Abstract
Transglutaminase-2 (TG2) is a critical crosslinking enzyme in the extracellular matrix (ECM) and tumor microenvironment (TME). While its expression has been linked to colorectal cancer (CRC), its functional role in the processes that drive disease appears to be context-dependant. There is now considerable evidence of a role for microRNAs (miRs) in the development and progression of cancer, including metastasis. A cell model of metastatic colon adenocarcinoma was used to investigate the contribution of miRs to the differential expression of TG2, and functional effects on inflammatory and invasive behaviour. The impact of TG2 in CRC was analysed in human colorectal tumour specimens, and by manipulations in SW480 and SW620 cells. Effects on invasive behaviour were measured using transwell invasion assays, and cytokine production assessed by ELISA. TG2 was identified as a target for miR-19 by in silico analysis, which was confirmed experimentally. Functional effects were evaluated by overexpression of pre-miR-19a in SW480 cells. Expression of TG2 correlated inversely with invasive behaviour, with knockdown in SW480 cells leading to enhanced invasion, and overexpression in SW620 cells the opposite. TG2 expression was observed in CRC primary tumours but lost in liver metastases. Finally miR-19 overexpression and subsequent decreased TG2 expression was linked to chromosome-13 amplification events, leading to altered invasive behaviour in CRC cells.
Keywords: Colorectal, transglutaminase-2, micro-RNA, invasion, metastasis
INTRODUCTION
Colorectal Cancer (CRC) is the fourth most common malignancy worldwide, and the third most common malignant cause of mortality in the western world [1, 2]. Although advances in screening and treatment have improved life expectancy in recent decades, prognosis remains significantly poorer in later stages when disease has spread to lymph nodes and distant metastatic sites [3]. Understanding and preventing this invasive progression would therefore significantly benefit patient outcome worldwide.
Transglutaminase-2 (TG2) activity has been linked to multiple biological processes associated with tumour development and progression, such as cell adhesion, motility, invasion, apoptosis, chemoresistance and epithelial-mesenchymal transition [4, 5]. The most ubiquitous member of the transglutaminase family of protein cross-linking enzymes, TG2 has been observed in various cancer tissues and cell lines, with activity linked to disease progression and metastasis in tumours with a diverse range of origins [6-10]. TG2 has been identified as a potential marker of CRC cancer progression by immunohistochemical analysis, following previous work demonstrating differential expression of TG2 in CRC cell lines with different metastatic potential [11-13]. However, published studies aiming to identify a definitive role for TG2 in cancer cell biology have demonstrated sometimes contradictory functional roles, such as promoting or inhibiting apoptosis. TG2 therefore appears to act in a context-dependant manner which may relate to cellular location and the availability of its many identified protein substrates [14], or to the balance between different isotypes of the enzyme which have been shown to have opposing consequences on cell behaviour [15].
MicroRNAs (miRNAs) are a family of short, non-coding, single-stranded RNAs, which inhibit the function of multiple target genes by binding to their 3′ untranslated region, leading to direct degradation of target mRNA or inhibiting translation [16]. A wide body of work now links miRNA expression to CRC by altering the expression of oncogenic and tumour suppressive genes [17, 18]. Further, miRNA deregulation is strongly linked to disease progression, with changes in miRNAs linked to metastasis [19, 20]. These “metastaMirs” are attractive therapeutic targets for treating metastatic CRC, as each miRNA influences the expression of multiple proteins downstream which may contribute to the development of the complex, multifactorial metastatic phenotype [21].
Since TG2 has multiple cell substrates and plays a critical role in cancer cell behaviour, its expression is carefully controlled. As well as translational regulation, TG2 abundance is also controlled through the SUMO pathway [22], and enzymatic function is dependent on the presence of calcium and inhibited by GTP [23]. To date, few studies have examined miRNA regulation of TG2, despite both TG2 and miRNAs being closely linked to cancer progression. In this study we investigated the differential expression of TG2 in colon cancer cell lines and tissue sections taken from primary and metastatic tumours, examined how TG2 expression affected invasive characteristics and inflammatory mediators synthesised by these cells, and finally determined how miRNA regulation alters these functional properties.
MATERIALS AND METHODS
Cell lines and reagents
The primary adenocarcinoma cell line SW480 was obtained from the European Collection of Cell Culture (Salisbury, UK), along with the patient-matched lymph-node metastasis-derived line SW620. Cells were cultured and passaged according to supplied information. SiRNA targeted against TG2 was obtained from Invitrogen, and transfected into cells using Hyperfect reagent (Qiagen) according to the manufacturer’s recommendations. TG2 expression plasmid (pLPCX-TG2) and the active site mutant (pLPCX-C277S) plasmid were used as previously described [22], along with an empty vector control (pcDNA3.1),and transfected into cells using Lipofectamine LTX (Invitrogen) according to the manufacturer’s instructions. Pre-MiR-19a plasmid (Genecoepia, Rockville MD) and a corresponding scrambled plasmid control (SCC) were transfected into SW480 cells using Fugene 6 (Roche) according to manufacturer’s recommendations (SW480/miR19A, and SW480/SCC). Stable transfection was achieved by selecting resistant clones using puromycin (1μg/mL), cell sorting by FACS for the IRES-driven GFP reporter, and after expansion used at early passage (<10). 24h prior to experiments, cells were also transfected with a miRNA-19A mimic or corresponding scramble control (Qiagen) using Hyperfect (Qiagen) according to manufacturer’s instructions. After 24h incubation, cells were trypsinized and used for experimental testing.
Matrigel invasion assay
Invasion assays were performed using 8μM transwell plates (Corning). Matrigel (BD Biosciences) was diluted 1:3 in serum-free medium and allowed to dry in the upper chamber of the wells. 100,000 cells were then added to the upper chamber in serum-free medium, and complete medium was added to the lower chamber as a chemoattractant. After 24 hours, cells invading the matrigel were released by trypsinisation, and counted using a CASY TTC counter (Roche Innovatis).
Western blot
Western blotting was performed to assess cellular expression of TG2, and actin expression used to confirm equal protein loading. Cells were lysed in PBS + 1% NP-40, and briefly sonicated before centrifugation to remove insoluble material. Alternatively, in some experiments protein extracts were prepared in 1% SDS following Trizol treatment according to the manufacturer’s protocol (Ambion). Total protein content of these preparations was assessed by BCA assay (Thermo Scientific), equal quantities of protein loaded onto SDS-PAGE gels for electrophoresis, and transferred to nitrocellulose membrane (Amersham). Membranes were blocked with 5% non-fat milk in TBS+0.5% Tween, then probed with appropriate primary antibodies; TG2 (AbCam, clone CUB7402, 1:2000), and actin to confirm equal protein loading (Santa Cruz, 1:2000). Bound proteins were detected using HRP-labelled secondary antibodies (Santa Cruz, 1:2000), and ECL chemiluminescent substrate (Thermo scientific).
TG2 activity assay
The assay for TG2 activity was performed as previously described [24]. Based on incorporation of the TG2 substrate monodanslycadaverin (bio-MDC, Cambridge Bioscience), cells were incubated for 1h with the substrate in the presence of 200mM CaCl2, fixed in 4% paraformaldehyde, and permeabilised with 0.1% Triton X-100 (Sigma). Biotinylated substrate was revealed using streptavidin-FITC (BD Pharmingen, 1:150), and TG2 protein co-stained using the antibody clone CUB7402 (AbCam, 1:100) and detected by anti-mouse Alexa-594 antibody (BD Biosciences, 1:300). Cells were counterstained with the nuclear stain DAPI (1:1000, Invitrogen) before mounting in Slow-fade medium (Invitrogen) and visualised under a fluorescent microscope. Mean corrected total cell fluorescence was calculated from fluorescence intensities obtained in ImageJ, using the equation Integrated Density – (Area of cell * background).
Flow cytometry and immunoassay
The expression of TG2 was assessed by flow cytometry in order to compare surface expression and intracellular expression. Cells were trypsinized from culture dishes, washed 3 times in PBS and suspended in flow buffer (PBS + 1% FCS, 0.05% sodium azide). Membrane permeabilisation was performed where necessary using Fix-perm reagents (EBiosciences, according to manufacturer’s instructions). For both intracellular and cell-surface staining, the primary TG2 antibody CUB7402 was used (1:100, Abcam), and detection performed using anti-mouse FITC-conjugated secondary antibody (1:300, Sigma). IL-8 production was measured using a commercial ELISA assay (R&D Systems).
Immunohistochemical and miRNA quantification from CRC patients
Immunohistochemical staining was performed on formalin-fixed specimens from patients undergoing resections for CRC at the University Hospital Southampton as part of an NIHR portfolio study (UK CRN ID6067). Tumour specimens were snap-frozen in liquid nitrogen within 10 minutes of surgery and stored in a designated UK Human Tissue Act-approved tumour bank. Samples were selected from three clinically distinct groups; a) colonic tissue from early stage disease (stage I/II), b) colonic tissue from late stage disease (lymph node involvement, stage III/IV), c) liver tissue from CRC metastatic disease (stage IV). Antigen retrieval was performed by microwave citrate method, and staining using the antibody clone CUB7402 (AbCam, 1:800). Semi-quantitative scoring of TG2 levels on whole tissue sections was performed independently and in a blinded manner by a specialist pathologist (GT) and a further investigator (AM). A modified 3-point scoring method was used: 1) low/negative staining (<10% positivity), 2) focal/patchy staining (10-50% positivity), 3) strong diffuse staining (>50% positive). All patients provided informed consent in accordance with the Helsinki protocol and the study was approved by the regional research ethics committee.
MicroRNA analysis
Prediction of miRNA targets for TG2 was performed using four target prediction algorithms: Targetscan (http;//www.targetscan.org; release 5.1); miRanda (http://www.microrna.org; 2010 release); miR Walk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/); and DIANA – microT (http://diana.cslab.ece.ntua.gr/microT/; v3.0).For quantification of miRNA levels in patient samples, laser-capturemicrodissection (LCM) was performed on frozen human tissue specimens. Sections were fixed in 75% ethanol, stained with 1% cresyl violet and dehydrated with ethanol before air drying. Microdissection was performed on the Leica AS microdissection platform, and captured CRC tissue collected directly into lysis buffer prior to RNA isolation (RNAaqueous microprep kit, Ambion). 10 randomly selected specimens were analysed from 2 clinically defined groups; a) primary CRC tumour tissue, b) patient-matched liver tissue from CRC metastasis. The expression of miRNAs was performed using TaqMan™ assays (Applied Biosystems) according to the manufacturer’s instructions, and normalised to U6 expression. Expression of miRNA was calculated relative to the endogenous reference gene U6B using the formula 2-ΔΔCT. MiRNA expression in cell lines was obtained from microarray data published previously [31], which is available in the EBI database (http://www.ebi.ac.uk/arrayexpress/experiments/; accession number E-MEXP-3270).
SNP6 array hybridization, data extraction and analysis
DNA was isolated from cell lines using the Qiagen DNeasy method prior to being purified, amplified, labeled and hybridized to the Affymetrix SNP6 platform (Affymetrix, Santa Clara, CA) as previously described [25]. The data was aligned (Build 36.3) and analysed by two independent researchers using Partek Genomics Suite (Partek Inc, Missouri, USA). Copy number alterations (CNAs) were defined as a deviation of 50 consecutive probes from a normal value of 2 (±0.3), within a consecutive genomic window of 50 Kilobases. The 270 HapMap Reference baseline (Affymetrix) was used as a control and germline copy number variants were excluded based on the Database of Genomic Variants (http://projects.tcag.ca/variation/). The allele ratio was calculated for each sample using the HapMap Allele Reference baseline (Affymetrix) and in the absence of paired normal DNA; copy number neutral loss of heterozygosity (CNNLOH) was defined as a region greater than 20Mb, extending to a telomere. We also analysed copy number data from 437 Colon Adenocarcinoma cases from the Cancer Genome Atlas data COAD data set (https://tcga-data.nci.nih.gov/tcga/) using the UCSC Cancer Genomics Browser(https://genome-cancer.soe.ucsc.edu/) to identify recurrent regions of copy number gain and loss that include our miRNAs of interest.
TG2 3′UTR luciferase reporter assay
TG2-3′UTR wild-type and TG2-3′UTR mutant vectors were generated by GenScript Inc (NJ, USA). A 750bp region of the TGM2 gene 3′UTR containing the single predicted miR-19a binding site was synthesized and was sub-cloned into the pRL-TK plasmid vector (Promega, UK) downstream of the Renilla-Luc gene at the Xba1 site. Insert orientation was in the same sense as the luciferase reporter in pRL-TK. The mutant vector was generated by changing the sequence TTTGCACA to TTTATTGA. Reporter genes were transfected into SW480/SCC and SW480/miRNA-19a lines using Fugene 6, and luciferase activity quantified using the Dual-luciferase reporter system (Promega) to collect the activity of firefly (PGL3 vector control) and renilla (TG2-3′UTR) measured in the same sample. SW480/SCC and SW480/miR-19a cells were plated at 4000 cells per well in 100ul DMEM in a CulturPlate-96 microplate (PerkinElmer, MA USA). 24 hours after plating, cells were transfected with 30nM Pre-miRs, 10ng PGL3 and 500ng of TG2-3′UTR (wild type or mutant) vectors per well. Light produced was measured using a plate reader at 2 second intervals and activity was calculated as renilla activity per light unit of firefly activity. 3′UTR renilla activity was normalised to firefly activity, and results presented as the difference between the wild-type and mutant vectors.
Statistical analysis
Statistically significant differences between experimental conditions were assessed using student’s t-test, and paired t-test where appropriate. Alternatively, where multiple comparisons were necessary, ANOVA with Bonferroni post-hoc test was used. All analyses were performed in GraphPad Prism, and p values < 0.05 were considered statistically significant. All experiments were performed a minimum of 3 times.
RESULTS
TG2 expression is decreased in SW620 cells compared to SW480 cells
It has been reported previously that differential TG2 levels are observed in the SW model of metastasis, with high levels in the primary colon adenocarcinoma SW480 line, and significantly reduced levels in the patient-matched SW620 lymph-node-derived line [11]. We confirmed these observations at protein level by Western blot (supplementary figure 1A), showing considerable reduction of the full-length 79kB isoform of TG2 in SW620 cells. Decreased protein expression of this full-length transcript in SW620 cells was matched by a decrease in mRNA transcript level (supplementary figure 1B). Interestingly, protein expression of the 55kD exon-10-truncated splice variant of TG2 (TG2-E10) was actually relatively higher in SW620 cells (supplementary figure 1A). This is in contrast to a decrease in mRNA transcript level (supplementary figure 1B), indicating differential regulation of the splice variants.
Reduced TG2 is associated with increased invasiveness in SW480/SW620 cells
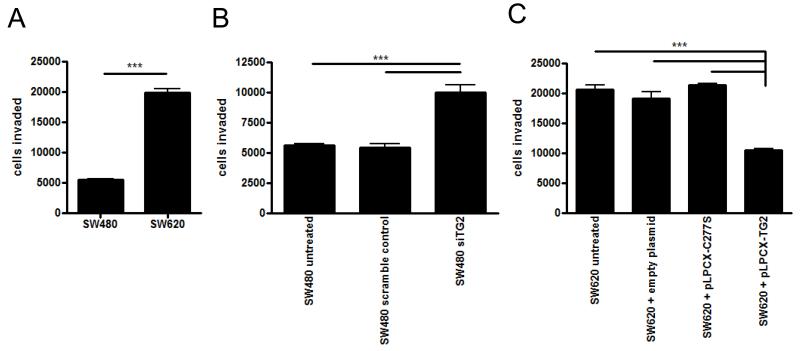
Because TG2 levels were significantly reduced in SW620 cells when compared to SW480, and SW620 cells are more invasive [12, 26], we hypothesised that TG2 may be inversely correlated to invasive potential. In an invasion assay using Matrigel as a substrate, SW620 cells, as expected, displayed increased invasive behaviour (figure 1A), with counts of invading cells reaching 20,000 compared to 5,500 for SW480 cells. This was not due to significant differences in cell proliferation within the lower chamber as differences were not observed in an MTT proliferation assay (data not shown), nor were any differences accounted for by apoptosis using Annexin V staining (data not shown). When SW480 cells were treated with siRNA to TG2, the cells became significantly more invasive, showing a 100% increase in the number of invading cells when compared to cells either untreated or treated with a scrambled, control siRNA (p < 0.0001, figure 1B). In contrast, overexpression of TG2 in SW620 cells using a plasmid encoding TG2 significantly decreased the number of invading cells by around 50%, compared to untreated, empty vector, or cells transfected with the cross-linking-deficient TG2 plasmid C277S (p < 0.0001, figure 1C). Thus TG2 cross-linking activity appears to restrict invasive behaviour of SW cells, whereas loss of TG2 in SW620 cells facilitates invasion.
Figure 1. TG2 is involved in the invasive ability of SW cells.
The number of cells invading through matrigel substrate towards an FCS chemoattractant in 24 hours was compared for SW480 cells and SW620 cells (A), in the presence or absence of TG2 siRNA to knock down TG2 expression, or scrambled siRNA as a control, in SW480 cells (B), and following the transfection of a TG2 expression plasmid or the catalytically inactive expression plasmid C277S into SW620 cells (C). Statistical significance is indicated by the asterisks, *** = p < 0.0001.
TG2 activity in SW cells is localised intracellularly
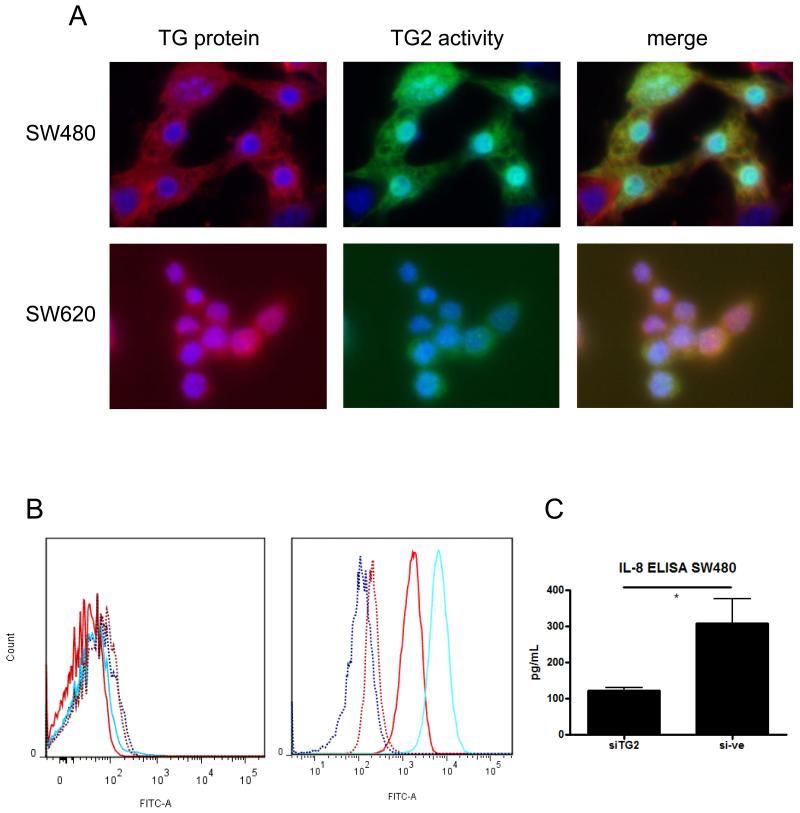
Since TG2 activity decreased invasiveness of SW cells, we wished to define its cellular localisation, because cell-surface TG2 may interact directly with the cell-surface and extracellular matrix proteins [27, 28], whereas intracellular TG2 has a cell signalling function [24, 29, 30]. Imaging of TG2 by immunofluorescent microscopy showed that TG2 protein was expressed throughout the cytoplasm in both SW480 and SW620 cells (figure 2A, red). Furthermore, cytoplasmic TG2 was catalytically active as assessed using a fluorescent TG2 substrate (figure 2A, green). The staining for protein and activity were co-localised (Figure 2A, merge). Expression of both TG2 protein and cross-linking activity was higher in SW480 cells compared to SW620 cells (mean CTCF of 33.3×105compared to 18.3×105 for protein, 19×105 compared to 9.3×105 for activity, respectively), consistent with previous data. Significant TG2 was also observed localised to the nucleus (figure 2A). By comparing nuclear expression to cytoplasmic expression, we observed that 17%/24% of the cellular TG2 protein/activity was localised to the nucleus of SW480 cells, and 11% of both protein/activity was localised to the nucleus of SW620 cells. The absence of cell surface TG2 data was confirmed by flow cytometry. Cells stained for cell surface expression of TG2 showed very limited staining, and no differences were observed between SW480 and SW620 cells (figure 2B). By contrast, when cells were permeabilised prior to staining, strong expression of TG2 was observed, with >99% cells expressing TG2, and with considerably stronger staining in SW480 cells compared to SW620 cells.
Figure 2. TG2 is distributed throughout the cell cytoplasm, and is not localised to the cell surface in SW cells.
The cellular distribution of TG2 was assessed by measuring the presence of TG2 protein in fixed cells by immunolocalisation with the antibody clone 7402 coupled to Alexa 492-conjugated secondary antibody (A, red stain), and co-localising protein to TG2 activity visualised using the biotinylated MDC-substrate assay coupled to a FITC-streptavidin secondary antibody (A, green). DAPI counterstaining (blue) indicates the cell nucleus (A). The presence of cell-surface TG2 was also determined by flow cytometry (B), using TG2 staining of whole live cells with the antibody clone 7402 (left panel), compared with cells permeabilised to assess intracellular levels (right panel). SW480 cells are displayed as blue lines, and SW620 cells as red lines. The dotted lines represent unstained controls. IL-8 production by SW480 cells in the presence or absence of siRNA to TG2 was analysed by ELISA (C). * = p < 0.05.
TG2 activity influences inflammatory profile of SW cells
Since intracellular TG2 is known to drive pro-inflammatory signalling [9, 23, 29], we also measured IL-8 production by SW480 and SW620 cells. IL-8 was not detected in SW620 supernatants, but was clearly produced by SW480 cells. Using TG2 specific siRNA, TG2 levels could be reduced from 300pg/ml to 100pg/ml compared to control (figure 2C, p < 0.05). We next compared the expression of cytokines in the two cell lines, and observed that the production of pro-inflammatory cytokines by SW480 cells was generally higher when compared to SW620 cells, including IL-8 and TNF-α (supplementary figure 2A, p < 0.05). However, although TG2 siRNA reduced IL-8 and TNF-α mRNA expression, the differences were not significantly different (supplementary figure 2B), and we could not detect expression of TNF-α in cell supernatants (data not shown). Since pro-inflammatory signalling pathways are also linked to the release of enzymes that can break down tissue, we also investigated whether Matrix Metalloproteinase (MMP) production was influenced by TG2. As expected, the more invasive SW620 cells expressed higher levels of MMP mRNA, notably significantly higher levels of MMP-7 (p < 0.05, supplementary figure 3A). Interestingly, SW620 cells also expressed significantly lower mRNA of one member of this enzyme family, MMP-14 (p < 0.05, supplementary figure 3A). However, when we compared MMP mRNA expression in SW480 cells treated with and without siRNA to TG2, no significant changes to MMP mRNA expression were observed (supplementary figure 3B), indicating that TG2-linked invasion was not directly related to enhanced MMP production.
TG2 levels are downregulated in metastatic tumours compared to primary tumours
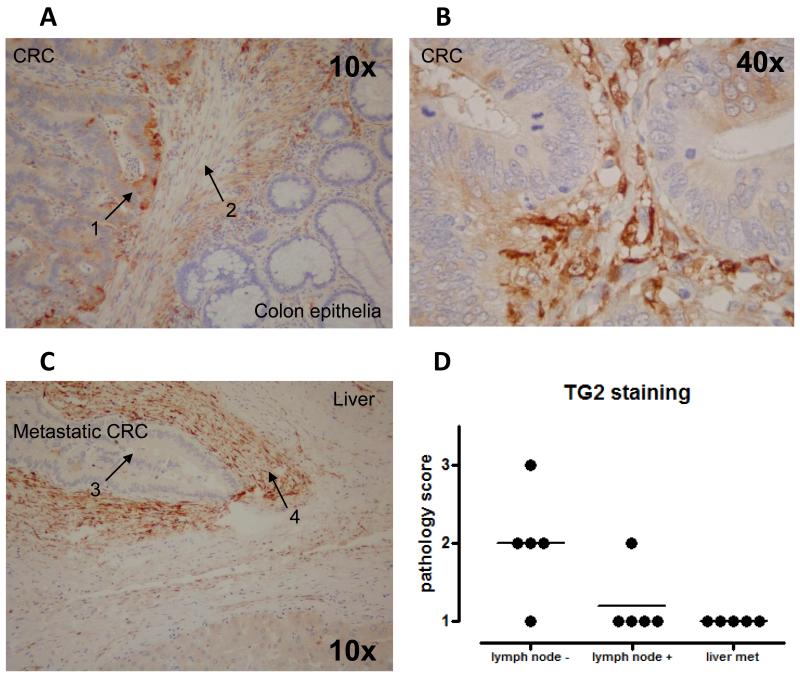
Because our data in the SW model demonstrated downregulation of TG2 in the metastatic SW620 cell line, and correlation of TG2 levels with tumour invasiveness, we investigated the expression of TG2 in human CRC sections, comparing primary tumours taken from patients grouped according to confirmed lymph node involvement, and from liver metastases. Staining for TG2 was detected in primary tumour sections (figure 3A and 3B), mainly at the invasive front (figure 3A), but not in liver metastases (figure 3C). Scoring by two independent investigators quantified this differential expression pattern (figure 3D), with TG2 expression found to negatively correlate with tumour stage. However, in these sections the most intense staining was detected in the tumour stroma (figure 3A-C), with cells surrounding the cancerous cells appearing to produce significant amounts of TG2, both at the primary and metastatic sites. No TG2 staining was observed in normal epithelia (data not shown).
Figure 3. TG2 is expressed in human CRC specimens, but lost in liver metastases.
TG2 was stained in CRC tumour sections using antibody clone 7402. Panel A shows a representative image showing staining in primary CRCs, with arrows indicating tumour (1) and stroma (2). Panel B shows TG2 staining at higher magnification in primary CRC, and panel C illustrates a representative image of a liver metastasis, with arrows showing tumour cells (3) and stroma (4). In both cases, strong stromal expression of TG2 is observed. As illustrated following scoring by a senior pathologist, TG2 expression is lost in sections taken from liver metastases (panel D).
TG2 is a predicted miRNA target
To better understand the differential protein expression of TG2 splice variants in SW cells (supplementary figure 1A), we next examined the mRNA expression of TG2 splice variants by RT-PCR. In SW480 cells, expression of the mRNA for all of the TG2 splice variants assessed was higher in SW480 compared to SW620 cells, at differences ranging from 100-2000-fold (p < 0.0001, supplementary figure 1B). The protein and mRNA expression of full-length TG2 therefore appear to correlate, but the protein level of the 55kD splice variant does not correlate to the mRNA expression of TG2-E10. There is increasing evidence that miRNAs are intimately involved in the metastatic progression of colon cancer [18, 20, 31, 32]. MiRNAs bind to the 3′-UTR of their target genes, and the TG2 splice variants are 3′-truncated [15, 33]. We therefore examined the possibility that TG2 may be a target for miRNA regulation, and explain why expression of TG2 splice variants is different at mRNA and protein level. Potential miRNA regulators of TG2 were identified by in silico analysis of the 3′UTR using a panel of 4 target prediction algorithms. These identified only a single miRNA, miRNA-19a/b, predicted to bind to the 3′UTR of TG2 across all platforms. Binding of miRNA-19a/b was predicted to occur at a conserved UUUGCACA sequence at position 1588-1595 of the 3′-UTR (supplementary figure 4A), suggesting miRNA-19a/b may represent a potential regulatory miRNA for TG2.
MiRNA-19 is upregulated in metastatic tumours compared to primary tumours
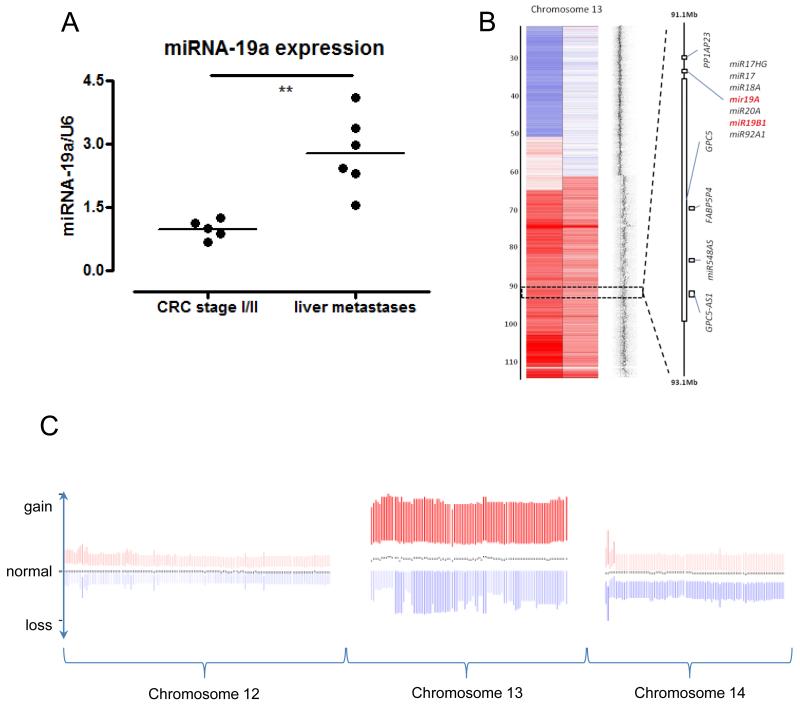
The level of miRNA-19 in sections taken from patients with CRC was assessed using LCM, in order to isolate epithelial and stromal expression (Supplementary figure 4B). MiRNA microarray profiling showed that miRNA-19a/b expression was significantly different in tumour epithelia when compared to normal epithelia (p < 0.05, supplementary figure 4C). MiRNA-19a/b expression was not significantly different in tumour stroma compared to normal stroma (Supplementary figure 4D). Noticably, both epithelial and stromal analyses showed several specimens with high expression of miRNA-19; interestingly, however, these did not correlate to the same patients for epithelial/stromal expression. Our data indicated that differences in TG2 expression were observed between primary tumour specimens and liver metastases. We therefore analysed miRNA-19a using TaqMan™ in LCM samples to compare these two groups. MiRNA-19a expression was significantly up-regulated in sections taken from liver metastases compared to sections taken from primary tumours (Figure 4A, p < 0.01).
Figure 4. MicroRNA-19a is upregulated in metastatic cells through instability of chromosome 13.
The expression of miRNA-19a was assessed by TaqMan in RNA isolated from sections of primary CRC tissue compared to liver metastases (A). Copy number heatmap from SNP6 data of chromosome 13 forSW620 and SW480 to the left and right, respectively. normal copy number, gain and loss are shown in grey, red and blue respectively, Data show duplication/ amplification of distal 13q including the miRNA-19a and miRNA-19b1 loci (B). Relative copy number for chromosomes 12, 13 and 14 positioned from left to right from the COAD data set (C). The mean profile for each chromosome is shown by the black dots, indicating copy number at different points along each chromosome from 437 patients with CRC. The colour intensity is proportional to the deviation from the median, and the shift upwards towards red indicates increased chromosome 13 copy number in relation to chromosomes 12 and 14.
MiRNA-19 is upregulated in SW620 cells compared to SW480 cells, and its genomic locus is amplified in CRC
To test if overexpression of miRNA-19 could be a mechanism for TG2 downregulation, we first established the levels of miRNA-19a and miRNA-19b in SW620 cells compared to SW480 cells. MicroRNA microarray profiling of SW620 and SW480 cells demonstrated a 2.6-fold increase of miRNA-19a and a 3-fold increase in miRNA-19b in SW620 cells compared to SW480 cells (normalised values of 274.49 vs 105.45, and 3082.57 vs 1026.25 for miRNA-19a and miRNA-19b, p = 0.01, and <0.0001, respectively [31]), subsequently validated by qPCR analysis, confirming observations from other groups [20, 34]. Deregulation of miRNAs has been attributed to genomic copy number changes [35], and miRNAs have been noted to be over-represented in regions of genomic gain in CRC [36], consequently we next examined if copy number changes could account for the upregulation of miR-19a and b in the SW cell lines. SW480 and SW620 cells were analysed using a Genome-Wide Human SNP Array 6.0 with the hapmap 270.422 data set as reference. MiR-19a and miR-19b1 are located on chromosome 13 and both were found to be gained in both cell lines (figure 4B). By contrast, miRNA-19b2 showed normal copy number in both cell lines from its locus on chromosome X (data not shown). To clarify if the miR-19a/b loci are subject to copy number change in primary human CRC, we next examined the Cancer Genome Atlas (TCGA) dataset. In the data available from 437 human CRCs, both miR-19a and miR-19b were subject to amplification through recurrent chromosome 13 gains. However, this was due to recurrent gains in chromosome 13, rather than any focal copy number alterations at the specific miR-19 loci (figure 4C). The data therefore indicate that advanced CRC frequently gains an additional copy of the whole chromosome 13. Further analysis of the TCGA dataset reveals that this amplification in CRC patients occurs in later stages of disease; no significant differences are observed when comparing patients with stage I, II or III disease, but significant differences (p < 0.05) are seen when comparing stage III and IV disease (data not shown).
MiRNA-19 directly targets TG2 and alters the invasive behaviour of SW cells
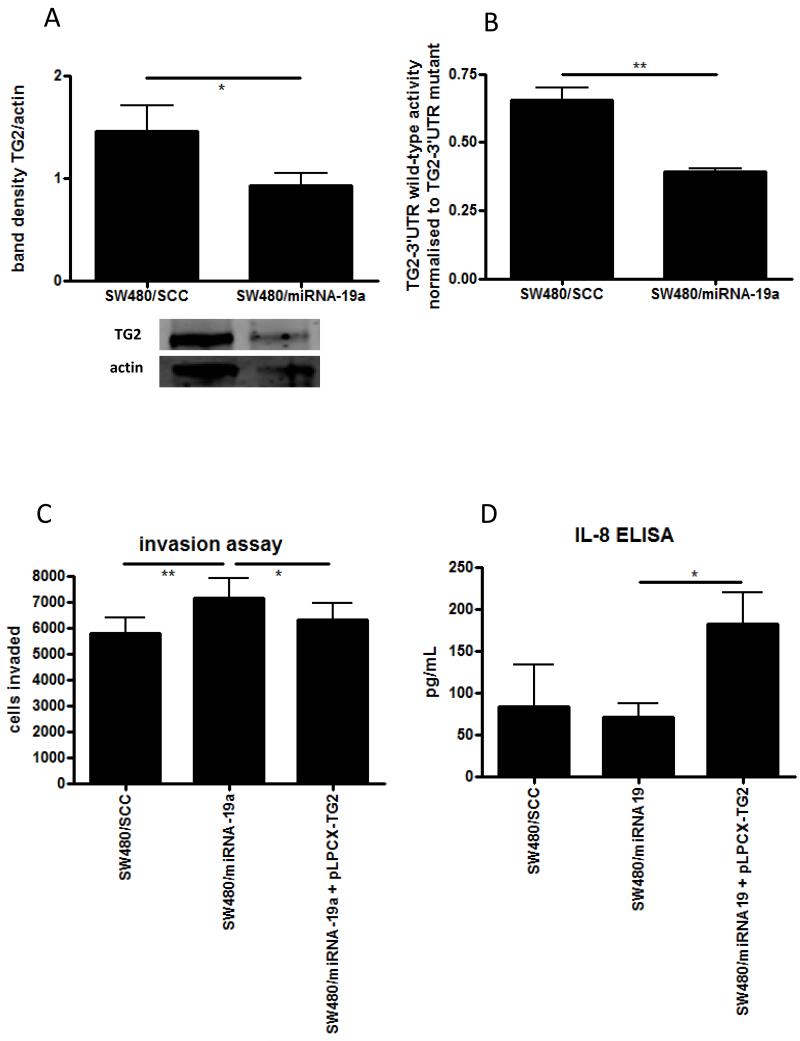
To confirm our in silico prediction of miRNA-19 targeting TG2, we manipulated miRNA-19a levels in SW480 cells by establishing stable cell lines over-expressing a scrambled plasmid control (SW480/SCC), a miRNA-19a expression plasmid (SW480/miR-19a), and by transient transfections with molecular miRNA mimics. Down-regulation of TG2 was observed by Western blot in SW480 cells manipulated to over-express miRNA-19a (figure 5A), which was statistically significant when assessed by densitometry (p < 0.05). The direct binding of miR-19a to the 3′UTR of TG2 was assessed using a luciferase reporter assay, which showed a reduction of 3′UTR activity by almost 50% in the SW480/miR-19a cells (p < 0.01, figure 5B). To evaluate the functional consequences of this, invasion assays were performed on these stable cell lines. SW480 cells over-expressing miRNA-19a showed enhanced invasiveness when compared to control cells (Figure 5C, p < 0.01). Thus elevating the levels of miRNA-19a has a similar effect to reducing TG2 using siRNA. To confirm that TG2 was the target for miRNA-19a in this model, we transfected SW480/19a cells with the TG2 expression plasmid, lacking the 3′-UTR binding site for miRNA-19a (pLPCX-TG2). Restoring TG2 in this way reduced invasion of SW480/miRNA-19a cells to a level similar to that observed in 480/SCC cells in a statistically significant manner (Figure 5C, p < 0.05). Finally, IL-8 production from the stable SW480 cell lines was assessed by ELISA. Although slightly lower IL-8 production was seen from SW480/miRNA-19a cells compared to SW480/SCC cells, this was not significant (Figure 5D). However, transfection with the pLPCX-TG2 plasmid significantly increased the production of IL-8 (p < 0.05), further illustrating the functional effect of TG2 lacking the 3′-UTR miRNA binding site.
Figure 5. MicroRNA-19 promotes changes in CRC cell behaviour through TG2.
The effect of of miRNA-19a over-expression in SW480 cells was assessed following stable transfection with a miRNA-19a expression plasmid (480/miRNA-19a), compared to a control plasmid (SCC). Western blotting was performed to observe the effect of miRNA-19a on TG2 protein levels (A) and quantified by densitometry, and direct targeting of TG2 by miRNA-19a was assessed using a 3′UTR luciferase reporter gene (B). The effect of miRNA-19a over-expression on invasion was assessed in the SW480 stable lines using the matrigel invasion assay (C). The effects of miRNA-19a on invasion were established to be through TG2 by co-transfecting the cells with a TG2 expression plasmid lacking the 3′-UTR miRNA binding sequence. The effect of miRNA-19a on inflammatory signalling was assessed using IL-8 ELISA on supernatents from SW480 stable cell lines (D). * = p <0.05, ** = p < 0.01.
DISCUSSION
There is a widespread body of literature spanning the past two decades illustrating that TG2 is involved with many cellular processes linked to tumour development and progression including chemoresistance, adhesion, migration, invasion and EMT, and TG2 has been found in tumour cells from a variety of origins. The primary role of TG2 is as a protein cross-linking enzyme, linking glutamine and lysine residues, and this can eventually lead to the formation of protein aggregates [37]. A wide range of TG2 substrates have been reported, which includes both intra- and extra-cellular proteins, implying a role for TG2 both inside and outside of the cell. The range of cell behaviours linked to TG2 and sometimes contradictory results in studies of TG2 activity suggest a role that is isoform-, context- and cell-type dependant [14].
It is interesting that we observe an inverse relationship between TG2 and invasion in vitro in the SW cell model. This finding supports studies describing a correlation between metastatic potential and TG2 expression in CRC cells [12], but runs contrary to reports in several other cell models [8, 27, 38, 39]. In a a similar manner to studies showing that TG2 can function in a pro- or anti-apoptotic manner [14], the cell-, isoform- and context-dependence of TG2 makes interpretation of such apparently contradictory results difficult. In an important study, it was demonstrated that the truncated form of TG2 promoted cell death, in contrast to the full-length protein which promoted cell survival [32]. Whilst we observe a down-regulation of full-length protein in SW620 cells compared to SW480 cells, we actually observe a small upregulation of the truncated protein (supplementary figure 1). If cell behaviour is linked to the balance between TG2 isoforms, this shift could well be critical - it is noteable that the smaller form is truncated at the 3′-end, making it likely that miR-19 will not inhibit transcription of the truncated protein. Since this truncation also removes the GTP binding site which inhibits cross-linking function, this isoform is highly active and may compensate for the miR-19-induced reduction of the full-length protein in the metastatic cell line, maintaining critical functionality such as adhesive and migratory behaviour.
Interestingly, in several previous studies where TG2 has been shown to promote invasive behaviour, this activity was not dependent on cross-linking activity, and active site mutated TG2 was also able to promote invasion [8, 39]. In contrast, when we transfected the active site mutated TG2 into SW620 cells, no effect on cell invasion was observed. Given the clear inhibition of invasive behaviour we observe when we transfect active TG2, we conclude that in the SW model system, TG2 inhibits invasive behaviour in a cross-linking dependent manner. Since promotion of invasion by TG2 in other model systems is not dependent on cross-linking it likely involves signal transduction mediated by interaction with integrins/FAK at the cell membrane/ECM boundary [8, 26-28, 40]. Our observation that SW cells lack cell surface expression of TG2 supports a role for TG2 in restricting invasion in early stage CRC through this separate mechanism. We did not specifically examine secretion of TG2 in this study, but experiments to examine whether TG2 released directly into the matrix would be informative, since modification of ECM by TG2 is known to restrict invasion [41]. In early CRC, it may be the case that cancer progression is driven by cross-linked, stiffened ECM [42], linking TG2 expression to poor prognosis as proposed by previous reports [13], but at the expense of rapid invasion, a feature that is reversed as the tumour progresses.
Whilst our data indicated a lack of cell surface expression of TG2, we observed extensive staining in the cytoplasm and nucleus. In these cell compartments, TG2 cross-linking activity is limited under physiologically normal conditions due to high nucleotide and low calcium conditions. However, intracellular cross-linking is known to occur as a consequence of cell stress –for example, activity is upregulated by ROS [23, 43]. One of the consequences of this response is the up-regulation of pro-inflammatory signalling pathways such as NF-κβ which has been reported in both inflammatory and tumour cell models [24, 29, 30], and we identified that IL-8 secretion from SW480 cells is inhibited by silencing TG2. IL-8 is known to play a significant role in CRC, and has been proposed as a marker of disease progression, but to our knowledge this is the first study linking IL-8 to TG2 in CRC models [44, 45]. Since SW620 cells are derived from an advanced stage, invasive, metastatic tumour, and IL-8 secretion was undetectable from these cells, it may be that stress-linked pro-inflammatory signalling through pathways like NF-κβ promotes progression of early CRC, whereas inhibition of this signalling in advanced disease promotes evasion of the immune system in advanced CRC. Further work would to clarify this would be extremely informative.
High TG2 levels inhibited invasive behaviour of SW cells, but silencing of TG2 did not significantly alter MMP expression (supplementary figure 3B), although MMP expression tended to be higher in SW620 cells. This may be a consequence of the experimental system; culturing cells in plates - as we did in the present study when analysing MMP gene expression - induces significantly different responses in tumour cells when compared to the 3D environment cells experience in vivo, or within the matrigel layer of the invasion assay [46, 47]. Actin is reported to be an intracellular TG2 substrate [48], so it is also feasible that intracellular TG2 could play a role in CRC cell invasion in the cytoskeletal remodelling involved during cell mobility and invasion. Further experiments to examine TG2 secretion, MMP expression and cytoskeletal changes by SW cells in a 3D model will be extremely informative.
The down-regulation of TG2 in metastatic SW620 cells and in sections taken from liver metastases illustrates the context-dependence of using TG2 as either a marker of disease or as a therapeutic target in CRC. The negative relationship between TG2 expression and invasive potential has been reported previously in the SW model [12], and the inverse relationship between TG2 and metastasis observed in other studies [49-51]. It will therefore be interesting to examine whether this biphasic model of TG2 involvement in cancer progression is a general phenomenon, as it would have significant implications for the targeting of TG2 therapeutically. Our observation of significant TG2 expression in the stroma of both primary and metastatic CRC, despite down-regulation of TG2 in metastatic cells, suggests that the majority of TG2 is produced as a defensive response rather than by the tumour [41, 52, 53]. Further clarification of the cellular source of TG2 in CRC is important, as the differential impact of TG2 activity in cancer cells and in the surrounding tissue complicates the use of TG2 as a therapeutic target. Identifying pathways that specifically regulate TG2 expressed in cancer cells may therefore offer a promising alternative approach.
Examining the role of miRNAs in regulating TG2 was a consequence of our data showing differential expression of two splice variants of TG2 at transcript and protein level; indeed, over the course of the study TG2 protein levels were observed to vary significantly. Examining putative miRNA binding sites revealed that TG2 is a predicted target for miRNA-19, which has two closely related members miRNA-19a and miRNA-19b within the miRNA17-92 cluster. This adds to previous studies identifying regulatory roles for miRNA-1285, miRNA-181a and miRNA-218 in regulating TG2 [54, 55], and since TG2 plays an important role in inflammatory disease [22, 24], multiple miRNA pathways may therefore have an important role in regulating innate immune responses as well as cancer cell behaviour linked to TG2 activity. It is interesting that we did not see a significant change in IL-8 in SW480/miR-19a cells compared to SW480/SCC control cells. This would be expected given that we observed that IL-8 production is inhibited by silencing TG2 in SW480 cells, TG2 is suppressed by miR-19, and we also observe that transfection of the TG2 plasmid into SW480/miR-19a cells upregulates IL-8. This could be the consequence of the multiple pathways that converge on NF-kB in cancer cells, for example NF-kB activation can be inhibited by blocking K-Ras activity in SW620 cells [56]. However, this data may also simply represent technical differences in manipulating TG2 using siRNA - which is highly efficient in our model - compared to manipulating TG2 using miRNA-19, which alters expression by less than 50% (figure 5A).
Multiple studies have demonstrated that miRNA-19 is upregulated in CRC patients, notably at the invasive front of the tumour, and also in SW620 cells when compared to SW480 cells [31, 32, 34]. [32, 34], strongly implicating these miRNAs in disease progression. We focussed on miRNA-19a, demonstrating upregulation in sections taken from liver metastases when compared to primary CRC sections, and in metastatic SW620 cells compared to primary SW480 cells. Overexpressing miRNA-19a led to a reduction in TG2 expression in SW480 cells, with consequent increased invasive behaviour. We therefore propose that the miRNA-19/TG2 axis can be added to the growing list of miRNA-regulated pathways that are linked to metastasis. Moreover, we identified that both in the SW cell model and in patients with CRC, overexpression of miRNA-19a/b is linked to chromosomal instability on chromosome 13, at the locus encoding a series of miRNAs including miRNA-19a and miRNA-b1. Amplification of chromosome 13 is observed frequently in CRC, despite encoding relatively few genes linked to oncogenic pathways [57], and these observations may provide a mechanistic link between CRC and instability at this locus, via miRNAs and TG2.
Identifying miRNA regulation of TG2 in CRC cells may provide a more targeted pathway to therapeutic intervention, given the presence of TG2 in both tumour and stroma. Further work is required to establish the precise mechanisms by which TG2 acts to influence cell invasion, and how invasion, inflammation and metastasis interact to drive the disease process. If TG2 is indeed a stress response, the role of ROS and calcium may be critical as they promote TG2 activity. We were not able to alter TG2 expression using ROS inhibitors, and defective calcium signalling is an established feature of CRC [58]. Continuing to investigate a pathway linking stress, inflammatory signalling and invasion has the potential to provide useful insights into the mechanisms driving an increasing burden on the world’s health.
Supplementary Material
Implications: Chromosome-13 amplification in advanced colorectal cancer contributes to invasion and metastasis by upregulating miR-19, which targets transglutaminase-2.
Acknowledgments
Financial support:
Cancer Research UK and the Royal College of Surgeons of England; Wessex Medical Research
Footnotes
Disclosures:
None
REFERENCES
- 1.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59(6):366–78. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 2.Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet. 2005;365(9454):153–65. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 3.Soreide K, Berg M, Skudal BS, Nedreboe BS. Advances in the understanding and treatment of colorectal cancer. Discov Med. 2011;12(66):393–404. [PubMed] [Google Scholar]
- 4.Li B, Cerione RA, Antonyak M. Tissue transglutaminase and its role in human cancer progression. Adv Enzymol Relat Areas Mol Biol. 2011;78:247–93. doi: 10.1002/9781118105771.ch6. [DOI] [PubMed] [Google Scholar]
- 5.Eckert RL, Kaartinen MT, Nurminskaya M, Belkin AM, Colak G, Johnson GV, et al. Transglutaminase regulation of cell function. Physiol Rev. 2014;94(2):383–417. doi: 10.1152/physrev.00019.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta K, Fok J, Miller FR, Koul D, Sahin AA. Prognostic significance of tissue transglutaminase in drug resistant and metastatic breast cancer. Clin Cancer Res. 2004;10(23):8068–76. doi: 10.1158/1078-0432.CCR-04-1107. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Mi W, Cai J, Ying W, Liu F, Lu H, et al. Quantitative proteomic signature of liver cancer cells: tissue transglutaminase 2 could be a novel protein candidate of human hepatocellular carcinoma. J Proteome Res. 2008;7(9):3847–59. doi: 10.1021/pr800153s. [DOI] [PubMed] [Google Scholar]
- 8.Verma A, Wang H, Manavathi B, Fok JY, Mann AP, Kumar R, et al. Increased expression of tissue transglutaminase in pancreatic ductal adenocarcinoma and its implications in drug resistance and metastasis. Cancer Res. 2006;66(21):10525–33. doi: 10.1158/0008-5472.CAN-06-2387. [DOI] [PubMed] [Google Scholar]
- 9.Verma A, Guha S, Diagaradjane P, Kunnumakkara AB, Sanguino AM, Lopez-Berestein G, et al. Therapeutic significance of elevated tissue transglutaminase expression in pancreatic cancer. Clin Cancer Res. 2008;14(8):2476–83. doi: 10.1158/1078-0432.CCR-07-4529. [DOI] [PubMed] [Google Scholar]
- 10.Lentini A, Abbruzzese A, Provenzano B, Tabolacci C, Beninati S. Transglutaminases: key regulators of cancer metastasis. Amino Acids. 2013;44(1):25–32. doi: 10.1007/s00726-012-1229-7. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda K. Induction of tissue transglutaminase expression by propionate and n-butyrate in colon cancer cell lines. J Nutr Biochem. 1999;10(7):397–404. doi: 10.1016/s0955-2863(99)00018-2. [DOI] [PubMed] [Google Scholar]
- 12.Zirvi KA, Keogh JP, Slomiany A, Slomiany BL. Transglutaminase activity in human colorectal carcinomas of differing metastatic potential. Cancer Lett. 1991;60(1):85–92. doi: 10.1016/0304-3835(91)90052-j. [DOI] [PubMed] [Google Scholar]
- 13.Miyoshi N, Ishii H, Mimori K, Tanaka F, Hitora T, Tei M, et al. TGM2 is a novel marker for prognosis and therapeutic target in colorectal cancer. Ann Surg Oncol. 2010;17(4):967–72. doi: 10.1245/s10434-009-0865-y. [DOI] [PubMed] [Google Scholar]
- 14.Chhabra A, Verma A, Mehta K. Tissue transglutaminase promotes or suppresses tumors depending on cell context. Anticancer Res. 2009;29(6):1909–19. [PubMed] [Google Scholar]
- 15.Antonyak MA, Jansen JM, Miller AM, Ly TK, Endo M, Cerione RA. Two isoforms of tissue transglutaminase mediate opposing cellular fates. Proc Natl Acad Sci U S A. 2006;103(49):18609–14. doi: 10.1073/pnas.0604844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W, Coller J. What comes first: translational repression or mRNA degradation? The deepening mystery of microRNA function. Cell Res. 2012;22(9):1322–4. doi: 10.1038/cr.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schetter AJ, Harris CC. Alterations of microRNAs contribute to colon carcinogenesis. Semin Oncol. 2011;38(6):734–42. doi: 10.1053/j.seminoncol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18(3):244–52. doi: 10.1097/PPO.0b013e318258b78f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang KH, Miller N, Kheirelseid EA, Lemetre C, Ball GR, Smith MJ, et al. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26(11):1415–22. doi: 10.1007/s00384-011-1279-4. [DOI] [PubMed] [Google Scholar]
- 20.de Krijger I, Mekenkamp LJ, Punt CJ, Nagtegaal ID. MicroRNAs in colorectal cancer metastasis. J Pathol. 2011;224(4):438–47. doi: 10.1002/path.2922. [DOI] [PubMed] [Google Scholar]
- 21.Mirnezami AH, Pickard K, Zhang L, Primrose JN, Packham G. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. Eur J Surg Oncol. 2009;35(4):339–47. doi: 10.1016/j.ejso.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Luciani A, Villella VR, Vasaturo A, Giardino I, Raia V, Pettoello-Mantovani M, et al. SUMOylation of tissue transglutaminase as link between oxidative stress and inflammation. J Immunol. 2009;183(4):2775–84. doi: 10.4049/jimmunol.0900993. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Lesort M, Guttmann RP, Johnson GV. Modulation of the in situ activity of tissue transglutaminase by calcium and GTP. J Biol Chem. 1998;273(4):2288–95. doi: 10.1074/jbc.273.4.2288. [DOI] [PubMed] [Google Scholar]
- 24.Maiuri L, Luciani A, Giardino I, Raia V, Villella VR, D’Apolito M, et al. Tissue transglutaminase activation modulates inflammation in cystic fibrosis via PPARgamma down-regulation. J Immunol. 2008;180(11):7697–705. doi: 10.4049/jimmunol.180.11.7697. [DOI] [PubMed] [Google Scholar]
- 25.Parker H, Rose-Zerilli MJ, Parker A, Chaplin T, Wade R, Gardiner A, et al. 13q deletion anatomy and disease progression in patients with chronic lymphocytic leukemia. Leukemia. 2011;25(3):489–97. doi: 10.1038/leu.2010.288. [DOI] [PubMed] [Google Scholar]
- 26.Kim HR, Wheeler MA, Wilson CM, Iida J, Eng D, Simpson MA, et al. Hyaluronan facilitates invasion of colon carcinoma cells in vitro via interaction with CD44. Cancer Res. 2004;64(13):4569–76. doi: 10.1158/0008-5472.CAN-04-0202. [DOI] [PubMed] [Google Scholar]
- 27.Priglinger SG, Alge CS, Neubauer AS, Kristin N, Hirneiss C, Eibl K, et al. TGF-beta2-induced cell surface tissue transglutaminase increases adhesion and migration of RPE cells on fibronectin through the gelatin-binding domain. Invest Ophthalmol Vis Sci. 2004;45(3):955–63. doi: 10.1167/iovs.03-0210. [DOI] [PubMed] [Google Scholar]
- 28.Mangala LS, Fok JY, Zorrilla-Calancha IR, Verma A, Mehta K. Tissue transglutaminase expression promotes cell attachment, invasion and survival in breast cancer cells. Oncogene. 2007;26(17):2459–70. doi: 10.1038/sj.onc.1210035. [DOI] [PubMed] [Google Scholar]
- 29.Verma A, Guha S, Wang H, Fok JY, Koul D, Abbruzzese J, et al. Tissue transglutaminase regulates focal adhesion kinase/AKT activation by modulating PTEN expression in pancreatic cancer cells. Clin Cancer Res. 2008;14(7):1997–2005. doi: 10.1158/1078-0432.CCR-07-1533. [DOI] [PubMed] [Google Scholar]
- 30.Mann AP, Verma A, Sethi G, Manavathi B, Wang H, Fok JY, et al. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-kappaB in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66(17):8788–95. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73(21):6435–47. doi: 10.1158/0008-5472.CAN-12-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahlert C, Klupp F, Brand K, Lasitschka F, Diederichs S, Kirchberg J, et al. Invasion front-specific expression and prognostic significance of microRNA in colorectal liver metastases. Cancer Sci. 2011;102(10):1799–807. doi: 10.1111/j.1349-7006.2011.02023.x. [DOI] [PubMed] [Google Scholar]
- 33.Lai TS, Liu Y, Li W, Greenberg CS. Identification of two GTP-independent alternatively spliced forms of tissue transglutaminase in human leukocytes, vascular smooth muscle, and endothelial cells. FASEB J. 2007;21(14):4131–43. doi: 10.1096/fj.06-7598com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374. doi: 10.1186/1471-2407-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamy P, Andersen CL, Dyrskjot L, Torring N, Orntoft T, Wiuf C. Are microRNAs located in genomic regions associated with cancer? Br J Cancer. 2006;95(10):1415–8. doi: 10.1038/sj.bjc.6603381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12(9):863–75. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 38.Kumar A, Xu J, Brady S, Gao H, Yu D, Reuben J, et al. Tissue transglutaminase promotes drug resistance and invasion by inducing mesenchymal transition in mammary epithelial cells. PLoS One. 2010;5(10):e13390. doi: 10.1371/journal.pone.0013390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang JY, Mangala LS, Fok JY, Lin YG, Merritt WM, Spannuth WA, et al. Clinical and biological significance of tissue transglutaminase in ovarian carcinoma. Cancer Res. 2008;68(14):5849–58. doi: 10.1158/0008-5472.CAN-07-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herman JF, Mangala LS, Mehta K. Implications of increased tissue transglutaminase (TG2) expression in drug-resistant breast cancer (MCF-7) cells. Oncogene. 2006;25(21):3049–58. doi: 10.1038/sj.onc.1209324. [DOI] [PubMed] [Google Scholar]
- 41.Mangala LS, Arun B, Sahin AA, Mehta K. Tissue transglutaminase-induced alterations in extracellular matrix inhibit tumor invasion. Mol Cancer. 2005;4:33. doi: 10.1186/1476-4598-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luciani A, Villella VR, Vasaturo A, Giardino I, Pettoello-Mantovani M, Guido S, et al. Lysosomal accumulation of gliadin p31-43 peptide induces oxidative stress and tissue transglutaminase-mediated PPARgamma downregulation in intestinal epithelial cells and coeliac mucosa. Gut. 2010;59(3):311–9. doi: 10.1136/gut.2009.183608. [DOI] [PubMed] [Google Scholar]
- 44.Bunger S, Haug U, Kelly FM, Klempt-Giessing K, Cartwright A, Posorski N, et al. Toward standardized high-throughput serum diagnostics: multiplex-protein array identifies IL-8 and VEGF as serum markers for colon cancer. J Biomol Screen. 2011;16(9):1018–26. doi: 10.1177/1087057111414894. [DOI] [PubMed] [Google Scholar]
- 45.Lee YS, Choi I, Ning Y, Kim NY, Khatchadourian V, Yang D, et al. Interleukin-8 and its receptor CXCR2 in the tumour microenvironment promote colon cancer growth, progression and metastasis. Br J Cancer. 2012;106(11):1833–41. doi: 10.1038/bjc.2012.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker EL, Lu J, Yu D, Bonnecaze RT, Zaman MH. Cancer cell stiffness: integrated roles of three-dimensional matrix stiffness and transforming potential. Biophys J. 2010;99(7):2048–57. doi: 10.1016/j.bpj.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker EL, Bonnecaze RT, Zaman MH. Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys J. 2009;97(4):1013–21. doi: 10.1016/j.bpj.2009.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemes Z, Jr., Adany R, Balazs M, Boross P, Fesus L. Identification of cytoplasmic actin as an abundant glutaminyl substrate for tissue transglutaminase in HL-60 and U937 cells undergoing apoptosis. J Biol Chem. 1997;272(33):20577–83. doi: 10.1074/jbc.272.33.20577. [DOI] [PubMed] [Google Scholar]
- 49.Barnes RN, Bungay PJ, Elliott BM, Walton PL, Griffin M. Alterations in the distribution and activity of transglutaminase during tumour growth and metastasis. Carcinogenesis. 1985;6(3):459–63. doi: 10.1093/carcin/6.3.459. [DOI] [PubMed] [Google Scholar]
- 50.Knight CR, Rees RC, Griffin M. Apoptosis: a potential role for cytosolic transglutaminase and its importance in tumour progression. Biochim Biophys Acta. 1991;1096(4):312–8. doi: 10.1016/0925-4439(91)90067-j. [DOI] [PubMed] [Google Scholar]
- 51.Hager H, Jensen PH, Hamilton-Dutoit S, Neilsen MS, Birckbichler P, Gliemann J. Expression of tissue transglutaminase in human bladder carcinoma. J Pathol. 1997;183(4):398–403. doi: 10.1002/(SICI)1096-9896(199712)183:4<398::AID-PATH947>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 52.Jones RA, Kotsakis P, Johnson TS, Chau DY, Ali S, Melino G, et al. Matrix changes induced by transglutaminase 2 lead to inhibition of angiogenesis and tumor growth. Cell Death Differ. 2006;13(9):1442–53. doi: 10.1038/sj.cdd.4401816. [DOI] [PubMed] [Google Scholar]
- 53.Haroon ZA, Lai TS, Hettasch JM, Lindberg RA, Dewhirst MW, Greenberg CS. Tissue transglutaminase is expressed as a host response to tumor invasion and inhibits tumor growth. Lab Invest. 1999;79(12):1679–86. [PubMed] [Google Scholar]
- 54.Hidaka H, Seki N, Yoshino H, Yamasaki T, Yamada Y, Nohata N, et al. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012;3(1):44–57. doi: 10.18632/oncotarget.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eom S, Kim Y, Kim M, Park D, Lee H, Lee YS, et al. Transglutaminase II/microRNA-218/-181a loop regulates positive feedback relationship between allergic inflammation and tumor metastasis. J Biol Chem. 2014;289(43):29483–505. doi: 10.1074/jbc.M114.603480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin G, Tang Z, Ye YB, Chen Q. NF-kappaB activity is downregulated by KRAS knockdown in SW620 cells via the RAS-ERK-IkappaBalpha pathway. Oncol Rep. 2012;27(5):1527–34. doi: 10.3892/or.2012.1669. [DOI] [PubMed] [Google Scholar]
- 57.Neklason DW, Tuohy TM, Stevens J, Otterud B, Baird L, Kerber RA, et al. Colorectal adenomas and cancer link to chromosome 13q22.1-13q31.3 in a large family with excess colorectal cancer. J Med Genet. 2010;47(10):692–9. doi: 10.1136/jmg.2009.076091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saidak Z, Mentaverri R, Brown EM. The role of the calcium-sensing receptor in the development and progression of cancer. Endocr Rev. 2009;30(2):178–95. doi: 10.1210/er.2008-0041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.