Abstract
Transcription-regulating long non-coding RNAs (lncRNAs) have the potential to control the site-specific expression of thousands of target genes. Previously, we showed that Evf2, the first described ultraconserved lncRNA, increases the association of transcriptional activators (DLX homeodomain proteins) with key DNA enhancers but represses gene expression. In this report, mass spectrometry shows that the Evf2-DLX1 ribonucleoprotein (RNP) contains the SWI/SNF-related chromatin remodelers Brahma-related gene 1 (BRG1, SMARCA4) and Brahma-associated factor (BAF170, SMARCC2) in the developing mouse forebrain. Evf2 RNA colocalizes with BRG1 in nuclear clouds and increases BRG1 association with key DNA regulatory enhancers in the developing forebrain. While BRG1 directly interacts with DLX1 and Evf2 through distinct binding sites, Evf2 directly inhibits BRG1 ATPase and chromatin remodeling activities. In vitro studies show that both RNA-BRG1 binding and RNA inhibition of BRG1 ATPase/remodeling activity are promiscuous, suggesting that context is a crucial factor in RNA-dependent chromatin remodeling inhibition. Together, these experiments support a model in which RNAs convert an active enhancer to a repressed enhancer by directly inhibiting chromatin remodeling activity, and address the apparent paradox of RNA-mediated stabilization of transcriptional activators at enhancers with a repressive outcome. The importance of BRG1/RNA and BRG1/homeodomain interactions in neurodevelopmental disorders is underscored by the finding that mutations in Coffin–Siris syndrome, a human intellectual disability disorder, localize to the BRG1 RNA-binding and DLX1-binding domains.
KEY WORDS: Long non-coding RNA, Chromatin remodeling, Homeodomain proteins, Evf2, Dlx6os1, BRG1, SMARCA4
Summary: In the developing mouse forebrain, the lncRNA Evf2 forms a complex with DLX1, BRG1 and BAF170. BRG1 directly interacts with Dlx1 and Evf2, the latter inhibiting its ATPase and chromatin remodeling activity.
INTRODUCTION
Genome-wide studies revealed that the majority of RNA transcripts in mammalian cells do not encode proteins [non-coding RNAs (ncRNAs)]. Long non-coding RNAs (lncRNAs) have emerged as a class of molecules with highly diverse structures and functions, including roles in transcriptional regulation (Geisler and Coller, 2013; Yang et al., 2014). Reports of single trans-acting lncRNAs controlling gene expression in cell lines (Lanz et al., 1999; Feng et al., 2006; Rinn et al., 2007) or in mice (Bond et al., 2009; Gabory et al., 2009; Rapicavoli et al., 2011) have been expanded by the genomic era to include thousands of lncRNAs with the potential for transcriptional effects. Families of enhancer RNAs (eRNAs; Ørom et al., 2010), ultraconserved RNAs [ucRNAs; also known as transcribed ultraconserved RNAs (t-UCRs); Calin et al., 2007] and opposite strand transcripts [OS; also called natural antisense transcripts (NATs); Yelin et al., 2003] have been identified. The biological significance of lncRNAs has increased with reports of roles in disease processes as diverse as lung cancer (Gutschner et al., 2013), pain response (Zhao et al., 2013) and microbial susceptibility (Gomez et al., 2013).
Transcription-regulating lncRNAs are not only functionally but also mechanistically diverse, regulating gene expression through trans and cis effects. Studies on Evf2, the first lncRNA member of larger classes of ucRNAs/t-UCRs, show that Evf2 (Dlx6os1, Dlx6as) is transcribed on the opposite strand, antisense to Dlx6, and therefore also belongs to the class of OS/NATs (Feng et al., 2006). Although antisense transcription predicted that Evf2 would have cis-regulatory effects, multiple experiments suggest that Evf2 exhibits both trans and cis effects (Feng et al., 2006; Bond et al., 2009; Berghoff et al., 2013). Chromatin immunoprecipitation (ChIP) experiments show that Evf2 increases the binding of transcriptional activators (DLX1/2; Zerucha et al., 2000) and the repressor methyl-CpG binding protein 2 (MECP2; Nan et al., 1997) to key Dlx5/6 enhancers (Zerucha et al., 2000) with a repressive outcome (Bond et al., 2009). Genetic epistasis experiments support a model in which Evf2 regulates Dlx5/6 gene expression by modulating the antagonistic interactions between DLX1/2 and MECP2, and regulating Dlx5/6 ultraconserved enhancer site-specific methylation (Berghoff et al., 2013). However, beyond complex formation with DLX proteins, Evf2-ribonucleoprotein (RNP)-containing complexes have not been characterized.
In these studies, we further investigate the mechanism of Evf2 transcriptional control, and focus on mouse E13.5 ganglionic eminence (GE), the site of sonic hedgehog activation of Dlx1/2/5/6 and Evf2 gene expression (Kohtz et al., 1998; Feng et al., 2006). Using mass spectrometry to sequence Evf2/DLX RNP complexes in vivo, we identify direct interactions between Evf2 and BRG1/BAFs, which are components of a SWI/SNF-related chromatin remodeling complex (Wang et al., 1996; Phelan et al., 1999; Kasten et al., 2011; Staahl and Crabtree, 2013), and between BRG1 (SMARCA4) and the DLX1 homeodomain protein. While Evf2 increases the association of BRG1 with key Dlx5/6 DNA regulatory enhancers in the developing forebrain, Evf2 also inhibits BRG1 ATPase and chromatin remodeling activity in vitro. Evf2/BRG1, Evf2-BAF170 (SMARCC2), and Evf2-BAF155 (SMARCC1) interactions are competed by specific ribohomopolymers but not tRNA, suggesting that RNA structure and length, but not sequence complexity, determine RNA binding. Together, these data support the contention that lncRNA-mediated transcriptional repression occurs through direct inhibition of chromatin remodeling, a mechanism distinct from recruitment of histone modification enzymes (Rinn et al., 2007; Nagano et al., 2008; Pandey et al., 2008; Zhao et al., 2008). Given that the BRG1 RNA-binding domain and one of the BRG1 DLX1-binding domains are mutated in the human intellectual disorder Coffin–Siris syndrome (CSS; Tsurusaki et al., 2012, 2014), characterization of RNA-mediated chromatin remodeling might be important for understanding neurodevelopmental disorders.
RESULTS
Evf2/DLX1 complexes contain SWI/SNF-related chromatin remodelers
Previous results show that Evf2 forms nuclear complexes with DLX homeodomain proteins (Feng et al., 2006) and that association of DLX1/2 with key Dlx5/6 DNA regulatory sequences decreases in mice lacking Evf2 (Evf2TS/TS) (Bond et al., 2009). Evf2TS/TS mice were generated by insertion of a triple polyA transcription STOP sequence (TS) in the 5′ end of Evf2, successfully preventing Evf2 transcription without disrupting expression of the adjacent Evf1 transcript (Bond et al., 2009). As described above, the E13.5 mouse GE is the site of sonic hedgehog-induced Dlx1/2/5/6 and Evf1/2 activation during forebrain development (Kohtz et al., 1998, 2001; Feng et al., 2006). In order to study Evf2 RNA/DLX protein complexes in E13.5 GE, we used anti-DLX immunoaffinity purification followed by mass spectrometry sequencing. We cross-linked a well-characterized anti-pan-DLX antibody (Kohtz et al., 2001; Feng et al., 2004, 2006; Bond et al., 2009) to cyanogen bromide-activated Sepharose beads, purified complexes from wild-type (Evf2+/+) and Evf2TS/TS E13.5 GE nuclear extracts, and compared the identity of DLX-bound proteins in the presence and absence of Evf2 RNA by mass spectrometry sequencing (Washburn et al., 2001).
DLX1 is the only DLX family member identified in both Evf2+/+ and Evf2TS/TS extracts. Members of SWI/SNF-related chromatin remodeling complexes are identified in Evf2+/+ nuclear extracts (Fig. 1A). DLX1-bound complexes from Evf2+/+ nuclear extracts contain the following eight proteins with the potential to affect chromatin remodeling: BRG1, BAF170, ARID1A (predicted), SNF2L (SMARCA1) and SNF2H (SMARCA5) (mammalian ISWI homologs), BAZ1A and BAZ1B (bromodomain adjacent to zinc finger proteins) and polybromo 1 (for a complete list of proteins see supplementary material Table S1). Total BRG1 and BAF170 protein levels are the same in Evf2+/+ and Evf2TS/TS nuclear extracts (Fig. 1B), supporting that mass spectrometry differences in BRG1 and BAF170 from Evf2+/+ and Evf2TS/TS DLX complexes do not result from Evf2 regulation of BRG1 or BAF170 protein production or stability. The endogenous DLX1-BRG1 complex in Evf2+/+ E13.5 GE nuclear extracts is further confirmed by co-immunoprecipitation of BRG1 with anti-DLX antibody (Fig. 1C). Although immunoprecipitation is performed in the presence of protease inhibitors, BRG1 might be cleaved during the immunoprecipitation process, as multiple bands are detected after immunoprecipitation with anti-BRG1 and anti-DLX (Fig. 1C).
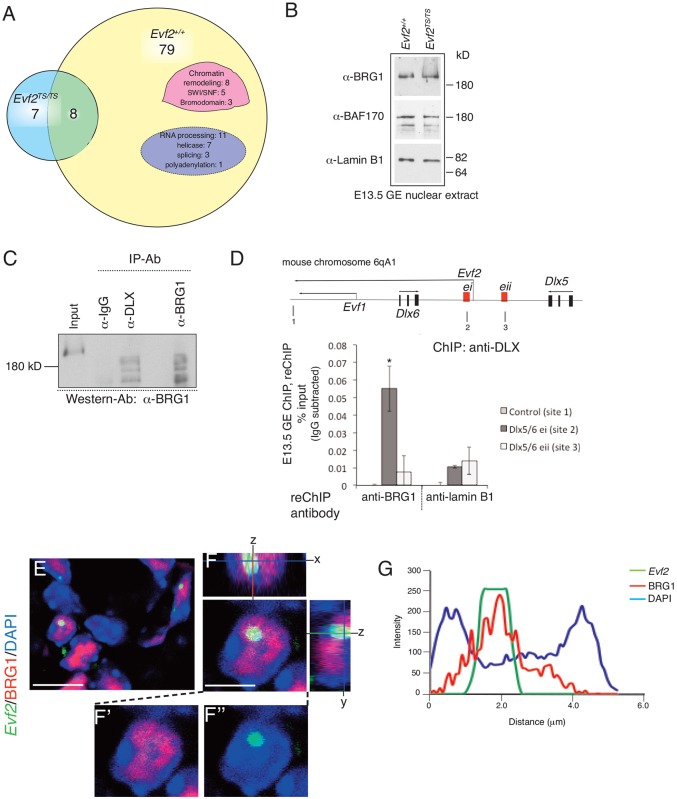
Fig. 1.
Identification of Evf2/DLX1/BRG1 complexes in mouse embryonic brain. (A) Venn diagram summarizing the results from mass spectrometry sequencing of E13.5 ganglionic eminence (GE) brain protein complexes. Proteins from E13.5 GE nuclear extracts were affinity purified on an anti-DLX column and sequenced. Among a total of 87 Evf2+/+ proteins, 79 were unique to Evf2+/+, of which eight were associated with CRs and 11 with RNA processing. Among a total of 15 Evf2TS/TS proteins, seven were unique to Evf2TS/TS. Eight proteins were common to Evf2+/+ and Evf2TS/TS. (B) Western blot analysis of Evf2+/+ and Evf2TS/TS nuclear extracts shows equal amounts of BRG1, BAF170 and lamin B proteins. (C) Co-immunoprecipitation of DLX/BRG1 complexes from Evf2+/+ E13.5 GE nuclear extracts. Immunoprecipitation with the antibodies indicated (IP-Ab) was followed by western blot analysis and probing with anti-BRG1 antibody. Input (10%) is loaded in the first lane; 25% of the anti-BRG1 immunoprecipitated protein is loaded in the last lane. (D) ChIP, followed by reChIP, defines a co-complex of DLX/BRG1 localized to the Dlx5/6 ultraconserved enhancer (ei). E13.5 GE chromatin was immunoprecipitated first with anti-DLX, and eluted complexes were then immunoprecipitated with anti-BRG1, anti-lamin B or anti-IgG. Percent input values are obtained after subtraction of IgG values. Primer sites (1-3) are indicated in the schematic. *P<0.05 (Student's two-tailed t-test) for BRG1, but not lamin B. Error bars indicate s.e.m. Chromatin was isolated from a pool of ∼20 GEs, and experiments were duplicated. (E-F″) Visualization of RNA/protein clouds in E13.5 GE. Evf2 fluorescent RNA in situ hybridization (FISH; green) and immunofluorescent detection of BRG1 protein (red) are visualized by confocal microscopy. Nuclei are stained with DAPI (blue). Confocal z-stack images show Evf2/BRG1 colocalization in yellow. (G) Fluorescent intensity profiles across the nucleus of a neuronal precursor in E13.5 GE (Evf2 RNA, green; BRG1 protein, red; nuclei, blue). In this nucleus, the highest peaks from Evf2 and BRG1 coincide, showing enrichment. Additional scans are shown in supplementary material Fig. S1, indicating Evf2/BRG1 correlation in the majority of nuclei, as well as some heterogeneity. Scale bars: 10 µm in E; 4 µm in F.
BRG1 is an essential component of SWI/SNF-related chromatin remodeling complexes that contain different combinations of BAFs, depending on cell type and state of differentiation (Lessard et al., 2007). BRG1 is an ATPase that can remodel nucleosome positioning along the DNA, even in the absence of other BAFs, in vitro (Workman and Kingston, 1992; Phelan et al., 1999). BRG1-BAFs regulate gene expression crucial for neural progenitor differentiation (Lessard et al., 2007; Yoo and Crabtree, 2009). In addition, Brg1 null mice show decreased Dlx1 gene expression in the developing ventral telencephalon (Lessard et al., 2007). These studies support a biological role of BRG1 in regulating gene expression in embryonic ventral telencephalic interneuron precursors, and led to further analysis of DLX-BRG1 complexes in E13.5 GE.
Protein complexes identified in nuclear extract lysates represent soluble but not necessarily chromatin-bound complexes. Given our previous experiments showing that Evf2 increases DLX binding to Dlx5/6 enhancers ei and eii (Bond et al., 2009), we next tested whether DLX-bound Dlx5/6 ei and eii complexes also contain BRG1. We performed ChIP using anti-DLX, followed by elution, and then reChIP with anti-BRG1 (Fig. 1D). DLX-BRG1 co-complexes are detected at the ultraconserved Dlx5/6 ei enhancer (site 2, but not sites 1 or 3; Fig. 1D). The ability to detect DLX-BRG1 complexes at Dlx5/6 ei but not eii could be due to differences in DLX and BRG1 enrichment at ei and eii, combined with the low efficiency of reChIP. Together, these data support the presence of both soluble and enhancer-bound DLX1-BRG1 complexes in E13.5 GE. The presence of DLX-BRG1 complexes at the Dlx5/6 ei enhancer supports a functional role for BRG1 in regulating enhancer activity.
BRG1 colocalizes with Evf2 RNA clouds
Fluorescent RNA in situ hybridization (FISH) previously showed that Evf2 forms nuclear RNA clouds in E13.5 GE neuronal progenitors, colocalizing with DLX homeodomain proteins (Feng et al., 2006). Evf2 RNA clouds appear similar to those reported for lncRNAs involved in imprinting, namely Xist and Kcnq1ot1 (Redrup et al., 2009; Brockdorff, 2011). Fluorescent RNA in situ hybridization and immunolocalization (FISH-immuno), followed by confocal microscopy, shows that BRG1 colocalizes with Evf2 in E13.5 GE nuclear RNA clouds (Fig. 1E-F″). Confocal z-stack analysis shows that BRG1 is scattered within the Evf2 RNA cloud (Fig. 1F). Intensity plots show enrichment of BRG1 within Evf2 clouds for the majority of nuclei (21/25) expressing Evf2 (Fig. 1G; supplementary material Fig. S1). This supports the idea that Evf2-BRG1 interactions occur within RNA clouds, but also suggests that there is heterogeneity in the relative distribution of Evf2 and BRG1 among E13.5 GE interneuron precursors.
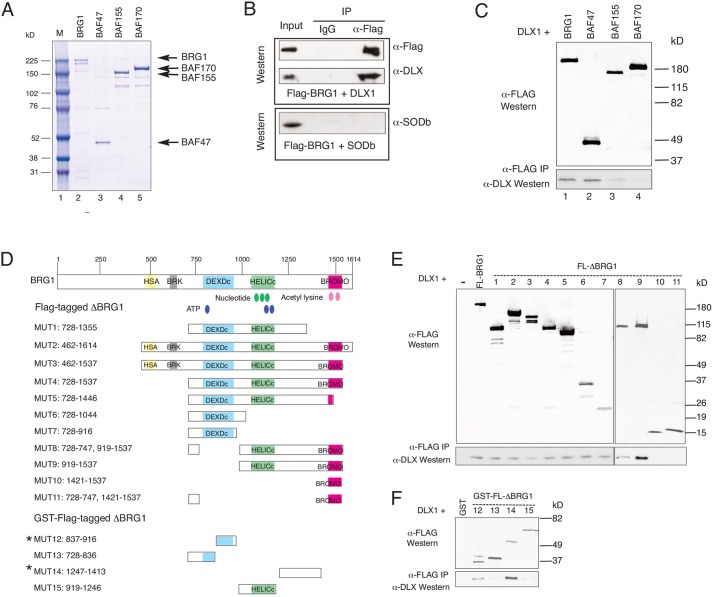
BRG1 contains two distinct DLX1-binding domains that correspond to sites of mutations found in CSS and overlap with the SNF2 ATP-coupling domain
Isolation of DLX1 complexes containing BRG1/BAF170 raises the possibility of direct interactions between DLX1 and BRG1/BAF170. Co-immunoprecipitation experiments using Flag-tagged BRG1 (Fig. 2A) and His-tagged DLX1 indicate that BRG1 directly binds DLX1, but not the control His-tagged protein SODb (Fig. 2B). In addition, BAF47 (SMARCB1; Wang et al., 1996), but not BAF170 or BAF155 (low-intensity band) bind DLX1 (Fig. 2C). Mass spectrometry results identified DLX1-BRG1 complexes in nuclear extracts containing Evf2 (Evf2+/+) but not in its absence (Evf2TS/TS) (Fig. 1A), raising the possibility that DLX1-BRG1 interactions might result from direct or indirect effects of Evf2. However, direct interactions between DLX1 and BRG1 in the absence of Evf2 (Fig. 2B) show that DLX1-BRG1 binding does not require the presence of Evf2 in vitro. Because we are dealing with such small amounts of protein (derived from embryonic brain GE nuclear extract lysates), we predict that the mass spectrometry results reflect enrichment rather than absolute numbers. Therefore, the mass spectrometry data indicating that BRG1 bound to DLX1 is detected in Evf2+/+, but not Evf2TS/TS, supports enrichment in the presence of Evf2 but does not exclude the possibility that some amount of BRG1 is bound to DLX1 in Evf2TS/TS. Together, these data support the formation of DLX1-BRG1-BAF170 complexes, stabilized by direct interactions between DLX1 and BRG1, and DLX1 and BAF47.
Fig. 2.
BRG1 binds DLX1 through two distinct binding domains. (A) Coomassie stain of Flag-tagged proteins isolated from baculovirus-infected insect cells. M, prestained size marker. (B-F) Direct interactions between DLX1 and BRG1 or BAF47. Flag-tagged and GST recombinant fusion proteins (8 pmoles) were: BRG1, FL-Δ-BRG1 (MUT1-15), BAF47, BAF155 and BAF170. His-tagged proteins (14 pmoles) were: DLX1, SODb. (B) DLX1 and BRG1 direct interactions. Flag-BRG1 is incubated with His-tagged DLX1 or His-tagged SODb, and immunoprecipitated (IP) with anti-IgG or anti-Flag-conjugated agarose. Western blot of IP complexes with anti-Flag, anti-DLX or anti-SODb indicates that DLX1 directly binds to BRG1. (C) DLX1 directly interacts with BAF47, but not BAF155 or BAF170. His-tagged DLX1 is incubated with Flag-tagged proteins, immunoprecipitated with anti-Flag and probed with anti-DLX by western blot. (D) Schematic of Flag-tagged BRG1 recombinant proteins. MUT 12-14 are GST fusions. Known functional domains include nucleotide-, ATP- and acetyl lysine-binding domains (green, blue and pink ovals) and HSA (helicase/SANT associated, β-actin binding), BRK (chromodomain/helicase shared domain with unknown function), DEXDc (DEAD-like helicase), HELICc (helicase) and BROMO (bromodomain). (E,F) Co-immunoprecipitation experiments with DLX1 and Flag-tagged BRG1 FL-Δ-BRG1 MUT1-15; anti-Flag detects BRG1 input, while anti-DLX1 detects DLX1 protein bound to BRG1.
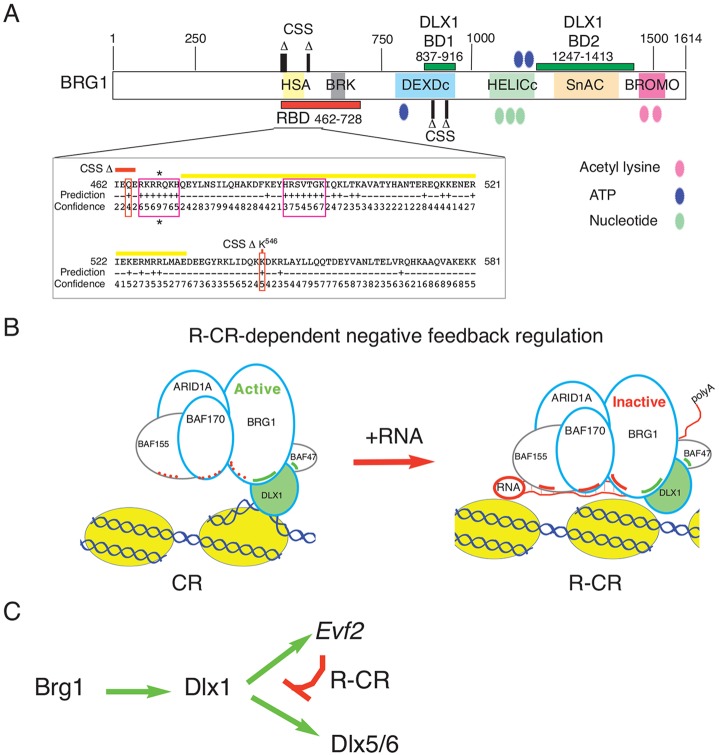
In order to define further the mechanism of BRG1-DLX1 interactions, we performed co-immunoprecipitation experiments with recombinant DLX1 and BRG1 deletion mutants (Fig. 2D-F). Two regions of BRG1 are sufficient to bind DLX1 [BRG1 837-916 (MUT12) and 1247-1413 (MUT14)]. BRG1 DLX1-binding domain 1 (837-916) localizes to a region known to contain mutations in CSS (Tsurusaki et al., 2012, 2014); BRG1 DLX1-binding domain 2 (1247-1413) is highly conserved and overlaps the SNF2 ATP-coupling domain (SnAC; see Fig. 6A), a region known to be required for yeast SNF2 remodeling and histone binding (Sen et al., 2013). Thus, DLX1-BRG1 interactions have the potential to play a role in a neurodevelopmental disorder.
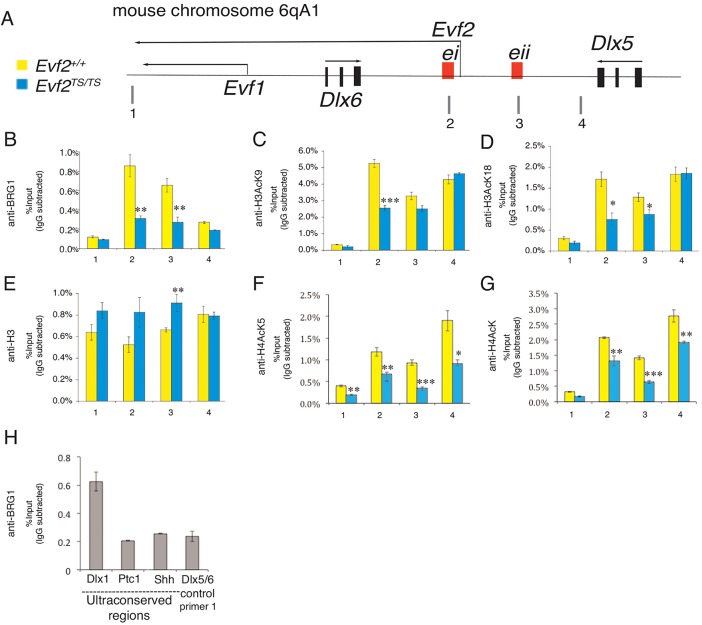
Evf2 increases BRG1 binding to key Dlx5/6 enhancers with changes in histone H3 and H4 lysine acetylation
Previous results show that Evf2 increases the association of DLX1/2 with the Dlx5/6 ei and eii intergenic enhancers (Bond et al., 2009). We next used ChIP to determine whether Evf2 affects BRG1 binding in the Dlx5/6 region. E13.5 GE chromatin from Evf2+/+ and Evf2TS/TS were compared for BRG1 occupancy (Fig. 3B). In the absence of Evf2, BRG1 binding to Dlx5/6 ei and eii decreases. BRG1 binding to site 1 (located downstream of Evf1), representing the background level of BRG1 binding, and to site 4 (a Dlx5/6 intergenic site) remain unaffected. Given the known interaction of the BRG1 bromodomain with acetylated lysines (Hassan et al., 2002), we next compared Evf2+/+ and Evf2TS/TS chromatin for the levels of histone lysine acetylation in the Dlx5/6 intergenic enhancers. ChIP using site-specific anti-H3 acetylated lysine antibodies on Evf2TS/TS E13.5 GE shows that H3AcK9 (Fig. 3C) and H3AcK18 (Fig. 3D) are significantly reduced at ei, and H3AcK18 to a lesser extent at eii, when compared with Evf2+/+. Decreased H3 lysine acetylation is not a result of decreased nucleosome density, as total H3 does not decrease at ei or eii (Fig. 3E). In Evf2TS/TS chromatin, H4AcK5 decreases at four sites (Fig. 3F), while total H4AcK (Fig. 3G) decreases at three intergenic sites (sites 2-4).
Fig. 3.
Evf2 stabilizes BRG1 binding to key DNA regulatory enhancers in the Dlx5/6 intergenic region. (A) Mouse chromosome 6qA1 (Dlx5/6 region) showing relative location of q-PCR primers (1-4), Dlx5/6 enhancers (the ultraconserved enhancer ei and the conserved enhancer eii, red boxes) and Evf2, Evf1, Dlx5, Dlx6 transcripts (black arrows). Dlx5 and Dlx6 protein coding exons (black boxes). (B-G) ChIP of Evf2+/+ (yellow bars) and Evf2TS/TS (blue bars) E13.5 GE tissue is followed by q-PCR using primers at sites 1-4. Antibodies for ChIP are indicated on the y-axis. Primer 1, a site downstream of the 3′ end of Evf2; primers 2-4 are Dlx5/6 intergenic sites: primer 2, ei; primer 3, eii (Zerucha et al., 2000); primer 4, an adjacent intergenic region with no known regulatory role. *P<0.05, **P<0.01, ***P<0.001 (Student's two-tailed t-test), n=10 embryos each genotype; error bars indicate s.e.m. (H) Anti-BRG1 ChIP from E13.5 GE. q-PCR shows enrichment of BRG1 at a subset of UCR sites: the Dlx1 UCR site, but not the Ptc1 or Shh UCRs.
We next asked whether BRG1 binds to other transcribed ultraconserved regions in E13.5 GE. Using the ultraconserved region (UCR) database (Woolfe et al., 2005), which reported over 1300 mouse UCRs, we searched for ucRNAs expressed in the developing brain (supplementary material Table S2). We identified three ucRNAs (Dlx1UR, Ptc1UR and ShhUR; supplementary material Fig. S2) that are expressed in E13.5 GE and transcribed on the opposite strand of genes in the SHH pathway (Marigo et al., 1996; Stone et al., 1996; Kohtz et al., 1998; Kohtz and Berghoff, 2010). ChIP shows that BRG1 is enriched in the Dlx1 UCR, but not in the Ptc1 (Ptch1) or Shh UCRs (Fig. 3H), supporting the idea that BRG1 binds a subset of transcribed UCRs in vivo.
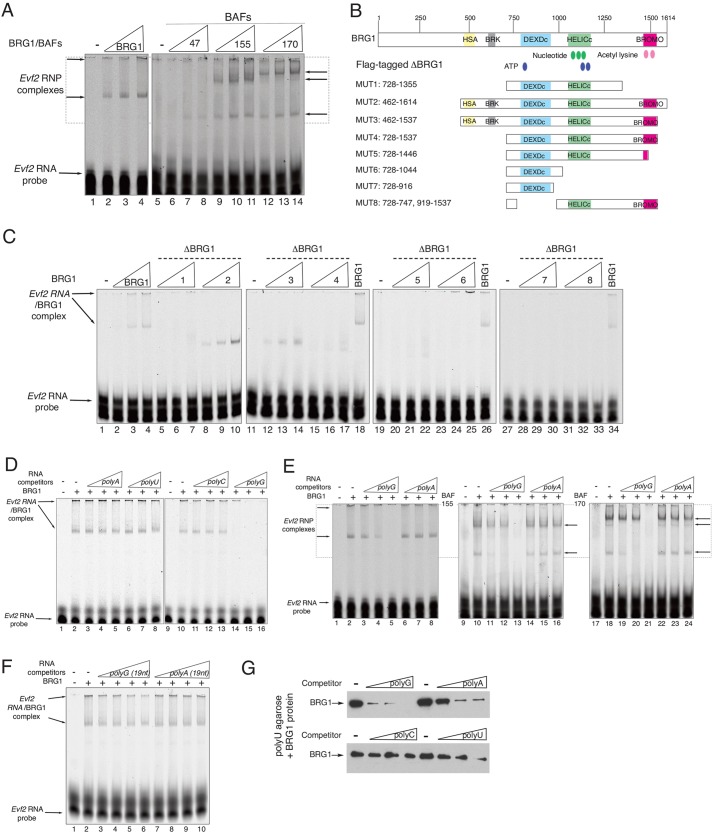
Promiscuous binding of RNAs to BRG1
We previously reported that Evf2 forms a complex with DLX proteins in nuclear clouds, in a soluble RNP from nuclear extracts, and increases DLX association with Dlx5/6 ei and eii (Feng et al., 2006; Bond et al., 2009). However, direct interactions between Evf2 RNA and DLX protein were not detected, suggesting involvement of an unknown RNA-binding protein. Complexes of DLX1 and BRG1 in vivo, direct interactions between DLX1 and BRG1 in vitro, and association between Evf2 and BRG1 in RNA clouds led us to test whether BRG1 directly binds Evf2. RNA electrophoretic mobility shift assays (REMSAs) show that recombinant BRG1 directly binds Evf2 (Fig. 4A). The Evf2 RNA probe in REMSAs contains 115 bases spanning the Evf2 UCR (blue boxed region in supplementary material Fig. S3A), a subregion of the Evf2 transcription-regulating region (red boxed region in supplementary material Fig. S3A) as previously defined by Feng et al. (2006).
Fig. 4.
Characterization of direct interactions between Evf2 RNA and BRG1/BAFs reveals promiscuous binding. RNA electrophoretic mobility shift assays (REMSAs) using near-infrared (NIR)-labeled Evf2 RNA (ultraconserved sequence; supplementary material Fig. S3, blue dashed box) and recombinant Flag-tagged BRG1 and BAF proteins. (A) Evf2 RNA probe (0.15 pmoles) is loaded in each lane with increasing concentrations of proteins: 2, 4, 8 picomoles; −, probe alone. (B) Flag-tagged BRG1 mutant proteins tested for binding to Evf2 by REMSA in C. (C) Lanes 1-34 contain Evf2 RNA probe with FL-BRG1 or FL-Δ-BRG1 (MUT1-8) added in increasing concentrations: 0.25, 1, 4 picomoles. (D) Evf2 RNA and BRG1 complexes are incubated with unlabeled ribohomopolymers in increasing concentrations (0.625, 2.5, 10 µg) in competitive REMSAs. All lanes contain NIR-labeled Evf2 RNA probe. BRG1 is added at 8 picomoles. (E) BRG1 (lanes 1-8), BAF155 (lanes 9-16) and BAF170 (lanes 17-24) complexes are incubated with unlabeled ribohomopolymers in increasing concentrations (0.008, 0.03, 0.125 µg) in competitive REMSAs. All lanes contain NIR-labeled Evf2 RNA probe. BRG1, BAF155 or BAF170 is added at 8 picomoles. (F) Evf2 RNA and BRG1 complexes are incubated with unlabeled ribohomopolymers (19 nt) in increasing concentrations (3.75, 7.5, 15.0, 30.0 picomoles) in competitive REMSAs. All lanes contain NIR-labeled Evf2 RNA probe. BRG1 is added at 8 picomoles. (G) Ribohomopolymer competition in BRG1-polyU-agarose binding assays. Western blot analysis is shown of BRG1 bound to polyU-agarose. BRG1 recombinant protein is incubated with polyU-agarose in the absence of RNA competitor (−), and in the presence of increasing amounts (0.008, 0.03, 0.125 µg) of polyG200-500nt, polyA200-500nt, polyC200-500nt or polyU200-500nt. Ribohomopolymer binding from highest to lowest affinity: polyG>polyA>polyU>polyC.
We next examined whether other members of the SWI/SNF complex bind Evf2 RNA. We find that BAF155 and BAF170, but not BAF47, directly bind Evf2 (Fig. 4A). We then used BRG1 deletion mutants to define the RNA-binding domain (RBD) in BRG1 to a region (462-728) containing the HSA and BRK domains (Fig. 4B,C).
In order to determine the sequence specificity of Evf2 RNA-BRG1 binding, we used ribohomopolymers in competitive REMSAs with Evf2 and BRG1, BAF155 or BAF170 (Fig. 4D-F). Evf2-BRG1 interactions can be competed by polyG RNA, but not polyA, polyU or polyC RNAs (Fig. 4D). Similarly, Evf2-BAF170 (Fig. 4E) and Evf2-BAF155 interactions can be competed by polyG200-500nt but not polyA200-500nt RNA. Shorter polyG19nt cannot compete for Evf2-binding sites in BRG1 (Fig. 4F). The range of concentrations tested for polyG200-500nt is 0.05-146 picomoles (Fig. 4D,E), and the lowest competing concentration is 0.18-0.44 pmoles (Fig. 4E, lane 4). Even at the highest concentration of polyG19nt tested (30 pmoles; Fig. 4F, lane 6), it does not compete.
In order to test whether BRG1 binds directly to other ribohomopolymers, we performed ribohomopolymer competitions in BRG1/polyU-agarose binding assays (Fig. 4G). BRG1 directly binds polyU-agarose (– competitor lanes); polyG, polyA and polyU RNAs, but not polyC RNA, compete for BRG1 binding to polyU-agarose. In order to further characterize the RNA binding properties of BRG1, we tested BRG1 binding to the following sequence complex RNAs: artificial pGEM RNA transcript, Dlx1UR and 28S RNA. All three RNAs outcompete Evf2-BRG1 binding (supplementary material Fig. S3B). Together, these data suggest that RNA binding by BRG1 is promiscuous, as it binds a subset of ribohomopolymers of a particular minimum length.
RNA inhibition of BRG1 ATPase activity and chromatin remodeling activity in vitro
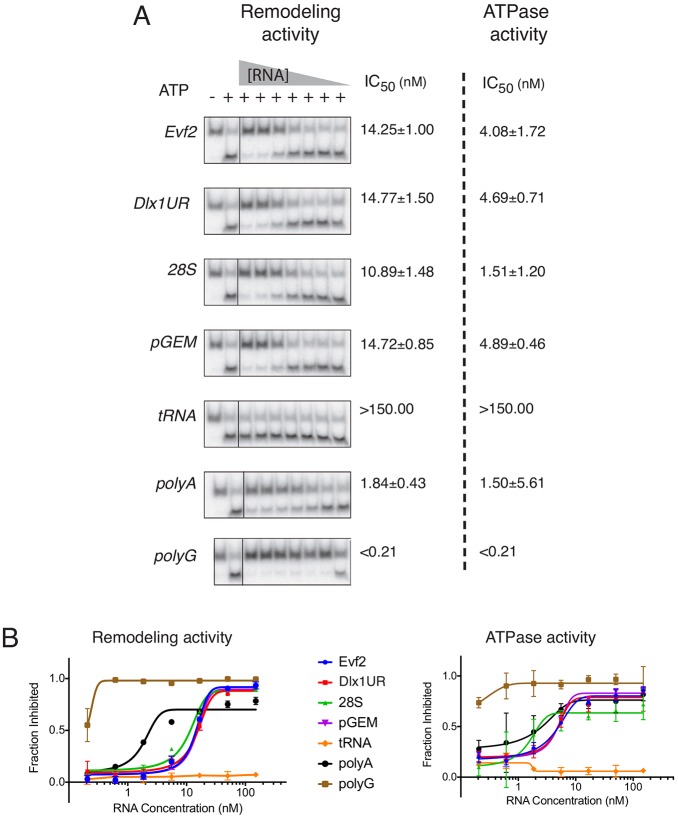
In vitro chromatin remodeling assays show that the SWI/SNF-related chromatin remodelers displace purified nucleosomes on a DNA template (Workman and Kingston, 1992; Phelan et al., 1999; Whitehouse et al., 1999). The ATPase subunit BRG1 is sufficient to displace nucleosomes in vitro. Direct interactions between BRG1, BAF170 and BAF155 and RNAs raise the possibility that RNAs directly influence chromatin remodeling. Therefore, we tested the effects of Evf2 and Dlx1UR lncRNAs, 28S, pGEM (artificial transcript), tRNA, and the highest-affinity binding ribohomopolymers polyG and polyA on BRG1-mediated ATPase activity and chromatin remodeling activity in vitro (Fig. 5). Sequence complex RNAs (Evf2622, 28S630, Dlx1UR534 and pGEM535) are transcribed in vitro to generate similar length RNAs. With the exception of tRNA, the tested RNAs inhibit BRG1 ATPase and remodeling activities (Fig. 5).
Fig. 5.
RNA inhibits BRG1 chromatin remodeling activity through inhibition of ATPase activity. BRG1 ATPase activity and chromatin remodeling activity are measured in the presence of seven different RNAs: Evf2, Dlx1UR, 28S, pGEM, tRNA, polyA, polyG. The IC50 (concentration of RNAs generating 50% inhibition of each activity) is indicated for each RNA. (A) BRG1 chromatin remodeling activity is assayed by nucleosome redistribution and restriction enzyme cleavage. Shown is acrylamide gel electrophoresis resolution of cut and uncut nucleosomes. BRG1 (100 nM) is incubated with trace nucleosomes (in 20 mM ATP, 3 mM MgCl2) and decreasing concentrations of RNAs: 150, 50, 16.67, 5.56, 1.85, 0.62, 0.21 nM. BRG1 ATPase activity is determined by thin layer chromatography analysis of reactions in which BRG1 (100 nM) is incubated with saturating nucleosomes (in 2 µM ATP, 3 mM MgCl2) and decreasing concentrations of RNAs as above. (B) Quantification of remodeling and ATPase assays; three independent experiments (error bars indicate s.e.m.).
Support that RNA-dependent inhibition of remodeling occurs through ATPase inhibition is based on the finding that IC50 (the concentration of RNA that inhibits ATPase or remodeling activity by 50%) values in the ATPase assay are lower than those observed in the remodeling assays (Fig. 5). The IC50 values for polyA RNA (1.50 µM) and polyG RNA (<0.21 µM) in ATPase inhibition are lower than the average IC50 for sequence complex RNAs (3.79 µM). The IC50 values for polyA RNA (1.84 µM) and polyG RNA (<0.21 µM) in remodeling inhibition are also lower than the average IC50 for sequence complex RNAs (13.66 µM). polyG RNA binds BRG1 with the highest affinity in both Evf2/REMSA competition (Fig. 4D) and polyU-agarose competition assays (Fig. 4G), supporting the contention that RNA-dependent inhibition of BRG1 ATPase and remodeling activity occurs through direct binding. Together, these data support RNA promiscuity in inhibiting the ATPase and remodeling activity of BRG1.
DISCUSSION
Histone-modifying enzymes and chromatin remodeling complexes constitute two major classes of proteins involved in regulating chromatin so that genes can be actively expressed or repressed. The SWI/SNF (yeast) and BAF (human) chromatin remodeling complexes recognize specific histone modifications and, in an ATP-dependent manner, reposition or eject nucleosomes (Workman and Kingston, 1992; Wang et al., 1996; Phelan et al., 1999; Clapier and Cairns, 2009; Kasten et al., 2011). lncRNAs are thought to repress gene expression through recruitment of histone-modifying enzymes (Rinn et al., 2007; Nagano et al., 2008; Pandey et al., 2008; Zhao et al., 2008), but a direct role for lncRNAs in chromatin remodeling has not been reported. However, experiments in Arabidopsis suggest that polV-produced lncRNAs interact with the lncRNA-binding protein ID2 and associate with SWI/SNF-related remodelers to silence gene expression through effects on nucleosome positioning (Zhu et al., 2013).
RNA binding properties of BRG
In this study, we propose that lncRNA-mediated transcriptional repression occurs through direct lncRNA-BRG1 binding and inhibition of BRG1 ATPase and chromatin remodeling activities (Fig. 5A,B and Fig. 6A,B). In vitro binding assays show that BRG1-RNA interactions are promiscuous and that the length of polyG ribohomopolymers is important (Fig. 4D-F). There are many RNA-binding proteins that exhibit non-specific or promiscuous RNA binding properties in vitro, but specific functional roles in the context of additional proteins or in vivo. Non-specific RNA binding has been described for factors involved in a wide range of RNA-dependent biological processes involving mRNA and microRNA processing, and promiscuous binding for lncRNA-polycomb repressor complex 2 (PRC2) interactions, where lncRNA repression is proposed to occur through a scanning model (Davidovich et al., 2013). In addition, early work on hnRNP-mediated RNA annealing activities and ribozyme catalysis led to the proposal that non-specific RNA-binding proteins may function as RNA chaperones, guiding RNA secondary or tertiary structures (Takagaki et al., 1992; Herschlag et al., 1994; Portman and Dreyfuss, 1994). It is of interest to note that preferential binding to specific ribohomopolymers is shared between BRG1/BAFs and members of the hnRNP family (Pinol-Roma et al., 1988) of RNA-binding proteins, some of which exhibit RNA chaperone activity. The known property of G-containing RNAs to form tetramers (Kim et al., 1991), combined with high-affinity polyG binding to BRG1 and BAFs (Fig. 4D,E), raises the possibility that RNA conformation might play a role in RNA-dependent inhibition of remodeling.
Fig. 6.
A model for transcriptional repression through RNA-mediated inhibition of chromatin remodeling. (A) BRG1 deletion analysis identifies two BRG1 DLX1-binding domains: DLX1-BD1 at 837-916 and DLX1-BD2 at 1247-1413 (dark green). The proposed RNA-binding domain (RBD) is at 462-728 (red). Sequence is shown for the N-terminal region of the RBD (462-581, BRG1-N-RBD), which displays higher confidence levels of RNA binding, as predicted by the BINDN+ program (Wang and Brown, 2006), than the C-terminal RBD (582-728). BRG1-N-RBD contains two stretches of potential RNA-binding residues (boxed in pink), one of which overlaps with the HSA region (yellow). *R469 has the highest confidence level prediction (+9) for RNA binding. CSS Δ indicates mutations identified in Coffin–Siris syndrome (CSS) by human exome sequencing (Tsurusaki et al., 2012). The CSS mutations (boxed in red) that are predicted to be involved in RNA binding are Q464 and K546. Two CSS mutations are found in BRG1 DLX1-BD1. (B) Model for RNA-dependent transcriptional repression. (Left) The Dlx5/6 active enhancer is bound by the DLX1-active chromatin remodeler (CR). CR components identified by mass spectrometry in E13.5 GE extracts are outlined in blue [BRG1, BAF170, ARID1A (predicted)]; additional known CRs are outlined in gray (BAF155, BAF47). The DLX1 homeodomain transcription factor (green, outlined in blue) binds DNA in a sequence-specific manner and binds BRG1 and BAF47 proteins (green lines). Nucleosomes (yellow spheres) are wound by DNA. The red dotted lines represent BRG1, BAF170 and BAF155 RNA-binding potential. (Right) Once the enhancer is activated, Evf2 (RNA, red) expression is activated. Sequence complex regions of the Evf2 RNA bind BRG1, BAF170 and BAF155 (red lines), stabilizing R-CR complex association with DNA, inhibiting BRG1 ATPase activity and remodeling activity, and repressing enhancer activity. RNA transcription and/or RNA retention may contribute to dynamic and/or precise modulation of enhancer activity. BRG1 (Brahma-related gene 1) is the ATPase remodeling component of the CR; BAFs, BRG1-associated factors 170, 155, 47; DLX1 is a homeodomain-containing, sequence-specific DNA-binding protein that acts as a transcription factor crucial for Evf2 and Dlx5/6 expression. (C) In the embryonic brain, Brg1 activates Dlx1 gene expression. Dlx1 activates Dlx5, Dlx6 and Evf2. Evf2 forms a complex with chromatin remodelers (R-CR), inhibits BRG1 remodeling activity, and attenuates Dlx1 activation of Dlx5/6.
Model for an RNA-bound chromatin remodeler (R-CR) in negative-feedback inhibition
We initially set out to answer the following: how does RNA-dependent recruitment of activating factors result in transcriptional repression? Previous work showed that Evf2 increases binding of both activators and repressors to the Dlx5/6 enhancer (Bond et al., 2009; Berghoff et al., 2013), leading to the idea that Evf2-mediated transcriptional repression results from the cumulative effects of antagonism between DLX1/2 and MECP2. In this work, mass spectrometry analysis of immunoaffinity purified DLX complexes in Evf2+/+ nuclear extracts (supplementary material Table S1) does not detect MECP2. This is consistent with a model in which DLX1/2 antagonism of MECP2 is competitive and occurs on mutually exclusive alleles (Bond et al., 2009; Berghoff et al., 2013). However, BRG1-RNA binding, and RNA-dependent direct inhibition of BRG1 ATPase and remodeling activity, support a role beyond that of mediating antagonism between transcription factors. Furthermore, Evf2 not only increases BRG1 and DLX1 association at the Dlx5/6 enhancers, but also increases histone acetylation (H3AcK9, H3AcK18, and H4AcK5 within Dlx5/6 ei and eii; Fig. 3). Given that a role for H4 acetylation-mediated inhibition of ISWI remodeling activity has been reported (Shogren-Knaak et al., 2006), it is possible that, in the presence of Evf2, increased histone lysine acetylation might further stabilize BRG1/BAF association through bromodomain interactions (Hassan et al., 2002) and contribute to remodeling inhibition.
These data support a model of lncRNA-dependent transcriptional repression at enhancers, distinct from polycomb repressor complex recruitment (Fig. 6B). In this model, DLX1-BRG1-BAFs [the chromatin remodeler (CR)] form an active complex at Dlx5/6 intergenic enhancers, activating adjacent gene expression (Dlx5, Dlx6 and Evf2). This initial recruitment occurs independently of Evf2, as Evf2 activation occurs after DLX1 binding. This is supported by genetic experiments in embryonic mouse brain: the requirement of BRG1 in Dlx1 activation (Lessard et al., 2007) and the requirement of Dlx1/2 in Dlx5/6/Evf2 activation (Anderson et al., 1997b; Berghoff et al., 2013). Once Evf2 expression is activated (Fig. 6B, red arrow), Evf2 binds to BRG1/BAFs (R-CR), stabilizes the association with Dlx5/6 ei and eii, and directly inhibits BRG1 remodeling activity through inhibition of ATPase activity. The R-CR converts an active enhancer bound by DLX1-BRG1-BAFs to an RNA-dependent repressed enhancer, supporting the idea that lncRNA activation functions in a negative-feedback mechanism to attenuate DLX1 activation.
In the proposed model, the lncRNA inhibits BRG1 ATPase and remodeling activity but the R-CR is retained at the enhancer, allowing subsequent gene reactivation. Evf2 RNA loss does not change total H3 at the Dlx5/6 ei, arguing against nucleosome ejection (Fig. 3E). It will be important to determine whether RNA binding is actually dynamic (CR↔R-CR) or is an endpoint, as shown in the model (Fig. 6B,C). A dynamic model is attractive in that RNA-bound activators are temporarily stabilized in order to achieve precise or rapid regulation. However, other possibilities that could achieve precise gene regulation include sequestration of positive factors, allele-specific effects, and transient and/or indirect effects of the RNA. The heterogeneity of individual profiles in the relationship between Evf2 nuclear clouds and BRG1 enrichment supports a complex mechanism, requiring future experiments to distinguish between dynamic, sequestration and allele-specific roles of the R-CR.
A recent report shows that the Myheart (Mhrt; myosin heavy chain-associated RNA transcript) lncRNA binds to BRG1 through the helicase domain, repressing gene expression through direct competition of BRG1-DNA binding (Han et al., 2014). Given that the BRG1 RBD defined in our work is distinct from that of MHRT (Fig. 6A), it will be important to determine the relative contributions of promiscuous RNA binding/remodeling inhibition and direct RNA-DNA competition in BRG1-lncRNA-mediated transcriptional repression. In addition to BRG1-lncRNA interactions in chromatin, SWI/SNF complexes have recently been shown to function in the assembly of nuclear bodies containing the lncRNA NEAT1 (Kawaguchi et al., 2015), thereby expanding the role of BRG1-lncRNA interactions in the nucleus.
Although direct interactions between Evf2, DLX1 and SWI/SNF, and binding of BRG1-DLX1 to Dlx5/6 enhancers, support a direct role of Evf2 RNA in BRG1 transcriptional regulation, it is also possible that the Evf2 RNA cloud plays a role as a ‘sink’ for proteins or other RNAs. Although it remains to be determined how many proteins, in addition to BRG1 and DLX, localize within the Evf2 RNA cloud, mass spectrometry of DLX-bound proteins predicts the presence of a large number of proteins with diverse functions. In light of SWI/SNF regulation of nuclear body assembly (Kawaguchi et al., 2015), it will be important to determine whether BRG1 and/or other DLX-bound proteins identified by mass spectrometry play a role in Evf2 RNA cloud formation, and whether Evf2 directly or indirectly affects the subnuclear organization/sequestering of factors involved in transcriptional regulation.
Biological significance of BRG1 RNA-binding and DLX1-binding domains
Several lines of evidence support the biological significance of Evf2 and DLX1 interactions during embryonic forebrain development. DLX1/2 regulate Dlx5/6 gene expression, and belong to a family of homeodomain transcription factors known for their roles in interneuron migration and differentiation (Panganiban and Rubenstein, 2002). Mice lacking Dlx1, 2, 5 and/or 6, either alone or in combination, exhibit a range of neuronal, craniofacial and limb defects (Anderson et al., 1997a,b; Depew et al., 1999; Pleasure et al., 2000; Merlo et al., 2002; Robledo et al., 2002; Cobos et al., 2005; Wang et al., 2010). In accordance with its role in regulating Dlx5/6 genes, mice lacking Evf2 display synaptic defects in the adult hippocampus (Bond et al., 2009).
The biological significance of CRs is highlighted by the recent identification of multiple CR mutations in human patients with autism, autism spectrum disorders (ASDs), schizophrenia and intellectual disability (reviewed by Staahl and Crabtree, 2013; Vogel-Ciernia and Wood, 2014). CSS is characterized by growth deficiency, intellectual disability, microcephaly, coarse facial features and nail defects; 20/23 CSS patients were reported to have a mutation in one of six SWI/SNF subunits, including BRG1 (Tsurusaki et al., 2012, 2014). Two BRG1 CSS deletion/mutations map within the N-terminal RBD domain (red boxes, Fig. 6A), while another two are in DLX1-binding domain 1 (837-916), raising the possibility that R-CR interactions might be involved (Fig. 6A). It is interesting that previous reports have linked disruption of Dlx5/6-dependent control of GABAergic interneuron development/function with autism. Loss of MECP2 in Dlx5/6-expressing GABAergic interneurons results in many behaviors associated with Rett syndrome, an ASD in humans (Nan et al., 1997; Chao et al., 2010). Loss of MECP2 increases Evf2 and Dlx5 expression in E13.5 GE (Berghoff et al., 2013) and Dlx5/6 expression in postnatal cortex (Horike et al., 2005). Loss of Evf2 destabilizes MECP2 binding to Dlx5/6 ei and eii, also resulting in increased Dlx5 and Dlx6 expression in E13.5 GE (Bond et al., 2009). Dlx1/2 antagonize MECP2 repression of Dlx5 (Berghoff et al., 2013). A Dlx5/6 ei SNP that disrupts DLX1/2 binding was identified in an autistic proband (Poitras et al., 2010). Given the global DNA-binding properties of MECP2, it has been difficult to envision how this may cause such specific neurological phenotypes, as in Rett syndrome. A similar problem stems from CR mutations, as CRs also control transcription globally. Together, these data raise an important question as to whether R-CRs provide the precise gene regulation necessary for higher order brain function and, when disrupted (as in MECP2 and CR mutations), how they might cause the subtle defects found in specific human neurological disorders.
MATERIALS AND METHODS
Primer and sequence data
Details of primers and sequence information for all RNAs are provided in the supplementary material Methods.
RNP isolation and protein identification
DLX immunoaffinity purification of complexes was performed from nuclear extracts (Dignam et al., 1983) using E13.5 GE of Evf2+/+ or Evf2TS/TS mice (Bond et al., 2009) or Swiss-Webster timed pregnant dams (Taconic) and anti-DLX antibody (Feng et al., 2006). Differential mass spectrometry and multidimensional protein identification technology were performed on DLX immunoaffinity purified complexes as previously described (Washburn et al., 2001). Supplementary material Table S1 lists mass spectrometry proteins identified in DLX-bound complexes from Evf2+/+ and Evf2TS/TS nuclear extract lysates. Raw data from mass spectrometry analysis can be accessed at: http://fields.scripps.edu/published/evf/. The mass spectrometry data have been submitted to Dryad and can be accessed under the doi: 10.5061/dryad.82gn0. The institutional IACUC committee approved all animal procedures.
Fluorescent in situ hybridization and immunohistochemistry colabeling (FISH-immuno)
FISH on E13.5 GE sections was performed using digoxigenin antisense Evf2 RNA probe as previously described (Feng et al., 2006) and 1:500 dilution of rabbit polyclonal anti-BRG1 (Wang et al., 1996), with tyramide amplification (Invitrogen). Details of Evf2 RNA and BRG1 protein colocalization are included in the supplementary material Methods.
Chromatin immunoprecipitation (ChIP)
E13.5 GE chromatin (25 µg) was diluted 1:10 in RIPA buffer (10 mM Tris pH 7.6, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100) with protease inhibitors (Roche), precleared by incubating with protein G agarose (Roche), and incubated with 2 or 5 µg ChIP-verified antibodies overnight at 4°C. The immunoprecipitated DNA was purified using the Qiaquick PCR Purification Kit (Qiagen). Immunoprecipitated DNA diluted 1:20 (in double-distilled water), primers, and Perfecta SYBR Green Fast Mix (Quanta Biosciences) was combined to 20 µl total reaction volume in a MicroAmp Fast Optical 96-well Reaction Plate (Life Technologies). Quantification of the immunoprecipitated material was performed in the Fast 7500 Real-Time PCR System (Life Technologies). For ChIP/reChIP, the ChIP protocol described above was used to perform anti-DLX ChIP, followed by elution and re-immunoprecipitation with anti-BRG1, anti-lamin B1 or anti-IgG as described (Truax and Greer, 2012). Percent input values obtained for IgG were subtracted from anti-BRG1 and anti-lamin B1 values. Statistical analysis employed an unpaired Student's t-test with equal variance. A detailed protocol for ChIP/reCHIP is included in the supplementary material Methods.
RNP interactions
The REMSA assembly reactions were carried out as described (Thomson et al., 1999) with several modifications. The RNA probe was labeled using near-infrared (NIR) in vitro transcription as described (Kohn et al., 2010). Flag-tagged BRG1 and BAF47, BAF155 and BAF170 proteins were purified from SF9 insect cells according to the Invitrogen Bac-to-Bac baculovirus expression system. The recombinant proteins were incubated with 0.15 pmoles Evf2 NIR-labeled probe in 10 μl reactions for 30 min at room temperature. For all competition experiments, protein and competitor RNA were pre-incubated for 10 min at room temperature before adding probe. Evf2, 28S, pGEM and Dlx1UR competitor RNAs were generated by in vitro transcription as described in the supplementary material Methods. The ribohomopolymers were obtained from Sigma (polyA, polyG) or Midland (polyC, polyU). The 19 nt RNA oligos were obtained from IDT DNA. 5 μg tRNA and 0.5 μl RNasin (Promega) were included in all the REMSA reactions. Pre-electrophoresis of 4% native polyacrylamide gels was performed for 20 min, REMSA reactions loaded and electrophoresed at 200 V for 40 min, and data visualized in the Odyssey Infrared Imager (LI-COR Biosciences). Comparisons of BRG1 binding to different ribohomopolymers were performed using a polyU-agarose binding assay, in which 8 picomoles of BRG1 was preincubated with the different soluble ribohomopolymers mentioned above. Further details of REMSAs and polyU-agarose binding assays are provided in the supplementary material Methods and Table S3.
BRG1 ATPase and remodeling assays
For protein purification and nucleosome assembly, Flag-epitope BRG1 and BRG1 mutants were purified from SF9 cells using baculovirus expression as previously described (Phelan et al., 1999). Recombinant histones were purified and reconstituted into octamers as described previously (Luger et al., 1999). Mononucleosomes were assembled onto a 237-bp DNA fragment containing a central 601 positioning sequence that had been modified to contain an internal PstI cleavage site. Conditions for RNA inhibition of BRG1 ATPase and remodeling activities are detailed in the supplementary material Methods and Table S3.
Supplementary Material
Acknowledgements
We thank Dr Weidong Wang (NIH) for the kind gift of rabbit anti-BRG1 antibody (multiple shipments); Dr Robin Park (Scripps) for the Venn diagram in Fig. 1; and Dr Michael VanGompel (present address UC Davis) for help with screening the UCR database (supplementary material Fig. S2).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
I.C. performed experiments (Figs 1, 3 and 4; supplementary data) and helped in manuscript preparation. D.E.L. established conditions for isolating embryonic brain RNPs, and RNA protein REMSAs. J.C. and R.E.K. contributed the BRG1 ATPase and remodeling experiments (Fig. 5). H.L. performed the BRG1-DLX1 direct interaction experiments (Fig. 2). K.S. performed reChIP experiments (Fig. 1). S.C. performed experiments in Figs 1 and 3. B.S.C. performed the screen of ucRNAs in E13.5 GE (Fig. 3; supplementary data). J.T. and J.R.Y. contributed mass spectroscopy results (Fig. 1; supplementary data). J.D.K. conceived experiments, proposed the model (Fig. 6) and prepared the manuscript.
Funding
This work was initially funded by the American Reinvestment and Recovery Act and the National Institute of Child Health and Human Development (NICHD) [R01HD056504 (2009-2011) to J.D.K.]; and presently by the National Institute of Mental Health (NIMH) [R01MH094653 to J.D.K.]; and the National Institute of General Medical Sciences (NIGMS) [5R37 GM048405-23 to R.E.K. and P41 GM103533 to J.R.Y.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.126318/-/DC1
References
- Anderson S. A., Eisenstat D. D., Shi L. and Rubenstein J. L. R. (1997a). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474-476. 10.1126/science.278.5337.474 [DOI] [PubMed] [Google Scholar]
- Anderson S. A., Qiu M., Bulfone A., Eisenstat D. D., Meneses J., Pedersen R. and Rubenstein J. L. R. (1997b). Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19, 27-37. 10.1016/S0896-6273(00)80345-1 [DOI] [PubMed] [Google Scholar]
- Berghoff E. G., Clark M. F., Chen S., Cajigas I., Leib D. E. and Kohtz J. D. (2013). Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140, 4407-4416. 10.1242/dev.099390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond A. M., VanGompel M. J. W., Sametsky E. A., Clark M. F., Savage J. C., Disterhoft J. F. and Kohtz J. D. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020-1027. 10.1038/nn.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. (2011). Chromosome silencing mechanisms in X-chromosome inactivation: unknown unknowns. Development 138, 5057-5065. 10.1242/dev.065276 [DOI] [PubMed] [Google Scholar]
- Calin G. A., Liu C.-G., Ferracin M., Hyslop T., Spizzo R., Sevignani C., Fabbri M., Cimmino A., Lee E. J., Wojcik S. E. et al. (2007). Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215-229. 10.1016/j.ccr.2007.07.027 [DOI] [PubMed] [Google Scholar]
- Chao H.-T., Chen H., Samaco R. C., Xue M., Chahrour M., Yoo J., Neul J. L., Gong S., Lu H.-C., Heintz N. et al. (2010). Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature 468, 263-269. 10.1038/nature09582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C. R. and Cairns B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273-304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Cobos I., Calcagnotto M. E., Vilaythong A. J., Thwin M. T., Noebels J. L., Baraban S. C. and Rubenstein J. L. R. (2005). Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat. Neurosci. 8, 1059-1068. 10.1038/nn1499 [DOI] [PubMed] [Google Scholar]
- Davidovich C., Zheng L., Goodrich K. J. and Cech T. R. (2013). Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 20, 1250-1257. 10.1038/nsmb.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew M. J., Liu J. K., Long J. E., Presley R., Meneses J. J., Pedersen R. A. and Rubenstein J. L. (1999). Dlx5 regulates regional development of the branchial arches and sensory capsules. Development 126, 3831-3846. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M. and Roeder R. G. (1983). Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475-1489. 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., White B., Tyurina O. V., Guner B., Larson T., Lee H. Y., Karlstrom R. O. and Kohtz J. D. (2004). Synergistic and antagonistic roles of the Sonic hedgehog N- and C-terminal lipids. Development 131, 4357-4370. 10.1242/dev.01301 [DOI] [PubMed] [Google Scholar]
- Feng J., Bi C., Clark B. S., Mady R., Shah P. and Kohtz J. D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470-1484. 10.1101/gad.1416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A., Ripoche M.-A., Le Digarcher A., Watrin F., Ziyyat A., Forne T., Jammes H., Ainscough J. F. X., Surani M. A., Journot L. et al. (2009). H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 136, 3413-3421. 10.1242/dev.036061 [DOI] [PubMed] [Google Scholar]
- Geisler S. and Coller J. (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14, 699-712. 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J. A., Wapinski O. L., Yang Y. W., Bureau J.-F., Gopinath S., Monack D. M., Chang H. Y., Brahic M. and Kirkegaard K. (2013). The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152, 743-754. 10.1016/j.cell.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Hammerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. et al. (2013). The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180-1189. 10.1158/0008-5472.CAN-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Li W., Lin C.-H., Yang J., Shang C., Nurnberg S. T., Jin K. K., Xu W., Lin C.-Y., Lin C.-J. et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102-106. 10.1038/nature13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A. H., Prochasson P., Neely K. E., Galasinski S. C., Chandy M., Carrozza M. J. and Workman J. L. (2002). Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111, 369-379. 10.1016/S0092-8674(02)01005-X [DOI] [PubMed] [Google Scholar]
- Herschlag D., Khosla M., Tsuchihashi Z. and Karpel R. L. (1994). An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 13, 2913-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike S., Cai S., Miyano M., Cheng J. F. and Kohwi-Shigematsu T. (2005). Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37, 31-40. [DOI] [PubMed] [Google Scholar]
- Kasten M. M., Clapier C. R. and Cairns B. R. (2011). SnapShot: chromatin remodeling: SWI/SNF. Cell 144, 310-310.e311. 10.1016/j.cell.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Kawaguchi T., Tanigawa A., Naganuma T., Ohkawa Y., Souquere S., Pierron G. and Hirose T. (2015). SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. USA 112, 4304-4309. 10.1073/pnas.1423819112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Cheong C. and Moore P. B. (1991). Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature 351, 331-332. 10.1038/351331a0 [DOI] [PubMed] [Google Scholar]
- Kohn M., Lederer M., Wachter K. and Huttelmaier S. (2010). Near-infrared (NIR) dye-labeled RNAs identify binding of ZBP1 to the noncoding Y3-RNA. RNA 16, 1420-1428. 10.1261/rna.2152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz J. D. and Berghoff E. G. (2010). Regulatory long non-coding RNAs and neuronal disorders. Physiol. Behav. 100, 250-254. 10.1016/j.physbeh.2010.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz J. D., Baker D. P., Corte G. and Fishell G. (1998). Regionalization within the mammalian telencephalon is mediated by changes in responsiveness to Sonic Hedgehog. Development 125, 5079-5089. [DOI] [PubMed] [Google Scholar]
- Kohtz J. D., Lee H. Y., Gaiano N., Segal J., Ng E., Larson T., Baker D. P., Garber E. A., Williams K. P. and Fishell G. (2001). N-terminal fatty-acylation of sonic hedgehog enhances the induction of rodent ventral forebrain neurons. Development 128, 2351-2363. [DOI] [PubMed] [Google Scholar]
- Lanz R. B., McKenna N. J., Onate S. A., Albrecht U., Wong J., Tsai S. Y., Tsai M.-J. and O'Malley B. W. (1999). A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97, 17-27. 10.1016/S0092-8674(00)80711-4 [DOI] [PubMed] [Google Scholar]
- Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A. and Crabtree G. R. (2007). An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201-215. 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T. J. and Richmond T. J. (1999). Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1-16. 10.1016/j.neuron.2007.06.019 [DOI] [PubMed] [Google Scholar]
- Marigo V., Davey R. A., Zuo Y., Cunningham J. M. and Tabin C. J. (1996). Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176-179. 10.1038/384176a0 [DOI] [PubMed] [Google Scholar]
- Merlo G. R., Paleari L., Mantero S., Genova F., Beverdam A., Palmisano G. L., Barbieri O. and Levi G. (2002). Mouse model of split hand/foot malformation type I. Genesis 33, 97-101. 10.1002/gene.10098 [DOI] [PubMed] [Google Scholar]
- Nagano T., Mitchell J. A., Sanz L. A., Pauler F. M., Ferguson-Smith A. C., Feil R. and Fraser P. (2008). The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717-1720. 10.1126/science.1163802 [DOI] [PubMed] [Google Scholar]
- Nan X., Campoy F. J. and Bird A. (1997). MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88, 471-481. 10.1016/S0092-8674(00)81887-5 [DOI] [PubMed] [Google Scholar]
- Ørom U. A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46-58. 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R. R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-DiNardo D. and Kanduri C. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232-246. 10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- Panganiban G. and Rubenstein J. L. (2002). Developmental functions of the Distal-less/Dlx homeobox genes. Development 129, 4371-4386. [DOI] [PubMed] [Google Scholar]
- Phelan M. L., Sif S., Narlikar G. J. and Kingston R. E. (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247-253. 10.1016/S1097-2765(00)80315-9 [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Choi Y. D., Matunis M. J. and Dreyfuss G. (1988). Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 2, 215-227. 10.1101/gad.2.2.215 [DOI] [PubMed] [Google Scholar]
- Pleasure S. J., Anderson S., Hevner R., Bagri A., Marin O., Lowenstein D. H. and Rubenstein J. L. R. (2000). Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron 28, 727-740. 10.1016/S0896-6273(00)00149-5 [DOI] [PubMed] [Google Scholar]
- Poitras L., Yu M., Lesage-Pelletier C., MacDonald R. B., Gagne J.-P., Hatch G., Kelly I., Hamilton S. P., Rubenstein J. L. R., Poirier G. G. et al. (2010). An SNP in an ultraconserved regulatory element affects Dlx5/Dlx6 regulation in the forebrain. Development 137, 3089-3097. 10.1242/dev.051052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman D. S. and Dreyfuss G. (1994). RNA annealing activities in HeLa nuclei. EMBO J. 13, 213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli N. A., Poth E. M., Zhu H. and Blackshaw S. (2011). The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 6, 32 10.1186/1749-8104-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrup L., Branco M. R., Perdeaux E. R., Krueger C., Lewis A., Santos F., Nagano T., Cobb B. S., Fraser P. and Reik W. (2009). The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136, 525-530. 10.1242/dev.031328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., Goodnough L. H., Helms J. A., Farnham P. J., Segal E. et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311-1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo R. F., Rajan L., Li X. and Lufkin T. (2002). The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 16, 1089-1101. 10.1101/gad.988402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P., Vivas P., Dechassa M. L., Mooney A. M., Poirier M. G. and Bartholomew B. (2013). The SnAC domain of SWI/SNF is a histone anchor required for remodeling. Mol. Cell. Biol. 33, 360-370. 10.1128/MCB.00922-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M. J., Davie J. R. and Peterson C. L. (2006). Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844-847. 10.1126/science.1124000 [DOI] [PubMed] [Google Scholar]
- Staahl B. T. and Crabtree G. R. (2013). Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr. Opin. Neurobiol. 23, 903-913. 10.1016/j.conb.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H. et al. (1996). The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129-134. 10.1038/384129a0 [DOI] [PubMed] [Google Scholar]
- Takagaki Y., MacDonald C. C., Shenk T. and Manley J. L. (1992). The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. USA 89, 1403-1407. 10.1073/pnas.89.4.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A. M., Rogers J. T., Walker C. E., Staton J. M. and Leedman P. J. (1999). Optimized RNA gel-shift and UV cross-linking assays for characterization of cytoplasmic RNA-protein interactions. BioTechniques 27, 1032-1039, 1042. [DOI] [PubMed] [Google Scholar]
- Truax A. D. and Greer S. F. (2012). ChIP and re-ChIP assays: investigating interactions between regulatory proteins, histone modifications, and the DNA sequences to which they bind. Meth. Mol. Biol. 809, 175-188. 10.1007/978-1-61779-376-9_12 [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y., Okamoto N., Ohashi H., Kosho T., Imai Y., Hibi-Ko Y., Kaname T., Naritomi K., Kawame H., Wakui K. et al. (2012). Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 44, 376-378. 10.1038/ng.2219 [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y., Okamoto N., Ohashi H., Mizuno S., Matsumoto N., Makita Y., Fukuda M., Isidor B., Perrier J., Aggarwal S. et al. (2014). Coffin-Siris syndrome is a SWI/SNF complex disorder. Clin. Genet. 85, 548-554. 10.1111/cge.12225 [DOI] [PubMed] [Google Scholar]
- Vogel-Ciernia A. and Wood M. A. (2014). Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology 80, 18-27. 10.1016/j.neuropharm.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. and Brown S. J. (2006). BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 34, W243-W248. 10.1093/nar/gkl298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cote J., Xue Y., Zhou S., Khavari P. A., Biggar S. R., Muchardt C., Kalpana G. V., Goff S. P., Yaniv M. et al. (1996). Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15, 5370-5382. [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dye C. A., Sohal V., Long J. E., Estrada R. C., Roztocil T., Lufkin T., Deisseroth K., Baraban S. C. and Rubenstein J. L. R. (2010). Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J. Neurosci. 30, 5334-5345. 10.1523/JNEUROSCI.5963-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. P., Wolters D. and Yates J. R. III (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242-247. 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Flaus A., Cairns B. R., White M. F., Workman J. L. and Owen-Hughes T. (1999). Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400, 784-787. 10.1038/23506 [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., Smith S. F., North P., Callaway H., Kelly K. et al. (2005). Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J. L. and Kingston R. E. (1992). Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science 258, 1780-1784. 10.1126/science.1465613 [DOI] [PubMed] [Google Scholar]
- Yang L., Froberg J. E. and Lee J. T. (2014). Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 39, 35-43. 10.1016/j.tibs.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R., Dahary D., Sorek R., Levanon E. Y., Goldstein O., Shoshan A., Diber A., Biton S., Tamir Y., Khosravi R. et al. (2003). Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21, 379-386. 10.1038/nbt808 [DOI] [PubMed] [Google Scholar]
- Yoo A. S. and Crabtree G. R. (2009). ATP-dependent chromatin remodeling in neural development. Curr. Opin. Neurobiol. 19, 120-126. 10.1016/j.conb.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerucha T., Stuhmer T., Hatch G., Park B. K., Long Q., Yu G., Gambarotta A., Schultz J. R., Rubenstein J. L. and Ekker M. (2000). A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 20, 709-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sun B. K., Erwin J. A., Song J.-J. and Lee J. T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750-756. 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Tang Z., Zhang H., Atianjoh F. E., Zhao J.-Y., Liang L., Wang W., Guan X., Kao S.-C., Tiwari V. et al. (2013). A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 16, 1024-1031. 10.1038/nn.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Rowley M. J., Böhmdorfer G. and Wierzbicki A. T. (2013). A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell 49, 298-309. 10.1016/j.molcel.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.