Abstract
The inflammasome activates caspase-1 and the release of interleukin-1β (IL-1β) and IL-18, and several inflammasomes protect against intestinal inflammation and colitis-associated colon cancer (CAC) in animal models. The absent in melanoma 2 (AIM2) inflammasome is activated by double-stranded DNA, and AIM2 expression is reduced in several types of cancer, but the mechanism by which AIM2 restricts tumor growth remains unclear. We found that Aim2-deficient mice had greater tumor load than Asc-deficient mice in the azoxymethane/dextran sodium sulfate (AOM/DSS) model of colorectal cancer. Tumor burden was also higher in Aim2−/−/ApcMin/+ than in APCMin/+ mice. The effects of AIM2 on CAC were independent of inflammasome activation and IL-1β and were primarily mediated by a non–bone marrow source of AIM2. In resting cells, AIM2 physically interacted with and limited activation of DNA-dependent protein kinase (DNA-PK), a PI3K-related family member that promotes Akt phosphorylation, whereas loss of AIM2 promoted DNA-PK–mediated Akt activation. AIM2 reduced Akt activation and tumor burden in colorectal cancer models, while an Akt inhibitor reduced tumor load in Aim2−/− mice. These findings suggest that Akt inhibitors could be used to treat AIM2-deficient human cancers.
Colon cancer is one of the leading causes of cancer-related deaths worldwide1. Although sporadic tumorigenesis accounts for the majority of these cases, colon cancer can result from prolonged inflammatory bowel disease (IBD; e.g., ulcerative colitis and Crohn’s disease). IBD pathogenesis is multifaceted and not fully understood, but it is largely associated with the chronic infiltration of immune cells, production of inflammatory cytokines and activation of the NF-κB family of transcription factors that promote tumorigenesis during colitis-associated colon cancer (CAC) (reviewed in ref. 2).
The NLR (nucleotide binding domain and leucine-rich-repeat-containing or NOD-like receptor) family of proteins has received much attention in IBD and CAC because of the genetic association of NOD2 with Crohn’s disease3,4. In particular, the inflammasome complex comprising NLRP3 and its adaptor, apoptosis-associated speck-like protein containing a carboxy-terminal CARD (ASC), serves as a protective factor in models of colitis and CAC by limiting inflammation and promoting epithelial cell repair primarily through production of IL-18 (refs. 5–8). Moreover, loss of IL-18 or the IL-18 receptor results in increased susceptibility to CAC tumorigenesis9. Similarly, the NLRP6 inflammasome also limits colitis and CAC through IL-18 production10 and by altering the intestinal microbiome11,12. Other reports imply an alternative role for the NLRP6 and NLRC4 inflammasomes in regulating intestinal cell apoptosis13,14, collectively indicating that multiple inflammasomes are required for limiting intestinal inflammation and colon cancer.
Absent in melanoma 2 (AIM2) is a pyrin-HIN protein that binds cytosolic double-stranded DNA and partners with the common inflammasome adaptor ASC to activate the inflammasome15,16. Although the role of AIM2 in inflammasome activation is well accepted, its role in tumorigenesis is less clear. AIM2 was originally proposed as a tumor suppressor gene in melanomas17, and AIM2 mutations are found in human colorectal tumors18,19, with reduced AIM2 expression being associated with poor prognosis in patients20. Furthermore, AIM2 suppresses cellular proliferation, but the mechanism is unknown21–23. Here, we report a previously unrecognized inflammasome-independent role for AIM2 in non–bone marrow cells in restricting tumorigenesis during colitis-associated and sporadic colon cancer through suppression of Akt, a master regulator of cellular survival.
RESULTS
AIM2 does not affect inflammation and inflammasome activation in colitis models
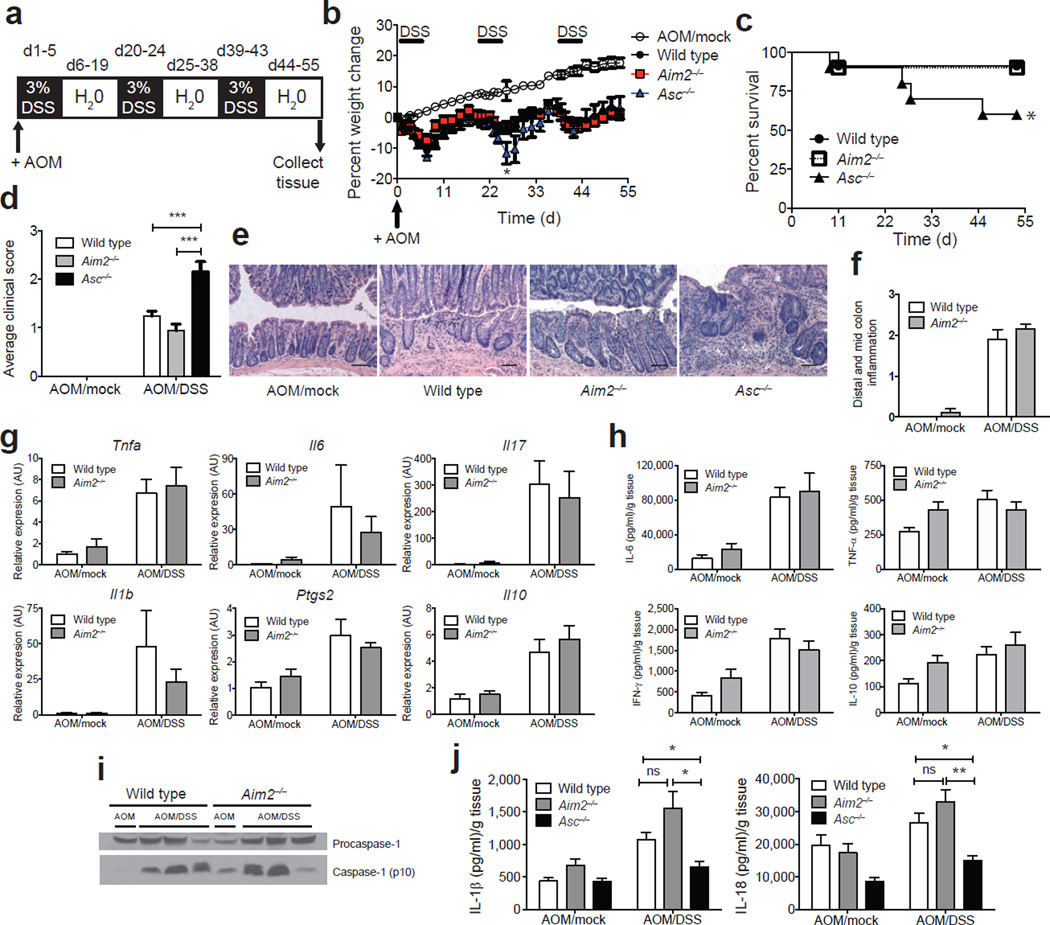
To assess the role of AIM2 in CAC, we generated Aim2-deficient mice (termed Aim2−/−) on the C57BL/6 genetic background (Supplementary Fig. 1a) and verified that Aim2−/− bone marrow–derived macrophages (BMDM) failed to produce IL-1β following LPS priming and poly(dA:dT) stimulation (Supplementary Fig. 1b). This is identical to what is observed with other strains of Aim2−/− mice15,16. Aim2−/− mice did not display alterations in immune cell frequencies or numbers of lymphoid organs (Supplementary Fig. 2). To study the role of AIM2 in CAC, wild-type, Aim2−/− and Asc−/− mice were intraperitoneally (i.p.) injected with azoxymethane (AOM) and then treated with three rounds of 3% dextran sodium sulfate (DSS) in drinking water24 (Fig. 1a). This model has been used to show the protective effect of multiple inflammasome components, including NLRP3, ASC, NLRC4, NLRP6 and caspase-1, against CAC5–7,10,12–14. The mice were assessed for weight loss, survival and clinical scores, which included a semiquantitative assessment of stool consistency and rectal bleeding7. The inflammasome-deficient Asc−/− mice showed greater weight loss, morbidity and clinical scores than Aim2−/− mice (Fig. 1b–d, Supplementary Fig. 3a), as previously reported5–7. Increased disease severity in Asc−/− mice was accompanied by increased infiltration of inflammatory cells (Fig. 1e) and proinflammatory cytokines in the colon5–7. In contrast, disease severity in Aim2−/− mice was indistinguishable from that in wild-type controls (Fig. 1b–d). Colon histopathology (blindly scored by a pathologist) revealed no difference in the inflammatory infiltrate between Aim2−/− and wild-type colons (Fig. 1e,f). Colonic cytokine mRNA expression (Fig. 1g) and cytokine production by colon explants (Fig. 1h) were also similar between Aim2−/− and wild-type mice. We next assessed the status of inflammasome activation during CAC in wild-type as compared to Aim2−/− mice. Similar protein expression of mature caspase-1 p10 subunit (Fig. 1i, Supplementary Fig. 4a) and concentrations of IL-1β and IL-18 (Fig. 1j) were detectable in the colons and colon explants, respectively, from AOM/DSS-treated wild-type and Aim2−/− mice. In contrast, IL-1β and IL-18 production was lower in Asc−/− colons. Because AIM2 associates with ASC to form an inflammasome15,16, it was somewhat unexpected that the outcomes for Asc−/− and Aim2−/− mice were distinct. However, intact inflammasome activation in Aim2−/− mice during CAC is likely due to the presence of other inflammasome-forming sensors, such as NLRP3 and NLRP6, which use ASC as an adaptor molecule and may compensate in the absence of AIM2 (refs. 5–7,11).
Figure 1.
AIM2 is distinct from ASC during colitis-associated colon cancer. (a) Induction procedure for the AOM/DSS model of CAC. (b–d) Weight loss (b), % survival (d, days) (c) and average clinical scores (d) in Aim2−/− (n = 25), Asc−/− (n = 11) and wild-type (n = 30) mice following induction of CAC using AOM/DSS. Mock wild-type animals (n = 10) received a single i.p. injection of AOM and were given regular drinking water instead of DSS. Error bars, s.e.m. (e) H&E staining of colons from AOM/DSS-treated wild-type, Aim2−/− and Asc−/− mice and (f) semiquantitative scoring of inflammation in colons from AOM/DSS-treated wild-type and Aim2−/− mice (n = 6 for WT AOM/Mock and Aim2−/− AOM/DSS, and n = 5 for WT AOM/DSS and Aim2−/− AOM/Mock). Images were taken at 200× magnification; scale bars, 100 µm. (g) Inflammatory cytokine mRNA (AU, arbitrary units) (n = 3 for WT AOM/Mock and Aim2−/− AOM/Mock, and n = 6 for WT AOM/DSS and Aim2−/− AOM/DSS) and (h) protein expression in wild-type and Aim2−/− colons (n = 5 for WT AOM/Mock and Aim2−/− AOM/Mock, n = 10 for WT AOM/DSS and n = 8 for Aim2−/− AOM/DSS). (i) Caspase-1 activation in wild-type and Aim2−/− colons and (j) IL-1β and IL-18 production in wild-type, Aim2−/− and Asc−/− colons (n = 5 for WT AOM/Mock, Aim2−/− AOM/Mock and Asc−/− AOM/Mock, and n = 10 for WT AOM/DSS, n = 8 for Aim2−/− AOM/DSS and n = 5 for Asc−/− AOM/DSS). Error bars, s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001; b,d,j, one-way ANOVA (Tukey's multiple-comparisons test); c, log-rank test.
Loss of intestinal barrier integrity promotes intestinal inflammation, and mice deficient in inflammasome components display increased intestinal permeability following the induction of experimental colitis and CAC5,6,10,13. However, intestinal permeability was not elevated in AOM/DSS-treated Aim2−/− mice compared to wild-type mice (Supplementary Fig. 3b), consistent with a lack of exaggerated inflammation in these mice. Additionally, weight change and colon length did not differ between Aim2−/− and wild-type mice following induction of acute DSS-induced colitis (Supplementary Fig. 5a–c). Moreover, wild-type and Aim2−/− mice showed similar morbidity and mortality during an extended time course after DSS treatment (Supplementary Fig. 5d–g) and produced similar levels of IL-1β and IL-18 after chronic DSS exposure (Supplementary Fig. 5h), suggesting that the inflammasome is intact in Aim2−/− mice during chronic colitis. As a positive control, IL-1β and IL-18 production was reduced in the colons of Asc−/− mice during chronic colitis (Supplementary Fig. 5h), as previously reported5–7. Finally, we examined another colitis model, which is driven by the transfer of naive CD4+ T cells25. The absence of AIM2 in transferred T cells did not alter disease severity (Supplementary Fig. 5i–k). Thus, absence of AIM2 did not affect the severity of colitis in different animal models.
AIM2 protects against precancerous polyp formation in colitis-associated colon cancer
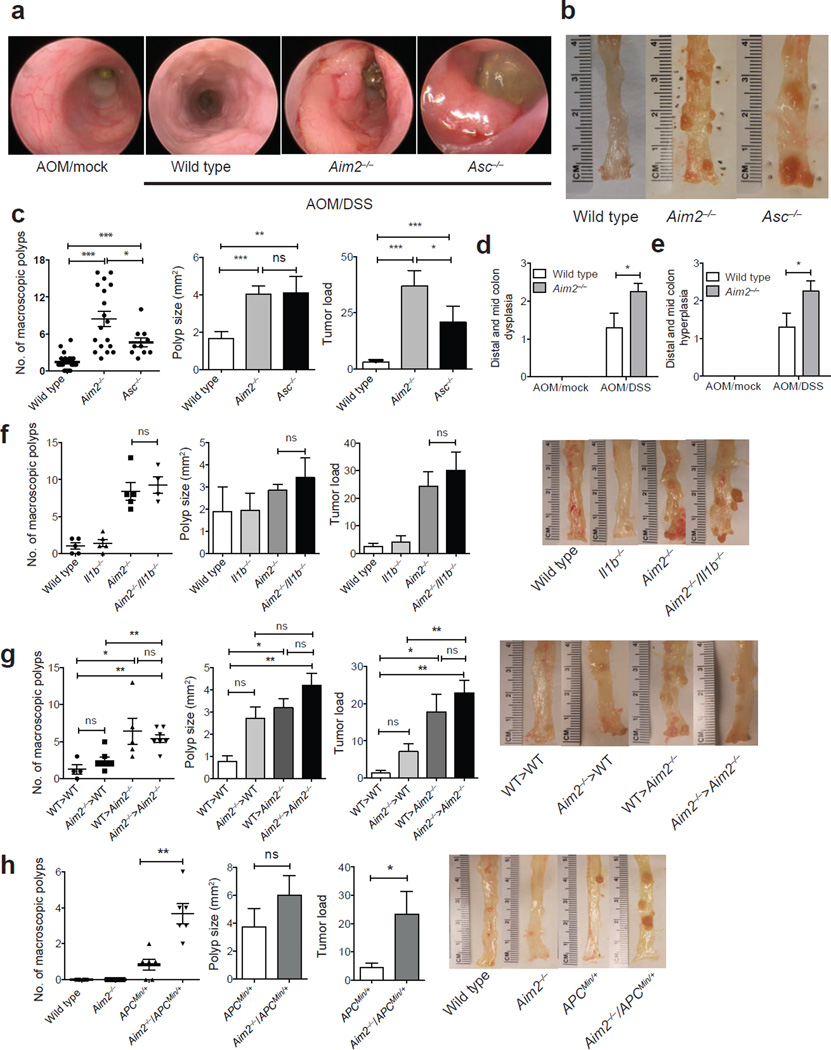
We next examined the impact of AIM2 on colon polyps, a measurement of precancerous growth. High-resolution endoscopy showed that Aim2−/− mice exposed to AOM/DSS had greater numbers of polyps compared to wild-type mice (Fig. 2a), and macroscopic polyp counts and tumor load assessments indicated that Aim2−/− mice had greater numbers of polyps and higher tumor load than Asc−/− and control mice (Fig. 2b,c). Colon histopathology from Aim2−/− mice revealed increased crypt dysplasia and hyperplasia compared to wild-type controls (Fig. 2d,e and Supplementary Fig. 6), even though the mice were similar to wild-type animals with respect to inflammatory immune infiltrates in the colon (Fig. 1f). Thus, Aim2 deficiency resulted in increased hyperplasia, dysplasia and polyps, without attendant increases in inflammation. In contrast, Asc deficiency increased polyp formation to a lesser extent than Aim2 deficiency, but did increase inflammation5–7. Because Aim2−/− colon explants exhibited a slight, but statistically nonsignificant, increase in IL-1β production (Fig. 1j), and IL-1β can promote tumorigenesis (reviewed in ref. 26), we generated Aim2−/−/Il1b−/− doubly deficient mice to determine whether IL-1β was responsible for the increased CAC in Aim2−/− mice. Consistent with a previous report using mice lacking the IL-1β receptor during CAC9, AOM/DSS-treated Il1b−/− mice had numbers of colon polyps and tumor loads similar to those of wild-type mice (Fig. 2f). However, Aim2−/−/Il1b−/− and Aim2−/− mice did not differ in terms of colon polyp numbers and tumor load (Fig. 2f), indicating that IL-1β was not responsible for the increased tumorigenesis in Aim2−/− mice. Thus, loss of AIM2 promoted tumorigenesis through a mechanism distinct from inflammasome regulation.
Figure 2.
AIM2 protects against colon tumorigenesis. (a,b) Mini-endoscopy (a) and representative images (b) of colons from AOM/DSS-treated wild-type (n = 5), Asc−/− (n = 3) and Aim2−/− mice (n = 5). (c) Macroscopic polyp counts, average polyp size per mouse and tumor load (n = 21 for WT, n = 18 for Aim2−/− and n = 10 for Asc−/−). (d,e) Semiquantitative scoring of colon hyperplasia (d) and dysplasia (e) in AOM/DSS-treated wild-type and Aim2−/− mice (n = 6 for WT AOM/Mock and Aim2−/− AOM/DSS, and n = 5 for WT AOM/DSS and Aim2−/− AOM/Mock). (f) Colon polyp counts, average polyp size per mouse and tumor load in AOM/DSS-treated wild-type, Aim2−/−, Il1b−/− and Aim2−/−/Il1b−/− mice, with representative images of colons in the right panel (n = 5 for wild-type, Aim2−/− and Il1b−/−, and n = 4 for Aim2−/−/Il1b−/−). (g) Colon polyp counts, average polyp size per mouse and tumor load in AOM/DSS-treated wild-type and Aim2−/− radiation bone marrow chimeras, with representative images of colons in the right panel (n = 4 for WT>WT, n = 5 for WT>Aim2−/− and n = 7 for Aim2−/−>WT and Aim2−/−>Aim2−/−). (h) Colon polyp counts, average polyp size per mouse and tumor load in ApcMin/+ and Aim2−/−/ApcMin/+ mice representative images of colons in the right panel (n = 6 mice/group). Each symbol represents one animal. Error bars, s.e.m. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001; c,g, one-way ANOVA (Tukey's multiple-comparisons test); d,e,f,h, unpaired t-test.
A non–bone marrow source of AIM2 protects against precancerous polyp formation
We next determined the cellular source responsible for the enhanced tumorigenesis in the absence of AIM2 by generating radiation bone marrow–chimeric mice. Wild-type and Aim2−/− mice were exposed to whole-body irradiation (8 Gy) to deplete the hematopoietic-progenitor compartment. Irradiated recipients were then reconstituted with either wild-type or Aim2−/− bone marrow, and CAC was induced using AOM/DSS. Wild-type mice reconstituted with Aim2−/− bone marrow (Aim2−/−>WT) exhibited a trend of increased tumor size and burden as compared to wild-type control mice (WT>WT), although it did not reach statistical significance. An additional statistical analysis by t-test showed that the polyp size was significantly larger in Aim2−/−>WT (P = 0.0273) compared to WT>WT mice, but the tumor load was not different (P = 0.073). By contrast, Aim2−/− recipient mice reconstituted with wild-type bone marrow (WT>Aim2−/−) were indistinguishable from Aim2−/− control mice (Aim2−/−>Aim2−/−) and had increased colon polyp numbers and tumor loads as compared to WT>WT controls (Fig. 2g). These results suggest that Aim2 expression in radioresistant, non–bone marrow cells was primarily associated with the restriction of tumorigenesis during CAC.
Because AIM2 suppressed colon tumorigenesis without affecting inflammasome function or limiting inflammation, and AIM2 has been implicated in human colon cancer18–20, we hypothesized that AIM2 would perform a similar protective role in a non–colitis-driven spontaneous model of colon cancer. To address this hypothesis, we generated Aim2−/− mice that contain a heterozygous mutation in the adenomatous polyposis coli (Apc) gene (Aim2−/−/ApcMin/+). Mutations in human APC are responsible for the hereditary form of colon cancer known as familial adenomatous polyposis (FAP), and mice with this mutation develop spontaneous intestinal, and to a lesser extent colorectal, tumors27,28. Aim2−/−/ApcMin/+ doubly deficient mice had greater numbers of colon tumors and higher tumor loads than singly deficient ApcMin/+ mice, the majority of which only had 0 or 1 colon polyp as previously described27,28 (Fig. 2h), demonstrating that AIM2 also restricts sporadic colon tumorigenesis.
AIM2 inhibits Akt activation to restrict tumorigenesis
To explore the mechanism by which AIM2 impacts tumorigenesis, we explored multiple pathways that might be affected in Aim2−/− colon tissue and cells. Loss of AIM2 in colorectal cancer lines has been associated with reduced HLA-DRA and HLA-DRB expression in vitro29, which could dampen antitumor immunity. However, I-Ab mRNA levels in wild-type and Aim2−/− colon polyps or adjacent tissue did not differ (Supplementary Fig. 7a). Mitogen-activated protein kinase (MAPK) pathways are major regulators of cellular proliferation and apoptosis (reviewed in ref. 30), but no consistent differences were detected between wild-type and Aim2−/− mice (Supplementary Fig. 7b). The NF-κB family of transcription factors is responsible for initiating inflammation and linking inflammation to cancer development (reviewed in ref. 2); however, activation of p65 and IκBα was not elevated in the colons of AOM/DSS-treated wild-type and Aim2−/− mice (Supplementary Fig. 7c). In addition, activated STAT3, which acts in concert with NF-κB to promote tumorigenesis (reviewed in ref. 31), was not elevated in Aim2−/− mice (Supplementary Fig. 7d).
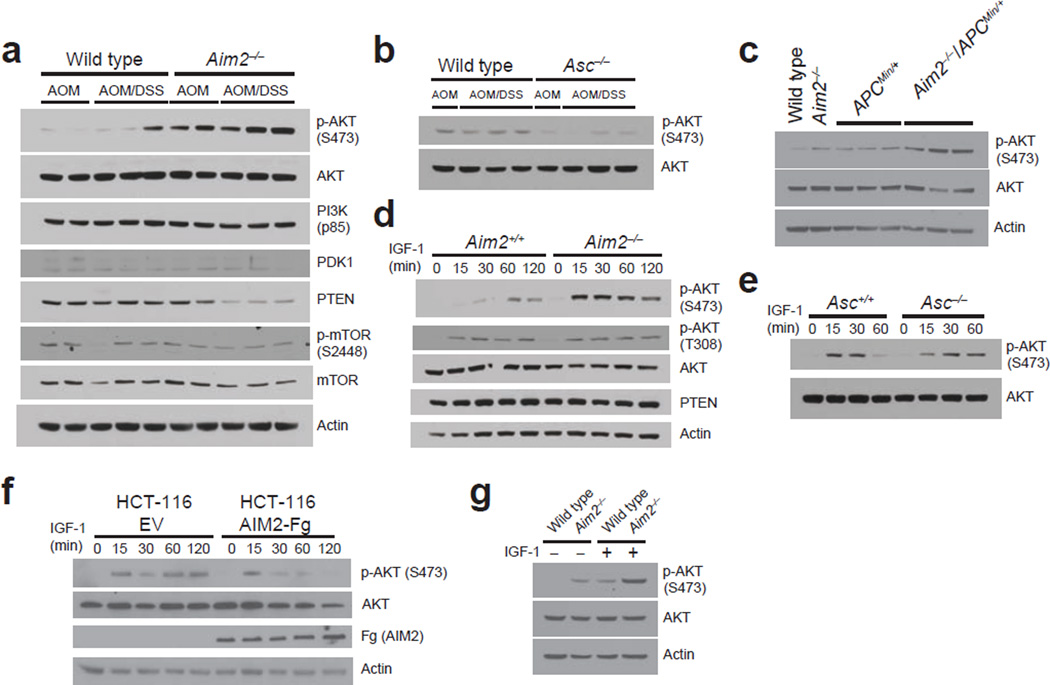
We next examined the phosphatidylinositol 3-kinase (PI3K)–Akt pathway of cellular survival because it is frequently mutated in human colorectal cancer32. Akt activation occurs through a series of events that involve the phosphorylation of Akt at threonine 308 (Thr308) by the 3-phosphoinositide–dependent protein kinase 1 (PDK1)33 and phosphorylation of serine 473 (Ser473) by the mammalian target of rapamycin (mTOR)-rictor complex34. Additionally, DNA-dependent protein kinase (DNA-PK) can directly or indirectly phosphorylate Akt Ser473 (refs. 35–37). Activated Akt promotes tumorigenesis by modulating cell cycle progression, apoptosis and cell motility. By contrast, the tumor suppressor phosphatase and tensin homology deleted on chromosome 10 (PTEN) inhibits the actions of PI3K38. Because Akt signaling is important in CAC39, we assessed Akt activation in Aim2−/− mice. Similar expression of total Akt (Fig. 3a,b) was found in the colons of wild-type, Aim2−/− and Asc−/− mice treated with AOM/DSS, but increased Ser473 p-Akt and reduced PTEN were found in Aim2−/− colons (Fig. 3a, Supplementary Fig. 4b,f). By contrast, slightly reduced levels of p-Akt were found in Asc−/− colons (Fig. 3b, Supplementary Fig. 4g). A similar elevation in p-Akt was detected in the colons of Aim2−/−/ApcMin/+ as compared to ApcMin/+ control mice (Fig. 3c, Supplementary Fig. 4h), demonstrating that AIM2 regulates Akt activity in models of both CAC and sporadic colon cancer.
Figure 3.
AIM2 negatively regulates Akt activity in vitro and in vivo. (a,b) Western blot analyses of PI3K pathway members in the colons of AOM/DSS-treated mice (representative of n = 6 mice/group). (c) Western blot analyses of Akt phosphorylation in colon homogenates from 12-week-old ApcMin/+ and Aim2−/−/ApcMin/+ mice (representative of n = 6 mice/group). (d–g) Western blot analyses of Akt phosphorylation in IGF-1–simulated MEFs from Aim2+/+ and Aim2−/− littermates (d), Asc+/+ and Asc−/− littermates (e), AIM2–3xFlag- or EV-transfected HCT-116 cells (f), and wild-type and Aim2−/− organoids (g). Actin (β-actin) and total Akt were used as loading controls. Results are representative of 1 of 3 experiments.
We next assessed whether AIM2 can directly regulate Akt in vitro. AIM2 is widely expressed throughout immune tissue and cells17 (Supplementary Fig. 8a). Human intestinal tissue17 and colon epithelial cells also express AIM2 (Supplementary Fig. 8b), and AIM2 mutations are present in colon adenomas and several human colon carcinoma cell lines, implying a role for AIM2 in epithelial cells18–20. To examine Akt activation in vitro, we treated cells with insulin-like growth factor-1 (IGF-1), which activates PI3K and Akt and is elevated in colon cancer40. IGF-1–treated Aim2−/− mouse embryonic fibroblasts (MEFs) had increased Ser473 (but not Thr308) p-Akt compared to Aim2+/+ controls (Fig. 3d, Supplementary Fig. 4i,j). This difference in p-Akt was not observed in Asc−/− MEFs (Fig. 3e, Supplementary Fig. 4l), indicating the different roles of AIM2 and ASC in regulating Akt activation.
We confirmed that AIM2 regulated Akt activation in colon cancer or colon epithelial cells through two approaches. First, the human colon carcinoma cell line HCT-116, which lacks functional AIM2 (ref. 19), was reconstituted by transfection with an AIM2-bearing vector Akt activation was lower in AIM2-expressing HCT-116 cells than in empty vector–transfected control cells (Fig. 3f, Supplementary Fig. 4m), indicating that AIM2 reduced Akt activation. Second, primary non-transformed colon epithelial organoid cultures were generated using a combination of growth factors and signaling molecules in a Matrigel system as previously reported (Supplementary Fig. 9)41–43. We assessed Akt activation in these three-dimensional organoid cultures isolated from wild-type and Aim2−/− colonic crypts and found increased p-Akt in Aim2−/− organoids as compared to wild-type control organoids upon IGF-1 stimulation. The basal level of p-Akt was also elevated in Aim2−/− organoids, likely due to the required exposure of epidermal growth factor for propagation of these primary cells (Fig. 3g, Supplementary Fig. 4n). These results suggest that AIM2 suppresses Akt activity in non–bone marrow cells, including colonic epithelial cells. In contrast to results from colonic epithelial cells, p-Akt did not differ in wild-type and Aim2−/− BMDM upon stimulation with LPS or IGF-1 (Supplementary Fig. 10a,b). Thus, AIM2 does not affect Akt activation in macrophages, a cell type well-characterized as expressing a functional AIM2 inflammasome.
Inhibition of Akt activation in Aim2−/−mice reduces tumor load in vivo
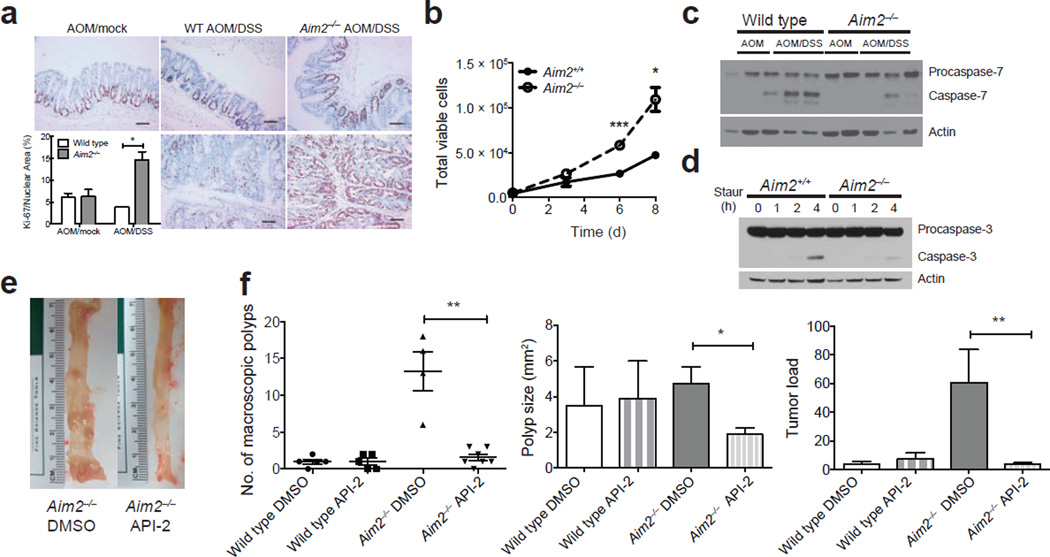
Consistent with the role of Akt in cell proliferation and survival, staining for Ki-67, a marker of proliferating cells, was increased in colonic crypts from AOM/DSS-treated Aim2−/− mice (Fig. 4a). Proliferation was also increased in Aim2−/− MEFs as compared to Aim+/+ controls (Fig. 4b). Moreover, in Aim2−/− colons, apoptotic caspase-7 cleavage was lower than in wild-type colons (Fig. 4c, Supplementary Fig. 4o), and Aim2−/− MEFs had lower apoptotic caspase-3 cleavage than controls following staurosporine treatment (Fig. 4d, Supplementary Fig. 4p).
Figure 4.
AIM2 restricts cell proliferation and promotes apoptosis. (a) Colon Ki-67 staining in AOM/DSS-treated wild-type and Aim2−/− mice (n = 6 for WT AOM/Mock and Aim2−/− AOM/DSS, and n = 5 for WT AOM/DSS and Aim2−/− AOM/Mock). 200× magnification; scale bars, 100 µm; error bars, s.e.m. (b) Cell numbers of Aim2+/+ and Aim2−/− MEFs (d, days). Error bars, s.e.m., from triplicate samples per group. (c) Western blot analyses of activated caspase-7 and procaspase-7 in colons from AOM/DSS-treated wild-type and Aim2−/− mice (n = 6 mice/group). (d) Western blot analyses of activated caspase-3 and procaspase-3 in staurosporine (Staur)-treated Aim2+/+ and Aim2−/− MEFs (h, hours). Results are representative of 1 of 3 experiments. (e) Representative images of colons from DMSO- or Akt inhibitor (API-2)-treated Aim2−/− mice. (f) Number of macroscopic colon polyps, average polyp size and tumor load in API-2-treated Aim2−/− mice. Each symbol represents one animal. Error bars denote standard error. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001. a,b, unpaired t-test; f, Mann-Whitney U-test (due to the non-normal distribution of the data sets and small sample size).
To determine whether activated Akt observed in the absence of AIM2 was required for increased tumorigenesis in Aim2−/− mice, AOM/DSS-treated wild-type and Aim2−/− mice were given a total of six i.p. injections (corresponding to the end of the second and third rounds of DSS) of a highly specific Akt inhibitor (API-2) previously described to target overactive Akt in cancer cells both in vitro and in vivo44. On day 55 of the AOM/DSS model, API-2–treated Aim2−/− mice had decreased colon polyp numbers, smaller polyps and reduced tumor loads (Fig. 4e,f) as compared to vehicle-treated Aim2−/− mice following AOM/DSS exposure. API-2 treatment did not reduce tumorigenesis in AOM/DSS-treated wild-type mice (Fig. 4f), which showed low numbers of colon polyps and little to no detectable p-Akt (shown earlier in Fig. 3a). These results suggest activated Akt promotes tumorigenesis in Aim2−/− mice, and AIM2 likely restricts tumorigenesis via limiting Akt activation.
AIM2 interacts with DNA-PK to regulate Akt activation
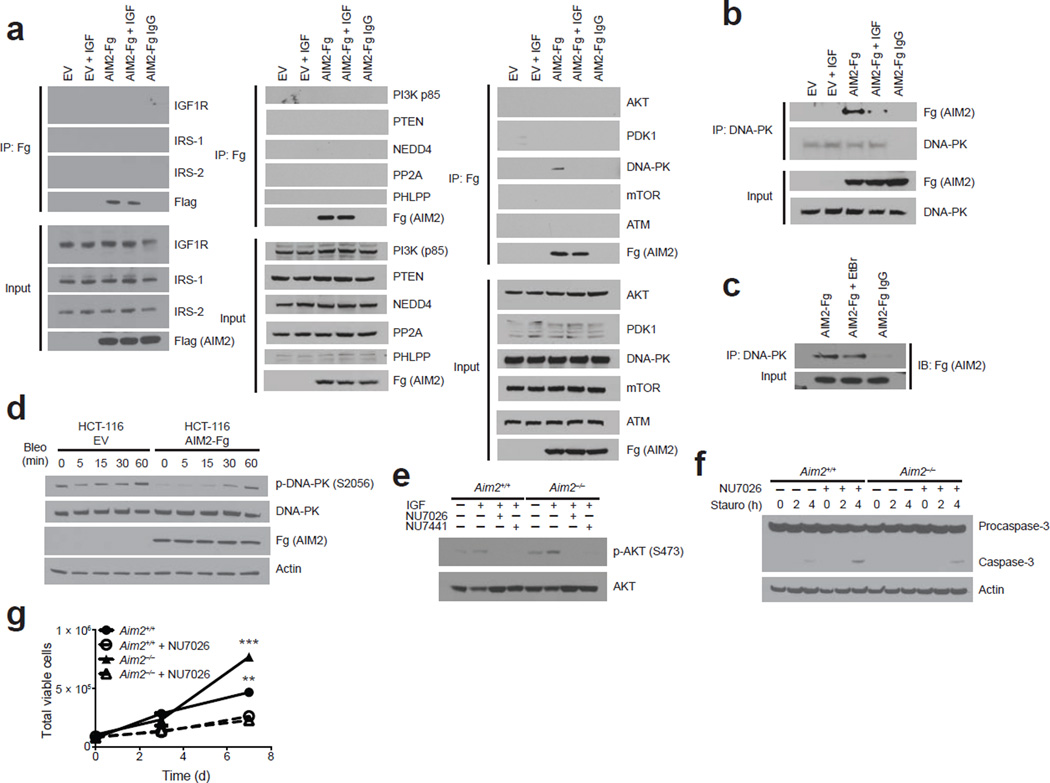
Akt signaling is controlled by kinases (including insulin receptor substrate-1/2 (IRS1/2), PI3K, 3-phosphoinositide dependent protein kinase-1 (PDK1), mTOR and DNA-PK) and phosphatases (including PTEN, protein phosphatase 2A (PP2A) and PH domain leucine-rich repeat protein phosphatase (PHLPP)) that are often hyperactive or inactivated, respectively, in cancer32. To further explore the mechanism by which AIM2 inhibits Akt activation, we examined whether AIM2 interacts with these kinases/phosphatases. The presence of p-Akt Ser473, but not Thr308, hyperphosphorylation in Aim2−/− MEFs (Fig. 3d) suggests that AIM2 targets an Akt Ser473–specific kinase or phosphatase. Enzymes in the mTOR or DNA-PK pathway are known to phosphorylate Akt at Ser473. Because specific antibodies against endogenous AIM2 are not available, we overexpressed a 3xFLAG-tagged human AIM2 (AIM2-3xFLAG) in HEK293T cells and performed co-immunoprecipitation experiments with endogenous Akt-PI3K components. AIM2 did not associate with Akt, IGF1 receptor (IGF1R), IRS1, IRS2, PI3K, PDK1, mTOR, ATM, the major PI3K-related phosphatases (that is, PTEN, PP2A and PHLPP) or the PTEN-specific ubiquitin ligase NEDD4 (Fig. 5a). However, AIM2 did associate with DNA-PK (Fig. 5a), and this was confirmed with reciprocal co-immunoprecipitations (Fig. 5b). DNA-PK consists of a 470-kDa catalytic subunit (DNA-PKcs) and two 73- and 80-kDa DNA-binding subunits (Ku70 and Ku80)45, and DNA-PK can promote cellular survival via Akt activation35,37. Elevated DNA-PK activity and expression has been reported in colorectal tumors46, and DNA-PK promotes c-Myc oncoprotein stability47. As both AIM2 and DNA-PK bind DNA, their interaction could be a consequence of nonspecific DNA bridging. To address this possibility, immunoprecipitates were treated with the DNA-intercalating agent ethidium bromide (EtBr) to disrupt DNA interactions that could promote indirect associations. The AIM2–DNA-PKcs association was maintained in the presence of EtBr (Fig. 5c), indicating that this association is not an artifact of their binding to DNA.
Figure 5.
AIM2 associates with DNA-PK and restricts its activity and Akt phosphorylation. (a) Immunoprecipitation of AIM2 and immunoblots of PI3K members and (b) reciprocal immunoprecipitation of DNA-PKcs and immunoblot of AIM2 in AIM2-Flag (Fg)- or empty vector (EV)-expressing HEK293T cells treated with IGF-1 (IGF) (c) Co-immunoprecipitation of DNA-PKcs and AIM2 in the presence of ethidium bromide (EtBr) in HEK293T cells. (d) Western blot analysis of phosphorylated DNA-PK (p-DNA-PK) in bleocin-treated AIM2-expressing HCT-116 cells. (e) Western blot analysis of p-Akt in 10 µM NU7026- or NU7441-treated Aim2+/+ and Aim2−/− MEFs. (f) Western blot analysis of activated caspase-3 in staurosporine (Stauro)-treated Aim2+/+ and Aim2−/− MEFs treated with NU7026 (5 µM; h, hours). (g) Cell number of Aim2+/+ and Aim2−/− MEFs cultured with 1 µM NU7026 (d, days). Error bars, s.e.m., from triplicate samples per group. Results are representative of 1 of 3 independent experiments (a,b,c,d,e,g) and representative of 1 of 2 independent experiments (f). **P < 0.01, ***P < 0.001. One-way ANOVA (Tukey's multiple-comparisons test).
The AIM2-DNA-PKcs interaction was detected in resting cells, but lost upon IGF-1 stimulation (Fig. 5a,b), suggesting that the interaction occurred in the resting state but these proteins dissociated from each other upon cellular activation, allowing DNA-PKcs to promote Akt activation. To address this possibility, we tested whether AIM2 could directly prevent DNA-PK activation by using AIM2-complemented HCT-116 cells treated with bleocin, which induces potent DNA-PK phosphorylation45. Indeed, AIM2 overexpression decreased phosphorylation of DNA-PK (p-DNA-PK; Fig. 5d, Supplementary Fig. 4q), demonstrating that AIM2 can limit DNA-PK activation. To assess the link between AIM2, DNA-PK and Akt activation, we tested two highly-specific DNA-PKcs inhibitors, NU7026 and NU7441 (ref. 48), in Aim2−/− and control cells. Akt Ser473 phosphorylation was higher in IGF-1-treated Aim2−/− cells as compared to IGF-1-treated wild-type controls, but this difference was not observed with either NU7026 or NU7441 treatment (Fig. 5e, Supplementary Fig. 4r). Additionally, p-Akt activation is known to have prosurvival effects. Reduced apoptosis and increased viability in Aim2−/− cells as compared to wild-type cells was reversed by DNA-PK inhibitors (Fig. 5f, Supplementary Fig. 4s and Fig. 5g). These results suggest DNA-PK is required for elevated Akt activation in Aim2−/− cells.
DISCUSSION
The innate immune DNA sensor AIM2 has been implicated as a potential tumor suppressor, although the mechanism for this effect remained unclear. Here, we assessed the role of the AIM2 inflammasome in the regulation of CAC, but instead found that AIM2 attenuates tumorigenesis through a mechanism independent of inflammasome activation and inflammation. In colon epithelial cells, AIM2 limits the activation of Akt, a master regulator of cellular survival often overexpressed in multiple human cancers. AIM2 in resting cells physically associates with and limits activation of DNA-PK, a PI3K-related family member that promotes Akt. Conversely, loss of AIM2 promotes DNA-PK–mediated Akt hyperactivation, thereby promoting proliferation and reducing apoptosis, two hallmarks of cancer development.
AIM2 mutations are found in human tumors18,19, reduced AIM2 expression has been associated with poor prognosis in patients20 and AIM2 suppresses proliferation21–23. Our findings indicate that loss of AIM2 results in activation of Akt, and suppression of Akt can reduce tumorigenesis in Aim2-deficeint animals. Thus, pharmacological targeting of Akt may be beneficial in individuals with AIM2-deficent tumors. This work implicates AIM2 as having a previously unrecognized role in the dual regulation of innate immune networks and signaling pathways that are critical to cellular survival and tumorigenesis. Although this study focused on colorectal cancer models, loss or mutations in APC in mice are also associated with intestinal tumorigenesis27,28. Furthermore, AIM2 is implicated in human melanoma17, and thus further work is needed to investigate whether targeting Akt is beneficial in other cancers associated with AIM2 deficiency.
ONLINE METHODS
Experimental animals
All animal protocols were approved by the University of North Carolina Chapel Hill (UNC) Institutional Animal Care and Use Committee (IACUC) in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animal numbers were empirically determined to optimize numbers necessary for statistical significance based on our previous reports utilizing these disease models7 (minimum of 4–7 animals/group for each type of analysis) while adhering to IACUC polices on maximum numbers of animals used in research. These numbers were also determined based on breeding efficiency, animal housing space and handling limitations. No randomization method was used due to the experimental setup, ad libitum administration of DSS and littermate groups. All experiments were performed under specific pathogen-free conditions using 6- to 8-week-old male mice. Animals that exhibited health concerns not related to the study conditions (for example, fight wounds and dermatitis) were excluded from the analysis. Wild-type (WT) C57BL/6 mice were obtained from the Jackson Laboratories (Bar Harbor, ME) and maintained at UNC Chapel Hill for more than 9 generations. Asc−/− mice on the C57BL/6 background have been described elsewhere49.
The Aim2 knockout (Aim2−/−) mice were generated by Ingenious Targeting Laboratory (Ronkonkoma, NY) on the C57BL/6 background via targeted replacement of the Aim2 coding region with a neomycin resistance gene through homologous recombination (depicted in Supplementary Fig. 1a). Primer sequences used for PCR-based Aim2−/− mouse genotyping are as follows: neomycin (KO) forward, GGAACTTCGCTAGACTAGTACGCGTG; neomycin (KO) reverse, CAACATTGTACAGATTGAGCAGG; Aim2 (WT) forward, GATGGAGAGTGAGTACCGGGAAATGCTGTT; and Aim2 (WT) reverse, TCTGCAAGTAGATTGGAGACAGACTCTGGTGA, resulting in a 450-bp band for the wild type and a 250-bp band for the targeted Aim2 knockout when PCR analysis is performed on the genomic DNA (Supplementary Fig. 1a).
The ApcMin/+ mice on the C57BL/6 background were purchased from Jackson Laboratories (Bar Harbor, ME) and maintained at UNC Chapel Hill. Aim2−/− mice were crossed with ApcMin/+ mice to generate Aim2−/−/ApcMin/+ mice.
Experimental colitis and colitis-associated colon cancer
For induction of colitis, mice were given 4–5% dextran sulfate sodium salt (DSS) (MP Biomedical, LLC) dissolved in drinking water for 5 consecutive days followed by regular drinking water for 6 or 10 days (Supplementary Fig. 5a,d)24. To induce colitis-associated colon cancer (CAC), animals were given a single i.p. injection of the mutagen azoxymethane (AOM, Sigma-Aldrich) (10 mg/kg body weight) in combination with three cycles of 3% DSS in drinking water for 5 days followed by regular drinking water for 14 days24 (Fig. 1a). Animals were killed following completion of either model or when moribund.
Animals were blindly monitored for weight loss daily during DSS treatment and once every 2 days during regular water administration to assess disease progression. In addition, clinical score assessments were blindly performed after administration of each round of DSS to evaluate stool consistency, rectal bleeding and stool blood measurements using Hemoccult Immunochemical Fecal Occult (Beckman Coulter) as previously described7.
For induction of chronic T cell–mediated colitis, spleens from healthy wild-type and Aim2−/− mice were isolated, gently dissociated and passed through a 70-µm cell strainer to generate single-cell suspension, and red blood cells were lysed using ACK buffer. Naive CD4+ T cells were purified by negative selection using the MACS Naïve CD4+ T Cell Isolation Kit (Miltenyl Biotech) and i.p. injected at 5 × 105 cells/mouse into recipient 8- to 10-week-old T and B cell–deficient Rag1−/− mice as previously described50. Animals were weighed weekly and killed at 8 weeks post-T cell transfer. Spleens were removed, total lymphocytes were isolated and the expansion of colitogenic CD4+T cells (and the absence of CD8+ T cells) was verified by flow cytometry.
Macroscopic polyp assessment
Colon tumorigenesis was visualized in situ using mini-endoscopy7. Animals were fasted overnight before endoscopy to minimize fecal obstruction during visualization, and a trained gastroenterologist (M.M.) performed endoscopy on the animals under oxygen-regulated isoflurane sedation on day 48 of the AOM/DSS model using a Coloview system (Karl Storz Veterinary Endoscope). The trained gastroenterologist was blinded to the genotype and experimental condition of the animals.
Upon completion of the AOM/DSS model of CAC, the mice were killed and colons were removed, flushed with PBS and opened longitudinally for visual polyp count by a trained investigator blinded to the genotype and experimental condition of the animals7. Tumor loads were determined as the sum of the diameters of all colon tumors present per mouse24. For spontaneous colon cancer assessment, ApcMin/+ and Aim2−/−/ApcMin/+ mice were killed at 12 weeks of age and colons were processed and assessed for macroscopic polyps as described above.
Assessment of intestinal permeability
AOM/DSS-treated wild-type and Aim2−/− mice were fasted for 3.5 h and fed FITC-dextran (MW 4,000, Sigma-Aldrich) at 60 mg/100 g body weight via oral gavage. Animals were killed 3.5 h later, and blood was removed via heart puncture. Fluorescence of serum FITC-dextran was measured using a plate reader at excitation 520 and emission 490 nm.
Generation of radiation bone marrow chimeras
Radiation bone marrow chimeric mice were generated as previously described7. Briefly, 10- to 12-week-old wild-type CD45.1+ and Aim2−/− CD45.2+ mice were lethally irradiated (8 Gy) and i.v. injected with 107 bone marrow cells isolated from either wild-type CD45+ or Aim2−/− CD45.2+ mice 24 h post-irradiation. Recipient mice were given 2 mg/ml neomycin in drinking water for 14 days, and bone marrow was allowed to reconstitute an additional 56 days before administration of AOM/DSS. The recipient mice were bled at 49 days post-bone marrow transplant, and the levels of peripheral blood lymphocyte CD45.1 verses CD45.2 was measured in order to determine the bone marrow reconstitution efficiency, which was 89–98% (data not shown).
Inhibition of Akt during colitis-associated colon cancer
Wild-type and Aim2−/− mice were given an i.p. injection of API-2 (1 mg/kg body weight in 10% DMSO) or 10% DMSO vehicle control on days 22, 25, 28, 41, 44 and 47 of the AOM/DSS model (3 injections during the second and third rounds of 3% DSS administration). Animals were killed at day 55 post-AOM/DSS, and macroscopic polyps were visualized as described above.
Histopathology
For assessment of tissue pathology, colons were affixed to cards in Swiss-roll fashion, fixed in 10% neutral-buffered formalin and routinely paraffin embedded and processed. Hematoxylin and eosin (H&E) staining was performed on 5-micron-thick colon sections, and inflammation, hyperplasia and dysplasia were scored by a Board-certified veterinary pathologist (A.B.R.) blinded to the genotype and experimental condition of each animal as previously described7. Additional sections were stained by immunohistochemistry for the proliferation marker Ki-67 (Cell Signaling Technology) and counterstained using 3,3′-diaminobenzidine (DAB) followed by hematoxylin counterstain. The percentage of Ki-67+ nuclei was calculated using ImageJ software.
qPCR
Inflammatory cytokine expression was assessed during CAC by washing colons with PBS and homogenizing the distal-most section for total RNA isolation using RNeasy Blood & Tissue kit (Qiagen), according to the manufacturer’s instructions. cDNA was generated using Superscript III (Invitrogen) according to the manufacturer’s instructions, and real-time PCR was performed for mouse Tnfa (Mm00443258), Il6 (Mm00446190), Il1b (Mm01336189), Il17a (Mm01189488), Ptgs2 (Mm00478374) and Il10 (Mm00439614) gene expression using mouse Taqman Gene Expression Assays (Applied Biosystems) and for I-Ab using SYBR green master mix (primer sequence: Reverse, CTGTCGTAGCGCACGTACT; Forward, GGCGAGTGCTACTTCACCA. The PCR results were normalized to Gapdh expression and were quantified by the ΔΔCT method.
Colon explant cultures for inflammatory cytokine production
Colon inflammatory cytokine secretion was assessed during CAC by flushing colons from AOM/DSS-treated mice repeatedly with PBS containing penicillin/streptomycin, and the distal-most 1-cm2 colon sections were cultured for 15 h in RPMI media containing 2× penicillin/streptomycin. Supernatants from these cultures were removed, cleared of debris by centrifugation and assessed for mouse IL-6, TNF, IFN-γ and IL-10 by Bio-Plex cytokine assay (Bio-Rad) and IL-1β (BD Biosciences) and IL-18 (eBioscience) by ELISA.
Western blotting
Protein expression of caspase-1, caspase-7, STAT3, MAPK, NF-κB and PI3K pathway members was assessed during CAC by homogenizing the distal-most section of the colon in 1% RIPA buffer (Boston Bioproducts) containing Complete Protease Inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (phosSTOP, Roche) cocktail. Cell lysates were cleared by centrifugation, and the protein concentration was determined by BCA protein assay (ThermoScientific). A total of 30 µg of protein was mixed in SDS loading dye (Novex) containing 20 mg/ml DTT reducing agent, boiled for 5 min, separated by SDS-PAGE using 4–12% Bis-Tris gels (Novex) and wet transferred to nitrocellulose membranes (Roche). Nitrocellulose membranes were blocked for 1 h with 5% non-fat milk and incubated overnight with primary antibodies, washed 5 times with TBS-T and incubated for 1–2 h at room temperature with the appropriate goat anti-rabbit or bovine anti-goat HRP-conjugated secondary antibodies (Santa Cruz Biotechnology and Sigma-Aldrich).
In vitro cell lines and primary cells were lysed in 1% RIPA or 1% NP-40 containing protease and phosphatase inhibitors (Roche), and then cellular lysates were cleared of cellular debris by centrifugation and were subjected to SDS-PAGE and western blotting as described above.
The cell lysates were immunoblotted using the following primary antibodies: anti-IL-1β (cat. no. AF401-NA) (R&D Systems); anti-PTEN (cat. no. 611500) (BD Bioscience); anti-caspase-1 (M-20) (cat. no. sc-514), anti-ERK1/2 (C-16) (cat. no. sc-093), anti-PHLPPL (T-17) (cat. no. sc-137663), and actin-HRP (C-11) (cat. no. sc-1615), anti-IRS1 (C-20) (cat. no. sc-559), anti-JNK (C-17) (cat. no. sc-474) (Santa Cruz Biotechnology); anti-pERK1/2 (T202/Y204) (cat. no. 4370), anti-p38 (cat. no. 9212), anti-pp38 (T180/Y182) (cat. no. 9216), anti-pJNK (T183/Y185) (cat. no. 9251), anti-p65 (D14E12), anti-pp65 (S536) (cat. no. 93H1), anti-IκBα (44D4) (cat. no. 4812), anti-pIκBα (S32) (14D4) (cat. no. 2859), anti-STAT3 (cat. no. 79D7), anti-pSTAT3 (Y705) (cat. no. 9131), anti-IGFR (D23H3) (cat. no. 9750), anti-IRS2 (L1326) (cat. no. 3089), anti-PI3K p85 (19H8) (cat. no. 4257), anti-mTOR (cat. no. 2972), anti-pmTOR (S2448) (cat. no. 2971), anti-pPDK1(cat. no. 3062), anti-pAkt (Thr308) (cat. no. 2965), anti-pAkt (Ser473) (cat. no. 4060), anti-Akt (cat. no. 9272), anti-caspase-3 (cat. no. 9662), anti-cleaved caspase-7 (Asp198) (cat. no. D6H1), anti-PP2A (81G5) (cat. no. 2041), anti-NEDD4 (cat. no. 2740) and anti-ATM (D2E2) (cat. no. 2873) (Cell Signaling Technology); anti-Flag-HRP (M2) (cat. no. A8592); anti-Flag (cat. no. F7425), anti-DNA-PKcs (cat. no. SAB4502385), (Sigma-Aldrich); and anti-pDNA-PK (S2056) (cat. no. ab18192) (Abcam). All primary antibodies were used at 1:1,000. Secondary antibodies include the following: goat anti-rabbit HRP (111-035-144), goat anti-mouse HRP (115-035-146) and donkey anti-goat HRP (805-035-180); all secondary antibodies were used at 1:40,000. Protein bands were quantified by densitometric analysis using ImageJ software.
Primary cell generation
Mouse embryonic fibroblasts (MEFs) were generated by mating an Aim2 heterozygous (Aim2+/−) breeder pair or an Asc heterozygous (Asc+/−) breeder pair and isolating the embryonic day 13.5 (E13.5) embryos from the female. The genotype of the Aim2+/+ and Aim2−/− littermates (or Asc+/+ and Asc−/− littermates) were confirmed by PCR, and the head and internal organs were removed from the embryos. The remaining tissue was minced, digested with 0.05% trypsin EDTA (Gibco) in the presence of DNase (Sigma-Aldrich) and the resulting cells were plated to confluency in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) containing 10% FBS (Sigma-Aldrich), penicillin/streptomycin, 4.5 g/L D-glucose, 4mM L-glutamine and 110 mg/L sodium pyruvate. A total of three independently generated Aim2+/+ and Aim2−/− littermate MEFs were used for this study, and all MEFs were non-immortalized and below passage 6.
Bone marrow–derived macrophages were generated by flushing bone marrow from the femurs and tibias of wild-type, Asc−/− and Aim2−/− mice. Red blood cells were lysed using ACK buffer, and the remaining bone marrow cells were differentiated into macrophages by culturing for 6–7 days in DMEM with GM-CSF-containing L929-conditioned media.
Highly proliferative 3D spheroids (termed organoids) of colonic epithelial cells were cultured in vitro according to previous publications41–43. Briefly, colon crypts were isolated from mice by soaking the excised colon in a chelating buffer (2.0 mM EDTA and 0.5 mM DTT) for 75 min followed by vigorous shaking. A total of 50,000 crypts were suspended in 300 µL of cold Matrigel (Corning Life Sciences) on ice, and the 15 µL of the suspension was added to the center of each well of a 24-well plate. The plate was inverted and incubated at 37 °C for 10 min to solidify Matrigel. A total of 0.5 ml of advanced DMEM/F12 medium containing Wnt-3A-containing L929-conditioned medium, R-spondin 2-containing HEK293-conditioned medium (a kind gift from J. Whitsett at Cincinnati Children’s Hospital), Noggin-containing HEK293-conditioned medium (provided by N.L.A) (volumetric ratio of 3:1:1:1), 50 ng/ml EGF (Biovision), 10 µM Y27632 ROCK inhibitor (LC Laboratories), 1mM N-acetyl-L-cysteine (Sigma-Aldrich), GlutaMAX, 10 mM HEPES, penicillin/streptomycin and 5 µg/ml gentamicin. The medium was changed every 48 h, and the organoids were passaged every 5 days at a 1:5 split ratio. Medium was removed from the well, and trypsin-EDTA (0.05%) was added. After incubation at 37 °C for 5 min, organoids were dissociated into small fragments by pipetting. Cells were then washed, embedded in fresh Matrigel and plated into 24-well plate. Day 5 differentiated organoids were used in experiments.
Cell culture
The human colorectal carcinoma HCT-116 cell line (American Type Culture Collection (ATCC)# CCL-247) was cultured in McCoy’s 5A media with 10% FBS and penicillin streptomycin. The embryonic kidney epithelial cell line HEK293T (ATCC# 3216) was cultured in DMEM containing 10% FBS (Sigma-Aldrich), penicillin/streptomycin, 4.5 g/L D-glucose, 4 mM L-glutamine and 110 mg/L sodium pyruvate. All cell lines were maintained at 37 °C with 5% CO2. Cells were routinely checked for mycoplasma by the Lineberger Cancer Center Tissue Culture Facility at UNC Chapel Hill and were authenticated by ATCC prior to purchase.
Co-immunoprecipitation
AIM2-Flag and DNA-PK co-immunoprecipitation experiments were carried out in 293T cells that were transfected with 1 µg/ml of AIM2-Flag or empty vector (pCIG2-IRES-PURO) using Lipofectamine reagent according to manufacturer’s instructions. Cells were stimulated as indicated at 48 h post-transfection and lysed in NP-40 lysis buffer containing Complete protease inhibitor cocktail (Roche). Cell lysates were centrifuged to remove insoluble material, pre-cleared with 10% zysorbin (Novex) and incubated overnight at 4°C with rotation with either anti-Flag antibody (a kind gift from M.A. Accavitti-Loper at University of Alabama, Birmingham) or anti-DNA-PKcs (Sigma-Aldrich). Ethidium bromide was added during the overnight antibody incubation at a concentration of 50 µg/ml where indicated. The lysates were incubated with Protein A/G Ultra-link resin (Thermo Scientific) for 2 h at 4 °C with rotation followed by 4 washes in NP-40 lysis buffer. For anti-Flag immunoprecipitations, protein was eluted using 3xFlag peptide (Sigma-Aldrich). For anti-DNA-PKcs immunoprecipitations, protein was eluted by boiling beads in SDS loading dye (Novex) containing 20 mg/ml DTT.
AIM2 constructs
pCIG2-PURO-hAIM2-3x-Flag was generated using traditional cloning methods. Briefly: two primers encoding a glycine linker and a 3x-Flag tag (GGGGDYKDDDDKGADYKDDDDKEFDYKDDDDK) were annealed and extended to generate a dsDNA fragment (GCACCATAAAGGTTATTAAGGCCAAAAAAAAAACAGGCGGTGGCGGTGATTATAAAGATGATGATGATAAAGGGGCAGACTACAAGG and atatGGATCCTCACTTGTCGTCGTCGTCCTTGTAGTCGAATTCCTTGTCGTCGTCGTCCTTGTAGTCTGCCCCTTTATC), which served as the reverse primer for the PCR amplification with an AIM2 forward primer (ggggGCTAGCACCATGGAGAGTAAATACAAGGAGATACTCTTGCTAACAGGC). The 3x-Flag tagged AIM2 amplicon was cloned into the NheI/BamHI site of pCIG2-IRES-PURO, a variant of pCIG2-IRES-eGFP51. TRC Lentiviral Human AIM2 shRNA constructs were purchased form Open Biosystems as bacterial glycerol stocks. Plasmids were purified using the Nucleobond Xtra Maxi Plus Kit (Macherey Nagel). AIM2-Flag expressing lentiviruses were generated in 293FT cells as previously described51.
AIM2 inflammasome activation
Wild-type and Aim2−/− macrophages were plated overnight, primed with 500 µg/ml LPS (Peprotech) for 3 h and stimulated for 6 h with 1 µg/ml poly(dA:dT) (Invivogen) combined with Lipofectamine 2000 transfection reagent (Invitrogen), according to the manufacturer’s instructions, for IL-1β assessment as previously described15.
PI3K activation
MEFs and macrophages were plated overnight and stimulated with 100 ng/ml of insulin-like growth factor-1 (IGF-1, R&D Systems) or 500 µg/ml lipopolysaccharide (LPS) to assess Akt activation by western blot. HCT-116 cells were plated overnight and transfected with 1 µg/ml of EV or AIM2-Flag using Lipofectamine 2000 according to the manufacturer’s instructions. HCT-116 cells were stimulated with IGF-1 (100 ng/ml) 48 h post-transfection. Day 5–differentiated primary organoids were generated as described above and stimulated with 100 ng/ml IGF-1 for 20 min.
Cell death assay
Aim2+/+ and Aim2−/− MEFs were plated overnight and stimulated with 0.5 µM staurosporine (Sigma-Aldrich) for the indicated times. Cells were washed and processed for cleaved caspase-3 expression by western blot analysis.
DNA-PKcs ihibitors
When indicated, MEFs were pre-treated for 30 min or 1 h with the indicated concentration of the DNA-PKcs inhibitors NU7026 (Cayman Chemical) or NU7441 (Tocris), or cultured with a single dose of NU7026 for proliferation assays.
Flow cytometry
Spleen, thymus and inguinal lymph nodes were isolated from naive Aim2+/+, Aim2+/− and Aim2−/− animals; tissue was gently dissociated and passed through a 70-µm cell strainer to generate single cell suspension; and red blood cells were lysed using ACK buffer. Cells were counted and stained using the following antibodies: anti-CD11b-FITC (cat. no. 11-0112, used at 1:200), anti-CD4-PerCpCy5.5 (cat. no. 45-0042, used at 1:80), anti-CD8-eFluor 450 (cat. no. 48-0083, used at 1:80), anti-CD11c-APC (cat. no. 17-0114, used at 1:80), anti-TCRβ-Alexa 700 (cat. no. 47-5961, used at 1:80), anti-F480-PE (cat. no. 12-4801, used at 1:80), anti-NK1.1-PECy7 (cat. no. 25-5941, used at 1:80) (eBioscience) and anti-B220-APC Cy7 (cat. no. 565371, used at 1:200) (BD Biosciences). Peripheral blood lymphocytes isolated from radiation bone marrow chimeric mice were stained with anti-CD45.1-PE (BD Biosciences, cat. no. 553776, used at 1:1,000) and anti-CD45.2-V500 (BD Biosciences, cat. no. 562130, used at 1:1,000). Flow cytometry was performed using an LSRII (BD Biosciences), and the resulting data were analyzed using FlowJo (TreeStar).
Statistical analysis
All results are presented as the mean ± s.e.m. (SEM). Significance between two groups was assessed by the Student’s two-tailed t-test. The non-parametric Mann-Whitney U-test was used to assess significance between two means of data sets lacking a normal distribution and having a small sample size. Data sets consisting of more than 2 groups were analyzed by analysis of variance (ANOVA) with Tukey-Kramer HSD post test for multiple comparisons if significance was determined. The product limit method of Kaplan and Meier was used for generating the survival curves, which were compared by using the log-rank test. A P value that was less than 0.05 was considered statistically significant for all data sets. All statistical analysis was performed using GraphPad Prism software. Samples that were greater than 3 s.d. from the mean were excluded as outliers.
Supplementary Material
ACKNOWLEDGMENTS
We thank E.H. Guthrie for technical assistance; I.C. Allen from the Department of Biomedical Sciences and Pathobiology at Virginia Tech for technical advice regarding the CAC model. P.K. Lund and S. Ding from the Department of Cell Biology and Physiology at UNC for their helpful discussions and technical advice pertaining to the ApcMin/+ model of spontaneous intestinal cancer; the Center for Gastrointestinal Biology and Disease (CGIBD) at UNC for providing core and technical support; and the Department of Microbiology and Immunology Flow Cytometry Core Facility at UNC for providing core and technical support. This work was supported by National Institute of Health (NIH) grants RO1-CA156330 (National Cancer Institute), U19-AI067798 (National Institute of Allergy and Infectious Disease/NIAID), R37-AI029564 (NIAID) and P01 DK094779 (National Institute of Diabetes and Digestive and Kidney Diseases/NIDDK) (J.P.-Y.T.). J.E.W. was supported in part by the NIH postdoctoral fellowship F32-K088417-01 (NIDDK) and in part by the American Cancer Society, PF-13-401-01-TBE. This work was also supported by the NIH grants R01 EY024556 (National Eye Institute) (N.L.A. and Y.W.) and R15 DK098754 (NIDDK) (B.K.D.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.E.W. and A.S.P. contributed equally to this manuscript. J.E.W., A.S.P. and J.P.-Y.T designed the experiments and wrote the manuscript with input from B.K.D. and D.A.R. J.E.W., A.S.P. and L.C. performed most of the analyses. A.S.P. generated the AIM2 constructs and performed lentivirus transductions. W.-C.C. assisted in generating primary fibroblasts. M.S. performed the qPCR cytokine experiments. A.A.K, A.D.T. and W.J.B. assisted in the CAC and APCmin/+ animal studies. A.B.R. performed the histopathological scoring. L.C. performed the clinical scoring. Y.W. and N.L.A. developed and generated the primary organoid cultures. B.R.B. and Y.O. performed the flow cytometric analysis. M.M. and C.J. performed and oversaw the mini-endoscopy. All contributing authors have agreed to submission of this manuscript for publication.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Weir HK, et al. Annual report to the nation on the status of cancer, 1975–2000, featuring the uses of surveillance data for cancer prevention and control. J. Natl. Cancer Inst. 2003;95:1276–1299. doi: 10.1093/jnci/djg040. [DOI] [PubMed] [Google Scholar]
- 2.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 4.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 5.Dupaul-Chicoine J, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen IC, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J. Immunol. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salcedo R, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J. Exp. Med. 2010;207:1625–1636. doi: 10.1084/jem.20100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu B, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc. Natl. Acad. Sci. USA. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Normand S, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. USA. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc. Natl. Acad. Sci. USA. 2010;107:21635–21640. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeYoung KL, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 18.Woerner SM, et al. Microsatellite instability of selective target genes in HNPCC-associated colon adenomas. Oncogene. 2005;24:2525–2535. doi: 10.1038/sj.onc.1208456. [DOI] [PubMed] [Google Scholar]
- 19.Woerner SM, et al. The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Genes Chromosom. Cancer. 2007;46:1080–1089. doi: 10.1002/gcc.20493. [DOI] [PubMed] [Google Scholar]
- 20.Dihlmann S, et al. Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. Int. J. Cancer. 2014 doi: 10.1002/ijc.28891. [DOI] [PubMed] [Google Scholar]
- 21.Choubey D, Walter S, Geng Y, Xin H. Cytoplasmic localization of the interferon-inducible protein that is encoded by the AIM2 (absent in melanoma) gene from the 200-gene family. FEBS Lett. 2000;474:38–42. doi: 10.1016/s0014-5793(00)01571-4. [DOI] [PubMed] [Google Scholar]
- 22.Chen IF, et al. AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol. Cancer Ther. 2006;5:1–7. doi: 10.1158/1535-7163.MCT-05-0310. [DOI] [PubMed] [Google Scholar]
- 23.Patsos G, Germann A, Gebert J, Dihlmann S. Restoration of absent in melanoma 2 (AIM2) induces G2/M cell cycle arrest and promotes invasion of colorectal cancer cells. Int. J. Cancer. 2010;126:1838–1849. doi: 10.1002/ijc.24905. [DOI] [PubMed] [Google Scholar]
- 24.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 25.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 26.Dinarello CA. Why not treat human cancer with interleukin-1 blockade? Cancer Metastasis Rev. 2010;29:317–329. doi: 10.1007/s10555-010-9229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 28.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Li L, Gretz N, Gebert J, Dihlmann S. Absent in Melanoma 2 (AIM2) is an important mediator of interferon-dependent and -independent HLA-DRA and HLA-DRB gene expression in colorectal cancers. Oncogene. 2012;31:1242–1253. doi: 10.1038/onc.2011.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 31.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons DW, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 33.Stephens L, et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 35.Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, et al. Protein phosphatase 2A and DNA-dependent protein kinase are involved in mediating rapamycin-induced Akt phosphorylation. J. Biol. Chem. 2013;288:13215–13224. doi: 10.1074/jbc.M113.463679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu D, Huang J, Basu A. Protein kinase Ce activates protein kinase B/Akt via DNA-PK to protect against tumor necrosis factor-α-induced cell death. J. Biol. Chem. 2006;281:22799–22807. doi: 10.1074/jbc.M603390200. [DOI] [PubMed] [Google Scholar]
- 38.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, et al. Effect of adenovirus-mediated PTEN gene on ulcerative colitis-associated colorectal cancer. Int. J. Colorectal Dis. 2013;28:1107–1115. doi: 10.1007/s00384-013-1678-9. [DOI] [PubMed] [Google Scholar]
- 40.Ma J, et al. Prospective study of colorectal cancer risk in men and plasma levels of insulin-like growth factor (IGF)-I and IGF-binding protein-3. J. Natl. Cancer Inst. 1999;91:620–625. doi: 10.1093/jnci/91.7.620. [DOI] [PubMed] [Google Scholar]
- 41.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, et al. Capture and 3D culture of colonic crypts and colonoids in a microarray platform. Lab Chip. 2013;13:4625–4634. doi: 10.1039/c3lc50813g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64:4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 45.Roberts SA, et al. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosoi Y, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int. J. Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- 47.An J, et al. DNA-dependent protein kinase catalytic subunit modulates the stability of c-Myc oncoprotein. Mol. Cancer. 2008;7:32. doi: 10.1186/1476-4598-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper A, et al. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 49.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 50.Ostanin DV, et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G135–G146. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrucelli A, Umashankar M, Zagallo P, Rak M, Goodrum F. Interactions between proteins encoded within the human cytomegalovirus UL133–UL138 locus. J. Virol. 2012;86:8653–8662. doi: 10.1128/JVI.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.