Abstract
Genetically modified pigs are commonly created via somatic cell nuclear transfer (SCNT). Treatment of reconstructed embryos with histone deacetylase inhibitors (HDACi) immediately after activation improves cloning efficiency. The objective of this experiment was to evaluate the transcriptome of SCNT embryos treated with suberoylanilide hydroxamic acid (SAHA), 4-iodo-SAHA (ISAHA), or Scriptaid as compared to untreated SCNT, in vitro–fertilized (IVF), and in vivo (IVV) blastocyst-stage embryos. SAHA (10 μM) had the highest level of blastocyst development at 43.9%, and all treatments except 10 μM ISAHA had the same percentage of blastocyst development as Scriptaid (p<0.05). Two treatments, 1.0 μM ISAHA and 1.0 μM SAHA, had higher mean cell number than No HDACi treatment (p<0.021). Embryo transfers performed with 10 μM SAHA- and 1 μM ISAHA-treated embryos resulted in the birth of healthy piglets. GenBank accession numbers from up- and downregulated transcripts were loaded into the Database for Annotation, Visualization and Integrated Discovery to identify enriched biological themes. HDACi treatment yielded the highest enrichment for transcripts within the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway, lysosome. The mean intensity of LysoTracker was lower in IVV embryos compared to IVF and SCNT embryos (p<0.0001). SAHA and ISAHA can successfully be used to create healthy piglets from SCNT.
Introduction
Low cloning efficiency is thought to be caused, at least partially, by inadequate nuclear remodeling and reprogramming of the donor nucleus (Whitworth and Prather, 2010). Although embryos can now be efficiently modified by zygote injection of CRISPR/Cas9 and TALEN RNA (Lillico et al., 2013; Whitworth et al., 2014), the production of offspring by somatic cell nuclear transfer (SCNT) is still required to create more elaborate site-specific genetic modifications requiring a targeting vector. In species like pigs that lack authentic embryonic stem cell (ESC) lines, SCNT is the most commonly used method to create genetically modified animals. Genetically modified pigs are powerful tools for agricultural (Prather et al., 2013; Whitworth et al., 2014) and biomedical models (Jensen et al., 2010; Renner et al., 2010; Rogers et al., 2008; Ross et al., 2012), as well as in xenotransplantation research (Kolber-Simonds et al., 2004; Lai et al., 2002; Zeyland et al., 2014).
The efficiency of SCNT has greatly improved with the introduction of histone deacetylase inhibitors (HDACi) treatment of reconstructed embryos after fusion of the donor cell to the cytoplasm and activation. In mice, trichostatin A (TSA), a hydroxamic acid-containing HDACi that specifically inhibits class I and IIa/b HDACs, improved nuclear remodeling of SCNT embryos and subsequent development to term (Ding et al., 2008; Kishigami et al., 2007). Mixed results were observed with the use of TSA in pigs. Development of cloned embryos to the blastocyst stage was improved in one study (Li et al., 2008), but resulted in low development of pig embryos in a second study (Zhao et al., 2010). Because of the inconsistency of the use of TSA in pig SCNT, the effectiveness of other HDACi was examined. Scriptaid [6-(1,3-dioxo-1H, 3H-benzo[de]isoquinolin-2-yl)-hexanoic acid hydroxyamide], another potent class I and class IIa/b HDACi, was highly effective in improving nuclear transfer efficiency in pigs, as treating reconstructed SCNT embryos with Scriptaid for 14–16 h postfusion and activation resulted in an increase in cloning efficiency of inbred NIH miniature pigs from 0% to 1.3%. This change reflected an increase in live piglet number from zero (nine embryo transfers) to 17 (10 embryo transfers) (Zhao et al., 2009). Scriptaid was also used to improve cloning efficiency in a difficult-to-clone adult ear fibroblast line and improved the liveborn rate from 0% to 3.7% (Zhao et al., 2010).
Another study examined gene expression at the blastocyst stage in SCNT embryos treated and not treated with Scriptaid after fusion and activation (Whitworth et al., 2011). In this microarray-based study, genes were identified as being different between SCNT blasocyst stage embryos (No HDACi) and in vivo blastocyst-stage embryos (IVV), as well as being different from the donor cell line used to construct the SCNT embryos. It was hypothesized that Scriptaid treatment was changing the transcriptional profile of the SCNT embryos to be more “in vivo–like” and thus also improving cloning efficiency. Interestingly, Scriptaid treatment did not alter gene expression in half of the misregulated transcripts and only returned gene expression to normal in vivo levels in three of the 14 closely evaluated transcripts (Whitworth et al., 2011).
In the mouse, two other inhibitors of class I and II a/b HDAC, suberoylanilide hydroxamic acid (SAHA) and oxamflatin, were shown to increase overall cloning efficiencies, but only SAHA increased blastocyst rates above non-HDACi treatment (Ono et al., 2010). Another class I and IIa HDACi, valproic acid, showed no improvements in development, suggesting inhibition of the class IIB HDACs (HDAC6 and HDAC10) may be of particular importance in the mouse; but valproic acid treatment in SCNT pig embryos did improve cloning efficiencies (Huang et al., 2011). Also in the pig, oxamflatin treatment did increase both the number of embryos that developed to blastocyst stage and total number of nuclei in oxamflatin-treated blastocysts when compared to the nontreated control, thus successfully increasing cloning efficiencies from 0.9% to 3.2% (Park et al., 2012). Another study compared oxamflatin directly to Scriptaid and showed a significant increase in the number of live pigs born when using oxamflatin (Mao et al., 2015). SAHA has yet to be tested for its ability to improve SCNT in the pig.
The exact mechanism for the improvements in nuclear remodeling by HDACi after SCNT are yet to be elucidated. Inhibition of deacetylases results in an increase in the global acetylation of histones (Zhao et al., 2010). It is thought that increased acetylation results in a change in the chromatin structure such that proteins like RNA polymerases can gain access to the DNA and begin transcription (Van Thuan et al., 2009). Here the effectiveness of HDACi, SAHA, and its hydrophobic derivitive 4-iodo-SAHA (ISAHA) was compared to Scriptaid treatment. Illumina deep sequencing was performed on normal IVV-derived blastocyst-stage embryos as well as embryos derived from in vitro fertilization (IVF), SCNT not treated with HDACi, and SCNT treated with three different HDACi. The goal of this experiment was to use this transcriptome data to identify a mechanism for the observed improvements in pig SCNT after HDACi treatment.
Materials and Methods
Ethical guidelines
All animal procedures were performed with an approved University of Missouri Institutional Animal Care and Use (ACUC) protocol.
Chemicals
All chemicals for embryo culture were purchased from Sigma-Aldrich Company (St. Louis, MO, USA) unless otherwise mentioned. SAHA and ISAHA were purchased from Cayman Chemical (Ann Arbor, MI, USA).
Animals, donor cell line, and oocytes
The recipient gilts used for embryo transfer were all from the University of Missouri swine herd that consists of large white landrace crosses from Newsham Genetics (West Des Moines, IA, USA). Oocytes were purchased from Applied Reproductive Technologies (A.R.T., Madison, WI, USA) and matured and prepared as described previously (Whitworth et al., 2009). Briefly, cumulus–oocyte complexes (COC) were collected from sow ovaries and shipped in maturation medium [Tissue Culture Medium-199 (TCM-199) with 2.9 mM HEPES, 5 μg/mL insulin, 10 ng/mL epidermal growth factor (EGF), 0.5 μg/mL pure follicle-stimulating hormone (p-FSH), 0.91 mM pyruvate, 0.5 mM cysteine, 10% porcine follicular fluid, and 25 ng/mL gentamicin] at 38.5°C. Upon arrival, COCs were transferred to fresh maturation medium and cultured for a total of 40 h. COCs were then vortexed for 4 min in 0.1% polyvinylalcohol (PVA) and 0.5 μg/mL hyaluronidase in a 0.3 M mannitol, TL-HEPES buffered-based medium. Metaphase II (MII) oocytes with a clearly visible extruded polar body and evenly distributed cytoplasm were placed in manipulation medium with 13.3 μM cytochalasin B and used for SCNT as described previously (Lai and Prather, 2003). The donor cell line was female and was genetically modified for xenotransplantation purposes for the National Swine Resource and Research Center (http://nsrrc.missouri.edu/ NSRRC:0025). The donor cells were thawed and cultured for 3–4 days before SCNT in 50% TCM-199 medium and 50% Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, and 5 ng/mL basic fibroblast growth factor (bFGF).
Treatment groups for blastocyst-stage embryo collection
BLIVV
Gilts were artificially inseminated on days 0 and 1 of the estrous cycle. Blastocyst-stage embryos were collected by midventral laparotomy on day 6 of gestation.
BLIVF
Oocytes were fertilized in vitro as previously described (Abeydeera et al., 1998). After IVF, oocytes were washed three times and placed in culture in Porcine Zygote Medium 3 (Yoshioka et al., 2002) supplemented with 1.69mM arginine (MU1) (Bauer et al., 2010). Embryos were cultured in 5% CO2 in air for 18 h and transferred to a humidified low-oxygen tension incubator 5% CO2 and 5% O2 for a total of 7 days.
SCNT
Reconstructed one-cell stage embryos were fused/activated in a fusion medium (0.3M mannitol, 0.1mM CaCl2, 0.1 mM MgCl2, 0.5 mM HEPES) with two DC pulses (1-sec interval) at 1.2 kV/cm for 30 μsec by using a BTX Electro Cell Manipulator (Harvard Apparatus). Immediately after fusion, HDACi-treated embryos were placed in one of three treatments: 0.5 μM Scriptaid, 1 μM ISAHA, or 10 μM SAHA in MU1. Preliminary studies also used 10 μM ISAHA and 1 μM SAHA. Nontreated control SCNT embryos were placed into directly into MU1. Embryos were cultured with HDACi for 14–16 h in 5% CO2 in air. After 14–16 h, embryos were washed and placed into fresh MU1 and transferred to a humidified, low-oxygen tension incubator (90% N2, 5% CO2, and 5% O2) at 38.5°C for a total of 7 days. IVF embryos were cultured similarly.
Blastocyst-stage development and total cell number
Blastocyst-stage embryo development was calculated by determining the total number of blastocyst-stage embryos at day 7 of development and comparing that to the total number of embryos in culture for that treatment (percent blastocyst). All zona pellucidae were removed by using a physiological saline lowered to a pH of 1.79. Zona-free embryos were then incubated with bisbenzimide (Hoechst 33342, Sigma, St. Louis, MO, USA) at 37°C for 30min at a concentration of 3ng/mL. Embryos were washed and mounted on slides under coverslips and visualized by using ultraviolet (UV) microscopy. The total number of nuclei were counted for each treatment and reported as a total cell number. Embryos with greater than 12 nuclei each were included in the analysis. There was a total of five biological replicates. The statistical analysis on blastocyst development and total cell number was performed by using Proc GLM procedure in SAS V9.1, and means were compared by using a protected least significant difference (LSD).
Embryo transfer and development to term
To determine if embryos treated with an HDACi (0.5 μM Scriptaid, 1 μM ISAHA, or 10 μM SAHA) could develop to term, embryo transfers were performed surgically on day 0 or 1 of estrus into a recipient gilt. Reconstructed one-cell stage embryos derived by NT were removed from HDACi treatment and transferred to recipient gilts within 3h. The number of reconstructed embryos transferred ranged from 184 to 254. On days 115–117 of pregnancy, piglets were delivered by cesarian section. Piglets were syringe fed colostrum, and health was monitored every 2h during the first 24h. Piglet survival 1 week (7 days) and 6 weeks (42 days) was compared by using Proc GLM procedure in SAS V9.1. Several healthy pigs were transferred to other universities between the ages of 38 and 136 days or were euthanized for experimental reasons other than poor health. The age of the pig when it was removed from University of Missouri facilities was used for the above comparison. A comparison of normal at the time of death versus abnormal at the time was statistically analyzed by the chi-squared method by using the Proc Freq procedure in SAS V9.1.
Sample collection
Blastocyst-stage embryos with a clear inner cell mass (ICM), trophectoderm, and blastocoel cavity were collected for both RNA analysis by deep sequencing and real-time PCR and for fixation to evaluate lysosomal intensity and localization as well as immunohistochemistry. For RNA analysis, blastocyst-stage embryos were counted and removed from culture medium. Zonae pellucidae were removed by rapid treatment with low pH phosphate-buffered saline (PBS) with 0.1% PVA and washed in diethylpyrocarbonate (DEPC)-treated 0.1% PVA in PBS. Embryos were pooled into groups of 10 embryos and snap frozen. Embryos were stored at −80°C until RNA isolation.
RNA isolation and amplification
Total RNA was isolated from each 10-embryo pool by using the AllPrep DNA/RNA Micro Kit (Qiagen, Germantown, MD, USA) and following the manufacturer's instructions. Of the 12 μL of eluted RNA, 5 μL were amplified with the Ovation RNA-Seq System V2 (Nugen, San Carlos, CA, USA). Concentrations of RNA/cDNA were determined by using a Nanodrop spectrophotometry system (Waltham, MA, USA). Amplified cDNA (1.2 μg) was then delivered to the MU DNA core.
Library preparation and deep sequencing
Amplified cDNA was re-evaluated for concentration and quality by using a BioAnalyzer 2100 (Agilent, Santa Clara, CA, USA). Amplified cDNA was diluted to a concentration of 10ng/mL and fragmented by using a Biorupter UCD-200TM (Diagenode, Liege, Belgium). The Biorupter settings used the energy setting on high, and the fragmentation time was 7.5min twice with on/off=0.5/0.5. One microgram of the fragmented DNA was used to generate the sequencing libraries by using Illumina's TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA, USA). The gel-free protocol was followed during the preparation process. The concentration of the final library was measured by using a Qubit dsDNA HS Array Kit (Life Technologies, Carlsbad, CA, USA), and the library size was also analyzed by using the Agilent Bioanalyzer 2100's High Sensitivity DNA Chips.
Libraries were sequenced on the Illumina HiSeq 2000 by using standard recipes (Recipe Version 1.3.26, HSC 1.5.15.1, RTA 1.13.48). Six samples to a lane were barcoded, multiplexed, and loaded at an 8 pM concentration. A single read collecting 100 bases and six bases of an index read were performed with Illumina TruSeq SBS version 3 chemistry. Demultiplexing and fastq files were generated by using CASAVA 1.8.2 (Illumina).
Raw sequencing data for IVV (n=4), IVF (n=4), SCNT (No HDACi) (n=4), SCNT (ISAHA) (n=4), SCNT (SAHA) (n=4), and SCNT (Scriptaid) (n=3) samples were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) and are available under accession numbers SAMN03330032–SAMN03330054
Read alignment and bioinformatics
Fastq files were then aligned to a custom database by the University of Missouri Informatics Research Core Facility as described previously (Isom et al., 2010). Briefly, the custom database was made from the 1,262,922 sequences [mostly expressed sequence tags (ESTs)] in the Swine UniGene Build #37 from NCBI (ftp://ftp.ncbi.nih.gov/repository/UniGene/Sus_scrofa), and another group of ESTs that were not included in the UniGene build: (1) 2064 ESTs from MII oocytes (GenBank acc. nos. GT640367–GT642429) and (2) 426 ESTs from reproductive tissues sequenced from the 5′ direction (GT640447–GT640620) (Green et al., 2006; Whitworth et al., 2004). All sequences were trimmed of low-complexity regions by using the program SeqClean (http://compbio.dfci.harvard.edu/tgi/software), and each of the UniGene clusters was assembled by using the CAP3 sequence assembly program to reduce redundancy and to extend length (Huang and Madan, 1999). The publicly available swine UniGene set from NCBI uses the longest member of the EST cluster to represent the group instead of a consensus contig. The other two sequence sources were then aligned to these clusters by using BLAST (Altschul et al., 1990). Reads were then normalized as described previously (Bauer et al., 2010). Briefly, the normalization factor for each biological replicate was calculated by identifying the sample with the largest number of alignable reads and then taking a ratio of the total number of reads for the other samples. Means were then calculated for the four biological replicates for each treatment group.
Differential expression as determined by EdgeR
The Bioconductor package EdgeR (3.0.7) was used to perform differential expression analyses of read counts from the RNA-seq data (Robinson et al., 2010). EdgeR implements a statistical method based on a generalized linear model (GLM), which compares reads counts from each aligned transcript. Transcripts were considered differentially expressed that had a p value less than 0.05 and a false discovery rate (FDR) of 0.05. Fifteen pairwise comparisons were performed including: Set 1, all treatments compared to IVV [IVV vs. IVF, IVV vs. SCNT (No HDACi), IVV vs. SCNT (Scriptaid), IVV vs. SCNT (SAHA), IVV vs. SCNT (ISAHA)]; set 2, all treatments compared to IVF [IVF vs. IVV (same as above), IVF vs. SCNT (No HDACi), IVF vs. SCNT (Scriptaid), IVF vs. SCNT (SAHA), IVF vs. SCNT (ISAHA); set 3, all SCNT (No HDACi) compared to HDACi-treated groups (SCNT vs. sCNT (Scriptaid), SCNT vs. SCNT (SAHA), SCNT vs. SCNT (ISAHA)]; and set 4, HDACi-treated SCNT groups compared to each other [SCNT (Scriptaid) vs. SCNT (SAHA), SCNT (Scriptaid) vs. SCNT (SAHA), and SCNT (SAHA) vs. SCNT (ISAHA)].
Due to the large number of the comparisons, the focus of analysis will be on comparisons set 1 (compared to IVV), set 2 (comparison between SCNT), and set 4 (comparison between HDACi treatments)). Normalized read counts, fold changes, and up- and downregulated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways that are significantly different between comparisons are available as supplementary Excel files 1–4. All of the normalized read counts aligned to the custom genome are also available in Supplementary Excel file 1 (Supplementary data are available at http://animalsciences.missouri.edu/faculty/prather/).
Database for Annotation, Visualization, and Integrated Discovery (DAVID)
To determine which biological themes were affected by the above treatments in the blastocyst-stage embryos, GenBank accessions for the up- and downregulated genes greater than 1.5-fold change identified between the pairwise comparisons (p<0.05) were uploaded into DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/tools.jsp) (Dennis et al., 2003). This software extracts biological features and meanings associated with large gene lists. The Homo sapiens genome was used as the background gene list, which allowed for identification of gene families that were enriched in the up- or downregulated groups. The enriched functional annotation terms were identified and listed according to their enrichment p values (also known as EASE score) and fold enrichment score by DAVID. The top three significant KEGG pathways and Gene Ontologies were reported for each comparison.
Evaluation of histone-related transcripts and pluripotency markers by deep sequencing
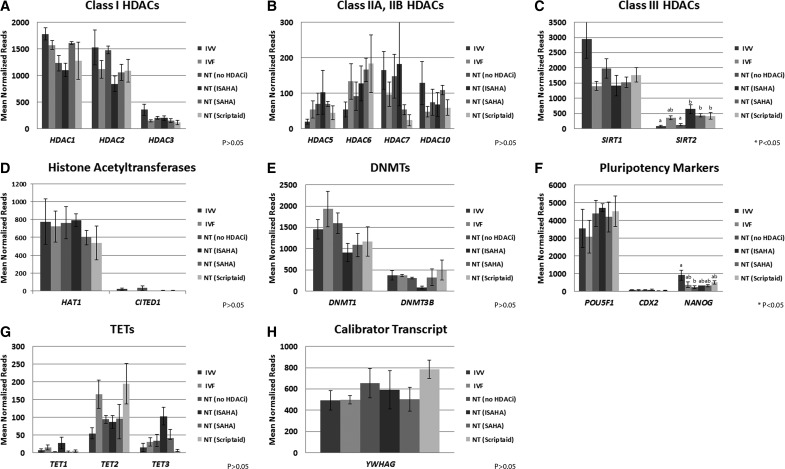
Histone-related transcripts and pluripotency markers including class I HDACs (HDAC1, HDAC2, HDAC3, HDAC8) class 2A HDAC (HDAC4, HDAC5, HDAC7, HDAC9), class 2B HDACs (HDAC6 and HDAC10), class 3 HDACs (SIRT1 and SIRT2), 2a and 2b histone acetyltransferases (HAT1 and CITED), DNA methyltransferases (DNMT1 and DNMT3B), (ten-eleven translocation (Tet) methylcytosine dioxygenases (TET1, TET2, and TET3) (Lee et al., 2014), and pluripotency markers (POU5F1, CDX2, and NANOG) were identified within the deep sequencing data set and evaluated for the number of normalize reads. Significant differences were determined as above by using EdgeR.
Deep sequencing validation by real-time PCR
Deep Sequencing results were validated by relative real-time PCR of lysosomal pathway transcripts, cathepsin K (CTSK), cathepsin A (CTSA), legumain (LGMN) hexosaminidase A (HEXA), hexosaminidase B (HEXB), and sphingomyelin phosphodiesterase 1 (SMPD1). Primers and annotations are listed in Table 1. Primers were ordered from Integrated DNA Technologies (IDT, Coralville, IA, USA). Real-time PCR parameters have been described previously (Whitworth et al., 2011). Primer efficiency was performed on each primer set by using a serial dilution of cDNA from the calibrator sample, also described previously (Whitworth et al., 2005). Three calibrator transcripts were tested for equal expression among the six treatments including: Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), actin, beta (ACTB), and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein and gamma polypeptide (YWHAG). Expression of GAPDH and ACTB was significantly different among treatments (p<0.0001 and p<0.0385, respectively).
Table 1.
Transcript Names, Primer Sequences, GenBank Accessions, and Annotations for Genes Analyzed by Real-Time PCR
| Gene name | Primer sequence 5′ to 3′ | GenBank Accession | Annotation |
|---|---|---|---|
| Lysosomal markers | |||
| CTSK | AGG TTG TAC TAC TGC TGC CTG TGA | NM_214302 | Sus scrofa cathepsin K (CTSK), mRNA |
| AAC GCC GAG AGA TTT CAT CCA CCT | |||
| CTSA | GGA GTC CCA GAA GGA TCC CA | NM_001243629 | Sus scrofa cathepsin A (CTSA), mRNA |
| GGG GAC TCG AGG TAC AAC AC | |||
| HEXA | GGT GGA TTT CAC CTG CTG GA | NM_001123221.1 | Sus scrofa hexosaminidase A (alpha polypeptide) (HEXA), mRNA |
| GAC AAT GCC CAG TAG CGT CT | |||
| HEXB | ACT ATG GCC TCG AGC AAG TG | NM_213921.1 | Sus scrofa hexosaminidase B (beta polypeptide) (HEXB), mRNA |
| CAG CTA TTC CTC GCC TGA CC | |||
| LGMN | CTG AAC GAG ACC ATC CAC TAC A | XM_001927082.4 | PREDICTED: Sus scrofa legumain (LGMN), mRNA |
| TTC GTC ATA GTA ACA GGC GTA AGA | |||
| SMPD1 | TGA ACA GGT ACG AGA ACA CCG | XM_003482522.1 | PREDICTED: Sus scrofa sphingomyelin phosphodiesterase 1, acid lysosomal (SMPD1), mRNA |
| AAC CGG GAT TCA GGC TGA TG | |||
| Calibrator transcript | |||
| YWHAG | TCCATCACTGAGGAAAACTGCTAA | XM_003124396.4 | PREDICTED: Sus scrofa tyrosine 3-monooxygenase/tryptophan 5-monooxygenase), activation protein, gamma polypeptide transcript variant X1, mRNA |
| TTTTTCCAACTCCGTGTTTCTCTA | |||
Real-time PCR was performed on the six treatments with the calibrator transcript YWHAG (NM_012479), which showed expression not to be different among treatments when measured by real-time PCR and read counts by deep sequencing (Fig. 1H) (p=0.159, p>0.05, respectively). The relative Ct (2−ΔΔCt) method was used to determine expression of candidate transcripts with the reference cDNA used as a calibrator sample. Bio-Rad iQ SYBR Green Supermix was used by following the recommended protocol from the manufacturer. A three-step protocol was used with a 60°C annealing temperature followed by a dissociation curve in the MyiQ single-color real-time PCR detection system (Bio-Rad, Hercules, CA, USA). All four biological replicates were tested in triplicate resulting in 12 Ct measurements/treatment. The statistical analysis on relative gene expression was performed by using Proc GLM procedure in SAS version 9.1 and means were compared by using a protected LSD. Data were log transformed before statistical analysis if not normally distributed as evaluated by Proc Univariate in SAS.
FIG. 1.
Mean normalized reads from deep sequencing from IVV, IVF, SCNT (No HDACi), SCNT (ISAHA), SCNT (SAHA), and SCNT (Scriptaid) blastocyst-stage embryos. Transcripts were subdivided into the following groups: Class I (A), Class IIA, IIB (B), and Class III (C) HDACs, histone acetyltransferase (HAT) activity (D), DNA methyltransferases (DNMTs) (E), pluripotency markers (F), TETs (G), and a calibrator transcript used for real-time PCR, YWHAG (H). Significant differences are indicated by * p value and superscripts a, b, c, d. NT=SCNT.
Lysosomal and cathepsin K localization
For lysosomal analysis and immunohistochemistry, zona pellucidae were also removed and embryos were incubated with 10mM LysoTracker Red DND-99 diluted in HEPES-buffered Tyrode's lactate medium ([TL-HEPES, 114 mM NaCl, 3.2 mM KCl, 25mM NaHCO3, 0.4 mM NaH2PO4H2O, 10 mM sodium lactate, 2 mM CaCl2, 0.5 mM MgCl·6H2O, 0.25 mM pyruvate, 0.6% bovine serum albumin (BSA), fraction V (Hagen et al., 1991)] for 30 min at 37°C. After LysoTracker treatment, embryos were washed three times in TL-HEPES, fixed in 4.0% paraformaldehyde for 30 min, and washed and stored in TL-HEPES at 4°C until immunohistochemistry (IHC). Three replicates with two to five embryos/replicate were collected for each treatment. IHC was performed with antibody for mouse monoclonal anti-cathepsin K (ab66237; Abcam Cambridge, MA, USA).
After fixation and storage, embryos were permeabilized with 0.1% Triton X and 0.1% Tween 20 in TL-HEPES for 30 min. All washes and incubations were performed in this same buffer. Embryos were then blocked in 1% BSA in permeabilization buffer for 1 h. Embryos were incubated in either primary antibody, CTSK (1:100), or without antibody at 4°C overnight. Embryos were washed three times and incubated with secondary rabbit anti-mouse immunoglublin G fluorescein isothiocyanate (IgG FITC) (IgG H&L) (1:500, ab6724; Abcam) for 1 h at 37°C. Embryos were washed three times and mounted in Flouromount G (Sigma, St. Louis, MO, USA) on glass slides under coverslips. Negative controls consisted of embryos treated with the same method in the absence of LysoTracker and primary antibody. Fluorescence images were captured by using a Nikon Eclipse Ti (Nikon, Melville, NY) with a 20× objective and the same exposure time for all treatments. Images were processed and mean intensities for the area around the LysoTracker-treated embryos were calculated in the NIS-Elements BR version 3.2 (Nikon) program. Background intensity for each replicate was subtracted from the mean intensity, and arbitrary units were imported into SAS V9.4. Significant differences were determined by using the Proc GLM procedure and means were separated by t grouping. Localization patterns were analyzed for CTSK, but mean intensities were not examined. Images were compiled in Adobe Photoshop CS3 (San Jose, CA, USA) without any manipulation.
Results
Blastocyst development and total cell number
Data from from five replicates were collected to compare percent blastocyst development between the treatments. There was not a significant difference in blastocyst development among any of the SCNT embryos regardless of treatment except for the highest concentration of ISAHA (10 μM), which signficantly decreased development rates (p<0.005; Table 2). Treatment with 10 μM SAHA had the numerically highest level of blastocyst development at 43.9%. Two treaments, 1.0 μM ISAHA and 1.0 μM SAHA, had significantly higher mean cell numbers compared to No HDACi treatment (p<0.021; Table 2). The treatments with the numerically highest percent blastocyst development (10 μM SAHA) and numerically highest mean total cell number (1 μM ISAHA) as well as the industry standard (0.5 μM Scriptaid) and No HDACi treatment were chosen for subsquent gene expression and lysosomal localization experiments.
Table 2.
Blastocyst-Stage Embryo Development Data and Mean Total Cell Number Data on Somatic Cell Nuclear Transfer Embryos Treated With ISAHA, SAHA, or Scriptaid Postactivation and Fusion
| Treatment | Percent blastocyst development | Total blastocyst | Number of embryos cultured | Mean total cell number±SEM | Total blastocysts examined | Replicates |
|---|---|---|---|---|---|---|
| No HDACi | 31.9a | 43 | 135 | 30.1±6.0a | 36 | 5 |
| 1.0 μM ISAHA | 34.2a | 26 | 76 | 37.9±3.1b | 20 | 5 |
| 10 μM ISAHA | 17.6b | 13 | 74 | 33.4±2.8ab | 13 | 5 |
| 1.0 μM SAHA | 32.6a | 47 | 144 | 37.6±2.3b | 32 | 5 |
| 10 μM SAHA | 43.9a | 47 | 107 | 35.7±1.7ab | 31 | 5 |
| 0.5 μM Scriptaid | 40.5a | 62 | 153 | 31.9±1.7ab | 39 | 5 |
Corresponds to significant differences between means as compared by least significant difference (LSD).
ISAHA, 4-iodo-suberoylanilide hydroxamic acid; SAHA, suberoylanilide hydroxamic acid; SEM, standard error of the mean; HDACi, histone deacetylase inhibitor.
Production of live offspring after treatment with HDACi
Only the HDACi treatments were evaluated for term development. It has been previously shown that No HDACi treatment results in low numbers of live offspring (Zhao et al., 2009; Zhao et al., 2010); the resulting piglets from this experiment were needed as xenotransplantation models, therefore embryos from the No HDACi treatment were not used for embryo transfer. SCNT embryos treated with 1 μM ISAHA, 10 μM SAHA, or 0.5 μM Scriptaid were transferred to recipient gilts on days 0 or 1 of estrus. All three treatments resulted in the birth of live pigs. The average litter size for 1 μM ISAHA, 10 μM SAHA, or 0.5 μM Scriptaid was 3.3, 2.5, and 4.5 with pregnancy rates of 75%, 100%, or 66.7%, respectively (Table 3). Of the nine piglets born after Scriptaid treatment, three died within 24h due to abnormalities (abnormal bladder, kidneys and liver, or macroglossia); only one piglet lived to 6 weeks of age, but this pig was euthanized due to poor leg and spinal structure. Of the 10 piglets born after ISAHA treatment, two died within 24h, one due to failure to thrive and one for unknown reasons. Four of the ISAHA pigs lived over 6 weeks. Of the five piglets born after SAHA treatment, two died within 24h, one due to macroglossia and contracted tendons and one due to muscular failure. Two of the SAHA pigs lived beyond 6 weeks. Summary of ages and abnormalities are in Table S1 (Supplementary Data are available at www.liebertpub.com/cell/).
Table 3.
Embryo Transfer Data
| Treatment | Embryos transferred | Number of recipients | Number pregnant | Pregnancy rate | Number piglets |
|---|---|---|---|---|---|
| 1.0 μM ISAHA | 858 | 4 | 3 | 75% | 10 |
| 10.0 μM SAHA | 395 | 2 | 2 | 100% | 5 |
| 0.5 μM Scriptaid | 616 | 3 | 2 | 66.7% | 9 |
ISAHA, 4-iodo-suberoylanilide hydroxamic acid; SAHA, suberoylanilide hydroxamic acid.
Piglet survivability to 1 week and 6 weeks was compared between the treatment groups. There was no significant difference between the treatments on piglet survivability to 1 week or 6 weeks (p>0.60 and p>0.43, respectively). Normal piglets from the ISAHA and SAHA groups were transferred to other institutions for use in xenotransplantation studies. A comparison between pigs that were normal at the time of death/transfer and the pigs that were abnormal at the time of death showed a statisical trend toward more ISAHA- and SAHA-treated pigs being normal at the time of death, zero (Scriptaid) compared to four (ISAHA) and two (SAHA) (p>0.08).
Analysis of RNA-seq reads
RNA-seq resulted in an average of 26,525,857 raw reads for each treatment replicate. A mean of 91.6% passed the low-complexity filter and resulted in a mean of 24,271,650 reads being aligned to the custom genome. The mean quality score for each replicate was 35.0 with a cutoff of 30. One treatment, Scriptaid replicate 2, resulted in a low number of raw reads after two sequencing attempts. The reads obtained also did not align to the custom swine genome and were therefore removed from the analysis. The quality control data for each treatment and replicate as well as the normalization factors used are detailed in Table S2.
Differential gene expression and DAVID analysis
Fifteen pairwise comparisons (details are described in Materials and Methods) were performed including four sets of comparions: Set 1, all treatments compared to IVV; set 2, all treatments compared to IVF; set 3, all SCNT (No HDACi) compared to HDACi-treated groups; and set 4, HDACi-treated SCNT groups compared to each other.
Comparison of set 1
All treatments were compared to IVV.
Gene expression
IVV vs. IVF, IVV vs. SCNT (No HDACi), IVV vs. SCNT (Scriptaid), IVV vs. SCNT (SAHA), and IVV vs. SCNT (ISAHA) were compared and genes with greater than a 1.5-fold change from IVV and a significant p value and FDR were considered significantly different (p<0.05). A detailed list of differentially expressed transcripts is available in a supplementary Excel file (Supplementary File 2). There were 560 upregulated and 224 downregulated transcripts in IVF when compared to IVV, resulting in the highest number of differentially expressed transcripts between these comparisons. There were 359 upregulated and 94 downregulated transcripts when comparing SCNT (No HDACi) blastocyst-stage embryos to IVV, resulting in the fewest number of differentially expressed transcripts between the comparisons. Among the HDACi-treated embryos, the SCNT (Scriptaid) had the fewest differentially expressed transcripts, with 394 upregulated and 182 downregulated. SCNT (SAHA) had the highest number of differentially expressed transcripts, with 525 upregulated and 245 downregulated. SCNT (ISAHA) had 412 upregulated and 237 downregulated transcripts when compared to IVV.
KEGG Pathways
GenBank accession numbers from transcripts that were significantly different from IVV (>1.5-fold and p<0.05) were loaded in DAVID and both up- and downregulated enriched biological pathways were identified (Table 4). For upregulated transcripts, the lysosome pathway was enriched in all of the comparisons except IVV vs. SCNT (No HDACi). Focal adhesion and the insulin signaling pathway were also upregulated in IVF compared to IVV. Glycine, serine, and threonine metabolism and the p53 signaling pathways were upregulated in the SCNT (No HDACi) blastocyst-stage embryos. In addition to the lysosomal pathway, the steroid biosynthesis pathway was also upregulated in SCNT embryos treated with HDACi. Only one downregulated pathway was identified in the IVF blastocyst-stage embryos, systemic lupus erythematosus. This pathway included three histone transcripts: H2A histone family, member X (H2AFX), histone cluster 1, H2ag; histone cluster 1 (HIST1H2AG) and H4l; and histone cluster 1 (HIST1H4K). The acute myeloid leukemia pathway was downregulated in the SAHA treatment and included transcripts BCL2-associated agonist of cell death (BAD), runt-related transcription factor 1 (RUNX1), and runt-related transcription factor 1, translocated to, 1 (cyclin D-related) (RUNX1T1). There were no downregulated KEGG pathways identified in the SCNT (No HDACi), Scriptaid, or ISAHA embryos when compared to IVV. A detailed list of enriched biological pathways and the associated transcripts with GenBank accession numbers can be found in Supplementary Excel File 2.
Table 4.
Enriched Biological Themes Identified in DAVID
| Category | Entry | Enriched KEGG pathways | Fold enrichment | p value | Associated genes |
|---|---|---|---|---|---|
| Upregulated | |||||
| IVF | hsa04142 | Lysosome | 4.41 | 1.25E-08 | CTSK, CTSA, CTSZ, HEXA, HEXB |
| hsa04510 | Focal adhesion | 2.68 | 3.39E-05 | ACTB, GLB1, GLA, SGSH, ABCB9 | |
| hsa04910 | Insulin signaling pathway | 2.95 | 1.65E-04 | CBLB, MKNK2, FASN, FBP1, INSR | |
| SCNT (no HDACi) | hsa00260 | Glycine, serine and threonine metabolism | 6.37 | 2.35E-02 | PSAT1, CBS, PHGDH, PSPH |
| hsa04115 | p53 signaling pathway | 3.63 | 4.68E-02 | FAS, CCND1, SERPINE1, SESN2,ZMAT3 | |
| Scriptaid | hsa04142 | Lysosome | 5.27 | 2.2E-07 | ASAH1, SGSH, ARSA, CTSK, HEXA, HEXB, LGMN |
| hsa00100 | Steroid biosynthesis | 15.86 | 2.6E-06 | DHCR24, CYP51A1P1,EBP, FDFT1, SQLE | |
| hsa00900 | Terpenoid backbone biosynthesis | 12.84 | 0.00046 | HMGCR, HMGCS1, ACAT1, IDI1, MVK | |
| SAHA | hsa04142 | Lysosome | 5.23 | 1.82E-10 | CTSK, CTSA, HEXA,HEXB, LGMN |
| hsa00100 | Steroid biosynthesis | 14.09 | 5.98E-08 | DHCR24, CYP51A1, EBP, FDFT1, HSD17B7 | |
| hsa00531 | Glycosaminoglycan degradation | 10.14 | 6.79E-06 | DHCR24, CYP51A1, EBP, FDFT1, HSD17B7 | |
| ISAHA | hsa04142 | Lysosome | 5.24 | 8.7E-08 | ATP6V0D1, ASAH1, CTSA, CTSH, CTSK |
| hsa00100 | Steroid biosynthesis | 16.97 | 1.6E-07 | DHCR24, CYP51A1, EBP, FDFT1, LSS | |
| hsa00531 | Glycosaminoglycan degradation | 10.30 | 0.00021 | SGSH, GLB1, IDS, HEXA, HEXB | |
| Downregulated | |||||
| IVF | hsa05322 | Systemic lupus erythematosus | 3.67 | 0.04465 | H2AFX, HIST1H2AG, HIST1H4K, HLA-DPA1, SNRPD1 |
| SCNT (No HDACi) | No KEGG pathways identified | ||||
| Scriptaid | No KEGG pathwaysi | ||||
| ISAHA | No KEGG pathways identified | ||||
| SAHA | hsa05221 | Acute myeloid leukemia | 6.54 | 0.00649 | BAD, RUNX1, RUNX1T1, KIT, MYC |
DAVID, Database for Annotation, Visualization, and Integrated Discovery; KEGG, Kyoto Encyclopedia of Genes and Genomes; IVF, in vitro fertilization; SCNT, somatic cell nuclear transfer; HDACi, histone deacetylase inhibitor; SAHA, suberoylanilide hydroxamic acid; ISAHA, 4-iodo-suberoylanilide hydroxamic acid.
Comparison of set 2
All treatments were compared to IVF, IVF vs. IVV (same as above), IVF vs. SCNT (No HDACi), IVF vs. SCNT (Scriptaid), IVF vs. SCNT (SAHA), and IVF vs. SCNT (ISAHA). Due to the large number of comarisons, this analysis was not the focus of this experiment, but the list of differentially expressed transcripts identified after EdgeR analysis is available as Supplementary File 3. Additionally, normalized reads values for each treatment for each treatment are also available as Supplementary Excel File 1).
Comparison of set 3
All SCNT (No HDACi) compared to HDACi-treated groups, SCNT vs. SCNT (Scriptaid), SCNT vs. SCNT (SAHA), SCNT vs. SCNT (ISAHA), and SCNT (Scriptaid) vs. SCNT (SAHA).
Gene expression
The SCNT vs. SCNT (Scriptaid) comparison resulted in the least number of differentially expressed transcripts and included 52 upregulated and 66 downregulated. The SCNT vs. SCNT (SAHA) comparison resulted in 64 upregulated and 81 downregulated transcripts. The SCNT vs. SCNT (ISAHA) comparison had the highest number of differentially expressed transcripts, including 82 upregulated and 120 downregulated.
KEGG Pathways
GenBank accession numbers from transcripts that were significantly different from SCNT (No HDACi) (>1.5-fold and p<0.05) were loaded into DAVID and both up- and downregulated enriched biological pathways were identified (Table 4). For upregulated transcripts, the lysosome pathway was enriched in all three of the HDACi-treated blastocyst-stage embryos, with an upregulation of CTSK, HEXA, and LGMN genes. ISAHA treatment also had an upregulation of the glycosphingolipid biosynthesis pathway and another glycan degradation pathway. There were no downregulated pathways identified in any of the HDACi-treated groups. KEGG pathways and associated transcripts are listed in Table 4. A detailed list of enriched biological pathways and the associated transcripts with GenBank accession numbers can be found in Supplementary Excel File 4.
Comparison of set 4
HDACi-treated SCNT groups were compared to each other. SCNT-Scriptaid vs. SCNT-SAHA, SCNT-SAHA vs. SCNT-ISAHA, and SCNT-Scriptaid vs. ISAHA resulted in only three, one, and one differentially expressed transcripts, respectively (p<0.05). Normalized read counts and fold changes are reported in Table 5 with details listed in Supplementary Excel File 1. DAVID analysis was not performed due to the small size of the gene lists. Gene lists are shown in Supplementary File 5.
Table 5.
Differential Expression Identified by RNA-seq in Blastocyst-Stage SCNT Embryos Treated with HDACi, Scriptaid, ISAHA, and SAHA after Fusion and Activation
| Comparison with statistical significance | Mean normalized reads Scriptaid | Mean normalized Reads SAHA | Mean normalized Reads ISAHA | Fold change (Log) | EdgeR p value | FDR | GenBank accession no. | Annotation |
|---|---|---|---|---|---|---|---|---|
| Scriptaid/SAHA | 529.5 | 8.5 | 93.2 | 6.0 | 2.62E-06 | 0.024 | XM_003355640.2 | PREDICTED: Sus scrofa histone deacetylase 7-like (LOC100627559) |
| Scriptaid/SAHA | 3764.8 | 21687.5 | 17554.5 | –2.5 | 9.94E-08 | 0.002 | NM_001244086.1 | Sus scrofa eukaryotic translation initiation factor 3, subunit L (EIF3L), mRNA |
| Scriptaid/SAHA | 0.0 | 60.9 | 8.4 | n/c | 4.58E-06 | 0.028 | NM_001243765.1 | Homo sapiens RAB GTPase activating protein 1 variant 4, mRNA |
| SAHA/ISAHA | 2.7 | 0.0 | 108.9 | n/c | 2.66E-06 | 0.049 | XM_001928419.2 | PREDICTED: Sus scrofa TruB pseudouridine (psi) synthase homolog 2 (Escherichia coli) (TRUB2) |
| Scriptaid/ISAHA | 153.0 | 0.0 | 19.5 | n/c | 7.88E-07 | 0.015 | XM_003130920.2 | PREDICTED: Sus scrofa stAR-related lipid transfer transfer protein 13-like (LOC100155651) |
SCNT, somatic cell nuclear transfer; HDACi, histone deacetylase inhibitor; ISAHA, 4-iodo-suberoylanilide hydroxamic acid; SAHA, suberoylanilide hydroxamic acid; FDR, false discovery rate; n/c, not calculated as log fold change cannot be determined if one sample does not contain normalized reads.
Differential gene expression of HDAC-related transcripts and pluripotency markers
Normalized read counts for several HDAC related transcripts, including class I, IIA and IIB, III HDACs, histone acetyltransferase (HAT1 and CITED1), DNA methyltransferases (DNMT1 and DNMT3B), Tet methylcytosine dioxygenase (TET1, TET2, and TET3), as well as pluripotency markers (POU5F1, CDX2, and NANOG) were examined among the six treatment groups. Interestingly, none of the class I, IIA, or IIB HDAC were significantly different by the blastocyst stage, even in the HDACi-treated embryos. HDAC1 and HDAC2 were highly expressed in all blastocyst-stage treatments, with 1781 and 1528 mean normalized reads in IVV. HDACs 3, 5, 7, and 10 were also equally expressed between treatments, but had lower expression ranges from 20 reads to 358 reads when compared to HDAC1 and HDAC2. Expression of HDAC4, HDAC8, and HDAC9 was either not detected or extremely low in blastocyst-stage embryos. The class III HDAC SIRT2 was significantly lower in IVV embryos compared to IVF, SCNT (ISAHA), SCNT (SAHA), and SCNT (Scriptaid), but was not significantly different than SCNT (No HDACi). SIRT1 was equally expressed in all treatments.
There were also no significant differences among the HATs, DNMTs, or TETs examined. Among the pluripotency markers, only NANOG was significantly different between IVV and SCNT (No HDACi). HDACi treatment increased expression of NANOG in SCNT embryos. The calibrator transcript YWHAG was also evaluated and resulted in equal expression as measured by real-time PCR (p=0.159) and by number of normalized reads across all treatments (p>0.05). Normalized reads for HDAC related transcripts and pluripotency markers are shown in Figure 1.
Deep sequencing validation by real-time PCR
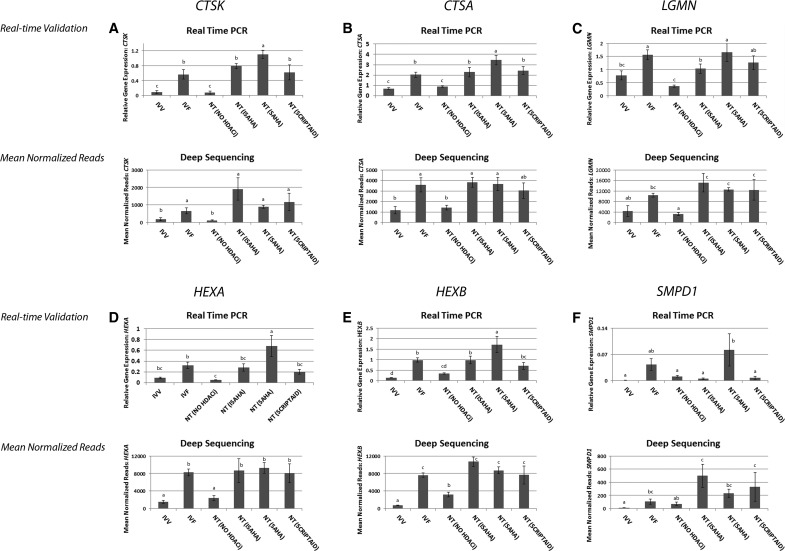
Validation of deep sequencing and the DAVID identified upregulation of the lysosomal pathway was performed by real-time PCR of six transcripts (CTSK, CTSA, LGMN, HEXA, HEXB, SMPD1) in addition to the calibrator transcript YWHAG. Expression of the calibrator transcript, YWHAG, was not different across all treatments (Fig. 1H). Deep sequencing showed that IVV and SCNT embryos had low expression of the lysosomal transcripts, but expression of lysosomal transcripts was increased in IVF and SCNT (HDACi-treated) embryos (Fig. 2). CTSK, CTSA, LGMN, HEXA, and HEXB showed a similar pattern of expression by real-time PCR with the exception of SAHA-treated SCNT blastocysts, which had increased expression above IVV and SCNT and also significantly higher expression over SCNT embryos treated with ISAHA and Scriptaid (p values, p<0.00001, p<0.00001, p<0.0008, p<0.0001, p<0.00001, p<0.019, respectively). SMPD1 was also expressed at low levels in IVV and NT embryos, but only had a significant increase in expression in SCNT (SAHA) (p<0.018). Deep sequencing showed an increase in expression of SMPD1 in all the HDACi-treated embryos as well as IVF, similar to the other transcripts (Fig. 2).
FIG. 2.
Real-time PCR validated the upregulation of lysosomal transcripts in the SCNT blastocyst-stage embryos treated with HDACi observed by deep sequencing. A side-by-side comparison of deep sequencing results and real-time PCR are shown in panels A, B, C, D, E, and F for transcripts CTSK, CTSA, LGMN, HEXA, HEXB and SMPD1, respectively. Significant differences with a p value<0.05 are indicated by superscripts a, b, c, d. NT=SCNT.
Lysosome localization and intensity
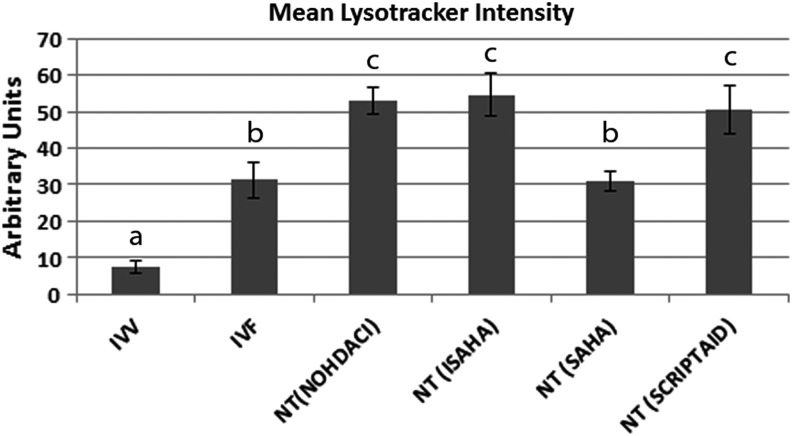
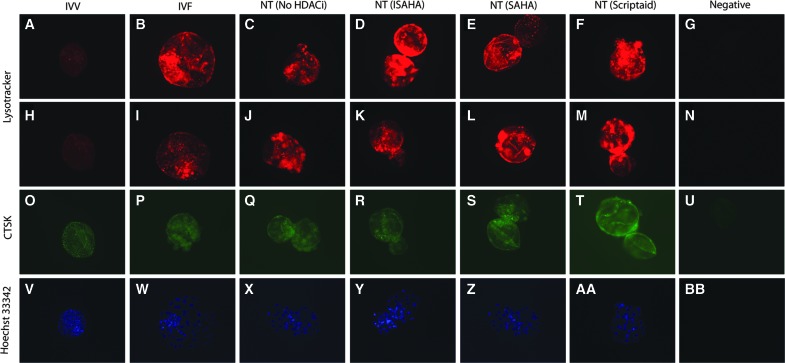
Lysosomal mean intensity was significantly lower in IVV blastocyst-stage embryos when compared to IVF, SCNT (No HDACi), SCNT (ISAHA), SCNT (SAHA), and SCNT (Scriptaid) (p<0.0001). SCNT (No HDACi), SCNT (ISAHA), and SCNT (Scriptaid) had the highest level of LysoTracker intensity. IVF and SCNT (SAHA) had an intermediate level of LysoTracker intensity (Fig. 3). The localization pattern of lysosomes in all of in vitro–cultured blastocyst-stage embryos revealed a large number of punctate areas of intensity around the entire embryo (Fig. 4). IVV embryos had very little staining of lysosomes, with only a few areas of punctate intensity around the embryo (Fig. 4A, H). There were no obvious differences in the localization patterns of the lysosomes between the SCNT embryos treated with HDACi.
FIG. 3.
The mean intensity from IVV, IVF, NT (No HDACi), NT (ISAHA), NT (SAHA), and NT (Scriptaid) blastocyst-stage embryos after incubation with LysoTracker. The mean intensity of the entire area of the embryo was used to for this calculation. Significant differences with a p value<0.05 are indicated by superscripts a, b, c. NT=SCNT.
FIG. 4.
Lysosomes were localized by LysoTracker in IVV, IVF, SCNT (No HDACi), SCNT (ISAHA), SCNT (SAHA), and SCNT (Scriptaid) blastocyst-stage embryos. Two representative embryos from each treatment are shown in panels A–M. Representative negative controls are shown in panels G and M. CTSK localization is shown in panels O–T with a representative negative control is shown in panel U. Total cell number was determined by staining with the DNA Hoechst 33342 and counting nuclei. A representative embryo for each treatment is shown in panels V–AA with a negative control shown in panel BB. NT=SCNT.
CTSK localization
CTSK localization was also different in IVV blastocyst-stage embryos when compared to in vitro–derived blastocysts (Fig. 4O–T). In the IVV embryos, CTSK appeared as punctate speckles around the entire embryo. In the other in vitro groups, CTSK did have some areas of localized CTSK expression, but CTSK appeared to be diffuse in the cytoplasm. There were no obvious differences in CTSK localization in the NT embryos treated with HDACi.
Discussion
Treatment of reconstructed SCNT embryos with SAHA and ISAHA postfusion and activation can successfully make healthy pigs from a donor cell line that had a particularly high rate of postnatal mortality when using Scriptaid. Previous studies showed that HDACi after activation and fusion resulted in some changes in gene expression, but did not create an SCNT blastocyst-stage embryo with a similar gene expression profile as a normal IVV embryo (Whitworth et al., 2011). Multiple studies have shown changes in gene expression in the “usual suspects” (i.e., histone deacetylases, DNA methyltransferases, and pluripotency markers) when using HDACi, but the mechanism of how HDACi truly improve cloning efficiency may actually lie in the embryo's ability or need for proper expression of lysosomal transcripts.
It is clear that HDACi changes the acetylation levels in the treated embryos. Scriptaid treatment increased acetylation levels of histone 4 at lysine 8 (AcH4K8) in the pronuclear SCNT embryo to a similar level observed in IVF embryos (Zhao et al., 2010). Sodium butyrate treatment also resulted in global acetylated histone 3 at lysine 12 (AcH3K12) profile in SCNT embryos that were similar to IVF (Liu et al., 2012). Treatment of SCNT embryos with a recently evaluated HDACi, CBHA (m-carboxycinnamic acid bishydroxamide) resulted in a rapid rise in acetylation of histone 3 lysine 9 (AcH3K9), lysine 18 (AcH3K18), and histone 4 lysine 16 (AcH4K16) immediately after treatment, but this increase in acetylation only persisted to the blastocyst stage in AcH3K18 (Song et al., 2014).
Inhibition of HDACs during reprogramming increases acetylation and changes gene expression, but a clear pattern of which changes are important has not been established. Most comparative studies focused on changes in gene expression between IVF and cultured embryos and SCNT embryos with and without HDACi, thus confounding the results with in vitro culture conditions. This study also compared IVF and SCNT embryos cultured to the blastocyst stage, but the transcriptome and lysosome localization and intensity of IVV embryos was also determined so that normal development could also be evaluated.
This study found no signficant differences in gene expression among class I or class IIa/b HDACs when analyzed by deep sequencing; thus, it appears that regulation of these HDACs was normalized among treatments by the blastocyst stage. Liu et al. (2012) did see a significant change in HDAC2 transcript expression after sodium butyrate treatment. Gene expression of the the Class III HDACs, SIRT2, which is not inhibited by Scriptaid, SAHA, or ISAHA, was significantly increased at the blastoycyst stage in HDACi-treated embryos. SIRT1 was not affected by treatment, but highly expressed in all of the blastocyst-stage embryos, which is in contrast to a previous report that showed SIRT 1–3 were expressed at a low level in blastocyst-stage embryos when analyzed by real-time PCR (Kwak et al., 2012).
We did not observe a significant change in any transcripts associated with histone acetyltransferase activity (HAT1 or CITED1), DNA methyltransferases (DNMT1 or DNMT3A), or Tet methylcytosine dioxygenases (TET1, TET2, and TET3). The pluripotency marker NANOG did have signifcantly lower expression in IVF embryos compared to IVV embryos, but there was no change in SCNT embryos as a result of HDACi. POU5F1 and CDX2 were expressed equally among treatments. Other groups have reported changes in gene expressoin of some of these transcripts as a result of in vitro culture or HDACi treatment as measured by real-time PCR. This observation may have not been found in transcriptome sequencing because the EdgeR analysis was quite stringent with a p value<0.05, FDR<0.05, and differerences had to be greater than 1.5-fold to be considered significant.
Lysosomes are membrane-bound organelles that contain acid hydrolases that break down cellular debris, including proteins, nucleic acids, carbohydrates, and lipids in a localized acidic environment. The lysosomal acid hydrolases include proteases (e.g., cathepsins, napsin, legumain, tripeptidyl pepetidase I), glycosidases (e.g., galactosidase, iduronidase, hexaminidase), sulfatases, lipases, and nucleases. The lysosome was the most upregulated KEGG pathway in SCNT embryos in response to HDACi compared to SCNT, as well as IVF compared to IVV, indicating both HDACi treatment and in vitro culture are having an effect. This upregulation included acid hydrolases (CTSK, CTSA, LGMN), glycosidaes (HEXA, HEXB), and a sphingomyelinase (SMPD1). Interestingly, IVV and untreated SCNT embryos had similar gene expression levels of lysosomal transcripts when compared to IVF and SCNT embyros treated with HDACi.
This suggests that HDACi treatment results in SCNT embryos that are more like IVF embryos than normal IVV embryos, and SCNT embryos that are not treated with HDACi have a gene expression pattern that is more similar to IVV embryos. It seems that embryos that are the result of fertilization and not nuclear reprogramming would have a similar transcriptional profile, but this does not seem to be the case. Our laboratory has reported multiple incidences of SCNT embryos being more similar to IVV embryos than to IVF embryos. When examining TRIM28 transcript expression, IVV and SCNT blastocyst-stage embryos had a similar expression level when compared to IVF (Hamm et al., 2014). Additionally, methylation profiles were also similar between IVV and untreated SCNT blastocysts when compared to IVF (Bonk et al., 2008). It should also be noted that gene expression as measured by real-time PCR detected an even higher expression of the lysosomal transcripts in the SCNT embryo treated with SAHA when compared to Scriptaid and ISAHA. It is unclear why these differences were observed as the calibrator transcript YWHAG was expressed equally among all treatments.
The lysosomes were identified in all of the treatment groups and compared at a constant exposure time. The IVV blastocyst-stage embryos had both low numbers of stained lysosomes and a low level of intensity when compared to the in vitro–cultured embryos. In a mouse study, lysosomal localization was examined in in vivo–ovulated and IVF and cultured preimplantation embryos and showed an increase in localization from the one-cell stage to the morula stage with a slight decrease by the blastocyst stage (Tsukamoto et al., 2013). The lysosomes of in vitro pig blastocysts from all of the treatments in this study had a similar staining pattern as the in vitro–cultured mouse blastocyst-stage embryos, but unfortunately there were no in vivo–cultured mouse embryos for comparison. Additionally, the changes in gene expression in the lysosomal transcripts did not correspond to a change in lysosomal intensity or localization pattern in the IVF or HDACi-treated SCNT blastocyst-stage embryos that could be measured by LysoTracker.
Embryos in all of the SCNT groups were derived from a female donor cell line, but the IVV and IVF embryos were pools of mixed-gender embryos. It has been shown that male and female bovine embryos can have different levels of gene expression of hydrolases and other proteolytic enzymes (Bermejo-Alvarez et al., 2010), but the differences observed as a result of HDACi treatment in this study were all from female embryos.
The cysteine protease cathepsins (CTS) and their inhibitors (CST) have also been shown to play an important role in tissue remodeling at the fetal maternal interface. Cathepsin B (CTSB) expression increases between days 25 and 30 of gestation in the chorionic epithelium in response to progesterone (Song et al., 2010). Cathepsin L1 (CTSL1) was localized to chorionic epithelia that form areolae, which absorb secretions from uterine glands. Message for the cysteine endopeptidase LGMN and its inhibitor CST6 localized with strong intensity in both the luminal epithelium (LE) and glandular epithelium (GE) as well as in the chorionic epithelium by day 30 of pregnancy in pigs (Shim et al., 2013). In SCNT pregnancies, both LGMN and CST6 were misregulated in the endometrium underlying the placenta compared to IVV pregnancies. These observations reflect the importance of proper acid hydrolase expression at the fetal maternal interface. Expression data from the Roslin Institute further validated the importance of lysosomal enzymes in day 50 placentas, which showed high expression of CTSA, CTSB, CTSD, LGMN, and HEXA. CTSK was also expressed, but at a lower level (Freeman et al., 2012).
Gene expression of blastocyst-stage embryos observed in this study illustrates that gene expression of lysosomal enzymes occurs aberrantly in both IVF and SCNT embryos treated with HDACi when compared to IVV or untreated SCNT embryos. A change at the protein level as measured by lysosome intensity and changes in CTSK localization was observed in all of the in vitro–cultured embryos. Increased expression of lysosomal transcripts illustrate that HDACi-treated and IVF embryos may have an increased need or ability to break down cellular debris and turn over protein. Previous work showed that poor-quality bovine embryos had an increased cathepsin B activity and addition of the cysteine peptidase inhibitor E-64 improved blastocyst rates and decreased apoptosis (Balboula et al., 2010). E-64 also improved development and reduced apoptosis of bovine SCNT embryos (Min et al., 2014). In this study, cathepsin B was upregulated in all of the in vitro–cultured embryos when compared to IVV, but not in response to HDACi.
In conclusion, treatment of reconstructed one-cell-stage pig embryos with HDACi, SAHA, or ISAHA can produce healthy offspring. Gene expression at the blastocyst stage between the three inhibitors Scriptaid, SAHA, and ISAHA was very similar as measured by transcriptome sequencing. The use of HDACi in SCNT embryos resulted in very few changes in gene expression of histone-related transcripts and pluripotency markers when compared to IVV, IVF, and untreated SCNT embryos. Interestingly, the greatest changes in gene expression occurred in lysosomal transcripts, thus indicating an increased ability or need for cellular turnover in these embryos.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the following people for their invaluable assistance with this project, including Mykel Anderson, Lonnie Dowell, Jason Dowell, Keith Giroux, Tricia Meyer, Jennifer Hamm, Dr. Jeffrey Whyte, Dr. Bethany Redel, Dr. Clifton Murphy, and Dr. Mike Linville. High-throughput sequencing services were performed at the University of Missouri DNA Core Facility with the help of Nathan Bivens and Karen Bromert.
This project was supported by the National Swine Resource and Research Center via funding from the National Institutes of Health (U42 OD011140 to RSP) and Food for the 21st Century at the University of Missouri.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Abeydeera L.R., Wang W.H., Prather R.S., and Day B.N. (1998). Maturation in vitro of pig oocytes in protein-free culture media: Fertilization and subsequent embryo development in vitro. Biol. Reprod. 58, 1316–1320 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., and Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Balboula A.Z., Yamanaka K., Sakatani M., Hegab A.O., Zaabel S.M., and Takahashi M. (2010). Intracellular cathepsin B activity is inversely correlated with the quality and developmental competence of bovine preimplantation embryos. Mol. Reprod. Dev. 77, 1031–1039 [DOI] [PubMed] [Google Scholar]
- Bauer B.K., Isom S.C., Spate L.D., Whitworth K.M., Spollen W.G., Blake S.M., Springer G.K., Murphy C.N., and Prather R.S. (2010). Transcriptional profiling by deep sequencing identifies differences in mRNA transcript abundance in in vivo-derived versus in vitro-cultured porcine blastocyst stage embryos. Biol. Reprod. 83, 791–798 [DOI] [PubMed] [Google Scholar]
- Bermejo-Alvarez P., Rizos D., Rath D., Lonergan P., and Gutierrez-Adan A. (2010). Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc. Natl. Acad. Sci. USA 107, 3394–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonk A.J., Li R., Lai L., Hao Y., Liu Z., Samuel M., Fergason E.A., Whitworth K.M., Murphy C.N., Antoniou E., and Prather R.S. (2008). Aberrant DNA methylation in porcine in vitro-, parthenogenetic-, and somatic cell nuclear transfer-produced blastocysts. Mol. Reprod. Dev. 75, 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., and Lempicki R.A. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- Ding X., Wang Y., Zhang D., Guo Z., and Zhang Y. (2008). Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology 70, 622–630 [DOI] [PubMed] [Google Scholar]
- Freeman T.C., Ivens A., Baillie J.K., Beraldi D., Barnett M.W., Dorward D., Downing A., Fairbairn L., Kapetanovic R., Raza S., Tomoiu A., Alberio R., Wu C., Su A.I., Summers K.M., Tuggle C.K., Archibald A.L., and Hume D.A. (2012). A gene expression atlas of the domestic pig. BMC Biol. 10, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J.A., Kim J.G., Whitworth K.M., Agca C., and Prather R.S. (2006). The use of microarrays to define functionally-related genes that are differentially expressed in the cycling pig uterus. Soc. Reprod. Fertil. Suppl. 62, 163–176 [PubMed] [Google Scholar]
- Hagen D.R., Prather R.S., Sims M.M., and First N.L. (1991). Development of one-cell porcine embryos to the blastocyst stage in simple media. J. Anim. Sci. 69, 1147–1150 [DOI] [PubMed] [Google Scholar]
- Hamm J., Tessanne K., Murphy C.N., and Prather R.S. (2014). Transcriptional regulators TRIM28, SETDB1, and TP53 are aberrantly expressed in porcine embryos produced by in vitro fertilization in comparison to in vivo- and somatic-cell nuclear transfer-derived embryos. Mol. Reprod. Dev. 81, 552–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., and Madan A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Tang X., Xie W., Zhou Y., Li D., Yao C., Zhou Y., Zhu J., Lai L., Ouyang H., and Pang D. (2011). Histone deacetylase inhibitor significantly improved the cloning efficiency of porcine somatic cell nuclear transfer embryos. Cell. Reprogram. 13, 513–520 [DOI] [PubMed] [Google Scholar]
- Isom S.C., Spollen W.G., Blake S.M., Bauer B.K., Springer G.K., and Prather R.S. (2010). Transcriptional profiling of day 12 porcine embryonic disc and trophectoderm samples using ultra-deep sequencing technologies. Mol. Reprod. Dev. 77, 812–819 [DOI] [PubMed] [Google Scholar]
- Jensen T.W., Mazur M.J., Pettigew J.E., Perez-Mendoza V.G., Zachary J., and Schook L.B. (2010). A cloned pig model for examining atherosclerosis induced by high fat, high cholesterol diets. Anim. Biotechnol. 21, 179–187 [DOI] [PubMed] [Google Scholar]
- Kishigami S., Bui H.T., Wakayama S., Tokunaga K., Van Thuan N., Hikichi T., Mizutani E., Ohta H., Suetsugu R., Sata T., and Wakayama T. (2007). Successful mouse cloning of an outbred strain by trichostatin A treatment after somatic nuclear transfer. J. Reprod. Dev. 53, 165–170 [DOI] [PubMed] [Google Scholar]
- Kolber-Simonds D., Lai L., Watt S.R., Denaro M., Arn S., Augenstein M.L., Betthauser J., Carter D.B., Greenstein J.L., Hao Y., Im G.S., Liu Z., Mell G.D., Murphy C.N., Park K.W., Rieke A., Ryan D.J., Sachs D.H., Forsberg E.J., Prather R.S., and Hawley R.J. (2004). Production of alpha-1,3-galactosyltransferase null pigs by means of nuclear transfer with fibroblasts bearing loss of heterozygosity mutations. Proc. Natl. Acad. Sci. USA 101, 7335–7340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S.S., Cheong S.A., Yoon J.D., Jeon Y., and Hyun S.H. (2012). Expression patterns of sirtuin genes in porcine preimplantation embryos and effects of sirtuin inhibitors on in vitro embryonic development after parthenogenetic activation and in vitro fertilization. Theriogenology 78, 1597–1610 [DOI] [PubMed] [Google Scholar]
- Lai L., and Prather R.S. (2003). Creating genetically modified pigs by using nuclear transfer. Reprod Biol Endocrinol 1, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L., Kolber-Simonds D., Park K.W., Cheong H.T., Greenstein J.L., Im G.S., Samuel M., Bonk A., Rieke A., Day B.N., Murphy C.N., Carter D.B., Hawley R.J., and Prather R.S. (2002). Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295, 1089–1092 [DOI] [PubMed] [Google Scholar]
- Lee K., Hamm J., Whitworth K., Spate L., Park K.W., Murphy C.N., and Prather R.S. (2014). Dynamics of TET family expression in porcine preimplantation embryos is related to zygotic genome activation and required for the maintenance of NANOG. Dev. Biol. 386, 86–95 [DOI] [PubMed] [Google Scholar]
- Li X., Kato Y., Tsuji Y., and Tsunoda Y. (2008). The effects of trichostatin A on mRNA expression of chromatin structure-, DNA methylation-, and development-related genes in cloned mouse blastocysts. Cloning Stem Cells 10, 133–142 [DOI] [PubMed] [Google Scholar]
- Lillico S.G., Proudfoot C., Carlson D.F., Stverakova D., Neil C., Blain C., King T.J., Ritchie W.A., Tan W., Mileham A.J., McLaren D.G., Fahrenkrug S.C., and Whitelaw C.B. (2013). Live pigs produced from genome edited zygotes. Sci. Rep. 3, 2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu Y., Gao F., Song G., Wen J., Guan J., Yin Y., Ma X., Tang B., and Li Z. (2012). Embryonic development and gene expression of porcine SCNT embryos treated with sodium butyrate. J. Exp. Zool. B Mol. Dev. Evol. 318, 224–234 [DOI] [PubMed] [Google Scholar]
- Mao J., Zhao M.T., Whitworth K.M., Spate L.D., Walters E.M., O'Gorman C., Lee K., Samuel M.S., Murphy C.N., Wells K., Rivera R.M., and Prather R.S. (2015). Oxamflatin treatment enhances cloned porcine embryo development and nuclear reprogramming. Cell. Reprogram. 17, 28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.H., Song B.S., Yeon J.Y., Kim J.W., Bae J.H., Park S.Y., Lee Y.H., Chang K.T., and Koo D.B. (2014). A cathepsin B inhibitor, E-64, improves the preimplantation development of bovine somatic cell nuclear transfer embryos. J. Reprod. Dev. 60, 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Li C., Mizutani E., Terashita Y., Yamagata K., and Wakayama T. (2010). Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol. Reprod. 83, 929–937 [DOI] [PubMed] [Google Scholar]
- Park S.J., Park H.J., Koo O.J., Choi W.J., Moon J.H., Kwon D.K., Kang J.T., Kim S., Choi J.Y., Jang G., and Lee B.C. (2012). Oxamflatin improves developmental competence of porcine somatic cell nuclear transfer embryos. Cell. Reprogram. 14, 398–406 [DOI] [PubMed] [Google Scholar]
- Prather R.S., Rowland R.R., Ewen C., Trible B., Kerrigan M., Bawa B., Teson J.M., Mao J., Lee K., Samuel M.S., Whitworth K.M., Murphy C.N., Egen T., and Green J.A. (2013). An intact sialoadhesin (Sn/SIGLEC1/CD169) is not required for attachment/internalization of the porcine reproductive and respiratory syndrome virus. J. Virol. 87, 9538–9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner S., Fehlings C., Herbach N., Hofmann A., von Waldthausen D.C., Kessler B., Ulrichs K., Chodnevskaja I., Moskalenko V., Amselgruber W., Goke B., Pfeifer A., Wanke R., and Wolf E. (2010). Glucose intolerance and reduced proliferation of pancreatic beta-cells in transgenic pigs with impaired glucose-dependent insulinotropic polypeptide function. Diabetes 59, 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C.S., Hao Y., Rokhlina T., Samuel M., Stoltz D.A., Li Y., Petroff E., Vermeer D.W., Kabel A.C., Yan Z., Spate L., Wax D., Murphy C.N., Rieke A., Whitworth K., Linville M.L., Korte S.W., Engelhardt J.F., Welsh M.J., and Prather R.S. (2008). Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer. J. Clin. Invest. 118, 1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J.W., Fernandez de Castro J.P., Zhao J., Samuel M., Walters E., Rios C., Bray-Ward P., Jones B.W., Marc R.E., Wang W., Zhou L., Noel J.M., McCall M.A., DeMarco P.J., Prather R.S., and Kaplan H.J. (2012). Generation of an inbred miniature pig model of retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 53, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J., Seo H., Choi Y., Yoo I., Lee C.K., Hyun S.H., Lee E., and Ka H. (2013). Analysis of legumain and cystatin 6 expression at the maternal-fetal interface in pigs. Mol. Reprod. Dev. 80, 570–580 [DOI] [PubMed] [Google Scholar]
- Song G., Bailey D.W., Dunlap K.A., Burghardt R.C., Spencer T.E., Bazer F.W., and Johnson G.A. (2010). Cathepsin B, cathepsin L, and cystatin C in the porcine uterus and placenta: Potential roles in endometrial/placental remodeling and in fluid-phase transport of proteins secreted by uterine epithelia across placental areolae. Biol. Reprod. 82, 854–864 [DOI] [PubMed] [Google Scholar]
- Song Y., Hai T., Wang Y., Guo R., Li W., Wang L., and Zhou Q. (2014). Epigenetic reprogramming, gene expression and in vitro development of porcine SCNT embryos are significantly improved by a histone deacetylase inhibitor–m-carboxycinnamic acid bishydroxamide (CBHA). Protein Cell 5, 382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S., Hara T., Yamamoto A., Ohta Y., Wada A., Ishida Y., Kito S., Nishikawa T., Minami N., Sato K., and Kokubo T. (2013). Functional analysis of lysosomes during mouse preimplantation embryo development. J. Reprod. Dev. 59, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Thuan N., Bui H.T., Kim J.H., Hikichi T., Wakayama S., Kishigami S., Mizutani E., and Wakayama T. (2009). The histone deacetylase inhibitor scriptaid enhances nascent mRNA production and rescues full-term development in cloned inbred mice. Reproduction 138, 309–317 [DOI] [PubMed] [Google Scholar]
- Whitworth K., Springer G.K., Forrester L.J., Spollen W.G., Ries J., Lamberson W.R., Bivens N., Murphy C.N., Mathialagan N., Green J.A., and Prather R.S. (2004). Developmental expression of 2489 gene clusters during pig embryogenesis: An expressed sequence tag project. Biol. Reprod. 71, 1230–1243 [DOI] [PubMed] [Google Scholar]
- Whitworth K.M., and Prather R.S. (2010). Somatic cell nuclear transfer efficiency: How can it be improved through nuclear remodeling and reprogramming? Mol. Reprod. Dev. 77, 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth K.M., Agca C., Kim J.G., Patel R.V., Springer G.K., Bivens N.J., Forrester L.J., Mathialagan N., Green J.A., and Prather R.S. (2005). Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol. Reprod. 72, 1437–1451 [DOI] [PubMed] [Google Scholar]
- Whitworth K.M., Li R., Spate L.D., Wax D.M., Rieke A., Whyte J.J., Manandhar G., Sutovsky M., Green J.A., Sutovsky P., and Prather R.S. (2009). Method of oocyte activation affects cloning efficiency in pigs. Mol. Reprod. Dev. 76, 490–500 [DOI] [PubMed] [Google Scholar]
- Whitworth K.M., Zhao J., Spate L.D., Li R., and Prather R.S. (2011). Scriptaid corrects gene expression of a few aberrantly reprogrammed transcripts in nuclear transfer pig blastocyst stage embryos. Cell. Reprogram. 13, 191–204 [DOI] [PubMed] [Google Scholar]
- Whitworth K.M., Lee K., Benne J.A., Beaton B.P., Spate L.D., Murphy S.L., Samuel M.S., Mao J., O'Gorman C., Walters E.M., Murphy C.N., Driver J., Mileham A., McLaren D., Wells K.D., and Prather R.S. (2014). Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol. Reprod. 91, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Suzuki C., Tanaka A., Anas I.M., and Iwamura S. (2002). Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol. Reprod. 66, 112–119 [DOI] [PubMed] [Google Scholar]
- Zeyland J., Lipinski D., and Slomski R. (2014). The current state of xenotransplantation. J. Appl. Genet. 56, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Ross J.W., Hao Y., Spate L.D., Walters E.M., Samuel M.S., Rieke A., Murphy C.N., and Prather R.S. (2009). Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol. Reprod. 81, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hao Y., Ross J.W., Spate L.D., Walters E.M., Samuel M.S., Rieke A., Murphy C.N., and Prather R.S. (2010). Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell. Reprogram. 12, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.