Abstract
Brain-derived neurotrophic factor (BDNF) has been shown to be important for neuronal survival and synaptic plasticity in the hippocampus in non-human animals. The Val66Met polymorphism in the BDNF gene, involving a valine (Val) to methionine (Met) substitution at codon 66, has been associated with lower BDNF secretion in vitro. However, there have been mixed results regarding associations between either circulating BDNF or the BDNF Val66Met polymorphism with hippocampal volume and memory in humans. The current study examined the association of BDNF genotype and plasma BDNF with hippocampal volume and memory in two large independent cohorts of middle-aged and older adults (both cognitively normal and early-stage dementia). Sample sizes ranged from 123 to 649. Measures of the BDNF genotype, plasma BDNF, MRI-based hippocampal volume and memory performance were obtained from the Knight Alzheimer Disease Research Center (ADRC) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI). There were no significant differences between BDNF Met+ and Met- groups on either hippocampal volume or memory in either cohort. In addition, plasma BDNF was not significantly associated with either hippocampal volume or memory in either cohort. Neither age, cognitive status nor gender moderated any of the relationships. Overall, current findings suggest that BDNF genotype and plasma BDNF may not be robust predictors for variance in hippocampal volume and memory in middle age and older adult cohorts.
Introduction
Healthy and pathological aging, particularly Alzheimer’s disease (AD), are associated with shrinkage of the hippocampus and memory decline (Allen, Bruss, Brown, & Damasio, 2005; Barnes et al., 2009; C. Jack et al., 1997; Martin & Gorenstein, 2010; Raz et al., 2005; Raz, Ghisletta, Rodrigue, Kennedy, & Lindenberger, 2010; Zelinski & Burnight, 1997). Multiple environmental, physiological and genetic factors may influence these age effects. Recent work has suggested that brain derived neurotrophic factor (BDNF) may be a contributing factor. BDNF is a neurotrophin that plays an important role in neurogenesis, neuronal survival and synaptic plasticity (Barnabé-Heider & Miller, 2003; Beck, Lindholm, Castrén, & Wree, 1994; Burke et al., 1994; Kojima et al., 2001; J. Lee, Duan, & Mattson, 2002; Bai Lu, 2003). BDNF is highly expressed in the hippocampus (Murer et al., 1999; Murer, Yan, & Raisman-Vozari, 2001), and has been implicated in hippocampal plasticity and in facilitating hippocampus-dependent memory functions (B Lu & Gottschalk, 2000; Ma, Wang, Wu, Wei, & Lee, 1998; Tyler, Alonso, Bramham, & Pozzo-Miller, 2002).
In humans, a Val66Met polymorphism (rs6265) has been identified in the BDNF gene. This single nucleotide polymorphism (SNP) substitutes valine (Val) for methionine (Met) at codon 66. This substitution interferes with intracellular trafficking of BDNF and activity-dependent BDNF secretion (Bath & Lee, 2006; Chen et al., 2004; Egan et al., 2003) in non-human animals. A number of studies have observed smaller hippocampal volumes (Bueller et al., 2006; Frodl et al., 2007; Pezawas et al., 2004; Schofield et al., 2009; but see Richter-Schmidinger et al., 2011; Szeszko et al., 2005) and lower episodic memory performance (Dempster et al., 2005; Egan et al., 2003; Goldberg et al., 2008; Hariri et al., 2003; Ho et al., 2006; Kennedy et al., 2014; Schofield et al., 2009; but see Richter-Schmidinger et al., 2011) in healthy young adult human Met allele carriers.
In contrast, past studies have generally not observed significant associations between the BDNF genotype and hippocampal volume in middle-aged and older adult cohorts (Benjamin et al., 2010; Karnik, Wang, Barch, Morris, & Csernansky, 2010; Miyajima et al., 2008; but see Pezawas et al., 2004). In addition, findings have been inconsistent in regards to episodic memory performance in this age range with some observations of significantly lower performance in Met carriers (Kennedy et al., 2014; Miyajima et al., 2008; Raz, Rodrigue, Kennedy, & Land, 2009), and other reports of no significant differences between Val/Val and Met carrier groups (Benjamin et al., 2010; Harris et al., 2006; Karnik et al., 2010; Tsai et al., 2008). Previous studies have used various methods for determining hippocampal volume, including manual tracing (Bueller et al., 2006; Frodl et al., 2007; Richter-Schmidinger et al., 2011; Szeszko et al., 2005), point counting (Benjamin et al., 2010; Miyajima et al., 2008) and voxel-based morphometry (Pezawas et al., 2004; Schofield et al., 2009). In terms of episodic memory, a variety of measures have been used, but the most commonly used test is the Logical Memory subtest from the Weschler Memory Scales (Dempster et al., 2005; Egan et al., 2003; Harris et al., 2006; Ho et al., 2006; Karnik et al., 2010). For both hippocampal volume and memory, no consistent pattern has been observed for different methods used and whether or not findings with BDNF genotype are significant. Moreover, sample sizes do not appear to systematically differ between studies finding significant differences and those that did not.
In regards to pathological aging, it appears that the BDNF genotype may not confer a greater risk for Alzheimer’s disease (e.g., Bian, Zhang, Zhang, & Zhao, 2005; Combarros, Infante, Llorca, & Berciano, 2004; Desai, Nebes, DeKosky, & Kamboh, 2005; Forlenza et al., 2010; Yu et al., 2008; Zhang et al., 2006; but see Fehér, Juhász, Rimanóczy, Kálmán, & Janka, 2009; Matsushita et al., 2005; Tsai, Hong, Liu, Liu, & Liou, 2006). However, the presence of the Met allele has been associated with a greater risk for cognitive decline in individuals with mild cognitive impairment (MCI;Forlenza et al., 2010), which is considered a potential prodromal stage of AD (Flicker, Ferris, & Reisberg, 1991).
Past work has also examined associations of circulating BDNF (i.e., in serum, plasma or cerebrospinal fluid) with hippocampal volume and memory. A few studies have examined associations between serum BDNF and hippocampal volume or memory in small cohorts of young to middle-aged controls for psychiatric populations, and findings have been mixed (Dias et al., 2009; Eker et al., 2010; Rizos et al., 2011). Notably, advancing age (Erickson et al., 2010; Li et al., 2009; Lommatzsch et al., 2005; Shimada et al., 2014; Ziegenhorn et al., 2007) and Alzheimer disease (Forlenza et al., 2010; Laske et al., 2007; Lee et al., 2009; Li et al., 2009; Yasutake, Kuroda, Yanagawa, Okamura, & Yoneda, 2006; Yu et al., 2008; but see Angelucci et al., 2010; Laske et al., 2006; O’Bryant et al., 2009) have generally been associated with lower circulating BDNF. While positive relationships between circulating BDNF and hippocampal volume in healthy older adults (Erickson et al., 2010) have been reported, other studies have not observed an association (Driscoll et al., 2012). Investigations of the association between circulating BDNF and memory in healthy older adults have demonstrated varying results with reports of positive (Erickson et al., 2010; Komulainen et al., 2008; Li et al., 2009), negative (Forlenza et al., 2010), and non-significant (Driscoll et al., 2012; Gunstad et al., 2008; O’Bryant et al., 2011) correlations. There has also been some limited work demonstrating a positive relationship between circulating BDNF and memory performance in MCI (Yu et al., 2008) and combined cohorts of healthy and MCI individuals (Shimada et al., 2014), but a negative relationship in AD (Forlenza et al., 2010) individuals.
Inconsistent findings in the BDNF and aging literature may relate to a reduced penetrance of BDNF genotype with age. Age-related pathological changes in brain structure and function may take place independent of genetic influence, which then may reduce genetic contribution to variance in hippocampal volume and memory (Erickson, Miller, & Roecklein, 2012). Mixed findings may also be related to limited sample sizes in some investigations that may have increased the likelihood of type II errors, assessment of the memory domain with only one measure of memory, and/or the gender composition of the cohorts. The primary goal of the current study was to assess the associations of the BDNF genotype and plasma BDNF with hippocampal volume and memory in cognitively normal and very mild to mildly demented adults. The present study advances the existing literature by: (1) examining the relationship in two independent cohorts; (2) examining both BDNF genotype and plasma BDNF within the same cohorts; (3) examining both hippocampal volume and memory within the same cohorts; and (4) use of composite estimates of the memory domain rather than analyses with single tasks.
Methods
Participants
Alzheimer’s Disease Neuroimaing Initiative (ADNI)
Data for one cohort used in the current study were obtained from the ADNI database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org.
Participants were classified using the Clinical Dementia Rating (CDR) scale, a reliable and validated protocol for staging dementia (Morris, 1993). CDR 0 corresponds to cognitively normal, whereas CDR 0.5 and 1 indicate very mild and mild dementia, respectively. 536 ADNI participants had both BDNF genotype (67% Val/Val, 30% Val/Met, 2% Met/Met) data and baseline MRI scans that passed quality control (see Table 1 for demographic information). Among these 536 participants, 355 participants had both baseline plasma BDNF data and baseline MRI scans that passed quality control (see Table 1 for demographic information). For memory analyses, data from the 12-month visit were used in the current report because the 12-month visit included a greater number of participants with available data. 649 participants had both BDNF genotype (66% Val/Val, 31% Val/Met, 3% Met/Met) and episodic memory data (see Table 2 for demographic information). 488 participants had both 12-month visit plasma BDNF and psychometric data (see Table 2 for demographic information).
Table 1.
Descriptive statistics for the MRI analyses (ADNI)
| Cognitively Normal | Early-Stage Dementia | |||
|---|---|---|---|---|
|
| ||||
| Genotype | Plasma | Genotype | Plasma | |
| N | 169 | 45 | 367 | 310 |
| Age, years (mean (SD)) | 75 (5) | 74 (6) | 74 (7)+ | 74 (7) |
| Female, n (%) | 81 (48) | 22 (49) | 150 (41) | 120 (39) |
| Education, years (mean (SD)) | 16 (3) | 6 (3) | 15 (3)* | 16 (3) |
| APOEε4+, n (%) | 44 (26) | 5 (11) | 227 (62)** | 187 (60)** |
| ε2/ε2, n (%) | 2 (1) | 0 (0) | 0 (0) | 0 (0) |
| ε2/ε3, n (%) | 22 (13) | 13 (29) | 10 (3) | 9 (3) |
| ε2/ε4, n (%) | 1 (1) | 1 (2) | 6 (2) | 6 (2) |
| ε3/ε3, n (%) | 101 (60) | 27 (60) | 130 (35) | 114 (37) |
| ε3/ε4, n (%) | 41 (24) | 4 (9) | 165 (45) | 134 (43) |
| ε4/ε4, n (%) | 2 (1) | 0 (0) | 56 (15) | 47 (15) |
| CDR 0.5, n (%) | n/a | n/a | 314 (86) | 281 (91) |
| MMSE (mean (SD)) | 29 (1) | 29 (1) | 26 (3)** | 26 (2)** |
| BDNF met+, n (%) | 52 (31) | 15 (33) | 124 (34) | 110 (35) |
| Val/Val, n (%) | 117 (69) | 30 (67) | 243 (66) | 200 (65) |
| Val/Met, n (%) | 48 (28) | 15 (33) | 115 (31) | 101 (33) |
| Met/Met, n (%) | 4 (2) | 0 (0) | 9 (2) | 9 (3) |
| Log plasma BDNF (mean(SD)) | n/a | .32 (.34) | n/a | .27 (.39) |
| Hippocampus, mm3 (mean (SD)) | 7291 (846) | 7452 (805) | 6072 (1037)** | 6195 (1035)** |
Note:
p<.1;
p<.05;
p<.001 for cognitively normal (CDR=0) compared to early-stage dementia (CDR=0.5/1).
Table 2.
Descriptive statistics for the cognitive analyses (ADNI)
| Cognitively Normal | Early Stage Dementia | |||
|---|---|---|---|---|
|
| ||||
| Genotype | Plasma | Genotype | Plasma | |
| N | 189 | 60 | 460 | 388 |
| Age, years (mean (SD)) | 77 (5) | 76 (6) | 76 (7) | 76 (7) |
| Female, n (%) | 90 (48) | 30 (50) | 168 (37)* | 134 (35)* |
| Education, years (mean (SD)) | 16 (3) | 15 (3) | 16 (3) | 16 (3) |
| APOE ε4+, n (%) | 51 (27) | 7 (12) | 266 (58)** | 222 (57)** |
| ε2/ε2, n (%) | 2 (1) | 0 (0) | 0 (0) | 0 (0) |
| ε2/ε3, n (%) | 27 (14) | 16 (27) | 14 (3) | 11 (3) |
| ε2/ε4, n (%) | 2 (1) | 1 (2) | 10 (2) | 8 (2) |
| ε3/ε3, n (%) | 109 (58) | 37 (62) | 180 (39) | 155 (40) |
| ε3/ε4, n (%) | 46 (24) | 6 (10) | 192 (42) | 160 (41) |
| ε4/ε4, n (%) | 3 (2) | 0 (0) | 64 (14) | 54 (14) |
| CDR 0.5, n (%) | n/a | n/a | 349 (76) | 311 (80) |
| MMSE (mean (SD)) | 29 (1) | 29 (1) | 25 (4)** | 25 (4)** |
| BDNF met+, n (%) | 58 (31) | 17 (28) | 160 (35) | 132 (34) |
| Val/Val, n (%) | 131 (69) | 43 (72) | 300 (65) | 256 (66) |
| Val/Met, n (%) | 54 (29) | 16 (27) | 145 (32) | 121 (31) |
| Met/Met, n (%) | 4 (2) | 1 (2) | 15 (3) | 11 (3) |
| Log plasma BDNF (mean(SD)) | n/a | .36 (.32) | n/a | .25 (.40)* |
| WMS-R Logical Memory - immediate (mean (SD)) | 15 (4) | 14 (4) | 7 (4)** | 7 (4)** |
| WMS-R Logical Memory - delayed (mean (SD)) | 13 (4) | 13 (5) | 4 (4)** | 4 (4)** |
| RAVLT - immediate (mean (SD)) | 44 (10) | 42 (9) | 27 (10)** | 27 (10)** |
| RAVLT - delayed (mean (SD)) | 8 (4) | 7 (4) | 2 (3)** | 2 (3)** |
| Standardized memory composite (mean (SD)) | 4.00 (2.48) | 5.20 (2.79) | −1.64 (2.63)** | −.80 (2.89)** |
Note:
p<.1;
p<.05;
p<.001 for cognitively normal (CDR=0) compared to early-stage dementia (CDR=0.5/1).
WMS-R = Wechsler Memory Scale – Revised; RAVLT = Rey Auditory Verbal Learning Test. WMS-R Logical Memory – immediate: Range 0–25; High score = good; WMS-R Logical Memory – delayed: Range 0–25; High score = good; RAVLT – immediate: Range 0–75; High score = good; RAVLT – delayed: Range 0–15; High score = good.
Washington University Knight Alzheimer Disease Research Center (Knight ADRC)
A second cohort of participants was recruited from the Knight ADRC. Participants were 45 to 100 years of age and were screened for neurological or systemic illnesses (e.g., Huntington’s Disease, Parkinson’s Disease). In order to compare with ADNI, participants who were CDR 1 or below were selected for analysis. 302 participants had both BDNF genotype (69% Val/Val, 30% Val/Met, 2% Met/Met) data and MRI scans that passed quality control (see Table 3 for demographic information). Among these 302 participants, 142 had plasma BDNF data within two years of their MRI scan (see Table 3 for demographic information). In addition, 331 participants had both BDNF genotype (66% Val/Val, 32% Val/Met, 2% Met/Met) and data from all three memory tests (see Memory Assessment for more detail; see Table 4 for demographic information). Furthermore, 123 participants had both plasma BDNF and data from all three memory tests. All plasma data were within two years of the memory assessment (see Table 4 for demographic information). BDNF genotype, MRI and memory data from a subset of the CDR 0 participants have been previously reported in Karnik et al., 2010. The Human Research Protection Office at Washington University approved the study and written informed consent was obtained from all participants.
Table 3.
Descriptive statistics for the MRI analyses (ADRC)
| Cognitively Normal | Early Stage Dementia | |||
|---|---|---|---|---|
|
| ||||
| Genotype | Plasma | Genotype | Plasma | |
| N | 233 | 121 | 69 | 21 |
| Age, years (mean (SD)) | 70 (10) | 71 (7) | 77 (6)** | 75 (5)* |
| Female, n (%) | 151 (65) | 79 (65) | 41 (59) | 12 (57) |
| Education, years (mean (SD)) | 16 (3) | 15 (3) | 14 (3)** | 14 (3)* |
| APOE ε4+, n (%) | 75 (32) | 32 (26) | 39 (57)** | 14 (67)** |
| ε2/ε2, n (%) | 2 (1) | 1 (1) | 0 (0) | 0 (0) |
| ε2/ε3, n (%) | 33 (14) | 19 (16) | 1 (1) | 0 (0) |
| ε2/ε4, n (%) | 8 (3) | 3 (2) | 1 (1) | 1 (5) |
| ε3/ε3, n (%) | 123 (53) | 69 (57) | 29 (42) | 7 (33) |
| ε3/ε4, n (%) | 60 (26) | 26 (21) | 36 (52) | 12 (57) |
| ε4/ε4, n (%) | 7 (3) | 3 (2) | 2 (3) | 1 (5) |
| CDR 0.5, n (%) | n/a | n/a | 46 (67) | 16 (76) |
| MMSE (mean(SD)) | 29 (1) | 29 (1) | 25 (4)** | 27 (3)* |
| BDNF met+, n (%) | 76 (33) | 38 (31) | 19 (28) | 6 (29) |
| Val/Val, n (%) | 157 (67) | 83 (69) | 50 (72) | 15 (71) |
| Val/Met, n (%) | 71 (30) | 34 (28) | 19 (28) | 6 (29) |
| Met/Met, n (%) | 5 (2) | 4 (3) | 0 (0) | 0 (0) |
| Log plasma BDNF (mean(SD)) | n/a | −.43 (.53) | n/a | −.49 (.60) |
| Hippocampus, mm3 (mean (SD)) | 7310 (1027) | 7287 (988) | 5915 (1033)** | 6024 (1083)** |
Note:
p<.1;
p<.05;
p<.001 for cognitively normal (CDR=0) compared to early-stage dementia (CDR=0.5/1).
Table 4.
Descriptive statistics for the cognitive analyses (ADRC)
| Cognitively Normal | Early Stage Dementia | |||
|---|---|---|---|---|
|
| ||||
| Genotype | Plasma | Genotype | Plasma | |
| N | 233 | 95 | 98 | 28 |
| Age, years (mean (SD)) | 75 (8) | 72 (6) | 79 (8)** | 74 (6) |
| Female, n (%) | 141 (61) | 59 (62) | 50 (51) | 14 (50) |
| Education, years (mean (SD)) | 15 (3) | 15 (3) | 15 (3)* | 15 (3) |
| APOE ε4+, n (%) | 68 (29) | 30 (32) | 48 (49)** | 14 (50)+ |
| ε2/ε2, n (%) | 1 (0) | 1 (1) | 0 (0) | 0 (0) |
| ε2/ε3, n (%) | 30 (13) | 14 (15) | 10 (10) | 2 (7) |
| ε2/ε4, n (%) | 7 (3) | 3 (3) | 5 (5) | 1 (4) |
| ε3/ε3, n (%) | 134 (58) | 50 (53) | 40 (41) | 12 (43) |
| ε3/ε4, n (%) | 55 (24) | 25 (26) | 36 (37) | 10 (36) |
| ε4/ε4, n (%) | 6 (3) | 2 (2) | 7 (7) | 3 (11) |
| CDR 0.5, n (%) | n/a | n/a | 71 (72) | 25 (89) |
| MMSE (mean (SD)) | 29 (1) | 29 (1) | 26 (3)** | 27 (2)* |
| BDNF met+, n (%) | 77 (33) | 31 (33) | 35 (36) | 10 (36) |
| Val/Val, n (%) | 156 (67) | 64 (67) | 63 (64) | 18 (64) |
| Val/Met, n (%) | 72 (31) | 29 (31) | 33 (34) | 10 (36) |
| Met/Met, n (%) | 5 (2) | 2 (2) | 2 (2) | 0 (0) |
| Log plasma BDNF, (mean(SD)) | n/a | −.58 (.40) | n/a | −.73 (.37)+ |
| WMS-R Logical Memory - immediate (mean (SD)) | 14 (4) | 13 (4) | 8 (5)** | 9 (5)** |
| WMS-R Logical Memory - delayed (mean (SD)) | 13 (4) | 12 (4) | 6 (5)** | 6 (5)** |
| WMS Associate Learning (mean (SD)) | 15 (4) | 14 (3) | 10 (4)** | 10 (3)** |
| Free and Cued Selective Reminding Test (mean (SD)) | 30 (6) | 30 (6) | 17 (10)** | 18 (8)** |
| Standardized memory composite (mean (SD)) | 1.44 (2.56) | 1.11 (2.60) | −3.43 (3.25)** | −3.75 (3.22)** |
Note:
p<.1;
p<.05;
p<.001 for cognitively normal (CDR=0) compared to early-stage dementia (CDR=0.5/1).
WMS-R = Wechsler Memory Scale – Revised; RAVLT = Rey Auditory Verbal Learning Test. WMS-R Logical Memory – immediate: Range 0–25; High score = good; WMS-R Logical Memory – delayed: Range 0–25; High score = good; WMS Associate Learning: Range 0–21; High score = good; Free and Cued Selective Reminding Test: Range 0–48; High score = good.
Genotyping
ADNI
Detailed procedures for genotyping, including for BDNF Val66Met SNP (rs6265), can be found in previous reports (Saykin et al., 2010; Shen et al., 2010). Briefly, 7mL of blood was taken in EDTA tubes, and genomic DNA was extracted using the QIAamp DNA Blood Maxi Kit (Qiagen, Inc., Valencia, CA). Next, to exclude for any degraded DNA samples, 50ng of genomic DNA was qualitatively analyzed with a 1% Tris-acetate-EDTA agarose gel. Samples were then analyzed using the Illumina Human-610-Quad BeadChip (Illumina, Inc., San Diego, CA). The APOE SNPs (rs429358 and rs7412) were not available on the Illumina Human610-Quad BeadChip. Thus, these SNPs were genotyped separately by polymerase chain reaction amplification and HhaI restriction enzyme digestion. The digested products were ran on a 4% Metaphor Gel and visualized by ethidium bromide staining to analyze the APOE genotype (Potkin et al., 2009; Saykin et al., 2010).
ADRC
Detailed procedures for genotyping in this cohort can be found in previus reports (Cruchaga et al., 2012, 2013). Briefly these DNA samples were genotyped with the Illumina 610 or the Omniexpress chip. Prior to analysis all samples and genotypes underwent stringent QC (Cruchaga et al., 2012). The genotype data for the BDNF val66Met SNP (rs6265) was extracted from the QC’d data. APOE genotyping for these samples was performed using Taqman assays as previously described (Cruchaga et al., 2010).
Plasma BDNF
ADNI
All plasma-based biomarker data were downloaded from the ADNI website (www.loni.ucla.edu/ADNI/) in October 2011. Detailed methods are posted on http://www.adni-info.org/Scientists/ADNIStudyProcedures.aspx. Briefly, blood samples were collected in the morning after an overnight fast, centrifuged to prepare plasma, and frozen on dry ice. Samples underwent an additional freeze-thaw cycle prior to measurement of BDNF. BDNF concentration was analyzed using the multiplex immunoassay panel, which is based upon Luminex’s xMAP Technology by Rules Based Medicine (RBM, Austin, TX). QC was performed on all samples.
ADRC
Similar procedures were performed for the ADRC samples, with the exception that plasma samples underwent only a single free-thaw cycle prior to analysis.
MR Acquisition
ADNI
All MRI data were downloaded from the ADNI website (www.loni.ucla.edu/ADNI/) in October 2011. Detailed methods are posted on http://www.adni-info.org/Scientists/ADNIStudyProcedures.aspx and in a previous report (C. Jack et al., 2008). Imaging was performed using 1.5T scanners (GE, Siemens or Phillips), and T1-weighted sagittal 3D MP-RAGE scans (TR=2400 ms, flip angle=8°, TI=1000 ms, 0.94 × 0.94 × 1.2 mm resolution) were acquired for each participant.
ADRC
Imaging was performed using either a Siemens Vision 1.5T scanner (n=101 for genotype analyses; n=39 for plasma analyses) or a Siemens Trio 3T scanner (n=201 for genotype analyses; n=103 for plasma analyses). For the Vision 1.5 scans, two to four T1-weighted sagittal MP-RAGE scans (TR=9.7 ms, flip angle=10°, TI=20 ms, 1 × 1 × 1.25 mm resolution) were acquired for each participant. For the Trio 3T scans, up to two T1-weighted sagittal MP-RAGE scans (TR=2400ms, flip angle=8°, TI=1000 ms, 1 × 1 × 1 mm resolution) were acquired for each participant.
Regional Volumetry
Hippocampal volume estimates were obtained using the Freesurfer image analysis suite. Total hippocampal volume estimated from the Freesurfer includes roughly the cornu ammonis subfields, dentate gyrus and subiculum. For the ADNI cohort, hippocampal volume estimates were obtained using Freesurfer v4.3. For the ADRC cohort, the Vision 1.5T scans were processed using Freesurfer v5.0, whereas the Trio 3T scans were processed using Freesurfer imaging analysis suite v5.1. The technical details of these procedures are described in prior publications (Dale, Fischl, & Sereno, 1999; Dale & Sereno, 1993; Fischl & Dale, 2000; Fischl, Liu, & Dale, 2001; Fischl et al., 2002; Fischl, Sereno, & Dale, 1999; Fischl, Sereno, Tootell, & Dale, 1999; Fischl et al., 2004). Briefly, this processing includes motion correction and averaging of multiple volumetric T1 weighted images, removal of non-brain tissue using a hybrid watershed/surface deformation procedure, automated Talairach transformation, segmentation of the subcortical white matter and deep gray matter volumetric structures intensity normalization, tessellation of the gray matter white matter boundary, automated topology correction, and surface deformation following intensity gradients to optimally place the gray/white and gray/cerebrospinal fluid borders at the location where the greatest shift in intensity defines the transition to the other tissue class. Once the cortical models are complete, the cerebral cortex is parcellated into units based on gyral and sulcal structure (Desikan et al., 2006; Fischl et al., 2004). Neuroanatomical labels are applied to each voxel based on a probabilistic atlas derived from a manually labeled training set that included older adults (Desikan et al., 2006). This procedure generates anatomical labeling and regional volume estimates with a high correspondence to manually generated labels and its delineation of the hippocampus has been validated in normal aging, mild cognitive impairment, and AD (Cherbuin, Anstey, Réglade-Meslin, & Sachdev, 2009; Fischl et al., 2002; Sánchez-Benavides et al., 2010; Tae, Kim, Lee, Nam, & Kim, 2008).
While there is evidence of reliability of Freesurfer-derived estimates of volume across scanner upgrades, different manufacturers, and number of MP-RAGE acquisitions, variation in field strength and Freesurfer version may introduce slight bias (e.g., Fennema-Notestine et al., 2007; Gronenschild et al., 2012; Han et al., 2006; Jovicich et al., 2009). To address potential biases, scanner type/Freesurfer version was included as a covariate in the analyses if it showed significant associations with the predictor or outcome variables.
As there were no a priori hypotheses regarding laterality effects, hippocampal volumes were summed across hemispheres. Estimated total intracranial volume (ICV; Buckner et al., 2004) was used to adjust hippocampal volumes for body size differences via a formula based on the analyses of covariance approach: Adjusted volume=raw volume-(b x (ICV – mean ICV)), where b is the slope of the regression of the ROI volume on ICV (Jack et al., 1989; Mathalon, Sullivan, Rawles, & Pfefferbaum, 1993). Adjusted hippocampal volume was used as the dependent variable in analyses for both the ADNI and the ADRC cohorts.
Memory Assessment
ADNI
All psychometric data were downloaded from the ADNI website (www.loni.ucla.edu/ADNI/) in October 2011. Detailed methods are posted on http://www.adni-info.org/Scientists/ADNIStudyProcedures.aspx. The episodic memory variables included immediate and delayed recall from the Rey Auditory Verbal Learning Test (Rey, 1964), and immediate and delayed recall from the Wechsler Memory Scale-Revised (WMS-R) Logical Memory subtest (Wechsler, 1987). A composite measure of memory was created by standardizing scores from each task (a total of 4 variables) and averaging the standardized scores.
ADRC
The episodic memory variables included the free recall score from the Free and Cued Selective Reminding Test (Grober, Buschke, Crystal, Bang, & Dresner, 1988), immediate and delayed recall from the WMS-R Logical Memory subtest (Wechsler, 1987), and WMS Associate Learning (Wechsler & Stone, 1973). A composite measure of memory was created by standardizing scores for each task (a total of 4 variables) and averaging the standardized scores.
The psychometric batteries for the ADNI and the ADRC cohorts did not include the exact same set of tests for assessing episodic memory. However, both cohorts included the immediate and delayed recall from the WMS-R Logical Memory subtest. All other memory measures were significantly correlated with this subtest (rs>.5, ps<.0001), suggesting that these tests are measures of a unitary aspect of episodic memory.
Analytical Approach
Outliers
Univariate outliers were defined as values 3.5 standard deviations from the mean of the cohort. For the ADNI cohort, one individual in the Genotype-MRI analyses had an outlier data point. For the ADRC cohort, one individual in the Plasma-Memory analyses had an outlier data point. Unless otherwise specified in the Results section, results were unchanged when outliers were removed.
Covariates
To determine the necessity of the inclusion of age (years), education (years), gender (coded as female=0, male=1), APOE genotype (coded as e4 non-carrier=0, e4 carrier=1), and CDR status (coded as CDR 0=0, CDR>0=1) as covariates in the analyses, the zero-order correlations (or t-tests for dichotomous variables) were examined between these variables and the predictors/outcomes. Pearson chi-square test was used to examine two categorical variables (e.g., BDNF genotype versus CDR status). Each analysis controlled for the covariates that were significantly associated with the predictor and/or outcome variables. In addition, the plasma BDNF-MRI assessment interval and scanner type (1.5T vs. 3T) were considered as additional covariates for ADRC MRI analyses. The mean interval between plasma BDNF and MRI assessments was +/− 4.2 months (SD=5.9) for the ADRC cohort and +/− 0.4 months (SD=1.4) for the ADNI cohort. Similarly, the plasma BDNF-memory assessment interval was considered as an additional covariate for ADRC memory analyses. The mean interval between plasma BDNF and memory assessments was +/− 2.3 months (SD=1.7) for the ADRC cohort and +/− 0.0 months (SD=0.6) for the ADNI cohort.
Statistical Analyses
A series of hierarchical regressions were conducted to address the primary questions regarding the main effects of BDNF genotype and plasma BDNF, and the moderating role of CDR on BDNF effects. The dependent variable was either hippocampal volume or memory performance. The predictor variable was either BDNF genotype, which was coded as 0 (Val/Val homozygotes) or 1 (Met allele carriers), or log-transformed plasma BDNF, which was a continuous variable. To correct for non-normality of residuals, a log transformation was applied to plasma BDNF prior to all analyses. In the regression analyses, covariates were entered in the first step. BDNF (i.e., genotype or plasma BDNF) was entered in the second step, and BDNF x CDR was entered in the last step.
The current study included 77 comparisons, including post-hoc analyses. Therefore, a false discovery rate correction was used to adjust all p-values that were below .1 to correct for multiple comparisons. Adjusted p-values were calculated by: (1) ranking the p-value of each comparison from smallest to largest; (2) multiplying the p-value by the total number of comparisons; and (3) dividing the value obtained from step 2 by the rank of p-value obtained from step 1.
Results
BDNF Genotype Analyses
Hippocampus
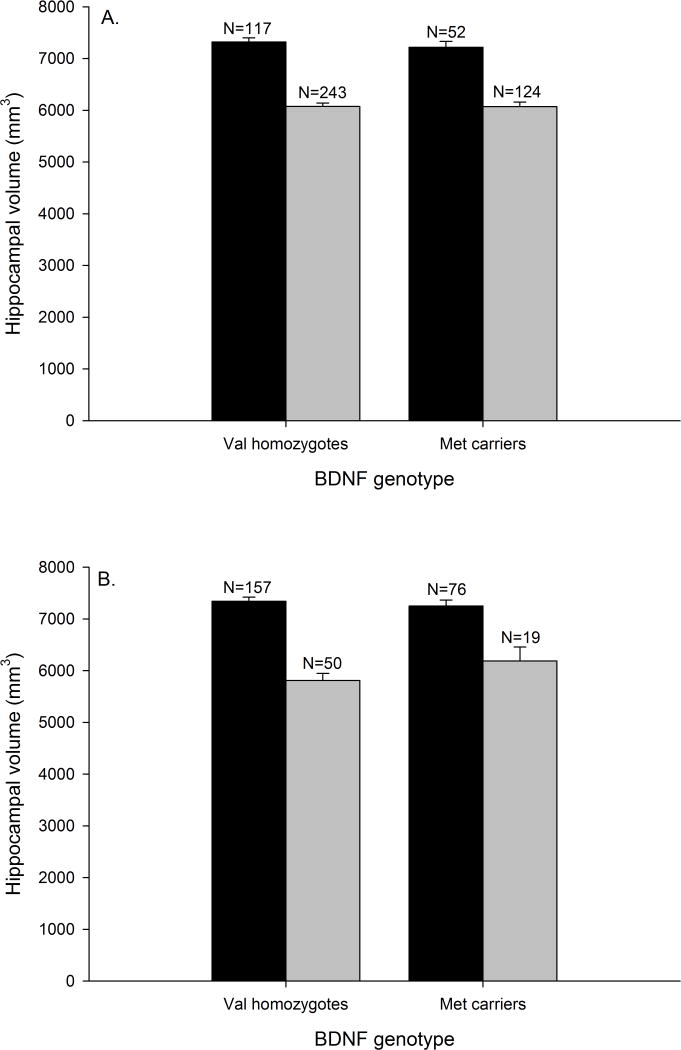
BDNF genotype was not significantly associated with hippocampal volume in either the ADNI or ADRC cohorts without or with covariates (i.e., ADNI: age, gender, APOE genotype, CDR status; ADRC: age, education, gender, APOE genotype, scanner type, CDR status) included (see Table 5, and Fig. 1A and 1B, respectively). Furthermore, the BDNF genotype x CDR interaction was not significant in either the ADNI or ADRC cohorts without or with covariates. Thus, there was not a significant difference in the magnitude of the effect between the cognitively normal and the early-stage dementia groups (see Table 5).
Table 5.
BDNF genotype and plasma BDNF analyses statistics
| Hippocampus | Memory | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Genotype | Plasma | Genotype | Plasma | |||||
|
| ||||||||
| ADNI | ADRC | ADNI | ADRC | ADNI | ADRC | ADNI | ADRC | |
| Unadjusted | ||||||||
| R-sq | .001 | .001 | .000 | .020 | .001 | .001 | .003 | .000 |
| F | .448 | .199 | .048 | 2.848 | .832 | .186 | 1.333 | .025 |
| Beta | −.029 | .026 | .012 | −.141 | −.036 | −.024 | .055 | .014 |
| p-value | .504 | .655 | .826 | .094*** | .362 | .667 | .249 | .876 |
|
| ||||||||
| Adjusted | ||||||||
| R-sq | .000 | .000 | .001 | .002 | .000 | .000 | .000 | .001 |
| F | .203 | .182 | .601 | .375 | .057 | .001 | .015 | .232 |
| Beta | −.016 | .017 | −.035 | −.053 | −.006 | −.001 | .005 | −.033 |
| p-value | .653 | .670 | .439 | .541 | .811 | .978 | .901 | .631 |
|
| ||||||||
| Unadjusted | ||||||||
| BDNFxCDR | ||||||||
| R-sq | .000 | .006 | .000 | .000 | .001 | .002 | .000 | .045 |
| F | .249 | 2.216 | .051 | .027 | .839 | 1.151 | .212 | 9.025 |
| Beta | .037 | .096 | .036 | −.014 | −.052 | .065 | .059 | .244 |
| p-value | .618 | .138 | .821 | .869 | .360 | .284 | .645 | .003** |
|
| ||||||||
| Adjusted | ||||||||
| BDNFxCDR | ||||||||
| R-sq | .000 | .001 | .000 | .000 | .003 | .003 | .000 | .054 |
| F | .251 | .869 | .161 | .086 | 3.561 | 1.760 | .343 | 13.930 |
| Beta | .034 | .048 | −.058 | −.021 | −.103 | .078 | .072 | .271 |
| p-value | .617 | .352 | .689 | .769 | .060+ | .186 | .559 | .000* |
False discovery rate adjusted p=.023;
False discovery rate adjusted p=.116;
False discovery rate adjusted p=.724;
False discovery rate adjusted p=.924
Figure 1.
BDNF genotype and hippocampal volume. A) ADNI cohort (CDR=0: F(1,167)=.520, p=.472; CDR>1: F(1,365)=.001, p=.973); B) ADRC cohort (CDR=0: F(1,231)=.400, p=.528; CDR>1: F(1,67)=1.822, p=.182). Black bars = Cognitively normal, and gray bars = Early-stage dementia. Data represent group means for hippocampal volume, and error bars are standard error of the mean.
Memory
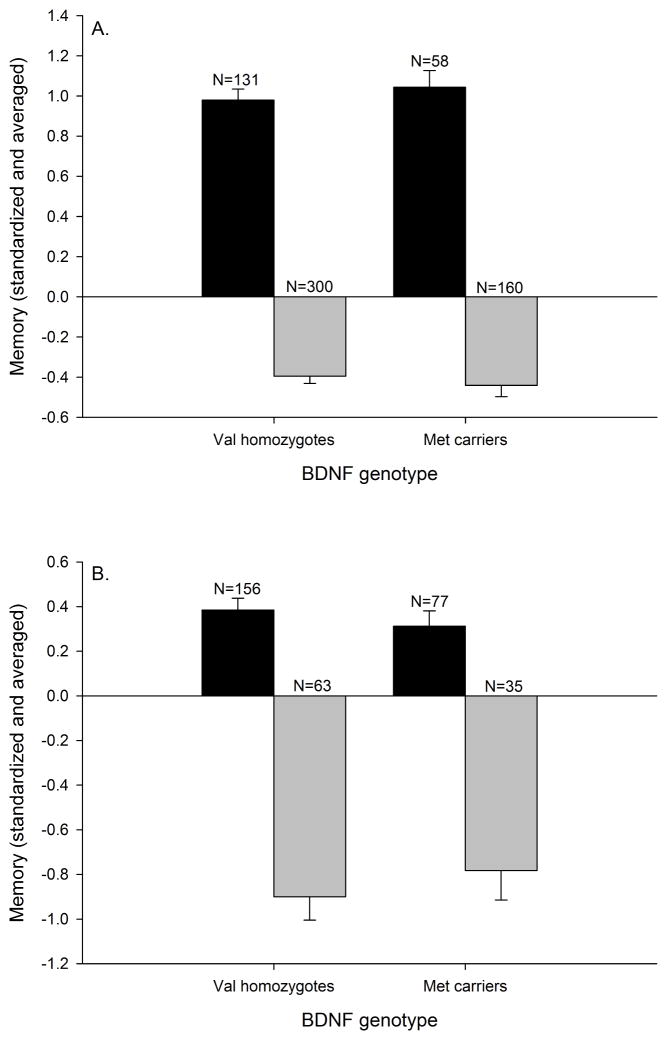
BDNF genotype was not significantly associated with memory in either the ADNI or ADRC cohorts without or with covariates (i.e., ADNI: education, gender, APOE genotype, CDR status; ADRC: age, education, gender, APOE genotype, CDR status) included (see Table 5, and Fig. 2A and 2B, respectively). Furthermore, the BDNF genotype x CDR interaction was not significant in the ADNI cohort without covariates. There was a non-significant trend after controlling for covariates, but this trend was absent after adjusting for multiple comparisons. The BDNF genotype x CDR interaction was not significant in the ADRC cohort without or with covariates. Thus, there was not a significant difference in the magnitude of the effect between the cognitively normal and the early-stage dementia groups (see Table 5).
Figure 2.
BDNF genotype and memory. A) ADNI cohort (CDR=0: F(1,187)=.424, p=.516; CDR>1: F(1,458)=.515, p=.473); B) ADRC cohort (CDR=0: F(1,231)=.647, p=.472; CDR>1: F(1,96)=.464, p=.498). Black bars = Cognitively normal, and gray bars = Early-stage dementia. Data represent group means for the standardized memory composite, and error bars are standard error of the mean.
Plasma BDNF Analyses
Hippocampus
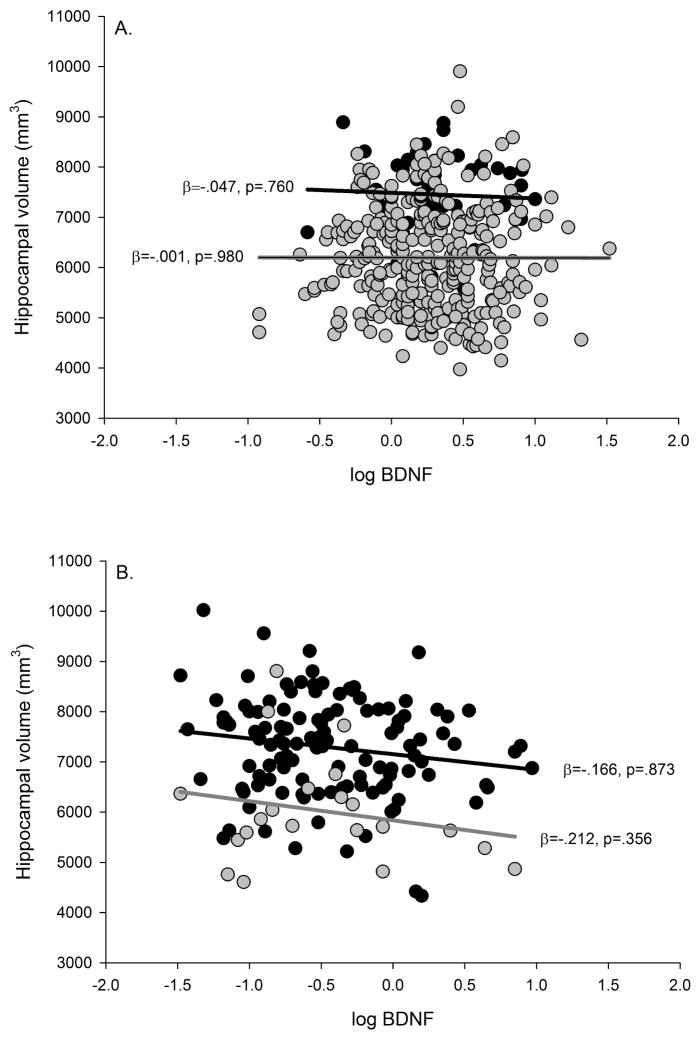
Plasma BDNF was not significantly associated with hippocampal volume in the ADNI cohorts without or with covariates (i.e., age, gender, APOE genotype, CDR status) included. In the ADRC cohort, there was a non-significant trend for an association in the model without covariates, but the trend was absent after adjusting for multiple comparisons, and after controlling for covariates (i.e., age, education, gender, APOE genotype, scanner type, CDR status, time interval between plasma BDNF assessment and MR scans) (see Table 5, Fig. 3A and 3B, respectively). Furthermore, the plasma BDNF x CDR interaction was not significant in either the ADNI or ADRC cohorts without or with covariates. Thus, there was not a significant difference in the magnitude of the effect between the cognitively normal and the early-stage dementia groups (see Table 5).
Figure 3.
Plasma BDNF and hippocampal volume. A) ADNI cohort; B) ADRC cohort. Black circles = Cognitively normal, and gray circles = Early-stage dementia. The black line represents the regression line for the cognitively normal group, and the gray line represents the regression line for the early-stage dementia group. P-values are adjusted FDR corrected.
Memory
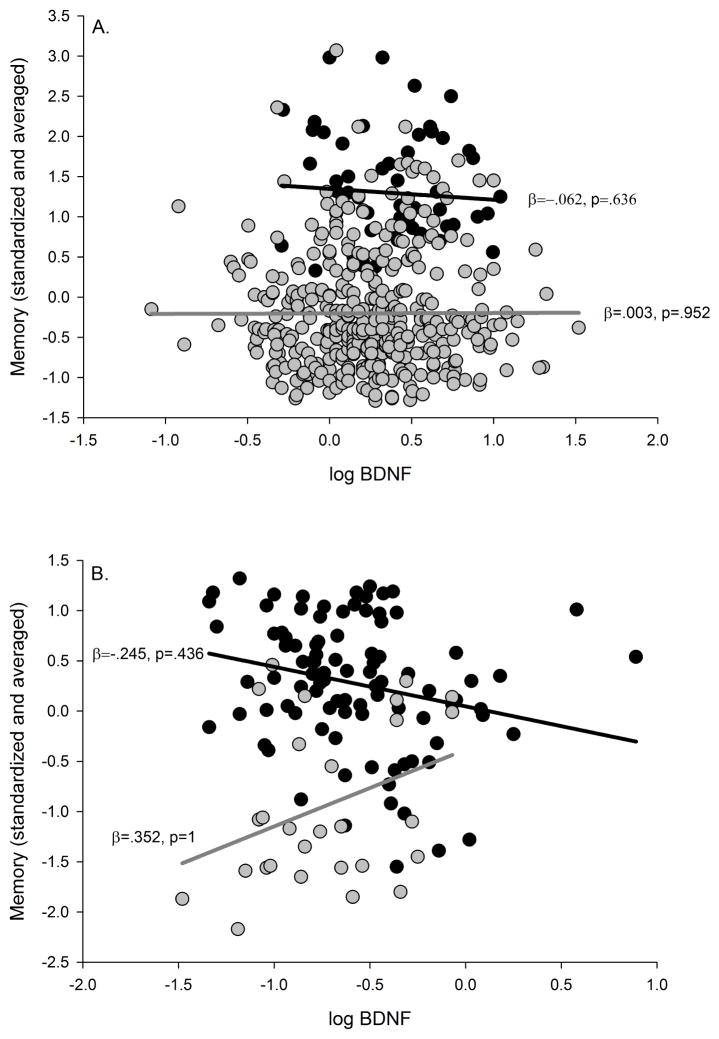
Plasma BDNF was not significantly associated with memory in either the ADNI or ADRC cohorts without or with covariates (i.e., ADNI: education, gender, APOE genotype, CDR status; ADRC: age, education, gender, CDR status) included (see Table 5, Fig. 4A and 4B, respectively). Furthermore, the plasma BDNF x CDR interaction was not significant in the ADNI cohort without or with covariates. Thus, there was not a significant difference in the magnitude of the effect between the cognitively normal and the early-stage dementia groups in the ADNI cohort. The plasma BDNF x CDR interaction was significant in the ADRC cohort without and with covariates. However, the BDNF x CDR interaction without covariates became non-significant after adjusting for multiple comparisons (see Table 5). There was a significant negative association between plasma BDNF and memory in the cognitively normal group and a non-significant positive trend in the early dementia group, but both associations became non-significant after adjusting for multiple comparisons.
Figure 4.
Plasma BDNF and memory. A) ADNI cohort; B) ADRC cohort. Black circles = Cognitively normal, and gray circles = Early-stage dementia. The black line represents the regression line for the cognitively normal group and the gray line represents the regression line for the early-stage dementia group. P-values are adjusted FDR corrected.
Post-hoc Analyses: Associate Learning (ADRC cohort)
There were an additional 319 individuals with scores for the WMS Associate Learning task and BDNF genotype data (see Supplementary Table 1 for demographic information). In addition, there were an additional 66 individuals with scores from the WMS Associate Learning task and plasma BDNF data (see Supplementary Table 1 for demographic information). In addition, Raz and colleagues (2009) observed a significant effect of the BDNF genotype on an associative memory task. Thus, we performed post-hoc analyses controlling for covariates with these increased sample sizes.
BDNF Genotype
BDNF genotype was not significantly associated with associate learning with covariates (i.e., age, education, gender, APOE genotype, CDR status) included. Furthermore, the BDNF genotype x CDR interaction was not significant with covariates (see Supplementary Table 2).
Plasma BDNF
Plasma BDNF was not significantly associated with associate learning with covariates (i.e., age, education, gender, CDR status) included. Furthermore, the plasma BDNF x CDR interaction was not significant with covariates (see Supplementary Table 2).
Post-hoc Analyses: BDNF x Age Interactions
Erickson and colleagues (2012) speculated that there may be lower penetrance of the BDNF genotype effects with advancing age in the context of age effects on brain structure and age-related diseases. Additionally, previous studies have found that circulating BDNF decreases with age in healthy adults (Erickson et al., 2010; Lommatzsch et al., 2005; Ziegenhorn et al., 2007). Thus, we examined whether there were interactive effects between BDNF and age on hippocampal volume and/or episodic memory performance. Hierarchical regressions were conducted to address these questions. In all analyses, the covariates were entered in the first step. Age, CDR status and BDNF (i.e., genotype or plasma) were entered in the second step. The BDNF x age interaction was entered in the third step. The BDNF x CDR and age x CDR interactions were entered in the fourth step. The 3-way interaction was entered in the last step.
BDNF Genotype
The BDNF genotype x age interaction was not significant in the ADNI cohort for hippocampal volume. The BDNF benotype x age interaction was a non-significant trend in the ADRC sample, but the trend was absent after adjusting for multiple comparisons. In addition, the BDNF genotype x age x CDR interaction was not significant in either the ADNI or ADRC cohorts for hippocampal volume.
The BDNF genotype x age interaction was not significant in either the ADNI or ADRC cohorts for memory performance. In addition, the BDNF genotype x age x CDR interaction was not significant in either the ADNI or ADRC cohorts for memory performance (see Table 6).
Table 6.
Post-hoc analyses statistics
| Hippocampus | Memory | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Genotype | Plasma | Genotype | Plasma | |||||
|
| ||||||||
| ADNI | ADRC | ADNI | ADRC | ADNI | ADRC | ADNI | ADRC | |
| BDNFxAge | ||||||||
| R-sq | .002 | .006 | .004 | .008 | .000 | .004 | .000 | .004 |
| F | 1.520 | 3.662 | 1.817 | 1.945 | .409 | 2.401 | .094 | .868 |
| Beta | .052 | −.090 | .062 | .093 | −.021 | −.074 | .011 | .063 |
| p-value | .218 | .057** | .179 | .166 | .523 | .122 | .759 | .353 |
|
| ||||||||
| BDNFxAgexCDR | ||||||||
| R-sq | .001 | .002 | .000 | .005 | .000 | .000 | .002 | .001 |
| F | .635 | 1.277 | .143 | 1.165 | .035 | .237 | 1.591 | .136 |
| Beta | −.081 | −.060 | −.076 | −.111 | .015 | .029 | −.190 | .030 |
| p-value | .426 | .259 | .706 | .282 | .851 | .627 | .208 | .713 |
|
| ||||||||
| BDNFxGender | ||||||||
| R-sq | .003 | .002 | .000 | .006 | .000 | .000 | .001 | .001 |
| F | 2.524 | 1.481 | .175 | 1.564 | .012 | .269 | .600 | .195 |
| Beta | −.099 | .068 | −.031 | −.100 | −.005 | .032 | −.050 | .039 |
| p-value | .113 | .225 | .676 | .213 | .911 | .604 | .439 | .660 |
|
| ||||||||
| BDNFxGenderxCDR | ||||||||
| R-sq | .002 | .002 | .000 | .002 | .001 | .000 | .004 | .010 |
| F | 1.331 | 1.271 | .004 | .379 | .788 | .012 | 2.895 | 2.611 |
| Beta | −.137 | .074 | .016 | −.052 | −.084 | .010 | −.375 | −.163 |
| p-value | .249 | .261 | .947 | .539 | .375 | .914 | .090* | .109 |
False discovery rate adjusted p=.770;
False discovery rate adjusted p=1
Plasma BDNF
The plasma BDNF x age interaction was not significant in either the ADNI or ADRC cohorts for hippocampal volume. In addition, the plasma BDNF x age x CDR interaction was not significant in either the ADNI or ADRC cohorts for hippocampal volume (See Table 6).
The plasma BDNF x age interaction was not significant in either the ADNI or ADRC cohorts for memory performance. In addition, the plasma BDNF x age x CDR interaction was not significant in either the ADNI or ADRC cohorts for memory performance (see Table 6).
Post-hoc Analyses: BDNF x Gender Interactions
Gender differences have been observed in serum BDNF levels (Shimada et al., 2014), in associations between plasma BDNF and episodic memory performance (Komulainen et al., 2008, and in BDNF genotype-related risk for AD (Fukomoto et al., 2010). Thus, we examined whether there were interactive effects between BDNF and gender on hippocampal volume and/or episodic memory performance using hierarchical regressions. In all analyses, the covariates were entered in the first step. Gender, CDR status and BDNF (i.e., genotype or plasma) were entered in the second step. BDNF x gender was entered in the third step to examine interactive effects between BDNF and gender. BDNF x CDR and gender x CDR were entered in the fourth step. The 3-way interaction was entered in the last step.
BDNF genotype
The BDNF genotype x gender interaction was not significant in either the ADNI or ADRC cohorts for hippocampal volume. In addition, The BDNF genotype x gender x CDR interaction was not significant in either the ADNI or ADRC cohorts for hippocampal volume (See Table 6).
The BDNF genotype x gender interaction was not significant in either the ADNI or ADRC cohorts for memory performance. In addition, the BDNF genotype x gender x CDR interaction was not significant in either the ADNI or ADRC cohorts for memory performance (see Table 6).
Plasma BDNF
The plasma BDNF x gender interaction was not significant in either the ADNI or ADRC cohorts for hippocampal volume. In addition, the plasma BDNF x gender x CDR interaction was not significant in either the ADNI or ADRC cohorts for hippocampal volume (See Table 6).
The plasma BDNF x gender interaction was not significant in either the ADNI or ADRC cohorts for memory performance. In addition, the plasma BDNF x gender x CDR interaction was a non-significant trend in the ADNI, but the trend was lost after adjusting for multiple comparisons. The plasma BDNF x gender x CDR interaction was not significant in the ADRC cohort. This non-significant interaction became a non-significant trend when an outlier was removed, but the trend was lost after adjusting for multiple comparisons (see Table 6).
Discussion
The current literature on the effects of BDNF, including the Val66Met polymorphism and circulating BDNF, on hippocampal volume and memory in humans has yielded mixed results. In the current study, neither BDNF genotype nor plasma BDNF were significantly associated with either hippocampal volume or memory in cognitively normal and early-stage dementia adults. We performed post-hoc testing to further explore what factors may have contributed to the null findings.
Several studies in young adults have observed significant differences in hippocampal volume and memory based on BDNF genotype (Bueller et al., 2006; Egan et al., 2003; Frodl et al., 2007; Hariri et al., 2003; Schofield et al., 2009). A potential reason for the current null findings in the cognitively normal cohorts may be the age of the participants. Past studies in healthy middle-aged and older adult cohorts have also failed to find a difference for hippocampal volume (Benjamin et al., 2010; Karnik et al., 2010; Miyajima et al., 2008; but see Pezawas et al., 2004). While two past studies including middle-aged and older adult cohorts observed worse memory performance for Met carriers (Miyajima et al., 2008; Raz et al., 2009), several others failed to find significant group differences (Benjamin et al., 2010; Harris et al., 2006; Karnik et al., 2010; Raz et al., 2008; Tsai et al., 2008). Thus, incorporating current results, the preponderance of investigations suggests minimal effects of the BDNF genotype on memory performance at later age ranges. Indeed, Erickson and colleagues (2012) have speculated that there may be lower penetrance of the BDNF genotype with advancing age, and other factors may have a stronger influence on brain structure and cognition. Furthermore, it has been suggested that the Met allele may be protective in older adults as there have been indications of better performance in older adult Met carriers in other cognitive domains (Erickson et al., 2012; Harris et al., 2006). While post-hoc analyses did not reveal significant interactive effects of BDNF genotype and age on either hippocampal volume or memory in either cohort after correcting for multiple comparisons, this could relate to the lack of younger adults (e.g., no participants <45 years of age) in the analyses.
There is some indication that Met homozygosity is associated with the lowest memory performance (Egan et al., 2003; Schofield et al., 2009), and so the relative proportion of this group in analyses may influence the degree to which significant results are observed. However, it has been estimated that only about 4% of the US population has the Met/Met genotype (Shimizu et al., 2004), and most studies combine the Val/Met and Met/Met groups. In studies with healthy middle-aged and older adults, the proportion of the Met/Met genotype has varied between 0% to 4% with no apparent differences in terms of whether significant BDNF effects are observed (Karnik et al., 2010; Miyajima et al., 2008; Raz et al., 2009). The current cohorts did not have sufficient Met/Met homozygotes to perform a post-hoc analysis (ADNI hippocampus: 2%; ADNI memory: 3%; ADRC hippocampus: 2%; ADRC memory: 2%). Future investigations could attempt to include a large number of Met/Met homozygotes to examine dose-dependent relationships with brain structure and function.
There were also no significant associations between plasma BDNF and hippocampal volume or memory in cognitively normal individuals in either cohort after correcting for multiple comparisons. Circulating BDNF is reduced with aging (e.g., Erickson et al., 2010; Lommatzsch et al., 2005; Ziegenhorn et al., 2007), and BDNF-induced long-term potentiation (LTP) is impaired in aged compared to younger rats (Gooney, Messaoudi, Maher, Bramham, & Lynch, 2004). Thus, there could be differing associations of circulating BDNF with hippocampal volume and memory with aging. The current study observed a significant negative association between age and plasma BDNF in the ADNI – hippocampus cohort only. Furthermore, we did not observe a significant interactive effect between plasma BDNF and age in either cohort. Again, interactive effects may be observed with the inclusion of a young adult group. Notably, conflicting findings have been observed in young adult cohorts as well, although these were predominately relatively small control cohorts (Dias et al., 2009; Eker et al., 2010; Rizos et al., 2011; Ruiz de Azua et al., 2013).
Previous studies have found no significant difference in circulating BDNF between Val homozygote and Met carriers in healthy (Jiang, Wang, Liu, Zhang & Chen, 2009; Li et al., 2009; Terracciano et al., 2009; Zhou et al., 2011) and demented (Li et al., 2009; Yu et al., 2008) middle-aged and older adults. Similarly, in post-hoc analyses there were no significant differences in plasma BDNF level between Val homozygote and Met carriers in our largest cognitively normal (ADRC – hippocampus) and demented (ADNI – memory) samples. These findings suggest that BDNF genotype may not strongly contribute to concentration of circulating BDNF in humans, and that findings with BDNF genotype may be different from those with circulating BDNF.
The role of BDNF in AD risk and pathophysiology has recently received attention (see Diniz & Teixeira, 2011 for a review). Thus far, the majority of previous studies have found no significant differences between AD individuals and healthy controls in terms of BDNF genotype and/or allele frequencies (e.g., Dias et al., 2009; Tsai et al., 2004; Zhang et al., 2006; but see Fukumoto et al., 2010). On the other hand, prior studies have reported a lower circulating BDNF level in AD individuals compared to healthy controls (e.g., Yasutake et al., 2006; Yu et al., 2008; Forlenza et al., 2010), though some work has observed higher BDNF levels in AD individuals (Angelucci et al., 2010; Laske et al., 2006) or no group differences (O’Bryant et al., 2009). In the current study, there were indications of lower plasma BDNF in early-stage dementia (see Tables). However, there were no significant associations of BDNF with hippocampus or memory in the AD groups in either cohort after correcting for multiple comparisons in the current study. These finding are inconsistent with one study that observed associations between circulating BDNF and memory in a cohort with mild cognitive impairment (Yu et al., 2008). This study consisted of a cohort of Chinese Han, and there is evidence of genotype frequency differences between Asian and European populations (e.g., Petryshen et al., 2010; Shimizu, Hashimoto, & Iyo, 2004). Thus, the cohort in Yu and colleagues (2008) had a higher proportion of Met homozygotes than our cohorts, and this may have contributed to the discrepant results. Higher BDNF has also been associated with slower cognitive decline in AD (Laske et al., 2011), and presence of the Met-allele has been associated with faster progression in individuals with mild cognitive impairment (Forlenza et al., 2010) in relatively small cohorts. Thus, further longitudinal work in larger cohorts may be warranted.
The current study measured circulating BDNF in plasma. It is possible that the particular sample analyzed (whole blood, serum, plasma or platelets) could contribute to conflicting findings (Lommatzsch et al., 2005; Trajkovska et al., 2007). However, both plasma and serum BDNF studies have varied in terms of whether or not significant associations with memory or hippocampus are observed in middle-aged to older adult cohorts (Driscoll et al., 2012; Erickson et al., 2010; Gunstad et al., 2008; Kamulainen et al., 2008; O’Bryant et al., 2011; Yu et al., 2008). In addition, positive associations of both serum and plasma BDNF with brain tissue concentrations in the prefrontal cortex and hippocampus have been observed (Elfving et al., 2010; Karege, Schwald, & Cisse, 2002; Klein et al., 2011; Sartorius et al., 2009).
Another potential reason for the null findings in terms of both BDNF genotype and circulating BDNF may be due to the presence of moderators. Multiple factors including stress (Elzinga et al., 2011; Grassi-Oliveira, Stein, Lopes, Teixeira, & Bauer, 2008; Kauer-Sant’Anna et al., 2007), physical exercise (Ferris, Williams, & Shen, 2007; Gold et al., 2003; Seifert et al., 2010; Zoladz et al., 2008), anti-depressant medications (Chen, Dowlatshahi, MacQueen, Wang, & Young, 2001; Shimizu et al., 2003), weight, gender and menstrual cycle phase (Lommatzsch et al., 2005; Shimada et al., 2014) may influence circulating BDNF concentrations. In addition, gender, exercise and vascular factors (e.g., hypertension) may interact with BDNF genotype (Fukumoto et al., 2010; Kim et al., 2011; Raz et al., 2008, 2009). Furthermore, previous studies have shown an interactive effect between BDNF genotype and early childhood adversity on circulating BDNF in individuals with a history of depression or anxiety (Elzinga et al., 2011) and on hippocampal volume (Carballedo et al., 2013; Gatt et al., 2009). Based on these studies, individuals with Met allele(s) and higher levels of childhood stress may have smaller hippocampal volume or memory performance than those with Met allele(s) but without childhood adversity. The current cohorts did not have data on early life stress experiences. It is possible that there was a mixture of individuals with high and low level of childhood adversity, contributing to null finding when these individuals are examined together. The current study did examine the interactive effect of BDNF and gender, and post-hoc analyses in the current cohorts did not reveal any significant moderating influence of gender. Future investigation could systematically examine the moderating effects of lifestyle and health factors to further elucidate the relationship between BDNF and brain structure and function.
Sample size is another issue to be considered as a contributing factor to the lack of significant effects. The current sample sizes for the BDNF genotype and hippocampus analyses were larger than those in the existing literature (Ns=36–173) (Benjamin et al., 2010; Bueller et al., 2006; Frodl et al., 2007; Karnik et al., 2010; Pezawas et al., 2004; Richter-Schmidinger et al., 2011; Schofield et al., 2009; Szeszko et al., 2005), and the BDNF genotype-memory samples were similar or larger than all but one of the existing studies (Ns=28–475) (Benjamin et al., 2010; Dempster et al., 2005; Egan et al., 2003; Goldberg et al., 2008; Hariri et al., 2003; Hashimoto et al., 2008; Ho et al., 2006; Karnik et al., 2010; Kennedy et al., 2014; Raz et al., 2008 and 2009; Richter-Schmidinger et al., 2011; Schofield et al., 2009; Strauss et al., 2004; Tsai et al., 2008; Yu et al., 2008; but see Miyajima et al., 2008; N=722). In addition, the current sample sizes for the plasma BDNF and hippocampus analyses were similar or larger than in the existing literature (Ns=20–142) (Driscoll et al., 2012; Eker et al., 2010; Erickson et al., 2010; Rizos et al., 2011), and the plasma BDNF-memory cohorts were similar or larger than all but two of the existing studies (Ns=35–429) (Dias et al., 2009; Driscoll et al., 2012; Gunstad et al., 2008; Li et al., 2009; O’Bryant et al., 2010; Ruiz de Azua et al., 2013; Yu et al., 2008; but see Komulainen et al., 2008 (N=1389); Shimada et al., 2014 (N=4463). Importantly, significant associations were not observed in either of the two current cohorts, and the observed beta weights were in the small range (Cohen, Manion, & Morrison, 2007). Prior significant associations were observed in both large (e.g., Komulainen et al., 2008; Shimada et al., 2014) and small (e.g., Bueller et al., 2006; Hariri et al., 2003) sample studies. In a post-hoc analysis with the ADRC cohort, we were able to include additional participants to examine the effects of either BDNF genotype (n=647) or plasma BDNF (n=189) on the Associate Learning task. However, no significant effects were observed. It appears unlikely that sample size was the major limiting issue in the current study.
Furthermore, the ADNI cohort consisted of three racial groups (White, Black, and Other races), and the ADRC cohort consisted of two racial groups (White and Black) (see Supplementary Tables 3 and 4). Previous studies have demonstrated differences in BDNF allele frequency among different racial populations (e.g., Petryshen et al., 2010; Shimizu et al., 2004). Some cohorts of the current study also showed frequency and/or plasma BDNF differences across racial groups. Thus, we examined BDNF effects in the largest racial group (Whites) and observed no differences in terms of results (data not shown).
There are some limitations in the current study that need to be addressed. First, for the ADRC cohort the scanner field strength and the Freesurfer version varied within the cohort. Although this variation was adjusted for in analyses, this may not have been sufficient to overcome any noise that was introduced into the data. In addition, different memory tests were administered across cohorts, which could have resulted in differing estimates of the episodic memory construct. However, our results also suggest that the failure to find an effect in one cohort was not specific to the particular set of memory tasks used for that cohort, but instead a more general phenonemenon observed across cohorts.
Overall, the current study did not find a significant BDNF effect, in terms of either genotype or plasma, on either hippocampal volume or memory. These null results were observed in two independent cohorts with sample sizes similar or larger than those in the existing literature. Thus, our findings suggest that Val66Met BDNF polymorphism and plasma BDNF may not be robust predictors for variance in hippocampal volume or memory in middle age and older adult cohorts. However, longitudinal investigations, and the incorporation of other brain regions and cognitive domains as well as potential moderators, may reveal protective roles of BDNF.
Supplementary Material
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; ; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Rev December 5, 2013 Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
This work was also supported by NIH grants P50 AG05861, P01 AG03991, P01 AG026276. This work was supported by a grant to Washington University from Pfizer. We thank the Clinical Core of the Knight Alzheimer Disease Research Center for participant assessments, the Genetics Core for BDNF and APOE genotyping, and the Biomarker Core of the Adult Children Study for plasma collection and processing.
Footnotes
The authors have no conflicts of interest related to this work.
References
- Allen JS, Bruss J, Brown CK, Damasio H. Normal neuroanatomical variation due to age: the major lobes and a parcellation of the temporal region. Neurobiology of Aging. 2005;26(9):1245–60. doi: 10.1016/j.neurobiolaging.2005.05.023. discussion 1279–82. [DOI] [PubMed] [Google Scholar]
- Angelucci F, Spalletta G, Iulio F, Ciaramella a, Salani F, Varsi a, Bossu P. Alzheimers Disease (AD) and Mild Cognitive Impairment (MCI) Patients are Characterized by Increased BDNF Serum Levels. Current Alzheimer Research. 2010;7(1):15–20. doi: 10.2174/156720510790274473. [DOI] [PubMed] [Google Scholar]
- Barnabé-Heider F, Miller FD. Endogenously produced neurotrophins regulate survival and differentiation of cortical progenitors via distinct signaling pathways. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2003;23(12):5149–60. doi: 10.1523/JNEUROSCI.23-12-05149.2003. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12832539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Bartlett JW, van de Pol La, Loy CT, Scahill RI, Frost C, Fox NC. A meta-analysis of hippocampal atrophy rates in Alzheimer’s disease. Neurobiology of Aging. 2009;30(11):1711–23. doi: 10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath KG, Lee FS. Variant BDNF (Val66Met) impact on brain structure and function. Cognitive, Affective & Behavioral Neuroscience. 2006;6(1):79–85. doi: 10.3758/cabn.6.1.79. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2735856&tool=pmcentrez&rendertype=abstract. [DOI] [PubMed] [Google Scholar]
- Beck T, Lindholm D, Castrén E, Wree a. Brain-derived neurotrophic factor protects against ischemic cell damage in rat hippocampus. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 1994;14(4):689–92. doi: 10.1038/jcbfm.1994.86. [DOI] [PubMed] [Google Scholar]
- Benjamin S, McQuoid DR, Potter GG, Payne ME, MacFall JR, Steffens DC, Taylor WD. The brain-derived neurotrophic factor Val66Met polymorphism, hippocampal volume, and cognitive function in geriatric depression. The American Journal of Geriatric Psychiatry: Official Journal of the American Association for Geriatric Psychiatry. 2010;18(4):323–31. doi: 10.1097/JGP.0b013e3181cabd2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian JT, Zhang JW, Zhang ZX, Zhao HL. Association analysis of brain-derived neurotrophic factor (BDNF) gene 196 A/G polymorphism with Alzheimer’s disease (AD) in mainland Chinese. Neuroscience Letters. 2005;387(1):11–6. doi: 10.1016/j.neulet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. NeuroImage. 2004;23(2):724–38. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Bueller Ja, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, Zubieta JK. BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biological Psychiatry. 2006;59(9):812–5. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Burke MA, Mobley WC, Cho J, Wiegand SJ, Lindsay RM, Mufson EJ, Kordower JH. Loss of developing cholinergic basal forebrain neurons following excitotoxic lesions of the hippocampus: rescue by neurotrophins. Experimental Neurology. 1994;130(2):178–95. doi: 10.1006/exnr.1994.1197. [DOI] [PubMed] [Google Scholar]
- Carballedo A, Morris D, Zill P, Fahey C, Reinhold E, Meisenzahl E, Frodl T. Brain-derived neurotrophic factor Val66Met polymorphism and early life adversity affect hippocampal volume. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2013;162B(2):183–90. doi: 10.1002/ajmg.b.32130. [DOI] [PubMed] [Google Scholar]
- Chen B, Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biological Psychiatry. 2001;50(4):260–5. doi: 10.1016/s0006-3223(01)01083-6. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11522260. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2004;24(18):4401–11. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbuin N, Anstey KJ, Réglade-Meslin C, Sachdev PS. In vivo hippocampal measurement and memory: a comparison of manual tracing and automated segmentation in a large community-based sample. PloS One. 2009;4(4):e5265. doi: 10.1371/journal.pone.0005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L, Manion L, Morrison K. Research Methods in Education. 6. New York: Routledge; 2007. [Google Scholar]
- Combarros O, Infante J, Llorca J, Berciano J. Polymorphism at codon 66 of the brain-derived neurotrophic factor gene is not associated with sporadic Alzheimer’s disease. Dementia and Geriatric Cognitive Disorders. 2004;18(1):55–8. doi: 10.1159/000077736. [DOI] [PubMed] [Google Scholar]
- Dale A, Fischl B, Sereno M. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. Retrieved from http://www.sciencedirect.com/science/article/pii/S1053811998903950. [DOI] [PubMed] [Google Scholar]
- Dale A, Sereno M. Improved localizadon of cortical activity by combining eeg and meg with mri cortical surface reconstruction: A linear approach. Journal of Cognitive Neuroscience. 1993;5(2):162–176. doi: 10.1162/jocn.1993.5.2.162. Retrieved from http://www.mitpressjournals.org/doi/abs/10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Dempster E, Toulopoulou T, McDonald C, Bramon E, Walshe M, Filbey F, Collier Da. Association between BDNF val66 met genotype and episodic memory. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2005;134B(1):73–5. doi: 10.1002/ajmg.b.30150. [DOI] [PubMed] [Google Scholar]
- Desai P, Nebes R, DeKosky ST, Kamboh MI. Investigation of the effect of brain-derived neurotrophic factor (BDNF) polymorphisms on the risk of late-onset Alzheimer’s disease (AD) and quantitative measures of AD progression. Neuroscience Letters. 2005;379(3):229–34. doi: 10.1016/j.neulet.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–80. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dias VV, Brissos S, Frey BN, Andreazza AC, Cardoso C, Kapczinski F. Cognitive function and serum levels of brain-derived neurotrophic factor in patients with bipolar disorder. Bipolar Disorders. 2009;11(6):663–71. doi: 10.1111/j.1399-5618.2009.00733.x. [DOI] [PubMed] [Google Scholar]
- Diniz B, Teixeira A. Brain-derived neurotrophic factor and Alzheimer’s disease: physiopathology and beyond. Neuromolecular Medicine. 2011;13(4):217–222. doi: 10.1007/s12017-011-8154-x. [DOI] [PubMed] [Google Scholar]
- Driscoll I, Martin B, An Y, Maudsley S, Ferrucci L, Mattson MP, Resnick SM. Plasma BDNF is associated with age-related white matter atrophy but not with cognitive function in older, non-demented adults. PloS One. 2012;7(4):e35217. doi: 10.1371/journal.pone.0035217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–69. doi: 10.1016/s0092-8674(03)00035-7. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12553913. [DOI] [PubMed] [Google Scholar]
- Eker C, Kitis O, Taneli F, Eker OD, Ozan E, Yucel K, Gonul AS. Correlation of serum BDNF levels with hippocampal volumes in first episode, medication-free depressed patients. European Archives of Psychiatry and Clinical Neuroscience. 2010;260(7):527–33. doi: 10.1007/s00406-010-0110-5. [DOI] [PubMed] [Google Scholar]
- Elfving B, Plougmann P, Muller H, Mathe A, Rosenberg R, Wegener G. Inverse correlation of brain and blood BDNF levels in a genetic rat model of depression. The International Journal of Neuropsychopharmacology. 2010;13(5):563–572. doi: 10.1017/S1461145709990721. [DOI] [PubMed] [Google Scholar]
- Elzinga BM, Molendijk ML, Oude Voshaar RC, Bus Baa, Prickaerts J, Spinhoven P, Penninx BJWH. The impact of childhood abuse and recent stress on serum brain-derived neurotrophic factor and the moderating role of BDNF Val66Met. Psychopharmacology. 2011;214(1):319–28. doi: 10.1007/s00213-010-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KI, Miller DL, Roecklein Ka. The aging hippocampus: interactions between exercise, depression, and BDNF. The Neuroscientist: A Review Journal Bringing Neurobiology, Neurology and Psychiatry. 2012;18(1):82–97. doi: 10.1177/1073858410397054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KI, Prakash RS, Voss MW, Chaddock L, Heo S, McLaren M, Kramer AF. Brain-Derived Neurotrophic Factor Is Associated with Age-Related Decline in Hippocampal Volume. The Journal of Neuroscience. 2010;30(15):5368–5375. doi: 10.1523/JNEUROSCI.6251-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér A, Juhász A, Rimanóczy A, Kálmán J, Janka Z. Association between BDNF Val66Met polymorphism and Alzheimer disease, dementia with Lewy bodies, and Pick disease. Alzheimer Disease and Associated Disorders. 2009;23(3):224–8. doi: 10.1097/WAD.0b013e318199dd7d. [DOI] [PubMed] [Google Scholar]
- Fennema-Notestine C, Gamst AC, Quinn BT, Pacheco J, Jernigan TL, Thal L, Gollub RL. Feasibility of multi-site clinical structural neuroimaging studies of aging using legacy data. Neuroinformatics. 2007;5(4):235–45. doi: 10.1007/s12021-007-9003-9. [DOI] [PubMed] [Google Scholar]
- Ferris LT, Williams JS, Shen CL. The effect of acute exercise on serum brain-derived neurotrophic factor levels and cognitive function. Medicine and Science in Sports and Exercise. 2007;39(4):728–34. doi: 10.1249/mss.0b013e31802f04c7. [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale A. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(20):11050–5. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale A. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Transactions on Medical Imaging. 2001;20(1):70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–55. doi: 10.1016/s0896-6273(02)00569-x. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11832223. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno M, Dale A. Cortical surface-based analysis: II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. Retrieved from http://www.sciencedirect.com/science/article/pii/S1053811998903962. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno M, Tootell R, Dale A. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human Brain Mapping. 1999;8(4):272–84. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10619420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat D, Dale A. Automatically Parcellating the Human Cerebral Cortex. Cerebral Cortex. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Flicker C, Ferris SH, Reisberg B. Mild cognitive impairment in the elderly: predictors of dementia. Neurology. 1991;41(7):1006–9. doi: 10.1212/wnl.41.7.1006. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/2067629. [DOI] [PubMed] [Google Scholar]
- Forlenza OV, Diniz BS, Teixeira AL, Ojopi EB, Talib LL, Mendonça VA, Gattaz WF. Effect of brain-derived neurotrophic factor Val66Met polymorphism and serum levels on the progression of mild cognitive impairment. The World Journal of Biological Psychiatry: The Official Journal of the World Federation of Societies of Biological Psychiatry. 2010;11(6):774–80. doi: 10.3109/15622971003797241. [DOI] [PubMed] [Google Scholar]
- Frodl T, Schüle C, Schmitt G, Born C, Baghai T, Zill P, Meisenzahl EM. Association of the brain-derived neurotrophic factor Val66Met polymorphism with reduced hippocampal volumes in major depression. Archives of General Psychiatry. 2007;64(4):410–6. doi: 10.1001/archpsyc.64.4.410. [DOI] [PubMed] [Google Scholar]
- Fukumoto N, Fujii T, Combarros O, Kamboh MI, Tsai SJ, Matsushita S, Kunugi H. Sexually dimorphic effect of the Val66Met polymorphism of BDNF on susceptibility to Alzheimer’s disease: New data and meta-analysis. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2010;153B(1):235–42. doi: 10.1002/ajmg.b.30986. [DOI] [PubMed] [Google Scholar]
- Gatt JM, Nemeroff CB, Dobson-Stone C, Paul RH, Bryant Ra, Schofield PR, Williams LM. Interactions between BDNF Val66Met polymorphism and early life stress predict brain and arousal pathways to syndromal depression and anxiety. Molecular Psychiatry. 2009;14(7):681–95. doi: 10.1038/mp.2008.143. [DOI] [PubMed] [Google Scholar]
- Gold SM, Schulz KH, Hartmann S, Mladek M, Lang UE, Hellweg R, Heesen C. Basal serum levels and reactivity of nerve growth factor and brain-derived neurotrophic factor to standardized acute exercise in multiple sclerosis and controls. Journal of Neuroimmunology. 2003;138(1–2):99–105. doi: 10.1016/S0165-5728(03)00121-8. [DOI] [PubMed] [Google Scholar]
- Goldberg TE, Iudicello J, Russo C, Elvevåg B, Straub R, Egan MF, Weinberger DR. BDNF Val66Met polymorphism significantly affects d’ in verbal recognition memory at short and long delays. Biological Psychology. 2008;77(1):20–4. doi: 10.1016/j.biopsycho.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Gooney M, Messaoudi E, Maher FO, Bramham CR, Lynch Ma. BDNF-induced LTP in dentate gyrus is impaired with age: analysis of changes in cell signaling events. Neurobiology of Aging. 2004;25(10):1323–31. doi: 10.1016/j.neurobiolaging.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Grassi-Oliveira R, Stein LM, Lopes RP, Teixeira AL, Bauer ME. Low plasma brain-derived neurotrophic factor and childhood physical neglect are associated with verbal memory impairment in major depression--a preliminary report. Biological Psychiatry. 2008;64(4):281–5. doi: 10.1016/j.biopsych.2008.02.023. [DOI] [PubMed] [Google Scholar]
- Grober E, Buschke H, Crystal H, Bang S, Dresner R. Screening for dementia by memory testing. Neurology. 1988;3 doi: 10.1212/wnl.38.6.900. [DOI] [PubMed] [Google Scholar]
- Gronenschild EHBM, Habets P, Jacobs HIL, Mengelers R, Rozendaal N, van Os J, Marcelis M. The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements. PloS One. 2012;7(6):e38234. doi: 10.1371/journal.pone.0038234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunstad J, Benitez A, Smith J, Glickman E, Spitznagel MB, Alexander T, Murray L. Serum brain-derived neurotrophic factor is associated with cognitive function in healthy older adults. Journal of Geriatric Psychiatry and Neurology. 2008;21(3):166–70. doi: 10.1177/0891988708316860. [DOI] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. NeuroImage. 2006;32(1):180–94. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2003;23(17):6690–4. doi: 10.1523/JNEUROSCI.23-17-06690.2003. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12890761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SE, Fox H, Wright aF, Hayward C, Starr JM, Whalley LJ, Deary IJ. The brain-derived neurotrophic factor Val66Met polymorphism is associated with age-related change in reasoning skills. Molecular Psychiatry. 2006;11(5):505–13. doi: 10.1038/sj.mp.4001799. [DOI] [PubMed] [Google Scholar]
- Ho B, Milev P, O’Leary D, Librant A, Andreasen N, Wassink T. Cognitive and magnetic resonance imaging brain morphometric correlates of brain-derived neurotrophic factor Val66Met gene polymorphism in patients with schizophrenia and healthy volunteers. Archives of …. 2006;63(7):731–740. doi: 10.1001/archpsyc.63.7.731.Cognitive. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C, Bernstein M, Fox N, Thompson P, Alexander G, Harvey D, Weiner M. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CJ, Twomey C, Zinsmeister A, Sharbrough F, Petersen R, Cascino G. Anterior temporal lobes and hippocampal formations: normative volumetric measurements from MR images in young adults. Radiology. 1989;172(2):549–554. doi: 10.1148/radiology.172.2.2748838. Retrieved from http://pubs.rsna.org/doi/abs/10.1148/radiology.172.2.2748838. [DOI] [PubMed] [Google Scholar]
- Jack C, Petersen R, Xu Y, Waring S, O’Brien P, Tangalos E, Kokmen E. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology. 1997;49(3):786–794. doi: 10.1212/wnl.49.3.786. Retrieved from http://www.neurology.org/content/49/3/786.short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, Fischl B. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. NeuroImage. 2009;46(1):177–92. doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karege F, Schwald M, Cisse M. Postnatal developmental profile of brain-derived neurotrophic factor in rat brain and platelets. Neuroscience Letters. 2002;328(3):261–264. doi: 10.1016/S0304-3940(02)00529-3. [DOI] [PubMed] [Google Scholar]
- Karnik MS, Wang L, Barch DM, Morris JC, Csernansky JG. BDNF polymorphism rs6265 and hippocampal structure and memory performance in healthy control subjects. Psychiatry Research. 2010;178(2):425–9. doi: 10.1016/j.psychres.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer-Sant’Anna M, Tramontina J, Andreazza AC, Cereser K, da Costa S, Santin A, Kapczinski F. Traumatic life events in bipolar disorder: impact on BDNF levels and psychopathology. Bipolar Disorders. 2007;9(Suppl 1)(4):128–35. doi: 10.1111/j.1399-5618.2007.00478.x. [DOI] [PubMed] [Google Scholar]
- Kennedy KM, Reese ED, Horn MM, Sizemore AN, Unni AK, Meerbrey ME, Rodrigue KM. BDNF val66met polymorphism affects aging of multiple types of memory. Brain Research. 2014:1–14. doi: 10.1016/j.brainres.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Stewart R, Bae KY, Kim SW, Yang SJ, Park KH, Yoon JS. Role of BDNF val66met polymorphism on the association between physical activity and incident dementia. Neurobiology of Aging. 2011;32(3):551.e5–12. doi: 10.1016/j.neurobiolaging.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Klein A, Williamson R, Santini M, Clemmensen C, Ettrup A, Rios M, Aznar S. Blood BDNF concentrations reflect brain-tissue BDNF levels across species. The International Journal of Neuropsychopharmacology. 2011;14(3):347–353. doi: 10.1017/S1461145710000738. [DOI] [PubMed] [Google Scholar]
- Kojima M, Takei N, Numakawa T, Ishikawa Y, Suzuki S, Matsumoto T, Hatanaka H. Biological characterization and optical imaging of brain-derived neurotrophic factor-green fluorescent protein suggest an activity-dependent local release of brain-derived neurotrophic factor in neurites of cultured hippocampal neurons. Journal of Neuroscience Research. 2001;64(1):1–10. doi: 10.1002/jnr.1080. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11276045. [DOI] [PubMed] [Google Scholar]
- Komulainen P, Pedersen M, Hänninen T, Bruunsgaard H, Lakka Ta, Kivipelto M, Rauramaa R. BDNF is a novel marker of cognitive function in ageing women: the DR’s EXTRA Study. Neurobiology of Learning and Memory. 2008;90(4):596–603. doi: 10.1016/j.nlm.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Laske C, Stellos K, Hoffmann N, Stransky E, Straten G, Eschweiler G, Leyhe T. Higher BDNF serum levels predict slower cognitive decline in Alzheimer’s disease patients. The International Journal of Neuropsychopharmacology. 2011;14(3):399–404. doi: 10.1017/S1461145710001008. [DOI] [PubMed] [Google Scholar]
- Laske C, Stransky E, Leyhe T, Eschweiler GW, Maetzler W, Wittorf A, Schott K. BDNF serum and CSF concentrations in Alzheimer’s disease, normal pressure hydrocephalus and healthy controls. Journal of Psychiatric Research. 2007;41(5):387–94. doi: 10.1016/j.jpsychires.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Laske C, Stransky E, Leyhe T, Eschweiler GW, Wittorf a, Richartz E, Schott K. Stage-dependent BDNF serum concentrations in Alzheimer’s disease. Journal of Neural Transmission (Vienna, Austria: 1996) 2006;113(9):1217–24. doi: 10.1007/s00702-005-0397-y. [DOI] [PubMed] [Google Scholar]
- Lee J, Duan W, Mattson MP. Evidence that brain-derived neurotrophic factor is required for basal neurogenesis and mediates, in part, the enhancement of neurogenesis by dietary restriction in the hippocampus of adult mice. Journal of Neurochemistry. 2002;82(6):1367–75. doi: 10.1046/j.1471-4159.2002.01085.x. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12354284. [DOI] [PubMed] [Google Scholar]
- Lee JG, Shin BS, You YS, Kim JE, Yoon SW, Jeon DW, Kim YH. Decreased serum brain-derived neurotrophic factor levels in elderly korean with dementia. Psychiatry Investigation. 2009;6(4):299–305. doi: 10.4306/pi.2009.6.4.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Peskind ER, Millard SP, Chi P, Sokal I, Yu CE, Montine TJ. Cerebrospinal fluid concentration of brain-derived neurotrophic factor and cognitive function in non-demented subjects. PloS One. 2009;4(5):e5424. doi: 10.1371/journal.pone.0005424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommatzsch M, Zingler D, Schuhbaeck K, Schloetcke K, Zingler C, Schuff-Werner P, Virchow JC. The impact of age, weight and gender on BDNF levels in human platelets and plasma. Neurobiology of Aging. 2005;26(1):115–23. doi: 10.1016/j.neurobiolaging.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Lu B. BDNF and activity-dependent synaptic modulation. Learning & Memory (Cold Spring Harbor, NY) 2003;10(2):86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Gottschalk W. Modulation of hippocampal synaptic transmission and plasticity by neurotrophins. Progress in Brain Research. 2000;128:231–41. doi: 10.1016/S0079-6123(00)28020-5. [DOI] [PubMed] [Google Scholar]
- Ma YL, Wang HL, Wu HC, Wei CL, Lee EH. Brain-derived neurotrophic factor antisense oligonucleotide impairs memory retention and inhibits long-term potentiation in rats. Neuroscience. 1998;82(4):957–67. doi: 10.1016/s0306-4522(97)00325-4. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9466420. [DOI] [PubMed] [Google Scholar]
- Martin J, Gorenstein M. Brocklehurst’s Textbook of Geriatric Medicine and Gerontology. Elsevier; 2010. Brocklehurst’s Textbook of Geriatric Medicine and Gerontology; pp. 170–177. [DOI] [Google Scholar]
- Mathalon DH, Sullivan EV, Rawles JM, Pfefferbaum A. Correction for head size in brain-imaging measurements. Psychiatry Research: Neuroimaging. 1993;50(2):121–139. doi: 10.1016/0925-4927(93)90016-B. [DOI] [PubMed] [Google Scholar]
- Matsushita S, Arai H, Matsui T, Yuzuriha T, Urakami K, Masaki T, Higuchi S. Brain-derived neurotrophic factor gene polymorphisms and Alzheimer’s disease. Journal of Neural Transmission (Vienna, Austria: 1996) 2005;112(5):703–11. doi: 10.1007/s00702-004-0210-3. [DOI] [PubMed] [Google Scholar]
- Miyajima F, Ollier W, Mayes a, Jackson a, Thacker N, Rabbitt P, Payton a. Brain-derived neurotrophic factor polymorphism Val66Met influences cognitive abilities in the elderly. Genes, Brain, and Behavior. 2008;7(4):411–7. doi: 10.1111/j.1601-183X.2007.00363.x. [DOI] [PubMed] [Google Scholar]
- Morris J. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43(11):2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Murer M, Boissiere F, Yan Q, Hunot S, Villares J, Faucheux B, Raisman-Vozari R. An immunohistochemical study of the distribution of brain-derived neurotrophic factor in the adult human brain, with particular reference to Alzheimer’s disease. Neuroscience. 1999;88(4):1015–1032. doi: 10.1016/S0306-4522(98)00219-X. [DOI] [PubMed] [Google Scholar]
- Murer M, Yan Q, Raisman-Vozari R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer’s disease and Parkinson’s disease. Progress in Neurobiology. 2001;63(1):71–124. doi: 10.1016/S0301-0082(00)00014-9. [DOI] [PubMed] [Google Scholar]
- O’Bryant SE, Hobson VL, Hall JR, Barber RC, Zhang S, Johnson L, Diaz-Arrastia R. Serum brain-derived neurotrophic factor levels are specifically associated with memory performance among Alzheimer’s disease cases. Dementia and Geriatric Cognitive Disorders. 2011;31(1):31–6. doi: 10.1159/000321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bryant SEO, Hobson V, Hall JR, Waring SC, Chan W, Massman P Texas Alzheimer’s Research Consortium. Brain-derived neurotrophic factor levels in Alzheimer’s disease. Journal of Alzheimer’s Disease. 2009;17(2):337–341. doi: 10.3233/JAD-2009-1051.Brain-Derived. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryshen T, Sabeti P, Aldinger K, Fry B, Fan J, Schaffner S, Sklar P. Population genetic study of the brain-derived neurotrophic factor (BDNF) gene. Molecular Psychiatry. 2010;15(8):810–815. doi: 10.1038/mp.2009.24.Population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Verchinski Ba, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2004;24(45):10099–102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner Ja, Kruggel F, Fallon JH, Macciardi F. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PloS One. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N, Dahle CL, Rodrigue KM, Kennedy KM, Land SJ, Jacobs BS. Brain-derived neurotrophic factor Val66Met and blood glucose: a synergistic effect on memory. Frontiers in Human Neuroscience. 2008;2(October):12. doi: 10.3389/neuro.09.012.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N, Ghisletta P, Rodrigue KM, Kennedy KM, Lindenberger U. Trajectories of brain aging in middle-aged and older adults: regional and individual differences. NeuroImage. 2010;51(2):501–11. doi: 10.1016/j.neuroimage.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N, Lindenberger U, Rodrigue KM, Kennedy KM, Head D, Williamson A, Acker JD. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cerebral Cortex (New York, NY: 1991) 2005;15(11):1676–89. doi: 10.1093/cercor/bhi044. [DOI] [PubMed] [Google Scholar]
- Raz N, Rodrigue K, Kennedy K, Land S. Genetic and vascular modifiers of age-sensitive cognitive skills: effects of COMT, BDNF, ApoE, and hypertension. Neuropsychology. 2009;23(1):105–116. doi: 10.1037/a0013487.Genetic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey A. L’examen clinique en psychologie. Paris: Presses Universitaires de France; 1964. [Google Scholar]
- Richter-Schmidinger T, Alexopoulos P, Horn M, Maus S, Reichel M, Rhein C, Kornhuber J. Influence of brain-derived neurotrophic-factor and apolipoprotein E genetic variants on hippocampal volume and memory performance in healthy young adults. Journal of Neural Transmission (Vienna, Austria: 1996) 2011;118(2):249–57. doi: 10.1007/s00702-010-0539-8. [DOI] [PubMed] [Google Scholar]
- Rizos EN, Papathanasiou M, Michalopoulou PG, Mazioti a, Douzenis a, Kastania a, Lykouras L. Association of serum BDNF levels with hippocampal volumes in first psychotic episode drug-naive schizophrenic patients. Schizophrenia Research. 2011;129(2–3):201–4. doi: 10.1016/j.schres.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Ruiz de Azua S, Matute C, Stertz L, Mosquera F, Palomino A, de la Rosa I, González-Pinto A. Plasma brain-derived neurotrophic factor levels, learning capacity and cognition in patients with first episode psychosis. BMC Psychiatry. 2013;13:27. doi: 10.1186/1471-244X-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Benavides G, Gómez-Ansón B, Sainz A, Vives Y, Delfino M, Peña-Casanova J. Manual validation of FreeSurfer’s automated hippocampal segmentation in normal aging, mild cognitive impairment, and Alzheimer Disease subjects. Psychiatry Research. 2010;181(3):219–25. doi: 10.1016/j.pscychresns.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Sartorius A, Hellweg R, Litzke J, Vogt M, Dormann C, Vollmayr B, Gass P. Correlations and discrepancies between serum and brain tissue levels of neurotrophins after electroconvulsive treatment in rats. Pharmacopsychiatry. 2009;42(6):270–276. doi: 10.1055/s-0029-1224162. [DOI] [PubMed] [Google Scholar]
- Saykin A, Shen L, Foroud T, Potkin S, Swaminathan S, Kim S, Weiner M. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s & Dementia. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013.Alzheimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PR, Williams LM, Paul RH, Gatt JM, Brown K, Luty A, Gordon E. Disturbances in selective information processing associated with the BDNF Val66Met polymorphism: evidence from cognition, the P300 and fronto-hippocampal systems. Biological Psychology. 2009;80(2):176–88. doi: 10.1016/j.biopsycho.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Seifert T, Brassard P, Wissenberg M, Rasmussen P, Nordby P, Stallknecht B, Secher NH. Endurance training enhances BDNF release from the human brain. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2010;298(2):R372–7. doi: 10.1152/ajpregu.00525.2009. [DOI] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Saykin AJ. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. NeuroImage. 2010;53(3):1051–63. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Makizako H, Doi T, Yoshida D, Tsutsumimoto K, Anan Y, Suzuki T. A large, cross-sectional observational study of serum BDNF, cognitive function, and mild cognitive impairment in the elderly. Frontiers in Aging Neuroscience. 2014;6(April):69. doi: 10.3389/fnagi.2014.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu E, Hashimoto K, Iyo M. Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: the possibility to explain ethnic mental traits. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2004;126B(1):122–3. doi: 10.1002/ajmg.b.20118. [DOI] [PubMed] [Google Scholar]
- Shimizu E, Hashimoto K, Okamura N, Koike K, Komatsu N, Kumakiri C, Iyo M. Alterations of serum levels of brain-derived neurotrophic factor (BDNF) in depressed patients with or without antidepressants. Biological Psychiatry. 2003;54(1):70–75. doi: 10.1016/S0006-3223(03)00181-1. [DOI] [PubMed] [Google Scholar]
- Szeszko PR, Lipsky R, Mentschel C, Robinson D, Gunduz-Bruce H, Sevy S, Malhotra aK. Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Molecular Psychiatry. 2005;10(7):631–6. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- Tae WS, Kim SS, Lee KU, Nam EC, Kim KW. Validation of hippocampal volumes measured using a manual method and two automated methods (FreeSurfer and IBASPM) in chronic major depressive disorder. Neuroradiology. 2008;50(7):569–81. doi: 10.1007/s00234-008-0383-9. [DOI] [PubMed] [Google Scholar]
- Trajkovska V, Marcussen AB, Vinberg M, Hartvig P, Aznar S, Knudsen GM. Measurements of brain-derived neurotrophic factor: methodological aspects and demographical data. Brain Research Bulletin. 2007;73(1–3):143–9. doi: 10.1016/j.brainresbull.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Gau Y-Ta, Liu ME, Hsieh CH, Liou YJ, Hong CJ. Association study of brain-derived neurotrophic factor and apolipoprotein E polymorphisms and cognitive function in aged males without dementia. Neuroscience Letters. 2008;433(2):158–62. doi: 10.1016/j.neulet.2007.12.057. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Hong CJ, Liu HC, Liu TY, Hsu LE, Lin CH. Association analysis of brain-derived neurotrophic factor Val66Met polymorphisms with Alzheimer’s disease and age of onset. Neuropsychobiology. 2004;49(1):10–2. doi: 10.1159/000075332. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Hong CJ, Liu HC, Liu TY, Liou YJ. The brain-derived neurotrophic factor gene as a possible susceptibility candidate for Alzheimer’s disease in a chinese population. Dementia and Geriatric Cognitive Disorders. 2006;21(3):139–43. doi: 10.1159/000090673. [DOI] [PubMed] [Google Scholar]
- Tyler WJ, Alonso M, Bramham CR, Pozzo-Miller LD. From acquisition to consolidation: on the role of brain-derived neurotrophic factor signaling in hippocampal-dependent learning. Learning & Memory (Cold Spring Harbor, NY) 2002;9(5):224–37. doi: 10.1101/lm.51202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. Manual: Wechsler Memory Scale-Revised. San Antonio, Texas: Psychological Corporation; 1987. [Google Scholar]
- Wechsler D, Stone C. Manual: Wechsler Memory Scale. New York: Psychological Corporation; 1973. [Google Scholar]
- Yasutake C, Kuroda K, Yanagawa T, Okamura T, Yoneda H. Serum BDNF, TNF-alpha and IL-1beta levels in dementia patients: comparison between Alzheimer’s disease and vascular dementia. European Archives of Psychiatry and Clinical Neuroscience. 2006;256(7):402–6. doi: 10.1007/s00406-006-0652-8. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhang Z, Shi Y, Bai F, Xie C, Qian Y, Deng L. Association study of the decreased serum BDNF concentrations in amnestic mild cognitive impairment and the Val66Met polymorphism in Chinese Han. The Journal of Clinical …. 2008;69(7):1104–1111. doi: 10.4088/jcp.v69n0710. Retrieved from http://europepmc.org/abstract/MED/18505307. [DOI] [PubMed] [Google Scholar]
- Zelinski EM, Burnight KP. Sixteen-year longitudinal and time lag changes in memory and cognition in older adults. Psychology and Aging. 1997;12(3):503–13. doi: 10.1037//0882-7974.12.3.503. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9308097. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ozbay F, Lappalainen J, Kranzler H, van Dyck C, Charney D, Gelernter J. Brain derived neurotrophic factor (BDNF) gene variants and Alzheimer’s disease, affective disorders, posttraumatic stress disorder, schizophrenia, and substance. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2006;141B(4):387–393. doi: 10.1002/ajmg.b.30332.Brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhorn Aa, Schulte-Herbrüggen O, Danker-Hopfe H, Malbranc M, Hartung HD, Anders D, Hellweg R. Serum neurotrophins--a study on the time course and influencing factors in a large old age sample. Neurobiology of Aging. 2007;28(9):1436–45. doi: 10.1016/j.neurobiolaging.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Zoladz Ja, Pilc a, Majerczak J, Grandys M, Zapart-Bukowska J, Duda K. Endurance training increases plasma brain-derived neurotrophic factor concentration in young healthy men. Journal of Physiology and Pharmacology: An Official Journal of the Polish Physiological Society. 2008;59(Suppl 7):119–32. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/19258661. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.