Abstract
Nicotinamide N-methyltransferase (Nnmt) methylates nicotinamide, a form of vitamin B3, to produce N1-methylnicotinamide (MNAM). Nnmt is an emerging metabolic regulator in adipocytes but its role in the liver, a tissue with the strongest Nnmt expression, is not known. In spite of its overall high expression, here we find that hepatic expression of Nnmt is highly variable and correlates with multiple metabolic parameters in mice and in humans. Further, we find that suppression of hepatic Nnmt expression in vivo alters glucose and cholesterol metabolism and that the metabolic effects of Nnmt in the liver are mediated by its product MNAM. Supplementation of high fat diet with MNAM decreases serum and liver cholesterol and liver triglycerides levels in mice. Mechanistically, increasing Nnmt expression or MNAM levels stabilizes sirtuin 1 protein, an effect, which is required for their metabolic benefits. In summary, we describe a novel regulatory pathway for vitamin B3 that could provide a new opportunity for metabolic disease therapy.
Introduction
NAD+ acts as a redox cofactor for more than 200 enzymatic reactions and also serves as a co-substrate for the sirtuins, a family of NAD+-dependent deacetylases1–3. Sirtuin 1 (Sirt1), the most studied member of the family, has emerged as an important regulator of nutrient metabolism. Sirt1 regulates gluconeogenesis through deacetylation of the peroxisome proliferator-activated receptor gamma coactivator 1 alpha (Pgc1α), forkhead box O 1 (FoxO1) and CREB-regulated transcription coactivator 2 (Crtc2), among other factors4–9. Sirt1 also suppresses lipogenesis and cholesterol synthesis partly through direct deacetylation and inhibition of the master regulators of lipid homeostasis, sterol regulatory element binding proteins 1 and 2 (Srebp-1 and −2)10–12. Absence of Sirt1 in the liver accelerates the metabolic disturbances of HFD13–15. Conversely, increased Sirt1 activity generally confers a metabolically beneficial profile. Transgenic overexpression of Sirt1 or increased availability of the Sirt1 substrate, NAD+, ameliorates many of the metabolic consequences of diet-induced obesity (DIO) in rodents, such as glucose intolerance, cholesterol and fat deposition in the liver, endoplasmic reticulum stress and inflammation although adverse effects were also reported16–21. The activity of Sirt1 itself is regulated through multiple mechanisms, which include interaction with other proteins and posttranslational modifications, both areas of current research22–24. In addition, Sirt1 is regulated by the availability of its substrate, NAD+ and may act as a sensor for metabolic adaptation to nutritional states. Thus enzymes in NAD+ metabolic pathways may regulate metabolism through Sirt125,26. Nnmt is one such enzyme, shown recently to regulate adipose tissue energy expenditure partly through global changes in histone methylation and increased NAD+ content27. Nnmt methylates nicotinamide (NAM) to MNAM using the universal methyl donor S-Adenosyl methionine (SAM) and producing S-adenosyl homocysteine (SAH)28–30. MNAM is further oxidized by aldehyde oxidase in the liver to two related compounds, 1-methyl-2-pyridone-5-carboxamide and 1-methyl-4-pyridone-3-carboxamide and all three are excreted in the urine31. Liver is the tissue with the strongest Nnmt expression but its role in this tissue is not well understood. In this study we examined the role of Nnmt in the liver and demonstrate that Nnmt regulates glucose, lipid and cholesterol metabolism by stabilizing Sirt1.
Results
Nnmt expression correlates with metabolic parameters
It has been recently shown that adipose tissue Nnmt expression is increased in mouse models of obesity27. For this reason, we examined the regulation of liver Nnmt expression by diets and in metabolic disease models. Nnmt expression was higher in the livers of db/db mice compared with controls as previously shown27 (Supplementary Fig. 1a). Nnmt expression was lower in the livers of ketogenic diet-fed mice (KD) and higher in the livers of calorically restricted mice (CR) compared with chow-fed mice. High-fat diet (HFD) feeding did not change liver Nnmt (Supplementary Fig. 1b). Consistent with that, MNAM content of the liver was not changed by HFD compared with chow (chow 2.88 ± 0.44 vs HFD 3.19 ± 0.35 pmol/mg wet weight, n = 8/group, data are mean ± s.e.m). Fasting and re-feeding experiments had no effect on liver Nnmt expression in C57BL6/J mice (Supplementary Fig. 1c).
Liver Nnmt expression varies widely (∼100-fold) among mouse inbred strains (Supplementary Fig. 2). We took advantage of this property and correlated liver Nnmt expression with metabolic phenotypes deposited in the Hybrid Mouse Diversity Panel (HMDP) database, which aims at finding novel genetic variation influencing metabolic diseases32. Liver Nnmt expression correlates inversely with high-density lipoprotein (HDL), total cholesterol, triglycerides (TGs), free fatty acids and other parameters (Supplementary Table 1). This correlation prompted us to examine the role of Nnmt in metabolic homeostasis.
Nnmt regulates glucose and cholesterol metabolism
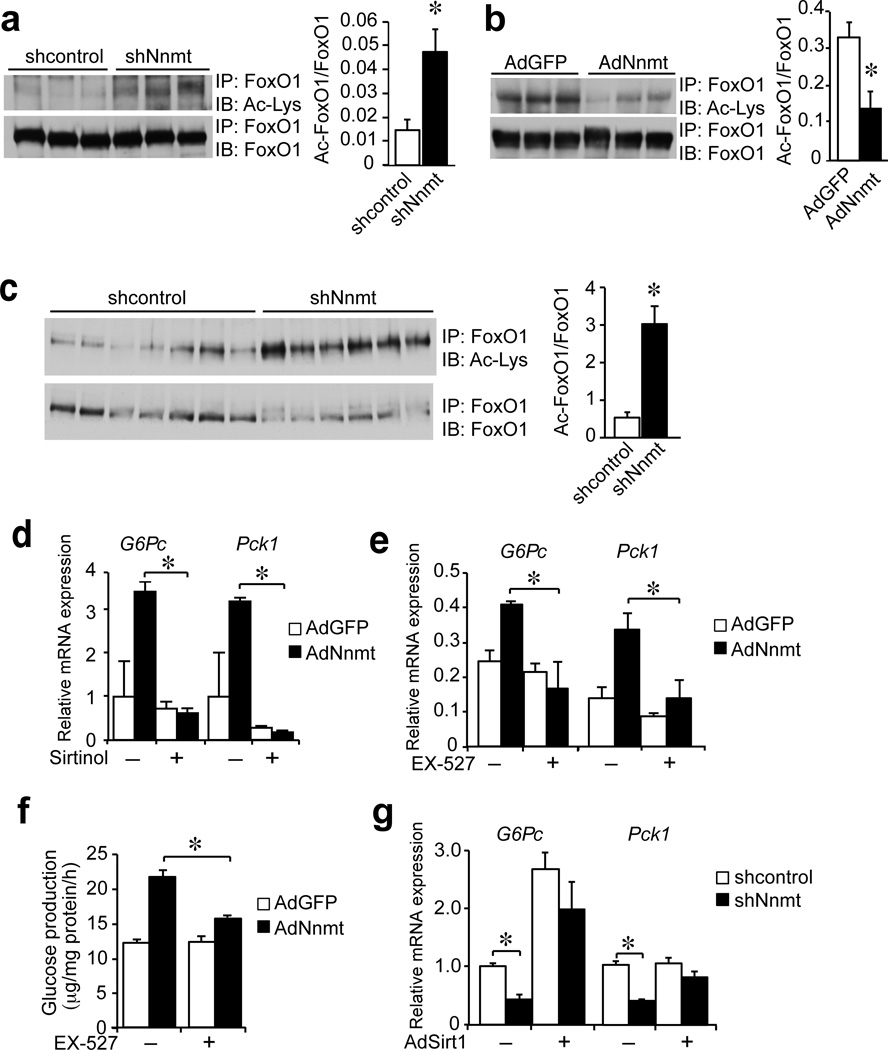
To establish a causal role for Nnmt regulating some of these metabolic parameters we manipulated Nnmt expression through adenoviral knockdown and overexpression in mouse primary hepatocytes and in vivo. We initially tested the role of Nnmt in gluconeogenesis, as this was the first reported metabolic pathway regulated by Sirt14. Primary hepatocytes with Nnmt knockdown (Fig. 1a) had significantly lower hepatocyte glucose output (50%) and significantly lower expression of both glucose-6-phosphatase catalytic (G6pc) (20%) and phosphoenolpyruvate carboxykinase 1 cytosolic (Pck1) (40%) compared with control hepatocytes (Fig. 1b). In contrast, primary hepatocytes with Nnmt overexpression (Fig. 1c) had significantly higher glucose output (1.4-fold), 3-fold higher expression of G6pc and 4-fold higher expression of Pck1 compared with control hepatocytes (Fig. 1d). These data indicate that Nnmt is a positive regulator of gluconeogenesis in primary hepatocytes.
Figure 1. Nnmt regulates gluconeogenesis and cholesterol metabolism.
(a) Nnmt protein expression in control (shcontrol) and Nnmt knockdown (shNnmt) hepatocytes. (b) Hepatocyte glucose output and mRNA expression of G6pc, Pck1 and Sirt1 in control (shcontrol) and Nnmt knockdown (shNnmt) hepatocytes (representative of 3 independent experiments performed in triplicates). (c) Nnmt protein expression in control (AdGFP) and Nnmt overexpressing (AdNnmt) hepatocytes. (d) Hepatocyte glucose output and mRNA expression of G6pc, Pck1 and Sirt1 in control (AdGFP) and Nnmt overexpressing (AdNnmt) hepatocytes (representative of >3 independent experiments performed in triplicates). (e) Overnight fasting blood glucose and insulin in mice with Nnmt knockdown (shcontrol n = 8, shNnmt n = 8). (f) Pyruvate tolerance test in mice with Nnmt knockdown (shcontrol n = 7, shNnmt n = 5). AUC = area under the curve. (g) Expression of gluconeogenic genes in the liver of Nnmt knockdown mice (shcontrol n = 8, shNnmt n = 8). (h) Serum and liver cholesterol in ad libitum fed Nnmt knockdown mice (shcontrol n = 8, shNnmt n = 7). (i) Expression of genes in cholesterol metabolism in Nnmt knockdown mice (shcontrol n = 8, shNnmt n = 7). Data are presented as mean ± s.e.m. Statistical significance was tested by unpaired Student’s t-test, *P < 0.05).
To test the role of Nnmt in vivo, we knocked down Nnmt in the liver of C57BL6/J mice using adenovirus. Nnmt knockdown mice had significantly lower overnight fasting glucose levels compared with control mice, whereas fasting insulin did not change (Fig. 1e). Pyruvate conversion to glucose was significantly lower in mice with Nnmt knockdown (50%), suggesting reduced gluconeogenesis in vivo (Fig. 1f). Expression of G6pc and fructose bisphosphatase 1 (Fbp1) was lower in the livers from Nnmt knockdown mice compared with control mice, while Pck1 and pyruvate carboxylase (Pcx) expression was not changed (Fig. 1g). Serum and hepatic TGs levels did not differ (data not shown) but serum and liver cholesterol levels were significantly higher in ad libitum-fed Nnmt knockdown mice compared with controls (Fig. 1h). Expression of Srebf2, the first two genes in the cholesterol synthesis pathway Hmgcr and Hmgcs1, and expression of several receptors and transporters in the cholesterol pathway, Abcg5, Scarb1, and Abcb11 were also higher (1.3–2 fold) in the livers of Nnmt knockdown mice compared with controls (Fig. 1i). Altogether, our results show that Nnmt is a novel regulator of hepatic glucose and cholesterol metabolism in vivo.
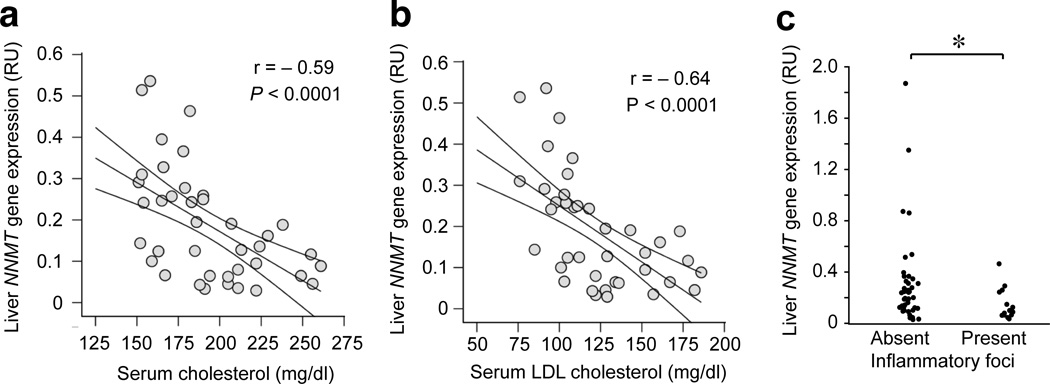
To explore the potential role of NNMT in human physiology we examined the correlation between liver NNMT expression and multiple metabolic parameters in a cohort of obese non-diabetic individuals (Supplementary Table 2). Liver NNMT correlated positively with glucose infusion rate (P = 0.03) suggesting enhanced glucose disposal (Table 1). We found significant inverse correlations between liver NNMT expression, total cholesterol (P < 0.0001) and low-density lipoprotein (LDL) cholesterol levels (P < 0.0001), fasting TG levels (P < 0.03) and cortisol levels (P < 0.045) (Table 1). The most robust correlations were observed between NNMT and cholesterol (total and LDL) (Fig. 2a,b). The coefficients of determination, R2 = 0.348 for total cholesterol and R2 = 0.41 for LDL cholesterol, suggest that variation in human liver NNMT expression can account for as much as 35–40% of the variation in the serum cholesterol of our subjects. In addition, liver inflammation decreased with increasing NNMT expression (Fig. 2c). These results implicate liver NNMT in human physiology.
Table 1.
Correlation among liver NNMT gene expression and selected metabolic parameters.
| n = 51 | r | P |
|---|---|---|
| Age (years) | – 0.21 | 0.15 |
| BMI (kg/m2) | – 0.08 | 0.5 |
| Fasting glucose (mg/dl) | 0.18 | 0.25 |
| Total cholesterol (mg/dl) | – 0.54 | <0.0001 |
| HDL cholesterol (mg/dl) | 0.21 | 0.15 |
| LDL cholesterol (mg/dl) | – 0.57 | <0.0001 |
| Fasting triglycerides (mg/dl) | – 0.31 | 0.03 |
| Glucose infusion rate (mg/kg/min) | 0.32 | 0.03 |
| Cortisol (µg/dl) | – 0.29 | 0.045 |
Pearson’s correlation coefficient (r) and P values for associations between liver NNMT expression and selected metabolic parameters in the subjects (n = 51). The significant associations persisted after excluding subjects taking hypolipidemic drugs (n = 10).
Figure 2. Human liver NNMT expression correlates with serum cholesterol and liver inflammation.
(a) Negative correlation between human liver NNMT expression and total cholesterol (n = 41, r = – 0.59, P < 0.0001). Subjects on hypolipidemic drugs (n = 10) were excluded from the correlation. (b) Negative correlation between human liver NNMT expression and LDL cholesterol (n = 41, r = –0.64, P < 0.0001). Subjects on hypolipidemic drugs (n = 10) were excluded from the correlation. (c) Liver NNMT expression according to liver lobular activity (index of inflammation) (n = 53, *P = 0.0367). RU = relative units.
Sirt1 is required for the metabolic actions of Nnmt
To explore whether Sirt1 is involved in the metabolic effects of Nnmt, we assessed the acetylation status of FoxO1, a well-regarded target of Sirt1 and thus used it as readout of Sirt1 activity. Acetylation of FoxO1 was significantly higher in primary hepatocytes with Nnmt knockdown in vitro compared with controls, indicating decreased Sirt1 activity (Fig. 3a). The opposite was also true as we found that FoxO1 acetylation was significantly lower in primary hepatocytes overexpressing Nnmt in vitro compared with controls (Fig. 3b). The livers of mice with Nnmt knockdown had significantly higher FoxO1 acetylation compared with control mice (>4-fold), suggesting impaired Sirt1 activity in vivo (Fig. 3c).
Figure 3. Sirt1 is necessary and sufficient for the metabolic effects of Nnmt.
(a) Immunobloting of acetylated, total FoxO1 and their ratio in control and Nnmt knockdown primary hepatocytes (triplicate determination). (b) Immunobloting of acetylated, total FoxO1 and their ratio in control and Nnmt overexpressing primary hepatocytes (triplicate determination). (c) Immunoblotting of acetylated and total FoxO1 and their ratio in the livers of control and Nnmt knockdown mice (shcontrol n = 6, shNnmt n = 7). (d) Expression of G6pc and Pck1 in control and Nnmt overexpressing primary hepatocytes treated with the Sirt1 inhibitor sirtinol (representative of 2 independent experiments performed in triplicates). (e) Expression of G6pc and Pck1 in control and Nnmt overexpressing primary hepatocytes treated with the Sirt1 inhibitor Ex-527 (performed in triplicates). (f) Effect of EX-527 on glucose production in control and Nnmt overexpressing primary hepatocytes (triplicate determination). (g) Expression of G6pc and Pck1 in control and Nnmt knockdown primary hepatocytes with Sirt1 overexpression (AdSirt1) (triplicate determination). Data are presented as mean ± s.e.m. Statistical significance was tested by unpaired Student’s t-test, *P < 0.05).
To test the causal involvement of Sirt1 in mediating the effects of Nnmt, we treated primary hepatocytes with the Sirt1 inhibitors sirtinol and EX-527. G6pc and Pck1 expression was significantly lower in inhibitor-treated hepatocytes compared with untreated hepatocytes overexpressing Nnmt (Fig. 3d,e). Pretreatment with the specific Sirt1 inhibitor EX-527 significantly lowered glucose production in inhibitor-treated hepatocytes compared with untreated hepatocytes overexpressing Nnmt (Fig. 3f). We also speculated that increasing Sirt1 expression should rescue the effects of Nnmt knockdown on the gluconeogenic gene expression. As expected, adenoviral overexpression of Sirt1 rescued the suppression of G6pc and Pck1 expression caused by Nnmt knockdown, showing that Sirt1 is both necessary and sufficient to mediate the effects of Nnmt on gluconeogenesis (Fig. 3g).
Nnmt regulates Sirt1 stability
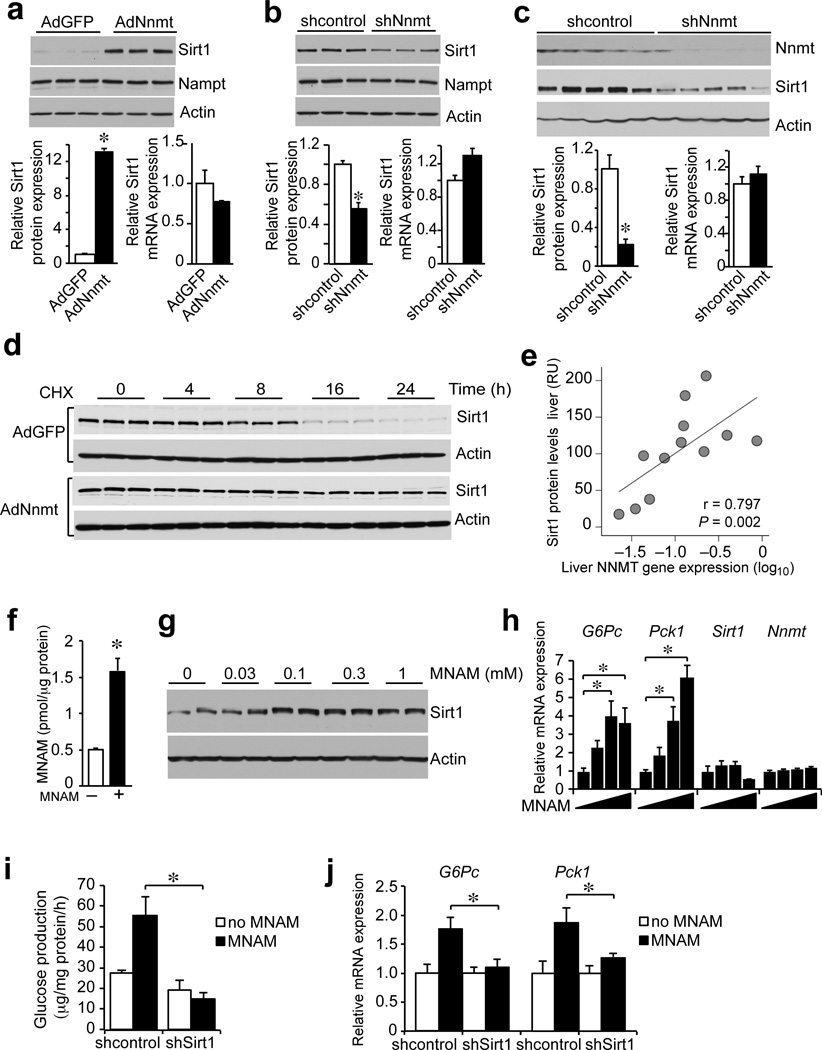
To investigate how Nnmt affects Sirt1 activity and metabolic pathways, we measured the content of Nnmt substrates and products, SAM, SAH, NAM, MNAM and the Sirt1 substrate, NAD+. SAM/SAH ratio, intracellular NAD+ and NAM content did not change in primary hepatocytes with Nnmt knockdown in vitro and in the livers of mice with Nnmt knockdown in vivo compared with controls (Supplementary Fig. 3a,b,c). Only the product of Nnmt, MNAM showed changes consistent with changes in Nnmt expression. Intracellular MNAM content was significantly higher (∼2.5-fold) in primary hepatocytes overexpressing Nnmt in vitro compared with controls (Supplementary Fig. 3d). MNAM content was below the detection limit in primary hepatocytes with Nnmt knockdown in vitro and was lower (30%) in the livers of mice with Nnmt knockdown in vivo compared with controls (Supplementary Fig. 3d).
As neither the SAM/SAH ratio nor NAD+ content, previously shown to be higher in adipocytes from Nnmt knockdown mice27, were changed in hepatocytes, we considered alternative ways Nnmt might be regulating Sirt1-dependent pathways. In particular, we examined whether Nnmt affects Sirt1 itself and noticed that expression of Sirt1 protein was significantly higher (>10-fold) in primary hepatocytes overexpressing Nnmt in vitro (Fig. 4a) and significantly lower (50%) in primary hepatocytes with Nnmt knockdown in vitro compared with controls (Fig. 4b). As an additional control, expression of Nampt, the enzyme regulating the limiting step in the nicotinamide salvage pathway, was not affected by changes in Nnmt expression (Fig. 4a,b). Sirt1 protein was also significantly lower in the livers of Nnmt knockdown mice compared with control mice (Fig. 4c). In the same experiments, Sirt1 mRNA expression was not changed, suggesting that Nnmt might be regulating the stability of Sirt1 protein. We measured Sirt1 half-life in primary hepatocytes treated with cyclohexamide, an inhibitor of protein translation. Sirt1 half-life was longer in Nnmt overexpressing hepatocytes (>25 h) compared with control (AdGFP) hepatocytes (10h) (Fig. 4d). Importantly, in human liver biopsies, SIRT1 protein correlates positively with NNMT expression (r = 0.797, n = 12) (Fig. 4e).
Figure 4. Nnmt regulates Sirt1 protein stability through its product MNAM.
(a) Sirt1 protein and mRNA expression in control and Nnmt overexpressing primary hepatocytes (representative of 3 independent experiments performed in triplicates). (b) Sirt1 protein and mRNA expression in control and Nnmt knockdown primary hepatocytes (representative of 3 independent experiments performed in triplicates). (c) Sirt1 protein and mRNA expression in livers of control and Nnmt knockdown mice (shcontrol n = 8, shNnmt n = 8). (d) Time course of Sirt1 protein degradation in cyclohexamide-treated control and Nnmt overexpressing hepatocytes (triplicate determination). (e) Correlation between Nnmt mRNA and Sirt1 protein expression from human liver biopsies (n = 12, P = 0.797). (f) Supplementation of media with 0.1 mM MNAM increases intracellular MNAM concentration (triplicate determination). (g) Dose-dependent effect of MNAM (0–1 mM) on Sirt1 protein expression in primary hepatocytes (representative of 2 independent experiments). (h) Dose-dependent effect of MNAM (0–0.3 mM) on G6pc ,Pck1, Sirt1 and Nnmt expression in primary hepatocytes (representative of 3 independent experiments performed in triplicates). (i) Glucose production in primary hepatocytes supplemented with MNAM and infected with either control or Sirt1 knockdown adenovirus (triplicate determination). (j) G6pc and Pck1 expression in primary hepatocytes supplemented with MNAM and infected with either control or Sirt1 knockdown adenovirus (triplicate determination). Data are presented as mean ± s.e.m. Statistical significance was tested by unpaired Student’s t-test. *P < 0.05. RU = relative units.
Regulation of Sirt1 protein stability is not well understood. Recent studies implicate the proteasome in the degradation of Sirt1 through ubiquitin-dependent and independent pathways and through caspase-mediated degradation33–35. We treated primary hepatocytes with the proteasome inhibitor MG132 for up to 20 h because toxicity ensues at later time. Thus expression of Sirt1 protein was modestly higher (∼2-fold) compared to untreated hepatocytes and comparable to that seen with Nnmt overexpression over the same time course (Supplementary Fig. 4a). In addition, the difference in Sirt1 protein expression between control and NNMT overexpressing hepatocytes was much smaller in MG132-treated cells consistent with the idea that Nnmt inhibits proteasome degradation of Sirt1. We treated cells with MG132 to accumulate ubiquitinated proteins and show that Nnmt overexpression lowers Sirt1 ubiquitination (Supplementary Fig. 4b) but does not affect general proteasome activity (Supplementary Fig. 4c).
MNAM mediates the metabolic function of Nnmt
Since Nnmt is an enzyme, we examined whether its enzymatic activity is required for the increased Sirt1 protein expression. To generate enzymatically inactive Nnmt, we modeled NAM into the available X-ray structure of mouse Nnmt and selected to independently mutate tyrosine 20 (Y20) and alanine 198 (A198) to tryptophan (Supplementary Fig. 5a)36–38, predicting that increasing the bulkiness of the side chain would disrupt NAM binding and thus Nnmt activity. We expressed wild type (wt) Nnmt (Nnmt-wt) and the mutants Nnmt-Y20W and Nnmt-A198W in a heterologous cell line and tested the immunoprecipitated proteins for their ability to methylate NAM. In contrast to wt Nnmt, neither Nnmt mutant was able to methylate NAM in the presence of its co-substrate SAM, despite similar amounts of Nnmt protein in the extracts (Supplementary Fig. 5b,c). Next, we co-transfected Sirt1 with either the wt or the Nnmt mutants to test their ability to increase Sirt1 protein expression and confirmed that Nnmt activity is required for this effect (Supplementary Fig. 5d).
These observations led us to test MNAM, the product of Nnmt. Although traditionally considered an inactive or toxic metabolite of terminal nicotinamide clearance, recent publications suggest that, at pharmacological doses, MNAM has biological activity and affects endothelial cell function39–41. Addition of MNAM to the media of primary hepatocytes resulted in 3-fold higher intracellular MNAM content compared with controls (Fig. 4f). Similar to Nnmt overexpression, MNAM treated hepatocytes showed a dose-dependent increase in Sirt1 protein expression compared with controls (Fig. 4g), and in G6pc and Pck1 mRNA expression without affecting Sirt1 or Nnmt mRNA expression (Fig. 4h). Consistent with the higher G6pc and Pck1 expression, MNAM-treated hepatocytes had higher glucose production (2-fold) compared with controls and these changes were abolished by Sirt1 knockdown (Fig. 4i,j). MNAM-treated cells had lower ubiquitinated Sirt1 protein without affecting general proteasome activity (Supplementary Fig. 5e,f) and MNAM does not directly affect Sirt1 activity in vitro (Supplementary Fig. 5g).
MNAM improves the metabolic profile of HFD-fed mice
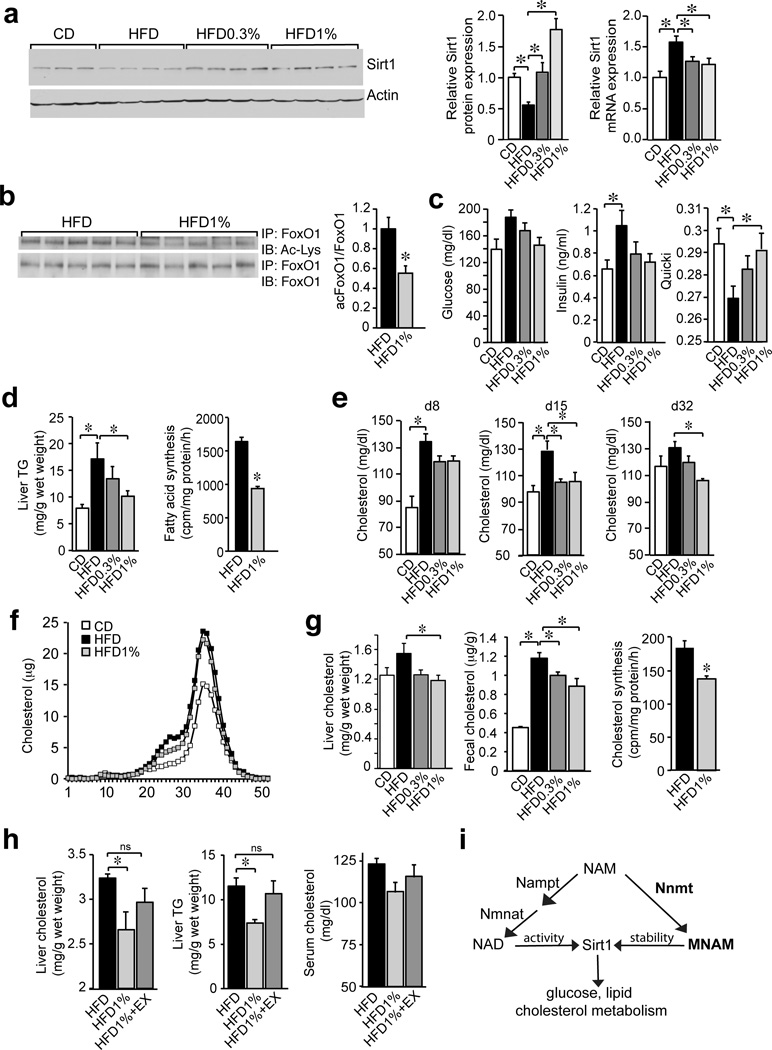
Increasing Sirt1 expression improves the metabolic profile of diet-induced obese (DIO) mice16,17,42. Since the small metabolite MNAM mediates the metabolic activity of Nnmt, we expected that in vivo treatment with MNAM would increase liver Sirt1 protein and improve the metabolic profile of mice fed a HFD. To test this idea, we fed several cohorts of wt C57BL6/J mice a HFD supplemented with different doses of MNAM (0.3% and 1%) for several weeks. MNAM content was 4-fold higher in the livers of MNAM-supplemented (HFD1%) compared with control mice (HFD) indicating that it is orally bioavailable (Supplementary Fig. 6a). HFD-fed mice gained an additional 7 grams of body weight after 8 weeks compared with mice fed low-fat control diet (CD) but MNAM supplementation had no effect on body weight gain (Supplementary Fig. 6b). Liver Sirt1 protein expression was significantly lower (50%) in HFD-fed compared with CD-fed mice (Fig. 5a). MNAM-supplemented mice had higher liver Sirt1 protein expression compared with mice fed HFD alone and beyond the levels seen on CD (Fig. 5a). The mRNA expression of Sirt1 followed the opposite pattern from the protein, suggesting that HFD regulates Sirt1 protein stability and MNAM reverses this effect (Fig. 5a).
Figure 5. MNAM improves cholesterol and lipid homeostasis in DIO mice.
(a) Liver Sirt1, Actin and Sirt1 mRNA from CD, HFD, HFD0.3% and HFD1% fed mice (b) Liver acetylated and total FoxO1 from HFD and HFD1% fed mice. (c) Blood glucose (4 h fast), insulin, and quantitative insulin sensitivity index (QUICKI) of mice fed CD, HFD, HFD0.3% and HFD1%. (d) Liver TGs content of mice fed CD, HFD, HFD0.3%, HFD1% (n = 5/group) and fatty acid synthesis in primary hepatocytes from mice fed HFD and HFD1% (n = 6/group, representative of 2 independent experiments). (e) Serum cholesterol (4 h fast) on days 8, 15 and 32 from mice fed CD, HFD, HFD0.3% and HFD1%. (f) Cholesterol content of lipoprotein fractions from mice fed CD, HFD and HFD1%. (g) Liver and fecal cholesterol of mice fed CD, HFD, HFD0.3% and HFD1%. Cholesterol synthesis in primary hepatocytes from mice fed HFD and HFD1% (n = 6/group, representative of 3 independent experiments). (h) Effect of pharmacological Sirt1 inhibition on serum and liver cholesterol and liver TGs levels in mice fed a HFD, HFD1%, HFD1%+EX (n = 5/group). (i) Schematic representation of the proposed pathway. One-way ANOVA followed by posthoc Dunnett’s with control set to HFD: a,c,d,e,g and h. Unpaired t-test: b,d (right panel), g (right panel); n = 7 for CD, n = 8 for HFD, HFD0.3%, HFD1% in a,b,c,e,g (left panel); f pooled sample from 8 mice/group; g (middle panel) technical triplicate of pooled samples. Data are presented as mean ± s.e.m, *P < 0.05.
Consistent with higher Sirt1 protein expression, liver FoxO1 acetylation was significantly lower in the livers of MNAM-supplemented mice (HFD1%) compared with control mice (HFD) (Fig. 5b). MNAM prevented the changes in fasting glucose and insulin caused by HFD, however this effect was transient and persisted only for one week (Fig. 5c). More sustained effects were seen on lipids and cholesterol. Serum TGs were not affected by HFD and MNAM treatment throughout the course of the studies (not shown) but MNAM-treated mice (HFD1%) had significantly lower liver TGs (60%) compared with control mice (HFD) (Fig. 5d) and the pattern of gene expression suggested that fatty acid synthesis is inhibited by MNAM (Supplementary Fig. 6c). Indeed, ex vivo hepatocytes isolated from mice treated with MNAM (HFD1%) had significantly lower fatty acid synthesis (58%) compared to control hepatocytes (HFD) (Fig. 5d). HFD-fed mice had significantly higher serum cholesterol compared to control mice (CD) and MNAM supplementation of HFD prevented this increase for several weeks (Fig. 5e), although by 8-weeks the effect was no longer apparent (not shown).
We also analyzed the lipoprotein profile to determine size of the particles carrying cholesterol. Plasma from HFD-fed mice had higher cholesterol content in HDL fractions (30–40) and in intermediate fractions (20–30) compared with CD-fed mice (Fig. 5f). MNAM-treated mice (HFD1%) had lower cholesterol in the intermediate fractions (20–30) compared with HFD-fed mice, suggesting a selective effect on the more atherogenic lipoprotein particles (Fig. 5f). After 8 weeks on HFD, liver cholesterol was significantly lower (75%) in MNAM-treated mice (HFD vs HFD1%) (Fig. 5g). Fecal cholesterol output was also lower in MNAM-treated mice compared to HFD alone, suggesting that the lower liver cholesterol was due to the decreased synthesis and not increased cholesterol clearance (Fig. 5g). As expected, cholesterol synthesis was lower (75%) in hepatocytes isolated from MNAM-treated mice (HFD1%) compared with control hepatocytes (HFD) (Fig. 5g).
Gene expression survey of cholesterol metabolic pathways was consistent with the liver cholesterol profile. Expression of the master cholesterol regulators Srebf2 and Nr1h3 (encoding LXRα) and several of their target genes in cholesterol synthesis (Srebf2, Hmgcs1) and transport (Ldlr, Abca1, Abcg8) were significantly higher in the livers of HFD-fed compared with CD-fed mice and MNAM supplementation of HFD prevented these changes in a dose-dependent manner (Supplementary Fig. 6d,e). Consistent with the improved metabolic profile of the liver, MNAM-fed mice had significantly lower liver expression of the proinflammatory cytokines Tnf and Il6 compared with HFD-fed mice (Supplementary Fig. 6f). To test if the metabolic effects of MNAM require Sirt1 activity in vivo, we fed mice HFD supplemented with MNAM in combination with the Sirt1 inhibitor EX-527. Sirt1 inhibition blocked the beneficial effects of MNAM on liver cholesterol and liver TGs. Changes in serum cholesterol showed similar trends but did not reach significance (Fig. 5h). Our data indicate that pharmacological doses of MNAM improve the metabolic dysregulation caused by HFD feeding. A schematic representation of the proposed pathway shows NAM increasing Sirt1 activity through NAD+ biosynthesis and Sirt1 protein through Nnmt and MNAM (Fig. 5i).
Discussion
Nnmt has recently emerged as a novel metabolic regulator27. Adipose tissue Nnmt expression correlates positively with adiposity in mice and antisense oligonucleotide knockdown of Nnmt protected against DIO27. In humans, adipose tissue NNMT expression and its product MNAM correlate positively with insulin resistance43. The function of Nnmt in the liver, a site of major Nnmt expression, is not well investigated. Dietary regulation of liver Nnmt expression shows some interesting patterns. Ketogenic diet feeding significantly suppresses liver Nnmt expression, which may contribute to the increased serum and liver cholesterol in this dietary model44. Of considerable interest is the increased Nnmt expression in C57BL6 mice under caloric restriction, as this would promote Sirt1 protein stability, which mediates many of the metabolic effects of caloric restriction16. Other strains of mice show a much more impressive response. Caloric restriction of the outbred ICR mice increases liver Nnmt expression 36.5-fold45. The contribution of Nnmt to the metabolic benefits of caloric restriction should be addressed in future experiments. Our data do not support the idea that downregulation of Nnmt expression contributes to the pathogenesis of obesity, at least in the models tested, as NNMT expression was either unchanged or even increased in the livers of HFD-fed and db/db mice, respectively.
Liver Nnmt is a major variable expression locus in mice46 and it correlates inversely with serum cholesterol, TGs and free fatty acid levels among other markers. Remarkably, we observed similar associations in a human obese cohort with significant inverse correlation between liver NNMT expression and serum total cholesterol, LDL cholesterol, TG levels and other parameters. The most significant associations are between NNMT and LDL cholesterol accounting for up to 40% of the variation in LDL cholesterol in our cohort and could potentially explain a large fraction of the variation in cholesterol in other human populations as well. Altogether, associations from mice and humans suggest that increased liver NNMT expression correlates with an improved metabolic profile, in contrast to its expression in adipose tissue27,43. We speculate that the significant variation in liver NNMT expression underlies the natural variation in metabolic traits seen among mice and humans and individuals with lower liver NNMT expression could be predisposed to the deleterious consequences of nutrient excess.
We tested the causal role of Nnmt in metabolic regulation by adenoviral knockdown and overexpression in vitro and in vivo in mice and showed that Nnmt regulates hepatic glucose, lipid and cholesterol metabolism. The mechanism of action underlying this regulation by Nnmt in the liver is completely different from adipose tissue. Nnmt is not a major methyltransferase in the liver and does not change methyl donor balance in hepatocytes in contrast to adipocytes and cancer cells27,47. This is not surprising, as liver expresses other major methyltransferases, which account for the majority of the methyl donor flux in this tissue48. Instead, we found that the metabolic effects of Nnmt are mediated by its product MNAM. Both Nnmt and MNAM increase Sirt1 protein expression independent of its mRNA levels, while Sirt1 is necessary for the metabolic effects of Nnmt and MNAM. Our results are consistent with a large body of literature implicating Sirt1 in the regulation of gluconeogenesis and cholesterol synthesis4,6,7,11.
The stability of Sirt1 itself is poorly understood and alternative pathways have been proposed34,35,49. Our data suggest that Nnmt and MNAM regulate the ubiquitin-proteasome degradation of Sirt1. Whether MNAM binds directly to Sirt1 itself or another protein involved in Sirt1 degradation is not known. Very recently, Mdm2 has been proposed as the Sirt1 E3-ligase and represents a potential candidate50. MNAM has long been considered an inactive metabolite. Recent studies show that it possesses anti-inflammatory and antithrombotic activity with incompletely understood mechanism of action, and also increases C.elegans life span through mitohormetic effects39,40,51. Our results define a novel intracellular mechanism for the metabolic effects of Nnmt through the production of an active endogenous metabolite MNAM that could be applicable to other cell types where Nnmt is strongly expressed, such as adipose tissue, endothelium, cancer cells, and possibly on cells not expressing Nnmt through paracrine effects of secreted MNAM.
We anticipated beneficial effects of dietary MNAM supplementation in mice fed a HFD because MNAM increased Sirt1 protein expression in vivo16–18. In line with the known metabolic effects of Sirt1, MNAM significantly lowered liver cholesterol and TG levels, while also suppressing fatty acid and cholesterol synthesis and the expression of lipogenic and cholesterol synthesis genes. MNAM supplementation also suppressed the expression of several target genes of Nr1h3 (encoding Lxrα) and Nr1h4 (encoding Fxr) in cholesterol and bile acid transport. It has been previously shown that Sirt1 deacetylates and activates Lxrα and Fxr52,53, therefore the reduction in their target genes is likely indirect and secondary to the reduced availability of their ligands due to the suppression of cholesterol synthesis.
We also observed systemic effects of MNAM with significant lowering of serum cholesterol and changes in lipoprotein profile. Mouse lipoprotein metabolism differs from humans, as most of the cholesterol is transported in HDL and not LDL. MNAM supplementation produced a selective decrease in larger lipoprotein particles and not HDL, suggesting that MNAM or its derivatives could be used to decrease LDL levels in humans. Serum TG levels did not change in our experiments. However, it should be noted that HFD and C57BL6 mice are not ideal models of triglyceridemia and a TG-lowering effect of MNAM has been reported in a rat model in the context of a high fructose diet41.
Methylation of NAM by Nnmt is a major pathway for the clearance of excess vitamin B3 from the body. Here we ascribe it a novel and unexpected function in Sirt1-dependent metabolic regulation. Thus in addition to well-established NAD+ biosynthetic pathways, which stimulate Sirt1 activity by increasing NAD+ levels, vitamin B3 clearance pathways also regulate hepatic nutrient metabolism through MNAM-mediated Sirt1 protein stabilization.
Online Methods
Materials
NAM, cyclohexamide, MG132, 8-Br-cAMP, Percoll and dexamethasone were obtained from Sigma-Aldrich (St.Louis, MO). MNAM was from Sigma-Aldrich (St.Louis, MO) and TCI America (Portland, OR). Nnmt chicken polyclonal antibody (29–288–23087) was from Genway (San Diego, CA). Nnmt rabbit antibody (ARP42281) was from Aviva Systems Biology (San Diego, CA). Sirt1 antibody (2028S) was from Cell Signaling (Danvers, MA). Actin mouse monoclonal antibody (ab3280) was from Abcam (Cambridge, MA). FoxO1antibody (sc-11350) was from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti Acetyl-lysine antibody (9441S) was from Cell Signaling (Danvers, MA). Secondary antibodies goat anti-chicken (703-035–155), donkey anti-Mouse (715-035–150) and donkey anti-Rabbit (711-035–152) were from Jackson Immunoresearch (West Grove, PA). Glucose was measured with the One Touch Ultra Mini glucose meter (Lifescan).
Mouse purchase
Eight week old C57BL6/J male mice were purchased from Jackson labs (Bar Harbor, ME). Mice were group housed with ad libitum access to food except where indicated. Water was available at all times. All procedures were approved by the Beth Israel Deaconess Medical Center IACUC.
Primary hepatocyte Isolation and culture
Primary hepatocytes were isolated from 8-week old C5BL6/J male mice by the collagenase perfusion method. Hepatocyte preparation media were from Invitrogen (Carlsbad, CA). Viable hepatocytes were enriched by Percoll gradient centrifugation. The hepatocytes were resuspended in Williams’ E Medium containing 10% FBS and seeded on collagen coated plates at a density of 800,000/ml. After a 4-hour attachment period the medium was switched to Williams’ E medium without serum containing insulin, transferrin and selenium (ITS) from Invitrogen and changed daily.
Adenovirus construction
Nnmt cDNA was cloned from the liver of C57BL6 mice and a Flag epitope was introduced to the N-terminus. Nnmt cDNA was cloned into the adenovirus vector pAd/CMV/V5-Dest (Invitrogen). ShRNA against Nnmt was obtained from the Broad Institute (Cambridge, MA) and cloned into the pAd/PL-Dest (Invitrogen). Control viruses contained either GFP or irrelevant shRNA. Adenoviruses were generated by the Harvard Gene Therapy Initiative Research Vector Core. The titers of the purified viruses ranged from 5–20 × 1010 pfu/ml.
Adenovirus infection
Primary hepatocytes were infected by overnight exposure to adenovirus (unless otherwise indicated) in William’s E medium at MOI 10. For Nnmt overexpression experiments, the hepatocytes were typically cultured for 48 hours. For Nnmt knockdown, the hepatocytes were typically cultured for 96 hours to allow for the degradation of the endogenous protein. For Sirt1 knockdown, hepatocytes were infected with adenovirus (MOI 30) overnight. At the same time, the hepatocytes were treated with 0.1 mM MNAM added to the media. After 48 hours the cells were used for gene expression assay or glucose production experiments. For EX-527 treatment, hepatocytes were incubated with 30 µM of EX-527 overnight prior to harvest.
Immunoblotting
Hepatocytes or livers were homogenized in RIPA buffer with protease, phosphate inhibitors and deacetylase inhibitors and lysed for 30 minutes at 4°C, followed by centrifugation for 15 minutes. Protein concentration of the supernatant was measured by the Bradford assay (Bio-Rad). 10 µg of total protein was analyzed by SDS-PAGE (Criterion gels, Bio-Rad) and transferred to nitrocellulose for 1 hr at 100V. The membrane was blocked with 5% BSA in TBST for 1 hour at room temperature and then incubated with the primary antibody overnight at 4°C. The next day, the membrane was washed 3 times with TBST and incubated with HRP-conjugated secondary antibody for 45 minutes at room temperature, followed by 3 more washes in TBST. Antibody binding was detected using chemiluminescence reagents from Pierce (Rockford, IL).
Glucose production
Primary mouse hepatocytes were plated on collagen I-coated 12-well plates (0.4 × 106/well). 4 hours later, the cells were infected with virus (MOI 10) overnight. For Nnmt over-expressing cells, media containing virus were washed away and switched to fresh maintenance media in the morning and then switched to William’s E media containing 0.5 mM cAMP and 0.1 µM dexamethasone at the end of the day. 18 hours later, the media were removed and the cells were rinsed with PBS twice. Then, 450 µl of glucose production buffer (consisting of glucose-free DMEM pH 7.4 without phenol red, 20 mM sodium pyruvate, 2mM L-glutamine and 15 mM Hepes) were added to the cell. 6 hours later, media were collected and the amount of glucose in the media was determined by the glucose assay kit (Sigma Aldrich). For Nnmt knockdown, cells were cultured for 3 more days after the cells were infected with virus. Then the cells were treated with William’s E media containing 0.5 mM cAMP and 0.1 mM dexamethasone for 18 hours and subsequently 450 µl of glucose production buffer. Media were collected at 6 hours and measured for glucose content as described in Nnmt overexpressing cells.
G6pc and Pck1 induction
18 hours before harvest, hepatocytes were washed once with PBS and switched to Williams' E medium containing 0.5 mM 8-Br-cAMP and 100 nM dexamethasone. After the treatment, hepatocytes were washed once with PBS and harvested for further analysis.
RNA extraction, cDNA synthesis and Quantitative PCR
RNA was extracted according to manufacturers instructions with the RNeasy kit from Qiagen (Valencia, CA). cDNA was prepared with the RT-for-PCR kit from Clontech (Mountain View, CA) or Quantitect Reverse Transcription kit (Qiagen). QPCR was performed with the SybrGreen Master Mix (Applied Biosciences) in a 7900HT cycler (Applied Biosciences). Cyclophilin or TBP was used for normalization. Sequences of the primers are available upon request.
Adenovirus infection of mice
Eight week old wt C57BL6/J male mice were injected via tail vein with a total viral load of 3×109 pfu/mouse in 100 µl of sterile PBS/3% glycerol. Mice were randomly allocated with respect to the virus. 7–10 days after infection mice were deeply anesthetized by ketamine/xylazine. Tissues were harvested and rapidly frozen in liquid nitrogen until further processing.
Pyruvate tolerance test
Ten days after virus-injection, mice were fasted overnight and injected intraperitoneally with pyruvate (2 g/kg body weight). Serum glucose was measured at 0, 15, 30, 60, 90 and 120 minutes after the injection.
Human subject recruitment
The human subject cohort comprised of 53 morbidly obese subjects (9 men and 44 women) at the Endocrinology Service of the Hospital Universitari de Girona Dr. Josep Trueta (Girona, Spain). Pre-established inclusion criteria: All subjects were of Caucasian origin. The subjects reported a stable body weight three months preceding the study, were free of any infections one month before and had no systemic disease. Pre-established exclusion criteria: Subjects with liver disease, specifically HCV infection and tumor disease, and subjects with thyroid dysfunction were excluded by biochemical work-up. A small number of participants were under hypolipidemic treatment [fibrates (n = 3) and statins (n = 7)]. Samples from all the subjects were obtained during elective gastric by-pass surgery. Liver samples were collected in RNA later, fragmented and immediately flash-frozen in liquid nitrogen before storage at −80°C.
Liver biopsies were analyzed by a single expert pathologist. The liver samples were stained with hematoxylin and eosin, Masson’s trichrome and reticulin. Non-alcoholic fatty liver disease (NAFLD) scoring system for was used to assess steatosis, hepatocellular ballooning, fibrosis and lobular inflammation 54. Lobular inflammation was graded as absent (0 inflammatory foci in a 200 x field) or present (>1 inflammatory foci in a 200 x field). The study has an 80% power to detect significant correlations (Spearman’s coefficient of at least 0.3) between variables in bilateral tests. The study was also powered to detect a significant difference of at least 0.5 SD units between groups. Two subjects with abnormally low total and LDL cholesterol levels were excluded from the metabolite correlations. Statistical analysis was performed in SPSS v.21. All subjects gave written informed consent, validated and approved by the ethical committee of the Hospital Universitari Dr. Josep Trueta (Comitè d’Ètica d’Investigació Clínica).
Analytical determinations (Human)
Serum glucose concentrations were measured in duplicate by the glucose oxidase method using a Beckman glucose analyser II (Beckman Instruments, Brea, California). Duplicate samples were used for serum insulin determination by the immunoradiometric assay (Medgenix Diagnostics, Fleunes, Belgium). The coefficients of variation (intra-assay) were 5.2% at a concentration of 10 mU/l and 3.4% at 130 mU/l. The coefficients of variation (inter-assay) were 6.9 and 4.5% at 14 and 89 mU/l, respectively. Total serum cholesterol was measured by an enzymatic, colorimetric method through the cholesterol esterase/cholesterol oxidase/peroxidase reaction (Cobas CHOL2). HDL cholesterol was quantified by a homogeneous enzymatic colorimetric assay through the cholesterol esterase/cholesterol oxidase/peroxidase reaction (Cobas HDLC3). Total serum TGs were measured by an enzymatic, colorimetric method with glycerol phosphate oxidase and peroxidase (Cobas TRIGL). LDL cholesterol was calculated using the Friedewald formula. Cortisol was etermined by routine laboratory test.
Euglycemic hyperinsulinemic clamp (Human)
Insulin action was determined by the euglycemic hyperinsulinemic clamp. After an overnight fast, two catheters were inserted into an antecubital vein, one for each arm, used to administer constant infusions of glucose and insulin, and to obtain arterialized venous blood samples. A 2-h euglycemic hyperinsulinemic clamp was initiated by a two-step primed infusion of insulin (80 mU/m2/min for 5 min, 60 mU/m2/min for 5 min) immediately followed by a continuous infusion of insulin at a rate of 40 mU/m2/min (regular insulin (Actrapid, Novo Nordisk, NJ)). Glucose infusion began at minute 4 at an initial perfusion rate of 2 mg/kg/min being then adjusted to maintain plasma glucose concentration at 4.9–5.5 mmol/L. Blood samples were collected every 5 minutes for determination of plasma glucose and insulin. Insulin sensitivity was assessed as the mean glucose infusion rate during the last 40 min. In the stationary equilibrium, the amount of glucose administered (M) equals the glucose taken by the body tissues and is a measure of overall insulin sensitivity.
Gene expression analyses (Human)
RNA was prepared from liver biopsies using the RNeasy Lipid Tissue Mini Kit (QIAgen; Gaithersburg, MD). The integrity of each RNA sample was evaluated by the Agilent Bioanalyzer® (Agilent Technologies; Palo Alto, CA). Total RNA was quantified spectrophotometrically (GeneQuant, GE Health Care; Piscataway, NJ) or with the bioanalyzer. Three µg of RNA were reverse transcribed to cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems; Darmstadt, Germany) according to the manufacturer’s protocol.
Gene expression was measured by real time quantitative PCR using the LightCycler® 480 Real-Time PCR System (Roche Diagnostics; Barcelona, Spain), using TaqMan® technology suitable for relative gene expression quantification. The reaction was performed in a final volume of 12 µl following the manufacturer’s protocol. The cycle program consisted of an initial denaturation for 10 min at 95°C followed by 40 cycles of 15 s denaturing phase at 92°C, 1 min annealing and extension phase at 60°C. Cyclophilin A. The pre-validated TaqMan® primer/probe set for cyclophilin was used as the endogenous control for the target genes in each reaction: cyclophilin A (PPIA; Hs99999904_m1, RefSeq. NM_021130.3). Pre-validated SybrGreen primers for human Nnmt were purchased from Roche Diagnostics (Barcelona, Spain). Primers sequences are as follows: NNMT forward: 5’-AGCTGGAGAAGTGGCTGAAG-3’ and NNMT reverse: 5’-TGGACCCTTGACTCTGTTCC-3’.
The Second Derivative Maximum Method was used for the determination of the crossing points (Cp). A Cp value was obtained for each amplification curve and ΔCp value was first calculated by subtracting the Cp value for human PPIA cDNA from the Cp value for each sample and transcript. Fold changes compared with the endogenous control were then determined by calculating 2−ΔCp, so gene expression results are expressed in all cases as expression ratio relative to PPIA gene expression, according to manufacturers’ guidelines.
FoxO1 acetylation
Hepatocytes (8×105) were plated into 6-well plates and infected with a control or Nnmt overexpressing adenovirus overnight. Next day the cells were infected again with the virus expressing FoxO1. 24 hours later, the cells were harvested in 200 µl high salt lysis buffer (1% Triton, 500 mM NaCl, 50 mM Tris-HCl pH 7.4 and inhibitors). After 30min incubation on ice, lysates were centrifuged at full speed for 30 minutes at 4°C. The supernatant was diluted with 2 volumes of dilution buffer (50 mM Tris-HCl pH7.4 with inhibitors) to bring down the concentration of Triton and salt. 250 µg of total protein from each sample was incubated with 2 µg of FoxO1 primary antibody overnight at 4°C. The next day 40 µl of protein A/G slurry was added for another 4 hours. The beads were then washed 3 times with wash buffer (0.3% Triton, 150 mM NaCl, 50 mM Tris-HCl) After the final wash, the beads were eluted with 40 µl l 2X sample buffer and boiled at 95 degree for 5 minutes. 20 µl of the eluate was resolved by SDS-PAGE gel and probed with the indicated antibodies. For liver endogenous FoxO1 acetylation assay, liver pieces were lysed and immunoprecipitated as described above. Briefly, 4 mg of total proteins were incubated with 2 µg of primary antibody overnight and then 40 µl of protein A/G slurry for 4 hours. The beads were then washed with wash buffer and eluted with 50µ 2X sample buffer. 20 µl of the eluate was loaded for western blotting.
HPLC methods
Hepatocytes infected with adenoviruses as described and treated with 1 mM of nicotinamide for 24 hours. Media were collected, centrifuged for 5 min and injected into HPLC for NAM and MNAM analysis. Hepatocytes were harvested in ice-cold 80% methanol and incubated at –80°C for 15 min. After centrifugation, the supernatant was dried under vacuum, resuspended in dH2O and analyzed by HPLC. 6-methylnicotinamide was included as an internal standard to account for sample loss during extraction. The metabolites were separated on Atlantis T3 column (4.6 mm id, 100 mm length) using the Breeze System (Waters, MA) protected by a 20 mm guard column. Metabolites were eluted isocratically in 20 mM KH2PO4 pH 3, 5% methanol, 2 mM sodium heptanesulfonic acid at 0.7 ml/min flow rate. NAM, MNAM were detected by UV absorption at 260 nm and identified by its retention time compared to standards.
Sirt1 degradation
Primary hepatocytes were infected with a control or Nnmt overexpressing adenovirus overnight and cultured for additional 24 hours. The cells were then treated with 10 µg/ml cyclohexamide and harvested at different time points in RIPA buffer with inhibitors. 10 µg of total protein from each sample were resolved by SDS-PAGE gel and probed with the indicated antibodies.
HEK293T transfection
HEK293T were originally purchased from ATCC and maintained in DMEM-HG containing 10% FBS. Cells were transfected with 0.05 µg plasmid encoding HA-Sirt1 along with 0.5 µg plasmid encoding either GFP, Flag-Nnmt-wt, Flag-Nnmt-Y20W or Flag-Nnmt-A198W with Lipofectamine 2000 (Invitrogen). 48 hours after the transfection, cells were rinsed with PBS and harvested in RIPA buffer. 10 µg of total protein from each sample was resolved by SDS-PAGE gel and probed with the indicated antibodies.
Proteasome assay
HEK293T cells were lysed on ice for 30 minutes in the lysis buffer provided in the 20S Proteasome assay kit (Cayman Chemicals, Ann Arbor, MI) and spun for 30 minutes. The supernatant was transferred to a new tube and incubated with increasing concentrations of MNAM for 15 min at room temperature. The proteasome activity of the supernatant was measured following manufacturer’s instructions. The proteasome inhibitor MG132 was used as a positive control.
MNAM treatment
Primary hepatocytes were plated as described. After a 5 hour attachment period, hepatocytes were treated with different concentrations of MNAM. After 24 hours, fresh media were added (containing NAM) and the incubation continued for another 18 hours before harvesting in RIPA buffer. 10 µg of total protein from each sample were resolved by SDS-PAGE gel and probed with the indicated antibodies.
Sirt1 activity assay
Direct effect of MNAM on Sirt1 activity was measured by the Sirt1 Direct Fluorescent Screening Assay Kit (Cayman Chemicals, An Arbor, MI) following manufacturer’s recommendations.
HFD experiments
7–8 week old wt C57BL6/J male mice were purchased from Jackson labs. After one-week acclimatization, mice were fed HFD-supplemented with MNAM. Individual mice were randomly allocated to each experimental group. The only criterion was a matched group mean weight. Mice were group-housed with food and water available ad libitum. Irradiated HFD (RD12451) and control diet (RD12450K) were purchased from Research Diets (New Brunswick, NJ). Diet was grounded in a blender and powdered MNAM was mixed with the diet to 0.3% v/v (HFD0.3%) and 1% v/v (HFD1%). For EX-527 feeding experiment, mice were fed with 0.01% EX-527 (approximately 10 mg/kg/day) mixed with HFD1%. Fresh diet was prepared weekly. At the end of the study mice were fasted for 6 hours, anesthetized by ketamine and xylazine and sacrificed by transcardial puncture. Tissues were dissected and frozen in liquid nitrogen until further processing.
Cholesterol and fatty acid synthesis
Mice were fed with HFD or HFD1% MNAM for two weeks. Primary hepatocytes from these mice were plated in collagen I coated 12-well plates (0.4 × 106/well). After 4 h attachment period, hepatocytes were incubated with maintenance media containing 0.2 mCi/ml [3]H2O. 18 hours later, media were removed, the cells harvested in 0.8 ml of 8N KOH and transferred to screw-cap tubes. An equal volume of ethanol was added and the tubes were heated at 90°C for 2 h with vortexing every 15 minutes. The mixture was transferred into 10 ml plastic tubes and nonsaponifiable lipids (cholesterol) were extracted two times with 2 ml of petroleum ether. Aliquots of the pooled extract were dried under the chemical hood. The 3H radioactivity was determined by scintillation counting and used to calculate the rate of cholesterol synthesis. The aqueous phase was acidified with 1 volume of 6 N HCl and saponifiable lipids were extracted two times with 2 ml of petroleum ether. The 3H radioactivity was determined by scintillation counting and used to calculate the rate of fatty acid synthesis.
Fecal lipid extraction
One gram of feces from each group of mice was pulverized and transferred to 15ml tube containing 2.5 ml water and incubated at 4°C overnight. 5ml Folch reagent (chloroform: methanol 2:1) was added to each tube and the contents were vortexed, followed by addition of 1 ml of 0.05% H2SO4 and vortexed again. After 30 minutes incubation at room temperature, the tubes were centrifuged for 5 minutes at 2000 rpm. The chloroform phase (lower phase) was transferred into a glass tube. The samples were extracted again with 5ml Folch reagent and the chloroform phases were pooled. The chloroform extract was vacuum dried and dissolved in butanol: Triton X-100: methanol (0.6:0.267:0.133). Cholesterol was measured with the enzymatic assay from Stanbio (Boerne, Texas).
Statistics
Typically, 8 mice were randomly allocated to each experimental group. The only criterion was a matched group mean weight. Based on our experience, we can detect a 50% difference in mean expression of multiple genes using this group size. Exclusion criteria for mouse studies were weight loss in excess of 20%, grossly abnormal organ morphology. Investigators were not blinded with respect to the experimental groups during animal studies. Statistical differences were evaluated in JMP Pro v.10 (SAS). Data are presented as mean and standard error (s.e.m.). Pairwise differences we tested by Student’s t-test. Multiple comparisons were tested by one-way ANOVA followed by posthoc Dunnett’s with control. Some data were log transformed to satisfy normal distribution and equal variance between cohorts. Significance was set at P < 0.05. Statistical outliers were identified in JMP Pro and excluded from the data.
Supplementary Material
Acknowledgements
We would like to thank F.F. Liu, D. Adams, A. M. Real, M.J. Lee for technical assistance and J. S. Flier for advice. We thank B. M. Spiegelman and P. Puigserver (Dana Farber Cancer Center, Boston, MA) for the donation of the Sirt1 and FoxO1 adenoviruses, D. E. Cohen (Brigham and Women’s Hospital, Boston, MA) for lipoprotein analysis and R. Ortiz (University Hospital of Girona, Spain) for the pathology evaluation of the human liver biopsies. This work was supported by grants from Takeda, the US National Institute of Health DK028082 (E.M.F.), DK083694 (P.P.), Boston Area Diabetes Endocrinology Research Center (BADERC) grant DK057521 (P.P.), and by grants FIS-PI11/00214 and FIS-PI12/02631 to J.M.F.R. from the Instituto de Salud Carlos III and Fondo Europeo de Desarrollo Regional (FEDER), Spain.
Footnotes
Author Contributions
S.H., Y.K., I.T. and P.P. designed, performed and interpreted experiments. X.W., D.P., Y.L. and J.M.A. performed experiments. E.M.F. interpreted experiments. J.M.M.N and J.M.F.R. performed and interpreted the human experiments. S.H., I.T., P.P and E.M.F. corrected the manuscript. P.P. conceived and managed the project, and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008;28:115–130. doi: 10.1146/annurev.nutr.28.061807.155443. [DOI] [PubMed] [Google Scholar]
- 4.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 5.Nemoto S, Fergusson MM, Finkel T. Science. Vol. 306. New York, N.Y: 2004. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway; pp. 2105–2108. [DOI] [PubMed] [Google Scholar]
- 6.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erion DM, et al. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponugoti B, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker AK, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu F, et al. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R-H, et al. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. The Journal of Clinical Investigation. 2011 doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 17.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantó C, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiang L, et al. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell Metab. 2011;14:758–767. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J-E, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 23.Zhao W, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim E-J, Kho J-H, Kang M-R, Um S-J. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Molecular Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 25.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. The Journal of Biological Chemistry. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 26.Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 2012;8:287–296. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- 27.Kraus D, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang-Lee YA, et al. Metabolic effects of nicotinamide administration in rats. J. Nutr. 1983;113:215–221. doi: 10.1093/jn/113.2.215. [DOI] [PubMed] [Google Scholar]
- 29.Scheller T, Orgacka H, Szumlanski CL, Weinshilboum RM. Mouse liver nicotinamide N-methyltransferase pharmacogenetics: biochemical properties and variation in activity among inbred strains. Pharmacogenetics. 1996;6:43–53. doi: 10.1097/00008571-199602000-00003. [DOI] [PubMed] [Google Scholar]
- 30.Yan L, Otterness DM, Craddock TL, Weinshilboum RM. Mouse liver nicotinamide N-methyltransferase: cDNA cloning, expression, and nucleotide sequence polymorphisms. Biochem. Pharmacol. 1997;54:1139–1149. doi: 10.1016/s0006-2952(97)00325-0. [DOI] [PubMed] [Google Scholar]
- 31.Felsted RL, Chaykin S. N1-Methylnicotinamide Oxidation in a Number of Mammals. J. Biol. Chem. 1967;242:1274–1279. [PubMed] [Google Scholar]
- 32.Ghazalpour A, et al. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm. Genome. 2012;23:680–692. doi: 10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 34.Gao Z, et al. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J. Biol. Chem. 2011;286:22227–22234. doi: 10.1074/jbc.M111.228874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong S, et al. The REGγ Proteasome Regulates Hepatic Lipid Metabolism through Inhibition of Autophagy. Cell Metabolism. 2013;18:380–391. doi: 10.1016/j.cmet.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 37.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michel F. Sanner. Python: A Programming Language for Software Integration and Development. J.Mol. Graphics Mod. 1999;17:55–61. [PubMed] [Google Scholar]
- 39.Chlopicki S, et al. 1-Methylnicotinamide (MNA), a primary metabolite of nicotinamide, exerts anti-thrombotic activity mediated by a cyclooxygenase-2/prostacyclin pathway. Br. J. Pharmacol. 2007;152:230–239. doi: 10.1038/sj.bjp.0707383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryniarski K, Biedron R, Jakubowski A, Chlopicki S, Marcinkiewicz J. Anti-inflammatory effect of 1-methylnicotinamide in contact hypersensitivity to oxazolone in mice; involvement of prostacyclin. Eur. J. Pharmacol. 2008;578:332–338. doi: 10.1016/j.ejphar.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Bartuś M, et al. 1-Methylnicotinamide (MNA) prevents endothelial dysfunction in hypertriglyceridemic and diabetic rats. Pharmacol Rep. 2008;60:127–138. [PubMed] [Google Scholar]
- 42.Li Y, et al. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. 2011;25:1664–1679. doi: 10.1096/fj.10-173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kannt A, et al. Association of nicotinamide-N-methyltransferase mRNA expression in human adipose tissue and the plasma concentration of its product, 1-methylnicotinamide, with insulin resistance. Diabetologia. 2015 doi: 10.1007/s00125-014-3490-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy AR, et al. A high-fat, ketogenic diet induces a unique metabolic state in mice. American journal of physiology. 2007;292:E1724–E39. doi: 10.1152/ajpendo.00717.2006. [DOI] [PubMed] [Google Scholar]
- 45.Estep PW, Warner JB, Bulyk ML. Short-term calorie restriction in male mice feminizes gene expression and alters key regulators of conserved aging regulatory pathways. PLoS ONE. 2009;4:e5242. doi: 10.1371/journal.pone.0005242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 47.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mato JM, Martínez-Chantar ML, Lu SC. Methionine metabolism and liver disease. Annu. Rev. Nutr. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- 49.Chalkiadaki A, Guarente L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012;16:180–188. doi: 10.1016/j.cmet.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng L, et al. Ubiquitinated Sirtuin 1 (SIRT1) Function Is Modulated during DNA Damage-Induced Cell Death and Survival. J. Biol. Chem. 2015 doi: 10.1074/jbc.M114.612796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmeisser K, et al. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat Chem Biol. 2013;9:693–700. doi: 10.1038/nchembio.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 53.Kemper JK, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Method-Only References
- 54.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.