Abstract
Monocytes represent a class of immune cells that play a key role in the innate and adaptive immune response against infections. One mechanism employed by monocytes for sensing foreign antigens is via toll-like receptors (TLRs)—transmembrane proteins that distinguish classes of foreign pathogens, for example, bacteria (TLR4, 5, and 9) vs. fungi (TLR2) vs. viruses (TLR3, 7, and 8). Binding of antigens activates a signaling cascade through TLR receptors that culminate in secretion of inflammatory cytokines. Detection of these cytokines can provide valuable clinical data for drug developers and disease investigations, but this usually requires a large sample volume and can be technically inefficient with traditional techniques such as flow cytometry, enzyme-linked immunosorbent assay, or luminex. This paper describes an approach whereby antibody arrays for capturing cells and secreted cytokines are encapsulated within a microfluidic device that can be reconfigured to operate in serial or parallel mode. In serial mode, the device represents one long channel that may be perfused with a small volume of minimally processed blood. Once monocytes are captured onto antibody spots imprinted into the floor of the device, the straight channel is reconfigured to form nine individually perfusable chambers. To prove this concept, the microfluidic platform was used to capture monocytes from minimally processed human blood in serial mode and then to stimulate monocytes with different TLR agonists in parallel mode. Three cytokines, tumor necrosis factor-α, interleukin (IL)-6, and IL-10, were detected using anti-cytokine antibody arrays integrated into each of the six chambers. We foresee further use of this device in applications such as pediatric immunology or drug/vaccine testing where it is important to balance small sample volume with the need for high information content.
INTRODUCTION
Upon entry into the human body, a pathogen is met by white blood cells and chemical messengers collectively known as the innate immune system.1 One of the key players in this defensive system to limit the spread of infections is the monocyte. Monocytes are leukocytes that circulate in the blood and can mature into macrophages or dendritic cells and trigger inflammation via secretion of signaling molecules such as cytokines.2,3 To activate cytokine secretion pathways, monocytes possess a group of receptors known as toll-like receptors (TLRs) that recognize common components of foreign pathogens and induce a variety of responses including production of cytokines such as interleukin (IL)-6, IL-10, and tumor necrosis factor (TNF)-α.4,5
Cytokine responses of monocytes to stimulation with TLR ligands have been utilized in various biomedical applications such as the testing of contaminants in drugs and treatments.6–8 For instance, as one of the safety requirements for FDA approval, all biological drugs and treatments in the U.S. must be tested for and meet acceptable levels of endotoxins and pyrogens.9 Hazardous levels of these toxins can lead to fever, inflammation, hepatotoxicity, and/or septic shock in patients and thus warrant notable concern.10,11 Currently, one of the methods used to check for contamination with endotoxins and pyrogens is the monocyte activation test.12 In this assay, monocytes are exposed to the reagent being tested and their cytokines are captured, measured, and compared to controls to determine the level of endotoxicity and pyrogenicity present in the sample.7,13 This test is capable of detecting all known relevant pyrogens to humans and offers a valuable tool to drug developers. In addition, there has been a growing repertoire of clinical studies also characterizing the monocyte cytokine secretion response to TLR but in patients with various diseases such as hepatitis C,14,15 endometrial cancer,16 autism,17 and primary biliary cirrhosis.18,19 Altered cytokine release profiles have been observed in the monocytes of these patients compared to negative healthy controls and have improved our understanding of the relationships between the innate immune system and certain diseases. Thus, characterization of the monocyte TLR response offers considerable clinical value.
The conventional method of monocyte activation and cytokine profiling is primarily comprised of two different immunological techniques: ELISA (enzyme-linked immunosorbent assay) and detection of cytokines using multiplexed immunoassay (e.g., luminex). These two conventional (macroscale) approaches require multiple handling steps, several pieces of equipment, and milliliter quantities of blood. Prior to each experiment, monocytes need to be purified, for example, by using magnetic separation column18 and then transferred into multi-well plates for cytokine profiling by ELISA or multiplex bead assay. While robust and sensitive, ELISA and multiplex assays have some drawbacks. In both of these techniques, cells are stimulated for a given period of time (often 24–48 h) after which media is collected for cytokine profiling. Some of the secreted cytokines have a short half-life due to proteolytic degradation, or they may be taken up by the cells during incubation period while other secreted cytokines may become complexed with many different molecules such as autoantibodies.20–22 Combining short half-life with the fact that certain cytokines are naturally produced at low concentrations may mean that assessing the concentration of various cytokines at the end of a long incubation period, as called for by ELISA-type approaches, may not reveal the presence of important but under-represented cytokines.23 A detection method that continuously captures and protects cytokines may thus be a more efficient and accurate alternative as is demonstrated by reports that have used this approach.23,24
Microfluidics combined with antibody (Ab) capture microarrays may resolve some of the challenges noted above while helping minimize the number of processing steps, processing time, the amount of reagents, and the equipment needed to achieve readout. Examples of recent studies employing a microfluidic platform include studies demonstrating rapid microfiltration and trapping of monocytes followed by in situ stimulation for TNF-α detection25 as well as studies demonstrating SPR-based detection of secreted cytokines from captured monocytes.26 These novel devices, however, did not allow for multiplexed stimulation and cytokine profiling of monocytes and may not be optimal to clinical researchers who are interested in profiling cytokines in response to a range of stimulants while using only a small sample volume. Previously, our laboratory used microarrays of cell- and cytokine-specific Ab spots to capture T-cells subsets in the proximity of cytokine sensors.27–30 In these past studies, we employed relatively simple microfluidic devices. The goal of the present study was to combine Ab microarrays for cell capture and cytokine analysis with reconfigurable microfluidics that can toggle between parallel and serial flow. Operating in serial mode, our device was used to capture monocytes from a small volume of (45 μl) of minimally processed blood. Switching the device to parallel mode created up to 9 individually perfusable chambers each containing ∼1000 monocytes as well as Ab spots for detection of three types of cytokines—IL6, IL10, and TNF-α. Operating the device in a parallel mode allowed testing of monocyte responses to 5 different TLR ligands while also incorporating a negative control experiment. Our reconfigurable microfluidic device allows the simplification of monocyte capture and cytokine profiling steps while requiring only 45 μl of blood. In the future, we intend to leverage the advantages of this microfluidic device towards assessing the immune response of infants under various stimulation conditions.
MATERIALS AND METHOD
Materials
Phosphate-buffered saline (PBS, 10×) without Ca2+ and Mg2+ salts, 4% paraformaldehyde (PFA), TWEEN 20, Triton-X 100, poly(ethyleneglycol) diacrylate (PEG-DA) (MW 700), 2-hydroxy-2-methyl-propiophenone (photoinitiator), anhydrous toluene (99.9%), and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (Saint Louis, MO). 3-acryloxypropyl trichlorosilane was purchased from Gelest, Inc. (Morrisville, PA). Purified mouse anti-human CD14 (322A-1 My4) antibodies were obtained from Beckman-Coulter (Fullerton, CA). Purified mouse anti-human IL-6 (clone 6708), IL-10 (clone 127107), TNF-α (clone 28401), and goat anti-human biotinylated polyclonal antibodies for IL-6, IL-10, and TNF-α were purchased from R&D Systems (Minneapolis, MN). Human recombinant IL-6, IL-10, and TNF-α were also purchased from R&D. The isotype controls mouse IgG2a and biotinylated mouse IgG2a antibodies were purchased from Serotec Antibodies (Raleigh, NC). Antibodies used for phenotyping of monocytes were anti-CD36-FITC (CB38) from BD Biosciences and anti-CD14-PE/Cy7 (61D3) from Abcam (Eugene, OR). The TLR agonists, poly I:C, flagellin, and CpG-B, were purchased from Invivogen (San Diego, CA). TLR agonists, LPS and LTA, were purchased from Sigma-Aldrich. Cell culture medium RPMI 1640 1× with L-glutamine was purchased from VWR (Radnor, PA). Fetal bovine serum (FBS), penicillin/streptomycin (PS), and 4,6-diamidino-2-phenylindole (DAPI) were purchased from Invitrogen (Carlsbad, CA). Saint-Gobain formula 3350 tygon tubing (1/32 in. ID), BD precision glide needles (20 gauge), butterfly needles (23 gauge), heparin-coated Vacutainer collection tubes for blood draws, and lymphoprep were purchased from Fisher Scientific (Waltham, MA). SU-8 negative photoresist and developer were purchased from Microchem (Newton, MA). 4 in. silicon wafers were purchased from University Wafer (South Boston, MA). Photomasks were purchased from CAD/Art Services (Bandon, OR). Sylgard 184 poly-(dimethylsiloxane) (PDMS) and curing agent were purchased from Ellsworth Adhesives (Concord, CA).
Fabrication of reconfigurable microfluidic devices
Standard soft lithography (SU-8) procedures were used to fabricate the reconfigurable two-layer microfluidic device using a procedure similar to previously described methods.31 The design for each layer was drawn in AutoCAD (Autodesk, Inc., San Rafael, CA) and the photomask was printed by CAD/Art services. As shown in Figure 1, the device could be operated in serial and parallel modes. In serial mode, the device had one straight channel 50 mm in length and 2.1 mm in width. In parallel mode, upon release of vacuum, the device was converted into 9 parallel chambers: each chamber 1.95 mm in length and 2.1 mm in width. The height of parallel chambers was 50 μm, while the height of a serial chamber was ∼100 μm. In brief, photoresist was spun-coated onto a silicon wafer and UV-patterned with a mask to create a mold with 50 μm height features. The spin speeds and exposure times were selected based on manufacturer's settings. Subsequently, PDMS was mixed in a 1:10 ratio of curing agent to elastomer and poured into each mold. Approximately 25–30 ml of PDMS mixture was used for the top layer while roughly 1–2 ml was used for the bottom layer to make it easily lifted. Both pieces were baked for approximately 1.5 h at 70 °C. After curing, a hole was punched through the control layer to create a vacuum insert. To prevent solution from leaking in-between layers, PDMS layers were irreversibly bound together.32 Each layer was plasma treated with a hand-held corona gun (Rotaloc, Littleton, CO) and manually aligned together by eye. They were then held together and baked at 70 °C for 10 min to complete the bonding. Holes were punched through both devices to form inlets and outlets. Prior to experiments, the device was rendered hydrophilic via oxidation in an oxygen plasma chamber (YES-R3, San Jose, CA) to prevent formation of air bubbles during channel filling.33
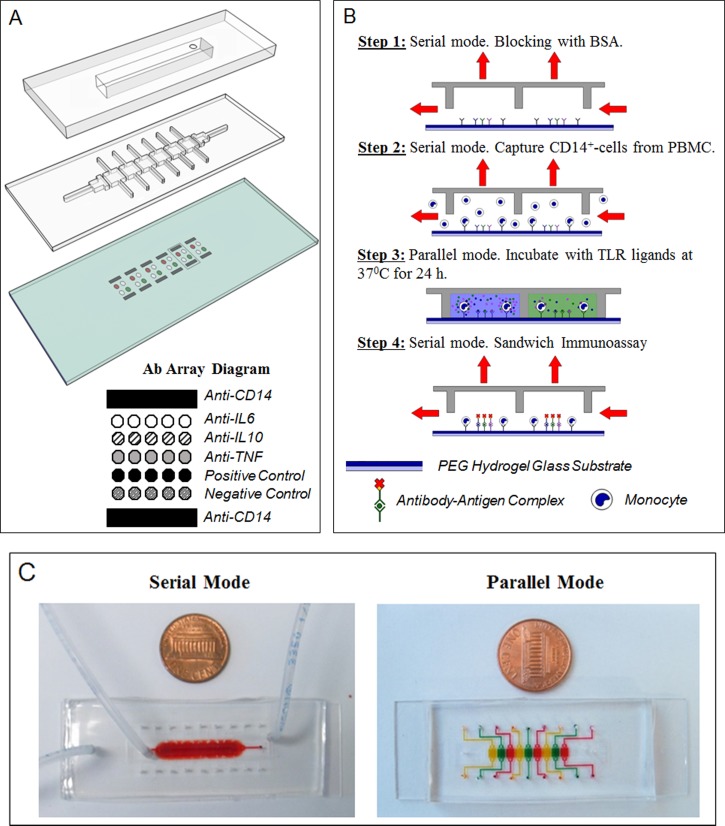
FIG. 1.
Microfluidic platform layout and operation. (a) The microfluidic platform consists of a two-layer PDMS device mounted on top of an antibody imprinted PEG substrate. A top control layer has a vacuum insertable chamber which once activated lifts up the bottom layer. The two layers work together to permit toggling between serial and parallel flow of solution. The hydrogel slide contains 6 set of antibody arrays containing IgG towards monocytes and cytokines. They were spaced based on the dimensions of each channel to allow each set of array to be comfortably enclosed by each respective PDMS chamber. Anti-CD14 cell capture spots were printed as interlaced spots to form a wide band of antibody spots adhesive to cells. Cytokine capture spots were printed between cell capture bands and consisted of IgG against IL-6, IL-10, TNF-α, biotinylated, and non-biotinylated isotype controls. (b) Overview of device operation. Step 1: Roof was lifted to create one channel (serial mode). The device was primed and incubated with BSA. Step 2: Mononuclear cells were infused into the device, with monocytes becoming captured on anti-CD14 Ab spots. Step 3: Roof was lowered creating individually addressable compartments. Each compartment contained the same number of cell capture and cytokine detection Ab spots. Each compartment was infused with a different TLR agonist to induce cytokine production from cells. Step 4: After incubation for 24 h, the roof was raised again. A mixture of biotinylated detection Abs and streptavidin conjugate were infused into the channel for immunostaining of cytokine capture spots. (c) Pictures of the device configured to operate in serial and parallel mode. Food dye was used to highlight that in serial mode, the device represents one straight channel, whereas in parallel mode it becomes 6 individually addressable compartments.
Hydrogel-coated substrates with imprinted Ab arrays
Standard microscope slides were plasma cleaned and modified with an acrylated silane as previously described.27 A mixture of PEG-diacrylate (700 MW) and 2% photoinitiator was spun coated at 800 rpm for 4 s onto the functionalized glass substrate, followed by UV exposure (365 nm, 60 mW/cm2) for 30 s to covalently attach a PEG hydrogel layer. The PEG slide was then placed in a lyophilizer (Dry Ice Benchtop Freeze Dry System, Labconco, Kansas City, MO) for 24 h to dehydrate, enhancing the physical adsorption of subsequent protein printing.28 Prior to printing, purified anti-human CD14, IL-6, IL-10, TNF-α, negative control, and biotinylated positive control IgG were each dissolved in 1× PBS at 0.2 mg/ml supplemented with 0.5% v/v BSA and 0.005% v/v tween20. Ab spots were robotically imprinted into the hydrogel surface via a Spotbot3 printer (Sunnyvale, CA) to create Ab microarrays with a merged band of cell capture spots flanking distinct cytokine detection spots (Figure 1(a)). A 150 μm pin was used to print cell capture and cytokine detection spots with 75 μm and 225 μm center to center spacing, respectively. There were 19 interlacing spots (per band) used for monocyte capture and 4 cytokine sensing Ab spots for each cytokine. Interlaced printing of cell capture spots formed a band rather than distinct spots to increase overall surface area for cell capture. Cell capture spots were also double-printed to maximize protein adsorption of cell capture antibodies. Cytokine capture antibody spots were printed in quadruplicates for each cytokine.
Capture and stimulation of cells inside a microfluidic device
As a proof of concept, a microfluidic device was used to analyze monocyte production of IL-6, IL-10, and TNF-α for the various TLR stimuli. Blood was collected from healthy adult donors through venipuncture under sterile conditions with informed consent and approval of the Institutional Review Board of the University of California at Davis (Protocol No. 222894–6). Peripheral blood mononuclear cells (PBMCs) were purified using a ficoll hypaque gradient. Prior to addition of cells, the microfluidic device was first aligned manually onto an antibody microarray PEG substrate by eye and held together with an acrylic clamp. Figures 1(b) and 1(c) depict the experimental overview. The top layer was actuated with vacuum to rinse the channels with PBS in serial. Subsequently, 1% BSA in PBS was incubated inside for 1 h to block regions surrounding antibody spots from nonspecific attachment of cells and proteins. Following another PBS rinse, PBMCs at 1 × 106 cells/ml were infused into the flow channel at 3 μl/min for cell capture. After ∼15 min, unbound cells were washed out and the vacuum was released to reconfigure the device back to parallel mode. This infusion time was found optimal in terms of allowing for capture of sufficient number of cells (∼500 cells per CD14 capture band) with limited nonspecific binding. Cell numbers were estimated by observing microfluidic devices through a microscope. Different stimuli were introduced into each channel. 5 different TLR agonists: 2.5 μg/ml poly I:C, 50 ng/ml flagellin, 3 μM CpG-B, 500 ng/ml LPS, and 500 ng/ml LTA dissolved in RPMI 1640 with 10% FBS and 1% PS were tested. Cells were also incubated in the same media inside a different channel without stimulant as a negative control. Finally, the device was transferred to a 37 °C and 5% CO2 controlled humidified incubator.
After 24 h, the device was taken out and washed in parallel with PBS to prevent downstream contamination of cytokines. The device was switched to serial mode to begin detection of captured cytokines. First, biotinylated anti-cytokine antibodies (5μg/ml) were incubated inside for 60 min. Second, channels were rinsed with PBS and incubated with streptavidin conjugated to Alexa 546 for 30 min. The device was rinsed with PBS to complete the immunostaining. The whole apparatus was then taken apart in PBS and rinsed with de-ionized water to remove any salt crystals. Fluorescence of captured cytokines was measured using a microarray scanner (GenePix 4000B, Molecular Devices, Sunnyvale, CA) and analyzed with GenePix Pro software as described previously.27 To assess the limit of detection and linear range, we challenged Ab arrays with varying concentrations of recombinant, IL-6, IL-10, and TNF-α. Recombinant cytokines were diluted to concentrations ranging from 0.01 ng–100 ng/ml and were infused into the microfluidic device arranged in a parallel mode. After 30 min, the chambers of the microfluidic device were thoroughly washed and then incubated with biotinylated Ab followed by streptavidin-Alexa546. After rinsing, the microfluidic devices were disassembled and glass substrates containing fluorescent Ab spots were analyzed using commercial microarray scanner (Genepix 4000B, Molecular Devices, Sunnyvale, CA). For cell capture purity confirmation experiments, captured monocytes were incubated with anti-CD36-FITC (1:5) and anti-CD14-PE/Cy7 (1:5) for 2 h. Cells were subsequently rinsed with PBS and fixed with 4% PFA in PBS for 15 min. Finally, the cells were permeabilized with 0.05% Triton-X for 5 min and incubated with DAPI for 10 min. Brightfield and fluorescence images of labeled cells were taken on a Zeiss LSM 510 confocal microscope.
RESULTS AND DISCUSSION
This paper describes the integration of reconfigurable microfluidics with Ab arrays for in situ capture of monocytes, treatment with multiple TLR ligands followed by detection of secreted cytokines. Reconfiguring the device between parallel and serial flow allowed exposure of cells to multiple stimulants and detection of multiple secreted cytokines based on the infusion of a small volume of minimally processed blood.
Design and operation of reconfigurable microfluidic device
The microfluidic platform consists of a thick control layer mounted on top of a thin flow layer (Figure 1(a)). The two layers work together to allow toggling of solution flow between serial and parallel directions. The bottom layer functions as the flow layer and contains 6 channels (2100 μm × 1950 μm × 50 μm each) and a set of inlet and outlet flowing perpendicularly. When placed on top of an Ab imprinted hydrogel surface, each channel forms a cup enclosing a volume of ∼204 nl and a set of Ab array spots. When left non-altered (no vacuum), the bottom layer is in its “parallel” configuration with each chamber being disconnected from the other. Inlets and outlets were punched at the ends of each channel to allow each channel to be individually addressable (Figure 1(c)). During incubation of channels with different stimulants, this configuration seals off any convective mixing between channels. After incubation, this configuration also allows each chamber to be washed simultaneously and independently, preventing any downstream contamination of cytokines that may occur once serial flow is reactivated.
The top layer functions as the layer that controls the direction of reagent flow through the device and contains a vacuum insert. When a vacuum is applied, it pulls up the roof of the bottom layer via negative pressure to connect all neighboring anterior channels to each other and to a perpendicular set of inlet and outlet. In this “serial” configuration, solution can be flown through all chambers perpendicularly to allow uniform treatment of channels with a sample or reagent source (Figure 1(c)). As shown by Cheng et al., shear stress is one of the most important parameters in the panning of monocytes from blood suspensions for high specificity, reduced pre-activation, and density of capture.34 A serial arrangement, thus, ensures that uniform shear stress occurs across all chambers during monocyte panning and that antibody cell capture spots are exposed to a shared sample source and homogenous seeding conditions. To revert back to parallel mode, vacuum was removed.
Capture of pure monocytes in reconfigurable microfluidic device
PBMC solution consists of a heterogenous mixture of mononuclear leukocytes purified from whole blood. Monocytes represent only ∼5%–10% of the total leukocyte population and are typically purified via a negative selection kit with the formation of antibodies tetramers targeted against markers on undesired cells or a positive selection kit with magnetic beads targeted against CD14 antigens of desired cells. Purity ratios using the negative selection method, however, typically only reach up to ∼75%–85%.18 Studies examining TLR-activated cytokine secretion in monocytes have found that the presence of these other cells left over from negative selection can sharply skew cytokine release profiles.18 This is likely due to common pattern recognition systems found among other mononuclear cells that can react to similar TLR agonists. Thus, to attain representative cytokine release profiles, a high purity isolation method is required. As for positive magnetic bead isolation, this method has been reported to achieve >95% monocyte purity but requires the usage of an expensive magnetic bead separator or separation columns and like negative selection, contains additional long incubations steps.
Microfluidic devices combined with capture of cells on Ab-modified surfaces require only a small sample volume and allow precise shear stress control in the process of isolating desired leukocyte subsets.34–38 In our study, monocyte-specific CD14 Abs were printed onto a dehydrated PEG hydrogel surface and then integrated to enable precise control of washing conditions. We chose to use CD14 clone My4 because of previous reports in the literature suggesting superior performance of this clone compared to other CD14 and CD36 clones.34 CD14 My4 was reported as most suitable for capturing monocytes while minimizing contamination of CD14+/C36– neutrophils. Using a flow rate of 3 μl/min and a shear stress of 0.3 dyne/cm2, we observed a cell capture density of 0.529 cells/mm2 within ∼15 min on the cell capture sites.
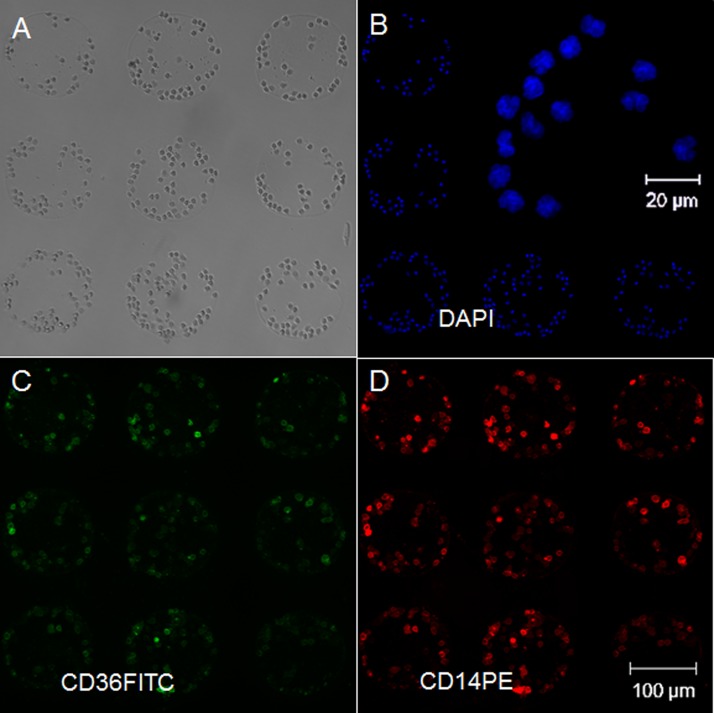
To confirm purity, captured cells were stained with both anti-CD36 FITC and anti-CD14 PE/Cy7 IgG and counted under the microscope. Among the leukocytes in PBMC, only monocytes (significantly), B-cells (weakly), and neutrophils (weakly) express CD14.39,40 Monocytes express CD36, while B-cells and neutrophils do not,41 therefore, immunostaining with anti-CD36 FITC, was used to identify captured cells as monocytes. Depending on the user's technique and patient's sample, neutrophils represent a contaminating fraction of up to 20% of the total cells in PBMC solution.42 Fluorescent images were obtained at 10× magnification using DAPI, fluorescein, and Cy7 excitation/emission filters. As shown in Figure 2, using our method, monocytes were purified on well-defined spotted arrays with ∼97% of captured cells staining for both CD14 PE/CY7 and CD36 FITC. A close-up image of DAPI staining as shown in Figure 2(b) reveals the characteristic lobed shaped monocyte nuclei. Furthermore, CD14+/CD36− cells were not observed on the spots, suggesting that neutrophil and B-cell contamination did not present a problem.
FIG. 2.
Confirming purity of captured cells by immunofluorescent staining. (a) Brightfield image of cells captured on anti-CD14. (b) Cells' nuclei were stained with DAPI. (c) CD14 surface antigens were stained with an antibody conjugated to a PE/Cy7 fluorophore. (d) CD36 surface antigens were stained with anti-CD36 FITC. Overall staining reveals high phenotypic purities of ∼97% CD14+/CD36+ monocyte cell capture.
Overall, our microfluidic monocyte isolation method provides purity similar to that of a standard positive selection protocol for monocytes (97% and 95% for microfluidic and macroscale separation, respectively). However, the microfluidic device described herein decreases the time for cell isolation, allows to the user to minimize the amount of reagents used, and eliminates the need for several different pieces of equipment. Furthermore, purified cells are captured directly inside cytokine profiling chambers and thus will remain at baseline until targeted stimulation.
Stimulation of monocytes with TLR agonists and detection of cytokine release in the microdevice
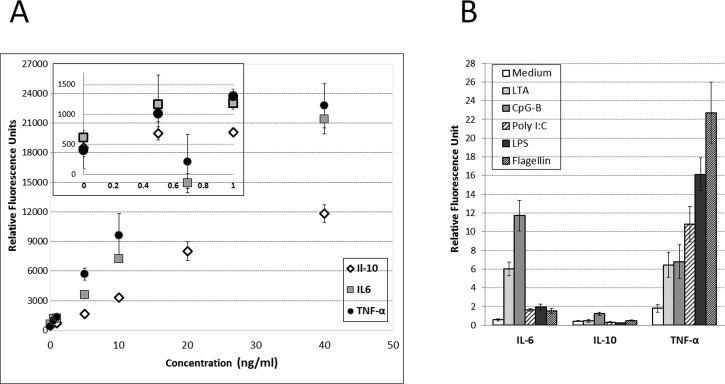
Prior to carrying out cytokine secretion studies, we wanted to characterize responses of Ab microarrays to our cytokines of interest—IL-6, IL-10, and TNF-α. Microfluidic chambers with imprinted Ab arrays were challenged with different cytokine concentration and then stained with fluorescently labeled secondary Abs (Figure 3(a)). Limit of detection, calculated as 3× standard deviation of zero concentration of cytokine, was determined to be 143 pg/ml, 177 pg/ml, and 109 pg/ml for IL-6, IL-10, and TNF-α, respectively. How do these detection limits compare to the levels of cytokines expected to be present in the microchambers? Our team has previously characterized release of TNF-α and IL-6 from monocytes stimulated with LPS.17 Based on these previous results, the sensitivity of Ab arrays described here is sufficient for detecting cytokine production from stimulated monocytes. As noted previously, the microfluidic device may be toggled between serial and parallel perfusion modes. The perfusion of blood and capture of monocytes occurred in serial mode after which the device was reconfigured to create multiple separate compartments for TLR stimulation studies. The TLR agonists tested were LPS (TLR4), LTA (TLR2), CpG-B (TLR9), Flagellin (TLR5), and poly I:C (TLR3). One additional chamber was infused with media without stimulants and served as negative control.
FIG. 3.
Quantification of captured cytokines from calibration curves. Fluorescently labeled captured cytokines were scanned with a microarray scanner and analyzed with Genepix Pro 6.0 to attain relative fluorescence intensity units (RFU). (a) A calibration curve was constructed, plotting relative fluorescence intensity against ng/ml. These curves were attained by incubating known concentrations of recombinant cytokines and immunostaining them in the same microfluidic platform. (b) Captured RFU signal in each channel was normalized by cell number to attain a cytokine release profile of 3 different cytokines from monocytes in 6 different cell conditions.
To quantify cytokine secretions, the microfluidic device was reconfigured once again creating one straight channel that was perfused with solution of biotinylated Abs followed by fluorescent streptavidin. The data presented in Figure 3(b) show that, as expected, cells in the negative control group did not produce appreciable quantities of cytokines. CpG-B stimulated cells (11.71 ± 1.646 RFU/cell) and LTA stimulated cells (6.016 ± 0.731 RFU/cell) produced highest level of of IL-6. Cells stimulated with CpG-B, also, released the most IL-10 (1.220 ± 0.164 RFU/cell). As for TNF-α secretion, the highest levels of this cytokine were observed from monocytes stimulated with flagellin (22.70 ± 3.275 RFU/cell). The results in Figure 3(b) highlight the fact that the microfluidic platform described here may be used to detect several cytokines from purified monocytes stimulated with several different TLR ligands. While there are other platforms with which to measure cytokine levels as well as platforms such as ELISpot that measures single cell secretion of a particular cytokine, the device described herein allows the measurement of cellular immune function from whatever population of cells are of interest in a systematic way, while incorporating multiple simultaneous exposures/perturbations using a very small blood sample.
CONCLUSION
We have developed a robust microfluidic device that facilitates the capture of a specific leukocyte subset (monocytes in our case) using a small volume of minimally processed blood. Further, the design of this device allows the functional assessment of cytokine production following simultaneous exposure to multiple stimulants in parallel. We envision this device to be utilized in future clinical applications to assess immune competency such as monocyte-specific cytokine signatures following interaction with TLR agonists in a pediatric population where only a small volume of blood is available. This device could also be adapted for studies involving small animals where sample volume is limited allowing the animals to remain alive for longitudinal studies. In the future, we propose to enhance this platform to directly capture monocytes from small volume samples of whole blood for early testing of innate immune function. Further, this strategy would extend beyond monocytes and could be adapted to analyze whatever cell population is desired.
ACKNOWLEDGMENTS
We would like to thank the JBJ foundation and the Autism Research Institute for financial support. Additional funding came from “Research Investment in Science and Engineering” project at UC Davis. We thank Dr. Chiamvimonvat for the help with fluorescence microscopy. UC Davis School of Medicine Capital Equipment Grant provided funding for the fluorescence microscope instrument. Scanning of the microarrays was performed at the Expression Analysis Facility of UC Davis.
References
- 1. Parihar A., Eubank T. D., and Doseff A. I., J. Innate Immun. 2(3), 204–215 (2010). 10.1159/000296507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furth R. V., Res. Immun. 149(7–8), 719–720 (1998). 10.1016/S0923-2494(99)80045-5 [DOI] [PubMed] [Google Scholar]
- 3. Chapuis F., Rosenzwajg M., Yagello M., Ekman M., Biberfeld P., and Gluckman J. C., Eur. J. Immunol. 27(2), 431–441 (1997). 10.1002/eji.1830270213 [DOI] [PubMed] [Google Scholar]
- 4. Takeda L. and Akira S., Semin. Immunol. 16(1), 3–9 (2004). 10.1016/j.smim.2003.10.003 [DOI] [PubMed] [Google Scholar]
- 5. Takeda K. and Akira S., J. Dermatol. Sci. 34(2), 73–82 (2004). 10.1016/j.jdermsci.2003.10.002 [DOI] [PubMed] [Google Scholar]
- 6. Hasiwa N., Daneshian M., Bruegger P., Fennrich S., Hochadel A. et al. , Altex 30(2), 169–208 (2013). 10.14573/altex.2013.2.169 [DOI] [PubMed] [Google Scholar]
- 7. Perdomo-Morales R., Pardo-Ruiz Z., Spreitzer I., Lagarto A., and Montag T., Altex 28(3), 227–235 (2011). 10.14573/altex.2011.3.227 [DOI] [PubMed] [Google Scholar]
- 8. Ding J. L. and Ho B., Trends Biotechnol. 19(8), 277–281 (2001). 10.1016/S0167-7799(01)01694-8 [DOI] [PubMed] [Google Scholar]
- 9.Food and Drug Administration et al. , Guidance for Industry: Pyrogen and Endotoxins Testing: Questions and Answers, 2012.
- 10. R. Culbertson, Jr. and Osburn B. I., Vet. Sci. Commun. 4(1), 3–14 (1980). 10.1007/BF02278476 [DOI] [Google Scholar]
- 11. Hartung T., Aaberge I., Berthold S., Carlin G., Charton E. et al. , Altern. Lab. Anim. 29(2), 99–123 (2001). [DOI] [PubMed] [Google Scholar]
- 12. Gaines Das R. E., Brugger P., Patel M., Mistry Y., and Poole S., J. Immunol. Methods 288(1–2), 165–177 (2004). 10.1016/j.jim.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 13. Schindler S., Von Aulock S., Daneshian M., and Hartung T., Altex 26(4), 265–277 (2009). [DOI] [PubMed] [Google Scholar]
- 14. Martin-Blondel G., Gales A., Bernad J., Cuzin L., Delobel P. et al. , J. Viral Hepat. 16(7), 485–491 (2009). 10.1111/j.1365-2893.2009.01094.x [DOI] [PubMed] [Google Scholar]
- 15. Villacres M. C., Literat O., DeGiacomo M., Du W., Frederick T. et al. , J. Viral Hepat. 15(2), 137–144 (2008) 10.1111/j.1365-2893.2007.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brooks N., Stojanovska L., Grant P., Apostolopoulos V., McDonald C. F. et al. , Int. J. Gynecol. Cancer 22(9), 1500–1508 (2012) 10.1097/IGC.0b013e3182249273. [DOI] [PubMed] [Google Scholar]
- 17. Enstrom A. M., Onore C. E., Van de Water J. A., and Ashwood P., Brain Behav. Immun. 24(1), 64–71 (2010). 10.1016/j.bbi.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mao T. K., Lian Z. X., Selmi C., Ichiki Y. et al. , Hepatology 42(4), 802–808 (2005). 10.1002/hep.20859 [DOI] [PubMed] [Google Scholar]
- 19. Zhao J., Zhao S., Zhao G., Liang L., Guo X. et al. , Scand. J. Gastroenterol. 46(4), 485–494 (2011). 10.3109/00365521.2010.539624 [DOI] [PubMed] [Google Scholar]
- 20. Kouwenhoven M., Ozenci V., Teleshova N., Hussein Y., Huang Y. M. et al. , Clin. Diagn. Lab. Immunol. 8(6), 1248–1257 (2001) 10.1128/CDLI.8.6.1248-1257.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Panicker G., Meadows K. S., Lee D. R., Risenbaum R., and Unger E. R., Cytokine 37(2), 176–179 (2007). 10.1016/j.cyto.2007.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barnes A., Lancelet 352(9124), 324–325 (1998). 10.1016/S0140-6736(05)60303-0 [DOI] [PubMed] [Google Scholar]
- 23. Finkelman F. D. and Morris S. C., Int. Immunol. 11(11), 1811–1818 (1999). 10.1093/intimm/11.11.1811 [DOI] [PubMed] [Google Scholar]
- 24. Meierhoff G., Ott P. A., Lehmann P. V., and Schloot N. C., Diabetes Metab. Res. Rev. 18(5), 367–380 (2002). 10.1002/dmrr.320 [DOI] [PubMed] [Google Scholar]
- 25. Huang N. T., Chen W., Oh B. R., Cornell T. T., Shanley T. P. et al. , Lab Chip 12(20), 4093–4101 (2012). 10.1039/c2lc40619e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen W., Huang N. T., Li X., Yu Z. T., Kurabayashi K. et al. , Front. Oncol. 3:98 (2013) 10.3389/fonc.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu H., Stybayeva G., Silangcruz J., Yan J., Ramanculov E. et al. , Anal Chem. 81(19), 8150–8156 (2009). 10.1021/ac901390j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu H., Stybayeva G., Macal M., Ramanculov E., George M. D. et al. , Lab Chip 8(12), 2197–2205 (2008). 10.1039/b810244a [DOI] [PubMed] [Google Scholar]
- 29. Stybayeva G., Mudanyali O., Seo S., Silangcruz J., Macal M. et al. , Anal Chem. 82, 3736–3744 (2010). 10.1021/ac100142a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen A., Vu T., Stybayeva G., Pan T., and Revzin A., Biomicrofluidics 7(2), 24105 (2013). 10.1063/1.4795423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kwa T., Zhou Q., Gao Y., Rahimian A., Kwon L. et al. , Lab Chip 14(10), 1695–1704 (2014). 10.1039/c4lc00037d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haubert K., Drier T., and Beebe D., Lab Chip 6, 1548–1549 (2006). 10.1039/b610567j [DOI] [PubMed] [Google Scholar]
- 33. Duffy D. C., McDonald J. C., Schueller O. J., and Whitesides G. M., Anal. Chem. 70(23), 4974–4984 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- 34. Cheng X., Gupta A., Chen C., Tompkins R. G., Rodriguez W. et al. , Lab Chip 9(10), 1357–1364 (2009). 10.1039/b818813k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sin A., Murthy S. K., Revzin A., Tompkins R. G., and Toner M., Biotechnol. Bioeng. 91(17), 816–826 (2005). 10.1002/bit.20556 [DOI] [PubMed] [Google Scholar]
- 36. Li P., Gao Y., and Pappas D., Anal. Chem. 84(19), 8140–8148 (2012). 10.1021/ac302002a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sekine K., Revzin A., Tompkins R. G., and Toner M., J. Immunol. Methods 313(1–2), 96–109 (2006). 10.1016/j.jim.2006.03.017 [DOI] [PubMed] [Google Scholar]
- 38. Zhu H., Macal M., Jones C. N., George M. D., Dandekar S. et al. , Anal. Chim. Acta 608(2), 186–196 (2008). 10.1016/j.aca.2007.12.021 [DOI] [PubMed] [Google Scholar]
- 39. Antal-Szalmas P., Strijp J. A., Weersink A. J., Verhoef J. et al. , J. Leukocyte Biol. 61(6), 721–728 (1997). [DOI] [PubMed] [Google Scholar]
- 40. Ziegler-Heitbrock H. W., Pechumer H., Petersmann I., Durieux J. J. et al. , Eur. J. Immunol. 24(8), 1937–1940 (1994). 10.1002/eji.1830240835 [DOI] [PubMed] [Google Scholar]
- 41. Huh H. Y., Pearce S. F., Yesner L. M., Schindler J. L. et al. , Blood 87(5), 2020–2028 (1996). [PubMed] [Google Scholar]
- 42. Bolen C. R., Uduman M., and Kleinstein S. H., BMC Bioinf. 12:258 (2011) 10.1186/1471-2105-12-258. [DOI] [PMC free article] [PubMed] [Google Scholar]