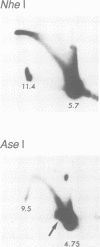
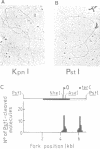
Abstract
Numerous bacterial replicons remain poorly characterized due to difficulties in localization of the replication origin. We have circumvented this problem in the characterization and fine mapping of the origin of plasmid pAM beta 1 by exploiting the Bacillus subtilis termination signal, terC. In terC-containing derivatives, theta-form molecules with two invariant endpoints accumulate. The endpoints, which correspond to plasmid origin and terC, were mapped with single-nucleotide precision. Analysis of the replication intermediates of wild-type molecules by two-dimensional gel electrophoresis confirmed the location of the plasmid origin. Our results demonstrate that pAM beta 1 replication proceeds unidirectionally by a theta mechanism. This work confirms the use of termination signals to localize origins, suggests that termination in B. subtilis occurs by a mechanism similar to that of Escherichia coli and establishes that in addition to rolling circle replicating plasmids, Gram positive bacteria harbour plasmids which replicate by a theta mechanism.
Full text
PDF
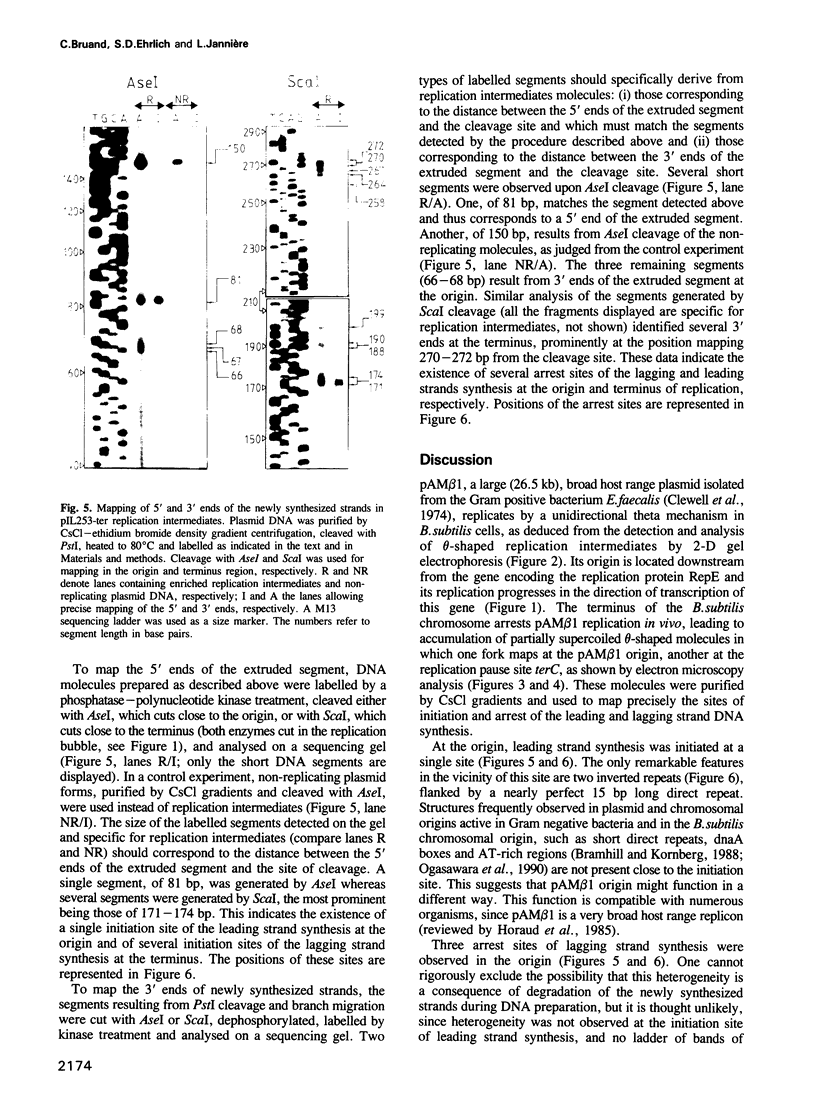
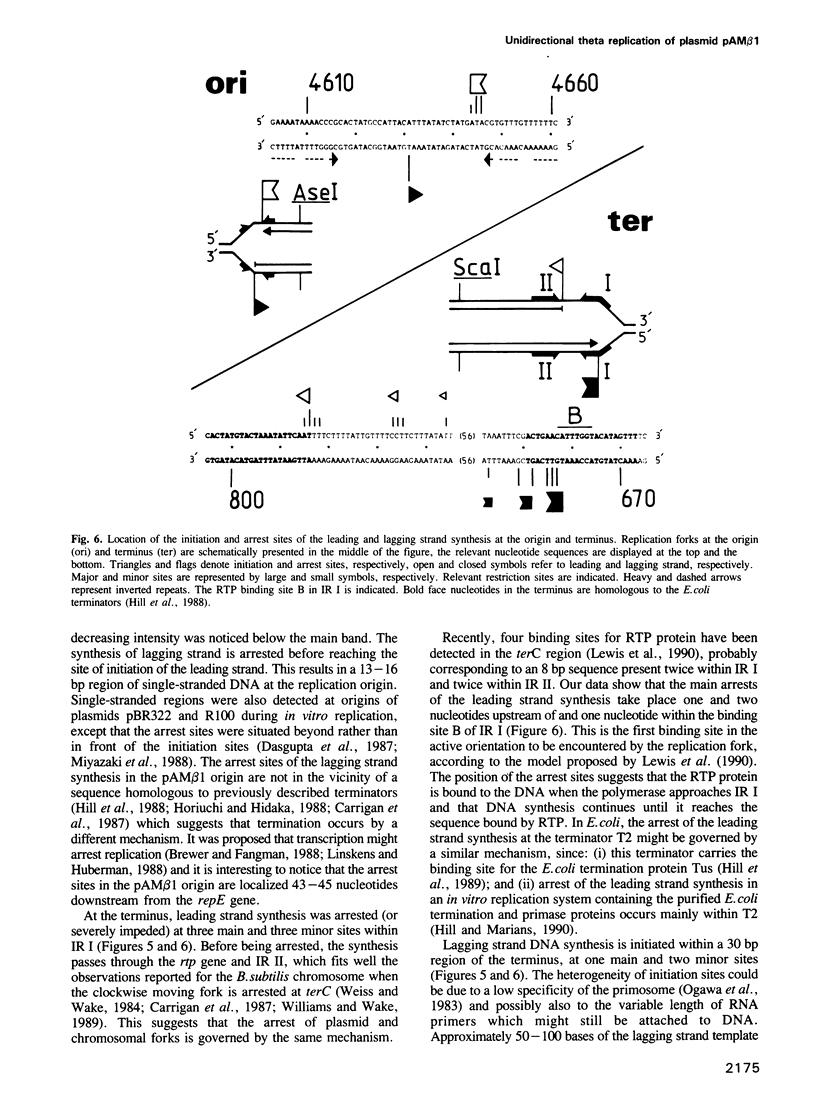


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Jansz H. S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Brantl S., Nowak A., Behnke D., Alonso J. C. Revision of the nucleotide sequence of the Streptococcus pyogenes plasmid pSM19035 repS gene. Nucleic Acids Res. 1989 Dec 11;17(23):10110–10110. doi: 10.1093/nar/17.23.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort G., Magot M., Ionesco H., Sebald M. Characterization and transferability of Clostridium perfringens plasmids. Plasmid. 1977 Nov;1(1):52–66. doi: 10.1016/0147-619x(77)90008-7. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988 Nov 18;55(4):637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Carrigan C. M., Haarsma J. A., Smith M. T., Wake R. G. Sequence features of the replication terminus of the Bacillus subtilis chromosome. Nucleic Acids Res. 1987 Oct 26;15(20):8501–8509. doi: 10.1093/nar/15.20.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S., Masukata H., Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987 Dec 24;51(6):1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- Garnier T., Cole S. T. Identification and molecular genetic analysis of replication functions of the bacteriocinogenic plasmid pIP404 from Clostridium perfringens. Plasmid. 1988 Mar;19(2):151–160. doi: 10.1016/0147-619x(88)90053-4. [DOI] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Replication origin of a single-stranded DNA plasmid pC194. EMBO J. 1989 Sep;8(9):2711–2716. doi: 10.1002/j.1460-2075.1989.tb08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon K. S., McKay L. L. Restriction enzyme analysis of lactose and bacteriocin plasmids from Streptococcus lactis subsp. diacetylactis WM4 and cloning of BclI fragments coding for bacteriocin production. Appl Environ Microbiol. 1987 May;53(5):1171–1174. doi: 10.1128/aem.53.5.1171-1174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann R., Neugebauer K., Pirkl E., Zentgraf H., Schaller H. Conversion of bacteriophage fd into an efficient single-stranded DNA vector system. Mol Gen Genet. 1980 Jan;177(2):231–242. doi: 10.1007/BF00267434. [DOI] [PubMed] [Google Scholar]
- Hill T. M., Marians K. J. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2481–2485. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. M., Pelletier A. J., Tecklenburg M. L., Kuempel P. L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988 Nov 4;55(3):459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Hill T. M., Tecklenburg M. L., Pelletier A. J., Kuempel P. L. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1593–1597. doi: 10.1073/pnas.86.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horaud T., Le Bouguenec C., Pepper K. Molecular genetics of resistance to macrolides, lincosamides and streptogramin B (MLS) in streptococci. J Antimicrob Chemother. 1985 Jul;16 (Suppl A):111–135. doi: 10.1093/jac/16.suppl_a.111. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Hidaka M., Akiyama M., Nishitani H., Sekiguchi M. Replication intermediate of a hybrid plasmid carrying the replication terminus (ter) site of R 6K as revealed by agarose gel electrophoresis. Mol Gen Genet. 1987 Dec;210(3):394–398. doi: 10.1007/BF00327188. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Hidaka M. Core sequence of two separable terminus sites of the R6K plasmid that exhibit polar inhibition of replication is a 20 bp inverted repeat. Cell. 1988 Aug 12;54(4):515–523. doi: 10.1016/0092-8674(88)90073-6. [DOI] [PubMed] [Google Scholar]
- Horodniceanu T., Bouanchaud D. H., Bieth G., Chabbert Y. A. R plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1976 Nov;10(5):795–801. doi: 10.1128/aac.10.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aiba S. Isolation and characterization of antibiotic resistance plasmids from thermophilic bacilli and construction of deletion plasmids. J Bacteriol. 1981 Jun;146(3):1091–1097. doi: 10.1128/jb.146.3.1091-1097.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Ishikawa H., Aiba S. Complete nucleotide sequence of the low copy number plasmid pRAT11 and replication control by the RepA protein in Bacillus subtilis. Mol Gen Genet. 1986 Oct;205(1):90–96. doi: 10.1007/BF02428036. [DOI] [PubMed] [Google Scholar]
- Jannière L., Bruand C., Ehrlich S. D. Structurally stable Bacillus subtilis cloning vectors. Gene. 1990 Mar 1;87(1):53–61. doi: 10.1016/0378-1119(90)90495-d. [DOI] [PubMed] [Google Scholar]
- Lewis P. J., Ralston G. B., Christopherson R. I., Wake R. G. Identification of the replication terminator protein binding sites in the terminus region of the Bacillus subtilis chromosome and stoichiometry of the binding. J Mol Biol. 1990 Jul 5;214(1):73–84. doi: 10.1016/0022-2836(90)90147-E. [DOI] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malke H. Genetics of resistance to macrolide antibiotics and lincomycin in natural isolates of Streptococcus pyogenes. Mol Gen Genet. 1974;135(4):349–367. doi: 10.1007/BF00271149. [DOI] [PubMed] [Google Scholar]
- Miyazaki C., Kawai Y., Ohtsubo H., Ohtsubo E. Unidirectional replication of plasmid R100. J Mol Biol. 1988 Nov 20;204(2):331–343. doi: 10.1016/0022-2836(88)90580-3. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Arai K., Okazaki T. Site selection and structure of DNA-linked RNA primers synthesized by the primosome in phage phi X174 DNA replication in vitro. J Biol Chem. 1983 Nov 10;258(21):13353–13358. [PubMed] [Google Scholar]
- Pelletier A. J., Hill T. M., Kuempel P. L. Termination sites T1 and T2 from the Escherichia coli chromosome inhibit DNA replication in ColE1-derived plasmids. J Bacteriol. 1989 Mar;171(3):1739–1741. doi: 10.1128/jb.171.3.1739-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich P. M., Haykinson MYa, Arutyunova L. S., Yomantas YuV, Stepanov A. I. The structure and source of plasmid DNA determine the cloning properties of vectors for Bacillus subtilis. Basic Life Sci. 1985;30:635–656. doi: 10.1007/978-1-4613-2447-8_44. [DOI] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Smith M. T., Aynsley C., Wake R. G. Cloning and localization of the Bacillus subtilis chromosome replication terminus, terC. Gene. 1985;38(1-3):9–17. doi: 10.1016/0378-1119(85)90198-2. [DOI] [PubMed] [Google Scholar]
- Smith M. T., Wake R. G. DNA sequence requirements for replication fork arrest at terC in Bacillus subtilis. J Bacteriol. 1988 Sep;170(9):4083–4090. doi: 10.1128/jb.170.9.4083-4090.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozhamannan S., Dabert P., Moretto V., Ehrlich S. D., Gruss A. Plus-origin mapping of single-stranded DNA plasmid pE194 and nick site homologies with other plasmids. J Bacteriol. 1990 Aug;172(8):4543–4548. doi: 10.1128/jb.172.8.4543-4548.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinfield T. J., Oultram J. D., Thompson D. E., Brehm J. K., Minton N. P. Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAM beta 1. Gene. 1990 Mar 1;87(1):79–90. [PubMed] [Google Scholar]
- Tanaka T., Sakaguchi K. Construction of a recombinant plasmid composed of B. subtilis leucine genes and a B. subtilis (natto) plasmid: its use as cloning vehicle in B. subtilis 168. Mol Gen Genet. 1978 Oct 24;165(3):269–276. doi: 10.1007/BF00332526. [DOI] [PubMed] [Google Scholar]
- Weiss A. S., Wake R. G. A unique DNA intermediate associated with termination of chromosome replication in Bacillus subtilis. Cell. 1984 Dec;39(3 Pt 2):683–689. doi: 10.1016/0092-8674(84)90475-6. [DOI] [PubMed] [Google Scholar]
- Westmoreland B. C., Szybalski W., Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969 Mar 21;163(3873):1343–1348. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]
- Williams N. K., Wake R. G. Sequence limits of DNA strands in the arrest replication fork at the Bacillus subtilis chromosome terminus. Nucleic Acids Res. 1989 Dec 11;17(23):9947–9956. doi: 10.1093/nar/17.23.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N. D., Boeke J. D. The filamentous phage (Ff) as vectors for recombinant DNA--a review. Gene. 1982 Jul-Aug;19(1):1–10. doi: 10.1016/0378-1119(82)90183-4. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]