Abstract
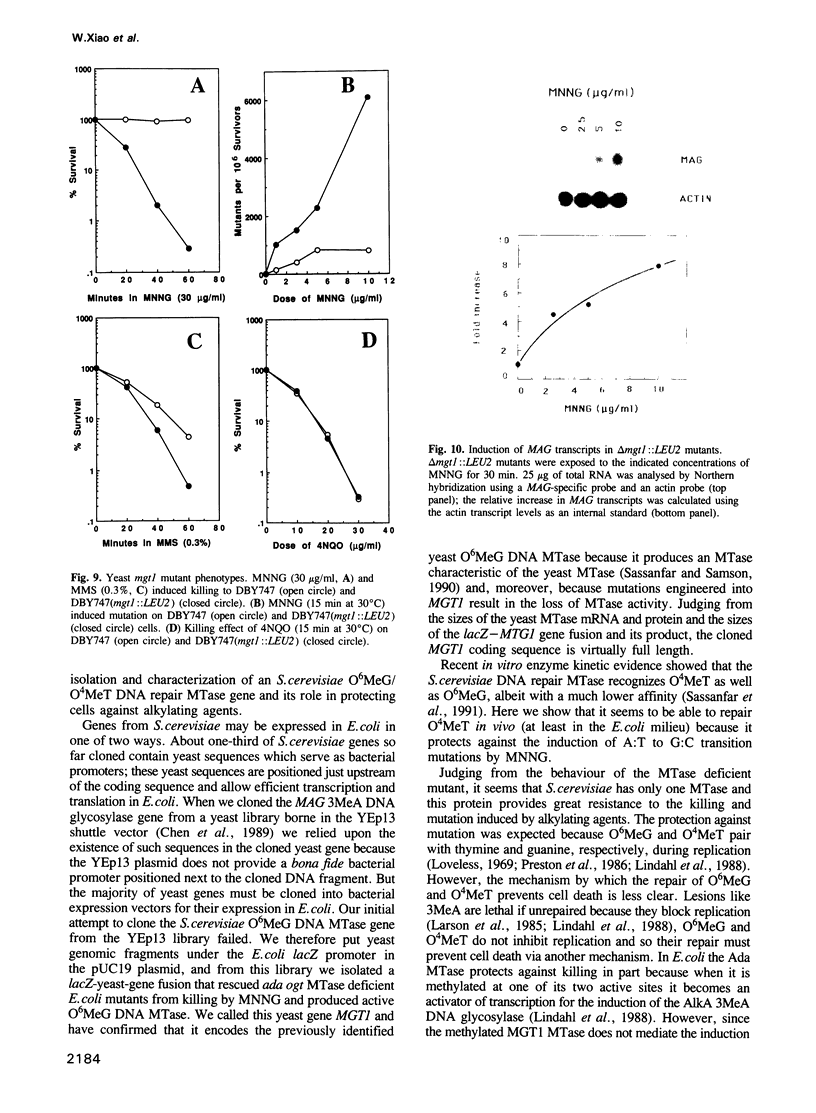
We previously identified and characterized biochemically an O6-methylguanine (O6MeG) DNA repair methyltransferase (MTase) in the yeast Saccharomyces cerevisiae and showed that it recognizes both O6MeG and O4-methylthymine (O4MeT) in vitro. Here we characterize the cloned S. cerevisiae O6MeG DNA MTase gene (MGT1) and determine its in vivo role in protecting yeast from DNA alkylation damage. We isolated a yeast DNA fragment that suppressed alkylation-induced killing and mutation in Escherichia coli ada ogt MTase deficient mutants and produced in these cells a protein similar to the yeast MTase. The cloned yeast fragment was mapped to chromosome IV and DNA sequencing identified an open reading frame, designated MGT1, which encodes a 188 amino acid protein with a molecular weight of 21,500 daltons. An 88 amino acid stretch of the MGT1 protein displays remarkable homology with four bacterial MTases and the human DNA MTase. S.cerevisiae mutants bearing an insertion in the MGT1 gene lacked DNA MTase activity and were very sensitive to alkylation induced killing and mutation. MGT1 transcript levels are not increased in response to DNA alkylation damage, nor is the MGT1 MTase involved in the regulation of the yeast 3-methyladenine DNA glycosylase gene (MAG). Expression of the MGT1 gene in E.coli prevented the induction by alkylating agents of both G:C to A:T and A:T to G:C transition mutations indicating that this eukaryotic MTase repairs both O6MeG and O4MeT in vivo.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Beranek D. T., Weis C. C., Swenson D. H. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980 Jul;1(7):595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- Berdal K. G., Bjørås M., Bjelland S., Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990 Dec;9(13):4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignami M., Lane D. P. O6-methylguanine in the SV40 origin of replication inhibits binding but increases unwinding by viral large T antigen. Nucleic Acids Res. 1990 Jul 11;18(13):3785–3793. doi: 10.1093/nar/18.13.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples C. G., Miller J. H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Papathanasiou M. A., Hollander M. C., Yarosh D. B. Expression of the O6-methylguanine-DNA methyltransferase gene MGMT in MER+ and MER- human tumor cells. Cancer Res. 1990 Dec 15;50(24):7908–7911. [PubMed] [Google Scholar]
- Friedberg E. C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1988 Mar;52(1):70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosina G., Abbondandolo A. The current evidence for an adaptive response to alkylating agents in mammalian cells, with special reference to experiments with in vitro cell cultures. Mutat Res. 1985 Sep;154(2):85–100. doi: 10.1016/0165-1110(85)90021-1. [DOI] [PubMed] [Google Scholar]
- Goth-Goldstein R., Johnson P. L. Repair of alkylation damage in Saccharomyces cerevisiae. Mol Gen Genet. 1990 May;221(3):353–357. doi: 10.1007/BF00259399. [DOI] [PubMed] [Google Scholar]
- Green D. A., Deutsch W. A. Repair of alkylated DNA: Drosophila have DNA methyltransferases but not DNA glycosylases. Mol Gen Genet. 1983;192(3):322–325. doi: 10.1007/BF00392169. [DOI] [PubMed] [Google Scholar]
- Hadden C. T., Foote R. S., Mitra S. Adaptive response of Bacillus subtilis to N-methyl-N'-nitro-N-nitrosoguanidine. J Bacteriol. 1983 Feb;153(2):756–762. doi: 10.1128/jb.153.2.756-762.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Brésil H., Serres M., Martel-Planche G., Wild C. P., Montesano R. Modulation of O6-methylguanine-DNA methyltransferase in rat and hamster liver after treatment with dimethylnitrosamine. Cancer Res. 1990 Sep 1;50(17):5426–5430. [PubMed] [Google Scholar]
- Harris A. L., Karran P., Lindahl T. O6-Methylguanine-DNA methyltransferase of human lymphoid cells: structural and kinetic properties and absence in repair-deficient cells. Cancer Res. 1983 Jul;43(7):3247–3252. [PubMed] [Google Scholar]
- Hayakawa H., Koike G., Sekiguchi M. Expression and cloning of complementary DNA for a human enzyme that repairs O6-methylguanine in DNA. J Mol Biol. 1990 Jun 20;213(4):739–747. doi: 10.1016/S0022-2836(05)80260-8. [DOI] [PubMed] [Google Scholar]
- Huisman O., Raymond W., Froehlich K. U., Errada P., Kleckner N., Botstein D., Hoyt M. A. A Tn10-lacZ-kanR-URA3 gene fusion transposon for insertion mutagenesis and fusion analysis of yeast and bacterial genes. Genetics. 1987 Jun;116(2):191–199. doi: 10.1093/genetics/116.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike G., Maki H., Takeya H., Hayakawa H., Sekiguchi M. Purification, structure, and biochemical properties of human O6-methylguanine-DNA methyltransferase. J Biol Chem. 1990 Sep 5;265(25):14754–14762. [PubMed] [Google Scholar]
- Larson K., Sahm J., Shenkar R., Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985 Jun-Jul;150(1-2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- Laval F. Induction of proteins involved in the repair of alkylated bases in mammalian cells by DNA-damaging agents. Mutat Res. 1990 Nov-Dec;233(1-2):211–218. doi: 10.1016/0027-5107(90)90164-y. [DOI] [PubMed] [Google Scholar]
- Laval F., Laval J. Adaptive response in mammalian cells: crossreactivity of different pretreatments on cytotoxicity as contrasted to mutagenicity. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1062–1066. doi: 10.1073/pnas.81.4.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Loveless A. Possible relevance of O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969 Jul 12;223(5202):206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- Maga J. A., McEntee K. Response of S. cerevisiae to N-methyl-N'-nitro-N-nitrosoguanidine: mutagenesis, survival and DDR gene expression. Mol Gen Genet. 1985;200(2):313–321. doi: 10.1007/BF00425442. [DOI] [PubMed] [Google Scholar]
- McKnight G. L., McConaughy B. L. Selection of functional cDNAs by complementation in yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4412–4416. doi: 10.1073/pnas.80.14.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano R., Brésil H., Margison G. P. Increased excision of O6-methylguanine from rat liver DNA after chronic administration of dimethylnitrosamine. Cancer Res. 1979 May;39(5):1798–1802. [PubMed] [Google Scholar]
- Montesano R., Brésil H., Planche-Martel G., Margison G. P., Pegg A. E. Effect of chronic treatment of rats with dimethylnitrosamine on the removal of O6-methylguanine from DNA. Cancer Res. 1980 Feb;40(2):452–458. [PubMed] [Google Scholar]
- Morohoshi F., Hayashi K., Munakata N. Bacillus subtilis ada operon encodes two DNA alkyltransferases. Nucleic Acids Res. 1990 Sep 25;18(18):5473–5480. doi: 10.1093/nar/18.18.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Hayashi K., Munakata N. Bacillus subtilis gene coding for constitutive O6-methylguanine-DNA alkyltransferase. Nucleic Acids Res. 1989 Aug 25;17(16):6531–6543. doi: 10.1093/nar/17.16.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuru Y., Nemoto N., Nakagawa K., Masahito P., Ishikawa T. O6-methylguanine DNA methyltransferase activity in liver from various fish species. Carcinogenesis. 1987 Aug;8(8):1123–1127. doi: 10.1093/carcin/8.8.1123. [DOI] [PubMed] [Google Scholar]
- O'Connor T. R., Laval F. Isolation and structure of a cDNA expressing a mammalian 3-methyladenine-DNA glycosylase. EMBO J. 1990 Oct;9(10):3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A. E., Dolan M. E. Properties and assay of mammalian O6-alkylguanine-DNA alkyltransferase. Pharmacol Ther. 1987;34(2):167–179. doi: 10.1016/0163-7258(87)90010-6. [DOI] [PubMed] [Google Scholar]
- Potter P. M., Wilkinson M. C., Fitton J., Carr F. J., Brennand J., Cooper D. P., Margison G. P. Characterisation and nucleotide sequence of ogt, the O6-alkylguanine-DNA-alkyltransferase gene of E. coli. Nucleic Acids Res. 1987 Nov 25;15(22):9177–9193. doi: 10.1093/nar/15.22.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston B. D., Singer B., Loeb L. A. Mutagenic potential of O4-methylthymine in vivo determined by an enzymatic approach to site-specific mutagenesis. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck G. W., Coons S., Carroll P., Samson L. A second DNA methyltransferase repair enzyme in Escherichia coli. Proc Natl Acad Sci U S A. 1988 May;85(9):3039–3043. doi: 10.1073/pnas.85.9.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck G. W., Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991 Mar;173(6):2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck G. W., Smith C. M., Goad D. L., Samson L. Characterization of the major DNA repair methyltransferase activity in unadapted Escherichia coli and identification of a similar activity in Salmonella typhimurium. J Bacteriol. 1989 Sep;171(9):4563–4568. doi: 10.1128/jb.171.9.4563-4568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B., Spurr N., Karran P. cDNA cloning and chromosomal assignment of the human O6-methylguanine-DNA methyltransferase. cDNA expression in Escherichia coli and gene expression in human cells. J Biol Chem. 1990 Jun 5;265(16):9563–9569. [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Samson L., Thomale J., Rajewsky M. F. Alternative pathways for the in vivo repair of O6-alkylguanine and O4-alkylthymine in Escherichia coli: the adaptive response and nucleotide excision repair. EMBO J. 1988 Jul;7(7):2261–2267. doi: 10.1002/j.1460-2075.1988.tb03066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Dosanjh M. K., Essigmann J. M., Samson L. Relative efficiencies of the bacterial, yeast, and human DNA methyltransferases for the repair of O6-methylguanine and O4-methylthymine. Suggestive evidence for O4-methylthymine repair by eukaryotic methyltransferases. J Biol Chem. 1991 Feb 15;266(5):2767–2771. [PubMed] [Google Scholar]
- Sassanfar M., Samson L. Identification and preliminary characterization of an O6-methylguanine DNA repair methyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990 Jan 5;265(1):20–25. [PubMed] [Google Scholar]
- Shevell D. E., Abou-Zamzam A. M., Demple B., Walker G. C. Construction of an Escherichia coli K-12 ada deletion by gene replacement in a recD strain reveals a second methyltransferase that repairs alkylated DNA. J Bacteriol. 1988 Jul;170(7):3294–3296. doi: 10.1128/jb.170.7.3294-3296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K., Shiota S., Collier J., Foote R. S., Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci U S A. 1990 Jan;87(2):686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Vaughan P., Sedgwick B., Hall J., Gannon J., Lindahl T. Environmental mutagens that induce the adaptive response to alkylating agents in Escherichia coli. Carcinogenesis. 1991 Feb;12(2):263–268. doi: 10.1093/carcin/12.2.263. [DOI] [PubMed] [Google Scholar]
- Waldstein E. A., Cao E. H., Setlow R. B. Adaptive resynthesis of O6-methylguanine-accepting protein can explain the differences between mammalian cells proficient and deficient in methyl excision repair. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5117–5121. doi: 10.1073/pnas.79.17.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- von Hofe E., Kennedy A. R. In vitro induction of O6-methylguanine-DNA methyltransferase in C3H/10T1/2 cells by X-rays is inhibited by nitrogen. Carcinogenesis. 1988 Apr;9(4):679–681. doi: 10.1093/carcin/9.4.679. [DOI] [PubMed] [Google Scholar]
- von Wronski M. A., Shiota S., Tano K., Mitra S., Bigner D. D., Brent T. P. Structural and immunological comparison of indigenous human O6-methylguanine-DNA methyltransferase with that encoded by a cloned cDNA. J Biol Chem. 1991 Jan 15;266(2):1064–1070. [PubMed] [Google Scholar]