Abstract
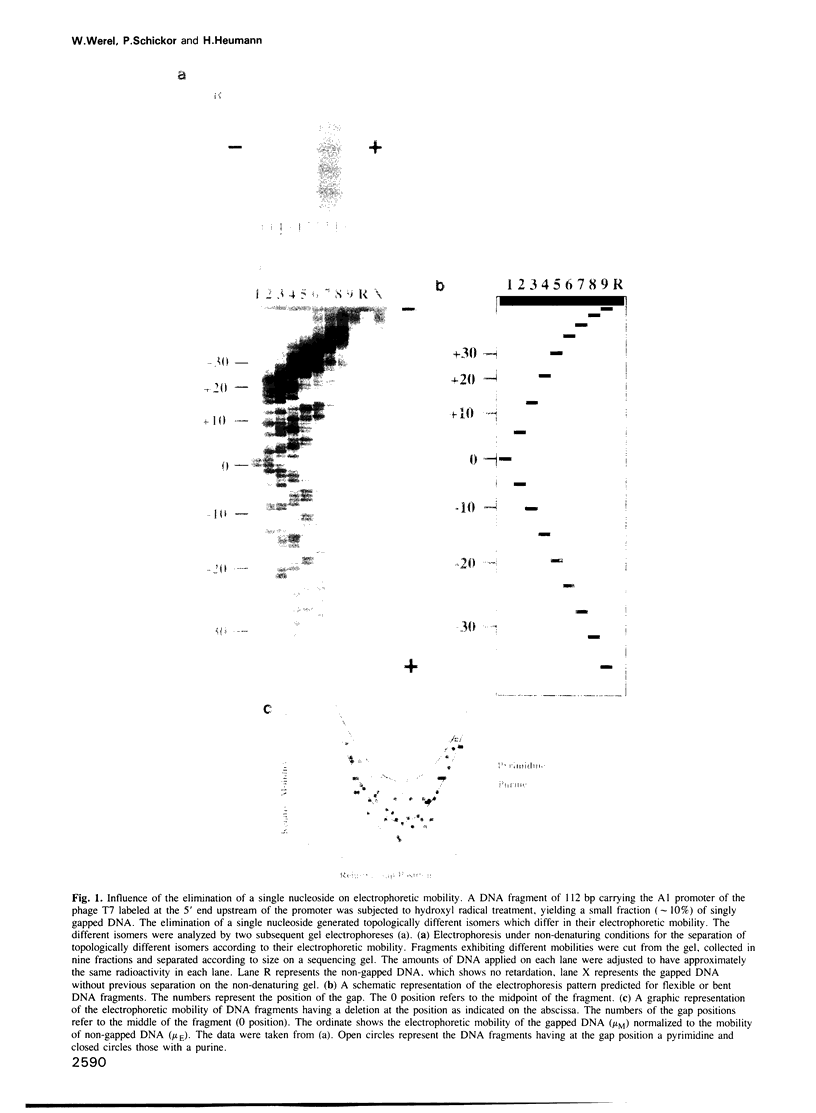
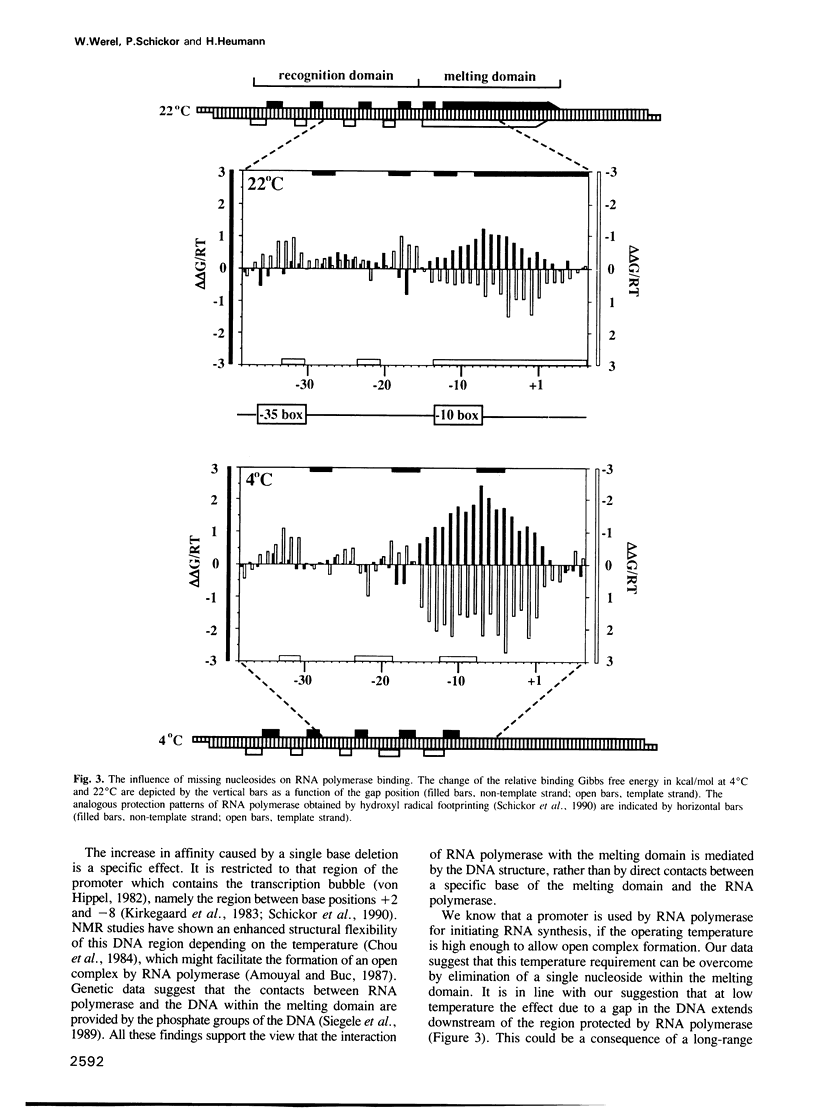
Two types of mechanisms are discussed for the formation of active protein-DNA complexes: contacts with specific bases and interaction via specific DNA structures within the cognate DNA. We have studied the effect of a single nucleoside deletion on the interaction of Escherichia coli RNA polymerase with a strong promoter. This study reveals three patterns of interaction which can be attributed to different sites of the promoter, (i) direct base contact with the template strand in the '-35 region' (the 'recognition domain'), (ii) a DNA structure dependent interaction in the '-10 region' (the 'melting domain'), and (iii) an interaction which is based on a defined spatial relationship between the two domains of a promoter, namely the 'recognition domain' and the 'melting domain'.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Ayers D. G., Auble D. T., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Role of the spacer DNA in functional complex formation. J Mol Biol. 1989 Jun 20;207(4):749–756. doi: 10.1016/0022-2836(89)90241-6. [DOI] [PubMed] [Google Scholar]
- Aymami J., Coll M., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. Molecular structure of nicked DNA: a substrate for DNA repair enzymes. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2526–2530. doi: 10.1073/pnas.87.7.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G., Beato M. Hydroxyl radical interference: a new method for the study of protein-DNA interactions. Nucleic Acids Res. 1989 Feb 25;17(4):1783–1783. doi: 10.1093/nar/17.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. H., Wemmer D. E., Hare D. R., Reid B. R. Sequence-specific recognition of DNA: NMR studies of the imino protons of a synthetic RNA polymerase promoter. Biochemistry. 1984 May 8;23(10):2257–2262. doi: 10.1021/bi00305a026. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Ampe C., Steitz T. A., Crothers D. M. Molecular characterization of the GCN4-DNA complex. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Overexpression and purification of the sigma subunit of Escherichia coli RNA polymerase. Gene. 1983 Dec;26(2-3):109–118. doi: 10.1016/0378-1119(83)90180-4. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry. 1989 Nov 28;28(24):9521–9527. doi: 10.1021/bi00450a041. [DOI] [PubMed] [Google Scholar]
- Heumann H., Metzger W., Niehörster M. Visualization of intermediary transcription states in the complex between Escherichia coli DNA-dependent RNA polymerases and a promoter-carrying DNA fragment using the gel retardation method. Eur J Biochem. 1986 Aug 1;158(3):575–579. doi: 10.1111/j.1432-1033.1986.tb09793.x. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S., Frisch H. L. Why does the electrophoretic mobility of DNA in gels vary with the length of the molecule? Biopolymers. 1982 May;21(5):995–997. doi: 10.1002/bip.360210511. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger W., Schickor P., Heumann H. A cinematographic view of Escherichia coli RNA polymerase translocation. EMBO J. 1989 Sep;8(9):2745–2754. doi: 10.1002/j.1460-2075.1989.tb08416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickor P., Metzger W., Werel W., Lederer H., Heumann H. Topography of intermediates in transcription initiation of E.coli. EMBO J. 1990 Jul;9(7):2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreck R., Zorbas H., Winnacker E. L., Baeuerle P. A. The NF-kappa B transcription factor induces DNA bending which is modulated by its 65-kD subunit. Nucleic Acids Res. 1990 Nov 25;18(22):6497–6502. doi: 10.1093/nar/18.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer G. E., Price M. A., Tullius T. D. Use of the hydroxyl radical and gel electrophoresis to study DNA structure. Electrophoresis. 1989 May-Jun;10(5-6):397–404. doi: 10.1002/elps.1150100518. [DOI] [PubMed] [Google Scholar]
- Siegele D. A., Hu J. C., Walter W. A., Gross C. A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989 Apr 20;206(4):591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zillig W., Zechel K., Halbwachs H. J. A new method of large scale preparation of highly purified DNA-dependent RNA-polymerase from E. coli. Hoppe Seylers Z Physiol Chem. 1970 Feb;351(2):221–224. doi: 10.1515/bchm2.1970.351.1.221. [DOI] [PubMed] [Google Scholar]