Abstract
Camptothecin (CAM), a complex pentacyclic pyrroloqinoline alkaloid, is the starting material for CAM-type drugs that are well-known antitumor plant drugs. Although many chemical and biological research efforts have been performed to produce CAM, a few attempts have been made to uncover the enzymatic mechanism involved in the biosynthesis of CAM. Enzyme-catalyzed oxidoreduction reactions are ubiquitously presented in living organisms, especially in the biosynthetic pathway of most secondary metabolites such as CAM. Due to a lack of its reduction partner, most catalytic oxidation steps involved in the biosynthesis of CAM have not been established. In the present study, an NADPH-cytochrome P450 reductase (CPR) encoding gene CamCPR was cloned from Camptotheca acuminata, a CAM-producing plant. The full length of CamCPR cDNA contained an open reading frame of 2127-bp nucleotides, corresponding to 708-amino acid residues. CamCPR showed 70 ~ 85% identities to other characterized plant CPRs and it was categorized to the group II of CPRs on the basis of the results of multiple sequence alignment of the N-terminal hydrophobic regions. The intact and truncate CamCPRs with N- or C-terminal His6-tag were heterologously overexpressed in Escherichia coli. The recombinant enzymes showed NADPH-dependent reductase activity toward a chemical substrate ferricyanide and a protein substrate cytochrome c. The N-terminal His6-tagged CamCPR showed 18- ~ 30-fold reduction activity higher than the C-terminal His6-tagged CamCPR, which supported a reported conclusion, i.e., the last C-terminal tryptophan of CPRs plays an important role in the discrimination between NADPH and NADH. Co-expression of CamCPR and a P450 monooxygenase, CYP73A25, a cinnamate 4-hydroxylase from cotton, and the following catalytic formation of p-coumaric acid suggested that CamCPR transforms electrons from NADPH to the heme center of P450 to support its oxidation reaction. Quantitative real-time PCR analysis showed that CamCPR was expressed in the roots, stems, and leaves of C. acuminata seedlings. The relative transcript level of CamCPR in leaves was 2.2-fold higher than that of roots and the stems showed 1.5-fold transcript level higher than the roots. The functional characterization of CamCPR will be helpful to disclose the mysterious mechanisms of the biosynthesis of CAM. The present study established a platform to characterize the P450 enzymes involved in the growth, development, and metabolism of eukaryotic organisms.
Introduction
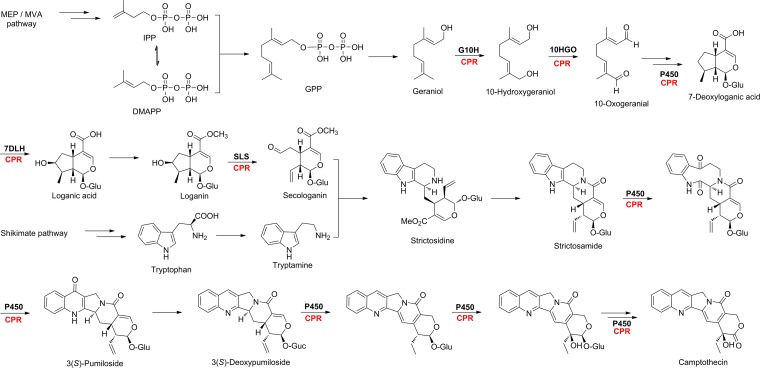
Camptothecin (CAM, Fig 1), a complex pentacyclic pyrroloqinoline alkaloid, was identified firstly from a China native tree Camptotheca acuminata [1]. CAM was verified to induce protein-linked DNA breakage via mammalian DNA topoisomerase I [2]. Since then CAM-type drugs, including 10-hydroxycamptothecin, topotecan, irinotecan and SN-38 were used as efficient anticancer drugs against a broad band of tumor types such as small lung cancer and refractory ovarian cancer [3,4]. These CAM-type drugs were chemically derived from CAM, although a small amount of 10-hydroxycamptothecin could be isolated from plant resources [5]. CAM itself was produced mainly by C. acuminata in China and Nothapodytes foetida in India [6], though several plant species of the Asterid clade, including Icacinaceae (Pyrenacantha klaineana and Merrilliodendron megacrapum), Rubiaceae (Ophiorrhiza pumila and O. mungos), Apocynaceae (Ervatamia heyneana) and Gelsemiaceae (Mostuea brunonis) have been reported to produce CAM [7]. In recent years, the heavy demand for CAM-type drugs has resulted in destructive harvesting of these plants in China and India. Many research efforts have been performed to uncover the mystery of the CAM biosynthesis, which will be helpful to increase the production amount of CAM via plant molecular breeding, metabolic engineering, etc. [8,9,10]. However, the knowledge of the biosynthetic machinery for CAM is yet to be fully deciphered and identified [10].
Fig 1. Putative biosynthetic pathway of CAM [11–20].
The terpenoid precursor secologanin was biosynthesized from MVA and / or MEP pathway through more than five steps of enzymatic oxidation. The amine precursor tryptamine was originated from shikimate pathway to tryptophan and followed by decarboxylation reaction. More than five cytochrome P450 enzymes were predicted to be involved in the conversion from strictosidine to CAM. The possible cytochrome P450 enzymes were highlighted in bold and its reduction partner CPR was in red. The other enzymes involved in the biosynthesis of CAM were omitted for clarity.
CAM was proposed to be derived from strictosidine, a common key intermediate of the monoterpenoid indole alkaloids [11] (Fig 1). Catalyzed by strictosidine synthase, strictosidine was proven to be synthesized from tryptamine and secologanin via the Pictect-Spengler condensation reaction [11] (Fig 1). Subsequent intramolecular cyclization of strictosidine yielded strictosamide, a penultimate precursor of CAM formation in C. acuminata [12] (Fig 1). Strictosamide was converted to CAM via a series of oxidation, rearrangement, cyclization, deglycosylation, dehydrogenation, and hydroxylation reactions (Fig 1). Among them, pumiloside and deoxypumiloside had been thought to be biogenetic intermediates in the formation of CAM from strictosamide [13,14] (Fig 1). Tryptamine, the amine precursor of strictosidine, was proven to be decarboxylation product of tryptophan from shikimate pathway [15] (Fig 1). Secologanin, the monoterpenoid precursor of stictosidine, was believed to be derived from geranyl diphosphate that is biosynthesized from mevalonate and/or non-mevalonate pathways, through a series of hydroxylation, epoxidation, oxidation, glycosylation, methylation, and cyclization steps [16,17,18,19,20] (Fig 1).
Most of the above mentioned reaction steps (Fig 1), including hydroxylation, oxidation, carbon-carbon bond scission, and dehydrogenation reactions, were thought to be catalyzed by various cytochrome P450 monooxygenases (P450s), one of the largest superfamily of enzymes [21]. The catalytic activities of most if not all eukaryotic P450s depend on their reduction partner, cytochrome P450 reductase (CPR, E.C.1.6.2.4). As the electrons donors, CPRs transfer two electrons from NADPH through the FAD and FMN cofactors into the central heme iron of P450s [22]. CPRs were proven to be presented in most living organisms, for instance, yeasts, plants and animals [23–30]. These CPRs were functionally characterized to donate electrons to P450s to support the oxidation reactions catalyzed by P450s [23–30]. Herein we report the molecular cloning, heterologous overexpression, and functional characterization of CamCPR, an NADPH-dependent CPR encoding gene from C. acuminata. The present study will facilitate the cloning and functional characterization of the P450s and thus accelerate the deciphering of CAM biosynthetic mysteries.
Materials and Methods
Plant materials and seedling growth
The fully matured seeds of C. acuminata were collected from matured C. acuminata trees located in the campus (104°4′12″ E, 30°37′59″ N) of Chengdu Institute of Biology of the Chinese Academy of Science, Chengdu, China. The seeds were washed with 5% Triton X-100 for 3 min, rinsed 8 ~ 10 times with sterile water, immersed in 70% EtOH for 1 min, soaked in 1% NaOCl for 3 min, and then re-rinsed 8 ~ 10 times with sterile water. The surface sterile seeds were transferred to half-strength MS medium solidified with 0.35% Phytagel (Sigma-Aldrich Co. LLC., MO) in Erlenmeyer flasks and grown in continuous darkness for 7 days at 25°C. The seedlings were transferred to a growth chamber and grown under a 16-h photoperiod provided by cool white fluorescent light (40 μmol m-2 s-l). The 20-day-seedlings were collected, frozen, and stored at -80°C.
Total RNA isolation
The total RNA was isolated from the seedlings of C. acuminata by using the TransZol UP Kit (Beijing TransGen Biotech Co., Ltd., China), according to the manufacturer’s manual. Briefly, the seedlings were ground into fine powder within liquid nitrogen and the total RNA was extracted using the TransZol UP Kit. The RNA pellets were dissolved in diethylpyrocarbonate (DEPC)-treated water. The quality and quantity of the total RNA were determined by the ratio of OD260 and OD280 recorded from UV-1100D spectrophotometer (Shanghai Mapada Instruments Co., Ltd., China). The total RNA was stored at -80°C for further usage.
Degenerate primer design and reverse transcription—polymerase chain reaction (RT-PCR) amplification of the core amplicon of CamCPR
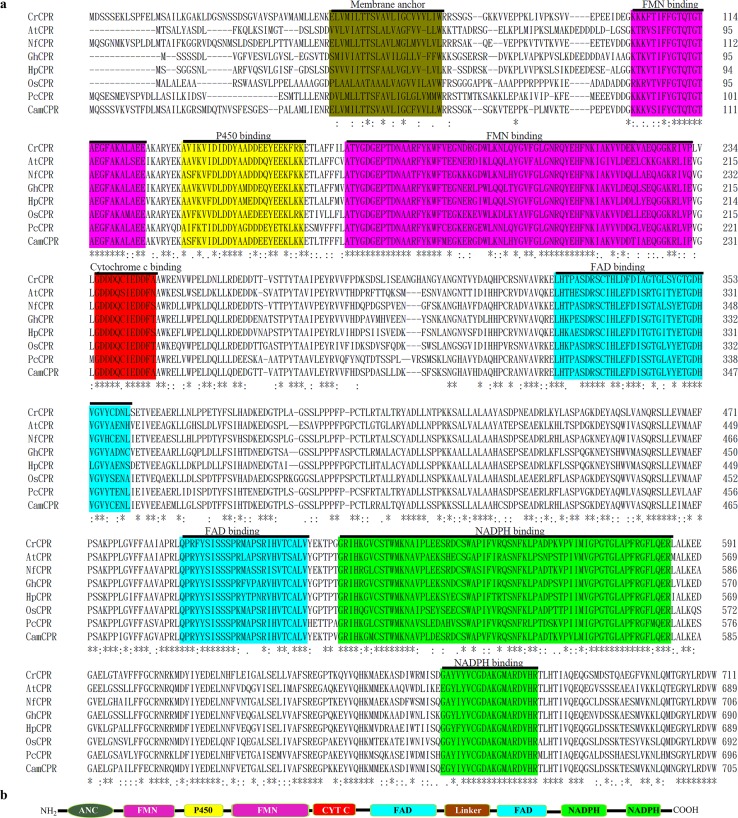
The amino acid residue sequences of the functionally characterized plant CPRs were retrieved from the GenBank database to perform a multiple sequences alignment by using the Clustal Omega multiple alignment tool to identify the conserved amino acid residues for a homology cloning strategy to clone the possible CPR encoding gene from C. acuminata (Fig 2). All characterized plant CPRs were proved to be membrane proteins, containing an N-terminal membrane anchor domain (Fig 2). Five highly conserved domains, including FMN-, FAD-, NADPH-, P450-, and cytochrome c-binding domains, were found in the amino acid residue sequence and structure of plant CPRs (Fig 2). Degenerate primers (Table 1) were designed on the basis of conserved amino acid residues of the FMN- and NADPH-binding sites of characterized plant CPRs (Fig 2). The primers were synthesized and purified by Sangon Biotech (Shanghai) Co., Ltd. Using the template total RNA and the degenerate primers, the CamCPR partial DNA was amplified by RT-PCR with One Step RNA PCR Kit (Tiangen Biotech (Beijing) Co., Ltd, China) following the standard RT-PCR program: 1 cycle of 50°C for 30 min, 1 cycle of 94°C for 2 min, 35 cycles of 94°C for 60 s, 43°C for 30 s, and 65°C for 2 min followed by a final extension at 65°C for 10 min in a thermal cycler (Eppendorf AG, Hamburg, Germany). The amplified PCR product was gel-purified and ligated into the pGM-T vector (Tiangen Biotech (Beijing) Co., Ltd, China). The constructs were transformed into E. coli DH5α competent cells and sequenced in both directions in Sangon Biotech (Shanghai) Co., Ltd. The nucleotide sequence was analyzed by using the similarity search BLAST program.
Fig 2. Multiple sequence alignment of the characterized plant CPRs and the deduced amino acid sequence of CamCPR using Clustal Omega multiple alignment tool (a) and schematic representation of the key domains of CamCPR (b).
The conserved membrane anchor, FMN-, FAD-, P450-, cytochrome c-, and NADPH-binding domains were highlighted in different colors. The identical amino acid residues within each domain were highlighted in the same color. The cytochrome P450 reductases shown here were CrCPR from Catharanthus roseus (X69791), AtCPR from Arabidopsis thaliana (NM_118585), NfCPR from Nothapodytes foetida (EU604540); GhCPR from Gossypium hirsutum (FJ719368), HpCPR from Hybrid poplar (AF302496), OsCPR from Oryza sativa (AF302496), and PcCPR from Petroselinum crispum (AF024635). ANC, membrane anchor sequence; FMN, flavin mononucleotide binding domain; P450, cytochrome P450 binding domain; FAD, flavin adenine dinucleotide binding domain; NADPH, nicotine amide dinucleotide phosphate binding domain.
Table 1. List of primers used in the study.
| Primer code | Sequence (5′-3′) | Direction | Application |
|---|---|---|---|
| CamCPR-F | CA(A/G)AC(C/T)GGIACIGC(C/T)GA(A/G) G | Forward | Degenerate primers |
| CamCPR-R | TT(A/G)GC(A/G)TCACC(A/G)CAIACATA | Reverse | |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | Forward | 5′-RACE |
| 5GSP1 | TCATACTGCCTGTTGCCGAGAC | Reverse | |
| 5GSP2 | CCATCTCCGTATGTAGCTAAGAAG | Reverse | |
| 3GSP1 | TACCGTCGTTCCACTAGTGATTT | Forward | 3′-RACE |
| 3RACE Outer Primer | TACCGTCGTTCCACTAGTGATTT | Reverse | |
| CamCPR-Full-F | CGGTAAGATGCAATCGAGTTCG | Forward | Full length cloning |
| CamCPR-Full-R | GAATTGGCAGCGAAGGTGAGTA | Reverse | |
| His6-CamCPR-F | GGAATTCCATATGCAATCGAGTTCGG | Forward | Overexpression |
| His6-tCamCPR-F | GGAATTCCATATGTCGTCAGGAAAGTC | Forward | |
| His6-CamCPR-R | CCGCTCGAGTCACCACACATCACGC | Reverse | |
| CamCPR-His6-F | CATGCCATGGCATCGAGTTCGGTTAAG | Forward | |
| tCamCPR-His6-F | GGAATTCCATATGTCGTCAGGAAAGTC | Forward | |
| CamCPR-His6-R | CCCAAGCTTCCACACATCACGCAAATACC | Reverse | |
| rtCamCPR-F | TGAAACTGGTGCACTCTCTGAG | Forward | Quantitative real-time PCR analysis |
| rtCamCPR-R | CAGTCTTGGAGCTGTCTAGAGATC | Reverse | |
| rtActin-F | TTCCTCATGCCATCCTCCGTCTT | Forward | |
| rtActin-R | CAGCGATACCTGAGAACATAGTGG | Reverse |
Rapid amplification of cDNA ends (RACE) of CamCPR
To get the complete open reading frame of CamCPR by using RACE methods, gene-specific primers (GSPs, Table 1) were designed on the basis of the nucleotide sequence of the core amplicon of CamCPR. The 5′- and 3′-ends of CamCPR were obtained by using 5′-RACE system for Rapid Amplification of cDNA Ends, Version 2.0 (Invitrogen, Shanghai) and 3′-Full RACE core set with PrimeScript RTase (Takara Biotechnology (Dalian) Co., Ltd, China), respectively, according to the manufacture’s corresponding instructions. Briefly, 5GSP1 was used as primer and the total RNA as template to get the first strand cDNA through S. N. A. P. purification. The purified cDNA was tailored with dCTP catalyzed by terminal transferase TdT. PCR amplification of the dC-tailored cDNA was performed using 5GSP2 and AAP as primers to afford the 5′-end of CamCPR, following the cycling conditions: 1 cycle of 94°C for 3 min, 35 cycles of 94°C for 30 s, 50°C for 1 min, 72°C for 2 min followed by a final extension at 72°C for 10 min. Meanwhile, 3′-RACE adaptor was used as primer and the total RNA as template to afford the first strand cDNA of the 3′-end of CamCPR. Subsequent PCR amplification was performed using 3GSP1 and 3′-RACE outer primer to obtain the 3′-end of CamCPR, following the cycling conditions: 1 cycle of 94°C for 3 min, 35 cycles of 94°C for 30 s, 50°C for 1 min, 72°C for 2 min followed by a final extension at 72°C for 10 min. The amplification products of both 5′- and 3′-RACE PCR were gel-purified, ligated into the pGM-T vector, and transformed into E. coli DH5α competent cells, respectively, following the procedure mentioned above. The nucleotide sequences of the 5′- and 3′-ends of CamCPR were sequenced in Sangon Biotech (Shanghai) Co., Ltd.
CamCPR full-length cDNA cloning
The full-length cDNA of CamCPR was generated on the basis of the above mentioned sequences of the core amplicon fragments, 5′- and 3′-RACE products. Two specific primers, CamCPR-Full-F and CamCPR-Full-R (Table 1), were designed and synthesized to clone the full-length of CamCPR from the total RNA of C. acuminata. Subsequently RT-PCR amplification using the above mentioned primers and total RNA as template afforded the desired PCR products. The RT-PCR conditions were 1 cycle of 50°C for 30 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 65°C for 3 min followed by a final extension at 65°C for 10 min. The RT-PCR products were gel-purified, ligated into the pGM-T vector, and transformed into E. coli DH5α competent cells, following the procedure mentioned above. The nucleotide sequences of the full length of CamCPR were sequenced in Sangon Biotech (Shanghai) Co., Ltd.
Bioinformatics analyses of CamCPR and its encoding protein
The full length nucleotide sequence of CamCPR was set as a query to search the nucleotide database of NCBI (http://www.ncbi.nlm.nih.gov/) by using BLAST program. The open reading frame (ORF) of CamCPR was predicted by using Translate tool (http://www.expasy.ch/tools/dna.html/). The properties of the deduced amino acid sequences of CamCPR were estimated by using the ExPASy ProtParam tool (http://www.expasy.ch/tools/protparam.html/). The transmembrane domain and N-terminal signal peptide of CamCPR were predicted by using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and SignalP (http://www.cbs.dtu.dk/services/SignalP/), respectively. The subcellular location of CamCPR was predicted by using WoLF PSORT (http://wolfpsort.org/). Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) was used for multiple sequence alignment. Structurally and functionally important regions were identified in the deduced amino acid sequence of CamCPR by Conseq services (http://consurf.tau.ac.il/). To assess the evolutionary relationships between the CamCPR and other CPR homologs from different plants species, the CamCPR was set as a query to search the database of NCBI by using BLASTp searches and the amino acid sequences of characterized CPRs were retrieved from NCBI. Then the amino acid residue sequences of CPRs were aligned using the ClustalW program (http://www.ebi.ac.uk). A phylogenetic tree was constructed by neighbour-joining method using MEGA 5 software [31]. Bootstrap analysis with 1,000 replicates was also conducted in order to obtain confidence levels for the branches.
Heterologous overexpression of CamCPR in E. coli
Different forward and reverse primers (Table 1) with various endonucleases restriction sites were designed and synthesized to amplify the expected nucleotide sequence of CamCPR by PCR using a HiFi Taq DNA polymerase (Sangon Biotech (Shanghai) Co., Ltd., China). It should be noted that two forward primers (Table 1), His6-tCamCPR-F for N-terimal His6-tag and tCamCPR-His6-F for C-terminal His6-tag, were designed to truncate the N-terminal 69 amino acid residues of CamCPR to afford the truncated CamCPRs (tCamCPR), according to a previous report [18]. The PCR conditions used were: 1 cycle of 94°C for 3 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min followed by a final extension at 72°C for 10 min. The PCR products were gel-purified, digested with the corresponding endonucleases, and subcloned into the corresponding vectors (pET-28a for intact CamCPR and pET-30a for tCamCPR) digested with the same endonucleases to afford the expression constructs. The E. coli BL21(DE3) component cells were transformed with the expression constructs to afford the recombinant strains.
For protein expression, a single colony of recombinant strain was inoculated into 5 mL of Luria–Bertani (LB) broth containing 50 μg/mL of kanamycin and incubated overnight at 37°C, 200 rpm in a shaking incubator. An aliquot culture (1 mL) was inoculated into 500 mL of Terrific Broth (TB) medium supplemented with 50 μg/mL of kanamycin and incubated at 37°C and 200 rpm. When the optical density (A600 nm) of the culture reached 0.8, the destination protein overexpression was induced by adding 1mM of isopropyl β-D-1-thiogalactopyranoside (IPTG; Sangon, Shanghai, China) into the culture. The culture was incubated at 28°C for another 16 h. Cells were harvested by centrifugation at 4000 rpm for 15 min at 4°C, washed twice with MOPS buffer (100 mM MOPS, 10% glycerol, 0.2 mM DTT, 1 mM EDTA, adjusted to pH 7.3 with NaOH, 4°C), re-suspended in the same buffer containing 1 mg/mL lysozyme and 1 mM phenylmethylsulfonyl fluoride (PMSF), and kept at 4°C for 30 min. The suspended cells were sonicated on ice-bath followed by centrifugation at 15,000 rpm for 30 min at 4°C. For purification of the membrane proteins, one more step was applied before the affinity chromatography. The supernatant was mixed with Triton X-100 (10%) to solubilize the membrane proteins with stirring gently on ice for 2 h. The mixture was centrifuged at 15,000 rpm for 30 min at 4°C and the resulting supernatant was incubated with nickel-nitrilotriacetic acid resin (Sangon) for 30 min. The mixtures of membrane protein or soluble protein and resin were loaded on to a gravity flow column and then eluted with MOPS buffer containing different concentration of imidazole (10, 50, and 250 mM). Purified protein was desalted by dialysis membranes (Sangon) with Tris-HCl buffer (50 mM Tris-HCl, pH 7.4). The purified protein was stored with Tris-HCl buffer containing 20% glycerol at -20°C. The purified protein samples were analyzed on 10% SDS-PAGE and their concentrations were estimated by using the ε 280 nm calculated from ExPASY ProParam as follows: His6-CamCPR and CamCPR-His6 (ε 280 nm = 90,020 M-1 cm-1), His6-tCamCPR and tCamCPR-His6 (ε 280 nm = 84,395 M-1 cm-1).
For the bacterial co-expression system used for P450 monooxygenase activity assay, the tCamCPR was inserted into NdeI / XhoI sites of pETDUET-1 (Novagen, Madison, USA), and the native ORF of CYP73A25 was inserted into the SalI / NotI sites of either empty pETDUET-1 or pETDUET-1 harboring tCamCPR. The constructs were transferred into E. coli BL21(DE3) component cells. The recombinant strain was overexpressed in TB medium with 100 μg/mL of ampicillin as described above. The bacterial cells were collected by centrifugation at 4000 rpm for 5 min at 4°C, washed twice with chill MOPS buffer, re-suspended in the same buffer containing 1 mg/mL lysozyme, and kept at 4°C for 30 min. After sonication on ice-bath, the cell lysate was centrifuged at 12,000 rpm for 10 min, and the supernatant was subjected to P450 activity assay. Overexpression of CYP73A25 and tCamCPR protein was confirmed by SDS-PAGE analysis. The reduction activity of tCamCPR was validated by cytochrome c reducing activity assay.
Enzymatic activity of CamCPR toward protein substrate
Cytochrome c reducing activity of CamCPR was assayed as described previously with minor modifications [32]. The reduction was monitored by the increase of absorbance at 550 nm, at 25°C. Briefly, 500 μL of the reaction mixture contained 25 μM cytochrome c in 50 mM Tris-HCl buffer (pH 7.4) and 1 μg protein. The reaction was initiated by adding 25 μM NADPH. The time dependent absorption change at 550 nm was recorded on a UV-1100D spectrophotometer. An absorption coefficient of 21 mM-1 cm-1 for equine heart cytochrome c was used for quantification. To determine kinetic parameters for cytochrome c, 100 μM NADPH was added to the reaction mixtures containing various concentrations of cytochrome c. The kinetic parameters for NADPH were measured by using 75 μM cytochrome c with varying NADPH concentrations. The kinetic constants K m and V max were calculated with nonlinear regression analysis using Origin 8.0 software (OriginLab Corporation, Northampton, USA). All concentration points were assayed in duplicate.
Enzymatic activity of CamCPR toward chemical substrate
According to the reported procedure [23], the ability of CamCPR to reduce chemical substrate ferricyanide was determined using 6.25 ~ 200 μM of substrate and 10 nM CamCPR in 50 mM phosphate buffer (pH 8.0). The change in absorbance at 420 nm for ferricyanide substrate was measured following the addition of varying NADPH concentrations. The reduction rates were calculated using extinction coefficients of 1.02 mM-1cm-1 for ferricyanide. The kinetic parameters were determined as mentioned above.
CamCPR supported heterogenous P450 monooxygenase activity
A functionally characterized P450 monooxygenase from cotton, CYP73A25 [24], was used to validate the function of CamCPR as a cytochrome P450 reductase. Overexpression of CYP73A25 and tCamCPR were performed as mentioned above. The cinnamic acid 4-hydroxylase activity was assayed in 50 mM phosphate buffer (pH 7.4) containing 1 mM trans-cinnamic acid, 1 mM glucose-6-phosphate and 1 mM glucose-6-phosphate dehydrogenase. The reaction was initiated by adding 2 mM NADPH and incubated at 30°C for 2 h. To quench the reaction, an equal volume of chill CH3OH was added to the reaction mixture. The formation of p-coumaric acid was determined by an HPLC equipped with an Altima C18 analytical column (250 mm×4.6 mm, 5 μm). The mobile phase consisted of buffer A (CH3OH) and buffer B (H2O), followed a linear gradient from buffer A/buffer B (20:80) to 95% buffer A over 22 min, from 95 to 20% buffer A over 2 min and continued at 20% buffer A for an additional 2 min, at a flow rate of 1 mL/min at 35°C, and monitored by a DAD detector.
Molecular modelling and 3-D structure prediction of CamCPR
The crystal structure of Rattus norvagicus CPR (RnCPR, PDB ID: c1j9zB) was resolved in high resolution. Here it was used as template to build the 3-D structure of CamCPR by using the toolkit from PHYRE2 server (Protein Homology/analogY Recognition Engine V 2.0, http://www.sbg.bio.ic.ac.uk/phyre2/html/). Based on the 3-D structure of RnCPR, the structurally, evolutionarily and functionally important regions of CamCPR were identified by using ConSurf software (http://consurf.tau.ac.il/) from the deduced amino acid residue sequence of CamCPR. The stereo-chemical and topological analyses of the modelled CamCPR were performed by using Ramachandran plotting obtained from Procheck module of the SAVES server (http://services.mbi.ucla.edu/SAVES/) and PDBSum (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html), respectively.
Quantitative real-time PCR
The C. acuminata seedlings were grown under identical conditions and three different individuals were set as biological replicates. A pair of specific primers, rtCamCPR-F and rtCamCPR-R, was designed for real-time PCR of CamCPR (Table 1), according to the instructions of real-time PCR system. Also as a control, a pair of primers for the reference gene actin was synthesized too (Table 1). To quantitate the tissue-specific expression of CamCPR, the fresh C. acuminata seedlings of 20 days were collected to extract the total RNA using the TransZol UP Kit as described before. According to the manufacturer’s instructions (Tiangen), the total RNA solution was pre-incubated at 72°C for 5 min to reduce secondary structures when used. Using the TIANScript cDNA synthesis kit (Tiangen), the cDNA (in a volume of 20 μL) was synthesized from 2 μg of the pre-incubated total RNA and the oligo (dT) primer. The above prepared cDNA of each sample was used as template for SYBR green PCR amplification to quantitate the tissue-specific expression of the target genes. The SYBR green PCR amplification was performed using the PIKOREAL24 real-time PCR System (Thermo Scientific, USA) according to the manufacturer’s instructions. Briefly, a total volume of 25 μL SYBR green PCR amplification solution contained 0.25 μL cDNA template, 0.2 μM of the primers, and 12.5 μL of AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China). The PCR amplification was performed under the following cycling conditions: 1 cycle of 95°C for 5 min; 60 cycles of 95°C for 10 s, 58°C for 30 s, 65°C for 30 s. Each sample and the negative control without template were conducted in triplicate. To test for primer dimers and unspecific products, the melting curves of the PCR products were recorded from 60°C to 95°C on the PIKOREAL24 real-time PCR System after the PCR amplification. The real-time PCR amplification curves and the following PCR products melting curves indicated that the primers were specific for CamCPR. The relative transcript levels of CamCPR in different tissues were calculated using the corresponding transcript levels of the reference gene encoding actin in the same tissue as control. And then the mRNA amount of CamCPR in roots was set as control to compare the expression levels of CamCPR in different tissues.
Results
Molecular cloning of the full length cDNA encoding CamCPR
A 1673-bp fragment of the CamCPR gene was obtained from RT-PCR using the above mentioned primers with total RNA as template. Subsequent 5´- and 3´- flanking regions were obtained by using RACE strategy. An 865- and a 659-bp fragment, respectively, were obtained from 5´- and 3´-RACE experiments. By overlapping the nucleotide sequences of the three fragments, the full-length sequence of CamCPR was generated, which was confirmed by molecular cloning and subsequent DNA sequencing.
The full nucleotide sequence of CamCPR was 2606 bp and contained a 2127-bp ORF encoding for a 708-amino acid protein (Fig 2). The ORF was flanked by a 135-bp 5′-untranslated region (UTR) and a 344-bp 3′-UTR. The nucleotide sequence of the full-length cDNA of CamCPR was deposited in NCBI GenBank under the accession number KP162177.
Bioinformatics properties of CamCPR
Using the ExPASy online tools, the theoretical isoelectric point of the deduced CamCPR was predicted to be 5.3. The molecular weight of CamCPR was 78.8 kDa. The deduced CamCPR contained the hallmarks of plant CPRs, including the membrane anchor, FMN-, FAD-, P450-, cytochrome c-, and NADPH-binding domains (Fig 2). A hydrophobic transmembrane region consisting of 19 amino acid residues was predicted at the N-terminus of CamCPR, which may function as an anchor involved in its location on the membrane of the endoplasmic reticulum. No obvious signal peptide cleavage site was predicted in CamCPR. Secondary structure prediction indicated that CamCPR consisted of α-helixes (35.77%), β-sheets (10.11%), and loops (54.12%).
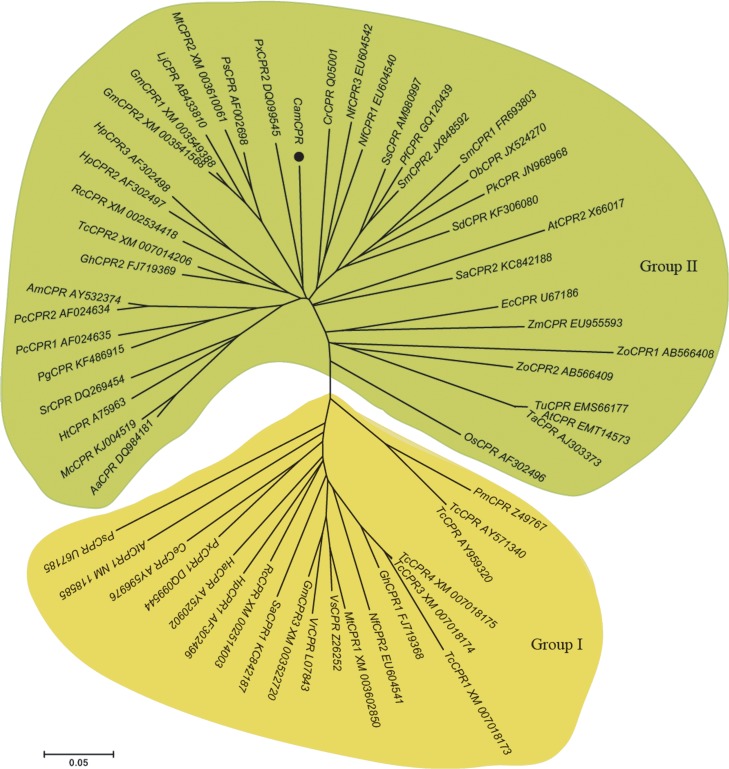
Similarity search showed that CamCPR shares 70 ~ 85% identities with characterized plant CPRs. To gain insights into the evolutionary relationship among CPRs from different plants, 50 CPRs from 33 dicotyls and 9 CPRs from 6 monocotyls were selected from NCBI database to align with CamCPR. A phylogenetic tree was generated by neighbor-joining method using MEGA 5 software (Fig 3). Plant CPRs were classified into groups I and II, based on the N-terminal hydrophobic regions [25]. CamCPR was phylogenetically grouped with AtCPR2, CrCPR and NfCPR1 of group II, which suggested a close evolutionary relationship within these enzymes.
Fig 3. Phylogenetic tree of the cytochrome P450 reductase family members.
Homologous sequences were obtained using the National Center for Biotechnology Information search engine (http://www.ncbi.nim.bih.gov/). The GenBank accession numbers for the sequences were as follows: CrCPR (Catharanthus roseus, X69791); AtCPR1 (Arabidopsis thaliana, NM_118585); AtCPR2 (Arabidopsis thaliana, X66017); NfCPR1 (Nothapodytes foetida, EU604540); NfCPR2 (Nothapodytes foetida, EU604541); NfCPR3 (Nothapodytes foetida, EU604542); GhCPR1 (Gossypium hirsutum, FJ719368); GhCPR2 (Gossypium hirsutum, FJ719369); HpCPR1 (Hybrid poplar, AF302496); HpCPR2 (Hybrid poplar, AF302497); HpCPR3 (Hybrid poplar, AF302498); OsCPR (Oryza sativa Japonica, AF302496); PcCPR1 (Petroselinum crispum, AF024634); PcCPR2 (Petroselinum crispum, AF024635); EcCPR (Eschscholzia californica, U67186); HtCPR (Helianthus tuberosus, A75963); VrCPR (Vigna radiate, L07843); PsmCPR (Papaver somniferum, U67185); PstCPR (Pisum sativum, AF002698); VsCPR (Vicia sativa, Z26252); AtCPR (Aegilops tauschii, EMT14573); AmCPR (Ammi majus, AY532374); AaCPR (Artemisia annua, DQ984181); CeCPR (Centaurium erythraea, AY596976); GmCPR1 (Glycine max, XM_003549388); GmCPR2 (Glycine max, XM_003541568); GmCPR3 (Glycine max, XM_003522720); HaCPR (Hypericum androsaemum, AY520902); LjCPR (Lotus japonicas, AB433810); McCPR (Matricaria chamomilla, KJ004519); MtCPR1 (Medicago truncatula, XM_003602850); MtCPR2 (Medicago truncatula, XM_003610061); ObCPR (Ocimum basilicum, JX524270); PgCPR (Panax ginseng, KF486915); PfCPR (Perilla frutescens, GQ120439); PxCPR1 (Petunia x hybrid, DQ099544); PxCPR2 (Petunia x hybrid, DQ099545); PkCPR (Picrorhiza kurrooa, JN968968); PmCPR (Pseudotsuga menziesii, Z49767); RcCPR1 (Ricinus communis, XM_002514003); RcCPR2 (Ricinus communis, XM_002534418); SmCPR1 (Salvia miltiorrhiza, FR693803); SmCPR2 (Salvia miltiorrhiza, JX848592); SaCPR1 (Santalum album, KC842187); SaCPR2 (Santalum album, KC842188); SdCPR (Scoparia dulcis, KF306080); SsCPR (Solenostemon scutellarioides, AM980997); SrCPR (Stevia rebaudiana, DQ269454); TchCPR (Taxus chinensis, AY959320); TcsCPR (Taxus cuspidate, AY571340); TcCPR1 (Theobroma cacao, XM_007018173); TcCPR2 (Theobroma cacao, XM_007014206); TcCPR3 (Theobroma cacao, XM_007018174); TcCPR4 (Theobroma cacao, XM_007018175); TaCPR (Triticum aestivum, AJ303373); TuCPR (Triticum urartu, EMS66177); ZmCPR (Zea mays, EU955593); ZoCPR1 (Zingiber officinale, AB566408); ZoCPR2 (Zingiber officinale, AB566409); Phylogenetic analysis was performed using the MEGA package and neighbor-joining program (http://www.megasoftware.net). The scale bar indicates the phylogenetic distance calculated according to the number of differences.
Heterologous overexpression and catalytic parameters of recombinant CamCPR
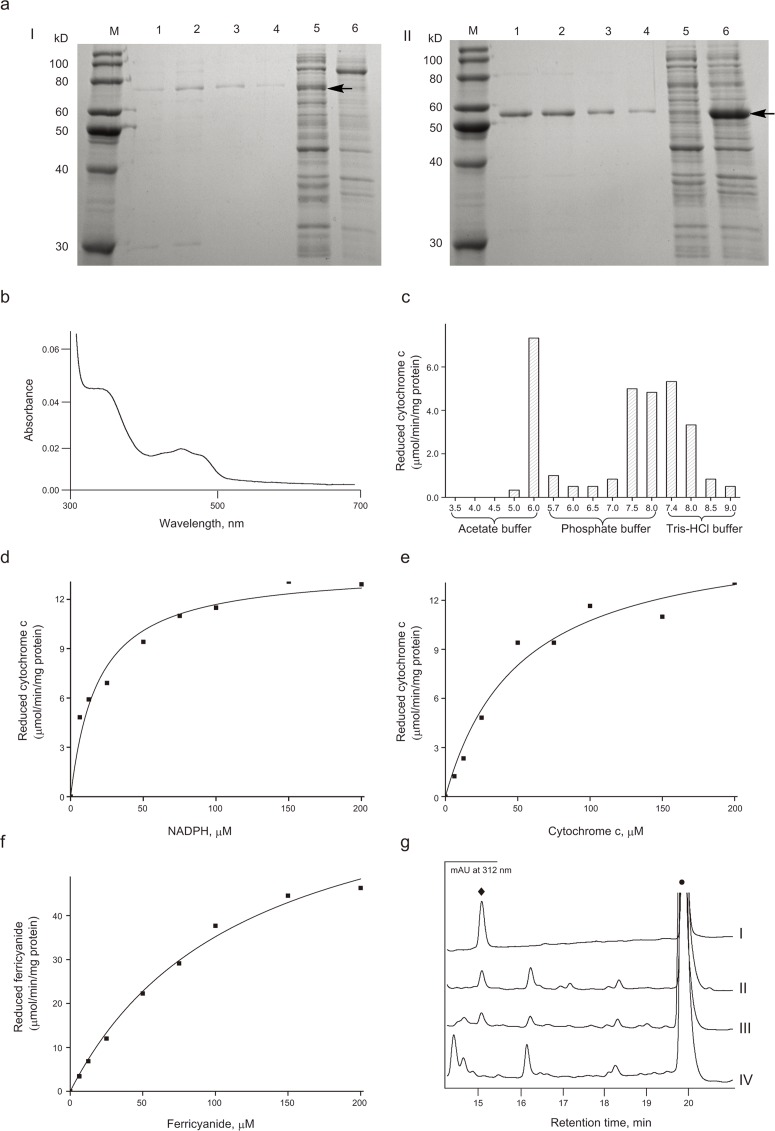
According to the previous report on CrCPR from C. roseus [18], the N-terminal 69 amino acid residues of CamCPR were truncated to afford tCamCPR. The full-length ORF of CamCPR and tCamCPR were subcloned into pET-28a or pET-30a vectors and heterologously overexpressed in E. coli BL21(DE3) as a fusion protein with N- or C-terminal His6-tag (Fig 4a). Similar to other plant CPRs [24], the recombinant tCamCPR showed characteristic absorbance bands of a flavoprotein (Fig 4b). The reaction buffers played an important role in the cytochrome c reducing activities catalyzed with CamCPR (Fig 4c). The CamCPR showed the highest reduction activity towards cytochrome c when the reduction assay was performed in the acetate buffer with pH = 6.0. However no reduction activity was observed when it was assayed in the same acetate buffer system with pH < 4.5. Meanwhile when the reduction assays were performed in the phosphate buffer system with pH = 6.0, the CamCPR showed weaker reduction activity. However, the reduction activity of CamCPR showed constant between pH = 7.4–8.0 whether the reaction buffer is phosphate or tris-HCl buffer system. The following assays were performed in 50 mM tris-HCl buffer at pH 7.4, although the optimal cytochrome c reducing activity was found in acetate buffer at pH 6.0 (Fig 4c).
Fig 4. Overexpression, purification, and characterization of recombinant CamCPR.
a, SDS-PAGE analyses of CamCPR (I) and tCamCPR (II). M, protein ruler; Lanes 1–4, purified enzyme; Lane 5 in I, cell lysate; Lane 5 in II, whole cell; Lane 6 in I, whole cell; Lane 6 in II, cell lysate. The target band was indicated with an arrowhead. b, the UV spectrum of tCamCPR was measured in 50 mM tris-HCl buffer (pH 7.4). c, the effects of different buffers with various pH on the cytochrome c reducing activity of tCamCPR. d–f, the steady-state kinetic constants of recombinant CamCPR. g, HPLC-DAD analyses of the reaction mixture of CamCPR supported cinnamic acid 4-hydrxoylase activity. Panel I, the authentic p-coumaric acid (♦) and trans-cinnamic acid (•); The HPLC traces of the whole reaction containing the cell lysates (panel II), the cells (panel III), and the boiled cells (panel IV) of the recombinant tCamCPR and CYP73A25 as catalyst.
The recombinant CamCPRs were assayed for NADPH- or NADH- dependent cytochrome c reduction activities, respectively (Table 2). NADPH was efficiently served as the electron donor and NADH could not be recognized by CamCPR, which means that the activity of CamCPR is dependent on NADPH (Table 2). However, the requirement of CPRs for electron acceptors was relatively less specific. Cytochrome c and ferricyanide [K3Fe(CN)6] can serve as the electron acceptors (Fig 4e and 4f). The kinetic parameters K m and V max of tCamCPR for NADPH, cytochrome c, and K3Fe(CN)6 were determined, respectively. The K m and V max of tCamCPR were 18.7 ± 3.6 μM and 13.8 ± 0.6 μmol/min/mg protein for NADPH (Fig 4d), 51.4 ± 12.6 μM and 16.3 ± 1.5 μmol/min/mg protein for cytochrome c (Fig 4e), and 119.3 ± 17.2 μM and 77.2 ± 4.7 μmol/min/mg protein for K3Fe(CN)6 (Fig 4f).
Table 2. Specific activities of various CamCPRs toward cytochrome c (25 μM), in the presence of 25 μM of NADPH or NADH.
Values are presented as mean ± SE. ND, not detected.
| Specific activity (μM/min/mg protein) | ||
|---|---|---|
| NADPH | NADH | |
| His6-CamCPR | 1.071±0.038 | ND |
| His6-tCamCPR | 3.258±0.021 | ND |
| CamCPR-His6 | 0.038±0.006 | ND |
| tCamCPR-His6 | 0.185±0.011 | ND |
CamCPR supported heterogenous P450 monooxygenase activity
The tCamCPR was inserted into pETDUET-1, followed by the insertion of the whole ORF of CYP73A25 from the plasmid pETDUET-1-CYP73A25-GhCPR [24]. The construct was overexpressed in E. coli BL21(DE3), induced by IPTG. The cell lysate was incubated with trans-cinnamic acid, the native substrate of CYP73A25. Comparison the HPLC-DAD traces of the whole reaction with cell lysate as catalyst (panel II, Fig 4g) and the whole reaction with whole cell as catalyst (panel III, Fig 4g) with that of the authentic standards (panel I, Fig 4g) and the whole reaction with boiled cells as control (panel IV, Fig 4g) demonstrated the formation of p-coumaric acid, the desired product of cinnamate 4-hydroxylase, which confirmed that tCamCPR functionally supports CYP73A25 to convert trans-cinnamic acid to form p-coumaric acid, i.e., 4-hydroxycinnamic acid.
Prediction of three-dimensional structure of CamCPR
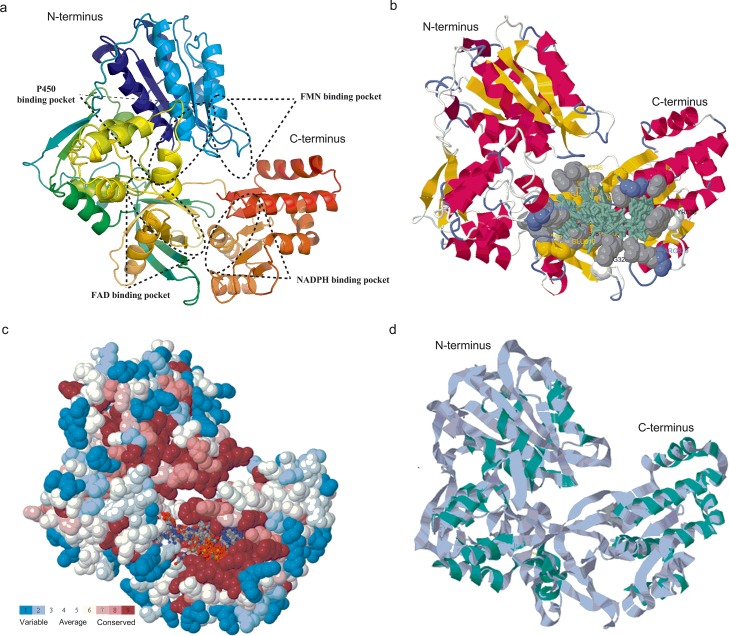
Based on the crystal structure of RnCPR (PDB ID: c1j9zB), the 3-D structure of CamCPR was predicted and constructed using different bioinformatics software (Fig 5). The presence of the conserved FMN-, FAD-, NADPH-, and P450-binding domains was also deduced from the predicted 3-D structure of CamCPR (Fig 5a). As depicted in Fig 5b, the amino acid residues involved in the binding of ligands such as FAD and NADPH were also predicted using the 3DLigandSite tool. Analyses of the evolutionary conservation of CamCPR surface amino acids indicated that many amino acid residues were conserved in CPRs. The amino acid residues with high scores were highlighted in red and were found to be the functional and structural residues of CamCPR by the bioinformatics tool of ConSeq servers (Fig 5c). Superimposition of the 3-D structures of CamCPR with the template RnCPR showed that all major domains aligned at same coordinates (Fig 5d), which indicated that CPRs were highly conserved reductases in eukaryotic organisms. The predicted structure of CamCPR was further refined by employing the bioinformatics tools from KoBaMIN web server and then the stereo-chemical qualities of the energy refined model of CamCPR was validated by the bioinformatics tools of the PROCHECK server. The results using Ramachandran plotting analysis showed that 91.6% amino acid residues of CamCPR located in the most favourable region, 6.1% residues in the additional allowed region, 1.5% in the generously allowed region, and 0.7% in the disallowed region. The relatively low percentage of amino acid residues in the disallowed regions suggested that the 3-D structure of CamCPR was validated.
Fig 5. The predicted 3-D structure of CamCPR.
a, schematic FMN-, FAD-, NADPH-, and P450-binding domains of CamCPR, built by the toolkit from PHYRE2 server; b, the amino acid residues involved in the binding of ligands (highlighted in greenish) such as FAD and NADPH, predicted by the 3DLigandSite tool; c, functional and structural residues of CamCPR with high scores were highlighted in red; and d, superimposition of CamCPR with the template RnCPR.
Quantitative analysis of tissue-specific expression of CamCPR
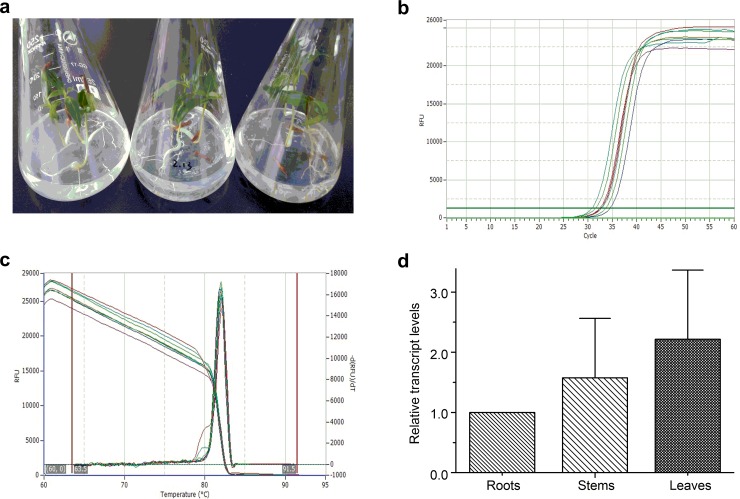
The real-time PCR amplification of CamCPR was performed to evaluate its expression in different tissues (Fig 6). The results showed that CamCPR was expressed in all tissues of C. auminata seedlings. However, the transcript level of CamCPR in stems was 1.5-fold higher than that of the roots. The leaves showed 2.2-fold higher transcript level than the roots.
Fig 6. Quantitative real-time PCR of CamCPR.
a, the 20-day-old C. acuminata seedlings; b, the real-time PCR amplification curves of CamCPR using rtCamCPR-F and rtCamCPR-R primers (Table 1); c, the PCR products melting curves of CamCPR; and d, the relative transcript levels of CamCPR in different tissues. The relative transcript level of CamCPR in the roots was set as control. Values are reported as means with standard error bars of three independent biological samples.
Discussion
CPRs, the reduction partner of eukaryotic cytochrome P450s, transfer electrons from electron donors such as NADPH to the central heme iron of P450s to support their oxidation reactions [22]. CPRs and P450s constitute multicomponent redox enzyme systems [33], which plays a pivotal role in the growth, development, and metabolism of eukaryotic organisms [26]. Generally speaking, each enzymatic oxidation reaction is catalyzed by a special P450 enzyme, which means that there are many P450s presented in eukaryotic organisms. Many CPRs have been identified and functionally characterized from living organisms. It was reported that only one CPR encoding gene presented in yeasts and animals [24,29,30]. The CPR serves as a versatile redox partner to communicate with different P450s and support P450s function in yeasts and animals. Meanwhile plants contain one, two or three paralogs of CPRs with different molecular weights, subcellular localizations and regulatory mechanisms [23,24,25,26,27,28]. Each plant has at least one constitutively expressed CPR to support its growth, development, and metabolism. Some species have inducible CPR that was suggested to support the plants to defend against environmental stresses. For the plants that have only one CPR encoding gene, it was suggested that the CPR is also involved in other biological processes. For C. acuminata, one CPR was obtained in this study using the homology cloning strategy, which was consistent with the transcriptome data analysis of C. acuminata (Fig 7). Four gene fragments encoding putative CPRs, caa_locus_6894, caa_locus_12198, caa_locus_112450, and caa_locus_37170, were retrieved from the Medicinal Plant Genomics Resource consortium (http://medicinalplantgenomics.msu.edu). These gene fragments were deduced to encode 187-, 94-, 58-, and 39-amino acid residues, respectively. The deduced amino acid residue sequence of each putative CPRs was set as query to search NCBI using BLAST program. The caa_locus_37170 was ruled out because it was annotated as a hypothetic protein. The multiple amino acid residues alignment of CamCPR, caa_locus_6894, caa_locus_12198, and caa_locus_112450 showed that the deduced amino acid residues from the transcriptome sequencing are part of CamCPR (Fig 7). The three putative CPRs segments, caa_locus_6894, caa_locus_12198, and caa_locus_112450, were suggested to be presented in young bark, immature leaf, callus, root culture, young flower, immature fruit, mature fruit, leaf, cotylendones, whole seedlings, young leaf, mature leaf, immature bark, and entire root, which was consistent with the real-time quantitative analysis of CamCPR expression. The mRNA of CamCPR was detected in all tissues of C. acuminata seedlings, indicating that the CamCPR is constitutively expressed in C. acuminata to support P450s oxidation reactions.
Fig 7. Amino acid residues alignment of CamCPR, caa_locus_6894, caa_locus_12198, and caa_locus_112450 using Clustal Omega multiple aligment tool.
The identical amino acid residues between CamCPR and caa_locus_6894 were highlighted in red, between CamCPR and caa_locus_12198 were in green, and between CamCPR and caa_locus_112450 were in blue.
It was found that the interacting domains of P450s and CPRs are highly conserved [34]. These domains were presented in CamCPR (Fig 2), which was evidenced by its cytochrome c reducing activity and its electrons supporting to CYP73A25, a functional P450 from cotton, to hydroxylate cinnamic acid to form 4-hydroxycinnamic acid. CPRs from vascular plants were clustered into two major phylogenetic groups based on the N-terminal hydrophobic regions [25]. CamCPR was categorized in group II CPRs, neighboring with AtCPR2, CrCPR and NfCPR1 (Fig 3).
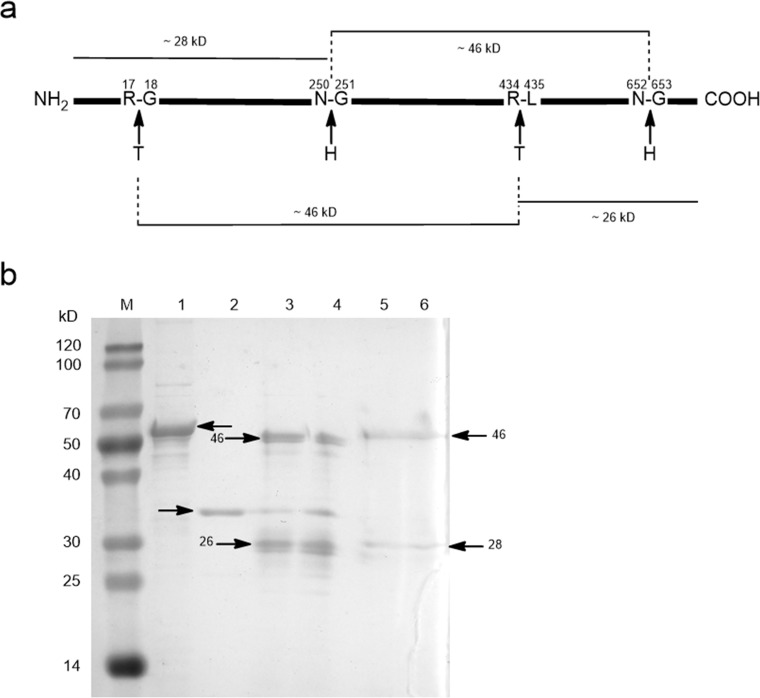
Tryptophan is the last C-terminal amino acid residue of most CPRs, for instance, W705 in CamCPR (Fig 2a). The tryptophan located in the nicotinamide binding site of CPRs, which plays a very important role in the discrimination between NADPH and NADH [35,36]. This amino acid residue serves as a trigger for releasing oxidized nicotinamide [35,36]. The intact and truncate CamCPRs with N- or C-terminal His6-tag were overexpressed and purified to homogeneity. The cytochrome c reducing activity assays showed that the recombinant protein with free last tryptophan residue showed 18- ~ 30-fold higher reduction activity than the fusion enzymes with C-terminal His6-tag (Table 2), which supported a previous conclusion that the free tryptophan is highly conserved and very important in CPRs, together with those mutation experiments [32,33]. It should be noted that the observed molecular weight of tCamCPR from SDS-PAGE analysis (panel II, Fig 4a) is smaller than its theoretical molecular weight. However the tCamCPR can be separated by Ni-NTA affinity resin, indicating an entire N-terminus of tCamCPR. Also the functional characterization suggested that the last tryptophan (W660 of tCamCPR) is present in tCamCPR. Enzymatic digestion and chemical degradation of tCamCPR were performed to confirm it (Fig 8). Thrombin was used to digest tCamCPR, according to the reported procedure [37]. Two main peptide fragments with ~ 46 and 26 kD were predicted (Fig 8a) and detected by SDS-PAGE analysis (Lanes 3 and 4, Fig 8b). The amide bond between Asn and Gly can be cleaved selectively by hydroxylamine [38]. Two cleavage sites were present in tCamCPR (Fig 8a) and the two predicted peptide fragments with ~ 28 and 46 kD (Fig 8a) were also detected by SDS-PAGE analysis (Lanes 5 and 6, Fig 8b). The results showed that tCamCPR is intact. Perhaps the abnormal gel shifting of tCamCPR is due to its amino acid residue composition [39].
Fig 8. Enzymatic digestion and chemical degradation of tCamCPR.
The predicted cleavage sites of tCamCPR (a) and the detected peptide fragments by SDS-PAGE analysis (b). T, thrombin; H, Hydroxylamine; M, protein ruler; Lane 1, tCamCPR; Lane 2, thrombin; Enzyme digestion products of tCamCPR with 1 U / 50 μL (Lane 3) and 2 U / 50 μL (Lane 4) of thrombin were used [37]; Lanes 5 and 6, chemical degradation products of tCamCPR by hydroxylamine [38].
Metabolic engineering is a highly efficient alternative for production of pharmaceutically important natural products in heterologous hosts via biotechnology such as microbial fermentation. However, an important prerequisite for any attempt at metabolic engineering is the detailed knowledge of the underlying biosynthetic and regulatory pathways in plants [27]. The present study and the other previous reports have shown that CPRs are highly conserved and CPRs from different plants can at least partially support the oxidation ability of P450s from different species [36]. According to the ‘‘share your parts” principle [36], CamCPR can be used as an ideal bio-brick in synthetic biology approaches to re-design or develop entirely different combinations of existing biological systems to produce CAM.
Conclusion
In summary, an NADPH-cytochrome P450 reductase CamCPR encoding gene was cloned from C. acuminata. CamCPR showed 70 ~ 85% identities to other characterized plant CPRs. The intact and truncate CamCPR with N- or C- His6-tag were overexpressed in E. coli and purified to homogeneity. CamCPRs showed NADPH-dependent reductase activity toward chemical and protein substrates. CamCPR can support the oxidation activity of a heterogenous P450 from cotton. The characterization and identification of CamCPR not only extends the plant CPRs family, but also facilitates the future functional studies of the enzyme interacting with other components of the cytochrome P450 enzyme systems. Additionally, the cloning and functional characterization of CamCPR will be helpful to uncover the mysterious mechanisms of the biosynthesis of CAM. The present study established a platform to characterize the P450 enzymes involved in the development, growth, and metabolism of plants
Data Availability
All relevant data are within the paper.
Funding Statement
The Science and Technology Project for Outstanding Youths in Life Science (KSCX2-EW-Q-6) from the Chinese Academy of Sciences (www.cas.cn) and the project 21172216 from the National Natural Science Foundation of China (www.nsfc.gov.cn). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wall ME, Wani MC, Cook CE, Palmer KH, McPhail AT, Sim GA (1966) Plant antitumor agents. I. The isolation and structure of camptothecin, a novel alkaloidal leukemia and tumor inhibitor from Camptotheca acuminata . J Am Chem Soc 88: 3887–3890. 5916382 [Google Scholar]
- 2. Hsiang YH, Hertzberg R, Hecht S, Liu LF (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem 260: 14873–14878. [PubMed] [Google Scholar]
- 3. Sirikantaramas S, Asano T, Sudo H, Yamazaki M, Saito K (2007) Camptothecin: therapeutic potential and biotechnology. Curr Pharm Biotechnol 8: 196–202. [DOI] [PubMed] [Google Scholar]
- 4. Demain AL, Vaishnav P (2011) Natural products for cancer chemotherapy. Microbiol Biotechnol 4: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thomas CJ, Rahier NJ, Hecht SM (2004) Camptothecin: current perspectives. Bioorg Med Chem 12: 1585–1604. [DOI] [PubMed] [Google Scholar]
- 6. Lorence A, Nessler CL (2004) Camptothecin, over four decades of surprising findings. Phytochemistry 65: 2735–2749. [DOI] [PubMed] [Google Scholar]
- 7. Shaanker RU, Ramesha BT, Ravikanth G, Gunaga R, Vasudeva R, Ganeshaiah KN (2008) Chemical profiling of Nothapodytes nimmoniana for camptothecin, an important anticancer alkaloid: towards the development of a sustainable production system in: Ramawat KG, Mérillon JM (Eds.) Bioactive molecules and medicinal plants. Springer-Verlag, Berlin and Heidelberg, pp. 197–213. [Google Scholar]
- 8. Kutchan TM (1995) Alkaloid biosynthesis—The basis for metabolic engineering of medicinal plants. Plant Cell 7: 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peng ZH, Hu JJ, Li HS (2010) Application of biotechnology in research of camptothecin. Prog Modern Biomed 10: 3175–3177. [Google Scholar]
- 10. Shen SH, Liu JY, Hu JQ, Li XH, Wang LL (2011) Advances in studies on biosynthetic pathways of camptothecin and their synthases. Chin Trad Herbal Drugs 42: 1862–1868. [Google Scholar]
- 11. Stöckigt J, Barleben L, Panjikar S, Loris EA (2008) 3D-Structure and function of strictosidine synthase—the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol Biochem 46: 340–355. 10.1016/j.plaphy.2007.12.011 [DOI] [PubMed] [Google Scholar]
- 12. Hutchinson CR, Heckendorf AH, Straughn JL, Daddona PE, Cane DE (1979) Biosynthesis of camptothecin. 3. Definition of strictosamide as the penultimate biosynthetic precursor assisted by 13C and 2H NMR spectroscopy. J Am Chem Soc 101: 3358–3369. [Google Scholar]
- 13. Aimi N, Nishimura M, Miwa A, Hoshino H, Sakai S, Haginiwa J (1989) Pumiloside and deoxypumiloside; plausible intermediates of camptothecin biosynthesis. Tetrahedron Lett 30: 4991–4994. [Google Scholar]
- 14. Kitajima M, Yoshida S, Yamagata K, Nakamura M, Takayama H, Saito K, et al. (2002) Camptothecin-related alkaloids from hairy roots of Ophiorrhiza pumila . Tetrahedron 58: 9169–9178. [Google Scholar]
- 15. López-Meyer M, Nessler CL (1997) Tryptophan decarboxylase is encoded by two autonomously regulated genes in Camptotheca acuminata which are differentially expressed during development and stress. Plant J 11: 1167–1175. [DOI] [PubMed] [Google Scholar]
- 16. Collu G, Unver N, Peltenburg-Looman AMG, van der Heijden R, Verpoorte R, Memelink J (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508: 215–220. [DOI] [PubMed] [Google Scholar]
- 17. Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, et al. (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492: 138–142. 10.1038/nature11692 [DOI] [PubMed] [Google Scholar]
- 18. Irmler S, Schröder G, St-Pierre B, Crouch NP, Hotze M, Schmidt J, et al. (2000) Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J 24: 797–804. [DOI] [PubMed] [Google Scholar]
- 19. Murata J, Roepke J, Gordon H, De Luca V (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524–542. 10.1105/tpc.107.056630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simkin AJ, Miettinen K, Claudel P, Burlat V, Guirimand G, Courdavault V, et al. (2013) Characterization of the plastidial geraniol synthase from Madagascar periwinkle which initiates the monoterpenoid branch of the alkaloid pathway in internal phloem associated parenchyma. Phytochemistry 85: 36–43. 10.1016/j.phytochem.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 21. http://drnelson.uthsc.edu/cytochromeP450.html.
- 22. http://www.p450.kvl.dk/p450rel.shtml.
- 23. Lee GY, Kim HM, Ma SH, Park SH, Joung YH, Yun CH (2014) Heterologous expression and functional characterization of the NADPH-cytochrome P450 reductase from Capsicum annuum . Plant Physiol Biochem 82: 116–122 and references therein. 10.1016/j.plaphy.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 24. Yang CQ, Lu S, Mao YB, Wang LJ, Chen XY (2010) Characterization of two NADPH: cytochrome P450 reductases from cotton (Gossypium hirsutum). Phytochemistry 71: 27–35 and references therein. 10.1016/j.phytochem.2009.09.026 [DOI] [PubMed] [Google Scholar]
- 25. Ro DK, Ehlting J, Douglas CJ (2002) Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductase from hybrid poplar. Plant Physiol 130: 1837–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rana S, Latto SK, Dhar N, Razdan S, Bhat WW, Dhar RS, et al. (2013) NADPH-cytochrome P450 reductase: molecular cloning and functional characterization of two paralogs from Withania somnifera (L.) Dunal. PLoS ONE 8: e57068 10.1371/journal.pone.0057068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhat WW, Rana S, Dhar N, Razdan S, Pandith SA, Vishwakarma R, et al. (2014) An inducible NADPH-cytochrome P450 reductase from Picrorhiza kurrooa–an imperative redox partner of cytochrome P450 enzymes. Funct Integr Genomics 14: 381–399 and references therein. 10.1007/s10142-014-0362-7 [DOI] [PubMed] [Google Scholar]
- 28. Huang FC, Sung PH, Do YY, Huang PL (2012) Differential expression and functional characterization of the NADPH cytochrome P450 reductase genes from Nothapodytes foetida . Plant Sci 190: 16–23 and references therein. 10.1016/j.plantsci.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 29. Kaewpa D, Boonsuepsakul S, Rongnoparut P (2007) Functional expression of mosquito NADPH-cytochrome P450 reductase in Escherichia coli . J Econ Entomol 100: 946–953 and references therein. [DOI] [PubMed] [Google Scholar]
- 30. Girhard M, Tieves F, Weber E, Smit MS, Urlacher VB (2013) Cytochrome P450 reductase from Candida apicola: versatile redox partner for bacterial P450s. Appl Microbiol Biotechnol 97: 1625–1635 and references therein. 10.1007/s00253-012-4026-z [DOI] [PubMed] [Google Scholar]
- 31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guengerich FP, Martin MV, Sohl CD, Cheng Q (2009) Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc 4: 1245–1251. 10.1038/nprot.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang M, Roberts DL, Paschke R, Shea TM, Masters BSS, Kim JJP (1997) Three-dimensional structure of NADPH–cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA 94: 8411–8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jensen K, Møller BL (2010) Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 71: 132–141. 10.1016/j.phytochem.2009.10.017 [DOI] [PubMed] [Google Scholar]
- 35. Elmore CL, Porter TD (2002) Modification of the nucleotide cofactor-binding site of cytochrome P-450 reductase to enhance turnover with NADH in vivo. J Biol Chem 277: 48960–48964. [DOI] [PubMed] [Google Scholar]
- 36. Murataliev MB, Feyereisen R, Walker FA (2004) Electron transfer by diflavin reductases. Biochim Biophys Acta 1698: 1–26. [DOI] [PubMed] [Google Scholar]
- 37. Chang JY (1985) Thrombin specificity: Requirement for apolar amino acids adjacent to the thrombin cleavage site of polypeptide substrate. Eur J Biochem 151: 217–224. [DOI] [PubMed] [Google Scholar]
- 38. Simpson RJ (2003) Peptide mapping and sequence analysis of gel-resolved proteins, Chapter 7 In: Simpson (Ed.) Proteins and Proteomics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA. [Google Scholar]
- 39. Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci USA 106: 1760–1765. 10.1073/pnas.0813167106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.