Abstract
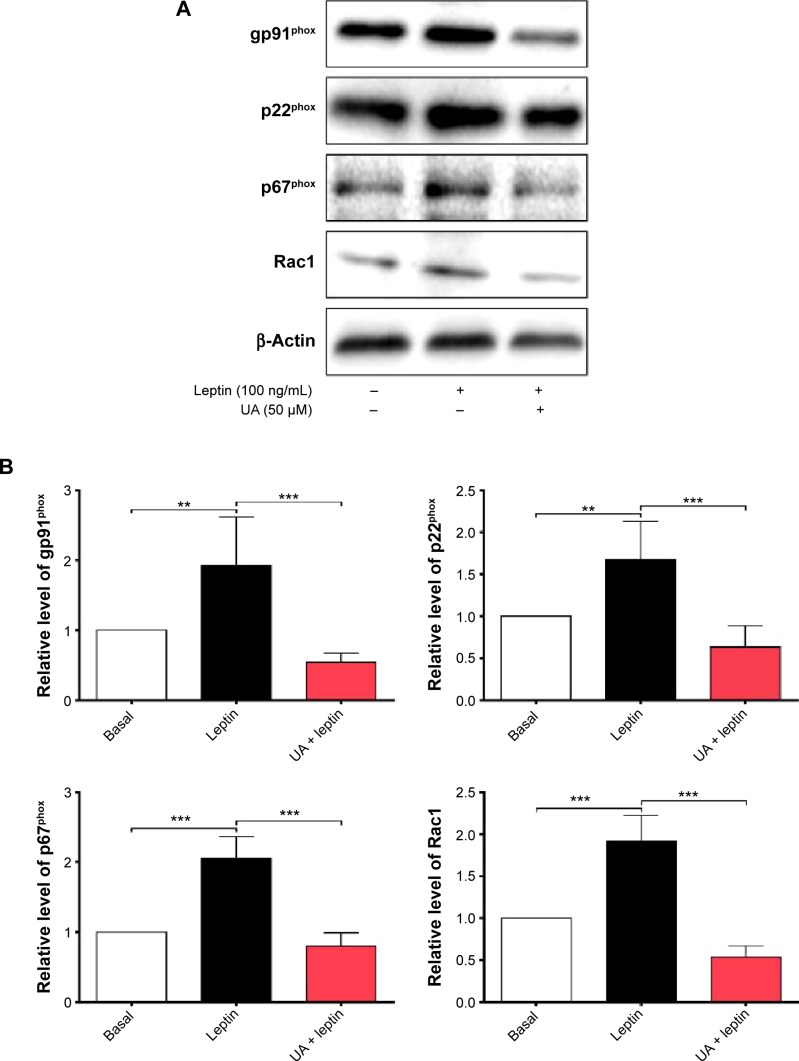
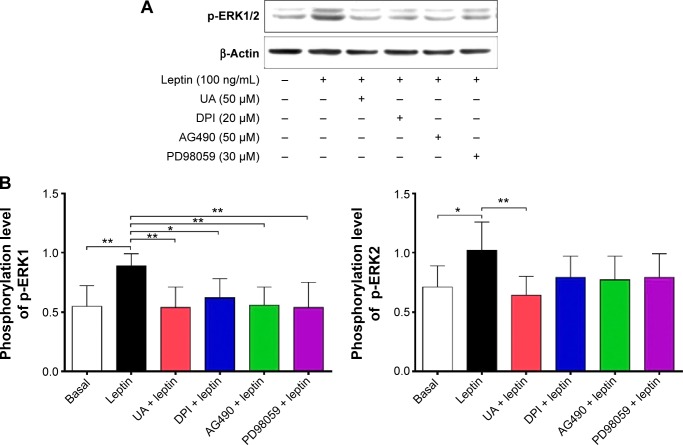
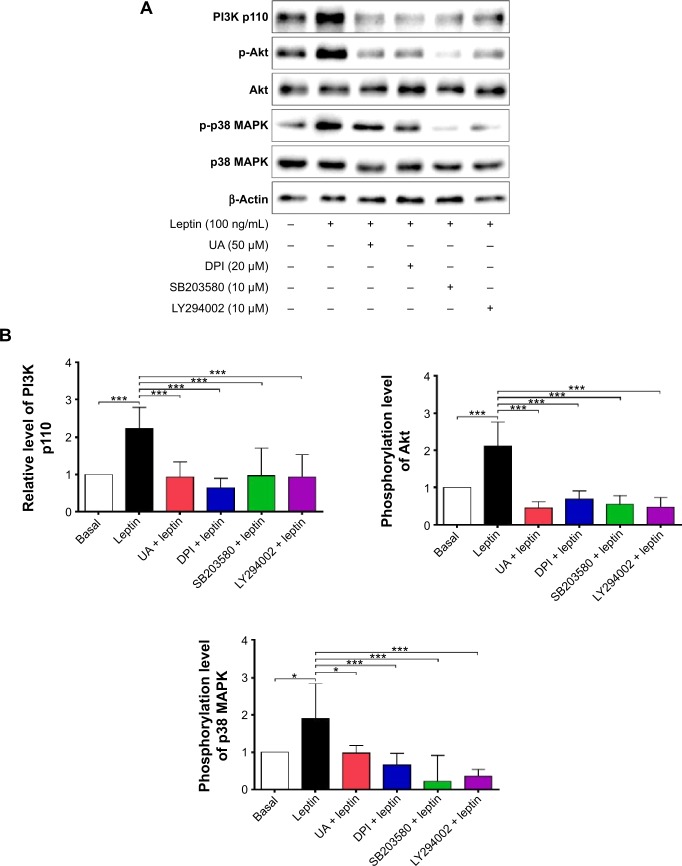
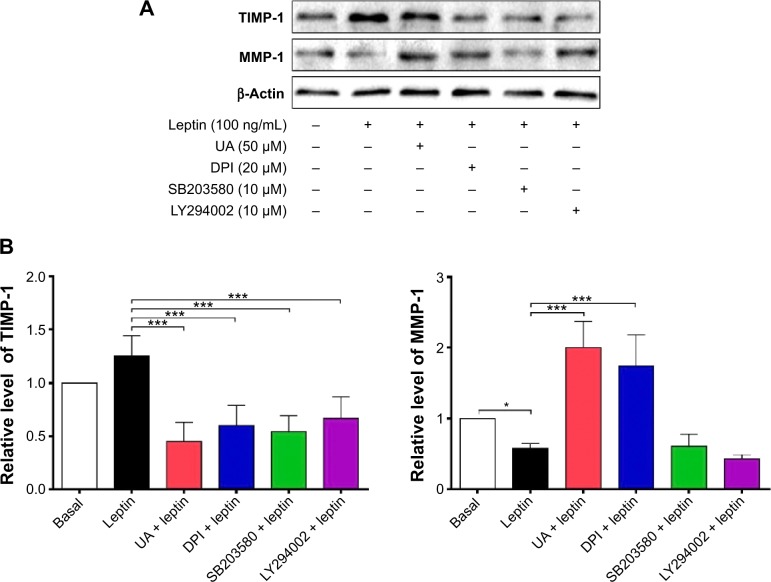
NADPH oxidases (NOXs) are a predominant mediator of redox homeostasis in hepatic stellate cells (HSCs), and oxidative stress plays an important role in the pathogenesis of liver fibrosis. Ursolic acid (UA) is a pentacyclic triterpenoid with various pharmacological activities, but the molecular targets and underlying mechanisms for its antifibrotic effect in the liver remain elusive. This study aimed to computationally predict the molecular interactome and mechanistically investigate the antifibrotic effect of UA on oxidative stress, with a focus on NOX4 activity and cross-linked signaling pathways in human HSCs and rat liver. Drug–drug interaction via chemical–protein interactome tool, a server that can predict drug–drug interaction via chemical–protein interactome, was used to predict the molecular targets of UA, and Database for Annotation, Visualization, and Integrated Discovery was employed to analyze the signaling pathways of the predicted targets of UA. The bioinformatic data showed that there were 611 molecular proteins possibly interacting with UA and that there were over 49 functional clusters responding to UA. The subsequential benchmarking data showed that UA significantly reduced the accumulation of type I collagen in HSCs in rat liver, increased the expression level of MMP-1, but decreased the expression level of TIMP-1 in HSC-T6 cells. UA also remarkably reduced the gene expression level of type I collagen in HSC-T6 cells. Furthermore, UA remarkably attenuated oxidative stress via negative regulation of NOX4 activity and expression in HSC-T6 cells. The employment of specific chemical inhibitors, SB203580, LY294002, PD98059, and AG490, demonstrated the involvement of ERK, PI3K/Akt, and p38 MAPK signaling pathways in the regulatory effect of UA on NOX4 activity and expression. Collectively, the antifibrotic effect of UA is partially due to the oxidative stress attenuating effect through manipulating NOX4 activity and expression. The results suggest that UA may act as a promising antifibrotic agent. More studies are warranted to evaluate the safety and efficacy of UA in the treatment of liver fibrosis.
Keywords: ursolic acid, liver fibrosis, NADPH oxidase, ROS, DDI-CPI, DAVID
Introduction
Liver fibrosis remains the major cause of morbidity and mortality worldwide. It causes several notorious complications, including ascites, portal hypertension, encephalopathy, and liver failure, and it accelerates the risk of hepatocellular carcinoma, placing a substantial burden to individual, society, and health care system.1,2 The liver fibrosis-driven chronic liver disease and cirrhosis cause 36,427 deaths and with a ratio of 11.5/100,000 in 2013 in USA.1 This is a complicated etiology of hepatic fibrosis. Viral infection is the most common contributing factor to liver fibrosis affecting 1%–2% of the US population,3 and liver fibrosis resultant cirrhosis is projected to reach 45% of those infected with hepatitis C virus patients in 2030.4 Hepatitis B virus is the main type of hepatitis virus in the People’s Republic of China with a substantial contribution to the incidence of liver fibrosis.5,6 Moreover, nonalcoholic fatty liver disease and nonalcoholic steatohepatitis are also important causes of liver fibrosis, and over 20% of patients with nonalcoholic steatohepatitis progress to cirrhosis worldwide.7 In addition, other etiologies of liver fibrosis and its late-stage liver injury include alcohol-induced disease, drug-induced toxicity, other liver infections (eg, schistosomiasis), immune-mediated liver diseases (eg, autoimmune hepatitis), metabolic disorders (eg, lipid, glycogen, or metal storage disorders), and cholestasis (eg, secondary biliary cirrhosis).
To date, the molecular mechanisms that underlie the development of liver fibrosis have been extensively investigated, including epithelial to mesenchymal transition and inflammatory response.8–10 Notably, increasing evidence suggests that oxidative stress has been implicated in the pathogenesis of liver fibrosis, with the intracellular accumulation of excessive reactive oxygen species (ROS), and the compelling evidence shows the involvement of ROS in the development of liver fibrosis with a regulatory role in a variety of cellular processes.7,10–13 However, the specific targets and signaling pathways have not been fully mapped yet, and the sources of ROS have not yet been conclusively determined in the pathogenesis of liver fibrosis. There are several enzymatic sources of ROS, including NADPH oxidase (NOX) family of oxidoreductases, nitric oxide synthases, xanthine oxidase, and cytochrome P450, of which NOX is the most important ROS-generating enzyme in both plants and animals, especially in mammals. In the liver, NOX has been involved in fibrogenesis. In particular, NOX4 is one of seven NOX isoforms and is the most widely distributed isoform.11 It has been reported that a functionally active form of NOX is expressed in hepatic stellate cells (HSCs) and that NOX-generated ROS serve as a second messenger for profibrogenic factor signal transduction in HSCs. However, regulation of NOX in HSCs remains largely elusive.
Recently, it has been proposed that antioxidants hold promise as potential antifibrotic therapies due to the ROS-attenuating effect.12,13 Ursolic acid (UA), a natural pentacyclic triterpenoid carboxylic acid, is the major component of certain traditional medicinal herbs and possesses a wide range of biological functions, such as antioxidative, anti-inflammatory, and anticancer activities.14 UA exerts a protective effect against ethanol-induced toxicity in isolated rat hepatocytes and ethanol-mediated experimental liver damage in rats.15,16 Our previous studies showed that UA significantly inhibited the proliferation of HSCs and induced apoptosis in vitro.17 Moreover, Steinkamp-Fenske et al18 showed that UA downregulated the expression level of NOX4 and suppressed ROS generation in human endothelial cells. However, there is a lack of study on the antifibrotic effect of UA, and the underlying mechanisms of the antifibrotic effect of UA are largely unknown.
Therefore, the present study aimed to evaluate the molecular targets of UA and analyze the molecular interactome of UA using computational and bioinformatic approaches and, subsequently, examine the antifibrotic effect of UA and delineate the underlying mechanisms in HSCs and Sprague Dawley rats.
Materials and methods
Prediction of the interactome of UA and pathway analysis by molecular docking and bioinformatic approach
The prediction of interactome of UA was performed using the drug–drug interaction via chemical–protein interactome (DDI-CPI) tool (http://cpi.bio-x.cn/ddi/) as previously described.19 DDI-CPI is a web-based server that can be used to predict DDI via CPI.20,21 In brief, protein targets were obtained from a third-party protein structure database named PDBbind (http://sw16.im.med.umich.edu/databases/pdbbind/index.jsp), which was based on the contents of Protein Data Bank (PDB).22 There are a total of 1,780 PDB entries of human proteins available in PDBbind, and a total of 301 nonredundant PDBs corresponding to 353 ligand-binding pockets were identified from it, with 86% of which have resolutions <2.5 Å. The docking boxes for each of the pockets were defined by expanding the circumscribed cube of the pocket with a margin of 8 Å at six directions (up, down, front, back, left, and right). For the preparation of the UA, the 2D structure of the UA was downloaded from PubChem. The hydrogen and Gasteiger charge were added, and the PDB file was converted into mol2 format using VEGA ZZ. The docking program AutoDock 4.2 was used to dock the prepared UA molecule into all 353 pockets, generating a score vector of 353 dimensions. Z′-scores were then calculated using the methodologies we applied previously.20,23,24 Here, an empirical threshold of −0.6 of the Z′-score was set to indicate that the binding of UA toward this target was likely to be true.
Pathway analysis by Database for Annotation, Visualization, and Integrated Discovery
Following the computational target prediction, the Database for Annotation, Visualization, and Integrated Discovery version 6.7 (DAVID, http://david.abcc.ncifcrf.gov/) was employed to interpret the biological function of the potential targets of UA derived from DDI-CPI.19–21 The protein IDs of these targets from UniProtKB, NCBI, and other sources were converted into gene lists by using the Gene ID Conversion Tool in DAVID. The DAVID database adds biological function annotation including gene ontology, pathway, protein–protein interactions, functional groups of genes (ie, clustering), and disease association derived from main public data sources. The genes of interest were visualized using BioCarta and Kyoto encyclopedia of Genes and Genomes (KEGG) pathway maps. The high classification stringency was selected for functional annotation clustering. Enrichment scores and Fisher’s exact test P-values (and corresponding false discovery rate) were then calculated to identify which functional-related gene groups were significantly enriched in the target list. These significant enriched gene groups could provide clues on how UA interacts with molecular targets in a systematic way.
Chemicals and reagents
Recombinant rat leptin was purchased from Peprotech Inc. (Rocky Hill, NJ, USA). UA, diphenyleneiodonium (DPI), dimethyl sulfoxide (DMSO), Thiazolyl blue tetrazolium bromide (MTT) and protease inhibitor and phosphatase inhibitor cocktails were purchased from Sigma-Aldrich Co., (St Louis, MO, USA). Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum, and AG490 were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Antioxidant N-acetyl-l-cysteine (NAC) was purchased from Solarbio Inc. (Beijing, People’s Republic of China). PD98059, an extracellular signal-regulated kinase (ERK) inhibitor, was purchased from Promega Corporation (Fitchburg, WI, USA). SB203580, a p38 mitogen-activated protein kinase (p38 MAPK) inhibitor, LY294002, a phosphoinositide 3-kinase (PI3K) inhibitor, and the ROS assay kit (which contains 2′,7′-dichlorofluorescin diacetate [DCF-DA]) were purchased from Beyotime Inc. (Jiangsu, People’s Republic of China). p47phox antibody was purchased from Bioworld Inc. (St Louis Park, MN, USA); gp91phox, p22phox, and Rac1 antibodies were purchased from Abcam Inc. (Cambridge, MA, USA). p67phox, PI3K (p110α), protein kinase B (Akt), phosphorylated (p)-Akt, p-ERK1/2, p-p38 MAPK, and p38 MAPK antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). TIMP metallopeptidase inhibitor 1 (TIMP-1) and matrix metalloproteinase-1 (MMP-1) antibodies were purchased from PL Laboratories Inc. (Port Moody, British Columbia, Canada). The α-smooth muscle actin (α-SMA) antibody was purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA), and the β-Actin antibody and goat anti-rabbit secondary antibodies were purchased from Zhongshan Golden Bridge Biotechnology Co., Ltd. (Beijing, People’s Republic of China).
Cell culture and treatment
The immortalized rat liver stellate cell line (HSC-T6) was kindly provided by Dr L-M Xu, Shanghai University of Chinese Traditional Medicine (Shanghai, People’s Republic of China). These cells exhibit the phenotype that most closely resembles primary rat stellate cells.25 HSC-T6 cells were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin (120 µg/mL), and streptomycin (100 µg/mL) at a humidified atmosphere of 5% CO2 at 37°C. The culture medium was replaced with serum-free DMEM (serum starvation) for 24 hours before the start of the experiments.
Animals and experimental design
A total of 32 male Sprague Dawley rats (weighing between 180 g and 200 g) were experimented in the present study. All animals were housed in plastic cages containing wood shaving and cotton bedding and maintained in a room at 22°C–25°C with a 12-hour day/night cycle and had free access to standard laboratory diet and water. The animal study protocol was approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Nanchang University. Dimethylnitrosamine (DMN) and UA were freshly dissolved in saline at predetermined concentrations.
A liver fibrosis model was established in rats by administering 1% DMN at a dosage of 1 mL/kg body weight by intraperitoneal injection for 3 consecutive days per week for 4 weeks. At week 5, the DMN-treated rats with liver fibrosis were randomly divided into three groups with or without UA treatment for another 4-week treatment. The UA groups were administered by intraperitoneal injections at dosages of 20 mg/kg (UA-2) and 40 mg/kg (UA-3) per day. The DMN group received DMN alone for 8 weeks. The vehicle group was treated with a volume of saline that was equivalent to the volume used per day in UA groups. The rats were euthanized by saline perfusion and exsanguinated via the inferior vena cava 1 day after the last treatment. The liver and blood samples were collected and stored at −80°C for further analysis. During the period of experiment, if the weight loses 10%, mice will be euthanized. Also, the mice will be immediately euthanatized if other symptoms such as pale mucous membranes, shivering, failure to respond to stimuli, piloerection, matted hair coat, soiled anogenital/vent area, and vocalization are observed.
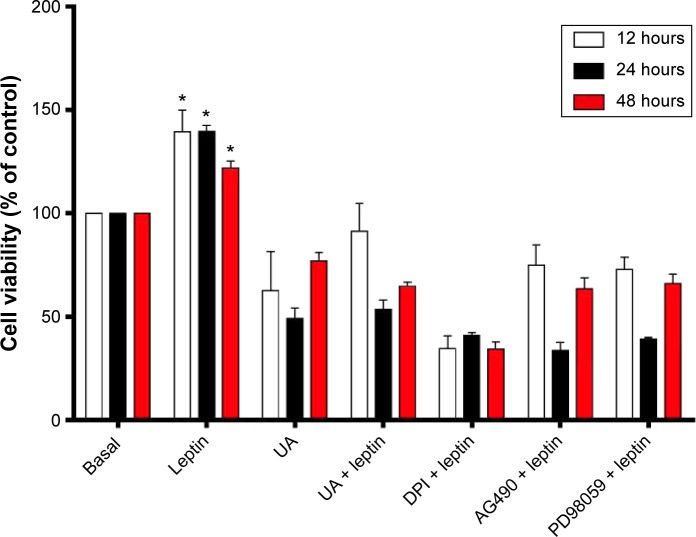
MTT assay
To examine the effect of UA on the proliferation of HSC-T6 cells, the MTT assay was performed. Briefly, HSC-T6 cells were seeded in to 96-well plates with medium containing leptin at a confluence of ~60% following a 24-hour starvation. The cells were treated with UA (50 µM), AG490 (50 µM), or DPI (20 µM) for 12 hours, 24 hours, or 48 hours, respectively. After the treatment, a volume of 10 µL of thiazolyl blue tetrazolium bromide (MTT) (5 mg/mL) was added and incubated for an additional 4 hours. Then, the medium was carefully aspirated, and the 100 µL of DMSO was added to dissolve the formazan particle. The absorption intensity was measured using a microplate reader (Bio-Rad 550; Bio-Rad Laboratories Inc., Hercules, CA, USA) at 490 nm.
Histological evaluation
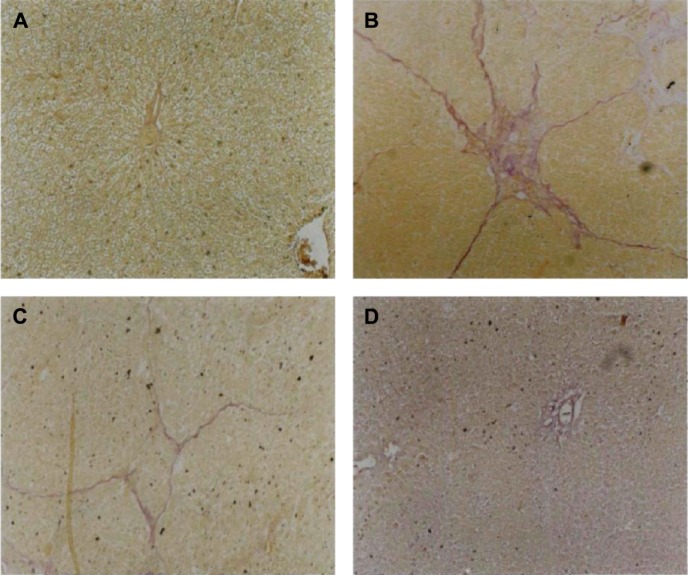
To examine the effect of UA on the general characterization, inflammatory cell infiltration, and disposition of connective tissue in rat liver, the hematoxylin and eosin and van Gieson staining were performed.
Determination of serum maleic dialdehyde and superoxide dismutase levels
In order to assess the antioxidative effect of UA in rats, the blood level of maleic dialdehyde (MDA) and superoxide dismutase (SOD) activity were examined using the thiobarbituric acid method.
Measurement of intracellular ROS level
To further examine the antioxidative effect of UA in HSC-T6 cells, the intracellular ROS level was determined using flow cytometry by the oxidation-sensitive probe DCF-DA as previously described.26,27 DCF-DA, a ROS probe that undergoes intracellular deacetylation followed by ROS-mediated oxidation to a fluorescent DCF, was used to measure ROS generation in the cytoplasm and cellular organelles.28,29 HSC-T6 cells were treated with UA (50 µM), NAC (10 mM), DPI (20 µM), or AG490 (50 µM) in the absence or presence of leptin. After the treatment, cells were incubated with DCF-DA (10 µM) for 20 minutes. Then, cellular fluorescence intensity was measured using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA) at excitation and emission wavelengths of 488 nm and 525 nm, respectively. The intracellular ROS level was calculated as a percentage of DCF fluorescence intensity relative to the vehicle controls (untreated HSC-T6 cells).
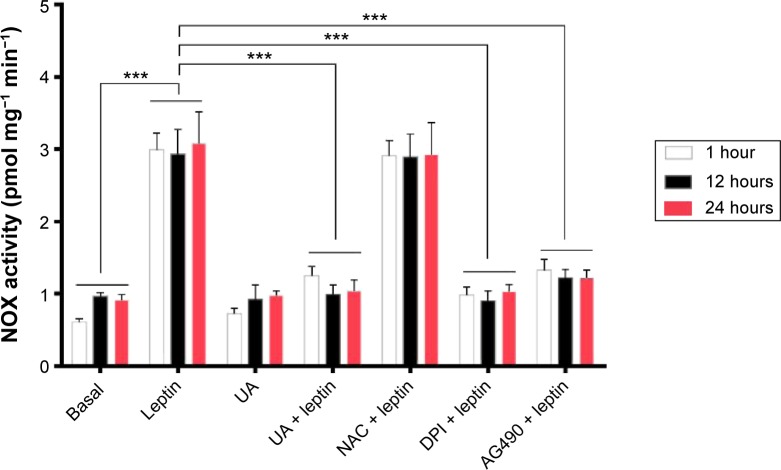
Determination of NOX activity
Following the examination of ROS generation in HSC-T6 cells, the NOX activity was determined to address the regulating effect of UA on the enzymatic source of ROS in HSC-T6 cells as previously described with some modifications.27,30,31 Briefly, HSC-T6 cells were incubated in the culture medium in the absence or presence of leptin and treated with UA (50 µM), NAC (10 mM), DPI (20 µM), or AG490 (50 µM). After the treatment, the cells were harvested by trypsinization, pelleted by centrifugation at 2,500× g for 5 minutes at 4°C and resuspended in PBS. Subsequently, the cells were incubated with 250 µM of NADPH. NADPH consumption was monitored by the decrease in absorbance at λ=340 nm for 10 minutes. For the specific analysis of NOX activity, the rate of NADPH consumption specifically inhibited by DPI was measured by pretreated with 10 µM of DPI for 30 minutes. An aliquot of cells was lysed by adding sodium dodecyl sulfate, and the protein concentration of the cell lysate was determined. The absorption extinction coefficient used to calculate the amount of NADPH consumed was 6.22 mM−1 cm−1. The data of NOX activity were expressed as picomol per liter of substrate per minute per milligram of protein.
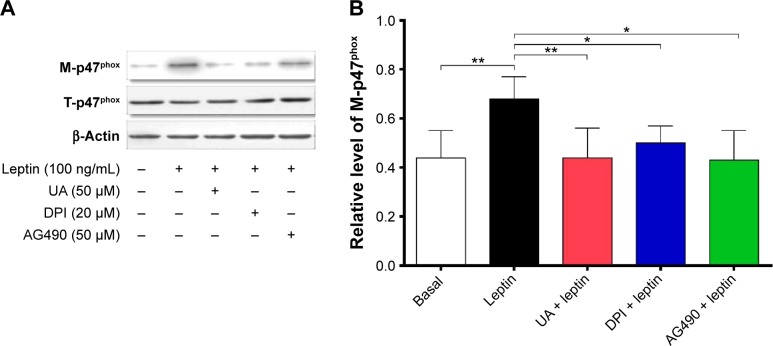
Western blotting analysis
Whole-cell extracts were obtained using Triton lysis buffer that contained protease inhibitor and phosphatase inhibitor cocktails. Liver extracts were obtained in modified radioimmunoprecipitation buffer. The proteins were loaded and separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Then, the membrane was blocked at room temperature for 1 hour with 5% nonfat milk in Tris-buffered saline with Tween (TBST), followed by the incubation with indicated primary antibodies overnight at 4°C. Next, the membranes were washed and incubated with the corresponding secondary antibody conjugated to horseradish peroxidase (HRP) at room temperature for 1 hour. Visualization was performed using Bio-Rad ChemiDoc™ XRS system (Hercules, CA, USA) with enhanced chemiluminescence substrate, and the blots were analyzed using Image Lab 3.0 (Bio-Rad Laboratories Inc., Hercules, CA, USA). Protein level was normalized to the matching densitometric value of internal control.
Reverse transcription polymerase chain reaction
Total RNA was extracted using TRNzol A+ total reagent (Tiangen Biotech, Beijing, People’s Republic of China) and subject to reverse transcription with dT15-oligonucleotide and Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Promega Corporation, Fitchburg, WI, USA). The primers for type I collagen and β-Actin used in the reverse transcription polymerase chain reaction are shown in Table 1. The mRNA levels of the type I collagen gene were normalized to the β-Actin mRNA level. The number of amplification cycles was 30, and the specific amplicons were analyzed by 1% agarose gel electrophoresis and visualized with ethidium bromide.
Table 1.
Primer sequences for type I collagen and β-Actin
| Gene | Sequence | Amplicon length (bp) |
|---|---|---|
| Type I Collagen | Sense: 5′-GGGGCAAGACAGTCATCGAA-3′ | 144 |
| Antisense: 5′-GGATGGAGGGAGTTTACACGAA-3′ | ||
| β-Actin | Sense: 5′-TCAGGTCATCACTATCGGCAAT-3′ | 432 |
| Antisense: 5′-AAAGAAAGGGTGTAAAACGCA-3′ |
Abbreviation: bp, base pair.
Statistical analysis
The results are expressed as the mean ± standard deviation. Differences between groups were compared using one-way analysis of variance followed by the Tukey’s test. Data with a skewed distribution or heterogeneity of variance were analyzed by the Kruskal–Wallis nonparametric test followed by a Nemenyi test. A P-value <0.05 was considered to be statistically significant.
Results
UA likely interacts with a number of important functional proteins
First, we computationally predicted the molecular targets of UA using our web-based DDI-CPI tool. There were 611 proteins that possibly interacted with UA (Table 2), including those involved in MAPK signaling pathway (FGFR2, TRAF2, FGFR1, HRAS, GRB2, MAPKAPK3, MAPKAPK2, AKT1, CDC42, TNFRSF1A, CASP3, RAC1, PRKACA, PAK1, TRAF6, AKT2, EGFR, PRKCA, MAP2K1, BRAF, TGFBR1, TP53, RAF1, MAPK10, PRKCB, MAPK1, DUSP3, RPS6KA1, MAPK14, MAPK3, PLA2G2A, MAPK9, and MAPK8), apoptosis (PIK3CG, TRAF2, XIAP, TP53, BCL2L1, AKT1, IRAK4, TNFRSF1A, CASP3, CASP7, BCL2, CASP8, PRKACA, and AKT2), energy metabolism (PPARA, PDPK1, PPARD, CHKB, RXRA, PPARG, FABP3, FABP4, FABP7, MMP-1, PCK1, NR1H3, NAMPT, CD38, NT5M, PNP, and NNMT), and cell proliferation (YWHAZ, TP53, TTK, CDK6, CHEK1, RB1, CHEK2, SFN, WEE1, CDK2, HDAC2, PLK1, GSK3B, MDM2, and ABL1). Furthermore, as shown in Table 3, our DAVID analysis showed that there were 49 functional clusters that were identified to be significantly enriched (enrichment score >3) in the target list derived from molecular docking calculations, including energy metabolism, signal transduction, vascular regulation, and carbohydrate metabolism. Furthermore, there were 76 KEGG pathways significantly enriched in the target list (Table 4), such as MAPK, p53, and mTOR signaling pathways.
Table 2.
Predicted molecular targets of UA
| PDB ID | Class | Target name | Function | Docking score |
|---|---|---|---|---|
| 2Q8G | PD | (Pyruvate dehydrogenase [lipoamide]) kinase isozyme 1, mitochondrial | Inhibits the mitochondrial pyruvate dehydrogenase complex by phosphorylation of the E1 α subunit, thus contributing to the regulation of glucose metabolism. | −7.3 |
| 1Y8O | PD | (Pyruvate dehydrogenase [lipoamide]) kinase isozyme 3, mitochondrial | Inhibits the mitochondrial pyruvate dehydrogenase complex by phosphorylation of the E1 α subunit, thus contributing to the regulation of glucose metabolism. | −7.7 |
| 2ZKJ | PD | (Pyruvate dehydrogenase [lipoamide]) kinase isozyme 4, mitochondrial | Inhibits the mitochondrial pyruvate dehydrogenase complex by phosphorylation of the E1 α subunit, thus contributing to the regulation of glucose metabolism. | −9.1 |
| 3IQU | PD | 14-3-3 protein σ | AP implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner. When bound to KRT17, regulates protein synthesis and epithelial cell growth by stimulating Akt/mTOR pathway (by similarity). p53-regulated inhibitor of G2/M progression. | −7.3 |
| 1QJA | PD | 14-3-3 protein ζ/δ | AP implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner. | −8.4 |
| 1OLS | PD | 2-Oxoisovalerate dehydrogenase subunit α, mitochondrial | The branched-chain α-keto dehydrogenase complex catalyzes the overall conversion of α-keto acids to acyl-CoA and CO2. It contains multiple copies of three enzymatic components: branched-chain α-keto acid decarboxylase (E1), lipoamide acyltransferase (E2), and lipoamide dehydrogenase (E3). | −3.2 |
| 2R4F, 2Q6B_2, 2Q6B, 3CCZ | PK | 3-Hydroxy-3-methylglutaryl-CoA reductase | Transmembrane glycoprotein that is the rate-limiting enzyme in cholesterol biosynthesis as well as in the biosynthesis of nonsterol isoprenoids that are essential for normal cell function, including ubiquinone and geranylgeranyl proteins. | −7.7 to −6 |
| 2O23, 1U7T | PK | 3-Hydroxyacyl-CoA dehydrogenase type 2 | Functions in mitochondrial tRNA maturation. Part of mitochondrial ribonuclease P, an enzyme composed of MRPP1/TRMT10C, MRPP2/HSD17B10, and MRPP3/KIAA0391, which cleaves tRNA molecules in their 5′-ends. By interacting with intracellular amyloid-β, it may contribute to the neuronal dysfunction associated with AD. | −9.4 to −7.5 |
| 1W1G | PD | 3-Phosphoinositide-dependent protein kinase 1 | Serine/threonine kinase that acts as a master kinase, phosphorylating and activating a subgroup of the AGC family of protein kinases. Its targets include: protein kinase B (PKB/AKT1, PKB/AKT2, PKB/AKT3), p70 ribosomal protein S6 kinase (RPS6KB1), p90 ribosomal protein S6 kinase (RPS6KA1, RPS6KA2, and RPS6KA3), cyclic AMP-dependent protein kinase (PRKACA), PKC (PRKCD and PRKCZ), serum- and GC-inducible kinase (SGK1, SGK2, and SGK3), PAK1, protein kinase PKN (PKN1 and PKN2). Plays a central role in the transduction of signals from INS by providing the activating phosphorylation to PKB/AKT1, thus propagating the signal to downstream targets controlling cell proliferation and survival as well as glucose and amino acid uptake and storage. Negatively regulates the TGF-β-induced signaling by: modulating the association of SMAD3 and SMAD7 with TGF-β receptor, phosphorylating SMAD2, SMAD3, SMAD4, and SMAD7; and preventing the nuclear translocation of SMAD3 and SMAD4 and the translocation of SMAD7 from the nucleus to the cytoplasm in response to TGF-β. Activates PPARG transcriptional activity and promotes adipocyte differentiation. Activates the NF-κB pathway via phosphorylation of IKKB. The tyrosine phosphorylated form is crucial for the regulation of focal adhesions by angiotensin-2. Controls proliferation, survival, and growth of developing pancreatic cells. Participates in the regulation of Ca2+ entry and Ca2+-activated K+ channels of mast cells. Essential for the motility of vascular ECs and is involved in the regulation of their chemotaxis. Plays a critical role in cardiac homeostasis by serving as a dual effector for cell survival and β-adrenergic response. Plays an important role during thymocyte development by regulating the expression of key nutrient receptors on the surface of pre-T-cells and mediating Notch-induced cell growth and proliferative responses. Provides negative feedback inhibition to TLR-mediated NF-κB activation in macrophages. Isoform 3 is catalytically inactive. | −6.5 |
| 1Q91 | PD | 5′(3′)-Deoxyribonucleotidase, mitochondrial | Dephosphorylates specifically the 5′ and 2′ (3′)-phosphates of uracil and thymine deoxyribonucleotides and so protects mitochondrial DNA replication from excess dTTP. Has only marginal activity toward dIMP and dGMP. | −9.4 |
| 2RJP | PD | A disintegrin and metalloproteinase with thrombospondin motifs 4 | Cleaves aggrecan, a cartilage proteoglycan, and may be involved in its turnover. May play an important role in the destruction of aggrecan in arthritic diseases. Could also be a critical factor in the exacerbation of neurodegeneration in AD. Cleaves aggrecan at the “392-Glu-|-Ala-393” site. | −6.9 |
| 3HYG | PD | A disintegrin and metalloproteinase with thrombospondin motifs 5 | Cleaves aggrecan, a cartilage proteoglycan, and may be involved in its turnover. May play an important role in the destruction of aggrecan in arthritic diseases. May play a role inproteolytic processing mostly during the peri-implantation period. | −6.7 |
| 3JRX | PD | Acetyl-CoA carboxylase 2 | ACC-β may be involved in the provision of malonyl-CoA or in the regulation of fatty acid oxidation, rather than fatty acid biosynthesis. Carries out three functions: biotin carboxyl carrier protein, biotin carboxylase, and carboxyl transferase. | −8.9 |
| 3EQR | PD | Activated CDC42 kinase 1 | Nonreceptor tyrosine-protein and serine/threonine–protein kinase that is implicated in cell spreading and migration, cell survival, cell growth, and proliferation. Transduces extracellular signals to cytosolic and nuclear effectors. Phosphorylates AKT1, AR, MCF2, WASL, and WWOX. Implicated in trafficking and clathrin-mediated endocytosis through binding to EGFR and clathrin. Binds to both poly- and monoubiquitin and regulates ligand-induced degradation of EGFR, thereby contributing to the accumulation of EGFR at the limiting membrane of early endosomes. Downstream effector of CDC42 which mediates CDC42-dependent cell migration via phosphorylation of BCAR1. May be involved both in adult synaptic function and plasticity and in brain development. Activates AKT1 by phosphorylating it on “Tyr-176”. Phosphorylates AR on “Tyr-267” and “Tyr-363”, thereby promoting its recruitment to androgen-responsive enhancers. Phosphorylates WWOX on “Tyr-287”. Phosphorylates MCF2, thereby enhancing its activity as a GEF toward Rho family proteins. Contributes to the control of AXL receptor levels. Confers metastatic properties on cancer cells and promotes tumor growth by negatively regulating tumor suppressor such as WWOX and positively regulating pro-survival factors such as AKT1 and AR. | −7.2 |
| 2I6A | PD | Adenosine kinase | ATP-dependent phosphorylation of adenosine and other related nucleoside analogs to monophosphate derivatives. Serves as a potential regulator of concentrations of extracellular adenosine and intracellular adenine nucleotides. | −2.9 |
| 3EML | PD | Adenosine receptor A2a | Receptor for adenosine. The activity of this receptor is mediated by G proteins that activate adenylyl cyclase. | −9 |
| 2PGJ | PD | ADP-ribosyl cyclase 1 | Synthesizes cyclic ADP-ribose, a second messenger for glucose-induced INS secretion. Also has cADPr hydrolase activity. Also moonlights as a receptor in cells of the immune system. | −9.3 |
| 1PY1 | PD | ADP-ribosylation factor-binding protein GGA1 | Plays a role in protein sorting and trafficking between the TGN and endosomes. Mediates the ARF-dependent recruitment of clathrin to the TGN and binds ubiquitinated proteins and membrane cargo molecules with a cytosolic AC-LL motif. | −6.5 |
| 2EXG | PD | Afadin | Belongs to an adhesion system, probably together with the E-cadherin–catenin system, which plays a role in the organization of homotypic, interneuronal, and heterotypic cell–cell AJs. Nectin- and actin-filament-binding protein that connects nectin to the actin cytoskeleton. | −6.9 |
| 1HSO, 1U3T, 1U3T_2, 1HSO_2 | PK | Alcohol dehydrogenase 1A | −8.5 to −7.2 | |
| 1U3U, 1U3U_2, 1U3V_2, 1U3V, 1HSZ | PK | Alcohol dehydrogenase 1B | N/A | −8.3 to −7.7 |
| 1HT0, 1U3W, 1U3W_2 | PK | Alcohol dehydrogenase 1C | N/A | −8.5 to −7.3 |
| 3COS | PK | Alcohol dehydrogenase 4 | −8.6 | |
| 1D1T, 1D1S, 1AGN | PK | Alcohol dehydrogenase class 4 µ/σ chain | Could function in retinol oxidation for the synthesis of retinoic acid, a hormone important for cellular differentiation. Medium-chain (octanol) and aromatic (m-nitrobenzaldehyde) compounds are the best substrates. Ethanol is not a good substrate but at the high ethanol concentrations reached in the digestive tract, it plays a role in the ethanol oxidation and contributes to the first-pass ethanol metabolism. | −8.5 to −7.7 |
| 2FZE, 3QJ5, 3QJ5_2, 2FZW | PK | Alcohol dehydrogenase class 3 | Class 3 ADH is remarkably ineffective in oxidizing ethanol, but it readily catalyzes the oxidation of long-chain primary alcohols and the oxidation of S-(hydroxymethyl) glutathione. | −9.1 to −8.2 |
| 3SZB | PK | Aldehyde dehydrogenase, dimeric NADP-preferring | ALDHs play a major role in the detoxification of alcohol-derived acetaldehyde. They are involved in the metabolism of corticosteroids, biogenic amines, neurotransmitters, and lipid peroxidation. This protein preferentially oxidizes aromatic aldehyde substrates. It may play a role in the oxidation of toxicaldehydes. | 5.2 |
| 3N80, 2VLE, 3INJ, 1O04 | PK | Aldehyde dehydrogenase, mitochondrial | N/A | −8.7 to −7.4 |
| 2XBA | PD | ALK tyrosine kinase receptor | Neuronal orphan receptor tyrosine kinase that is essentially and transiently expressed in specific regions of the central and peripheral nervous systems and plays an important role in the genesis and differentiation of the nervous system. Transduces signals from ligands at the cell surface, through specific activation of the MAPK pathway. Phosphorylates almost exclusively at the first tyrosine of the Y-x-x-x-Y-Y motif. Following activation by ligand, ALK induces tyrosine phosphorylation of CBL, FRS2, IRS1, and SHC1 as well as of the MAPKs MAPK1/ERK2 and MAPK3/ERK1. Acts as a receptor for ligands PTN, a secreted growth factor, and MDK, a PTN-related factor, thus participating in PTN and MDK signal transduction. PTN binding induces MAPK pathway activation, which is important for the anti-apoptotic signaling of PTN and regulation of cell proliferation. MDK binding induces phosphorylation of the ALK target IRS1, activates MAPKs and PI3K, resulting also in cell proliferation induction. Drives NF-κB activation, probably through IRS1 and the activation of the AKT serine/threonine kinase. Recruitment of IRS1 to activated ALK and the activation of NF-κB are essential for the autocrine growth and survival signaling of MDK. | −7.2 |
| 3MPH, 3HIG, 3HI7, 3HIG_2 | PK | Amiloride-sensitive amine oxidase (copper-containing) | Catalyzes the degradation of compounds such as putrescine, histamine, spermine, and spermidine, substances involved in allergic and immune responses, cell proliferation, tissue differentiation, tumor formation, and possibly apoptosis. Placental DAO is thought to play a role in the regulation of the female reproductive function. | −9.6 to 13.4 |
| 2BXR_2, 2BXR, 2Z5X, 2Z5Y, 2Z5Y_2, 2Z5X_2 | PK | Amine oxidase (flavin-containing) A | Catalyzes the oxidative deamination of biogenic and xenobiotic amines and has important functions in the metabolism of neuroactive and vasoactive amines in the CNS and peripheral tissues. MAOA preferentially oxidizes biogenic amines such as 5-HT, norepinephrine, and epinephrine. | −10.3 to −1.9 |
| 2V5Z, 2XFN, 1S3E, 2XFN_2, 2V5Z_2, 1S3E_2 | PK | Amine oxidase (flavin-containing) B | Catalyzes the oxidative deamination of biogenic and xenobiotic amines and has important functions in the metabolism of neuroactive and vasoactive amines in the CNS and peripheral tissues. MAOB preferentially degrades benzylamine and phenylethylamine. | −6.9 to −1.6 |
| 3B66 | PD | ANDR | Steroid hormone receptors are ligand-activated transcription factors that regulate eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Transcription factor activity is modulated by bound coactivator and corepressor proteins. Transcription activation is downregulated by NR0B2. Activated, but not phosphorylated, by HIPK3 and ZIPK/DAPK3. | −5.3 |
| 2OO8 | PD | Angiopoietin-1 receptor | Tyrosine-protein kinase that acts as cell-surface receptor for ANGPT1, ANGPT2, and ANGPT4 and regulates angiogenesis, EC survival, proliferation, migration, adhesion and cell spreading, reorganization of the actin cytoskeleton, but also maintenance of vascular quiescence. Has anti-inflammatory effects by preventing the leakage of proinflammatory plasma proteins and leukocytes from blood vessels. Required for normal angiogenesis and heart development during embryogenesis. Required for postnatal hematopoiesis. After birth, activates or inhibits angiogenesis, depending on the context. Inhibits angiogenesis and promotes vascular stability in quiescent vessels, where ECs have tight contacts. In quiescent vessels, ANGPT1 oligomers recruit TEK to cell–cell contacts, forming complexes with TEK molecules from adjoining cells, and this leads to preferential activation of phosphatidylinositol 3-kinase and the AKT1 signaling cascades. In migrating ECs that lack cell–cell adhesions, ANGT1 recruits TEK to contact with the ECM, leading to the formation of focal adhesion complexes, activation of PTK2/FAK and of the downstream kinases MAPK1/ERK2 and MAPK3/ERK1, and ultimately to the stimulation of sprouting angiogenesis. ANGPT1 signaling triggers receptor dimerization and autophosphorylation at specific tyrosine residues that then serve as binding sites for scaffold proteins and effectors. Signaling is modulated by ANGPT2 that has lower affinity for TEK, can promote TEK autophosphorylation in the absence of ANGPT1, but inhibits ANGPT1-mediated signaling by competing for the same binding site. Signaling is also modulated by formation of heterodimers with TIE1 and by proteolytic processing that gives rise to a soluble TEK extracellular domain. The soluble extracellular domain modulates signaling by functioning as decoy receptor for angiopoietins. TEK phosphorylates DOK2, GRB7, GRB14, PIK3R1; SHC1, and TIE1. | −8 |
| 1O86 | PD | Angiotensin-converting enzyme | Converts angiotensin-I to angiotensin-2 by release of the terminal His-Leu; this results in an increase of the vasoconstrictor activity of angiotensin. Also able to inactivate bradykinin, a potent vasodilator. Has also a glycosidase activity that releases GPI-anchored proteins from the membrane by cleaving the mannose linkage in the GPI moiety. | −9.7 |
| 1JJ7 | PK | Antigen peptide transporter 1 | Involved in the transport of antigens from the cytoplasm to the endoplasmic reticulum for association with MHC class I molecules. Also acts as a molecular scaffold for the final stage of MHC class I folding, namely the binding of peptide. Nascent MHC class I molecules associate with TAP via tapasin. Inhibited by the covalent attachment of herpes simplex virus ICP47 protein, which blocks the peptide-binding site of TAP. Inhibited by human cytomegalo virus US6 glycoprotein, which binds to the lumenal side of the TAP complex and inhibits peptide translocation by specifically blocking ATP binding to TAP1 and prevents the conformational rearrangement of TAP induced by peptide binding. Inhibited by human adenovirus E3-19K glycoprotein, which binds the TAP complex and acts as a tapasin inhibitor, preventing MHC class I/TAP association. Expression of TAP1 is downregulated by human Epstein–Barr virus vIL-10 protein, thereby affecting the transport of peptides into the endoplasmic reticulum and subsequent peptide loading by MHC class I molecules. | −6.6 |
| 1AZX | PD | Antithrombin-III | Most important serine protease inhibitor in plasma that regulates the blood coagulation cascade. AT-III inhibits thrombin, matriptase-3/TMPRSS7 as well as factors IXa, Xa, and XIa. Its inhibitory activity is greatly enhanced in the presence of heparin. | −6.2 |
| 3L81 | PD | AP-4 complex subunit mu-1 | Subunit of novel type of clathrin- or nonclathrin-associated protein coat involved in targeting proteins from the TGN to the endosomal–lysosomal system. | −6 |
| 3KIV | PD | Apolipoprotein(a) | Apo(a) is the main constituent of Lp(a). It has serine proteinase activity and is able of autoproteolysis. Inhibits tissue-type plasminogen activator 1. Lp(a) may be a ligand for megalin/Gp 330. | −6.1 |
| 2O22 | PD | Apoptosis regulator Bcl-2 | Suppresses apoptosis in a variety of cell systems including factor-dependent lympho hematopoietic and neural cells. Regulates cell death by controlling the mitochondrial membrane permeability. Appears to function in a feedback loop system with caspases. Inhibits caspase activity either by preventing the release of cytochrome c from the mitochondria and by binding to the APAF-1. | −7.5 |
| 2AEB | PD | Arginase-1 | N/A | −6.7 |
| 1PQ3 | PD | Arginase-2, mitochondrial | May play a role in the regulation of extra-urea cycle arginine metabolism and also in downregulation of NO synthesis. Extrahepatic arginase functions to regulate L-arginine bioavailability to NO synthase. Since NO synthase is found in the penile corpus cavernosum smooth muscle, the clitoral corpus cavernosum and the vagina, arginase II plays a role in both male and female sexual arousal. It is therefore a potential target for the treatment of male and female sexual arousal disorders. | −8.1 |
| 3H82 | PK | Aryl hydrocarbon receptor nuclear translocator | Required for activity of the Ah (dioxin) receptor. This protein is required for the ligand-binding subunit to translocate from the cytosol to the nucleus after ligand binding. The complex then initiates transcription of genes involved in the activation of PAH procarcinogens. The heterodimer with HIF1A or EPAS1/HIF2A functions as a transcriptional regulator of the adaptive response to hypoxia. | −5.8 |
| 2PQT, 2IJA | PK | Arylamine N-acetyltransferase 1 | Participates in the detoxification of a plethora of hydrazine and arylamine drugs. Catalyzes the N- or O-acetylation of various arylamine and heterocyclic amine substrates and is able to bioactivate several known carcinogens. | −8.2 to −6 |
| 2PFR | PK | Arylamine N-acetyltransferase 2 | Participates in the detoxification of a plethora of hydrazine and arylamine drugs. Catalyzes the N- or O-acetylation of various arylamine and heterocyclic amine substrates and is able to bioactivate several known carcinogens. | −7 |
| 1E2S | PK | Arylsulfatase A | Hydrolyzes cerebroside sulfate. | −6.8 |
| 2O4H | PD | Aspartoacylase | Catalyzes the deacetylation of NAA to produce acetate and L-aspartate. NAA occurs in high concentration in brain and its hydrolysis. NAA plays a significant part in the maintenance of intact white matter. In other tissues, it acts as a scavenger of NAA from body fluids. | −7.2 |
| 4AYT, 4AYW, 4AYX | PK | ATP-binding cassette subfamily B member 10, mitochondrial | May mediate critical mitochondrial transport functions related to heme biosynthesis (by similarity). | −7.6 to −6.7 |
| 3NHA, 3NH9 | PK | ATP-binding cassette subfamily B member 6, mitochondrial | Binds heme and porphyrins and functions in their ATP-dependent uptake into the mitochondria. Plays a crucial role in heme synthesis. | −7.8 to −6.7 |
| 3LLM | PD | ATP-dependent RNA helicase A | Unwinds double-stranded DNA and RNA in a 3′ to 5′ direction. Alteration of secondary structure may subsequently influence interactions with proteins or other nucleic acids. Functions as a transcriptional activator. Component of the CRD-mediated complex that promotes MYC mRNA stability. Involved with LARP6 in the stabilization of type I collagen mRNAs for CO1A1 and CO1A2. | −6.7 |
| 3FDN | PD | Aurora kinase A | Mitotic serine/threonine kinases that contribute to the regulation of cell-cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, SAC, and cytokinesis. Required for initial activation of CDK1 at centrosomes. Phosphorylates numerous target proteins, including ARHGEF2, BORA, BRCA1, CDC25B, DLGP5, HDAC6, KIF2A, LATS2, NDEL1, PARD3, PPP1R2, PLK1, RASSF1, TACC3, p53/TP53, and TPX2. Regulates KIF2A tubulin depolymerase activity. Required for normal axon formation. Plays a role in microtubule remodeling during neurite extension. Important for microtubule formation and/or stabilization. Also acts as a key regulatory component of the p53/TP53 pathway, and particularly the checkpoint-response pathways critical for oncogenic transformation of cells, by phosphorylating and stabilizating p53/TP53. Phosphorylates its own inhibitors, the PP1 isoforms, to inhibit their activity. Necessary for proper cilia disassembly prior to mitosis. | −7.7 |
| 2KE1 | PD | Autoimmune regulator | Transcriptional regulator that binds to DNA as a dimmer or as a tetramer, but not as a monomer. Binds to G-doublets in an A/T-rich environment; the preferred motif is a tandem repeat of 5′-. ATTGGTTA-3′ combined with a 5′-TTATTA-3′ box. Binds to nucleosomes (by similarity). Binds to chromatin and interacts selectively with histone H3 that is not methylated at “Lys-4”, not phosphorylated at “Thr-3”, and not methylated at “Arg-2”. Functions as a sensor of histone H3 modifications that are important for the epigenetic regulation of gene expression. Functions as a transcriptional activator and promotes the expression of otherwise tissue-specific self-antigens in the thymus, which is important for self-tolerance and the avoidance of autoimmune reactions. | −7 |
| 2I3H | PD | Baculoviral IAP repeat-containing protein 7 | Apoptotic regulator capable of exerting proapoptotic and anti-apoptotic activities and plays crucial roles in apoptosis, cell proliferation, and cell-cycle control. Its anti-apoptotic activity is mediated through the inhibition of CASP3, CASP7, and CASP9 as well as by its E3 ubiquitin-protein ligase activity. As it is a weak caspase inhibitor, its anti-apoptotic activity is thought to be due to its ability to ubiquitinate DIABLO/SMAC targeting it for degradation, thereby promoting cell survival. May contribute to caspase inhibition, by blocking the ability of DIABLO/SMAC to disrupt XIAP/BIRC4–caspase interactions. Protects against apoptosis induced by TNF or by chemical agents such as adriamycin, etoposide, or staurosporine. Suppression of apoptosisis mediated by activation of MAPK8/JNK1, and possibly also of MAPK9/JNK2. This activation depends on TAB1 and NR2C2/TAK1. In vitro, inhibits CASP3 and proteolytic activation of pro-CASP9. Isoform 1 blocks staurosporine-induced apoptosis. Isoform 2 blocks etoposide-induced apoptosis. Isoform 2 protects against NK cell killing whereas isoform 1 augments killing. | −7.1 |
| 3LBZ | PD | B-cell lymphoma 6 protein | Transcriptional repressor that is required for germinal center formation and antibody affinity maturation. Probably plays an important role in lymphoma genesis. | −6.5 |
| 2YXJ | PD | Bcl-2-like protein 1 | Potent inhibitor of cell death. Inhibits activation of caspases (by similarity). Appears to regulate cell death by blocking the VDAC by binding to it and preventing the release of the caspase activator, CYC1, from the mitochondrial membrane. Also acts as a regulator of G2 checkpoint and progression to cytokinesis during mitosis. Isoform Bcl-X(S) promotes apoptosis. | −8.8 |
| 3I28, 1ZD3, 3ANS | PK | Bifunctional epoxide hydrolase 2 | Bifunctional enzyme. The C-terminal domain has epoxidehydrolase activity and acts on epoxides (alkene oxides, oxiranes) and arene oxides. Plays a role in xenobiotic metabolism by degrading potentially toxic epoxides. Also determines steady-state levels of physiological mediators. The N-terminal domain has lipid phosphatase activity, with the highest activity toward threo-9,10-phosphonooxy-hydroxy-octadecanoic acid, followed by erythro-9,10-phosphonooxy-hydroxy-octadecanoic acid, 12-phosphonooxy-octadec-9Z-enoic acid, 12-phosphonooxy-octadec-9E-enoic acid, and p-nitrophenyl phosphate. | −10.3 to −9.6 |
| 2W3O | PD | Bifunctional polynucleotide phosphatase/kinase | Plays a key role in the repair of DNA damage, functioning as part of both the NHEJ and BER pathways. Through its two catalytic activities, PNK ensures that DNA termini are compatible with extension and ligation either by removing 3′-phosphates from or by phosphorylating 5′-hydroxyl groups on the ribose sugar of the DNA backbone. | −5.9 |
| 1PKX | PD | Bifunctional purine biosynthesis protein PURH | Bifunctional enzyme that catalyzes two steps in purine biosynthesis. | −6.4 |
| 3OKI | PD | Bile acid receptor | Ligand-activated transcription factor. Receptor for bile acids such as chenodeoxycholic acid, lithocholic acid, and deoxycholic acid. Represses the transcription of the CYP7A1 through the induction of NR0B2 or FGF19 expression, via two distinct mechanisms. Activates the IBABP. Activates the transcription of bile salt export pump ABCB11 by directly recruiting histone methyltransferase CARM1 to this locus. | −6.2 |
| 3F3Y_2, 3F3Y, 1EFH | PK | Bile salt sulfotransferase | Sulfotransferase that utilizes PAPS as sulfonate donor to catalyze the sulfonation of steroids and bile acids in the liver and adrenal glands. | −8.7 to −6.1 |
| 1XSC | PD | Bis(5′-nucleosyl)-tetraphosphatase (asymmetrical) | A symmetrically hydrolyzes Ap4A to yield AMP and ATP. Plays a major role in maintaining homeostasis. | −6.5 |
| 3K0K | PD | Breast cancer type 1 susceptibility protein | E3 ubiquitin-protein ligase that specifically mediates the formation of “Lys-6”-linked polyubiquitin chains and plays a central role in DNA repair by facilitating cellular responses to DNA damage. It is unclear whether it also mediates the formation of other types of polyubiquitin chains. The E3 ubiquitin-protein ligase activity is required for its tumor suppressor function. The BRCA1-BARD1 heterodimer coordinates a diverse range of cellular pathways such as DNA damage repair, ubiquitination, and transcriptional regulation to maintain genomic stability. Regulates centrosomal microtubule nucleation. Required for normal cell-cycle progression from G2 to mitosis. Required for appropriate cell-cycle arrests after ionizing irradiation in both the S phase and the G2 phase of the cell-cycle. Involved in transcriptional regulation of P21 in response to DNA damage. Required for FANCD2 targeting to sites of DNA damage. May function as a transcriptional regulator. Inhibits lipid synthesis by binding to inactive phosphorylated ACACA and preventing its dephosphorylation. Contributes to HRR via its direct interaction with PALB2, fine-tunes recombinational repair partly through its modulatory role in the PALB2-dependent loading of BRCA2-RAD51 repair machinery at DNA breaks. Component of the BRCA1-RBBP8 complex which regulates CHEK1 activation and controls cell-cycle G2/M checkpoints on DNA damage via BRCA1-mediated ubiquitination of RBBP8. | −6.8 |
| 3P5O | PD | Bromodomain-containing protein 4 | Plays a role in a process governing chromosomal dynamics during mitosis (by similarity). | −7.7 |
| 3N7S | PD | CGRP type 1 receptor | Receptor for CGRP together with RAMP1 and receptor for adrenomedullin together with RAMP3 (by similarity). Receptor for adrenomedullin together with RAMP2. The activity of this receptor is mediated by G proteins that activates adenylyl cyclase. | −8.7 |
| 3AGM | PD | cAMP-dependent protein kinase catalytic subunit α | Phosphorylates a large number of substrates in the cytoplasm and the nucleus. Regulates the abundance of compartmentalized pools of its regulatory subunits through phosphorylation of PJA2 that binds and ubiquitinates these subunits, leading to their subsequent proteolysis. Phosphorylates CDC25B, ABL1, NFKB1, CLDN3, PSMC5/RPT6, PJA2, RYR2, RORA, TRPC1, and VASP. RORA is activated by phosphorylation. Required for glucose-mediated adipogenic differentiation increase and osteogenic differentiation inhibition from osteoblasts. Involved in the regulation of platelets in response to thrombin and collagen; maintains circulating platelets in a resting state by phosphorylating proteins in numerous platelet inhibitory pathways when in complex with NF-κB (NFKB1 and NFKB2) and I-κB-α (NFKBIA), but thrombin and collagen disrupt these complexes and free active PRKACA stimulates platelets and leads to platelet aggregation by phosphorylating VASP. Prevents the antiproliferative and anti-invasive effects of α-difluoromethylornithine in breast cancer cells when activated. RYR2 channel activity is potentiated by phosphorylation in presence of luminal Ca2+, leading to reduced amplitude and increased frequency of SOICR characterized by an increased rate of Ca2+ release and propagation velocity of spontaneous Ca2+ waves, despite reduced wave amplitude and resting cytosolic Ca2+. TRPC1 activation by phosphorylation promotes Ca2+ influx, essential for the increase in permeability induced by thrombin in confluent endothelial monolayers. PSMC5/RPT6 activation by phosphorylation stimulates proteasome. Regulates negatively TJs in ovarian cancer cells via CLDN3 phosphorylation. NFKB1 phosphorylation promotes NF-κB p50-p50 DNA binding. Involved in embryonic development by downregulating the Hh signaling pathway that determines embryo pattern formation and morphogenesis. Isoform 2 phosphorylates and activates ABL1 in sperm flagellum to promote spermatozoa capacitation. Prevents meiosis redumption in prophase-arrested oocytes via CDC25B inactivation by phosphorylation. May also regulate REM sleep in the PPT. | −8.2 |
| 2QYK | PD | cAMP-specific 3′,5′-cyclic phosphodiesterase 4A | Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes. | −8.4 |
| 3FRG | PD | cAMP-specific 3′,5′-cyclic phosphodiesterase 4B | Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes. May be involved in mediating CNS effects of therapeutic agents ranging from antidepressants to antiasthmatic and anti-inflammatory agents. | −8.3 |
| 1Y2K | PD | cAMP-specific 3′,5′-cyclic phosphodiesterase 4D | Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes. | −8.6 |
| 2FOY | PD | Carbonic anhydrase 1 | Reversible hydration of carbon dioxide. Can hydrate scyanamide to urea. | −8.5 |
| 3CZV | PD | Carbonic anhydrase 13 | Reversible hydration of carbon dioxide. | −7 |
| 2FOU | PD | Carbonic anhydrase 2 | Essential for bone resorption and osteoclast differentiation (by similarity). Reversible hydration of carbon dioxide. Can hydrate cyanamide to urea. Involved in the regulation of fluid secretion into the anterior chamber of the eye. | −6.6 |
| 3FW3 | PD | Carbonic anhydrase 4 | Reversible hydration of carbon dioxide. May stimulate the sodium/bicarbonate transporter activity of SLC4A4 that acts in pH homeostasis. It is essential for acid overload removal from the retina and retina epithelium, and acid release in the choriocapillaris in the choroid. | −6.6 |
| 2PFG_2, 2PFG, 3BHJ_3, 3BHJ, 3BHJ_2, 1WMA | PK | Carbonyl reductase (NADPH) 1 | NADPH-dependent reductase with broad substrate specificity. Catalyzes the reduction of a wide variety of carbonyl compounds including quinones, prostaglandins, menadione, plus various xenobiotics. Catalyzes the reduction of the antitumor anthracyclines doxorubicin and daunorubicin to the cardiotoxic compounds doxorubicinol and daunorubicinol. Can convert prostaglandin E2 to prostaglandin F2-α. Can bind glutathione, which explains its higher affinity for glutathione-conjugated substrates. Catalyzes the reduction of S-nitrosoglutathione. | −9.3 to −7.4 |
| 2HRB | PK | Carbonyl reductase (NADPH) 3 | Has low NADPH-dependent oxidoreductase activity toward 4-benzoylpyridine and menadione (in vitro). | −8.9 |
| 2V77 | PD | Carboxypeptidase A1 | Carboxypeptidase that catalyzes the release of a C-terminal amino acid, but has little or no action with -Asp, -Glu, -Arg, -Lys, or -Pro. | −7 |
| 2PCU | PD | Carboxypeptidase A4 | Metalloprotease that could be involved in the histone hyperacetylation pathway. | −7.5 |
| 3D67 | PD | Carboxypeptidase B2 | Cleaves C-terminal arginine or lysine residues from biologically active peptides such as kinins or anaphylatoxins in the circulation, thereby regulating their activities. Downregulates fibrinolysis by removing C-terminal lysine residues from fibrin that has already been partially degraded by plasmin. | −7.5 |
| 3H30 | PD | Casein kinase II subunit α | Catalytic subunit of a constitutively active serine/threonine–protein kinase complex that phosphorylates a large number of substrates containing acidic residues C-terminal to the phosphorylated serine or threonine. Regulates numerous cellular processes, such as cell-cycle progression, apoptosis, and transcription as well as viral infection. May act as a regulatory node that integrates and coordinates numerous signals, leading to an appropriate cellular response. During mitosis, functions as a component of the p53/TP53-dependent SAC that maintains cyclin-B-CDK1 activity and G2 arrest in response to spindle damage. Also required for p53/TP53-mediated apoptosis, phosphorylating “Ser-392” of p53/TP53 following UV irradiation. Can also negatively regulate apoptosis. Phosphorylates the caspases CASP9 and CASP2 and the apoptotic regulator NOL3. Phosphorylation protects CASP9 from cleavage and activation by CASP8 and inhibits the dimerization of CASP2 and activation of CASP8. Regulates transcription by direct phosphorylation of RNA polymerases I, II, III, and IV. Also phosphorylates and regulates numerous transcription factors including NF-κB, STAT1, CREB1, IRF1, IRF2, ATF1, SRF, MAX, JUN, FOS, MYC, and MYB. Phosphorylates Hsp90 and its co-chaperones FKBP4 and CDC37, which is essential for chaperone function. Regulates Wnt signaling by phosphorylating CTNNB1 and the transcription factor LEF1. Acts as an ectokinase that phosphorylates several extracellular proteins. During viral infection, phosphorylates various proteins involved in the viral life cycles of EBV, HSV, HBV, HCV, HIV, CMV and HPV. | −7.6 |
| 3E3B | PD | Casein kinase II subunit α′ | Catalytic subunit of a constitutively active serine/threonine–protein kinase complex that phosphorylates a large number of substrates containing acidic residues C-terminal to the phosphorylated serine or threonine. Regulates numerous cellular processes, such as cell-cycle progression, apoptosis, and transcription as well as viral infection. May act as a regulatory node which integrates and coordinates numerous signals, leading to an appropriate cellular response. During mitosis, functions as a component of the p53/TP53-dependent SAC that maintains cyclin-B-CDK1 activity and G2 arrest in response to spindle damage. Also required for p53/TP53-mediatedapoptosis, phosphorylating “Ser-392” of p53/TP53 following UV irradiation. Can also negatively regulate apoptosis. Phosphorylates the caspases CASP9 and CASP2 and the apoptotic regulator NOL3. Phosphorylation protects CASP9 from cleavage and activation by CASP8, and inhibits the dimerization of CASP2 and activation of CASP8. Regulates transcription by direct phosphorylation of RNA polymerases I, II, III and IV. Also phosphorylates and regulates numerous transcription factors including NF-κB, STAT1, CREB1, IRF1, IRF2, ATF1, SRF, MAX, JUN, FOS, MYC, and MYB. Phosphorylates Hsp90 and its co-chaperones FKBP4 and CDC37, which is essential for chaperone function. Regulates Wnt signaling by phosphorylating CTNNB1 and the transcription factor LEF1. Acts as an ectokinase that phosphorylates several extracellular proteins. During viral infection, phosphorylates various proteins involved in the viral life cycles of EBV, HSV, HBV, HCV, HIV, CMV, and HPV. | -8.1 |
| 1RWX | PD | Caspase-1 | Thiol protease that cleaves IL-1 β between an Asp and an Ala, releasing the mature cytokine that is involved in a variety of inflammatory processes. Important for defense against pathogens. Cleaves and activates SREBPs. Can also promote apoptosis. | −7.2 |
| 2H5I | PD | Caspase-3 | Involved in the activation cascade of caspases responsible for apoptosis execution. At the onset of apoptosis, it proteolytically cleaves PARP at a “216-Asp-|-Gly-217” bond. Cleaves and activates SREBPs between the basic helix-loop-helix leucine zipper domain and the membrane attachment domain. Cleaves and activates caspase-6, -7 and -9. Involved in the cleavage of huntingtin. Triggers cell adhesion in sympathetic neurons through RET cleavage. | −8.4 |
| 2QL9 | PD | Caspase-7 | Involved in the activation cascade of caspases responsible for apoptosis execution. Cleaves and activates SREBPs. Proteolytically cleaves PARP at a “216-Asp-|-Gly-217” bond. Overexpression promotes programmed cell death. | −8.3 |
| 1QTN | PD | Caspase-8 | Most upstream protease of the activation cascade of caspases responsible for the TNFRSF6/FAS-mediated and TNFRSF1A-induced cell death. Binding to the adapter molecule, FADD recruits it to either receptor. The resulting aggregate called DISC performs CASP8 proteolytic activation. The active dimeric enzyme is then liberated from the DISC and is free to activate downstream apoptotic proteases. Proteolytic fragments of the N-terminal propeptide (termed CAP3, CAP5, and CAP6) are likely retained in the DISC. Cleaves and activates CASP3, CASP4, CASP6, CASP7, CASP9, and CASP10. May participate in the GZMB apoptotic pathways. Cleaves ADPRT. Hydrolyzes the small-molecule substrate, Ac-Asp-Glu-Val-Asp AMC. Likely target for the cow pox virus CRMA death inhibitory protein. Isoforms 5–8 lack the catalytic site and may interfere with the pro-apoptotic activity of the complex. | −9.3 |
| 3BWM_2, 3BWY_2, 3BWM, 3BWY, 3A7E, 3A7E_2 | PK | Catechol O-methyltransferase | Catalyzes the O-methylation, and thereby the inactivation, of catecholamine neurotransmitters and catechol hormones. Also shortens the biological half-lives of certain neuroactive drugs like L-DOPA, α-methyl DOPA, and isoproterenol. | −7 to −6.4 |
| 1GMY | PD | Cathepsin B | Thiol protease that is believed to participate in intracellular degradation and turnover of proteins. Has also been implicated in tumor invasion and metastasis. | −7.1 |
| 1LYB | PD | Cathepsin D | Acid protease active in intracellular protein breakdown. Involved in the pathogenesis of several diseases such as breast cancer and possibly AD. | −7.8 |
| 1T32 | PD | Cathepsin G | Serine protease with trypsin- and chymotrypsin-like specificity. Cleaves complement C3. Has antibacterial activity against the Gram-negative bacterium Peudomonas aeruginosa, antibacterial activity is inhibited by LPS from P. aeruginosa, Z-Gly-Leu-Phe-CH2Cl, and phenylmethylsulfonyl fluoride. | −7.1 |
| 3KWZ | PD | Cathepsin K | Closely involved in osteoclastic bone resorption and may participate partially in the disorder of bone remodeling. Displays potent endoprotease activity against fibrinogen at acid pH. May play an important role in ECM degradation. | −7.6 |
| 3HHA | PD | Cathepsin L1 | Important for the overall degradation of proteins in lysosomes. | −7.9 |
| 3OVX | PD | Cathepsin S | Thiol protease. Key protease responsible for the removal of the invariant chain from MHC class II molecules. The bond specificity of this proteinase is in part similar to the specificities of cathepsin L and cathepsin N. | −7.1 |
| 1A4R | PD | Cell division control protein 42 homolog | Plasma membrane-associated small GTPase that cycles between an active GTP-bound and an inactive GDP-bound state. Inactive state binds to a variety of effector proteins to regulate cellular responses. Involved in epithelial cell polarization processes. Regulates the bipolar attachment of spindle microtubules to kinetochores before chromosome congression in metaphase. Plays a role in the extension and maintenance of the formation of thin, actin-rich surface projections called filopodia. Mediates CDC42-dependent cell migration. | −7.6 |
| 2FR3 | PD | Cellular retinoic acid-binding protein 2 | Transports retinoic acid to the nucleus. Regulates the access of retinoic acid to the nuclear retinoic acid receptors. | −7.4 |
| 2VUK | PD | Cellular tumor antigen p53 | Acts as a tumor suppressor in many tumor types; induces growth arrest or apoptosis depending on the physiological circumstances and cell type. Involved in cell-cycle regulation as a trans-activator that acts to negatively regulate cell division by controlling a set of genes required for this process. One of the activated genes is an inhibitor of CDKs. Apoptosis induction seems to be mediated either by stimulation of BAX and FAS antigen expression or by repression of Bcl-2 expression. Induces the transcription of lincRNA-p21 and lincRNA-Mkln1. LincRNA-p21 participates in TP53-dependent transcriptional repression, leading to apoptosis and seem to have to effect on cell-cycle regulation. Implicated in Notch signaling crossover. Prevents CDK7 kinase activity when associated with CAK complex in response to DNA damage, thus stopping cell-cycle progression. Isoform 2 enhances the transactivation activity of isoform 1 from some but not all TP53-inducible promoters. Isoform 4 suppresses transactivation activity and impairs growth suppression mediated by isoform 1. Isoform 7 inhibits isoform 1-mediated apoptosis. | −6.3 |
| 3ITU | PD | cGMP-dependent 3′,5′-cyclic phosphodiesterase | Cyclic nucleotide phosphodiesterase with a dual-specificity for the second messengers cAMP and cGMP, which are key regulators of many important physiological processes. | −8.6 |
| 1SO2 | PD | cGMP-inhibited 3′,5′-cyclic phosphodiesterase B | Cyclic nucleotide phosphodiesterase with a dual-specificity for the second messengers cAMP and cGMP, which are key regulators of many important physiological processes. May play a role in fat metabolism. Regulates cAMP binding of RAPGEF3. Through simultaneous binding to RAPGEF3 and PIK3R6 assembles a signaling complex in which the PI3K γ complex is activated by RAPGEF3 and which is involved in angiogenesis. | −9.7 |
| 1XOZ | PD | cGMP-specific 3′,5′-cyclic phosphodiesterase | Plays a role in signal transduction by regulating the intracellular concentration of cyclic nucleotides. This phosphodiesterase catalyzes the specific hydrolysis of cGMP to 5′-GMP. | −9.7 |
| 1WB0 | PD | Chitotriosidase-1 | Degrades chitin, chitotriose, and chitobiose. May participate in the defense against nematodes and other pathogens. Isoform 3 has no enzymatic activity. | −9.2 |
| 3G15 | PD | Choline kinase α | Has a key role in phospholipid biosynthesis and may contribute to tumor cell growth. Catalyzes the first step in phosphatidylcholine biosynthesis. Contributes to phosphatidylethanolamine biosynthesis. Phosphorylates choline and ethanolamine. Has higher activity with choline. | −8.9 |
| 3FEG | PD | Choline/ethanolamine kinase | Has a key role in phospholipid biosynthesis. Catalyzes the first step in phosphatidylethanolamine biosynthesis. Phosphorylates ethanolamine and can also act on choline (in vitro). Has higher activity with ethanolamine. May not significantly contribute to in vivo phosphatidylcholine biosynthesis. | −9 |
| 2WIJ | PD | Cholinesterase | Esterase with broad substrate specificity. Contributes to the inactivation of the neurotransmitter acetylcholine. Can degrade neurotoxic organophosphate esters. | −10.8 |
| 3N7O | PD | Chymase | Major secreted protease of mast cells with suspected roles in vasoactive peptide generation, ECM degradation, and regulation of gland secretion. | −7.8 |
| 3BHO | PD | Cleavage and polyadenylation specificity factor subunit 5 | Component of the CFIm complex that plays a key role in pre-mRNA 3′-processing. Involved in association with CPSF6 or CPSF7 in pre-MRNA 3′-end poly(A) site cleavage and poly(A) addition. NUDT21/CPSF5 binds to cleavage and polyadenylation RNA substrates. The homodimer mediates simultaneous sequence-specific recognition of two 5′-UGUA-3′ elements within the pre-mRNA. Binds to, but does not hydrolyze mono- and di-adenosine nucleotides. May have a role in mRNA export. | −9.8 |
| 3LC3 | PD | Coagulation factor IX | Factor IX is a vitamin K-dependent plasma protein that participates in the intrinsic pathway of blood coagulation by converting factor X to its active form in the presence of Ca2+ ions, phospholipids, and factor VIIIa. | −7.3 |
| 2BZ6 | PD | Coagulation factor VII | Initiates the extrinsic pathway of blood coagulation. Serine protease that circulates in the blood in a zymogen form. Factor VII is converted to factor VIIa by factors Xa, XIIa, IXa, or thrombin by minor proteolysis. In the presence of tissue factor and calcium ions, factor VIIa then converts factor X to factor Xa by limited proteolysis. Factor VIIa will also convert factor IX to factor IXa in the presence of tissue factor and calcium ions. | −8 |
| 3HNB | PD | Coagulation factor VIII | Factor VIII, along with calcium ions and phospholipid, acts as a cofactor for factor IXa when it converts factor X to the activated form, factor Xa. | −5.5 |
| 2JKH | PD | Coagulation factor X | Factor Xa is a vitamin K-dependent glycoprotein that converts prothrombin to thrombin in the presence of factor Va, calcium ions, and phospholipid during blood clotting. | −8 |
| 3BG8 | PD | Coagulation factor XI | Factor XI triggers the middle phase of the intrinsic pathway of blood coagulation by activating factor IX. | −7.6 |
| 830C | PD | Collagenase 3 | Degrades type I collagen. Does not act on gelatin or casein. Could have a role in tumoral process. | −7.6 |
| 2JG8 | PD | Complement C1q subcomponent subunit A | C1q associates with the proenzymes C1r and C1s to yield C1, the first component of the serum complement system. The collagen-like regions of C1q interact with the Ca2+-dependent C1r(2)-C1s(2) proenzyme complex, and efficient activation of C1 takes place on interaction of the globular heads of C1q with the Fc regions of IgG or IgM antibody present in immune complexes. | −7 |
| 1BIO | PD | Complement factor D | Factor D cleaves factor B when the latter is complexed with factor C3b, activating the C3bbb complex, which then becomes the C3 convertase of the alternate pathway. Its function is homologous to that of C1s in the classic pathway. | −6 |
| 2KMX | PD | Copper-transporting ATPase 1 | May supply copper to copper-requiring proteins within the secretory pathway, when localized in the TGN. Under conditions of elevated extracellular copper, it relocalized to the plasma membrane where it functions in the efflux of copper from cells. | −6.5 |
| 3ODU | PD | C-X-C chemokine receptor type 4 | Receptor for the C-X-C chemokine CXCL12/SDF-1 that transduces a signal by increasing intracellular calcium ion levels and enhancing MAPK1/MAPK3 activation. Acts as a receptor for extracellular ubiquitin, leading to enhanced intracellular calcium ions concentrations and reduced cellular cAMP levels. Involved in hematopoiesis and in cardiac ventricular septum formation. Also plays an essential role in vascularization of the gastrointestinal tract, probably by regulating vascular branching and/or remodeling processes in ECs. Involved in cerebellar development. In the CNS, could mediate hippocampal-neuron survival. Acts as a co-receptor (CD4 being the primary receptor) for HIV-1 X4 isolates and as a primary receptor for some HIV-2 isolates. Promotes Env-mediated fusion of the virus. | −9.9 |
| 2R3I | PD | CDK2 | Serine/threonine–protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis. Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, and EZH2. Interacts with cyclins A, B1, B3, D, or E. Triggers duplication of centrosomes and DNA. Acts at the G1 –S transition to promote the E2F transcriptional program and the initiation of DNA synthesis and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in hESCs. Activity of CDK2 is maximal during S phase and G2 ; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1 –S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1 (by similarity). Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1 –S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress toward mitosis. In response to DNA damage, DSB repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociation from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1 –S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the NO-mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1 –S transition. CTNNB1 phosphorylation regulates INS internalization. |
−6.9 |
| 1UNG | PD | CDK5 | Proline-directed serine/threonine–protein kinase essential for neuronal cell-cycle arrest and differentiation may be involved in apoptotic cell death in neuronal diseases by triggering abortive cell-cycle re-entry. Interacts with D1- and D3-type G1 cyclins. Phosphorylates SRC, NOS3, VIM/vimentin, p35/CDK5R1, MEF2A, SIPA1L1, SH3GLB1, PXN, PAK1, MCAM/MUC18, SEPT5, SYN1, DNM1, AMPH, SYNJ1, CDK16, RAC1, RHOA, CDC42, TONEBP/NFAT5, MAPT/TAU, MAP1B, histone H1, p53/TP53, HDAC1, APEX1, PTK2/FAK1, huntingtin/HTT, ATM, MAP2, NEFH, and NEFM. Regulates several neuronal development and physiological processes including neuronal survival, migration and differentiation, axonal and neurite growth, synaptogenesis, OLs differentiation, synaptic plasticity, and neurotransmission, by phosphorylating key proteins. Activated by interaction with CDK5R1(p35) and ATP6V0D1(p39), especially in postmitotic neurons, and promotes CDK5R1(p35) expression in an autostimulation loop. Phosphorylates many downstream substrates such as Rho and Ras family small GTPases (eg, PAK1, RAC1, RHOA, CDC42) or microtubule- binding proteins (eg, MAPT/TAU, MAP2, MAP1B), and modulates actin dynamics to regulate neurite growth and/or spine morphogenesis. Phosphorylates also exocytosis-associated proteins such as MCAM/MUC18, SEPT5, SYN1, and PCTAIRE 1/CDK16 as well as endocytosis-associated proteins such as DNM1, AMPH, and SYNJ1 at synaptic terminals. In the mature CNS, regulates neurotransmitter movements by phosphorylating substrates associated with neurotransmitter release and synapse plasticity; synaptic vesicle exocytosis, vesicles fusion with the presynaptic membrane, and endocytosis. Promotes cell survival by activating anti-apoptotic proteins BCL2 and STAT3, and negatively regulating of JNK3/MAPK10 activity. Phosphorylation of p53/TP53 in response to genotoxic and oxidative stresses enhances its stabilization by preventing ubiquitin ligase- mediated proteasomal degradation, and induces transactivation of p53/TP53 target genes, thus regulating apoptosis. Phosphorylation of p35/CDK5R1 enhances its stabilization by preventing calpain-mediated proteolysis producing p25/CDK5R1 and avoiding ubiquitin ligase-mediated proteasomal degradation. During aberrant cell-cycle activity and DNA damage, p25/CDK5 activity elicits cell-cycle activity and double-strand DNA breaks that precedes neuronal death by deregulating HDAC1. DNA damage-triggered phosphorylation of huntingtin/HTT in nuclei of neurons protects neurons against polyglutamine expansion as well as DNA damage-mediated toxicity. Phosphorylation of PXN reduces its interaction with PTK2/FAK1 in MCFAs during differentiation of OLs. Negative regulator of Wnt/β-catenin signaling pathway. Activator of the GAIT (IFN- γ-activated inhibitor of translation) pathway, which suppresses expression of a posttranscriptional regulon of proinflammatory genes in myeloid cells; phosphorylates the linker domain of glutamyl-prolyl tRNA synthetase (EPRS) in a IFN-γ-dependent manner, the initial event in assembly of the GAIT complex. Phosphorylation of SH3GLB1 is required for autophagy induction in starved neurons. Phosphorylation of TONEBP/NFAT5 in response to osmotic stress mediates its rapid nuclear localization. MEF2 is inactivated by phosphorylation in nucleus in response to neurotoxin, thus leading to neuronal apoptosis. APEX1AP-endodeoxyribonuclease is repressed by phosphorylation, resulting in accumulation of DNA damage and contributing to neuronal death. NOS3 phosphorylation downregulates NOS3-derived nitrite (NO) levels. SRC phosphorylation mediates its ubiquitin-dependent degradation and thus leads to cytoskeletal reorganization. May regulate EC migration and angiogenesis via the modulation of lamellipodia formation. Involved in dendritic spine morphogenesis by mediating the EFNA1-EPHA4 signaling. | −7.7 |
| 1XO2 | PD | CDK6 | Serine/threonine–protein kinase involved in the control of the cell cycle and differentiation; promotes G1 –S transition. Phosphorylates pRB/RB1 and NPM1. Interacts with D-type G1 cyclins during interphase at G1 to form a pRB/RB1 kinase and controls the entrance into the cell cycle. Involved in initiation and maintenance of cell-cycle exit during cell differentiation; prevents cell proliferation and regulates negatively cell differentiation, but is required for the proliferation of specific cell types (eg, erythroid and hematopoietic cells). Essential for cell proliferation within the dentate gyrus of the hippocampus and the subventricular zone of the lateral ventricles. Required during thymocyte development. Promotes the production of newborn neurons, probably by modulating G1 length. Promotes, at least in astrocytes, changes in patterns of gene expression, changes in the actin cytoskeleton including loss of stress fibers, and enhances motility during cell differentiation. Prevents myeloid differentiation by interfering with RUNX1 and reducing its transcription transactivation activity, but promotes proliferation of normal myeloid progenitors. Delays senescence. Promotes the proliferation of β-cells in pancreatic islets of Langerhans. | −7.5 |
| 2PZE | PD | Cystic fibrosis transmembrane conductance regulator | Involved in the transport of chloride ions. May regulate bicarbonate secretion and salvage in epithelial cells by regulating the SLC4A7 transporter. | −6.5 |
| 1MQ0 | PK | Cytidine deaminase | This enzyme scavenges exogenous and endogenous cytidine and 2′-dC for UMP synthesis. | −7.9 |
| 2CIA | PD | Cytoplasmic protein NCK2 | AP that associates with tyrosine-phosphorylated growth factor receptors or their cellular substrates. Maintains low levels of EIF2S1 phosphorylation by promoting its dephosphorylation by PP1. Plays a role in ELK1-dependent transcriptional activation in response to activated R as signaling. | −7 |
| 2ZI5_2, 2NO7, 3IPX, 2ZI5, 1P5Z, 2NO7_2 | PK | dC kinase | Required for the phosphorylation of the deoxyribonucleosides dC, dG, and dA. Has broad substrate specificity and does not display selectivity based on the chirality of the substrate. It is also an essential enzyme for the phosphorylation of numerous nucleoside analogs widely employed as antiviral and chemotherapeutic agents. | −9.7 to −5.4 |
| 2HQU | PD | Deoxyuridine 5′-triphosphate nucleotidohydrolase, mitochondrial | This enzyme is involved in nucleotide metabolism: it produces dUMP, the immediate precursor of thymidine nucleotides and it decreases the intracellular concentration of dUTP so that uracil cannot be incorporated into DNA. | −8.1 |
| 1KMV | PD | Dihydrofolate reductase | Key enzyme in folate metabolism. Contributes to the de novo mitochondrial thymidylate biosynthesis pathway. Catalyzes an essential reaction for de novo glycine and purine synthesis and for DNA precursor synthesis. Binds its own mRNA and that of DHFRL1. | −7.2 |
| 2G63 | PD | Dipeptidyl peptidase 4 | Cell surface glycoprotein receptor involved in the costimulatory signal essential for TCR-mediated T-cell activation. Acts as a positive regulator of T-cell coactivation, by binding at least ADA, CAV1, IGF2R, and PTPRC. Its binding to CAV1 and CARD11 induces T-cell proliferation and NF-κB activation in a TCR/CD3-dependent manner. Its interaction with ADA also regulates lymphocyte–epithelial cell adhesion. Involved in association with FAP in the pericellular proteolysis of the ECM, the migration and invasion of ECs into the ECM. May be involved in the promotion of lymphatic adhesion, migration, and tube formation of Ecs. When overexpressed, enhanced cell proliferation, a process inhibited by GPC3. Acts also as a serine exopeptidase with a dipeptidyl peptidase activity that regulates various physiological processes by cleaving peptides in the circulation, including many chemokines, mitogenic growth factors, neuropeptides, and peptide hormones. Removes N-terminal dipeptides sequentially from polypeptides having unsubstituted N-termini provided that the penultimate residue is proline. |
−8.7 |
| 2DDF | PD | Disintegrin and metalloproteinase domain-containing protein 17 | Cleaves the membrane-bound precursor of TNF-α to its mature soluble form. Responsible for the proteolytical release of soluble JAM3 from ECs surface. Responsible for the proteolytic release of several other cell-surface proteins, including p75 TNF-receptor, IL-1 receptor type II, p55TNF-receptor, transforming growth factor-α, L-selectin, GH receptor, MUC1, and the APP. Also involved in the activation of Notch pathway (by similarity). | −6.6 |
| 2OQS | PD | Disks large homolog 1 | Essential multidomain scaffolding protein required for normal development (by similarity). Recruits channels, receptors, and signaling molecules to discrete plasma membrane domains in polarized cells. May play a role in AJ assembly, signal transduction, cell proliferation, synaptogenesis, and lymphocyte activation. Regulates the excitability of cardiac myocytes by modulating the functional expression of Kv4 channels. Functional regulator of Kv1.5 channel. | −7.2 |
| 1FAO | PD | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3-phosphoinositide | May act as a B-cell-associated adapter that regulates B-cell antigen receptor-signaling downstream of PI3K. | −6.5 |
| 3E8N | PD | Dual specificity MAPK1 | Dual specificity protein kinase that acts as an essential component of the MAP kinase signal transduction pathway. Binding of extracellular ligands such as growth factors, cytokines, and hormones to their cell-surface receptors activates Ras and this initiates RAF1 activation. RAF1 then further activates the dual-specificity protein kinases MAP2K1/MEK1 and MAP2K2/MEK2. Both MAP2K1/MEK1 and MAP2K2/MEK2 function specifically in the MAPK/ERK cascade and catalyze the concomitant phosphorylation of a threonine and a tyrosine residue in a Thr-Glu-Tyr sequence located in the extracellular signal-regulated kinases MAPK3/ERK1 and MAPK1/ERK2, leading to their activation and further transduction of the signal within the MAPK/ERK cascade. Depending on the cellular context, this pathway mediates diverse biological functions such as cell growth, adhesion, survival and differentiation, predominantly through the regulation of transcription, metabolism, and cytoskeletal rearrangements. One target of the MAPK/ERK cascade is PPARG, a nuclear receptor that promotes differentiation and apoptosis. MAP2K1/MEK1 has been shown to export PPARG from the nucleus. The MAPK/ERK cascade is also involved in the regulation of endosomal dynamics, including lysosome processing and endosome cycling through the PNRC as well as in the fragmentation of the Golgi apparatus during mitosis. | −7.6 |
| 3HMP | PD | Dual specificity protein kinase TTK | Phosphorylates proteins on serine, threonine, and tyrosine. Probably associated with cell proliferation. Essential for chromosome alignment by enhancing AURKB activity (via direct CDCA8 phosphorylation) at the centromere, and for the mitotic checkpoint. | −8 |
| 3F81 | PD | Dual specificity protein phosphatase 3 | Shows activity both for tyrosine-protein phosphate and serine-protein phosphate but displays a strong preference toward phosphotyrosines. Specifically dephosphorylates and inactivates ERK1 and ERK2. | −8 |
| 3ANQ | PD | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | May play a role in a signaling pathway regulating nuclear functions of cell proliferation. Phosphorylates serine, threonine, and tyrosine residues in its sequence and in exogenous substrates. | −8.7 |
| 3LBL | PD | E3 ubiquitin-protein ligase Mdm2 | E3 ubiquitin-protein ligase that mediates ubiquitination of p53/TP53, leading to its degradation by the proteasome. Inhibits p53/TP53- and p73/TP73-mediated cell-cycle arrest and apoptosis by binding its transcriptional activation domain. Also acts as an ubiquitin ligase E3 toward itself and ARRB1. Permits the nuclear export of p53/TP53. Promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma RB1 protein. Inhibits DAXX-mediated apoptosis by inducing its ubiquitination and degradation. Component of the TRIM28/KAP1-MDM2-p53/TP53 complex involved in stabilizing p53/TP53. Also component of the TRIM28/KAP1-ERBB4-MDM2 complex that links growth factor and DNA damage response pathways. Mediates ubiquitination and subsequent proteasome degradation of DYRK2 in nucleus. Ubiquitinates IGF1R and promotes it to proteasomal degradation. | −8.5 |
| 3NY3 | PD | E3 ubiquitin-protein ligase UBR2 | E3 ubiquitin-protein ligase that is a component of the N-end rule pathway. Recognizes and binds to proteins bearing specific N-terminal residues that are destabilizing according to the N-end rule, leading to their ubiquitination and subsequent degradation. Plays a critical role in chromatin inactivation and chromosome-wide transcriptional silencing during meiosis via ubiquitination of histone H2A. Binds leucine and is a negative regulator of the leucine-mTOR signaling pathway, thereby controlling cell growth. | −7.3 |
| 3HL5 | PD | E3 ubiquitin-protein ligase XIAP | Multifunctional protein that not only regulates caspases and apoptosis but also modulates inflammatory signaling and immunity, copper homeostasis, mitogenic kinase signaling, cell proliferation as well as cell invasion and metastasis. Acts as a direct caspase inhibitor. Directly binds to the active site pocket of CASP3 and CASP7 and obstructs substrate entry. Inactivates CASP9 by keeping it in a monomeric, inactive state. Acts as an E3 ubiquitin-protein ligase regulating NF-κB signaling and the target proteins for its E3 ubiquitin-protein ligase activity include: RIPK1, CASP3, CASP7, CASP8, CASP9, MAP3K2/MEKK2, DIABLO/SMAC, AIFM1, CCS, and BIRC5/survivin. Ubiquitinion of CCS leads to enhancement of its chaperone activity toward its physiologic target, SOD1, rather than proteasomal degradation. Ubiquitinion of MAP3K2/MEKK2 and AIFM1 does not lead to proteasomal degradation. Plays a role in copper homeostasis by ubiquitinating COMMD1 and promoting its proteasomal degradation. Can also function as E3 ubiquitin-protein ligase of the NEDD8 conjugation pathway, targeting effector caspases for neddylation and inactivation. Regulates the BMP signaling pathway and the SMAD- and MAP3K7/TAK1-dependent pathways leading to NF-κB and JNK activation. Acts as an important regulator of innate immune signaling via regulation of NLRs. Protects cells from spontaneous formation of the ripoptosome, a large multiprotein complex that has the capability to kill cancer cells in a caspase-dependent and caspase-independent manner. Suppresses ripoptosome formation by ubiquitinating RIPK1 and CASP8. Acts as a positive regulator of Wnt signaling and ubiquitinates TLE1, TLE2, TLE3, TLE4, and AES. Ubiquitination of TLE3 results in inhibition of its interaction with TCF7L2/TCF4, thereby allowing efficient recruitment and binding of the transcriptional coactivator β-catenin to TCF7L2/TCF4 that is required to initiate a Wnt-specific transcriptional program. | −6.8 |
| 1JOC | PD | Early endosome antigen 1 | Binds phospholipid vesicles containing phosphatidylinositol 3-phosphate and participates in endosomal trafficking. | −6.1 |
| 1X9D | PD | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-α-mannosidase | Involved in glycoprotein quality control targeting of misfolded glycoproteins for degradation. It primarily trims a single α-1,2-linked mannose residue from Man(9)GlcNAc(2) to produce Man(8)GlcNAc(2), but at high enzyme concentrations, as found in the ERQC components, it further trims the carbohydrates to Man(5-6) GlcNAc(2). | −8.7 |
| 3DWB | PD | Endothelin-converting enzyme 1 | Converts big endothelin-1 to endothelin-1. | −10.1 |
| 1H1H | PD | Eosinophil cationic protein | Cytotoxin and helminthotoxin with low-efficiency ribonuclease activity. Possesses a wide variety of biological activities. Exhibits antibacterial activity, including cytoplasmic membrane depolarization of preferentially Gram-negative, but also Gram-positive strains. Promotes Escherichia coli outer membrane detachment, alteration of the overall cell shape and partial loss of cell content. | −7 |
| 2VWX | PD | Ephrin type-B receptor 4 | Receptor tyrosine kinase that binds promiscuously transmembrane ephrin-B family ligands residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. The signaling pathway downstream of the receptor is referred to as forward signaling while the signaling pathway downstream of the ephrin ligand is referred to as reverse signaling. Together with its cognate ligand/functional ligand EFNB2 plays a central role in heart morphogenesis and angiogenesis through regulation of cell adhesion and cell migration. EPHB4-mediated forward signaling controls cellular repulsion and segregation form EFNB2-expressing cells. Plays also a role in postnatal blood vessel remodeling, morphogenesis and permeability and is thus important in the context of tumor angiogenesis. | −8.5 |
| 2RGP | PD | EGFR | Receptor tyrosine kinase-binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses. Known ligands include EGF, TGFA/TGF-α, amphiregulin, epigen/EPGN, BTC/β cellulin, epiregulin/EREG and HBEGF/heparin-binding EGF. Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits APs like GRB2 that in turn activates complex downstream signaling cascades. Activates at least four major downstream signaling cascades including the Ras-RAF-MEK-ERK, PI3K-AKT, PLCγ-PKC, and STATs modules. May also activate the NF-κB signaling cascade. Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling. Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/β-catenin. Isoform 2 may act as an antagonist of EGF action. | −7.9 |
| 1I5R | PD | Estradiol 17-β-dehydrogenase 1 | Favors the reduction of estrogens and androgens. Also has 20-α-HSD activity. Uses preferentially NADH. | −10.5 |
| 1XPC | PD | ER | Nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic ERE sequence or association with other DNA-binding transcription factors, such as AP-1/c-JUN, c-FOS, ATF-2, Sp1, and Sp3, to mediate ERE-independent signaling. Ligand binding induces a conformational change allowing subsequent or combinatorial association with multiprotein coactivator complexes through LXXLL motifs of their respective components. Mutual transrepression occurs between the ER and NF-κB in a cell-type specific manner. Decreases NF-κB DNA-binding activity and inhibits NF-κB-mediated transcription from the IL6 promoter and displaces RELA/p65 and associated coregulators from the promoter. Recruited to the NF-κB response element of the CCL2 and IL8 promoters and can displace CREBBP. Present with NF-κB components RELA/p65 and NFKB1/p50 on ERE sequences. Can also act synergistically with NF-κB to activate transcription involving respective recruitment adjacent response elements; the function involves CREBBP. Can activate the transcriptional activity of TFF1. Also mediates membrane-initiated estrogen signaling involving various kinase cascades. Isoform 3 is involved in activation of NOS3 and endothelial NO production. Isoforms lacking one or several functional domains are thought to modulate transcriptional activity by competitive ligand or DNA binding and/or heterodimerization with the full length receptor. Isoform 3 can bind to ERE and inhibit isoform 1. | −6.7 |
| 1YYE | PD | ER β | Nuclear hormone receptor. Binds estrogens with an affinity similar to that of ESR1 and activates expression of reporter genes containing EREs in an estrogen-dependent manner. Isoform β-cx lacks ligand-binding ability and has no or only very low binding activity resulting in the loss of ligand-dependent transactivation ability. DNA binding by ESR1 and ESR2 is rapidly lost at 37°C in the absence of ligand while in the presence of 17 β-estradioland 4-hydroxy-tamoxifen loss in DNA binding at elevated temperature is more gradual. | −4.9 |
| 1G3M, 1G3M_2, 1HY3 | PK | Estrogen sulfotransferase | Sulfotransferase that utilizes PAPS as sulfonate donor to catalyze the sulfate conjugation of estradiol and estrone. May play a role in the regulation of ER activity by metabolizing free estradiol. Maximally sulfates β-estradiol and estrone at concentrations of 20 nM. Also sulfates dehydroepiandrosterone, pregnenolone, ethinylestradiol, equalenin, diethylstilbesterol, and 1-naphthol, at significantly higher concentrations; however, cortisol, testosterone, and dopamine are not sulfated. | −7.8 to −6.4 |
| 2E2R | PD | Estrogen-related receptor γ | Orphan receptor that acts as transcription activator in the absence of bound ligand. Binds specifically to an ERE and activates reporter genes controlled by ERE (by similarity). | −5.6 |
| 2W97 | PD | Eukaryotic translation initiation factor 4E | Its translation stimulation activity is repressed by binding to the CYFIP1-FMR1 complex (by similarity). Recognizes and binds the 7-methyl guanosine-containing mRNA cap during an early step in the initiation of protein synthesis and facilitates ribosome binding by inducing the unwinding of the mRNAs secondary structures. Component of the CYFIP1-EIF4E-FMR1 complex that binds to the mRNA cap and mediates translational repression. In the CYFIP1-EIF4E-FMR1 complex, this subunit mediates the binding to them RNA cap. | −7.7 |
| 3N45 | PD | Farnesyl pyrophosphate synthase | Key enzyme in isoprenoid biosynthesis that catalyzes the formation of FPP, a precursor for several classes of essential metabolites including sterols, dolichols, carotenoids, and ubiquinones. FPP also serves as substrate for protein farnesylation and geranylgeranylation. Catalyzes the sequential condensation of isopentenyl pyrophosphate with the allylic pyrophosphates, dimethylallyl pyrophosphate, and then with the resultant geranylpyrophosphate to the ultimate product farnesyl pyrophosphate. | −8.8 |
| 2NNQ | PD | FABP, adipocyte | Lipid transport protein in adipocytes. Binds both long-chain fatty acids and retinoic acid. Delivers long-chain fatty acids and retinoic acid to their cognate receptors in the nucleus (by similarity). | −5.9 |
| 1FDQ | PD | FABP, brain | B-FABP could be involved in the transport of a so far unknown hydrophobic ligand with potential morphogenic activity during CNS development. It is required for the establishment of the radial glial fiber system in developing brain, a system that is necessary for the migration of immature neurons to establish cortical layers (by similarity). | −6.3 |
| 1HMR | PD | FABP, heart | FABP is thought to play a role in the intracellular transport of long-chain fatty acids and their acyl-CoA esters. | −6.7 |
| 3C4F | PD | Fibroblast growth factor receptor 1 | Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of embryonic development, cell proliferation, differentiation, and migration. Required for normal mesoderm patterning and correct axial organization during embryonic development, normal skeletogenesis, and normal development of the GnRH neuronal system. Phosphorylates PLCG1, FRS2, GAB1, and SHB. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules DAG and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1, and SOS1 and mediates activation of Ras, MAPK1/ERK2, MAPK3/ERK1, and the MAPK signaling pathway as well as of the AKT1 signaling pathway. Promotes phosphorylation of SHC1, STAT1, and PTPN11/SHP2. In the nucleus, enhances RPS6KA1 and CREB1 activity and contributes to the regulation of transcription. FGFR1 signaling is downregulated by IL-17RD/SEF and by FGFR1 ubiquitination, internalization, and degradation. | −8.3 |
| 3B2T | PD | Fibroblast growth factor receptor 2 | Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation, migration, and apoptosis and in the regulation of embryonic development. Required for normal embryonic patterning, trophoblast function, limb bud development, lung morphogenesis, osteogenesis, and skin development. Plays an essential role in the regulation of osteoblast differentiation, proliferation and apoptosis, and is required for normal skeleton development. Promotes cell proliferation in keratinocytes and immature osteoblasts, and promotes apoptosis in differentiated osteoblasts. Phosphorylates PLCG1, FRS2, and PAK4. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules DAG and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1, and SOS1 and mediates activation of Ras, MAPK1/ERK2, MAPK3/ERK1, and the MAPK signaling pathway as well as of the AKT1 signaling pathway. FGFR2 signaling is downregulated by ubiquitination, internalization, and degradation. Mutations that lead to constitutive kinase activation or impair normal FGFR2 maturation, internalization, and degradation lead to aberrant signaling. Over-expressed FGFR2 promotes activation of STAT1. | −9 |
| 3BZ3 | PD | Focal adhesion kinase 1 | Nonreceptor PTK that plays an essential role in regulating cell migration, adhesion, spreading, reorganization of the actin cytoskeleton, formation and disassembly of focal adhesions and cell protrusions, cell-cycle progression, cell proliferation, and apoptosis. Required for early embryonic development and placenta development. Required for embryonic angiogenesis, normal cardiomyocyte migration and proliferation, and normal heart development. Regulates axon growth and neuronal cell migration, axon branching and synapse formation; required for normal development of the nervous system. Plays a role in osteogenesis and differentiation of osteoblasts. Functions in integrin signal transduction, but also in signaling downstream of numerous growth factor receptors, (GPCRs), EPHA2, netrin receptors, and LDL receptors. Forms multisubunit signaling complexes with SRC and SRC family members upon activation; this leads to the phosphorylation of additional tyrosine residues, creating binding sites for scaffold proteins, effectors, and substrates. Regulates numerous signaling pathways. Promotes activation of PI3K and the AKT1signaling cascade. Promotes activation of MAPK1/ERK2, MAPK3/ERK1, and the MAPK signaling cascade. Promotes localized and transient activation of GEFs and GAPs, and thereby modulates the activity of Rho family GTPases. Signaling via CAS family members mediates activation of RAC1. Recruits the ubiquitin ligase MDM2 to P53/TP53 in the nucleus, and thereby regulates P53/TP53 activity, P53/TP53 ubiquitination, and proteasomal degradation. Phosphorylates SRC; this increases SRC kinase activity. Phosphorylates ACTN1, ARHGEF7, GRB7, RET, and WASL. Promotes phosphorylation of PXN and STAT1; most likely PXN and STAT1 are phosphorylated by a SRC family kinase that is recruited to autophosphorylated PTK2/FAK1, rather than by PTK2/FAK1 itself. Promotes phosphorylation of BCAR1; GIT2 and SHC1; this requires both SRC and PTK2/FAK1. Promotes phosphorylation of BMX and PIK3R1. Isoform 6 (FRNK) does not contain a kinase domain and inhibits PTK2/FAK1 phosphorylation and signaling. Its enhanced expression can attenuate the nuclear accumulation of LPXN and limit its ability to enhance SRF-dependent gene transcription. | −7.6 |
| 2JJK | PD | Fructose-1,6-bisphosphatase 1 | −7 | |
| 3NYN | PD | G protein-coupled receptor kinase 6 | Specifically phosphorylates the activated forms of G protein-coupled receptors. Such receptor phosphorylation initiates β-arrestin-mediated receptor desensitization, internalization, and signaling events leading to their desensitization. Seems to be involved in the desensitization of D2-like dopamine receptors instriatum and chemokine receptor CXCR4 that is critical for CXCL12-induced cell chemotaxis (By similarity). Phosphorylates RHO (in vitro) and a non G-protein-coupled receptor: LRP6 during Wnt signaling (in vitro). | −6.6 |
| 3OY8 | PD | Galectin-1 | May regulate apoptosis, cell proliferation, and cell differentiation. Binds β-galactoside and a wide array of complex carbohydrates. Inhibits CD45 protein phosphatase activity and therefore the dephosphorylation of LYN kinase. | −6.4 |
| 2XG3 | PD | Galectin-3 | Galactose-specific lectin that binds IgE. May mediate with the α-3, β-1 integrin the stimulation by CSPG4 of ECs migration. Together with DMBT1, required for terminal differentiation of columnar epithelial cells during early embryogenesis (by similarity). In the nucleus: acts as a pre-mRNA splicing factor. Involved in acute inflammatory responses including neutrophil activation and adhesion, chemoattraction of monocytes macrophages, opsonization of apoptotic neutrophils, and activation of mast cells. | −6.3 |
| 2Q80 | PD | Geranylgeranyl pyrophosphate synthase | Catalyzes the trans-addition of the three molecules of IPP onto DMAPP to form geranylgeranyl pyrophosphate, an important precursor of carotenoids and geranylated proteins. | −8.7 |
| 1NHZ, 3K22_2, 3K22, 3E7C | PK | GC receptor | Receptor for GCs. Has a dual mode of action: as a transcription factor that binds to GREs and as a modulator of other transcription factors. Affects inflammatory responses, cellular proliferation, and differentiation in target tissues. Could act as a coactivator for STAT5-dependent transcription upon GH stimulation and could reveal an essential role of hepatic GR in the control of body growth. Involved in chromatin remodeling. Plays a significant role in transactivation. Involved in nuclear translocation (by similarity). | −7 to −5.7 |
| 2BHL, 2BH9, 2BH9_2, 1QKI, 1QKI_2 | PK | Glucose-6-phosphate 1-dehydrogenase | Produces pentose sugars for nucleic acid synthesis and main producer of NADPH reducing power. | −9.4 to −7.9 |
| 2V3D | PD | Glucosylceramidase | N/A | −8.6 |
| 3BI1 | PD | Glutamate carboxypeptidase 2 | Has both folate hydrolase and NAALADase activity. Has a preference for tri-α-glutamate peptides. In the intestine, required for the uptake of folate. In the brain, modulates excitatory neurotransmission through the hydrolysis of the neuropeptide, NAAG, thereby releasing glutamate. Isoform PSM-4 and isoform PSM-5 would appear to be physiologically irrelevant. Involved in prostate tumor progression. Also exhibits a dipeptidyl-peptidase IV type activity. In vitro, cleaves Gly-Pro-AMC. | −9.5 |
| 2AFW | PD | Glutaminyl-peptide cyclotransferase | Responsible for the biosynthesis of pyroglutamyl peptides. Has a bias against acidic and tryptophan residues adjacent to the N-terminal glutaminyl residue and a lack of importance of chain length after the second residue. Also catalyzes N-terminal pyroglutamate formation. In vitro, catalyzes pyroglutamate formation of N-terminally truncated form of APP amyloid-β peptides [Glu-3]-β-amyloid. May be involved in the N-terminal pyroglutamate formation of several amyloid-related plaque-forming peptides. | −8.3 |
| 1K3L, 1K3Y, 3KTL, 1YDK | PK | Glutathione S-transferase A1 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. | −8.5 to −7.3 |
| 2VCT, 4ACS, 2WJU | PK | Glutathione S-transferase A2 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. | −9.6 to −8.3 |
| 2VCV, 2VCV_2, 1TDI | PK | Glutathione S-transferase A3 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Catalyzes isomerization reactions that contribute to the biosynthesis of steroid hormones. Efficiently catalyze obligatory double-bond isomerizations of δ(5)-androstene-3,17-dione and δ(5)-pregnene- 3,20-dione precursors to testosterone and progesterone, respectively. | −11.4 to −8.4 |
| 3IK7 | PK | Glutathione S-transferase A4 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. This isozyme has a high catalytic efficiency with 4-hydroxyalkenals such as 4-HNE. | −8 |
| 3RPN, 1YZX | PK | Glutathione S-transferase κ1 | Significant glutathione conjugating activity is found only with the model substrate, CDNB. | −8.5 to −8.0 |
| 1XWK, 1YJ6, 1XW6 | PK | Glutathione S-transferase µ1 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. | −7.7 to −7.1 |
| 1HNA, 3GUR, 2C4J, 1XW5 | PK | Glutathione S-transferase µ2 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. | −7.9 to −7.0 |
| 1EEM, 3LFL, 3VLN | PK | Glutathione S-transferase ω1 | Exhibits glutathione-dependent thiol transferase and dehydroascorbate reductase activities. Has S-(phenacyl) glutathione reductase activity. Has also glutathione S-transferase activity. Participates in the biotransformation of inorganic arsenic and reduces MMA and dimethylarsonic acid. | −7.7 to −7.3 |
| 3QAG, 3Q19 | PK | Glutathione S-transferase ω2 | Exhibits glutathione-dependent thiol transferase activity. Has high dehydroascorbate reductase activity and may contribute to the recycling of ascorbic acid. Participates in the biotransformation of inorganic arsenic and reduces MMA. | −7.9 to −7.7 |
| 3CSH_2, 2A2R, 3GUS, 3CSH, 3GUS_2 | PK | Glutathione S-transferase P | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Regulates negatively CDK5 activity via p25/p35 translocation to prevent neurodegeneration. | −8.1 to −7.9 |
| 2C3Q | PK | Glutathione S-transferase θ1 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Acts on 1,2-epoxy-3-(4-nitrophenoxy) propane, phenethylisothiocyanate 4-nitrobenzyl chloride and 4-nitrophenethyl bromide. Displays glutathione peroxidase activity with cumene hydroperoxide. | −6.8 |
| 3JDW | PD | Glycine amidinotransferase, mitochondrial | Catalyzes the biosynthesis of guanidinoacetate, the immediate precursor of creatine. Creatine plays a vital role in energy metabolism in muscle tissues. May play a role in embryonic and CNS development. May be involved in the response to heart failure by elevating local creatine synthesis. | −6.9 |
| 2AZT, 1R74 | PK | Glycine N-methyltransferase | Catalyzes the methylation of glycine by using S-AdoMet to form N-methylglycine (sarcosine) with the concomitant production of S-adenosyl homocysteine (AdoHcy). Possible crucial role in the regulation of tissue concentration of AdoMet and of metabolism of methionine. | −7.6 to −7.2 |
| 1L5R | PD | Glycogen phosphorylase, liver form | Phosphorylase is an important allosteric enzyme in carbohydrate metabolism. Enzymes from different sources differ in their regulatory mechanisms and in their natural substrates. However, all known phosphorylases share catalytic and structural properties. | −8.5 |
| 1Q5K | PD | Glycogen synthase kinase-3 β | Constitutively active protein kinase that acts as a negative regulator in the hormonal control of glucose homeostasis, Wnt signaling, and regulation of transcription factors and microtubules, by phosphorylating and inactivating glycogensynthase (GYS1 or GYS2), EIF2B, CTNNB1/β-catenin, APC, AXIN1, DPYSL2/CRMP2, JUN, NFATC1/NFATC, MAPT/TAU, and MACF1. Requires primed phosphorylation of the majority of its substrates. In skeletal muscle, contributes to INS regulation of glycogen synthesis by phosphorylating and inhibiting GYS1 activity and hence glycogen synthesis. May also mediate the development of INS resistance by regulating activation of transcription factors. Regulates protein synthesis by controlling the activity of initiation factor 2B (EIF2BE/EIF2B5) in the same manner as glycogen synthase. In Wnt signaling, GSK3B forms a multimeric complex with APC, AXIN1, and CTNNB1/β-catenin and phosphorylates the N-terminus of CTNNB1, leading to its degradation mediated by ubiquitin/proteasomes. Phosphorylates JUN at sites proximal to its DNA-binding domain, thereby reducing its affinity for DNA. Phosphorylates NFATC1/NFATC on conserved serine residues promoting NFATC1/NFATC nuclear export, shutting off NFATC1/NFATC gene regulation, and thereby opposing the action of calcineurin. Phosphorylates MAPT/TAU on “Thr-548”, decreasing significantly MAPT/TAU ability to bind and stabilize microtubules. MAPT/TAU is the principal component of neurofibrillary tangles in AD. Plays an important role in ERBB2-dependent stabilization of microtubules at the cell cortex. Phosphorylates MACF1, inhibiting its binding to microtubules which is critical for its role in bulge stem cell migration and skin wound repair. Probably regulates NF-κB (NFKB1) at the transcriptional level and is required for the NF-κB-mediated anti-apoptotic response to TNF-α (TNF/TNFA). Negatively regulates replication in pancreatic β-cells, resulting in apoptosis, loss of β-cells and diabetes. Phosphorylates MUC1 in breast cancer cells, decreasing the interaction of MUC1 with CTNNB1/β-catenin. Is necessary for the establishment of neuronal polarity and axonout growth. Phosphorylates MARK2, leading to inhibit its activity. Phosphorylates SIK1 at “Thr-182”, leading to sustain its activity. Phosphorylates ZC3HAV1 that enhances its antiviral activity. | −8.9 |
| 2EUK | PD | Glycolipid transfer protein | Accelerates the intermembrane transfer of various glycolipids. Catalyzes the transfer of various glycosphingolipids between membranes but does not catalyze the transfer of phospholipids. May be involved in the intracellular translocation of glucosylceramides. | −9.7 |
| 1ORF | PK | Granzyme A | This enzyme is necessary for target cell lysis in cell-mediated immune responses. It cleaves after Lys or Arg. Cleaves APEX1 after “Lys-31” and destroys its oxidative repair activity. Involved in apoptosis. | −7.7 |
| 1IAU | PD | Granzyme B | This enzyme is necessary for target cell lysis in cell-mediated immune responses. It cleaves after Asp. Seems to be linked to an activation cascade of caspases (aspartate-specific cysteine proteases) responsible for apoptosis execution. Cleaves caspase-3, -7, -9, and 10 to give rise to active enzymes mediating apoptosis. | −7.5 |
| 2W0Z | PD | Growth factor receptor-bound protein 2 | AP that provides a critical link between cell surface growth factor receptors and the Ras signaling pathway. Isoform 2 does not bind to phosphorylated EGFR but inhibits EGF-induced transactivation of a Ras-responsive element. Isoform 2 acts as a dominant negative protein over GRB2 and, by suppressing proliferative signals, may trigger active programmed cell death. | −6.4 |
| 1CLU | PD | GTPase HRas | Ras proteins bind GDP/GTP and possess intrinsic GTPase activity. | −7.3 |
| 2XTN | PD | GTPase IMAP family member 2 | N/A | −7.3 |
| 3ORH | PK | Guanidinoacetate N-methyltransferase | N/A | −7.4 |
| 2G83 | PD | Guanine nucleotide-binding protein G(i) subunit α-1 | Guanine nucleotide-binding proteins (G proteins) are involved as modulators or transducers in various transmembrane signaling systems. The G(i) proteins are involved in hormonal regulation of adenylate cyclase: they inhibit the cyclase in response to β-adrenergic stimuli. The inactive GDP-bound form prevents the association of RGS14 with centrosomes and is required for the translocation of RGS14 from the cytoplasm to the plasma membrane. May play a role in cell division. | −8.3 |
| 2J9L | PD | H+/Cl− exchange transporter 5 | Proton-coupled chloride transporter. Functions as antiport system and exchanges chloride ions against protons. Important for normal acidification of the endosome lumen. May play an important role in renal tubular function. | −6.9 |
| 2KBS | PD | Harmonin | Required for normal development and maintenance of cochlear hair cell bundles. Anchoring/scaffolding protein that is a part of the functional network formed by USH1C, USH1G, CDH23, and MYO7A, which mediates mechanotransduction in cochlear hair cells. Required for normal hearing (by similarity). | −6.1 |
| 3EKO | PD | Heat shock protein HSP 90-α | Molecular chaperone that promotes the maturation, structural maintenance, and proper regulation of specific target proteins involved for instance in cell-cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle, and chaperone function. | −7 |
| 3NMQ | PD | Heat shock protein HSP 90-β | Molecular chaperone that promotes the maturation, structural maintenance, and proper regulation of specific target proteins involved for instance in cell-cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle, and chaperone function. | −8 |
| 2CVD | PD | Hematopoietic prostaglandin D synthase | Bifunctional enzyme that catalyzes both the conversion of PGH2 to PGD2, a prostaglandin involved in smooth muscle contraction/relaxation and a potent inhibitor of platelet aggregation, and the conjugation of glutathione with a wide range of aryl halides and organic isothiocyanates. Also exhibits low glutathione-peroxidase activity toward cumene hydroperoxide. | −8.2 |
| 3F66 | PD | Hepatocyte growth factor receptor | Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to hepatocyte growth factor/HGF ligand. Regulates many physiological processes including proliferation, scattering, morphogenesis, and survival. Ligand binding at the cell surface induces autophosphorylation of MET on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with the PI3K subunit PIK3R1, PLCG1, SRC, GRB2, STAT3, or the adapter GAB1. Recruitment of these downstream effectors by MET leads to the activation of several signaling cascades including the Ras-ERK, PI3K-AKT, or PLCγ-PKC. The Ras-ERK activation is associated with the morphogenetic effects while PI3K/AKT coordinates prosurvival effects. During embryonic development, MET signaling plays a role in gastrulation, development and migration of muscles and neuronal precursors, angiogenesis, and kidney formation. In adults, participates in wound healing as well as organ regeneration and tissue remodeling. Promotes also differentiation and proliferation of hematopoietic cells. Acts as a receptor for Listeria internalin in lB, mediating entry of the pathogen into cells. | −9.9 |
| 1ZKL | PD | High affinity cAMP-specific 3′,5′-cyclic phosphodiesterase 7A | Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes. May have a role in muscle signal transduction. | −8.5 |
| 3ECN | PD | High affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8A | Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes. May be involved in maintaining basal levels of the cyclic nucleotide and/or in the cAMP regulation of germ cell development. | −8.1 |
| 2HD1 | PD | High affinity cGMP-specific 3′,5′-cyclic phosphodiesterase 9A | Hydrolyzes the second messenger cGMP, which is a key regulator of many important physiological processes. | −9.5 |
| 2AOT, 2AOT_2, 1JQD, 1JQE, 1JQD_2 | PK | Histamine N-methyltransferase | Inactivates histamine by N-methylation. Plays an important role in degrading histamine and in regulating the airway response to histamine. | −9.8 to −8.2 |
| 3IOI | PD | Histo-blood group ABO system transferase | This protein is the basis of the ABO blood group system. The histo-blood group ABO involves three carbohydrate antigens: A, B, and H. A, B, and AB individuals express a glycosyl transferase activity that converts the H antigen to the A antigen (by addition of UDP-GalNAc) or to the B antigen (by addition of UDP-Gal), whereas O individuals lack such activity. | −8.7 |
| 1WUG | PD | HAT KAT2B | Functions as a HAT to promote transcriptional activation. Has significant HAT activity with core histones (H3 and H4) and also with nucleosome core particles. Inhibits cell-cycle progression and counteracts the mitogenic activity of the adenoviral oncoprotein E1A. In case of HIV-1 infection, it is recruited by the viral protein Tat. Regulates Tat’s transactivating activity and may help inducing chromatin remodeling of proviral genes. | −7.5 |
| 3MAX | PD | HDAC2 | Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3, and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell-cycle progression, and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Forms transcriptional repressor complexes by associating with MAD, SIN3, YY1, and N-COR. Interacts in the late S phase of DNA replication with DNMT1 in the other transcriptional repressor complex composed of DNMT1, DMAP1, PCNA, CAF1. Deacetylates TSHZ3 and regulates its transcriptional repressor activity. Component of a RCOR-GFI-KDM1A-HDAC complex that suppresses, via HDAC recruitment, a number of genes implicated in multilineage blood cell development. | −7.2 |
| 2VQM | PD | HDAC4 | Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3, and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell-cycle progression, and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Involved in muscle maturation via its interaction with the myocyte enhancer factors such as MEF2A, MEF2C, and MEF2D. | −8.4 |
| 3C10 | PD | HDAC7 | Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3, and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell-cycle progression, and developmental events. HDACs act via the formation of large multiprotein complexes. Involved in muscle maturation by repressing transcription of myocyte enhancer factors such as MEF2A, MEF2B, and MEF2C. During muscle differentiation, it shuttles into the cytoplasm, allowing the expression of myocyte enhancer factors (by similarity). May be involved in EBV latency, possibly by repressing the viral BZLF1 gene. | −6.8 |
| 3MZ6 | PD | HDAC8 | Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3, and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell-cycle progression, and developmental events. HDACs act via the formation of large multiprotein complexes. May play a role in smooth muscle cell contractility. | −6.8 |
| 3MO5 | PD | Histone–lysine N-methyltransferase EHMT1 | Histone methyltransferase that specifically mono- and dimethylates “Lys-9” of histone H3 (H3K9me1 and H3K9me2, respectively) in euchromatin. H3K9me represents a specific tag for epigenetic transcriptional repression by recruiting HP1 proteins to methylated histones. Also weakly methylates “Lys-27” of histone H3 (H3K27me). Also required for DNA methylation, the histone methyltransferase activity is not required for DNA methylation, suggesting that these two activities function independently. Probably targeted to histone H3 by different DNA-binding proteins like E2F6, MGA, MAX, and/or DP1. During G0 phase, it probably contributes to silencing of MYC- and E2F-responsive genes, suggesting a role in G0 –G transition in cell cycle. In addition to the histone methyltransferase activity, also methylates nonhistone proteins: mediates dimethylation of “Lys-373” of p53/TP53. | -8.4 |
| 3K5K | PD | Histone–lysine N-methyltransferase EHMT2 | Histone methyltransferase that specifically mono- and dimethylates “Lys-9” of histone H3 (H3K9me1 and H3K9me2, respectively) in euchromatin. H3K9me represents a specific tag for epigenetic transcriptional repression by recruiting HP1 proteins to methylated histones. Also mediates monomethylation of “Lys-56” of histone H3 (H3K56me1) in G1 phase, leading to promote interaction between histone H3 and PCNA and regulating DNA replication. Also weakly methylates “Lys-27” of histone H3 (H3K27me). Also required for DNA methylation, the histone methyltransferase activity is not required for DNA methylation, suggesting that these two activities function independently. Probably targeted to histone H3 by different DNA-binding proteins like E2F6, MGA, MAX, and/or DP1. May also methylate histone H1. In addition to the histone methyltransferase activity, also methylates non-histone proteins: mediates dimethylation of “Lys-373” of p53/TP53. Also methylates CDYL, WIZ, ACIN1, DNMT1, HDAC1, ERCC6, KLF12 and itself. | -8.4 |
| 3LQJ | PD | Histone–lysine N-methyltransferase MLL | Histone methyltransferase that plays an essential role in early development and hematopoiesis. Catalytic subunit of the MLL1-MLL complex, a multiprotein complex that mediates both methylation of “Lys-4” of histone H3 (H3K4me) complex and acetylation of “Lys-16” of histone H4 (H4K16ac). In the MLL1-MLL complex, it specifically mediates H3K4me, a specific tag for epigenetic transcriptional activation. Has weak methyltransferase activity by itself and requires other component of the MLL1-MLL complex to obtain full methyltransferase activity. Has no activity toward histone H3 phosphorylated on “Thr-3”, less activity toward H3 dimethylated on “Arg-8” or “Lys-9”, while it has higher activity toward H3 acetylated on “Lys-9”. Required for transcriptional activation of HOXA9. Promotes PPP1R15A-inducedapoptosis. | −8.4 |
| 1X7Q | PD | HLA class I histocompatibility antigen, A-11 α chain | Involved in the presentation of foreign antigens to the immune system. | −8.1 |
| 3D25 | PD | HLA class I histocompatibility antigen, A-2 α chain | Involved in the presentation of foreign antigens to the immune system. | −8.2 |
| 1XR9 | PD | HLA class I histocompatibility antigen, B-15 α chain | Involved in the presentation of foreign antigens to the immune system. | −8.6 |
| 3BZE | PD | HLA class I histocompatibility antigen, α chain E | Preferably binds to a peptide derived from the signal sequence of most HLA-A, -B, -C, and -G molecules. | −8.9 |
| 1HMP, 1BZY | PK | Hypoxanthine–guanine phosphoribosyl transferase | Converts guanine to guanosine monophosphate, and hypoxanthine to inosine monophosphate. Transfers the 5-phosphoribosyl group from 5-phosphoribosylpyro phosphate onto the purine. Plays a central role in the generation of purine nucleotides through the purine salvage pathway. | −7.3 to −5.9 |
| 2W0X | PD | Hypoxia-inducible factor 1-α inhibitor | Hydroxylates HIF-1 α at “Asp-803” in the CAD. Functions as an oxygen sensor and, under normoxic conditions, the hydroxylation prevents interaction of HIF-1 with transcriptional coactivators including Cbp/p300-interacting transactivator. Involved in transcriptional repression through interaction with HIF1A, VHL, and HDACs. Hydroxylates specific Asn residues within ARDs of NFKB1, NFKBIA, NOTCH1, ASB4, PPP1R12A, and several other ARD-containing proteins. Also hydroxylates Asp and His residues within ARDs of ANK1 and TNKS2, respectively. Negatively regulates NOTCH1 activity, accelerating myogenic differentiation. Positively regulates ASB4 activity, promoting vascular differentiation. | −7.5 |
| 1D6V | PD | Ig κ chain C region | N/A | −8 |
| 1GAF | PD | Ig γ-1 chain C region | N/A | −6.6 |
| 3EKN | PD | INS receptor | Receptor tyrosine kinase that mediates the pleiotropic actions of INS. Binding of INS leads to phosphorylation of several intracellular substrates, including, IRSs (IRS1, 2, 3, 4), SHC, GAB1, CBL, and other signaling intermediates. Each of these phosphorylated proteins serve as docking proteins for other signaling proteins that contain Src-homology-2 domains (SH2 domain) that specifically recognize different phosphotyrosines residues, including the p85 regulatory subunit of PI3K and SHP2. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway, which is responsible for most of the metabolic actions of INS, and the Ras-MAPK pathway, which regulates expression of some genes and cooperates with the PI3K pathway to control cell growth and differentiation. Binding of the SH2 domains of PI3K to phosphotyrosines on IRS1 leads to the activation of PI3K and the generation of PIP3, a lipid second messenger, which activates several PIP3-dependent serine/threonine kinases, such as PDPK1 and subsequently AKT/PKB. The net effect of this pathway is to produce a translocation of the glucose transporter SLC2A4/GLUT4 from cytoplasmic vesicles to the cell membrane to facilitate glucose transport. Moreover, upon INS stimulation, activated AKT/PKB is responsible for: anti-apoptotic effect of INS by inducing phosphorylation of BAD; regulates the expression of gluconeogenic and lipogenic enzymes by controlling the activity of the winged helix or forkhead (FOX) class of transcription factors. Another pathway regulated by PI3K-AKT/PKB activation is mTORC1 signaling pathway that regulates cell growth and metabolism and integrates signals from INS. AKT mediates INS-stimulated protein synthesis by phosphorylating, TSC2 thereby activating mTORC1 pathway. The Ras/RAF/MAP2K/MAPK pathway is mainly involved in mediating cell growth, survival, and cellular differentiation of INS. Phosphorylated IRS1 recruits GRB2-SOS complex, which triggers the activation of the Ras/RAF/MAP2K/MAPK pathway. In addition to binding INS, the INS receptor can bind IGFs (IGFI and IGFII). Isoform Short has a higher affinity for IGFII binding. When present in a hybrid receptor with IGF1R, binds IGF1. PubMed:12138094 shows that hybrid receptors composed of IGF1R and INSR isoform Long are activated with a high affinity by IGF1, with low affinity by IGF2 and not significantly activated by INS, and that hybrid receptors composed of IGF1R and INSR isoform Short are activated by IGF1, IGF2, and INS. In contrast, PubMed:16831875 shows that hybrid receptors composed of IGF1R and INSR isoform Long and hybrid receptors composed of IGF1R and INSR isoform Short have similar binding characteristics, both bind IGF1 and have a low affinity for INS. | −9.2 |
| 3E4A | PD | INS-degrading enzyme | Plays a role in the cellular breakdown of INS, IAPP, glucagon, bradykinin, kallidin, and other peptides, and thereby plays a role in intercellular peptide signaling. Degrades amyloid formed by APP and IAPP. May play a role in the degradation and clearance of naturally secreted amyloid β-protein by neurons and microglia. | −9.5 |
| 2OJ9 | PD | IGF1 receptor | Receptor tyrosine kinase that mediates actions of IGF1. Binds IGF1 with high affinity and IGF2 and INS with a lower affinity. The activated IGF1R is involved in cell growth and survival control. IGF1R is crucial for tumor transformation and survival of malignant cell. Ligand binding activates the receptor kinase, leading to receptor autophosphorylation and tyrosines phosphorylation of multiple substrates that function as signaling APs including, the IRSs (IRS1/2), Shc, and 14-3-3 proteins. Phosphorylation of IRS proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway and the Ras-MAPK pathway. The result of activating the MAPK pathway is increased cellular proliferation, whereas activating the PI3K pathway inhibits apoptosis and stimulates protein synthesis. Phosphorylated IRS1 can activate the 85 kDa regulatory subunit of PI3K (PIK3R1), leading to activation of several downstream substrates, including protein AKT/PKB. AKT phosphorylation, in turn, enhances protein synthesis through mTOR activation and triggers the antiapoptotic effects of IGFIR through phosphorylation and inactivation of BAD. In parallel to PI3K-driven signaling, recruitment of Grb2/SOS by phosphorylated IRS1 or Shc leads to recruitment of Ras and activation of the Ras-MAPK pathway. In addition to these two main signaling pathways, IGF1R signals also through the Janus kinase/signal transducer and activator of transcription pathway (JAK/STAT). Phosphorylation of JAK proteins can lead to phosphorylation/activation of STAT proteins. In particular, activation of STAT3 may be essential for the transforming activity of IGF1R. The JAK/STAT pathway activates gene transcription and may be responsible for the transforming activity. JNKs can also be activated by the IGF1R. IGF1 exerts inhibiting activities on JNK activation via phosphorylation and inhibition of MAP3K5/ASK1, which is able to directly associate with the IGF1R. When present in a hybrid receptor with INSR, binds IGF1. PubMed:12138094 shows that hybrid receptors composed of IGF1R and INSR isoform Long are activated with a high affinity by IGF1, with low affinity by IGF2 and not significantly activated by INS, and that hybrid receptors composed of IGF1R and INSR isoform Short are activated by IGF1, IGF2, and INS. In contrast, PubMed:16831875 shows that hybrid receptors composed of IGF1R and INSR isoform Long and hybrid receptors composed of IGF1R and INSR isoform Short have similar binding characteristics, both bind IGF1 and have a low affinity for INS. | −7.4 |
| 1IMX | PD | IGFI | The IGFs, isolated from plasma, are structurally and functionally related to INS but have a much higher growth-promoting activity. May be a physiologicalregulator of [1-14C]-2-deoxy-D-glucose (2DG) transport and glycogen synthesis in osteoblasts. Stimulates glucose transport in rat bone-derived osteoblastic (PyMS) cells and is effective at much lower concentrations than INS, not only regarding glycogen and DNA synthesis but also with regard to enhancing glucose uptake. | −6.6 |
| 2ICA | PD | Integrin α-L | Integrin α-L/β-2 is a receptor for ICAM1, ICAM2, ICAM3, and ICAM4. It is involved in a variety of immune phenomena including leukocyte-EC interaction, cytotoxic T-cell mediated killing, and antibody-dependent killing by granulocytes and monocytes. | −7.9 |
| 2OIC | PD | IL-1 receptor-associated kinase 4 | Serine/threonine–protein kinase that plays a critical role in initiating innate immune response against foreign pathogens. Involved in TLR and IL-1R signaling pathways. Is rapidly recruited by MYD88 to the receptor-signaling complex upon TLR activation to form the Myddosome together with IRAK2. Phosphorylates initially IRAK1, thus stimulating the kinase activity and intensive autophosphorylation of IRAK1. Phosphorylates E3 ubiquitin ligases pellino proteins (PELI1, PELI2, and PELI3) to promote pellino-mediated polyubiquitination of IRAK1. Then, the ubiquitin-binding domain of IKBKG/NEMO binds to polyubiquitinated IRAK1 bringing together the IRAK1-MAP3K7/TAK1-TRAF6 complex and the NEMO-IKKA-IKKB complex. In turn, MAP3K7/TAK1 activates IKKs (CHUK/IKKA and IKBKB/IKKB) leading to NF-κB nuclear translocation and activation. Alternatively, phosphorylates TIRAP to promote its ubiquitination and subsequent degradation. Phosphorylates NCF1 and regulates NADPH oxidase activation after LPS stimulation suggesting a similar mechanism during microbial infections. | −8.7 |
| 1M48 | PD | IL-2 | Produced by T-cells in response to antigenic or mitogenic stimulation, this protein is required for T-cell proliferation and other activities crucial to regulation of the immune response. Can stimulate B-cells, monocytes, lymphokine-activated killer cells, NK cells, and glioma cells. | −6.9 |
| 966C | PD | Interstitial collagenase | Cleaves collagens of types I, II, and III at one site in the helical domain. Also cleaves collagens of types VII and X. In case of HIV infection, interacts and cleaves the secreted viral Tat protein, leading to a decrease in neuronal Tat’s mediated neurotoxicity. | −7.9 |
| 2PSX | PD | Kallikrein-5 | May be involved in desquamation. | −6.8 |
| 3NC9 | PD | Ketohexokinase | N/A | −8.5 |
| 2PG2 | PD | Kinesin-like protein KIF11 | Motor protein required for establishing a bipolar spindle. Blocking of KIF11 prevents centrosome migration and arrest cells in mitosis with monoastral microtubule arrays. | −9.7 |
| 3FVS, 3FVX, 3FVX_2, 3FVU, 3FVU_2 | PK | Kynurenine–oxoglutarate transaminase 1 | Catalyzes the irreversible transamination of the L-tryptophan metabolite L-kynurenine to form KA. Metabolizes the cysteine conjugates of certain halogenated alkenes and alkanes to form reactive metabolites. Catalyzes the β-elimination of S-conjugates and Se-conjugates of L-(seleno) cysteine, resulting in the cleavage of the C-S or C-Se bond. | −11.2 to −10.4 |
| 1QIN | PD | Lactoylglutathione lyase | Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione. Involved in the regulation of TNF-induced transcriptional activity of NF-κB. | −8.6 |
| 3.00E+73 | PD | LanC-like protein 1 | May play a role in EPS8 signaling. Binds glutathion. | −7.4 |
| 3F70 | PD | Lethal(3)malignant brain tumor-like protein 2 | Putative PcG protein. PcG proteins maintain the transcriptionally repressive state of genes, probably via a modification of chromatin, rendering it heritably changed in its expressibility. Its association with a chromatin-remodeling complex suggests that it may contribute to prevent expression of genes that trigger the cell into mitosis. Binds to monomethylated and dimethylated “Lys-20” on histone H4. Binds histone H3 peptides that are monomethylated or dimethylated on “Lys-4”, “Lys-9”, or “Lys-27”. | −6.2 |
| 3FUN | PD | Leukotriene A4 hydrolase | Epoxide hydrolase that catalyzes the final step in the biosynthesis of the proinflammatory mediator leukotriene B4. Has also aminopeptidase activity. | −8.8 |
| 2H7C, 1MX1 | PK | Liver carboxylesterase 1 | Involved in the detoxification of xenobiotics and in the activation of ester and amide prodrugs. Hydrolyzes aromatic and aliphatic esters, but has no catalytic activity toward amides or a fatty acyl-CoA ester. Hydrolyzes the methyl ester group of cocaine to form benzoylecgonine. Catalyzes the transesterification of cocaine to form cocaethylene. Displays fatty acid ethyl ester synthase activity, catalyzing the ethyl esterification of oleic acid to ethyloleate. | −7.2 to −3.0 |
| 3NJY | PD | Lysine-specific demethylase 4A | Histone demethylase that specifically demethylates “Lys-9” and “Lys-36” residues of histone H3, thereby playing a central role in histone code. Does not demethylate “Lys-4”, “Lys-27” of histone H3, nor “Lys-20” of histone H4. Demethylates trimethylated “Lys-9” and “Lys-36” residues of H3, while it has no activity on mono and dimethylated residues. Demethylation of Lys residue generates formaldehyde and succinate. Participates in transcriptional repression of ASCL2 and E2F-responsive promoters via the recruitment of HDACs and NCOR1, respectively. Isoform 2: Crucial for muscle differentiation, promotes transcriptional activation of the Myog gene by directing the removal of repressive chromatin marks at its promoter. Lacks the N-terminal demethylase domain. | −6.5 |
| 3GL6 | PD | Lysine-specific demethylase 5A | Histone demethylase that specifically demethylates “Lys-4” residue of histone H3, thereby playing a central role in histone code. Does not demethylate “Lys-9”, “Lys-27”, “Lys-36”, “Lys-79” of histone H3, or “Lys-20” of histone H4. Demethylates trimethylated and imethylated but not monomethylated “Lys” of H3. May stimulate transcription mediated by nuclear receptors. May be involved in transcriptional regulation of Hox proteins during cell differentiation. May participate in transcriptional repression of cytokines such as CXCL12. | −6.7 |
| 2UXX | PD | Lysine-specific histone demethylase 1A | Histone demethylase that demethylates both “Lys-4” (H3K4me) and “Lys-9” (H3K9me) of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Acts as a corepressor by mediating demethylation of H3K4me, a specific tag for epigenetic transcriptional activation. Demethylates both mono-(H3K4me1) and dimethylated (H3K4me2) H3K4me. May play a role in the repression of neuronal genes. Alone, it is unable to demethylate H3K4me on nucleosomes and requires the presence of RCOR1/CoREST to achieve such activity. Also acts as a coactivator of ANDR-dependent transcription, by being recruited to ANDR target genes and mediating demethylation of H3K9me, a specific tag for epigenetic transcriptional repression. The presence of PRKCB in ANDR-containing complexes, which mediates phosphorylation of “Thr-6” of histone H3 (H3T6ph), a specific tag that prevents demethylation H3K4me, and prevents H3K4me demethylase activity of KDM1A. Demethylates dimethylated “Lys-370” of p53/TP53, which prevents interaction of p53/TP53 with TP53BP1 and represses p53/TP53-mediated transcriptional activation. Demethylates and stabilizes the DNA methylase DNMT1. Required for gastrulation during embryogenesis. Component of a RCOR-GFI-KDM1A-HDAC complex that suppresses, via HDAC recruitment, a number of genes implicated in multilineage blood cell development. | −9.8 |
| 3BL9 | PD | m7GpppX diphosphatase | Necessary for the complete degradation of mRNAs, both in normal mRNA turnover and in NMD. Removes the 7-methyl guanine cap structure from mRNA fragments shorter than ten nucleotides that are produced by 3′–.5′ exosome-mediated mRNA decay. Releases m7GMP. Can also degrade m7GDP to m7GMP. Has no activity toward mRNA molecules longer than 25 nucleotides. | −10 |
| 2I1M | PD | Macrophage CSF1 receptor | Tyrosine-protein kinase that acts as cell-surface receptor for CSF1 and IL34 and plays an essential role in the regulation of survival, proliferation and differentiation of hematopoietic precursor cells, especially mononuclear phagocytes, such as macrophages and monocytes. Promotes the release of proinflammatory chemokines in response to IL34 and CSF1, and thereby plays an important role in innate immunity and in inflammatory processes. Plays an important role in the regulation of osteoclast proliferation and differentiation, the regulation of bone resorption, and is required for normal bone and tooth development. Required for normal male and female fertility, and for normal development of milk ducts and acinar structures in the mammary gland during pregnancy. Promotes reorganization of the actin cytoskeleton, regulates formation of membrane ruffles, cell adhesion and cell migration, and promotes cancer cell invasion. Activates several signaling pathways in response to ligand binding. Phosphorylates PIK3R1, PLCG2, GRB2, SLA2, and CBL. Activation of PLCG2 leads to the production of the cellular signaling molecules, such as DAG and inositol 1,4,5-trisphosphate, that then lead to the activation of PKC family members, especially PRKCD. Phosphorylation of PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase, leads to activation of the AKT1 signaling pathway. Activated CSF1R also mediates activation of the MAP kinases MAPK1/ERK2 and/or MAPK3/ERK1, and of the SRC family kinases SRC, FYN, and YES1. Activated CSF1R transmits signals both via proteins that directly interact with phosphorylated tyrosine residues in its intracellular domain and via APs, such as GRB2. Promotes activation of STAT family members STAT3, STAT5A, and/or STAT5B. Promotes tyrosine phosphorylation of SHC1 and INPP5D/SHIP-1. Receptor signaling is downregulated by protein phosphatases, such as INPP5D/SHIP-1, that dephosphorylate the receptor and its downstream effectors and by rapid internalization of the activated receptor. | −8.1 |
| 3F17 | PD | Macrophage metalloelastase | May be involved in tissue injury and remodeling. Has significant elastolytic activity. Can accept large and small amino acids at the P1′ site, but has a preference for leucine. Aromatic or hydrophobic residues are preferred at the P1 site, with small hydrophobic residues (preferably alanine) occupying P3. | −6.4 |
| 3IJG | PD | Macrophage migration inhibitory factor | Proinflammatory cytokine. Involved in the innate immune response to bacterial pathogens. The expression of MIF at sites of inflammation suggests a role as mediator in regulating the function of macrophages in host defense. Counteracts the anti-inflammatory activity of GCs. Has phenyl pyruvate tautomerase and dopachrome tautomerase activity (in vitro), but the physiological substrate is not known. It is not clear whether the tautomerase activity has any physiological relevance, and whether it is important for cytokine activity. | −7 |
| 1FW1 | PK | Maleylacetoacetate isomerase | Bifunctional enzyme showing minimal glutathione-conjugating activity with ethacrynic acid and 7-chloro-4-nitrobenz-2-oxa-1,3-diazole and maleylacetoacetate isomerase activity. Has also low glutathione peroxidase activity with tert-butyl and cumene hydroperoxides. Is able to catalyze the glutathione-dependent oxygenation of dichloroacetic acid to glyoxylic acid. | −5.8 |
| 3L4Y | PD | Maltase-glucoamylase, intestinal | May serve as an alternate pathway for starch digestion when luminal α-amylase activity is reduced because of immaturity or malnutrition. May play a unique role in the digestion of malted dietary oligosaccharides used in food manufacturing. | −7.6 |
| 3M2W | PD | MAPK 2 | Stress-activated serine/threonine–protein kinase involved in cytokines production, endocytosis, reorganization of the cytoskeleton, cell migration, cell-cycle control, chromatin remodeling, DNA damage response, and transcriptional regulation. Following stress, it is phosphorylated and activated by MAP kinase p38-α/MAPK14, leading to phosphorylation of substrates. Phosphorylates serine in the peptide sequence, Hyd-X-R-X(2)-S, where Hyd is a large hydrophobic residue. Phosphorylates ALOX5, CDC25B, CDC25C, ELAVL1, HNRNPA0, HSF1, HSP27/HSPB1, KRT18, KRT20, LIMK1, LSP1, PABPC1, PARN, PDE4A, RCSD1, RPS6KA3, TAB3, and TTP/ZFP36. Mediates phosphorylation of HSP27/HSPB1 in response to stress, leading to dissociate HSP27/HSPB1 from large sHsps oligomers and impair their chaperone activities and ability to protect against oxidative stress effectively. Involved in inflammatory response by regulating TNF and IL6 production posttranscriptionally: acts by phosphorylating ARE-binding proteins ELAVL1, HNRNPA0, PABPC1, and TTP/ZFP36, leading to regulate the stability and translation of TNF and IL6 mRNAs. Phosphorylation of TTP/ZFP36, a major posttranscriptional regulator of TNF, promotes its binding to 14-3-3 proteins and reduces its ARE mRNA affinity, leading to inhibition of dependent degradation of ARE-containing transcript. Also involved in late G2/M checkpoint following DNA damage through a process of posttranscriptional mRNA stabilization: following DNA damage, it relocalizes from nucleus to cytoplasm and phosphorylates HNRNPA0 and PARN, leading to stabilize GADD45A mRNA. Involved in TLR signaling pathway in DCs: required for acute TLR-induced macropinocytosis by phosphorylating and activating RPS6KA3. | −6.9 |
| 3FHR | PD | MAPK 3 | Stress-activated serine/threonine–protein kinase involved in cytokines production, endocytosis, cell migration, chromatin remodeling, and transcriptional regulation. Following stress, it is phosphorylated and activated by MAP kinase p38-α/MAPK14, leading to phosphorylation of substrates. Phosphorylates serine in the peptide sequence, Hyd-X-R-X(2)-S, where Hyd is a large hydrophobic residue. MAPKAPK2 and MAPKAPK3 share the same function and substrate specificity, but MAPKAPK3 kinase activity and level in protein expression are lower compared to MAPKAPK2. Phosphorylates HSP27/HSPB1, KRT18, KRT20, RCSD1, RPS6KA3, TAB3, and TTP/ZFP36. Mediates phosphorylation of HSP27/HSPB1 in response to stress, leading to dissociate HSP27/HSPB1 from large sHsps oligomers and impair their chaperone activities and ability to protect against oxidative stress effectively. Involved in inflammatory response by regulating TNF and IL6 production posttranscriptionally: acts by phosphorylating ARE-binding proteins, such as TTP/ZFP36, leading to regulate the stability and translation of TNF and IL6 mRNAs. Phosphorylation of TTP/ZFP36, a major posttranscriptional regulator of TNF, promotes its binding to 14-3-3 proteins and reduces its ARE mRNA affinity, leading to inhibition of dependent degradation of ARE-containing transcript. Involved in TLR signaling pathway in DCs: required for acute TLR-induced macropinocytosis by phosphorylating and activating RPS6KA3. Also acts as a modulator of polycomb-mediated repression. | −6.9 |
| 3G0E | PD | Mast/stem cell growth factor receptor kit | Tyrosine-protein kinase that acts as cell-surface receptor for the cytokine KITLG/SCF and plays an essential role in the regulation of cell survival and proliferation, hematopoiesis, stem cell maintenance, gametogenesis, mast cell development, migration and function, and in melanogenesis. In response to KITLG/SCF binding, KIT can activate several signaling pathways. Phosphorylates PIK3R1, PLCG1, SH2B2/APS, and CBL. Activates the AKT1 signaling pathway by phosphorylation of PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase. Activated KIT also transmits signals via GRB2 and activation of Ras, RAF1, and the MAP kinases MAPK1/ERK2 and/or MAPK3/ERK1. Promotes activation of STAT family members STAT1, STAT3, STAT5A, and STAT5B. Activation of PLCG1 leads to the production of the cellular signaling molecules DAG and inositol 1,4,5-trisphosphate. KIT signaling is modulated by protein phosphatases and by rapid internalization and degradation of the receptor. Activated KIT promotes phosphorylation of the protein phosphatases PTPN6/SHP-1and PTPRU, and of the transcription factors STAT1, STAT3, STAT5A, and STAT5B. Promotes phosphorylation of PIK3R1, CBL, CRK (isoform Crk-II), LYN, MAPK1/ERK2, and/or MAPK3/ERK1, PLCG1, SRC, and SHC1. | −7.1 |
| 1MMQ | PD | Matrilysin | Degrades casein, gelatins of types I, III, IV, and V, and fibronectin. Activates procollagenase. | −7.4 |
| 1RM8 | PD | Matrix metalloproteinase 16 | Endopeptidase that degrades various components of the extracellular matrix, such as collagen type III and fibronectin. Activates progelatinase A. Involved in the matrix remodeling of blood vessels. Isoform Short cleaves fibronectin and also type III collagen, but at lower rate. It has no effect on type I, II, IV, and V collagens. However, upon interaction with CSPG4, it may be involved in degradation and invasion of type I collagen by melanoma cells. | −7.6 |
| 2JSD | PD | Matrix metalloproteinase 20 | Degrades amelogenin, the major protein component of the enamel matrix and two of the macromolecules characterizing the cartilage extracellular matrix: aggrecan and the COMP. May play a central role in tooth enamel formation. | −7 |
| 2OVX | PD | Matrix metalloproteinase 9 | May play an essential role in local proteolysis of the extracellular matrix and in leukocyte migration. Could play a role in bone osteoclastic resorption. Cleaves KiSS1 at a Gly-|-Leubond. Cleaves type IV and V collagens into large C-terminal three quarter fragments and shorter N-terminal one quarter fragments. Degrades fibronectin but not laminin or Pz-peptide. | −7.5 |
| 3K05 | PD | Mediator of DNA damage checkpoint protein 1 | Required for checkpoint-mediated cell-cycle arrest in response to DNA damage within both the S phase and G2/M phases of the cell cycle. May serve as a scaffold for the recruitment of DNA repair and signal transduction proteins to discrete foci of DNA damage marked by “Ser-139” phosphorylation of histone H2AFX. Also required for downstream events subsequent to the recruitment of these proteins. These include phosphorylation and activation of the ATM, CHEK1, and CHEK2 kinases, and stabilization of TP53 and apoptosis. ATM and CHEK2 may also be activated independently by a parallel pathway mediated by TP53BP1. | −6.3 |
| 2NQ6 | PD | Methionine aminopeptidase 1 | Removes the N-terminal methionine from nascent proteins. Required for normal progression through the cell cycle. | −8.6 |
| 2OAZ | PD | Methionine aminopeptidase 2 | Removes the N-terminal methionine from nascent proteins. The catalytic activity of human METAP2 toward Met-Val peptides is consistently two orders of magnitude higher than that of METAP1, suggesting that it is responsible for processing proteins containing N-terminal Met-Val and Met-Thr sequences in vivo. Protects eukaryotic initiation factor EIF2S1 from translation-inhibiting phosphorylation by inhibitory kinases such as EIF2AK2/PKR and EIF2AK1/HCR. Plays a critical role in the regulation of protein synthesis. | −8.1 |
| 1PME | PD | MAPK1 | Serine/threonine kinase that acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the two MAPKs that play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival, and differentiation through the regulation of transcription, translation, and cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. Approximately 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis, and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the PNRC; as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4, or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3, or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1, or PPARG), regulators of translation (such as EIF4EBP1), and a variety of other signaling-related molecules (such as ARHGEF2, DCC, FRS2, or GRB10). Protein kinases (such as RAF1, RPS6KA1/RSK1, RPS6KA3/RSK2, RPS6KA2/RSK3, RPS6KA6/RSK4, SYK, MKNK1/MNK1, MKNK2/MNK2, RPS6KA5/MSK1, RPS6KA4/MSK2, MAPKAPK3, or MAPKAPK5) and phosphatases (such as DUSP1, DUSP4, DUSP6, or DUSP16) are other substrates that enable the propagation the MAPK/ERK signal to additional cytosolic and nuclear targets, thereby extending the specificity of the cascade. Acts as a transcriptional repressor. Binds to a [GC]AAA[GC] consensus sequence. Repress the expression of interferon γ-induced genes. Seems to bind to the promoter of CCL5, DMP1, IFIH1, IFITM1, IRF7, IRF9, LAMP3, OAS1, OAS2, OAS3, and STAT1. Transcriptional activity is independent of kinase activity. | −8.2 |
| 3DA6 | PD | MAPK10 | Serine/threonine–protein kinase involved in various processes such as neuronal proliferation, differentiation, migration, and programmed cell death. Extracellular stimuli such as proinflammatory cytokines or physical stress stimulate the SAP/JNK signaling pathway. In this cascade, two dual specificity kinases MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK10/JNK3. In turn, MAPK10/JNK3 phosphorylates a number of transcription factors, primarily components of AP-1 such as JUN and ATF2 and thus regulates AP-1 transcriptional activity. Plays regulatory roles in the signaling pathways during neuronal apoptosis. Phosphorylates the neuronal microtubule regulator STMN2. Acts in the regulation of the β-APP/APP signaling during neuronal differentiation by phosphorylating APP. Participates also in neurite growth in spiral ganglion neurons. | −7.1 |
| 2QD9 | PD | MAPK14 | Serine/threonine kinase that acts as an essential component of the MAP kinase signal transduction pathway. MAPK14 is one of the four p38 MAPKs that play an important role in the cascades of cellular responses evoked by extracellular stimuli such as proinflammatory cytokines or physical stress, leading to direct activation of transcription factors. Accordingly, p38 MAPKs phosphorylate a broad range of proteins, and it has been estimated that they may hav ~200–300 substrates each. Some of the targets are downstream kinases, which are activated through phosphorylation and further phosphorylate additional targets. RPS6KA5/MSK1 and RPS6KA4/MSK2 can directly phosphorylate and activate transcription factors such as CREB1, ATF1, the NF-κB isoform RELA/NFKB3, STAT1, and STAT3, but can also phosphorylate histone H3 and the nucleosomal protein HMGN1. RPS6KA5/MSK1 and RPS6KA4/MSK2 play important roles in the rapid induction of immediate-early genes in response to stress or mitogenic stimuli, either by inducing chromatin remodeling or by recruiting the transcription machinery. On the other hand, two other kinase targets, MAPKAPK2/MK2 and MAPKAPK3/MK3, participate in the control of gene expression mostly at the posttranscriptional level, by phosphorylating ZFP36 (tristetraprolin) and ELAVL1, and by regulating EEF2K, which is important for the elongation of mRNA during translation. MKNK1/MNK1 and MKNK2/MNK2, two other kinases activated by p38 MAPKs, regulate protein synthesis by phosphorylating the initiation factor EIF4E2. MAPK14 interacts also with casein kinase II, leading to its activation through autophosphorylation and further phosphorylation of TP53/p53. In the cytoplasm, the p38 MAPK pathway is an important regulator of protein turnover. For example, CFLAR is an inhibitor of TNF-induced apoptosis whose proteasome-mediated degradation is regulated by p38 MAPK phosphorylation. In a similar way, MAPK14 phosphorylates the ubiquitin ligase SIAH2, regulating its activity toward EGLN3. MAPK14 may also inhibit the lysosomal degradation pathway of autophagy by interfering with the intracellular trafficking of the transmembrane protein ATG9. Another function of MAPK14 is to regulate the endocytosis of membrane receptors by different mechanisms that impinge on the small GTPase RAB5A. In addition, clathrin-mediated EGFR internalization induced by inflammatory cytokines and UV irradiation depends on MAPK14-mediated phosphorylation of EGFR itself as well as of RAB5A effectors. Ectodomain shedding of transmembrane proteins is regulated by p38 MAPKs as well. In response to inflammatory stimuli, p38 MAPKs phosphorylate the membrane-associated metalloprotease ADAM17. Such phosphorylation is required forADAM17-mediated ectodomain shedding of TGF-α family ligands, which results in the activation of EGFR signaling and cell proliferation. Another p38 MAPK substrate is FGFR1. FGFR1 can be translocated from the extracellular space into the cytosol and nucleus of target cells and regulates processes such as rRNA synthesis and cell growth. FGFR1 translocation requires p38 MAPK activation. In the nucleus, many transcription factors are phosphorylated and activated by p38 MAPKs in response to different stimuli. Classic examples include ATF1, ATF2, ATF6, ELK1, PTPRH, DDIT3, TP53/p53, MEF2C, and MEF2A. The p38 MAPKs are emerging as important modulators of gene expression by regulating chromatin modifiers and remodelers. The promoters of several genes involved in the inflammatory response, such as IL6, IL8, and IL-12B, display a p38 MAPK-dependent enrichment of histone H3 phosphorylation on “Ser-10” (H3S10ph) in LPS-stimulated myeloid cells. This phosphorylation enhances the accessibility of the cryptic NF-κB-binding sites marking promoters for increased NF-κB recruitment. Phosphorylates CDC25B and CDC25C that are required for binding to 14-3-3 proteins and lead to initiation of a G2 delay after ultraviolet radiation. Phosphorylates TIAR following DNA damage, releasing TIAR from GADD45A mRNA and preventing mRNA degradation. The p38 MAPKs may also have kinase-independent roles, which are thought to be due to the binding to targets in the absence of phosphorylation. Protein GlcNAcylation catalyzed by the OGT is regulated by MAPK14, and, although OGT does not seem to be phosphorylated by MAPK14, their interaction increases upon MAPK14 activation induced by glucose deprivation. This interaction may regulate OGT activity by recruiting it to specific targets such as neurofilament H, stimulating its GlcNAcylation. Required in mid-fetal development for the growth of embryo-derived blood vessels in the labyrinth layer of the placenta. Also plays an essential role in developmental and stress-induced erythropoiesis, through regulation of EPO gene expression. Isoform MXI2 activation is stimulated by mitogens and oxidative stress and only poorly phosphorylates ELK1 and ATF2. Isoform EXIP may play a role in the early onset of apoptosis. Phosphorylates S100A9 at “Thr-113”. | −8.4 |
| 2ZOQ | PD | MAPK3 | Serine/threonine kinase that acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the two MAPKs that play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival, and differentiation through the regulation of transcription, translation, and cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. Approximately 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis, and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the PNRC as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4, or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3, or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1, or PPARG), regulators of translation (such as EIF4EBP1), and a variety of other signaling-related molecules (like ARHGEF2, FRS2, or GRB10). Protein kinases (such as RAF1, RPS6KA1/RSK1, RPS6KA3/RSK2, RPS6KA2/RSK3, RPS6KA6/RSK4, SYK, MKNK1/MNK1, MKNK2/MNK2, RPS6KA5/MSK1, RPS6KA4/MSK2, MAPKAPK3, or MAPKAPK5) and phosphatases (such as DUSP1, DUSP4, DUSP6, or DUSP16) are other substrates that enable the propagation the MAPK/ERK signal to additional cytosolic and nuclear targets, thereby extending the specificity of the cascade. | −6.7 |
| 3ELJ | PD | MAPK8 | Serine/threonine–protein kinase involved in various processes such as cell proliferation, differentiation, migration, transformation, and programmed cell death. Extracellular stimuli such as proinflammatory cytokines or physical stress stimulate the SAP/JNK signaling pathway. In this cascade, two dual specificity kinases MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK8/JNK1. In turn, MAPK8/JNK1 phosphorylates a number of transcription factors, primarily components of AP-1 such as JUN, JDP2, and ATF2 and thus regulates AP-1 transcriptional activity. Phosphorylates the replication licensing factor CDT1, inhibiting the interaction between CDT1 and the histone H4 acetylase HBO1 to replication origins. Loss of this interaction abrogates the acetylation required for replication initiation. Promotes stressed cell apoptosis by phosphorylating key regulatory factors including p53/TP53 and Yes-associates protein YAP1. In T-cells, MAPK8 and MAPK9 are required for polarized differentiation of T-helper cells into Th1 cells. Contributes to the survival of erythroid cells by phosphorylating the antagonist of cell death BAD upon EPO stimulation. Mediates starvation-induced BCL2 phosphorylation, BCL2 dissociation from BECN1, and thus activation of autophagy. Phosphorylates STMN2 and hence regulates microtubule dynamics, controlling neurite elongation in cortical neurons. In the developing brain, through its cytoplasmic activity on STMN2, negatively regulates the rate of exit from multipolar stage and of radial migration from the ventricular zone. Phosphorylates several other substrates including HSF4, the deacetylase SIRT1, ELK1, or the E3 ligase ITCH. JNK1 isoforms display different binding patterns: β-1 preferentially binds to c-Jun, whereas α-1, α-2, and β-2 have a similar low level of binding to both c-Jun or ATF2. However, there is no correlation between binding and phosphorylation, which is achieved at about the same efficiency by all isoforms. | −6.9 |
| 3E7O | PD | MAPK9 | Serine/threonine–protein kinase involved in various processes such as cell proliferation, differentiation, migration, transformation, and programmed cell death. Extracellular stimuli such as proinflammatory cytokines or physical stress stimulate the SAP/JNK signaling pathway. In this cascade, two dual specificity kinases MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK9/JNK2. In turn, MAPK9/JNK2 phosphorylates a number of transcription factors, primarily components of AP-1 such as JUN and ATF2 and thus regulates AP-1 transcriptional activity. In response to oxidative or ribotoxic stresses, inhibits rRNA synthesis by phosphorylating and inactivating the RNA polymerase 1-specific transcription initiation factor RRN3. Promotes stressed cell apoptosis by phosphorylating key regulatory factors including TP53 and YAP1. In T-cells, MAPK8 and MAPK9 are required for polarized differentiation of T-helper cells into Th1 cells. Upon TCR stimulation, is activated by CARMA1, BCL10, MAP2K7, and MAP3K7/TAK1 to regulate JUN protein levels. Plays an important role in the osmotic stress-induced epithelial tight-junctions disruption. When activated, promotes β-catenin/CTNNB1 degradation and inhibits the canonical Wnt signaling pathway. Participates also in neurite growth in spiral ganglion neurons. MAPK9 isoforms display different binding patterns: α-1 and α-2 preferentially bind to JUN, whereas β-1 and β-2 bind to ATF2. However, there is no correlation between binding and phosphorylation, which is achieved at about the same efficiency by all isoforms. JUNB is not a substrate for JNK2α-2, and JUND binds only weakly to it. | −8.1 |
| 3DTC | PD | MAPK9 | Serine/threonine kinase that acts as an essential component of the MAP kinase signal transduction pathway. Plays an important role in the cascades of cellular responses evoked by changes in the environment. Once activated, acts as an upstream activator of the MKK/JNK signal transduction cascade through the phosphorylation of MAP2K4/MKK4 and MAP2K7/MKK7 which in turn activate the JNKs. The MKK/JNK signaling pathway regulates stress response via activator protein-1 (JUN) and GATA4 transcription factors. Plays also a role in mitochondrial death signaling pathway, including the release cytochrome c, leading to apoptosis. | −7.1 |
| 2CBZ | PK | Multidrug resistance-associated protein 1 | Mediates export of organic anions and drugs from the cytoplasm. Mediates ATP-dependent transport of glutathione and glutathione conjugates, leukotriene C4, estradiol-17-β-o-glucuronide, methotrexate, antiviral drugs and other xenobiotics. Confers resistance to anticancer drugs. Hydrolyzes ATP with low efficiency. | −6.8 |
| 3I4A | PD | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | Hydrolyzes NG,NG′-dimethyl-L-arginine (ADMA) and NG-monomethyl-L-arginine, which act as inhibitors of NOS. Has therefore a role in the regulation of NO generation. | −5.9 |
| 3FED | PD | N-acetylated-α-linked acidic dipeptidase 2 | Has NAALADase activity. Also exhibits a dipeptidyl-peptidase IV type activity. Inactivate the peptide neurotransmitter N-acetylaspartyl glutamate. | −8.7 |
| 1KBQ_2, 1H69, 1KBQ, 1D4A | PK | NAD(P)H dehydrogenase (quinine) 1 | The enzyme apparently serves as a quinone reductase in connection with conjugation reactions of hydroquinones involved in detoxification pathways as well as in biosynthetic processes such as the vitamin K-dependent γ-carboxylation of glutamate residues in prothrombin synthesis. | −8.7 to −7.3 |
| 1GZ4 | PD | NAD-dependent malic enzyme, mitochondrial | N/A | −8.7 |
| 2NYR | PD | NAD-dependent protein deacylase sirtuin-5, mitochondrial | NAD-dependent lysine demalonylase and desuccinylase that specifically removes malonyl and succinyl groups on target proteins. Activates CPS1 and contributes to the regulation of blood ammonia levels during prolonged fasting: acts by mediating desuccinylation of CPS1, thereby increasing CPS1 activity in response to elevated NAD levels during fasting. Has weak NAD-dependent protein deacetylase activity; however, this activity may not be physiologically relevant in vivo. Deacetylates cytochrome c (CYCS) and a number of other proteins in vitro. | −10 |
| 3QE2, 3QE2_2, 3QE2_3, 3QFS_2, 3QFT, 3QFT_2, 3QFS | PK | NADPH-cytochrome P450 reductase | This enzyme is required for electron transfer from NADP to cytochrome P450 in microsomes. It can also provide electron transfer to heme oxygenase and cytochrome B5. | −10.0 to −9.0 |
| 3GZN | PD | NEDD8-activating enzyme E1 regulatory subunit | Regulatory subunit of the dimeric UBA3-NAE1 E1 enzyme. E1 activates NEDD8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a NEDD8-UBA3 thioester and free AMP. E1 finally transfers NEDD8 to the catalytic cysteine of UBE2M. Necessary for cell-cycle progression through the S–M checkpoint. Overexpression of NAE1 causes apoptosis through deregulation of NEDD8 conjugation. | −8.2 |
| 1R1H | PD | Neprilysin | Thermolysin-like specificity, but is almost confined on acting on polypeptides of up to 30 amino acids. Biologically important in the destruction of opioid peptides such as Met-and Leu-enkephalins by cleavage of a Gly-Phe bond. Able to cleave angiotensin-1, angiotensin-2 and angiotensins 1–9. Involved in the degradation of ANF. Displays UV-inducible elastase activity toward skin preelastic and elastic fibers. | −8.9 |
| 3I97 | PD | Neuropilin-1 | The membrane-bound isoform 1 is a receptor involved in the development of the cardiovascular system, in angiogenesis, in the formation of certain neuronal circuits and in organogenesis outside the nervous system. It mediates the chemo-repulsant activity of semaphorins. It binds to semaphorin 3A, the PLGF-2 isoform of PGF, and the VEGF-165 isoform of VEGF and VEGF-B. Coexpression with KDR results in increased VEGF-165 binding to KDR as well as increased chemotaxis. It may regulate VEGF-induced angiogenesis. The soluble isoform 2 binds VEGF-165 and appears to inhibit its binding to cells. It may also induce apoptosis by sequestering VEGF-165. May bind as well various members of this maphorin family. Its expression has an adverse effect on blood vessel number and integrity. | −5.8 |
| 1ZS0 | PD | Neutrophil collagenase | Can degrade fibrillar type I, II, and III collagens. | −7.5 |
| 1H6H | PD | Neutrophil cytosol factor 4 | Component of the NADPH-oxidase, a multicomponent enzyme system responsible for the oxidative burst in which electrons are transported from NADPH to molecular oxygen, generating reactive oxidant intermediates. It may be important for the assembly and/or activation of the NADPH-oxidase complex. | −8.6 |
| 1H1B | PD | Neutrophil elastase | Modifies the functions of NK cells, monocytes, and granulocytes. Inhibits C5a-dependent neutrophil enzyme release and chemotaxis. | −7 |
| 3DSZ | PD | Neutrophil gelatinase-associated lipocalin | Iron-trafficking protein involved in multiple processes such as apoptosis, innate immunity, and renal development. Binds iron through association with 2,5-DHBA, a side rophore that shares structural similarities with bacterial enterobactin, and delivers or removes iron from the cell, depending on the context. Iron-bound form (holo-24p3) is internalized following binding to the SLC22A17 (24p3R) receptor, leading to release of iron and subsequent increase of intracellular iron concentration. In contrast, association of the iron-free form (apo-24p3) with the SLC22A17 (24p3R) receptor is followed by association with an intracellular side rophore, iron chelation, and iron transfer to the extracellular medium, thereby reducing intracellular iron concentration. Involved in apoptosis due to IL3 deprivation: iron-loaded form increases intracellular iron concentration without promoting apoptosis, while iron-free form decreases intracellular iron levels, inducing expression of the proapoptotic protein BCL2L11/BIM, resulting in apoptosis. Involved in innate immunity, possibly by sequestrating iron, leading to limit bacterial growth. | −9.2 |
| 2IIP, 3ROD, 3ROD_2 | PK | Nicotinamide N-methyltransferase | Catalyzes the N-methylation of nicotinamide and other pyridines to form pyridinium ions. This activity is important for biotransformation of many drugs and xenobiotic compounds. | −6.3 to 14.0 |
| 3DKJ | PD | Nicotinamide phosphoribosyl transferase | Catalyzes the condensation of nicotinamide with 5-phosphoribosyl-1-pyrophosphate to yield nicotinamide mononucleotide, an intermediate in the biosynthesis of NAD. It is the rate-limiting component in the mammalian NAD biosynthesis pathway (by similarity). | −6.8 |
| 3F9Y | PD | N-Lysine methyltransferase SETD8 | Protein-lysine N-methyltransferase that monomethylates both histones and nonhistone proteins. Specifically monomethylates “Lys-20” of histone H4 (H4K20me1). H4K20me1 is enriched during mitosis and represents a specific tag for epigenetic transcriptional repression. Mainly functions in euchromatin regions, thereby playing a central role in the silencing of euchromatic genes. Required for cell proliferation, probably by contributing to the maintenance of proper higher order structure of DNA during mitosis. Involved in chromosome condensation and proper cytokinesis. Nucleosomes are preferred as substrate compared to free histones. Mediates monomethylation of p53/TP53 at “Lys-382”, leading to repress p53/TP53-target genes. | −7.8 |
| 3NYX 2BZZ | PD PD | Nonreceptor tyrosine-protein kinase TYK2 Nonsecretory ribonuclease | Probably involved in intracellular signal transduction by being involved in the initiation of type I IFN signaling. Phosphorylates the interferon-α/β receptor α chain. This is a nonsecretory ribonuclease. It is a pyrimidine-specific nuclease with a slight preference for U. Cytotoxin and helminthotoxin. Selectively chemotactic for DCs. Possesses a wide variety of biological activities. | −7 −8.4 |
| 1H2T | PD | Nuclear cap-binding protein subunit 1 | Component of the CBC, which binds cotranscriptionally to the 5′-cap of pre-mRNAs and is involved in various processes such as pre-mRNA splicing, translation regulation, NMD, RNA-mediated genesilencing (RNAi) by miRNAs, and mRNA export. The CBC is involved in mRNA export from the nucleus via its interaction with ALYREF/THOC4/ALY, leading to the recruitment of the mRNA export machinery to the 5′− end of mRNA and to mRNA export in a 5′–3′ direction through the nuclear pore. The CBC is also involved in mediating U snRNA and intron less mRNAs export from the nucleus. The CBC is essential for a pioneer round of mRNA translation, before steady-state translation when the CBC is replaced by cytoplasmic cap-binding protein eIF4E. The pioneer round of mRNA translation mediated by the CBC plays a central role in NMD, NMD only taking place in mRNAs bound to the CBC, but not one IF4E-bound mRNAs. The CBC enhances NMD in mRNAs containing at least one EJC via its interaction with UPF1, promoting the interaction between UPF1 and UPF2. The CBC is also involved in “failsafe” NMD, which is independent of the EJC, while it does not participate in SMD. During cell proliferation, the CBC complex is also involved in miRNAs biogenesis via its interaction with SRRT/ARS2 and is required for miRNA-mediated RNA interference. The CBC also acts as a negative regulator of PARN, thereby acting as an inhibitor of mRNA deadenylation. In the CBC, NCBP1/CBP80 does not bind directly capped RNAs (m7GpppG-capped RNA) but is required to stabilize the movement of the N-terminal loop of NCBP2/CBP20 and lock the CBC into a high affinity cap-binding state with the cap structure. | −7.3 |
| 1M13, 1NRL, 3HVL | PD | Nuclear receptor subfamily 1 group I member 2 | Nuclear receptor that binds and is activated by variety of endogenous and xenobiotic compounds. Transcription factor that activates the transcription of multiple genes involved in the metabolism and secretion of potentially harmful xenobiotics, drugs, and endogenous compounds. Activated by the antibiotic rifampicin and various plant metabolites, such as hyperforin, guggulipid, colupulone, and isoflavones. Response to specific ligands is species specific. Activated by naturally occurring steroids, such as pregnenolone and progesterone. Binds to a response element in the promoters of the CYP3A4 and ABCB1/MDR1 genes. | −9.4 to −8.1 |
| 1XV9, 1XVP | PK | Nuclear receptor subfamily 1 group I member 3 | Binds and transactivates the RAREs that control expression of the retinoic acid receptor β2 and alcohol dehydrogenase 3 genes. Transactivates both the phenobarbital responsive element module of the human CYP2B6 gene and the CYP3A4 xenobiotic response element. | −6.8 to −6.5 |
| 1OAI | PD | Nuclear RNA export factor 1 | Involved in the nuclear export of mRNA species bearing retroviral CTEs and in the export of mRNA from the nucleus to the cytoplasm. The NXF1–NXT1 heterodimer is involved in the export of HSP70 mRNA in conjunction with ALYREF/THOC4 and THOC5. | −6.3 |
| 1UCN | PD | Nucleoside diphosphate kinase A | Major role in the synthesis of nucleoside triphosphates other than ATP. Possesses nucleoside-diphosphate kinase, serine/threonine-specific protein kinase, geranyl and farnesylpyrophosphate kinase, histidine protein kinase, and 3′–5′exonuclease activities. Involved in cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor endocytosis, and gene expression. Required for neural development including neural patterning and cell fate determination. | −5.8 |
| 3BBB | PD | Nucleoside diphosphate kinase B | Major role in the synthesis of nucleoside triphosphates other than ATP. Negatively regulates Rho activity by interacting with AKAP13/LBC. Acts as a transcriptional activator of the MYC gene; binds DNA nonspecifically (PubMed:8392752). Exhibits histidine protein kinase activity. | −8.9 |
| 2ON3 | PD | Ornithine decarboxylase | N/A | −7.8 |
| 3IPQ | PD | Oxysterols receptor LXR-α | Orphan receptor. Interaction with RXR shifts RXR fromits role as a silent DNA-binding partner to an active ligand-binding subunit in mediating retinoid responses through target genes defined by LXRES. LXRES are DR4-type response elements characterized by direct repeats of two similar hexanuclotide half-sites spaced by four nucleotides. Plays an important role in the regulation of cholesterol homeostasis, regulating cholesterol uptake through MYLIP-dependent ubiquitination of LDLR, VLDLR, and LRP8 (by similarity). | −7.4 |
| 3KFC | PD | Oxysterols receptor LXR-β | Orphan receptor. Binds preferentially to double-stranded oligonucleotide direct repeats having the consensus half-site sequence 5′-AGGTCA-3′ and 4-nt spacing (DR-4). Regulates cholesterol uptake through MYLIP-dependent ubiquitination of LDLR, VLDLR, and LRP8 (by similarity). | −7.1 |
| 1U33 | PD | Pancreatic α-amylase | N/A | −9.6 |
| 2QZ4 | PK | Paraplegin | Putative ATP-dependent zinc metalloprotease. | −7 |
| 1YND | PD | Peptidyl-prolyl cis–trans isomerase A | PPIases accelerate the folding of proteins. It catalyzes the cis–trans isomerization of proline imidic peptide bonds in oligopeptides. | −7.3 |
| 1FKB | PD | Peptidyl-prolyl cis–trans isomerase FKBP1A | Keeps in an inactive conformation TGFBR1, the TGF-β type I serine/threonine kinase receptor, preventing TGF-β receptor activation in absence of ligand. Recruits SMAD7 to ACVR1B which prevents the association of SMAD2 and SMAD3 with the activin receptor complex, thereby blocking the activin signal. May modulate the RYR1 calcium channel activity. PPIases accelerate the folding of proteins. It catalyzes the cis–trans isomerization of proline imidic peptide bonds in oligopeptides. | −7.9 |
| 1PBK | PD | Peptidyl-prolyl cis–trans isomerase FKBP3 | FKBPs and rapamycin-binding proteins constitute a family of receptors for the two immunosuppressants that inhibit T-cell proliferation by arresting two distinct cytoplasmic signal transmission pathways. PPIases accelerate the folding of proteins. | −8 |
| 3I6C | PD | Peptidyl-prolyl cis–trans isomerase NIMA-interacting 1 | Essential PPIase that regulates mitosis presumably by interacting with NIMA and attenuating its mitosis-promoting activity. Displays a preference for an acidic residue N-terminal to the isomerized proline bond. Catalyzes pSer/Thr-Pro cis–trans isomerizations. Downregulates kinase activity of BTK. Can transactivate multiple oncogenes and induce centrosome amplification, chromosome instability, and cell transformation. Required for the efficient dephosphorylation and recycling of RAF1 after mitogen activation. | −5.6 |
| 1IKT | PD | Peroxisomal multifunctional enzyme type 2 | Bifunctional enzyme acting on the peroxisomal β-oxidation pathway for fatty acids. Catalyzes the formation of 3-ketoacyl-CoA intermediates from both straight-chain and 2-methyl-branched-chain fatty acids. | −10 |
| 1FCH | PD | Peroxisomal targeting signal 1 receptor | Binds to the C-terminal PTS1-type tripeptide peroxisomal targeting signal (SKL-type) and plays an essential role in peroxisomal protein import. | −6.9 |
| 3KDU, 2ZNN, 2P54 | PK | Peroxisome proliferator-activated receptor α | Ligand-activated transcription factor. Key regulator of lipid metabolism. Activated by the endogenous ligand 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Activated by oleylethanolamide, a naturally occurring lipid that regulates satiety (by similarity). Receptor for peroxisome proliferators such as hypolipidemic drugs and fatty acids. Regulates the peroxisomal β-oxidation pathway of fatty acids. Functions as transcription activator for the ACOX1 and P450 genes. Transactivation activity requires heterodimerization with RXRA and is antagonized by NR2C2. | −7.2 to −5.8 |
| 3B1M, 3LMP, 3U9Q, 3V9V, 3H0A | PK | PPARG | Receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Once activated by a ligand, the receptor binds to a promoter element in the gene for acyl-CoA oxidase and activates its transcription. It therefore controls the peroxisomal β-oxidation pathway of fatty acids. Key regulator of adipocyte differentiation and glucose homeostasis. Acts as a critical regulator of gut homeostasis by suppressing NF-κB-mediated proinflammatory responses. | −8.3 to −6.1 |
| 2AWH, 3TKM, 3GZ9 | PK | Peroxisome proliferator-activated receptor δ | Ligand-activated transcription factor. Receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Has a preference for poly-unsaturated fatty acids, such as γ-linoleic acid and eicosapentanoic acid. Once activated by a ligand, the receptor binds to promoter elements of target genes. Regulates the peroxisomal β-oxidation pathway of fatty acids. Functions as transcription activator for the acyl-CoA oxidase gene. Decreases expression of NPC1L1 once activated by a ligand. | −7.6 to −6.4 |
| 3HFV | PD | Phenylalanine–tRNA ligase, mitochondrial | Catalyzes direct attachment of p-Tyr (Tyr) to tRNAPhe. Permits also, with a lower efficiency, the attachment of m-Tyr to tRNAPhe, thereby opening the way for delivery of the misacylated tRNA to the ribosome and incorporation of ROS-damaged amino acid into proteins. | −8.8 |
| 2G8N, 3KQS, 2G72, 2G72_2, 2G8N_2, 3KQS_2 | PK | Phenylethanolamine N-methyltransferase | Converts noradrenaline to adrenaline. | −6.7 to 19.9 |
| 1D5R | PD | PIP3-phosphatase and dual-specificity protein phosphatase PTEN | Tumor suppressor. Acts as a dual-specificity protein phosphatase, dephosphorylating tyrosine-, serine-and threonine-phosphorylated proteins. Also acts as a lipid phosphatase, removing the phosphate in the D3 position of the inositol ring from PIP3, phosphatidylinositol 3,4-diphosphate, phosphatidylinositol 3-phosphate, and inositol 1,3,4,5-tetrakisphosphate with order of substrate preference in vitro PtdIns(3,4,5)P3 . PtdIns(3,4)P2 . PtdIns3P . Ins(1,3,4,5)P4. The lipid phosphatase activity is critical for its tumor suppressor function. Antagonizes the PI3K-AKT/PKB signaling pathway by dephosphorylating phosphoinositides, and thereby modulating cell-cycle progression and cell survival. The unphosphorylated form cooperates with AIP1 to suppress AKT1 activation. Dephosphorylates tyrosine-phosphorylated focal adhesion kinase and inhibits cell migration and integrin-mediated cell spreading and focal adhesion formation. Plays a role as a key modulator of the AKT-mTOR signaling pathway controlling the tempo of the process of newborn neurons integration during adult neurogenesis, including correct neuron positioning, dendritic development, and synapse formation. May be a negative regulator of INS signaling and glucose metabolism in adipose tissue. The nuclear monoubiquitinated form possesses greater apoptotic potential, whereas the cytoplasmic nonubiquitinated form induces less tumor suppressive ability. | −6.9 |
| 3LJ3 | PD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit γ isoform | PI3K that phosphorylates PtdIns(4,5)P2 (phosphatidylinositol 4,5-bisphosphate) to generate PIP3. PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, activating signaling cascades involved in cell growth, survival, proliferation, motility, and morphology. Links GPCR activation to PIP3 production. Involved in immune, inflammatory, and allergic responses. Modulates leukocyte chemo taxis to inflammatory sites and in response to chemo attractant agents. May control leukocyte polarization and migration by regulating the spatial accumulation of PIP3 and by regulating the organization of F-actin formation and integrin-based adhesion at the leading edge. Controls motility of DCs. Together with PIK3CD is involved in NK cell development and migration toward the sites of inflammation. Participates in T-lymphocyte migration. Regulates T-lymphocyte proliferation and cytokine production. Together with PIK3CD participates in T-lymphocyte development. Required for B-lymphocyte development and signaling. Together with PIK3CD participates in neutrophil respiratory burst. Together with PIK3CD is involved in neutrophil chemotaxis and extra vasation. Together with PIK3CB promotes platelet aggregation and thrombosis. Regulates α-IIb/β-3 integrins (ITGA2B/ITGB3) adhesive function in platelets downstream of P2Y12 through a lipid kinase activity-independent mechanism. May have also a lipid kinase activity-dependent function in platelet aggregation. Involved in endothelial progenitor cell migration. Negative regulator of cardiac contractility. Modulates cardiac contractility by anchoring PKA and PDE3B activation, reducing cAMP levels. Regulates cardiac contractility also by promoting β-adrenergic receptor internalization by binding to ADRBK1 and by nonmuscle tropomyosin phosphorylation. Also exhibits has serine/threonine protein kinase activity: both lipid and protein kinase activities are required for β-adrenergic receptor endocytosis. May also have a scaffolding role in modulating cardiac contractility. Contributes to cardiac hypertrophy under pathological stress. Through simultaneous binding of PDE3B to RAPGEF3 and PIK3R6 is assembled in a signaling complex in which the PI3K γ complex is activated by RAPGEF3 and which is involved in angiogenesis. | −9.2 |
| 1NHX | PD | PEP carboxykinase, cytosolic (GTP) | Catalyzes the conversion of OAA to PEP, the rate-limiting step in the metabolic pathway that produces glucose from lactate and other precursors derived from the citric acid cycle. | −7.1 |
| 1KVO | PD | Phospholipase A2, membrane associated | Thought to participate in the regulation of the phospholipid metabolism in biomembranes including eicosanoid biosynthesis. Catalyzes the calcium-dependent hydrolysis of the 2-acyl groups in 3-sn-phosphoglycerides. | −8 |
| 3EBB | PD | Phospholipase A2-activating protein | Involved in the maintenance of ubiquitin levels (by similarity). | −8.6 |
| 3ACL | PD | Pirin | Possible transcriptional coregulator. May contribute to the regulation of cellular processes via its interaction with BCL3. May be required for efficient terminal myeloid maturation of hematopoietic cells. May play a role in the regulation of cell migration. May promote apoptosis when overexpressed. Has quercetin 2,3-dioxygenase activity (in vitro). | −8.1 |
| 1CEA | PD | Plasminogen | Plasmin dissolves the fibrin of blood clots and acts as a proteolytic factor in a variety of other processes including embryonic development, tissue remodeling, tumor invasion, and inflammation. In ovulation, weakens the walls of the Graafian follicle. It activates the urokinase-type plasminogen activator, collagenases and several complement zymogens, such as C1 and C5. Cleavage of fibronectin and laminin leads to cell detachment and apoptosis. Also cleaves fibrin, thrombospondin, and von Willebrand factor. Its role in tissue remodeling and tumor invasion may be modulated by CSPG4. Binds to cells. Angiostatin is an angiogenesis inhibitor that blocks neovascularization and growth of experimental primary and metastatic tumors in vivo. | −8.4 |
| 1A7C | PD | Plasminogen activator inhibitor 1 | Serine protease inhibitor. This inhibitor acts as “bait” for tissue plasminogen activator, urokinase, protein C, and matriptase-3/TMPRSS7. Its rapid interaction with PLAT may function as a major control point in the regulation of fibrinolysis. | −7.2 |
| 2I5F | PD | Pleckstrin | Major PKC substrate of platelets. | −6.6 |
| 3KJD | PD | PARP 2 | Involved in the BER pathway, by catalyzing the poly(ADP-ribosyl)ation of a limited number of acceptor proteins involved in chromatin architecture and in DNA metabolism. This modification follows DNA damages and appears as an obligatory step in a detection/signaling pathway, leading to the reparation of DNA strand breaks. | −7.3 |
| 3C4H | PD | PARP 3 | Involved in the BER pathway, by catalyzing the poly(ADP-ribosyl)ation of a limited number of acceptor proteins involved in chromatin architecture and in DNA metabolism. This modification follows DNA damages and appears as an obligatory step in a detection/signaling pathway, leading to the reparation of DNA strand breaks. May link the DNA damage surveillance network to the mitotic fidelity checkpoint. Negatively influences the G1/S cell-cycle progression without interfering with centrosome duplication. Binds DNA. May be involved in the regulation of PRC2 and PRC3 complex-dependent gene silencing. | −10.4 |
| 3CTR | PD | Poly(A)-specific ribonuclease PARN | 3′-Exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Interacts with both the 3′-endpoly(A) tail and the 5′-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails. Involved in NMD, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AREs in their 3′-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization. | −6.2 |
| 3GJW | PD | PARP 1 | Involved in the BER pathway, by catalyzing the poly(ADP-ribosyl)ation of a limited number of acceptor proteins involved in chromatin architecture and in DNA metabolism. This modification follows DNA damages and appears as an obligatory step in a detection/signaling pathway leading to the reparation of DNA strand breaks. Mediates the poly(ADP-ribosyl)ation of APLF and CHFR. Positively regulates the transcription of MTUS1 and negatively regulates the transcription of MTUS2/TIP150. With EEF1A1 and TXK, forms a complex that acts as a Th1 cell-specific transcription factor and binds the promoter of IFN-γ to directly regulate its transcription, and is thus involved importantly in Th1 cytokine production. | −11.3 |
| 3K27 | PD | Polycomb protein EED | PcG protein. Component of the PRC2/EED-EZH2 complex, which methylates “Lys-9” and “Lys-27” of histone H3, leading to transcriptional repression of the affected target gene. Also recognizes “Lys-26” trimethylated histone H1 with the effect of inhibiting PRC2 complex methyltransferase activity on nucleosomal histone H3 “Lys-27”, whereas H3 “Lys-27” recognition has the opposite effect, enabling the propagation of this repressive mark. The PRC2/EED-EZH2 complex may also serve as a recruiting platform for DNA methyltransferases, thereby linking two epigenetic repression systems. Genes repressed by the PRC2/EED-EZH2 complex include HOXC8, HOXA9, MYT1, and CDKN2A. | −6.7 |
| 1SQN | PD | Progesterone receptor | The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. PRB is involved activation of c-SRC/MAPK signaling on hormone stimulation. Isoform A is inactive in stimulating c-Src/MAPK signaling on hormone stimulation. | −6 |
| 2R03 | PD | Programmed cell death 6-interacting protein | Class E VPS protein involved in concentration and sorting of cargo proteins of the MVB for incorporation into ILVs that are generated by invagination and scission from the limiting membrane of the endosome. Binds to the phospholipid LBPA which is abundant in MVBs internal membranes. The MVB pathway appears to require the sequential function of ESCRT-O, -I, -II, and -III complexes. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis and enveloped virus budding (HIV-1 and other lentiviruses). Appears to be an adapter for a subset of ESCRT-III proteins, such as CHMP4, to function at distinct membranes. Required for completion of cytokinesis. Involved in HIV-1 virus budding. Can replace TSG101 it its role of supporting HIV-1 release; this function implies the interaction with CHMP4B. May play a role in the regulation of both apoptosis and cell proliferation. | −7.8 |
| 3DDU | PD | Prolyl endopeptidase | Cleaves peptide bonds on the C-terminal side of prolyl residues within peptides that are up tô30 amino acids long. | −8.2 |
| 3.00E+16 | PD | Prostasin | Possesses a trypsin-like cleavage specificity with a preference for poly-basic substrates. Stimulates ENaC activity through activating cleavage of the γ subunits (SCNN1G). | −6.9 |
| 1ND5 | PD | Prostatic acid phosphatase | A nonspecific tyrosine phosphatase that dephosphorylates a diverse number of substrates under acidic conditions (pH 4–6) including alkyl, aryl, and acyl orthophosphate monoesters and phosphorylated proteins. Has lipid phosphatase activity and inactivates lysophosphatidic acid in seminal plasma. Isoform 2: the cellular form also has ecto-5′-nucleotidase activity in DRG neurons. Generates adenosine from AMP that acts as a pain suppressor. Acts as a tumor suppressor of prostate cancer through dephosphorylation of ERBB2 and deactivation of MAPK-mediated signaling. | −8.2 |
| 1LD8 | PD | Protein farnesyl transferase subunit β | Catalyzes the transfer of a farnesyl moiety from farnesyl pyrophosphate to a cysteine at the fourth position from the C-terminus of several proteins. The β subunit is responsible for peptide binding. | −10.3 |
| 1ZRZ | PD | PKC iota type | Calcium- and DAG -independent serine/threonine–protein kinase, that plays a general protective role against apoptotic stimuli, is involved in NF-κB activation, cell survival, differentiation and polarity, and contributes to the regulation of microtubule dynamics in the early secretory pathway. Is necessary for BCR-ABL oncogene-mediated resistance to apoptotic drug in leukemia cells, protecting leukemia cells against drug-induced apoptosis. In cultured neurons, prevents amyloid β protein-induced apoptosis by interrupting cell death process at a very early step. In glioblastoma cells, may function downstream of phosphatidylinositol 3-kinase (PI[3]K) and PDPK1 in the promotion of cell survival by phosphorylating and inhibiting the pro-apoptotic factor BAD. Can form a protein complex in NSCLC cells with PARD6A and ECT2 and regulate ECT2 oncogenic activity by phosphorylation, which in turn promotes transformed growth and invasion. In response to NGF, acts downstream of SRC to phosphorylate and activate IRAK1, allowing the subsequent activation of NF-κB and neuronal cell survival. Functions in the organization of the apical domain in epithelial cells by phosphorylating EZR. This step is crucial for activation and normal distribution of EZR at the early stages of intestinal epithelial cell differentiation. Forms a protein complex with LLGL1 and PARD6B independently of PARD3 to regulate epithelial cell polarity. Plays a role in microtubule dynamics in the early secretory pathway through interaction with RAB2A and GAPDH and recruitment to VTCs. In HCAECs, is activated by saturated fatty acids and mediates lipid-induced apoptosis. | −8.8 |
| 1XJD | PD | PKC θ type | Calcium-independent, phospholipid- and DAG-dependent serine/threonine–protein kinase that mediates nonredundant functions in TCR signaling, including T-cells activation, proliferation, differentiation, and survival, by mediating activation of multiple transcription factors such as NF-κB, JUN, NFATC1, and NFATC2. In TCR-CD3/CD28-co-stimulated T-cells, is required for the activation of NF-κB and JUN, which in turn is essential for IL2 production, and participates to the calcium-dependent NFATC1 and NFATC2 transactivation. Mediates the activation of the canonical NF-κB pathway (NFKB1) by direct phosphorylation of CARD11 on several serine residues, inducing CARD11 association with lipid rafts and recruitment of the BCL10-MALT1 complex, which then activates IKK complex, resulting in nuclear translocation and activation of NFKB1. May also play an indirect role in activation of the noncanonical NF-κB (NFKB2) pathway. In the signaling pathway leading to JUN activation, acts by phosphorylating the mediator STK39/SPAK and may not act through MAP kinases signaling. Plays a critical role in TCR/CD28-induced NFATC1 and NFATC2 transactivation by participating in the regulation of reduced inositol 1,4,5-trisphosphate generation and intracellular calcium mobilization. After costimulation of T-cells through CD28 can phosphorylate CBLB and is required for the ubiquitination and subsequent degradation of CBLB, which is a prerequisite for the activation of TCR. During T-cells differentiation, plays an important role in the development of Th2 cells following immune and inflammatory responses, and, in the development of inflammatory autoimmune diseases, is necessary for the activation of IL-17-producing Th17 cells. May play a minor role in Th1 response. Upon TCR stimulation, mediates T-cell protective survival signal by phosphorylating BAD, thus protecting T-cells from BAD-induced apoptosis, and by upregulating BCL-X(L)/BCL2L1 levels through NF-κB and JUN pathways. In platelets, regulates signal transduction downstream of the ITGA2B, CD36/GP4, F2R/PAR1 and F2RL3/PAR4 receptors, playing a positive role in “outside-in” signaling and granule secretion signal transduction. May relay signals from the activated ITGA2B receptor by regulating the uncoupling of WASP and WIPF1, thereby permitting the regulation of actin filament nucleation and branching activity of the Arp2/3 complex. May mediate inhibitory effects of free fatty acids on insulin signaling by phosphorylating IRS1, which in turn blocks IRS1 tyrosine phosphorylation and downstream activation of the PI3K/AKT pathway. Phosphorylates MSN in the presence of phosphatidylglycerol or phosphatidylinositol. Phosphorylates PDPK1 at “Ser-504” and “Ser-532” and negatively regulates its ability to phosphorylate PKB/AKT1. | −7.9 |
| 3IW4 | PD | PKC α type | Calcium-activated, phospholipid- and DAG-dependent serine/threonine–protein kinase that is involved in positive and negative regulation of cell proliferation, apoptosis, differentiation, migration and adhesion, tumorigenesis, cardiac hypertrophy, angiogenesis, platelet function, and inflammation, by directly phosphorylating targets such as RAF1, BCL2, CSPG4, TNNT2/CTNT, or activating signaling cascade involving MAPK1/3 (ERK1/2) and RAP1GAP. Involved in cell proliferation and cell growth arrest by positive and negative regulation of the cell cycle. Can promote cell growth by phosphorylating and activating RAF1, which mediates the activation of the MAPK/ERK signaling cascade, and/or by upregulating CDKN1A, which facilitates active CDK complex formation in glioma cells. In intestinal cells stimulated by the phorbol ester PMA, can trigger a cell-cycle arrest program which is associated with the accumulation of the hyperphosphorylated growth-suppressive form of RB1 and induction of the CDK inhibitors CDKN1A and CDKN1B. Exhibits anti-apoptotic function in glioma cells and protects them from apoptosis by suppressing the p53/TP53-mediated activation of IGFBP3, and in leukemia cells mediates anti-apoptotic action by phosphorylating BCL2. During macrophage differentiation induced by macrophage CSF1, is translocated to the nucleus and is associated with macrophage development. After wounding, translocates from focal contacts to lamellipodia and participates in the modulation of desmosomal adhesion. Plays a role in cell motility by phosphorylating CSPG4, which induces association of CSPG4 with extensive lamellipodia at the cell periphery and polarization of the cell accompanied by increases in cell motility. Is highly expressed in a number of cancer cells where it can act as a tumor promoter and is implicated in malignant phenotypes of several tumors such as gliomas and breast cancers. Negatively regulates myocardial contractility and positively regulates angiogenesis, platelet aggregation, and thrombus formation in arteries. Mediates hypertrophic growth of neonatal cardiomyocytes, in part through a MAPK1/3 (ERK1/2)-dependent signaling pathway, and upon PMA treatment, is required to induce cardiomyocyte hypertrophy up to heart failure and death, by increasing protein synthesis, protein–DNA ratio, and cell surface area. Regulates cardiomyocyte function by phosphorylating cardiac troponin T (TNNT2/CTNT), which induces significant reduction in actomyosin ATPase activity, myofilament calcium sensitivity, and myocardial contractility. In angiogenesis, is required for full EC migration, adhesion to VTN, and VEGFA-dependent regulation of kinase activation, and vascular tube formation. Involved in the stabilization of VEGFA mRNA at posttranscriptional level and mediates VEGFA-induced cell proliferation. In the regulation of calcium-induced platelet aggregation, mediates signals from the CD36/GP4 receptor for granule release, and activates the integrin heterodimer ITGA2B-ITGB3 through the RAP1GAP pathway for adhesion. During response to LPSs, may regulate selective LPS-induced macrophage functions involved in host defense and inflammation. But in some inflammatory responses, may negatively regulate NF-κB-induced genes, through IL-1A-dependent induction of NF-κB inhibitor α (NFKBIA/IKBA). Upon stimulation with TPA, phosphorylates EIF4G1, which modulates EIF4G1 binding to MKNK1 and may be involved in the regulation of EIF4E phosphorylation. Phosphorylates KIT, leading to inhibition of KIT activity. | −7.3 |
| 2I0E | PD | PKC β type | Calcium-activated, phospholipid- and DAG-dependent serine/threonine–protein kinase involved in various cellular processes such as regulation of the BCR signalosome, oxidative stress-induced apoptosis, ANDR-dependent transcription regulation, INS signaling, and ECs proliferation. Plays a key role in B-cell activation by regulating BCR-induced NF-κB activation. Mediates the activation of the canonical NF-κB pathway (NFKB1) by direct phosphorylation of CARD11/CARMA1 at “Ser-559”, “Ser-644”, and “Ser-652”. Phosphorylation induces CARD11/CARMA1 association with lipid rafts and recruitment of the BCL10-MALT1complex as well as MAP3K7/TAK1, which then activates IKK complex, resulting in nuclear translocation and activation of NFKB1. Plays a direct role in the negative feedback regulation of the BCR signaling, by downmodulating BTK function via direct phosphorylation of BTK at “Ser-180”, which results in the alteration of BTK plasma membrane localization and in turn inhibition of BTK activity. Involved in apoptosis following oxidative damage: in case of oxidative conditions, specifically phosphorylates “Ser-36” of isoform p66Shc of SHC1, leading to mitochondrial accumulation of p66Shc, where p66Shc acts as a ROS producer. Acts as a coactivator of ANDR-dependent transcription, by being recruited to ANDR target genes and specifically mediating phosphorylation of “Thr-6” of histone H3 (H3T6ph), a specific tag for epigenetic transcriptional activation that prevents demethylation of histone H3 “Lys-4” (H3K4me) by LSD1/KDM1A. In INS signaling, may function downstream of IRS1 in muscle cells and mediate INS-dependent DNA synthesis through the RAF1-MAPK/ERK signaling cascade. May participate in the regulation of glucose transport in adipocytes by negatively modulating the INS-stimulated translocation of the glucose transporter SLC2A4/GLUT4. Under high glucose in pancreatic β-cells, is probably involved in the inhibition of the INS gene transcription, via regulation of MYC expression. In ECs, activation of PRKCB induces increased phosphorylation of RB1, increased VEGFA-induced cell proliferation, and inhibits PI3K/AKT-dependent NO synthase (NOS3/eNOS) regulation by INS, which causes endothelial dysfunction. Also involved intriglyceride homeostasis (by similarity). | −9.1 |
| 1MFG | PD | Protein LAP2 | Acts as an adapter for the receptor ERBB2, in epithelia. By binding the unphosphorylated “Tyr-1248” of receptor ERBB2, it may contribute to stabilize this unphosphorylated state. | −6.6 |
| 3FEA | PD | Protein Mdm4 | Inhibits p53/TP53- and TP73/p73-mediated cell-cycle arrest and apoptosis by binding its transcriptional activation domain. Inhibits degradation of MDM2. Can reverse MDM2-targeted degradation of TP53 while maintaining suppression of TP53 transactivation and apoptotic functions. | −6.3 |
| 2WOR | PD | Protein S100-A7 | N/A | −6.7 |
| 3HCM | PD | Protein S100-B | Weakly binds calcium but binds zinc very tightly distinct binding sites with different affinities exist for both ions on each monomer. Physiological concentrations of potassium ion antagonize the binding of both divalent cations, especially affecting high-affinity calcium-binding sites. Binds to and initiates the activation of STK38 by releasing autoinhibitory intra molecular interactions within the kinase. Interaction with AGER after myocardial infarction may play a role in myocyte apoptosis by activating ERK1/2 and p53/TP53 signaling pathways (by similarity). Could assist ATAD3A cytoplasmic processing, preventing aggregation, and favoring mitochondrial localization. | −6.9 |
| 2Q3Z | PD | Protein–glutamine γ-glutamyltransferase 2 | Catalyzes the cross-linking of proteins and the conjugation of polyamines to proteins. | −8.1 |
| 1VJJ | PD | Protein–glutamine γ-glutamyltransferase E | Catalyzes the calcium-dependent formation of isopeptide cross-links between glutamine and lysine residues in various proteins as well as the conjugation of polyamines to proteins. Involved in the formation of the CE, a specialized component consisting of covalent cross-links of proteins beneath the plasma membrane of terminally differentiated keratinocytes. Catalyzes small proline-rich proteins (SPRR1 and SPRR2) and LOR cross-linking to form small interchain oligomers, which are further cross-linked by TGM1 onto the growing CE scaffold (by similarity). In hair follicles, involved in cross-linking structural proteins to hardening the inner root sheath. | −9 |
| 3FZS | PD | PTK 2-β | Nonreceptor PTK that regulates reorganization of the actin cytoskeleton, cell polarization, cell migration, adhesion, spreading, and bone remodeling. Plays a role in the regulation of the humoral immune response and is required for normal levels of marginal B-cells in the spleen and normal migration of splenic B-cells. Required for normal macrophage polarization and migration toward sites of inflammation. Regulates cytoskeleton rearrangement and cell spreading in T-cells and contributes to the regulation of T-cell responses. Promotes osteoclastic bone resorption; this requires both PTK2B/PYK2 and SRC. May inhibit differentiation and activity of osteoprogenitor cells. Functions in signaling downstream of integrin and collagen receptors, immune receptors, GPCRs, and cytokine, chemokine, and growth factor receptors; and mediates responses to cellular stress. Forms multisubunit signaling complexes with SRC and its family members upon activation; this leads to the phosphorylation of additional tyrosine residues, creating binding sites for scaffold proteins, effectors, and substrates. Regulates numerous signaling pathways. Promotes activation of phosphatidylinositol 3-kinase and of the AKT1 signaling cascade. Promotes activation of NOS3. Regulates production of the cellular messenger cGMP. Promotes activation of the MAP kinase signaling cascade, including activation of MAPK1/ERK2, MAPK3/ERK1, and MAPK8/JNK1. Promotes activation of Rho family GTPases, such as RHOA and RAC1. Recruits the ubiquitin ligase MDM2 to P53/TP53 in the nucleus, and thereby regulates P53/TP53 activity, P53/TP53 ubiquitination and proteasomal degradation. Acts as a scaffold, binding to both PDPK1 and SRC, thereby allowing SRC to phosphorylate PDPK1 at “Tyr-9”, “Tyr-373”, and “Tyr-376”. Promotes phosphorylation of NMDA receptors by SRC family members, and thereby contributes to the regulation of NMDA receptor ion channel activity and intracellular Ca2+ levels. May also regulate potassium ion transport by phosphorylation of potassium channel subunits. Phosphorylates SRC; this increases SRC kinase activity. Phosphorylates ASAP1, NPHP1, KCNA2, and SHC1. Promotes phosphorylation of ASAP2, RHOU, and PXN; this requires both SRC and PTK2/PYK2. | −8.3 |
| 2BVR | PD | Prothrombin | Thrombin, which cleaves bonds after Arg and Lys, converts fibrinogen to fibrin and activates factors V, VII, VIII, XIII, and, in complex with thrombomodulin, protein C. Functions in blood homeostasis, inflammation, and wound healing. | −8 |
| 2X2L | PD | Proto-oncogene tyrosine-protein kinase receptor Ret | Receptor tyrosine-protein kinase involved in numerous cellular mechanisms including cell proliferation, neuronal navigation, cell migration, and cell differentiation upon binding with glial cell derived neuro trophic factor family ligands. Phosphorylates PTK2/FAK1. Regulates both cell death/survival balance and positional information. Required for the molecular mechanisms orchestration during intestine organogenesis; involved in the development of enteric nervous system and renal organogenesis during embryonic life, and promotes the formation of Peyer’s patch-like structures, a major component of the gut-associated lymphoid tissue. Modulates cell adhesion via its cleavage by caspase in sympathetic neurons and mediates cell migration in an integrin (eg, ITGB1 and ITGB3)-dependent manner. Involved in the development of the neural crest. Active in the absence of ligand, triggering apoptosis through a mechanism that requires receptor intracellular caspase cleavage. Acts as a dependence receptor; in the presence of the ligand GDNF in somatotrophs (within pituitary), promotes survival and downregulates GH production, but triggers apoptosisin absence of GDNF. Regulates nociceptor survival and size. Triggers the differentiation of RA mechanoreceptors. Mediator of several diseases such as neuroendocrine cancers; these diseases are characterized by aberrant integrins-regulated cell migration. | −6.5 |
| 1O4A | PD | Proto-oncogene tyrosine-protein kinase Src | Nonreceptor PTK that is activated following engagement of many different classes of cellular receptors including immune response receptors, integrins and other adhesion receptors, receptor PTKs, G protein-coupled receptors as well as cytokine receptors. Participates in signaling pathways that control a diverse spectrum of biological activities including gene transcription, immune response, cell adhesion, cell-cycle progression, apoptosis, migration, and transformation. Due to functional redundancy between members of the SRC kinase family, identification of the specific role of each SRC kinase is very difficult. SRC appears to be one of the primary kinases activated following engagement of receptors and plays a role in the activation of other PTK families. Receptor clustering or dimerization leads to recruitment of SRC to the receptor complexes where it phosphorylates the tyrosine residues within the receptor cytoplasmic domains. Plays an important role in the regulation of cytoskeletal organization through phosphorylation of specific substrates such as AFAP1. Phosphorylation of AFAP1 allows the SRC SH2 domain to bind AFAP1 and to localize to actin filaments. Cytoskeletal reorganization is also controlled through the phosphorylation of cortactin (CTTN). When cells adhere via focal adhesions to the extracellular matrix, signals are transmitted by integrins into the cell resulting in tyrosine phosphorylation of a number of focal adhesion proteins, including PTK2/FAK1 and PXN. In addition to phosphorylating focal adhesion proteins, SRC is also active at the sites of cell–cell contact AJs and phosphorylates substrates such as β-catenin (CTNNB1), δ-catenin (CTNND1), and plakoglobin (JUP). Another type of cell–cell junction, the gap junction, is also a target for SRC, which phosphorylates connexin-43 (GJA1). SRC is implicated in regulation of pre-mRNA-processing and phosphorylates RNA-binding proteins such as KHDRBS1. Also plays a role in PDGF-mediated tyrosine phosphorylation of both STAT1 and STAT3, leading to increased DNA-binding activity of these transcription factors. Involved in the Ras pathway through phosphorylation of RASA1 and RASGRF1. Plays a role in EGF-mediated calcium-activated chloride channel activation. Required for EGFR internalization through phosphorylation of clathrin heavy chains (CLTC and CLTCL1) at “Tyr-1477”. Involved in β-arrestin (ARRB1 and ARRB2) desensitization through phosphorylation and activation of ADRBK1, leading to β-arrestin phosphorylation and internalization. Has a critical role in the stimulation of the CDK20/MAPK3 MAPK cascade by EGF. Might be involved not only in mediating the transduction of mitogenic signals at the level of the plasma membrane but also in controlling progression through the cell cycle via interaction with regulatory proteins in the nucleus. Plays an important role in osteoclastic bone resorption in conjunction with PTK2B/PYK2. Both the formation of a SRC-PTK2B/PYK2 complex and SRC kinase activity are necessary for this function. Recruited to activated integrins by PTK2B/PYK2, thereby phosphorylating CBL, which in turn induces the activation and recruitment of phosphatidylinositol 3-kinase to the cell membrane in a signaling pathway that is critical for osteoclast function. Promotes energy production in osteoclasts by activating mitochondrial cytochrome C oxidase. Phosphorylates DDR2 ontyrosine residues, thereby promoting its subsequent autophosphorylation. Phosphorylates RUNX3 and COX2 on tyrosine residues, TNK2 on “Tyr-284” and CBL on “Tyr-731”. Enhances DDX58/RIG-I-elicited antiviral signaling. Phosphorylates PDPK1 at “Tyr-9”, “Tyr-373”, and “Tyr-376”. | −6 |
| 2OS9 | PD | Pulmonary surfactant-associated protein D | Contributes to the lung’s defense against inhaled microorganisms. May participate in the extracellular reorganization or turnover of pulmonary surfactant. Binds strongly maltose residues and to a lesser extent other α-glucosylmoieties. | −7.2 |
| 3BGS | PD | Purine nucleoside phosphorylase | The purine nucleoside phosphorylases catalyze the phosphorolytic breakdown of the N-glycosidic bond in the β-(deoxy)ribonucleoside molecules, with the formation of the corresponding free purine bases and pentose-1-phosphate. | −7.5 |
| 2VPE | PD | Pygopus homolog 1 | Involved in signal transduction through the Wnt pathway. | −7 |
| 3EXE | PD | Pyruvate dehydrogenase E1 component subunit α, somatic form, mitochondrial | The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO2 It contains multiple copies of three enzymatic components: pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2), and lipoamide dehydrogenase (E3). | −4.7 |
| 1T5A | PD | Pyruvate kinase isozymes M1/M2 | Glycolytic enzyme that catalyzes the transfer of a phosphoryl group from PEP to ADP, generating ATP. Stimulates POU5F1-mediated transcriptional activation. Plays a general role in caspase-independent cell death of tumor cells. The ratio between the highly active tetrameric form and nearly inactive dimeric form determines whether glucose carbons are channeled to biosynthetic processes or used for glycolytic ATP production. The transition between the two forms contributes to the control of glycolysis and is important for tumor cell proliferation and survival. | −8.2 |
| 2UVM | PD | RAC-α serine/threonine–protein kinase | AKT1 is one of three closely related serine/threonine–protein kinases (AKT1, AKT2, and AKT3) called the AKT kinase, and which regulate many processes including metabolism, proliferation, cell survival, growth, and angiogenesis. This is mediated through serine and/or threonine phosphorylation of a range of downstream substrates. Over 100 substrate candidates have been reported so far, but for most of them, no isoform specificity has been reported. AKT is responsible of the regulation of glucose uptake by mediating INS-induced translocation of the SLC2A4/GLUT4 glucose transporter to the cell surface. Phosphorylation of PTPN1 at “Ser-50” negatively modulates its phosphatase activity preventing dephosphorylation of the INS receptor and the attenuation of INS signaling. Phosphorylation of TBC1D4 triggers the binding of this effector to inhibitory 14-3-3 proteins, which is required for INS-stimulated glucose transport. AKT regulates also the storage of glucose in the form of glycogen by phosphorylating GSK3A at “Ser-21” and GSK3B at “Ser-9”, resulting in inhibition of its kinase activity. Phosphorylation of GSK3 isoforms by AKT is also thought to be one mechanism by which cell proliferation is driven. AKT regulates also cell survival via the phosphorylation of MAP3K5 (apoptosis signal-related kinase). Phosphorylation of “Ser-83” decreases MAP3K5 kinase activity stimulated by oxidative stress, and thereby prevents apoptosis. AKT mediates INS-stimulated protein synthesis by phosphorylating TSC2 at “Ser-939” and “Thr-1462”, thereby activating mTORC1 signaling and leading to both phosphorylation of 4E-BP1 and in activation of RPS6KB1. AKT is involved in the phosphorylation of members of the FOXO factors (Forkhead family of transcription factors), leading to binding of 14-3-3 proteins and cytoplasmic localization. In particular, FOXO1 is phosphorylated at “Thr-24”, “Ser-256”, and “Ser-319”. FOXO3 and FOXO4 are phosphorylated on equivalent sites. AKT has an important role in the regulation of NF-κB-dependent gene transcription and positively regulates the activity of CREB1 (cAMP-response element-binding protein). The phosphorylation of CREB1 induces the binding of accessory proteins that are necessary for the transcription of pro-survival genes such as BCL2 and MCL1. AKT phosphorylates “Ser-454” on ATP citrate lyase (ACLY), thereby potentially regulating ACLY activity and fatty acid synthesis. Activates the 3B isoform of cyclic nucleotide phosphodiesterase (PDE3B) via phosphorylation of “Ser-273”, resulting in reduced cyclic AMP levels and inhibition of lipolysis. Phosphorylates PIKFYVE on “Ser-318”, which results in increased PI(3)P-5 activity. The Rho GTPase-activating protein DLC1 is another substrate and its phosphorylation is implicated in the regulation cell proliferation and cell growth. AKT plays a role as key modulator of the AKT-mTOR signaling pathway controlling the tempo of the process of newborn neurons integration during adult neurogenesis, including correct neuron positioning, dendritic development, and synapse formation. Signals downstream of phosphatidylinositol 3-kinase (PI(3)K) to mediate the effects of various growth factors such as PDGF, EGF, INS, and IGFI. AKT mediates the antiapoptotic effects of IGFI. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. May be involved in the regulation of the placental development. Phosphorylates STK4/MST1 at “Thr-120” and “Thr-387” leading to inhibition of its: kinase activity, nuclear translocation, autophosphorylation, and ability to phosphorylate FOXO3. Phosphorylates STK3/MST2 at “Thr-117” and “Thr-384” leading to inhibition of its: cleavage, kinase activity, autophosphorylation at Thr-180, binding to RASSF1, and nuclear translocation. Phosphorylates SRPK2 and enhances its kinase activity toward SRSF2 and ACIN1 and promotes its nuclear translocation. Phosphorylates RAF1 at “Ser-259” and negatively regulates its activity. Phosphorylation of BAD stimulates its pro-apoptotic activity. AKT1-specific substrates have been recently identified, including PALLD, which phosphorylation modulates cytoskeletal or ganization and cell motility; PHB, playing an important role in cell metabolism and proliferation; and CDKN1A, for which phosphorylation at “Thr-145” induces its release from CDK2 and cytoplasmic relocalization. These recent findings indicate that the AKT1 isoform has a more specific rolein cell motility and proliferation. Phosphorylates CLK2, thereby controlling cell survival to ionizing radiation. | −6.7 |
| 2JDO | PD | RAC-β serine/threonine–protein kinase | AKT2 is one of three closely related serine/threonine–protein kinases (AKT1, AKT2, and AKT3) called the AKT kinase, and which regulate many processes including metabolism, proliferation, cell survival, growth, and angiogenesis. This is mediated through serine and/or threonine phosphorylation of a range of downstream substrates. Over 100 substrate candidates have been reported so far, but for most of them, no isoform specificity has been reported. AKT is responsible of the regulation of glucose uptake by mediating INS-induced translocation of the SLC2A4/GLUT4 glucose transporter to the cell surface. Phosphorylation of PTPN1 at “Ser-50” negatively modulates its phosphatase activity preventing dephosphorylation of the INS receptor and the attenuation of INS signaling. Phosphorylation of TBC1D4 triggers the binding of this effector to inhibitory 14-3-3 proteins, which is required for INS-stimulated glucose transport. AKT regulates also the storage of glucose in the form of glycogen by phosphorylating GSK3A at “Ser-21” and GSK3B at “Ser-9”, resulting in inhibition of its kinase activity. Phosphorylation of GSK3 isoforms by AKT is also thought to be one mechanism by which cell proliferation is driven. AKT regulates also cell survival via the phosphorylation of MAP3K5 (apoptosis signal-related kinase). Phosphorylation of “Ser-83” decreases MAP3K5 kinase activity stimulated by oxidative stress, and thereby prevents apoptosis. AKT mediates INS-stimulated protein synthesis by phosphorylating TSC2 at “Ser-939” and “Thr-1462”, thereby activating mTORC1 signaling and leading to both phosphorylation of 4E-BP1 and in activation of RPS6KB1. AKT is involved in the phosphorylation of members of the FOXO factors (Forkhead family of transcription factors), leading to binding of 14-3-3 proteins and cytoplasmic localization. In particular, FOXO1 is phosphorylated at “Thr-24”, “Ser-256”, and “Ser-319”. FOXO3 and FOXO4 are phosphorylated on equivalent sites. AKT has an important role in the regulation of NF-κB-dependent gene transcription and positively regulates the activity of CREB1 (cyclic AMP [cAMP]-response element-binding protein). The phosphorylation of CREB1 induces the binding of accessory proteins that are necessary for the transcription of prosurvival genes such as BCL2 and MCL1. AKT phosphorylates “Ser-454” on ACLY, thereby potentially regulating ACLY activity and fatty acid synthesis. Activates the 3B isoform of cyclic nucleotide phosphodiesterase (PDE3B) via phosphorylation of “Ser-273”, resulting in reduced cyclic AMP levels and inhibition of lipolysis. Phosphorylates PIKFYVE on “Ser-318”, which results in increased PI(3)P-5 activity. The Rho GTPase-activating protein DLC1 is another substrate and its phosphorylation is implicated in the regulation cell proliferation and cell growth. AKT plays a role as key modulator of the AKT-mTOR signaling pathway controlling the tempo of the process of newborn neurons integration during adult neurogenesis, including correct neuron positioning, dendritic development, and synapse formation. Signals downstream of phosphatidylinositol 3-kinase (PI[3]K) to mediate the effects of various growth factors such as PDGF, EGF, INS, and IGFI. AKT mediates the antiapoptotic effects of IGF-I. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. May be involved in the regulation of the placental development. One of the few specific substrates of AKT2 identified recently is PITX2. Phosphorylation of PITX2 impairs its association with the CCND1 mRNA-stabilizing complex thus shortening the half-life of CCND1. AKT2 seems also to be the principal isoform responsible of the regulation of glucose uptake. AKT2 is also specifically involved in skeletal muscle differentiation. Downregulation by RNA interference reduces the expression of the phosphorylated form of BAD, resulting in the induction of caspase-dependent apoptosis. Phosphorylates CLK2 on “Thr-343”. | −8.4 |
| 1RYF | PD | Ras-related C3 botulinum toxin substrate 1 | Plasma membrane-associated small GTPase that cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular responses such as secretory processes, phagocytosis of apoptotic cells, epithelial cell polarization, and growth factor-induced formation of membrane ruffles. Rac1 p21/rho GDI heterodimer is the active component of the cytosolic factor sigma1, which is involved in stimulation of the NADPH oxidase activity in macrophages (by similarity). Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. Stimulates PKN2 kinase activity. In concert with RAB7A, plays a role in regulating the formation of RBs in osteoclasts. In glioma cells, promotes cell migration and invasion. Isoform B has an accelerated GEF-independent GDP/GTP exchange and an impaired GTP hydrolysis, which is restored partially by GTPase-activating proteins. It is able to bind to the GTPase-binding domain of PAK but not full-length PAK in a GTP-dependent manner, suggesting that the insertion does not completely abolish effector interaction. | −7.8 |
| 2R4B | PD | Receptor tyrosine-protein kinase ERBB4 | Tyrosine-protein kinase that plays an essential role as cell surface receptor for neuregulins and EGF family members and regulates development of the heart, CNS, mammary gland, gene transcription, and cell proliferation, differentiation, migration, and apoptosis. Required for normal cardiac muscle differentiation during embryonic development, and for postnatal cardiomyocyte proliferation. Required for normal development of the embryonic CNS, especially for normal neural crest cell migration and normal axon guidance. Required for mammary gland differentiation, induction of milk proteins and lactation. Acts as cell-surface receptor for the neuregulins NRG1, NRG2, NRG3, and NRG4 and the EGF family members BTC, EREG, and HBEGF. Ligand-binding triggers receptor dimerization and autophosphorylation at specific tyrosine residues that then serve as binding sites for scaffold proteins and effectors. Ligand specificity and signaling is modulated by alternative splicing, proteolytic processing, and the formation of heterodimers with other ERBB family members, thereby creating multiple combinations of intracellular phosphotyrosines that trigger ligand and context-specific cellular responses. Mediates phosphorylation of SHC1 and activation of the MAP kinases MAPK1/ERK2 and MAPK3/ERK1. Isoforms JM-A CYT-1 and JM-B CYT-1 phosphorylate PIK3R1, leading to the activation of phosphatidylinositol 3-kinase and AKT1 and protect cells against apoptosis. Isoforms JM-A CYT-1 and JM-B CYT-1 mediate reorganization of the actin cytoskeleton and promote cell migration in response to NRG1. Isoforms JM-A CYT-2 and JM-B CYT-2 lack the phosphotyrosine that mediates interaction with PIK3R1 and hence do not phosphorylate PIK3R1, do not protect cells against apoptosis, and do not promote reorganization of the actin cytoskeleton and cell migration. Proteolytic processing of isoforms JM-A CYT-1 and JM-A CYT-2 gives rise to the corresponding soluble intracellular domains (4ICD) that translocate to the nucleus, promote nuclear import of STAT5A, activation of STAT5A, mammary epithelium differentiation, cell proliferation, and activation of gene expression. The ERBB4-soluble intracellular domains (4ICD) colocalize with STAT5A at the CSN2 promoter to regulate transcription of milk proteins during lactation. The ERBB4-soluble intracellular domains can also translocate to mitochondria and promote apoptosis. | −5.9 |
| 2H03 | PD | Receptor-type tyrosine-protein phosphatase β | Plays an important role in blood vessel remodeling and angiogenesis. Not necessary for the initial formation of blood vessels, but is essential for their maintenance and remodeling. Can induce dephosphorylation of TEK/TIE2, CDH5/VE-cadherin, and KDR/VEGFR-2. Regulates angiopoietin-TIE2 signaling in ECs. Acts as a negative regulator of TIE2, and controls TIE2-driven EC proliferation, which in turn affects blood vessel remodeling during embryonic development and determines blood vessel size during perinatal growth. Essential for the maintenance of EC contact integrity and for the adhesive function of VE-cadherin in ECs and this requires the presence of plakoglobin (by similarity). | −6.8 |
| 1HRN | PD | Renin | Renin is a highly specific endopeptidase, with the only known function to generate angiotensin I from angiotensinogen in the plasma, initiating a cascade of reactions that produce an elevation of blood pressure and increased sodium retention by the kidney. | −9.9 |
| 3HX3 | PD | Retinaldehyde-binding protein 1 | Soluble retinoid carrier is essential for proper function of both rod and cone photoreceptors. Participates in the regeneration of active 11-cis-retinol and 11-cis-retinaldehyde, from the inactive 11-trans products of the RHO photocycle and in the de novo synthesis of these retinoids from 11-trans metabolic precursors. The cycling of retinoids between photoreceptor and adjacent pigment epithelium cells is known as the “visual cycle”. | −7.3 |
| 1GUX | PD | Retinoblastoma-associated protein | Key regulator of entry into cell division that acts as a tumor suppressor. Promotes G0–G1 transition when phosphorylated by CDK3/cyclin-C. Acts as a transcription repressor of E2F1 target genes. The under phosphorylated, active form of RB1 interacts with E2F1 and represses its transcription activity, leading to cell-cycle arrest. Directly involved in heterochromatin formation by maintaining overall chromatin structure and, in particular, that of constitutive heterochromatin by stabilizing histonemethylation. Recruits and targets histone methyltransferases SUV39H1, SUV420H1, and SUV420H2, leading to epigenetic transcriptional repression. Controls histone H4 “Lys-20” trimethylation. Inhibits the intrinsic kinase activity of TAF1. Mediates transcriptional repression by SMARCA4/BRG1 by recruiting a HDAC complex to the c-FOS promoter. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex (by similarity). In case of viral infections, interactions with SV40 large T antigen, HPV E7 protein or adenovirus E1A protein induce the disassembly of RB1-E2F1 complex, thereby disrupting RB1’s activity. | −6.2 |
| 1MZN, 2P1T | PK | Retinoic acid receptor RXRα | Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RAR/RXR heterodimers bind to the RAREs composed of tandem 5′-AGGTCA-3′ sites known as DR1–DR5. The high-affinity ligand for RXRs is 9-cis retinoic acid. RXRA serves as a common heterodimeric partner for a number of nuclear receptors. The RXR/RAR heterodimers bind to the RAREs composed of tandem 5′-AGGTCA-3′ sites known as DR1–DR5. In the absence of ligand, the RXR-RAR heterodimers associate with a multiprotein complex containing transcription corepressors that induce histone acetylation, chromatin condensation, and transcriptional suppression. On ligand binding, the corepressors dissociate from the receptors and associate with the coactivators, leading to transcriptional activation. The RXRα/PPARα heterodimer is required for PPARA transcriptional activity on fatty acid oxidation genes such as ACOX1 and the P450 system genes. | −6.2 to −4.6 |
| 1XAP | PD | Retinoic acid receptor β | Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RXR/RAR heterodimers bind to the RAREs composed of tandem 5′-AGGTCA-3′ sites known as DR1–DR5. In the absence or presence of hormone ligand, acts mainly as an activator of gene expression due to weak binding to corepressors. In concert with RARG, required for skeletal growth, matrix homeostasis, and growth plate function. | −7.6 |
| 1FCY | PD | Retinoic acid receptor γ | Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid and regulate gene expression in various biological processes. The RAR/RXR heterodimers bind to the RAREs composed of tandem 5′-AGGTCA-3′ sites known as DR1–DR5. In the absence of ligand, acts mainly as an activator of gene expression due to weak binding to corepressors. Required for limb bud development. In concert with RARA or RARB, required for skeletal growth, matrix homeostasis, and growth plate function (by similarity). | −6.9 |
| 1RBP | PD | Retinol-binding protein 4 | Delivers retinol from the liver stores to the peripheral tissues. In plasma, the RBP–retinol complex interacts with transthyretin, this prevents its loss by filtration through the kidney glomeruli. | −7.6 |
| 2P0D | PD | Rho GTPase-activating protein 9 | GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state. Has a substantial GAP activity toward CDC42 and RAC1 and less toward RHOA. Has a role in regulating adhesion of hematopoietic cells to the extracellular matrix. | −6.4 |
| 2ETR | PD | Rho-associated protein kinase 1 | Protein kinase that is a key regulator of actin cytoskeleton and cell polarity. Involved in regulation of smooth muscle contraction, actin cytoskeleton organization, stress fiber and focal adhesion formation, neurite retraction, cell adhesion and motility via phosphorylation of DAPK3, GFAP, LIMK1, LIMK2, MYL9/MLC2, PFN1, and PPP1R12A. Phosphorylates FHOD1 and acts synergistically with it to promote SRC-dependent nonapoptotic plasma membrane blebbing. Phosphorylates JIP3 and regulates the recruitment of JNK to JIP3 upon UVB-induced stress. Acts as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability. Acts as a negative regulator of VEGF-induced angiogenic EC activation. Required for centrosome positioning and centrosome-dependent exit from mitosis. Plays a role in terminal erythroid differentiation. May regulate closure of the eyelids and ventral body wall by inducing the assembly of actomyosin bundles. Promotes keratinocyte terminal differentiation. | −9 |
| 2Z7R | PD | Ribosomal protein S6 kinase α-1 | Serine/threonine–protein kinase that acts downstream of ERK (MAPK1/ERK2 and MAPK3/ERK1) signaling and mediates mitogenic and stress-induced activation of the transcription factors CREB1, ETV1/ER81, and NR4A1/NUR77, regulates translation through RPS6 and EIF4B phosphorylation, and mediates cellular proliferation, survival, and differentiation by modulating mTOR signaling and repressing pro-apoptotic function of BAD and DAPK1. In fibroblast, is required for EGF-stimulated phosphorylation of CREB1, which results in the subsequent transcriptional activation of several immediate-early genes. In response to mitogenic stimulation (EGF and PMA), phosphorylates and activates NR4A1/NUR77 and ETV1/ER81 transcription factors and the cofactor CREBBP. Upon INS-derived signal, acts indirectly on the transcription regulation of several genes by phosphorylating GSK3B at “Ser-9” and inhibiting its activity. Phosphorylates RPS6 in response to serum or EGF via an mTOR-independent mechanism and promotes translation initiation by facilitating assembly of the preinitiation complex. In response to INS, phosphorylates EIF4B, enhancing EIF4B affinity for the EIF3 complex and stimulating cap-dependent translation. Is involved in the mTOR nutrient-sensing pathway by directly phosphorylating TSC2 at “Ser-1798”, which potently inhibits TSC2 ability to suppress mTOR signaling and mediates phosphorylation of RPTOR, which regulates mTORC1 activity and may promote rapamycin-sensitive signaling independently of the PI3K/AKT pathway. Mediates cell survival by phosphorylating the pro-apoptotic proteins BAD and DAPK1 and suppressing their pro-apoptotic function. Promotes the survival of hepatic stellate cells by phosphorylating CEBPB in response to the hepatotoxin CCl4. Is involved in cell-cycle regulation by phosphorylating the CDK inhibitor CDKN1B, which promotes CDKN1B association with 14-3-3 proteins and prevents its translocation to the nucleus and inhibition of G1 progression. | −7.7 |
| 3TEM, 1SG0, 1XI2, 1XI2_2, 3TEM_2, 1SG0_2 | PK | Ribosyldihydronicotinamide dehydrogenase (quinine) | The enzyme apparently serves as a quinone reductase in connection with conjugation reactions of hydroquinones involved in detoxification pathways as well as in biosynthetic processes such as the vitamin K-dependent γ-carboxylation of glutamate residues in prothrombin synthesis. | −7.9 to -7.2 |
| 3H0W | PD | S-AdoMet decarboxylase proenzyme | N/A | −9.1 |
| 2OBV | PK | S-AdoMet synthase isoform type 1 | Catalyzes the formation of S-adenosyl methionine from methionine and ATP. | −7.5 |
| 1O5E | PD | Serine protease hepsin | Plays an essential role in cell growth and maintenance of cell morphology. May mediate the activating cleavage of HGF and MST1/HGFL. | −7.5 |
| 2JOA | PD | Serine protease HTRA1 | Serine protease with a variety of targets, including extracellular matrix proteins such as fibronectin. HTRA1-generated fibronectin fragments further induce synovial cells to upregulate MMP-1 and MMP-3 production. May also degrade proteoglycans, such as aggrecan, decorin, and fibromodulin. Through cleavage of proteoglycans, may release soluble FGF-glycosaminogly can complexes that promote the range and intensity of FGF signals in the extracellular space. Regulates the availability of IGFs by cleaving IGF-binding proteins. Inhibits signaling mediated by TGF-β family members. This activity requires the integrity of the catalytic site, although it is unclear whether TGF-β proteins are themselves degraded. By acting on TGF-β signaling, may regulate many physiological processes, including retinal angiogenesis and neuronal survival and maturation during development. Intracellularly, degrades TSC2, leading to the activation of TSC2 downstream targets. | −7.3 |
| 3C4C | PD | Serine/threonine–protein kinase B-RAF | Involved in the transduction of mitogenic signals from the cell membrane to the nucleus. May play a role in the postsynaptic responses of hippocampal neuron. | −9.5 |
| 3PA3 | PD | Serine/threonine–protein kinase CHK1 | Serine/threonine–protein kinase which is required for checkpoint-mediated cell-cycle arrest and activation of DNA repair in response to the presence of DNA damage or unreplicated DNA. May also negatively regulate cell-cycle progression during unperturbed cell cycles. This regulation is achieved by a number of mechanisms that together help to preserve the integrity of the genome. Recognizes the substrate consensus sequence [R-X-X-S/T]. Binds to and phosphorylates CDC25A, CDC25B, and CDC25C. Phosphorylation of CDC25A at “Ser-178” and “Thr-507” and phosphorylation of CDC25C at “Ser-216” create binding sites for 14-3-3 proteins which inhibit CDC25A and CDC25C. Phosphorylation of CDC25A at “Ser-76”, “Ser-124”, “Ser-178”, “Ser-279”, and “Ser-293” promotes proteolysis of CDC25A. Phosphorylation of CDC25A at “Ser-76” primes the protein for subsequent phosphorylation at “Ser-79”, “Ser-82”, and “Ser-88” by NEK11, which is required for polyubiquitination and degradation of CDCD25A. Inhibition of CDC25 leads to increased inhibitory tyrosine phosphorylation of CDK-cyclin complexes and blocks cell-cycle progression. Also phosphorylates NEK6. Binds to and phosphorylates RAD51 at “Thr-309”, which promotes the release of RAD51 from BRCA2 and enhances the association of RAD51 with chromatin, thereby promoting DNA repair by homologous recombination. Phosphorylates multiple sites within the C-terminus of TP53, which promotes activation of TP53 by acetylation and promotes cell-cycle arrest and suppression of cellular proliferation. Also promotes repair of DNA cross-links through phosphorylation of FANCE. Binds to and phosphorylates TLK1 at “Ser-743”, which prevents the TLK1-dependent phosphorylation of the chromatin assembly factor ASF1A. This may enhance chromatin assembly both in the presence or absence of DNA damage. May also play a role in replication fork maintenance through regulation of PCNA. May regulate the transcription of genes that regulate cell-cycle progression through the phosphorylation of histones. Phosphorylates histone H3.1 (to form H3T11ph), which leads to epigenetic inhibition of a subset of genes. May also phosphorylate RB1 to promote its interaction with the E2F family of transcription factors and subsequent cell-cycle arrest. Isoform 2: Endogenous repressor of isoform 1, interacts with, and antagonizes CHK1 to promote the S to G2 M phase transition. | −6.8 |
| 2W0J | PD | Serine/threonine–protein kinase CHK2 | Serine/threonine–protein kinase that is required for checkpoint-mediated cell-cycle arrest, activation of DNA repair and apoptosis in response to the presence of DNA DSBs. May also negatively regulate cell-cycle progression during unperturbed cell cycles. Following activation, phosphorylates numerous effectors preferentially at the consensus sequence [L-X-R-X-X-S/T]. Regulates cell-cycle checkpoint arrest through phosphorylation of CDC25A, CDC25B, and CDC25C, inhibiting their activity. Inhibition of CDC25 phosphatase activity leads to increased inhibitory tyrosine phosphorylation of CDK-cyclin complexes and blocks cell-cycle progression. May also phosphorylate NEK6 which is involved in G2/M cell-cycle arrest. Regulates DNA repair through phosphorylation of BRCA2, enhancing the association of RAD51 with chromatin that promotes DNA repair by homologous recombination. Also stimulates the transcription of genes involved in DNA repair (including BRCA2) through the phosphorylation and activation of the transcription factor FOXM1. Regulates apoptosis through the phosphorylation of p53/TP53, MDM4, and PML. Phosphorylation of p53/TP53 at “Ser-20” by CHEK2 may alleviate inhibition by MDM2, leading to accumulation of active p53/TP53. Phosphorylation of MDM4 may also reduce degradation of p53/TP53. Also controls the transcription of pro-apoptotic genes through phosphorylation of the transcription factor E2F1. Tumor suppressor, it may also have a DNA damage-independent function in mitotic spindle assembly by phosphorylating BRCA1. Its absence may be a cause of the chromosomal instability observed in some cancer cells. | −7.1 |
| 2XKD | PD | Serine/threonine–protein kinase NEK2 | Protein kinase that is involved in the control of centrosome separation and bipolar spindle formation in mitotic cells and chromatin condensation in meiotic cells. Regulates centrosome separation (essential for the formation of bipolar spindles and high-fidelity chromosome separation) by phosphorylating centrosomal proteins such as CROCC, CEP250, and NINL, resulting in their displacement from the centrosomes. Regulates kinetochore microtubule attachment stability in mitosis via phosphorylation of NDC80. Involved in regulation of mitotic checkpoint protein complex via phosphorylation of CDC20 and MAD2L1. Plays an active role in chromatin condensation during the first meiotic division through phosphorylation of HMGA2. Phosphorylates: PPP1CC, SGOL1, NECAB3, and NPM1. Essential for localization of MAD2L1 to kinetochore and MAPK1 and NPM1 to the centrosome. Isoform 1 phosphorylates and activates NEK11 in G1/S-arrestedcells. Isoform 2, which is not present in the nucleolus, does not. | −7.7 |
| 3FXZ | PD | Serine/threonine–protein kinase PAK 1 | The activated kinase acts on a variety of targets. Likely to be the GTPase effector that links the Rho-related GTPases to the JNK MAP kinase pathway. Activated by CDC42 and RAC1. Involved in dissolution of stress fibers and reorganization of focal complexes. Involved in regulation of microtubule biogenesis through phosphorylation of TBCB. DVL1 and PAK1 form aternary complex with MUSK which is important for MUSK-dependent regulation of AChR clustering during the formation of the NMJ. Activity is inhibited in cells undergoing apoptosis, potentially due to binding of CDC2L1 and CDC2L2. Phosphorylates MYL9/MLC2. Phosphorylates RAF1 at “Ser-338” and “Ser-339” resulting in: activation of RAF1, stimulation of RAF1 translocation to mitochondria, phosphorylation of BAD by RAF1, and RAF1 binding to BCL2. | −7.7 |
| 2X4Z | PD | Serine/threonine–protein kinase PAK 4 | Serine/threonine protein kinase that plays a role in a variety of different signaling pathways including cytoskeleton regulation, cell migration, growth, proliferation, or cell survival. Activation by various effectors including growth factor receptors or active CDC42 and RAC1 results in a conformational change and a subsequent autophosphorylation on several serine and/or threonine residues. Phosphorylates and inactivates the protein phosphatase SSH1, leading to increased inhibitory phosphorylation of the actin binding/depolymerizing factor cofilin. Decreased cofilin activity may lead to stabilization of actin filaments. Phosphorylates LIMK1, a kinase that also inhibits the activity of cofilin. Phosphorylates integrin β5/ITGB5 and thus regulates cell motility. Phosphorylates ARHGEF2 and activates the downstream target RHOA that plays a role in the regulation of assembly of focal adhesions and actin stress fibers. Stimulates cell survival by phosphorylating the BCL2 antagonist of cell death BAD. Alternatively, inhibits apoptosis by preventing caspase-8 binding to death domain receptors in a kinase-independent manner. Plays a role in cell-cycle progression by controlling levels of the cell-cycle regulatory protein CDKN1A and by phosphorylating RAN. | −8.3 |
| 3F2A | PD | Serine/threonine–protein kinase PIM1 | Proto-oncogene with serine/threonine kinase activity involved in cell survival and cell proliferation and thus providing a selective advantage in tumorigenesis. Exerts its oncogenic activity through: the regulation of MYC transcriptional activity, the regulation of cell-cycle progression, and by phosphorylation and inhibition of proapoptotic proteins (BAD, MAP3K5, FOXO3). Phosphorylation of MYC leads to an increase of MYC protein stability, and thereby an increase of transcriptional activity. The stabilization of MYC exerced by PIM1 might explain partly the strong synergism between these two oncogenes in tumorigenesis. Mediates survival signaling through phosphorylation of BAD, which induces release of the anti-apoptotic protein Bcl-X(L)/BCL2L1. Phosphorylation of MAP3K5, an other proapoptotic protein, by PIM1, significantly decreases MAP3K5 kinase activity and inhibits MAP3K5-mediated phosphorylation of JNK and JNK/p38MAPK subsequently reducing caspase-3 activation and cell apoptosis. Stimulates cell-cycle progression at the G1–S and G2–M transitions by phosphorylation of CDC25A and CDC25C. Phosphorylation of CDKN1A, a regulator of cell-cycle progression at G1, results in the relocation of CDKN1A to the cytoplasm and enhanced CDKN1A protein stability. Promotes cell-cycle progression and tumorigenesis by downregulating expression of a regulator of cell-cycle progression, CDKN1B, at both transcriptional and posttranslational levels. Phosphorylation of CDKN1B induces binding of 14-3-3-proteins, permits nuclear export, and inhibits proteasome-dependent degradation. May affect the structure or silencing of chromatin by phosphorylating HP1 γ/CBX3. Acts also as a regulator of homing and migration of bone marrow cells involving functional interaction with the CXCL12-CXCR4 signaling axis. | −8.1 |
| 3FVH | PD | Serine/threonine–protein kinase PLK1 | Serine/threonine–protein kinase that performs several important functions throughout M phase of the cell cycle, including the regulation of centrosome maturation and spindle assembly, the removal of cohesins from chromosome arms, the inactivation of an aphase-promoting complex/cyclosome (APC/C) inhibitors, and the regulation of mitotic exit and cytokinesis. Polo-like kinase proteins act by binding and phosphorylating proteins are that already phosphorylated on a specific motif recognized by the POLO box domains. Phosphorylates BORA, BUB1B/BUBR1, CCNB1, CDC25C, CEP55, ECT2, ERCC6L, FBXO5/EMI1, FOXM1, KIF20A/MKLP2, MLF1IP, NEDD1, NINL, NPM1, NUDC, PKMYT1/MYT1, PLK1S1/KIZ, PPP1R12A/MYPT1, PRC1, RACGAP1/CYK4, SGOL1, STAG2/SA2, TEX14, TOPORS, p73/TP73, TPT1, and WEE1. Plays a key role in centrosome functions and the assembly of bipolar spindles by phosphorylating PLK1S1/KIZ, NEDD1, and NINL. NEDD1 phosphorylation promotes subsequent targeting of the γ-tubulin ring complex (gTuRC) to the centrosome, an important step for spindle formation. Phosphorylation of NINL component of the centrosome leads to NINL dissociation from other centrosomal proteins. Involved in mitosis exit and cytokinesis by phosphorylating CEP55, ECT2, KIF20A/MKLP2, MLF1IP, PRC1, and RACGAP1. Recruited at the central spindle by phosphorylating and docking PRC1 and KIF20A/MKLP2; creates its own docking sites on PRC1 and KIF20A/MKLP2 by mediating phosphorylation of sites subsequently recognized by the POLO box domains. Phosphorylates RACGAP1, thereby creating a docking site for the Rho GTP exchange factor ECT2 that is essential for the cleavage furrow formation. Promotes the central spindle recruitment of ECT2. Plays a central role in G2–M transition of mitotic cell cycle by phosphorylating CCNB1, CDC25C, FOXM1, MLF1IP, PKMYT1/MYT1, PPP1R12A/MYPT1, and WEE1. Part of a regulatory circuit that promotes the activation of CDK1 by phosphorylating the positive regulator CDC25C and inhibiting the negative regulators WEE1 and PKMYT1/MYT1. Also acts by mediating phosphorylation of cyclin-B1 (CCNB1) on centrosomes in prophase. Phosphorylates FOXM1, a key mitotic transcription regulator, leading to enhance FOXM1 transcriptional activity. Involved in kinetochore functions and sister chromatide cohesion by phosphorylating BUB1B/BUBR1, FBXO5/EMI1, and STAG2/SA2. PLK1 is high on nonattached kinetochores suggesting a role of PLK1 in kinetochore attachment or in SAC regulation. Required for kinetochore localization of BUB1B. Regulates the dissociation of cohesin from chromosomes by phosphorylating cohesin subunits such as STAG2/SA2. Phosphorylates SGOL1: required for spindle pole localization of isoform 3 of SGOL1 and plays a role in regulating its centriole cohesion function. Mediates phosphorylation of FBXO5/EMI1, a negative regulator of the APC/C during prophase, leading to FBXO5/EMI1 ubiquitination and degradation by the proteasome. Acts as a negative regulator of p53 family members: phosphorylates TOPORS, leading to inhibit the sumoylation of p53/TP53 and simultaneously enhance the ubiquitination and subsequent degradation of p53/TP53. Phosphorylates the transactivation domain of the transcription factor p73/TP73, leading to inhibit p73/TP73-mediated transcriptional activation and pro-apoptotic functions. Phosphorylates BORA, and thereby promotes the degradation of BORA. Contributes to the regulation of AURKA function. Also required for recovery after DNA damage checkpoint and entry into mitosis. | −8.2 |
| 3HDM | PD | Serine/threonine–protein kinase SGK1 | Serine/threonine–protein kinase that is involved in the regulation of a wide variety of ion channels, membrane transporters, cellular enzymes, transcription factors, neuronal excitability, cell growth, proliferation, survival, migration, and apoptosis. Plays an important role in cellular stress response. Contributes to regulation of renal Na+ retention, renal K+ elimination, salt appetite, gastric acid secretion, intestinal Na+/H+ exchange and nutrient transport, INS-dependent salt sensitivity of blood pressure, salt sensitivity of peripheral glucose uptake, cardiac repolarization, and memory consolidation. Upregulates Na+ channels: SCNN1A/ENAC, SCN5A, and ASIC1/ACCN2; K+ channels: KCNJ1/ROMK1, KCNA1-5, KCNQ1-5, and KCNE1; epithelial Ca2+ channels: TRPV5 and TRPV6; chloride channels: BSND, CLCN2, and CFTR; glutamate transporters: SLC1A3/EAAT1, SLC1A2/EAAT2, SLC1A1/EAAT3, SLC1A6/EAAT4, and SLC1A7/EAAT5; amino acid transporters: SLC1A5/ASCT2, SLC38A1/SN1, and SLC6A19; creatine transporter: SLC6A8; Na+/dicarboxylate cotransporter: SLC13A2/NADC1; Na+-dependent phosphate cotransporter: SLC34A2/NAPI-2B; and glutamate receptor: GRIK2/GLUR6. Upregulates carriers: SLC9A3/NHE3, SLC12A1/NKCC2, SLC12A3/NCC, SLC5A3/SMIT, SLC2A1/GLUT1, SLC5A1/SGLT1, and SLC15A2/PEPT2. Regulates enzymes: GSK3A/B, PMM2, and Na+/K+ ATPase, and transcription factors: CTNNB1 and nuclear factor NF-κB. Stimulates sodium transport into epithelial cells by enhancing the stability and expression of SCNN1A/ENAC. This is achieved by phosphorylating the NEDD4L ubiquitin E3 ligase, promoting its interaction with 14-3-3 proteins, thereby preventing it from binding to SCNN1A/ENAC and targeting it for degradation. Regulates SOCE by stimulating ORAI1 and STIM1. Regulates KCNJ1/ROMK1directly via its phosphorylation or indirectly via increased interaction with SLC9A3R2/NHERF2. Phosphorylates MDM2 and activates MDM2-dependent ubiquitination of p53/TP53. Phosphorylates MAPT/TAU and mediates microtubule depolymerization and neurite formation in hippocampal neurons. Phosphorylates SLC2A4/GLUT4 and upregulates its activity. Phosphorylates APBB1/FE65 and promotes its localization to the nucleus. Phosphorylates MAPK1/ERK2 and activates it by enhancing its interaction with MAP2K1/MEK1 and MAP2K2/MEK2. Phosphorylates FBXW7 and plays an inhibitory role in the NOTCH1 signaling. Phosphorylates FOXO1 resulting in its relocalization from the nucleus to the cytoplasm. Phosphorylates FOXO3, promoting its exit from the nucleus and interference with FOXO3-dependent transcription. Phosphorylates BRAF and MAP3K3/MEKK3 and inhibits their activity. Phosphorylates SLC9A3/NHE3 in response to dexamethasone, resulting in its activation and increased localization at the cell membrane. Phosphorylates CREB1. Necessary for vascular remodeling during angiogenesis. Sustained high levels and activity may contribute to conditions such as hypertension and diabetic nephropathy. Isoform 2 exhibited a greater effect on cell plasma membrane expression of SCNN1A/ENAC and Na+ transport than isoform 1. | −7.2 |
| 2IE4 | PD | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A α isoform | The PR65 subunit of protein phosphatase 2A serves as a scaffolding molecule to coordinate the assembly of the catalytic subunit and a variable regulatory B subunit. Required for proper chromosome segregation and for centromeric localization of SGOL1 in mitosis. | −7.6 |
| 3E7A | PD | Serine/threonine-protein phosphatase PP1-α catalytic subunit | Protein phosphatase that associates with over 200 regulatory proteins to form highly specific holoenzymes which dephosphorylate hundreds of biological targets. PP1 is essential for cell division and participates in the regulation of glycogen metabolism, muscle contractility, and protein synthesis. Involved in regulation of ionic conductances and long-term synaptic plasticity. May play an important role in dephosphorylating substrates such as the postsynaptic density-associated Ca2+/calmodulin-dependent protein kinase II. Component of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure and cell-cycle progression during the transition from mitosis into interphase. Regulates NEK2 function in terms of kinase activity and centrosome number and splitting, both in the presence and absence of radiation-induced DNA damage. Regulator of neural tube and optic fissure closure, and ENCCs migration during development. | −6.8 |
| 1IT6 | PD | Serine/threonine-protein phosphatase PP1-γ catalytic subunit | Protein phosphatase that associates with over 200 regulatory proteins to form highly specific holoenzymes which dephosphorylate hundreds of biological targets. PP1 is essential for cell division and participates in the regulation of glycogen metabolism, muscle contractility, and protein synthesis. Dephosphorylates RPS6KB1 Involved in regulation of ionic conductances and long-term synaptic plasticity. May play an important role in dephosphorylating substrates such as the postsynaptic density-associated Ca2+/calmodulin-dependent protein kinase II. Component of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure and cell-cycle progression during the transition from mitosis into interphase. | −7.6 |
| 1J04, 3R9A, 1H0C | PK | Serine–pyruvate aminotransferase | N/A | −7.2 to −6.8 |
| 3A73, 3A73_2, 3SQJ_7, 3SQJ, 3A73_4, 3A73_8, 3SQJ_8, 3SQJ_4, 3SQJ_3, 3A73_6, 3A73_5, 3A73_7, 3SQJ_6, 3SQJ_2, 3SQJ_5, 3A73_3 | PK | Serum albumin | Serum albumin, the main protein of plasma, has a good binding capacity for water, Ca2+, Na+, K+, fatty acids, hormones, bilirubin, and drugs. Its main function is the regulation of the colloidal osmotic pressure of blood. Major zinc transporter in plasma, typically binds approximately 80% of all plasma zinc. | −9.5 to –6.1 |
| 2W08 | PD | Serum amyloid P-component | Can interact with DNA and histones and may scavenge nuclear material released from damaged circulating cells. May also function as a calcium-dependent lectin. | −6.3 |
| 2VYT | PD | Sex comb on midleg-like protein 2 | Putative PcG protein. PcG proteins act by forming multiprotein complexes, which are required to maintain the transcriptionally repressive state of homeotic genes throughout development (by similarity). | −7.4 |
| 1KDK | PD | Sex hormone-binding globulin | Functions as an androgen transport protein, but may also be involved in receptor-mediated processes. Each dimer binds one molecule of steroid. Specific for 5-α-dihydrotestosterone, testosterone, and 17-β-estradiol. Regulates the plasma metabolic clearance rate of steroid hormones by controlling their plasma concentration. | −6.3 |
| 2ZG1 | PD | Sialic acid-binding Ig-like lectin 5 | Putative adhesion molecule that mediates sialic-acid dependent binding to cells. Binds equally to α-2,3-linked and α-2,6-linked sialic acid. The sialic acid recognition site maybe masked by cis interactions with sialic acids on the same cell surface. | −6.5 |
| 1K27 | PD | MTA phosphorylase | Catalyzes the reversible phosphorylation of MTA to adenine and 5-methylthioribose-1-phosphate. Involved in the breakdown of MTA, a major by-product of polyamine biosynthesis. Responsible for the first step in the methionine salvage pathway after MTA has been generated from S-AdoMet. Has broad substrate specificity with 6-aminopurine nucleosides as preferred substrates. | −7.2 |
| 1XK5 | PD | Snurportin-1 | Functions as an UsnRNP-specific nuclear import adapter. Involved in the m3G-cap-dependent nuclear import of UsnRNPs. Binds specifically to the terminal m3G-cap UsnRNAs. | −8.1 |
| 3IVV | PD | Speckle-type POZ protein | Inhibits IPF1/PDX1 transactivation of established target promoters, such as INS, may be by recruiting a repressor complex (by similarity). In complex with CUL3, involved in ubiquitination of BMI1, H2AFY, and DAXX, and probably also in ubiquitination and proteasomal degradation of Gli2 or Gli3. | −7.1 |
| 1EZF | PD | Squalene synthase | N/A | −9.4 |
| 3OMC | PD | Staphylococcal nuclease domain-containing protein 1 | Functions as a bridging factor between STAT6 and the basal transcription factor. Plays a role in PIM1 regulation of MYB activity. Functions as a transcriptional coactivator for the EBNA2. | −7.7 |
| 1ELR | PD | Stress-induced-phosphoprotein 1 | Mediates the association of the molecular chaperones HSC70 and HSP90 (HSPCA and HSPCB). | −6.5 |
| 1HY7 | PD | Stromelysin-1 | Can degrade fibronectin, laminin, gelatins of type I, III, IV, and V; type III, IV, X, and IX collagens, and cartilage proteoglycans. Activates procollagenase. | −8.2 |
| 2W8Q | PK | Succinate-semialdehyde dehydrogenase, mitochondrial | Catalyzes one step in the degradation of the inhibitory neurotransmitter GABA. | −2.8 |
| 3U3O_2, 1Z28, 2D06, 1LS6_2, 1LS6, 3U3O | PK | Sulfotransferase 1A1 | Sulfotransferase that utilizes PAPS as sulfonate donor to catalyze the sulfate conjugation of catecholamines, phenolic drugs, and neurotransmitters. Has also estrogen sulfotransferase activity. Responsible for the sulfonation and activation of minoxidil. It mediates the metabolic activation of carcinogenic N-hydroxyarylamines to DNA-binding products and so it could participate as modulating factor of cancer risk. | −6.4 to −5.5 |
| 1Z29 | PK | Sulfotransferase 1A2 | Sulfotransferase that utilizes PAPS as sulfonate donor to catalyze the sulfate conjugation of catecholamines, phenolic drugs, and neurotransmitters. Is also responsible for the sulfonation and activation of minoxidil. Mediates the metabolic activation of carcinogenic N-hydroxyarylamines to DNA-binding products and could so participate as modulating factor of cancer risk. | −6.7 |
| 3BFX | PK | Sulfotransferase 1C2 | Sulfotransferase that utilizes PAPS as sulfonate donor to catalyze the sulfate conjugation of drugs, xenobiotic compounds, hormones, and neurotransmitters. May be involved in the activation of carcinogenic hyroxylamines. Shows activity toward p-nitrophenol and N-hydroxy-2-acetylamino-fluorene (N-OH-2AAF). | −7.7 |
| 2H8K, 2REO | PK | Sulfotransferase 1C3 | Sulfotransferase that utilizes PAPS as sulfonate donor and has low sulphotransferase activity toward various substrates with alcohol groups (in vitro). May catalyze the sulfate conjugation of xenobiotic compounds and endogenous substrates. | −6.9 to −5.3 |
| 2Z5F, 3CKL, 3CKL_2, 1Q20_2, 1Q22_2, 1Q22, 1Q1Z, 1Q20 | PK | Sulfotransferase family cytosolic 1B member 1 | Sulfotransferase that utilizes PAPS as sulfonate donor to catalyze the sulfate conjugation of many hormones, neurotransmitters, drugs, and xenobiotic compounds. Sulfonation increases the water solubility of most compounds, and therefore their renal excretion, but it can also result in bioactivation to form active metabolites. Sulfates dopamine, small phenols such as 1-naphthol and p-nitrophenol and thyroid hormones, including 3,3′-diiodothyronine, triidothyronine, reverse triiodothyronine, and thyroxine. | −10.4 to −4.9 |
| 2GV6 | PD | Suppressor of tumorigenicity 14 protein | Degrades extracellular matrix. Proposed to play a role in breast cancer invasion and metastasis. Exhibits trypsin-like activity as defined by cleavage of synthetic substrates with Argor Lys as the P1 site. | −8.1 |
| 1OBX | PD | Syntenin-1 | Seems to function as an AP. In AJs, may function to couple syndecans to cytoskeletal proteins or signaling components. Seems to couple transcription factor SOX4 to the IL5 receptor (IL5RA). May also play a role in vesicular trafficking. Seems to be required for the targeting of TGFA to the cell surface in the early secretory pathway. | −7.5 |
| 3KR8 | PD | Tankyrase-2 | Poly-ADP-ribosyl transferase involved in various processes such as Wnt signaling pathway, telomere length, and vesicle trafficking. Acts as an activator of the Wnt signaling pathway by mediating poly-ADP-ribosylation of AXIN1 and AXIN2, two key components of the β-catenin destruction complex: poly-ADP-ribosylated target proteins are recognized by RNF146, which mediates their ubiquitination and subsequent degradation. Also mediates poly-ADP-ribosylation of BLZF1 and CASC3, followed by recruitment of RNF146 and subsequent ubiquitination. Mediates poly-ADP-ribosylation of TERF1, thereby contributing to the regulation of telomere length. May also regulate vesicle trafficking and modulate the subcellular distribution of SLC2A4/GLUT4-vesicles. | −6.9 |
| 3HMM | PD | TGF-β receptor type 1 | Transmembrane serine/threonine kinase forming with the TGF-β type II serine/threonine kinase receptor, TGFBR2, thenon-promiscuous receptor for the TGF-β cytokines TGFB1, TGFB2, and TGFB3. Transduces the TGFB1, TGFB2, and TGFB3 signals from the cell surface to the cytoplasm and is thus regulating a plethora of physiological and pathological processes including cell-cycle arrest in epithelial and hematopoietic cells, control of mesenchymal cell proliferation and differentiation, wound healing, extracellular matrix production, immunosuppression, and carcinogenesis. The formation of the receptor complex composed of two TGFBR1 and two TGFBR2 molecules symmetrically bound to the cytokine dimer results in the phosphorylation and the activation of TGFBR1 by the constitutively active TGFBR2. Activated TGFBR1 phosphorylates SMAD2 that dissociates from the receptor and interacts with SMAD4. The SMAD2-SMAD4 complex is subsequently translocated to the nucleus where it modulates the transcription of the TGF-β-regulated genes. This constitutes the canonical SMAD-dependent TGF-β signaling cascade. Also involved in noncanonical, SMAD-independent TGF-β signaling pathways. For instance, TGFBR1 induces TRAF6 autoubiquitination which in turn results in MAP3K7 ubiquitination and activation to trigger apoptosis. Also regulates epithelial to mesenchymal transition through a SMAD-independent signaling pathway through PARD6A phosphorylation and activation | −7.5 |
| 2BZG, 2H11 | PK | Thiopurine S-methyltransferase | Catalyzes the S-methylation of thiopurine drugs such as 6-mercaptopurine. | −6.3 to −6.2 |
| 1UOU | PD | Thymidine phosphorylase | May have a role in maintaining the integrity of the blood vessels. Has growth promoting activity on ECs, angiogenic activity in vivo and chemotactic activity on ECs in vitro. Catalyzes the reversible phosphorolysis of thymidine. The produced molecules are then utilized as carbon and energy sources or in the rescue of pyrimidine bases for nucleotide synthesis. | −6.1 |
| 1I00 | PD | Thymidylate synthase | Contributes to the de novo mitochondrial thymidylate biosynthesis pathway. | −9.1 |
| 3JZB | PD | Thyroid hormone receptor α | Nuclear hormone receptor. High-affinity receptor for triiodothyronine. | −5.9 |
| 1N46 | PD | Thyroid hormone receptor β | High-affinity receptor for triiodothyronine. | −6.8 |
| 2XN6_2, 2XN7_2, 2XN7, 2CEO, 2XN6 | PK | Thyroxine-binding globulin | Major thyroid hormone transport protein in serum. | −9.7 to −6.9 |
| 3CYY | PD | TJ protein ZO-1 | The N-terminal may be involved in transducing a signal required for TJ assembly, while the C-terminal may have specific properties of TJs. The α domain might be involved in stabilizing junctions. Plays a role in the regulation of cell migration by targeting CDC42BPB to the leading edge of migrating cells. | −8.5 |
| 1A5H | PD | Tissue-type plasminogen activator | Converts the abundant, but inactive, zymogen plasminogen to plasmin by hydrolyzing a single Arg-Val bond in plasminogen. By controlling plasmin-mediated proteolysis, it plays an important role in tissue remodeling and degradation, in cell migration and many other physiopathological events. Plays a direct role in facilitating neuronal migration. | −7.6 |
| 3KZE | PD | T-lymphoma invasion and metastasis-inducing protein 1 | Modulates the activity of RHO-like proteins and connects extracellular signals to cytoskeletal activities. Acts as a GDP-dissociation stimulator protein that stimulates the GDP-GTP exchange activity of RHO-like GTPases and activates them. Activates RAC1, CDC42, and to a lesser extent RHOA. | −7.4 |
| 1QSC | PD | TNF receptor-associated factor 2 | Regulates activation of NF-κB and JNK and plays a central role in the regulation of cell survival and apoptosis. Required for normal antibody isotype switching from IgM to IgG. Has E3 ubiquitin-protein ligase activity and promotes “Lys-63”-linked ubiquitination of target proteins, such as BIRC3, RIPK1, and TICAM1. Is an essential constituent of several E3 ubiquitin-protein ligase complexes, where it promotes the ubiquitination of target proteins by bringing them into contact with other E3 ubiquitin ligases. Regulates BIRC2 and BIRC3 protein levels by inhibiting their autoubiquitination and subsequent degradation; this does not depend on the TRAF2 RING-type zinc finger domain. | −6.3 |
| 1LB6 | PD | TNF receptor-associated factor 6 | E3 ubiquitin ligase that, together with UBE2N and UBE2V1, mediates the synthesis of “Lys-63”-linked-polyubiquitinchains conjugated to proteins, such as IKBKG, AKT1, and AKT2. Also mediates ubiquitination of free/unanchored polyubiquitin chain that leads to MAP3K7 activation. Leads to the activation of NF-κB and JUN. May be essential for the formation of functional osteoclasts. Seems to also play a role in DCs maturation and/or activation. Represses c-Myb-mediated transactivation in B-lymphocytes. AP that seems to play a role in signal transduction initiated via TNF, IL-1, and IL-17 receptors. Regulates osteoclast differentiation by mediating the activation of AP complex 1 (AP-1) and NF-κB, in response to RANK-L stimulation. | −7.1 |
| 2CE9 | PD | Transducin-like enhancer protein 1 | Transcriptional corepressor that binds to a number of transcription factors. Inhibits NF-κB-regulated gene expression. Inhibits the transcriptional activation mediated by FOXA2 and by CTNNB1 and TCF family members in Wnt signaling. The effects of full-length TLE family members may be modulated by association with dominant-negative AES. Unusual function as coactivator for ESRRG. | −7.8 |
| 3HU3 | PD | tER ATPase | Necessary for the fragmentation of Golgi stacks during mitosis and for their reassembly after mitosis. Involved in the formation of the tER. The transfer of membranes from the endoplasmic reticulum to the Golgi apparatus occurs via 50–70 nm transition vesicles that derive from part-rough, part-smooth transitional elements of the tER. Vesicle budding from the tER is an ATP-dependent process. The ternary complex containing UFD1L, VCP, and NPLOC4 binds ubiquitinated proteins and is necessary for the export of misfolded proteins from the ER to the cytoplasm, where they are degraded by the proteasome. The NPLOC4-UFD1L-VCP complex regulates spindle disassembly at the end of mitosis and is necessary for the formation of a closed nuclear envelope. Regulates E3 ubiquitin-protein ligase activity of RNF19A (by similarity). Component of the VCP/p97-AMFR/gp78 complex that participates in the final step of the sterol-mediated ubiquitination and ERAD of HMGCR. Also involved in DNA damage response: recruited to DSBs sites in a RNF8- and RNF168-dependent manner and promotes the recruitment of TP53BP1 at DNA damage sites. |
−10.1 |
| 3HJ0 | PD | Transthyretin | Thyroid hormone-binding protein. Probably transports thyroxine from the blood stream to the brain. | −6.9 |
| 1NJS | PD | Trifunctional purine biosynthetic protein adenosine-3 | N/A | −6.7 |
| 1HTI | PD | Triosephosphate isomerase | N/A | −7.5 |
| 3CKK | PK | tRNA [guanine-N(7)-]-methyltransferase | Catalyzes the formation of N(7)-methylguanine at position 46 (m7G46) in tRNA. | −6.8 |
| 1IH0 | PD | Troponin C, slow skeletal and cardiac muscles | Troponin is the central regulatory protein of striated muscle contraction. Tn consists of three components: Tn-I, which is the inhibitor of actomyosin ATPase; Tn-T, which contains the binding site for tropomyosin; and Tn-C. The binding of calcium to Tn-C abolishes the inhibitory action of Tn on actin filaments. | −6.6 |
| 1H4W | PD | Trypsin-3 | Digestive protease specialized for the degradation of trypsin inhibitors. In the ileum, may be involved in defensing processing, including DEFA5. | −6.9 |
| 2FPZ | PD | Tryptase β-2 | Tryptase is the major neutral protease present in mast cells and is secreted upon the coupled activation-degranulation response of this cell type. Has an immunoprotective role during bacterial infection. Required to efficiently combat Klebsiella pneumoniae infection (by similarity). | −8.2 |
| 3HF6 | PD | Tryptophan 5-hydroxylase 1 | N/A | −10.6 |
| 1FT4 | PD | TNF receptor super family member 1A | Receptor for TNFSF2/TNF-α and homotrimeric TNFSF1/lymphotoxin-α. The adapter molecule FADD recruits caspase-8 to the activated receptor. The resulting DISC performs caspase-8 proteolytic activation that initiates the subsequent cascade of caspases (aspartate-specific cysteine proteases) mediating apoptosis. Contributes to the induction of noncytocidal TNF effects including anti-viral state and activation of the acid sphingomyelinase. | −6.8 |
| 3OBQ | PD | Tumor susceptibility gene 101 protein | Component of the ESCRT-I complex, a regulator of vesicular trafficking process. Binds to ubiquitinated cargo proteins and is required for the sorting of endocytic ubiquitinated cargos into MVBs. Mediates the association between the ESCRT-0 and ESCRT-I complex. Required for completion of cytokinesis; the function requires CEP55. May be involved in cell growth and differentiation. Acts as a negative growth regulator. Involved in the budding of many viruses through an interaction with viral proteins that contain a late-budding motif P-[ST]-A-P. This interaction is essential for viral particle budding of numerous retroviruses. | −6.5 |
| 2HZI | PD | Tyrosine-protein kinase ABL1 | Nonreceptor tyrosine-protein kinase that plays a role in many key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, and DNA damage response and apoptosis. Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like WASF3 (involved in branch formation); ANXA1 (involved in membrane anchoring); DBN1, DBNL, CTTN, RAPH1, and ENAH (involved in signaling); or MAPT and PXN (microtubule-binding proteins). Phosphorylation of WASF3 is critical for the stimulation of lamellipodia formation and cell migration. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as BCAR1, CRK, CRKL, DOK1, EFS, or NEDD9. Phosphorylates multiple receptor tyrosine kinases and more particularly promotes endocytosis of EGFR, facilitates the formation of neuro muscular synapses through MUSK, inhibits PDGFRB-mediated chemotaxis, and modulates the endocytosis of activated BCR complexes. Other substrates that are involved in endocytosis regulation are the caveolin (CAV1) and RIN1. Moreover, ABL1 regulates the CBL family of ubiquitin ligases that drive receptor downregulation and actin remodeling. Phosphorylation of CBL leads to increased EGFR stability. Involved in late-stage autophagy by regulating positively the trafficking and function of lysosomal components. ABL1 targets to mitochondria in response to oxidative stress, and thereby mediates mitochondrial dysfunction and cell death. ABL1 is also translocated in the nucleus where it has DNA-binding activity and is involved in DNA damage response and apoptosis. Many substrates are known mediators of DNA repair: DDB1, DDB2, ERCC3, ERCC6, RAD9A, RAD51, RAD52, and WRN. Activates the proapoptotic pathway when the DNA damage is too severe to be repaired. Phosphorylates TP73, a primary regulator for this type of damage-induced apoptosis. Phosphorylates PSMA7 that leads to an inhibition of proteasomal activity and cell cycle transition blocks. ABL1 acts also as a regulator of multiple pathological signaling cascades during infection. Several known tyrosine-phosphorylated microbial proteins have been identified as ABL1 substrates. This is the case of A36R of vaccinia virus, Tir (translocated intimin receptor) of pathogenic E. coli and possibly Citrobacter, CagA (cytotoxin-associated gene A) of Helicobacter pylori, or AnkA (ankyrin repeat-containing protein A) of Anaplasma phagocytophilum. Pathogens can highjack ABL1 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1. | −7.5 |
| 3GEN | PD | Tyrosine-protein kinase BTK | Nonreceptor tyrosine kinase in dispensable for B lymphocyte development, differentiation, and signaling. Binding of antigen to the B-cell antigen receptor triggers signaling that ultimately leads to B-cell activation. After BCR engagement and activation at the plasma membrane, phosphorylates PLCG2 at several sites, igniting the downstream signaling pathway through calcium mobilization, followed by activation of the PKC family members. PLCG2 phosphorylation is performed in close cooperation with the AP B-cell linker protein BLNK. BTK acts as a platform to bring together a diverse array of signaling proteins and is implicated in cytokine receptor signaling pathways. Plays an important role in the function of immune cells of innate as well as adaptive immunity, as a component of the TLRs pathway. The TLR pathway acts as a primary surveillance system for the detection of pathogens and is crucial to the activation of host defense. Especially, is a critical molecule in regulating TLR9 activation in splenic B-cells. Within the TLR pathway, induces tyrosine phosphorylation of TIRAP which leads to TIRAP degradation. BTK plays also a critical role in transcription regulation. Induces the activity of NF-κB, which is involved in regulating the expression of hundreds of genes. BTK is involved in the signaling pathway linking TLR8 and TLR9 to NF-κB. Transiently phosphorylates transcription factor GTF2I on tyrosine residues in response to BCR. GTF2I then translocates to the nucleus to bind regulatory enhancer elements to modulate gene expression. ARID3A and NFAT are other transcriptional target of BTK. BTK is required for the formation of functional ARID3A DNA-binding complexes. There is however no evidence that BTK itself binds directly to DNA. BTK has a dual role in the regulation of apoptosis. | −7.5 |
| 2DQ7 | PD | Tyrosine-protein kinase FYN | Non-receptor tyrosine-protein kinase that plays a role in many biological processes including regulation of cell growth and survival, cell adhesion, integrin-mediated signaling, cytoskeletal remodeling, cell motility, immune response and axon guidance. Inactive FYN is phosphorylated on its C-terminal tail within the catalytic domain. Following activation by PKA, the protein subsequently associates with PTK2/FAK1, allowing PTK2/FAK1phosphorylation, activation and targeting to focal adhesions. Involved in the regulation of cell adhesion and motility through phosphorylation of CTNNB1 (β-catenin) and CTNND1 (δ-catenin). Regulates cytoskeletal remodeling by phosphorylating several proteins including the actin regulator WAS and the microtubule-associated proteins MAP2 and MAPT. Promotes cell survival by phosphorylating AGAP2/PIKE-A and preventing its apoptotic cleavage. Participates in signal transduction pathways that regulate the integrity of the glomerular slit diaphragm (an essential part of the glomerular filter of the kidney) by phosphorylating several slit diaphragm components including NPHS1, KIRREL and TRPC6. Plays a role in neural processes by phosphorylating DPYSL2, a multifunctional AP within the CNS, ARHGAP32, a regulator for Rho family GTPases implicated in various neural functions, and SNCA, a small pre-synaptic protein. Participates in the downstream signaling pathways that lead to T-cell differentiation and proliferation following TCR stimulation. Also participates in negative feedback regulation of TCR signaling through phosphorylation of PAG1, thereby promoting interaction between PAG1 and CSK and recruitment of CSK to lipid rafts. CSK maintains LCK and FYN in an inactive form. Promotes CD28-induced phosphorylation of VAV1. | −8.9 |
| 2HK5 | PD | Tyrosine-protein kinase HCK | Nonreceptor tyrosine-protein kinase found in hematopoietic cells that transmits signals from cell surface receptors and plays an important role in the regulation of innate immune responses, including neutrophil, monocyte, macrophage and mast cell functions, phagocytosis, cell survival and proliferation, and cell adhesion and migration. Acts downstream of receptors that bind the Fc region of immunoglobulins, such as FCGR1A and FCGR2A, but also CSF3R, PLAUR, the receptors for IFN-γ, IL2, IL6, and IL8, and integrins, such as ITGB1 and ITGB2. During the phagocytic process, mediates mobilization of secretory lysosomes, degranulation, and activation of NADPH oxidase to bring about the respiratory burst. Plays a role in the release of inflammatory molecules. Promotes reorganization of the actin cytoskeleton and actin polymerization, formation of podosomes and cell protrusions. Inhibits TP73-mediated transcription activation and TP73-mediated apoptosis. Phosphorylates CBL in response to activation of immunoglobulin γ Fc region receptors. Phosphorylates ADAM15, BCR, ELMO1, FCGR2A, GAB1, GAB2, RAPGEF1, STAT5B, TP73, VAV1, and WAS. | −8 |
| 3MIY | PD | Tyrosine-protein kinase ITK/TSK | Tyrosine kinase that plays an essential role in regulation of the adaptive immune response. Regulates the development, function, and differentiation of conventional T-cells and nonconventional NKT-cells. When APCs activate TCR, a series of phosphorylation lead to the recruitment of ITK to the cell membrane, in the vicinity of the stimulated TCR receptor, where it is phosphorylated by LCK. Phosphorylation leads to ITK autophosphorylation and full activation. Once activated, phosphorylates PLCG1, leading to the activation of this lipase and subsequent cleavage of its substrates. In turn, the endoplasmic reticulum releases calcium ions in the cytoplasm and the NFAT translocates into the nucleus to perform its transcriptional duty. Phosphorylates 2 essential APs: the linker for activation of T-cells/LAT protein and LCP2. Then, a large number of signaling molecules such as VAV1 are recruited and ultimately lead to lymphokine production, T-cell proliferation and differentiation. | −6 |
| 3EYG | PD | Tyrosine-protein kinase JAK1 | Tyrosine kinase of the nonreceptor type, involved in the IFN-α/β/γ signal pathway. Kinase partner for the IL2 receptor. | −7.3 |
| 3.00E + 64 | PD | Tyrosine-protein kinase JAK2 | Nonreceptor tyrosine kinase involved in various processes such as cell growth, development, differentiation, or histone modifications. Mediates essential signaling events in both innate and adaptive immunity. In the cytoplasm, plays a pivotal role in signal transduction via its association with type I receptors such as GHR, PRLR, LEPR, EPOR, and THPO; or type II receptors including IFN-α, IFN-β, IFN-γ, and multiple ILs. Following ligand binding to cell surface receptors, phosphorylates specific tyrosine residues on the cytoplasmic tails of the receptor, creating docking sites for STAT proteins. Subsequently, phosphorylates the STAT proteins once they are recruited to the receptor. Phosphorylated STATs then form homodimer or heterodimers and translocate to the nucleus to activate gene transcription. For example, cell stimulation with EPO during erythropoiesis leads to JAK2 autophosphorylation, activation, and its association with EPOR that becomes phosphorylated in its cytoplasmic domain. Then, STAT5 (STAT5A or STAT5B) is recruited, phosphorylated, and activated by JAK2. Once activated, dimerized STAT5 translocates into the nucleus and promotes the transcription of several essential genes involved in the modulation of erythropoiesis. In addition, JAK2 mediates angiotensin-2-induced ARHGEF1 phosphorylation. Plays a role in cell cycle by phosphorylating CDKN1B. Cooperates with TEC through reciprocal phosphorylation to mediate cytokine-driven activation of FOS transcription. In the nucleus, plays a key role in chromatin by specifically mediating phosphorylation of “Tyr-41” of histone H3 (H3Y41ph), a specific tag that promotes exclusion of CBX5 (HP1α) from chromatin. | −7.4 |
| 3PJC | PD | Tyrosine-protein kinase JAK3 | Nonreceptor tyrosine kinase involved in various processes such as cell growth, development, or differentiation. Mediates essential signaling events in both innate and adaptive immunity and plays a crucial role in hematopoiesis during the development of T-cells. In the cytoplasm, plays a pivotal role in signal transduction via its association with type I receptors sharing the common γ subunits such as IL-2R, IL-4R, IL-7R, IL-9R, IL-15R, and IL-21R. Following ligand binding to cell surface receptors, phosphorylates specific tyrosine residues on the cytoplasmic tails of the receptor, creating docking sites for STAT proteins. Subsequently, phosphorylates the STAT proteins once they are recruited to the receptor. Phosphorylated STATs then form homodimer or heterodimers and translocate to the nucleus to activate gene transcription. For example, upon IL-2R activation by IL2, JAK1, and JAK3 molecules bind to IL-2R β (IL-2RB) and γ chain (IL-2RG) subunits inducing the tyrosine phosphorylation of both receptor subunits on their cytoplasmic domain. Then, STAT5A and STAT5B are recruited, phosphorylated, and activated by JAK1 and JAK3. Once activated, dimerized STAT5 translocates to the nucleus and promotes the transcription of specific target genes in acytokine-specific fashion. | −10 |
| 1LKK | PD | Tyrosine-protein kinase LCK | Nonreceptor tyrosine-protein kinase that plays an essential role in the selection and maturation of developing T-cells in the thymus and in the function of mature T-cells. Plays a key role in T-cell antigen receptor-linked signal transduction pathways. Constitutively associated with the cytoplasmic portions of the CD4 and CD8 surface receptors. Association of the TCR with a peptide antigen-bound MHC facilitates the interaction of CD4 and CD8 with MHC class II and class I molecules, respectively, thereby recruiting the associated LCK protein to the vicinity of the TCR/CD3 complex. LCK then phosphorylates tyrosines residues within the ITAMs of the cytoplasmic tails of the TCR-γ chains and CD3 subunits, initiating the TCR/CD3 signaling pathway. Once stimulated, the TCR recruits the tyrosine kinase ZAP70, which becomes phosphorylated and activated by LCK. Following this, a large number of signaling molecules are recruited, ultimately leading to lymphokine production. LCK also contributes to signaling by other receptor molecules. Associates directly with the cytoplasmic tail of CD2, which leads to hyperphosphorylation and activation of LCK. Also plays a role in the IL2 receptor-linked signaling pathway that controls the T-cell proliferative response. Binding of IL2 to its receptor results in increased activity of LCK. Is expressed at all stages of thymocyte development and is required for the regulation of maturation events that are governed by both pre-TCR and mature α βTCR. Phosphorylates other substrates including RUNX3, PTK2B/PYK2, the microtubule-associated protein MAPT, RHOH, or TYROBP. | −6.4 |
| 3A4O | PD | Tyrosine-protein kinase LYN | Nonreceptor tyrosine-protein kinase that transmits signals from cell surface receptors and plays an important role in the regulation of innate and adaptive immune responses, hematopoiesis, responses to growth factors and cytokines, integrin signaling, but also responses to DNA damage and genotoxic agents. Functions primarily as negative regulator, but can also function as activator, depending on the context. Required not only for the initiation of the B-cell response, but also for its downregulation and termination. Plays an important role in the regulation of B-cell differentiation, proliferation, survival, and apoptosis and is important for immune self-tolerance. Acts downstream of several immune receptors, including the BCR, CD79A, CD79B, CD5, CD19, CD22, FCER1, FCGR2, FCGR1A, TLR2, and TLR4. Plays a role in the inflammatory response to bacterial LPS. Mediates the responses to cytokines and growth factors in hematopoietic progenitors, platelets, erythrocytes, and in mature myeloid cells, such as DCs, neutrophils, and eosinophils. Acts downstream of EPOR, KIT, MPL, the chemokine receptor CXCR4 as well as the receptors for IL3, IL5, and CSF2. Plays an important role in integrin signaling. Regulates cell proliferation, survival, differentiation, migration, adhesion, degranulation, and cytokine release. Downregulates signaling pathways by phosphorylation of ITIMs, which then serve as binding sites for phosphatases, such as PTPN6/SHP-1, PTPN11/SHP-2, and INPP5D/SHIP-1, which modulate signaling by dephosphorylation of kinases and their substrates. Phosphorylates LIME1 in response to CD22 activation. Phosphorylates BTK, CBL, CD5, CD19, CD72, CD79A, CD79B, CSF2RB, DOK1, HCLS1, LILRB3/PIR-B, MS4A2/FCER1B, PTK2B/PYK2, SYK, and TEC. Promotes phosphorylation of SIRPA, PTPN6/SHP-1, PTPN11/SHP-2, and INPP5D/SHIP-1. Mediates phosphorylation of the BCR-ABL fusion protein. Required for rapid phosphorylation of FER in response to FCER1 activation. Mediates KIT phosphorylation. Acts as an effector of EPOR in controlling KIT expression and may play a role in erythroid differentiation during the switch between proliferation and maturation. Depending on the context, activates or inhibits several signaling cascades. Regulates phosphatidylinositol 3-kinase activity and AKT1 activation. Regulates activation of the MAP kinase signaling cascade, including activation of MAP2K1/MEK1, MAPK1/ERK2, MAPK3/ERK1, MAPK8/JNK1, and MAPK9/JNK2. Mediates activation of STAT5A and/or STAT5B. Phosphorylates LPXN on “Tyr-72”. | −8.8 |
| 3BPR | PD | Tyrosine-protein kinase MER | Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to several ligands including LGALS3, TUB, TULP1, or GAS6. Regulates many physiological processes including cell survival, migration, differentiation, and phagocytosis of apoptotic cells (efferocytosis). Ligand binding at the cell surface induces autophosphorylation of MERTK on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with GRB2 or PLCG2 and induces phosphorylation of MAPK1, MAPK2, FAK/PTK2, or RAC1. MERTK signaling plays a role in various processes such as macrophage clearance of apoptotic cells, platelet aggregation, cytoskeleton reorganization, and engulfment. Functions in the RPE as a regulator of rod outer segments fragments phagocytosis. Plays also an important role in inhibition of TLR-mediated innate immune response by activating STAT1, which selectively induces production of suppressors of cytokine signaling SOCS1 and SOCS3. | −8.1 |
| 1XBB | PD | Tyrosine-protein kinase SYK | Nonreceptor tyrosine kinase that mediates signal transduction downstream of a variety of transmembrane receptors including classic immunoreceptors like the BCR. Regulates several biological processes including innate and adaptive immunity, cell adhesion, osteoclast maturation, platelet activation, and vascular development. Assembles into signaling complexes with activated receptors at the plasma membrane via interaction between its SH2 domains and the receptor tyrosine-phosphorylated ITAM domains. The association with the receptor can also be indirect and mediated by APs containing ITAM or partial hemITAM domains. The phosphorylation of the ITAM domains is generally mediated by SRC subfamily kinases upon engagement of the receptor. More rarely signal transduction via SYK could be ITAM independent. Direct downstream effectors phosphorylated by SYK include VAV1, PLCG1, PI-3-kinase, LCP2, and BLNK. Initially identified as essential in BCR signaling, it is necessary for the maturation of B-cells most probably at the pro-B to pre-B transition. Activated upon BCR engagement, it phosphorylates and activates BLNK, an adapter linking the activated BCR to downstream signaling adapters and effectors. It also phosphorylates and activates PLCG1 and the PKC signaling pathway. It also phosphorylates BTK and regulates its activity in B-cell antigen receptor-coupled signaling. Besides its function, downstream of BCR plays also a role in TCR signaling. Plays also a crucial role in the innate immune response to fungal, bacterial, and viral pathogens. It is for instance activated by the membrane lectin CLEC7A. Upon stimulation by fungal proteins, CLEC7A together with SYK activates immune cells inducing the production of ROS. Also activates the inflammasome and NF-κB-mediated transcription of chemokines and cytokines in presence of pathogens. Regulates neutrophil degranulation and phagocytosis through activation of the MAPK signaling cascade. Also mediates the activation of DCs by cell necrosis stimuli. Also involved in mast cells activation. Also functions downstream of receptors mediating cell adhesion. Relays for instance, integrin-mediated neutrophils and macrophages activation and P-selectin receptor/SELPG-mediated recruitment of leukocytes to inflammatory loci. Plays also a role in nonimmune processes. It is for instance involved in vascular development where it may regulate blood and lymphatic vascular separation. It is also required for osteoclast development and function. Functions in the activation of platelets by collagen, mediating PLCG2 phosphorylation and activation. May be coupled to the collagen receptor by the ITAM domain-containing FCER1G. Also activated by the membrane lectin CLEC1B that is required for activation of platelets by PDPN/podoplanin. Involved in platelet adhesion being activated by ITGB3 engaged by fibrinogen. | −7.8 |
| 1U59 | PD | Tyrosine-protein kinase ZAP-70 | Tyrosine kinase that plays an essential role in regulation of the adaptive immune response. Regulates motility, adhesion, and cytokine expression of mature T-cells as well as thymocyte development. Contributes also to the development and activation of primary B-lymphocytes. When APCs activate TCR, a series of phosphorylations lead to the recruitment of ZAP70 to the doubly phosphorylated TCR component CD247/CD3Z through ITAM motif at the plasma membrane. This recruitment serves to localization to the stimulated TCR and to relieve its autoinhibited conformation. Release of ZAP70 active conformation is further stabilized by phosphorylation mediated by LCK. Subsequently, ZAP70 phosphorylates at least two essential APs: LAT and LCP2. In turn, a large number of signaling molecules are recruited and ultimately lead to lymphokine production, T-cell proliferation, and differentiation. Furthermore, ZAP70 controls cytoskeleton modifications, adhesion, and mobility of T-lymphocytes, thus ensuring correct delivery of effectors to the APC. ZAP70 is also required for TCR-CD247/CD3Z internalization and degradation through interaction with the E3 ubiquitin-protein ligase CBL and APs SLA and SLA2. Thus, ZAP70 regulates both T-cell activation–switch on and switch off–by modulating TCR expression at the T-cell surface. During thymocyte development, ZAP70 promotes survival and cell-cycle progression of developing thymocytes before positive selection (when cells are still CD4/CD8 double negative). Also, ZAP70-dependent signaling pathway may also contribute to primary B-cells formation and activation through BCR. | −8.5 |
| 2F71 | PD | Tyrosine-protein phosphatase nonreceptor type 1 | Tyrosine-protein phosphatase that acts as a regulator of endoplasmic reticulum unfolded protein response. Mediates dephosphorylation of EIF2AK3/p-ERK; inactivating the protein kinase activity of EIF2AK3/p-ERK. May play an important role in CKII- and p60c-src-induced signal transduction cascades. May regulate the EFNA5-EPHA3 signaling pathway which modulates cell reorganization and cell–cell repulsion. | −7.3 |
| 3O5X | PD | Tyrosine-protein phosphatase nonreceptor type 11 | Acts downstream of various receptor and cytoplasmic PTKs to participate in the signal transduction from the cell surface to the nucleus. Dephosphorylates ROCK2 at Tyr-722 resulting in stimulation of its RhoA-binding activity. | −7.7 |
| 3FCI | PD | Uracil-DNA glycosylase | Excises uracil residues from the DNA which can arise as a result of misincorporation of dUMP residues by DNA polymerase or deamination of cytosine | −7.9 |
| 3MI2 | PD | Uridine 5′-monophosphate synthase | N/A | −6.3 |
| 3MHW | PD | Urokinase-type plasminogen activator | Specifically cleave the zymogen plasminogen to form the active enzyme plasmin. | −8.3 |
| 3EWH | PD | Vascular endothelial growth factor receptor 2 | Tyrosine-protein kinase that acts as a cell-surface receptor for VEGFA, VEGFC, and VEGFD. Plays an essential role in the regulation of angiogenesis, vascular development, vascular permeability, and embryonic hematopoiesis. Promotes proliferation, survival, migration, and differentiation of ECs. Promotes reorganization of the actin cytoskeleton. Isoforms lacking a transmembrane domain, such as isoforms 2 and 3, may function as decoy receptors for VEGFA, VEGFC, and/or VEGFD. Isoform 2 plays an important role as negative regulator of VEGFA- and VEGFC-mediated lymph angiogenesis by limiting the amount of free VEGFA and/or VEGFC and preventing their binding to FLT4. Modulates FLT1 and FLT4 signaling by forming heterodimers. Binding of vascular growth factors to isoform 1 leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules DAG and inositol 1,4,5-trisphosphate and the activation of PKC. Mediates activation of MAPK1/ERK2, MAPK3/ERK1, and the MAPK signaling pathway as well as of the AKT1 signaling pathway. Mediates phosphorylation of PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase, reorganization of the actin cytoskeleton and activation of PTK2/FAK1. Required for VEGFA-mediated induction of NOS2 and NOS3, leading to the production of the signaling molecule NO by ECs. Phosphorylates PLCG1. Promotes phosphorylation of FYN, NCK1, NOS3, PIK3R1, PTK2/FAK1, and SRC. | −8.1 |
| 1DB1 | PD | Vitamin D3 receptor | Nuclear hormone receptor. Transcription factor that mediates the action of vitamin D3 by controlling the expression of hormone-sensitive genes. Regulates transcription of hormone-sensitive genes via its association with the WINAC complex, a chromatin-remodeling complex. Recruited to promoters via its interaction with the WINAC complex subunit BAZ1B/WSTF, which mediates the interaction with acetylated histones, an essential step for VDR–promoter association. Plays a central role in calcium homeostasis. | −9.1 |
| 2H6Q | PD | WD repeat-containing protein 5 | Contributes to histone modification. May position the N-terminus of histone H3 for efficient trimethylation at “Lys-4”. As part of the MLL1-MLL complex, it is involved in methylation and dimethylation at “Lys-4” of histone H3. H3 “Lys-4” methylation represents a specific tag for epigenetic transcriptional activation. As part of the NSL complex, it may be involved in acetylation of nucleosomal histone H4 on several lysine residues. May regulate osteoblasts differentiation. | −7.9 |
| 2IN6 | PD | Wee1-like protein kinase | Acts as a negative regulator of entry into mitosis (G2–M transition) by protecting the nucleus from cytoplasmically activated cyclin B1-complexed CDK1 before the onset of mitosis by mediating phosphorylation of CDK1 on “Tyr-15”. Specifically phosphorylates and inactivates cyclin B1-complexed CDK1 reaching a maximum during G2 phase and a minimum as cells enter M phase. Phosphorylation of cyclin B1-CDK1 occurs exclusively on “Tyr-15” and phosphorylation of monomeric CDK1 does not occur. Its activity increases during S and G2 phases and decreases at M phase when it is hyperphosphorylated. A correlated decrease in protein level occurs at M/G1 phase, probably due to its degradation. | −7.7 |
| 2E1Q, 2E1Q_3, 2E1Q_2 | PK | Xanthine dehydrogenase/oxidase | Key enzyme in purine degradation. Catalyzes the oxidation of hypoxanthine to xanthine. Catalyzes the oxidation of xanthine to uric acid. Contributes to the generation of ROS. Has also low oxidase activity toward aldehydes (in vitro). | −9.3 to 14.2 |
| 2RR4 | PD | Zinc finger CW-type PWWP domain protein 1 | N/A | −7.2 |
| 3KQ0 | PK | α-1-Acid glycoprotein 1 | Functions as transport protein in the blood stream. Binds various ligands in the interior of its β-barrel domain. Also binds synthetic drugs and influences their distribution and availability in the body. Appears to function in modulating the activity of the immune system during the acute-phase reaction. | −9.2 |
| 3APV, 3APU, 3APW | PK | α-1-Acid glycoprotein 2 | Functions as transport protein in the blood stream. Binds various hydrophobic ligands in the interior of its β-barrel domain. Also binds synthetic drugs and influences their distribution and availability. Appears to function in modulating the activity of the immune system during the acute-phase reaction. | −8.7 to −6.6 |
Notes: The prediction of targets of UA was performed using the DDI-CPI tool (http://cpi.bio-x.cn/ddi/)20,21 that is a web-based server used to predict drug–drug interaction via chemical–protein interactome. The protein targets were obtained from a third-party protein structure database named PDBBind (http://sw16.im.med.umich.edu/databases/pdbbind/index.jsp),20,21 which was based on the contents of PDB.
Abbreviations: AC-LL, acidic cluster-dileucine; ACLY, ATP citrate lyase; AD, Alzheimer’s disease; ADH, alcohol dehydrogenase; AJ, adherens junction; ALDH, aldehyde dehydrogenase; ANDR, androgen receptor; ANF, atrial natriuretic factor; AP, adapter protein; APAF, apoptosis-activating factor; APC/C, anaphase-promoting complex/cyclosome; APCs, antigen-presenting cells; APP, amyloid precursor protein; AT, antithrombin; BCR, B-cell receptor; CAD, C-terminal transactivation domain; CBC, cap-binding complex; CDK, cyclin-dependent kinase; CDNB, 1-chloro-2,4-dinitrobenzene; CE, cornified envelope; CFIm, cleavage factor Im; CGRP, calcitonin-gene-related peptide; CNS, central nervous system; CoA, coenzyme; COMP, cartilage oligomeric matrix protein; CPI, chemical–protein interactome; CSF1, colony-stimulating factor; 1; CSF1R, colony-stimulating factor 1 receptor; CTRs, constitutive transport elements; CYP7A1, cholesterol 7-α-hydroxylase gene; dA, deoxyadenosine; DAG, diacylglycerol; dC, deoxycytidine; DCs, dendritic cells; DDI, drug–drug interaction; dG, deoxyguanosine; DHBA, dihydroxybenzoic acid; DISC, death-inducing signaling complex; DRG, dorsal root ganglion; DSBs, double-strand breaks; ECs, endothelial cells; EBNA2, Epstein–Barr virus nuclear antigen 2; EBV, Epstein–Barr virus; ECM, extracellular matrix; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; EJC, exon junction complex; ENaC, epithelial sodium channel; ENCCs, enteric neural crest cell; EPO, erythropoietin; EPOR, erythropoietin receptor; ER, estrogen receptor; ERAD, endoplasmic reticulum-associated degradation; ERE, estrogen response element; ERQC, ER quality control compartment; FABP, fatty acid-binding protein; FKBPs, FK506-binding proteins; FPP, farnesyl diphosphate; GABA, γ-aminobutyric acid; GAPs, GTPase-activating proteins; GC, glucocorticoids; GEF, guanine nucleotide exchange factor; GH, growth hormone; GHR, growth hormone receptor; GnRH, gonadotropin-releasing hormone; GPCRs, G-protein coupled receptors; GRE, glucocorticoid response element; HAT, histone acetyltransferase; HCAECs, human coronary artery endothelial cells; HDAC, histone deacetylase; hESCs, human embryonic stem cells; Hh, Hedgehog; HNE, hydroxynonenal; HRR, homologous recombination repair; HSF4, heat shock factor protein 4; HT, hydroxytryptamine; IBABP, intestinal bile acid-binding protein; IGF, insulin-like growth factor; IL, interleukin; INS, insulin; IRS, insulin receptor substrate; ITAMs, immunoreceptor tyrosine-based activation motifs; ITIM, immunoreceptor tyrosine-based inhibitory motifs; ILVs, intralumenal vesicles; JNK, c-Jun N-terminal kinase; KA, kynurenic acid; LBPA, lysobisphosphatidic acid; LEPR, leptin receptor; lincRNA-p21, long intergenic noncoding RNA p21; Lp(a), lipoprotein(a); LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MCFA, matrix-cell focal adhesions; MDK, midkine; m3G, trimethylguanosine; MHC, major histocompatibility complex; miRNAs, microRNAs; MMA, monomethylarsonic acid; MSN, moesin; MTA, S-methyl-5′-thioadenosine; MVB, multivesicular body; NAA, N-acetylaspartic acid; NAAG, N-aceylaspartyl glutamate; NAALADase, N-acetylated-α-linked-acidic dipeptidase; NFAT, nuclear factor of activated T-cells; NGF, nerve growth factor; NHEJ, nonhomologous end-joining; NK, natural killer; NLRs, nod-like receptors; NMD, nonsense-mediated mRNA decay; NMJ, neuromuscular junction; NO, nitric oxide; NSCLC, non-small-cell lung cancer; NSL, nonspecific lethal; OAA, oxaloacetate; OLs, oligodendrocytes; PAK, p21-activated kinase; PALLD, paladin; PAPS, 3′-phospho-5′-adenylylsulfate; PARP, poly(ADP-ribose) polymerase; PcG, polycomb group; PDB, Protein Data Bank; PDGF, platelet-derived growth factor; PEP, phosphoenolpyruvate; PHB, prohibitin; PI3K, phosphoinositide-3-kinase; PIP3, phosphatidylinositol-3,4,5-triphosphate; PKA, protein kinase A; PKC, protein kinase C; PNRC, perinuclear recycling compartment; PPARG, peroxisome proliferator-activated receptor γ; PP1, protein phosphatase type 1; PPT, pedunculopontine tegmental; PRB, progesterone receptor isoform B; PRLR, prolactin receptor; PTK, protein-tyrosine kinase; PTN, pleiotrophin; PXN, paxillin; RA, rapidly adapting; RAREs, retinoic acid response elements; RBs, ruffled borders; REM, rapid eye movement; RHO, rhodopsin; ROS, reactive oxygen species; RPE, retinal pigment epithelium; SAC, spindle assembly checkpoint; SAP, stress-activated protein kinase; sHsps, small heat shock protein; SMD, Staufen-mediated mRNA decay; SOCE, store-operated Ca2+ entry; SOICR, store overload-induced Ca2+ release; SREBPs, sterol regulatory element-binding proteins; SRF, serum response factor; STAT, signal transducers and activators of transcription; TCR, T-cell receptor; tEr, transitional endoplasmic reticulum; TGN, trans-Golgi network; Th1, T-helper 1; Th2, T-helper 2; THPO, thrombopoietin; TJ, tight junction; TLR, toll-like receptor; TNF, tumor necrosis factor; TPA, 12-O-tetradecanoylphorbol-13-acetate; UA, ursolic acid; VDAC, voltage-dependent anion channel; VEGF, vascular endothelial growth factor; VTCs, vesicular tubular clusters; VTN, vitronectin.
Table 3.
The top enriched clusters (enrichment score >3) by the DAVID database for the target list of UA derived from molecular docking calculations
| Category | Term | Count | % | P-value | Genes | Fold enrichment | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|
| Annotation cluster 1 | Enrichment score: 31.55 | |||||||
| UP_SEQ_FEATURE | ATP binding site | 85 | 18.20 | 1.71E-43 | SPG7, TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAT1A, MAP3K9, PAK4, PRKACA, PAK1, INSR, AKT2, SYK, PRKCA, EGFR, RET, SGK1, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PRKCQ, NME2, NME1, MAPK3, LCK, MAPK9, MAPK8, FGFR2, CHKA, FGFR1, ERBB4, NEK2, CHKB, ERBB2, MAPKAPK3, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, ZAP70, CSF1R, PDK1, ITK, MAP2K1, HCK, PDK3, TGFBR1, PDK4, MET, RAF1, MAPK10, KDR, KHK, RPS6KA1, PLK1, FYN, MAPK14, GSK3B, DYRK1A, GRK6, JAK1, JAK3, TNK2, MERTK, ABL1 | 6.42 | 9.33E-41 | 2.87E-40 |
| SP_PIR_KEYWORDS | Kinase | 89 | 19.06 | 5.29E-39 | TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, INSR, AKT2, SYK, PIK3CG, PRKCA, EGFR, RET, SGK1, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PRKCQ, PNKP, NME2, NME1, ADK, MAPK3, LCK, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, CDK5R1, ERBB4, NEK2, CHKB, ERBB2, DCK, MAPKAPK3, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, ZAP70, CSF1R, PDK1, ITK, MAP2K1, HCK, PDK3, TGFBR1, PDK4, MET, RAF1, MAPK10, KDR, KHK, RPS6KA1, PLK1, FYN, MAPK14, GSK3B, DYRK1A, GRK6, JAK1, JAK3, TNK2, MERTK, ABL1, PLAU | 5.33 | 1.40E-36 | 7.68E-36 |
| INTERPRO | Protein kinase, ATP binding site | 77 | 16.49 | 1.08E-37 | TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, INSR, AKT2, SYK, PRKCA, EGFR, SGK1, RET, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PRKCQ, MAPK3, LCK, MAPK9, MAPK8, FGFR2, FGFR1, ERBB4, NEK2, ERBB2, MAPKAPK3, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, ZAP70, CSF1R, ITK, MAP2K1, HCK, TGFBR1, MET, RAF1, MAPK10, KDR, RPS6KA1, FYN, PLK1, MAPK14, GSK3B, DYRK1A, GRK6, JAK1, ABCC1, JAK3, TNK2, MERTK, ABL1 | 6.05 | 9.68E-35 | 1.68E-34 |
| UP_SEQ_FEATURE | Protein kinase | 72 | 15.42 | 5.38E-36 | TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, INSR, AKT2, SYK, PRKCA, EGFR, SGK1, RET, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, MAPK1, PRKCQ, MAPK3, LCK, MAPK9, MAPK8, FGFR2, FGFR1, ERBB4, NEK2, ERBB2, MAPKAPK3, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, ZAP70, CSF1R, ITK, MAP2K1, TGFBR1, HCK, MET, RAF1, MAPK10, KDR, FYN, PLK1, MAPK14, GSK3B, DYRK1A, GRK6, TNK2, MERTK, ABL1 | 6.28 | 2.20E-33 | 9.04E-33 |
| INTERPRO | Protein kinase | 76 | 16.27 | 2.09E-35 | TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, INSR, AKT2, SYK, PRKCA, EGFR, SGK1, RET, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PRKCQ, MAPK3, LCK, MAPK9, MAPK8, FGFR2, FGFR1, ERBB4, NEK2, ERBB2, MAPKAPK3, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, ZAP70, CSF1R, ITK, MAP2K1, HCK, TGFBR1, MET, RAF1, MAPK10, KDR, RPS6KA1, FYN, PLK1, MAPK14, GSK3B, DYRK1A, GRK6, JAK1, JAK3, TNK2, MERTK, ABL1 | 5.71 | 9.38E-33 | 3.26E-32 |
| UP_SEQ_FEATURE | Nucleotide phosphate-binding region | 96 | 20.56 | 1.27E-32 | SPG7, TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAT1A, MAP3K9, PAK4, ABCB10, PRKACA, PAK1, INSR, AKT2, SYK, PRKCA, EGFR, RET, SGK1, KIF11, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PNKP, PRKCQ, LCK, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, MAPKAPK3, DCK, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, TAP1, ZAP70, CSF1R, PDK1, ITK, DHX9, MAP2K1, HCK, PDK3, TGFBR1, PDK4, MET, RAF1, ACACB, MAPK10, ABCB6, KDR, GART, KHK, RPS6KA1, VCP, PLK1, FYN, MAPK14, GSK3B, DYRK1A, UBA3, GRK6, JAK1, TNK2, JAK3, MERTK, ABL1, CLCN5 | 4.08 | 4.17E-30 | 2.14E-29 |
| GOTERM_BP_FAT | Protein amino acid phosphorylation | 88 | 18.84 | 1.18E-29 | TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, APP, CSNK2A1, CXCR4, MAP3K9, PAK4, PRKACA, PAK1, BRD4, INSR, AKT2, SYK, PIK3CG, PRKCA, EGFR, RET, SGK1, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PRKCQ, LCK, F2, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CDK5R1, ERBB4, NEK2, ERBB2, MAPKAPK3, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, BCL2, TEK, ZAP70, CSF1R, PDK1, ITK, MAP2K1, HCK, PDK3, TGFBR1, PDK4, MET, BIRC7, RAF1, MAPK10, KDR, PTPN11, RPS6KA1, PLK1, FYN, MAPK14, GSK3B, DYRK1A, GRK6, JAK1, JAK3, TNK2, MERTK, ABL1 | 4.00 | 3.79E-26 | 2.13E-26 |
| GOTERM_MF_FAT | Protein kinase activity | 82 | 17.56 | 7.32E-27 | NRP1, TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, INSR, AKT2, SYK, PRKCA, EGFR, RET, SGK1, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PRKCQ, MAPK3, LCK, MAPK9, MAPK8, FGFR2, FGFR1, CDK5R1, ERBB4, NEK2, ERBB2, MAPKAPK3, FKBP1A, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, ZAP70, CSF1R, PDK1, ITK, MAP2K1, HCK, PDK3, TGFBR1, PDK4, MET, RAF1, MAPK10, KDR, RPS6KA1, FYN, PLK1, MAPK14, GSK3B, DYRK1A, GRK6, JAK1, JAK3, TNK2, MERTK, ABL1 | 3.90 | 6.19E-24 | 1.13E-23 |
| GOTERM_BP_FAT | Phosphorylation | 90 | 19.27 | 2.98E-25 | TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, APP, CSNK2A1, CXCR4, MAP3K9, PAK4, PRKACA, PAK1, BRD4, INSR, AKT2, SYK, PIK3CG, PRKCA, EGFR, RET, SGK1, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PRKCQ, PNKP, NME2, NME1, LCK, F2, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CDK5R1, ERBB4, NEK2, ERBB2, MAPKAPK3, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, BCL2, TEK, ZAP70, CSF1R, PDK1, ITK, MAP2K1, HCK, PDK3, TGFBR1, PDK4, MET, BIRC7, RAF1, MAPK10, KDR, PTPN11, RPS6KA1, PLK1, FYN, MAPK14, GSK3B, DYRK1A, GRK6, JAK1, TNK2, JAK3, MERTK, ABL1 | 3.41 | 3.20E-22 | 5.41E-22 |
| GOTERM_BP_FAT | Phosphate metabolic process | 99 | 21.20 | 1.53E-24 | TTK, AURKA, BTK, CSNK2A2, APP, CSNK2A1, MAP3K9, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, PPP1CC, WEE1, MAPK1, PNKP, PPP1CA, NME2, NME1, F2, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CDK5R1, ERBB4, NEK2, ERBB2, MAPKAPK3, CHEK1, ADRBK1, MAPKAPK2, CHEK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CSF1R, ITK, MAP2K1, TGFBR1, MET, KDR, PTPN11, PLK1, DYRK1A, GRK6, TNK2, PTPN1, MERTK, ABL1, PTEN, AKT1, PDPK1, CXCR4, PAK4, PAK1, BRD4, AKT2, PRKCA, PPP2R1A, SGK1, LYN, PRKCI, CDK6, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, LCK, KIT, IGF1R, DAPP1, PTK2B, BCL2, PPP2CA, PTPRB, PDK1, HCK, PDK3, PDK4, BIRC7, RAF1, MAPK10, DUSP3, RPS6KA1, FYN, MAPK14, GSK3B, JAK1, JAK3 | 3.09 | 1.23E-21 | 2.78E-21 |
| GOTERM_BP_FAT | Phosphorus metabolic process | 99 | 21.20 | 1.53E-24 | TTK, AURKA, BTK, CSNK2A2, APP, CSNK2A1, MAP3K9, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, PPP1CC, WEE1, MAPK1, PNKP, PPP1CA, NME2, NME1, F2, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CDK5R1, ERBB4, NEK2, ERBB2, MAPKAPK3, CHEK1, ADRBK1, MAPKAPK2, CHEK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CSF1R, ITK, MAP2K1, TGFBR1, MET, KDR, PTPN11, PLK1, DYRK1A, GRK6, TNK2, PTPN1, MERTK, ABL1, PTEN, AKT1, PDPK1, CXCR4, PAK4, PAK1, BRD4, AKT2, PRKCA, PPP2R1A, SGK1, LYN, PRKCI, CDK6, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, LCK, KIT, IGF1R, DAPP1, PTK2B, BCL2, PPP2CA, PTPRB, PDK1, HCK, PDK3, PDK4, BIRC7, RAF1, MAPK10, DUSP3, RPS6KA1, FYN, MAPK14, GSK3B, JAK1, JAK3 | 3.09 | 1.23E-21 | 2.78E-21 |
| Annotation cluster 2 | Enrichment score: 23.53 | |||||||
| SP_PIR_KEYWORDS | Tyrosine protein kinase | 36 | 7.71 | 1.26E-29 | FGFR2, FGFR1, ERBB4, ERBB2, TTK, KIT, EPHB4, SRC, BTK, IGF1R, PTK2, PTK2B, TEK, ZAP70, INSR, SYK, CSF1R, EGFR, ITK, RET, MAP2K1, LYN, HCK, MET, ALK, WEE1, KDR, TYK2, FYN, DYRK1A, LCK, JAK1, JAK3, TNK2, MERTK, ABL1 | 13.36 | 1.33E-27 | 1.83E-26 |
| INTERPRO | Tyrosine protein kinase, active site | 32 | 6.85 | 9.13E-25 | FGFR2, FGFR1, ERBB4, ERBB2, KIT, EPHB4, SRC, BTK, IGF1R, PTK2, PTK2B, TEK, ZAP70, INSR, SYK, CSF1R, EGFR, ITK, RET, LYN, HCK, MET, ALK, KDR, TYK2, FYN, LCK, JAK1, JAK3, TNK2, MERTK, ABL1 | 11.79 | 2.73E-22 | 1.42E-21 |
| INTERPRO | Tyrosine protein kinase | 34 | 7.28 | 3.37E-24 | FGFR2, FGFR1, ERBB4, ERBB2, KIT, EPHB4, SRC, BTK, IRAK4, IGF1R, PTK2, PTK2B, MAP3K9, TEK, ZAP70, INSR, SYK, CSF1R, EGFR, ITK, RET, LYN, HCK, MET, ALK, KDR, TYK2, FYN, LCK, JAK1, JAK3, TNK2, MERTK, ABL1 | 10.30 | 7.57E-22 | 5.26E-21 |
| SMART | TyrKc | 34 | 7.28 | 1.43E-22 | FGFR2, FGFR1, ERBB4, ERBB2, KIT, EPHB4, SRC, BTK, IRAK4, IGF1R, PTK2, PTK2B, MAP3K9, TEK, ZAP70, INSR, SYK, CSF1R, EGFR, ITK, RET, LYN, HCK, MET, ALK, KDR, TYK2, FYN, LCK, JAK1, JAK3, TNK2, MERTK, ABL1 | 8.93 | 2.41E-20 | 1.74E-19 |
| GOTERM_MF_FAT | Protein tyrosine kinase activity | 38 | 8.14 | 3.94E-20 | FGFR2, FGFR1, NRP1, ERBB4, ERBB2, TTK, KIT, EPHB4, SRC, BTK, IRAK4, IGF1R, PTK2, PTK2B, TEK, ZAP70, INSR, SYK, CSF1R, EGFR, ITK, RET, MAP2K1, LYN, HCK, MET, ALK, WEE1, KDR, TYK2, FYN, DYRK1A, LCK, JAK1, JAK3, TNK2, MERTK, ABL1 | 6.60 | 1.67E-17 | 6.10E-17 |
| Annotation cluster 3 | Enrichment score: 16.58 | |||||||
| UP_SEQ_FEATURE | Nucleotide phosphate- binding region: ATP | 96 | 20.56 | 1.27E-32 | SPG7, TTK, AURKA, BTK, CSNK2A2, AKT1, PDPK1, CSNK2A1, MAT1A, MAP3K9, PAK4, ABCB10, PRKACA, PAK1, INSR, AKT2, SYK, PRKCA, EGFR, RET, SGK1, KIF11, LYN, BRAF, ROCK1, PIM1, PRKCI, CDK6, ALK, CDK5, WEE1, CDK2, PRKCB, TYK2, MAPK1, PNKP, PRKCQ, LCK, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, MAPKAPK3, DCK, ADRBK1, CHEK1, MAPKAPK2, KIT, CHEK2, EPHB4, SRC, IRAK4, IGF1R, PTK2, PTK2B, TEK, TAP1, ZAP70, CSF1R, PDK1, ITK, DHX9, MAP2K1, HCK, PDK3, TGFBR1, PDK4, MET, RAF1, ACACB, MAPK10, ABCB6, KDR, GART, KHK, RPS6KA1, VCP, PLK1, FYN, MAPK14, GSK3B, DYRK1A, UBA3, GRK6, JAK1, TNK2, JAK3, MERTK, ABL1, CLCN5 | 4.08 | 4.17E-30 | 2.14E-29 |
| SP_PIR_KEYWORDS | ATP binding | 107 | 22.91 | 7.66E-29 | IDE, TTK, AURKA, BTK, CSNK2A2, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, WEE1, MAPK1, PNKP, NME2, NME1, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CSF1R, DHX9, ITK, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, KHK, RPS6KA1, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 3.32 | 6.74E-27 | 1.11E-25 |
| SP_PIR_KEYWORDS | Nucleotide binding | 118 | 25.27 | 1.72E-26 | HRAS, IDE, TTK, AURKA, BTK, CSNK2A2, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, NUDT2, RET, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, PDE5A, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, GNAI1, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, RAC1, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, CDC42, PDPK1, NT5M, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, GIMAP2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, PCK1, KHK, PDE2A, RPS6KA1, PYGL, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 2.88 | 1.14E-24 | 2.50E-23 |
| GOTERM_MF_FAT | ATP binding | 110 | 23.56 | 4.31E-15 | IDE, TTK, AURKA, BTK, CSNK2A2, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, KHK, RPS6KA1, FYN, PYGL, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 2.15 | 5.23E-13 | 6.69E-12 |
| GOTERM_MF_FAT | Nucleoside binding | 116 | 24.84 | 6.62E-15 | IDE, TTK, AURKA, PNP, BTK, CSNK2A2, KDM1A, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, PDE5A, MAPK3, MAPK9, MAPK8, FGFR2, XDH, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CDA, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, PCK1, KHK, RPS6KA1, PYGL, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 2.08 | 6.26E-13 | 1.03E-11 |
| GOTERM_MF_FAT | Adenyl ribonucleotide binding | 110 | 23.56 | 1.07E-14 | IDE, TTK, AURKA, BTK, CSNK2A2, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, KHK, RPS6KA1, FYN, PYGL, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 2.12 | 9.11E-13 | 1.67E-11 |
| GOTERM_MF_FAT | Purine nucleoside binding | 114 | 24.41 | 2.63E-14 | IDE, TTK, AURKA, BTK, CSNK2A2, KDM1A, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, PDE5A, MAPK3, MAPK9, MAPK8, FGFR2, XDH, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, PCK1, KHK, RPS6KA1, PYGL, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 2.05 | 2.02E-12 | 4.07E-11 |
| GOTERM_MF_FAT | Adenyl nucleotide binding | 112 | 23.98 | 5.87E-14 | IDE, TTK, AURKA, BTK, CSNK2A2, KDM1A, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, MAPK3, MAPK9, MAPK8, FGFR2, XDH, FGFR1, CHKA, ERBB4, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, KHK, RPS6KA1, FYN, PYGL, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 2.05 | 3.82E-12 | 9.08E-11 |
| GOTERM_MF_FAT | Nucleotide binding | 137 | 29.34 | 4.05E-12 | NCBP2, HRAS, HMGCR, IDE, TTK, AURKA, BTK, CSNK2A2, KDM1A, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, NUDT2, ROCK1, BRAF, PIM1, PPARGC1A, WEE1, POR, PNKP, MAPK1, NME2, NME1, PDE5A, MAPK3, MAPK9, MAPK8, XDH, FGFR2, FGFR1, CHKA, HSD17B10, ME2, ERBB4, GNAI1, NEK2, FARS2, CHKB, ERBB2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, RAC1, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, NXF1, ACACB, ABCB6, KDR, GART, ATP7A, DHFR, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, PARP1, UBE2E1, CLCN5, HSP90AB1, SPG7, AKT1, CDC42, PDPK1, PARN, NT5M, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, PRKCI, TP53, CFTR, CDK6, CBR3, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, G6PD, ADK, LCK, DCK, ADH5, KIT, TYMS, IGF1R, PTK2B, ADH4, TAP1, TGM2, GIMAP2, PDK1, HCK, PDK3, PDK4, SIRT5, RAF1, MAPK10, PCK1, KHK, RPS6KA1, PYGL, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 1.76 | 2.28E-10 | 6.26E-09 |
| GOTERM_MF_FAT | Ribonucleotide binding | 118 | 25.27 | 9.49E-12 | HRAS, IDE, TTK, AURKA, BTK, CSNK2A2, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, NUDT2, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, PDE5A, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, GNAI1, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, RAC1, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, CDC42, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, GIMAP2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, PCK1, KHK, RPS6KA1, PYGL, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 1.85 | 4.72E-10 | 1.47E-08 |
| GOTERM_MF_FAT | Purine ribonucleotide binding | 118 | 25.27 | 9.49E-12 | HRAS, IDE, TTK, AURKA, BTK, CSNK2A2, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, NUDT2, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, PDE5A, MAPK3, MAPK9, MAPK8, FGFR2, FGFR1, CHKA, ERBB4, GNAI1, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, RAC1, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, CDC42, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, GIMAP2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, PCK1, KHK, RPS6KA1, PYGL, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 1.85 | 4.72E-10 | 1.47E-08 |
| GOTERM_MF_FAT | Purine nucleotide binding | 120 | 25.70 | 3.44E-11 | HRAS, IDE, TTK, AURKA, BTK, CSNK2A2, KDM1A, CSNK2A1, MAT1A, MAP3K9, ABCB10, PRKACA, INSR, SYK, PIK3CG, EGFR, RET, NUDT2, BRAF, ROCK1, PIM1, WEE1, PNKP, MAPK1, NME2, NME1, PDE5A, MAPK3, MAPK9, MAPK8, FGFR2, XDH, FGFR1, CHKA, ERBB4, GNAI1, NEK2, CHKB, ERBB2, FARS2, MAPKAPK3, CHEK1, ADRBK1, CHEK2, MAPKAPK2, EPHB4, SRC, IRAK4, PTK2, TEK, RAC1, ZAP70, CSF1R, ITK, DHX9, MAP2K1, TGFBR1, MET, ACACB, ABCB6, KDR, GART, ATP7A, VCP, PLK1, UBA3, DYRK1A, GRK6, TNK2, MERTK, ABL1, CLCN5, UBE2E1, HSP90AB1, SPG7, AKT1, CDC42, PDPK1, PAK4, PAK1, AKT2, PRKCA, SGK1, HSP90AA1, KIF11, LYN, TP53, PRKCI, CDK6, CFTR, ALK, CDK5, CDK2, PRKCB, TYK2, PRKCQ, ADK, LCK, DCK, KIT, IGF1R, PTK2B, TAP1, TGM2, GIMAP2, PDK1, HCK, PDK3, PDK4, RAF1, MAPK10, PCK1, KHK, RPS6KA1, PYGL, FYN, MAPK14, GSK3B, JAK1, ABCC1, JAK3 | 1.81 | 1.45E-09 | 5.32E-08 |
| Annotation cluster 4 | Enrichment score: 16.40 | |||||||
| UP_SEQ_FEATURE | DNA-binding region: nuclear receptor | 20 | 4.28 | 2.07E-19 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 18.19 | 5.65E-17 | 3.48E-16 |
| UP_SEQ_FEATURE | Zinc finger region: NR C4 type | 20 | 4.28 | 2.07E-19 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 18.19 | 5.65E-17 | 3.48E-16 |
| INTERPRO | Zinc finger, nuclear hormone receptor type | 20 | 4.28 | 4.10E-18 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 15.54 | 7.36E-16 | 6.39E-15 |
| INTERPRO | Nuclear hormone receptor, ligand binding, core | 20 | 4.28 | 1.08E-17 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 14.90 | 1.61E-15 | 1.68E-14 |
| INTERPRO | Nuclear hormone receptor, ligand binding | 20 | 4.28 | 1.08E-17 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 14.90 | 1.61E-15 | 1.68E-14 |
| SMART | ZnF_C4 | 20 | 4.28 | 4.44E-17 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 13.47 | 3.73E-15 | 5.40E-14 |
| SMART | HOLI | 20 | 4.28 | 1.16E-16 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 12.91 | 6.22E-15 | 1.33E-13 |
| INTERPRO | Steroid hormone receptor | 19 | 4.07 | 1.59E-16 | PPARA, PPARD, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 14.45 | 1.42E-14 | 1.78E-13 |
| GOTERM_MF_FAT | Steroid hormone receptor activity | 20 | 4.28 | 8.16E-16 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 11.78 | 1.31E-13 | 1.20E-12 |
| INTERPRO | Zinc finger, NHR/GATA- type | 19 | 4.07 | 8.68E-16 | PPARA, PPARD, AR, RARG, THRA, THRB, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 13.32 | 7.25E-14 | 1.39E-12 |
| GOTERM_MF_FAT | Ligand-dependent nuclear receptor activity | 20 | 4.28 | 3.24E-14 | PPARA, PPARD, AR, RARG, THRA, THRB, RXRA, PPARG, ESRRG, ESR1, NR3C1, ESR2, NR1H2, PGR, VDR, NR1I3, NR1I2, RARB, NR1H4, NR1H3 | 9.95 | 2.29E-12 | 5.01E-11 |
| Annotation cluster 5 | Enrichment score: 15.83 | |||||||
| SP_PIR_KEYWORDS | Protease | 60 | 12.85 | 5.06E-25 | SPG7, METAP1, METAP2, MMP-9, IDE, MMP-8, MMP-7, MMP-3, MMP-1, MMP-20, APP, CASP3, LPA, HTRA1, CASP7, CASP8, CPA1, CFD, CASP1, TPSB2, DPP4, F11, F10, GZMA, ELANE, F9, GZMB, MMP-16, CTSS, F7, MMP-13, MMP-12, NAALAD2, CTSK, FOLH1, BACE2, ST14, BACE1, F2, CTSD, ADAM17, CTSB, ADAMTS5, CTSG, ADAMTS4, PREP, MME, ACE, ECE1, REN, PRSS3, PLAT, CPA4, HPN, KLK5, PLG, LTA4H, CMA1, CPB2, PLAU | 5.11 | 2.43E-23 | 7.34E-22 |
| GOTERM_MF_FAT | Endopeptidase activity | 51 | 10.92 | 1.01E-16 | SPG7, MMP-9, IDE, MMP-8, MMP-7, MMP-3, MMP-1, CASP3, MMP-20, LPA, HTRA1, CASP7, CASP8, SERPINE1, CFD, CASP1, TPSB2, DPP4, F11, F10, GZMA, ELANE, F9, GZMB, MMP-16, CTSS, F7, MMP-13, MMP-12, CTSK, BACE2, ST14, F2, BACE1, CTSD, ADAM17, CTSB, ADAMTS5, CTSG, ADAMTS4, PREP, MME, ECE1, REN, PRSS3, PLAT, HPN, KLK5, PLG, CMA1, PLAU | 3.92 | 3.13E-14 | 1.67E-13 |
| GOTERM_MF_FAT | Peptidase activity | 62 | 13.28 | 2.65E-15 | SPG7, METAP1, METAP2, MMP-9, IDE, MMP-8, MMP-7, MMP-3, MMP-1, MMP-20, APP, CASP3, LPA, HTRA1, CASP7, CASP8, SERPINE1, CPA1, CFD, CASP1, TPSB2, DPP4, F11, F10, GZMA, ELANE, F9, GZMB, MMP-16, CTSS, F7, MMP-13, MMP-12, NAALAD2, CTSK, FOLH1, BACE2, ST14, BACE1, F2, CTSD, ADAM17, CTSB, CTSG, ADAMTS5, ADAMTS4, PREP, MME, ACE, ECE1, REN, PRSS3, PLAT, CPA4, HPN, KLK5, PLG, QPCT, LTA4H, CMA1, CPB2, PLAU | 3.12 | 3.76E-13 | 4.12E-12 |
| GOTERM_MF_FAT | Peptidase activity, acting on L-amino acid peptides | 60 | 12.85 | 4.84E-15 | SPG7, METAP1, METAP2, MMP-9, IDE, MMP-8, MMP-7, MMP-3, MMP-1, MMP-20, CASP3, LPA, HTRA1, CASP7, CASP8, SERPINE1, CPA1, CFD, CASP1, TPSB2, DPP4, F11, F10, GZMA, ELANE, F9, GZMB, MMP-16, CTSS, F7, MMP-13, MMP-12, NAALAD2, CTSK, FOLH1, BACE2, ST14, BACE1, F2, CTSD, ADAM17, CTSB, ADAMTS5, CTSG, ADAMTS4, PREP, MME, ACE, ECE1, REN, PRSS3, PLAT, CPA4, HPN, KLK5, PLG, LTA4H, CMA1, CPB2, PLAU | 3.15 | 5.17E-13 | 7.55E-12 |
| GOTERM_BP_FAT | Proteolysis | 76 | 16.27 | 1.10E-10 | METAP1, SPG7, METAP2, TSG101, MMP-9, IDE, MMP-8, MMP-7, MMP-3, C1QC, MMP-1, MMP-20, CASP3, LPA, HTRA1, CASP7, CASP8, CPA1, CASP1, CFD, TPSB2, DPP4, F11, F10, GZMA, ELANE, F9, MMP- 16, UBR2, GZMB, CTSS, F7, MMP-13, MMP-12, C1QA, PRKCQ, C1QB, NAALAD2, CTSK, FOLH1, BACE2, ST14, BACE1, F2, CTSD, ADAM17, MDM2, CTSB, CTSG, ADAMTS5, ADAMTS4, PREP, XIAP, MME, NEDD8, ACE, ECE1, REN, PRSS3, TRAF6, SPOP, PLAT, CPA4, HPN, KLK5, PLG, BRCA1, NAE1, QPCT, VCP, UBA3, CMA1, LTA4H, CPB2, PLAU, UBE2E1 | 2.19 | 1.32E-08 | 2.00E-07 |
| Annotation cluster 6 | Enrichment score: 14.64 | |||||||
| SP_PIR_KEYWORDS | Serine/threonine protein kinase | 44 | 9.42 | 3.20E-17 | NEK2, MAPKAPK3, TTK, CHEK1, ADRBK1, AURKA, MAPKAPK2, CHEK2, CSNK2A2, IRAK4, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, AKT2, PRKCA, SGK1, MAP2K1, ROCK1, BRAF, TGFBR1, PIM1, PRKCI, RAF1, CDK6, MAPK10, CDK5, WEE1, CDK2, PRKCB, MAPK1, PRKCQ, RPS6KA1, PLK1, MAPK14, GSK3B, DYRK1A, MAPK3, GRK6, MAPK9, MAPK8 | 4.76 | 1.20E-15 | 4.64E-14 |
| INTERPRO | Serine/threonine protein kinase | 38 | 8.14 | 2.09E-16 | NEK2, MAPKAPK3, TTK, CHEK1, ADRBK1, AURKA, MAPKAPK2, CHEK2, CSNK2A2, AKT1, PDPK1, CSNK2A1, PAK4, PRKACA, PAK1, AKT2, PRKCA, SGK1, MAP2K1, ROCK1, PIM1, PRKCI, CDK6, MAPK10, CDK5, CDK2, PRKCB, MAPK1, PRKCQ, RPS6KA1, PLK1, MAPK14, GSK3B, DYRK1A, MAPK3, GRK6, MAPK9, MAPK8 | 5.25 | 2.49E-14 | 3.44E-13 |
| INTERPRO | Serine/threonine protein kinase, active site | 44 | 9.42 | 4.17E-16 | NEK2, MAPKAPK3, TTK, CHEK1, ADRBK1, AURKA, MAPKAPK2, CHEK2, CSNK2A2, IRAK4, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, AKT2, PRKCA, SGK1, MAP2K1, ROCK1, BRAF, TGFBR1, PIM1, PRKCI, RAF1, CDK6, MAPK10, CDK5, WEE1, CDK2, PRKCB, MAPK1, PRKCQ, RPS6KA1, PLK1, MAPK14, GSK3B, DYRK1A, MAPK3, GRK6, MAPK9, MAPK8 | 4.44 | 4.43E-14 | 6.88E-13 |
| INTERPRO | Serine/threonine protein kinase related | 44 | 9.42 | 6.21E-16 | NEK2, MAPKAPK3, TTK, CHEK1, ADRBK1, AURKA, MAPKAPK2, CHEK2, CSNK2A2, IRAK4, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, AKT2, PRKCA, SGK1, MAP2K1, ROCK1, BRAF, TGFBR1, PIM1, PRKCI, RAF1, CDK6, MAPK10, CDK5, WEE1, CDK2, PRKCB, MAPK1, PRKCQ, RPS6KA1, PLK1, MAPK14, GSK3B, DYRK1A, MAPK3, GRK6, MAPK9, MAPK8 | 4.38 | 5.98E-14 | 1.03E-12 |
| SMART | S_TKc | 38 | 8.14 | 1.09E-14 | NEK2, MAPKAPK3, TTK, CHEK1, ADRBK1, AURKA, MAPKAPK2, CHEK2, CSNK2A2, AKT1, PDPK1, CSNK2A1, PAK4, PRKACA, PAK1, AKT2, PRKCA, SGK1, MAP2K1, ROCK1, PIM1, PRKCI, CDK6, MAPK10, CDK5, CDK2, PRKCB, MAPK1, PRKCQ, RPS6KA1, PLK1, MAPK14, GSK3B, DYRK1A, MAPK3, GRK6, MAPK9, MAPK8 | 4.55 | 4.57E-13 | 1.32E-11 |
| GOTERM_MF_FAT | Protein serine/threonine kinase activity | 47 | 10.06 | 7.51E-12 | CDK5R1, NEK2, MAPKAPK3, TTK, CHEK1, FKBP1A, ADRBK1, AURKA, MAPKAPK2, CHEK2, CSNK2A2, IRAK4, AKT1, PDPK1, CSNK2A1, MAP3K9, PAK4, PRKACA, PAK1, AKT2, EGFR, PRKCA, SGK1, MAP2K1, ROCK1, BRAF, TGFBR1, PIM1, PRKCI, RAF1, CDK6, MAPK10, CDK5, WEE1, CDK2, PRKCB, MAPK1, PRKCQ, RPS6KA1, PLK1, MAPK14, GSK3B, DYRK1A, MAPK3, GRK6, MAPK9, MAPK8 | 3.15 | 3.97E-10 | 1.16E-08 |
| Annotation cluster 7 | Enrichment score: 14.56 | |||||||
| GOTERM_BP_FAT | Regulation of programmed cell death | 74 | 15.85 | 2.10E-15 | HRAS, MMP-9, PTEN, BTK, AKT1, CASP3, APP, FNTA, TIAM1, CASP8, RARB, CASP1, NQO1, PIK3CG, EGFR, PRKCA, PPP2R1A, NUDT2, RARG, BRAF, ROCK1, RXRA, PIM1, ESR1, TP53, PRKCI, ESR2, F7, CDK5, MAPK1, CD38, NME2, NME1, LCK, F2, MAPK9, ADAM17, MAPK8, CTSB, GSTP1, TRAF2, CDK5R1, YWHAZ, XIAP, ADORA2A, ERBB2, BCL2L1, KIT, NR3C1, SFN, CHEK2, SRC, MIF, VDR, IGF1R, ALB, BCL2, PPP2CA, RAC1, TGM2, BCL6, GLO1, TRAF6, LGALS1, TGFBR1, BIRC7, IGF1, PLG, BRCA1, NAE1, ATP7A, VCP, GSK3B, ABL1, IL2 | 2.76 | 7.55E-13 | 3.83E-12 |
| GOTERM_BP_FAT | Regulation of cell death | 74 | 15.85 | 2.52E-15 | HRAS, MMP-9, PTEN, BTK, AKT1, CASP3, APP, FNTA, TIAM1, CASP8, RARB, CASP1, NQO1, PIK3CG, EGFR, PRKCA, PPP2R1A, NUDT2, RARG, BRAF, ROCK1, RXRA, PIM1, ESR1, TP53, PRKCI, ESR2, F7, CDK5, MAPK1, CD38, NME2, NME1, LCK, F2, MAPK9, ADAM17, MAPK8, CTSB, GSTP1, TRAF2, CDK5R1, YWHAZ, XIAP, ADORA2A, ERBB2, BCL2L1, KIT, NR3C1, SFN, CHEK2, SRC, MIF, VDR, IGF1R, ALB, BCL2, PPP2CA, RAC1, TGM2, BCL6, GLO1, TRAF6, LGALS1, TGFBR1, BIRC7, IGF1, PLG, BRCA1, NAE1, ATP7A, VCP, GSK3B, ABL1, IL2 | 2.75 | 8.23E-13 | 4.63E-12 |
| GOTERM_BP_FAT | Regulation of apoptosis | 73 | 15.63 | 4.05E-15 | HRAS, MMP-9, PTEN, BTK, AKT1, CASP3, APP, FNTA, TIAM1, CASP8, RARB, CASP1, NQO1, PIK3CG, PRKCA, EGFR, PPP2R1A, NUDT2, RARG, BRAF, ROCK1, RXRA, PIM1, ESR1, TP53, PRKCI, ESR2, F7, CDK5, MAPK1, CD38, NME2, NME1, LCK, F2, MAPK9, ADAM17, MAPK8, CTSB, GSTP1, TRAF2, CDK5R1, YWHAZ, XIAP, ADORA2A, ERBB2, BCL2L1, NR3C1, SFN, CHEK2, SRC, MIF, VDR, IGF1R, ALB, BCL2, PPP2CA, RAC1, TGM2, BCL6, GLO1, TRAF6, LGALS1, TGFBR1, BIRC7, IGF1, PLG, BRCA1, NAE1, ATP7A, VCP, GSK3B, ABL1, IL2 | 2.75 | 1.17E-12 | 7.25E-12 |
| Annotation cluster 8 | Enrichment score: 10.94 | |||||||
| GOTERM_BP_FAT | Regulation of phosphorylation | 49 | 10.49 | 1.72E-12 | HMGCR, TTK, PTEN, AKT1, CDC42, APP, CASP3, PDPK1, CXCR4, MAP3K9, PRKACA, PAK1, INSR, SYK, PRKCA, EGFR, PPP2R1A, LYN, ELANE, PIM1, RB1, CDK5, NCK2, F2, ADAM17, TRAF2, CDK5R1, ADORA2A, ERBB2, CHEK1, FKBP1A, SFN, KIT, PIN1, PTK2B, PPP2CA, BCL2, RAC1, TRAF6, MAP2K1, TGFBR1, MET, BIRC7, IGF1, PTPN11, ATP7A, FABP4, TNK2, IL2 | 3.19 | 3.25E-10 | 3.12E-09 |
| GOTERM_BP_FAT | Regulation of phosphorus metabolic process | 50 | 10.71 | 2.04E-12 | HMGCR, TTK, PTEN, AKT1, CDC42, APP, CASP3, PDPK1, CXCR4, MAP3K9, PRKACA, PAK1, INSR, SYK, PRKCA, EGFR, PPP2R1A, LYN, ELANE, PIM1, RB1, CDK5, NCK2, F2, ADAM17, TRAF2, CDK5R1, ADORA2A, ERBB2, CHEK1, FKBP1A, SFN, KIT, PIN1, PTK2B, PPP2CA, BCL2, RAC1, TRAF6, MAP2K1, PLEK, TGFBR1, MET, BIRC7, IGF1, PTPN11, ATP7A, FABP4, TNK2, IL2 | 3.13 | 3.65E-10 | 3.70E-09 |
| GOTERM_BP_FAT | Regulation of phosphate metabolic process | 50 | 10.71 | 2.04E-12 | HMGCR, TTK, PTEN, AKT1, CDC42, APP, CASP3, PDPK1, CXCR4, MAP3K9, PRKACA, PAK1, INSR, SYK, PRKCA, EGFR, PPP2R1A, LYN, ELANE, PIM1, RB1, CDK5, NCK2, F2, ADAM17, TRAF2, CDK5R1, ADORA2A, ERBB2, CHEK1, FKBP1A, SFN, KIT, PIN1, PTK2B, PPP2CA, BCL2, RAC1, TRAF6, MAP2K1, PLEK, TGFBR1, MET, BIRC7, IGF1, PTPN11, ATP7A, FABP4, TNK2, IL2 | 3.13 | 3.65E-10 | 3.70E-09 |
| GOTERM_BP_FAT | Regulation of transferase activity | 41 | 8.78 | 3.67E-11 | TRAF2, CDK5R1, HMGCR, ADORA2A, ERBB2, PPARG, CHEK1, SFN, KIT, PTEN, AKT1, CDC42, PDPK1, CASP3, APP, CXCR4, PTK2B, MAP3K9, PPP2CA, RAC1, PRKACA, PAK1, TRAF6, INSR, SYK, EGFR, PRKCA, PPP2R1A, MAP2K1, TGFBR1, ELANE, MET, PIM1, BIRC7, RB1, CDK5, PTPN11, NCK2, FABP4, ADAM17, IL2 | 3.34 | 4.93E-09 | 6.67E-08 |
| GOTERM_BP_FAT | Regulation of kinase activity | 40 | 8.56531 | 4.06E-11 | TRAF2, CDK5R1, HMGCR, ADORA2A, ERBB2, CHEK1, SFN, KIT, PTEN, AKT1, CDC42, CASP3, APP, PDPK1, CXCR4, PTK2B, MAP3K9, PPP2CA, RAC1, PRKACA, PAK1, TRAF6, INSR, SYK, EGFR, PRKCA, PPP2R1A, MAP2K1, TGFBR1, ELANE, MET, PIM1, BIRC7, RB1, CDK5, PTPN11, NCK2, FABP4, ADAM17, IL2 | 3.40 | 5.24E-09 | 7.37E-08 |
| GOTERM_BP_FAT | Regulation of protein kinase activity | 38 | 8.145 | 2.24E-10 | TRAF2, CDK5R1, HMGCR, ADORA2A, ERBB2, CHEK1, SFN, KIT, PTEN, AKT1, CASP3, APP, PDPK1, CXCR4, PTK2B, MAP3K9, PPP2CA, PRKACA, PAK1, TRAF6, INSR, SYK, EGFR, PRKCA, PPP2R1A, MAP2K1, TGFBR1, ELANE, MET, PIM1, BIRC7, RB1, CDK5, PTPN11, NCK2, FABP4, ADAM17, IL2 | 3.34 | 2.33E-08 | 4.07E-07 |
| Annotation cluster 9 | Enrichment score: 10.39 | |||||||
| SP_PIR_KEYWORDS | Serine proteinase | 17 | 3.64 | 9.89E-14 | PLAT, F11, PREP, HPN, F10, GZMA, F9, GZMB, F7, PLG, LPA, PRSS3, F2, CMA1, TPSB2, CTSG, PLAU | 12.97 | 2.18E-12 | 1.44E-10 |
| UP_SEQ_FEATURE | Charge relay system | 28 | 6.00 | 3.26E-13 | PREP, HMGCR, LPA, HTRA1, BCHE, PRSS3, CFD, TPSB2, DPP4, F11, PLAT, HPN, F10, CES1, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, NAALAD2, FOLH1, ST14, F2, CMA1, CTSG, PLAU | 5.85 | 4.45E-11 | 5.48E-10 |
| SP_PIR_KEYWORDS | Serine protease | 23 | 4.93 | 1.74E-12 | PLAT, F11, PREP, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, LPA, HTRA1, ST14, F2, PRSS3, CMA1, TPSB2, CFD, DPP4, CTSG, PLAU | 6.97 | 3.68E-11 | 2.53E-09 |
| UP_SEQ_FEATURE | Peptidase S1 | 20 | 4.28 | 9.94E-12 | PLAT, F11, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, LPA, ST14, F2, PRSS3, CMA1, TPSB2, CFD, CTSG, PLAU | 7.72 | 1.08E-09 | 1.67E-08 |
| INTERPRO | Peptidase S1 and S6, chymotrypsin/Hap | 21 | 4.50 | 6.51E-11 | PLAT, F11, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, LPA, HTRA1, ST14, F2, PRSS3, CMA1, TPSB2, CFD, CTSG, PLAU | 6.47 | 4.50E-09 | 1.01E-07 |
| INTERPRO | Peptidase S1A, chymotrypsin | 20 | 4.28 | 6.91E-11 | PLAT, F11, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, LPA, ST14, F2, PRSS3, CMA1, TPSB2, CFD, CTSG, PLAU | 6.87 | 4.43E-09 | 1.08E-07 |
| INTERPRO | Peptidase S1/S6, chymotrypsin/Hap, active site | 20 | 4.28 | 8.22E-11 | PLAT, F11, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, LPA, ST14, F2, PRSS3, CMA1, TPSB2, CFD, CTSG, PLAU | 6.81 | 4.92E-09 | 1.28E-07 |
| GOTERM_MF_FAT | Serine-type endopeptidase activity | 25 | 5.35 | 5.61E-10 | PREP, MMP-8, LPA, HTRA1, PRSS3, SERPINE1, CFD, TPSB2, DPP4, F11, PLAT, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, F2, ST14, CMA1, CTSG, PLAU | 4.68 | 2.26E-08 | 8.67E-07 |
| SMART | Tryp_SPc | 21 | 4.50 | 6.36E-10 | PLAT, F11, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, LPA, HTRA1, ST14, F2, PRSS3, CMA1, TPSB2, CFD, CTSG, PLAU | 5.61 | 2.14E-08 | 7.74E-07 |
| GOTERM_MF_FAT | Serine-type peptidase activity | 26 | 5.57 | 2.29E-09 | PREP, MMP-8, LPA, HTRA1, PRSS3, SERPINE1, CFD, TPSB2, DPP4, F11, PLAT, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, NAALAD2, ST14, F2, CMA1, CTSG, PLAU | 4.21 | 8.44E-08 | 3.55E-06 |
| GOTERM_MF_FAT | Serine hydrolase activity | 26 | 5.57 | 2.90E-09 | PREP, MMP-8, LPA, HTRA1, PRSS3, SERPINE1, CFD, TPSB2, DPP4, F11, PLAT, HPN, F10, GZMA, KLK5, ELANE, F9, GZMB, F7, PLG, NAALAD2, ST14, F2, CMA1, CTSG, PLAU | 4.17 | 1.02E-07 | 4.49E-06 |
| Annotation cluster 10 | Enrichment score: 9.02 | |||||||
| GOTERM_BP_FAT | Negative regulation of programmed cell death | 38 | 8.4 | 6.92E-10 | YWHAZ, HRAS, XIAP, ADORA2A, ERBB2, BCL2L1, KIT, PTEN, MIF, AKT1, IGF1R, CASP3, FNTA, ALB, BCL2, TGM2, GLO1, BCL6, TRAF6, PIK3CG, EGFR, ROCK1, BRAF, TGFBR1, PIM1, BIRC7, PRKCI, TP53, ESR1, IGF1, F7, ESR2, NME2, NME1, GSK3B, ADAM17, MAPK8, GSTP1, IL2 | 3.21 | 6.76E-08 | 1.26E-06 |
| GOTERM_BP_FAT | Negative regulation of cell death | 38 | 8.14 | 7.49E-10 | YWHAZ, HRAS, XIAP, ADORA2A, ERBB2, BCL2L1, KIT, PTEN, MIF, AKT1, IGF1R, CASP3, FNTA, ALB, BCL2, TGM2, GLO1, BCL6, TRAF6, PIK3CG, EGFR, ROCK1, BRAF, TGFBR1, PIM1, BIRC7, PRKCI, TP53, ESR1, IGF1, F7, ESR2, NME2, NME1, GSK3B, ADAM17, MAPK8, GSTP1, IL2 | 3.20 | 7.10E-08 | 1.36E-06 |
| GOTERM_BP_FAT | Negative regulation of apoptosis | 37 | 7.92 | 1.71E-09 | YWHAZ, HRAS, XIAP, ADORA2A, ERBB2, BCL2L1, PTEN, MIF, AKT1, IGF1R, CASP3, FNTA, ALB, BCL2, TGM2, GLO1, BCL6, TRAF6, EGFR, PIK3CG, ROCK1, BRAF, TGFBR1, PIM1, BIRC7, PRKCI, TP53, ESR1, IGF1, F7, ESR2, NME2, NME1, GSK3B, ADAM17, MAPK8, GSTP1, IL2 | 3.17 | 1.31E-07 | 3.11E-06 |
| Annotation cluster 11 | Enrichment score: 8.99 | |||||||
| GOTERM_BP_FAT | Positive regulation of apoptosis | 42 | 8.99 | 8.49E-10 | TRAF2, CDK5R1, ADORA2A, MMP-9, BCL2L1, CHEK2, SFN, NR3C1, PTEN, SRC, BTK, AKT1, VDR, APP, CASP3, TIAM1, PPP2CA, BCL2, CASP8, RAC1, TGM2, BCL6, RARB, CASP1, TRAF6, NQO1, PRKCA, PPP2R1A, NUDT2, RARG, TGFBR1, RXRA, TP53, CDK5, BRCA1, PLG, MAPK1, CD38, LCK, MAPK9, MAPK8, ABL1 | 2.96 | 7.82E-08 | 1.54E-06 |
| GOTERM_BP_FAT | Positive regulation of programmed cell death | 42 | 8.99 | 1.05E-09 | TRAF2, CDK5R1, ADORA2A, MMP-9, BCL2L1, CHEK2, SFN, NR3C1, PTEN, SRC, BTK, AKT1, VDR, APP, CASP3, TIAM1, PPP2CA, BCL2, CASP8, RAC1, TGM2, BCL6, RARB, CASP1, TRAF6, NQO1, PRKCA, PPP2R1A, NUDT2, RARG, TGFBR1, RXRA, TP53, CDK5, BRCA1, PLG, MAPK1, CD38, LCK, MAPK9, MAPK8, ABL1 | 2.94 | 8.87E-08 | 1.90E-06 |
| GOTERM_BP_FAT | Positive regulation of cell death | 42 | 8.99 | 1.20E-09 | TRAF2, CDK5R1, ADORA2A, MMP-9, BCL2L1, CHEK2, SFN, NR3C1, PTEN, SRC, BTK, AKT1, VDR, APP, CASP3, TIAM1, PPP2CA, BCL2, CASP8, RAC1, TGM2, BCL6, RARB, CASP1, TRAF6, NQO1, PRKCA, PPP2R1A, NUDT2, RARG, TGFBR1, RXRA, TP53, CDK5, BRCA1, PLG, MAPK1, CD38, LCK, MAPK9, MAPK8, ABL1 | 2.93 | 9.91E-08 | 2.18E-06 |
| Annotation cluster 12 | Enrichment score: 8.97 | |||||||
| GOTERM_BP_FAT | Regulation of cell motion | 28 | 6.00 | 1.90E-10 | NRP1, ERBB4, MMP-9, KIT, PTEN, AKT1, IGF1R, ACE, PTK2B, CXCR4, BCL2, TEK, RAC1, BCL6, INSR, EGFR, F10, LYN, MAP2K1, TGFBR1, IGF1, F7, CDK5, PLG, KDR, MAPK1, ADAM17, HDAC7 | 4.40 | 2.04E-08 | 3.45E-07 |
| GOTERM_BP_FAT | Regulation of locomotion | 27 | 5.78 | 8.53E-10 | ERBB4, ADORA2A, MMP-9, KIT, PTEN, AKT1, IGF1R, ACE, PTK2B, CXCR4, BCL2, TEK, RAC1, INSR, PRKCA, EGFR, F10, MAP2K1, ELANE, IGF1, F7, CDK5, PLG, KDR, MAPK1, ADAM17, HDAC7 | 4.27 | 7.64E-08 | 1.55E-06 |
| GOTERM_BP_FAT | Regulation of cell migration | 24 | 5.14 | 7.40E-09 | EGFR, F10, ERBB4, MAP2K1, MMP-9, IGF1, KIT, F7, CDK5, PTEN, PLG, KDR, AKT1, MAPK1, IGF1R, ACE, CXCR4, PTK2B, BCL2, TEK, RAC1, ADAM17, INSR, HDAC7 | 4.31 | 5.08E-07 | 1.34E-05 |
| Annotation cluster 13 | Enrichment score: 8.68 | |||||||
| UP_SEQ_FEATURE | GST N-terminal | 12 | 2.57 | 4.56E-13 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTZ1, GSTT1, GSTO2, GSTO1, HPGDS, GSTP1 | 23.39 | 5.74E-11 | 7.65E-10 |
| INTERPRO | Glutathione S-transferase, N-terminal | 12 | 2.57 | 3.81E-12 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTZ1, GSTT1, GSTO2, GSTO1, HPGDS, GSTP1 | 19.50 | 2.85E-10 | 5.94E-09 |
| GOTERM_MF_FAT | Glutathione transferase activity | 12 | 2.57 | 9.51E-12 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTK1, GSTZ1, GSTT1, GSTO2, GSTO1, GSTP1 | 17.31 | 4.47E-10 | 1.47E-08 |
| UP_SEQ_FEATURE | GST C-terminal | 12 | 2.57 | 1.92E-10 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTZ1, GSTT1, GSTO2, GSTO1, HPGDS, GSTP1 | 14.88 | 1.96E-08 | 3.22E-07 |
| INTERPRO | Glutathione S-transferase, C-terminal | 10 | 2.14 | 2.08E-09 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTZ1, GSTT1, GSTO1, GSTP1 | 17.02 | 1.17E-07 | 3.24E-06 |
| INTERPRO | Glutathione S-transferase, C-terminal-like | 11 | 2.36 | 3.25E-09 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTZ1, GSTT1, GSTO2, GSTO1, GSTP1 | 13.56 | 1.71E-07 | 5.06E-06 |
| INTERPRO | Glutathione S-transferase/chloride channel, C-terminal | 11 | 2.36 | 4.75E-09 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTZ1, GSTT1, GSTO2, GSTO1, GSTP1 | 13.11 | 2.37E-07 | 7.40E-06 |
| KEGG_PATHWAY | Glutathione metabolism | 14 | 3.00 | 1.57E-05 | GSTA1, GSTA2, ODC1, GSTA3, GSTA4, GSTT1, GSTM1, GSTM2, G6PD, GSTK1, GSTZ1, GSTO2, GSTO1, GSTP1 | 4.18 | 9.77E-05 | 0.018816 |
| INTERPRO | Thioredoxin fold | 11 | 2.356 | 5.18E-04 | GSTM1, GSTA1, GSTM2, GSTA2, GSTA3, GSTA4, GSTZ1, GSTT1, GSTO2, GSTO1, GSTP1 | 3.90 | 0.01217529 | 0.804529 |
| Annotation cluster 14 | Enrichment score: 7.42 | |||||||
| GOTERM_BP_FAT | Positive regulation of cell motion | 19 | 4.07 | 2.63E-09 | EGFR, F10, ERBB4, LYN, MAP2K1, MMP-9, TGFBR1, IGF1, KIT, F7, KDR, IGF1R, MAPK1, PTK2B, BCL2, ADAM17, BCL6, INSR, HDAC7 | 5.88 | 1.97E-07 | 4.77E-06 |
| GOTERM_BP_FAT | Positive regulation of locomotion | 17 | 3.64 | 1.14E-07 | PRKCA, EGFR, F10, ERBB4, MAP2K1, MMP-9, IGF1, F7, KIT, KDR, IGF1R, MAPK1, PTK2B, BCL2, ADAM17, INSR, HDAC7 | 5.26 | 5.58E-06 | 2.07E-04 |
| GOTERM_BP_FAT | Positive regulation of cell migration | 16 | 3.43 | 1.85E-07 | EGFR, F10, ERBB4, MAP2K1, MMP-9, IGF1, F7, KIT, KDR, IGF1R, MAPK1, PTK2B, BCL2, ADAM17, INSR, HDAC7 | 5.45 | 8.90E-06 | 3.36E-04 |
| Annotation cluster 15 | Enrichment score: 7.15 | |||||||
| SP_PIR_KEYWORDS | SH2 domain | 17 | 3.64 | 1.08E-08 | ITK, LYN, GRB2, HCK, SRC, PTPN11, BTK, TYK2, NCK2, DAPP1, FYN, LCK, ZAP70, JAK1, JAK3, ABL1, SYK | 6.31 | 1.89E-07 | 1.56E-05 |
| INTERPRO | SH2 domain | 17 | 3.64 | 7.50E-08 | ITK, LYN, GRB2, HCK, SRC, PTPN11, BTK, TYK2, NCK2, DAPP1, FYN, LCK, ZAP70, JAK1, JAK3, ABL1, SYK | 5.48 | 3.06E-06 | 1.17E-04 |
| SMART | SH2 domain | 17 | 3.64 | 4.51E-07 | ITK, LYN, GRB2, HCK, SRC, PTPN11, BTK, TYK2, NCK2, DAPP1, FYN, LCK, ZAP70, JAK1, JAK3, ABL1, SYK | 4.75 | 1.26E-05 | 5.49E-04 |
| Annotation cluster 16 | Enrichment score: 7.12 | |||||||
| GOTERM_BP_FAT | Cell migration | 31 | 6.64 | 8.87E-09 | PPARD, CDK5R1, NRP1, HMGCR, PEX5, KIT, PTEN, SRC, PTK2, GAB2, CXCR4, PTK2B, PRSS3, RAC1, SFTPD, SYK, PRKCA, PLAT, RET, ROCK1, TGFBR1, MET, ELANE, ESR2, CDK5, KDR, NCK2, FYN, ADAM17, SDCBP, PLAU | 3.41 | 5.84E-07 | 1.61E-05 |
| GOTERM_BP_FAT | Cell motility | 31 | 6.64 | 9.98E-08 | PPARD, CDK5R1, NRP1, HMGCR, PEX5, KIT, PTEN, SRC, PTK2, GAB2, CXCR4, PTK2B, PRSS3, RAC1, SFTPD, SYK, PRKCA, PLAT, RET, ROCK1, TGFBR1, MET, ELANE, ESR2, CDK5, KDR, NCK2, FYN, ADAM17, SDCBP, PLAU | 3.06 | 4.95E-06 | 1.81E-04 |
| GOTERM_BP_FAT | Localization of cell | 31 | 6.64 | 9.98E-08 | PPARD, CDK5R1, NRP1, HMGCR, PEX5, KIT, PTEN, SRC, PTK2, GAB2, CXCR4, PTK2B, PRSS3, RAC1, SFTPD, SYK, PRKCA, PLAT, RET, ROCK1, TGFBR1, MET, ELANE, ESR2, CDK5, KDR, NCK2, FYN, ADAM17, SDCBP, PLAU | 3.06 | 4.95E-06 | 1.81E-04 |
| GOTERM_BP_FAT | Cell motion | 39 | 8.35 | 3.80E-07 | ITGAL, PPARD, CDK5R1, NRP1, HMGCR, ERBB2, PEX5, KIT, PTEN, SRC, APP, PTK2, GAB2, CXCR4, PTK2B, PAK4, PRSS3, RAC1, SFTPD, SYK, PLAT, PRKCA, RET, MAP2K1, ROCK1, TGFBR1, ELANE, MET, IGF1, ESR2, CDK5, KDR, NCK2, FYN, MAPK14, ADAM17, SDCBP, MAPK8, PLAU | 2.49 | 1.68E-05 | 6.90E-04 |
| Annotation cluster 17 | Enrichment score: 7.03 | |||||||
| UP_SEQ_FEATURE | Short sequence motif: cysteine switch | 12 | 2.57 | 4.58E-09 | MMP-20, MMP-9, MMP-8, MMP-7, ADAM17, MMP-16, MMP-3, MMP-13, ADAMTS5, MMP-12, MMP-1, ADAMTS4 | 11.42 | 4.41E-07 | 7.69E-06 |
| INTERPRO | Peptidase M, neutral zinc metallopeptidases, zinc- binding site | 16 | 3.43 | 6.40E-08 | MMP-9, MMP-8, MMP-7, MME, MMP-16, MMP-3, MMP-13, MMP-1, MMP-12, ACE, MMP-20, ECE1, ADAM17, LTA4H, ADAMTS5, ADAMTS4 | 5.96 | 2.74E-06 | 9.97E-05 |
| GOTERM_MF_FAT | Metalloendopeptidase activity | 16 | 3.43 | 2.72E-06 | SPG7, MMP-9, IDE, MMP-8, MMP-7, MME, MMP-16, MMP-3, MMP-13, MMP-1, MMP-12, MMP-20, ECE1, ADAM17, ADAMTS5, ADAMTS4 | 4.44 | 7.42E-05 | 0.004202 |
| Annotation cluster 18 | Enrichment score: 6.95 | |||||||
| GOTERM_BP_FAT | Positive regulation of kinase activity | 27 | 5.78 | 4.35E-08 | TRAF2, ADORA2A, ERBB2, KIT, AKT1, CDC42, PDPK1, PTK2B, CXCR4, MAP3K9, RAC1, PRKACA, PAK1, TRAF6, INSR, SYK, EGFR, MAP2K1, TGFBR1, MET, ELANE, BIRC7, PIM1, CDK5, PTPN11, ADAM17, IL2 | 3.55 | 2.46E-06 | 7.90E-05 |
| GOTERM_BP_FAT | Positive regulation of transferase activity | 27 | 5.78 | 9.47E-08 | TRAF2, ADORA2A, ERBB2, KIT, AKT1, CDC42, PDPK1, PTK2B, CXCR4, MAP3K9, RAC1, PRKACA, PAK1, TRAF6, INSR, SYK, EGFR, MAP2K1, TGFBR1, MET, ELANE, BIRC7, PIM1, CDK5, PTPN11, ADAM17, IL2 | 3.41 | 4.77E-06 | 1.72E-04 |
| GOTERM_BP_FAT | Positive regulation of protein kinase activity | 25 | 5.35 | 3.34E-07 | TRAF2, ADORA2A, ERBB2, KIT, AKT1, PDPK1, PTK2B, CXCR4, MAP3K9, PRKACA, PAK1, TRAF6, INSR, SYK, EGFR, MAP2K1, TGFBR1, MET, ELANE, BIRC7, PIM1, CDK5, PTPN11, ADAM17, IL2 | 3.40 | 1.49E-05 | 6.05E-04 |
| Annotation cluster 19 | Enrichment score: 6.74 | |||||||
| GOTERM_BP_FAT | Positive regulation of biosynthetic process | 52 | 11.13 | 5.71E-08 | HSP90AB1, PPARA, HRAS, THRA, THRB, PPARG, AKT1, APP, FNTA, RARB, KDM5A, DDAH1, INSR, AKT2, SYK, EGFR, AR, RARG, HSP90AA1, RXRA, ELANE, TP53, RB1, ESR2, PPARGC1A, CDK2, TNKS2, PRKCQ, MAPK1, NCOA1, NCOA2, F2, ADORA2A, ARNT, NR1H2, IGF1R, TNFRSF1A, PTK2B, TRAF6, NR1H4, NR1H3, BCKDHA, EPAS1, TGFBR1, ESRRG, IGF1, BRCA1, HDAC4, HDAC2, MAPK14, JAK3, IL2 | 2.27 | 3.07E-06 | 1.04E-04 |
| GOTERM_BP_FAT | Positive regulation of nitrogen compound metabolic process | 49 | 10.49 | 9.00E-08 | HSP90AB1, PPARA, NCBP1, HRAS, THRA, THRB, PPARG, AKT1, APP, RARB, KDM5A, DDAH1, INSR, EGFR, AR, RARG, HSP90AA1, RXRA, TP53, CD40, ESR2, RB1, PPARGC1A, CDK2, TNKS2, MAPK1, NCOA1, NCOA2, ADORA2A, COMT, HPRT1, ARNT, NR1H2, IGF1R, TNFRSF1A, NR1H4, NR1H3, BCKDHA, MAP2K1, EPAS1, TGFBR1, ESRRG, IGF1, BRCA1, HDAC4, HDAC2, MAPK14, JAK3, IL2 | 2.31 | 4.61E-06 | 1.63E-04 |
| GOTERM_BP_FAT | Positive regulation of cellular biosynthetic process | 50 | 10.71 | 2.34E-07 | HSP90AB1, PPARA, HRAS, THRA, THRB, PPARG, AKT1, APP, RARB, KDM5A, DDAH1, INSR, AKT2, SYK, EGFR, AR, RARG, HSP90AA1, RXRA, ELANE, TP53, RB1, ESR2, PPARGC1A, CDK2, TNKS2, PRKCQ, MAPK1, NCOA1, NCOA2, ADORA2A, ARNT, NR1H2, IGF1R, TNFRSF1A, PTK2B, TRAF6, NR1H4, NR1H3, BCKDHA, EPAS1, TGFBR1, ESRRG, IGF1, BRCA1, HDAC4, HDAC2, MAPK14, JAK3, IL2 | 2.21 | 1.11E-05 | 4.24E-04 |
| GOTERM_BP_FAT | Positive regulation of macromolecule biosynthetic process | 47 | 10.06 | 9.04E-07 | PPARA, HRAS, THRA, THRB, PPARG, ARNT, AKT1, NR1H2, IGF1R, TNFRSF1A, APP, FNTA, PTK2B, RARB, KDM5A, TRAF6, INSR, NR1H4, AKT2, SYK, NR1H3, BCKDHA, AR, RARG, EPAS1, TGFBR1, RXRA, ELANE, ESRRG, TP53, IGF1, RB1, ESR2, PPARGC1A, BRCA1, CDK2, TNKS2, HDAC4, MAPK1, PRKCQ, NCOA1, HDAC2, NCOA2, MAPK14, F2, JAK3, IL2 | 2.18 | 3.73E-05 | 0.001641 |
| Annotation cluster 20 | Enrichment score: 6.14 | |||||||
| UP_SEQ_FEATURE | AGC-kinase C-terminal | 12 | 2.57 | 7.32E-08 | PRKCA, AKT1, PRKCQ, SGK1, ROCK1, RPS6KA1, GRK6, PRKCI, PRKACA, ADRBK1, PRKCB, AKT2 | 8.93 | 5.99E-06 | 1.23E-04 |
| INTERPRO | AGC-kinase, C-terminal | 12 | 2.57 | 2.81E-07 | PRKCA, AKT1, PRKCQ, SGK1, ROCK1, RPS6KA1, GRK6, PRKCI, PRKACA, ADRBK1, PRKCB, AKT2 | 7.80 | 1.05E-05 | 4.38E-04 |
| SMART | S_TK_X | 12 | 2.57 | 1.07E-06 | PRKCA, AKT1, PRKCQ, SGK1, ROCK1, RPS6KA1, GRK6, PRKCI, PRKACA, ADRBK1, PRKCB, AKT2 | 6.76 | 2.25E-05 | 0.001302 |
| INTERPRO | Protein kinase, C-terminal | 9 | 1.93 | 1.22E-05 | PRKCA, AKT1, PRKCQ, SGK1, ROCK1, RPS6KA1, PRKCI, PRKCB, AKT2 | 8.04 | 3.31E-04 | 0.018952 |
| Annotation cluster 21 | Enrichment score: 5.96 | |||||||
| INTERPRO | 3′,5′-Cyclic nucleotide phosphodiesterase | 9 | 1.93 | 5.13E-08 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 15.32 | 2.30E-06 | 8.00E-05 |
| INTERPRO | Metal-dependent phosphohydrolase, HD region | 9 | 1.93 | 1.72E-07 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 13.41 | 6.73E-06 | 2.68E-04 |
| UP_SEQ_FEATURE | Metal ion-binding site: divalent metal cation 2 | 9 | 1.93 | 2.38E-07 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 13.16 | 1.86E-05 | 4.00E-04 |
| UP_SEQ_FEATURE | Metal ion-binding site: divalent metal cation 1 | 9 | 1.93 | 2.38E-07 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 13.16 | 1.86E-05 | 4.00E-04 |
| SMART | HDc | 9 | 1.93 | 4.89E-07 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 11.62 | 1.17E-05 | 5.95E-04 |
| GOTERM_MF_FAT | 3′,5′-Cyclic-nucleotide phosphodiesterase activity | 9 | 1.93 | 8.68E-07 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 10.82 | 2.53E-05 | 0.001342 |
| GOTERM_MF_FAT | Cyclic-nucleotide phosphodiesterase activity | 9 | 1.93 | 1.24E-06 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 10.39 | 3.49E-05 | 0.001915 |
| GOTERM_MF_FAT | Phosphoric diester hydrolase activity | 9 | 1.93 | 0.008435 | PDE2A, PDE7A, PDE4A, PDE4B, PDE5A, PDE3B, PDE9A, PDE4D, PDE8A | 3.09 | 0.08777958 | 12.27639 |
| Annotation cluster 22 | Enrichment score: 5.80 | |||||||
| UP_SEQ_FEATURE | Short sequence motif: cysteine switch | 12 | 2.57 | 4.58E-09 | MMP-20, MMP-9, MMP-8, MMP-7, ADAM17, MMP-16, MMP-3, MMP-13, ADAMTS5, MMP-12, MMP-1, ADAMTS4 | 11.42 | 4.41E-07 | 7.69E-06 |
| INTERPRO | Peptidoglycan binding like | 9 | 1.93 | 3.26E-08 | MMP-20, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 16.09 | 1.54E-06 | 5.07E-05 |
| UP_SEQ_FEATURE | Hemopexin-like 4 domain | 8 | 1.71 | 6.08E-07 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 14.88 | 4.15E-05 | 0.001021 |
| UP_SEQ_FEATURE | Hemopexin-like 3 domain | 8 | 1.71 | 6.08E-07 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 14.88 | 4.15E-05 | 0.001021 |
| UP_SEQ_FEATURE | Hemopexin-like 1 domain | 8 | 1.71 | 8.56E-07 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 14.24 | 5.39E-05 | 0.001437 |
| UP_SEQ_FEATURE | Hemopexin-like 2 domain | 8 | 1.71 | 8.56E-07 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 14.24 | 5.39E-05 | 0.001437 |
| INTERPRO | Peptidase, metallopeptidases | 9 | 1.93 | 8.89E-07 | MMP-20, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 11.09 | 3.07E-05 | 0.001385 |
| INTERPRO | Hemopexin/matrixin, repeat | 8 | 1.71 | 2.10E-06 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 12.43 | 6.73E-05 | 0.003271 |
| INTERPRO | Hemopexin/matrixin, conserved site | 8 | 1.71 | 2.10E-06 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 12.43 | 6.73E-05 | 0.003271 |
| INTERPRO | Hemopexin/matrixin | 8 | 1.71 | 2.10E-06 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 12.43 | 6.73E-05 | 0.003271 |
| INTERPRO | Peptidase M10A and M12B, matrixin and adamalysin | 10 | 2.14 | 2.31E-06 | MMP-20, MMP-9, MMP-8, MMP-7, ADAM17, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 8.31 | 7.15E-05 | 0.003597 |
| SMART | ZnMc | 9 | 1.93 | 2.48E-06 | MMP-20, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 9.62 | 4.63E-05 | 0.003017 |
| SMART | HX | 8 | 1.71 | 5.21E-06 | MMP-20, MMP-9, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 10.78 | 8.76E-05 | 0.006349 |
| INTERPRO | Peptidase M10A, matrix metallopeotidase | 7 | 1.50 | 6.39E-06 | MMP-20, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 13.90 | 1.85E-04 | 0.009958 |
| PIR_SUPERFAMILY | Peptidase_M10A_matrix | 7 | 1.50 | 1.06E-04 | MMP-20, MMP-8, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 8.41 | 0.00874563 | 0.144244 |
| PIR_SUPERFAMILY | Matrix metalloproteinase, stromelysin type | 6 | 1.28 | 2.87E-04 | MMP-20, MMP-8, MMP-16, MMP-3, MMP-13, MMP-1 | 9.27 | 0.01875986 | 0.388252 |
| Annotation cluster 23 | Enrichment score: 5.73 | |||||||
| UP_SEQ_FEATURE | Metal ion-binding site: calcium 3; via carbonyl oxygen | 8 | 1.71 | 4.23E-07 | PRKCA, MMP-9, MMP-8, MMP-3, MMP-13, MMP-12, MMP-1, PRKCB | 15.59 | 3.01E-05 | 7.11E-04 |
| UP_SEQ_FEATURE | Metal ion-binding site: calcium 1 | 10 | 2.14 | 1.15E-06 | PRKCA, APCS, MMP-9, MMP-8, MMP-7, MMP-3, MMP-13, MMP-12, MMP-1, PRKCB | 9.09 | 6.70E-05 | 0.001923 |
| UP_SEQ_FEATURE | Metal ion-binding site: calcium 2; via carbonyl oxygen | 9 | 1.93 | 3.72E-06 | PRKCA, MMP-9, MMP-8, MMP-7, MMP-3, MMP-13, MMP-12, MMP-1, PRKCB | 9.44 | 1.90E-04 | 0.006252 |
| UP_SEQ_FEATURE | Metal ion-binding site: calcium 2 | 10 | 2.14 | 6.62E-06 | PRKCA, APCS, MMP-9, MMP-8, MMP-7, MMP-3, MMP-13, MMP-12, MMP-1, PRKCB | 7.44 | 3.19E-04 | 0.011118 |
| Annotation cluster 24 | Enrichment score: 5.69 | |||||||
| UP_SEQ_FEATURE | Metal ion-binding site: zinc 2; catalytic | 8 | 1.71 | 1.45E-08 | ACE, MMP-9, MMP-8, MMP-7, MMP-3, MMP-13, MMP-12, MMP-1 | 23.39 | 1.32E-06 | 2.43E-05 |
| INTERPRO | Peptidoglycan binding like | 9 | 1.93 | 3.26E-08 | MMP-20, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 16.09 | 1.54E-06 | 5.07E-05 |
| UP_SEQ_FEATURE | Metal ion-binding site: zinc 2; in inhibited form | 7 | 1.50 | 3.93E-08 | MMP-9, MMP-8, MMP-7, MMP-3, MMP-13, MMP-12, MMP-1 | 28.65 | 3.39E-06 | 6.60E-05 |
| SP_PIR_KEYWORDS | Metalloproteinase | 9 | 1.93 | 5.85E-08 | ACE, MMP-9, MMP-8, MMP-7, MME, MMP-3, MMP-13, MMP-12, MMP-1 | 15.45 | 9.65E-07 | 8.48E-05 |
| INTERPRO | Peptidase, metallopeptidases | 9 | 1.93 | 8.89E-07 | MMP-20, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 11.09 | 3.07E-05 | 0.001385 |
| SP_PIR_KEYWORDS | Collagen degradation | 7 | 1.50 | 1.93E-06 | MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-1 | 16.96 | 2.43E-05 | 0.002803 |
| SMART | ZnMc | 9 | 1.93 | 2.48E-06 | MMP-20, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-12, MMP-1 | 9.62 | 4.63E-05 | 0.003017 |
| GOTERM_BP_FAT | Multicellular organismal catabolic process | 8 | 1.71 | 1.52E-05 | ACE, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-1 | 9.33 | 4.29E-04 | 0.027575 |
| GOTERM_BP_FAT | Collagen catabolic process | 7 | 1.50 | 3.21E-05 | MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-1 | 10.62 | 7.95E-04 | 0.058172 |
| GOTERM_BP_FAT | Multicellular organismal macromolecule metabolic process | 8 | 1.71 | 5.27E-05 | ACE, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-1 | 7.83 | 0.00113118 | 0.095563 |
| GOTERM_BP_FAT | Multicellular organismal metabolic process | 8 | 1.71 | 1.74E-04 | ACE, MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-1 | 6.56 | 0.0030583 | 0.315166 |
| GOTERM_BP_FAT | Collagen metabolic process | 7 | 1.50 | 2.49E-04 | MMP-9, MMP-8, MMP-7, MMP-16, MMP-3, MMP-13, MMP-1 | 7.58 | 0.00404851 | 0.451323 |
| Annotation cluster 25 | Enrichment score: 5.36 | |||||||
| GOTERM_BP_FAT | Hemopoiesis | 24 | 5.14 | 3.31E-06 | LYN, PLEK, EPAS1, MMP-9, PPARG, TP53, CDK6, RB1, KIT, KDR, ATP7A, HDAC4, CDC42, ACE, G6PD, BCL2, LCK, CASP8, ZAP70, ADAM17, BCL6, TRAF6, IL2, SYK | 3.08 | 1.21E-04 | 0.006014 |
| GOTERM_BP_FAT | Immune system development | 26 | 5.57 | 4.69E-06 | MMP-9, UNG, PPARG, KIT, CDC42, ACE, BCL2, CASP8, ZAP70, BCL6, TRAF6, SYK, PLEK, LYN, EPAS1, TGFBR1, TP53, CDK6, RB1, KDR, ATP7A, HDAC4, G6PD, LCK, ADAM17, IL2 | 2.86 | 1.59E-04 | 0.00851 |
| GOTERM_BP_FAT | Hemopoietic or lymphoid organ development | 25 | 5.35 | 5.23E-06 | MMP-9, PPARG, KIT, CDC42, ACE, BCL2, CASP8, ZAP70, BCL6, TRAF6, SYK, PLEK, LYN, EPAS1, TGFBR1, TP53, CDK6, RB1, KDR, ATP7A, HDAC4, G6PD, LCK, ADAM17, IL2 | 2.92 | 1.74E-04 | 0.009499 |
| Annotation cluster 26 | Enrichment score: 5.32 | |||||||
| GOTERM_BP_FAT | Programmed cell death | 44 | 9.42 | 2.08E-06 | TRAF2, PPARD, HRAS, XIAP, ADORA2A, GJA1, BCL2L1, SFN, KIT, PTEN, AKT1, TNFRSF1A, APP, CASP3, ECE1, CXCR4, TIAM1, PTK2B, CASP7, BCL2, RAC1, CASP8, TGM2, PAK1, TRAF6, CASP1, SGK1, ROCK1, GZMA, LGALS1, TP53, BIRC7, RAF1, GZMB, CDK5, BRCA1, PLG, NAE1, VCP, F2, LCK, MAPK8, MDM4, PDCD6IP | 2.18 | 7.88E-05 | 0.003772 |
| GOTERM_BP_FAT | Cell death | 48 | 10.28 | 5.10E-06 | TRAF2, PPARD, HRAS, SPG7, XIAP, ADORA2A, GJA1, BCL2L1, SFN, KIT, HPRT1, PTEN, AKT1, TNFRSF1A, APP, CASP3, ECE1, CXCR4, TIAM1, PTK2B, CASP7, BCL2, RAC1, CASP8, TGM2, PAK1, TRAF6, CASP1, AR, SGK1, ROCK1, GZMA, LGALS1, TP53, BIRC7, RAF1, GZMB, CDK5, BRCA1, PLG, NAE1, VCP, LCK, F2, CTSD, MAPK8, MDM4, PDCD6IP | 2.02 | 1.71E-04 | 0.009255 |
| GOTERM_BP_FAT | Death | 48 | 10.28 | 6.06E-06 | TRAF2, PPARD, HRAS, SPG7, XIAP, ADORA2A, GJA1, BCL2L1, SFN, KIT, HPRT1, PTEN, AKT1, TNFRSF1A, APP, CASP3, ECE1, CXCR4, TIAM1, PTK2B, CASP7, BCL2, RAC1, CASP8, TGM2, PAK1, TRAF6, CASP1, AR, SGK1, ROCK1, GZMA, LGALS1, TP53, BIRC7, RAF1, GZMB, CDK5, BRCA1, PLG, NAE1, VCP, LCK, F2, CTSD, MAPK8, MDM4, PDCD6IP | 2.01 | 1.95E-04 | 0.011004 |
| GOTERM_BP_FAT | Apoptosis | 42 | 8.99 | 8.02E-06 | TRAF2, PPARD, HRAS, XIAP, ADORA2A, GJA1, BCL2L1, SFN, PTEN, AKT1, TNFRSF1A, APP, CASP3, ECE1, CXCR4, TIAM1, PTK2B, CASP7, BCL2, RAC1, CASP8, PAK1, CASP1, TRAF6, SGK1, ROCK1, GZMA, LGALS1, TP53, BIRC7, RAF1, GZMB, CDK5, BRCA1, PLG, NAE1, VCP, F2, LCK, MAPK8, MDM4, PDCD6IP | 2.12 | 2.46E-04 | 0.014551 |
| Annotation cluster 27 | Enrichment score: 5.27 | |||||||
| GOTERM_BP_FAT | Glucose metabolic process | 20 | 4.28 | 6.48E-07 | PDK1, RBP4, PPARD, ALDH5A1, PDK3, PDK4, FBP1, DLAT, PPP1CC, PPARGC1A, PDHB, PCK1, AKT1, PPP1CA, TPI1, G6PD, PYGL, MAPK14, GSK3B, PDHA1 | 3.96 | 2.75E-05 | 0.001176 |
| GOTERM_BP_FAT | Hexose metabolic process | 21 | 4.50 | 5.31E-06 | PDK1, RBP4, PPARD, ALDH5A1, PDK3, PDK4, FBP1, DLAT, PPP1CC, PPARGC1A, PDHB, PCK1, AKT1, KHK, PPP1CA, TPI1, G6PD, PYGL, MAPK14, GSK3B, PDHA1 | 3.32 | 1.75E-04 | 0.009641 |
| GOTERM_BP_FAT | Monosaccharide metabolic process | 21 | 4.50 | 4.51E-05 | PDK1, RBP4, PPARD, ALDH5A1, PDK3, PDK4, FBP1, DLAT, PPP1CC, PPARGC1A, PDHB, PCK1, AKT1, KHK, PPP1CA, TPI1, G6PD, PYGL, MAPK14, GSK3B, PDHA1 | 2.87 | 9.94E-04 | 0.081769 |
| Annotation cluster 28 | Enrichment score: 4.77 | |||||||
| UP_SEQ_FEATURE | Short sequence motif: TXY | 6 | 1.28 | 5.80E-06 | MAPK1, MAPK14, MAPK3, MAPK9, MAPK8, MAPK10 | 20.46 | 2.88E-04 | 0.009734 |
| INTERPRO | MAP kinase, conserved site | 6 | 1.28 | 1.12E-05 | MAPK1, MAPK14, MAPK3, MAPK9, MAPK8, MAPK10 | 17.87 | 3.13E-04 | 0.017402 |
| GOTERM_MF_FAT | MAP kinase activity | 6 | 1.28 | 7.48E-05 | MAPK1, MAPK14, MAPK3, MAPK9, MAPK8, MAPK10 | 12.36 | 0.00154179 | 0.11556 |
| Annotation cluster 29 | Enrichment score: 4.76 | |||||||
| GOTERM_BP_FAT | Blood coagulation | 15 | 3.21 | 5.98E-06 | PLAT, F11, F10, PLEK, ADORA2A, F8, F9, CD40, F7, PLG, LPA, SERPINE1, F2, SERPINC1, PLAU | 4.46 | 1.95E-04 | 0.01085 |
| GOTERM_BP_FAT | Coagulation | 15 | 3.21 | 5.98E-06 | PLAT, F11, F10, PLEK, ADORA2A, F8, F9, CD40, F7, PLG, LPA, SERPINE1, F2, SERPINC1, PLAU | 4.46 | 1.95E-04 | 0.01085 |
| GOTERM_BP_FAT | Hemostasis | 15 | 3.21 | 1.17E-05 | PLAT, F11, F10, PLEK, ADORA2A, F8, F9, CD40, F7, PLG, LPA, SERPINE1, F2, SERPINC1, PLAU | 4.21 | 3.47E-04 | 0.021303 |
| GOTERM_BP_FAT | Regulation of body fluid levels | 15 | 3.21 | 2.26E-04 | PLAT, F11, F10, PLEK, ADORA2A, F8, F9, CD40, F7, PLG, LPA, SERPINE1, F2, SERPINC1, PLAU | 3.23 | 0.00375601 | 0.410281 |
| Annotation cluster 30 | Enrichment score: 4.68 | |||||||
| UP_SEQ_FEATURE | Region of interest: S-adenosyl-L-methionine binding | 9 | 1.93 | 1.55E-06 | EHMT1, PNMT, METTL1, SETD8, GAMT, COMT, GNMT, EHMT2, NNMT | 10.52 | 8.76E-05 | 0.002605 |
| UP_SEQ_FEATURE | Binding site: S-adenosyl-L- methionine | 10 | 2.14 | 3.45E-06 | EHMT1, HNMT, PNMT, SETD8, GAMT, COMT, GNMT, EHMT2, TPMT, NNMT | 8.02 | 1.82E-04 | 0.005799 |
| SP_PIR_KEYWORDS | S-Adenosyl-L-methionine | 12 | 2.57 | 4.03E-05 | EHMT1, HNMT, PNMT, METTL1, SETD8, GAMT, COMT, GNMT, EHMT2, AMD1, TPMT, NNMT | 4.80 | 3.49E-04 | 0.05845 |
| SP_PIR_KEYWORDS | Methyltransferase | 12 | 2.57 | 8.74E-04 | TYMS, EHMT1, HNMT, PNMT, METTL1, SETD8, GAMT, COMT, GNMT, EHMT2, TPMT, NNMT | 3.39 | 0.00613378 | 1.260168 |
| Annotation cluster 31 | Enrichment score: 4.64 | |||||||
| GOTERM_BP_FAT | Regulation of carbohydrate metabolic process | 10 | 2.14 | 3.66E-06 | AKT1, HDAC4, PLEK, IGF1, NR3C1, INSR, PPARGC1A, NR1H4, ARNT, AKT2 | 7.78 | 1.31E-04 | 0.006647 |
| GOTERM_BP_FAT | Regulation of cellular carbohydrate metabolic process | 9 | 1.93 | 2.64E-05 | AKT1, HDAC4, PLEK, IGF1, NR3C1, INSR, PPARGC1A, ARNT, AKT2 | 7.18 | 6.86E-04 | 0.047923 |
| GOTERM_BP_FAT | Regulation of glucose metabolic process | 8 | 1.71 | 1.20E-04 | AKT1, HDAC4, IGF1, NR3C1, INSR, PPARGC1A, ARNT, AKT2 | 6.93 | 0.00225032 | 0.21798 |
| Annotation cluster 32 | Enrichment score: 4.32 | |||||||
| SP_PIR_KEYWORDS | Zinc | 98 | 20.99 | 2.10E-09 | THRA, THRB, S100A7, MMP-9, IDE, MMP-8, MMP-7, ADH1C, ADH1B, ADH1A, MMP-3, MMP-1, BTK, PGR, FNTB, HMHA1, APP, ASPA, RARB, CPA1, DDAH1, NQO2, RARG, ROCK1, BRAF, RXRA, UBR2, PPP1CC, PPP1CA, PYGO1, ADAMTS5, ADAMTS4, TRAF2, MME, NR1H2, VDR, ACE, ECE1, ALB, CDA, GLO1, TRAF6, NR1H4, NR1H3, CPA4, ITK, EHMT1, ESRRG, EHMT2, BRCA1, NR1I3, NR1I2, S100B, AIRE, HGS, PARP1, CPB2, PPARA, SPG7, PPARD, PPARG, ZCWPW1, MMP-20, KDM5A, PRKCA, AR, TP53, PRKCI, ESR1, MMP-16, ESR2, MMP-13, MMP-12, PRKCB, PRKCQ, FOLH1, NAALAD2, CA4, ADAM17, MDM2, PDE9A, MDM4, CA2, CA1, XIAP, CA13, ADH5, ADH7, EEA1, NR3C1, ADH4, PPP2CA, BCL6, L3MBTL2, BIRC7, SIRT5, RAF1, QPCT, LTA4H, KDM4A | 1.84 | 4.10E-08 | 3.04E-06 |
| GOTERM_MF_FAT | Transition metal ion binding | 123 | 26.34 | 0.00201 | THRA, THRB, S100A7, MMP-9, IDE, MMP-8, ADH1C, MMP-7, ADH1B, ADH1A, MMP-3, MMP-1, BTK, PGR, FNTB, HMHA1, APP, ASPA, FNTA, MAT1A, CPA1, RARB, DDAH1, NQO2, RARG, ROCK1, BRAF, RXRA, PIM1, F8, UBR2, PPP1CC, POR, PPP1CA, PDE5A, PYGO1, ADAMTS5, ADAMTS4, XDH, TRAF2, MME, NR1H2, ARG1, VDR, ACE, ECE1, ALB, ARG2, PIR, CDA, GLO1, TRAF6, NR1H4, NR1H3, ITK, CPA4, EHMT1, TGFBR1, ESRRG, ACACB, EHMT2, BRCA1, GART, ATP7A, HDAC4, NR1I3, NR1I2, S100B, AIRE, HGS, PTPN1, PARP1, ABL1, CPB2, ABO, PPARA, PPARD, SPG7, METAP1, METAP2, PPARG, ZCWPW1, MMP-20, HIF1AN, PDE8A, KDM5A, PRKCA, AR, ESR1, PRKCI, TP53, MMP-16, ESR2, MMP-13, MMP-12, PRKCB, PRKCQ, FOLH1, NAALAD2, ADAM17, CA4, MDM2, PDE9A, CA2, MDM4, CA1, TPH1, XIAP, CA13, ADH5, EEA1, ADH7, NR3C1, ADH4, PPP2CA, BCL6, L3MBTL2, BIRC7, SIRT5, RAF1, PCK1, QPCT, FYN, LTA4H, KDM4A | 1.27 | 0.02584329 | 3.062766 |
| GOTERM_MF_FAT | Zinc ion binding | 97 | 20.77 | 0.026629 | THRA, THRB, S100A7, MMP-9, IDE, MMP-8, MMP-7, ADH1C, ADH1B, ADH1A, MMP-3, MMP-1, BTK, PGR, FNTB, HMHA1, APP, ASPA, FNTA, RARB, CPA1, DDAH1, NQO2, RARG, ROCK1, BRAF, RXRA, UBR2, PDE5A, PYGO1, ADAMTS5, ADAMTS4, TRAF2, MME, NR1H2, VDR, ACE, ECE1, CDA, GLO1, TRAF6, NR1H4, NR1H3, CPA4, ITK, EHMT1, ESRRG, EHMT2, BRCA1, HDAC4, NR1I3, NR1I2, S100B, AIRE, HGS, PTPN1, PARP1, CPB2, PPARA, SPG7, PPARD, PPARG, ZCWPW1, MMP- 20, KDM5A, PRKCA, AR, TP53, PRKCI, ESR1, MMP-16, ESR2, MMP-13, MMP-12, PRKCB, PRKCQ, FOLH1, NAALAD2, CA4, ADAM17, MDM2, MDM4, CA2, CA1, XIAP, CA13, ADH5, ADH7, EEA1, NR3C1, ADH4, BCL6, L3MBTL2, BIRC7, SIRT5, RAF1, QPCT, LTA4H, KDM4A | 1.21 | 0.19883278 | 34.1204 |
| Annotation cluster 33 | Enrichment score: 4.22 | |||||||
| GOTERM_BP_FAT | Phenol metabolic process | 9 | 1.93 | 1.39E-05 | ATP7A, PNMT, EPAS1, SULT1B1, MAOA, SULT1A1, SULT1A2, COMT, HPRT1 | 7.80 | 3.95E-04 | 0.025154 |
| GOTERM_BP_FAT | Diol metabolic process | 8 | 1.71 | 9.89E-05 | ATP7A, PNMT, EPAS1, MAOA, SULT1A1, SULT1A2, COMT, HPRT1 | 7.14 | 0.00196673 | 0.179443 |
| GOTERM_BP_FAT | Catechol metabolic process | 8 | 1.71 | 9.89E-05 | ATP7A, PNMT, EPAS1, MAOA, SULT1A1, SULT1A2, COMT, HPRT1 | 7.14 | 0.00196673 | 0.179443 |
| GOTERM_BP_FAT | Catecholamine metabolic process | 8 | 1.71 | 9.89E-05 | ATP7A, PNMT, EPAS1, MAOA, SULT1A1, SULT1A2, COMT, HPRT1 | 7.14 | 0.00196673 | 0.179443 |
| Annotation cluster 34 | Enrichment score: 4.16 | |||||||
| PIR_SUPERFAMILY | Alcohol sulfotransferase | 7 | 1.50 | 1.65E-06 | SULT2A1, SULT1B1, SULT1A1, SULT1A2, SULT1C2, SULT1E1, SULT1C3 | 15.14 | 1.82E-04 | 0.002238 |
| UP_SEQ_FEATURE | Nucleotide phosphate- binding region: PAPS | 8 | 1.71 | 9.72E-06 | SULT2A1, SULT1B1, SULT1A1, SULT2B1, SULT1A2, SULT1C2, SULT1E1, SULT1C3 | 10.23 | 4.54E-04 | 0.016313 |
| INTERPRO | Sulfotransferase | 8 | 1.71 | 1.86E-05 | SULT2A1, SULT1B1, SULT1A1, SULT2B1, SULT1A2, SULT1C2, SULT1E1, SULT1C3 | 9.23 | 4.90E-04 | 0.028912 |
| GOTERM_MF_FAT | Sulfotransferase activity | 8 | 1.71 | 0.00138 | SULT2A1, SULT1B1, SULT1A1, SULT2B1, SULT1A2, SULT1C2, SULT1E1, SULT1C3 | 4.71 | 0.01896331 | 2.111955 |
| GOTERM_MF_FAT | Transferase activity, transferring sulfur- containing groups | 8 | 1.71 | 0.0037 | SULT2A1, SULT1B1, SULT1A1, SULT2B1, SULT1A2, SULT1C2, SULT1E1, SULT1C3 | 3.98 | 0.04321176 | 5.571159 |
| Annotation cluster 35 | Enrichment score: 4.11 | |||||||
| GOTERM_BP_FAT | Protein complex assembly | 36 | 7.71 | 2.61E-05 | HSD17B10, TRAF2, HRAS, GRB2, IDE, GJA1, PEX5, FKBP1A, HPRT1, SRC, MIF, TNFRSF1A, IGF1R, PTK2, PTK2B, RAC1, CASP8, TGM2, CDA, GNMT, INSR, SYK, PPP2R1A, HSP90AA1, APCS, MAP2K1, BRAF, ALDH5A1, TP53, CD40, CDK5, PPARGC1A, NCK2, VCP, MDM2, MDM4 | 2.16 | 6.89E-04 | 0.047358 |
| GOTERM_BP_FAT | Protein complex biogenesis | 36 | 7.71 | 2.61E-05 | HSD17B10, TRAF2, HRAS, GRB2, IDE, GJA1, PEX5, FKBP1A, HPRT1, SRC, MIF, TNFRSF1A, IGF1R, PTK2, PTK2B, RAC1, CASP8, TGM2, CDA, GNMT, INSR, SYK, PPP2R1A, HSP90AA1, APCS, MAP2K1, BRAF, ALDH5A1, TP53, CD40, CDK5, PPARGC1A, NCK2, VCP, MDM2, MDM4 | 2.16 | 6.89E-04 | 0.047358 |
| GOTERM_BP_FAT | Macromolecular complex assembly | 41 | 8.78 | 1.65E-04 | NCBP2, HSD17B10, TRAF2, NCBP1, HRAS, GRB2, IDE, GJA1, PEX5, FKBP1A, HPRT1, SRC, MIF, IGF1R, TNFRSF1A, PTK2, HIST1H4A, PTK2B, RAC1, CASP8, TGM2, CDA, GNMT, INSR, SYK, PPP2R1A, HSP90AA1, APCS, MAP2K1, BRAF, ALDH5A1, SNUPN, TP53, CD40, CDK5, PPARGC1A, NCK2, VCP, HIST1H3A, MDM2, MDM4, HIST1H3G | 1.87 | 0.00291459 | 0.298718 |
| GOTERM_BP_FAT | Macromolecular complex subunit organization | 42 | 9.00 | 3.20E-04 | NCBP2, HSD17B10, NCBP1, TRAF2, HRAS, GRB2, IDE, GJA1, PEX5, FKBP1A, HPRT1, SRC, MIF, IGF1R, TNFRSF1A, PTK2, HIST1H4A, PTK2B, RAC1, CASP8, TGM2, CDA, GNMT, INSR, SYK, PPP2R1A, HSP90AA1, APCS, MAP2K1, BRAF, ALDH5A1, SNUPN, TP53, CD40, CDK5, PPARGC1A, NCK2, VCP, HIST1H3A, PLA2G2A, MDM2, MDM4, HIST1H3G | 1.79 | 0.0048243 | 0.578406 |
| Annotation cluster 36 | Enrichment score: 4.00 | |||||||
| GOTERM_BP_FAT | Regulation of cell activation | 18 | 3.85 | 6.55E-05 | TRAF2, PLEK, ADORA2A, ERBB2, CD40, PNP, MIF, CD38, PRKCQ, NCK2, CASP3, LCK, SFTPD, ZAP70, BCL6, TRAF6, SYK, IL2 | 3.12 | 0.00136967 | 0.118797 |
| GOTERM_BP_FAT | Regulation of lymphocyte activation | 16 | 3.43 | 1.08E-04 | TRAF2, ADORA2A, ERBB2, CD40, PNP, PRKCQ, NCK2, CD38, CASP3, LCK, SFTPD, ZAP70, BCL6, TRAF6, SYK, IL2 | 3.28 | 0.00208753 | 0.196338 |
| GOTERM_BP_FAT | Regulation of leukocyte activation | 17 | 3.64 | 1.17E-04 | TRAF2, ADORA2A, ERBB2, CD40, PNP, MIF, PRKCQ, NCK2, CD38, CASP3, LCK, SFTPD, ZAP70, BCL6, TRAF6, SYK, IL2 | 3.11 | 0.00220533 | 0.212381 |
| GOTERM_BP_FAT | Regulation of T-cell activation | 14 | 3.00 | 1.22E-04 | TRAF2, ADORA2A, ERBB2, PNP, PRKCQ, NCK2, CASP3, LCK, SFTPD, ZAP70, BCL6, TRAF6, SYK, IL2 | 3.63 | 0.0022739 | 0.221543 |
| Annotation cluster 37 | Enrichment score: 3.97 | |||||||
| GOTERM_BP_FAT | Positive regulation of phosphorylation | 13 | 2.78 | 7.83E-05 | EGFR, PRKCA, LYN, TGFBR1, IGF1, TTK, BCL2, F2, ADAM17, TNK2, INSR, SYK, IL2 | 4.07 | 0.00161717 | 0.142086 |
| GOTERM_BP_FAT | Positive regulation of phosphate metabolic process | 13 | 2.78 | 1.06E-04 | EGFR, PRKCA, LYN, TGFBR1, IGF1, TTK, BCL2, F2, ADAM17, TNK2, INSR, SYK, IL2 | 3.94 | 0.00206093 | 0.191517 |
| GOTERM_BP_FAT | Positive regulation of phosphorus metabolic process | 13 | 2.78 | 1.06E-04 | EGFR, PRKCA, LYN, TGFBR1, IGF1, TTK, BCL2, F2, ADAM17, TNK2, INSR, SYK, IL2 | 3.94 | 0.00206093 | 0.191517 |
| GOTERM_BP_FAT | Positive regulation of protein amino acid phosphorylation | 12 | 2.57 | 1.56E-04 | PRKCA, LYN, BCL2, TGFBR1, F2, ADAM17, IGF1, TTK, TNK2, INSR, IL2, SYK | 4.09 | 0.00277529 | 0.282881 |
| Annotation cluster 38 | Enrichment score: 3.85 | |||||||
| GOTERM_BP_FAT | Regulation of establishment of protein localization | 15 | 3.21 | 4.31E-05 | RBP4, ADORA2A, TGFBR1, PRKCI, CD40, CDK5, PTPN11, AKT1, PRKCQ, PDPK1, GSK3B, PRKACA, TRAF6, CASP1, IL2 | 3.76 | 9.64E-04 | 0.078194 |
| GOTERM_BP_FAT | Regulation of protein localization | 16 | 3.43 | 4.84E-05 | RBP4, ADORA2A, TGFBR1, PRKCI, CD40, CDK5, PTPN11, AKT1, PRKCQ, PDPK1, GSK3B, BCL2, PRKACA, TRAF6, CASP1, IL2 | 3.52 | 0.00105367 | 0.087828 |
| GOTERM_BP_FAT | Regulation of protein transport | 12 | 2.57 | 0.00132 | PRKCQ, RBP4, ADORA2A, GSK3B, TGFBR1, PRKACA, CD40, TRAF6, CASP1, CDK5, IL2, PTPN11 | 3.19 | 0.0157027 | 2.369144 |
| Annotation cluster 39 | Enrichment score: 3.73 | |||||||
| INTERPRO | EGF receptor, L domain | 5 | 1.07 | 1.96E-05 | EGFR, IGF1R, ERBB4, ERBB2, INSR | 25.53 | 5.03E-04 | 0.030568 |
| INTERPRO | Furin-like cysteine-rich region | 5 | 1.07 | 1.96E-05 | EGFR, IGF1R, ERBB4, ERBB2, INSR | 25.53 | 5.03E-04 | 0.030568 |
| INTERPRO | Furin-like repeat | 5 | 1.07 | 0.001343 | EGFR, IGF1R, ERBB4, ERBB2, INSR | 9.93 | 0.02767984 | 2.072751 |
| SMART | FU | 5 | 1.07 | 0.002245 | EGFR, IGF1R, ERBB4, ERBB2, INSR | 8.61 | 0.03373603 | 2.698772 |
| Annotation cluster 40 | Enrichment score: 3.73 | |||||||
| GOTERM_BP_FAT | Cell projection organization | 30 | 6.42 | 1.26E-05 | FGFR1, CDK5R1, NRP1, ADORA2A, ERBB2, GJA1, HPRT1, PTEN, AKT1, CDC42, IGF1R, APP, PTK2, CXCR4, PTK2B, BCL2, RAC1, CDH23, EGFR, PRKCA, MAP2K1, ROCK1, PLEK, RXRA, CDK5, PTPN11, ATP7A, NCK2, S100B, SDCBP | 2.47 | 3.66E-04 | 0.022889 |
| GOTERM_BP_FAT | Neuron projection development | 23 | 4.93 | 3.91E-05 | PRKCA, EGFR, FGFR1, CDK5R1, NRP1, MAP2K1, ADORA2A, RXRA, ERBB2, GJA1, HPRT1, CDK5, PTEN, PTPN11, ATP7A, IGF1R, PTK2, APP, S100B, CXCR4, PTK2B, BCL2, RAC1 | 2.73 | 9.07E-04 | 0.070998 |
| GOTERM_BP_FAT | Neuron projection morphogenesis | 20 | 4.28 | 7.95E-05 | PRKCA, EGFR, CDK5R1, NRP1, MAP2K1, ADORA2A, RXRA, ERBB2, GJA1, HPRT1, CDK5, PTPN11, ATP7A, IGF1R, PTK2, APP, S100B, CXCR4, BCL2, RAC1 | 2.85 | 0.00163049 | 0.144174 |
| GOTERM_BP_FAT | Neuron development | 26 | 5.57 | 1.45E-04 | FGFR1, CDK5R1, NRP1, ADORA2A, ERBB2, GJA1, HPRT1, PTEN, IGF1R, APP, PTK2, PTK2B, CXCR4, BCL2, RAC1, CDH23, EGFR, PRKCA, RET, MAP2K1, RXRA, PRKCI, CDK5, PTPN11, ATP7A, S100B | 2.33 | 0.00262906 | 0.26354 |
| GOTERM_BP_FAT | Neuron differentiation | 30 | 6.42 | 2.81E-04 | FGFR1, CDK5R1, NRP1, ADORA2A, ERBB2, GJA1, PEX5, HPRT1, PTEN, CDC42, IGF1R, APP, PTK2, CXCR4, PTK2B, BCL2, RAC1, CDH23, EGFR, PRKCA, RET, MAP2K1, RXRA, TGFBR1, PRKCI, CDK5, PTPN11, ATP7A, PRKCQ, S100B | 2.08 | 0.00445775 | 0.50945 |
| GOTERM_BP_FAT | Cell morphogenesis | 26 | 5.57 | 3.08E-04 | CDK5R1, NRP1, ADORA2A, ERBB2, GJA1, HPRT1, IGF1R, CDC42, APP, PTK2, CXCR4, BCL2, RAC1, BCL6, DLG1, CDH23, EGFR, PRKCA, MAP2K1, RXRA, PRKCI, CDK5, PTPN11, ATP7A, S100B, ERBB2IP | 2.22 | 0.00471944 | 0.557892 |
| GOTERM_BP_FAT | Cell projection morphogenesis | 20 | 4.28 | 4.83E-04 | PRKCA, EGFR, CDK5R1, NRP1, MAP2K1, ADORA2A, RXRA, ERBB2, GJA1, HPRT1, CDK5, PTPN11, ATP7A, IGF1R, PTK2, APP, S100B, CXCR4, BCL2, RAC1 | 2.48 | 0.00684232 | 0.873863 |
| GOTERM_BP_FAT | Cell part morphogenesis | 20 | 4.28 | 8.26E-04 | PRKCA, EGFR, CDK5R1, NRP1, MAP2K1, ADORA2A, RXRA, ERBB2, GJA1, HPRT1, CDK5, PTPN11, ATP7A, IGF1R, PTK2, APP, S100B, CXCR4, BCL2, RAC1 | 2.37 | 0.01076448 | 1.488171 |
| GOTERM_BP_FAT | Cellular component morphogenesis | 26 | 5.57 | 0.001471 | CDK5R1, NRP1, ADORA2A, ERBB2, GJA1, HPRT1, IGF1R, CDC42, APP, PTK2, CXCR4, BCL2, RAC1, BCL6, DLG1, CDH23, EGFR, PRKCA, MAP2K1, RXRA, PRKCI, CDK5, PTPN11, ATP7A, S100B, ERBB2IP | 1.99 | 0.01716579 | 2.636383 |
| Annotation cluster 41 | Enrichment score: 3.64 | |||||||
| GOTERM_CC_FAT | Membrane-enclosed lumen | 83 | 17.77 | 9.64E-05 | NCBP2, NCBP1, TSG101, IDE, PEX5, XRCC1, AGXT, C1QC, PDHB, AKT1, KDM1A, PARN, APP, CASP3, CSNK2A1, CASP7, CASP8, PDHA1, BRD4, CFD, KDM5A, RARG, LYN, GATM, TP53, F8, TLE1, RB1, PPP1CC, PPARGC1A, WEE1, CDK2, MAPK1, PNKP, PPP1CA, MAPK3, MDM2, MAPK8, ME2, NEK2, FARS2, CHEK1, KIT, NR3C1, CHEK2, HIST1H4A, BCHE, ALB, MDC1, GSTK1, NUDT21, HSD17B4, PDK1, BCKDHA, DHX9, KAT2B, CES1, EPAS1, PDK3, PDK4, BCKDHB, IGF1, SIRT5, DLAT, NXF1, BRCA1, PCK1, HDAC4, DUSP3, HDAC2, RPS6KA1, VCP, PLK1, MAPK14, DYRK1A, ALDH2, KDM4A, ABL1, PARP1, PARP2, HDAC8, HDAC7, UBE2E1 | 1.50 | 0.00321839 | 0.13268 |
| GOTERM_CC_FAT | Organelle lumen | 81 | 17.34 | 1.45E-04 | NCBP2, NCBP1, TSG101, IDE, PEX5, XRCC1, AGXT, C1QC, PDHB, AKT1, KDM1A, PARN, APP, CASP3, CSNK2A1, CASP7, CASP8, PDHA1, BRD4, CFD, KDM5A, RARG, TP53, F8, TLE1, RB1, PPP1CC, PPARGC1A, WEE1, CDK2, MAPK1, PNKP, PPP1CA, MAPK3, MDM2, MAPK8, ME2, NEK2, FARS2, CHEK1, KIT, NR3C1, CHEK2, HIST1H4A, BCHE, ALB, MDC1, GSTK1, NUDT21, HSD17B4, PDK1, BCKDHA, DHX9, KAT2B, CES1, EPAS1, PDK3, PDK4, BCKDHB, IGF1, SIRT5, DLAT, NXF1, BRCA1, PCK1, HDAC4, DUSP3, HDAC2, RPS6KA1, VCP, PLK1, MAPK14, DYRK1A, ALDH2, KDM4A, ABL1, PARP1, PARP2, HDAC8, HDAC7, UBE2E1 | 1.50 | 0.00410352 | 0.199949 |
| GOTERM_CC_FAT | Intracellular organelle lumen | 76 | 16.27 | 8.68E-04 | NCBP2, NCBP1, TSG101, IDE, PEX5, XRCC1, AGXT, C1QC, PDHB, AKT1, KDM1A, PARN, CASP3, CSNK2A1, CASP7, CASP8, PDHA1, BRD4, KDM5A, RARG, TP53, TLE1, RB1, PPP1CC, PPARGC1A, WEE1, CDK2, MAPK1, PNKP, PPP1CA, MAPK3, MDM2, MAPK8, ME2, NEK2, FARS2, CHEK1, NR3C1, KIT, CHEK2, HIST1H4A, BCHE, MDC1, GSTK1, NUDT21, HSD17B4, PDK1, BCKDHA, DHX9, KAT2B, CES1, EPAS1, PDK3, PDK4, BCKDHB, SIRT5, DLAT, NXF1, BRCA1, PCK1, HDAC4, DUSP3, HDAC2, RPS6KA1, VCP, PLK1, MAPK14, DYRK1A, ALDH2, KDM4A, ABL1, PARP1, PARP2, HDAC8, HDAC7, UBE2E1 | 1.44 | 0.01759311 | 1.189132 |
| Annotation cluster 42 | Enrichment score: 3.60 | |||||||
| GOTERM_BP_FAT | Negative regulation of macromolecule biosynthetic process | 36 | 7.71 | 1.32E-04 | PPARA, PPARD, THRA, THRB, TSG101, PPARG, NR1H2, KDM1A, VDR, FNTA, EED, SFTPD, BCL6, RARB, NR1H4, PRKCA, RARG, EHMT1, RXRA, ELANE, TP53, SIRT5, TLE1, RB1, BRCA1, HDAC4, NR1I2, NCOA2, HDAC2, BACE2, UBA3, MDM2, FABP4, SETD8, MDM4, HDAC8 | 2.00 | 0.00243752 | 0.238856 |
| GOTERM_BP_FAT | Negative regulation of gene expression | 34 | 7.28 | 1.33E-04 | PPARA, PPARD, THRA, THRB, TSG101, PPARG, BCL2L1, NR1H2, KDM1A, VDR, SND1, EED, BCL6, RARB, INSR, NR1H4, RARG, EHMT1, RXRA, TP53, SIRT5, TLE1, RB1, BRCA1, HDAC4, NCOA2, HDAC2, NR1I2, UBA3, MDM2, FABP4, SETD8, MDM4, HDAC8 | 2.05 | 0.00244468 | 0.240933 |
| GOTERM_BP_FAT | Negative regulation of biosynthetic process | 37 | 7.92 | 1.53E-04 | PPARA, PPARD, THRA, THRB, TSG101, PPARG, NR1H2, KDM1A, VDR, FNTA, EED, SFTPD, BCL6, RARB, NR1H4, PRKCA, RARG, EHMT1, PLEK, RXRA, ELANE, TP53, SIRT5, TLE1, RB1, BRCA1, HDAC4, NR1I2, NCOA2, HDAC2, BACE2, UBA3, MDM2, FABP4, SETD8, MDM4, HDAC8 | 1.96 | 0.00272817 | 0.276544 |
| GOTERM_BP_FAT | Negative regulation of cellular biosynthetic process | 36 | 7.71 | 2.14E-04 | PPARA, PPARD, THRA, THRB, TSG101, PPARG, NR1H2, KDM1A, VDR, EED, SFTPD, BCL6, RARB, NR1H4, PRKCA, RARG, EHMT1, PLEK, RXRA, ELANE, TP53, SIRT5, TLE1, RB1, BRCA1, HDAC4, NR1I2, NCOA2, HDAC2, BACE2, UBA3, MDM2, FABP4, SETD8, MDM4, HDAC8 | 1.95 | 0.00359443 | 0.388594 |
| GOTERM_BP_FAT | Negative regulation of transcription | 31 | 6.64 | 2.76E-04 | PPARA, PPARD, THRA, THRB, TSG101, PPARG, NR1H2, KDM1A, VDR, EED, BCL6, RARB, NR1H4, RARG, EHMT1, RXRA, TP53, SIRT5, TLE1, RB1, BRCA1, HDAC4, NCOA2, HDAC2, NR1I2, UBA3, MDM2, FABP4, SETD8, MDM4, HDAC8 | 2.05 | 0.00440251 | 0.500666 |
| GOTERM_BP_FAT | Negative regulation of nitrogen compound metabolic process | 33 | 7.07 | 4.87E-04 | PPARA, PPARD, THRA, THRB, TSG101, PPARG, COMT, NR1H2, VDR, KDM1A, EED, CDA, BCL6, RARB, NR1H4, RARG, EHMT1, RXRA, TP53, SIRT5, TLE1, RB1, BRCA1, HDAC4, NCOA2, HDAC2, NR1I2, UBA3, MDM2, FABP4, SETD8, MDM4, HDAC8 | 1.93 | 0.00683604 | 0.88072 |
| GOTERM_BP_FAT | Negative regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | 32 | 6.85 | 8.13E-04 | PPARA, PPARD, THRA, THRB, TSG101, PPARG, NR1H2, KDM1A, VDR, EED, CDA, BCL6, RARB, NR1H4, RARG, EHMT1, RXRA, TP53, SIRT5, TLE1, RB1, BRCA1, HDAC4, NCOA2, HDAC2, NR1I2, UBA3, MDM2, FABP4, SETD8, MDM4, HDAC8 | 1.90 | 0.0106896 | 1.465914 |
| Annotation cluster 43 | Enrichment score: 3.59 | |||||||
| GOTERM_BP_FAT | Positive regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | 42 | 8.99 | 1.87E-05 | NCBP1, PPARA, HRAS, THRA, THRB, ADORA2A, PPARG, ARNT, NR1H2, IGF1R, TNFRSF1A, APP, RARB, KDM5A, INSR, NR1H4, NR1H3, BCKDHA, AR, RARG, MAP2K1, EPAS1, TGFBR1, RXRA, ESRRG, TP53, IGF1, RB1, ESR2, CD40, PPARGC1A, BRCA1, CDK2, TNKS2, HDAC4, MAPK1, NCOA1, NCOA2, HDAC2, MAPK14, JAK3, IL2 | 2.04 | 5.12E-04 | 0.034005 |
| GOTERM_BP_FAT | Positive regulation of gene expression | 37 | 7.92 | 1.99E-04 | PPARA, THRA, THRB, PPARG, KIT, ARNT, NR1H2, TNFRSF1A, APP, RARB, KDM5A, NR1H4, NR1H3, BCKDHA, AR, RARG, EPAS1, TGFBR1, RXRA, ESRRG, TP53, ESR1, IGF1, RB1, ESR2, PPARGC1A, BRCA1, CDK2, HDAC4, MAPK1, NCOA1, NCOA2, HDAC2, MAPK14, MAPK9, JAK3, IL2 | 1.93 | 0.00342307 | 0.360451 |
| GOTERM_BP_FAT | Positive regulation of transcription from RNA polymerase II promoter | 27 | 5.78 | 2.41E-04 | PPARA, THRA, THRB, PPARG, ARNT, NR1H2, TNFRSF1A, APP, RARB, NR1H4, NR1H3, AR, RARG, EPAS1, RXRA, TP53, IGF1, RB1, ESR2, PPARGC1A, HDAC4, NCOA1, NCOA2, HDAC2, MAPK14, JAK3, IL2 | 2.21 | 0.00396061 | 0.437076 |
| GOTERM_BP_FAT | Positive regulation of RNA metabolic process | 32 | 6.85 | 2.87E-04 | PPARA, NCBP1, THRA, THRB, PPARG, ARNT, NR1H2, TNFRSF1A, APP, RARB, NR1H4, NR1H3, BCKDHA, AR, RARG, MAP2K1, EPAS1, RXRA, ESRRG, TP53, IGF1, RB1, ESR2, PPARGC1A, BRCA1, HDAC4, NCOA1, NCOA2, HDAC2, MAPK14, JAK3, IL2 | 2.02 | 0.00447801 | 0.519307 |
| GOTERM_BP_FAT | Positive regulation of transcription | 34 | 7.28 | 9.92E-04 | PPARA, THRA, THRB, PPARG, ARNT, NR1H2, TNFRSF1A, APP, RARB, KDM5A, NR1H4, NR1H3, BCKDHA, AR, RARG, EPAS1, RXRA, TGFBR1, ESRRG, TP53, IGF1, RB1, ESR2, PPARGC1A, BRCA1, CDK2, HDAC4, MAPK1, NCOA1, NCOA2, HDAC2, MAPK14, JAK3, IL2 | 1.83 | 0.01261501 | 1.785517 |
| GOTERM_BP_FAT | Positive regulation of transcription, DNA dependent | 30 | 6.42 | 0.001113 | PPARA, THRA, THRB, PPARG, ARNT, NR1H2, TNFRSF1A, APP, RARB, NR1H4, NR1H3, BCKDHA, AR, RARG, EPAS1, RXRA, ESRRG, TP53, IGF1, RB1, ESR2, PPARGC1A, BRCA1, HDAC4, NCOA1, NCOA2, HDAC2, MAPK14, JAK3, IL2 | 1.91 | 0.0137637 | 2.001185 |
| Annotation cluster 44 | Enrichment score: 3.57 | |||||||
| GOTERM_BP_FAT | Regulation of glucose metabolic process | 8 | 1.71 | 1.20E-04 | AKT1, HDAC4, IGF1, NR3C1, INSR, PPARGC1A, ARNT, AKT2 | 6.93 | 0.00225032 | 0.21798 |
| GOTERM_BP_FAT | Positive regulation of glucose metabolic process | 6 | 1.28 | 2.99E-04 | AKT1, IGF1, INSR, PPARGC1A, ARNT, AKT2 | 9.58 | 0.00461973 | 0.540924 |
| GOTERM_BP_FAT | Positive regulation of cellular carbohydrate metabolic process | 6 | 1.28 | 3.88E-04 | AKT1, IGF1, INSR, PPARGC1A, ARNT, AKT2 | 9.10 | 0.00566472 | 0.70135 |
| GOTERM_BP_FAT | Positive regulation of carbohydrate metabolic process | 6 | 1.28 | 3.88E-04 | AKT1, IGF1, INSR, PPARGC1A, ARNT, AKT2 | 9.10 | 0.00566472 | 0.70135 |
| Annotation cluster 45 | Enrichment score: 3.49 | |||||||
| GOTERM_BP_FAT | Positive regulation of cyclin-dependent prote in kinase activity during G1/S | 4 | 0.86 | 1.38E-04 | EGFR, AKT1, PIM1, ADAM17 | 30.33 | 0.00252379 | 0.25015 |
| GOTERM_BP_FAT | Regulation of cyclin- dependent protein kinase activity during G1/S | 4 | 0.86 | 1.38E-04 | EGFR, AKT1, PIM1, ADAM17 | 30.33 | 0.00252379 | 0.25015 |
| GOTERM_BP_FAT | Positive regulation of cyclin-dependent protein kinase activity | 4 | 0.86 | 0.00175 | EGFR, AKT1, PIM1, ADAM17 | 15.17 | 0.01988844 | 3.129 |
| Annotation cluster 46 | Enrichment score: 3.44 | |||||||
| GOTERM_BP_FAT | Regulation of lymphocyte proliferation | 12 | 2.57 | 8.23E-05 | CD38, PRKCQ, NCK2, CASP3, ERBB2, ZAP70, SFTPD, BCL6, CD40, PNP, IL2, SYK | 4.39 | 0.00166731 | 0.149307 |
| GOTERM_BP_FAT | Regulation of mononuclear cell proliferation | 12 | 2.57 | 9.20E-05 | CD38, PRKCQ, NCK2, CASP3, ERBB2, ZAP70, SFTPD, BCL6, CD40, PNP, IL2, SYK | 4.33 | 0.00183952 | 0.1668 |
| GOTERM_BP_FAT | Regulation of leukocyte proliferation | 12 | 2.57 | 9.20E-05 | CD38, PRKCQ, NCK2, CASP3, ERBB2, ZAP70, SFTPD, BCL6, CD40, PNP, IL2, SYK | 4.33 | 0.00183952 | 0.1668 |
| GOTERM_BP_FAT | Positive regulation of cell activation | 13 | 2.78 | 2.86E-04 | TRAF2, PLEK, CD40, PNP, PRKCQ, NCK2, CD38, LCK, ZAP70, BCL6, TRAF6, SYK, IL2 | 3.55 | 0.00449264 | 0.518482 |
| GOTERM_BP_FAT | Positive regulation of lymphocyte activation | 12 | 2.57 | 3.36E-04 | TRAF2, CD38, PRKCQ, NCK2, LCK, ZAP70, BCL6, CD40, TRAF6, PNP, IL2, SYK | 3.75 | 0.00504753 | 0.607989 |
| GOTERM_BP_FAT | Positive regulation of lymphocyte proliferation | 9 | 1.93 | 4.03E-04 | CD38, PRKCQ, NCK2, ZAP70, BCL6, CD40, PNP, IL2, SYK | 4.96 | 0.0058375 | 0.729273 |
| GOTERM_BP_FAT | Positive regulation of mononuclear cell proliferation | 9 | 1.93 | 4.57E-04 | CD38, PRKCQ, NCK2, ZAP70, BCL6, CD40, PNP, IL2, SYK | 4.87 | 0.00655668 | 0.826394 |
| GOTERM_BP_FAT | Positive regulation of leukocyte proliferation | 9 | 1.93 | 4.57E-04 | CD38, PRKCQ, NCK2, ZAP70, BCL6, CD40, PNP, IL2, SYK | 4.87 | 0.00655668 | 0.826394 |
| GOTERM_BP_FAT | Positive regulation of leukocyte activation | 12 | 2.57 | 7.21E-04 | TRAF2, CD38, PRKCQ, NCK2, LCK, ZAP70, BCL6, CD40, TRAF6, PNP, IL2, SYK | 3.43 | 0.00971853 | 1.300425 |
| GOTERM_BP_FAT | Regulation of T-cell proliferation | 9 | 1.93 | 9.16E-04 | PRKCQ, NCK2, CASP3, ERBB2, ZAP70, SFTPD, PNP, IL2, SYK | 4.40 | 0.01184284 | 1.650107 |
| GOTERM_BP_FAT | Positive regulation of T-cell activation | 9 | 1.93 | 0.003429 | TRAF2, PRKCQ, NCK2, LCK, ZAP70, TRAF6, PNP, IL2, SYK | 3.59 | 0.03453311 | 6.043888 |
| Annotation cluster 47 | Enrichment score: 3.30 | |||||||
| GOTERM_BP_FAT | Negative regulation of transferase activity | 12 | 2.57 | 3.07E-04 | PRKCA, AKT1, PPP2R1A, PDPK1, CASP3, HMGCR, ADORA2A, PPP2CA, PPARG, FABP4, RB1, SFN | 3.79 | 0.00471957 | 0.555258 |
| GOTERM_BP_FAT | Negative regulation of protein kinase activity | 11 | 2.36 | 5.57E-04 | PRKCA, AKT1, PPP2R1A, PDPK1, CASP3, HMGCR, ADORA2A, PPP2CA, FABP4, RB1, SFN | 3.84 | 0.00764671 | 1.006445 |
| GOTERM_BP_FAT | Negative regulation of kinase activity | 11 | 2.36 | 7.30E-04 | PRKCA, AKT1, PPP2R1A, PDPK1, CASP3, HMGCR, ADORA2A, PPP2CA, FABP4, RB1, SFN | 3.71 | 0.00979929 | 1.316686 |
| Annotation cluster 48 | Enrichment score: 3.14 | |||||||
| SP_PIR_KEYWORDS | SH3 domain | 16 | 3.43 | 1.94E-04 | ITK, LYN, GRB2, HCK, NCF4, SRC, BTK, NCK2, TJP1, FYN, MAP3K9, LCK, TNK2, ABL1, ARHGAP9, DLG1 | 3.14 | 0.0015486 | 0.280714 |
| UP_SEQ_FEATURE | SH3 domain | 14 | 3.00 | 4.16E-04 | ITK, LYN, HCK, NCF4, SRC, BTK, TJP1, FYN, MAP3K9, LCK, TNK2, ABL1, ARHGAP9, DLG1 | 3.24 | 0.0157035 | 0.695681 |
| INTERPRO | Src homology-3 domain | 16 | 3.43 | 9.74E-04 | ITK, LYN, GRB2, HCK, NCF4, SRC, BTK, NCK2, TJP1, FYN, MAP3K9, LCK, TNK2, ABL1, ARHGAP9, DLG1 | 2.69 | 0.0206159 | 1.506777 |
| SMART | SH3 domain | 16 | 3.43 | 0.003617 | ITK, LYN, GRB2, HCK, NCF4, SRC, BTK, NCK2, TJP1, FYN, MAP3K9, LCK, TNK2, ABL1, ARHGAP9, DLG1 | 2.33 | 0.04574947 | 4.316077 |
| Annotation cluster 49 | Enrichment score: 3.06 | |||||||
| GOTERM_BP_FAT | Mononuclear cell proliferation | 8 | 1.71 | 5.31E-04 | ACE, CXCR4, FYN, BCL2, TP53, FKBP1A, CD40, HPRT1 | 5.51 | 0.00731779 | 0.959108 |
| GOTERM_BP_FAT | Leukocyte proliferation | 8 | 1.71 | 5.31E-04 | ACE, CXCR4, FYN, BCL2, TP53, FKBP1A, CD40, HPRT1 | 5.51 | 0.00731779 | 0.959108 |
| GOTERM_BP_FAT | Lymphocyte proliferation | 7 | 1.50 | 0.002353 | CXCR4, FYN, BCL2, TP53, FKBP1A, CD40, HPRT1 | 5.06 | 0.02523826 | 4.185354 |
Abbreviations: DAVID, Database for Annotation, Visualization, and Integrated Discovery; UA, ursolic acid; FDR, false discovery rate.
Table 4.
The KEGG pathways by the DAVID database for the target list of UA derived from molecular docking calculations
| Category | Term | Count | % | P-value | Genes | Fold enrichment | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|
| KEGG pathway | Pathways in cancer | 58 | 12.42 | 4.97E-12 | HSP90AB1, HRAS, PPARD, MMP-9, PPARG, PTEN, MMP-1, AKT1, CDC42, CASP3, CASP8, RARB, AKT2, PRKCA, EGFR, PIK3CG, AR, RET, HSP90AA1, BRAF, RXRA, TP53, CDK6, RB1, CDK2, PRKCB, MAPK1, MAPK3, MDM2, MAPK9, MAPK8, GSTP1, FGFR2, TRAF2, FGFR1, XIAP, GRB2, ERBB2, BCL2L1, KIT, ARNT, IGF1R, PTK2, BCL2, RAC1, TRAF6, CSF1R, EPAS1, MAP2K1, TGFBR1, MET, RAF1, IGF1, MAPK10, HDAC2, GSK3B, JAK1, ABL1 | 2.64 | 7.75E-10 | 5.97E-09 |
| KEGG pathway | Prostate cancer | 28 | 6.00 | 7.42E-12 | FGFR2, HSP90AB1, FGFR1, HRAS, GRB2, ERBB2, PTEN, AKT1, IGF1R, PDPK1, BCL2, AKT2, EGFR, PIK3CG, AR, HSP90AA1, BRAF, MAP2K1, TP53, IGF1, RAF1, RB1, CDK2, MAPK1, GSK3B, MAPK3, MDM2, GSTP1 | 4.69 | 5.79E-10 | 8.92E-09 |
| KEGG pathway | Non-small-cell lung cancer | 20 | 4.28 | 6.18E-10 | PRKCA, EGFR, PIK3CG, HRAS, MAP2K1, BRAF, GRB2, RXRA, ERBB2, TP53, RAF1, CDK6, RB1, PRKCB, AKT1, MAPK1, PDPK1, MAPK3, RARB, AKT2 | 5.52 | 3.21E-08 | 7.43E-07 |
| KEGG pathway | ERBB signaling pathway | 25 | 5.35 | 1.01E-09 | HRAS, ERBB4, GRB2, ERBB2, SRC, AKT1, PTK2, PAK4, PAK1, AKT2, PRKCA, EGFR, PIK3CG, BRAF, MAP2K1, RAF1, MAPK10, PRKCB, NCK2, MAPK1, GSK3B, MAPK3, MAPK9, MAPK8, ABL1 | 4.29 | 3.95E-08 | 1.22E-06 |
| KEGG pathway | Fc epsilon RI signaling pathway | 23 | 4.93 | 3.27E-09 | PRKCA, PIK3CG, PDK1, HRAS, MAP2K1, LYN, GRB2, RAF1, MAPK10, PRKCB, BTK, AKT1, MAPK1, GAB2, FYN, MAPK14, MAPK3, RAC1, PLA2G2A, MAPK9, MAPK8, AKT2, SYK | 4.40 | 1.02E-07 | 3.93E-06 |
| KEGG pathway | Glioma | 20 | 4.28 | 1.18E-08 | PRKCA, EGFR, PIK3CG, HRAS, MAP2K1, BRAF, GRB2, TP53, IGF1, RAF1, CDK6, RB1, PTEN, PRKCB, AKT1, MAPK1, IGF1R, MAPK3, MDM2, AKT2 | 4.73 | 3.07E-07 | 1.42E-05 |
| KEGG pathway | Pancreatic cancer | 21 | 4.50 | 2.31E-08 | EGFR, PIK3CG, MAP2K1, BRAF, ERBB2, TGFBR1, TP53, RAF1, CDK6, RB1, BCL2L1, MAPK10, AKT1, CDC42, MAPK1, MAPK3, RAC1, MAPK9, JAK1, MAPK8, AKT2 | 4.35 | 5.16E-07 | 2.78E-05 |
| KEGG pathway | Progesterone-mediated oocyte maturation | 23 | 4.93 | 2.38E-08 | HSP90AB1, PIK3CG, HSP90AA1, MAP2K1, BRAF, GNAI1, IGF1, RAF1, PDE3B, MAPK10, CDK2, AKT1, PGR, MAPK1, IGF1R, RPS6KA1, PLK1, MAPK14, MAPK3, MAPK9, MAPK8, PRKACA, AKT2 | 3.99 | 4.65E-07 | 2.87E-05 |
| KEGG pathway | Drug metabolism | 19 | 4.07 | 5.60E-08 | GSTA1, GSTA2, GSTA3, GSTA4, MAOA, MAOB, ADH5, ADH1C, ADH1B, GSTT1, ADH1A, ADH7, ALDH3A1, GSTM1, GSTM2, ADH4, GSTK1, GSTZ1, GSTO2, GSTO1, GSTP1 | 4.57 | 9.71E-07 | 6.73E-05 |
| KEGG pathway | Focal adhesion | 36 | 7.71 | 9.22E-08 | HRAS, XIAP, GRB2, ERBB2, PTEN, SRC, AKT1, CDC42, IGF1R, PDPK1, PTK2, PAK4, BCL2, RAC1, PAK1, AKT2, EGFR, PIK3CG, PRKCA, MAP2K1, ROCK1, BRAF, MET, IGF1, RAF1, MAPK10, PPP1CC, PRKCB, KDR, MAPK1, PPP1CA, FYN, GSK3B, MAPK3, MAPK9, MAPK8 | 2.67 | 1.44E-06 | 1.11E-04 |
| KEGG pathway | Neurotrophin signaling pathway | 27 | 5.78 | 1.05E-07 | HRAS, YWHAZ, GRB2, MAPKAPK2, IRAK4, AKT1, CDC42, BCL2, RAC1, TRAF6, AKT2, PDK1, PIK3CG, BRAF, MAP2K1, TP53, RAF1, MAPK10, PTPN11, MAPK1, RPS6KA1, MAPK14, GSK3B, MAPK3, MAPK9, MAPK8, ABL1 | 3.25 | 1.49E-06 | 1.26E-04 |
| KEGG pathway | Insulin signaling pathway | 28 | 6.00 | 1.66E-07 | HRAS, GRB2, PDE3B, AKT1, PDPK1, PRKACA, INSR, AKT2, PIK3CG, MAP2K1, BRAF, PRKCI, FBP1, RAF1, ACACB, MAPK10, PPP1CC, PPARGC1A, PCK1, MAPK1, PPP1CA, EIF4E, PYGL, GSK3B, MAPK3, MAPK9, MAPK8, PTPN1 | 3.09 | 2.16E-06 | 1.99E-04 |
| KEGG pathway | Chronic myeloid leukemia | 20 | 4.28 | 2.62E-07 | PIK3CG, HRAS, BRAF, MAP2K1, GRB2, TGFBR1, TP53, RAF1, CDK6, RB1, BCL2L1, PTPN11, AKT1, MAPK1, HDAC2, GAB2, MAPK3, MDM2, ABL1, AKT2 | 3.98 | 3.14E-06 | 3.15E-04 |
| KEGG pathway | Colorectal cancer | 21 | 4.50 | 3.78E-07 | EGFR, PIK3CG, MAP2K1, BRAF, GRB2, TGFBR1, MET, TP53, RAF1, MAPK10, AKT1, MAPK1, IGF1R, CASP3, GSK3B, BCL2, MAPK3, RAC1, MAPK9, MAPK8, AKT2 | 3.73 | 4.21E-06 | 4.54E-04 |
| KEGG pathway | T-cell receptor signaling pathway | 24 | 5.14 | 4.36E-07 | PIK3CG, PDK1, ITK, HRAS, MAP2K1, GRB2, RAF1, AKT1, CDC42, MAPK1, NCK2, PRKCQ, FYN, MAPK14, GSK3B, PAK4, LCK, MAPK3, ZAP70, MAPK9, PAK1, IL2, AKT2, DLG1 | 3.31 | 4.54E-06 | 5.24E-04 |
| KEGG pathway | Melanoma | 19 | 4.07 | 5.39E-07 | EGFR, PIK3CG, FGFR1, HRAS, MAP2K1, BRAF, MET, TP53, IGF1, RAF1, CDK6, RB1, PTEN, AKT1, MAPK1, IGF1R, MAPK3, MDM2, AKT2 | 3.99 | 5.25E-06 | 6.48E-04 |
| KEGG pathway | Endometrial cancer | 16 | 3.43 | 8.09E-07 | EGFR, PIK3CG, HRAS, BRAF, MAP2K1, GRB2, ERBB2, TP53, RAF1, PTEN, AKT1, MAPK1, PDPK1, GSK3B, MAPK3, AKT2 | 4.59 | 7.42E-06 | 9.72E-04 |
| KEGG pathway | Metabolism of xenobiotics by cytochrome P450 | 17 | 3.64 | 1.12E-06 | GSTA1, GSTA2, GSTA3, GSTA4, ADH5, ADH1C, ADH1B, GSTT1, ADH1A, ADH7, ALDH3A1, GSTM1, GSTM2, ADH4, GSTK1, GSTZ1, GSTO2, GSTO1, GSTP1 | 4.23 | 9.69E-06 | 0.001345 |
| KEGG pathway | VEGF signaling pathway | 19 | 4.07 | 1.29E-06 | PRKCA, PIK3CG, HRAS, MAP2K1, MAPKAPK3, RAF1, MAPKAPK2, SRC, KDR, PRKCB, AKT1, CDC42, MAPK1, PTK2, MAPK14, MAPK3, RAC1, PLA2G2A, AKT2 | 3.78 | 1.06E-05 | 0.001555 |
| KEGG pathway | Bladder cancer | 14 | 3.00 | 1.84E-06 | EGFR, HRAS, BRAF, MAP2K1, ERBB2, MMP-9, TP53, RAF1, RB1, MMP-1, MAPK1, TYMP, MAPK3, MDM2 | 4.97 | 1.43E-05 | 0.00221 |
| KEGG pathway | Adherens junction | 19 | 4.07 | 1.95E-06 | EGFR, PTPRB, FGFR1, ERBB2, TGFBR1, MET, SRC, CSNK2A2, CDC42, MAPK1, IGF1R, TJP1, CSNK2A1, FYN, MAPK3, RAC1, PTPN1, INSR, MLLT4 | 3.68 | 1.45E-05 | 0.002349 |
| KEGG pathway | Renal cell carcinoma | 18 | 3.85 | 2.15E-06 | PIK3CG, HRAS, MAP2K1, BRAF, EPAS1, GRB2, MET, RAF1, ARNT, PTPN11, AKT1, CDC42, MAPK1, PAK4, MAPK3, RAC1, PAK1, AKT2 | 3.83 | 1.52E-05 | 0.002583 |
| KEGG pathway | Natural killer cell- mediated cytotoxicity | 25 | 5.35 | 5.63E-06 | ITGAL, HRAS, GRB2, CASP3, PTK2B, RAC1, ZAP70, PAK1, SYK, PRKCA, PIK3CG, BRAF, MAP2K1, HLA-A, RAF1, GZMB, HLA-B, HLA-E, HLA-G, PTPN11, PRKCB, MAPK1, FYN, LCK, MAPK3 | 2.80 | 3.82E-05 | 0.006771 |
| KEGG pathway | Chemokine signaling pathway | 30 | 6.42 | 1.36E-05 | HRAS, GRB2, GNAI1, ADRBK1, AKT1, CDC42, PTK2, CXCR4, TIAM1, PTK2B, RAC1, PRKACA, GNG2, PAK1, AKT2, PIK3CG, ITK, MAP2K1, ROCK1, BRAF, LYN, HCK, RAF1, PRKCB, MAPK1, GNB1, GSK3B, MAPK3, GRK6, JAK3 | 2.39 | 8.82E-05 | 0.016317 |
| KEGG pathway | Glutathione metabolism | 14 | 3.00 | 1.57E-05 | GSTA1, GSTA2, ODC1, GSTA3, GSTA4, GSTT1, GSTM1, GSTM2, G6PD, GSTK1, GSTZ1, GSTO2, GSTO1, GSTP1 | 4.18 | 9.77E-05 | 0.018816 |
| KEGG pathway | Complement and coagulation cascades | 16 | 3.43 | 3.59E-05 | PLAT, F11, F10, F8, F9, F7, C1QC, PLG, C1QA, C1QB, SERPINE1, F2, SERPINC1, CFD, CPB2, PLAU | 3.46 | 2.15E-04 | 0.043123 |
| KEGG pathway | B-cell receptor signaling pathway | 16 | 3.43 | 9.98E-05 | PIK3CG, HRAS, LYN, MAP2K1, GRB2, RAF1, PRKCB, BTK, AKT1, MAPK1, DAPP1, GSK3B, MAPK3, RAC1, AKT2, SYK | 3.18 | 5.76E-04 | 0.119869 |
| KEGG pathway | Adipocytokine signaling pathway | 15 | 3.21 | 1.03E-04 | TRAF2, PPARA, CHKB, RXRA, ACACB, MAPK10, PPARGC1A, PCK1, PTPN11, AKT1, PRKCQ, TNFRSF1A, MAPK9, MAPK8, AKT2 | 3.34 | 5.76E-04 | 0.124225 |
| KEGG pathway | Epithelial cell signaling in Helicobacter pylori infection | 15 | 3.21 | 1.22E-04 | EGFR, LYN, MET, MAPK10, SRC, PTPN11, CDC42, TJP1, CASP3, MAPK14, RAC1, ADAM17, MAPK9, MAPK8, PAK1 | 3.29 | 6.59E-04 | 0.147114 |
| KEGG pathway | Long-term depression | 15 | 3.21 | 1.44E-04 | PRKCA, PPP2R1A, HRAS, BRAF, LYN, MAP2K1, GNAI1, IGF1, RAF1, PRKCB, IGF1R, MAPK1, PPP2CA, MAPK3, PLA2G2A | 3.24 | 7.51E-04 | 0.173569 |
| KEGG pathway | Endocytosis | 27 | 5.78 | 1.89E-04 | FGFR2, HRAS, ERBB4, TSG101, ADRBK1, EEA1, KIT, SRC, IGF1R, CDC42, CXCR4, TRAF6, CSF1R, EGFR, RET, TGFBR1, MET, PRKCI, HLA-A, HLA-B, HLA-E, HLA-G, KDR, GRK6, HGS, MDM2, PDCD6IP | 2.19 | 9.52E-04 | 0.227192 |
| KEGG pathway | GnRH signaling pathway | 18 | 3.85 | 2.21E-04 | PRKCA, EGFR, HRAS, MAP2K1, GRB2, RAF1, MAPK10, SRC, PRKCB, CDC42, MAPK1, PTK2B, MAPK14, MAPK3, PLA2G2A, MAPK9, PRKACA, MAPK8 | 2.74 | 0.0010769 | 0.265376 |
| KEGG pathway | Acute myeloid leukemia | 13 | 2.78 | 3.55E-04 | PIK3CG, PPARD, HRAS, BRAF, MAP2K1, GRB2, PIM1, RAF1, KIT, AKT1, MAPK1, MAPK3, AKT2 | 3.34 | 0.0016786 | 0.426361 |
| KEGG pathway | Small-cell lung cancer | 16 | 3.43 | 3.73E-04 | PIK3CG, TRAF2, XIAP, RXRA, TP53, CDK6, RB1, BCL2L1, PTEN, CDK2, AKT1, PTK2, BCL2, RARB, TRAF6, AKT2 | 2.84 | 0.0017096 | 0.44736 |
| KEGG pathway | Thyroid cancer | 9 | 1.93 | 4.55E-04 | MAPK1, HRAS, RET, MAP2K1, BRAF, RXRA, MAPK3, PPARG, TP53 | 4.63 | 0.0020265 | 0.545693 |
| KEGG pathway | Glycolysis/gluconeogenesis | 13 | 2.78 | 4.94E-04 | ADH5, FBP1, ADH1C, ADH1B, ADH1A, ADH7, DLAT, PDHB, PCK1, ALDH3A1, ALDH7A1, TPI1, ADH4, ALDH2, PDHA1 | 3.23 | 0.0021381 | 0.592084 |
| KEGG pathway | Tyrosine metabolism | 11 | 2.36 | 5.00E-04 | PNMT, MAOA, MAOB, ADH5, ADH1C, ADH1B, ADH1A, ADH7, COMT, ALDH3A1, MIF, ADH4, GSTZ1 | 3.73 | 0.0021056 | 0.599243 |
| KEGG pathway | NOD-like receptor signaling pathway | 13 | 2.78 | 6.75E-04 | HSP90AB1, HSP90AA1, XIAP, MAPK10, MAPK1, ERBB2IP, MAPK14, MAPK3, CASP8, MAPK9, MAPK8, CASP1, TRAF6 | 3.13 | 0.002769 | 0.808763 |
| KEGG pathway | MAPK signaling pathway | 33 | 7.07 | 7.57E-04 | FGFR2, TRAF2, FGFR1, HRAS, GRB2, MAPKAPK3, MAPKAPK2, AKT1, CDC42, TNFRSF1A, CASP3, RAC1, PRKACA, PAK1, TRAF6, AKT2, EGFR, PRKCA, MAP2K1, BRAF, TGFBR1, TP53, RAF1, MAPK10, PRKCB, MAPK1, DUSP3, RPS6KA1, MAPK14, MAPK3, PLA2G2A, MAPK9, MAPK8 | 1.84 | 0.0030264 | 0.906871 |
| KEGG pathway | Aldosterone-regulated sodium reabsorption | 10 | 2.14 | 0.001243 | PRKCA, PIK3CG, MAPK1, PDPK1, SGK1, MAPK3, IGF1, SFN, INSR, PRKCB | 3.64 | 0.0048377 | 1.483839 |
| KEGG pathway | Fcγ R-mediated phagocytosis | 16 | 3.43 | 0.001417 | PRKCA, PIK3CG, LYN, MAP2K1, HCK, RAF1, PRKCB, AKT1, CDC42, MAPK1, GAB2, MAPK3, RAC1, PAK1, AKT2, SYK | 2.51 | 0.0053789 | 1.689783 |
| KEGG pathway | p53 signaling pathway | 13 | 2.78 | 0.001583 | TP53, IGF1, CHEK1, CDK6, CHEK2, SFN, PTEN, CDK2, CASP3, CASP8, SERPINE1, MDM2, MDM4 | 2.85 | 0.0058663 | 1.886424 |
| KEGG pathway | Prion diseases | 9 | 1.93 | 0.001753 | C1QA, C1QB, MAPK1, MAP2K1, FYN, MAPK3, STIP1, PRKACA, C1QC | 3.83 | 0.0063436 | 2.086826 |
| KEGG pathway | Axon guidance | 19 | 4.07 | 0.002119 | HRAS, NRP1, ROCK1, GNAI1, MET, EPHB4, CDK5, CDC42, NCK2, MAPK1, PTK2, CXCR4, FYN, GSK3B, PAK4, MAPK3, RAC1, PAK1, ABL1 | 2.20 | 0.0074937 | 2.51844 |
| KEGG pathway | Oocyte meiosis | 17 | 3.64 | 0.002402 | PPP2R1A, YWHAZ, AR, MAP2K1, IGF1, AURKA, PPP1CC, CDK2, PGR, MAPK1, IGF1R, PPP1CA, RPS6KA1, PLK1, PPP2CA, MAPK3, PRKACA | 2.30 | 0.0083009 | 2.849446 |
| KEGG pathway | Pyruvate metabolism | 9 | 1.93 | 0.004272 | ALDH7A1, ME2, ALDH2, GLO1, PDHA1, DLAT, ACACB, PDHB, PCK1 | 3.36 | 0.0144151 | 5.017313 |
| KEGG pathway | Apoptosis | 14 | 3.00 | 0.00474 | PIK3CG, TRAF2, XIAP, TP53, BCL2L1, AKT1, IRAK4, TNFRSF1A, CASP3, CASP7, BCL2, CASP8, PRKACA, AKT2 | 2.40 | 0.0156455 | 5.551863 |
| KEGG pathway | Leukocyte transendothelial migration | 17 | 3.64 | 0.004911 | PRKCA, PIK3CG, ITK, ITGAL, ROCK1, GNAI1, NCF4, MMP-9, PTPN11, PRKCB, CDC42, PTK2, PTK2B, CXCR4, MAPK14, RAC1, MLLT4 | 2.15 | 0.0158742 | 5.747625 |
| KEGG pathway | Long-term potentiation | 12 | 2.57 | 0.00495 | PRKCA, MAPK1, PPP1CA, HRAS, RPS6KA1, MAP2K1, BRAF, MAPK3, RAF1, PRKACA, PPP1CC, PRKCB | 2.63 | 0.0156745 | 5.791613 |
| KEGG pathway | PPAR signaling pathway | 12 | 2.57 | 0.005547 | PPARA, PDPK1, PPARD, CHKB, RXRA, PPARG, FABP3, FABP4, FABP7, MMP-1, PCK1, NR1H3 | 2.59 | 0.0172046 | 6.46863 |
| KEGG pathway | Gap junction | 14 | 3.00 | 0.005771 | PRKCA, EGFR, HRAS, MAP2K1, GRB2, GNAI1, GJA1, RAF1, SRC, PRKCB, MAPK1, TJP1, MAPK3, PRKACA | 2.35 | 0.0175475 | 6.721689 |
| KEGG pathway | Drug metabolism | 9 | 1.93 | 0.006762 | XDH, TYMP, UMPS, CES1, NAT1, NAT2, CDA, HPRT1, TPMT | 3.12 | 0.0201485 | 7.833234 |
| KEGG pathway | mTOR signaling pathway | 10 | 2.14 | 0.006809 | PIK3CG, AKT1, MAPK1, PDPK1, EIF4E, RPS6KA1, BRAF, MAPK3, IGF1, AKT2 | 2.87 | 0.0199102 | 7.886251 |
| KEGG pathway | Toll-like receptor signaling pathway | 15 | 3.21 | 0.006813 | PIK3CG, MAP2K1, CD40, MAPK10, AKT1, IRAK4, MAPK1, MAPK14, MAPK3, RAC1, CASP8, MAPK9, MAPK8, TRAF6, AKT2 | 2.21 | 0.0195569 | 7.890864 |
| KEGG pathway | Viral myocarditis | 12 | 2.57 | 0.00691 | EIF4G1, ITGAL, CASP3, FYN, CASP8, RAC1, HLA-A, HLA-B, CD40, HLA-E, ABL1, HLA-G | 2.52 | 0.0194747 | 7.998282 |
| KEGG pathway | Vascular smooth muscle contraction | 16 | 3.43 | 0.007143 | PRKCA, BRAF, MAP2K1, ROCK1, ADORA2A, RAF1, PPP1CC, PRKCB, PRKCQ, MAPK1, PPP1CA, MAPK3, PLA2G2A, PRKACA, CALCRL, RAMP1 | 2.13 | 0.019771 | 8.257365 |
| KEGG pathway | Primary immunodeficiency | 8 | 1.71 | 0.007398 | UNG, LCK, TAP1, ZAP70, AIRE, JAK3, CD40, BTK | 3.41 | 0.020118 | 8.540799 |
| KEGG pathway | Arginine and proline metabolism | 10 | 2.14 | 0.007733 | ODC1, ARG1, ALDH7A1, GATM, MAOA, ARG2, MAOB, ALDH2, GAMT, AMD1 | 2.81 | 0.0206644 | 8.91132 |
| KEGG pathway | Histidine metabolism | 7 | 1.50 | 0.010917 | ASPA, ALDH7A1, HNMT, MAOA, MAOB, ALDH2, ALDH3A1 | 3.60 | 0.028606 | 12.36319 |
| KEGG pathway | Purine metabolism | 19 | 4.07 | 0.01285 | XDH, NUDT2, DCK, PDE3B, PDE4D, HPRT1, PNP, GART, NME2, PDE2A, ATIC, NME1, NT5M, PDE7A, PDE4A, ADK, PDE4B, PDE5A, PDE9A, PDE8A | 1.85 | 0.0330669 | 14.40039 |
| KEGG pathway | Tight junction | 17 | 3.64 | 0.01618 | PRKCA, PPP2R1A, HRAS, GNAI1, PRKCI, PTEN, SRC, PRKCB, AKT1, CSNK2A2, CDC42, PRKCQ, TJP1, CSNK2A1, PPP2CA, MLLT4, AKT2 | 1.89 | 0.0408596 | 17.80881 |
| KEGG pathway | Renin–angiotensin system | 5 | 1.07 | 0.023312 | ACE, REN, MME, CMA1, CTSG | 4.39 | 0.0576238 | 24.69256 |
| KEGG pathway | Allograft rejection | 7 | 1.50 | 0.030239 | HLA-A, GZMB, HLA-B, CD40, HLA-E, HLA-G, IL2 | 2.90 | 0.0732134 | 30.86838 |
| KEGG pathway | Cell cycle | 15 | 3.21 | 0.038296 | YWHAZ, TP53, TTK, CDK6, CHEK1, RB1, CHEK2, SFN, WEE1, CDK2, HDAC2, PLK1, GSK3B, MDM2, ABL1 | 1.79 | 0.090792 | 37.46662 |
| KEGG pathway | Sulfur metabolism | 4 | 0.86 | 0.041478 | SULT1A1, SULT2B1, SULT1A2, SULT1E1 | 4.97 | 0.0966722 | 39.90887 |
| KEGG pathway | Fatty acid metabolism | 7 | 1.50 | 0.04772 | ALDH7A1, ADH4, CHKB, ADH1C, ALDH2, ADH5, ADH1B, ADH7, ADH1A | 2.61 | 0.1091442 | 44.44857 |
| KEGG pathway | Glycine, serine, and threonine metabolism | 6 | 1.28 | 0.052437 | GATM, MAOA, MAOB, GAMT, GNMT, AGXT | 2.89 | 0.117864 | 47.66781 |
| KEGG pathway | ALS | 8 | 1.71 | 0.060969 | TNFRSF1A, CASP3, MAPK14, BCL2, RAC1, TP53, BCL2L1, CASP1 | 2.25 | 0.1343859 | 53.06039 |
| KEGG pathway | Nicotinate and nicotinamide metabolism | 5 | 1.07 | 0.072217 | NAMPT, CD38, NT5M, PNP, NNMT | 3.11 | 0.155886 | 59.39099 |
| KEGG pathway | Caffeine metabolism | 3 | 0.64 | 0.074832 | XDH, NAT1, NAT2 | 6.39 | 0.1591473 | 60.74629 |
| KEGG pathway | Alzheimer’s disease | 17 | 3.64 | 0.076679 | HSD17B10, CDK5R1, IDE, MME, CDK5, NAE1, MAPK1, TNFRSF1A, APP, CASP3, BACE2, GSK3B, CASP7, MAPK3, CASP8, BACE1, ADAM17 | 1.56 | 0.1607853 | 61.67824 |
| KEGG pathway | Wnt signaling pathway | 16 | 3.43 | 0.078172 | PRKCA, PPP2R1A, PPARD, ROCK1, MMP-7, TP53, MAPK10, PRKCB, CSNK2A2, CSNK2A1, PPP2CA, GSK3B, RAC1, MAPK9, PRKACA, MAPK8 | 1.58 | 0.1616832 | 62.41638 |
| KEGG pathway | Regulation of actin cytoskeleton | 21 | 4.50 | 0.080954 | EGFR, FGFR2, PIK3CG, FGFR1, ITGAL, HRAS, ROCK1, BRAF, MAP2K1, RAF1, PPP1CC, CDC42, MAPK1, PPP1CA, PTK2, TIAM1, PAK4, F2, MAPK3, RAC1, PAK1 | 1.46 | 0.1650651 | 63.75741 |
| KEGG pathway | Dorso–ventral axis formation | 5 | 1.07 | 0.081647 | EGFR, MAPK1, MAP2K1, GRB2, MAPK3 | 2.98 | 0.164358 | 64.08485 |
| KEGG pathway | One carbon pool by folate | 4 | 0.86 | 0.086763 | TYMS, DHFR, ATIC, GART | 3.73 | 0.172031 | 66.4176 |
| KEGG pathway | Type II diabetes mellitus | 7 | 1.50 | 0.090845 | PIK3CG, MAPK1, MAPK3, MAPK9, MAPK8, MAPK10, INSR | 2.22 | 0.1775696 | 68.17856 |
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; DAVID, Database for Annotation, Visualization, and Integrated Discovery; UA, ursolic acid; FDR, false discovery rate; ALS, amyotrophic lateral sclerosis. it shows that UA exerts an ameliorating effect on DMN-induced liver fibrosis in rats through reducing the accumulation of collagen and suppressing the expression of α-SMA in rat HSCs.
Following the bioinformatical prediction of the molecular interactome of UA, we functionally and mechanistically examined the effect of UA on the liver fibrosis through the assessment of collagen accumulation and ROS generation in vitro and in vivo.
UA ameliorates DMN-induced hepatic fibrosis in rats
First, we established the model of liver fibrosis and examined the effect of UA on liver fibrosis in rats through examining the accumulation of collagen in rat HSCs. As shown in Figure 1A and B and Table 5, there is marked difference in the collagen disposition in rat HSCs. Compared to the vehicle group, rats that received 1% DMN treatment remarkably increased the accumulation of collagen in HSCs (Figure 1B). Administration of rats with UA at 20 mg/kg and 40 mg/kg decreased the collagen accumulation in rat HSCs (Figure 1C and D). Notably, high dose (40 mg/kg) of UA normalized the level of collagen in rat HSCs (Figure 1D).
Figure 1.
Effect of UA on DMN-induced hepatic fibrogenesis in rats.
Notes: Rats were administered with 1% DMN dissolved in saline (1 mL/kg body weight) via intraperitoneal injection for 3 consecutive days per week for 4 weeks to induce liver fibrosis. Representative photomicrographs of the liver histology from groups (n=5) treated with vehicle control (A), DMN alone (B), DMN +20 mg/kg UA (C), and DMN +40 mg/kg UA (D). Magnification: ×100.
Abbreviations: UA, ursolic acid; DMN, dimethylnitrosamine.
Table 5.
Effect of UA on rat liver collagen hyperplastic degree in experimental hepatic fibrotic rats (Mean ± SD, n=8)
| Group | Dose (mg kg−1) | N | − | + | ++ | +++ |
|---|---|---|---|---|---|---|
| Normal | wt | 8 | 8 | 0 | 0 | 0 |
| Model• | wt | 8 | 0 | 1 | 4 | 3 |
| UA-2•,★ | 20 | 8 | 1 | 4 | 2 | 1 |
| UA-3•,▲ | 40 | 8 | 1 | 5 | 2 | 0 |
Notes:
P<0.01 vs normal;
P<0.01,
P<0.05 vs model. An increased number of + symbols represent an increase in collagen hyperplastic degree with treatment. − represents without treatment.
Abbreviations: UA, ursolic acid; SD, standard deviation; wt, without treatment.
Furthermore, we examined the expression level of α-SMA, a marker of HSCs activation and fibrogenesis in rat liver. Treatment of rats with 1% DMN gave a remarkable increase (5.7-fold) in the expression lever of α-SMA in rat liver, compared to the vehicle control (Figure 2A and B, P<0.001). However, administration of rats with UA at 20 mg/kg and 40 mg/kg markedly decreased UA-induced expression level of α-SMA by 54.9% and 70.6% in rat liver, respectively (Figure 2A and B, P<0.001). Higher dose (40 mg/kg) of UA exhibited a more potent inhibitory effect on the expression level of α-SMA than that of low dose (20 mg/kg) of UA in rat liver (Figure 2A and B). Taken together,
Figure 2.
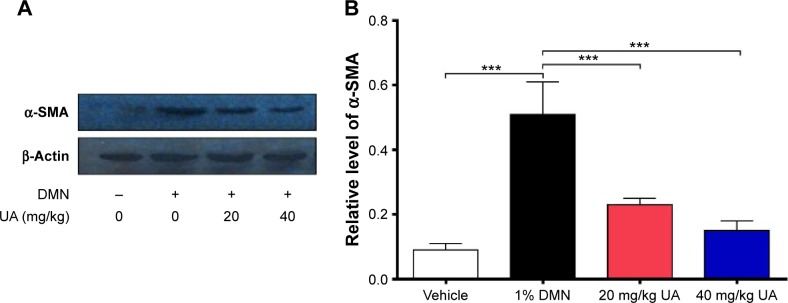
Effect of UA on the expression of α-SMA in rat liver.
Notes: Rats were administered with 1% DMN dissolved in saline (1 mL/kg body weight) via intraperitoneal injection for 3 consecutive days per week for 4 weeks to induce liver fibrosis. (A) Representative blot of α-SMA from groups treated with vehicle control, DMN alone, DMN +20 mg/kg UA, and DMN +40 mg/kg UA. (B) Bar graph showing the relative expression level of α-SMA in rat liver. β-Actin acts as an internal control. Data are expressed as mean ± SD (n=5). ***P<0.001. + and − represent with or without treatment, respectively.
Abbreviations: UA, ursolic acid; α-SMA, α-smooth muscle actin; DMN, dimethylnitrosamine; SD, standard deviation.
UA restores serum SOD level and suppresses serum MDA in DMN-induced liver fibrotic rats
Growing evidence shows that oxidative stress has been implicated in the pathogenesis of liver fibrosis.32 The antioxidative effect of UA was examined in rats treated with 1% DMN by determining the serum levels of SOD and MDA. SOD has a capability of detoxifying superoxide and protecting cells against oxidative stress, whereas MDA is a principal toxic product of lipid peroxidation under oxidative stress.33 As shown in Figure 3A, DMN-alone treated group exhibited low serum level of SOD, and there was a 29.4% decrease (P<0.001), compared to the vehicle group; whereas UA treatment remarkably increased the serum level of SOD. In comparison to the DMN group, there was a 1.6- and 2.0-fold rise in the serum level of SOD (P<0.001). On the contrary, there was an 8.4-fold increase in the serum level of MDA in DMN-alone treated group, compared to the vehicle group (Figure 3B, P<0.001). UA treatment suppressed DMN-induced elevation in the serum level of MDA. There was a 78.4% and 84.2% reduction in the serum level of MDA, compared to DMN group (Figure 3B, P<0.001). Taken together, the data show that UA treatment can attenuate oxidative stress through the regulation of serum level of SOD and MDA in DMN-induced liver fibrotic rats.
Figure 3.
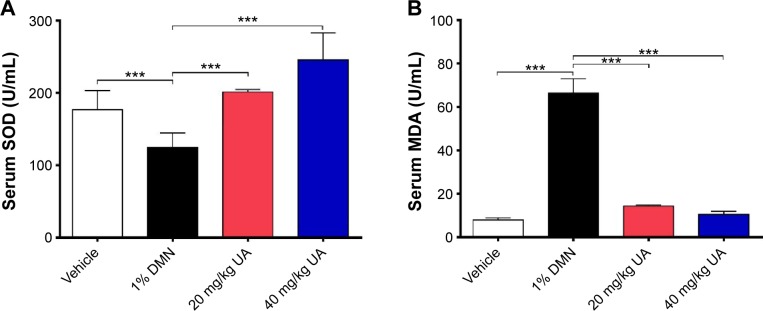
UA increased serum SOD level and decreased serum MDA level in liver fibrotic rats.
Notes: Rats were administered with 1% DMN dissolved in saline (1 mL/kg body weight) by intraperitoneal injection for 3 consecutive days per week for 4 weeks to induce liver fibrosis. Bar graphs showing the serum level of SOD (A) and MDA (B) in rats received treatment of vehicle control, DMN alone, DMN +20 mg/kg UA, and DMN +40 mg/kg UA. Data are expressed as mean ± SD (n=5). ***P<0.001.
Abbreviations: UA, ursolic acid; SOD, superoxide dismutase; MDA, maleic dialdehyde; DMN, dimethylnitrosamine; SD, standard deviation.
UA inhibits proliferation in HSC-T6 cells
Since we have observed the beneficial effects of UA in DMN-induced liver fibrotic rats, we further investigated the molecular mechanisms that underlie the antifibrotic effect of UA in vitro. We first examined the effect of UA (50 µM), DPI (20 µM, an NOX inhibitor), AG490 (50 µM, a specific and potent inhibitor of JAK2), and PD98059 (30 µM, a potent and selective inhibitor of MAP kinase) on the proliferation of HSC-T6 cells using MTT assay (Figure 4). Treatment of HSC-T6 cells with leptin (100 ng/mL) markedly induced cell proliferation over 48 hours (P<0.001). Treatment of HSC-T6 cells with UA, AG490, DPI, or PD98059 prevented leptin-induced proliferation over 48 hours (Figure 4). DPI exhibited the most potent preventive effect, and the data showed that UA exerted a comparable inhibitory effect to DPI to the cell proliferation. Taken together, the initial data suggest that ROS, JAK2, and MAPK/ERK signaling pathways are involved in the leptin-promoted proliferation in HSC-T6 cells.
Figure 4.
UA inhibits the proliferation of HSC-T6 cells.
Notes: Leptin (100 ng/mL) stimulated HSC-T6 cells proliferation at 12 hours, 24 hours, and 48 hours. Leptin-induced HSC-T6 cell proliferation was significantly suppressed by the pretreatment with UA (50 µM), AG490 (50 µM), DPI (20 µM), or PD98059 (30 µM). Data are expressed as mean ± SD from six independent experiments. The symbol *indicates the comparison between leptin and other treatments, *P<0.001.
Abbreviations: UA, ursolic acid; HSC, hepatic stellate cell; DPI, diphenyleneiodonium; SD, standard deviation.
UA suppresses intracellular ROS generation in HSC-T6 cells
We also examined the effect of UA on the ROS generation in the HSC-T6 cells using DCF-DA fluorescence (Figure 5A and B). Within 1 hour, leptin-stimulated HSC-T6 cells showed a significant increase in ROS production compared to the vehicle control group. There was a 3.4-fold elevation in the intracellular ROS level in leptin (100 ng/mL)-treated cells, compared to basal level (P<0.001; Figure 5A). The leptin-induced effect on ROS generation was abolished by pretreatment with UA (50 µM), NAC (10 mM, a ROS scavenger), DPI (20 µM), or AG490 (50 µM) in HSC-T6 cells (P<0.001; Figure 5A). Of note, UA suppressed leptin-induced ROS generation in a time-dependent manner over 24 hours, which was similar to DPI (Figure 5B). NAC potently scavenged intracellular ROS level within 1 hour; however, the ROS scavenging effect of NAC was gradually attenuated over 24 hours (Figure 5B). Taken together, the data suggest that UA reduces intracellular ROS level mainly through the inhibition of enzymatic source of ROS, rather than the scavenging of intracellular ROS.
Figure 5.
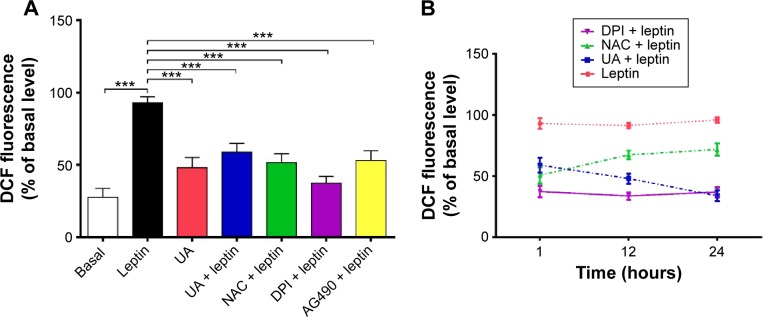
UA inhibits leptin-induced ROS production in HSC-T6 cells.
Notes: HSC-T6 cells were seeded into six-well plates and pretreated with vehicle control, UA (50 µM), NAC (10 mM), DPI (20 µM), or AG490 (50 µM) for 30 minutes. Then, the cells were stimulated by leptin (100 ng/mL), and the intracellular ROS level was measured using DCF-DA fluorescence probes (10 µM). (A) HSC-T6 cells were stimulated by leptin for 1 hour showing a significant induction in ROS production that was blocked by the pretreatment with UA, NAC, DPI, or AG490. (B) UA suppressed the leptin-induced ROS generation in a time-dependent manner and exhibited a similar inhibitory effect on ROS generation to DPI over 24 hours. Data are expressed as mean ± SD from six independent experiments. ***P<0.001.
Abbreviations: UA, ursolic acid; ROS, reactive oxygen species; HSC, hepatic stellate cell; NAC, N-acetyl-L-cysteine; DPI, diphenyleneiodonium; DCF-DA, 2′,7′-dichlorofluorescin diacetate; SD, standard deviation.
UA inhibits leptin-induced NOX activation in HSC-T6 cells
Since we observed the inhibitory effect of UA on intracellular ROS generation via the regulation of enzymatic source, we further examined the effect of UA on the activity of NOX in HSC-T6 cells. As shown in Figure 6, the NOX activity remarkably increased in the HSC-T6 cells when treated with leptin (100 ng/mL) for 1 hour, 12 hours, or 24 hours, compared to the basal level (P<0.001). UA (50 µM) markedly inhibited leptin-induced NOX activation after 1 hour, 12 hours, or 24 hours. UA’s inhibitory effect on NOX activity was similar to that of DPI (20 µM) and AG490 (50 µM). However, NAC showed no effect on leptin-induced NOX activation. Taken together, the data further showed that UA suppresses intracellular ROS production via the modulation of enzymatic source, NOX, in HSC-T6 cells.
Figure 6.
UA suppressed leptin-induced NOX activation in HSC-T6 cells.
Notes: Cells were pretreated for 30 minutes with UA (50 µM), NAC (10 mM), or AG490 (50 µM) and incubated for 1 hour, 12 hours, or 24 hours in the presence of leptin (100 ng/mL). Cells were detached by trypsinization and processed as described in the Materials and methods section. Data are expressed as mean ± SD from six independent experiments. ***P<0.01.
Abbreviations: UA, ursolic acid; NOX, NADPH oxidase; HSC, hepatic stellate cell; NAC, N-acetyl-L-cysteine; SD, standard deviation.
UA inhibits leptin-induced p47phox membrane translocation
p47phox translocation from cytoplasm to cell membrane occurs during NOX activation; thus, the translocation was examined through testing the content of the membrane-bound p47phox and the total cellular p47phox in HSC-T6 cells. There was a marked alteration in the leptin (100 ng/mL)-induced translocation of p47phox, whereas the total level of p47phox did not dramatically change in the presence of UA (50 µM), DPI (20 µM), or AG490 (50 µM) (Figure 7A and B). Stimulation of HSC-T6 cells with leptin for 30 minutes remarkably induced the translocation of p47phox from cytoplasm to cell membrane. There was a 1.5-fold increase in the membrane- bound p47phox in leptin-stimulated cells, compared to the control cells (P<0.01; Figure 7A and B). Pretreatment of cells with UA blocked the leptin-induced translocation of p47phox and normalized the membrane-bound p47phox. These effects were observed in DPI- and AG490-treated cells as well (P<0.05; Figure 7A and B). Collectively, UA blocks the translocation of p47phox from cytoplasm to cell membrane, contributing to the inhibitory effect of NOX activity and ROS generation in HSC-T6 cells.
Figure 7.
UA suppressed leptin-induced translocation of the p47phox from cytoplasm to cell membrane in HSC-T6 cells.
Notes: Cells were pretreated with UA (50 µM), DPI (20 µM), or AG490 (50 µM) for 30 minutes and then were stimulated by leptin (100 ng/mL) for another 30 minutes. The membrane bound and total level of p47phox were examined by Western blotting assay. (A) Representative blots of membrane (M-) p47phox and total (T-) p47phox in HSC-T6 cells. (B) Bar graph showing the relative level of M-p47phox in HSC-T6 cells. Data are expressed as mean ± SD from six independent experiments. β-Actin acts as an internal control. *P<0.05; **P<0.01.
Abbreviations: UA, ursolic acid; HSC, hepatic stellate cell; DPI, diphenyleneiodonium; SD, standard deviation.
UA inhibits the leptin-induced expression of gp91phox, p22phox, p67phox, and Rac1
To further investigate the effect of UA on the activity of NOX in HSC-T6 cells, we examined the effect of UA on the expression level of NOX subunits, including gp91phox, p22phox, p67phox, and Rac1, which are important to NOX activity. As shown in Figure 8A and B, after HSC-T6 cells were stimulated with lep-tin (100 ng/mL) for 12 hours, the expression level of gp91phox, p22phox, p67phox, and Rac1 was remarkably increased 1.9-, 1.7-, 2.1-, and 1.9-fold compared to the control group, respectively (P<0.01 or <0.001). Pretreatment of cells with UA (50 µM) markedly decreased the leptin-induced protein expression of gp91phox, p22phox, p67phox, and Rac1 by 71.9%, 62.5%, 61.0%, and 71.9%, compared to the leptin-treated cells, respectively (P<0.001). These results suggest that UA potently negatively regulate the expression level of NOX4 subunits, contributing to the reducing effect of ROS generation.
Figure 8.
Effect of UA on the expression of NOX subunits of HSC-T6 cells.
Notes: Cells were pretreated with UA (50 µM) for 30 minutes and then stimulated with leptin (100 ng/mL) for another 12 hours. The expression level of gp91phox, p22phox, p67phox, and Rac1 was examined using Western blotting assay. (A) Representative blots of gp91phox, p22phox, p67phox, and Rac1 in HSC-T6 cells. (B) Bar graphs showing the relative level of gp91phox, p22phox, p67phox, and Rac1 in HSC-T6 cells. Data are expressed as mean ± SD from six independent experiments. β-Actin acts as an internal control. **P<0.01; ***P<0.001.
Abbreviations: UA, ursolic acid; NOX, NADPH oxidase; HSC, hepatic stellate cell; SD, standard deviation.
UA inhibits ERK, PI3K/Akt, and p38 MAPK signaling pathways in HSC-T6 cells
NOX-derived ROS regulate signal transduction involved in liver fibrosis in HSCs. We evaluated the effect of UA on the modulation of PI3K/Akt, p38 MAPK, and ERK1/2 signaling pathways in HSC-T6 cells using Western blotting analysis. As shown in Figure 9A and B, 30-minute stimulation of HSC-T6 cells with 100 ng/mL leptin remarkably increased the level of p-ERK1 and p-ERK2 by 1.6- and 1.4-fold, compared to the control group, respectively (P<0.05 or 0.01). Pretreatment of cells with UA (50 µM) completely blocked the leptin-induced phosphorylation of ERK1/2 in HSC-T6 cells (P<0.01; Figure 9A and B). Incubation of HSC-T6 cells with DPI (20 µM), AG490 (50 µM), or PD98059 (30 µM) markedly inhibited leptin-induced phosphorylation of ERK1 (P<0.05 or 0.01; Figure 9A and B). There was a decrease in the level of p-ERK2 but without statistical significance when treated with DPI, AG490, or PD98059 (P<0.05; Figure 9A and B). Furthermore, the effect of UA on PI3K/Akt and p38 MAPK signaling pathways was examined in HSC-T6 cells. As shown in Figure 10A and B, stimulation of HSC-T6 cells with leptin markedly increased the level of p-p38 MAPK, PI3K, and p-Akt (P<0.05 or 0.001). However, pretreatment of cells with UA dramatically suppressed leptin-induced increase in the level of p-p38 MAPK, PI3K, and p-Akt (P<0.05 or 0.001; Figure 10A and B). Notably, DPI, SB203580, and LY294002 showed a similar effect to UA on leptin-induced elevation in the level of p-p38 MAPK, PI3K, and p-Akt (P<0.001; Figure 10A and B). Taken together, UA negatively regulates ERK, PI3K/Akt, and p38 MAPK signaling pathways in HSC-T6 cells, which contributes, at least in part, to the underlying mechanism of the antifibrotic effect of UA.
Figure 9.
UA inhibits ERK signaling pathway in HSC-T6 cells.
Notes: Cells were pretreated with UA (50 µM), DPI (20 µM), AG490 (50 µM), or PD98059 (30 µM) for 30 minutes and then stimulated with leptin (100 ng/mL) for 30 minutes. The phosphorylation level of ERK1/2 was examined using Western blotting assay. (A) Representative blots of p-ERK1/2 in HSC-T6 cells. (B) Bar graphs showing the relative level of p-ERK1 and p-ERK2 in HSC-T6 cells. Data are expressed as mean ± SD from six independent experiments. β-Actin acts as an internal control. *P<0.05; **P<0.01.
Abbreviations: UA, ursolic acid; ERK, extracellular signal-regulated kinase; HSC, hepatic stellate cell; DPI, diphenyleneiodonium; SD, standard deviation.
Figure 10.
UA inhibits p38 MAPK and PI3K/Akt signaling pathways in HSC-T6 cells.
Notes: Cells were pretreated with UA (50 µM), DPI (20 µM), SB203580 (10 µM), or LY294002 (10 µM) for 30 minutes and then stimulated with leptin (100 ng/mL) for 30 minutes. The level of p-p38 MAPK, p38 MAPK, PI3K, p-Akt, and Akt was examined using Western blotting assay. (A) Representative blots of p-p38 MAPK, p38 MAPK, PI3K, p-Akt, and Akt in HSC-T6 cells. (B) Bar graphs showing the relative level of p-p38 MAPK, p38 MAPK, PI3K, p-Akt, and Akt in HSC-T6 cells. Data are expressed as mean ± SD from six independent experiments. β-Actin acts as an internal control. *P<0.05; ***P<0.001. + and − represent with or without treatment, respectively.
Abbreviations: UA, ursolic acid; p38 MAPK, p38 mitogen-activated protein kinase; PI3K, phosphoinositide 3-kinase; HSC, hepatic stellate cell; DPI, diphenyleneiodonium; SD, standard deviation.
UA promotes the expression of MMP-1 but inhibits the expression of TIMP-1 and type I collagen in HSC-T6 cells
Finally, we examined the effect of UA on the protein expression of TIMP-1 and MMP-1 and mRNA expression of type I collagen in HSC-T6 cells. HSC-T6 cells were treated with leptin at 100 ng/mL for 24 hours, the TIMP-1 protein expression level and the type I collagen mRNA expression level markedly increased, but the MMP-1 protein expression level significantly decreased (Figures 11A and B and Figures 12A and B). Pretreatment of cells with UA (50 µM), DPI (20 µM), SB203580 (10 µM), or LY294002 (10 µM) inhibited leptin-promoted expression of TIMP-1 (P<0.001; Figure 11A and B), while UA and DPI remarkably increased leptin-suppressed expression level of MMP-1 (P<0.001; Figure 11A and B). SB203580 and LY294002 did not show significant regulatory effect on the expression of MMP-1 in HSC-T6 cells (P>0.05; Figure 11A and B). Moreover, pretreatment of HSC-T6 cells with UA, DPI, AG490, or PD98059 markedly blocked the leptin-induced mNRA expression of type I collagen (P<0.05 or <0.01; Figure 12A and B). Collectively, UA inhibits liver fibrosis with the involvement of ERK, PI3K/Akt, and p38 MAPK signaling pathways.
Figure 11.
Effect of UA on the expression of TIMP-1 and MMP-1 in HSC-T6 cells.
Notes: Cells were pretreated with UA (50 µM), DPI (20 µM), SB203580 (10 µM), or LY294002 (10 µM) for 30 minutes and then stimulated with leptin for another 24 hours. The expression level of TIMP-1 and MMP-1 was examined using Western blotting assays. (A) Representative blots of TIMP-1 and MMP-1 in HSC-T6 cells. (B) Bar graphs showing the relative level of TIMP-1 and MMP-1 in HSC-T6 cells. Data are expressed as mean ± SD from six independent experiments. β-Actin acts as an internal control. *P<0.05; ***P<0.001. + and − represent with or without treatment, respectively.
Abbreviations: UA, ursolic acid; TIMP-1, TIMP metallopeptidase inhibitor 1; MMP-1, matrix metalloproteinase-1; HSC, hepatic stellate cell; DPI, diphenyleneiodonium; SD, standard deviation.
Figure 12.
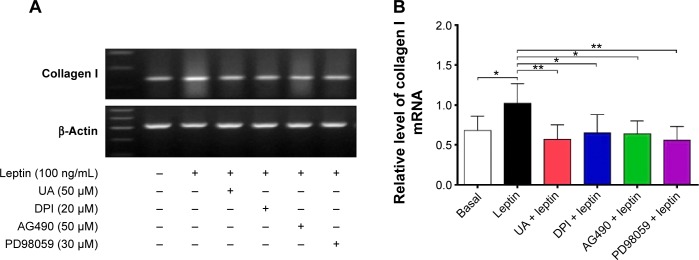
Effect of UA on the mRNA expression of collagen I in HSC-T6 cells.
Notes: Cells were pretreated with UA (50 µM), DPI (20 µM), AG490 (50 µM), or PD98059 (30 µM) for 30 minutes and then stimulated with leptin (100 ng/mL) for another 24 hours. The mRNA expression level of type I collagen I was examined using PCR assays. (A) Representative blots of type I collagen I in HSC-T6 cells. (B) Bar graph showing the relative level of type I collagen I in HSC-T6 cells. Data are expressed as mean ± SD from six independent experiments. β-Actin acts as an internal control. *P<0.05; **P<0.01. + and − represent with or without treatment, respectively.
Abbreviations: UA, ursolic acid; HSC, hepatic stellate cell; DPI, diphenyleneiodonium; PCR, polymerase chain reaction; SD, standard deviation.
Discussion
Liver fibrosis represents a major challenge with considerable morbidity and mortality worldwide, due to the complexity of etiology.34 Compelling evidence indicates that oxidative stress plays a causal role in the pathogenesis of liver fibrosis11,35,36 and that antioxidant may be a promising agent to treat liver fibrosis through the attenuation of oxidative stress.13,37 In the present study, we have predicted the molecular interactome of UA, and there are 611 molecular proteins possibly interacting with UA. The subsequential functional and mechanistic experiments show that UA reverses DMN-induced liver fibrosis through the attenuation of oxidative stress with the involvement of ERK, PI3K/Akt, and p38 MAPK signaling pathways in vitro and in vivo.
In recent years, there is a growing interest in the investigation of the beneficial effects and molecular mechanisms of UA that is a natural pentacyclic triterpenoid carboxylic acid and a major component of some traditional medicinal herbs.38 Increasing evidence shows that UA possesses a variety of bioactivities, including antioxidative, anti-inflammation, and anticancer activities, although the full spectrum of molecular targets has not yet been revealed.38,39 In the present study, the computational experiment provided an interactome of UA, with 611 potential molecular targets been predicted. The bioinformatic analysis showed various signaling pathways that may be able to explain the beneficial effects of UA. These results can provide a clue for us to investigate the beneficial effects and underlying mechanisms of UA.
Employment of computational and bioinformatic approaches have become a practical and valuable way to efficiently predict the molecular interactome of a chemical molecule. The DDI-CPI tool has been used to predict the potential targets, and DAVID has been employed to analyze the molecular targets and related signaling pathways that are regulated by UA. An approach using DDI-CPI server offers a fast, efficient, and inexpensive strategy to predict the potential targets, identify drug repositioning potential, and evaluate and determine adverse drug reactions of a chemical/drug via molecular docking of small compound across human proteome,19–21,24,40,41 although this web-based program has limitations that may affect the accuracy of the outcome.21 Our findings showed that UA may modulate a number of functional proteins and related signaling pathways. These proteins and signaling pathways have important roles in the regulation of redox homeostasis, cell proliferation, apoptosis, energy metabolism, xenobiotics metabolism, lipid and carbohydrate metabolism, and inflammatory response.
Oxidative stress plays a causal role in the development of liver fibrosis.11,35,36,42 It is caused by an increase in the generation of pro-oxidant molecules and/or a decrease in the antioxidants in a given cellular compartment, in which the pro-oxidants overweigh the capability of antioxidant systems, resulting in an imbalance in the amounts of oxidants and antioxidants. NOXs are multicomponent enzymes mediating the transfer of electrons from cytosolic NADPH to O2 to produce O2−,43–45 and they are the primary enzymatic source of ROS.46 NOXs are differentially expressed and distributed among the tissues and subject to regulation by various factors. Moreover, NOXs derived from ROS are also important signaling molecules under pathophysiological conditions. It is known that ROS is involved in a wide range of cellular processes,46–49 including host defence, inflammation, cellular signaling, gene expression, cellular death, cellular senescence, regulation of cell death, O2 sensing, biosynthesis, protein cross-linking, regulation of cellular redox potential, reduction of metal ions, regulation of matrix metalloproteinases, angiogenesis, and cross-link with the nitric oxide system. This role suggests that NOXs should be considered a potential pharmacological target for antifibrotic therapies.26,50
To date, seven members of the NOX family have been distinguished on the basis of the membrane spanning catalytic subunits of NOX or DUOX that they utilize to transfer electrons from NADPH to O2 and produce ROS. They are NOX1, 2, 3, 4, 5 as well as DUOX1- and 2-containing oxidases.46,51–54 The core structure of all NOX isoforms consists of six conserved transmembrane domains and a cytosolic C terminal. Increased activity and expression of NOX isoforms has been demonstrated in a wide variety of diseases and/or disorders, which has been marked by the excessive NOX-generated ROS resulted from upregulation of activity and/or expression of various NOX family members.55–57 In the last decade, extensive attention has been focused on the redox control and the underlying molecular mechanisms of NOX-dependent ROS generation. In addition, the regulation on NOX isoforms has also been studied.55–57 It has been shown that the regulation of NOX isoforms varies from transcriptional level to posttranslational level, although the underlying mechanisms of NOX regulation have not been fully revealed.55–57
NOX4, like the other NOX isoforms, its structure consists mainly of a six-transmembrane domain known as the gp9phox domain; NOX4 predominantly generates hydrogen peroxide rather than superoxide, although it is able to generate superoxide under specific conditions.35,42,58 Despite their extensive similarity in structure and enzymatic function, members of the NOX family differ in their mechanism of activation. In particular, NOX4 requires interaction with a second membrane-bound subunit, p22phox.54,59 NOX4 is a multiprotein complex that generates ROS in both phagocytic and nonphagocytic cells, regulating intracellular signaling.11,36 In the liver, NOX4 plays a central role in fibrogenesis.11 A functionally active form of NOX4 is expressed in HSCs, and the ROS derived from NOX4 act as a second messenger for pro-fibrogenic factor signal transduction in HSCs. HSCs activation is a critical event in the occurrence and development of fibrosis, because activated HSCs are the primary source of extracellular matrix in liver upon injury.2 This role suggests that NOX4 may act as a potential therapeutic target for antifibrotic therapies.26,50
The NOX subunits and regulatory protein are essential for the superoxide generating function and activity of NOX isoforms. The subunits and regulatory proteins required for NOX4 activation include membrane-bound and cytosolic proteins – p22phox, which is a membrane-bound protein helping to stabilize the NOX proteins and dock cytosolic factors, and p47phox, p67phox, the small GTPase Rac, and the modulatory p40phox, which are cytosolic proteins working together to activate the NOX enzymes.46,47,60 Rac1, a member of the Rho family of small GTPase proteins, regulates the activation of NOX4 and the production of ROS and plays an important role in regulating NOX activity.59 Over-expression of Rac1 in HSCs promotes liver injury and fibrosis in mice,61 and induces the phagocytosis of apoptotic bodies by HSCs.62 The level of ROS in cells and tissues is controlled by the tissue- and stimulus-specific expressions of NOX protein and their regulatory subunits and by the acute regulation via calcium, protein phosphorylation, guanine nucleotide exchange on Rac, and the assembly of regulatory subunits.54 Our results demonstrated that leptin induced an increase in the expression level of the NOX4 regulatory subunits, including gp91phox, p22phox, p67phox, and Rac1 in HSC-T6 cells and that pretreatment with UA blocked the leptin-induced increase in the expression level of gp91phox, p22phox, p67phox, and Rac1. Furthermore, pretreatment of UA inhibited the translocation of p47phox from cytoplasm to cell membrane. The negative regulatory effect of UA on the expression of NOX4 subunits explains the reduction of ROS generation in HSC-T6 cells and rats. Consequently, the UA-attenuated oxidative stress leads to a reversal of liver fibrosis and protection of HSC-T6 cells, evident from the decrease in the accumulation of type I collagen in HSCs and the expression of TIMP-1 and type I collagen at transcriptional or translational level, but increase the level of MMP-1.
Furthermore, increasing evidence shows that post-translational modification of NOX represents an important mechanism for NOX regulation. The phosphorylation of NOX subunits and their assembly into active complexes are mediated by various mechanisms, of which many of them are redox sensitive. It has been demonstrated that a number of stimuli and pathways are involved in the activation of NOX by phosphorylation. They comprise phospholipases (PLC/γ, PLD), arachidonic acid metabolites, GTP-binding proteins (Ras, Rac1/2), PKC, PI3K, MAPK, and nonreceptor protein tyrosine kinases.63,64 Previous studies showed that NOX-mediated ROS generation through the activation of Akt and MAPK phosphorylation, leading to an increase in AP-1-DNA-binding activity, promotion in the expression of type I collagen, TGF-β1, and other inflammatory cytokines26 and that NOX-mediated ROS production interplays with p38 MAPK50 and JAK1/2-STAT3/ERK signaling pathways in HSCs.65 In agreement with previous studies, the data clearly show that UA modulates the NOX activity and expression with the involvement of ERK, PI3K/Akt, and p38 MAPK signaling pathways in HSCs via the employment of specific chemical inhibitors. In aggregate, it suggests that targeting NOX4 represents a promising strategy for liver fibrosis treatment. Notably, recent studies have indicated that the NOX1/4 dual inhibitor GKT137831 can significantly inhibit the activation of HSCs and the development of liver fibrosis.12,66–68 This inhibitor is currently being investigated in Phase II clinical trials. Therefore, NOX-targeting drugs with low toxicity that are effective against fibrosis are expected to lead to a breakthrough in the treatment of liver fibrosis. However, there are many factors that may affect the therapeutic outcome of antifibrotic agents in the treatment of liver fibrosis.69 Thus, more well-designed studies are needed to investigate the therapeutic effect and underlying mechanism of the antifibrotic effect.
In summary, the present study has depicted a full spectrum of molecular targets and related signaling pathways that possibly respond to UA treatment. The benchmarking results have clearly shown that UA exhibits a potent antifibrotic effect in rats evident from the reduction in the accumulation of type I collagen in rat HSCs. The underlying molecular mechanism and beneficial effect of UA can be ascribed to the oxidative stress attenuating effect through negative regulation of NOX4 activity and expression with the involvement of ERK, PI3K/Akt, and p38 MAPK signaling pathways in HSCs. These results render UA as a promising antifibrotic agent via targeting NOX4, and further studies are warranted to evaluate the safety and efficacy of UA in the treatment of liver fibrosis.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Murphy SL, Xu J, Kochanek KD. Deaths: final data for 2010. Natl Vital Stat Rep. 2013;61(4):1–117. [PubMed] [Google Scholar]
- 2.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115(2):209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell AE, Colvin HM, Palmer Beasley R. Institute of Medicine recommendations for the prevention and control of hepatitis B and C. Hepatology. 2010;51(3):729–733. doi: 10.1002/hep.23561. [DOI] [PubMed] [Google Scholar]
- 4.Davis GL, Alter MJ, El-Serag H, Poynard T, Jennings LW. Aging of hepatitis C virus (HCV)-infected persons in the United States: a multiple cohort model of HCV prevalence and disease progression. Gastroenterology. 2010;138(2):e1–e6. doi: 10.1053/j.gastro.2009.09.067. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Fan D. Hepatitis B in China. Lancet. 2007;369(9573):1582–1583. doi: 10.1016/S0140-6736(07)60723-5. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Zhu H, Wu Y, et al. HIV, hepatitis B virus, and hepatitis C virus co-infection in patients in the China national free antiretroviral treatment program, 2010–2012: a retrospective observational cohort study. Lancet Infect Dis. 2014;14(11):1065–1072. doi: 10.1016/S1473-3099(14)70946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmison J, McCullough AJ. Pathogenesis of non-alcoholic steatohepatitis: human data. Clin Liver Dis. 2007;11(1):75–104. ix. doi: 10.1016/j.cld.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Brenner DA. Molecular pathogenesis of liver fibrosis. Trans Am Clin Climatol Assoc. 2009;120:361–368. [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134(6):1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novo E, Cannito S, Paternostro C, Bocca C, Miglietta A, Parola M. Cellular and molecular mechanisms in liver fibrogenesis. Arch Biochem Biophys. 2014;548:20–37. doi: 10.1016/j.abb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 11.De Minicis S, Brenner DA. NOX in liver fibrosis. Arch Biochem Biophys. 2007;462(2):266–272. doi: 10.1016/j.abb.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen-Naftaly M, Friedman SL. Current status of novel antifibrotic therapies in patients with chronic liver disease. Therap Adv Gastroenterol. 2011;4(6):391–417. doi: 10.1177/1756283X11413002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbloom J, Mendoza FA, Jimenez SA. Strategies for anti-fibrotic therapies. Biochim Biophys Acta. 2013;1832(7):1088–1103. doi: 10.1016/j.bbadis.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Dufour D, Pichette A, Mshvildadze V, et al. Antioxidant, anti-inflammatory and anticancer activities of methanolic extracts from Ledum groenlandicum Retzius. J Ethnopharmacol. 2007;111(1):22–28. doi: 10.1016/j.jep.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Saravanan R, Viswanathan P, Pugalendi KV. Protective effect of ursolic acid on ethanol-mediated experimental liver damage in rats. Life Sci. 2006;78(7):713–718. doi: 10.1016/j.lfs.2005.05.060. [DOI] [PubMed] [Google Scholar]
- 16.Saraswat B, Visen PK, Agarwal DP. Ursolic acid isolated from Eucalyptus tereticornis protects against ethanol toxicity in isolated rat hepatocytes. Phytother Res. 2000;14(3):163–166. doi: 10.1002/(sici)1099-1573(200005)14:3<163::aid-ptr588>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 17.Shen YM, Zhu X, Zhang KH, et al. Effect of ursolic acid on proliferation and apoptosis of hepatic stellate cells in vitro. Zhonghua Gan Zang Bing Za Zhi. 2008;16(4):298–301. [PubMed] [Google Scholar]
- 18.Steinkamp-Fenske K, Bollinger L, Völler N, et al. Ursolic acid from the Chinese herb danshen (Salvia miltiorrhiza L.) upregulates eNOS and downregulates Nox4 expression in human endothelial cells. Atherosclerosis. 2007;195(1):e104–e111. doi: 10.1016/j.atherosclerosis.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Shermann BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Luo H, Chen J, Shi L, et al. DRAR-CPI: a server for identifying drug repositioning potential and adverse drug reactions via the chemical-protein interactome. Nucleic Acids Res. 2011;39(Web Server issue):W492–W498. doi: 10.1093/nar/gkr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo H, Zhang P, Huang H, et al. DDI-CPI, a server that predicts drug-drug interactions through implementing the chemical-protein interactome. Nucleic Acids Res. 2014;42(Web Server issue):W46–W52. doi: 10.1093/nar/gku433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47(12):2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Luo H, Chen J, Xing Q, He L. SePreSA: a server for the prediction of populations susceptible to serious adverse drug reactions implementing the methodology of a chemical-protein interactome. Nucleic Acids Res. 2009;37(Web Server issue):W406–W412. doi: 10.1093/nar/gkp312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, Chen J, Shi L, Hudock MP, Wang K, He L. Identifying unexpected therapeutic targets via chemical-protein interactome. PLoS One. 2010;5(3):e9568. doi: 10.1371/journal.pone.0009568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel S, Piantedosi R, Frank J, et al. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41(6):882–893. [PubMed] [Google Scholar]
- 26.Bataller R, Schwabe RF, Choi YH, et al. NADPH oxidase signal trans-duces angiotensin II in hepatic stellate cells and is critical in hepatic fibrosis. J Clin Invest. 2003;112(9):1383–1394. doi: 10.1172/JCI18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrera B, Murillo MM, Alvarez-Barrientos A, Beltrán J, Fernández M, Fabregat I. Source of early reactive oxygen species in the apoptosis induced by transforming growth factor-β in fetal rat hepatocytes. Free Radic Biol Med. 2004;36(1):16–26. doi: 10.1016/j.freeradbiomed.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Joseph JA. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic Biol Med. 1999;27(5–6):612–616. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- 29.LeBel CP, Ischiropoulos H, Bondy SC. Evaluation of the probe 2′,7′-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem Res Toxicol. 1992;5(2):227–231. doi: 10.1021/tx00026a012. [DOI] [PubMed] [Google Scholar]
- 30.Proell V, Carmona-Cuenca I, Murillo MM, Huber H, Fabregat I, Mikulits W. TGF-β dependent regulation of oxygen radicals during transdifferentiation of activated hepatic stellate cells to myofibroblastoid cells. Comp Hepatol. 2007;6:1. doi: 10.1186/1476-5926-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou ZW, Xie XL, Zhou SF, Li CG. Mechanism of reversal of high glucose-induced endothelial nitric -none IIA in human endothelial cell line EA.hy926. Eur J Pharmacol. 2012;697(1–3):97–105. doi: 10.1016/j.ejphar.2012.09.051. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Valle V, Chavez-Tapia NC, Uribe M, Mendez-Sanchez N. Role of oxidative stress and molecular changes in liver fibrosis: a review. Curr Med Chem. 2012;19(28):4850–4860. doi: 10.2174/092986712803341520. [DOI] [PubMed] [Google Scholar]
- 33.Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci U S A. 2012;109(15):5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 35.Galli F, Battistoni A, Gambari R, et al. Working Group on Inflammation in Cystic Fibrosis Oxidative stress and antioxidant therapy in cystic fibrosis. Biochim Biophys Acta. 2012;1822(5):690–713. doi: 10.1016/j.bbadis.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Paik YH, Kim J, Aoyama T, De Minicis S, Bataller R, Brenner DA. Role of NADPH oxidases in liver fibrosis. Antioxid Redox Signal. 2014;20(17):2854–2872. doi: 10.1089/ars.2013.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maraldi T. Natural compounds as modulators of NADPH oxidases. Oxid Med Cell Longev. 2013;2013:271602. doi: 10.1155/2013/271602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda Y, Murakami A, Ohigashi H. Ursolic acid: an anti- and pro-inflammatory triterpenoid. Mol Nutr Food Res. 2008;52(1):26–42. doi: 10.1002/mnfr.200700389. [DOI] [PubMed] [Google Scholar]
- 39.Mazumder K, Tanaka K, Fukase K. Cytotoxic activity of ursolic acid derivatives obtained by isolation and oxidative derivatization. Molecules. 2013;18(8):8929–8944. doi: 10.3390/molecules18088929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, Wang KJ, Wang LS, et al. Chemical-protein interactome and its application in off-target identification. Interdiscip Sci. 2011;3(1):22–30. doi: 10.1007/s12539-011-0051-8. [DOI] [PubMed] [Google Scholar]
- 41.Yang L, Wang K, Chen J, et al. Exploring off-targets and off-systems for adverse drug reactions via chemical-protein interactome–clozapine-induced agranulocytosis as a case study. PLoS Comput Biol. 2011;7(3):e1002016. doi: 10.1371/journal.pcbi.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang F, Liu GS, Dusting GJ, Chan EC. NADPH oxidase-dependent redox signaling in TGF-beta-mediated fibrotic responses. Redox Biol. 2014;2:267–272. doi: 10.1016/j.redox.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babior BM, Lambeth JD, Nauseef W. The neutrophil NADPH oxidase. Arch Biochem Biophys. 2002;397(2):342–344. doi: 10.1006/abbi.2001.2642. [DOI] [PubMed] [Google Scholar]
- 44.Babior BM, Kipnes RS, Curnutte JT. Biological defense mechanisms. The production by leukocytes of superoxide, a potential bactericidal agent. J Clin Invest. 1973;52(3):741–744. doi: 10.1172/JCI107236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babior BM. The NADPH oxidase of endothelial cells. IUBMB Life. 2000;50(4–5):267–269. doi: 10.1080/713803730. [DOI] [PubMed] [Google Scholar]
- 46.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 47.Katsuyama M, Matsuno K, Yabe-Nishimura C. Physiological roles of NOX/NADPH oxidase, the superoxide-generating enzyme. J Clin Biochem Nutr. 2012;50(1):9–22. doi: 10.3164/jcbn.11-06SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lassegue B, Griendling KK. NADPH oxidases: functions and pathologies in the vasculature. Arterioscler Thromb Vasc Biol. 2010;30(4):653–661. doi: 10.1161/ATVBAHA.108.181610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283(25):16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adachi T, Togashi H, Suzuki A, et al. NAD(P)H oxidase plays a crucial role in PDGF-induced proliferation of hepatic stellate cells. Hepatology. 2005;41(6):1272–1281. doi: 10.1002/hep.20719. [DOI] [PubMed] [Google Scholar]
- 51.Ris-Stalpers C. Physiology and pathophysiology of the DUOXes. Antioxid Redox Signal. 2006;8(9–10):1563–1572. doi: 10.1089/ars.2006.8.1563. [DOI] [PubMed] [Google Scholar]
- 52.Kawahara T, Quinn MT, Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol. 2007;7:109. doi: 10.1186/1471-2148-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawahara T, Lambeth JD. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evol Biol. 2007;7:178. doi: 10.1186/1471-2148-7-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambeth JD, Kawahara T, Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic Biol Med. 2007;43(3):319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cave AC, Brewer AC, Narayanapanicker A, et al. NADPH oxidases in cardiovascular health and disease. Antioxid Redox Signal. 2006;8(5–6):691–728. doi: 10.1089/ars.2006.8.691. [DOI] [PubMed] [Google Scholar]
- 56.Streeter J, Thiel W, Brieger K, Miller FJ., Jr Opportunity Nox: the future of NADPH oxidases as therapeutic targets in cardiovascular disease. Cardiovasc Ther. 2012;31(3):125–137. doi: 10.1111/j.1755-5922.2011.00310.x. [DOI] [PubMed] [Google Scholar]
- 57.Drummond GR, Selemidis S, Griendling KK, Sobey CG. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat Rev Drug Discov. 2011;10(6):453–471. doi: 10.1038/nrd3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sancho P, Mainez J, Crosas-Molist E, et al. NADPH oxidase NOX4 mediates stellate cell activation and hepatocyte cell death during liver fibrosis development. PLoS One. 2012;7(9):e45285. doi: 10.1371/journal.pone.0045285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hordijk PL. Regulation of NADPH oxidases: the role of Rac proteins. Circ Res. 2006;98(4):453–462. doi: 10.1161/01.RES.0000204727.46710.5e. [DOI] [PubMed] [Google Scholar]
- 60.Katsuyama M. NOX/NADPH oxidase, the superoxide-generating enzyme: its transcriptional regulation and physiological roles. J Pharmacol Sci. 2010;114(2):134–146. doi: 10.1254/jphs.10r01cr. [DOI] [PubMed] [Google Scholar]
- 61.Choi SS, Sicklick JK, Ma Q, et al. Sustained activation of Rac1 in hepatic stellate cells promotes liver injury and fibrosis in mice. Hepatology. 2006;44(5):1267–1277. doi: 10.1002/hep.21375. [DOI] [PubMed] [Google Scholar]
- 62.Mehal W, Imaeda A. Cell death and fibrogenesis. Semin Liver Dis. 2010;30(3):226–231. doi: 10.1055/s-0030-1255352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamori T, Inanami O, Nagahata H, Kuwabara M. Phosphoinositide 3-kinase regulates the phosphorylation of NADPH oxidase component p47(phox) by controlling cPKC/PKCdelta but not Akt. Biochem Biophys Res Commun. 2004;316(3):720–730. doi: 10.1016/j.bbrc.2004.02.108. [DOI] [PubMed] [Google Scholar]
- 64.Kilpatrick LE, Sun S, Li H, Vary TC, Korchak HM. Regulation of TNF-induced oxygen radical production in human neutrophils: role of delta-PKC. J Leukoc Biol. 2010;87(1):153–164. doi: 10.1189/jlb.0408230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Minicis S, Seki E, Oesterreicher C, Schnabl B, Schwabe RF, Brenner DA. Reduced nicotinamide adenine dinucleotide phosphate oxidase mediates fibrotic and inflammatory effects of leptin on hepatic stellate cells. Hepatology. 2008;48(6):2016–2026. doi: 10.1002/hep.22560. [DOI] [PubMed] [Google Scholar]
- 66.Schuppan D, Kim YO. Evolving therapies for liver fibrosis. J Clin Invest. 2013;123(5):1887–1901. doi: 10.1172/JCI66028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang JX, Chen X, Serizawa N, et al. Liver fibrosis and hepatocyte apoptosis are attenuated by GKT137831, a novel NOX4/NOX1 inhibitor in vivo. Free Radic Biol Med. 2012;53(2):289–296. doi: 10.1016/j.freeradbiomed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Babalola O, Mamalis A, Lev-Tov H, Jagdeo J. NADPH oxidase enzymes in skin fibrosis: molecular targets and therapeutic agents. Arch Dermatol Res. 2014;306(4):313–330. doi: 10.1007/s00403-013-1416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guy J, Peters MG. Liver disease in women: the influence of gender on epidemiology, natural history, and patient outcomes. Gastroenterol Hepatol (N Y) 2013;9(10):633–639. [PMC free article] [PubMed] [Google Scholar]