Abstract
Replication of the mitochondrial genome depends on the single DNA polymerase (pol gamma). Mutations in the POLG gene, encoding the catalytic subunit of the human polymerase gamma, have been linked to a wide variety of mitochondrial disorders that show remarkable heterogeneity, with more than 200 sequence variants, often very rare, found in patients. The pathogenicity and dominance status of many such mutations remain, however, unclear. Remarkable structural and functional conservation of human POLG and its S. cerevisiae ortholog (Mip1p) led to the development of many successful yeast models, enabling to study the phenotype of putative pathogenic mutations. In a group of patients with suspicion of mitochondrial pathology, we identified five novel POLG sequence variants, four of which (p.Arg869Ter, p.Gln968Glu, p.Thr1053Argfs*6, and p.Val1106Ala), together with one previously known but uncharacterised variant (p.Arg309Cys), were amenable to modelling in yeast. Familial analysis indicated causal relationship of these variants with disease, consistent with autosomal recessive inheritance. To investigate the effect of these sequence changes on mtDNA replication, we obtained the corresponding yeast mip1 alleles (Arg265Cys, Arg672Ter, Arg770Glu, Thr809Ter, and Val863Ala, respectively) and tested their effect on mitochondrial genome stability and replication fidelity. For three of them (Arg265Cys, Arg672Ter, and Thr809Ter), we observed a strong, partially dominant phenotype of a complete loss of functional mtDNA, whereas the remaining two led to partial mtDNA depletion and significant increase in point mutation frequencies. These results show good correlation with the severity of symptoms observed in patients and allow to establish these variants as pathogenic mutations.
Electronic supplementary material
The online version of this article (doi:10.1007/s00439-015-1578-x) contains supplementary material, which is available to authorized users.
Introduction
Mitochondria arose from an α-proteobacterial symbiont in an endosymbiosis event approximately 1.5 billion of years ago (Dyall et al. 2004; Gray et al. 1999). These organelles are present in all aerobic eukaryotic cells, and are crucial for energy production, and many other processes, like Fe-S cluster synthesis, Ca2+ homeostasis, fatty acid oxidation and apoptosis (Aon et al. 2014; Kasahara and Scorrano 2014; Stehling et al. 2014).
As a legacy of their endosymbiotic origin, mitochondria are the only organelles in animal and fungal cells containing their own DNA (mtDNA). Unlike nuclear DNA (nDNA), there are hundreds or even thousands of copies of mtDNA in a single cell. Usually all the copies of mitochondrial DNA are identical (homoplasmy), but mitochondrial DNA molecules with different sequence variants (polymorphic or pathogenic) can coexist with wild-type mtDNA (heteroplasmy) (Aanen et al. 2014).
Human mitochondrial DNA is a circular double-stranded molecule encoding 13 proteins which are subunits of the respiratory chain complexes, 22 tRNAs and 2 rRNAs (Holt et al. 2014; Mishra and Chan 2014). Therefore, the vast majority of 1000–1500 proteins necessary for the proper functioning of the organelle and cellular regulation of mtDNA integrity and copy number are encoded by nDNA and transported into the mitochondrion. The reciprocal influence of nuclear gene expression and processes located in mitochondria is referred to as intergenomic communication (Spinazzola and Zeviani 2007).
Mitochondrial diseases are a group of heterogenic multiorgan disorders, particularly affecting tissues with high energy requirement, like muscles and nerves. They may be caused by mtDNA mutations, nuclear gene mutations (for example in genes encoding subunits of respiratory chain complexes), or by mutations in nuclear genes encoding proteins involved in mtDNA replication and stability, having a secondary effect on mitochondrial DNA amount and quality (Spinazzola and Zeviani 2007).
Replication of human mitochondrial DNA depends entirely on nuclear-encoded proteins. The only DNA polymerase present in human mitochondria is polymerase γ, a member of the type A family of DNA polymerases. It functions as a heterotrimer and consists of a catalytic subunit (POLG) encoded by the POLG gene (15q26.1), and two accessory subunits (POLG2) encoded by the POLG2 (17q23.3) gene. POLG is a 140 kDa protein composed of 1239 aa (the N-terminal 25 aa form the mitochondrial import signal and are cut off during import) and is capable of replicating DNA on its own; the interaction with POLG2 increases polymerase fidelity and processivity. Three domains can be distinguished in the catalytic subunit: the exonuclease domain with the proofreading activity (26–418 aa), the polymerase domain (756–1239 aa), and the linker region (419–755 aa) located between the two domains which is a platform for direct interaction with one of the accessory subunits (Graziewicz et al. 2006; Longley et al. 2005; Stumpf and Copeland 2011). An alternative localization of the POLG domain boundaries was proposed after the publication of the crystal structure of the Pol γ holoenzyme, in which the exonuclease domain encompasses amino acids 171–440, the linker region covers aa 476–785, and the polymerase domain is split into two and comprises amino acids 441–475 and 786–1239 (Lee et al. 2009). The exact domain sequence boundaries are still subject to dispute.
Mutations in the POLG gene may disrupt mtDNA replication, leading to the formation of multiple mtDNA deletions, to depletion (pathological decrease of the mtDNA copy number), or to a combination of these defects, which in turn disrupts the mitochondrial function and energy production (Stumpf and Copeland 2011). POLG mutations can, therefore, lead to a spectrum of mitochondrial diseases with dominant or recessive type of inheritance, which can be divided into three main groups: sporadic/familial progressive external ophthalmoplegia (PEO) (OMIM #157640 and #258450), adult-onset ataxia (OMIM #607459), and Alpers syndrome (OMIM #203700). Mitochondrial diseases attributed to mutations in POLG show a remarkable heterogeneity with, according to the Human DNA Polymerase Gamma Mutation Database (http://tools.niehs.nih.gov/polg/), more than 200 variants known to date, many of them rare.
The budding yeast is widely used as a genetic model system to study mtDNA stability and mutagenesis. Unlike most other microorganisms, S. cerevisiae can stably exist both in a haploid and a diploid state. Recessive mutations can be conveniently isolated and manifested in haploid strains, while complementation tests can be performed in the diploids. Heterozygous mutants allow the effects of dominant alleles to be observed. The most useful feature of these microorganisms in the study of mitochondrial function is that respiratory deficient mutants are viable and the presence of mtDNA is optional. The respiratory deficient phenotype can be easily observed as a formation of the so-called petite colonies on fermentative media, and lack of growth on respiratory-only media (Ephrussi and Slonimski 1955). S. cerevisiae is, therefore, an excellent model for the study of communication between nuclear and mitochondrial genetic systems. Due to the evolutionary conservation of some features of the mitochondrial system, like the mtDNA polymerase, they also may be used as a model for research on human mutations and their role in mtDNA replication (Barrientos 2003; Baruffini et al. 2007; Foury and Kucej 2002; Graziewicz et al. 2006; Longley et al. 2005; Stumpf and Copeland 2011; Lodi et al. 2015).
The yeast mitochondrial DNA polymerase (Mip1) is a nuclear-encoded protein, essential for the maintenance of the mitochondrial genome. It is orthologous to the human polymerase γ, and in spite of the evolutionary distance, its domain structure and amino acid sequence are essentially conserved (Foury 1989; Ropp and Copeland 1996). A yeast strain with its native mitochondrial DNA polymerase replaced by the human polymerase γ is viable and even maintains the functional mitochondrial genome essentially as efficiently as the wild type yeast (Qian et al. 2014). The Mip1 protein consists of four domains: the exonuclease-like domain, the linker region, the polymerase-like domain and the C-terminal domain. The polymerase and exonuclease domains are highly similar to their human equivalents. Unlike human Pol γ, Mip1 does not have any accessory subunits, and does not form a heterotrimer complex (Lucas et al. 2004; Yakubovskaya et al. 2006). Consequently, the linker region is not well conserved, and its sequence shows no significant similarity to that of the human ortholog. Another difference is the presence in Mip1 of a C-terminal domain, absent in human POLG (Viikov et al. 2011, 2012). In spite of those differences between yeast and human mtDNA polymerases, it is still possible to model many of the polymerase γ mutations found in patients with mitochondrial diseases in the yeast protein. As it is with POLG, even a single amino acid substitution in one of the conserved domains of Mip1 can lead to a severe mitochondrial deficiency phenotype. Combining comparable phenotypes with easy and fast analysis makes budding yeast an excellent model for examining pathologies of human polymerase γ (Barrientos 2003; Baruffini et al. 2006, 2007, 2011, 2014; Baruffini and Lodi 2010; Foury and Kucej 2002; Lodi et al. 2015; Stuart et al. 2006; Stricker et al. 2009; Stumpf et al. 2010; Stumpf and Copeland 2011, 2013, 2014; Szczepanowska and Foury 2010).
In search of new possibly pathogenic variants which can be modelled in S. cerevisiae, POLG sequence analysis was conducted for 60 patients with mitochondrial disease. Known well-characterised variants and novel sequence changes located in regions poorly conserved between human and yeast were rejected. We chose four previously undescribed variants for further study. The c.2604 C>T (p.Arg869Ter), c.2901C>G (p.Gln968Glu), c.3155_3156delCA (p.Thr1053Argfs*6) and c.3316 T>C (p.Val1106Ala) are located in the polymerase domain. In addition, one previously known substitution c.924C>T (p.Arg309Cys) located in the exonuclease domain was also included in the study. This mutation was reported once and only vaguely mentioned in the original work, additionally symptoms observed in both patients differ (Amiot et al. 2009). A novel nonsense mutation p.Arg869Ter in the polymerase domain serves as a control of the usefulness of our yeast model.
Materials and methods
Population analysis of novel POLG mutations
Analysis of the presence of the mtDNA deletions was conducted by long range PCR as described previously (Kierdaszuk et al. 2009). For patients with confirmed multiple mitochondrial DNA deletions, depletion or strong clinical indication, we performed POLG sequence analysis similar to (Filosto et al. 2003) (primer sequences are provided in Online Resource 1).
Reactions were performed in 25 µl final volume containing 0.16 µM of each of the forward and reverse primer, 0.4 mM of dNTPs, 1 U of Taq polymerase (A&A Biotechnology) and 100 ng of DNA. PCR conditions were 94° for 5 min, followed by 35 cycles of 94° for 1 min, appropriate annealing temperature (64° for POLG1_1, 6, 7, 8, 9, 10 C and 58° for POLG1_2, 3, 4, 5, 11, 12) for 1 min, 72° for 1 min and final elongation at 72° for 7 min. Sequencing was performed in the Laboratory of DNA Sequencing and Oligonucleotide Synthesis, Institute of Biochemistry and Biophysics, Polish Academy of Sciences. Sequences were compared with the POLG reference sequence (RefSeq NG_008218.1). The novel sequence variants were deposited in GenBank with accession numbers KP783508 (p.Gln968Glu), KP783509 (p.Arg290Cys), KP783511 (p.Arg869Ter), KP783513 (p.Val1106Ala), and KP783512 (p.Thr1053Argfs*6). We have also deposited sequence for p.Arg309Cys (KP783510) variant since it was not present in the GeneBank.
To estimate the frequency of the analysed POLG sequence variants in the Polish population, we designed and performed PCR–RFLP or high-resolution melting (HRM) tests on control DNA samples isolated from adults without neuromuscular disorders who gave their informed consent for genetic testing. HRM is a technique based on real-time PCR which utilizes differences in melting temperature of amplicons based on their nucleotide composition and enables to distinguish samples even with single-nucleotide substitution (Montgomery et al. 2010). Control samples for which test results were inconclusive were subsequently sequenced. In the case of p.Arg309Cys, a restriction site for the HhaI enzyme disappears in the POLG1_3 fragment, whereas for p.Thr1053Argfs*6 and p.Val1106Ala restriction sites for BaeGI and HincII, respectively, disappear in the POLG1_10 fragment. The digestion reactions were performed as follows: 5 µl of PCR product, 10 U of enzyme (New England Biolabs), 2 µl of buffer 2 (HhaI) or 3.1 (BaeGI and HincII), water up to 20 µl and incubated overnight in 37°.
To test the population frequencies of variants p.Arg869Ter and p.Gln968Glu, two pairs of primers for HRM test were designed (HRM869F/R and HRM968F/R, respectively). HRM reactions were prepared in 10 µl final volume containing MasterMix 1x (Roche), 3 mM (p.Arg869Ter) or 3.5 mM (p.Gln968Glu) MgCl2, 0.15 µM of each of the forward and reverse primer, and 60 ng of DNA. Initial denaturation for 10 min at 95° (4.8°/s) was followed by 45 cycles of 10 s at 95° (4.8°/s), 15 s at 60° (p.Arg869Ter) or 67° (p.Gln968Glu) (2.5°/s), 20 s at 72° (4.8°/s). Melting curves were determined as follows: 95° for 5 min (4.4°/s), 40° for 3 min (2.2°/s), 60° for 1 s (1°/s) and heating up to 95° (0.02°/s) with 25 acquisitions per 1°.
To verify reciprocal in trans localization of variants p.Arg309Cys and a second undescribed variant in patient AI2, we cloned a POLG fragment using primers H3POLG1_3F and S1POLG1_3R and pGEM-T Easy Vector into XL10-Gold chemocompetent E. coli (Stratagene).
Yeast strains, plasmids and media
S. cerevisiae strains used in the research were two derivatives of W303, differing in the mating type: W303-1B (Chiron et al. 2005) (MATaade2-1 leu2-3, 112 ura3-1 trp1-1 his3-11, 15 can1-100) and CW04 (Chiron et al. 2005) (MATαade2-1 leu2-3, 112 ura3-1 trp1-1 his3-11, 15 can1-100). Strain Δmip1 was derived from CW04 by deleting the MIP1 gene with the KanMX4 cassette (MATa ade2-1 leu2-3, 112 ura3-1 trp1-1 his3-11, 15 can1-100 mip1::KanMX4). D273-10B/51 (MATαade5, [rho0]) (Groudinsky et al. 1981) was used as a tester strain for the mitochondrial genome integrity.
The wild-type MIP1 gene with approximately 300 bp of upstream and downstream sequences was amplified using primers mip1l and mip1r (Online Resource 1) and cloned into centromeric plasmids YCplac33 (URA3) and YCplac111 (LEU2) (Gietz and Sugino 1988) using the SLIC method (Li and Elledge 2007). The resulting constructs were named YCplac33MIP1 and YCplac111MIP1, respectively. Before transformation, all the constructs were verified by sequencing. For overexpression of mutant alleles, new constructs based on the pRS425 vector (Christianson et al. 1992) were created, by subcloning mutated sequences using the SLIC method (Li and Elledge 2007). All the constructs were verified by sequencing.
The following media were used in this work: YP medium consisted of 1 % yeast extract (Bioshop) 2 % Peptone (Bioshop) and 40 mg/l adenine (Sigma). YP medium was supplemented with 2 % glucose (YPD) or 2 % glycerol (YPG). SC medium consisted of 6.7 g/l YNB without amino acids (Bioshop) supplemented with an adequate amount of drop-out mix (Formedium™) according to the manufacturer’s instructions. The SC media were supplemented with 2 % glucose or 2 % glycerol. For evaluating the petite frequency, YPDG medium was used: 1 % yeast extract (Bioshop), 2 % Peptone (Bioshop), 40 mg/l adenine, 2 % glycerol and 0.1 % glucose. YPAEG-ery and YPAEG-oli media were used to determine frequencies of point mutations in mutant strains; they consisted of: 1 % yeast extract (Bioshop) 2 % Peptone (Bioshop) and 40 mg/l adenine, 2 % glycerol and 25 mM potassium phosphate buffer (pH 6.5) and 3 g/l erythromycin (Sigma) or 3 mg/l oligomycin (Sigma), respectively. For solid media, 2 % agar (Bioshop) was added.
Construction of mip1 mutant strains
The mip1 mutant alleles were obtained by site-directed mutagenesis of the wild-type MIP1 gene cloned into YCplac111 (YCplac111MIP1), using the double primer method of Zheng et al. (2004). The following mutated alleles were created (Table 2): Arg265Cys, Arg672Ter, Arg770Glu, Arg770Gln, Thr809Ter, Val863Ala (see Online Resource 1 for primer sequences). The W303/Δmip1 diploid, obtained by mating W303-1B and Δmip1 strains, was transformed with both the wild-type YCplac33MIP1 and a mutated version of YCplac111mip1 plasmids. This provided a functional wild-type MIP1 allele, allowing to maintain mtDNA (provided by the W303-1B parental strain) in the Δmip1 background. This diploid was subsequently sporulated, and haploid spores bearing both plasmids and the genomic Δmip1 gene deletion were selected using replica plating (-Ura, -Leu, G418 selection media). Subsequently, the YCplac33MIP1 URA3 plasmid carrying the wild-type MIP1 gene was removed by counterselection on 5-FOA plates (Sikorski and Boeke 1991), leaving the mutated variant of Mip1p as the only polymerase γ in the cells.
Table 2.
Analysis of mtDNA stability and point mutations in mutant yeast strains carrying MIP1 alleles corresponding to the POLG variants found in human patients
| Variant in H. sapiens | Mutation in S. cerevisiae | Domain | Petite frequency (%) | OliR/108CFU | EryR/108CFU | |||
|---|---|---|---|---|---|---|---|---|
| Homoallelic | Heteroallelic with wt MIP1 | |||||||
| 30° | 37° | 30° | 37° | |||||
| – | wt MIP1 | – | 1.4 ± 0.8 | 2.8 ± 1.5 | 1.0 ± 0.7 | 1.3 ± 0.4 | 0.7 ± 0.3 | 0.5 ± 0.18 |
| Hemiallelic vector/MIP1 | 1.7 ± 0.8 | 2.5 ± 0.7 | ||||||
| p.Arg309Cys | Arg265Cys | exo | 100 ± 0** | 100 ± 0** | 16.2 ± 4.3** | 17.5 ± 4.6** | – | – |
| p.Arg869Ter | Arg672Ter | pol | 100 ± 0** | 100 ± 0** | 15.9 ± 4.0** | 18 ± 3.8** | – | – |
| p.Gln968Glu | Arg770Glu | pol | 5.6 ± 1.8* | 7 ± 2.9* | 2.7 ± 1.8 | 3.0 ± 1.7 | 15 ± 7.8* | 7.4 ± 1.4* |
| Arg770Gln (humanized) | 1.6 ± 1.0 | 2.0 ± 0.9 | 1.5 ± 1.2 | 1.7 ± 0.5 | 3.8 ± 1.8 | 1.6 ± 0.33 | ||
| p.Thr1053Argfs*6 | Thr809Ter | pol | 100 ± 0** | 100 ± 0** | 13.9 ± 5.8** | 16.0 ± 5.9** | – | – |
| p.Val1106Ala | Val863Ala | pol | 5.3 ± 1.7* | 4.7 ± 1.4* | 3.4 ± 1.7* | 2.9 ± 1.5 | 3.6 ± 2.7* | 3.4 ± 0.1* |
Statistical significance calculated compared to the wild-type Mip1 for homoallelic strains, except for Arg770Glu, where the humanized Arg770Gln variant was used as reference
For the heteroallelic strains, statistical significance was calculated compared to the hemiallelic vector/MIP1 strain
* p < 0.05, ** p < 0.01
When the amino acid in the human POLG sequence was different from the corresponding residue in yeast Mip1p, in addition to the allele with the wild-type yeast residue replaced with the mutated human POLG amino acid (putative pathological allele), a second strain was created, with the wild-type human residue introduced into the yeast sequence, resulting in the humanized allele. The activity of the mutated protein was then compared both to the wild-type yeast sequence, and to the humanized variant of Mip1p.
Determining the effect of mutations on oxidative growth phenotype
To define oxidative growth phenotype, 10 μl of 10−1, 10−2, 10−3 and 10−4 dilutions of liquid culture of each strain (starting from OD600 = 1) was spotted on YPD and YPG plates and incubated at 30° (normal temperature) or 37° (elevated temperature) for 3 days.
Determination of mtDNA stability in yeast mip1 mutants
Single colonies of strains obtained in the 5-FOA counterselection procedure were grown in liquid YPD medium at 30° or 37°. After 48 h of incubation, cells were diluted to OD600 = 1 and around 300–500 colonies were plated on YPDG medium. Colonies were incubated for 60 h at 30° and 37°. Subsequently, both petite and rho+ colonies were counted using the tetrazolium test (Ogur et al. 1957) (using 2 mg/ml of TTC).
For heteroallelic strains, appropriate selective SC medium with 2 % glucose instead of YPD was used for initial incubation. This was necessary to prevent the loss of plasmids in the preculture.
For each strain, at least 20 independent experiments were performed. Statistical significance of observed differences was evaluated using the T test (two-tailed, unequal variance) for p < 0.05 confidence levels.
To confirm that the respiratory deficient phenotype was due to changes in mtDNA, the petite strains were crossed to a wild-type rho0 tester strain (D273-10B/51), and the resulting diploids assayed for respiratory growth on YPG plates.
Detection of mtDNA in yeast petite cells
The presence of mtDNA in yeast strains displaying the petite phenotype was assessed by staining of DNA in fixed cells with the Hoechst 3324 dye (Invitrogen), followed by fluorescent microscopy imaging as described previously (Puchta et al. 2010).
RT-PCR for MIP1 mRNA in yeast
Total cellular RNA was extracted according to the method described previously (Schmitt et al. 1990). RNA sample was treated with DNAse (Roche) in the presence of RNAse inhibitor (Ribolock, Thermo) in the buffer recommended by the manufacturer. DNA-free RNA was then extracted with phenol/chloroform/octanol (25:24:1), precipitated from the aqueous phase by ethanol/sodium acetate (pH 5.2) and dissolved in water. 5 μg of DNA-free RNA was reverse transcribed by Maxima™ Reverse Transcriptase (Thermo Scientific) according to the manufacturer’s instructions. Reactions without reverse transcriptase (RT-) were used as controls to test for genomic DNA contamination. cDNA was then amplified by PCR using the rtMipL and rtMipshR or rtMipL and rtMiploR primer pairs (Online Resource 1).
Quantification of point mutation accumulation in yeast mtDNA
To evaluate point mutation frequencies, two independent series of experiments consisting of 10 repeats were conducted. Each colony was grown in 3 ml of YPG for 48 h at 30°. Subsequently, 6–8 × 107 colonies were plated on YPAEG-ery medium and incubated at 30° for 9 days. Simultaneously, for each YPG culture, 50 μl of a 10−5 dilution was plated on a YPG plate to establish the exact number of colony-forming units in the culture.
The mutation frequencies in Table 2 are shown as the mean count of EryR colonies per 108 CFU. The Mann–Whitney U test was used to establish statistical significance for this experiment.
For the OliR experiment, the same protocol was used, except that instead of 3 g/l erythromycin, 3 mg/l oligomycin was added to the medium and the number of colonies plated on YPAEG-oli plate was approximately 3–5 × 107 CFU.
Results
Novel POLG variants in mitochondrial disease patients
In a search for previously undescribed variants, POLG coding sequence analysis was conducted for 60 patients with clinical diagnosis of myopathy (OMIM # 251900) (n = 18), encephalomyopathy (n = 5), PEO (OMIM #157640 and #258450) (n = 4), PEO and myopathy (n = 3), encephalopathy (n = 3), polyneuropathy (n = 2), myoclonic epilepsy associated with ragged-red fibres (MERRF) (OMIM #545000) (n = 2), Kearns-Sayre syndrome (OMIM #530000) (n = 1), myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) (OMIM #540000) (n = 1), mitochondrial neurogastrointestinal encephalopathy (MNGIE) (OMIM #613662) (n = 1), sensory ataxic neuropathy, dysarthria and ophthalmoparesis (SANDO) (OMIM #607459) (n = 2), general suspicion of mitochondrial disease (n = 15) or Alpers syndrome (OMIM #203700) (n = 3). Sequencing of the POLG gene in these patients, followed by in silico translation, revealed 1 poorly uncharacterised and 5 previously unreported sequence changes located in the regions conserved between yeast and human (c.924 C>T/p.Arg309Cys and c.867 C>T/p.Arg290Cys—proband from family A, c.2604 C>T/p.Arg869Ter—proband from family B, c.2901 C>G/p.Gln968Glu—proband from family C, c.3155_3166delCA/p.Thr1053Argfs*6—proband from family D and c.3316 T>C/p.Val1106Ala—proband from family E). None of the novel variants were present in the Human Polymerase Gamma Mutation Database, NCBI dbSNP (Sherry 2001), the 1000 genomes project (Genomes Project Consortium 2012), or in the Exome Variant Server database (http://evs.gs.washington.edu/EVS/). These variants, along with the poorly characterised p.Arg309Cys, were therefore selected for further investigation, and their sequences deposited in GenBank (accession numbers are listed in “Materials and methods”).
Patients’ clinical presentation
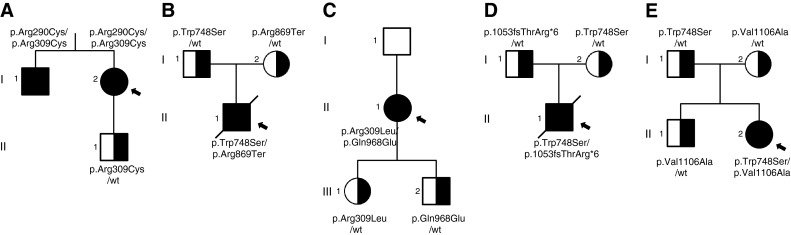
The results of sequence analysis, clinical and molecular evaluation of the four patients carrying previously unreported POLG variants and one with the known, but poorly characterised variant, are summarized in Table 1. Proband from family A (AI2) (Fig. 1a) is a 54-year-old woman with SANDO syndrome and positive family history (her brother presented similar symptoms). From the age of 31, she suffered from progressive external ophthalmoplegia, ptosis, dysarthria, weakness of upper and lower limbs and sensory ataxic neuropathy. Additionally, mental retardation was diagnosed. Nerve conduction studies indicated axonal sensory and motor neuropathy. MRI showed brain atrophy. Skeletal muscle biopsy disclosed ragged-red fibres on light microscopy examination. Analysis of mitochondrial DNA revealed multiple deletions in muscle tissue. Along with the p.Arg309Cys variant, patient AI2 also had another previously unknown sequence change c.867 C>T (p.Arg290Cys) for which, due to a low level of similarity in this position between POLG and Mip1 (Fig. 2c), creating a yeast model was impractical. According to PolyPhen-2 (Adzhubei et al. 2010), a software tool which predicts possible impact of a single amino acid substitution on the function of human proteins, both these variants found in patient AI2 are probably damaging, with a score of 1.00. SIFT Human Protein DB tool also predicts that both variants p.Arg290Cys and p.Arg309Cys are not tolerated (SeqRep = 0.92) (Ng and Henikoff 2001). The proband’s brother (AI1) carries both p.Arg309Cys and p.Arg290Cys variants present in the proband AI2.
Table 1.
Clinical symptoms, mtDNA and POLG sequence analysis in patients
| Proband | POLG sequence variantsa | Result of mtDNA analysis | Clinical diagnosis |
|---|---|---|---|
| AI2 | c.867 C>T (p.Arg290Cys)/c.924 C>T (p.Arg309Cys) | Multiple deletionsb | SANDO |
| BII1 | c.2243 G>C (p.Trp748Ser)/c.2604 C>T (p.Arg869Ter) | Depletionc | Alpers syndrome |
| CII1 | c.925 G>T (p.Arg309Leu)/c.2901 C>G (p.Gln968Glu) | Multiple deletionsb | PEO |
| DII1 | c.2243 G>C (p.Trp748Ser)/c.3155_3156delCA (p.Thr1053Argfs*6) | Depletionc | Alpers syndrome |
| EII2 | c.2243 G>C (p.Trp748Ser)/c.3316 T>C (p.Val1106Ala) | No lesions detected | SANDO |
Bold text indicates novel variants
aCompared to the POLG reference sequence (RefSeq NG_008218.1)
bMuscle tissue
cLiver tissue
Fig. 1.
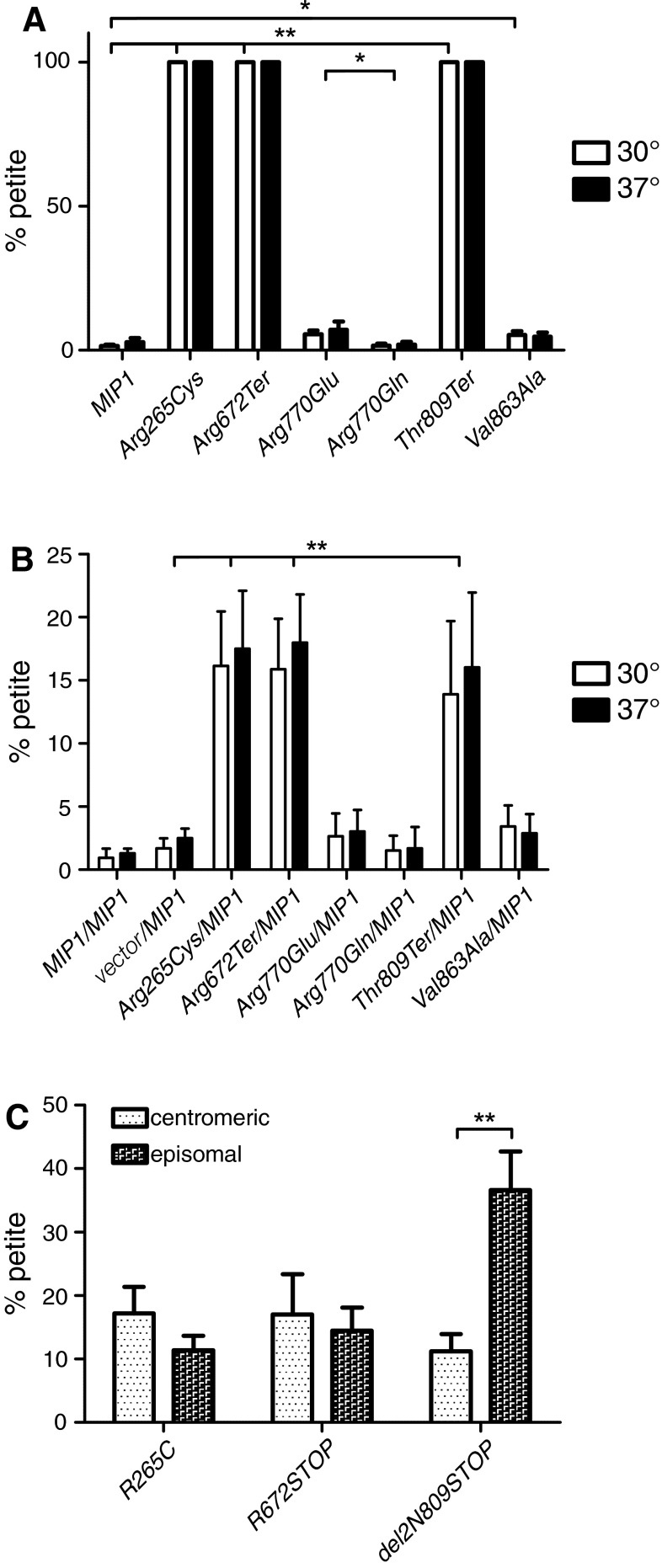
POLG variant distribution analysis in patients and their families. a Proband (I2) and her brother (I1) are both affected and have one known, but poorly characterised and one novel variant (p.Arg290Cys/p.Arg309Cys), whereas the proband’s healthy son (II1) is a carrier of the p.Arg309Cys variant. b Proband (II1) died at the age of 2.5 years due to liver failure caused by valproic acid administration. He carried heterozygous p.Trp748Ser mutation and heterozygous novel variant p.Arg869Ter. His parents (I1 and I2) are heterozygous carriers of either pathogenic mutation p.Trp748Ser (I1), or the novel variant p.Arg869Ter (I2). c Proband (II1) carries heterozygous pathogenic mutation p.Arg309Leu and heterozygous novel variant p.Gln968Glu. Her children (III1 and III2) are unaffected heterozygous carriers of pathogenic mutation p.Arg309Leu (III1), and the novel variant p.Gln968Glu (III2). The presence of p.Gln968Glu in the proband’s father (I1) was excluded, but due to the lack of biological material we were unable to perform the analysis for the presence of the p.Arg309Leu mutation. d The proband (II1) died at the age of 3.5 years after liver failure due to valproic acid administration. Both parents (I1 and I2) have either the novel p.Thr1053Argfs*6 (I1) variant or the known pathogenic mutation p.Trp748Ser (I2). e Proband (II2) carries heterozygous pathogenic mutation p.Trp748Ser together with heterozygous novel variant p.Val1106Ala. Her father (I1) is a carrier of the known pathogenic mutation p.Trp748Ser, whereas both her mother (I2) and brother (II1) have heterozygous novel variant p.Val1106Ala. All heterozygous family members are asymptomatic. Since the analysis of familial distribution of POLG variants present in families a–e indicates that all the probands are compound heterozygotes in trans, an autosomal recessive mode of inheritance is suggested
Fig. 2.
Modelling human POLG variants using the S. cerevisiae ortholog—Mip1p. a Schematic representation of POLG and Mip1 protein primary structure, with locations of the studied human POLG variants and the corresponding residues in S. cerevisiae Mip1p. The exonuclease domain is shown shaded light grey and the polymerase domain is shaded dark grey. Sequence of the linker region is not conserved. Blocks I, II, and III in the exonuclease domain, and A, B, and C in the polymerase domain are the regions of highest sequence conservation. b Alignment of the mitochondrial DNA polymerase amino acid sequences of selected vertebrate and yeast species in the regions of human variants selected for yeast modelling. Conserved residues (>0.5 consensus identity) are highlighted, arrows point to residues changed in the variant alleles under study. c Alignment of the mitochondrial DNA polymerase amino acid sequences of selected vertebrate and yeast species in the region around human Arg290 residue (marked with an asterisk), where lack of conservation between vertebrate and yeast sequences makes modelling of the Arg290Cys variant impractical
The proband from family B (BII1) was without any symptoms at infancy. At the age of 12 months, he became progressively ataxic and recurrent convulsions appeared, developing quickly to severe epileptic encephalopathy. Soon after introduction of anticonvulsant valproic acid administration, a fulminant liver failure was observed and the patient died at the age of 2.5 years. Severe mitochondrial depletion found in the liver tissue on autopsy (0.08 %, while the threshold for diagnosing depletion is set at <30 % of the normal mtDNA level) was in agreement with the clinical suspicion of Alpers syndrome. POLG sequencing revealed two in trans variants, one known pathogenic p.Trp748Ser substitution (Palin et al. 2010; Van Goethem et al. 2004), and a novel variant p.Arg869Ter.
Proband from family C (CII1) (Fig. 1c) is a 53-year-old woman with progressive external ophthalmoplegia and negative family history. At the age of 38, she observed impaired eye movements and ptosis. Nerve conduction studies were normal, and electromyography indicated myogenic changes. Cardiological assessment revealed no abnormalities. Skeletal muscle biopsy disclosed ragged-red fibres on light microscopy, and multiple abnormal mitochondria with paracrystalline inclusions on electron microscopy examination. Multiple mtDNA deletions were present in muscle tissue. POLG sequencing revealed a known p.Arg309Leu (Lamantea et al. 2002) mutation and a novel variant p.Gln968Glu, predicted to be not tolerated by SIFT (SeqRep = 0.97), and probably damaging with a score of 0.992 according to PolyPhen2.
In the proband from family D (DII1) (Fig. 1d), a psychomotor retardation and muscle hypotonia were observed since 2 months of age, and autistic behaviour was observed during early chilhood. At the age of 3 years, the patient was given valproic acid due to episodes of unconsciousness and abnormal EEG pattern and responded suddenly with an acute liver failure. He died 6 months later before the qualification for liver transplantation was completed. Severe depletion of mtDNA was found in the liver section taken at autopsy (3.4 %, ref. >30 %), as in the case of the patient BII1. In the POLG sequence, in addition to the known p.Trp748Ser mutation, a novel variant p.Thr1053Argfs*6 was found.
In spite of normal developmental milestones, the proband from family E (EII2) (Fig. 1e) started to have walking difficulties at the age of 13 years. Her condition deteriorated, and she developed ataxic gait and dysarthria. Two years later, she developed action-exacerbated myoclonus. When last seen at 15 years, she had no ophthalmoparesis. Nerve conduction studies showed sensory-motor polyneuropathy of lower limb nerves. We did not detect mtDNA deletions in a DNA sample isolated from peripheral blood. Since muscle tissue was not available, the decision to sequence POLG was undertaken based on the patient’s clinical presentation, and indicated that she was a compound heterozygote p.Trp748Ser/p.Val1106Ala. The novel variant (p.Val1106Ala) is designated as not tolerated (SIFT SeqRep = 0.98) and possibly damaging with a score of 0.85 (PolyPhen2). Interestingly, another amino acid change (valine to isoleucine) in the same position was described previously as probably pathogenic (Horvath et al. 2006).
Each of the novel variants coexisted in the patient with a known pathogenic recessive mutation, and all the patients’ parents were healthy; therefore, we suspected that all the variants, if pathogenic, were also recessive. For all the patients, DNA samples isolated from other members of their families were also available. To confirm in trans localization of novel versus known variants in patients AI2, BII1, CII1, DII1 and EII2, we performed family studies, and in one case cloning.
Familial and population study
To verify reciprocal localization of novel and known variants in patients, we acquired DNA samples from their family members, and amplified and subsequently sequenced the relevant POLG fragments (Fig. 1). In three families (B, D and E), the proband’s parents were each carrier of one of the two pathogenic/possibly pathogenic variants. In the case of proband AI2, the parents’ DNA was unavailable, but POLG sequence analysis of her son (AII1) proved him to be the carrier of the known, but poorly characterised p.Arg309Cys variant. Her symptomatic brother (AI1) had both variants p.Arg290Cys and p.Arg309Cys found in the proband. Additionally, we performed cloning of POLG alleles in patient AI2, which further confirmed that she was a compound heterozygote. The family of patient CII1 did not give consent to obtain their DNA from peripheral blood, but agreed to perform buccal swabs. Both her children are heterozygous carriers of the pathogenic (CIII1-p.Arg309Leu) or possibly pathogenic (CIII2-p.Gln968Glu) variant. The presence of the novel variant p.Gln968Glu in her father (CI1) was ruled out, but due to a low yield of DNA extraction, we were not able to confirm the presence of the p.Arg309Leu mutation. Thus, we confirmed that in all the affected individuals, the two variants were in trans.
The population frequency of all novel variants was estimated using PCR–RFLP or HRM tests. Because the population frequency of p.Arg309Cys was not previously assessed, we also conducted PCR–RFLP test for this variant. None of the analysed POLG sequence variants were found in 320 chromosomes of healthy adults, which means that the population frequency of each variant is significantly lower than 1 %.
The analysis of familial distribution and frequency in the general population suggests that the analysed POLG sequence changes are probably pathogenic and inherited in a recessive manner.
Modelling the putative pathogenic mutations in yeast
To gain some insights into the effects of the novel variants in the human polymerase γ using the well established and accessible S. cerevisiae model, we first aligned the amino acid sequences of human POLG and its yeast ortholog Mip1p, and identified the residues that were mutated in patient sequences (Fig. 2). Of the 5 residues, three (Arg309, Arg869, and Val1106) were conserved in the yeast Mip1p. The site of the frameshift mutation p.Thr1053Argfs*6 also corresponds to a block of a nearly complete amino acid sequence identity. The Gln968 residue, also in a conserved region, corresponds to Arg770 in the yeast protein. In this case, in addition to the mutated allele (with Glu in this position), we also created the humanized allele (Arg770Gln).
The mutant mip1 alleles were created by site-directed mutagenesis and introduced into the Δmip1 background on centromeric (low copy number) vectors, as described in “Materials and methods”. The plasmid shuffling strategy was used to maintain functional mtDNA owing to the presence of the wild-type MIP1 gene on an URA3 vector, which could subsequently be eliminated by 5-FOA counterselection (Sikorski and Boeke 1991).
Influence of mip1 mutations on respiratory growth phenotype in the yeast model
Growth on a non-fermentable carbon source (glycerol or ethanol) is the fastest and simplest method to examine respiratory function in S. cerevisiae strains. Among the five tested strains carrying yeast equivalents of putative pathogenic mutations, three exhibited a severe respiratory deficiency. Mutants Arg265Cys, Arg672Ter and Thr809Ter were unable to support growth on glycerol at 30° (Fig. 3). Growth of the remaining strains did not differ from the wild-type. Incubation at a restrictive temperature (37°) contributed to an even more severe phenotype of mutants which were unable to grow on glycerol (Arg265Cys, Arg672Ter, Thr809Ter), causing slower growth even on a medium with fermentable carbon source. The elevated temperature did not significantly affect the rest of the tested strains.
Fig. 3.
Respiratory growth of S. cerevisiae strains bearing the wild type and mutated alleles of MIP1. Fermentative growth was tested on YPD medium (YP with 2 % glucose) at 30° and 37°. Respiratory growth was tested by plating the strains on YPG medium (YP with 2 % glycerol) and incubation at 30° and 37°. Each strain was spotted in 10−1, 10−2, 10−3 and 10−4 dilutions (starting from OD600 = 1). The pictures were taken after 3 days of incubation. a Fermentative (YPD) and respiratory (YPG) growth at normal temperature (30°). All the strains grow on the fermentable carbon source (YPD), although the strains bearing the Arg265Cys, Arg672Ter and Thr809Ter variants show slightly slower growth. These strains are not able to grow on medium with the nonfermentable carbon source (YPG) (b). The same growth assay as in (a), but carried out at the elevated temperature (heat stress, 37°). The phenotype of strains bearing the Arg265Cys, Arg672Ter and Thr809Ter variants is even more pronounced at the elevated temperature, and the growth defect on the fermentable carbon source is more apparent. The strains that are not affected at normal temperature do not show a respiratory deficiency at 37° either
Determination of petite frequencies in strains with mip1 mutations
Petite mutants of S. cerevisiae are unable to grow on non-fermentable carbon sources and form only small anaerobic colonies in the presence of glucose. While this phenotype can result from a point mutation in mtDNA or a mutation in a nuclear gene, most often it is caused by a complete loss (rho0) or extensive deletions (rho−) of mtDNA. Petite colonies appear at a low (1–5 %) rate spontaneously in cultures grown on fermentable carbon sources. Mutations in nuclear genes encoding proteins involved in mitochondrial function often result in a marked increase in the frequency of petite generation (Contamine and Picard 2000; Lipinski et al. 2010).
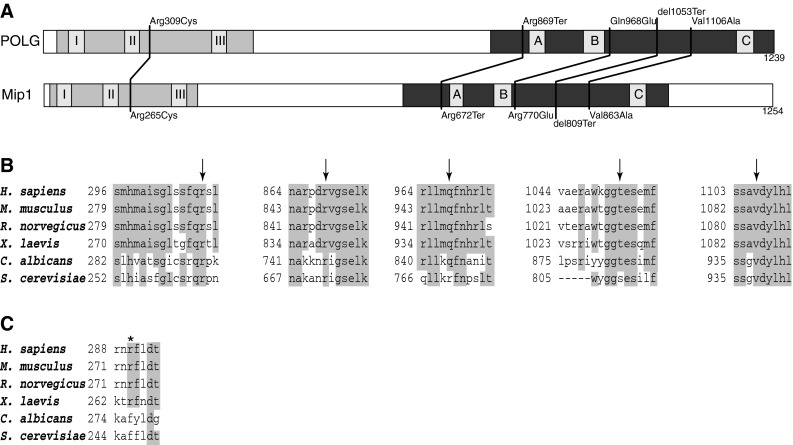
All the strains carrying mip1 alleles corresponding to the modelled human mutations show increased petite frequencies in comparison to the one with the wild-type MIP1 (Fig. 4a; Table 2). Furthermore, mutations Arg265Cys, Arg672Ter, Thr809Ter, which cause a complete respiratory deficiency, lead to a complete loss of functional mtDNA in the tested strains.
Fig. 4.
Mitochondrial genome stability in yeast strains carrying MIP1 alleles corresponding to the POLG variants found in human patients. a Frequencies of petite mutants in strains carrying the wild-type MIP1 or mutated mip1 alleles in the homoallelic setting (as the only MIP1 allele on a centromeric plasmid). Mutations Arg265Cys, Arg672Ter, and Thr809Ter lead to a complete loss of functional mtDNA (100 % petites) and the petite frequency in Mip1Val863Ala strain is three times higher than in the wild type strain. The Arg770Glu mutation significantly increases petite accumulation in comparison to either the wild type, or humanized (Arg770Gln) allele. There is no significant difference between the humanized Arg770Gln and the wild-type allele. b Frequencies of petite mutants in the pure wild-type (MIP1/MIP1), hemiallelic (vector/MIP1) and heteroallelic (MIP1/mip1) strains, with both alleles expressed from low copy number (centromeric) plasmids. In heteroallelic strains with Arg265Cys/MIP1, Arg672Ter/MIP1 and Thr809Ter/MIP1 alleles, petites accumulate at a significantly higher frequency than in the hemiallelic (vector/MIP1) strain; nevertheless, they still retain >80 % of functional mtDNA. Petite frequency in the Val863Ala/MIP1 shows a slight (but significant at p < 0.05) difference compared with the hemiallelic (vector/MIP1) at 30°, but not at 37° (not marked on the graph). The Arg770Glu/MIP1 heteroallelic strain shows no significant difference compared to either the hemiallelic (vector/MIP1) or the humanized strain. Only the strongly significant differences (p < 0.01) were marked with asterisks in this panel for clarity. c Frequencies of petite mutants resulting from the overexpression (on the multicopy pRS425 plasmid) of mutant alleles (in the background of the wild-type MIP1 on a low copy number vector). For Arg265Cys and Arg672Ter variants, no significant difference was observed between the low copy number and multicopy vectors, but for the Thr809Ter allele the petite frequency was significantly higher when the mutated allele was on a multicopy plasmid, suggesting an antimorphic character of the mutation. The T test (two-tailed, unequal variance) was used to assess the statistical significance, *p < 0.05, and **p < 0.01. ns not significant. Numerical data can be found in Table 2
For the Arg770Glu mutation, petite accumulation is significantly (p < 0.05) higher than in its humanized allele mip1Arg770Gln and in the wild-type version of the protein (5.6, 1.6, and 1.4 % at 30°, respectively). Furthermore, there is no significant difference between wild-type Mip1p (1.4 % petite) and humanized Arg770Gln (1.6 %). Petite frequency of the strain carrying the Val863Ala allele (5.3 %) is approximately three times higher than in the wild-type strain at 30°, and this difference is statistically significant (p < 0.05).
To confirm that the observed respiratory negative phenotype was related to the loss of functional mtDNA, petite colonies from the strains carrying the Arg265Cys, Arg672Ter and Thr809Ter variants (50 colonies each) were crossed to a wild-type rho0 tester, and the resulting diploids assayed for respiratory growth. This test confirmed that in all the petite colonies there was no functional mtDNA left (rho−/rho0). Due to the presence of nonfunctional residual mitochondrial genome fragments in rho− mtDNA, it is not practical to perform quantitative analysis of mtDNA levels in yeast cells using PCR-based methods. For the strains carrying the Arg265Cys, Arg672Ter and Thr809Ter variants, DNA staining with Hoechst dye followed by fluorescent microscopy imaging revealed that, unlike in the wild-type control, extranuclear DNA signal is mostly absent, suggesting that they are essentially rho0 (Online Resource 2).
The severe phenotype of the Arg265Cys, Arg672Ter and Thr809Ter variants raised the possibility that these mutations abolished the expression of the MIP1 gene, and thus were equivalent to transcriptional nullomorphs. We performed an RT-PCR assay on total RNA isolated from the strains carrying the Arg265Cys, Arg672Ter and Thr809Ter variants as the only MIP1 alleles, and found no apparent change compared to the wild-type control, thus confirming that these mutations do not interfere with the expression of the MIP1 gene, at least on mRNA level (Online Resource 3).
Determination of petite frequencies in heteroallelic strains
To determine the dominance/recessivity of the mutations, we assayed the frequency of petite formation in heteroallelic yeast strains bearing each of the mutant alleles as well as the wild-type MIP1 allele (Fig. 4b; Table 2). A hemiallelic strain containing the empty YCplac111 vector as well as the vector carrying the wild type MIP1 (vector/MIP1) was also used as a control to normalise the allele dosage and differentiate between antimorphy and haploinsufficiency. The petite frequency in the hemiallelic strain is slightly, but significantly (p < 0.05) higher than in the MIP1/MIP1 strain, and this effect is more pronounced at the elevated temperature (37°). MIP1 haploinsufficiency is evident when a diploid strain heterozygous for the Δmip1 allele is used (Baruffini et al. 2006). The haploid deletant strain with plasmid-borne wild-type and mutant MIP1 alleles used in this study is clearly less sensitive to haploinsufficiency.
In the Val863Ala/MIP1 strain, the petite frequency was slightly, yet significantly (p < 0.05) higher than in the vector/MIP1 strain at 30°. No increase in petite frequency could be observed at the elevated temperature (37°). Even though the difference at 30° was statistically significant, the petite frequency in this heteroallelic strain remained low (well below 5 %); the dominant negative effect of this allele is thus negligible.
The petite accumulation frequency for the heteroallelic strain Arg770Glu/MIP1 strain, as well as for the humanized Arg770Gln/MIP1, was not significantly elevated above the level observed in the hemiallelic vector/MIP1 control at either temperature; this mutation is, therefore, recessive.
Strains heteroallelic for the three mutations that showed the most severe phenotype (complete respiratory deficiency and loss of functional mtDNA) in the monoallelic setting, Arg265Cys/MIP1, Arg672Ter/MIP1, and Thr809Ter/MIP1, accumulated petites at a much higher frequency than the hemiallelic vector/MIP1 control strain (at both temperatures), although they still retained >80 % of functional mtDNA. These alleles thus have a partially dominant negative effect on the mtDNA replication process (Fig. 4b; Table 2). To verify whether this effect is dependent on allele dosage, we also constructed strains carrying the mutant alleles on multicopy (episomal) plasmids, and the wild-type allele on a centromeric plasmid (Fig. 4c). For Arg265Cys and Arg672Ter, no significant difference was observed between the low copy number and multicopy vectors, for the Thr809Ter allele; however, increasing the copy number of the mutated variant resulted in a significantly (p < 0.01) more pronounced phenotype of the heteroallelic strain, suggesting a true antimorphic character of this mutation.
Frequency of mtDNA point mutations in the yeast model
Two of the five mutated mip1 alleles, Arg770Glu and Val863Ala, showed only a partial decrease in overall mtDNA stability (measured by the frequency of petite colony formation), whereas the humanized Arg770Gln variant was indistinguishable from the wild type in that aspect. No effect on the respiratory growth was observed in these three strains. To test whether the replicative function of the mitochondrial DNA polymerase in these strains was not affected in a more subtle way, we performed experiments assessing the point mutation rates in mtDNA.
Quantification of point mutation accumulation in mtDNA is based on estimating the frequencies of spontaneous oligomycin- (OliR) and erythromycin-resistant (EryR) mutants that result from substitutions in mitochondrial genes encoding the ATPase subunits or the large subunit rRNA, respectively. This assay can indicate changes in the fidelity of mtDNA replication, such as defects in the proofreading activity of the mitochondrial DNA polymerase.
Two mutations (Arg770Glu, Val863Ala) and the humanized allele (Arg770Gln) were examined in comparison to the wild-type strain (Table 2; Fig. 5). The strain carrying the Val863Ala allele showed a slight, but still statistically significant (p < 0.05, Mann–Whitney U test) increase in the frequency of both OliR and EryR mutations. The effect of the Arg770Glu allele, on the other hand, was more pronounced, and significantly (p < 0.05) higher both in comparison with the wild-type MIP1, and with the Arg770Gln humanized variant (differences between the humanized allele and the wild type version were not significant). Interestingly, this substitution (as well as Val863Ala) is located in the polymerase domain (Table 2). It has to be noted, however, that the effect observed for this variant, while statistically significant (p < 0.05), is far less evident than that reported for an MIP1 allele devoid of the corrective exonuclease domain activity (exo−), where the EryR mutation frequency increased by 550-fold (Stumpf et al. 2010).
Fig. 5.
Replication fidelity, expressed as point mutation frequencies in yeast strains carrying MIP1 alleles corresponding to the POLG variants found in human patients. Oligomycin- and erythromycin-resistant mutant frequencies were calculated as a fraction of the rho + cells plated on the respiratory (YPG) medium for the mip1 alleles that did not cause a complete loss of mtDNA stability. a Frequencies of mutations (in mitochondrial genes encoding the ATPase subunits) resulting in oligomycin resistance. Black circles correspond to the antibiotic resistant colonies on individual plates. b Frequencies of mutations (in the mitochondrial gene encoding the large subunit rRNA) resulting in erythromycin resistance. Open circles correspond to the antibiotic resistant colonies on individual plates. Horizontal lines indicate the mean value of medians from two independent experimental repetitions (10 plates each). In both assays, the strain carrying the Val863Ala mutation showed a significant increase in the point mutation frequency. The effect of the Arg770Glu allele on mutation frequencies was even more pronounced, and significantly elevated either in comparison to the wild-type MIP1, or to the Arg770Gln humanized variant, whereas differences between the humanized variant and the wild-type allele were not statistically significant. Mann–Whitney U test was used to assess the statistical significance, *p < 0.05, and **p < 0.01. ns not significant. Numerical data can be found in Table 2
Discussion
To date, over 200 POLG sequence changes are known, and for the vast majority their pathogenicity remains unclear. Since most variants are classified as pathogenic only on the basis of their presence in the patient, there are only a few well-documented POLG mutations. Here, we present an in-depth analysis of 5 POLG variants—4 novel and 1 known but poorly characterised, found in patients with mitochondrial disorder symptoms: p.Arg309Cys, p.Arg869Ter, p.Gln968Glu, p.Thr1053Argfs*6 and p.Val1106Ala, combining familial and population studies with a functional study in a yeast model. Since the population frequency of all the analysed variants is lower than 1 %, it is highly probable that they are not polymorphic sequence changes but true pathogenic mutations.
As it is often the case for the severe recessive mitochondrial disorders caused by POLG mutations (like the Alpers syndrome) (Chan et al. 2009; Euro et al. 2011; Mancuso et al. 2004; Wiltshire et al. 2008; Wong et al. 2008), the affected individuals are compound heterozygotes. In our study, all the novel POLG variants in the patients coexist with another known mutation. Pedigree analysis, together with the results of the general population study, strongly suggests that p.Arg309Cys, p.Arg869Ter, p.Gln968Glu, p.Thr1053Argfs*6 and p.Val1106Ala are indeed pathogenic variants inherited in a recessive manner. To verify these assumptions, we used the yeast S. cerevisiae as a model for studying phenotypes of the 4 novel and 1 known but poorly characterised POLG mutations that occur in sequence regions conserved between yeast and human orthologs. The validity of using S. cerevisiae as a model in studying known human pathogenic POLG mutations was demonstrated in many cases (Barrientos 2003; Baruffini et al. 2006, 2007, 2011, 2014; Baruffini and Lodi 2010; Foury and Kucej 2002; Lodi et al. 2015; Stuart et al. 2006; Stumpf et al. 2010; Stumpf and Copeland 2011, 2013, 2014), including cases where no other evidence for causation was available (Stumpf et al. 2010), yet studies simultaneously reporting new human mutations in a patient cohort and analysing the phenotype of the corresponding yeast strains are still very rare (Baruffini et al. 2011). Furthermore, for all the modelled mutations, we observed phenotypes consistent with the clinical status of the human patients.
All the mutant strains in our yeast model display a phenotype that in some aspects differs significantly from the wild type. Arg770Glu and Val863Ala, corresponding to human p.Gln968Glu and p.Val1106Ala POLG variants, respectively, are located in the polymerase domain and show a rather mild defect of mtDNA stability (measured by a moderate increase in petite frequency). Additionally, in both these strains, we observed an increase in the frequency of point mutations (measured by the rates of spontaneous erythromycin and oligomycin resistant colony formation). This correlates with the observations made in patients—proband EII2 carrying the p.Val1106Ala variant has no detectable mtDNA lesions, in spite of polyneuropathy symptoms, whereas in the muscle tissue of proband CII1 carrying the p.Gln968Glu variant, multiple DNA deletions are evident, but the phenotype is still relatively mild (PEO). Correspondingly, the decrease in mtDNA stability and increase in the frequency of point mutations were more pronounced in the yeast strain carrying the Arg770Glu variant than in the one with the Val863Ala allele. The relevance of the relatively mild increase in the frequency of point mutations we observed for these variants in the yeast model for the human pathology is, however, not evident. In yeast, a mutant deficient in the exonuclease function of Mip1p (Exo−) shows an increase in mtDNA point mutations of about 550-fold (Stumpf et al. 2010) that is orders of magnitude larger than the effect of the Arg770Glu and Val863Ala alleles (about 15-fold at best). Also, increases in mtDNA mutation rate in the exonuclease deficient POLG mouse model (Trifunovic et al. 2004; Vermulst et al. 2007) lead to a premature ageing phenotype, but do not result in marked physiological effects in young animals. The effects of changes that involve mtDNA point mutation frequencies are likely to be cumulative, and thus difficult to compare between different model systems.
The other two polymerase domain mutations are Arg672Ter and Thr809Ter, corresponding to human variants p.Arg869Ter and p.Thr1053Argfs*6, respectively. These two alleles encode protein variants with a sequence truncated in the middle of the polymerase domain, due to the presence of a premature STOP codon. Expectedly, in the monoallelic yeast strains, we observe a total loss of Mip1 activity, resulting in a complete loss of functional mtDNA. Accordingly, compound heterozygote patients carrying these variants exhibited a severe Alpers syndrome (Table 1). In the yeast model, in the heteroallelic setting (with a mutated allele and the wild-type allele present in the same strain), they show a partially dominant effect, with a significant, albeit not complete, loss of mtDNA stability (Fig. 4b; Table 2) compared to a hemiallelic control. In the case of Thr809Ter, the partially dominant effect shows an allele dosage dependency (Fig. 4c), as increasing the copy number of the mutated allele by expressing it from a multicopy plasmid results in a further increase in petite colony formation, suggesting that this mutation is antimorphic.
While it is difficult to speculate about the exact molecular mechanism of the observed defect without in-depth biochemical experiments, there are several possible mechanisms that could explain the disruptive effect of these mutations, resulting in a partially dominant negative phenotype. The truncated proteins could disrupt replication by competing for the binding of substrates or other protein factors essential for replication. Similar dominant negative phenotypes of mip1 alleles modelling putative human pathological variants were observed in previous studies (Stumpf et al. 2010). Interestingly, in familial analysis of human p.Arg869Ter and p.Thr1053Argfs*6 variants, no mitochondrial disorder symptoms were reported for heterozygous carriers of these alleles. The yeast model system appears to be more sensitive to perturbations in the mtDNA maintenance, as the lack of dominant mitochondrial disease in humans for alleles showing partial dominance in the yeast model was noted for at least five different cases (Stumpf et al. 2010). This could be explained by the lower copy number of mtDNA molecules per cell in yeast (about 20–30 copies per cell, 50–100 fold less than in typical vertebrate cells), and by the generally less robust character of the S. cerevisiae mitochondrial genome system, evidenced by the spontaneous generation of petites, evident even in wild-type strains and exacerbated by many different genetic and environmental factors (Contamine and Picard 2000; Lipinski et al. 2010). Alternatively, the carriers may suffer from defects that are very mild compared to the severe pathology in affected compound heterozygotes, and thus are not evident in a typical familial study, or will develop a phenotype later in life. Low penetrance of pathologies such as migraines and neuropathies was suggested for simple heterozygous carriers of strongly pathogenic POLG variants (Tzoulis et al. 2006).
Interestingly, the only change in our analysis that is located in the exonuclease domain of the polymerase γ enzyme, p.Arg309Cys, corresponding to Arg265Cys in S. cerevisiae Mip1p, results in a yeast phenotype that is as severe as that of the truncated polymerase domain variants described in the previous paragraph, with a complete loss of functional mtDNA in the monoallelic strain and a significant increase in petite frequency in the heteroallelic setting (Table 2; Fig. 4). Such a strong phenotype is unexpected for a simple substitution in the exonuclease domain, but stands in agreement with an original report of a patient with homozygous p.Arg309Cys variant, resulting from parents consanguinity, having way more severe phenotype than patient AI2 who is a compound heterozygote. Unlike proband AI2, the p.Arg309Cys homozygote started showing symptoms during adolescence and besides PEO, ataxia, myopathy and dyshartia she also suffered from chronic intestinal pseudo-obstruction, epilepsy, strokes, hearing loss and pigmentary retinopathy, which led to her death at the age of 22 (Amiot et al. 2009). The phenotype of this variant is probably not related to an essential character of the mutated residue, as a different substitution in the same position; Arg265Leu results only in a slight increase of petite frequency in yeast (Stumpf et al. 2010) and the human p.Arg309Leu allele in compound heterozygotes [patient CII1 in this study and one patient described previously (Lamantea et al. 2002)] causes only a relatively mild PEO phenotype. A previously described mip1 mutant completely deficient in the exonuclease function (Exo−) shows a massive increase in mtDNA point mutations (about 550 fold), but only about 10 % petite frequency (Stumpf et al. 2010). The severe phenotype of Arg265Cys could be due to the presence of an additional cysteine residue which can form undesirable disulphide bonds with another residue in the protein, and thus seriously alter the protein conformation (Farnum et al. 2014).
To summarize, the yeast model proved to be a valuable tool for the initial assessment of the phenotypic effect of novel or poorly characterised POLG variants found in human patients. The phenotypes of strains with mutations introduced into orthologous positions in regions conserved between human POLG and S. cerevisiae Mip1p are in good agreement with clinical observations, as variants found in patients affected with the most severe forms of mitochondriopathy cause a complete loss of mtDNA stability in the monoallelic setting in yeast. The less severe variants corresponded to a partial loss of mtDNA stability (increase of petite formation rate) and/or increase in point mutation frequencies.
A recently developed alternative to orthologous modelling is based on the expression of human POLG as the only mtDNA polymerase in yeast cells devoid of the native MIP1 gene, and is termed “humanized yeast” (Qian et al. 2014). Both approaches have their merits and shortcomings, and should be viewed as complementary rather than exclusive. Orthologous modelling is obviously unavailable when the variants occur in sequence regions that are not conserved between human and yeast polymerase γ sequences, like in case of proband AI2 carrying a novel variant p.Arg290Cys in addition to p.Arg309Cys. Using the human mitochondrial polymerase would overcome this particular limitation, but given the substantial differences in the structure and replication of yeast and human mtDNA, phenotypes observed for the variants of the human enzyme could also be related to changes in its capability to function in a heterologous environment.
Yeast Mip1 exhibits amazing sensitivity when used to model putative pathogenic variants of the orthologous human mtDNA polymerase. In the monoallelic setting, even the slight phenotypic effects manifest themselves as observable changes in the mitochondrial genome stability and fidelity of replication. As phenotypic tests involving very large numbers of individual colonies are feasible and economical in yeast, statistical significance of these observations can be reliably assessed. For the severe defects that cause complete loss of functional mtDNA, heteroallelic strains can be easily constructed, and they provide valuable insights into the effects of these alleles, including dominance/recessiveness relationships that can be crucial for understanding the effects of corresponding human variants.
In conclusion, we proved that the mitochondrial DNA polymerase variants discovered in patients with mitochondrial disease, and modelled in yeast, have an influence on different aspects of the mitochondrial genome maintenance, and are thus likely to be bona fide pathogenic mutations. Our study also further confirms the utility of yeast Mip1p as a tool for studying human POLG variants.
Electronic supplementary material
Acknowledgments
We would like to state the authors’ contributions as follows: study design and supervision, data analysis: PG (yeast modelling), KT (human variants); genetic study coordination: MK, KT; clinical evaluation: AK, BK (family A), EP (families B, D), AL (family C), AKP, MN (family E); preliminary genetic analysis: MK, KT, DP-A, EB; POLG sequencing: MK (85 %), DL (15 %); population study: MK (variants: p.Arg869Ter, p.Thr1053Argfs*6, p.Val1106Ala), JV (variant p.Arg309Cys), DS, ML (variant: p.Gln968Glu); family distribution studies: MK; yeast experiments: JK, AK-G (design and supervision of the point mutation rate test), preparation of the manuscript: MK, JK, PG, KT, EB. We wish to thank Aleksander Chlebowski for his assistance with fluorescent microscopy. This work was supported by the EU Operational Programme Innovative Economy via the Foundation for Polish Science [TEAM/2010-6/6 to JK, PG and KT, VENTURES/2011-8/3 to MK], and by the National Science Center of Poland [NCN 2012/05/B/NZ2/01627 to DP-A].
Footnotes
M. Kaliszewska and J. Kruszewski are joint first authors.
Contributor Information
Paweł Golik, Phone: +48 22 592 32 34, Email: pgolik@igib.uw.edu.pl.
Katarzyna Tońska, Phone: +48 22 592 22 41, Email: kaska@igib.uw.edu.pl.
References
- Aanen DK, Spelbrink JN, Beekman M. What cost mitochondria? The maintenance of functional mitochondrial DNA within and across generations. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130438. doi: 10.1098/rstb.2013.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiot A, Tchikviladze M, Joly F, Slama A, Hatem DC, Jardel C, Messing B, Lombes A. Frequency of mitochondrial defects in patients with chronic intestinal pseudo-obstruction. Gastroenterology. 2009;137:101–109. doi: 10.1053/j.gastro.2009.03.054. [DOI] [PubMed] [Google Scholar]
- Aon MA, Bhatt N, Cortassa SC. Mitochondrial and cellular mechanisms for managing lipid excess. Front Physiol. 2014;5:282. doi: 10.3389/fphys.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A. Yeast models of human mitochondrial diseases. IUBMB Life. 2003;55:83–95. doi: 10.1002/tbmb.718540876. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Lodi T. Construction and validation of a yeast model system for studying in vivo the susceptibility to nucleoside analogues of DNA polymerase gamma allelic variants. Mitochondrion. 2010;10:183–187. doi: 10.1016/j.mito.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Lodi T, Dallabona C, Puglisi A, Zeviani M, Ferrero I. Genetic and chemical rescue of the Saccharomyces cerevisiae phenotype induced by mitochondrial DNA polymerase mutations associated with progressive external ophthalmoplegia in humans. Hum Mol Genet. 2006;15:2846–2855. doi: 10.1093/hmg/ddl219. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Ferrero I, Foury F. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with disease in human. Biochim Biophys Acta. 2007;1772:1225–1235. doi: 10.1016/j.bbadis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Horvath R, Dallabona C, Czermin B, Lamantea E, Bindoff L, Invernizzi F, Ferrero I, Zeviani M, Lodi T. Predicting the contribution of novel POLG mutations to human disease through analysis in yeast model. Mitochondrion. 2011;11:182–190. doi: 10.1016/j.mito.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Ferrari J, Dallabona C, Donnini C, Lodi T. Polymorphisms in DNA polymerase gamma affect the mtDNA stability and the NRTI-induced mitochondrial toxicity in Saccharomyces cerevisiae. Mitochondrion. 2014;20c:52–63. doi: 10.1016/j.mito.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SS, Naviaux RK, Basinger AA, Casas KA, Copeland WC. De novo mutation in POLG leads to haplotype insufficiency and Alpers syndrome. Mitochondrion. 2009;9:340–345. doi: 10.1016/j.mito.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiron S, Suleau A, Bonnefoy N. Mitochondrial translation: elongation factor tu is essential in fission yeast and depends on an exchange factor conserved in humans but not in budding yeast. Genetics. 2005;169:1891–1901. doi: 10.1534/genetics.104.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]
- Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/MMBR.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Ephrussi B, Slonimski PP. Subcellular units involved in the synthesis of respiratory enzymes in yeast. Nature. 1955;176:1207–1208. doi: 10.1038/1761207b0. [DOI] [PubMed] [Google Scholar]
- Euro L, Farnum GA, Palin E, Suomalainen A, Kaguni LS. Clustering of Alpers disease mutations and catalytic defects in biochemical variants reveal new features of molecular mechanism of the human mitochondrial replicase, Pol gamma. Nucleic Acids Res. 2011;39:9072–9084. doi: 10.1093/nar/gkr618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnum GA, Nurminen A, Kaguni LS. Mapping 136 pathogenic mutations into functional modules in human DNA polymerase gamma establishes predictive genotype-phenotype correlations for the complete spectrum of POLG syndromes. Biochim Biophys Acta. 2014;1837:1113–1121. doi: 10.1016/j.bbabio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filosto M, Mancuso M, Nishigaki Y, Pancrudo J, Harati Y, Gooch C, Mankodi A, Bayne L, Bonilla E, Shanske S, Hirano M, DiMauro S. Clinical and genetic heterogeneity in progressive external ophthalmoplegia due to mutations in polymerase gamma. Arch Neurol. 2003;60:1279–1284. doi: 10.1001/archneur.60.9.1279. [DOI] [PubMed] [Google Scholar]
- Foury F. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989;264:20552–20560. [PubMed] [Google Scholar]
- Foury F, Kucej M. Yeast mitochondrial biogenesis: a model system for humans? Curr Opin Chem Biol. 2002;6:106–111. doi: 10.1016/S1367-5931(01)00276-9. [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- Groudinsky O, Dujardin G, Slonimski PP. Long range control circuits within mitochondria and between nucleus and mitochondria. II. Genetic and biochemical analyses of suppressors which selectively alleviate the mitochondrial intron mutations. Mol Gen Genet. 1981;184:493–503. doi: 10.1007/BF00352529. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Speijer D, Kirkwood TB. The road to rack and ruin: selecting deleterious mitochondrial DNA variants. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130451. doi: 10.1098/rstb.2013.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath R, Hudson G, Ferrari G, Futterer N, Ahola S, Lamantea E, Prokisch H, Lochmuller H, McFarland R, Ramesh V, Klopstock T, Freisinger P, Salvi F, Mayr JA, Santer R, Tesarova M, Zeman J, Udd B, Taylor RW, Turnbull D, Hanna M, Fialho D, Suomalainen A, Zeviani M, Chinnery PF. Phenotypic spectrum associated with mutations of the mitochondrial polymerase gamma gene. Brain. 2006;129:1674–1684. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24:761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Kierdaszuk B, Jamrozik Z, Tonska K, Bartnik E, Kaliszewska M, Kaminska A, Kwiecinski H. Mitochondrial cytopathies: clinical, morphological and genetic characteristics. Neurol Neurochir Pol. 2009;43:216–227. [PubMed] [Google Scholar]
- Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi GP, Zeviani M. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Lipinski KA, Kaniak-Golik A, Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome- from genetics to evolution and systems biology. Biochim Biophys Acta. 2010;1797:1086–1098. doi: 10.1016/j.bbabio.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Lodi T, Dallabona C, Nolli C, Goffrini P, Donnini C, Baruffini E. DNA polymerase gamma and disease: what we have learned from yeast. Front Genet. 2015;6:106. doi: 10.3389/fgene.2015.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Lucas P, Lasserre JP, Plissonneau J, Castroviejo M. Absence of accessory subunit in the DNA polymerase gamma purified from yeast mitochondria. Mitochondrion. 2004;4:13–20. doi: 10.1016/j.mito.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Filosto M, Bellan M, Liguori R, Montagna P, Baruzzi A, DiMauro S, Carelli V. POLG mutations causing ophthalmoplegia, sensorimotor polyneuropathy, ataxia, and deafness. Neurology. 2004;62:316–318. doi: 10.1212/WNL.62.2.316. [DOI] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JL, Sanford LN, Wittwer CT. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn. 2010;10:219–240. doi: 10.1586/erm.09.84. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogur M, St. John R, Nagai S. Tetrazolium overlay technique for population studies of respiration deficiency in yeast. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- Palin EJ, Lesonen A, Farr CL, Euro L, Suomalainen A, Kaguni LS. Functional analysis of H. sapiens DNA polymerase gamma spacer mutation W748S with and without common variant E1143G. Biochim Biophys Acta. 2010;1802:545–551. doi: 10.1016/j.bbadis.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta O, Lubas M, Lipinski KA, Piatkowski J, Malecki M, Golik P. DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA. Genetics. 2010;184:959–973. doi: 10.1534/genetics.110.113969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Kachroo AH, Yellman CM, Marcotte EM, Johnson KA. Yeast cells expressing the human mitochondrial DNA polymerase reveal correlations between polymerase fidelity and human disease progression. J Biol Chem. 2014;289:5970–5985. doi: 10.1074/jbc.M113.526418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Zeviani M. Disorders of nuclear-mitochondrial intergenomic communication. Biosci Rep. 2007;27:39–51. doi: 10.1007/s10540-007-9036-1. [DOI] [PubMed] [Google Scholar]
- Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Stricker S, Pruss H, Horvath R, Baruffini E, Lodi T, Siebert E, Endres M, Zschenderlein R, Meisel A. A variable neurodegenerative phenotype with polymerase gamma mutation. J Neurol Neurosurg Psychiatry. 2009;80:1181–1182. doi: 10.1136/jnnp.2008.166066. [DOI] [PubMed] [Google Scholar]
- Stuart GR, Santos JH, Strand MK, Van Houten B, Copeland WC. Mitochondrial and nuclear DNA defects in Saccharomyces cerevisiae with mutations in DNA polymerase gamma associated with progressive external ophthalmoplegia. Hum Mol Genet. 2006;15:363–374. doi: 10.1093/hmg/ddi454. [DOI] [PubMed] [Google Scholar]
- Stumpf JD, Copeland WC. Mitochondrial DNA replication and disease: insights from DNA polymerase gamma mutations. Cell Mol Life Sci. 2011;68:219–233. doi: 10.1007/s00018-010-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf JD, Copeland WC. The exonuclease activity of the yeast mitochondrial DNA polymerase gamma suppresses mitochondrial DNA deletions between short direct repeats in Saccharomyces cerevisiae. Genetics. 2013;194:519–522. doi: 10.1534/genetics.113.150920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf JD, Copeland WC. MMS exposure promotes increased MtDNA mutagenesis in the presence of replication-defective disease-associated DNA polymerase gamma variants. PLoS Genet. 2014;10:e1004748. doi: 10.1371/journal.pgen.1004748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf JD, Bailey CM, Spell D, Stillwagon M, Anderson KS, Copeland WC. mip1 containing mutations associated with mitochondrial disease causes mutagenesis and depletion of mtDNA in Saccharomyces cerevisiae. Hum Mol Genet. 2010;19:2123–2133. doi: 10.1093/hmg/ddq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowska K, Foury F. A cluster of pathogenic mutations in the 3′–5′ exonuclease domain of DNA polymerase gamma defines a novel module coupling DNA synthesis and degradation. Hum Mol Genet. 2010;19:3516–3529. doi: 10.1093/hmg/ddq267. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Tzoulis C, Engelsen BA, Telstad W, Aasly J, Zeviani M, Winterthun S, Ferrari G, Aarseth JH, Bindoff LA. The spectrum of clinical disease caused by the A467T and W748S POLG mutations: a study of 26 cases. Brain. 2006;129:1685–1692. doi: 10.1093/brain/awl097. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Luoma P, Rantamaki M, Al Memar A, Kaakkola S, Hackman P, Krahe R, Lofgren A, Martin JJ, De Jonghe P, Suomalainen A, Udd B, Van Broeckhoven C. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–1257. doi: 10.1212/01.WNL.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- Viikov K, Valjamae P, Sedman J. Yeast mitochondrial DNA polymerase is a highly processive single-subunit enzyme. Mitochondrion. 2011;11:119–126. doi: 10.1016/j.mito.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Viikov K, Jasnovidova O, Tamm T, Sedman J. C-terminal extension of the yeast mitochondrial DNA polymerase determines the balance between synthesis and degradation. PLoS One. 2012;7:e33482. doi: 10.1371/journal.pone.0033482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire E, Davidzon G, DiMauro S, Akman HO, Sadleir L, Haas L, Zuccollo J, McEwen A, Thorburn DR. Juvenile Alpers disease. Arch Neurol. 2008;65:121–124. doi: 10.1001/archneurol.2007.14. [DOI] [PubMed] [Google Scholar]
- Wong LJ, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, Basinger AA, Burton BK, Swoboda K, Gilbert DL, Vanderver A, Saneto RP, Maranda B, Arnold G, Abdenur JE, Waters PJ, Copeland WC. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum Mutat. 2008;29:E150–E172. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J Biol Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.