Stem cell transplantation has emerged as a promising therapeutic strategy for treating many injuries and diseases including peripheral arterial disease,[1] cardiac disease,[2, 3] stroke,[4] peripheral nerve injury,[5] and spinal cord injury.[6] Cell transplantation by injection is a minimally invasive and clinically preferred strategy that delivers cells directly to the desired site of repair.[7] However, numerous studies in recent years have shown that injectable cell therapies result in low cell retention and engraftment, which is a major obstacle to clinical translation.[2, 3] Low cell retention is partly attributed to (i) mechanical forces during injection that damage the cell membrane, and (ii) the lack of a three-dimensional (3D) matrix to support cell viability post-injection.[2, 3, 8] Injectable hydrogels have been explored as a strategy to address both of these causes of low cell retention.[8–20] Pre-encapsulating cells in a shear-thinning, physical hydrogel can provide mechanical protection that prevents cell membrane damage during injection.[8] Weak hydrogels with shear moduli < 50 Pa were previously found to offer the best protection; however, these materials typically biodegrade too quickly (within one to two weeks) to support long-term cell survival in a 3D matrix.[8, 16, 21–23] Therefore, it has been challenging to design a hydrogel that can simultaneously address both causes of low cell retention in a single gel formulation. Toward this end, we aim to design an injectable hydrogel that undergoes two different physical crosslinking mechanisms: 1) weak, heterodimeric, molecular-recognition ex vivo to provide mechanical cell protection during injection and 2) additional crosslinking in vivo induced by thermo-responsive formation of a reinforcing network to support long-term cell survival within a 3D matrix.
While several naturally derived hydrogel materials have been used for injectable cell transplantation, including alginate,[8] collagen,[9, 10] fibrin,[11] hyaluronan,[12] Matrigel,[13] and decellularized extracellular matrices,[24] these materials can be subject to batch-to-batch variations.[25] To optimize the hydrogel flow properties, synthetic efforts have focused on the development of shear-thinning and self-healing hydrogels, which flow as a liquid during injection and recover back into a solid gel after delivery to retain the encapsulated cells at the desired site.[26] Currently, these hydrogel systems are mainly based on the self-assembly of peptides[14] and/or block copolymers.[22, 27] These hydrogels often require exposing cells to non-physiological conditions (e.g. high ionic strength, low pH, or low temperature) to induce the sol-gel transition and to achieve cell encapsulation.
Hydrogels based on heterodimeric molecular-recognition between protein motifs, including growth factor mediated hydrogels,[28] Dock-and-Lock hydrogels,[18, 19] leucine-zipper hydrogels,[15] and mixing-induced two-component hydrogels (MITCH),[16, 17] are appealing due to their ability to easily encapsulate cells by simply mixing together complementary polymers. However, these hydrogels with dynamic and weak physical crosslinks are very soft and subject to fast biodegradation after delivery. Therefore, an additional crosslinking step post-injection may be a promising method to increase the gel stiffness and decrease the degradation rate. While dual-stage hydrogel crosslinking has been reported, the design of these hydrogels to provide mechanical cell protection during injection and to support long-term in vivo cell viability and retention has not been demonstrated.[15, 19] When designing an in situ crosslinking strategy, we chose to avoid diffusible small molecules and chemical reactions that may have unwanted off-target effects. We also wanted to develop a crosslinking protocol that would be simple to surgically implement without need for extra equipment (e.g. UV lamps). Thermoresponsive, physical crosslinking offers mild network formation upon warming in situ due to entropically driven dehydration and collapse of polymer components, leading to non-cytotoxic encapsulation of cells.[29]
We hypothesized that a hydrogel that underwent weak, heterodimeric, molecular-recognition ex vivo and thermo-responsive crosslinking in situ would simultaneously address two of the major causes of transplanted cell death and significantly improve the retention of viable transplanted stem cells. To achieve this, we designed a novel hetero-arm star copolymer that was conjugated with both modular polypeptide domains and a thermo-responsive component to enable physical crosslinking processes before and after syringe injection. We termed this material Shear-thinning Hydrogel for Injectable Encapsulation and Long-term Delivery (SHIELD). Specifically, the molecular recognition takes place between a star-shaped peptide-polyethylene glycol (PEG) copolymer assembling with an engineered recombinant protein (C7) to form a weak, physical network ex vivo. The in situ crosslinking induces formation of a reinforcing double network via thermal phase transition of poly(N-isopropylacrylamide) (PNIPAM) chains conjugated to the PEG copolymer (Figure 1a).
Figure 1.
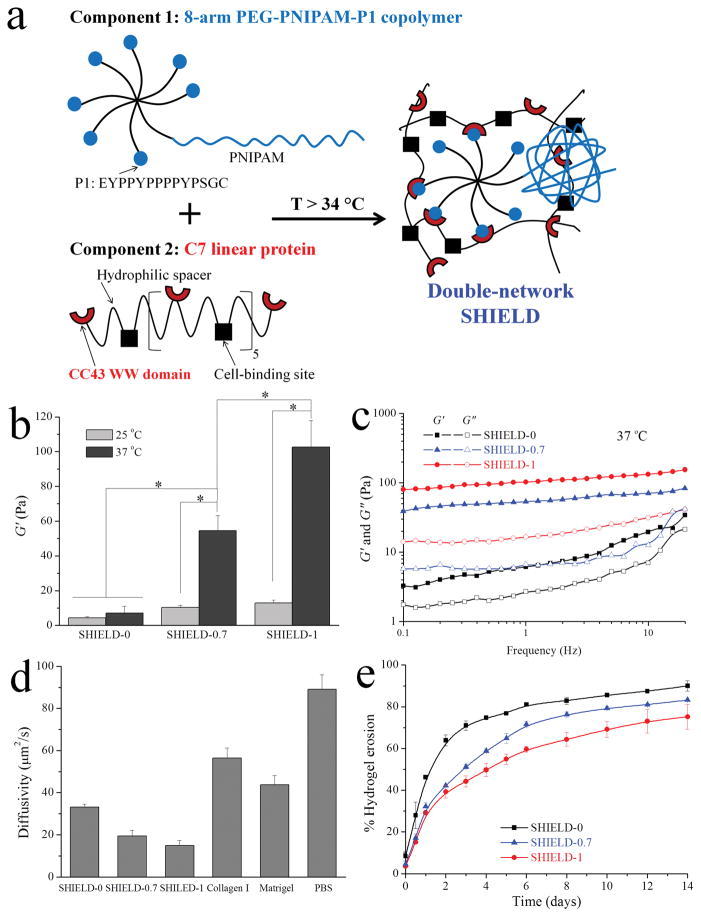
Schematic and material properties of Shear-thinning Hydrogel for Injectable Encapsulation and Long-term Delivery (SHIELD). (a) Component 1 is a 8-arm PEG with 1 arm conjugated with PNIPAM and the other 7 arms conjugated with proline-rich peptide (denoted as P1) domains. Component 2 is a recombinant C7 linear protein copolymer bearing CC43 WW (denoted as C) domains and RGD cell-binding domains connected by hydrophilic spacers. (b) Shear storage moduli (G′) of SHIELD-0, SHIELD-0.7, and SHIELD-1 (with 0, 0.7, and 1 wt% PNIPAM, respectively) at 25 or 37 °C and 1 Hz. * p < 0.05, n ≥ 3. (c) Storage (G′) and loss moduli (G″) of SHIELD as a function of frequency at 37 °C. (d) FRAP characterization of dextran (MW = 40 kDa) diffusivity within hydrogels and control media at 37 °C. All differences between any two groups are statistically significant, p < 0.05, n ≥ 3. (e) SHIELD erosion kinetics represented by cumulative fraction of the hydrogel material eroded into bulk PBS medium at 37 °C over 14 days. p < 0.05 between any two groups at the same time point after day 2, n ≥ 3.
The site-specific conjugation of both PNIPAM and P1 peptides to 8-arm PEG-vinyl sulfone (VS) was achieved using Michael-type addition between amines (present on PNIPAM) or thiols (present on P1 peptides) and VS, which allows for rapid and selective reaction within aqueous conditions. This two-step reaction yielded high degrees of conjugation (74% for PNIPAM and 90% for P1, NMR spectra in Figure S1, Supporting Information). PNIPAM peak integration of the 8-arm PEG-PNIPAM copolymer indicated that PNIPAM was conjugated to ~1 arm of the 8-arm PEG-VS with unreacted VS double bonds remaining on the other 7 arms. These double bonds completely disappeared after the second conjugation step, and tyrosine aromatic proton peaks from the P1 peptide appeared, indicating the successful synthesis of the 8-arm PEG-PNIPAM-P1 copolymer. Smooth GPC curves suggested that the copolymer retained a narrow molecular weight distribution similar to the original PEG-VS, with no traces of PNIPAM chain contamination (Figure S2, Supporting Information). The apparent weight-average molecular weight increased from 18,570 g/mol for 8-arm PEG-VS to 32,400 g/mol for the PEG-PNIPAM-P1 copolymer due to the conjugation of PNIPAM and P1 peptide chains. The exact molecular weights are likely somewhat different given the hetero-arm configuration of the synthesis products and the linear polymer configuration of the molecular weight standards.
In solution, the PEG-PNIPAM-P1 copolymer (10 wt% in phosphate-buffered saline (PBS)) showed typical viscous behavior of an unentangled polymer solution with loss moduli (G″) much higher than storage moduli (G′) before reaching a crossover point at high frequencies (Figure S3, Supporting Information). Similar rheological behavior was found for the linear C7 protein solution (10 wt% in PBS). Upon mixing this PEG-PNIPAM-P1 copolymer solution with the C7 (10 wt% in PBS), SHIELD-1 (1 wt% PNIPAM moiety) formed within seconds, and the G′ significantly increased to ~13 Pa at 25 °C, due to a hetero-assembled network formed by the hetero-dimeric, specific binding of C and P domains (Figure 1b, further statistical analysis provided in Figure S4, Supporting Information). All hydrogels demonstrated frequency sweep curves characteristic of elastic networks formed by physical crosslinking, with G′ consistently larger than G″, confirming gelation upon mixing of the two components (Figure 1c). At body temperature, the G′ of the network further increased an order of magnitude from ~13 Pa to ~100 Pa as a result of the formation of a secondary PNIPAM thermo-responsive, self-assembled network within the already existing hetero-assembled network (Figure 1b,c). The lower critical solution temperature (LCST) of SHIELD-1 was measured to be ~34 °C (Figure S5, Supporting Information). As a control, SHIELD-0 was created using C7 mixed with PEG-P1 copolymer with all eight arms of PEG-VS conjugated with P1 peptides (0 wt% PNIPAM). As expected, the stiffness of the SHIELD-0 control did not increase at body temperature (Figure S5, Supporting Information). By mixing SHIELD-0 with SHIELD-1 at a ratio of 30/70, we created SHIELD-0.7 (0.7 wt% PNIPAM) with an intermediate G′ of ~70 Pa, suggesting that the mechanical properties of this family of hydrogels can be easily tuned by controlling the extent of formation of the secondary PNIPAM network (Figures 1c).
To further characterize these double-network hydrogels, we measured the diffusivity of a 40 kDa dextran within the various formulations using fluorescence recovery after photobleaching (FRAP). Diffusivity is known to correlate with network mesh size. Increasing the PNIPAM secondary network density from 0 to 0.7 to 1 wt% (SHIELD-0, SHIELD-0.7, and SHIELD-1, respectively) resulted in significantly slower diffusion, indicating a smaller mesh size for the double-network hydrogels (Figure 1d). For comparison, all three hetero-assembled hydrogels had significantly lower diffusivities compared to Type I collagen and Matrigel, two common matrices for cell transplantation with similar storage moduli (G′ of ~25 Pa and ~90 Pa, respectively).[9, 30] In general, hydrogels with smaller mesh sizes, and hence decreased diffusivity, are expected to have slower biodegradation rates, which we hypothesized would improve long-term cell retention. Because the diffusivities of SHIELD are similar to normal tissues, such as muscle[31] and cartilage,[32] we also expected SHIELD to allow for sufficient nutrient exchange and paracrine signaling to support long-term cell viability. As with other physically crosslinked hydrogels, the loss of hydrogel material is expected to occur predominantly by surface erosion of the double network rather than internal degradation of the polymers.[21] Analysis of hydrogel erosion kinetics in bulk PBS medium showed that SHIELD-1, a double network hydrogel, eroded significantly more slowly than the SHIELD-0, a single network hydrogel (Figure 1e). These data are consistent with measurements of hydrogel moduli and diffusivity, as increasing the number of physical crosslinks per chain results in stiffer hydrogels, a smaller mesh size, decreased diffusivity, and decreased surface erosion.
We next evaluated the shear-thinning and cell-protective properties of the hydrogels during high-shear flow, such as that experienced during syringe needle injection. Linear viscosity measurements at alternating low and high shear rates showed that SHIELD-1 exhibited shear-thinning behavior, or thixotropy, with much lower viscosity at higher shear rates at both room and body temperatures (Figure 2a). The SHIELD-1 responded to high shear rates almost instantaneously (< 1 sec) with a sharp yielding transition, due to the inherently fast on-rate kinetics of C to P domain interactions.[33] At low shear rates, the networks reversibly and rapidly (< 2 sec) self-healed due to reformation of the physical network junctions. The response time of SHIELD-1 upon shear-thinning and self-healing is much shorter than other shear-thinning hydrogel systems based on protein-ligand interactions or peptide nanofiber entanglement, which can take minutes to hours due to passive re-entanglement of polymer chains or stepwise re-assembly into peptide nanofibers.[21, 26, 34] The rapid shear-thinning and self-healing kinetics are ideal for injectable applications, where the hydrogels must be able to flow under hand pressure to facilitate easy transplantation and able to recover the gel state immediately post-injection to remain localized at the desired site.[8, 35]
Figure 2.
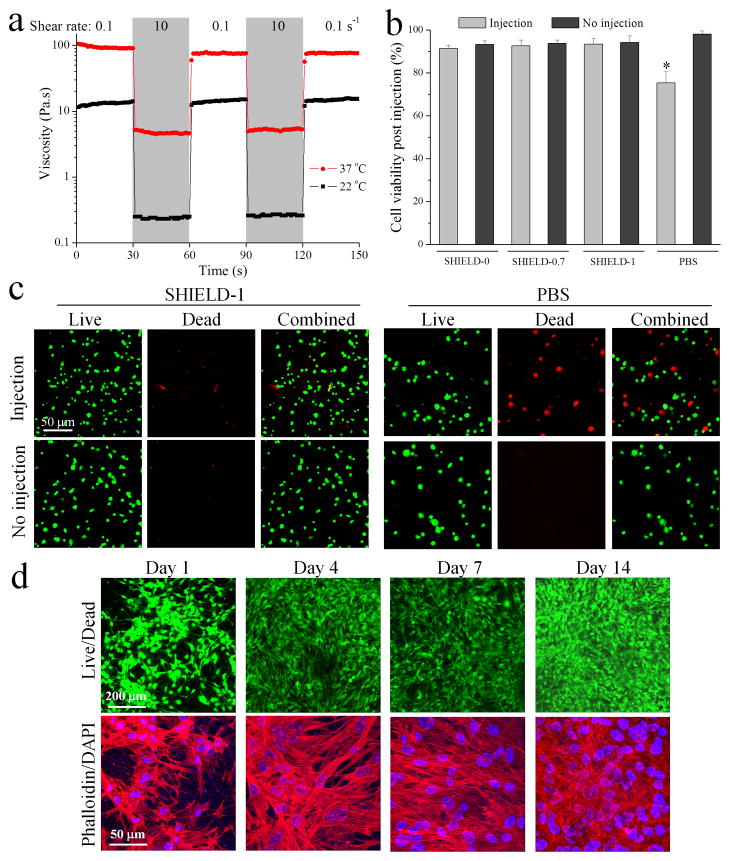
Cell protective properties of SHIELD. (a) Shear-thinning and self-healing of SHIELD-1 under alternating shear rates of 0.1 and 10 s−1 at 22 and 37 °C. (b) Acute hASC viability following in vitro injection through a 28-G syringe needle at 1.0 mL/min. * p < 0.05 compared to other groups, n = 5. (c) Fluorescence images of hASCs stained with LIVE/DEAD assay (green/red, respectively) within SHIELD-1 or PBS immediately post-injection. (d) Confocal 3D projection images of hASCs cultured within SHIELD-1 at days 1, 4, 7, and 14 post-injection stained with LIVE/DEAD assay (green/red, respectively, top row) and DAPI (blue) for cell nuclei and rhodamine phalloidin (red) for F-actin cytoskeleton (bottom row).
We then tested the cytocompatibility of SHIELD-1 and their potential to provide cell protection during injection using a preclinical model of human adipose-derived stem cell (hASC) transplantation. hASCs possess tremendous potential for multiple regenerative medicine therapies and are harvested through voluntary lipoaspiration of adult fat tissue to circumvent procurement and ethical concerns.[36] hASCs from consenting donors were encapsulated in the hydrogel within a 1-mL syringe barrel prior to ejection through a 28-gauge syringe needle using a syringe pump at a flow rate of 1 mL/min. Cells were immediately analyzed with a LIVE/DEAD assay to count the number of cells with intact or damaged membranes, respectively. Immediately after injection, 93 ± 4% of the hASCs were still alive within the hydrogel, statistically similar to non-injected controls (Figure 2b,c). In contrast, when the cells were injected in a saline solution (PBS), significantly more cells exhibited membrane damage, resulting in a viability of 69 ± 5% (Figure 2b,c). Similar levels of mechanical cell protection were exhibited by SHIELD-0 and SHIELD-0.7 (Figure 2b). These results suggest that the weak, hetero-dimeric, molecular-recognition network present in all three SHIELD variants provides significant cell protection from the damaging mechanical forces experienced during cell transplantation. These results are consistent with our previously published data that weak, shear-thinning alginate gels with shear moduli < 50 Pa protected cells from membrane damage during syringe-needle injection.8 Thus, the weak, primary SHIELD network formed ex vivo has appropriate mechanical properties to provide acute mechanical shielding during cell injection.
After in vitro injection, the hASCs within SHIELD-1 were brought to physiological temperature to induce formation of the thermo-responsive, secondary reinforcing network. These 3D cultures were maintained for 14 days post-injection and analyzed for cell viability, homogeneity, and morphology. All cultures remained proliferative with a homogeneous cell distribution, and minimal dead cells were observed within the hydrogels at all time points (Figure 2d, see Figure S6 for comparison images of cells cultured within SHIELD-0). Quantification of cell number after two weeks of culture suggests that proliferation within SHIELD-1 is statistically greater than that in SHIELD-0 (Figure S6, Supporting Information). Cells exhibited well-spread cytoskeletal morphologies with distinct actin filament networks within SHIELD-1. Together these data demonstrate that the double network hydrogels support cell encapsulation, delivery by injection, and cell proliferation and spreading in vitro.
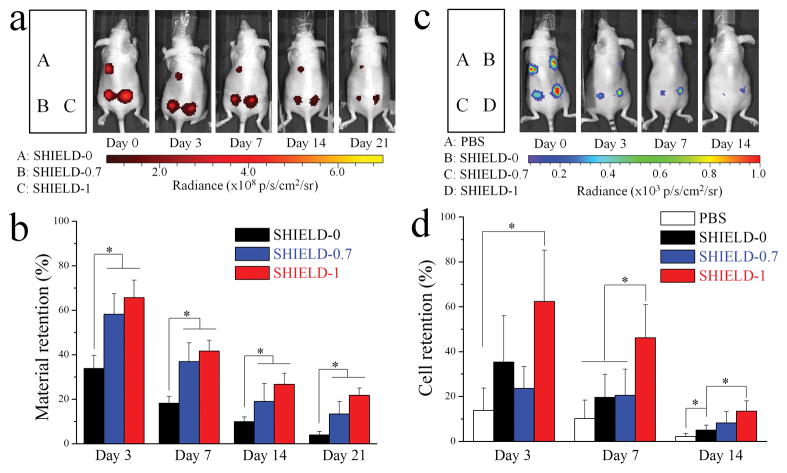
We next studied the ability of the hydrogels to improve cell transplantation efficiency in vivo. Immediately following encapsulation of hASCs within SHIELD-0, SHIELD-0.7, or SHIELD-1, they were injected subcutaneously into nude mice. Cell delivery in PBS was included as a control. All cell-embedded hydrogels were easily injectable under hand force through a 28-G needle and recovered as compact gel structures, which were visible and palpable as small nodules at the injection sites. Hydrogel material location and retention were tracked non-invasively by labeling the C7 polymer with a near-infrared fluorescence dye prior to injection (Figure 3a). At day 3, only ~30% of the SHIELD-0 material was retained (Figure 3b, further statistical analysis provided in Figure S7, Supporting Information). In contrast, ~60% of the SHIELD-1 and SHIELD-0.7 materials remained at day 3. This statistically significant trend was observed across all time points during the 3-week experiment. These data are consistent with our in vitro hydrogel erosion results and confirm that the PNIPAM network efficiently enhances material retention time in vivo.
Figure 3.
Material and cell retention after in vivo subcutaneous injection of hASCs within SHIELD-0, SHIELD-0.7, and SHIELD-1. (a) Fluorescence images of hydrogels conjugated with near-infrared dye at 0, 3, 7, 14, and 21 days post-injection. (b) Fluorescence imaging quantification of material retention relative to day 0. * p < 0.05, n = 5. (c) Bioluminescence images (BLI) of hASCs at 0, 3, 7, and 14 days post-injection. (d) BLI quantification of viable cell retention relative to day 0. * p < 0.05, n = 4.
To evaluate our hypothesis that decreased biodegradation rates would result in enhanced cell retention, viable transplanted cells were non-invasively quantified for 14 days post-injection using bioluminescence imaging (BLI). The hASCs were engineered to secrete firefly luciferase, which catalyzes emission of a bioluminescence signal upon reaction of D-luciferin.[37] By day 3, only ~13% of hASCs transplanted using PBS as a delivery vehicle were still metabolically active (Figure 3c,d, further statistical analysis provided in Figure S8, Supporting Information). In contrast, SHIELD-1 significantly enhanced cell retention, with ~60% viable cells at day 3. It should be noted that this in vivo evaluation does not take into account the acute cell-protective properties of the hydrogel. All intensities are reported relative to day 0 values post-injection, when presumably a significant percentage of PBS-delivered cells had already died due to acute membrane damage during injection. Therefore, these increases in cell numbers may be due to decreased cell apoptosis, decreased cell migration away from the injection site, and/or enhanced cell proliferation. At day 7, double-network SHIELD-1 improved cell retention 5-fold and 2.5-fold compared to PBS and single-network SHIELD-0, respectively. This effect was further enhanced at day 14, when SHIELD-1 cell retention was 6-fold and 3-fold higher than PBS and SHIELD-0, respectively. These results are consistent with our hypothesis and demonstrate that formation of a secondary, thermo-responsive network within an existing physical network can significantly promote long-term retention of metabolically active, transplanted cells at the target site.
Regardless of cell type, transplantation model, and methods to quantify cell retention, it has been widely found that only a small fraction of transplanted cells are retained acutely.[2, 4, 38] As a result, multiple cell injections are often required to achieve functional recovery, although the optimal cell number required for specific cell-based therapies remains to be determined.[39] The survival and retention time of the transplanted stem cells delivered within SHIELD-1 are significantly better than many other injectable, single-network hydrogels, which typically result in a substantial loss of 90 - 99% of transplanted cells in one to two weeks post-transplantation.[10, 12, 16, 20] For example, hASCs encapsulated and injected within a widely-used Type I collagen gel had < 5% retention at day 7 post-injection.[16] The survival of neural progenitor cells transplanted within a hyaluronan-heparin-collagen hydrogel into the cavity of stroked rat brain was modestly increased compared to culture media (cell survival at day 14 of 8% vs. 4%, hydrogel vs. culture media, respectively).[20] In another study, transplantation of cardiomyoblasts into ischemic heart resulted in < 5% survival when delivered within a collagen matrix and < 10% survival within a Matrigel/collagen matrix at day 14 post-injection.[10] The key role of hASCs in functional recovery has been hypothesized to be the secretion of paracrine factors that promote endogenous cell function.[40] Because SHIELD delivery significantly enhances hASC retention at the desired site, fewer transplanted cells and fewer cell injections will be needed to achieve an equivalent level of secreted paracrine factors and hence therapeutic efficacy. Thus SHIELD use will minimize the number of cells required for transplantation, decreasing the expense, time, and complexity of ex vivo expansion of patient-isolated cells. Furthermore, because of the highly tunable nature of the SHIELD system, the material degradation lifetime can be tailored to match the requirements for various endogenous regeneration processes.
In summary, we have developed a physically-crosslinked, double-network hydrogel to address the problem of post-transplantation cell death and to potentially eliminate a major bottleneck in regenerative therapy, as clinical outcome is contingent on the number of viable donor cells. We demonstrated that SHIELD undergoes two distinct crosslinking mechanisms that simultaneously address two of the major causes of transplanted cell death by providing (i) protection from the mechanically disruptive forces experienced during syringe-needle flow and (ii) a highly tunable 3D microenvironment that supports cell survival and retention post-injection. By minimizing the number of transplanted cells required for therapeutic efficacy, SHIELD use will decrease the cost, duration, and complexity of cell-based regenerative medicine therapies.
Experimental Section
Material synthesis
8-arm polyethylene glycol vinyl sulfone (8-arm PEG-VS) with nominal molecular weights of 20,000 g/mol were purchased from Nanocs (Boston, MA). Peptide P1 (EYPPYPPPPYPSGC, 1563 g/mol) was purchased through custom peptide synthesis from Genscript Corp (Piscataway, NJ, USA). All other chemicals were purchased from Sigma-Aldrich (Milwaukee, WI) unless otherwise noted. Amine-terminated PNIPAM was fractionated by dissolving in acetone and stepwise precipitation upon addition of hexane. The high molecular weight fraction had a weight-average molecular weight (Mw) of 10,650 g/mol with a polydispersity index (PDI) of 1.2 as determined by gel permeation chromatography (GPC). GPC was carried out at room temperature in tetrahydrofuran (THF) as the eluent at a flow rate of 1.0 mL/min using a Viscotek chromatograph and a Viscotek S3580 refractive index detector (Houston, TX) and standard monodisperse polystyrenes for calibration. A Michael-type addition of amine-terminated PNIPAM to 8-arm PEG-VS was conducted at an amine:VS ratio of 1:8 in a Schlenk tube at pH 9.5 and 0.5 M triethylamine at 25 °C for 24 h. Then the rest of the unreacted arms of PEG-VS were further reacted with excess P1 in the presence of tris(2-carboxyethyl)phosphine (TCEP). This PEG-PNIPAM-P1 copolymer solution was lyophilized, washed with chloroform to remove unreacted PEG, and then dialyzed (Molecular Weight Cut-Off = 15,000 g/mol) against deionized water (pH 7.4) to remove unreacted PNIPAM and P1. For comparison, PEG-P1 copolymer was synthesized by reacting 8-arm PEG-VS with excess P1 and purified as described above. The chemical structures were confirmed by 1H NMR spectrometry, acquired on a Varian Inova 500 MHz NMR spectrometer using deuterium oxide as a solvent (Figure S1, Supporting Information). 1H NMR (500 MHz, D2O) for PEG-VS: δ = 6.7, 6.3 (m, H2C=), 6.1 (d, =CH-SO2), 3.6 (PEG backbone protons); PEG-VS-PNIPAM: δ = 6.7, 6.3 (m, H2C=), 6.1 (d, =CH-SO2), 3.6 (PEG backbone protons), 1.8, 1.4 (PNIPAM backbone protons), 1.0 (s, -C(CH3)2); PEG-PNIPAM-P1: δ = 7.0, 6.7 (tyrosine aromatic protons), 3.6 (PEG backbone protons), 1.8, 1.4 (PNIPAM backbone protons), 1.0 (s, -C(CH3)2). The degree of end-group conversion was 74% for PNIPAM and 90% for P1. To determine the Mw and PDI, GPC was performed as described above. The C7 recombinant protein polymer (Table S1 for full amino acid sequence, Supporting Information) was cloned, synthesized, and purified as reported previously.[17] Briefly, the DNA sequence encoding the C7 linear protein block copolymer was cloned into the pET-15b vector (Novagen) and transformed into the BL21(DE3)pLysS Escherichia coli host strain (Life Technologies). The protein was expressed following isopropyl β-D-1-thiogalactopyranoside (IPTG) induction, purified by affinity chromatography via the specific binding of N-terminal polyhistidine tag to Ni-nitrilotriacetic acid resin (Qiagen), dialyzed against phosphate-buffered saline (PBS), and concentrated by diafiltration across Amicon Ultracel filter units (Millipore).
Hydrogel preparation
Each WW domain in C7 was treated as one C unit, and each pendant peptide group in the PEG-PNIPAM-P1 copolymer was treated as one P unit. Weight percentage of PNIPAM component was used to name various SHIELD formulations. SHIELD-1 (SHIELD with 1 wt% PNIPAM moiety) was formed by mixing C7 and PEG-PNIPAM-P1 copolymer to achieve a C:P ratio of 1:1 and a final concentration of 10% w/v in PBS (Figure 1a). Similarly, SHIELD-0 (SHIELD with 0 wt% PNIPAM moiety) was formed by mixing C7 and PEG-P1 copolymer to achieve the same final concentration of 10% w/v and C:P ratio of 1:1. PEG-PNIPAM-P1 and PEG-P1 copolymers were also mixed at a weight ratio of 70/30 and then mixed with C7 to prepare a 10% w/v gels named SHIELD-0.7, which contains 0.7 wt% PNIPAM.
Rheological characterization
Dynamic oscillatory rheology experiments were performed on a stress-controlled rheometer (AR-G2, TA instrument, New Castle, DE) using a 25-mm diameter cone-plate geometry (n ≥ 3). Samples were loaded immediately onto the rheometer after mixing and a humidity chamber was secured in place to prevent dehydration. Frequency sweeps from 0.1 – 20 Hz at 25 and 37 °C were performed at 5% constant strain to obtain storage moduli (G′) and loss moduli (G″). Shear-thinning and self-healing properties of the gel samples were characterized by measuring linear viscosity (η) under a time sweep mode at alternating low and high shear rates of 0.1 s−1 and 10 s−1, respectively, for 30 s each and a total of 150 s.
FRAP diffusivity measurement
FRAP measurement was performed using dextran (molecular weight = 40 kg/mol) conjugated to fluorescein isothiocyanate (FITC) (Invitrogen). Dextran was mixed individually with various SHIELD formulations (gel volume = 30 μL, concentration = 10% w/v, C:P ratio = 1:1) at a final dextran concentration of 4 mg/mL. The total fluorescence intensity was visualized at 37 °C using a Leica TCS SP5 confocal microscope at low light intensity. Photobleaching was then conducted by exposing a 100 × 100 μm spot in the field of view to a high intensity laser light. A series of images were taken every three seconds for five minutes to track the recovery of dextran fluorescence (n ≥ 3). Measurements of the control samples were performed using PBS, rat-tail Type I collagen (2 mg/mL, BD Biosciences), and growth-factor-reduced Matrigel (BD Biosciences) as diffusion media, prepared according to manufacturers’ protocols. The resulting fluorescence recovery profiles were modeled by Fickian diffusion according to a previously reported protocol to calculate dextran diffusivities.[42]
Hydrogel erosion kinetics
Various SHIELD formulations were formed in circular silicone molds (diameter = 4 mm, height = 2 mm) within a 24-well plate (n ≥ 3). The mixture was allowed to undergo sol-gel phase transition for 15 min at 37 °C in a humidified incubator. Then 1.5 mL of PBS was added to each well. Volumes of 100 μL were sampled from the PBS supernatant of each well and replenished with 100 μL of fresh PBS over a period of 14 days. Gel erosion kinetics were analyzed by using absorbance spectroscopy at 280 nm (SpectraMax M2 Spectrophotometer, Molecular Devices) to measure the amount of protein released into the supernatant at each time point. All values were normalized to that of the initial hydrogel directly disrupted and solubilized in PBS.
In vitro cell injection and quantification of viability
hASCs were obtained from de-identified human lipoaspirate from the flank and thigh regions by suction assisted liposuction. All tissue donors responded to an Informed Consent approved by the Stanford Institutional Review Board. hASCs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 IU/ml penicillin/streptomycin at 37 °C and 5% atmospheric CO2. Cells were expanded and passaged by trypsinization for subsequent use. All in vitro injection experiments were performed with 30-μl gel volume containing 5 × 104 cells. Cell suspension (5 μl) was first mixed with the 8-arm PEG-PNIPAM-P1 copolymer solution (20% w/v in PBS) and PBS before further mixing with C7 (10% w/v in PBS). The volumes of PEG-PNIPAM-P1 copolymer solution and C7 were adjusted to achieve a final C:P ratio of 1:1 at a total cell-laden hydrogel concentration of 10% w/v. For cell injection studies, the final mixing step with C7 was performed in the barrel of a 1-mL insulin syringe fitted with a 28 G needle. The mixture was allowed to gel for 5 min before injecting into a circular silicone mold (diameter = 4 mm, height = 2 mm) within a 24-well plate using a syringe pump (SP220I; World Precision Instruments) at a flow rate of 1 mL/min. Cell viability was determined using LIVE/DEAD viability/cytotoxicity kit (Invitrogen) immediately post-injection and at days 1, 4, 7, and 14 post-injection (n = 5), according to manufacturer’s instructions. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 solution in PBS, and stained with rhodamine phalloidin (1:300, Life Technologies) and 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/mL, Life Technologies). Images were collected using the Leica confocal microscope by creating z-stacks of greater than 200-μm depth with 2.4-μm intervals between slices in the middle of the hydrogel and then compressing into a maximum projection image. Cell number was quantified from confocal images at each time point.
In vivo transplantation and bioluminescence imaging
All experiments followed protocols approved by the Stanford Administrative Panel on Laboratory Animal Care. NIH guidelines for the care and use of laboratory animals (NIH Publication #85-23 Rev. 1985) were observed. To track the material retention, C7 protein was labeled with Cyanine5.5 NHS ester, which is a near-infrared emitting dye (Lumiprobe), according to the manufacturer’s protocol. To track the cell retention, hASCs were transduced with lentivirus particles expressing the firefly luciferase gene (Qiagen). For in vivo transplantation, athymic nude mice (25–30 g, male, Charles River Laboratories) were anesthetized with isoflurane, and hydrogel samples (30 μl total with 5 × 105 cells) were hand-injected subcutaneously via an insulin syringe with a 28 G needle. hASCsFluc+ resuspended at the same concentration in saline (30 μl) were also injected as controls. To minimize location specific bias, injection sites were randomized and rotated across the four injection sites per mouse. To monitor cell viability and distribution, bioluminescence imaging (BLI) was performed with an IVIS imaging system (Xenogen Corp.), and data was acquired with LivingImage™ software (Xenogen Corp.) on days 0, 3, 7, and 14. Before imaging, mice were anesthetized with 2% isoflurane/air. Reporter probe D-luciferin was administered via intraperitoneal injection at a dose of 350 mg/kg body weight. BLI was acquired at 5-min intervals with an exposure time of 30 sec. For each image acquisition, a gray scale body surface image was collected, followed by an overlay of the pseudo-colored image of photon counts from active luciferase within the mouse. Image acquisition continued until all samples had reached peak intensity (10–40 min). Signal intensity for each sample was quantified as total flux (photons/sec) within a region of interest at peak intensity (n = 5). Fluorescence images of Cyanine5.5 dye were also taken with an exposure time of 0.5 sec at each time point using the Cy5.5 filter sets (excitation: 673 nm, emission: 707 nm) and their intensities were quantified using the same software (n = 4). All values were normalized to day 0.
Statistical analysis
All data are presented as mean ± standard deviation. Statistical comparisons were performed by one-way analysis of variance (ANOVA) with Tukey post-hoc test on diffusivity and hydrogel erosion results. Two-way ANOVA with Tukey post-hoc test was performed on cell viability, cell number, shear moduli and in vivo results containing two independent variables. Values were considered to be significantly different when the p value was <0.05.
Supplementary Material
Acknowledgments
This study was supported by grants from NSF (DMR-0846363), NIH (R01-DK085720, DP2-OD006477), CIRM (RT2-01938), and Coulter Foundation (CP-2014-4). The authors thank Karen Dubbin for FRAP method optimization, Tyler Stukenbroeker and Prof. Robert Waymouth for use of GPC, and Andreina Parisi Amon, Allison Nauta, Benjamin Levi, and Prof. Michael Longaker for hASC isolation.
Footnotes
Supporting Information is available from the Wiley Online Library.
Contributor Information
Dr. Lei Cai, Department of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA
Ruby E. Dewi, Department of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA
Prof. Sarah C. Heilshorn, Email: heilshorn@stanford.edu, Department of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA. Department of Bioengineering, Stanford University, Stanford, CA 94305, USA
References
- 1.Raval Z, Losordo DW. Circ Res. 2013;112:1288. doi: 10.1161/CIRCRESAHA.113.300565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terrovitis JV, Smith RR, Marban E. Circ Res. 2010;106:479. doi: 10.1161/CIRCRESAHA.109.208991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Nat Biotechnol. 2007;25:1015. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 4.Bliss T, Guzman R, Daadi M, Steinberg GK. Stroke. 2007;38:817. doi: 10.1161/01.STR.0000247888.25985.62. [DOI] [PubMed] [Google Scholar]
- 5.Walsh S, Midha R. Neurosurgical focus. 2009;26:E2. doi: 10.3171/FOC.2009.26.2.E2. [DOI] [PubMed] [Google Scholar]
- 6.Macaya D, Spector M. Biomedical materials. 2012;7:012001. doi: 10.1088/1748-6041/7/1/012001. [DOI] [PubMed] [Google Scholar]; Cooke MJ, Vulic K, Shoichet MS. Soft Matter. 2010;6:4988. [Google Scholar]
- 7.Dib N, Khawaja H, Varner S, McCarthy M, Campbell A. J Cardiovasc Transl. 2011;4:177. doi: 10.1007/s12265-010-9253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguado BA, Mulyasasmita W, Su J, Lampe KJ, Heilshorn SC. Tissue Eng Part A. 2012;18:806. doi: 10.1089/ten.tea.2011.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMarco RL, Su J, Yan KS, Dewi R, Kuo CJ, Heilshorn SC. Integrative biology: quantitative biosciences from nano to macro. 2014;6:127. doi: 10.1039/c3ib40188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutschka I, Chen IY, Kofidis T, Arai T, von Degenfeld G, Sheikh AY, Hendry SL, Pearl J, Hoyt G, Sista R, Yang PC, Blau HM, Gambhir SS, Robbins RC. Circulation. 2006;114:I167. doi: 10.1161/CIRCULATIONAHA.105.001297. [DOI] [PubMed] [Google Scholar]
- 11.Christman KL, Vardanian AJ, Fang Q, Sievers RE, Fok HH, Lee RJ. J Am Coll Cardiol. 2004;44:654. doi: 10.1016/j.jacc.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 12.Liang Y, Walczak P, Bulte JWM. Biomaterials. 2013;34:5521. doi: 10.1016/j.biomaterials.2013.03.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Li W, Johnson S, Ingram D, Yoder M, Badylak S. Endothelium: journal of endothelial cell research. 2004;11:199. doi: 10.1080/10623320490512390. [DOI] [PubMed] [Google Scholar]
- 14.Bakota EL, Wang Y, Danesh FR, Hartgerink JD. Biomacromolecules. 2011;12:1651. doi: 10.1021/bm200035r. [DOI] [PMC free article] [PubMed] [Google Scholar]; Webber MJ, Tongers J, Renault MA, Roncalli JG, Losordo DW, Stupp SI. Acta Biomaterialia. 2010;6:3. doi: 10.1016/j.actbio.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, Lamm MS, Pochan DJ, Schneider JP. Proc Natl Acad Sci U S A. 2007;104:7791. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yan C, Altunbas A, Yucel T, Nagarkar RP, Schneider JP, Pochan DJ. Soft Matter. 2010;6:5143. doi: 10.1039/C0SM00642D. [DOI] [PMC free article] [PubMed] [Google Scholar]; Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 15.Glassman MJ, Chan J, Olsen BD. Advanced Functional Materials. 2013;23:1182. doi: 10.1002/adfm.201202034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parisi-Amon A, Mulyasasmita W, Chung C, Heilshorn SC. Adv Healthc Mater. 2013;2:428. doi: 10.1002/adhm.201200293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong Po Foo CT, Lee JS, Mulyasasmita W, Parisi-Amon A, Heilshorn SC. Proc Natl Acad Sci U S A. 2009;106:22067. doi: 10.1073/pnas.0904851106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mulyasasmita W, Cai L, Dewi RE, Jha A, Ullmann SD, Luong RH, Huang NF, Heilshorn SC. Journal of controlled release: official journal of the Controlled Release Society. 2014 doi: 10.1016/j.jconrel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu HD, Charati MB, Kim IL, Burdick JA. Biomaterials. 2012;33:2145. doi: 10.1016/j.biomaterials.2011.11.076. [DOI] [PubMed] [Google Scholar]
- 19.Lu HD, Soranno DE, Rodell CB, Kim IL, Burdick JA. Adv Healthc Mater. 2013;2:1028. doi: 10.1002/adhm.201200343. [DOI] [PubMed] [Google Scholar]
- 20.Zhong J, Chan A, Morad L, Kornblum HI, Fan G, Carmichael ST. Neurorehabil Neural Repair. 2010;24:636. doi: 10.1177/1545968310361958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen W, Zhang K, Kornfield JA, Tirrell DA. Nat Mater. 2006;5:153. doi: 10.1038/nmat1573. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Ni X, Leong KW. Journal of biomedical materials research Part A. 2003;65:196. doi: 10.1002/jbm.a.10444. [DOI] [PubMed] [Google Scholar]
- 23.Wheeldon IR, Barton SC, Banta S. Biomacromolecules. 2007;8:2990. doi: 10.1021/bm700858p. [DOI] [PubMed] [Google Scholar]; Anderson BC, Pandit NK, Mallapragada SK. J Control Release. 2001;70:157. doi: 10.1016/s0168-3659(00)00341-2. [DOI] [PubMed] [Google Scholar]
- 24.Singelyn JM, Sundaramurthy P, Johnson TD, Schup-Magoffin PJ, Hu DP, Faulk DM, Wang J, Mayle KM, Bartels K, Salvatore M, Kinsey AM, Demaria AN, Dib N, Christman KL. J Am Coll Cardiol. 2012;59:751. doi: 10.1016/j.jacc.2011.10.888. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robertson MJ, Dries-Devlin JL, Kren SM, Burchfield JS, Taylor DA. PLoS One. 2014;9:e90406. doi: 10.1371/journal.pone.0090406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kharkar PM, Kiick KL, Kloxin AM. Chem Soc Rev. 2013;42:7335. doi: 10.1039/c3cs60040h. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Y, Rodrigues J, Tomas H. Chemical Society Reviews. 2012;41:2193. doi: 10.1039/c1cs15203c. [DOI] [PubMed] [Google Scholar]; Nguyen MK, Alsberg E. Progress in Polymer Science. 2014;39:1235. doi: 10.1016/j.progpolymsci.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guvendiren M, Lu HD, Burdick JA. Soft Matter. 2012;8:260. [Google Scholar]
- 27.Yamaguchi H, Kobayashi Y, Kobayashi R, Takashima Y, Hashidzume A, Harada A. Nat Commun. 2012;3:603. doi: 10.1038/ncomms1617. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harada A, Kataoka K. Progress in Polymer Science. 2006;31:949. [Google Scholar]
- 28.Yamaguchi N, Zhang L, Chae BS, Palla CS, Furst EM, Kiick KL. J Am Chem Soc. 2007;129:3040. doi: 10.1021/ja0680358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klouda L, Mikos AG. Eur J Pharm Biopharm. 2008;68:34. doi: 10.1016/j.ejpb.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comolli N, Neuhuber B, Fischer I, Lowman A. Acta Biomater. 2009;5:1046. doi: 10.1016/j.actbio.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ohya S, Nakayama Y, Matsuda T. Journal of artificial organs: the official journal of the Japanese Society for Artificial Organs. 2004;7:181. doi: 10.1007/s10047-004-0265-9. [DOI] [PubMed] [Google Scholar]
- 30.Zuidema JM, Rivet CJ, Gilbert RJ, Morrison FA. Journal of Biomedical Materials Research Part B: Applied Biomaterials. 2014;102:1063. doi: 10.1002/jbm.b.33088. [DOI] [PubMed] [Google Scholar]
- 31.Gefen A, Cornelissen LH, Gawlitta D, Bader DL, Oomens CW. J Biomech. 2008;41:845. doi: 10.1016/j.jbiomech.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 32.Leddy H, Guilak F. Annals of Biomedical Engineering. 2003;31:753. doi: 10.1114/1.1581879. [DOI] [PubMed] [Google Scholar]
- 33.Mulyasasmita W, Lee JS, Heilshorn SC. Biomacromolecules. 2011;12:3406. doi: 10.1021/bm200959e. [DOI] [PMC free article] [PubMed] [Google Scholar]; Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Nature. 2005;437:579. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 34.Pham TTH, Skrzeszewska PJ, Werten MWT, Rombouts WH, Cohen Stuart MA, de Wolf FA, van der Gucht J. Soft Matter. 2013;9:6391. [Google Scholar]
- 35.Yan C, Mackay ME, Czymmek K, Nagarkar RP, Schneider JP, Pochan DJ. Langmuir. 2012;28:6076. doi: 10.1021/la2041746. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yan C, Pochan DJ. Chemical Society Reviews. 2010;39:3528. doi: 10.1039/b919449p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nauta A, Seidel C, Deveza L, Montoro D, Grova M, Ko SH, Hyun J, Gurtner GC, Longaker MT, Yang F. Molecular therapy: the journal of the American Society of Gene Therapy. 2013;21:445. doi: 10.1038/mt.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]; de Villiers JA, Houreld N, Abrahamse H. Stem cell reviews. 2009;5:256. doi: 10.1007/s12015-009-9084-y. [DOI] [PubMed] [Google Scholar]; Aguena M, Fanganiello RD, Tissiani LA, Ishiy FA, Atique R, Alonso N, Passos-Bueno MR. Stem cells international. 2012;2012:303610. doi: 10.1155/2012/303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Almeida PE, van Rappard JRM, Wu JC. American Journal of Physiology-Heart and Circulatory Physiology. 2011;301:H663. doi: 10.1152/ajpheart.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blocklet D, Toungouz M, Berkenboom G, Lambermont M, Unger P, Preumont N, Stoupel E, Egrise D, Degaute JP, Goldman M, Goldman S. Stem Cells. 2006;24:333. doi: 10.1634/stemcells.2005-0201. [DOI] [PubMed] [Google Scholar]; Teng CJ, Luo J, Chiu RC, Shum-Tim D. The Journal of thoracic and cardiovascular surgery. 2006;132:628. doi: 10.1016/j.jtcvs.2006.05.034. [DOI] [PubMed] [Google Scholar]; Hicks AU, Lappalainen RS, Narkilahti S, Suuronen R, Corbett D, Sivenius J, Hovatta O, Jolkkonen J. The European journal of neuroscience. 2009;29:562. doi: 10.1111/j.1460-9568.2008.06599.x. [DOI] [PubMed] [Google Scholar]
- 39.Hu X, Wang J, Chen J, Luo R, He A, Xie X, Li J. European journal of cardio-thoracic surgery: official journal of the European Association for Cardio-thoracic Surgery. 2007;31:438. doi: 10.1016/j.ejcts.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 40.Gimble JM, Katz AJ, Bunnell BA. Circ Res. 2007;100:1249. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu F, Mizuno H, Uysal CA, Cai X, Ogawa R, Hyakusoku H. Plastic and Reconstructive Surgery. 2008;121:50. doi: 10.1097/01.prs.0000293876.10700.b8. [DOI] [PubMed] [Google Scholar]; Garcia-Olmo D, Herreros D, Pascual I, Antonio Pascual J, Del-Valle E, Zorrilla J, De-La-Quintana P, Garcia-Arranz M, Pascual M. Diseases of the Colon & Rectum. 2009;52:79. doi: 10.1007/DCR.0b013e3181973487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.