Commission (UGC), India. T.S.K.P. is supported by DBT Program Support on Neuroproteomics (BT/01/COE/08/05) to IOB. This work was supported by an NIH roadmap grant for Technology Centers of Networks and Pathways (U54GM103520). The authors acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation and the Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with The Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation's Parkinson's disease Programs.
Abbreviations
- IL-1
Interleukin-1
- IL-33
Interleukin-33
- BCA
Bicinchoninic acid assay
- bRPLC
basic Reverse phase liquid chromatography
- HCD
Higher energy collision induced dissociation
- IAP
Immunoaffinity purification
- TEABC
Triethyl ammonium bicarbonate
- TPCK
Tosyl phenylalanyl chloromethyl ketone
- TiO2
Titanium dioxide
Footnotes
Conflict of interest: The authors have declared no conflict of interest.
References
- 1.Schmitz J, Owyang A, Oldham E, Song Y, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Carriere V, Roussel L, Ortega N, Lacorre DA, et al. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci U S A. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baekkevold ES, Roussigne M, Yamanaka T, Johansen FE, et al. Molecular characterization of NF-HEV, a nuclear factor preferentially expressed in human high endothelial venules. Am J Pathol. 2003;163:69–79. doi: 10.1016/S0002-9440(10)63631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moussion C, Ortega N, Girard JP. The IL-1-like cytokine IL-33 is constitutively expressed in the nucleus of endothelial cells and epithelial cells in vivo: a novel ‘alarmin’? PLoS One. 2008;3:e3331. doi: 10.1371/journal.pone.0003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cayrol C, Girard JP. The IL-1-like cytokine IL-33 is inactivated after maturation by caspase-1. Proc Natl Acad Sci U S A. 2009;106:9021–9026. doi: 10.1073/pnas.0812690106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meisel C, Bonhagen K, Lohning M, Coyle AJ, et al. Regulation and function of T1/ST2 expression on CD4+ T cells: induction of type 2 cytokine production by T1/ST2 cross-linking. J Immunol. 2001;166:3143–3150. doi: 10.4049/jimmunol.166.5.3143. [DOI] [PubMed] [Google Scholar]
- 7.Allakhverdi Z, Smith DE, Comeau MR, Delespesse G. Cutting edge: The ST2 ligand IL-33 potently activates and drives maturation of human mast cells. J Immunol. 2007;179:2051–2054. doi: 10.4049/jimmunol.179.4.2051. [DOI] [PubMed] [Google Scholar]
- 8.Besnard AG, Togbe D, Guillou N, Erard F, et al. IL-33-activated dendritic cells are critical for allergic airway inflammation. Eur J Immunol. 2011;41:1675–1686. doi: 10.1002/eji.201041033. [DOI] [PubMed] [Google Scholar]
- 9.Komai-Koma M, Gilchrist DS, McKenzie AN, Goodyear CS, et al. IL-33 activates B1 cells and exacerbates contact sensitivity. J Immunol. 2011;186:2584–2591. doi: 10.4049/jimmunol.1002103. [DOI] [PubMed] [Google Scholar]
- 10.Villarreal DO, Wise MC, Walters JN, Reuschel EL, et al. Alarmin IL-33 acts as an immunoadjuvant to enhance antigen-specific tumor immunity. Cancer Res. 2014;74:1789–1800. doi: 10.1158/0008-5472.CAN-13-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Q, Li G, Zhu Y, Liu L, et al. IL-33 synergizes with TCR and IL-12 signaling to promote the effector function of CD8+ T cells. Eur J Immunol. 2011;41:3351–3360. doi: 10.1002/eji.201141629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki K, Sanada S, Kudinova AY, Steinhauser ML, et al. Interleukin-33 prevents apoptosis and improves survival after experimental myocardial infarction through ST2 signaling. Circ Heart Fail. 2009;2:684–691. doi: 10.1161/CIRCHEARTFAILURE.109.873240. [DOI] [PubMed] [Google Scholar]
- 13.Funakoshi-Tago M, Tago K, Hayakawa M, Tominaga S, et al. TRAF6 is a critical signal transducer in IL-33 signaling pathway. Cell Signal. 2008;20:1679–1686. doi: 10.1016/j.cellsig.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Prefontaine D, Lajoie-Kadoch S, Foley S, Audusseau S, et al. Increased expression of IL-33 in severe asthma: evidence of expression by airway smooth muscle cells. J Immunol. 2009;183:5094–5103. doi: 10.4049/jimmunol.0802387. [DOI] [PubMed] [Google Scholar]
- 15.Guo Z, Wu J, Zhao J, Liu F, et al. IL-33 promotes airway remodeling and is a marker of asthma disease severity. J Asthma. 2014 doi: 10.3109/02770903.2014.921196. [DOI] [PubMed] [Google Scholar]
- 16.Savinko T, Matikainen S, Saarialho-Kere U, Lehto M, et al. IL-33 and ST2 in atopic dermatitis: expression profiles and modulation by triggering factors. J Invest Dermatol. 2012;132:1392–1400. doi: 10.1038/jid.2011.446. [DOI] [PubMed] [Google Scholar]
- 17.Imai Y, Yasuda K, Sakaguchi Y, Haneda T, et al. Skin-specific expression of IL-33 activates group 2 innate lymphoid cells and elicits atopic dermatitis-like inflammation in mice. Proc Natl Acad Sci U S A. 2013;110:13921–13926. doi: 10.1073/pnas.1307321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marvie P, Lisbonne M, L’Helgoualc’h A, Rauch M, et al. Interleukin-33 overexpression is associated with liver fibrosis in mice and humans. J Cell Mol Med. 2010;14:1726–1739. doi: 10.1111/j.1582-4934.2009.00801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rankin AL, Mumm JB, Murphy E, Turner S, et al. IL-33 induces IL-13-dependent cutaneous fibrosis. J Immunol. 2010;184:1526–1535. doi: 10.4049/jimmunol.0903306. [DOI] [PubMed] [Google Scholar]
- 20.McHedlidze T, Waldner M, Zopf S, Walker J, et al. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity. 2013;39:357–371. doi: 10.1016/j.immuni.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pushparaj PN, Li D, Komai-Koma M, Guabiraba R, et al. Interleukin-33 exacerbates acute colitis via interleukin-4 in mice. Immunology. 2013;140:70–77. doi: 10.1111/imm.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beltran CJ, Nunez LE, Diaz-Jimenez D, Farfan N, et al. Characterization of the novel ST2/IL-33 system in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1097–1107. doi: 10.1002/ibd.21175. [DOI] [PubMed] [Google Scholar]
- 23.Ross R, Ross XL, Ghadially H, Lahr T, et al. Mouse langerhans cells differentially express an activated T cell-attracting CC chemokine. J Invest Dermatol. 1999;113:991–998. doi: 10.1046/j.1523-1747.1999.00803.x. [DOI] [PubMed] [Google Scholar]
- 24.Hong J, Bae S, Jhun H, Lee S, et al. Identification of constitutively active interleukin 33 (IL-33) splice variant. J Biol Chem. 2011;286:20078–20086. doi: 10.1074/jbc.M111.219089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Grinchuk V, Urban JF, Jr, Bohl J, et al. Macrophages as IL-25/IL-33-responsive cells play an important role in the induction of type 2 immunityMacrophages as IL-25/IL-33-responsive cells play an important role in the induction of type 2 immunity. PLoS One. 2013;8:e59441. doi: 10.1371/journal.pone.0059441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 27.Rush J, Moritz A, Lee KA, Guo A, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 28.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 29.Selvan LD, Renuse S, Kaviyil JE, Sharma J, et al. Phosphoproteome of Cryptococcus neoformans. J Proteomics. 2014;97:287–295. doi: 10.1016/j.jprot.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 30.Vizcaino JA, Deutsch EW, Wang R, Csordas A, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S, Sato Y, Kawashima M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinformatics. 2011;13:15–24. doi: 10.1002/0471250953.bi1315s35. Unit 13. [DOI] [PubMed] [Google Scholar]
- 35.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Brint EK, Fitzgerald KA, Smith P, Coyle AJ, et al. Characterization of signaling pathways activated by the interleukin 1 (IL-1) receptor homologue T1/ST2. A role for Jun N-terminal kinase in IL-4 induction. J Biol Chem. 2002;277:49205–49211. doi: 10.1074/jbc.M209685200. [DOI] [PubMed] [Google Scholar]
- 37.Choi YS, Choi HJ, Min JK, Pyun BJ, et al. Interleukin-33 induces angiogenesis and vascular permeability through ST2/TRAF6-mediated endothelial nitric oxide production. Blood. 2009;114:3117–3126. doi: 10.1182/blood-2009-02-203372. [DOI] [PubMed] [Google Scholar]
- 38.Tare N, Li H, Morschauser A, Cote-Sierra J, et al. KU812 cells provide a novel in vitro model of the human IL-33/ST2L axis: functional responses and identification of signaling pathways. Exp Cell Res. 2010;316:2527–2537. doi: 10.1016/j.yexcr.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Yagami A, Orihara K, Morita H, Futamura K, et al. IL-33 mediates inflammatory responses in human lung tissue cells. J Immunol. 2010;185:5743–5750. doi: 10.4049/jimmunol.0903818. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y, Xia F, Hermance N, Mabb A, et al. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-kappaB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol Cell Biol. 2011;31:2774–2786. doi: 10.1128/MCB.01139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shim JH, Xiao C, Paschal AE, Bailey ST, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ben-Levy R, Leighton IA, Doza YN, Attwood P, et al. Identification of novel phosphorylation sites required for activation of MAPKAP kinase-2. EMBO J. 1995;14:5920–5930. doi: 10.1002/j.1460-2075.1995.tb00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engel K, Schultz H, Martin F, Kotlyarov A, et al. Constitutive activation of mitogen-activated protein kinase-activated protein kinase 2 by mutation of phosphorylation sites and an A-helix motif. J Biol Chem. 1995;270:27213–27221. doi: 10.1074/jbc.270.45.27213. [DOI] [PubMed] [Google Scholar]
- 44.Feng J, Ito M, Ichikawa K, Isaka N, et al. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem. 1999;274:37385–37390. doi: 10.1074/jbc.274.52.37385. [DOI] [PubMed] [Google Scholar]
- 45.Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, et al. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 47.Yndestad A, Marshall AK, Hodgkinson JD, Tham el L, et al. Modulation of interleukin signalling and gene expression in cardiac myocytes by endothelin-1. Int J Biochem Cell Biol. 2010;42:263–272. doi: 10.1016/j.biocel.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 48.Chow JY, Wong CK, Cheung PF, Lam CW. Intracellular signaling mechanisms regulating the activation of human eosinophils by the novel Th2 cytokine IL-33: implications for allergic inflammation. Cell Mol Immunol. 2010;7:26–34. doi: 10.1038/cmi.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Funakoshi-Tago M, Tago K, Sato Y, Tominaga S, Kasahara T. JAK2 is an important signal transducer in IL-33-induced NF-kappaB activation. Cell Signal. 2011;23:363–370. doi: 10.1016/j.cellsig.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Garton AJ, Tonks NK. PTP-PEST: a protein tyrosine phosphatase regulated by serine phosphorylation. EMBO J. 1994;13:3763–3771. doi: 10.1002/j.1460-2075.1994.tb06687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 52.Hordijk PL, ten Klooster JP, van der Kammen RA, Michiels F, et al. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 53.Kunda P, Paglini G, Quiroga S, Kosik K, Caceres A. Evidence for the involvement of Tiam1 in axon formation. J Neurosci. 2001;21:2361–2372. doi: 10.1523/JNEUROSCI.21-07-02361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satoh A, Nakanishi H, Obaishi H, Wada M, et al. Neurabin-II/spinophilin An actin filament-binding protein with one pdz domain localized at cadherin-based cell-cell adhesion sites. J Biol Chem. 1998;273:3470–3475. doi: 10.1074/jbc.273.6.3470. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto K, Murata H, Putranto EW, Kataoka K, et al. DOCK7 is a critical regulator of the RAGE-Cdc42 signaling axis that induces formation of dendritic pseudopodia in human cancer cells. Oncol Rep. 2013;29:1073–1079. doi: 10.3892/or.2012.2191. [DOI] [PubMed] [Google Scholar]
- 56.Vadlamudi RK, Li F, Adam L, Nguyen D, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 57.O’Neill LA, Bird TA, Gearing AJ, Saklatvala J. Interleukin-1 signal transduction Increased GTP binding and hydrolysis in membranes of a murine thymoma line (EL4) J Biol Chem. 1990;265:3146–3152. [PubMed] [Google Scholar]
- 58.Singh R, Wang B, Shirvaikar A, Khan S, et al. The IL-1 receptor and Rho directly associate to drive cell activation in inflammation. J Clin Invest. 1999;103:1561–1570. doi: 10.1172/JCI5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bagrodia S, Derijard B, Davis RJ, Cerione RA. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. Functional annotation of phosphoproteins. Molecular function and biological processes of phosphoproteins were obtained from Panther Classification System to calculate the distribution of molecular functions of phosphoproteins identified (A) Molecular function and (B) Biological process of IL-33-upregulated phosphoproteins. (C) Molecular function and (D) Biological process of IL-33-downregulated phosphoproteins.
Supporting Information Figure S2. Known molecules identified in IL-33 signaling pathway. Representative MS spectra of known phosphorylated proteins. A–D, phosphorylation status of Map kinases—Mapk1, Mapk8, Mapk14 and Map3k7, and transcription regulators—c-Jun (E) and Atf2 (F) were upregulated as evidenced by MS spectra showing the changes in the relative abundance of phosphopeptides.
Supporting Information Figure S3. MAPK signaling pathway. KEGG pathway analysis of proteins differentially phosphorylated by IL-33 using DAVID functional analysis identified the MAPK signaling pathway to be enriched. Proteins colored in red were found to be differentially phosphorylated in response to IL-33.
Supporting Information Table S1. A list of phosphoPSMs identified by MASCOT and SEQUEST.
Supporting Information Table S2. A list of phosphopeptides quantitated by SILAC.


