Abstract
Epigenetic regulation of gene expression is a central mechanism that governs cell stemness, determination, commitment, and differentiation. It has been recently found that PHF8, a major H4K20/H3K9 demethylase, plays a critical role in craniofacial and bone development. In this study, we hypothesize that PHF8 promotes osteoblastogenesis by epigenetically regulating the expression of a nuclear matrix protein, special AT-rich sequence-binding protein 2 (SATB2) that plays pivotal roles in skeletal patterning and osteoblast differentiation. Our results showed that expression levels of PHF8 and SATB2 in preosteoblasts and bone marrow stromal cells (BMSCs) increased simultaneously during osteogenic induction. Overexpressing PHF8 in these cells upregulated the expression of SATB2, Runx2, osterix, and bone matrix proteins. Conversely, knockdown of PHF8 reduced the expression of these genes. Furthermore, ChIP assays confirmed that PHF8 specifically bound to the transcription start site (TSS) of the SATB2 promoter, and the expression of H3K9me1 at the TSS region of SATB2 decreased in PHF8 overexpressed group. Implantation of the BMSCs overexpressing PHF8 with silk protein scaffolds promoted bone regeneration in critical-sized defects in mouse calvaria. Taken together, our results demonstrated that PHF8 epigenetically modulates SATB2 activity, triggering BMSCs osteogenic differentiation and facilitating bone formation and regeneration in biodegradable silk scaffolds.
Introduction
Over 500,000 surgeries correcting bone deformities and critical size defects occur each year, yet 50% of typical graft procedures fail.1,2 Consequently, the repair of massive bony defects remains challenging in the clinical setting. Tissue engineering provides a new method for bone regeneration. It has three parts: the source of seed cells, suitable scaffold material, and effective cytokines. The ideal scaffold should have good biodegradability, distinguishing mechanical properties, and low inflammatory response. As a natural material, silk scaffold (SS) has good histocompatibility and excellent slow-release function because of its special porous mesh membrane structure. Now, SS, which was used for bone tissue engineering scaffold, has been in the stage of clinical trials. Kim et al.3 used silk nanofiber membrane as scaffold to repair cranial defects and found completely new bone repair after 12 weeks and exhibited no inflammatory response. Our lab also used SSs to repair mouse skull defects and achieved good results.4–6 Stem cells play pivotal roles in bone tissue regeneration and bone wound repairing. We have previously explored the effects of bone marrow stromal cells (BMSCs) and induced pluripotent stem cells regulated by the osteogenic transcription factors in osteogenetic differentiation and bone regeneration.5–9 The differentiation of stem cells toward specific functional cell types is the process used to establish specific gene expression patterns, which is a result of elaborated control of activation/silencing of large numbers of genes. Initial regulation in coordinating the expression of osteogenic genes and orchestrating molecular mechanisms in osteoblastogenesis exists and epigenetic regulation of gene expression may serve this role.
Epigenetic modification does not involve changes in DNA sequence, but it causes alterations in DNA methylation and acetylation pattern that modifies the local chromatin structure and thereby serves to regulate expression of specific genes. Histone methylation played important roles in cellular proliferation and differentiation.10,11 PHF8 was a histone demethylase associated with X-linked mental retardation. PHF8 bound through its PHD domain to H3K4me3 nucleosomes and demethylated H3K9, H3K27, and H4K20 at the transcription start site (TSS) regions of active promoters12,13 and then regulated gene transcription.
PHF8 has been shown to be involved in various biological processes. It has been confirmed that PHF8 regulates many cell cycle genes14 and controls the expression of genes associated with cell adhesion and cytoskeleton organization such as RhoA, Rac1, and GSK3β.15 PHF8 was also shown to govern retinoic acid response in acute promyelocytic leukemia16 and affect cell migration and invasion in cancer.17 Besides, PHF8 regulates rRNA synthesis via its histone H3K9me1/2 demethylase activity.18,19
It has been recently found that PHF8 played a critical role in craniofacial and bone development.20 Using a zebrafish model Qi et al. found that PHF8 was mostly expressed in the head and jaw regions. Injection of a zPHF8 morpholino caused abnormalities in craniofacial organs and wild-type (but not catalytically inactive) zPHF8 showed significant rescue of craniofacial defects induced by zPHF8 morpholino. These important findings identified a critical role of zPHF8 in craniofacial development.
Special AT-rich sequence-binding protein 2 (SATB2) is a DNA-binding protein that regulates chromatin organization and gene expression.21 Similar to PHF8, SATB2 is also expressed in branchial arches and osteoblast-lineage cells. SATB2−/− mice exhibited defects in osteoblast differentiation and function, which consequently delayed bone formation and mineralization. SATB2−/− embryos showed multiple craniofacial defects including a significant truncation of the mandible and a cleft palate.22 Our previous studies demonstrated that SATB2 enhanced expressions of bone matrix proteins and osteogenic transcription factors in BMSCs and dental follicle cells, and played pivotal roles in bone regeneration, suggesting that SATB2 can be used as an osteogenic transcription factor to overcome hurdles in craniofacial regeneration.9 However, the profiles of the epigenetic regulation of the SATB2 expression during osteogenic differentiation of BMSCs, particularly in oral and craniofacial development are still largely unknown.
The studies described above indicate the similarities in gene expression and function between PHF8 and SATB2. In this study, we characterized the epigenetic regulation of PHF8 on SATB2 in BMSCs and the role of PHF8 in osteoblast differentiation and calvarial bone regeneration in mouse calvarial defects filled with BMSCs packed in SSs.
Materials and Methods
Cell culture
MC3T3-E1 murine preosteoblast cells were maintained in alpha minimum essential medium (α-MEM) with 10% fetal bovine serum and antibiotics. The MC3T3-E1 cells were then cultured in medium containing 50 mg/mL ascorbic acid (Sigma) and 5 mM β-glycerophosphate (Sigma) to induce osteogenic differentiation. BMSCs were obtained from the femurs and tibias of 4-week-old mice, were cultured in DMEM supplemented with 20% fetal bovine serum and antibiotics in cell culture dish with a diameter of 60 mm (Becton, Dickinson and Company) until they reached 80–90% confluence, and then passaged and maintained at α-MEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin as we described previously.23 To induce osteogenic differentiation of BMSCs, cells were switched to osteogenic medium containing 50 mg/mL ascorbic acid (Sigma), 10 nM dexamethasone (Sigma), and 5 mM β-glycerophosphate (Sigma) for 1, 3, 7, 10, 14, and 21 days, respectively.
Preparation of retroviral vectors and cell infection
The plasmids pBABE-control shRNA, pBABE-PHF8 shRNA, and pOZ-Flag-HA-PHF8 (mouse PHF8 cDNA) were a generous gift from Dr. Yang Shi's laboratory (Department of Pathology, Harvard Medical School). The pOZ-Flag-HA-PHF8 and packaging vectors pOZ-ENV and pOZ-Gagpol were co-transfected into HEK-293T cells using lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the supernatant filtered through a 0.45 mm filter (Millipore) was used to infect targeted cells with polybrene at a final concentration of 8 μg/mL. The empty retroviral vector pOZ-Flag-HA was also packaged and used as a control. pBABE-PHF8 shRNA and packing vector pCG-Gagpol and pCG-VSVG were co-transfected into HEK-293T cells to construct the viruses expressing PHF8 shRNA. Quantitative reverse transcription polymerase chain reaction (RT-PCR) assay was used for quantification of both genomic viral RNA after production and of targeted gene transcripts following transduction.
Cytoimmunofluorescence test
The glass coverslips were put in 24-well plate and the MC3T3-E1 cells were seeded in 24-well plate. The following day, cells were fixed with 4% paraformaldehyde, followed by permeabilization with 0.05% Triton X-100 (Sigma-Aldrich). Then, cells were blocked for 30 min with 10% normal donkey serum and incubated with PHF8 primary antibody (Abacm) overnight at 4°C. After three washes of 5 min each in phosphate-buffered saline (PBS), the cells were incubated in conjugated secondary antibody (Alexa 488 green; Invitrogen) and then mounted with ProLong® Gold Antifade Reagent with DAPI (Invitrogen). Images were collected using an Axiovert 405 M epifluorescence inverted light microscope (Lumen200; Prior Scientific, Inc.) and an Olympus DP 73 digital camera.
Real-time RT-PCR for mRNA analysis
Real-time RT-PCR for mRNA analysis was performed using SYBR Green Mastermix (Affymetrix) on a Bio-Rad iQ5 thermal cycler (Bio-Rad Laboratories). The relative expression level of the housekeeping gene GAPDH was used to normalize gene expression in each sample in different groups. The sequences of the primer for amplification of mouse ALP, BSP, OCN, Runx2, osterix, SATB2, PHF8, and GAPDH were shown in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/tea).
Western blot analysis
MC3T3-E1 and BMSCs were infected with viruses that express PHF8, empty vector, PHF8 shRNA, and control shRNA, respectively. Whole protein lysates were prepared as described previously.24 Antibodies for PHF8 (Abcam), SATB2, OSX, and Runx2 were obtained from Santa Cruz Biotechnologies. The second antibody was HRP-labeled goat anti-rabbit IgG (Santa Cruz). The proteins were visualized using electrogenerated chemiluminescence reagents from Pierce Biotechnology. Image J was used to qualify the protein expression.
ChIP analysis
The MC3T3-E1 cells and BMSCs from a 150 cm plate were cross-linked with 10% formaldehyde and quenched with 0.125 M glycine. Cells were washed with cold PBS and then lysed with cell lysis buffer complemented with protease inhibitor Cocktail II (Sigma). DNA fragmentation was performed with Vibra-Cell™ sonicator using 500 μL nuclear lysis buffer lysate with 20 cycles of 15 s ON and 45 s OFF. In qChIP, 100 μg of chromatin were incubated with 2 μg of PHF8 antibody (Abcam) or 2 μg of the H3K9me1 antibody (Abcam) at 4°C overnight. Twenty microliters of Protein A or G beads was added to each tube for 2 h at 4°C and the complex were then washed with low salt buffer, high salt buffer, LiCl buffer, and TE buffer each for 5 min. The immunoprecipitants were de-cross-linked at 62°C overnight. The immunoprecipitated DNA was dissolved in 30 μL H2O. One microliter of DNA was used for real-time polymerase chain reaction (PCR). The primers for qChIP were shown in Supplementary Table S2.
SS preparation and cell seeding
The silk fibroin scaffolds (disk-shaped, 4 mm diameter and 2 mm thick) were prepared as described previously.5,25 For cell seeding, cells were released from the culture substratum and concentrated to 2×107 cells/mL in serum-free medium. Then, BMSCs were seeded into the SSs by pipetting 0.5 mL of the cell suspension onto the materials. The BMSCs/SS construct was incubated overnight to allow cell attachment in vitro before implantation.
Animal surgery
The animal protocols used in this study were approved by the Institutional Animal Care and Use Committee at Tufts University/Tufts Medical Center (Approved Protocol #B2011-49). All mice were kept in a controlled temperature-and controlled room under a 12 h light, 12 h dark cycle.
Eight-week-old C57BL/6J mice were anesthetized, and a 4-mm-diameter calvarial critical-sized defect was created on each side of the calvarial bone using a dental bur attached to a slow-speed hand piece with minimal invasion of the Dura mater. The critical-sized defects in mice were randomly divided into six groups to receive the following implants (n=5 per group): (1) SS alone; (2) SS with BMSCs; (3) SS with PHF8-modified BMSCs; (4) SS with empty vector-modified BMSCs; (5) SS with PHF8 shRNA-modified BMSCs; (6) SS with control shRNA-modified BMSCs.
Quantitative real time reverse transcription polymerase chain reaction of newly formed tissue
The mice were sacrificed 5 weeks after surgery and the newly formed bone samples were frozen immediately in liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen) from the newly formed tissues and used for cDNA synthesis. Real-time PCR was used to detect the mRNA expressions of ALP, BSP, OCN, SATB2, Runx2, OSX, and PHF8.
Micro CT measurement
Five weeks after surgery, the morphology of the reconstructed cranium was assessed using a micro-CT system (μCT-40; Scanco Medical). The CT settings were used as follows: pixel matrix, 1024×1024; slice thickness, 20 mm. After scanning, the micro-CT images were segmented using a nominal threshold value of 300 as reported previously5 and a three dimensional histomorphometric analysis was performed automatically. The parameters of bone volume were used for comparison in this study.
Histomorphometric analysis and immunohistochemical staining
For histological examination, cranial specimens were fixed with 10% formalin and then decalcified with 10% ethylenediaminetetraacetic acid (pH 7.0) for 2 weeks. Samples were then dehydrated in ethanol and embedded in paraffin wax. Five-millimeter sections were cut and mounted on glass slides. Three randomly selected sections from each sample were stained with hematoxylin and eosin (H&E). H&E staining was performed using standard methods as previously described.26 The expressions of OCN were detected using immunohistochemical staining followed by counterstaining in hematoxylin with a Histostain SP kit (Invitrogen). H&E staining and immunostaining sections were photographed with an OLYMPUS BX53 microscope. Newly formed bone in H&E-stained sections was quantified in five sections of at least five different defects for each treatment at 200× magnification using Image Pro Plus software. New bone formation on each section was expressed as a percentage of the total area of the defect. The localization of OCN staining was studied on transverse sections of the cranium as previously described.26 All slides were coded to prevent the introduction of examiner bias.
Long bones, calvarial bone, periodontal ligament, heart, brain, kidney, liver, and muscle tissues were also collected from 8-week wild-type mice without surgery. Five-millimeter sections were cut and mounted on glass slides. The expressions of PHF8 were detected using immunohistochemical staining followed by counterstaining in hematoxylin with a Histostain SP kit (Invitrogen).
Statistical analysis
All results were expressed as mean±standard error of the mean of three or more independent experiments. One-way analysis of variance with post-hoc tests was used to test significance using the software SPSS 13.0. Values of p<0.05 were considered statistically significant.
Results
The cellular localization of PHF8 and the expression pattern of PHF8 in different tissues
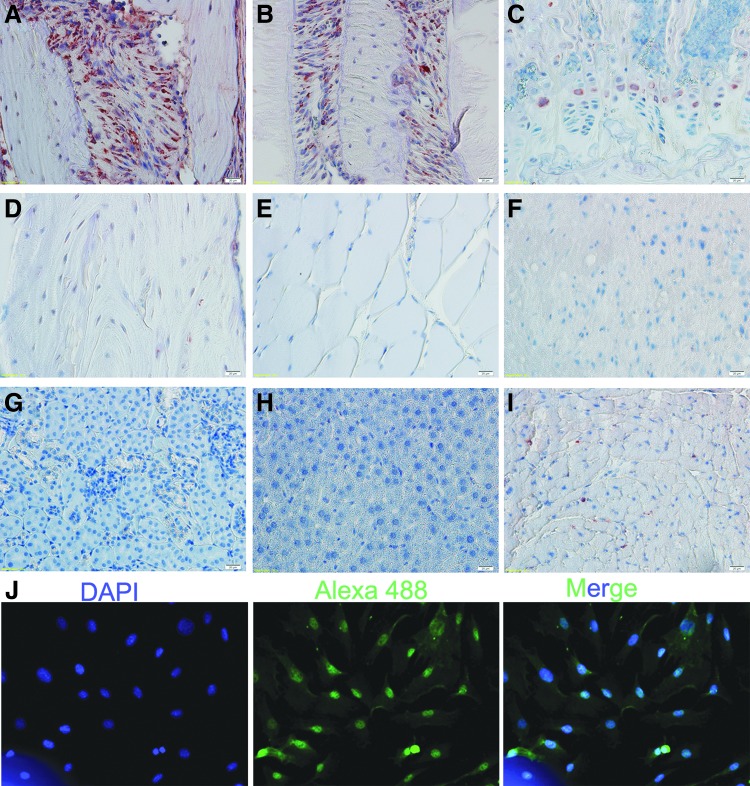
To detect the expression pattern of PHF8 in different tissues, we collected tissues from 8-week wild-type mice and used immunohistochemical staining to detect the PHF8 expression (Fig. 1A–I). We found that there was little PHF8 expression in kidney, liver, and muscle. In heart, only a few cells were PHF8-positive cells. In long bones, calvarial bones, and periodontal ligaments, there were more PHF8-positive cells than that in other tissues. Importantly, the PHF8-positive cells mainly distributed in the growth plate of long bone and the cranial suture. These results indicated that PHF8 might play an important role in bone formation and regeneration.
FIG. 1.
The localization of PHF8 and the expression pattern of PHF8 during osteogenic differentiation. Original magnification: 400×. (A–I) The expression pattern of PHF8 in different tissues detected by IHC. (A) Calvarial bone; (B) periodontal ligament; (C) growth plate; (D) long bone; (E) muscle; (F) brain; (G) kidney; (H) liver; (I) heart; (J) The cellular localization of PHF8. PHF8 protein was labeled with Alexa 488 green, and nuclei were labeled with DAPI (blue). Color images available online at www.liebertpub.com/tea
PHF8 is a histone demethylase and it exhibits its demethylase activity in the nucleus. In this study, we used immunocytofluorescent staining to detect the cellular localization of PHF8 and found that PHF8 expressions were localized in nucleus in MC3T3-E1 cells (Fig. 1J).
The expression of PHF8 increased during osteogenic differentiation
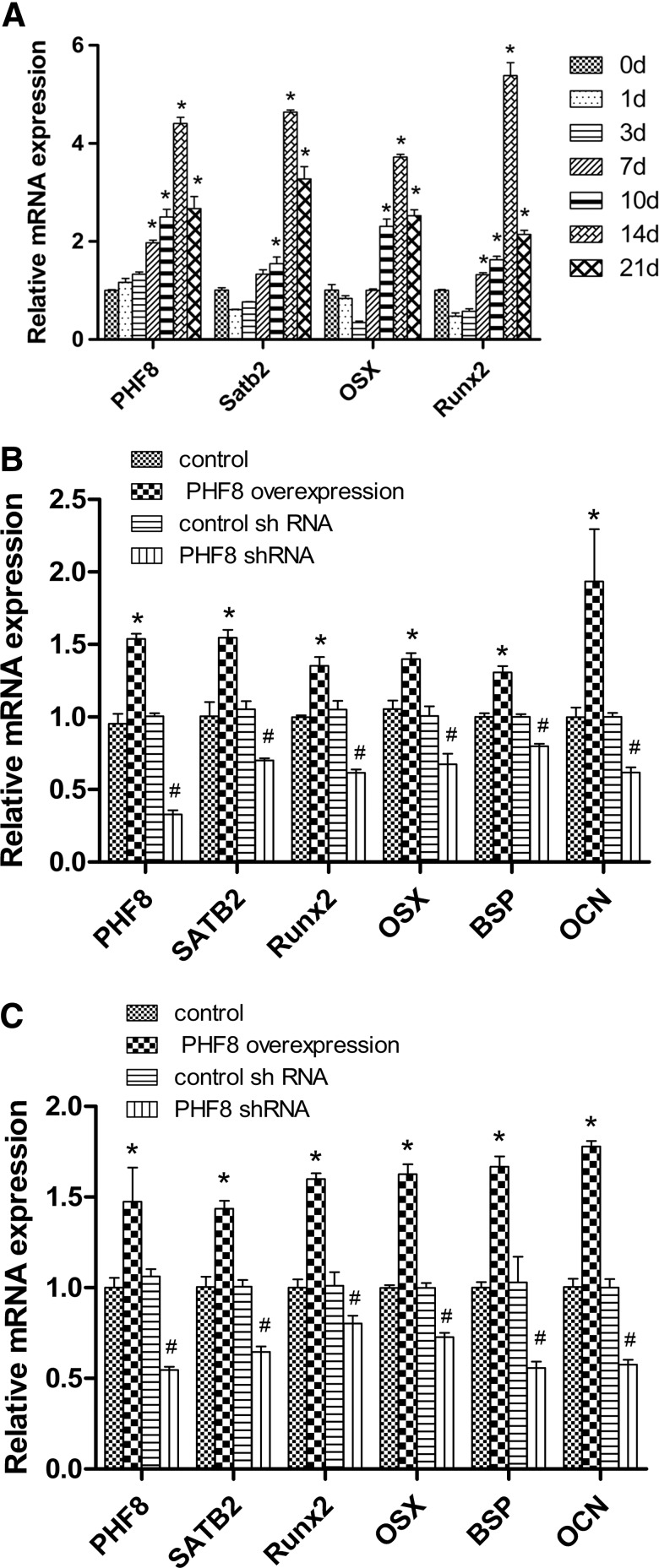
BMSCs were obtained from 4-week-old mice and treated with osteogenic medium for 1, 3, 7, 10, 14, and 21 days, respectively. RNA was extracted and real-time PCR was conducted to detect the gene expression of PHF8. Our results showed that the expression of both PHF8 and SATB2 increased during osteogenic differentiation (Fig. 2A).
FIG. 2.
PHF8 regulated the gene expression levels of the osteogenic markers. The expression of PHF8, SATB2, OSX, and Runx2 increased during osteogenic differentiation, *p<0.05 versus 0 day group (A). PHF8 cDNA overexpression and knockdown of PHF8 using shRNA, respectively, in MC3T3-E1 cells (B) and BMSCs (C). The gene expressions of SATB2, OSX, Runx2, BSP, and OCN were detected by realtime reverse transcription polymerase chain reaction. *p<0.05 versus control vector group; #p<0.05 versus control shRNA group. BMSCs, bone marrow stromal cells; SATB2, special AT-rich sequence-binding protein 2.
PHF8 promoted osteogenic differentiation of MC3T3-E1 cells and BMSCs
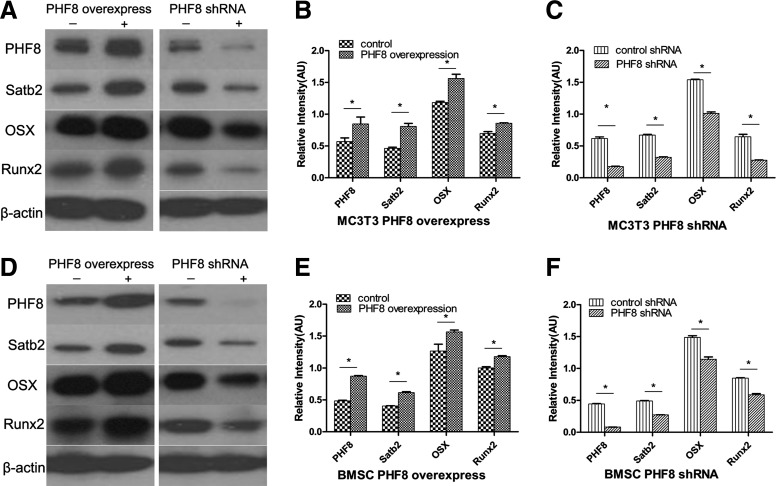
To determine whether PHF8 affects osteogenic differentiation, we overexpressed or knocked down PHF8 expression in MC3T3-E1 cells and BMSCs. The results showed that PHF8 overexpression upregulated the expression of all bone marker genes and knockdown of PHF8 using shRNA downregulated the expression of SATB2, OSX, Runx2, BSP, and OC in MC3T3-E1 cells and BMSCs (Figs. 2B, C and 3). These results provided the evidence that PHF8 promoted osteogenic differentiation of MC3T3-E1 cells and BMSCs.
FIG. 3.
PHF8 regulated bone transcription factors. The protein expressions of PHF8, Satb2, OSX, and Runx2 were detected by western blot. (A) PHF8 overexpression (left panel) and PHF8 suppression by shRNA (right panel) in MC3T3-E1 cells. (B, C) Quantification of protein levels. (D–F) The same experiments as (A–C) in BMSCs. *p<0.05.
PHF8 directly bound to the TSS region of SATB2
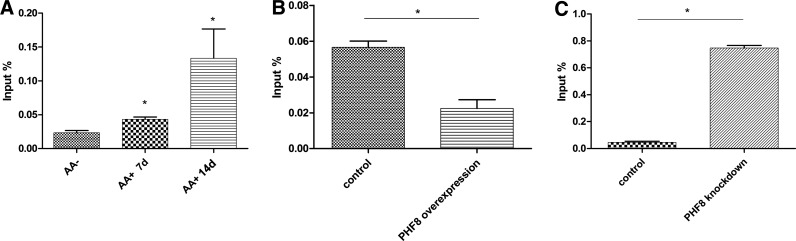
To detect the mechanism of how PHF8 regulates osteogenic differentiation, we performed ChIP analysis to determine whether PHF8 can directly bind to transcription factor SATB2. MC3T3-E1 cells were treated with osteogenic medium for 7 and 14 days, and then cross-linked. The DNA-protein complex was then incubated with PHF8 antibody and then DNA was extracted. The results showed that PHF8 bound to the TSS region of SATB2 gene (Fig. 4A). We also found that the binding of PHF8 to the TSS region of SATB2 increased when cells were treated with osteogenic medium (Fig. 4A).
FIG. 4.
ChIP-qPCR analyze the binding of PHF8 and H3K9me1 on Satb2. (A) MC3T3-E1 cells were treated with osteogenic medium for 7 and 14 days, and then cross-linked. The DNA-protein complex was then incubated with PHF8 antibody and then DNA was extracted, real-time PCR was used to detect the binding of PHF8 on SATB2. (B, C) PHF8 was overexpressed (B) or suppressed (C) in MC3T3-E1 cells, and then cross-linked. The DNA-protein complex was incubated with H3K9me1 antibody and then DNA was extracted, real-time PCR was used to detect the binding of H3K9me1 on SATB2. *p<0.05. PCR, polymerase chain reaction.
PHF8 regulated osteogenic differentiation via demethylating H3K9me1 at the promoter of SATB2
To determine whether PHF8 regulates SATB2 transcription through the H3K9me1 demethylase activity, we overexpressed or knocked down PHF8 expression in MC3T3-E1 cells undergoing osteogenic differentiation, and then performed ChIP analysis to detect the binding of H3K9me1 at the promoter of SATB2. Our results confirmed that overexpressing PHF8 downregulated the binding of H3K9me1 at the TSS region of SATB2, while knockdown of PHF8 using shRNA upregulated the binding of H3K9me1 (Fig. 4B, C). H3K9me1 can silence gene transcription and the demethylation of H3K9me1 causes the DNA to unwind from histone protein, and converting chromatin into a transcriptionally active conformation. Therefore, this result suggested that the binding of PHF8 to SATB2 and the resultant demethylation activity of PHF8 toward H3K9me1 at the promoter of SATB2 converted SATB2 into a transcriptional active conformation.
Enhancement of bone healing by transduction of PHF8 in mice
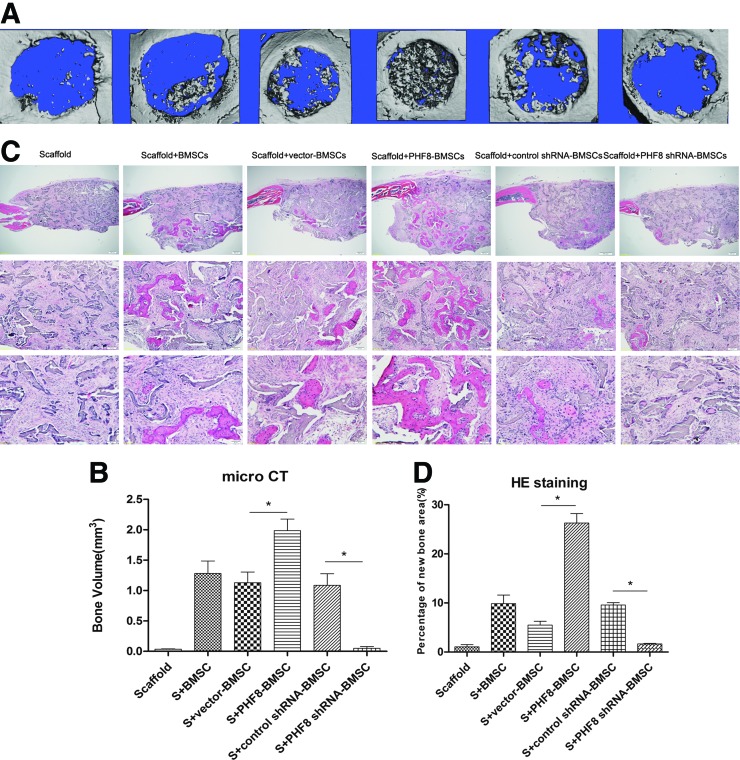
Critical size full thickness defects of 4 mm in diameter were made in both sides of the cranial bone and filled with a SS seeded with gene-modified BMSCs. Five weeks after surgery, micro CT was used to detect the morphology of the newly formed bone and evaluate new bone formation within the defects. As shown in Figure 5, larger, newly formed bone filled the defects of the PHF8-modified BMSCs group when compared with the empty vector-modified or unmodified groups, which showed the formation of scattered new bone in the defect sites (Fig. 5A). No obvious evidence of new bone formation was observed in the defects filled with scaffolds alone or PHF8 shRNA-modified BMSCs (Fig. 5A).
FIG. 5.
Micro CT and HE staining analysis. (A) Five weeks after surgery, micro CT results indicated the PHF8-modified BMSCs group showed the largest amounts of new bone formation. (B) HE staining demonstrated that PHF8-modified BMSCs group showed more new bone formation than other groups. Original magnification: top panel, 40×; middle panel, 200×; bottom panel, 400×. (C) Volume of newly formed bone detected by micro CT. (D) Percentage of new bone area in different treatment groups measured in HE staining sections. HE, hematoxylin and eosin; S, scaffold; *p<0.05. Color images available online at www.liebertpub.com/tea
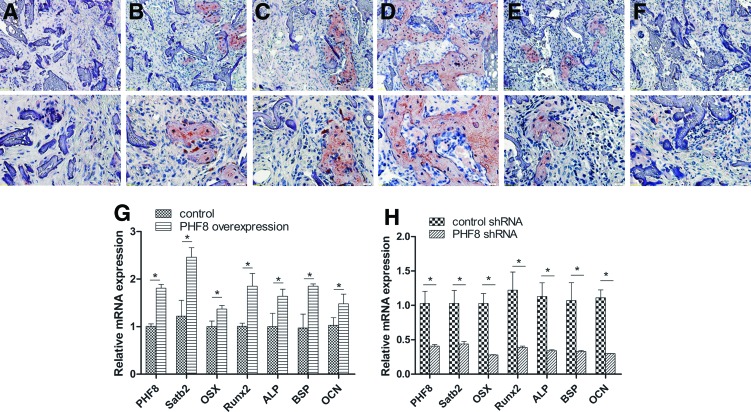
To quantify the new bone regeneration within the calvarial defects, the volume of regenerated bone (BV) were measured. As shown in Figure 5C, BV was significantly higher for the PHF8-modified BMSCs group when compared with all other groups. Histological evidence further supported the micro CT results, indicating that the specimens from the PHF8-modified BMSCs group showed the most extensive new bone formation (Fig. 5B). In contrast, a small amount of newly formed bone tissue was shown in the empty vector-modified BMSCs, unmodified BMSCs group, or control shRNA-modified group. Defects were filled with fibrous connective tissue and no obvious bone formation was found in the PHF8 shRNA-modified BMSCs group and scaffold alone group (Fig. 5B). The percentage of new bone area in the PHF8-modified BMSCs group was significantly higher than the other four groups 5 weeks postoperation (Fig. 5D). To further analyze the new bone mineralization, immunohistochemistry was performed to determine OCN expression levels. As shown in Figure 6, immunohistochemistry exhibited strong expression for OCN in areas of new bone formation within the defect region from samples treated with the PHF8-modified BMSCs, whereas OCN staining was weaker in empty vector-transduced BMSCs, control shRNA-modified BMSCs, and untransduced BMSCs groups. Moreover, no obvious positive staining was observed in the PHF8 shRNA-modified BMSCs group or scaffold alone group (Fig. 6A–F). Moreover, to further determine the effect of PHF8-modified BMSCs in new bone formation, total RNA was isolated from four the experimental groups (PHF8-modified BMSCs group; empty vector-modified BMSCs group; PHF8 shRNA-modified BMSCs group; control shRNA-modified BMSCs group), and quantitative real time reverse transcription polymerase chain reaction was performed to detect mRNA expression levels of ALP, BSP, OCN, SATB2, Runx2, and OSX. As shown in Figure 6, the ALP, BSP, OCN, SATB2, Runx2, and OSX mRNA expression showed a higher level in the defects with PHF8-modified BMSCs group than the control group 5 weeks postoperation (Fig. 6G), while knockdown of PHF8 expression using shRNA in BMSCs inhibited the expression of these genes (Fig. 6H). These results demonstrated that BMSCs modified with PHF8 and seeded on SSs enhanced cranial critical-size bone healing in mice.
FIG. 6.
Immunohistochemical and real-time PCR analysis of newly formed bones. (A–F) Immunohistochemical analysis of new bone formation in each group at 5 weeks postoperation (A–F); Original magnification: upper panel, 200×; lower panel, 400×). OCN staining demonstrates no obvious positive staining in scaffold alone (A) and PHF8 shRNA-modified BMSCs group (F), while a weak OCN staining in scaffold with BMSCs group (B), vector-modified BMSCs group (C) and control shRNA-modified BMSCs group (E), and a stronger OCN expression was observed in the new bone tissue from PHF8-modified BMSCs group (D). (G–H) Gene expression levels of PHF8, Satb2, ALP, OSX, Runx2, OCN, and BSP in the new bone of PHF8-modified BMSCs treatment group (G) and PHF8 shRNA treatment group (H). *p<0.05. Color images available online at www.liebertpub.com/tea
Discussion
Epigenetic regulation has been shown to play an important role in various biological processes. As one of the modes of epigenetic regulation and a mechanism for modifying chromatin structure, histone methylation was associated with the stimulation of several pathways known to be important for biological processes including neural development,27 vascular development,28,29 and bone regeneration.30,31 Our research aimed to characterize the role PHF8 played in osteogenic differentiation and bone regeneration.
In this study, we first detected the distribution of the PHF8 in different tissues in mice. We found that there were more PHF8-positive cells in the growth plate of the long bone and calvarial sutures than other tissues in 8-week mice. The growth plate is responsible for bone lengthening32 and sutures play an important role in bone repair and bone regeneration.33 Based on the distribution of PHF8 expression in these tissues, we hypothesized that PHF8 plays an important role in bone regeneration. When we treated BMSCs and preosteoblasts with osteogenic medium, we found that the gene expression of PHF8 and other osteogenic markers such as SATB2, Runx2, and OSX were upregulated compared with control groups. These results indicated that PHF8 might play an important role in osteogenic differentiation of BMSCs and MC3T3-E1.
BMSCs can differentiate into several kinds of cells including osteoblasts and play an important role in bone regeneration.34,35 Enhanced osteogenic differentiation of BMSCs induced by genetic modification could change the character of these cells. Such genetic modifications were frequently achieved by overexpression of regulatory proteins such as BMP2 and VEGF.36,37 In this study, we found that PHF8 overexpression increased the expression of SATB2, Runx2, OSX, ALP, BSP, and OCN while knockdown of PHF8 downregulated the expression of those genes in MC3T3-E1 cells and BMSCs. Our previous studies and those of other labs confirmed that SATB2 plays an important role in osteogenic differentiation and bone regeneration,5,38 and SATB2 regulates the expression of OSX during osteoblast differentiation.9 Given that SATB2 plays an important role in integrating genetic and epigenetic signaling39 and PHF8 overexpression upregulated the expression of SATB2 during osteogenic differentiation, we inferred that PHF8 might regulate SABT2 to activate osteogenic differentiation of BMSCs. To this end, we performed ChIP analysis to confirm the epigenetic regulation of PHF8 through SATB2.
Epigenetic regulation is largely controlled by histone modifications, which enable gene promoters to be accessible or inaccessible to transcription factors.40,41 One such important histone modification is histone H3 lysine monomethylation (H3K9me1) on gene promoters, which silences gene transcription.42 PHF8 is a histone demethylase that can demethylate H3K20 and H3K9me1/2,19 and it apparently would be localized in the nucleus of the cells. We first confirmed that PHF8 was mainly localized in the nucleus of the cells using immunocytofluorescent assays. Then, ChIP analysis found that PHF8 bound to the TSS region of SATB2. More interestingly, the occupancy of the PHF8 at the TSS region increased during osteogenic differentiation. These results indicated that PHF8 might promote osteogenic differentiation via regulation the expression of SATB2. Next, we performed another ChIP analysis and found that PHF8 overexpression decreased the binding of H3K9me1 to the promoter of SATB2. As PHF8 is an H3K9me1/2 demethylase, these results indicated that PHF8 could demethylate H3K9me1 at the promoter region of SATB2, thus causing the DNA to unwind from histone protein, and converting SATB2 into a transcriptionally active conformation. These in vitro data confirmed that PHF8 regulated the expression of SATB2 via its histone demethylation activity and then promoted osteogenic differentiation.
Tissue engineering has been widely used in bone research field, which is an especially good choice to repair large size bone defects by using resorbable scaffolds supplemented with regeneration-competent cells and growth factors.43 The biodegradability, distinguishing mechanical properties, and low inflammatory response of silk fibroin make it a promising scaffold for bone regeneration.6,44 Our previous research also found that silk fibroin was an ideal scaffold for bone repair.4,5 As a result, we choose silk fibroin as scaffolds in this research. Besides the scaffolds, the seeded cells and growth factors are key elements for forming tissue-engineered bone. As an important regulator, epigenetic pathways are able to play a crucial role in the control of gene activity during different stages of development and throughout life, which will lead to more effective ways to prevent and treat diseases affecting the oral and craniofacial region. PHF8 could demethylate H3K9me1 at the promoter of SATB2 and then promoted osteogenic differentiation in vitro. Therefore, we wanted to know whether PHF8 could promote bone healing in vivo. Critical size calvarial bone defects were formed in mice and then implanted with PHF8-modified BMSCs in the defects housed within the SSs. We found little new bone formation in the defects 2 weeks after surgery. However, when we used immunohistochemistry to detect the expression of OCN in cells in the defects, we found that there were more OCN-positive cells in PHF8-modified BMSCs implanted group (data not shown). These results indicated that PHF8 could promote osteogenic differentiation of BMSCs in vivo. Five weeks after surgery, we found substantially more new bone formation in PHF8-modified BMSCs group than other groups. However, little new bone was found in PHF8 shRNA-modified BMSCs treatment group. These results provided the evidence that PHF8 played an important role in bone-related wound healing. As an epigenetic regulator, PHF8 could demethylate H3K9me1 and converted SATB2 chromatin into a transcriptionally active conformation during osteogenic differentiation. We and other researchers have also shown that SATB2 acted as a “node” to transcriptionally regulate oral and craniofacial development and osteoblast differentiation, and promote bone tissue regeneration.5,9,38 PHF8 could be used as a useful epigenetic regulator in the tissue engineering field as it can promote BMSCs osteogenic differentiation and bone regeneration (Fig. 7). Further understanding of how the nuclear matrix gene SATB2, chromatin structure modifier PHF8, and transcriptional activity coordinate the regulation of multiple steps during osteoblastic differentiation of BMSCs will allow us to better optimize tissue regeneration strategies.
FIG. 7.
Schematic representation of epigenetic regulation by PHF8 in promoting BMSCs osteogenic differentiation and bone repair. PHF8 demethylate H3K9me1 at the promoter of SATB2 and activate SATB2 transcription. SATB2 then upregulate the expression of osteogenic factors such as OSX, ALP, BSP, OCN, and promote osteogenic differentiation of BMSCs and bone repair. Color images available online at www.liebertpub.com/tea
Conclusions
The data demonstrated that PHF8, an epigenetic modifier, regulated osteogenic differentiation of BMSCs via modulating H3K9me1 methylation of the master gene SATB2. Ex vivo gene therapy of PHF8-modified BMSCs within SSs effectively promoted bone regeneration in critical-sized calvarial defects. Although further studies are needed to optimize PHF8 levels during bone regeneration, the present results provide strong evidence for using PHF8 to repair bone defects.
Supplementary Material
Acknowledgments
We thank Jean Tang for her assistance in tissue sample preparation and histology, and Dana Murray for her help in the preparation of the article. We are grateful to the other members of the Chen lab for discussion and technical help. This project was supported by National Institutes of Health (NIH) grants R01DE16710 and R01DE21464, and the International Association of Dental Research (IADR) and the Academy of Osseointegration (AO) Innovation in Implant Science Award (to J.C.); Ministry of Education, Science and Technology (MEST) grant 2007-0054931 in South Korea (to S.H.K.); NIH contract grant P41 EB002520 (to D.L.K.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Boyce T., Edwards J., and Scarborough N. Allograft bone. The influence of processing on safety and performance. Orthop Clin North Am 30, 571, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Huang C., Das A., Barker D., Tholpady S., Wang T., Cui Q., et al. . Local delivery of FTY720 accelerates cranial allograft incorporation and bone formation. Cell Tissue Res 347, 553, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim K.H., Jeong L., Park H.N., Shin S.Y., Park W.H., Lee S.C., et al. . Biological efficacy of silk fibroin nanofiber membranes for guided bone regeneration. J Biotechnol 120, 327, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Karageorgiou V., Tomkins M., Fajardo R., Meinel L., Snyder B., Wade K., et al. . Porous silk fibroin 3-D scaffolds for delivery of bone morphogenetic protein-2 in vitro and in vivo. J Biomed Mater Res A 78, 324, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Ye J.H., Xu Y.J., Gao J., Yan S.G., Zhao J., Tu Q., et al. . Critical-size calvarial bone defects healing in a mouse model with silk scaffolds and SATB2-modified iPSCs. Biomaterials 32, 5065, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X., Zhao J., Wang S., Sun X., Zhang X., Chen J., et al. . Mandibular repair in rats with premineralized silk scaffolds and BMP-2-modified bMSCs. Biomaterials 30, 4522, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu B., Zhang J., Brewer E., Tu Q., Yu L., Tang J., et al. . Osterix enhances BMSC-associated osseointegration of implants. J Dent Res 88, 1003, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu Q., Zhang J., James L., Dickson J., Tang J., Yang P., et al. . Cbfa1/Runx2-deficiency delays bone wound healing and locally delivered Cbfa1/Runx2 promotes bone repair in animal models. Wound Repair Regen 15, 404, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Tu Q., Grosschedl R., Kim M.S., Griffin T., Drissi H., et al. . Roles of SATB2 in osteogenic differentiation and bone regeneration. Tissue Eng Part A 17, 1767, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., and Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15, 2343, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Klose R.J., Kallin E.M., and Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7, 715, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Kleine-Kohlbrecher D., Christensen J., Vandamme J., Abarrategui I., Bak M., Tommerup N., et al. . A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell 38, 165, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton J.R., Upadhyay A.K., Qi H.H., Zhang X., Shi Y., and Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17, 38, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim H.J., Dimova N.V., Tan M.K., Sigoillot F.D., King R.W., and Shi Y. The G2-M regulator, histone demethylase PHF8, is targeted for degradation by APCcdc20. Mol Cell Biol 33, 4166, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asensio-Juan E., Gallego C., and Martinez-Balbas M.A. The histone demethylase PHF8 is essential for cytoskeleton dynamics. Nucleic Acids Res 40, 9429, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arteaga M.F., Mikesch J.H., Qiu J., Christensen J., Helin K., Kogan S.C., et al. . The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell 23, 376, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjorkman M., Ostling P., Harma V., Virtanen J., Mpindi J.P., Rantala J., et al. . Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene 31, 3444, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Zhu Z., Wang Y., Li X., Xu L., Wang X., Sun T., et al. . PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res 20, 794, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Feng W., Yonezawa M., Ye J., Jenuwein T., and Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol 17, 445, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Qi H.H., Sarkissian M., Hu G.Q., Wang Z., Bhattacharjee A., Gordon D.B., et al. . Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alcamo E.A., Chirivella L., Dautzenberg M., Dobreva G., Farinas I., Grosschedl R., et al. . Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364, 2008 [DOI] [PubMed] [Google Scholar]
- 22.Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Farinas I., et al. . SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125, 971, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Yu L., Tu Q., Han Q., Zhang L., Sui L., Zheng L., et al. . Adiponectin regulates bone marrow mesenchymal stem cell niche through a unique signal transduction pathway: an approach for treating bone disease in diabetes. Stem Cells 33, 240, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valverde P., Tu Q., and Chen J. BSP and RANKL induce osteoclastogenesis and bone resorption synergistically. J Bone Miner Res 20, 1669, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Kim H.J., Kim U.J., Kim H.S., Li C., Wada M., Leisk G.G., et al. . Bone tissue engineering with premineralized silk scaffolds. Bone 42, 1226, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu Q., Valverde P., Li S., Zhang J., Yang P., and Chen J. Osterix overexpression in mesenchymal stem cells stimulates healing of critical-sized defects in murine calvarial bone. Tissue Eng 13, 2431, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zovkic I.B., and Sweatt J.D. Epigenetic mechanisms in learned fear: implications for PTSD. Neuropsychopharmacology 38, 77, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villeneuve L.M., Reddy M.A., Lanting L.L., Wang M., Meng L., and Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A 105, 9047, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clifford R.L., John A.E., Brightling C.E., and Knox A.J. Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. J Immunol 189, 819, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye L., Fan Z., Yu B., Chang J., Al Hezaimi K., Zhou X., et al. . Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eslaminejad M.B., Fani N., and Shahhoseini M. Epigenetic regulation of osteogenic and chondrogenic differentiation of mesenchymal stem cells in culture. Cell J 15, 1, 2013 [PMC free article] [PubMed] [Google Scholar]
- 32.Musumeci G., Castrogiovanni P., Loreto C., Castorina S., Pichler K., and Weinberg A.M. Post-traumatic caspase-3 expression in the adjacent areas of growth plate injury site: a morphological study. Int J Mol Sci 14, 15767, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao Y., Wang G., Wang Z., Wang C., Zhang H., and Liu C. Synergistic enhancement of new bone formation by recombinant human bone morphogenetic protein-2 and osteoprotegerin in trans-sutural distraction osteogenesis: a pilot study in dogs. J Oral Maxillofac Surg 69, e446, 2011 [DOI] [PubMed] [Google Scholar]
- 34.Song G., Habibovic P., Bao C., Hu J., van Blitterswijk C.A., Yuan H., et al. . The homing of bone marrow MSCs to non-osseous sites for ectopic bone formation induced by osteoinductive calcium phosphate. Biomaterials 34, 2167, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim C.T., Ren X., Afizah M.H., Tarigan-Panjaitan S., Yang Z., Wu Y., et al. . Repair of osteochondral defects with rehydrated freeze-dried oligo[poly(ethylene glycol) fumarate] hydrogels seeded with bone marrow mesenchymal stem cells in a porcine model. Tissue Eng Part A 19, 1852, 2013 [DOI] [PubMed] [Google Scholar]
- 36.Kasten P., Beverungen M., Lorenz H., Wieland J., Fehr M., and Geiger F. Comparison of platelet-rich plasma and VEGF-transfected mesenchymal stem cells on vascularization and bone formation in a critical-size bone defect. Cells Tissues Organs 196, 523, 2012 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Mostafa N.Z., Hsu C.Y., Rose L., Kucharki C., Yan J., et al. . Modification of human BMSC with nanoparticles of polymeric biomaterials and plasmid DNA for BMP-2 secretion. J Surg Res 183, 8, 2013 [DOI] [PubMed] [Google Scholar]
- 38.Yan S.G., Zhang J., Tu Q.S., Ye J.H., Luo E., Schuler M., et al. . Enhanced osseointegration of titanium implant through the local delivery of transcription factor SATB2. Biomaterials 32, 8676, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gyorgy A.B., Szemes M., de Juan Romero C., Tarabykin V., and Agoston D.V. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur J Neurosci 27, 865, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Jenuwein T., and Allis C.D. Translating the histone code. Science 293, 1074, 2001 [DOI] [PubMed] [Google Scholar]
- 41.Kouzarides T. Chromatin modifications and their function. Cell 128, 693, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Pinheiro I., Margueron R., Shukeir N., Eisold M., Fritzsch C., Richter F.M., et al. . Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948, 2012 [DOI] [PubMed] [Google Scholar]
- 43.Petite H., Viateau V., Bensaid W., Meunier A., de Pollak C., Bourguignon M., et al. . Tissue-engineered bone regeneration. Nat Biotechnol 18, 959, 2000 [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Kim H.J., Vunjak-Novakovic G., and Kaplan D.L. Stem cell-based tissue engineering with silk biomaterials. Biomaterials 27, 6064, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.