Abstract
Hypoxia selects the most aggressive and drug-resistant clones in solid malignancies. One of the pivotal transcription factors induced by hypoxia is Hif-1α. However, in serous ovarian cancer (SEOC), Hif-1α expression is not a prognostic biomarker. This study aims to assess the hypothesis that the serine-threonine kinase Nek6 functions as a downstream effector cooperating with Hif-1α in driving ovarian cancer aggressiveness. Nek6 was overexpressed and Hif-1α was silenced in A2780 cells. Nek6 was also stably silenced in Hey cells. The dependence of Nek6 expression on Hif-1α was assayed as a function of hypoxic growth conditions. Nek6 interaction with the cytoskeletal gateway of drug resistance was investigated with far western blot. The co-expression of NEK6, HIF1A, TUBB3 and GBP1 transcripts was quantified with qPCR in two cohorts of SEOC patients (346 locally treated patients and 344 from the TCGA dataset). Nek6 expression is induced by hypoxia in a Hif-1α dependent fashion. Nek6 directly interacts with GBP-1, thus being a component of the cytoskeletal gateway of drug resistance. Nek6 overexpression increases and silencing decreases the anchorage-independent growth of cultured cells. In SEOC patients, NEK6 expression is significantly correlated with HIF1A. Co-expression of NEK6, HIF1A, TUBB3 and GBP1 transcripts identifies a subset of SEOC patients characterized by poor outcome and drug resistance. This study demonstrates the functional relevance of Nek6 in the context of the adaptive response to hypoxia in SEOC. This finding may help identify a sub-population of patients at high risk of relapse to standard first-line chemotherapy.
Keywords: Ovarian cancer, Nek6, class III beta-tubulin, Hif-1-alpha, GBP-1
Introduction
Serous epithelial ovarian cancer (SEOC) is the most common type of ovarian cancer and the deadliest gynecologic malignancy. Due to the clinically indolent nature of the disease in the early stages, the majority of patients are diagnosed when surgery alone is not sufficient to eradicate the cancer. Combination chemotherapy with platinum/taxane remains a cornerstone of treatment for most patients, and duration of chemotherapy response is a proven predictor of outcome. Patients relapsing more than 12 months following chemotherapy are defined as chemo-sensitive and can expect a relatively favorable 5-year survival rate of 60% [1]. By contrast, women relapsing within 3 months will likely experience rapid fatal progression; this cohort has a 5-year survival rate of less than 5% [1]. Novel strategies aimed at circumventing the injurious effects of chemoresistance in the management of SEOC are desperately needed [2]. Elucidating the molecular mechanism(s) underlying chemoresistance is mandatory if progress on this front is to be realized.
It has already been established that the tumor microenvironment has a profound impact on cancer biology and response to treatments [3]. Cancers which are exposed to harsh microenvironments featured by low oxygen and diminished nutrient supply will activate adaptive, “survival” pathways that simultaneously provide, to patients’ detriment, protection from the therapeutic effects of chemotherapy. The expression of class III β-tubulin (encoded by the gene TUBB3) is a key event in this cascade [4] contributing to its power as a prognostic marker in ovarian cancer [5,6] and other solid malignancies [7]. Of note, class III β-tubulin is one of the genes regulated directly by Hif-1α (Hypoxia-inducible factor 1-alpha) [8], and it enhances the incorporation of GBP-1 (Interferon-induced guanylate-binding protein 1) GTPase into microtubules. GBP-1 facilitates the cytoskeletal incorporation of prosurvival kinases, protecting them from proteasome degradation and prolonging their pro-survival signaling [9]. The functional interaction between GBP-1, class III β-tubulin and the pro-survival kinases constitutes the so-called cytoskeletal gateway of drug resistance [9].
Nek6 (NIMA (Never In Mitosis Gene A)-Related Kinase 6) is a serine-threonine protein kinase required for progression through the metaphase portion of mitosis. Nek6 is elevated in malignant tumors and human cancer cell lines as compared with normal tissue and fibroblast cells and is believed to play a role in tumorigenesis [10].
The cytoskeletal gateway of drug resistance allows incorporation into the cytoskeleton of at least 19 protein kinases capable to interact with GBP-1 [9]. One of these 19 proteins is Nek6. Therefore, we aimed this study at investigating Nek6 expression in ovarian cancer and its functional connection with the adaptive response to hypoxia and the cytoskeletal gateway of drug resistance. The results support the hypothesis that Nek6 contributes to the aggressive phenotype of ovarian cancer via the functional circuit comprised of Hif-1α and class III β-tubulin induced by hypoxia.
Materials and methods
Cell cultures and growth experiments
A2780, Hey and SKOV-6 ovarian cancer cells have been previously described [11]. Cell lines were authenticated through short tandem repeat analysis and used within 15 passages from authentication. For normoxia, cells were grown in a fully humidified atmosphere of 5% CO2/95% air in RPMI medium complemented with fetal bovine serum and antibiotics. For hypoxia, experiments were performed using a sealed chamber (Billups-Rotenberg Inc., Del Mar, CA) filled with 93% nitrogen, 5% CO2, and 2% oxygen. Growth experiments were carried out as previously described [12]. Cell lines were routinely screened for mycoplasma infection using the Universal Mycoplasma Kit (ATCC, Manassas, VA) according to manufacturer’s instructions. All the reagents were purchased from Sigma-Aldrich (St. Louis, MO) if not otherwise specified. Growth inhibition experiments were performed as previously described [8,13].
Nek6 overexpression
Human NEK6 (NM_014397) was amplified from a human testis library by PCR with forward 5’-ATAAAGCTTATGGCAGGACAGCCCGGCCAC-3’ and reverse 5’-AAGAATTCTCAGGTGCTGGACATCCAGAT-3’ primers. Amplicon was then cloned in pUSE(+) expression vector (Upstate Biotechnology, Lake Placid, NY). The empty vector served as negative control. Stable transfection in A2780 cells was performed as previously described [14]. Briefly, after electroporation, cells were selected in the presence of G418 (1.5 μg/mL). When colonies appeared, individual clones were obtained at limiting dilution. Twenty-four clones were screened at the gene and protein level. The two clones with the highest Nek6 expression were selected for further analysis.
Nek6 and Hif-1α silencing
Inserts targeting NEK6 were designed using GeneScript software with the following oligonucleotides: siNEK6-F 5’-GATCCCGATCGAGCAGTGTGACTACTTCAAGAGAGTAGTCACACTGCTCGATCTTTTTTCCAAA-3’ and siNEK6-R 5’-AGCTTTTGGAAAAAAGATCGAGCAGTGTGACTACTCTCTTGAAGTAGTCACACTGCTCGATCGG-3’.
The inserts were subsequently cloned in BamHI/HindIII sites of the small interfering RNA expression vector pRNAT-U6.1/Neo (GeneScript, Piscataway, NJ). As a control, the SiC construct was built which expressed a small RNA not targeting any region of the human genome. For SiC, the annealed oligonucleotides were: SiC-F 5’-GATCCCGTTAGTCTGATATGGGATGGGCTTGATATCCGGCCCATCCCATATCAGACTAATTTTTTCCAAA-3’ and SiC-R 5’-AGCTTTTGGAAAAAATTAGTCTGATATGGGATGGGCCGGATATCAAGCCCATCCCATATCAGACTAACGG-3’. Cells were stably transfected as described above in Hey cells [12].
A2780 cells were transiently transfected with Hif-1α siRNA duplex whose sequence has been already reported [8]. After transfection, cells were cultured under hypoxic conditions for 48 h.
Real-time quantitative PCR and western blots
Total RNA was obtained from cultured cells using RNAeasy plus mini kit (Qiagen, Valencia, CA) according to the manufacturer’s directions. cDNA was prepared using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Real-time quantitative PCR was done using the iCycler iQ System (Bio-Rad) and the iQ SYBR Green Supermix (Bio-Rad) in a final volume of 25 µL, starting with a 3-min template denaturation step at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Amplifications were carried out using the following primers: NEK6 forward 5’-CGCGCCGTTCGTGCCCT-3’ and reverse 5’-GCCAAGCGAGCAGCGAAAAGACA-3’; TUBB3 forward 5’-GCGAGATGTACGAAGACGAC-3’ and reverse 5’-TTTAGACACTGCTGGCTTCG-3’. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize the possible variation in sample concentration, with forward 5’-CTGACCTGCCGTCTAGAAA-3’ and reverse 5’-CCACCATGGAGAAGGGTGG-3’; β-Actin (ACTB) replaced GAPDH for normalization for analyses in hypoxia, using the forward 5’-GCCGACAGGATGCAGAAGGAG-3’ and reverse 5’-CAGGATGGAGCCGCCGATC-3’.
The experiments were repeated two or more times and each time the samples were run in triplicates. The results were analyzed as described previously [15] using the Excel spreadsheet RelQuant (Bio-Rad).
The procedure of western blot has been already described [9]. The following antibodies were utilized: anti-Nek6 rabbit monoclonal antibody (1:5000, Abcam, Cambridge, UK); anti-Gapdh (1:5000, Abcam, Cambridge, UK); anti-β-actin (1:5000, Sigma-Aldrich), anti-class III β-tubulin polyclonal antibody (1:10000, Covance, Berkeley, CA); mouse monoclonal anti-Hif1-α (1:250, Novus Biologicals, Littleton, CO). After incubation with secondary horseradish peroxidase-conjugated antibodies (Bio-Rad), visualization was performed with the enhanced chemiluminescence-plus system (Amersham Biosciences, Buckinghamshire, UK) using a Versadoc imaging system (Bio-Rad).
Analysis of the GBP-1: Nek6 protein: protein interaction
Interaction between the two proteins was assessed with the far western blotting technique as previously described [16]. Briefly, recombinant GBP-1 and Nek6 were produced in E. Coli and freshly purified. Five µg of GBP-1 and carbonic anhydrase (CA) were blotted onto polyvinylidene difluoride membrane. Proteins were subjected to a denaturation process in protein binding buffer (100 mmol/L NaCl, 20 mmol/L Tris (pH 7.6), 0.5 mmol/L EDTA, 10% glycerol, 0.1% Tween 20, 2% milk powder, and 1 mmol/L DTT) containing decreasing amounts of guanidine-HCl from 6 to 0.1 mol/L. Renaturation of proteins was performed overnight at 4°C in protein binding buffer without guanidine-HCl. Membranes were blocked with 5% milk in PBST (PBS with 0.05% Tween 20) for 2 h at 22°C. Recombinant Nek6 was added to protein binding buffer and membranes were incubated 16 h at 4°C. Membranes were washed thrice in PBST and incubated with primary (anti-Nek6) and secondary (horseradish-conjugated) antibodies in PBST with 3% milk. Detection was done by Versadoc (Bio-Rad) after incubation in SuperSignal (Pierce, Rockford, IL). For coimmunoprecipitation experiments we used the same procedure we described previously [17], using for detection the above described anti-Nek6 antibody.
Chromatin immunoprecipitation (ChIP)-PCR assay
In silico analysis, using the MatInspector Professional software (Genomatix, Munich, Germany), indicated the presence of putative Hif-1α binding sites at 5’ flanking region of NEK6 gene (NM_014397), at 5’ of translational start site and at 3’ UTR of NEK6 gene.
ChIP was performed in A2780 cells, either in normoxia or hypoxia for 48 and 72 h, as previously described [8,18]. Briefly, immunoprecipitation was performed overnight with 10 μg of anti-Hif-1α monoclonal antibody (NB100-105, Novus Biologicals, Littleton CO) and anti-immunoglobulin (Ig) antibody as negative control (goat, Bio-Rad).
Genomic regions close to the putative Hif-1α binding sites at 5’ of translational start site (cagcgccaCGTGgggag -2947 bp to -2931 bp from ATG on plus strand and cctccccaCGTGgcgct -2946 bp to -2929 bp from ATG on the minus strand), were PCR amplified using the following primers: forward: 5’-cttattcccggtcccccttg-3’ and reverse: 5’-gtcactcaccagctagacc-3’; another site (aaaggggaCGTGgaagg -4121 bp to -4105 bp from ATG on the minus strand) was PCR amplified with primers forward: 5’-ccggggacgggcagt-3’ and reverse: 5’-ccacaccccaacagaaaccc-3’. Two additional putative Hif-1α binding sites, identified by MatInspector Professional software in the 3’UTR of NEK6 gene (tttccacaCGTGgactc +1354 bp to +1362 bp from stop codon on plus strand and cgagtccaCGTGtggaa +1355 bp to +1363 bp from stop codon on the minus strand), were PCR amplified using the following primers: forward: 5’-tgattcctgacacctgctgc-3’ and reverse: 5’agtaactcccagtcaccctgt-3’. In addition, negative control primers, amplifying a region of genomic DNA between the GAPDH gene and CNAP1 gene (accession number: BC028182), were used: forward: 5’-atggttgccactggggatct-3’ and reverse: 5’-tcgcaaagcctaggggaaga-3’.
Soft agar assay
For the soft agar assay in hypoxia vs. normoxia condition, respectively 8000 cells were layered in 300µl of culture medium on the bottom agar (final solution of 0.6% agar in growth media 1X RPMI 1640, 10% FBS) in six-well culture plates and top agar (final solution of 0.3% agar in growth media 1X RPMI 1640, 10% FBS) was added. To avoid agar drying growth media was added on 0.3% top agar. After 24 h from layering, cells was cultured in normoxia and hypoxia condition for 72 h, then colony numbers were counted.
Translational analysis
Formalin fixed paraffin embedded (FFPE) samples were obtained from ovarian cancer that had been preserved between 2000 and 2008 following a protocol approved by the Danbury Hospital IRB (prot. Number DH12/17). Patients gave explicit informed consent to be included in the study. All the enrolled patients underwent standard treatment including platinum/taxane combination. Samples were collected at the first surgery before any treatment. The procedure has been already described [13,19,20]. Briefly, FFPE samples were cut to 10 μm thickness and two tissue slices were put into a 1.5 ml tube. One milliliter of xylene was added for deparaffinization followed by mixing twice with a high speed vortex for 3 min at room temperature. Total RNA was then automatically extracted with the Qiacube using the Qiagen miRNeasy FFPE kit following manufacturers’ protocols. The RNA from the cell line A2780 was automatically extracted with the QIAcube using the Qiagen miRNeasy kit following manufacturer’s protocols. RNA quantity and the quality were assessed by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Analysis was carried out using the 48.48 dynamic array (Fluidigm, South San Francisco, CA) and a Biomark platform following the manufacturer’s protocol. Data were normalized using the geometric average of three housekeeping genes (GAPDH, TUBB and ACTB1) and the delta/delta Ct method [21]. As validation set the level 3 data of normalized RNA-seq coming from 344 patients were downloaded from the TCGA data portal. RNA-seq instead of microarray data were used for the absence of a validated probe for NEK6 [22] in the U133A chips used in the TCGA analysis. Clinical information was also downloaded and the two files merged using the unique 12 number identifier of each individual patient.
Statistical analysis
Cox and Kaplan-Meier analysis was performed as described previously [13]. Patients were categorized as positive or negative according to a cut off optimized using the R script S1 previously published [23]. A cumulative score for the expression of each of the four investigated factors was created by giving one or zero point if the factor is positive or negative, respectively. According a score 0 and 4 correspond to a patient not expressing or expressing the 4 factors, respectively. Correlation test was performed using the Pearson method and t-test or ANOVA were used to detect significant differences with a type I error rate of 5%. All the analyses were performed with the JMP9 software (SAS, Cary, NC).
Results
NEK6 expression in ovarian cancer and normal ovarian tissue
NEK6 and HIF1A (encoding for Nek6 and Hif-1α, respectively) transcript levels were investigated in 902 SEOC patients and in 51 normal ovarian controls using the GENT (Gene Expression across Normal and Tumor tissue) software. GENT is a database assembling normalized microarray data coming from over 40,000 samples [24]. In cancer tissues, expression levels of NEK6 were significantly higher than those identified in normal tissues (Figure 1A). Although this phenomenon was not observed for HIF1A (Figure 1B), NEK6 and HIF1A were significantly correlated (Ρ=0.243; CI 0.18-0.31; P<0.001). We also tested the correlation levels between HIF1A and its well-known targets such as SLC2A1 [25] (Ρ=0.241; CI 0.18-0.33; P<0.001) and CA9 [26] (Ρ=0.186; CI 0.12-0.25; P<0.001). The obtained ρ values were very similar to those obtained for NEK6. Women expressing higher levels of HIF1A in cancer tissues concomitantly expressed higher levels of NEK6 (Figure 1C). This correlation is consistent with the hypothesis that Nek6 is a downstream effector of Hif-1α in ovarian cancer.
Figure 1.

Dot plot showing expression of NEK6 (A) and HIF1A (B) in SEOC (n=902) and normal ovarian tissue (n=51). Expression values have been retrieved with the software GENT (http://medical-genome.kribb.re.kr/GENT/). Each dot represents a patient and line in the chart is the mean value. Difference between SEOC and normal tissue is significant for NEK6 (P<0.0001, t-test), but not for HIF1A (P=0.36, t-test). Dot plot showing expression of NEK6 in SEOC according to HIF1A (C). SEOC patients were categorized as high and low expressors of HIF1A according to the mean value shown in B. Patients with high expression of HIF1A exhibited a significant high expression of NEK6 (P<0.0001, t-test).
NEK6 is modulated by hypoxia in Hif-1α direct way
In order to assess whether Nek6 is modulated by hypoxia, A2780, Hey and SKOV-6 ovarian cancer cells were exposed to hypoxia for 48 and 72 hours. At the mRNA level, we noted a significant upregulation in hypoxia compared to normoxia in all the three cell lines (Figure 2A). Additionally, we observed a significant induction of Nek6 protein at both 48 hours and 72 hours of hypoxia in all cell lines examined (Figure 2B). To investigate the role of Hif-1α in mediating NEK6 expression, HIF-1α was knocked down using RNA interference (Figure 2C, 2D). A2780 cells were transfected either with a scrambled sequence (SiC) or with a short RNA specific for HIF-1α (SiHIF-1α). HIF-1α silencing suppressed the hypoxia-induced increase of NEK6 at the gene (Figure 2C) and protein level (Figure 2D), confirming that Hif-1α is required for this effect.
Figure 2.
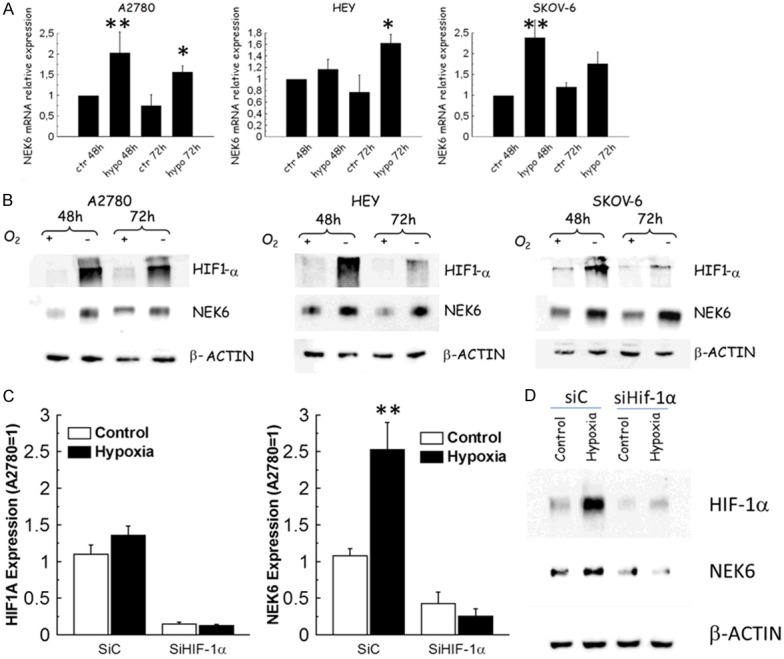
Expression analysis of NEK6 and HIF1A in a panel of ovarian cancer cells exposed to hypoxia for 48h and 72 h. In all the charts single and double asterisks indicate a P<0.05 or P<0.001, respectively. A. Bar charts indicate that hypoxia increases NEK6 expression in all the cell lines after 48 and 72h of hypoxia. Bar and error bars refer to mean and standard deviation of two independent experiments performed in triplicate samples. B. Representative western blot analysis obtained in whole cell lysates from the same cell lines. Under hypoxic condition Nek6 is induced both 48 h and 72 h in all cell lines examined. b-actin served as a loading control and Hif-1α as a marker of actual exposure to hypoxia. C. Bar chart reporting the effects of Hif-1α silencing in terms of HIF1A and NEK6 expression assessed through qPCR. Bar and error bars refer to mean and standard deviation of two independent experiments performed in triplicate samples. D. Western blot analysis for Hif-1α and Nek6 in A2780 cells transiently transfected with control silencing siC or with siHif-1α. β-actin was used as a loading control. The increase of expression of both Hif-1α and Nek6 upon 48 h of hypoxia is visible in siC transfected cells but not in those transfected with siHIF-1α. This figure demonstrates both the efficiency of Hif-1α silencing and dependence of Nek6 on Hif-1α. This experiment was repeated twice with similar results.
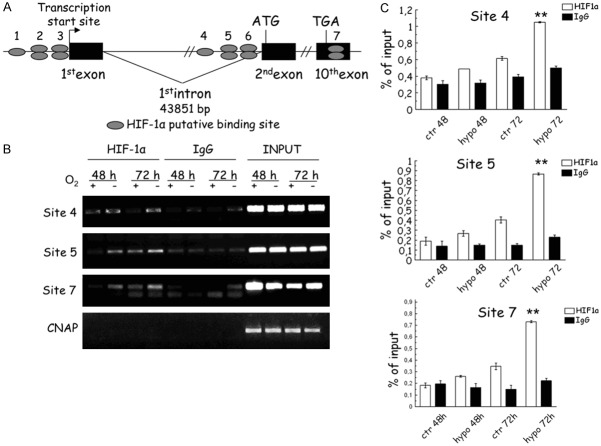
The presence of a direct binding of Hif-1α was assessed using ChIP. A bioinformatic analysis for the presence of HRE (Hypoxia Responsive Elements) motifs was conducted and seven putative sites were identified in both 5’ (six sites) and 3’ (1 site) flanking region of the NEK6 gene (Figure 3A). A clear Hif-1α binding was noticeable in two sites in the 5’ region upstream the ATG and in the 3’UTR of the NEK6 gene (Figure 3B) in A2780 cells. This 3’UTR location may be functionally very important for NEK6 regulation by Hif-1α, as demonstrated with other Hif-1a targets such as EPO [27,28] and TUBB3 [8] regulated by Hif-1α in the 3’UTR region. A functional relevance of these three binding sites is further supported by quantitative analysis through qPCR (Figure 3C), which demonstrated a significant Hif-1α binding after 72 h of hypoxia in all the three sites.
Figure 3.
Results of ChIP in A2780 cells. In A Schematic representation of the NEK6 gene and location of the seven putative Hif-1a binding sites In B ChIP-PCR analysis reveal engagement of Hif-1a in at least three sites (site 4, 5 and 7). In C Results of the ChIP-qPCR analysis, served to quantify the results (bar chart). Bar and error bars refer to mean and standard deviation of two independent experiments performed in triplicate samples. Double asterisks mean a significant effect (P<0.001) after 72 h of hypoxia.
Nek6 and the cytoskeletal gateway of response to hypoxia
We have previously demonstrated that hypoxia induces class III TUBB3 expression through Hif-1α [8], and we have described the role of the GBP-1 GTPase in hypoxia as a functional gateway to incorporate pro-survival kinases [9]. In order to verify if Nek6 is a component of the cytoskeletal gateway, we probed the interaction between Nek6 and GBP-1 by far western blot. Recombinant GBP-1 was spotted onto a PVDF membrane and recombinant Nek6 was incubated at 4 different concentrations from 1 to 5 µg. As negative control, carbonic anhydrase (CA) across the same concentration range was spotted in parallel lanes and probed with recombinant Nek6 in the same box. Anti-Nek6 antibody subsequently applied to the washed membrane revealed a clear, dose-dependent band of the expected 67 kD size (corresponding to GBP-1) in the Nek6 lanes but not the CA lanes, confirming specific binding of Nek6 to GBP-1 (Figure 4A). To further demonstrate Nek6: GBP-1 binding specificity, CA rather than recombinant Nek6 was on the membrane, and the experiment was repeated. As expected and displayed in Figure 4B, no bands were seen, thus demonstrating that the band seen in Figure 4A were specific. To further demonstrate the occurrence of the interaction, we performed a co-immunoprecipitation experiment. A protein lysate was prepared from scraped A2780 cells in basal conditions and after 72 h of hypoxia. GBP-1 was pulled down and detection of Nek6 was then performed. As negative control an anti-human IgG was used to pull down proteins. Results show the presence of the GBP-1: NEK6 interaction, more evident in hypoxia when the expression levels of Nek6 increases in A2780 (Figure 4C).
Figure 4.
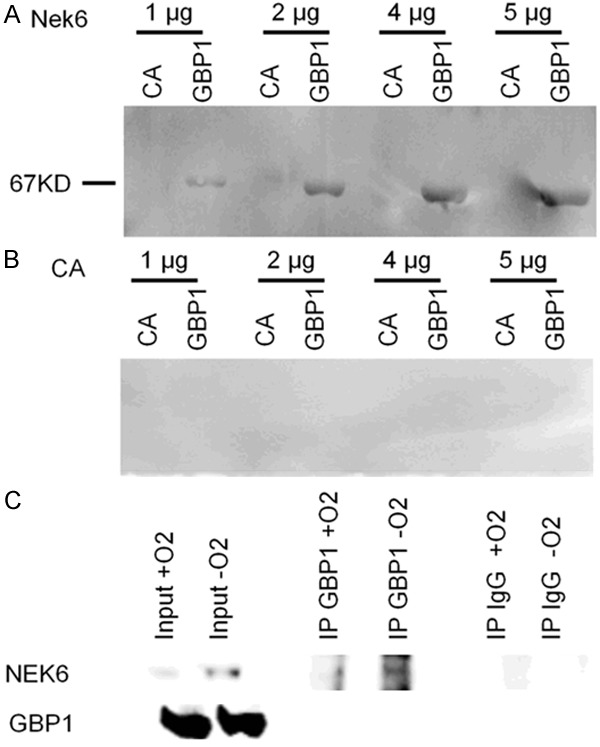
Representative far western blot showing the GBP-1: Nek6 interaction. In A GBP-1 was spotted onto the membrane. Carbonic anhydrase (CA) and recombinant Nek6 was incubated at 4 concentrations (from 1 to 5 µg). Interaction was revealed with an anti-Nek6 antibody. A clear band at the expected weight of GBP-1 (67kD) was seen at all the four concentrations with Nek6 but not with CA. In B the same experiment was repeated by spotting CA onto the membrane. No signal was detected with either Nek6 or CA at all concentrations. This experiment was repeated thrice with the same results. In C results of co-IP performed in A2780 cells in normoxic or hypoxic conditions (72 h). On the left input signal for both Nek6 and GBP-1; in the middle, pulldown of the GBP-1 protein and revelation with the anti-Nek6 identifies a signal which is higher in hypoxic conditions; on the right no signal is detected when the pulldown is performed with an anti-human-IgG. This experiment has been repeated thrice with similar results.
Nek6 expression promotes anchorage-independent growth
Anchorage independent growth is another hallmark of the activation of the class III β-tubulin pathway [29]. To investigate whether Nek6 can influence anchorage independent growth in ovarian cancer cell lines, we developed a functional “on/off” soft agar assay. There are significant risks of off targets effect when a gene is overexpressed or silenced. We believe that such risks are minimized when a combined “on/off” approach is utilized in multiple cellular models. The “on” state employed A2780 cells which are characterized by low basal expression of Nek6. We prepared a construct carrying the coding sequence, and two clones were selected after stable overexpression. Two clones transformed with the empty vector pUSE served as negative controls. A representative image of the plates is provided as Figure 5A. Overexpression of Nek6 was verified with qPCR (Figure 5B) and western blot (Figure 5C) and at the mRNA and protein level, respectively. The test was performed in normoxic and hypoxic conditions. Analysis was extended to the unselected pools from where the clones were selected. When Nek6 was overexpressed (in the pool and in the clones), a significant increase in the colony number was detected in both normoxia and hypoxia (Figure 5D). On the other hand, no significant differences were observed in the pool of A2780-pUSE and its clones.
Figure 5.
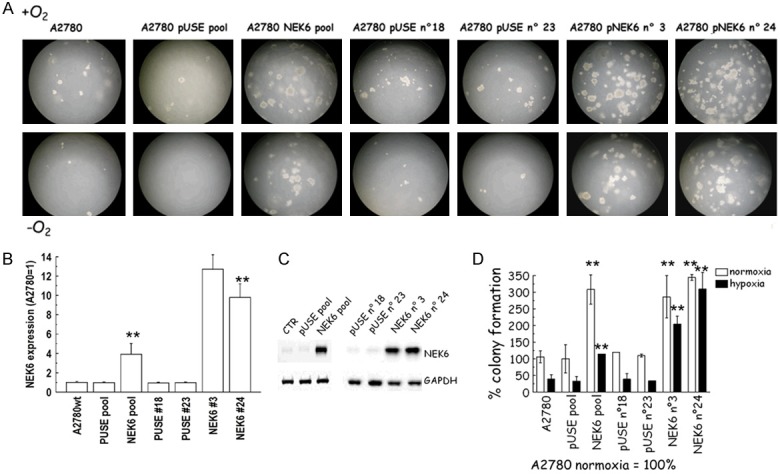
(A) Representative images of the anchorage independent clonogenic growth of A2780 cells obtained with Nek6 overexpression. (B-D) Nek6 over-expression in A2780 cells. A2780 cells were stably transfected with a construct carrying the NEK6 coding sequence and with the empty vector (pUSE). Two clones were selected from each pool population, qPCR analysis (B) and western blot (C) show Nek6 overexpression in A2780-NEK6, compared to A2780-pUSE control clones. (D) The soft agar assays represented in (A) were quantified by counting the number of colonies formed. The graph shows the results expressed as % of colony formation. Increase of number of colonies was significant in both hypoxia and normoxia in all the clones with Nek6 overexpression (double asterisks, P<0.001, ANOVA).
The “off” state utilized Hey cells which have high basal expression of Nek6. Cells were stably transfected with a DNA silencing construct (SiNEK6) using a sequence not targeting any region in the human genome (SiC) as a negative control. A representative image of the plates is provided in Figure 6A. Efficacy of silencing was verified at the mRNA (Figure 6B) and protein level (Figure 6C). Two clones were selected from SiNEK6 and SiC. Soft agar assay was performed as described above in the selected clones and in the pool. As expected, we observed a significant lower number of colonies in the pool of Hey-siNEK6 in both hypoxia and normoxia. In the clones we also observed a reduction in the number of colonies, which was significant only in hypoxia (Figure 6D).
Figure 6.
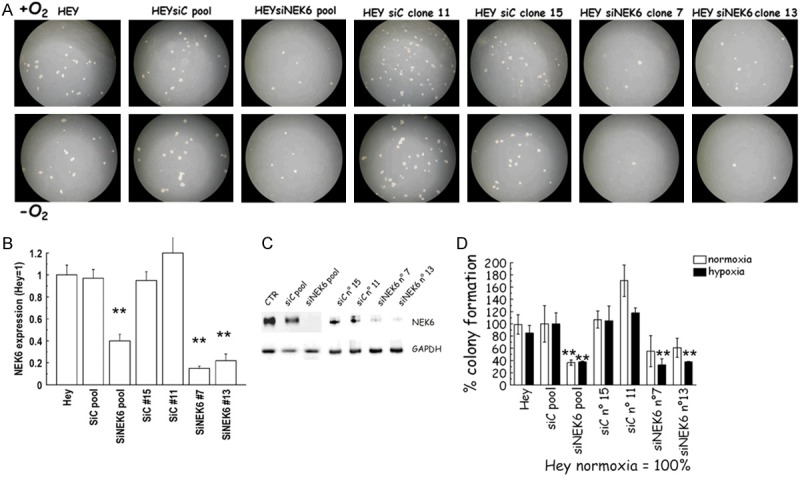
(A) Representative images of the anchorage independent clonogenic growth of Hey cells obtained with Nek6 stable genetic silencing. (B-D) Nek6 silencing in Hey cells. Hey cells were stably transfected with siNEK6 or a nonspecific oligonucleotide silencing vectors (SiC). Two clones were selected from each pool population, qPCR analysis (B) and western blot (C) show Nek6 silencing in Hey-siNEK6 clones, compared to Hey-SiC control clones. (D) The soft agar assays shown in A were quantified by counting the number of colonies formed. The graph shows the results expressed as % of colony formation. Decrease of number of colonies was significant in hypoxia whenever Nek6 was silenced (double asterisks, P<0.001, ANOVA).
Nek6 expression and sensitivity to cisplatin and paclitaxel
In order to assess the effect of the on/off approach on chemotherapy sensitivity, we performed a growth inhibition assay in the presence of paclitaxel or cisplatin for 72 h. All the results are summarized in Table 1. The “on” effects in terms of paclitaxel (Figure 7A) and cisplatin (Figure 7B) sensitivity were compared in A2780 cells across cell lines with NEK6 overexpression vs. cells transformed with the empty pUSE vector. As average, NEK6 overexpression was accompanied by a significant but modest increase in resistance to both paclitaxel (3 nM NEK6 vs. 2 nM PUSE) and cisplatin (400 nM NEK6 vs. 250 nM PUSE). Similarly, the “off” effects were investigated in Hey cells stably transformed with SiNEK6 or the SiC control. In this case we noticed that silencing produced a modest significant increase of paclitaxel (Figure 7C) and cisplatin (Figure 7D) sensitivity. In fact as average IC50 of paclitaxel was 7.8 nM in the SiC vs. 4.7 nM of the SiNEK6 group, while the values for cisplatin were 3100 nM in the SiC vs. 1400 nM in the SiNEK6 group, respectively. Altogether these findings suggest that Nek6 modulation is associated with a modest modulation of sensitivity to the first line chemotherapeutics used in ovarian cancer.
Table 1.
Results of the cytotoxicity assay. Each experiment has been repeated twice in triplicate and results are summarized as mean and standard deviation (SD) of IC50 (nM)
| Mean Paclitaxel | Mean Paclitaxel | SD Paclitaxel | Mean Cisplatin | Mean Cisplatin | SD Cisplatin | |
|---|---|---|---|---|---|---|
| A2780wt | 1.40 | 0.14 | 241.00 | 22.63 | ||
| NEK6_clone10 | 3.60 | 0.42 | 281.00 | 2.83 | ||
| NEK6_clone17 | 2.95 | 1.06 | 374.00 | 33.94 | ||
| NEK6_clone24 | 1.60 | 0.00 | 421.50 | 2.12 | ||
| NEK6_clone3 | 3.50 | 0.57 | 431.00 | 9.90 | ||
| NEK6_pool | 3.50 | 0.42 | 502.50 | 24.75 | ||
| PUSE_clone_18 | 1.98 | 0.18 | 226.50 | 0.71 | ||
| PUSE_clone_19 | 1.85 | 0.21 | 276.00 | 57.98 | ||
| PUSE_clone_21 | 2.63 | 0.11 | 219.00 | 155.56 | ||
| PUSE_clone_23 | 2.61 | 0.55 | 239.50 | 38.89 | ||
| PUSE_pool | 1.87 | 0.16 | 247.50 | 45.96 | ||
| Hey wt | 7.65 | 1.20 | 3365.00 | 162.63 | ||
| SiC_clone_11 | 8.80 | 0.14 | 2591.00 | 169.71 | ||
| SiC_clone_15 | 6.05 | 3.32 | 3549.50 | 579.12 | ||
| SiC_pool | 8.10 | 0.42 | 2783.50 | 94.05 | ||
| SiNEK6_clone23 | 2.90 | 0.28 | 1304.50 | 317.49 | ||
| SiNEK6_clone7 | 5.30 | 5.80 | 1133.50 | 60.10 | ||
| SiNEK6_pool | 6.15 | 0.35 | 1737.00 | 145.66 |
Figure 7.
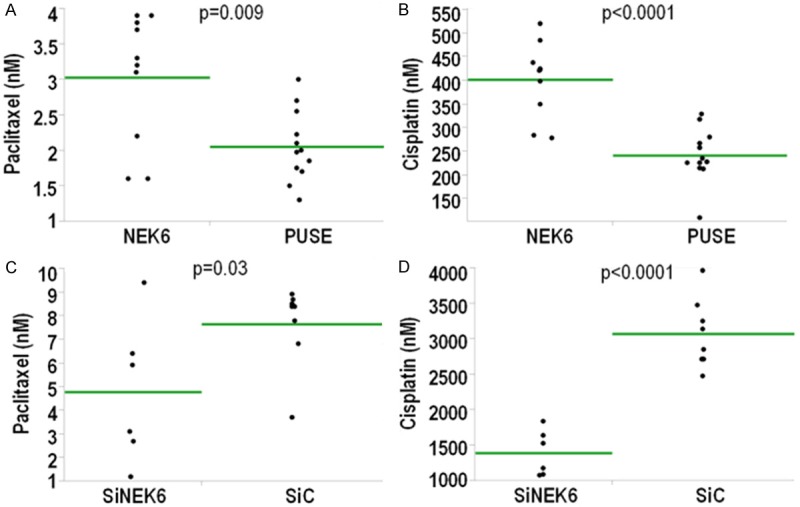
A-D. Effect of overexpression and silencing on chemotherapy sensitivity. The “on/off” effects of Nek6 overexpression/silencing were assessed in a panel of clones and in the pool. In A&B the “on” effects are shown (A2780) while in C&D the “off” effects (Hey). In A&C and in B&D data are for paclitaxel and cisplatin, respectively. Each dot corresponds to the IC50 calculated in a triplicate experiment. The green line corresponds to the average of each group and the p-value of testing the difference across the groups (t-test).
NEK6 in ovarian cancer patients cooperates with HIF1A, TUBB3 and GBP1
The findings presented above demonstrate that Nek6 is a component of the cytoskeletal gateway of drug resistance in ovarian cancer induced by hypoxia under the control of Hif-1α. By facilitating anchorage independent cancer cell colony growth, Nek6 is implicated as a potential mediator of ovarian cancer aggressiveness. We postulate that the aggressive phenotype should be most pronounced when all the components of the related molecular pathways are expressed together. To test this hypothesis, we analyzed the expression of HIF1A, NEK6, TUBB3 and GBP1 using qPCR in a clinical setting of 346 SEOC patients whose clinical features are summarized in Table 2. TUBB3 (Ρ=0.21, P=0.0002), GBP1 (Ρ=0.14, P=0.0014) and NEK6 (Ρ=0.31, P<0.0001) were all significantly correlated with HIF1A, with NEK6 showing the strongest correlation. Thus, the data from the clinical samples lend further support to those derived from cell culture indicating that Nek6 is regulated in hypoxia in a Hif-1α-dependent fashion in the context of a concerted survival pathway. We then analyzed each of the factors as an outcome predictor using the Kaplan-Meier method (Figure 8) and Cox hazard ratio model in univariate and multivariate analysis (combined with age and stage). Endpoint analysis was overall survival (OS). As shown in Table 3, both TUBB3 and NEK6 but not GBP1 and HIF1A were significant predictors in univariate and multivariate analysis. To test for a possible cooperative effect on prognosis of the four transcripts, a cumulative diagnostic score was computed for each patient giving one point for each of the transcripts overexpressed. The diagnostic score was calculated only for those patients in which all the four transcripts were detectable, thus reducing the sample size to 271 patients. Diagnostic score (range 0 to 4) proved predictive in univariate and multivariate Cox analysis (Table 3). Of note, patients with score 4 exhibited aggressive disease with a median OS time of 29 months (Figure 9A), a value very close to the 25 months documented in a large meta-analysis of patients treated with palliative care only [1]. Additionally, we categorized patients according to duration of response after first-line chemotherapy. Patients relapsing after one year were categorized as sensitive as suggested by international guidelines [30]; those relapsing before one year were categorized as resistant. Among patients with a total score of 0.71% were sensitive compared to just 50% among those with a total score of 4 (Figure 9B). This observation supports the hypothesis that the expression of NEK6 and its partners is biologically relevant in aggressive disease.
Table 2.
Clinical features of the analyzed setting of ovarian cancer patients
| Characteristics | CU* clinical setting | TCGA |
|---|---|---|
| Cases | 346 | 344 |
| Age, yrs | ||
| Median (range) | 60 (24-84) | 58 (30-87) |
| FIGO Stage (%) | ||
| I-II | 12 (3.5) | 19 (5.6) |
| III | 271 (78.3) | 276 (80.2) |
| IV | 63 (18.2) | 49 (14.2) |
| Histotype (%) | ||
| Papillary-serous | 346 (100) | 344 (100) |
| Ca 125 | ||
| Median (range) | 683 U/mL (6->10,000) | N.A |
| Status (%) | ||
| Dead | 232 (67.0) | 190 (59.6) |
| Alive | 114 (33.0) | 156 (40.4) |
| Median follow up (Alive) | 68 months | onths |
Catholic University clinical setting.
Figure 8.
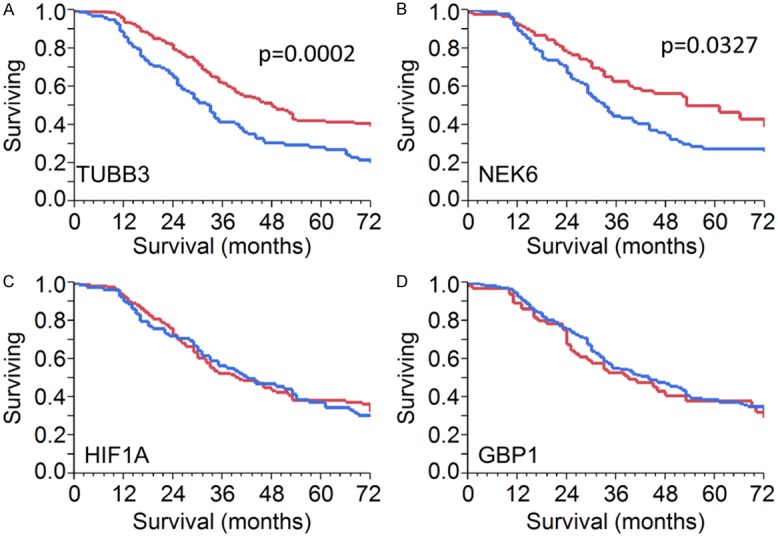
Analysis of expression of NEK6, HIF1A, GBP1 and TUBB3 in a first clinical setting of 346 SEOC patients. Kaplan-Meier survival curve for TUBB3 (A), NEK6 (B), HIF1A (C) and GBP1 (D). Difference was significant for TUBB3 and NEK6 but not for HIF1A and GBP1.
Table 3.
Univariate and multivariate Cox’s proportional hazard ratio in 346 ovarian cancer patients
| n (%) | HR | 95% CI | P-value | |
|---|---|---|---|---|
| Overall Survival (Univariate analysis) | ||||
| Age | 346/346 (100) | 2.30 | 1. 44-4.58 | 0.0083 |
| Ca125 | 296/346 (86) | 0.87 | 0.37-1.82 | 0.5562 |
| Stage | 346/346 (100) | 2.99 | 1.38-7.84 | 0.0038 |
| HIF1A | 341/346 (99) | 1.03 | 0.79-1.34 | 0.781 |
| GBP1 | 329/346 (95) | 0.88 | 0.67-1.15 | 0.3568 |
| TUBB3 | 318/346 (92) | 1.67 | 1.26-2.21 | 0.0004 |
| NEK6 | 277/346 (80) | 1.37 | 1.01-1.82 | 0.0366 |
| Score | 271/346 (78) | 2.28 | 1.29-4.09 | 0.006 |
| Overall Survival (Multivariate analysis) | ||||
| Age | 346/346 (100) | 2.27 | 1.16-4.52 | 0.0167 |
| Stage | 346/346 (100) | 2.87 | 1.32-7.52 | 0.0121 |
| TUBB3 | 318/346 (92) | 1.71 | 1.29-2.27 | 0.0003 |
| NEK6 | 277/346 (80) | 1.45 | 1.08-1.94 | 0.013 |
| Score | 271/346 (78) | 2.29 | 1.27-4.10 | 0.006 |
Figure 9.
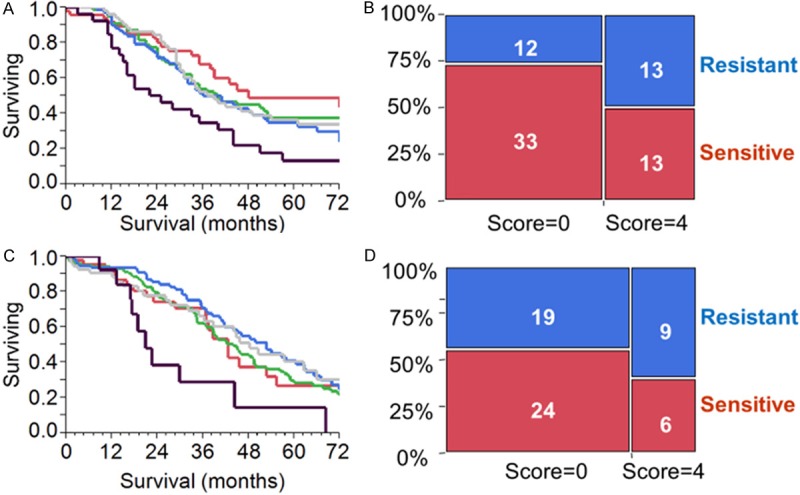
A-D. Analysis of the combined diagnostic score marking the concomitant expression of NEK6, HIF1A, GBP1 and TUBB3. For the expression of each transcript, one point was given. Score 0 and 4 are for patients expressing none or all the transcripts, respectively. A&B. Analysis in a first clinical cohort of 346 SEOC patients. Transcript expression was measured with qPCR. A. Kaplan-Meier analysis reporting the survival curve for patients with score 0 (red), score 1 (green), score 2 (yellow), score 3 (blue) and score 4 (black). Difference between score 0 and score 4 was significant (log-rank test, P=0.001). B. Proportional bar chart showing the number of resistant and sensitive SEOC patients according to score. Patients with score 4 exhibited a higher rate of resistance. C&D. Analysis in a second clinical cohort of 344 SEOC patients (TCGA). Transcript expression was measured with RNA-seq. C. Kaplan-Meier analysis reporting the survival curve for patients with score 0 (red), score 1 (green), score 2 (yellow), score 3 (blue) and score 4 (black). Difference between score 0 and score 4 was significant (log-rank test, P=0.03). D. Proportional bar chart showing the number of resistant and sensitive SEOC patients according to score. Patients with score 4 exhibited a higher rate of resistance.
To validate our results in a separate patient cohort, we downloaded the RNA-seq data of The Cancer Genome Atlas (TCGA) reporting normalized gene expression values for 344 patients (Table 2). We performed the same analysis described above obtaining a diagnostic score for each patient synthesizing the expression data of the four transcripts. The diagnostic scores, as shown in Figure 9C, were again significant in univariate (HR 2.22; CI 1.01-4.75; P=0.05) and multivariate analysis (HR 2.24; CI 1.03-5.04; P=0.04). When patients were grouped according to the duration of response after first-line chemotherapy, we noted, as in the first cohort, that patients with score of 0 were more likely to have sensitive disease compared to patients with score of 4 (56% vs. 40%, Figure 9D).
Discussion
Ovarian cancer is the most lethal gynecologic malignancy, and a full understanding of the mechanisms underlying resistance to first-line treatment is currently lacking. It has been reported that hypoxia activates an adaptive response which renders cancer cells derived from solid tumors less chemo and radiosensitive [3]. This response is mainly driven by the transcription factor Hif-1α, and several strategies of Hif-1α inhibition are undergoing active investigation in clinical trials [31]. By contrast to most other solid tumors and as demonstrated in this report and in previous analyses [32], Hif-1α alone is insufficient to predict outcome in ovarian cancer. We speculate that in this cancer type, only a portion of Hif-1α is active. In this sense, characterization of downstream mediators is required to properly classify ovarian cancer patients with respect to Hif-1α functional status. Here we report for the first time a functional link between Hif-1α and Nek6. Four lines of evidence support a regulation of Nek6 by Hif-1α: i) the presence of a significant correlation between HIF1A and NEK6 in two independent set of patients; ii) concomitant induction of HIF1A and NEK6 in three cellular models upon hypoxia; iii) presence in the 3`UTR of one HRE motif engaged by Hif-1α upon hypoxia; iv) Hif-1α silencing inhibits NEK6 upregulation. All these findings support that Nek6 is a downstream target of Hif-1α both in vitro and in ovarian cancer patients. This functional link is particularly compelling as earlier studies have reported Nek6 overexpression in liver, breast, colorectal, lung and laryngeal cancer [33]. Additionally, Takeno et al. reported higher levels of Nek6 protein in advanced gastric cancers as compared with early-stage samples [34]. Furthermore, Nek6 has been previously described as a factor capable of enhancing cancer aggressiveness in multiple solid tumors [35] and as a potent inhibitor of the p53-induced senescence [36]. In this context, the present investigation adds SEOC to the diseases in which Nek6 is a direct driver of biological aggressiveness. Similar to other kinases, we show also that Nek6 binds to GBP-1, a GTPase whose incorporation in the microtubules is facilitated by class III β-tubulin in cells subjected to microenvironmental stress [9].
As applicable to chemoresistance, class III β-tubulin disrupts microtubule polymerization and thereby inhibits the effectiveness of drugs such as paclitaxel which work via this molecular mechanism [37]. In some circumstances, however, class III β-tubulin expression has an opposite clinical effect; its expression is associated with chemosensitivity rather than chemoresistance. In breast cancer cells, for example, TUBB3 gene is regulated by estrogens [38] and high expression is an indicator of a disease which will be more sensitive to chemotherapy and aromatase inhibitors [39]. These facts suggest that, as reported above for Hif-1α, class III β-tubulin is a pure prognostic predictor of poor outcome only in the context of a multi-molecular concerted pathway [4]. These observations highlight the importance of the elucidation of upstream and downstream functional relationships in order to correctly classify patients for the expression of a given prognosticator. This phenomenon is particularly relevant for SEOC, which is a disease featured by extreme clonal heterogeneity without apparent driving mutations [40]. For these reasons, measurement of the expression of genes in isolation, without a clear understanding of functional interactions, can confound any attempt at predicting ovarian cancer aggressiveness.
The data presented in our study shows that expression levels of clinically relevant and functionally related transcripts NEK6, GBP1, HIF1A and TUBB3 can be combined to provide a diagnostic score which is predictive of aggressive disease. We discovered this phenomenon in a clinical cohort of 346 women, and we validated it in a separate cohort of 344 patients (TCGA study).
Elucidation of the linkages between genes induced by hypoxia and those involved in response to treatments may allow a more effective use of the anticancer strategies available for ovarian cancer. Recently, bevacizumab has been approved by EMEA (European Medicine Agency) for the first-line treatment of ovarian cancer patients [41]. Hypoxia resulting from bevacizumab treatment is believed to represent the most important functional circuit for resistance to drugs targeting VEGF (Vascular Endothelial Growth Factor) and angiogenesis [42]. A diagnostic score comprised of NEK6, HIF1A, GBP1 and TUBB3 expression levels could serve as a predictive marker and identify patients with high probability of resistance (and therefore contraindication) to bevacizumab, as element of the first-line chemotherapy in SEOC. Conversely, it may facilitate selection of patients for CRLX101, a nanoparticle assembly containing cyclodextrin-based polymer and camptothecin. This uniquely formulated drug downregulates VEGF (31) and may counter resistance to bevacizumab, a concept which is undergoing clinical trial in SEOC patients (NCT01652079).
In summary, this study provides evidence that Nek6 is a factor driven by hypoxia that cooperates with Hif-1α and the cytoskeletal gateway of drug resistance in mediating an aggressive phenotype in SEOC patients. The strength of our investigation comes to the fact that we were capable to translate the initial observations found in cell lines into two independent clinical set of patients, thus minimizing the risk that our findings are false positive results. A weakness of the investigation consists in the fact that we were not able to analyze Nek6 expression at the protein level in our patient cohort, due to the absence of a suitable anti-Nek6 antibody working in FFPE tissues. Nevertheless we believe our findings identify a novel functional circuit underlying biological aggressiveness in SEOC and may facilitate the personalization of the best first line treatment in this clinical setting.
Acknowledgements
This work was partially supported by a Grant from AIRC (Associazione Italiana Ricerca sul Cancro, IG11975), Associazione OPPO e le sue stanze ONLUS, by a liberal donation from Mr. and Mrs. Ruggles. This work is dedicated to the memory of Nancy Vivien Maiorano who passed away for cancer at the age of 48. Sponsors of the study did not have any role in the study conception and management.
Disclosure of conflict of interest
None.
Abbreviations
- SEOC
Serous Ovarian Cancer
- Hif-1α
Hypoxia-inducible factor 1-alpha
- GBP-1
Interferon-induced guanylate-binding protein 1
- Nek6
NIMA (Never In Mitosis Gene A)-Related Kinase 6
- GENT
Gene Expression across Normal and Tumor tissue
- SiC
Scrambled interfering control
- HRE
Hypoxia Responsive Elements
- ChIP
Chromatin Immuno-precipitation
- CA
Carbonic Anhydrase
- OS
Overall Survival
- HR
Hazard Ratio
- CI
Confidence Interval
- 3’UTR
3’Untranslated Region
- EMEA
European Medicine Agency
References
- 1.Kyrgiou M, Salanti G, Pavlidis N, Paraskevaidis E, Ioannidis JP. Survival benefits with diverse chemotherapy regimens for ovarian cancer: meta-analysis of multiple treatments. J Natl Cancer Inst. 2006;98:1655–1663. doi: 10.1093/jnci/djj443. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan S, Coward JI, Bast RC Jr, Berchuck A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R, Etemadmoghadam D, Friedlander M, Gabra H, Kaye SB, Lord CJ, Lengyel E, Levine DA, McNeish IA, Menon U, Mills GB, Nephew KP, Oza AM, Sood AK, Stronach EA, Walczak H, Bowtell DD, Balkwill FR. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariani M, Shahabi S, Sieber S, Scambia G, Ferlini C. Class III beta-tubulin (TUBB3): more than a biomarker in solid tumors? Curr Mol Med. 2011;11:726–731. doi: 10.2174/156652411798062368. [DOI] [PubMed] [Google Scholar]
- 5.Mozzetti S, Ferlini C, Concolino P, Filippetti F, Raspaglio G, Prislei S, Gallo D, Martinelli E, Ranelletti FO, Ferrandina G, Scambia G. Class III beta-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin Cancer Res. 2005;11:298–305. [PubMed] [Google Scholar]
- 6.Ferrandina G, Zannoni GF, Martinelli E, Paglia A, Gallotta V, Mozzetti S, Scambia G, Ferlini C. Class III beta-tubulin overexpression is a marker of poor clinical outcome in advanced ovarian cancer patients. Clin Cancer Res. 2006;12:2774–2779. doi: 10.1158/1078-0432.CCR-05-2715. [DOI] [PubMed] [Google Scholar]
- 7.Seve P, Dumontet C. Is class III beta-tubulin a predictive factor in patients receiving tubulin-binding agents? Lancet Oncol. 2008;9:168–175. doi: 10.1016/S1470-2045(08)70029-9. [DOI] [PubMed] [Google Scholar]
- 8.Raspaglio G, Filippetti F, Prislei S, Penci R, De Maria I, Cicchillitti L, Mozzetti S, Scambia G, Ferlini C. Hypoxia induces class III betatubulin gene expression by HIF-1alpha binding to its 3’ flanking region. Gene. 2008;409:100–108. doi: 10.1016/j.gene.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 9.De Donato M, Mariani M, Petrella L, Martinelli E, Zannoni GF, Vellone V, Ferrandina G, Shahabi S, Scambia G, Ferlini C. Class III beta-tubulin and the cytoskeletal gateway for drug resistance in ovarian cancer. J Cell Physiol. 2012;227:1034–1041. doi: 10.1002/jcp.22813. [DOI] [PubMed] [Google Scholar]
- 10.Kahn BB, Rossetti L, Lodish HF, Charron MJ. Decreased in vivo glucose uptake but normal expression of GLUT1 and GLUT4 in skeletal muscle of diabetic rats. J Clin Invest. 1991;87:2197–2206. doi: 10.1172/JCI115254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baranello C, Mariani M, Andreoli M, Fanelli M, Martinelli E, Ferrandina G, Scambia G, Shahabi S, Ferlini C. Adrenomedullin in ovarian cancer: foe in vitro and friend in vivo? PLoS One. 2012;7:e40678. doi: 10.1371/journal.pone.0040678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferlini C, Raspaglio G, Mozzetti S, Distefano M, Filippetti F, Martinelli E, Ferrandina G, Gallo D, Ranelletti FO, Scambia G. Bcl-2 down-regulation is a novel mechanism of paclitaxel resistance. Mol Pharmacol. 2003;64:51–58. doi: 10.1124/mol.64.1.51. [DOI] [PubMed] [Google Scholar]
- 13.Mariani M, Zannoni GF, Sioletic S, Sieber S, Martino C, Martinelli E, Coco C, Scambia G, Shahabi S, Ferlini C. Gender influences the Class III and V beta-tubulin ability to predict poor outcome in colorectal cancer. Clin Cancer Res. 2012;18:2964–75. doi: 10.1158/1078-0432.CCR-11-2318. [DOI] [PubMed] [Google Scholar]
- 14.Prislei S, Mozzetti S, Filippetti F, De Donato M, Raspaglio G, Cicchillitti L, Scambia G, Ferlini C. From plasma membrane to cytoskeleton: a novel function for semaphorin 6A. Mol Cancer Ther. 2008;7:233–241. doi: 10.1158/1535-7163.MCT-07-0390. [DOI] [PubMed] [Google Scholar]
- 15.Mozzetti S, Iantomasi R, De Maria I, Prislei S, Mariani M, Camperchioli A, Bartollino S, Gallo D, Scambia G, Ferlini C. Molecular mechanisms of patupilone resistance. Cancer Res. 2008;68:10197–10204. doi: 10.1158/0008-5472.CAN-08-2091. [DOI] [PubMed] [Google Scholar]
- 16.Ferlini C, Cicchillitti L, Raspaglio G, Bartollino S, Cimitan S, Bertucci C, Mozzetti S, Gallo D, Persico M, Fattorusso C, Campiani G, Scambia G. Paclitaxel directly binds to Bcl-2 and functionally mimics activity of Nur77. Cancer Res. 2009;69:6906–6914. doi: 10.1158/0008-5472.CAN-09-0540. [DOI] [PubMed] [Google Scholar]
- 17.Andreoli M, Persico M, Kumar A, Orteca N, Kumar V, Pepe A, Mahalingam S, Alegria AE, Petrella L, Sevciunaite L, Camperchioli A, Mariani M, Di Dato A, Novellino E, Scambia G, Malhotra SV, Ferlini C, Fattorusso C. Identification of the First Inhibitor of the GBP1:PIM1 Interaction. Implications for the Development of a New Class of Anticancer Agents against Paclitaxel Resistant Cancer Cells. J Med Chem. 2014;57:7916–7932. doi: 10.1021/jm5009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raspaglio G, Petrillo M, Martinelli E, Li Puma DD, Mariani M, De Donato M, Filippetti F, Mozzetti S, Prislei S, Zannoni GF, Scambia G, Ferlini C. Sox9 and Hif-2alpha regulate TUBB3 gene expression and affect ovarian cancer aggressiveness. Gene. 2014;542:173–181. doi: 10.1016/j.gene.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 19.Prislei S, Martinelli E, Mariani M, Raspaglio G, Sieber S, Ferrandina G, Shahabi S, Scambia G, Ferlini C. MiR-200c and HuR in ovarian cancer. BMC Cancer. 2013;13:72. doi: 10.1186/1471-2407-13-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariani M, He S, McHugh M, Andreoli M, Pandya D, Sieber S, Wu Z, Fiedler P, Shahabi S, Ferlini C. Integrated multidimensional analysis is required for accurate prognostic biomarkers in colorectal cancer. PLoS One. 2014;9:e101065. doi: 10.1371/journal.pone.0101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Airley R, Loncaster J, Davidson S, Bromley M, Roberts S, Patterson A, Hunter R, Stratford I, West C. Glucose transporter glut-1 expression correlates with tumor hypoxia and predicts metastasis-free survival in advanced carcinoma of the cervix. Clin Cancer Res. 2001;7:928–934. [PubMed] [Google Scholar]
- 26.Chia SK, Wykoff CC, Watson PH, Han C, Leek RD, Pastorek J, Gatter KC, Ratcliffe P, Harris AL. Prognostic significance of a novel hypoxia-regulated marker, carbonic anhydrase IX, in invasive breast carcinoma. J. Clin. Oncol. 2001;19:3660–3668. doi: 10.1200/JCO.2001.19.16.3660. [DOI] [PubMed] [Google Scholar]
- 27.Bunn HF, Gu J, Huang LE, Park JW, Zhu H. Erythropoietin: a model system for studying oxygen-dependent gene regulation. J Exp Biol. 1998;201:1197–1201. doi: 10.1242/jeb.201.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Elsner T, Ramirez JR, Sanz-Rodriguez F, Varela E, Bernabeu C, Botella LM. A cross-talk between hypoxia and TGF-beta orchestrates erythropoietin gene regulation through SP1 and Smads. J Mol Biol. 2004;336:9–24. doi: 10.1016/j.jmb.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 29.McCarroll JA, Gan PP, Liu M, Kavallaris M. betaIII-tubulin is a multifunctional protein involved in drug sensitivity and tumorigenesis in non-small cell lung cancer. Cancer Res. 2010;70:4995–5003. doi: 10.1158/0008-5472.CAN-09-4487. [DOI] [PubMed] [Google Scholar]
- 30.Parmar MK, Ledermann JA, Colombo N, du Bois A, Delaloye JF, Kristensen GB, Wheeler S, Swart AM, Qian W, Torri V, Floriani I, Jayson G, Lamont A, Trope C. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGOOVAR-2.2 trial. Lancet. 2003;361:2099–2106. doi: 10.1016/s0140-6736(03)13718-x. [DOI] [PubMed] [Google Scholar]
- 31.Monti E, Gariboldi MB. HIF-1 as a target for cancer chemotherapy, chemosensitization and chemoprevention. Curr Mol Pharmacol. 2011;4:62–77. doi: 10.2174/1874467211104010062. [DOI] [PubMed] [Google Scholar]
- 32.Birner P, Schindl M, Obermair A, Breitenecker G, Oberhuber G. Expression of hypoxia-inducible factor 1alpha in epithelial ovarian tumors: its impact on prognosis and on response to chemotherapy. Clin Cancer Res. 2001;7:1661–1668. [PubMed] [Google Scholar]
- 33.Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M, Boldorini R, Pallotti F, Viale G, Gishizky ML, Draetta GF, Di Fiore PP. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006;66:8147–8154. doi: 10.1158/0008-5472.CAN-05-3489. [DOI] [PubMed] [Google Scholar]
- 34.Takeno A, Takemasa I, Doki Y, Yamasaki M, Miyata H, Takiguchi S, Fujiwara Y, Matsubara K, Monden M. Integrative approach for differentially overexpressed genes in gastric cancer by combining large-scale gene expression profiling and network analysis. Br J Cancer. 2008;99:1307–1315. doi: 10.1038/sj.bjc.6604682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nassirpour R, Shao L, Flanagan P, Abrams T, Jallal B, Smeal T, Yin MJ. Nek6 mediates human cancer cell transformation and is a potential cancer therapeutic target. Mol Cancer Res. 2010;8:717–728. doi: 10.1158/1541-7786.MCR-09-0291. [DOI] [PubMed] [Google Scholar]
- 36.Jee HJ, Kim AJ, Song N, Kim HJ, Kim M, Koh H, Yun J. Nek6 overexpression antagonizes p53-induced senescence in human cancer cells. Cell Cycle. 2010;9:4703–4710. doi: 10.4161/cc.9.23.14059. [DOI] [PubMed] [Google Scholar]
- 37.Derry WB, Wilson L, Khan IA, Luduena RF, Jordan MA. Taxol differentially modulates the dynamics of microtubules assembled from unfractionated and purified beta-tubulin isotypes. Biochemistry. 1997;36:3554–3562. doi: 10.1021/bi962724m. [DOI] [PubMed] [Google Scholar]
- 38.Saussede-Aim J, Matera EL, Ferlini C, Dumontet C. Beta3-tubulin is induced by estradiol in human breast carcinoma cells through an estrogen-receptor dependent pathway. Cell Motil Cytoskeleton. 2009;66:378–388. doi: 10.1002/cm.20377. [DOI] [PubMed] [Google Scholar]
- 39.Galmarini CM, Treilleux I, Cardoso F, Bernard-Marty C, Durbecq V, Gancberg D, Bissery MC, Paesmans M, Larsimont D, Piccart MJ, Di Leo A, Dumontet C. Class III beta-tubulin isotype predicts response in advanced breast cancer patients randomly treated either with single-agent doxorubicin or docetaxel. Clin Cancer Res. 2008;14:4511–4516. doi: 10.1158/1078-0432.CCR-07-4741. [DOI] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia A, Singh H. Bevacizumab and ovarian cancer. Ther Adv Med Oncol. 2013;5:133–141. doi: 10.1177/1758834012467661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tejpar S, Prenen H, Mazzone M. Overcoming resistance to antiangiogenic therapies. Oncologist. 2012;17:1039–1050. doi: 10.1634/theoncologist.2012-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]