Abstract
The genus Streptobacillus (S.) remained monotypic for almost 90 years until two new species were recently described. The type species, S. moniliformis, is one of the two etiological agents of rat bite fever, an under-diagnosed, worldwide occurring zoonosis. In a polyphasic approach field isolates and reference strains of S. moniliformis, S. hongkongensis, S. felis as well as divergent isolates were characterized by comparison of molecular data (n = 29) and from the majority also by their physiological as well as proteomic properties (n = 22). Based on growth-independent physiological profiling using VITEK2-compact, API ZYM and the Micronaut system fastidious growth-related difficulties could be overcome and streptobacilli could definitively be typed despite generally few differences. While differing in their isolation sites and dates, S. moniliformis isolates were found to possess almost identical spectra in matrix-assisted laser desorption ionization—time of flight mass spectrometry and Fourier transform infrared spectroscopy. Spectroscopic methods facilitated differentiation of S. moniliformis, S. hongkongensis and S. felis as well as one divergent isolate. Sequencing of 16S rRNA gene as well as functional genes groEL, recA and gyrB revealed only little intraspecific variability, but generally proved suitable for interspecies discrimination between all three taxa and two groups of divergent isolates.
Introduction
For almost 90 years, the genus Streptobacillus [1] (S.; Leptotrichiaceae, Fusobacteriales) comprised the monotypic S. moniliformis, one of the two etiological organisms of rat bite fever (RBF) [2, 3]. Beside RBF S. moniliformis causes also Haverhill fever (HF), which represent two rarely observed syndromes of this worldwide occurring bacterial zoonosis [3]. The infection is predominantly transmitted through rat bites, scratches or by direct or indirect contact with rat urine [4–6]. Approximately 50–100% of wild rats carry S. moniliformis in their oro- or nasopharynx and shed the organism with saliva and urine [2, 7, 8]. Despite its wide distribution in natural hosts S. moniliformis is a rarely detected and most likely under-reported pathogen in humans and animals [3]. In the last few years Streptobacillus-like organisms have been observed beside S. moniliformis, from which S. hongkongensis [9] and S. felis [10] were recently described as new species causing quinsy and septic arthritis in humans and pneumonia in a cat, respectively. Furthermore, Streptobacillus spp. were reported from a canine oral microbiome project (sequence COT-370; [11]) as well as from Japanese rats [12]. The present study aimed to compare 29 field isolates and reference strains of S. moniliformis, S. hongkongensis, S. felis as well as two yet undescribed species from various geographic areas, host species and isolation sites with respect to phenotypic and molecular properties. In contrast to earlier investigations [13–15] a spectrum of methods was employed to overcome known diagnostic difficulties due to the fastidious growth of this microorganism.
Materials and Methods
Bacterial strains
The present study included 29 members of the genus Streptobacillus from different host species which cover isolations of S. moniliformis over the past 90 years from La Réunion near Africa, Asia, Australia, Europe and North America as well as type strains of S. moniliformis, S. hongkongensis and S. felis (Table 1). Eight field isolates and reference strains (“strains” hereafter) were associated with human infections; 17 were obtained from rodents, i.e. 12 originated from rats, four from mice and one strain was derived from a spinifex hopping mouse (Notomys alexis); three strains were isolated from turkeys and one strain was isolated from a cat. From seven Japanese rat strains there was only DNA available. The S. moniliformis strains associated with human disease were isolated from cases of RBF (n = 5), HF (n = 1) and unknown origin (n = 1) (Table 1).
Table 1. Field isolates and reference strains as well as origins, clinical symptoms and host species of Streptobacillus moniliformis, Streptobacillus spp. and Sebaldella termitidis used in this study.
| Strain no. | Strain designation | Species | Year of isolation | Host | Clinic / sample | Country | Strain reference |
|---|---|---|---|---|---|---|---|
| 1 | DSM 12112 T (= ATCC 14647 T ) | S. moniliformis | 1925 | Human | RBF | France | [1] |
| 2 | CIP 55–48 | S. moniliformis | 1947 | Mouse | Lymph adenitis | UK | |
| 3 | ATCC 27747 | S. moniliformis | 1964 | Turkey | Septic arthritis | USA | [16] |
| 4 | NCTC 10773 | S. moniliformis | 1971 | Human | Blood culture | UK | |
| 5 | NCTC 11194 | S. moniliformis | 1977 | Human | RBF | UK | |
| 6 | AHL 370–1 | Streptobacillus sp. | 1979 | Spinifex hopping mouse | Liver | Australia | [17] |
| 7 | IPDH 144/80 | S. moniliformis | 1980 | Turkey | Septic arthritis | Germany | |
| 8 | CIP 81–99 | S. moniliformis | 1981 | Human | Blood culture (wild rat bite) | France | |
| 9 | AHL 370–4 | S. moniliformis | 1982 | Mouse | Ear infection | Australia | |
| 10 | NCTC 11941 | S. moniliformis | 1983 | Human | Haverhill fever | UK | |
| 11 | IPDH 109/83 | S. moniliformis | 1983 | Turkey | Septic arthritis | Germany | |
| 12 | ATCC 49567 | S. moniliformis | 1989 | Mouse | Lymph adenitis | Germany | [18] |
| 13 | Kun 3 (RIVM) | S. moniliformis | 1991 | Rat | Healthy | The Netherlands | [19] |
| 14 | ATCC 49940 | S. moniliformis | 1992 | Rat | Otitis media | Germany | [20] |
| 15 | B10/15 | S. moniliformis | unknown | Wild rat | Unknown | The Netherlands | |
| 16 | A378/1 | S. moniliformis | 1995 | Wild rat | Vaginal swab | Germany | DKFZ strain collection |
| 17 | VA11257/2007 | S. moniliformis | 2007 | Human (farmer) | RBF, endocarditis | Germany | [21] |
| 18 | VK105/14 | S. moniliformis | 2008 | Domestic rat | Abscess | Germany | TiHo strain collection |
| 19 | B5/1 | S. moniliformis | 2009 | Laboratory mouse | After rat bite | Germany | DKFZ strain collection |
| 20 | Marseille | S. moniliformis | 2009 | Rat | RBF | La Réunion | [22] |
| 21 | IKC1 | S. moniliformis | Rat | Oral swab | Japan | AB330754, inactivated (DNA); [12] | |
| 22 | IKC5 | S. moniliformis | Rat | Oral swab | Japan | AB330755, inactivated (DNA); [12] | |
| 23 | IKB1 | S. moniliformis | Rat | Oral swab | Japan | AB330756, inactivated (DNA); [12] | |
| 24 | TSD4 | S. moniliformis | Rat | Oral swab | Japan | AB330757, inactivated (DNA); [12] | |
| 25 | OGS16 | Streptobacillus sp. | Rat | Oral swab | Japan | AB330758, inactivated (DNA); [12] | |
| 26 | KWG2 | Streptobacillus sp. | Rat | Oral swab | Japan | AB330759, inactivated (DNA); [12] | |
| 27 | KWG24 | Streptobacillus sp. | Rat | Oral swab | Japan | AB330760, inactivated (DNA); [12] | |
| 28 | 131000547 T (DSM 29248 T ) | S. felis | 2013 | Cat | Pneumonia | Germany | [10] |
| 29 | DSM 26322 T (HKU33 T ) | S. hongkongensis | 2014 | Human | Abscess | Hong Kong | [9] |
| 30 | NCTC 11300 T (ATCC 33386 T ) | Sebaldella termitidis | 1962 | Termite | Intestine |
T: type strain; ATCC: American Type Culture Collection, Rockville, USA; NCTC: National Collection of Type Cultures, London, UK; CIP: Collection Institut Pasteur, Paris, France; IPDH: Institut für Geflügelkrankheiten, Hannover, Germany; RIVM: Rijksinstituut voor Volksgezondheid en Milieuhygiene, Bilthoven, The Netherlands; AHL: Animal Health Laboratory, South Perth, Australia; ZfV: Zentralinstitut für Versuchstierzucht, Hannover, Germany; DKFZ: Deutsches Krebsforschungszentrum, Heidelberg, Germany; TiHo: Tierärztliche Hochschule Hannover, Germany; RBF: rat bite fever
Phenotypic characterization
Culture requirements
For the growth of Streptobacillus spp. Columbia agar supplemented with 5% sheep blood (Oxoid, Wesel, Germany; SBA) was incubated for 2–5 days at 37°C in a capnophilic atmosphere of 10% CO2. Liquid media (brain-heart infusion and peptone broth, supplemented with 20% bovine or horse serum [all Oxoid]) were used for streptobacillary growth and incubated for 2–7 days under the same conditions.
Biochemical properties
Extended biochemical profiling was carried out according to the manufacturer’s instructions using commercial fermentative test systems, i.e. Micronaut Strep2 and RPO (Merlin Diagnostika, Bornheim-Hersel, Germany; [23]), VITEK2-compact with the NHI card and API ZYM (both bioMeriéux, Nürtingen, Germany; Table 2).
Table 2. Physiological characteristics of field isolates and reference strains from Streptobacillus moniliformis and of reference strains from Streptobacillus felis 131000547T, Streptobacillus hongkongensis DSM 26322T and Sebaldella termitidis NCTC 11300T.
| Strain no. | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 28 | 29 | 30 |
| haemolysis on SBA | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - | + | + | - |
| tripeptidase* # | + | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + | - | + |
| prolin aminopeptidase* # | + | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + | - | - |
| hydroxyprolin aminopeptidase* # | + | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + | - | - |
| glycyltryptophan aminopeptidase* # | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - § | - |
| arginine aminopeptidase* # | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + | - | + | - | - |
| pyrase* # | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - § | - |
| neuraminidase* # | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + | w | + | - | + | + | - | - | - |
| arginine dihydrolase* # | + | + | + | + | + | + | + | + | - | + | - | + | + | + | + | - | + | + | + | + | - | + | - |
| glycylprolin aminopeptidase* # | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| asparatyl aminopeptidase* # | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| growth in SPMS medium* # | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - |
| urease* | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | + | - | - | - | + | - | - |
| p-nitrophenyl-β-D-glucuronide* | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - |
| esculine* | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - | - | + |
| phenylalanine arylamidase † | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - § | - § | - |
| L-pyrrolidonyl arylamidase † | + | - | - | + | - | - | + | + | + | + | + | + | + | - | + | + | + | - | + | + | - | - | - |
| phospahatase † | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + § | - |
| tyrosine arylamidase † | + | + | + | + | + | w | + | + | + | + | + | + | + | + | + | + | + | w | + | + | - § | - § | - |
| ala-phe-pro-arylamidase † | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - § | - § | + |
| phenylphosphonate † | - | w | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + |
| D-mannose † | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + |
| N-acetyl-D-glucosamine † | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | w | - | - | - | + | + |
| D-glucose † | + | w | - | + | - | - | - | - | - | + | - | - | - | - | - | - | + | - | - | + | - | + | + |
| alkaline phosphatase ‡ | w | - | w | w | - | - | w | - | - | + | - | w | w | - | - | - | - | - | - | - | + | + | + |
| esterase (C4) ‡ | w | w | w | w | w | + | w | - | w | + | w | w | + | w | - | w | - | - | - | - | + | w | - |
| esterase lipase (C8) ‡ | + | + | + | + | + | + | w | w | + | + | + | + | + | + | w | + | w | w | w | w | + | w | - |
| lipase (C14) ‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| leucine arylamidase ‡ | w | - | w | w | - | w | w | w | - | w | w | + | - | - | - | - | - | - | - | - | - | - | - |
| valine arylamidase ‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| cystine arylamidase ‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| trypsin ‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| α-chymotrypsin ‡ | + | w | w | + | + | + | - | w | w | w | w | + | + | w | + | + | + | + | + | + | - | - | - |
| acid phosphatase ‡ | w | - | - | - | - | + | w | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | + |
| naphthol-AS-BI-phosphohydrolase ‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | w | w |
| β-Glucuronidase ‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| α-Glucosidase ‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
Physiological characteristics were obtained by an individual panel of eleven# discriminatory reactions designed for the identification of Streptobacillus spp. (Micronaut Strep2 and RPO; all Merlin Diagnostika GmbH)*, VITEK2-compact with the NHI card†, API-ZYM‡ (both bioMeriéux) and haemolytic properties on Columbia agar with 5% sheep blood; for congruent results see text; +: positive;-: negative; w: weak reaction;
§ potential discriminatory character for species identification
Presumptive physiological characterization further employed standard microbiological procedures: Haemolytic properties of the bacteria were observed on SBA. Tests for catalase activity were carried out with 3% H2O2 on microscopic slides and those for cytochrome oxidase with the BBL DrySlide system (Becton Dickinson, Heidelberg, Germany).
Antimicrobial susceptibility testing
The antimicrobial susceptibility pattern was determined using minimal inhibitory concentrations (MIC) obtained by broth microdilution test (Merlin Diagnostika) as described earlier [24]. Following the adaptation of the read-out system by using SPMS culture medium containing cattle serum and gelatine, the commercially available Micronaut-S Campylobacter (all Merlin Diagnostika) was carried out (S1 Table). The test design contained the following 12 antimicrobial substances: azithromycin (AZM), ciprofloxacin (CIP), clindamycin (CLI), chloramphenicol (CMP), erythromycin (ERY), gentamicin (GEN), meropenem (MER), nalidixic acid (NAL), streptomycin (STR), trimethoprim/sulfamethoxazol (T/S), telithromycin (TEL), and tetracycline (TET). Results were interpreted according to Clinical and Laboratory Standards Institute criteria [25].
Haemagglutination
Screening for adhesive properties was performed for 15 strains (No. 1–15 according to Table 1) by previously described haemagglutination experiments using erythrocytes from 11 different host species [26]. In detail, red blood cells from humans (blood type AB, rhesus factor positive), BALB/c and C57B1/6J mice, rats, turkeys, guinea-pigs, hamsters, chickens, sheep, horses, pigs and cattle were included [26]. For slide agglutination experiments defibrinated blood samples were diluted 1:4 in phosphate buffered saline (PBS). Colonies from a 24 h culture of S. moniliformis were used in a turbidity of McFarland 6 in 150 μl PBS. Strong reactions were read as agglutination after gently mixing 15 μl bacterial suspensions with 10 μl diluted blood samples after 30 sec. Delayed reactions were read after 2 min incubation on ice. Haemagglutination was also assessed in microtiter plates by adding the bacterial suspension from the slide agglutination experiment to a final volume of 60 μl per well thereby yielding blood dilutions of 1:7 and 1:10. Sealed plates were incubated for 24 h at 4°C. Haemagglutination was detected by semiquantitative reading as strong (++) or moderate/weak (+) shape of coat-forming layers on the well wall, whereas negative reactions caused sedimentation of erythrocytes on the bottom. E. coli KK 158/1 and a setup without bacteria served as positive and negative controls, respectively [26].
Matrix assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)
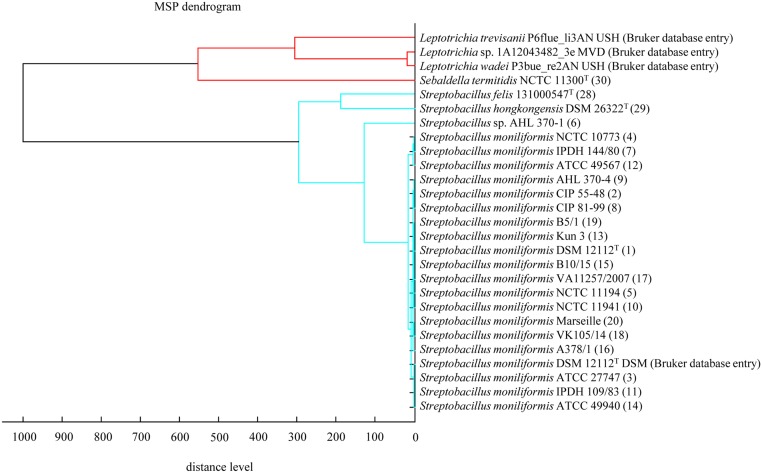
Bacteria were incubated for 24 h, subsequently selected from the SBA plates and subjected to steel targets according to manufacturer's instructions (BrukerBiotyper, BrukerDaltonics, Bremen, Germany). The viable bacteria were prepared using the direct smear method as well as an extraction protocol provided by the manufacturer. Briefly, freshly grown bacteria were harvested and diluted in ethanol, centrifuged (2.000 x g, 2 min), air dried and resuspended in aqueous volumes of 70% formic acid and acetonitrile followed by a vortex step. One microliter was directly transferred to the steel target. Analysis was performed on a MALDI-TOF MS Biotyper Version V3.3.1.0. The database used (DB 5627, BrukerDaltonics) comprised only one entry for S. moniliformis from strain DSM 12112T (= ATCC 14647T). A dendrogram including selected main spectra peak lists (MSP) of the family Leptotrichiaceae from the Bruker database and the S. moniliformis strains from this study is depicted in Fig 1.
Fig 1. Dendrogram including all main spectra peak lists (MSP) of the family Leptotrichiaceae available in the Bruker Taxonomy Database.
Spectra of Streptobacillus moniliformis field and reference strains, Streptobacillus hongkongensis DSM 26322T, Streptobacillus felis 131000547T and Sebaldella termitidis ATCC 33386T were measured using the direct transfer protocol. The dendrogram was generated using the BioTyper MSP Dendrogram Creation Standard Method (v1.4) of the MALDI Biotyper OC Software (v3.1, build 66). The database used (DB 5627, BrukerDaltonics) comprised a singular entry from S. moniliformis (DSM 12112T = ATCC 14647T); T: type strain, ATCC: American Type Culture Collection, Rockville, USA, NCTC: National Collection of Type Cultures, London, UK, DSM: Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany, IPDH: Institut für Geflügelkrankheiten, Hannover, Germany.
Fourier Transform-Infrared Spectroscopy (FT-IR)
Bacterial isolates were cultivated independently in 5–7 replicates at 37°C for five days in a capnophilic atmosphere of 10% CO2 on SBA. Harvesting of cells, preparation of bacterial films on zinc selenide plates, drying and handling was performed as described previously [27]. The dried bacterial films were used directly for examination by FT-IR. Infrared spectra were recorded for each sample in a transmission mode from 500 to 4000 cm-1 with an FT-IR spectrometer (Tensor27 with HTS-XT-module, BrukerOptics, Ettlingen, Germany). Acquisition and first analysis of data was carried out using OPUS Software (vers. 4.2, BrukerOptics). To get a first impression the IR spectra of all viable strains listed in Table 1 were compared by hierarchical cluster analysis [28, 29]. Therefore, the second derivation of the vector normalized spectra in the wave number range of 500–1400 cm-1 and 2800–3000 cm-1 were used for calculation with Ward’s algorithm (OPUS 4.2; [30]). Spectra identified as outliers were quashed. The wave numbers 550–1800 cm-1 and 2800–3200 cm-1 of second derivative spectra were selected and vector normalized. After a principal component analysis, the first 40 components were used for linear discriminant analysis with spectra grouped by isolate. The diagram obtained depicts the arrangement of isolates according to their spectral differences (Fig 2).
Fig 2. Linear discriminant analysis (LDA) of 201 infrared spectra of one strain each of Sebaldella termitidis NCTC 11300T, Streptobacillus hongkongensis DSM 26322T, Streptobacillus felis 131000547T, 19 Streptobacillus moniliformis field isolates and reference strains and Streptobacillus sp. isolate AHL 370–1 from a spinifex hopping mouse.
The wave numbers 550–1800 cm-1 and 2800–3200 cm-1 of second derivative spectra were selected and vector normalized. After a principal component analysis, the first 40 components were used for the LDA. In this LDA every isolate was defined as one group. Spectra of Sebaldella termitidis ATCC 33386T are represented by squares, Streptobacillus felis 131000547T by diamonds, Streptobacillus hongkongensis DSM 26322T by triangles, Streptobacillus sp. (AHL 370–1) by circles and Streptobacillus moniliformis field isolates and reference strains by dots. Ellipses contain 95% of all group spectra assuming a bivariate normal distribution.
Molecular characterization
Species specific PCR for S. moniliformis
Two PCRs for the detection of S. moniliformis based on the 16S rRNA gene were performed with minor modifications [12, 31]. Freshly cultured colonies were suspended in 180 μl sterile distilled water. After incubation for 15 min at 100°C the lysates were centrifuged at 12.000 x g for 10 min to remove cell debris. One microliter of the supernatant was used for subsequent PCR analysis according to Kimura et al. (primers S5: 5’-CATACTCGGAATAAGATGG-3’ and AS2: 5’-GCTTAGCTCCTCTTTGTAC-3’; PCR program as follows: x1 (95°C, 180 sec), x35 (95°C, 20 sec, 53°C, 60 sec, 72°C, 60 sec), x1 (72°C, 420 sec)) [12] and Nicklas (primers SbmF: 5’-GAGAGAGCTTTGCATCCT-3’ and SbmR: 5’-GTAACTTCAGGTGCAACT-3’; x1 (94°C, 240 sec), x35 (94°C, 60 sec; 50°C, 60 sec; 72°C, 60 sec), x1 (72°C, 420 sec)) (cited in [31]), respectively. All PCRs were carried out in a T3000 Thermocycler (Biometra, Göttingen, Germany). Amplicons of 269 and 1222 bp, respectively, were detected by electrophoresis in 1.5% agarose gel containing ethidium bromide (1 mg/ml), visualised in an UV transilluminator and photographed. DNA extracted from S. moniliformis strain NCTC 11941 served as positive control in all runs.
DNA sequence analysis and determination of guanine/cytosine (G/C) contents
DNA was extracted from a pure bacterial culture from all viable strains with a commercial kit according to the manufacturer’s instructions (MasterPure Complete DNA and RNA Purification Kit, Epicentre, distributed by Biozym Scientific, Hessisch Oldendorf, Germany) and DNA from all strains was subjected to genome sequencing. De novo assembly was performed with CLC Genomics Workbench, Version 7.5 (CLC Bio, Aarhus, Denmark). For automatic annotation we used the RAST Server: Rapid Annotations using Subsystems Technology [32]. Sequence analysis from non-published genomes and calculation of G/C contents were carried out with Geneious (v. 8.1.3; Biomatters, Auckland, NZ) [33]. Nucleotide sequences of partial 16S rRNA genes and of groEL, recA and gyrB genes were aligned by using MAFFT [34] (MAFFT v7.017, implemented in Geneious). Maximum likelihood phylogenies and trees were estimated (100 bootstrap replicates) and visualized with PhyML [35], using the HKY85 model [36].
Instead of weak DNA-DNA hybridization results for members of this genus [26] (data not shown) average nucleotide identity (ANI) was carried out according to the method described by Goris et al. [37] using the ezbiocloud platform (http://www.ezbiocloud.net/ezgenome/ani). According to Richter & Rosello-Mora [38] the cut-off for species boundary with this method is at 95–96%.
Results
Phenotypic characterization
Presumptive confirmation of bacterial strains
Streptobacilli grew well on SBA after 2–5 days of incubation at 37°C in 10% CO2. Colonies were tiny, drop-like, shiny, slightly convex, 0.1–0.4 mm in diameter. Some of the colonies showed a “fried-egg” appearance. Strains normally grew without haemolysis, but strains S. moniliformis ATCC 49940, S. felis 131000547T and S. hongkongensis DSM 26322T displayed alpha-haemolytic colonies as already described [10, 20, 39]. In liquid media streptobacillary growth could be detected after 2–7 days as “puff-ball” or “bread crumb-like” appearance. Gram staining revealed irregular Gram-negative pleomorphic, fusiform to filamentous, non-spore forming, non-encapsulated, non-acid-fast rods that were arranged in chains and clumps, sometimes displaying irregular, lateral bulbar swellings.
Biochemical studies
Despite its Gram-negative staining characteristics of S. moniliformis, a representative species specific panel of biochemical reactions was derived from preliminary testing with the Micronaut Strep2 (for streptococci and enterococci) and RPO (for Gram-positive bacteria) test systems. A panel of eleven relevant chemotaxonomic discriminatory parameters proved sufficient for the identification of S. moniliformis, which included besides principle growth characteristics and negative fermentative reactions during overnight incubation (data not shown) the following reactions (positive percentage): tripeptidase (95.2), proline aminopeptidase (95.2), arginine dihydrolase (85.7), arginine aminopeptidase (90.5), glycyltryptophan aminopeptidase (100), growth in SPMS medium (100), neuraminidase (90.5), glycylproline aminopeptidase (100), hydroxyproline aminopeptidase (95.2), pyrase (100) and asparatyl aminopeptidase (0) (Table 2). Further physiological tests obtained by Micronaut Strep2 and RPO gave congruent results for all S. moniliformis strains for hydrolysis of p-nitrophenyl-β-D-glucosamide, alkaline phosphatase and arginine hydrolysis (positive) as well as for tellurite, fermentation of cellobiose, arbutin, sorbitole, amygdaline and raffinose, chitin, H-asp-β-naphthylamide, p-nitrophenyl-α-D-galactopyranoside, p-nitrophenyl-β-D-galactopyranoside, p-nitrophenyl-β-D-fucopyranoside, p-nitrophenyl-β-D-glucopyranoside (negative). Neither the single strains of S. felis 131000547T and S. hongkongensis DSM 26322T nor strain AHL 370–1 could unambiguously be separated from S. moniliformis by the above mentioned physiological characters, although S. hongkongensis was the only glycyltryptophan aminopeptidase- and pyrase-negative strain under study. VITEK2-compact identified strains of S. moniliformis, S. felis 131000547T and S. hongkongensis DSM 26322T as Neisseria (N.) cinerea (93–99% confidence), N. elongata (87% confidence) or inconclusive between N. cinerea and N. elongata (NHI card profiles 0210000002, 0220000000, 0220000040, 0233000000, 0232000000, 0233000000, 0273000000, 0273200000, 0277220000, 0237100002, 0237220000). From the 30 different reactions all but eleven were congruently negative for arginine-arylamidase, γ-glutamyl-transferase, L-lysine-arylamidase, D-galactose, Ellman’s reagent, L-pyrrolidonyl-arylamidase, tyrosine-arylamidase, glycogen, D-maltose, sucrose, urease, β-galactopyranosidase indoxyl, ornithine-decarboxylase, α-arabinosidase, pyruvate, phosphoryl-choline, D-malate, maltotriose, L-glutamine, D-ribose, phenylphosphonate and D-xylose and positive for leucine-arylamidase and L-proline-arylamidase for all tested 22 strains of the genus Streptobacillus (nos. 1–20, 28, 29 according to Table 2). All S. moniliformis strains were phenylalanine-arylamidase-, tyrosine-arylamidase- and ala-phe-pro-arylamidase-positive in contrast to S. felis 131000547T and S. hongkongensis DSM 26322T. Furthermore, S. hongkongensis DSM 26322T was the only phosphatase-positive strain under investigation.
The API-ZYM test was carried out with 22 Streptobacillus strains and revealed consistent enzymatic pattern for lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosaminidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase (all negative). All Streptobacillus strains from this study were positive for esterase lipase (C8). Contrarily, S. hongkongensis DSM 26322T was the only naphthol-AS-BI-phosphohydrolase-positive strain and strain IPDH 144/80 was solely α-chymotrypsin-negative. All further differing biochemical test results are presented in Table 2. Presumptive physiological characterization revealed corresponding results for all strains for cytochrome oxidase, catalase, urease, nitrate reduction and indole production (all negative).
Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined for all 22 viable Streptobacillus strains tested in this study and results are presented in S1 Table. Congruent in vitro results could be obtained for all strains with respect to azithromycin (≤0.0625–2), clindamycin (≤0.125–0.25), chloramphenicol (≤0.5–4), meropenem (≤0.0625–1), telithromycin (≤0.125–4), tetracycline (≤0.125–1; all susceptible [MICs in mg/L]). A resistant phenotype was recorded for trimethoprim/sulfamethoxazol (≥8/152) for all strains except S. hongkongensis DSM 26322T. Some strains of S. moniliformis displayed at least resistance or intermediate resistance to ciprofloxacin (2), erythromycin (16), gentamicin (4–8), nalidixic acid (32–128) and streptomycin (4–32).
Haemagglutination
Adhesive properties were detected in all 12 S. moniliformis strains tested. Erythrocytes of 11 different vertebrate species were agglutinated with varying intensity. The slide agglutination test represented results for spontaneous agglutination and most intense reactions could be observed in erythrocytes from turkeys, humans, guinea-pigs and pigs. Red blood cells from rats and chickens showed a strong reaction (++). C57BL/6J mice, known to represent a highly susceptible mouse strain towards streptobacillosis [18], were less strongly agglutinated compared to erythrocytes from the more resistant BALB/c mice (mostly + in contrast to mostly ++).
Results from the haemagglutination in microtiter plates differed in some way (Table 3). Again, erythrocytes from turkeys, humans and pigs, but also from rats and C57BL/6J mice proved to show the strongest haemagglutination reactions. Agglutination with erythrocytes from chickens, guinea-pigs and BALB/c mice was weaker compared to the results from slide agglutination tests.
Table 3. Adhesive properties of selected S. moniliformis strains from this study.
| Erythrocytes from | DSM 12112T | NCTC 10773 | NCTC 11194 | CIP 81–99 | NCTC 11941 | ATCC 27747 | IPDH 144/80 | IPDH 109/83 | ATCC 49567 | CIP 55–48 | ATCC 49940 | Kun 3 (RIVM) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Turkey | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| turkey* | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | + | ++ |
| Human | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | + | ++ |
| human* | ++ | + | + | ++ | ++ | + | ++ | ++ | ++ | ++ | - | ++ |
| guinea pig | + | + | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | + | ++ |
| guinea pig* | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ |
| mouse (C57BL/6) | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + |
| mouse (C57BL/6)* | + | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + |
| Rat | ++ | ++ | ++ | ++ | ++ | ++ | + | + | ++ | ++ | + | ++ |
| rat* | ++ | ++ | + | ++ | ++ | + | + | + | + | ++ | + | ++ |
| Pig | ++ | ++ | + | ++ | ++ | ++ | ++ | - | ++ | ++ | ++ | ++ |
| pig* | ++ | ++ | + | ++ | ++ | ++ | + | - | ++ | ++ | ++ | ++ |
| mouse (BALB/c) | + | + | + | ++ | ++ | + | ++ | ++ | ++ | ++ | - | + |
| mouse (BALB/c)* | + | + | + | ++ | + | + | ++ | ++ | ++ | ++ | + | + |
| Chicken | + | ++ | ++ | + | + | ++ | + | ++ | + | + | + | ++ |
| chicken* | + | ++ | ++ | + | + | ++ | ++ | ++ | + | ++ | + | ++ |
| Cattle | ++ | + | + | ++ | ++ | ++ | ++ | + | + | ++ | + | ++ |
| cattle* | ++ | + | + | ++ | + | ++ | + | + | + | ++ | + | ++ |
| Sheep | ++ | - | + | + | + | + | - | ++ | + | + | ++ | ++ |
| sheep* | + | + | + | + | - | + | + | + | + | + | + | + |
| Hamster | + | + | + | + | + | + | ++ | + | + | ++ | + | + |
| hamster* | + | + | + | + | + | + | ++ | + | + | + | - | + |
| Horse | - | - | + | + | - | + | + | + | + | + | - | + |
| horse* | + | - | + | + | - | + | + | + | - | + | - | + |
Haemagglutination was tested in microtiter plates in the presence* and absence of 1% mannose; strong (++), moderate/weak (+) or no (-) reaction
In both experiments erythrocytes from cattle, sheep, hamsters and horses showed the weakest or even no agglutination. By adding mannose, a known agonist of a common adhesin receptor, no significant differences could be observed indicating mannose-resistant agglutination in all cases. No differences were observed between agglutination of erythrocytes from ‘original’ host species (from which respective strains were originally isolated) and other host red blood cells, but susceptibility was generally highest in species of potential hosts compared to non-host species.
MALDI-TOF MS
For MALDI-TOF MS, all 20 viable strains of S. moniliformis were identified to the species level with a score level between 2.0 and 2.4 using the direct smear and the extraction method for sample preparation. This was also true for strain AHL 370–1, which albeit clustered most distantly from all S. moniliformis strains. Streptobacillus felis 131000547T as well as S. hongkongensis DSM 26322T and Sebaldella termitidis ATCC 33386T could not be identified yielding only score levels between 1.3 and 1.5 (database DB 5627, BrukerDaltonics). Following the manual inclusion of respective spectra of these strains to the database these were most closely related to S. moniliformis. A dendrogram including selected main spectra peaks (MSP) of the family Leptotrichiaceae from the Bruker database as well as from type strains of S. felis, S. hongkongensis and Sebaldella termitidis is depicted in Fig 1.
FT-IR
The comparison of the infrared-spectra of 22 viable strains of Streptobacillus spp. as well as the closely related Sebaldella termitidis ATCC 33386T showed a clear separation into the three species S. moniliformis, S. felis and S. hongkongensis (Fig 2). Based on spectra within the genus Streptobacillus the spectral cloud derived from spectra of divergent strain AHL 370–1 could be delineated from all other strains (S1 Fig).
Molecular characterization
Species specific PCR for S. moniliformis
Amplification of the specific target sequences resulted in characteristic amplicon sizes of approximately 269 and 1222 bp employing the PCR assays according to Kimura et al. [12] and Nicklas (cited in Rohde et al. [31]), respectively. All 23 S. moniliformis strains, the four divergent strains AHL 370–1, OGS16, KWG2 and KWG24 and also S. felis 131000547T were found positive in both PCRs, whereas S. hongkongensis DSM 26322T resulted in a specific amplicon in the PCR according to Nicklas but not to Kimura et al. (data not shown).
Phylogenetic analysis and determination of G/C contents
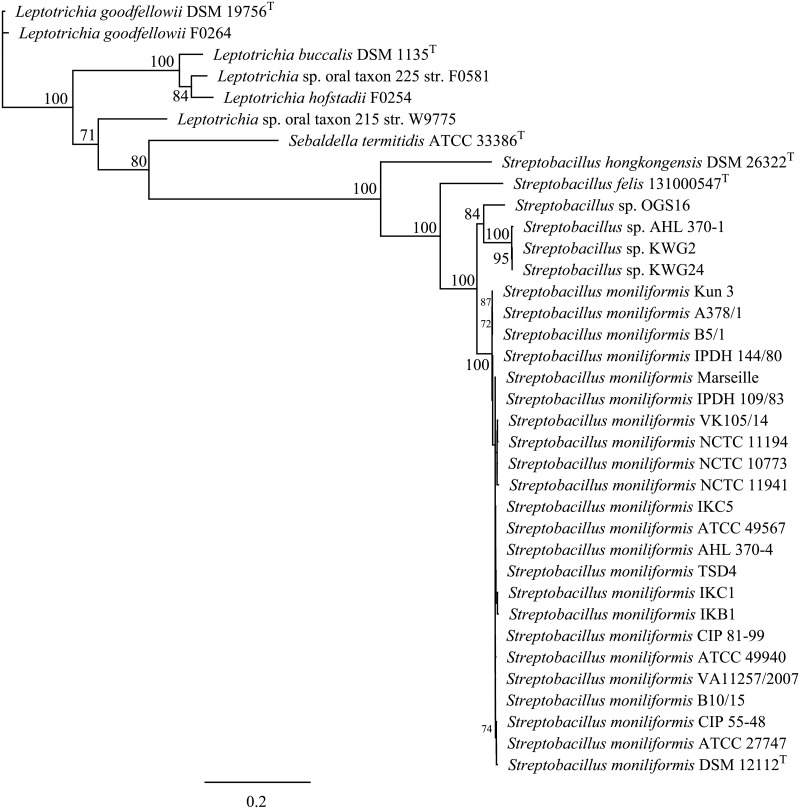
Alignment of sequences of the partial 16S rRNA gene (1482 bp) revealed a sequence homology of 99.8–100% for 23 S. moniliformis strains under study (Fig 3). Three (AHL 370–1, KWG2, KWG24) and one (OGS16) divergent strains, respectively, were clustering separately (≥ 90% bootstrap support) and displayed sequence homology of 97.97–98.58% to the type strain DSM 12112T. The 16S rRNA gene homology between S. moniliformis DSM 12112T compared to S. felis 131000547T was 97.11%, whereas the homology between the S. moniliformis type strain and S. hongkongensis DSM 26322T was 92.73%. The type strain of S. hongkongensis clustered closer with that of Sneathia sanguinegens than with S. moniliformis and S. felis 131000547T.
Fig 3. Maximum-likelihood tree showing the phylogenetic position within the family Leptotrichiaceae.
The tree was generated in Geneious using PhyML [35] and based on 16S rRNA gene sequences. GenBank accession numbers are KR001904-1922, KP657489, KP657490-KP657495, and HG421076. Numbers at branch nodes refer to bootstrap values (100 replicates). Bar: 0.06 nucleotide substitutions per side.
The outstanding position of the aforementioned four S. moniliformis strains as separate clusters was also supported by analysis of functional genes groEL, gyrB, and recA. The phylogenetic analysis based on concatenated sequences (4.632 bp) of these genes (groEL [1.602 bp], recA [1.047 bp], gyrB [1.983 bp]) (Fig 4) roughly revealed that strains AHL 370–1, KWG2 and KWG24 and also strain OGS16 clustered at two separate positions apart from the other members of the genus (100% bootstrap support), so that it was possible to clearly delineate S. moniliformis, divergent strain clusters 1 and 2, S. hongkongensis and S. felis.
Fig 4. Phylogenetic trees based on concatenated sequences (4632 nt) of groEL (1.602 nt), recA (1.047 nt) and gyrB (1.983 nt), sequences including type strains of all species of the family Leptotrichiaceae showing the phylogenetic relationship of Streptobacillus species from this study.
GenBank accession nos. are KR001923-1941 and KP657496-503 for groEL, KR001942-1960 and KP657504-511 for recA, KR001961-1979 and KP676101-108 for gyrB sequences. The tree was generated with the maximum-likelihood program PhyML (Substitution Model HKY85; number of bootstraps: 100) [35] after alignment of sequences with MAFFT v7.017, both implemented in Geneious. Numbers at nodes represent bootstrap values >70%. Bar: 0.2 substitutions per sequence position.
G/C contents of Streptobacillus spp. obtained from non-published genomes from all strains revealed 25.7–28.9% (S2 Table), which is quite in accordance with previously determined values based on melting point analyses [26]. Average nucleotide identity (ANI) analysis revealed an overall DNA-DNA relatedness of 98.51 to 99.3% (recipr. 98.17–99.41%) between the type strain DSM 12112T and all S. moniliformis strains under study, except for isolates AHL 370–1, KWG2, KWG24 and OGS16. ANI values below the cut-off for species boundary were calculated for Streptobacillus spp. isolates AHL 370–1, KWG2, KWG24 and OGS16 (88.21 to 89.1%; recipr. 87.89 to 89.87%). The average nucleotide identity between S. moniliformis type strain DSM 12112T and S. hongkongensis DSM 26322T and S. felis 131000547T was 74.04% (recipr. 75.03%) and 82.0% (recipr. 82.02%), respectively (S2 Table).
Nucleotide sequence accession numbers
Partial 16S rRNA sequences of 27 Streptobacillus strains have been submitted to the GenBank database under accession nos. KR001904-KR001922, HG421076, KP657489, KP657491-KP657495, KT311784, CP001779. The groEL, gyrB, and recA sequences have been submitted under accession nos. KR001923-KR001941, KP657496-KP657497, KP657499-KP657503, KT311785, CP001779 (groEL), KR001942-KR001960, KP676101-KP676102, KP676104-KP676108, KT311786, CP001779 (gyrB), and KR001961-KR001979, KP657504-KP657505, KP657507-KP657511, KT311787, CP001779 (recA), respectively.
Discussion
The aim of this study was a comparison of different strains of the genus Streptobacillus including representatives of S. moniliformis and two strains of the novel species S. hongkongensis and S. felis. Few studies have focused on strain diversity within the species S. moniliformis, the etiological agent of the worldwide occurring zoonosis RBF, and compared strains from different origins, also considering the occurrence of L-forms in this species [13–15, 26, 40, 41]. A similar broad spatiotemporal diversity of strains like in the present study never formed the basis for a comparison, also including novel members of the recently extended genus for the first time. Most previous studies revealed only little differences between strains with respect to their pheno- and genotype [15, 26]. Minor biochemical strain differences between studies were merely attributed to different preparations of culture media [12, 14, 15, 41–44] and might also be explained by difficulties due to the fastidious growth or the person reading the tests [26]. Even within the same study single physiological discrepancies were noted in repeated experiments with the same strain, thereby reflecting inconsistency in test results rather than discriminatory traits [26]. Protein profiles as observed by SDS-PAGE were found to be identical between S. moniliformis strains in a former study [13] and other chemotaxonomical investigations found a homologous fatty acid profile for all tested S. moniliformis strains of tetradecanoic acid (14:0), palmitic acid (16:0), octadecanoic acid with linoleic acid (18:2) and oleic acid (18:1), and stearic acid (18:0) [45–47].
However, comprehensive analyses of differences based on spectroscopic data of biomolecules as well as genotypic properties of strains were not yet published in international literature. The present study included 23 S. moniliformis strains from at least five different host species that cover isolations over the past 90 years from almost all subcontinents as well as the type strains of S. hongkongensis and S. felis and four divergent strains that presumably belong to two yet undescribed species. Classical biochemistry of most key reactions was in accordance with published results, although we also observed significant growth differences depending on the kind of carbohydrate preparation (data not shown). Therefore, standardized test systems for physiological typing of S. moniliformis were included. Earlier studies involved API 20E for assessing biochemical profiles [12]. As this test requires viable bacteria and as previously chosen culture conditions were not appropriate for the growth of S. moniliformis, published results of this method may be interpreted with caution. We have successfully employed commercially available systems API ZYM [6, 12, 14, 20] and VITEK2-compact (NHI profile) [9] and basically validated the Micronaut system for a novel application with Streptobacillus. All these systems use biochemical end-point measurements and are thus independent from bacterial growth though standardized and the Micronaut system earlier proved to be suitable for bacterial species and biotype discrimination [48]. The designed identification panel of this system could unequivocally discriminate members of the genus Streptobacillus from other bacterial species by considering growth characteristics (fastidious capnophilic growth, occurrence of L-forms, “puff-ball”- or “bread crumb”-like appearance in liquid media [2, 3, 12, 14, 15] and eleven biochemical reactions (Table 2). Briefly, five of these reactions were identical among all S. moniliformis as well as S. felis 131000547T. However, most other physiological tests showed too diverse reactions to discriminate species or biotypes, especially because the variability of S. hongkongensis, S. felis and the yet undescribed members could not or not sufficiently be evaluated by one or three strains each. Like in other studies, we could confirm major uniformity between strains of S. moniliformis for most physiological reactions. Nevertheless, minor discrepancies were observed, some of which were earlier reported [12, 14, 42–44] and can–in the case of API ZYM reactions–be explained by differences in semi quantitative reading of results, too [12, 14, 15, 41–43, 49].
Antimicrobial resistance profiles revealed that S. moniliformis is susceptible to all β-lactam antibiotics [14], and no β-lactamase activity could be demonstrated so far [50]. Penicillin G was repeatedly reported as the most active antimicrobial substance in in vitro tests, which further supports its use as the drug of choice in the treatment of RBF and HF, followed by tetracycline [2]. The majority of studies have performed antimicrobial susceptibility testing (AST) with rather old-fashioned methods without determining MIC values and not including all relevant actual agents and novel Streptobacillus species. In order to provide a more up-to-date picture we included recently isolated strains and carried out AST by broth microdilution with a commercially available test. The strains from this study were very similar in their resistance pattern and largely confirmed a generally good therapeutic basis. However, at least some isolates were resistant or intermediate resistant to ciprofloxacin, erythromycin, gentamicin, nalidixic acid and streptomycin. Interestingly and in contrast to Woo et al. [9], S. hongkongensis DSM 26322T turned out to be the only member of the genus under study that was trimethoprim/sulfamethoxazol-sensitive. Based on chemoresistance, biochemical and chemotaxonomic results from this and former studies other traits should be propagated, both for diagnostics as well as for strain comparisons in Streptobacillus. One of these applicable methods is the acquisition of MALDI-TOF and FT-IR spectra that were initially included on a large scale basis. By MALDI-TOF MS all tested strains of S. moniliformis could be assigned with high accuracy to species level. This was also true for representative spectra of S. hongkongensis and S. felis [10] and facilitated also discrimination of one of the undescribed Streptobacillus sp. (strain AHL 370–1). In contrast to MALDI-TOF MS, where the spectral information mirrors protein components, spectra generated by FT-IR include information from a broad variety of main component biomolecules [51]. Again, spectra from type strains of S. hongkongensis and S. felis were shown to be distinct, but closely adjacent to those from all other S. moniliformis strains from this study, whereas spectra from strain AHL 370–1 could be noted as a small outgroup nearby the S. moniliformis cluster (Fig 2). It remains to be elucidated, whether strains KWG2, KWG24 and OGS16, from which only DNA was available, will confirm spectral traits found here. Interestingly, despite displaying a unique profile in electrophoretic protein patterns [13] no antigenic differences could be observed for strain AHL 370–1 [52].
Based on highly uniform electrophoretic protein patterns it was hypothesized that S. moniliformis strains could be grouped with respect to host species, geographic origin, disease pattern and route of infection, i.e. isolates from cases of RBF and HF should be clearly assigned by protein profiling [13]. We and others have not found evidence for these assumptions [15, 26, 52]. On the other hand, especially differences of HF- versus RBF-strains are unlikely because rats represent the source of infection in both cases and disparities could better be explained by different gene expression following oral or parenteral infection [3] or simply by too few HF-strains under study. Additionally, the time between infection and strain isolation from the host following rat exposure is usually too short to facilitate adaptation of strains and expression of a different phenotype. A number of studies have, nevertheless, proven that strains isolated from susceptible host species were able to cause infection in rodents, thereby partially fulfilling Koch’s postulates [16, 18]. As hypothesized by Nolan et al. [50], no phylotypes of S. moniliformis were yet detected indicating that this species is rarely found in the environment outside of its natural hosts.
A number of studies have used 16S rRNA gene sequencing as a diagnostic tool for species determination of S. moniliformis in strains and clinical specimens [53–56]. Both PCR protocols targeting the 16S rRNA gene published as specific for S. moniliformis were suitable to detect all strains from humans, rats, mice, a spinifex hopping mouse and turkeys from this study and also S. felis [10]. In contrast, S. hongkongensis was only detected by the PCR described by Rohde et al. [31] suggesting that both PCR are merely genus rather than species-specific. In order to elucidate the sufficiency, usefulness and discriminatory power of marker genes for inter- as well as intraspecific analysis within the genus Streptobacillus, sequence data from all strains were analysed. Within the phylum Fusobacteria sequencing of 16S rDNA, 16S-23S rDNA internal transcribed spacer, gyrB, groEL, recA, rpoB, conserved indels and genes for group-specific proteins, 43-kDa outer membrane protein and zinc protease have been proposed for species identification or phylogenetic analysis [57–65] and eleven whole genome sequences were generated to date. We have used functional genes 16S rRNA, groEL, recA and gyrB, which unequivocally could discriminate the strains from this study to species level. Fueled by the work presented here and further studies we have identified two other candidate species of Streptobacillus, one of which will be described shortly, thereby significantly extending the knowledge of this former monotypic genus. A more detailed insight into some of these candidates was solely possible by comparison of further genes, which otherwise would have been missed solely by 16S rRNA gene sequencing. Interestingly, a previous study came to the conclusion that disparities in 16S rRNA might in fact be based on different co-evolution of strains in different rat host species as suggested by Kimura et al. (2008). Indeed were isolates directly or indirectly (mouse after rat bite) obtained from brown rats (Rattus norvegicus; strains no. 14, 16, 18, 19, 21–24) belonging to S. moniliformis, but for the remaining S. moniliformis strains the exact source rat species remained obscure. Variations of the 16S rRNA gene were mainly observed in the region at nucleotide positions 1 to 300 [12], which could also be confirmed in our study and involved four strains (AHL 370–1, KWG2, KWG24, OGS16 cf. Fig 3). Their outstanding position from S. moniliformis was also supported by analysis of other functional genes (cf. Fig 4), but these four strains represented in fact two separate clusters. Interestingly, we found that strain AHL 370–1 from the spinifex hopping mouse clustered identical with strains KWG2 and KWG24 and respective genomes were also indistinguishable as obtained by ANI.
We therefore believe that strains OGS16, KWG2 and KWG24 are not exclusively restricted to R. rattus. This assumption is, on the other hand, also supported by the complete absence of human isolates resembling the 16S rRNA pattern of these three strains despite the rarity of R. rattus in many regions worldwide. Further epidemiologically based studies are required to elucidate the true prevalence of these microorganisms in different rat as well as further rodent host species. However, at present there is indeed no evidence that these genetic differences are geographically related, because the yet undescribed species were found in close proximity in Japan [12] and strain AHL 370–1 from this study originated from Australia.
With respect to G/C contents our investigations revealed a nearly identical G/C content of 25.7–28.9 mol% in all investigated Streptobacillus strains. The G/C content of Sebaldella termitidis was only 33.38 mol% [26, 66], thereby deceeding the classical value for species discrimination of 10 mol% [67]. Results from the literature of S. hongkongensis revealed G/C content of 26.0 [9, 10] and S. felis had 26.4 mol%. Analogous results could also be confirmed by ANI. A value of 70% DDH was proposed by Wayne et al. (1987) [68] as a recommended standard for delineating species. Goris et al. (2007) [37] could demonstrate a close relationship between DDH values and ANI in that the recommended cut-off point of 70% for species discrimination corresponded to 95% ANI. The 95–96% species boundary is also supported by Richter & Rosselló-Móra [38], who have developed an alignment free interface to calculate ANI also used in this study, which should even work for taxonomic issues on a 20% partial genome of strains under study. In contrast to the highly homologous group of S. moniliformis strains not only S. hongkongensis and S. felis but again also AHL 370–1, KWG2, KWG24 and OGS16 had significantly lower ANI values thereby clearly pointing towards separate species apart from S. moniliformis.
Concerning pathogenicity one might suspect haemolysis as a possible marker of virulence [3]. Indeed were haemolytic strains of S. moniliformis, S. hongkongensis and S. felis involved in clinical disease in a rat (strain no. 14, ATCC49940) [15], a dog [7], a cat [39] and a human [10], but other clinical isolates, especially those causing severe or even fatal disease turned out to be non-haemolytic so that other virulence factors apparently play a more important role. These might include the extraordinary high amount of DNase in all strains, which was released even independently of bacterial growth [26]. However, another possible relationship was detected during haemagglutination experiments that could also be explained as trait of virulence: susceptibility of erythrocytes to haemagglutination was generally highest in species serving as potential hosts like humans, mice, rats, turkeys and guinea-pigs compared to non-host species suggesting a predisposing role for potential host species. No significant differences were observed between agglutination of erythrocytes from ‘original’ host species, i.e. from which respective strains were originally isolated and other host red blood cells, which again gives rise to the assumption that infectious strains do at least not rapidly adapt to their hosts following infection. However, these experiments unequivocally suggest the presence of adhesins, a mechanism involved in pathogenicity which is a prerequisite for the ‘successful’ infection of a host. Indeed, there appear to remain other factors besides adhesins as can be concluded from infections in hosts with missing host specificity. These might include not yet identified genetic factors at the host side which can be concluded from differences in susceptibility to infection like for instance the genetically diverse, highly susceptible C57BL/6J mice compared to BALB/c mice [18, 69].
Conclusion
Rat bite fever represents a significant public health threat that is under-diagnosed in humans and animal species. We have analyzed the largest-ever collection of strains with respect to time and geography of this rarely isolated microorganism. In a polyphasic approach we could show that some recently described as well as novel candidate species besides S. moniliformis could be differentiated, which are not restricted to 16S rRNA gene differences. It will be challenging to note the true prevalence of these novel members in human and animal infections. Systematical data for several of the strains from this study with respect to G/C contents as well as haemagglutination were assessed before [26], but never made available to the public at large, which will hopefully help in future research to further elucidate virulence properties of these neglected bacterial species. Growth characteristics together with traditional physiological methods and also standardized biochemistry are suitable for genus determination, but do not possess enough discriminatory power to sufficiently differentiate Streptobacillus spp. We have shown that the strains of S. moniliformis as well as putative and designated type strains of the novel species could be easily classified with modern genotypic and phenotypic methods including sequence analysis of selected functional genes as well as FT-IR and MALDI-TOF MS. The latter proved to have the potential to easily diagnose the novel species S. hongkongensis and S. felis and also at least one yet undescribed species. Further genetic studies on Streptobacillus should include investigations on possible virulence genes and differences in pathogenic mechanism between strains.
Supporting Information
The wave numbers 550–1800 cm-1 and 2800–3200 cm-1 of second derivative spectra were selected and vector normalized. After a principal component analysis, the first 40 components were used for the LDA. In this LDA every isolate was defined as one group. Spectra of Streptobacillus felis are represented by diamonds, Streptobacillus hongkongenis by triangles, Streptobacillus sp. (AHL 370–1) by circles and S. moniliformis by dots. Note that LDA axis 1 and 3 are shown, because axis 3 contains the differences between Streptobacillus sp. (AHL 370–1) and S. moniliformis.
(TIF)
Broth microdilution susceptibility testing was performed with the Merlin Micronaut system; results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) MIC criteria based on CLSI MIC interpretive standards for other non-Enterobacteriaceae and anaerobes [25]. AZM: azithromycin, CIP: ciprofloxacin, CLI: clindamycin, CMP: chloramphenicol, ERY: erythromycin, GEN: gentamicin, MER: meropenem, NAL: nalidixic acid, STR: streptomycin, T/S: trimethoprim/sulfamethoxazole, TEL: telithromycin, TET: tetracycline, R: resistant, I: intermediate susceptible, S: susceptible phenotype, MIC values in mg/L.
(DOCX)
(DOCX)
Acknowledgments
For excellent technical assistance we thank Ulrike Kling, Anna Mohr, Katharina Engel and Barbara Depner and Barbara Gamb for making even the most exotic manuscripts available. Stefanie P. Glaeser and Christoph Lämmler are acknowledged for comments on an earlier draft of the manuscript. Viola Spamer and Osama Sammra contributed in valuable pretrials. This study would not have been possible without the strain collection and the great interest of Michael Wullenweber in S. moniliformis. We are greatly acknowledging the support of Walter Geißdörfer (Erlangen), Judith Rohde (Hannover), Bernard La Scola (Marseille) and Koichi Imaoka (Tokyo) for providing strains or DNA of strains no. 17, 18, 19 and 21–28, respectively. Parts of this work belong to the doctoral thesis of Nicola Hofmann.
Data Availability
Data are available from GenBank: BankIt1841833 NCTC11941_16S KT311784; BankIt1841876 NCTC11941_groEL KT311785; BankIt1841876 NCTC11941_gyrB KT311786; BankIt1841876 NCTC11941_recA KT311787.
Funding Statement
The Hessian State Laboratory (Hessisches Landeslabor) is supported by Hessian Ministry for the Environment, Climate Change, Agriculture and Consumer Protection (HMUKLV). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Merlin Diagnostika provided support in the form of a salary for author KA, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Levaditi C, Nicolau S, Poincloux P. Sur le rôle étiologique de Streptobacillus moniliformis (nov. spec.) dans l'érythème polymorphe aigu septicémique. C R Acad Sci. 1925;180:1188–90. [Google Scholar]
- 2. Elliott SP. Rat bite fever and Streptobacillus moniliformis . Clin Microbiol Rev. 2007;20(1):13–22. Epub 2007/01/16. 10.1128/cmr.00016-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaastra W, Boot R, Ho HT, Lipman LJ. Rat bite fever. Vet Microbiol. 2009;133(3):211–28. Epub 2008/11/15. 10.1016/j.vetmic.2008.09.079 . [DOI] [PubMed] [Google Scholar]
- 4. Torres L, Lopez AI, Escobar S, Marne C, Marco ML, Perez M, et al. Bacteremia by Streptobacillus moniliformis: first case described in Spain. Eur J Clin Microbiol Infect Dis. 2003;22(4):258–60. Epub 2003/04/24. . [DOI] [PubMed] [Google Scholar]
- 5. Bleich A, Nicklas W. Zoonoses transmitted by mouse and rat maintained as laboratory or pet animals [in German]. Berl Münch Tierärztl Wochenschr. 2008;121(7–8):241–55. Epub 2008/08/21. . [PubMed] [Google Scholar]
- 6. Hayashimoto N, Yoshida H, Goto K, Takakura A. Isolation of Streptobacillus moniliformis from a pet rat. J Vet Med Sci. 2008;70(5):493–5. Epub 2008/06/06. . [DOI] [PubMed] [Google Scholar]
- 7. Ditchfield J, Lord LH, McKay KA. Streptobacillus moniliformis infection in a dog. Can Vet J. 1961;2(12):457–9. Epub 1961/12/01. [PMC free article] [PubMed] [Google Scholar]
- 8. Washburn RG. Streptobacillus moniliformis (rat-bite fever) In: Mandell GL, Bennett JE, Dolin RG, editors. Principles and practice of infectious diseases Vol 2 New York: Churchill Livingstone; 1995. p. 2084–6. [Google Scholar]
- 9. Woo PC, Wu AK, Tsang CC, Leung KW, Ngan AH, Curreem SO, et al. Streptobacillus hongkongensis sp. nov., isolated from patients with quinsy and septic arthritis in Hong Kong, and emended descriptions of the genus Streptobacillus and the species Streptobacillus moniliformis . Int J Syst Evol Microbiol. 2014. 10.1099/ijs.0.061242-0 . [DOI] [PubMed] [Google Scholar]
- 10. Eisenberg T, Glaeser S, Nicklas W, Mauder N, Contzen M, Aledelbi K, et al. Streptobacillus felis sp. nov. isolated from a cat with pneumonia. Int J Syst Evol Microbiol—accepted manuscript. 2015. [DOI] [PubMed] [Google Scholar]
- 11. Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, et al. The canine oral microbiome. PLoS One. 2012;7(4):e36067 Epub 2012/05/05. 10.1371/journal.pone.0036067 PONE-D-12-02767 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kimura M, Tanikawa T, Suzuki M, Koizumi N, Kamiyama T, Imaoka K, et al. Detection of Streptobacillus spp. in feral rats by specific polymerase chain reaction. Microbiol Immunol. 2008;52(1):9–15. Epub 2008/03/21. 10.1111/j.1348-0421.2008.00005.x . [DOI] [PubMed] [Google Scholar]
- 13. Costas M, Owen RJ. Numerical analysis of electrophoretic protein patterns of Streptobacillus moniliformis strains from human, murine and avian infections. J Med Microbiol. 1987;23(4):303–11. Epub 1987/06/01. . [DOI] [PubMed] [Google Scholar]
- 14. Edwards R, Finch RG. Characterisation and antibiotic susceptibilities of Streptobacillus moniliformis . J Med Microbiol. 1986;21(1):39–42. Epub 1986/02/01. . [DOI] [PubMed] [Google Scholar]
- 15. Wullenweber M. Streptobacillus moniliformis—a zoonotic pathogen. Taxonomic considerations, host species, diagnosis, therapy, geographical distribution. Lab Animal. 1995;29(1):1–15. Epub 1995/01/01. . [DOI] [PubMed] [Google Scholar]
- 16. Yamamoto R, Clark GT. Streptobacillus moniliformis infection in turkeys. Vet Rec. 1966;79(4):95–100. Epub 1966/07/23. . [DOI] [PubMed] [Google Scholar]
- 17. Hopkinson WI, Lloyd JM. Streptobacillus moniliformis septicaemia in spinifex hopping mice (Notomys alexis). Aust Vet J. 1981;57(11):533–4. Epub 1981/11/01. . [DOI] [PubMed] [Google Scholar]
- 18. Wullenweber M, Kaspareit-Rittinghausen J, Farouq M. Streptobacillus moniliformis epizootic in barrier-maintained C57BL/6J mice and susceptibility to infection of different strains of mice. Lab Anim Sci. 1990;40(6):608–12. Epub 1990/11/01. . [PubMed] [Google Scholar]
- 19. Boot R, Oosterhuis A, Thuis HC. PCR for the detection of Streptobacillus moniliformis . Lab Anim. 2002;36(2):200–8. Epub 2002/04/12. . [DOI] [PubMed] [Google Scholar]
- 20. Wullenweber M, Jonas C, Kunstyr I. Streptobacillus moniliformis isolated from otitis media of conventionally kept laboratory rats. J Exp Anim Sci. 1992;35(1):49–57. Epub 1992/03/01. . [PubMed] [Google Scholar]
- 21. Kondruweit M, Weyand M, Mahmoud FO, Geissdorfer W, Schoerner C, Ropers D, et al. Fulminant endocarditis caused by Streptobacillus moniliformis in a young man. J Thorac Cardiovasc Surg. 2007;134(6):1579–80. Epub 2007/11/21. 10.1016/j.jtcvs.2007.08.010 . [DOI] [PubMed] [Google Scholar]
- 22. Loridant S, Jaffar-Bandjee MC, La Scola B. Shell vial cell culture as a tool for Streptobacillus moniliformis "resuscitation". Am J Trop Med Hyg. 2011;84(2):306–7. Epub 2011/02/05. 10.4269/ajtmh.2011.10-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manafi M, Kneifel W, Bascomb S. Fluorogenic and chromogenic substrates used in bacterial diagnostics. Microbiological reviews. 1991;55(3):335–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wellinghausen N, Pietzcker T, Poppert S, Belak S, Fieser N, Bartel M, et al. Evaluation of the Merlin MICRONAUT system for rapid direct susceptibility testing of gram-positive cocci and gram-negative bacilli from positive blood cultures. J Clin Microbiol. 2007;45(3):789–95. 10.1128/JCM.01856-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anonymous. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document 2013;M100–S23. [Google Scholar]
- 26.Hofmann N. Phenotypical and molecular taxonomic investigations on the systematic status of Streptobacillus moniliformis, the agent of rat-bite-fever [in German]. Thesis Dr. rer. nat., Faculty of Biology, Leibniz Universität Hannover 1994.
- 27. Kuhm AE, Suter D, Felleisen R, Rau J. Identification of Yersinia enterocolitica at the species and subspecies levels by Fourier transform infrared spectroscopy. Appl Environ Microbiol. 2009;75(18):5809–13. 10.1128/AEM.00206-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Helm D, Labischinski H, Schallehn G, Naumann D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J Gen Microbiol. 1991;137(1):69–79. . [DOI] [PubMed] [Google Scholar]
- 29. Rau J, Perz R, Klittich G, Contzen M. Cereulide forming presumptive Bacillus cereus strains from food—differentiating analyses using cultural methods, LC-MS/MS, PCR, and infrared spectroscopy in consideration of thermotolerant isolates [in German]. Berl Münch Tierärztl Wochenschr. 2009;122(1–2):25–36. . [PubMed] [Google Scholar]
- 30. Ward JH. Hierarchical grouping to optimize an objective function. J Amer Statist Assoc 1963;58:236–44. [Google Scholar]
- 31. Rohde J, Rapsch C, Fehr M. Case report: Abscessation due to Streptobacillus moniliformis in a rat [in German]. Prakt Tierarzt. 2008;89(6):466–73. [Google Scholar]
- 32. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology. 2003;52(5):696–704. . [DOI] [PubMed] [Google Scholar]
- 36. Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of molecular evolution. 1985;22(2):160–74. . [DOI] [PubMed] [Google Scholar]
- 37. Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. 10.1099/ijs.0.64483-0 . [DOI] [PubMed] [Google Scholar]
- 38. Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106(45):19126–31. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eisenberg T, Nesseler A, Nicklas W, Spamer V, Seeger H, Zschöck M. Streptobacillus sp. isolated from a cat with pneumonia. J Clin Microbiol Case Reports. 2014;2014:1–7. 10.1099/jmmcr.0.000562 [DOI] [Google Scholar]
- 40. Roughgarden JW. Antimicrobial therapy of ratbite fever. A review. Archives of Internal Medicine. 1965;(116):39–54. [DOI] [PubMed] [Google Scholar]
- 41. Cohen RL, Wittler RG, Faber JE. Modified biochemical tests for characterization of L-phase variants of bacteria. Appl Microbiol. 1968;16(11):1655–62. Epub 1968/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lambe DW Jr, McPhedran AM, Mertz JA, Stewart P. Streptobacillus moniliformis isolated from a case of Haverhill fever: biochemical characterization and inhibitory effect of sodium polyanethol sulfonate. Am J Clin Pathol. 1973;60(6):854–60. Epub 1973/12/01. . [DOI] [PubMed] [Google Scholar]
- 43. Smith CD, Sampson CC. Studies of Streptobacillus moniliformis from a case of human rat-bite fever. Am J Med Technol. 1960;26:47–50. Epub 1960/01/01. . [PubMed] [Google Scholar]
- 44. Wittler RG, Cary SG. Genus Streptobacillus Levaditi In: Buchanan RE, Gibbons NE, editors. Bergey's manual of determinative bacteriology, 8th edition Baltimore: Williams & Wilkins Co; 1974. p. 378–81. [Google Scholar]
- 45. Pins MR, Holden JM, Yang JM, Madoff S, Ferraro MJ. Isolation of presumptive Streptobacillus moniliformis from abscesses associated with the female genital tract. Clin Infect Dis. 1996;22(3):471–6. Epub 1996/03/01. . [DOI] [PubMed] [Google Scholar]
- 46. Rowbotham TJ. Rapid identification of Streptobacillus moniliformis . Lancet. 1983;2(8349):567 Epub 1983/09/03. . [DOI] [PubMed] [Google Scholar]
- 47. Rygg M, Bruun CF. Rat bite fever (Streptobacillus moniliformis) with septicemia in a child. Scand J Infect Dis. 1992;24(4):535–40. Epub 1992/01/01. . [DOI] [PubMed] [Google Scholar]
- 48. Al Dahouk S, Scholz HC, Tomaso H, Bahn P, Gollner C, Karges W, et al. Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system. BMC Microbiol. 2010;10:269 Epub 2010/10/26. 10.1186/1471-2180-10-269 1471-2180-10-269 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Greenwood JR, Harvey SM. Streptobacillus moniliformis In: Dworkin M, Falkow M, Rosenberg SE, Schleifer KH, Stackebrandt E, editors. The Prokaryotes, a handbook on the biology of bacteria. volume 7, Proteobacteria. New York, NY: Springer; 2006. p. 983–5. [Google Scholar]
- 50. Nolan M, Gronow S, Lapidus A, Ivanova N, Copeland A, Lucas S, et al. Complete genome sequence of Streptobacillus moniliformis type strain (9901). Stand Genomic Sci. 2009;1(3):300–7. Epub 2009/01/01. 10.4056/sigs.48727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Naumann D. Infrared spectroscopy in microbiology In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. Chichester: John Wiley & Sons Ltd; 2000. p. 102–31. [Google Scholar]
- 52. Boot R, Bakker RH, Thuis H, Veenema JL, De Hoog H. An enzyme-linked immunosorbent assay (ELISA) for monitoring rodent colonies for Streptobacillus moniliformis antibodies. Lab Anim. 1993;27(4):350–7. Epub 1993/10/01. . [DOI] [PubMed] [Google Scholar]
- 53. Berger C, Altwegg M, Meyer A, Nadal D. Broad range polymerase chain reaction for diagnosis of rat-bite fever caused by Streptobacillus moniliformis . Pediatr Infect Dis J. 2001;20(12):1181–2. Epub 2001/12/12. . [DOI] [PubMed] [Google Scholar]
- 54. Glasman PJ, Thuraisingam A. Rat bite fever: a misnomer? BMJ Case Rep. 2009;2009 Epub 2009/01/01. 10.1136/bcr.04.2009.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakagomi D, Deguchi N, Yagasaki A, Harada K, Shibagaki N, Kimura M, et al. Rat-bite fever identified by polymerase chain reaction detection of Streptobacillus moniliformis DNA. J Dermatol. 2008;35(10):667–70. Epub 2008/11/20. 10.1111/j.1346-8138.2008.00541.x . [DOI] [PubMed] [Google Scholar]
- 56. Wallet F, Savage C, Loiez C, Renaux E, Pischedda P, Courcol RJ. Molecular diagnosis of arthritis due to Streptobacillus moniliformis . Diagn Microbiol Infect Dis. 2003;47(4):623–4. Epub 2004/01/09. . [DOI] [PubMed] [Google Scholar]
- 57. Woo PC, Wong SS, Teng JL, Leung KW, Ngan AH, Zhao DQ, et al. Leptotrichia hongkongensis sp. nov., a novel Leptotrichia species with the oral cavity as its natural reservoir. J Zhejiang Univ Sci B. 2010;11(6):391–401. Epub 2010/05/28. 10.1631/jzus.B1000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Conrads G, Claros MC, Citron DM, Tyrrell KL, Merriam V, Goldstein EJ. 16S-23S rDNA internal transcribed spacer sequences for analysis of the phylogenetic relationships among species of the genus Fusobacterium . Int J Syst Evol Microbiol. 2002;52(Pt 2):493–9. Epub 2002/04/05. . [DOI] [PubMed] [Google Scholar]
- 59. Sun D, Zhang H, Lv S, Wang H, Guo D. Identification of a 43-kDa outer membrane protein of Fusobacterium necrophorum that exhibits similarity with pore-forming proteins of other Fusobacterium species. Res Vet Sci. 2013;95(1):27–33. Epub 2013/02/26. 10.1016/j.rvsc.2013.01.016 . [DOI] [PubMed] [Google Scholar]
- 60. Kim HS, Lee DS, Chang YH, Kim MJ, Koh S, Kim J, et al. Application of rpoB and zinc protease gene for use in molecular discrimination of Fusobacterium nucleatum subspecies. J Clin Microbiol. 2010;48(2):545–53. Epub 2009/12/04. 10.1128/jcm.01631-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shah HN, Olsen I, Bernard K, Finegold SM, Gharbia S, Gupta RS. Approaches to the study of the systematics of anaerobic, gram-negative, non-sporeforming rods: current status and perspectives. Anaerobe. 2009;15(5):179–94. Epub 2009/08/22. 10.1016/j.anaerobe.2009.08.003 . [DOI] [PubMed] [Google Scholar]
- 62. Strauss J, White A, Ambrose C, McDonald J, Allen-Vercoe E. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe. 2008;14(6):301–9. Epub 2008/12/31. 10.1016/j.anaerobe.2008.12.003 . [DOI] [PubMed] [Google Scholar]
- 63. Jin J, Haga T, Shinjo T, Goto Y. Phylogenetic analysis of Fusobacterium necrophorum, Fusobacterium varium and Fusobacterium nucleatum based on gyrB gene sequences. J Vet Med Sci. 2004;66(10):1243–5. Epub 2004/11/06. . [DOI] [PubMed] [Google Scholar]
- 64. Jalava J, Eerola E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int J Syst Bacteriol. 1999;49 Pt 4:1375–9. Epub 1999/11/11. . [DOI] [PubMed] [Google Scholar]
- 65. Lawson PA, Gharbia SE, Shah HN, Clark DR, Collins MD. Intrageneric relationships of members of the genus Fusobacterium as determined by reverse transcriptase sequencing of small-subunit rRNA. Int J Syst Bacteriol. 1991;41(3):347–54. Epub 1991/07/01. . [DOI] [PubMed] [Google Scholar]
- 66. Harmon-Smith M, Celia L, Chertkov O, Lapidus A, Copeland A, Glavina Del Rio T, et al. Complete genome sequence of Sebaldella termitidis type strain (NCTC 11300). Stand Genomic Sci. 2010;2(2):220–7. Epub 2011/02/10. 10.4056/sigs.811799 ; PubMed Central PMCID: PMCPmc3035275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schleifer KH, Stackebrandt E. Molecular systematics of prokaryotes. Annual review of microbiology. 1983;37:143–87. 10.1146/annurev.mi.37.100183.001043 . [DOI] [PubMed] [Google Scholar]
- 68. Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 1987;37:463–4. [Google Scholar]
- 69. Wullenweber M, Hedrich HJ, Reetz IC. Susceptibility to streptobacillosis of mice is highly influenced by genetic factors. AALAS Bulletin. 1991;30:43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The wave numbers 550–1800 cm-1 and 2800–3200 cm-1 of second derivative spectra were selected and vector normalized. After a principal component analysis, the first 40 components were used for the LDA. In this LDA every isolate was defined as one group. Spectra of Streptobacillus felis are represented by diamonds, Streptobacillus hongkongenis by triangles, Streptobacillus sp. (AHL 370–1) by circles and S. moniliformis by dots. Note that LDA axis 1 and 3 are shown, because axis 3 contains the differences between Streptobacillus sp. (AHL 370–1) and S. moniliformis.
(TIF)
Broth microdilution susceptibility testing was performed with the Merlin Micronaut system; results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) MIC criteria based on CLSI MIC interpretive standards for other non-Enterobacteriaceae and anaerobes [25]. AZM: azithromycin, CIP: ciprofloxacin, CLI: clindamycin, CMP: chloramphenicol, ERY: erythromycin, GEN: gentamicin, MER: meropenem, NAL: nalidixic acid, STR: streptomycin, T/S: trimethoprim/sulfamethoxazole, TEL: telithromycin, TET: tetracycline, R: resistant, I: intermediate susceptible, S: susceptible phenotype, MIC values in mg/L.
(DOCX)
(DOCX)
Data Availability Statement
Data are available from GenBank: BankIt1841833 NCTC11941_16S KT311784; BankIt1841876 NCTC11941_groEL KT311785; BankIt1841876 NCTC11941_gyrB KT311786; BankIt1841876 NCTC11941_recA KT311787.