Abstract
Chimeric antigen receptor (CAR) T cell therapies have demonstrated durable and potentially curative therapeutic efficacy against B cell leukemia in clinical trials. A CAR strategy can target any tumor surface antigens as long as an antigen-binding receptor can be generated. New CARs which target solid tumors and have the potential to target multiple tumor types are needed. In this study, B7H6, a ligand for the NK cell activating receptor NKp30, was targeted to create a CAR which targets multiple tumor types. B7H6 is expressed on various primary human tumors, including leukemia, lymphoma, and gastrointestinal stromal tumors (GISTs), but it is not constitutively expressed on normal tissues. B7H6-specific CAR T cells have robust cellular cytotoxicity and IFN-γ secretion when co-cultured with B7H6+ tumor cells, and they exhibit little self-reactivity to immature dendritic cells (iDCs) or pro-inflammatory monocytes. In vivo, B7H6-specific CAR T cells greatly enhanced the survival of RMA/B7H6 lymphoma bearing mice. The long-term survivor mice were protected against a B7H6-deficient tumor re-challenge. This CAR therapy also decreased tumor burden in a murine ovarian cancer model. In conclusion, B7H6-specific CARs have the potential to treat B7H6+ hematologic and solid tumors.
Keywords: CAR, NK cell, adoptive T cell therapy, lymphoma, ovarian cancer
INTRODUCTION
Chimeric antigen receptor (CAR) T cell therapies have demonstrated durable and potentially curative therapeutic efficacy in various clinical trials.1–4 CARs generally consist of a single-chain variable fragment from antibody (scFv) for target recognition, fused with co-stimulation signaling domains, such as CD28 or 4-1BB, and a CD3ζ signaling domain to provide T cell activation signals.5 This approach can target almost any tumor surface antigens as long as a specific receptor can be generated. Additionally, antigen recognition by CARs is often independent of MHC expression, which enables a single CAR design to be used in patients regardless of MHC allele expression. Although CAR strategies can target almost all types of tumors, approaches that target multiple tumor types are rare and CARs that target molecules more selectively expressed on tumors are needed.
CAR designs utilizing NK cell activating receptors, such NKG2D and NKp30 provide a strategy which targets multiple tumor types yet maintains tumor selectivity over normal cells.6–8 Many NK cell activating receptors recognize stress induced ligands that are specifically expressed on tumor cells.9 For example, NKp30 recognizes B7H6, which is expressed on a diverse number of human tumor cell lines and on some primary human leukemia, lymphomas and gastrointestinal stromal tumors (GISTs).10–12 Moreover, data suggest that NK cells utilize NKp30 for tumor immune surveillance in clinical settings, such as in acute myeloid leukemia (AML) patients, a NCRdull phenotype on NK cells correlated with poor survival.13 In GIST patients immunostimulatory forms of NKp30 correlated with a better prognosis.12 Furthermore, B7H6 mRNA is not expressed in 48 normal tissues under steady state conditions.11 This suggests that NKp30-based CARs may target multiple tumor types. However, BAG-6, another ligand for NKp30, is expressed on iDCs and can trigger NK cell killing of iDCs,14, 15 and NKp30-based CARs mediated self-reactivity against PBMCs and iDCs.8
A potential approach to overcome the self-reactivity of NKp30-based CARs to PBMCs and iDCs is to create CARs targeting B7H6. In this study, we show that B7H6-specific CAR T cells mediate robust in vitro and in vivo activity against B7H6 expressing tumor cells with little activity against PBMCs or iDCs. Thus, a B7H6-specific CAR T cell therapy may be beneficial for a variety of patients with hematologic or solid tumors.
RESULTS
Construction and expression of B7H6-specific CARs and NKp30-based CARs
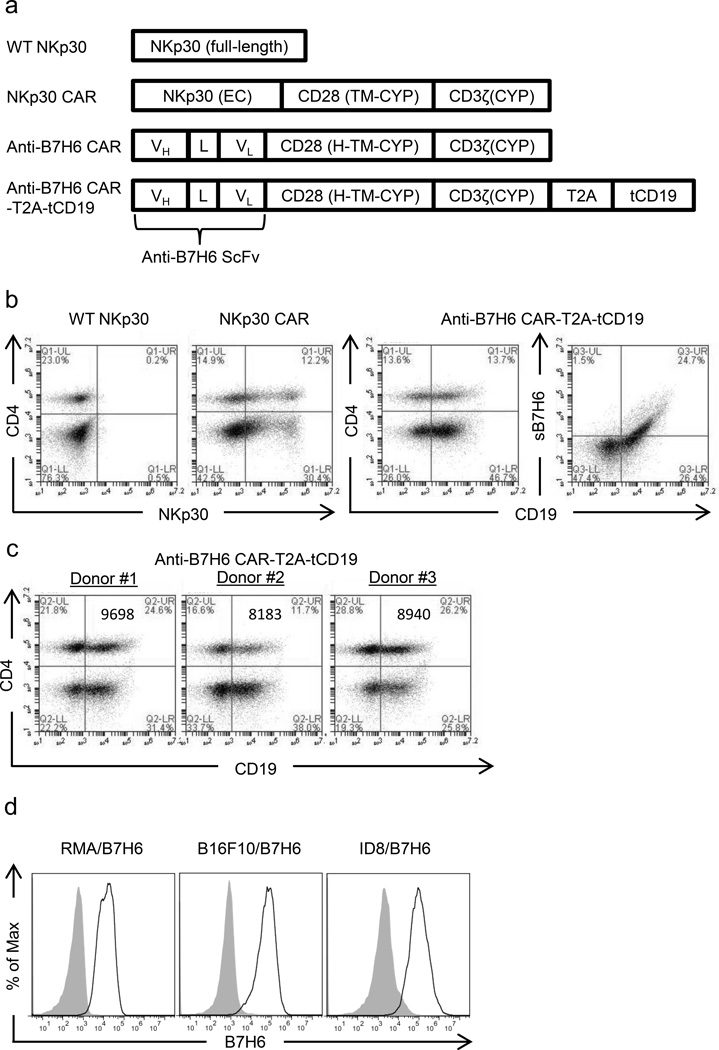
To generate a CAR specific to B7H6 but not other NKp30 ligands, a single chain variable fragment from an anti-B7H6 mAb (47.39) was constructed by linking heavy chain variable region and light chain variable region with a (Glycine4Serine3) linker. This anti-B7H6 scFv was fused with human CD28 hinge (H), transmembrane (TM), and cytoplasmic (CYP) domains, followed by a human CD3ζ CYP domain to create a B7H6-specific CAR (anti-B7H6 CAR) (Figure 1a). Wild type (WT) NKp30 and a NKp30-based CAR (NKp30 CAR) were used for comparison with the anti-B7H6 CAR.8 T cells express WT NKp30 poorly and no specific activity is expected from this CAR, so WT NKp30 transduced T cells were used as a transduction control. The NKp30 CAR contains human CD28 TM and CYP domains between the NKp30 extracellular (EC) and CD3ζ CYP domains (Figure 1a). These CARs can be expressed efficiently on the T cell surface and confer primary and CD28 costimulatory signals through CD3ζ CYP and CD28 CYP domains upon CAR binding to its ligand.8 In order to assess anti-B7H6 CAR expression and to facilitate sorting of CAR+ T cells, a retroviral vector with the anti-B7H6 CAR, a furin cleavage site containing T2A sequence, and a truncated human CD19 gene was also constructed (Figure 1a). Surface expression of anti-B7H6 CARs on transduced human T cells were analyzed by flow cytometry after staining T cells with soluble B7H6 or by using CD19 expression as a surrogate marker of the CAR expression (Figure 1b). Although there is potential for donor to donor variability in CAR expression, the expression of anti-B7H6 CAR on T cells from different human PBMC donors showed very similar patterns of expression (Figure 1c). NKp30 CAR and anti-B7H6 CARs can be expressed efficiently on human T cells, whereas WT NKp30 express poorly on T cells (Figure 1b), as previously shown.8
Figure 1. Design and expression of NKp30-based CAR (NKp30 CAR) and B7H6-specific CARs (anti-B7H6 CARs).
(a) WT NKp30 is the full length wild-type NKp30 gene. A NKp30 CAR was created by fusing NKp30 extracellular (EC) domain with human CD28 transmembrane (TM), and cytoplasmic (CYP) domains, followed by a human CD3ζ CYP domain. A B7H6-specific CAR was created by fusing anti-B7H6 scFv DNA with the human CD28 hinge (H), transmembrane (TM), and cytoplasmic (CYP) domains, followed by a human CD3ζ CYP domain DNA. The anti-B7H6 CAR-T2A-tCD19 construct was created by combining the anti-B7H6 CAR DNA with a T2A sequence containing a furin cleavage site and a truncated (t) human CD19 DNA sequence. (b) Human PBMCs were transduced with WT NKp30, NKp30 CAR, or anti-B7H6 CAR-T2A-tCD19 constructs. Transduced T cells were stained with anti-CD4 mAbs, anti-NKp30 mAbs, soluble B7H6 (sB7H6), and/or anti-CD19 mAbs. CD4- T cells are CD8+ T cells. The data are representative of data from 3 different human donors. (c) Anti-B7H6 CAR expression on T cells from different PBMC donors were analyzed. The values in the graph represent the mean fluorescent intensities of CD19 expression for each sample. (d) RMA/B7H6, B16F10/B7H6, and ID8/B7H6 were stained with anti-B7H6 mAbs followed by goat anti-msIgG Abs (open histograms) or with goat anti-msIgG Abs only (filled histograms). Data shown are representative of 3 independent experiments.
B7H6 expression on tumor cells
To identify the potential targets for anti-B7H6 CAR T cell therapy, we screened a panel of human tumor cell lines for B7H6 expression.8 B7H6 expression was found in several hematological malignancy cell lines, including lymphoma, leukemia, and multiple myeloma and in several solid tumor cell lines, including melanoma, breast cancer, and pancreatic cancer cell lines. A broader survey of B7H6 expression reported that it was found on 24 out of 119 human tumor cell lines.11 Since B7H6 gene is expressed in Homo Sapiens and Pongo abelii but not Mus musculus, we generated B7H6 expressing murine tumors from RMA T cell lymphoma, B16F10 melanoma, ID8 ovarian cancer cell lines, which we named as RMA/B7H6, B16F10/B7H6, and ID8/B7H6 for further in vitro and in vivo experiments (Figure 1c).
B7H6-specific CAR T cells mediate cytotoxicity against B7H6 expressing tumors
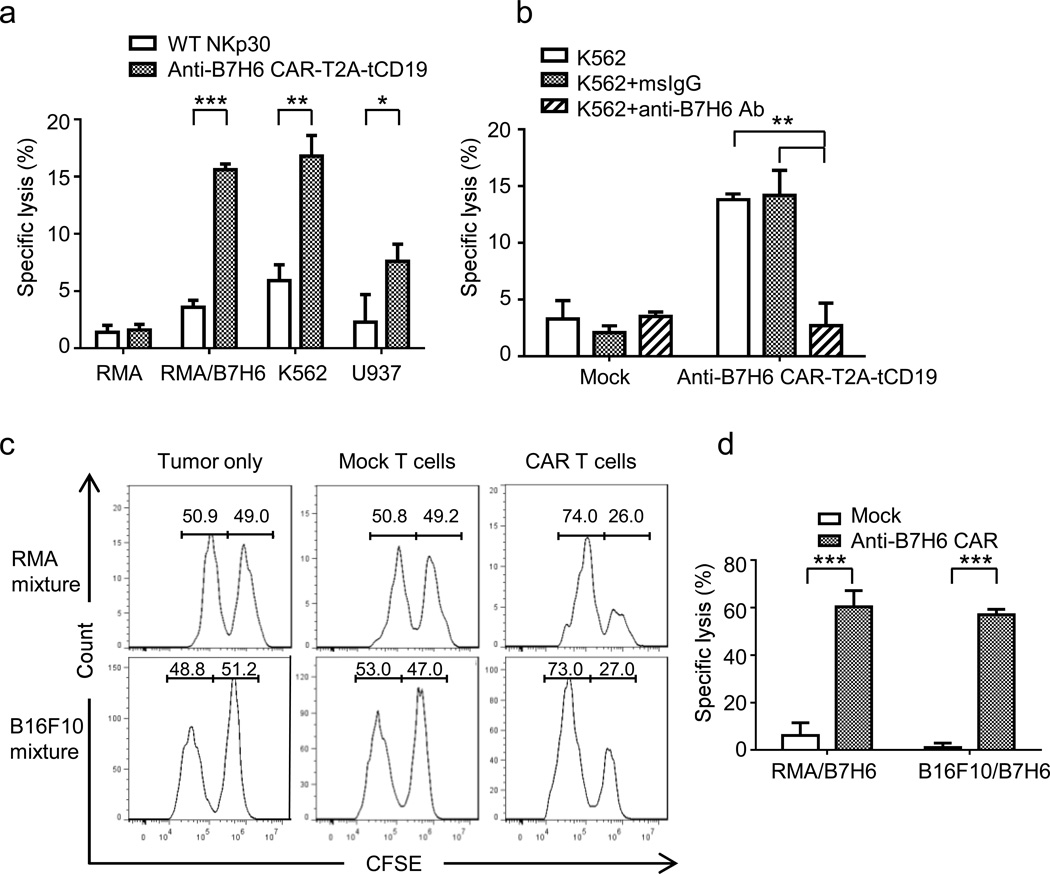
To test whether B7H6-specific CAR T cells mediate cytotoxicity, we co-cultured human anti-B7H6 CAR-T2A-tCD19 T cells with B7H6+ tumor cell lines (RMA/B7H6, K562, U937) or B7H6- tumor cell lines (RMA). We observed that human anti-B7H6 CAR T cells mediated robust cytotoxicity against B7H6+ tumors but not B7H6- tumor cells (Figure 2a). To confirm that cytotoxicity was dependent on B7H6, B7H6 was pre-blocked on K562 tumor cells with anti-B7H6 mAbs before incubating tumor cells with CAR T cells. Pre-blocking tumor cells with anti-B7H6 mAbs significantly reduced the cytotoxicity of anti-B7H6 CAR T cells to the cytotoxicity observed with Mock (untransduced) T cells (Figure 2b). We also observed that murine anti-B7H6 CAR T cells specifically killed B7H6+ but not B7H6- tumor cells when cultured with a 1:1 mixture of B7H6- and B7H6+ tumor cells (RMA and RMA/B7H6 or B16F10 and B16F10/B7H6) (Figure 2c & 2d). These data show that both human and murine anti-B7H6 CAR T cells mediate specific killing of B7H6+ tumor cells.
Figure 2. In vitro cytotoxicity of B7H6-specific CAR T cells on tumor cell lines.
(a) Human PBMCs transduced with WT NKp30 or anti-B7H6 CAR-T2A-tCD19 were co-cultured with B7H6- (RMA) or B7H6+ (RMA/B7H6, K562, U937) tumor cells at an E:T ratio 5:1 in a 6h LDH-release assay. The data are mean +SD of triplicates, and representative of 2 independent experiments. (b) K562 cells were pre-incubated with msIgG or anti-B7H6 mAbs. Mock (untransduced) or anti-B7H6 CAR-T2A-tCD19 human T cells were co-cultured with K562 at an E:T ratio 5:1 in a 6h LDH-release assay. (c) Mouse splenocytes untransduced (Mock) or transduced with the anti-B7H6 CAR were co-cultured with a 50% RMA + 50% RMA/B7H6 cell mixture or a 50% B16F10 + 50% B16F10/B7H6 cell mixture at an E:T ratio 5:1. RMA, B16F10 and RMA/B7H6, B16F10/B7H6 cells were labeled with 0.1uM (B7H6- cells) and 1uM CFSE (B7H6+ cells), respectively. After 8h, samples were harvested and analyzed by flow cytometry. Data shown were pre-gated on live cells and excluded T cells. (d) Specific lysis by Mock and anti-B7H6 CAR T cells against RMA/B7H6 and B16F10/B7H6 are shown as mean +SD. Data are representative of 2 independent experiments. An * indicates p < 0.05; ** indicates p < 0.005; *** indicates p < 0.0001.
B7H6-specific CAR T cells produce IFN-γ when cultured with B7H6 expressing tumors but not immature DCs or LPS, IL-1β, or TNF-α stimulated PBMCs
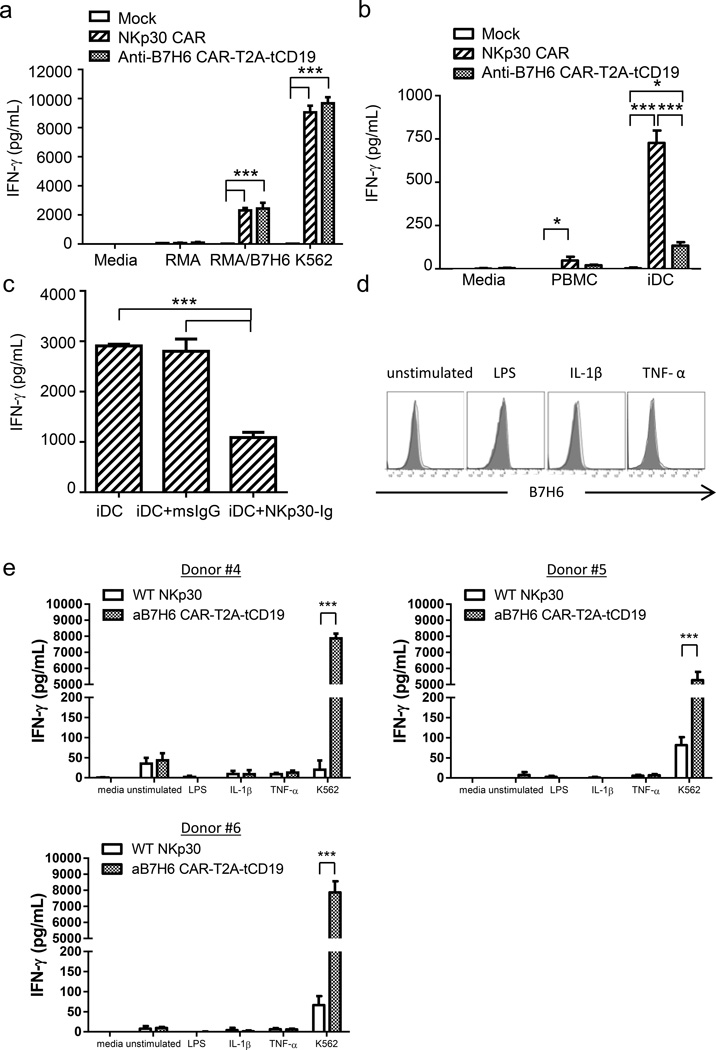
IFN-γ plays an important role in mediating adoptive T cell therapy efficacies.16–18 To examine whether human B7H6-specific CAR T cells produce IFN-γ upon recognition of B7H6+ tumors, Mock, NKp30 CAR, and anti-B7H6 CAR T cells were co-cultured with B7H6+ (RMA/B7H6, K562) or B7H6- (RMA) tumor cells. B7H6-specific CAR T cells mediated robust IFN-γ secretion (> 2000 pg/ml) when co-cultured with B7H6+ tumor cells (Figure 3a). The amount of IFN-γ secretion by anti-B7H6 CAR T cells was comparable to NKp30 CAR T cells. It is known that NK cells interact with iDCs and mediate iDC killing and IFN-γ secretion through NKp30 and BAG-6 interactions.15 Since anti-B7H6 CAR does not recognize NKp30 ligands other than B7H6, anti-B7H6 CAR T cells should exhibit little reactivity to autologous PBMCs or iDCs compared to NKp30 CAR T cells. Indeed, human anti-B7H6 CAR T cells exhibit little self-reactivity to autologous PBMCs or iDCs compared to NKp30 CAR T cells (Figure 3b). By pre-blocking the ligands on iDCs with NKp30-Ig, there was a significant decrease in the IFN-γ secretion by NKp30 CAR T cells in these co-cultures, confirming the self-reactivity of NKp30 CAR T cells to iDCs is contributed by NKp30 recognition of its ligands (Figure 3c). Besides tumor cells, B7H6 has been reported to be expressed on circulating pro-inflammatory monocytes in a group of patients suffering severe sepsis or on monocytes stimulated in vitro with Toll-like receptor (TLR) agonists or pro-inflammatory cytokines.19 However, we did not observe the 47.39 mAb which was used as the antigen binding fragment of the B7H6-specific CAR, recognizes B7H6 expression on lipopolysaccharide (LPS), interleukin-1β (IL-1β), or tumor necrosis factor-α (TNF-α) stimulated CD14+ monocytes (Figure 3d). Furthermore, co-culturing B7H6-specific CAR T cells with LPS, IL-1β, or TNF-α stimulated autologous PBMCs did not elicit specific IFN-γ secretion by CAR T cells. This phenomenon was consistent when CAR T cells were derived from different human donors (Figure 3e). Since there is little self-reactivity against iDCs and pro-inflammatory monocytes by B7H6-specific CAR T cells, these CAR T cells should provide a broad therapeutic window.
Figure 3. B7H6-specific CAR T cells produce IFN-γ in response to B7H6 expressing tumors but not immature DCs or LPS, IL-1β, or TNF-α stimulated PBMCs.
(a) Mock, NKp30 CAR, or anti-B7H6 CAR T cells were co-cultured with tumor cells at an E:T ratio 1:1. After 24h, cell-free conditioned media was harvested and IFN-γ concentration was determined by ELISA. The results are shown as mean + SD and are representative of 2 different PBMC donors. (b) Mock, NKp30 CAR, or anti-B7H6 CAR T cells were co-cultured with autologous PBMCs or autologous immature DCs at an E:T ratio 5:1 (105:2×104). Cell-free media was harvested 24h later and IFN-γ concentration was measured. The results are shown as mean + SD and are representative of 2 PBMC different donors. (c) Autologous iDCs were pre-incubated with msIgG or NKp30-Ig for 45min before they were co-cultured with NKp30 CAR T cells at an E:T ratio 5:1 (105:2×104). After 24h, cell-free culture media was harvested and IFN-γ concentration was determined by ELISA. The results are shown as mean + SD and are representative of 2 different donors. (d) Human PBMCs were cultured and stimulated with LPS, IL-1β, or TNF-α for 48h. Cells were harvested and stained with anti-CD14 mAbs and anti-B7H6 mAbs. Histograms shown are pre-gated on CD14+ monocytes. Filled histograms represent isotype controls and open histograms represent B7H6 expression. Data shown are representative of 3 different PBMC donors. (e) WT NKp30 or anti-B7H6 CAR T cells were co-cultured with unstimulated, LPS, IL-1β, or TNF-α stimulated autologous PBMCs for 24h. Cell free supernatant was harvested and IFN-γ concentration was determined by ELISA. The results of CAR T cells derived from three different donors are shown as mean + SD. An * indicates p < 0.05; ** indicates p < 0.005; *** indicates p < 0.0001.
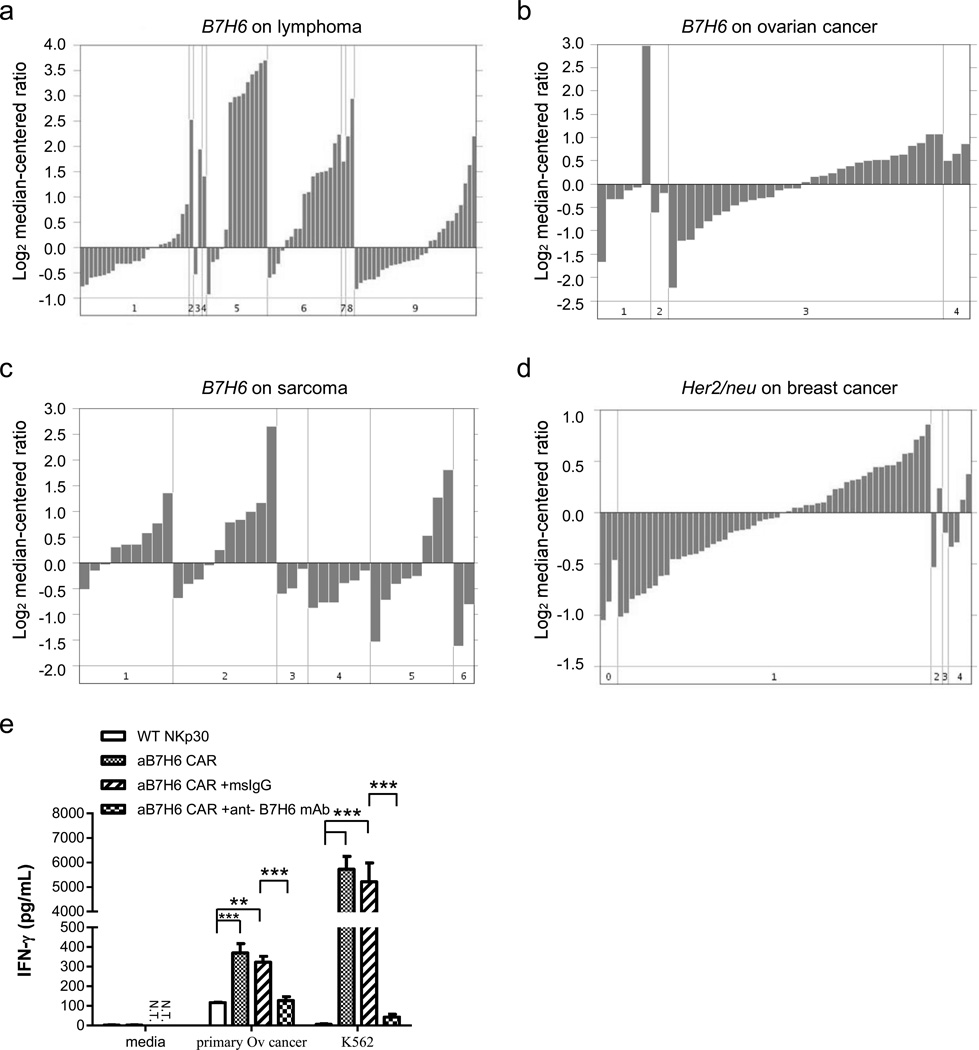
B7H6 mRNA expression is upregulated in multiple primary human tumor types, and B7H6-specific CAR T cells react to human primary ovarian cancer
B7H6 is expressed by some subsets of hematological malignancies, such as acute nonlymphoblastic leukemia (ANLL), acute lymphoblastic leukemia (ALL), and lymphomas.11 In order to identify more primary human tumor types that potentially express B7H6, we took a data mining approach to identify B7H6 expression in published microarray data from primary human tumor samples. By using the Oncomine database20 interface (https://www.oncomine.org) and searching with the keyword "DKFZp686O24166", an alias of the B7H6 gene, and further filtered the results by differential analysis filter, we identified several subsets of primary tumor types expressing high amounts of B7H6 mRNA. Table 1 summarizes the tumor types (those having from 15 to 285 different patient samples) that have more than 10% of the samples with high B7H6 mRNA expression (B7H6 mRNA amount > median gene mRNA amount in the particular microarray). Analysis showed that there was B7H6 mRNA expression in primary human lymphoma,21 ovarian cancer,22 and sarcomas23 (Figure 4a, 4b, 4c). Her2/neu expression, a known breast cancer target, in human breast cancers is also shown24 using a similar data analysis for comparison (Figure 4d). As expected, some subsets of leukemia and lymphoma had high B7H6 mRNA expression, and this correlated with published data on B7H6 protein expression.11 Furthermore, GIST, the most common mesenchymal tumor of the gastrointestinal tract, whose clinical prognosis is correlated with immunostimulatory NKp30 isoforms, was found to have B7H6 expression.12 When B7H6-specific CAR T cells were co-cultured with primary human ovarian cancers, specific IFN-γ production was observed, and pre-incubating cancer cells with anti-B7H6 mAbs before co-culturing with CAR T cells significantly abrogated the response (Figure 4e). These findings highlighted the potential to target multiple clinically relevant tumor types with B7H6-specific CAR T cell therapy.
Table 1.
B7H6 in Primary human tumor samples1
| Primary tumor type | Percent (ratio) of patient samples with B7H6 mRNA amount > median gene mRNA amount |
|---|---|
| Diffuse Large B Cell Lymphoma | 46.4% (39/84) |
| Acute Lymphoblastic Leukemia | 36.7% (11/30) |
| Ovarian Serous Surface Papillary Carcinoma | 51.6% (16/31) |
| Medulloblastoma | 29.1% (83/285) |
| Glioblastoma | 11.5% (3/26) |
| Gastrointestinal Stromal Tumor (GIST) | 53.3% (8/15) |
| Leiomyosarcoma | 30.8% (8/26) |
| Malignant Fibrous Histiocytoma | 21.1% (4/19) |
| Dermatofibrosarcoma Protuberans | 14.3% (3/21) |
| Clear Cell Renal Cell Carcinoma | 13.0% (21/161) |
| Invasive Ductal Breast Carcinoma | 10.3% (4/39) |
Summary of the tumor types (which have 15 to 285 different patient samples tested) having more than 10% of samples exhibiting high B7H6 mRNA expression (B7H6 mRNA amount > median gene mRNA amount in the particular microarray). Data are identified from the Oncomine database (https://www.oncomine.org)
Figure 4. B7H6 mRNA expression is upregulated in multiple types of primary human tumor samples, and B7H6-specific CAR T cells recognize human primary ovarian cancer.
B7H6 mRNA expression from various primary human tumor samples was analyzed from the Oncomine database (https://www.oncomine.org). (a) B7H6 mRNA expression in B cell lymphoma patient samples. Cancer subsets are as indicated: 1. Activated B-cell-Like Diffuse Large B-Cell Lymphoma. 2. B-Cell Neoplasm (Not Specified). 3. B-cell Non-Hodgkin's Lymphoma. 4. Burkitt's Lymphoma. 5. Diffuse Large B-Cell Lymphoma. 6. Germinal Center B-Cell-Like Diffuse Large B-Cell Lymphoma. 7. Grade 3 Follicular Lymphoma. 8. Immunoblastic Lymphoma. 9. Type 3 Diffuse Large B-Cell Lymphoma. (b) B7H6 mRNA expression in ovarian cancer patient samples. Cancer subsets are as indicated: 1. Ovarian Clear Cell Adenocarcinoma. 2. Ovarian Endometrioid Adenocarcinoma. 3. Ovarian Serous Surface Papillary Carcinoma. 4. Undifferentiated Ovarian Carcinoma. (c) B7H6 mRNA expression in various types of sarcoma samples. Cancer subsets are as indicated: 1. Gastrointestinal Stromal Tumor. 2. Leiomyosarcoma. 3. Liposarcoma. 4. Malignant Fibrous Histiocytoma. 5. Monophasic Synovial Sarcoma. 6. Schwannoma. (d) Her2/neu mRNA expression in breast cancer samples. Disease subsets are as indicated: 0. Not specified. 1. Ductal Breast Carcinoma. 2. Ductal Breast Carcinoma in Situ. 3. Fibroadenoma. 4. Lobular Breast Carcinoma. Note the scale difference in each figure. (e) WT NKp30 or B7H6-specific CAR T cells were co-cultured with a primary human ovarian cancer sample at an E:T ratio 1:1. For blocking experiments, cancer samples were pre-incubated with 5µg of msIgG or anti-B7H6 mAbs before co-culturing with T cells. Cell-free conditioned media was harvested after 24h and IFN-γ concentration was determined by ELISA. The results shown are mean + SD and are representative of T cells made from 2 different PBMC donors. An * indicates p < 0.05; ** indicates p < 0.005; *** indicates p < 0.0001. N.T. indicates not tested.
Anti-B7H6 CAR T cell therapy improved outcomes in murine lymphoma and ovarian cancer models
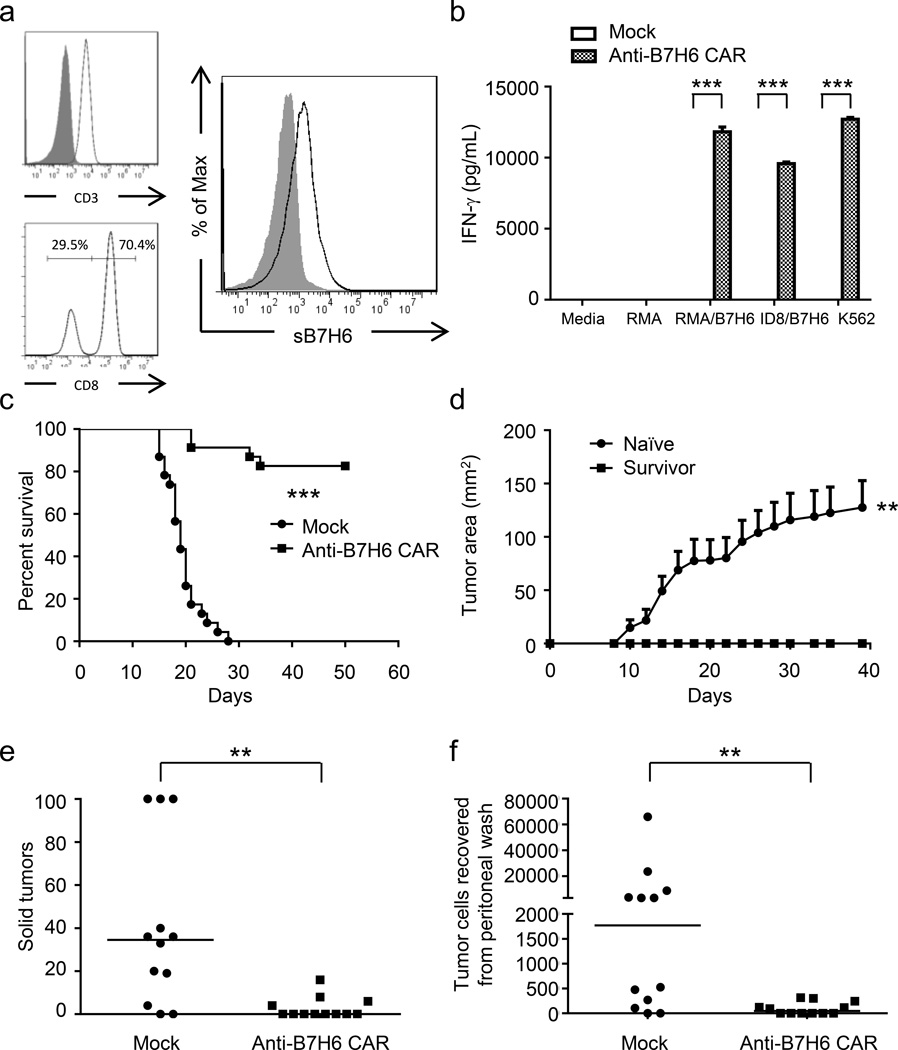
To study the therapeutic efficacy of anti-B7H6 CAR T cells in vivo, the B7H6-specific CAR was expressed in murine T cells. More than 99% of cells in the CAR cell preparations used for in vivo efficacy studies were CD3+ T cells (Figure 5a). This anti-B7H6 CAR can be expressed efficiently on murine T cells (Figure 5a), and these CAR T cells recognize B7H6 expressing tumors and secrete IFN-γ (Figure 5b). In a systemic T cell lymphoma model, B6 mice injected with RMA/B7H6 tumor cells i.v. on day 0 were treated with anti-B7H6 CAR T cells i.v. on days 5, 7, and 9. Anti-B7H6 CAR T cell therapy greatly enhanced the survival of tumor-bearing mice with 83% becoming long-term tumor-free survivors while all control mice had to be sacrificed by day 29 (Figure 5c). Furthermore, the long-term tumor-free survivors were protected upon re-challenge with B7H6-deficient RMA tumor cells (Figure 5d). In an ovarian cancer model, ID8/B7H6 tumor cells were injected into mice i.p. on day 0 and were treated with anti-B7H6 CAR T cells i.p. on day 7. Tumor burden was assessed on day 42 by counting the solid tumors on the peritoneal wall and determining the number of free tumor cells in the peritoneum wash. We observed that anti-B7H6 CAR T cell therapy greatly reduced the tumor burden of ID8/B7H6 tumor bearing mice (Figure 5e & 5f).
Figure 5. B7H6-specific CAR T cells promote survival of RMA/B7H6 lymphoma-bearing mice and decrease tumor burden in ID8/B7H6 ovarian cancer-bearing mice.
(a) Cell surface molecules on murine B7H6-specific CAR T cells. Expression of CD3, CD8, and the B7H6-specific CAR are shown. Filled histograms represent isotype controls and open histograms represent specific expression. More than 99% of cells in the cell preparation are CD3+ T cells. CD8- cells are CD4+ T cells. (b) Murine Mock T cells or anti-B7H6 CAR T cells were co-cultured with tumor cells at an E:T ratio 1:1 (RMA, RMA/B7H6, K562) or 4:1 (ID8/B7H6). After 24h, cell-free culture media was harvested and the IFN-γ concentration was measured by ELISA. The results are shown as mean + SD and are representative of 2 independent experiments. (c) B6 mice were injected with RMA/B7H6 (105 cells, i.v.) on day 0. Murine Mock T cells or anti-B7H6 CAR T cells were administered (5 × 106 cells, i.v.) on days 5, 7, and 9. Data are presented in Kaplan-Meier survival curves and are pooled from 3 independent experiments (n=23 for each group). (d) Naïve and survived mice (from Figure 5c) were challenged with 104 RMA tumor cells subcutaneously, and tumor area was measured every other day. Data shown are mean + SEM and are pooled from 2 independent experiments (n=7 for each group). (e & f) ID8/B7H6 (GFP+) ovarian cancer cells (5 × 106 cells, i.p.) were injected into B6 mice on day 0. Murine Mock T cells or anti-B7H6 CAR T cells were administered on day 7 (5 × 106 cells, i.p.). The total number of solid tumors (stopped counting at 100) for each mouse on the peritoneal wall was counted (e) and the number of GFP+ tumor cells in the peritoneal wash (f) were measured on day 42. Each dot represents data from an individual mouse. Horizontal bars represent median values. These data are combined from two independent experiments. An * indicates p < 0.05; ** indicates p < 0.005; *** indicates p < 0.0001.
DISCUSSION
In this study, we identified several types of primary human tumors including lymphoma, leukemia, ovarian cancer, breast cancer, renal cell carcinoma, various sarcomas and brain tumors as expressing high amount of B7H6 mRNA. The finding that B7H6 mRNA expression was upregulated in leukemia and GIST correlated well with clinical observations of NKp30-mediated immunosurveillance in these tumors.12, 13 These observations highlighted the potential broad tumor coverage of B7H6-specific CAR T cell therapy.
Self-reactivity is a major concern for CAR T cell therapy and can potentially cause unexpected severe adverse effects. The severity can range from manageable side effects, such as B cell depletion caused by anti-CD19 CAR T cells1–3 to acute mortality caused by anti-ERBB2 CAR T cells.25 In this study, we created B7H6-specific CARs which maintained NKp30-based CAR tumor targeting capacity but spared the reactivity against PBMCs and iDCs, so these CAR T cells should be effective against tumors without recognizing normal hematopoietic cells.
B7H6 was identified as a tumor-specific ligand for NKp30.11 Quantitative reverse transcription PCR on a normal tissue array showed no detectable expression in any of the 48 normal tissues assayed.11 Recently, a study showed that B7H6 protein may be transiently induced on monocytes and neutrophils upon TLR ligand stimulation or cytokine stimulation in vitro, and B7H6 was found on circulating pro-inflammatory monocytes in a group of patients suffering from severe sepsis conditions.19 However, we found little or no B7H6 upregulation on CD14+ monocytes upon LPS, IL-1β, or TNF-α stimulation. In addition, B7H6-specific CAR T cells did not react to these activated monocytes in PBMCs. Thus, there may be other reasons for the differences of the findings in this study compared to the previous one, but it suggests that simple activation by TLR agonists and inflammatory cytokines is not sufficient to trigger B7H6 expression on PBMCs.
The finding that B7H6 mRNA expression was upregulated in lymphoma, leukemia, and GIST correlated with previous published results.11, 12 The observation that B7H6 mRNA was upregulated in some subsets of ovarian cancers also correlated with the in vitro functional data. Several subsets of tumors identified to express high amount of B7H6 mRNA have a high clinical incidence, such as diffuse large B cell lymphoma which is the most common form of lymphoma in adults.26 GIST is the most common form of mesenchymal tumor of the gastrointestinal tract and expresses B7H6.27 These findings highlighted the potential of targeting B7H6 for tumor immunotherapy.
Mus musculus does not express B7H6.11 Therefore to investigate the therapeutic efficacy of B7H6-specific CAR T cells in immunocompetent mice, the human B7H6 gene was introduced into various murine tumor cell lines. Exogenously expressing B7H6 on tumor cell lines may lead to higher B7H6 expression on the cell surface than is found on primary human tumor cells. Human B7H6 may be immunogenic in mice, but we have not observed a selection for B7H6-loss variants in these models suggesting any immunogenicity is low. Although it is important to note these caveats, these B7H6 tumor models allow one to study B7H6-specific CAR T cells in animals with a competent immune system.
In conclusion, B7H6-specific CAR T cells produce cytokines and mediate cytotoxicity in the presence of B7H6+ tumor cells. B7H6-specific CAR T cell therapy mediated robust therapeutic efficacy in murine lymphoma and ovarian cancer models. This CAR can also lead to resistant against tumor rechallenge, which may be a consequence of a broader host immune memory against other tumor antigens. Expression data indicate that subsets of lymphoma, leukemia, ovarian cancers, brain tumors, breast cancers, renal cell carcinomas, and various sarcomas express B7H6. B7H6-specific CAR T cell therapy has the potential to treat B7H6+ hematological malignancies and solid tumors.
MATERIALS AND METHODS
Mice
C57BL/6 (B6) mice were purchased from the National Cancer Institute (Frederick, MD). Mice were 7–12 weeks old at the start of the experiments. All experiments were conducted under the approval of Dartmouth College's Institutional Animal Care and Use Committee.
Cell culture and cell lines
Bosc23, PT67, GP+E86, K562, U937 cell lines were obtained from American Type Culture Collection (Rockville, MD). An RMA subline RMA/B7H6, B16F10 subline B16F10/B7H6, and ID8 subline ID8/B7H6 were generated by transducing the parental cell lines with a dualtropic retrovirus containing the B7H6 gene (see below), followed by 1mg/mL G418 (Corning, Cambridge, MA) or 2ug/mL Puromycin (Sigma-Aldrich, Saint Louis, MO) selection for 7 days. Bosc23, PT67, GP+E86, and ID8/B7H6 cells were cultured in DMEM with a high glucose concentration (4.5g/L), supplemented with 10% heat-inactive FBS (Atlanta Biologicals, Lawrenceville, GA), 10mM HEPES, 0.1mM non-essential amino acids, 1mM sodium pyruvate, 100U/mL penicillin, 100ug/mL streptomycin, and 50uM 2-ME. Other cell lines were cultured in RPMI 1640 containing the same supplements as DMEM. When harvesting adhesion growing cells for staining or functional assay, PBS containing 10mM EDTA was used instead of Trypsin. Trypsin treatment cleaves B7H6 from cell surface (data not shown). Human DCs were generated as described previously.8 To test PBMCs activation with LPS, IL-1β, or TNF-α, human blood cells from cell cones obtained from leukapheresis cell donations (Dartmouth-Hitchcock Medical Center Blood Donor Center) were cultured in 24 well plates at a cell density 3×106 cells/well in complete RPMI 1640 at 37°C with 5% CO2 without or with following stimulation, LPS (1µg/mL; Sigma-Aldrich), IL-1β (1ng/mL; PeproTech, Rocky Hill, NJ), TNF-α (100ng/mL; PeproTech) for 48 h. Cells analyzed by flow cytometry, and CD14+ monocytes were gated for the analysis of B7H6 expression. De-identified primary ovarian cancer samples from patients were obtained at the time of primary debulking surgery performed at Dartmouth-Hitchcock Medical Center by the division of Gynecology Oncology. Informed consent was obtained prior to surgery. The patients were having primary debulking surgery with intent to remove all gross disease. Excess primary ovarian tumor tissue as well as metastatic intra-abdominal disease was separated from the resected specimens for research purposes prior to surgical pathology analysis. No tissue was removed solely for this research study. Cancer tissues were mechanically disrupted [Collagenase (catalog number: C5138; Sigma-Aldrich) treatment cleaves B7H6 from cell surface] and red blood cells in the samples were lysed with ACK lysis buffer (0.15M NH4Cl, 10mM KHCO3, 0.1mM EDTA, PH 7.4) before cell culture. Primary ovarian cancer cells were cultured for two days before used for functional assay.
Construction of B7H6-specific chimeric antigen receptor (CAR)
A B7H6-specific CAR was constructed by fusing the single change variable fragment (scFv) of an anti-B7H6 hybridoma,8 clone 47.39, to the human CD28 hinge-transmembrane-cytoplasmic domains [amino acid (aa) 135–220] and a CD3ζ endodomain [aa 52–164]. The construction of the anti-B7H6 scFv was done by fusing the variable region of heavy chain [VH, aa 1–134] and light chains [VL, aa 23–129] with a 15 amino acid glycine (G)-serine (S) linker (G4S)3 linker (3 repeats of GGGGS). The B7H6-specific CAR construct was then cloned into a retroviral vector pFB-neo (Stratagene, Palo Alto, CA). B7H6-specific CAR-T2A-tCD19 construct was created by fusing the B7H6-specific CAR to a Furin cleavage site-SGSG-T2A sequence (AGAGCCAAAAGGTCTGGCTCCGGTGAGGGCAGAGGAAGTCTTCTAACATGCGGTGACGTGGAGGAGAATCCCGGCCCT), followed by a truncated human CD19 [aa 1–327]. All PCRs were done by using high-fidelity DNA polymerase Phusion (New England BioLabs, Ipswich, MA).
Retrovirus production and transduction
Eighteen hours before transfection, Bosc23 or 293T cells were plated at a density of 3 × 106 cells/10cm plate in 10mL complete DMEM media. Transfection of CAR plasmids into Bosc23 cells was performed using Lipofectamine 2000 (Life Technology, Carlsbad, CA) following the manufacturer's protocols. Transfection of CAR plasmids and ψEco packaging plasmids (Gift from Patricia Ernst, Dartmouth College, Hanover, NH) into 293T cells was performed by using 20ug of CAR vectors, plus 10ug of ψEco packaging plasmids, with 9uL of X-fect (Clonetech, Palo Alto, CA) according to the manufacturer's protocols. Forty-eight to seventy-two hours after transfection, ecotropic viral supernatant was harvested and filtered with 0.45µm filter before storage at −80°C. Virus-producing PT67 cells were generated by cross-infection with ecotropic viruses collected from the transfected Bosc23 or 293T cells, three times, followed by G418 (1mg/mL) selection for 7 days. Virus-producing GP+E86 cells were generated by cross-infection using dualtropic virus produced by PT67 cells, followed by G418 (1mg/mL) selection for 7 days. Retroviral transduction of T cells was performed as previously described.8 For experiments using B7H6-specific CAR-T2A-tCD19 T cells, T cells were activated, transduced, and expanded. CD19+ cells were positively selected with anti-CD19-PE magnetic beads (STEMCELL, Vancouver, Canada) and expanded for 5 days before functional assays. Greater than 99% of cells in CAR T cell preparations derived from human PBMCs or murine splenocytes were CD3+ cells.
Production of soluble B7H6, DyLight 650 conjugated soluble B7H6, and NKp30-msIgG fusion protein
A soluble B7H6 (extracellular domain) was prepared by cloning of the B7H6 extracellular portion [aa 25–262] into pET15b expression vector (Novagen, Madison, WI). Soluble B7H6 was expressed as inclusion bodies in E.coli. Extraction of B7H6 inclusion bodies was done by using CelLytic™ B cell lysis reagent (Sigma-Aldrich). Dissolving and refolding of soluble B7H6 was done by using CelLytic™ IB inclusion body solubilization reagent (Sigma-Aldrich). Inclusion body extraction, dissolving, and refolding were done following the manufacture's protocols. Conjugation of soluble B7H6 to DyLight 650 NHS Ester (Thermo Fisher Scientific, Waltham, MA) was done according to the manufacturer's protocol. NKp30-msIgG fusion sequence was constructed by fusing the NKp30 extracellular portion [aa 1–135] with a murine IgG2a hinge-CH2-CH3 portion, and the construct was further cloned into a pFB-neo vector. Production of NKp30-msIgG fusion protein was performed by transiently transfecting the NKp30-msIgG plasmid into 293F cells (Life Technology) by 40Kd PEI (Polysciences Inc, Warrington, PA). Transfection was done by gently mixing 293F cells with DNA and PEI at a final concentration of 2 × 107/mL cells, 12.5ug/mL DNA, 25ug/mL PEI and shaking it on an orbital shaker at 37°C at 120 rpm for 3 hrs. After 3hrs, the whole mixture was diluted with Gibco® FreeStyle 293™ Expression Medium (Life Technology) at a 1:9 ratio to reach a final cell concentration 2×106 cells/mL. The culture was maintained in 37°C shaker at 120rpm for 4 days. Cell free culture supernatants were harvested and purified by HiTrap rProtein A HP column (GE healthcare, Waukesha, WI) following the manufacture's protocol.
Flow cytometry
Flow cytometry analysis of B7H6 expression on tumor cells were performed by staining cells with anti-B7H6 mAbs,8 followed by DyLight 649-conjugated goat anti-mouse IgG (clone: Poly4053, Biolegend, San Diego, CA). For staining of mouse cells, the samples were pre-incubated with FcR block antibody (anti-mouse CD16/CD32, 2.4G2; Bio X Cell, Lebanon, NH), and for staining of human cells, samples were pre-incubated with human gamma globulin (Cohn fraction, Sigma-Aldrich). Samples were analyzed by Accuri C6 cytometer (BD Biosciences, San Jose, CA) and data was analyzed by Accuri or FlowJo software (TreeStar Inc, Ashland, OR).
In vitro cytotoxicity assay
LDH release cytotoxicity assays were done with CytoTox 96 Non-Radioactive Cytotoxicity Assay Kit (Promega, Madison, WI) following the manufacturer’s protocol. T cells and tumor cells were co-cultured at an Effector cell to Target cell (E:T) ratio 5:1 (105:2×104) for 6h in 96-well U-bottom plates in triplicates. The specific lysis was calculated by the following equation: percentage of specific lysis = [(experimental release-effector spontaneous release-target spontaneous release) / target maximal release-target spontaneous release] × 100.
Flow cytotoxicity assays were done by labeling B7H6 negative and B7H6 positive tumor cell lines with 0.1uM or 1uM CFSE (Life Technology), respectively. B7H6 negative and B7H6 positive tumor cells were mixed at a 1:1 ratio before co-cultured with T cells at an E:T ratio of 5:1 (2×105:4×104) for 8h in triplicates. Samples were harvested and the ratio of B7H6 negative and B7H6 positive tumor cells were identified by flow cytometry. The specific lysis was calculated by the following equation: percentage of specific lysis = [1− (% of B7H6 positive tumor in B7H6-specific CAR or Mock T cell group × % of B7H6 negative tumor in tumor only group) / (% of B7H6 negative tumor in B7H6-specific CAR or Mock T cell group × % of B7H6 positive tumor in tumor only group)] × 100.
Cytokine production by T cells
T cells were co-cultured with suspension tumor cells, primary ovarian cancer cells, or LPS, IL-1β, or TNF-α stimulated PBMCs at an E:T ratio of 1:1 (105:105) in 96 well V-bottom plates or with adherent tumor cells at an E:T ratio of 4:1 (105:2.5×104) in 96 well flat-bottom plates in triplicates. Cell-free culture medium was harvested after 24h, the IFN-γ concentration was measured by ELISA using IFN-γ Duoset ELISA kits (R&D systems, Minneapolis, MN).
Treatment of tumor bearing mice with anti-B7H6 CAR T cells
As a systemic T cell lymphoma model, B6 mice were injected with 1 × 105 RMA/B7H6 cells via the tail vein in 400µL HBSS. Five, 7, and 9 days later, mice were treated with 5 × 106 Mock (untransduced) T cells or B7H6-specific CAR T cells i.v. The health of mice were monitored, and mice were sacrificed when moribund signs were observed. As an ovarian cancer model, B6 mice were injected with 5 × 106 ID8/B7H6 (GFP+) cells i.p. in 400µL HBSS. Seven days after tumor inoculation, mice were treated with 5 × 106 Mock T cells or B7H6-specific CAR T cells i.p. Forty-two days after initial tumor inoculation, tumor burden was measured by counting solid tumors on the peritoneal wall of each mouse (counting stopped at 100 tumors) and determining the number of suspension tumor cells (GFP+) in a peritoneal wash, as done previously.28
Statistical analysis
A student's t -test or ANOVA were used to analyze differences between groups. Kaplan-Meier survival curves were used to analyze survival benefit. Statistical analysis was done by GraphPad Prism software (GraphPad Software, San Diego, CA)
ACKNOWLEDGEMENTS
The authors thank the National Cancer Institute Biological Resource Branch for providing recombinant human IL-2 and the staff of the Center for Comparative Medicine and Research in Dartmouth College for providing animal care. This work was supported by a grant from the NIH CA130911 and funds from the Center for Synthetic Immunity.
Tong Zhang and Charles Sentman are inventors on a patent application covering the B7H6 CAR described in this study. This technology has been licensed by Cardio3 Biosciences. This work is managed in compliance with the policies of Dartmouth College.
Footnotes
CONFLICT OF INTEREST
Ming-Ru Wu and Leslie DeMars declare no potential conflict of interest.
REFERENCES
- 1.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95):95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5(177):177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadelain M, Brentjens R, Riviere I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3(4):388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T, Lemoi BA, Sentman CL. Chimeric NK-receptor-bearing T cells mediate antitumor immunotherapy. Blood. 2005;106(5):1544–1551. doi: 10.1182/blood-2004-11-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber A, Zhang T, DeMars LR, Conejo-Garcia J, Roby KF, Sentman CL. Chimeric NKG2D receptor-bearing T cells as immunotherapy for ovarian cancer. Cancer Res. 2007;67(10):5003–5008. doi: 10.1158/0008-5472.CAN-06-4047. [DOI] [PubMed] [Google Scholar]
- 8.Zhang T, Wu MR, Sentman CL. An NKp30-based chimeric antigen receptor promotes T cell effector functions and antitumor efficacy in vivo. J Immunol. 2012;189(5):2290–2299. doi: 10.4049/jimmunol.1103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 10.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190(10):1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206(7):1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delahaye NF, Rusakiewicz S, Martins I, Menard C, Roux S, Lyonnet L, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med. 2011;17(6):700–707. doi: 10.1038/nm.2366. [DOI] [PubMed] [Google Scholar]
- 13.Fauriat C, Just-Landi S, Mallet F, Arnoulet C, Sainty D, Olive D, et al. Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood. 2007;109(1):323–330. doi: 10.1182/blood-2005-08-027979. [DOI] [PubMed] [Google Scholar]
- 14.Ferlazzo G, Tsang ML, Moretta L, Melioli G, Steinman RM, Munz C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med. 2002;195(3):343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simhadri VR, Reiners KS, Hansen HP, Topolar D, Simhadri VL, Nohroudi K, et al. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS One. 2008;3(10):e3377. doi: 10.1371/journal.pone.0003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barth RJ, Jr, Mule JJ, Spiess PJ, Rosenberg SA. Interferon gamma and tumor necrosis factor have a role in tumor regressions mediated by murine CD8+ tumor-infiltrating lymphocytes. J Exp Med. 1991;173(3):647–658. doi: 10.1084/jem.173.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Z, Blankenstein T. CD4+ T cell--mediated tumor rejection involves inhibition of angiogenesis that is dependent on IFN gamma receptor expression by nonhematopoietic cells. Immunity. 2000;12(6):677–686. doi: 10.1016/s1074-7613(00)80218-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Barber A, Sentman CL. Chimeric NKG2D modified T cells inhibit systemic T-cell lymphoma growth in a manner involving multiple cytokines and cytotoxic pathways. Cancer Res. 2007;67(22):11029–11036. doi: 10.1158/0008-5472.CAN-07-2251. [DOI] [PubMed] [Google Scholar]
- 19.Matta J, Baratin M, Chiche L, Forel JM, Cognet C, Thomas G, et al. Induction of B7-H6, a ligand for the natural killer cell-activating receptor NKp30, in inflammatory conditions. Blood. 2013;122(3):394–404. doi: 10.1182/blood-2013-01-481705. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14(11):4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O'Connell JX, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359(9314):1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 24.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18(4):843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood. 2006;107(1):265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connolly EM, Gaffney E, Reynolds JV. Gastrointestinal stromal tumours. Br J Surg. 2003;90(10):1178–1186. doi: 10.1002/bjs.4352. [DOI] [PubMed] [Google Scholar]
- 28.Barber A, Zhang T, Sentman CL. Immunotherapy with chimeric NKG2D receptors leads to long-term tumor-free survival and development of host antitumor immunity in murine ovarian cancer. J Immunol. 2008;180(1):72–78. doi: 10.4049/jimmunol.180.1.72. [DOI] [PubMed] [Google Scholar]