Abstract
Hypermethylation of GPX3 (glutathione peroxidase 3) promoter has been identified in various solid tumors. However, the pattern of GPX3 promoter methylation in acute myeloid leukemia (AML) remains unknown. The current study was intended to investigate the clinical significance of GPX3 promoter methylation in de novo AML patients and further determine its role in regulating GPX3 expression. GPX3 promoter methylation status was detected in 181 de novo AML patients and 44 normal controls by real-time quantitative methylation-specific PCR and bisulfite sequencing PCR. Real-time quantitative PCR was carried out to assess GPX3 expression. GPX3 promoter was significantly methylated in AML patients compared with normal controls (P=0.022). The patients with GPX3 methylation presented significantly older age than those with GPX3 unmethylation (P=0.011). GPX3 methylated patients had significantly lower frequency of C/EBPA mutation and higher incidence of FLT3-ITD mutation (P=0.037 and 0.030, respectively). The non-M3 patients with GPX3 methylation had significantly lower overall survival than those with GPX3 unmethylation (P=0.036). No significant correlation was observed between GPX3 expression and its promoter methylation (R=0.110, P=0.284). However, GPX3 mRNA level was significantly increased after 5-aza-2’-deoxycytidine treatment in leukemic cell line THP1. Our data suggest that GPX3 methylation predicts adverse clinical outcome in non-M3 AML patients. Moreover, GPX3 expression is regulated by its promoter methylation in leukemic cell line THP1.
Keywords: GPX3, methylation, prognosis, regulation, acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is a clonal hematological malignancy with diverse clinical outcome, characterized by a block in differentiation of hematopoiesis and growth of a clonal population of neoplastic cells or blasts [1,2]. Genetic alterations play crucial roles not only in the pathogenesis but also in the prognosis of AML [3-5]. Recently, epigenetic modifications such as aberrant DNA methylation have been identified to contribute to the pathogenesis of AML [3]. Moreover, aberrant methylation of various oncogenes and/or tumor suppressor genes (TSGs) has been found as potential biomarkers for the prognosis of AML [6,7]. These give new insights into disease pathogenesis and provide opportunities for therapeutic advances.
GPX (glutathione peroxidase) family is composed of 8 members (GPX1-GPX8) with their roles in reducing redundant reactive oxygen species (ROS) against oxidative damages to host cells [8]. GPX3, locates on chromosome 5q23, accounts for nearly all the GPX activities in plasma [8]. Tumor suppressor function of GPX3 has been identified in quite a few tumors [9-11]. Accumulating studies have revealed the pattern of GPX3 promoter hypermethylation in a variety of cancers [12-19]. Moreover, the prognostic value of GPX3 promoter hypermethylation has also been revealed in several cancers [17-19]. However, little is known about the pattern of GPX3 promoter methylation and its clinical relevance in AML by far. The present study was aimed to investigate the clinical significance of GPX3 promoter methylation in de novo AML patients and further determine its role in the regulation of GPX3 expression.
Materials and methods
Patients
A total of 181 patients with a diagnosis of AML as well as 44 healthy donors were included into the study approved by the Institutional Review Board of the Affiliated People’s Hospital of Jiangsu University. The diagnosis and classification of the patients were based on the revised French-American-British (FAB) classification and the 2008 World Health Organization (WHO) criteria [20,21]. Treatment protocol for AML patients was described previously [22]. The parameters of AML patients were summarized in Table 1. Bone marrow (BM) specimens were collected form all the patients and healthy donors after written informed consents were obtained. BM mononuclear cells were extracted from BM specimens by gradient centrifugation using Lymphocyte Separation Medium (TBD sciences, Tianjin, China).
Table 1.
Association between GPX3 promoter methylation and clinical parameters in AML patients
| Patient’s parameters | Status of GPX3 promoter methylation | ||
|---|---|---|---|
|
| |||
| Unmethylated (n=134) | Methylated (n=47) | P value | |
| Sex, male/female | 77/57 | 31/16 | 0.388 |
| Median age, years (range) | 48 (3-93) | 59 (15-87) | 0.011 |
| Median WBC, ×109/L (range) | 16.4 (0.8-528.0) | 16.3 (0.9-185.4) | 0.742 |
| Median hemoglobin, g/L (range) | 74 (32-131) | 74.5 (33-138) | 0.834 |
| Median platelets, ×109/L (range) | 40 (3-264) | 40.5 (6-119) | 0.859 |
| BM blasts, % (range) | 43.5 (5.0-97.5) | 51.5 (3.0-94.5) | 0.259 |
| FAB | 0.541 | ||
| M0 | 1 (1%) | 0 (0%) | |
| M1 | 13 (10%) | 7 (15%) | |
| M2 | 49 (37%) | 17 (36%) | |
| M3 | 21 (16%) | 7 (15%) | |
| M4 | 27 (21%) | 12 (26%) | |
| M5 | 14 (10%) | 4 (8%) | |
| M6 | 9 (7%) | 0 (0%) | |
| WHO | 0.294 | ||
| AML with t(8;21) | 15 (11%) | 4 (9%) | |
| APL with t(15;17) | 21 (16%) | 7 (15%) | |
| AML with 11q23 | 1 (1%) | 1 (2%) | |
| ML without maturation | 10 (7%) | 7 (15%) | |
| AML with maturation | 36 (27%) | 13 (28%) | |
| Acute myelomonocytic leukemia | 26 (19%) | 13 (28%) | |
| Acute monoblastic/monocytic leukemia | 14 (10%) | 2 (4%) | |
| Acute erythroid leukemia | 9 (7%) | 0 (0%) | |
| No data | 2 (1%) | 0 (0%) | |
| Karyotype classification | 0.563 | ||
| Favorable | 36 (27%) | 11 (23%) | |
| Intermediate | 76 (57%) | 24 (57%) | |
| Poor | 15 (11%) | 8 (17%) | |
| No data | 7 (5%) | 4 (9%) | |
| Karyotype | 0.834 | ||
| Normal | 59 (44%) | 18 (38%) | |
| T(8;21) | 15 (11%) | 4 (9%) | |
| T(15;17) | 20 (15%) | 7 (15%) | |
| 11q23 | 1 (1%) | 1 (2%) | |
| Complex | 12 (9%) | 6 (13%) | |
| Others | 20 (15%) | 7 (15%) | |
| No data | 7 (5%) | 4 (9%) | |
| Gene Mutation | |||
| C/EBPA (+/-) | 24/97 | 3/43 | 0.037 |
| NPM1 (+/-) | 15/106 | 5/41 | 1.000 |
| FLT3-ITD (+/-) | 10/111 | 10/36 | 0.030 |
| c-KIT (+/-) | 6/115 | 2/44 | 1.000 |
| N/K RAS (+/-) | 12/109 | 6/40 | 0.581 |
| IDH1/2 (+/-) | 8/113 | 1/45 | 0.447 |
| DNMT3A (+/-) | 8/113 | 4/42 | 0.738 |
| U2AF1 (+/-) | 3/118 | 2/44 | 0.616 |
| CR (+/-) | 42/43 | 20/22 | 1.000 |
WBC: white blood cells; FAB: French-American-British classification; AML: acute myeloid leukemia; CR: complete remission.
Cell line, cell culture and 5-aza-dC treatment
Human leukemic cell line THP1 cells were cultured in IMDM medium containing 10% fetal calf serum and grown at 37°C in 5% CO2 humidified atmosphere. For demethylation studies, cells were incubated with a final concentration of 0 μM, 0.1 μM, 1 μM, 10 μM, and 50 μM 5-aza-2’-deoxycytidine (5-aza-dC) (Sigma-Aldrich, Steinheim, USA) for 72 h. All cells were cultured until harvested for the extraction of RNA and DNA.
RNA isolation, reverse transcription and RQ-PCR
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription reaction with 40 μL volume was composed of 5 × buffer 10 mM, 10 mM of dNTPs, 10 μM of random hexamers, 80 U of RNAsin, and 200 U of MMLV reverse transcriptase (MBI Fermentas, Hanover, USA). The reaction conditions were incubated for 10 min at 25°C, 60 min at 42°C, and then stored at -20°C.
Real-time quantitative PCR (RQ-PCR) was performed on a 7300 Thermo cycler (Applied Biosystems, CA, USA). The primer sequences for GPX3 expression were 5’-GCCGGGGACAAGAGAAGT-3’ (forward) and 5’-GAGGACGTATTTGCCAGCAT-3’ (reverse) [17]. The reaction system with 20 μL volume consisted of cDNA 20 ng, 0.8 μM of primers, 10 μM of AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA), and 0.4 μM of ROX Reference Dye 1 (Invitrogen, Carlsbad, CA, USA). The RQ-PCR reaction conditions were 95°C for 5 min, followed by 45 cycles at 95°C for 10 s, 63°C for 30 s, 72°C for 30 s, and 80°C for 30 s to collect fluorescence, finally followed by 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s. Both positive and negative controls were included in each assay. Relative GPX3 transcript levels were calculated by the formulas NGPX3=(EGPX3)ΔCT GPX3 (control-sample)÷(EABL)ΔCT ABL (control-sample) and E=10(-1/slope) (the slope referred to CT versus cDNA concentration plot).
DNA isolation, chemical modification and RQ-MSP
Genomic DNA was isolated using genomic DNA purification kit (Gentra, Minneapolis, MN, USA) and was modified using the CpGenome DNA Modification Kit (Chemicon, Ternecula, Canada) according to the manufacturer’s recommendations. The primer sequences for the methylated (M) GPX3 promoter were 5’-TATGTTATTGTCGTTTCGGGAC-3’ (forward) and 5’-GTCCGTCTAAAATATCCGACG-3’ (reverse), and for the unmethylated (U) GPX3 promoter were 5’-TTTATGTTATTGTTGTTTTGGGATG-3’ (forward) and 5’-ATCCATCTAAAATATCCAACACTCC-3’ (reverse) [15]. Real-time quantitative methylation-specific PCR (RQ-MSP) was performed for M-MSP reaction, which was composed of primers 0.8 μM, 10 μM of AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA), 0.4 μM of ROX Reference Dye 1 (Invitrogen, Carlsbad, CA, USA), and 20 ng of modified DNA. The program for amplification was 95°C for 5 min, 40 cycles for 10 s at 95°C, 30 s at 64°C, 72°C for 30 s, and 80°C for 30 s, finally a melting program of one cycle at 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60 °C for 15 s. U-MSP reaction using the same reagents was incubated for 95°C for 5 min, 40 cycles for 10 s at 95°C, 30 s at 58°C, and 30 s at 72°C followed by a final 7 min extension step at 72°C. Both positive and negative controls were included in each assay. The normalized ratio (NM-GPX3) calculated relative to the reference ALU was used to assess the level of GPX3 promoter methylation in samples. NM-GPX3 was calculated using the equation: NM-GPX3=(EM-GPX3)ΔCT M-GPX3 (control-sample)÷(EALU)ΔCT ALU (control-sample).
Bisulfite sequencing
The primer sequences for bisulfite modified GPX3 promoter were 5’-ATTTTGGAGTTAAAAGAGGAAG-3’ (forward) and 5’-CTACCTAATCCCTAACCACC-3’ (reverse). Bisulfite sequencing PCR (BSP) reaction system contained 1 × PCR buffer (KCl 0.25 mM), dNTP Mixture 6.25 μM, primers 0.5 μM, hot start DNA polymerase 0.75 U (Takara, Tokyo, Japan), and modified DNA 20 ng. The BSP was carried out at 98°C for 10 s, 40 cycles for 10 s at 98°C, 30 s at 56°C, 72°C for 30 s, and followed by a final 7 min extension step at 72°C. The PCR products were analyzed on 2% agarose gels. The PCR products were purified and cloned into pMD19-T Vector (Takara, Tokyo, Japan), then transfected into DH5A competent cells (Vazyme, Carlsbad, CA, USA). Eight clones from each sample were sequenced (BGI Tech Solutions Co., Shanghai, China).
Gene mutation detection
The detection of N/K-RAS, DNMT3A, U2AF1, IDH1/2, c-KIT, and NPM1 mutations were performed for PCR products using HRMA with the LightScanner platform (Idaho Technology Inc., Salt Lake City, Utah) [23-26]. All positive samples were confirmed by DNA direct sequencing. FLT3-ITD and C/EBPA mutations were detected by direct DNA sequencing [27].
Statistical analysis
SPSS 18.0 software package (SPSS, Chicago, IL) was applied to perform statistical analyses. Mann-Whitney’s U test was carried to compare the difference of continuous variables between two groups. Pearson Chi-square analysis or Fisher exact test was employed to compare the difference of categorical variables. Correlation analysis between GPX3 expression and its promoter methylation was performed by spearman rank correlation test. Kaplan-Meier analysis and multivariate analysis were used to analyze the impact of GPX3 expression on survival respectively. For all analyses, a two-tailed P<0.05 was defined as statistically significant.
Results
GPX3 methylation in normal controls and AML patients
According to RQ-MSP, GPX3 promoter was significantly methylated in AML patients (median 0.012, range 0.000-7.493) compared with normal controls (median 0.005, range 0.000-1.000) (P=0.022, Figure 1). The representative electrophoresis results of RQ-MSP products were shown in Figure 2.
Figure 1.

Relative methylation levels of GPX3 in normal controls and AML patients.
Figure 2.
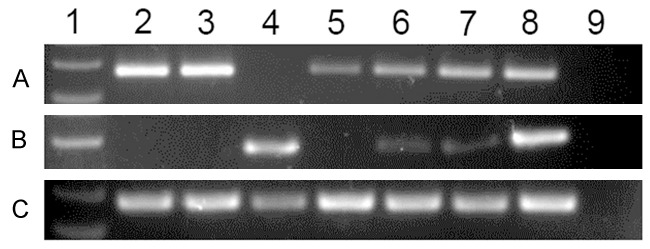
Electrophoresis results of RQ-PCR and RQ-MSP products in normal controls and AML patients. 1: Gene RulerTM 100 bp DNA ladder; 2, 3: controls; 4-7: AML patients; 8: positive control; 9: negative control. A: GPX3 expression; B: GPX3 methylation; C: GPX3 unmethylation.
Two controls and two GPX3 unmethylated AML patients as well as two GPX3 methylated AML patients were selected randomly to further investigate the GPX3 methylation density by BSP. Both controls and unmethylated AML patients presented almost fully unmethylated GPX3 promoter (Figure 3). While the two methylated AML patients presented higher density of GPX3 methylation (Figure 3).
Figure 3.
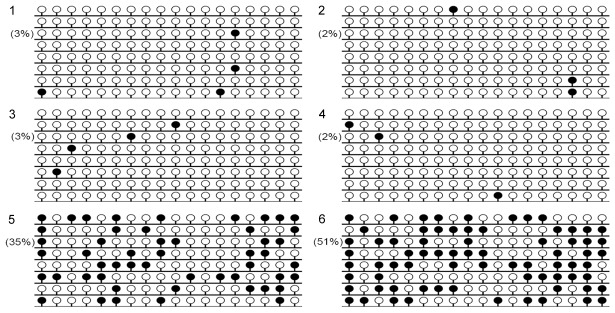
Methylation density of GPX3 promoter in normal controls and AML patients. White cycle: unmethylated CpG dinucleotide; Black cycle: methylated CpG dinucleotide. 1, 2: controls; 3, 4: unmethylated AML patients; 5, 6: methylated AML patients.
Association between GPX3 methylation and expression
GPX3 expression was detected in 97 AML patients with available mRNA. GPX3 mRNA level in AML patients ranged from 0.000 to 9.407 with a median level of 0.035. No significant correlation was observed between GPX3 expression and its promoter methylation (R=0.110, P=0.284).
Association between GPX3 methylation and clinical characteristics of AML patients
The level of methylated GPX3 promoter in controls was 0.034±0.150 (range 0.000-1.000). NM-GPX3 above the value of 0.184 (defined as the mean + SD) was set to define GPX3 promoter methylation in AML patients. Only 1 of 44 (2%) controls presented methylated GPX3 promoter. However, GPX3 promoter methylation was identified in 26% (47/181) of AML patients (P<0.001). According to the cutoff value, the whole AML patients were divided into two groups: GPX3 methylated and GPX3 unmethylated. There were no significant differences in sex, white blood cell, hemoglobin, platelets, and BM blasts between the methylated and unmethylated patients (P>0.05, Table 1). No significant difference was observed in the distribution of both FAB and WHO as well as karyotypic classifications between the patients with and without GPX3 methylation (P>0.05, Table 1). However, GPX3 methylated cases showed significantly older age than GPX3 unmethylated cases (P=0.011, Table 1). Significant differences were observed in the frequencies of both C/EBPA and FLT3-ITD mutations between GPX3 methylated and unmethylated cases. The methylated patients had significantly lower frequency of C/EBPA mutation and higher incidence of FLT3-ITD mutation (P=0.037 and 0.030, respectively, Table 1). Since the GPX3 gene locates at the chromosome 5, we further analyzed GPX3 methylation pattern in the patients with and without -5/5q-. No significant difference was found in the level of GPX3 methylation between the -5/5q- and non-(-5/5q-) cases (median 0.060 vs 0.010, P=0.211).
Association between GPX3 methylation and clinical outcome
Follow-up data were obtained in 127 patients. GPX3 methylated and unmethylated patients showed similar complete remission (CR) rate in whole AML (48% vs 49%, P=1.000, Table 1). Moreover, there were also no significant differences in CR rate between GPX3 methylated and unmethylated patients among both non-M3 AML [37% (13/35) vs 45% (34/75), P=0.535] and cytogenetically normal AML (CN-AML) [47% (8/17) vs 42% (23/55), P=1.000]. Survival analyses were performed in 121 patients with survival data ranging from 1 to 92 months with a median of 8 months. No significant differences were observed in overall survival (OS) between the methylated and unmethylated cases in both whole AML (median 4 vs 9 months, P=0.439) and CN-AML (median 3 vs 11 months, P=0.179). However, among non-M3 patients, GPX3 methylated patients had significantly lower OS than GPX3 unmethylated patients (median 3 vs 8 months, P=0.036, Figure 4). Moreover, multivariate analysis also confirmed the prognostic significance of GPX3 methylation in non-M3 patients (Table 2) but not in whole AML as well as CN-AML patients (data not shown).
Figure 4.
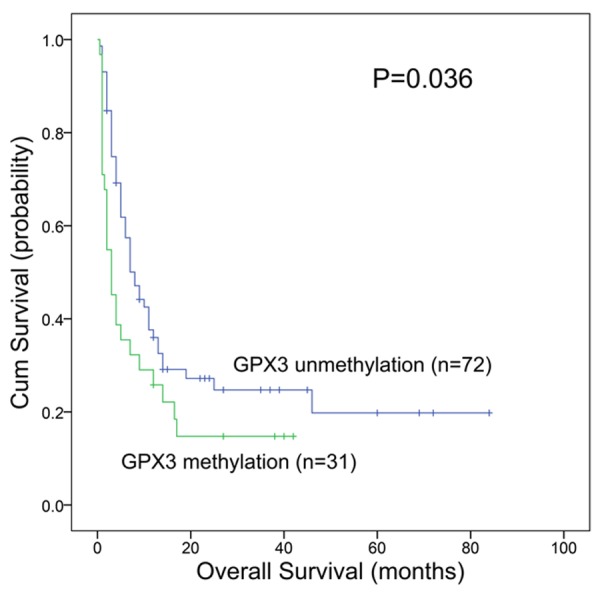
The impact of GPX3 methylation on overall survival of non-M3 AML patients.
Table 2.
Multivariate analysis of prognostic factors for overall survival in non-M3 AML patients
| Hazard ratio (95% CI) | P value | |
|---|---|---|
| Age | 2.344 (1.396-3.936) | 0.001 |
| WBC | 2.063 (1.242-3.425) | 0.005 |
| Karyotypic classification | 1.425 (1.003-2.025) | 0.048 |
| GPX3 methylation | 1.851 (1.051-3.262) | 0.033 |
| FLT3 mutation | 0.460 (0.209-1.013) | 0.054 |
| NPM1 mutation | 1.192 (0.505-2.811) | 0.688 |
| C/EBPA mutation | 0.999 (0.486-2.054) | 0.997 |
| c-KIT mutation | 0.362 (0.048-2.738) | 0.325 |
| N/K RAS mutation | 1.397 (0.647-3.021) | 0.395 |
| IDH1/2 mutation | 1.061 (0.406-2.770) | 0.904 |
| DNMT3A mutation | 1.112 (0.399-3.094) | 0.839 |
| U2AF1 mutation | 3.372 (1.269-8.961) | 0.015 |
Epigenetic mechanism regulating GPX3 expression in leukemic cell line
To determine the role of GPX3 promoter methylation in regulating GPX3 expression in AML, THP1 cell line was treated with 5-aza-dC. THP1 showed extremely low GPX3 mRNA level and fully methylated GPX3 promoter before 5-aza-dC treatment (Figure 5). GPX3 mRNA level was significantly increased after 5-aza-dC treatment in a dose-dependent manner, meanwhile, GPX3 promoter methylation level decreased (Figure 5).
Figure 5.
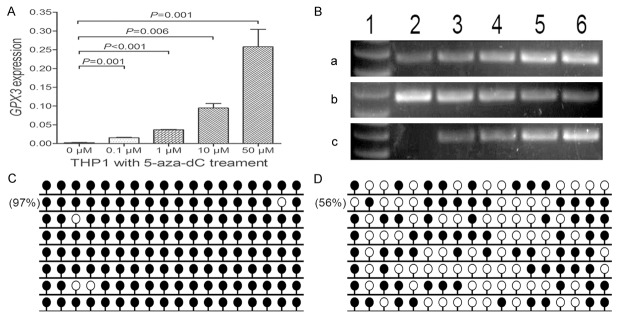
GPX3 expression and methylation in THP1 cell line before and after 5-aza-dC treatment. A: GPX3 relative expression levels. B: Electrophoresis results of RQ-PCR and RQ-MSP products. 1: Gene RulerTM 100 bp DNA ladder; 2: 0 μM; 3: 0.1 μM; 4: 1 μM; 5: 10 μM; 6: 50 μM. a: GPX3 expression; b: GPX3 methylation; c: GPX3 unmethylation. C: GPX3 methylation density before treatment. D: GPX3 methylation density after treatment (50 μM).
Discussion
Alterations in DNA methylation are frequent, early events in carcinogenesis [28]. Hypermethylation of TSGs in promoter-associated CpG islands is correlated with gene silencing, whereas hypomethylation in other regions is associated with genomic instability [29]. Moreover, DNA methylation of various TSGs has been identified as potential biomarkers for early detection, diagnosis, prognosis, therapeutic stratification, and post-therapeutic monitoring in a host of cancers [28]. GPX3 is one of these TSGs having been identified. Li et al demonstrated that GPX3 promoter methylation could serve as the potential biomarker for the early diagnosis in esophageal squamous cell carcinoma [30]. Peng et al disclosed the association of GPX3 promoter methylation with lymph node metastasis in gastric carcinomas [17]. Chen et al revealed that GPX3 promoter hypermethylation was associated with head and neck cancer (HNC) chemoresistance and acted as a potentially prognostic indicator for HNC patients treated with cisplatin-based chemotherapy [18]. Furthermore, Kaiser et al also indicated the prognostic significance of GPX3 promoter methylation in multiple myeloma [19].
In the current study, we investigated the status of GPX3 promoter methylation and indicated that GPX3 promoter hypermethylation was a frequent event in de novo AML patients. Although we did not observe the adverse impact of GPX3 methylation on CR in AML patients, our study by both Kaplan-Meier and multivariate analyses revealed the negatively prognostic value of GPX3 methylation among non-M3 AML patients. To the best of our knowledge, our investigation for the first time reported that GPX3 promoter methylation could provide helpful prognostic information in de novo AML patients. Recently, several gene mutations including IDH1/2, TET2, JAK2-V617F, and PML have been identified to be contributed to epigenetic modifications in myeloid malignancies [31]. However, our study did not observe the significant association between GPX3 promoter methylation and IDH1/2 gene mutations. Interestingly, we observed the significantly increased incidence of C/EBPA wild type and FLT3-ITD mutation in the methylated AML patients. Further studies are required to determine the underlying role of C/EBPA and FLT3-ITD mutations during the process of leukemogenesis caused by GPX3 promoter methylation.
Accumulating studies have revealed the association between GPX3 expression and its promoter methylation in a variety of cancers [13-19]. Moreover, GPX3 expression could be up-regulated after the treatment with 5-aza-dC in different cancer cell lines including human esophageal squamous cell carcinoma cell lines, cervical cancer cell lines, gastric carcinoma cell lines, and multiple myeloma cell lines [15,17,19,32]. Our investigation further confirmed the epigenetic mechanism in the regulation of GPX3 expression in leukemic cell line THP1. However, our study did not observe the significant association between GPX3 expression and its promoter methylation in the AML patients. These results suggested that other mechanism might be involved in the regulation of GPX3 expression in de novo AML patients. Further studies are needed to explore the underlying mechanism regulating GPX3 expression in de novo AML patients.
Taken together, our study indicates that GPX3 methylation is associated with C/EBPA wild type and FLT3-ITD mutation in de novo AML patients. In spite of the correlation, GPX3 methylation acts as an independent prognostic biomarker in non-M3 AML patients. Moreover, GPX3 expression is regulated by its promoter methylation in leukemic cell line THP1.
Acknowledgements
This study was supported by National Natural Science foundation of China (81270630, 81172592), Science and Technology Special Project in Clinical Medicine of Jiangsu Province (BL2012056), 333 Project of Jiangsu Province (BRA2013136), Science and Technology Infrastructure Program of Zhenjiang (SS2012003), Medical Key Talent Project of Zhenjiang, Social Development Foundation of Zhenjiang (SH2013042, SH2013082, SH2014044, SH2014086), and Jiangsu Government Scholarship for Overseas Studies.
Disclosure of conflict of interest
None.
References
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Smith M, Barnett M, Bassan R, Gatta G, Tondini C, Kern W. Adult acute myeloid leukaemia. Crit Rev Oncol Hematol. 2004;50:197–222. doi: 10.1016/j.critrevonc.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:497–529. doi: 10.1053/beha.2001.0152. [DOI] [PubMed] [Google Scholar]
- 5.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 6.Deneberg S. Epigenetics in myeloid malignancies. Methods Mol Biol. 2012;863:119–137. doi: 10.1007/978-1-61779-612-8_7. [DOI] [PubMed] [Google Scholar]
- 7.Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH, Kohlschmidt J, Mrozek K, Wu YZ, Bucci D, Curfman JP, Whitman SP, Eisfeld AK, Mendler JH, Schwind S, Becker H, Bar C, Carroll AJ, Baer MR, Wetzler M, Carter TH, Powell BL, Kolitz JE, Byrd JC, Plass C, Garzon R, Caligiuri MA, Stone RM, Volinia S, Bundschuh R, Bloomfield CD. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J. Clin. Oncol. 2014;32:548–556. doi: 10.1200/JCO.2013.50.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brigelius-Flohe R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830:3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Yu YP, Yu G, Tseng G, Cieply K, Nelson J, Defrances M, Zarnegar R, Michalopoulos G, Luo JH. Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 2007;67:8043–8050. doi: 10.1158/0008-5472.CAN-07-0648. [DOI] [PubMed] [Google Scholar]
- 10.Barrett CW, Ning W, Chen X, Smith JJ, Washington MK, Hill KE, Coburn LA, Peek RM, Chaturvedi R, Wilson KT, Burk RF, Williams CS. Tumor suppressor function of the plasma glutathione peroxidase gpx3 in colitis-associated carcinoma. Cancer Res. 2013;73:1245–1255. doi: 10.1158/0008-5472.CAN-12-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi X, Ng KT, Lian QZ, Liu XB, Li CX, Geng W, Ling CC, Ma YY, Yeung WH, Tu WW, Fan ST, Lo CM, Man K. Clinical significance and therapeutic value of glutathione peroxidase 3 (GPx3) in hepatocellular carcinoma. Oncotarget. 2014;5:11103–11120. doi: 10.18632/oncotarget.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohamed MM, Sabet S, Peng DF, Nouh MA, El-Shinawi M. Promoter hypermethylation and suppression of glutathione peroxidase 3 are associated with inflammatory breast carcinogenesis. Oxid Med Cell Longev. 2014;2014:787195. doi: 10.1155/2014/787195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min SY, Kim HS, Jung EJ, Jung EJ, Jee CD, Kim WH. Prognostic significance of glutathione peroxidase 1 (GPX1) down-regulation and correlation with aberrant promoter methylation in human gastric cancer. Anticancer Res. 2012;32:3169–3175. [PubMed] [Google Scholar]
- 14.Falck E, Karlsson S, Carlsson J, Helenius G, Karlsson M, Klinga-Levan K. Loss of glutathione peroxidase 3 expression is correlated with epigenetic mechanisms in endometrial adenocarcinoma. Cancer Cell Int. 2010;10:46. doi: 10.1186/1475-2867-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Wang Y, Li P, Zhu S, Wang J, Zhang S. Identification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinoma. Dig Dis Sci. 2011;56:681–688. doi: 10.1007/s10620-010-1369-0. [DOI] [PubMed] [Google Scholar]
- 16.Lee OJ, Schneider-Stock R, McChesney PA, Kuester D, Roessner A, Vieth M, Moskaluk CA, El-Rifai W. Hypermethylation and loss of expression of glutathione peroxidase-3 in Barrett’s tumorigenesis. Neoplasia. 2005;7:854–861. doi: 10.1593/neo.05328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng DF, Hu TL, Schneider BG, Chen Z, Xu ZK, El-Rifai W. Silencing of glutathione peroxidase 3 through DNA hypermethylation is associated with lymph node metastasis in gastric carcinomas. PLoS One. 2012;7:e46214. doi: 10.1371/journal.pone.0046214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Rao X, House MG, Nephew KP, Cullen KJ, Guo Z. GPx3 promoter hypermethylation is a frequent event in human cancer and is associated with tumorigenesis and chemotherapy response. Cancer Lett. 2011;309:37–45. doi: 10.1016/j.canlet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser MF, Johnson DC, Wu P, Walker BA, Brioli A, Mirabella F, Wardell CP, Melchor L, Davies FE, Morgan GJ. Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma. Blood. 2013;122:219–226. doi: 10.1182/blood-2013-03-487884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swerdlow SH CE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. France: IARC Press; 2008. [Google Scholar]
- 21.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Lin J, Yang J, Qian J, Qian W, Yao DM, Deng ZQ, Liu Q, Chen XX, Xie D, An C, Tang CY. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leuk Res. 2013;37:1642–1647. doi: 10.1016/j.leukres.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 23.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6:e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91:519–525. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Qian J, Sun A, Lin J, Xiao G, Yin J, Chen S, Wu D. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46:579–583. doi: 10.1016/j.clinbiochem.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 26.Qian J, Yao DM, Lin J, Qian W, Wang CZ, Chai HY, Yang J, Li Y, Deng ZQ, Ma JC, Chen XX. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2012;7:e45760. doi: 10.1371/journal.pone.0045760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen XM, Lin J, Yang J, Yao DM, Deng ZQ, Tang CY, Xiao GF, Yang L, Ma JC, Hu JB, Qian W, Qian J. Double CEBPA mutations are prognostically favorable in non-M3 acute myeloid leukemia patients with wild-type NPM1 and FLT3-ITD. Int J Clin Exp Pathol. 2014;7:6832–6840. [PMC free article] [PubMed] [Google Scholar]
- 28.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 29.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Zhou F, Jiang C, Wang Y, Lu Y, Yang F, Wang N, Yang H, Zheng Y, Zhang J. Identification of a DNA methylome profile of esophageal squamous cell carcinoma and potential plasma epigenetic biomarkers for early diagnosis. PLoS One. 2014;9:e103162. doi: 10.1371/journal.pone.0103162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Zheng Z, Yingji S, Kim H, Jin R, Renshu L, Lee DY, Roh MR, Yang S. Downregulation of glutathione peroxidase 3 is associated with lymph node metastasis and prognosis in cervical cancer. Oncol Rep. 2014;31:2587–2592. doi: 10.3892/or.2014.3152. [DOI] [PubMed] [Google Scholar]


