SUMMARY
Major features of transcription by human RNA Polymerase II (Pol II) remain poorly defined due to a lack of quantitative approaches for visualizing Pol II progress at nucleotide resolution. We developed a simple and powerful approach for performing native elongating transcript sequencing (NET-seq) in human cells that globally maps strand-specific Pol II density at nucleotide resolution. NET-seq exposes a mode of antisense transcription that originates downstream and converges on transcription from the canonical promoter. Convergent transcription is associated with a distinctive chromatin configuration and is characteristic of lower-expressed genes. Integration of NET-seq with genomic footprinting data reveals stereotypic Pol II pausing coincident with transcription factor occupancy. Finally, exons retained in mature transcripts display Pol II pausing signatures that differ markedly from skipped exons, indicating an intrinsic capacity for Pol II to recognize exons with different processing fates. Together, human NET-seq exposes the topography and regulatory complexity of human gene expression.
INTRODUCTION
High throughput sequencing analyses of transcription have discovered new classes of RNAs and new levels of regulatory complexity. Many of these results were obtained with two experimental strategies to measure RNA polymerase density genome-wide. The first, RNA polymerase II (Pol II) ChIP-seq or ChIP-chip, identifies DNA bound to RNA polymerase. The second set of approaches, global run-on sequencing (GRO-seq) and precision run-on sequencing (PRO-seq), restarts RNA polymerase in vitro with labeled nucleotides to purify and sequence nascent RNA (Core et al., 2008; Kwak et al., 2013). GRO-seq and Pol II ChIP detect strong transcriptional pauses ~50 bp downstream of many transcription start sites (Core et al., 2008; Kwak et al., 2013; Muse et al., 2007; Rahl et al., 2010; Zeitlinger et al., 2007), demonstrating that promoter-proximal pausing is more prevalent than initially observed (Core et al., 2008; Krumm et al., 1992; Kwak et al., 2013; Muse et al., 2007; Rahl et al., 2010; Rougvie and Lis, 1988; Strobl and Eick, 1992; Zeitlinger et al., 2007). Abundant unstable transcripts upstream of and antisense to promoters revealed that divergent transcription is a common feature of eukaryotic promoters (Core et al., 2008; Neil et al., 2009; Preker et al., 2008; Seila et al., 2008; Xu et al., 2009). Despite progress in understanding how these transcripts are terminated and degraded (Almada et al., 2013; Core et al., 2008; Kwak et al., 2013; Ntini et al., 2013; Preker et al., 2008), their roles remain unknown (Wu and Sharp, 2013). Finally, recent studies confirm that splicing is largely co-transcriptional and splicing outcome is kinetically tied to elongation rate (Bhatt et al., 2012; Dujardin et al., 2014; Fong et al., 2014; Ip et al., 2011; Krumm et al., 1992; la Mata et al., 2003; Roberts et al., 1998; Rougvie and Lis, 1988; Shukla et al., 2011; Strobl and Eick, 1992; Tilgner et al., 2012). However, it has been impossible to determine whether such kinetic coupling in human cells is mediated by pausing events genome-wide, due to the high resolution required to measure pausing on short human exons.
The strongly stereotyped locations of promoter-proximal pauses and divergent antisense transcription can be exposed by averaging Pol II density from many genes (metagene analysis) obtained at low resolution (Core et al., 2008; Neil et al., 2009; Preker et al., 2008; Rahl et al., 2010; Seila et al., 2008; Xu et al., 2009). Yet, the precise architecture of promoter-associated transcriptional activity and of pausing outside of promoter regions have been obscured by the resolution limitations of current methodologies, preventing deeper insight into the underlying regulatory mechanisms. Indeed, the interplay between chromatin structure, transcription factors and the transcription machinery is largely undefined. Pol II ChIP-seq is typically limited in its resolution to >200 bp resolution and lacks strand specificity. GRO-seq is similarly limited to ~50 bp resolution, and although PRO-seq has higher resolution, both run-on methods require transcription elongation complexes to resume polymerization in vitro, a variable process sensitive to the experimental conditions and the Pol II pausing state (Core et al., 2008; Weber et al., 2014). Recently, we showed that the extraordinary stability of the RNA-DNA-RNA polymerase ternary complex can be exploited to capture nascent RNA (Churchman and Weissman, 2011). Native elongating transcript sequencing (NET-seq) quantitatively purifies Pol II complexes and sequences the 3′ end of nascent RNA to reveal the strand-specific position of Pol II with single-nucleotide resolution. NET-seq detects all transcriptionally engaged Pol II, including productively transcribing Pol II, paused Pol II, and Pol II recovering from pausing (Churchman and Weissman, 2011).
Here we develop a NET-seq approach that quantitatively defines the full spectrum of transcriptional activity in a strand-specific manner and at nucleotide resolution in human cells. We find that many promoters display antisense transcription downstream of a promoter-proximal pause, resulting in convergent sense and antisense transcriptional activities that face one another in close proximity. Convergent transcription is associated with a distinct chromatin conformation and is a feature of lower-expressed genes, suggesting a possible regulatory role. NET-seq reveals that Pol II density profiles differ between retained exons, skipped exons, and introns in human cells, indicating generalized kinetic coupling of transcription and splicing. Human NET-seq is readily applicable to diverse cell types and provides a general strategy to study transcriptional complexity.
RESULTS
A Robust Human NET-seq Methodology
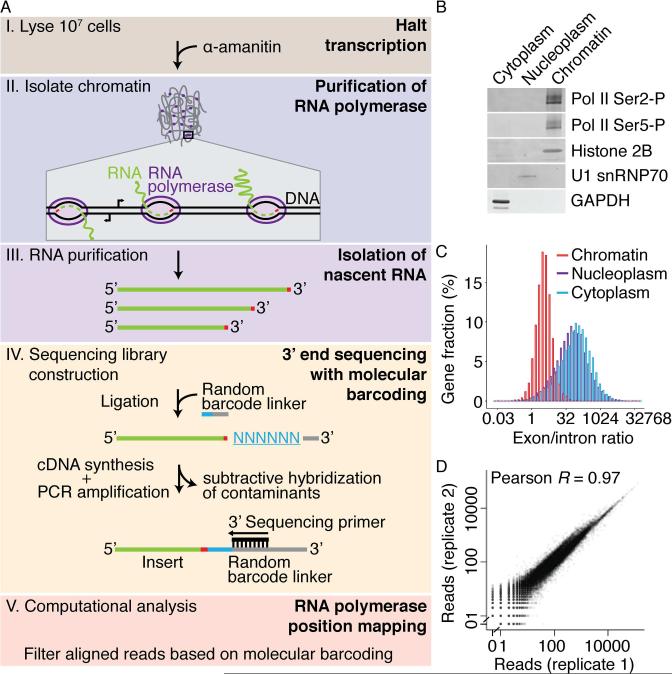
The first step of NET-seq purifies nascent RNA through its tight interaction with RNA polymerase. In yeast, this is achieved through an epitope-tagged Pol II subunit that enables highly quantitative purification and specific elution (Churchman and Weissman, 2011). In adapting NET-seq to human cells, we biochemically purify >99% of all engaged RNA polymerase in a highly specific manner that can be applied to any mammalian cell line or tissue without genetic modification (Figure 1A). This method avoids using Pol II antisera, which could bias the population of isolated Pol II complexes due to posttranslational modifications and epitope masking by heterogenous Pol II binding partners and structural conformations. Instead, human NET-seq exploits the high stability of the RNA-DNA-RNA polymerase ternary complex, even in the presence of high salt and urea (Cai and Luse, 1987; Wuarin and Schibler, 1994), to purify engaged RNA polymerase, along with its nascent RNA, through an association with chromatin after cellular fractionation into cytoplasm, nucleoplasm and chromatin (Bhatt et al., 2012; Pandya-Jones and Black, 2009; Wuarin and Schibler, 1994). To prevent transcriptional run-on during fractionation, lysate is kept at ≤4°C and α-amanitin, a potent transcriptional inhibitor (Lindell et al., 1970), is included in every step. Through optimization of current fractionation approaches, we identified buffers and washing conditions that cleanly purify >99% of elongating RNA polymerase II (C-terminal domain (CTD) Ser2-P, Ser5-P and the general CTD hyper-phosphorylated form of Pol II) in the chromatin fraction (See Experimental Procedures, Figure 1B and S1A). Western blot analyses of Pol II isoforms and factors with well-defined subcellular localizations verify the stringency of our fractionation conditions (Figure 1B).
Figure 1. A Robust and Simplified NET-seq Approach for Human Cells.
(A) Schematic view of the key steps of the human NET-seq approach. The transcription inhibitor, α-amanitin, is introduced at cell lysis and maintained through all purification steps. Engaged RNA polymerase is purified through the isolation of chromatin. The 3′ end of the co-purified nascent RNA (red) is ligated to a linker containing a mixed random hexameric sequence (blue) that serves as a molecular barcode. After cDNA synthesis, contaminant species are removed by hybridization. PCR amplification results in a DNA sequencing library with the sequencing primer binding site proximal to the random hexamer barcode. Finally, the 3′ ends of the sequenced nascent RNA are aligned to the human genome yielding RNA polymerase density at nucleotide resolution. Analysis of the molecular barcode allows reads arising from DNA library construction artifacts to be filtered out. (B) Representative western blot analysis of cellular fractions in HeLa S3 cells. Subcellular localization markers were also probed (Chromatin marker, Histone 2B; nucleoplasm marker, U1 snRNP70; cytoplasm marker, GAPDH). (C) Histograms of the size-normalized ratio of subcellular RNA-seq reads that map to exons versus introns for each gene. (D) Number of uniquely aligned reads per Pol II gene for two biological replicates from HeLa S3 cells (Pearson's correlation, R = 0.97). 0.5 pseudocounts were added to genes with zero counts in one of the replicates. The dataset with higher coverage was randomly downsampled to match the total number of reads of the other dataset. See also Figure S1 and Table S1.
To confirm that our purification strategy specifically isolates nascent RNA, we sequenced the RNA in each fraction. Unprocessed RNA species, such as intron-containing Pol II transcripts and spacer-containing Pol I transcripts (Figures 1C and S1B), are heavily enriched in the chromatin fraction. Importantly, the large majority of intron-containing RNAs observed in the nucleoplasm persist to the cytoplasm, indicating that these RNAs are products of intron-retaining alternative splicing and not nascent transcripts (Figure S1C). Together, these results demonstrate that RNA polymerase and nascent RNA are quantitatively purified through the isolation of chromatin.
The second step of the NET-seq approach requires sequencing the 3’ ends of the nascent RNA, which localizes Pol II genome-wide at nucleotide resolution (Churchman and Weissman, 2011; Ferrari et al. 2013; Weber et al., 2014). In large part, our yeast library construction protocol is used (Churchman and Weissman, 2012), with two important changes to account for the increased complexity of the human genome. First, we addressed reverse transcription (RT) artifacts that arise from the significant size of human nascent RNA. We found that reverse transcription frequently initiates within the RNA if there are stretches as short as six nucleotides of complementarity to the RT primer (Figure S1D). When the 3′ ends of the nascent RNA are ligated to a linker pool, consisting of a random hexamer at the 5′ end followed by a common sequence, ligation efficiency increases and mispriming events are dramatically reduced (Figure S1D). Furthermore, the hexamer serves as a molecular barcode for each molecule and enables the computational removal of reads arising from residual mispriming events and PCR duplicates. Second, we deplete abundant mature snRNAs, snoRNAs, rRNA and others through subtractive hybridization targeting their 3′ ends (Figure S1E, Table S1) to increase sequencing coverage for nascent transcripts. Finally, library construction steps are optimized to be highly efficient (>90%) and are continually monitored through quality controls to minimize bias. Together, our optimized library construction protocol faithfully converts the 3′ ends of nascent human RNA to a DNA sequencing library that allows the high fidelity mapping of strand-specific Pol II density.
To observe genome-wide transcriptional activity, a NET-seq library was prepared from HeLa S3 cells and sequenced to high coverage (768 million total reads, 360 million uniquely aligned). Each sequencing read was aligned to the human genome and the genomic location of the 3′ end of the nascent RNA was recorded to map RNA polymerase density with nucleotide resolution. As expected, we recovered nascent RNA from all three nuclear RNA polymerases (Pol I, Pol II, Pol III), as well as mature chromatin-associated RNAs, such as snRNAs, and splicing intermediates (Figures S1F and S1G). Here we focused our analysis on Pol II-synthesized RNAs, but our results suggest that the NET-seq approach is amenable to the study of other RNA polymerases. Importantly, comparison of a biological replicate library (175 million total reads, 83 million uniquely aligned) shows strong agreement, indicating the robustness of the approach (Pearson's coefficient, 0.97, Figure 1D). To demonstrate that NET-seq is easily adaptable to other cell lines, we applied our approach to HEK 293T cells and obtained data from two replicates with similar quality (replicate 1: 1.203 billion total reads, 555 million uniquely aligned; replicate 2: 358 million total reads, 135 million uniquely aligned, Figure S1H). From these analyses, we conclude that human NET-seq is capable of reproducibly monitoring transcriptional activity across the human genome and adaptation to new cell lines is straightforward.
NET-seq Reveals Transcriptional Activity at Nucleotide Resolution Genome-Wide
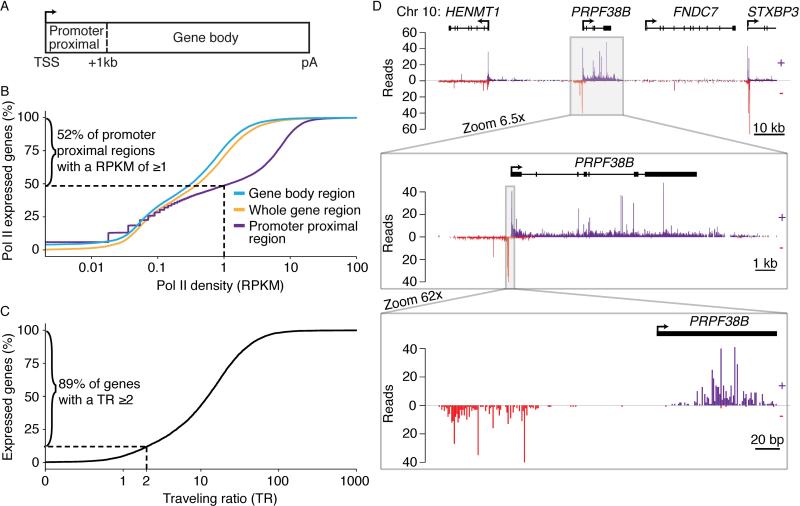
The resolution afforded by NET-seq and the sequencing coverage obtained provide an in-depth view of genome-wide transcriptional activity. In both HeLa S3 and HEK 293T NET-seq data, greater than 50% of genes have coverage >1 read per kb per million uniquely aligned reads (RPKM) to Pol II genes in promoter-proximal regions, conservatively defined as the region between the earliest annotated transcription start site and +1 kb (Figures 2A-B, S2C). When coverage is calculated across entire genes, the percentage decreases to less than 30% in both cell lines, due to the prevalence of promoter-proximal pausing (Figure 2A-B). Indeed, most (89% in HeLa S3 cells and 94% in HEK 293T cells) expressed genes display promoter-proximal pausing defined by a traveling ratio (coverage ratio between a narrow promoter-proximal region and the gene body) of ≥ 2, consistent with earlier observations in mouse embryonic stem cells (Figure 2C, Figure S2D) (Rahl et al., 2010). Furthermore, we detect unstable RNA production, antisense transcription upstream of many promoters (89% in HeLa S3 cells and 82% in HEK 293T cells), transcription downstream of many polyadenylation sites (95% in HeLa S3 cells and 88% in HEK 293T cells) and enhancer RNAs (Figure S2A,B,E,F).
Figure 2. NET-seq Reports on Transcription Globally and Locally.
(A) Schematic defining gene regions used in analysis of NET-seq data. (B) Distributions of the percent of expressed Pol II transcribed protein-coding genes (N=19108) with a given Pol II coverage for different gene regions as defined in Figure 2A. (C) Distributions of the percent of well expressed Pol II protein-coding genes (N = 8912) with a given traveling ratio. Well expressed Pol II genes are defined as those genes with an RPKM of 1 or greater in a tight promoter-proximal region (−30 bp to +300 bp of the TSS). Traveling ratio (TR) is defined as the RPKM of the tight promoter-proximal region divided by the RPKM of the gene body region. (D) Number of NET-seq reads at three zoom levels around the PRPF38B locus for HeLa S3 cells. Reads that aligned to the positive strand (+) are in violet and reads that aligned to the negative strand (-) are in red. The TSS and the direction of transcription is indicated by an arrow. Annotation of exonic and intronic regions are shown as boxes and lines, respectively. RPKM, reads per kb per million uniquely aligned reads at Pol II-transcribed genes.;TSS, transcription start site; pA, polyadenylation site. See also Figure S2.
NET-seq data describes transcriptional activity at many length scales. At the single gene level, strong signal is observed at promoter regions and across introns (Figure 2D, upper and middle panels). Signal variation across the gene body suggests that transcription elongation is discontinuous following release from promoter-proximal regions and that pausing is a general feature during productive Pol II transcription. Near transcription start sites (TSSs), NET-seq detects sense and antisense transcription of divergent promoters at single-nucleotide resolution, revealing that promoter-proximal pausing does not occur at only one position; instead there are narrow regions of high Pol II density (Figure 2D, lower panel). Together, NET-seq data uncover key features of human transcription activity, and the high resolution and the coverage of the data provide deeper insight into these complexities.
Widespread Convergent Transcription in Promoter-Proximal Regions
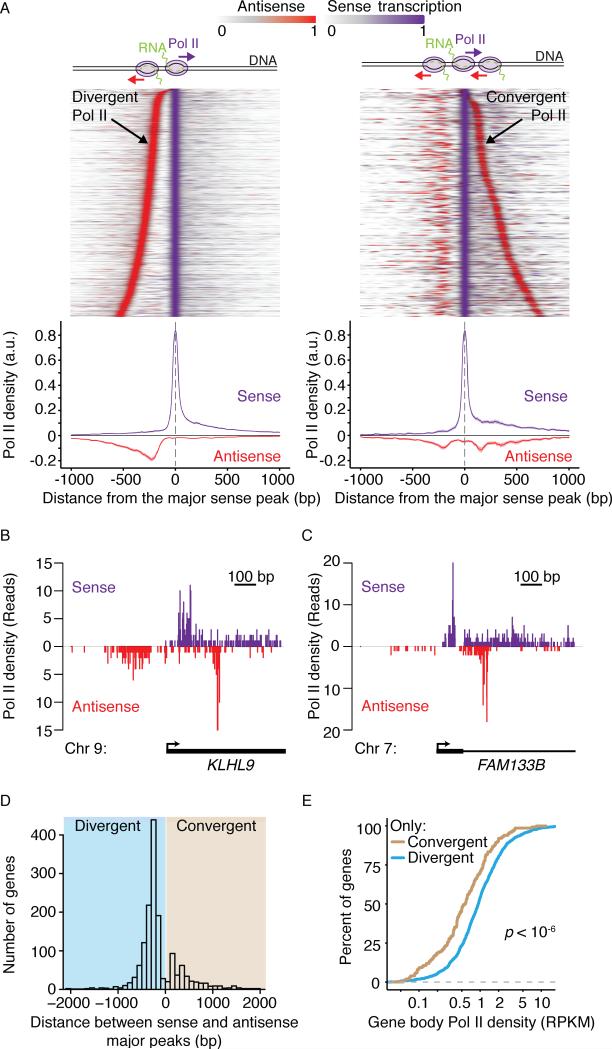
Several previous studies showed widespread divergent transcription at eukaryotic promoters (Churchman and Weissman, 2011; Core et al., 2008; Neil et al., 2009; Seila et al., 2008; Xu et al., 2009). We analyzed this phenomenon for a stringently defined set of Pol II transcribed genes that do not overlap other genes within 2.5 kb of the TSS and polyadenylation site and are longer than 2 kb to avoid misinterpreting transcription from other genes as antisense transcription (N = 3937 genes). Analysis of regions 2 kb upstream and downstream of transcription start sites with broad coverage (coefficient of variation > 0.5, N = 1488) reveals divergent transcription in 77% of promoter-proximal regions, consistent with other studies (Figure 3A, left panel) (Core et al., 2008; Seila et al., 2008). Surprisingly, close inspection of our data revealed an unappreciated form of antisense transcription near promoters. At 25% of promoter-proximal regions, we observe antisense transcription originating downstream of sense transcription (Figure 3A, right panel), which we term convergent transcription. Convergent transcription is clearly observed at single promoter regions (Figure 3B-C) and in most cases, such as near the KLHL9 promoter, convergent transcription is accompanied by divergent transcription (Figure 3B). However, it also occurs in the absence of divergent transcription (for example, FAM133B, Figure 3C). Furthermore, GRO-seq also detects these transcripts. A re-analysis of mouse embryonic stem cell data reveals convergent antisense transcription (Jonkers et al., 2014) (Figure S3A).
Figure 3. Convergent Transcription Observed at the Promoter-Proximal Regions of Lower-Expressed Genes.
(A) Promoters are classified depending on whether they contain a peak of convergent antisense Pol II transcription, as illustrated by the cartoon above the heat maps. A stringent set of promoter-proximal regions were selected for analysis to ensure that transcription arising from other transcription units would not bias classification (see Experimental Procedures). Heat maps of Pol II density are displayed for each class (Left, no convergent peak, N = 850 genes; Right, convergent peak, N = 310). NET-seq signal from each promoter region (+/- 2kb centered at the sense transcription peak) is normalized to vary between 0 and 1 and smoothed with a 50 bp sliding window average. Heat maps are sorted by the distance between the sense and antisense peaks. Mean Pol II density profile is displayed below the heat maps. Solid lines indicate the mean values and shading shows the 95% confidence interval. Sense transcription is shown in violet and antisense transcription is shown in red. (B-C) Examples of NET-seq reads in two promoter-proximal regions that display convergent Pol II transcription. (D) Histogram of distances between the major peak of Pol II density in the sense direction and the peaks in the antisense direction for all analyzed promoters. (E) Distributions of the percentage of genes with a given Pol II density in the gene body region, as defined in Figure 2A. Genes with only convergent transcription (yellow) or only divergent transcription (blue) in their promoter-proximal regions are compared. The p value is calculated by the Kolmogorov–Smirnov test. RPKM, reads per kb per million uniquely aligned reads at Pol II-transcribed genes. See also Figure S3.
To characterize the structural attributes on these modes of transcriptional activity, distances between sense and antisense peaks were determined for each promoter-proximal region (Figure 3D). A stereotypical distance (250 ± 50 bp) separates the sense and antisense peaks in divergent transcription, while the sense and antisense peaks in convergent transcription are also separated by a stereotypical distance (150 bp ± 50 bp), indicating that convergent antisense transcription is not simply the result of spurious antisense transcription initiation events across the promoter-proximal region (Figure 3D).
A Distinct Chromatin Structure Associated with Convergent Transcription
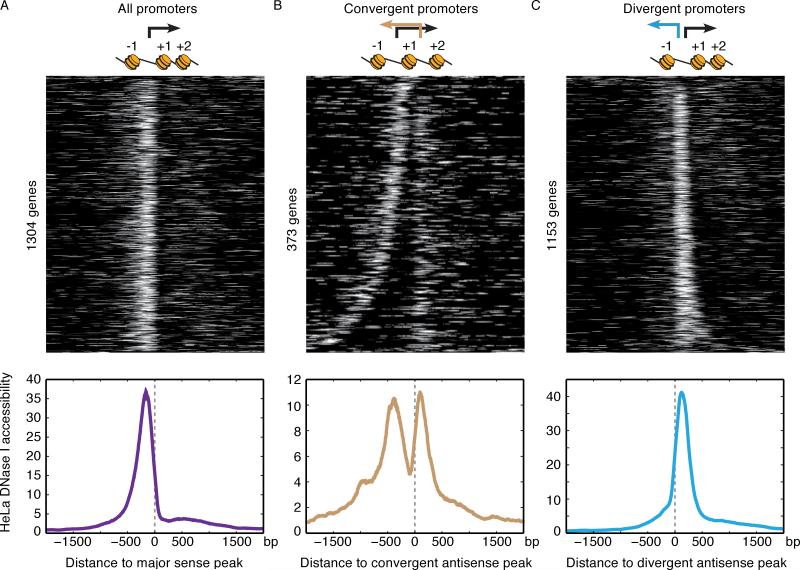
Many chromatin modifiers control antisense transcription (Churchman and Weissman, 2011; DeGennaro et al., 2013; Kim et al., 2012; Marquardt et al., 2014; Whitehouse et al., 2007), and we asked how promoter-proximal transcriptional activity relates to local chromatin structure. We used DNase-seq to map regions of open chromatin and highly positioned nucleosomes in the same HeLa S3 cells used for NET-seq (Thurman et al., 2012). We examined the distribution of DNase I accessibility relative to promoter-proximal peaks in NET-seq data (Figure 4). At genes that have a sense Pol II peak (representing promoter-proximally paused Pol II), we observe strong DNase I hypersensitivity upstream of the peak, determining the canonical promoter (Figure 4A), and reduced DNase sensitivity downstream of the peak corresponding to the +1 nucleosome. Thus promoter-proximal pausing occurs prior to the +1 nucleosome in mammalian cells. Comparison of DNase I data relative to the divergent antisense peak shows that this transcriptional activity originates from the 5′ side of the promoter hypersensitivity region, consistent with the model that divergent antisense transcription is a consequence of an open chromatin region (Seila et al., 2009) (Figure 4C). In contrast, genes with convergent transcription show two distinct peaks in DNase I hypersensitivity: a canonical promoter peak and a downstream peak located proximal to the convergent antisense peak (Figure 4B). Thus convergent antisense transcription likely originates locally. Furthermore, the dip between the two peaks of DNase I hypersensitivity likely represents the +1 nucleosome, consistent with the ~150 bp spacing between the sense and convergent antisense Pol II peaks (Figures 3D and 4B). These results indicate that convergent transcription reflects sense and antisense transcription that initiate locally and undergo promoter-proximal pausing flanking the +1 nucleosome.
Figure 4. Convergent Transcription is Associated With a Distinct Chromatin Structure.
Heatmaps showing DNase I accessibility in HeLa S3 cells surrounding all (A) promoters aligned to the sense NET-seq peak, (B) promoters that have convergent transcription aligned to the antisense convergent NET-seq peak and (C) promoters that have divergent transcription aligned to the antisense divergent peak. Below each heatmap is the mean DNase I accessibility profile of the region shown in heatmap. Above each heatmap are arrows showing the transcriptional activity observed in each promoter-proximal region. A cartoon displays the chromatin structure determined by analysis of the DNase-seq data.
Convergent Transcription is a Feature of Lower-expressed Genes
Convergent transcription can regulate gene expression through transcriptional interference mechanisms (Callen et al., 2004; Elledge and Davis, 1989; Gullerova and Proudfoot, 2012; Hobson et al., 2012; Martens et al. 2004; Prescott and Proudfoot, 2002; Shearwin et al., 2005). Thus, we considered whether promoter-proximal convergent transcription may be involved in release of Pol II from promoter-proximal pausing into productive elongation. We compared Pol II density within the gene body (+1 kb to the polyadenylation site, illustrated in Figure 2A) at genes that display only convergent transcription to genes that display only divergent transcription. Notably, genes with only convergent transcription near their promoters show consistently less transcription downstream of their promoter regions (Figure 3E) (1.8 fold less on average, Kolmogorov–Smirnov test, p<<10-6). Comparison of less stringently defined sets of genes, such as all genes with convergent transcription to all genes without convergent transcription, showed a similar effect (Figure S3B). In agreement with this observation, analysis of ENCODE HeLa S3 ChIP-seq data reveals that H3K79me2 histone marks, which correlates with transcription elongation, occur at significantly lower levels in the gene bodies of genes with convergent antisense transcription (Figure S3C) (Consortium, 2012; Guenther et al., 2007; Wozniak and Strahl, 2014). Thus convergent antisense transcription could interfere with productive transcription elongation or could be a consequence of less productive elongation. Either of these possibilities could be directly mediated by Pol II or by another factor, such as chromatin.
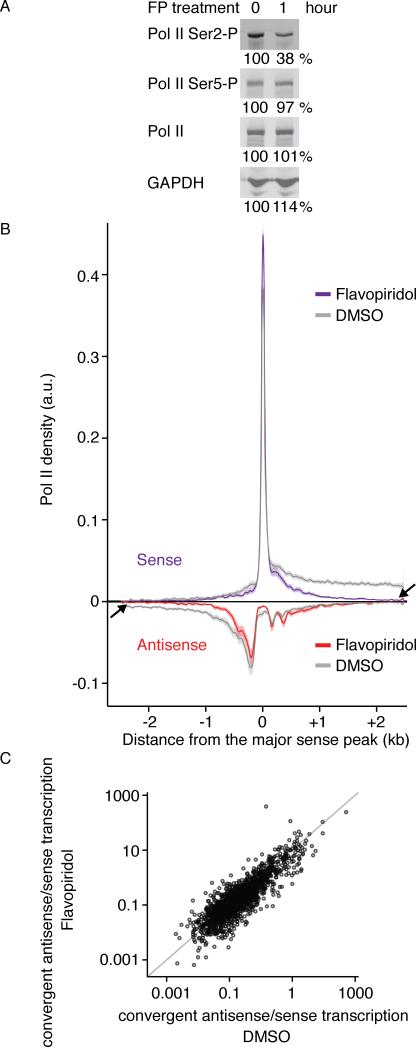
To test whether convergent antisense transcription is a consequence of reduced sense transcription elongation, we globally suppressed productive elongation by inhibiting positive transcription elongation factor b (P-TEFb). Most promoter-proximally paused Pol II are released through recruitment of P-TEFb that phosphorylates multiple proteins, including Ser2 residues of the Pol II CTD (Kim and Sharp, 2001; Peterlin and Price, 2006). Therefore, active P-TEFb greatly facilitates the transition to productive elongation, but does not affect transcription initiation (Lis et al., 2000; Peterlin and Price, 2006; Rahl et al., 2010). We performed NET-seq analysis on HeLa S3 cells exposed to the P-TEFb inhibitor flavopiridol (FP) (Chao, 2001) or DMSO alone. As expected, after 60 minutes FP reduced Pol II CTD Ser2 phosphorylation, but phosphorylation of Ser5 residues and overall Pol II levels remained unchanged (Figure 5A, S4). We generated NET-seq libraries from HeLa S3 cells after a one-hour FP treatment or DMSO control (FP treatment NET-seq dataset, 486 million total reads, 262 uniquely mapped reads; DMSO control NET-seq dataset, 491 million total reads, 263 million uniquely mapped). In agreement with previous studies, we observe a global decrease in Pol II density outside of promoter-proximal regions compared to the DMSO control (Figure 5B, arrows) (Flynn et al., 2011; Jonkers et al., 2014; Rahl et al., 2010). Thus FP treatment reduces productive elongation of most genes. To quantify the effect of FP treatment on convergent transcription, we calculated the ratio of convergent antisense to sense transcription at all promoter proximal regions. If convergent transcription were a simple consequence of lower expression, it should not only be increased proportionally to promoter-proximal sense transcription following FP treatment, but importantly it should appear in genes where it wasn't detected before. We observe that the convergent antisense to sense transcription ratio remains constant following FP treatment, indicating that sense and convergent antisense transcription levels covary, and we do not detect a new subpopulation of genes with convergent transcription in their promoter-proximal regions (Figure 5C). This result suggests that the lack of sense productive transcription elongation is not sufficient to induce convergent transcription. Thus, if convergent antisense transcription is not a simple consequence of low sense expression, then it may contribute to the cause.
Figure 5. P-TEFb Inhibition Proportionally Affects Levels of Sense Transcription and Convergent Antisense Transcription.
(A) Western blot analysis of whole cell extract of HeLa S3 cells with flavopiridol (FP) treatment (1 hr). The percentage at the bottom of each lane is the amount of the respective protein (as determined by image quantification) before and after FP treatment. GAPDH serves as a loading control. (B) Meta-gene analysis of NET-seq data from HeLa S3 cells treated with 1 μM FP (purple and red) or DMSO-control (gray) for one hour. Arrows indicate regions where transcription is affected by FP treatment. Genes that passed promoter classification criteria (described in Experimental Methods) in both datasets are included in the analysis (N = 304). NET-seq signal from each promoter region (+/- 2.5 kb centered at the TSS) are binned into 10 bp windows and normalized to vary between 0 and 1. Solid lines indicate the mean normalized Pol II density and shading shows the 95% confidence interval. (C) A scatter plot comparing the convergent to sense ratio after treatment with DMSO (control) and after FP treatment for a stringent subset of non-overlapping genes (N = 3937). The ratio is the sum of NET-seq signal on the antisense strand versus the sense strand across the 500 bp region after the TSS. TSS, transcription start site. See also Figure S4.
Impact of Transcription Factor Occupancy on Pol II Elongation
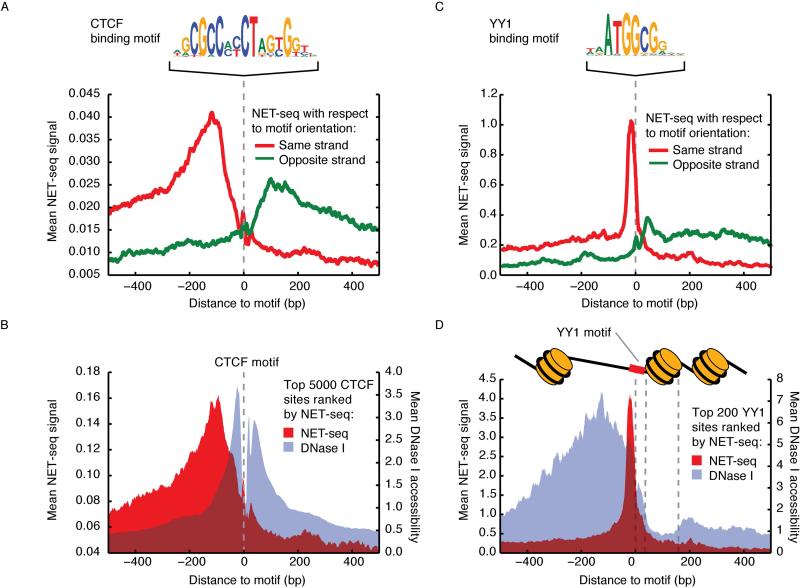
DNA-bound transcription factors (TFs) have the potential to obstruct elongating Pol II. To investigate the relationship between TF occupancy and Pol II progress, we expanded our DNase-seq data from HeLa S3 cells to genomic footprinting depth (269 million uniquely mapped genomic reads), enabling detailed mapping of the occupancy of TF recognition sites within DNase I hypersensitivity sites (DHSs). As CTCF is implicated in Pol II pausing in vitro and within the cell (Shukla et al., 2011), we quantified NET-seq signal and DNase-seq signal around CTCF recognition sites within DHSs on both strands. We observed higher Pol II density just upstream of the CTCF sites, suggesting that CTCF might represent a barrier to Pol II elongation genome-wide (Figure 6A-B). Interestingly, the NET-seq signal around these sites differs in magnitude for each strand, indicating that CTCF may pose strand-specific obstacles (Figure 6A).
Figure 6. Pol II Pausing Associated with Transcription Factor Occupancy.
(A) Average NET-seq signal around 16,339 CTCF motifs with accessible chromatin. In red is NET-seq signal oriented to the strand of the motif (Pol II transcription from left to right) and in green is NET-seq on the other strand (Pol II transcription from right to left). The CTCF binding motif is pictured above. The NET-seq data was smoothed by a 10 bp sliding window average. (B) Mean NET-seq (red) and DNase I cleavage (gray) signal (10 bp windowed averages) surrounding the top 5,000 CTCF motifs sorted by NET-seq signal. (C) Average NET-seq signal (smoothed by a 10 bp sliding window average) around 731 YY1 motifs with accessible chromatin. In red is NET-seq signal oriented to the strand of the motif (Pol II transcription from left to right) and in green is NET-seq on the other strand (Pol II transcription from right to left). The YY1 binding motif is pictured above. (D) Mean per-nucleotide NET-seq (red) and DNase I cleavage (gray) signal surrounding the top 200 YY1 motifs sorted by NET-seq signal. Both signals are presented as 10 bp windowed averages. Schematic of nucleosome positioning relative to YY1 inferred from DNase I accessibility is above plot.
As transcriptional pausing has been seen upstream of nucleosomes in yeast and Drosophila cells (Churchman and Weissman, 2011; Mavrich et al., 2008; Weber et al., 2014), we investigated Pol II density around YY1, a canonical promoter-centric transcription factor (Xi et al., 2007) thought to position +1 nucleosomes (Vierstra et al., 2013). Thus, we speculated that YY1 occupancy might impact Pol II elongation. Given that poly-zinc finger TFs engage DNA asymmetrically, we also speculated that any impact on Pol II might also be strand-specific. We observed a peak in NET-seq signal precisely at YY1 sites in DHSs, consistent with YY1-directed pausing (Figure 6C-D). Strikingly, this effect was highly directional, and is predominant when Pol II engages YY1 from the upstream direction (Figure 6D). These results indicate that TFs might directly regulate Pol II elongation in direction- or strand-specific ways.
Fine Structure of Pol II Pausing Along Constitutive and Alternative Exons
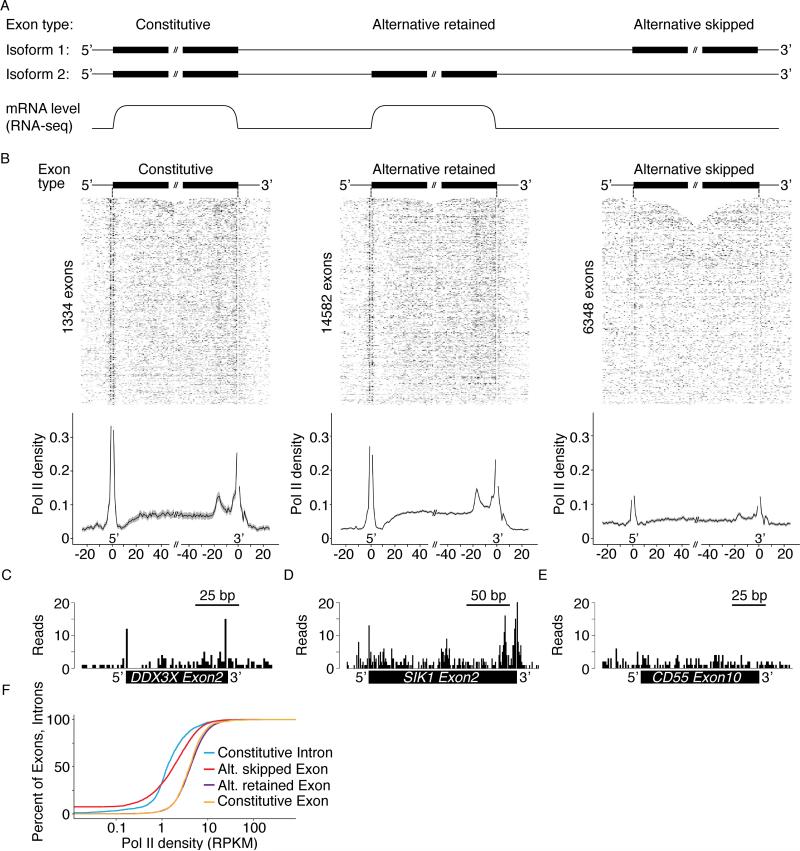
Alteration to transcription elongation rates affects splicing outcomes, which has lead to the proposal of the kinetic model of transcription and splicing coupling (Dujardin et al., 2014; Fong et al., 2014; Ip et al., 2011; la Mata et al., 2003; Roberts et al., 1998). However, the degree to which transcription rate is modulated locally around exons is unclear. Higher Pol II density at human exons versus introns was reported using Pol II ChIP-seq and ChIP-chip (Brodsky et al., 2005; Schwartz et al., 2009), but in another study, no significant difference was observed (Spies et al., 2009). Furthermore, the precise pattern across individual exons could not be resolved. In Drosophila cells, PRO-seq observed high Pol II density across exons, and detected a high enrichment of Pol II density at the 5′ ends (Kwak et al., 2013). We analyzed NET-seq data at constitutive exons and revealed significantly higher coverage than at introns in both HeLa S3 (2.4× higher) and HEK 293T cells (2.2× higher) (p-value << 10-15, Kolmogorov–Smirnov test), suggesting that transcription elongation is slower at exons in human cells (Figures 7A-B, F and S5B). Any contamination from processed mRNA would inflate these differences, however, our quality controls (Figure 1B-C) suggest that this is a small effect, if at all. Interestingly, NET-seq signal across exons is not flat: sharp increases in Pol II density occur in the few base pairs surrounding the 5′ and 3′ ends of constitutive exons, indicating strong Pol II pausing at exon boundaries (Figure 7B). Furthermore, a broader peak of RNA polymerase density is present ~17 nt before the 3′ end of exons. The general features of this pattern are observed at single exons, for example, exons 2 of the DDX3X and SIK1 genes (Figure 7C-D). Finally, we observe similar trends in the NET-seq data from HEK 293T cells (Figure S5B). This analysis suggests that exon borders impose a structured barrier to Pol II elongation.
Figure 7. Pol II Density Across Exons Reveals a Stereotypical Pausing Pattern that Depends on Splicing Outcome.
(A) Schematic of the classification of constitutive, alternative retained and alternative skipped exons based on annotated isoforms and detected levels in cytosolic RNA-seq data. (B) A stringent set of exons were selected for analysis from genes containing NET-seq signal of ≥ 1 RPKM (see Experimental Procedures). Heat maps and meta-exon analysis of HeLa S3 Pol II density across each type of exon as defined in Figure 7A (Left to right, constitutive exons, N = 1334, alternative retained, N = 14582 and alternative skipped, N = 6348). NET-seq signal from each exon (+/- 25 bp) is normalized to vary from 0 to 1. Solid lines on the meta-exon plots indicate the mean values and the gray shading represents the 95% confidence interval. The single nucleotide positions where splicing intermediates align (3′ ends of introns and exons) were entirely removed from analysis (see Experimental Procedures) and appear as a blank position in the figures. Raw NET-seq reads across the constitutive exon 2 within the DDX3X gene (C), alternative retained exon 2 within the SIK1 gene (D) and the alternative skipped exon 10 within the CD55 gene (E). (F) Distribution of the percent of exons or introns with a given Pol II density. RPKM, reads per kb per million Pol II uniquely aligned reads. See also Figure S5.
Most human exons can be alternatively spliced, with retained exons varying between cell types (Pan et al., 2008; Wang et al., 2008). We expanded our analysis to alternatively spliced exons and investigated whether transcriptional pausing varies at exons with different splicing outcomes. We focused our analysis on genes with an NET-seq RPKM of greater than 1 in the gene body (Figure S5A) and defined skipped exons as those undetected in the cytoplasmic RNA-seq data (Figure 7A). As for constitutive exons, retained alternative exons have higher Pol II density compared to the density across introns (Figure 7F). These exons also have a similar pausing pattern as constitutive exons, which is visible by meta-exon analysis and at the single exon (Figure 7B, D). Interestingly, Pol II density is lower at skipped exons than at alternative retained exons (Figure 7F). Strikingly, the Pol II density pattern is similarly shaped across skipped and retained exons, albeit significantly different in amplitude (Figure 7B, E). The residual pausing pattern at skipped exons could be due to the small number of retained exons that are misannotated as skipped. Finally, the same differences in the Pol II density patterns across retained and skipped exons are observed in HEK293T cells (Figure S5B). Together these data show that Pol II recognizes exon structures with different processing fates, suggesting that alternative splicing is kinetically coupled to transcription elongation genome-wide.
DISCUSSION
Here we demonstrate that human NET-seq provides complete, strand-specific maps of transcription at single-nucleotide resolution. NET-seq thereby defines transcriptional pausing sites and directly measures unstable transcripts. Finally, NET-seq instantaneously reports the transcription status of genes, in contrast to RNA-seq, which reports the balance between RNA synthesis and degradation.
Our work describes an unappreciated aspect of promoter-proximal transcription: the presence of convergent transcription at many human genes. Importantly, we show that convergent transcription is a hallmark of lower-expressed genes, suggesting a potential role in the regulation of promoter-proximal pausing. Prominent DNase I hypersensitivity sites flanking the convergent antisense peak indicate that promoter-proximal convergent transcription reflects initiation at a defined promoter located a characteristic distance from the canonical sense promoter.
Other than expression level, one small commonality was found between the genes with convergent transcription: the dinucleotide CC occurs slightly more frequently in regions displaying convergent transcription (12.4% ± 0.4% for convergent, 11.1% ± 0.2% for not convergent). Thus it appears that convergent transcription is a prevalent phenomenon that is not restricted to a specific class of genes. An intriguing possibility is that paused antisense Pol II directly blocks or clashes with sense transcription, as can occur in yeast (Prescott and Proudfoot, 2002). The sense and convergent antisense peaks are too far apart (~150 bp) to reflect direct contact of paused polymerases, but the DNase-seq data reveal that this distance likely represents the +1 nucleosome that is positioned between them. Interference could arise through positioning of the +1 nucleosome or indirect mechanisms such as transcription-induced changes in DNA topology, chromatin modifications or transcription factor occupancy. In any event, NET-seq data do not resolve whether sense and antisense transcription occur simultaneously as the approach requires averaging over a population of cells. Therefore, potential roles of convergent transcription during initiation, elongation and termination will have to be investigated within cell populations and at the single cell level.
Our study yields a global picture of how transcription elongation is altered at alternatively spliced exons in human cells. Changes in transcription elongation influence alternative splicing, which is thought to be mediated either by the differential recruitment of splicing factors (recruitment model) or by biasing kinetic competition between multiple splicing outcomes (kinetic model) (Bentley, 2014; Dujardin et al., 2013; Kornblihtt et al., 2004). Here, we show that alternative splicing outcomes in human cells are associated with Pol II exon density and strong pauses at the 5′ and 3′ ends, consistent with the kinetic model. What causes pauses at exons is an important question. Nucleosomes can influence transcriptional pausing (Churchman and Weissman, 2011; Hodges et al., 2009; Izban and Luse, 1991; Skene et al., 2014) and, importantly, nucleosome occupancy and histone modifications transition at exon boundaries according to splice site strength (Huff et al., 2010; Schwartz et al., 2009; Spies et al., 2009; Tilgner et al., 2009). DNA sequence and DNA methylation at exon boundaries could contribute to pausing, because sequence elements have been shown to cause transcriptional pausing (Gelfman et al., 2013; Herbert et al., 2006; Kassavetis and Chamberlin, 1981; Larson et al., 2014; Maizels, 1973; Vvedenskaya et al., 2014) Additionally, transcription factors could underlie pausing at retained exons, as is the case with CTCF binding at exon 5 of the CD45 gene (Shukla et al., 2011). The broad peak of Pol II density 15 bp from the 3′ end of the exon may reflect Pol II backtracking during the recovery from the strong pause at the 3′ end of the exon. Backtracking would produce small cleavage products consistent with the population of tiny RNAs that were previously identified in this region (Taft et al., 2009).
We expect adaptation of human NET-seq to any human cell type to be straightforward, resulting in a tool to illuminate a variety of biological processes. Future applications include high-resolution analyses of transcription regulation across cell types, responses to signaling pathways, and cellular differentiation.
EXPERIMENTAL PROCEDURES
Cell fractionation and RNA purification
Cell fractionation is performed as described by (Bhatt et al., 2012; Pandya-Jones and Black, 2009) and based on (Wuarin and Schibler, 1994) with modifications. All steps are conducted on ice or at 4°C and in the presence of 25 μM α-amanitin, 50 Units/ml SUPERaseIN and Protease inhibitors cOmplete. HeLa S3 cells and HEK 293T cells are grown in DMEM containing 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin to a confluency of 90%. Following lysis of 1×107 cells, the nuclei are washed with the nuclei wash buffer (0.1% Triton-X-100, 1 mM EDTA, in 1× PBS) to remove cytoplasmic remnants. Nuclei lysis is performed without MgCl2 (1% NP-40, 20 mM Hepes pH 7.5, 300 mM NaCl, 1 M Urea, 0.2 mM EDTA, 1 mM DTT). The success of cell fractionation is monitored by Western blot analysis and subcellular RNA-seq.
Sequencing library constructions
For NET-seq, the library preparation is performed as described by (Churchman and Weissman, 2011; 2012) with modifications. For 3′ RNA ligation, a pre-adenylated DNA linker with a mixed random hexameric barcode sequence at its 5′ end is used. cDNA containing the 3′ end sequences of a subset of mature and heavily sequenced snRNAs, snoRNAs, rRNAs and mitochondrial tRNAs are specifically depleted using biotinylated DNA oligos (Supplemental Table S1) as described by (Ingolia et al., 2012). For subcellular RNA-seq, the sequencing libraries are prepared as described in (Churchman and Weissman, 2012), with the ribosomal RNA removed using the Ribo-Zero Magnetic Kit (Epicentre). DNA libraries are sequenced by the NextSeq 500 and HiSeq 2000 Illumina platforms.
Processing and Alignment of Sequencing Reads
Reads are trimmed and aligned using STAR (v2.4.0) (Dobin et al., 2013). For NET-seq data, only the position matching the 5′ end of the sequencing read (after removal of the barcode), corresponding to the 3′ end of the nascent RNA fragment, is recorded with a python script using HTSeq package (Anders et al., 2014). Reverse transcription mispriming events are identified and removed when molecular barcode sequences match exactly to the genomic sequence adjacent to the aligned read. Reads that align to the same genomic position and contain identical barcodes are considered PCR duplication events and are removed. Splicing intermediates have 3′ hydroxyls and will enter NET-seq libraries and contribute to the reads aligning to the exact single nucleotide 3′ ends of introns and 3′ ends of exons (Figure S1G). Therefore, reads that map precisely at the exact single nucleotide ends of introns and exons are discarded and the single 1 bp genomic positions are not considered in subsequent analysis.
Annotation of Exons and Introns
Clear exonic regions are identified by determining the minimum overlapping exonic region of all isoforms that have an exon at that position. If the region is present in all isoforms, it is considered a constitutive exon, otherwise it is labeled alternative. Alternative skipped exons are classified by those alternative exons that are entirely undetected in the cytoplasm RNA-seq data and the rest of the alternative exons are classified as retained. Constitutive intronic regions are identified as the minimum intronic overlapping regions present in all isoforms.
NET-seq Exon Metagene and Heatmap Analysis
The set of exons included in the analysis are required to be within genes of an RPKM greater than 1 in gene bodies (defined in Figure 2A) and not overlapping any other annotated feature. They are required to begin and end at the same position in all isoforms that contain the exon. First and last exons of genes are removed from analysis. NET-seq signal across each exon +/- 25 bp is normalized to range between 0 and 1 so that each exon contributes to the analysis with the same weight. Precise single nucleotide genomic loci where splicing intermediates map (exact 3′ ends of introns and exons) are not included in the analysis and those locations are left blank in any plots.
Analysis of Promoter-Proximal Regions
Promoter-proximal regions were carefully selected for analysis to ensure that there is minimal contamination from transcription arising from other transcription units. Starting with genes that are Pol II protein-coding, non-overlapping within a region of 2.5kb upstream of the TSS and 2.5kb downstream of the polyA site, and longer than 2 kb, NET-seq data at promoter-proximal regions are required to have a coefficient of variation greater than 0.5 and have at least 40 positions covered in the sense strand. Within a 4 kb window surrounding the TSS, peaks were identified in the sense from these genes. If more than 40 bases on the antisense strand have NET-seq signal, peaks were also identified on the antisense strand. Promoters with a major or minor antisense peaks located downstream of the sense major peak are classified as displaying convergent transcription. Promoters with a major or minor antisense peaks located upstream of the sense major peak are classified as displaying divergent transcription.
Supplementary Material
Article Highlights.
Human NET-seq maps global RNA polymerase II (Pol II) density at high resolution
Widespread convergent transcription occurs near promoters of lower-expressed genes.
Strong Pol II pausing at sites of occupied transcription factors, including YY1 and CTCF.
NET-seq reveals pronounced Pol II pausing at the boundaries of retained exons.
ACKNOWLEDGMENTS
We thank F. Winston, J. Gray, M. Couvillion, S. Doris and E. Feinberg for critical comments on the manuscript; We thank J.Gray and A. Snavely for help with eRNA analysis; We thank F. Winston, K. Struhl, S. Buratowski, A. Ciuffi and A. Regev for advice and discussions; We thank M. Gebremeskel for tissue culture support; K. Waraska at the HMS Biopolymers Facility and Z. Herbert at the DFCI Molecular Biology Core Facilities for sequencing; and N. Pho and BD Kim at HMS Research Computing for computing support. This work was supported by US National Institutes of Health NHGRI grants R01HG007173 to L.S.C. and U54HG007010 to J.A.S., a Damon Runyon Dale F. Frey Award for Breakthrough Scientists (to L.S.C.), and a Burroughs Wellcome Fund Career Award at the Scientific Interface (to L.S.C). A.M. was supported by Long-Term Postdoctoral Fellowships of the Human Frontier Science Program (LT000314/2013L) and EMBO (ALTF858-2012). J.dI. was supported by the Swiss National Science Foundation Postdoctoral Fellowship. J.V. was supported by US National Science Foundation Graduate Research Fellowship under grant DGE-071824.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.M., J.dI. and L.S.C. designed the NET-seq experiments; A.M. established NET-seq and subcellular RNA-seq experimental protocols, with input from J.dI.; A.M. and S.P.M. carried out experiments; J.dI. developed a bioinformatics analysis pipeline for human NET-seq and subcellular RNA-seq, with input from A.M.; A.R., J.V., R.S. and J.A.S. generated and analyzed the DNase-seq data; J.dI., U.E. and L.S.C. analyzed NET-seq data; A.M., J.dl., J.A.S. and L.S.C. wrote the manuscript.
Accession Number
All NET-seq and RNA-seq data sets are available at GEO under the accession number GSE61332. DNase-seq data sets are available at ENCODE under the ENCODE DCC accession number ENCBS229UDI.
References
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high- throughput sequencing data. bioRxiv. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Mol Cell Biol. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong A-J, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Luse DS. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J Biol Chem. 1987;262:298–304. [PubMed] [Google Scholar]
- Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Chao SH. Flavopiridol Inactivates P-TEFb and Blocks Most RNA Polymerase II Transcription in Vivo. Journal of Biological Chemistry. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Native elongating transcript sequencing (NET- seq). Current Protocols in Molecular Biology / Edited by Frederick M Ausubel [Et Al] 2012 doi: 10.1002/0471142727.mb0414s98. Chapter 4, Unit4.14.1–Unit4.14.17. [DOI] [PubMed] [Google Scholar]
- Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro CM, Alver BH, Marguerat S, Stepanova E, Davis CP, Bähler J, Park PJ, Winston F. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol. 2013;33:4779–4792. doi: 10.1128/MCB.01068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, la Mata, de M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell. 2014;54:683–690. doi: 10.1016/j.molcel.2014.03.044. [DOI] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, Petrillo E, Buggiano V, Gómez Acuña LI, Fiszbein A, Godoy Herz MA, Nieto Moreno N, Munoz MJ, Alló M, et al. Transcriptional elongation and alternative splicing. Biochimica Et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Ferrari F, Plachetka A, Alekseyenko AA, Jung YL, Ozsolak F, Kharchenko PV, Park PJ, Kuroda MI. “Jump start and gain” model for dosage compensation in Drosophila based on direct sequencing of nascent transcripts. Cell Reports. 2013;5:629–636. doi: 10.1016/j.celrep.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proceedings of the National Academy of Sciences. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu X-D, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28:2663–2676. doi: 10.1101/gad.252106.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Cohen N, Yearim A, Ast G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013;23:789–799. doi: 10.1101/gr.143503.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat Struct Mol Biol. 2012;19:1193–1201. doi: 10.1038/nsmb.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal Fluctuations Govern the Transcription Dynamics of RNA Polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407–e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Chamberlin MJ. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J Biol Chem. 1981;256:2777–2786. [PubMed] [Google Scholar]
- Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem. 2001;276:12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Set3 HDAC Mediates Effectsof Overlapping Noncoding Transcription on Gene Induction Kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, la Mata, de M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Lindell TJ, Weinberg F, Morris PW, Roeder RG, Rutter WJ. Specific Inhibition of Nuclear RNA Polymerase II by agr-Amanitin. Science. 1970;170:447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Maizels NM. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci USA. 1973;70:3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Escalante-Chong R, Pho N, Wang J, Churchman LS, Springer M, Buratowski S. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014;157:1712–1723. doi: 10.1016/j.cell.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. Polyadenylation site–induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. Rna. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the Elongation Phase of Transcription with P TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, Mccuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Hernandez AE, Groudine M, Henikoff S, Espinosa JM. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife. 2014;3 doi: 10.7554/eLife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. Embo J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest ARR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigó R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, Guigó R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- Vierstra J, Wang H, John S, Sandstrom R, Stamatoyannopoulos JA. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nat Meth. 2013;11:66–72. doi: 10.1038/nmeth.2713. [DOI] [PubMed] [Google Scholar]
- Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. Nucleosomes Are Context-Specific, H2A.Z-Modulated Barriers to RNA Polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Wu X, Sharp PA. Divergent Transcription: A Driving Force for New Gene Origination? Cell. 2013;155:990–996. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Yu Y, Fu Y, Foley J, Halees A, Weng Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res. 2007;17:798–806. doi: 10.1101/gr.5754707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.