Abstract
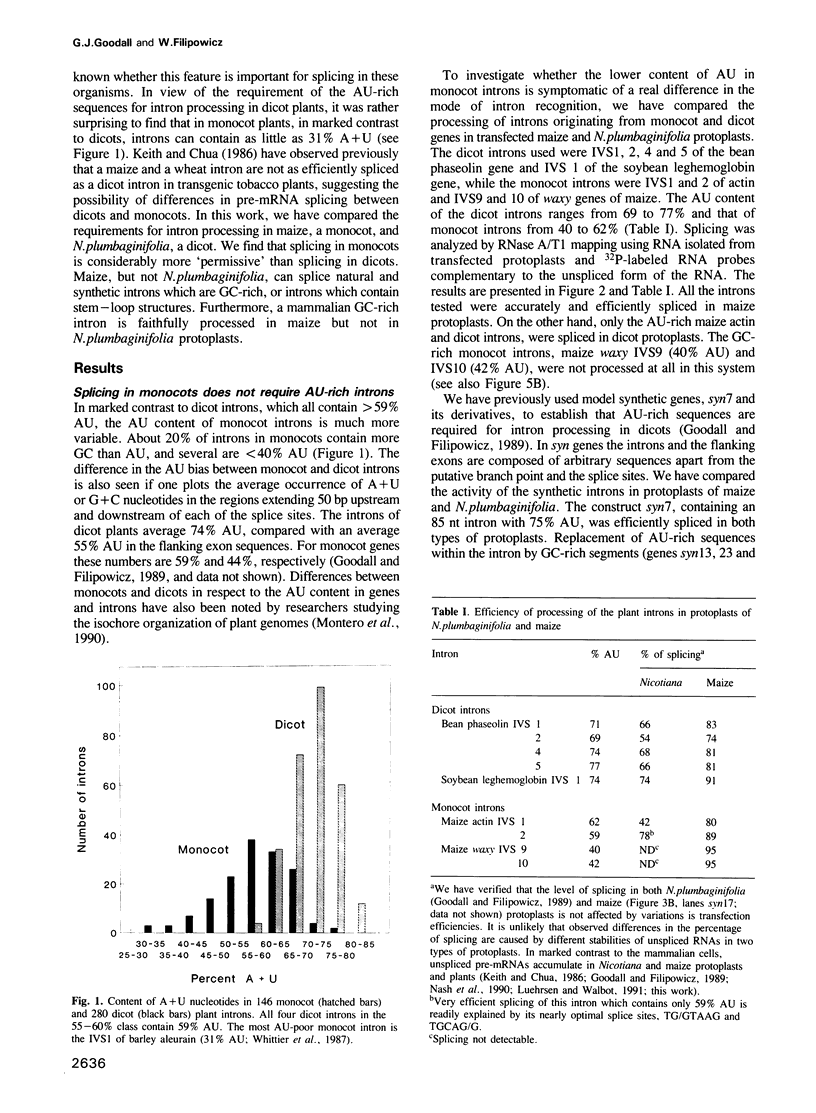
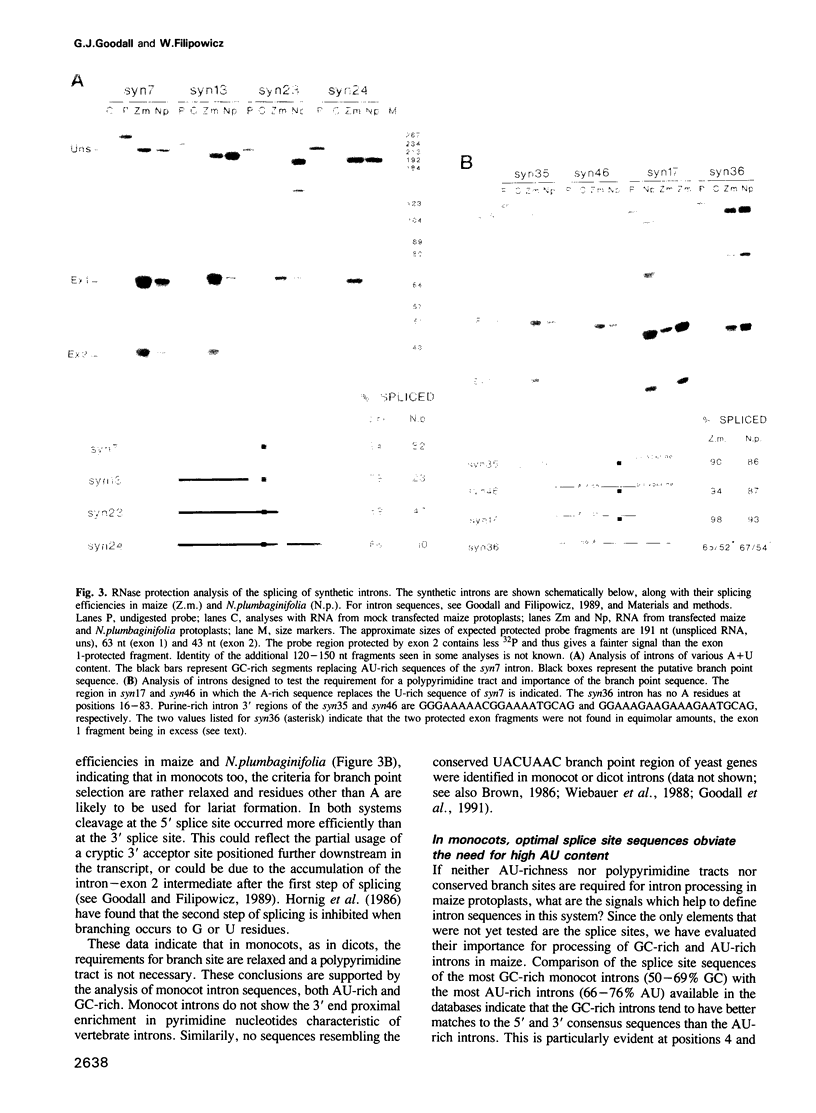
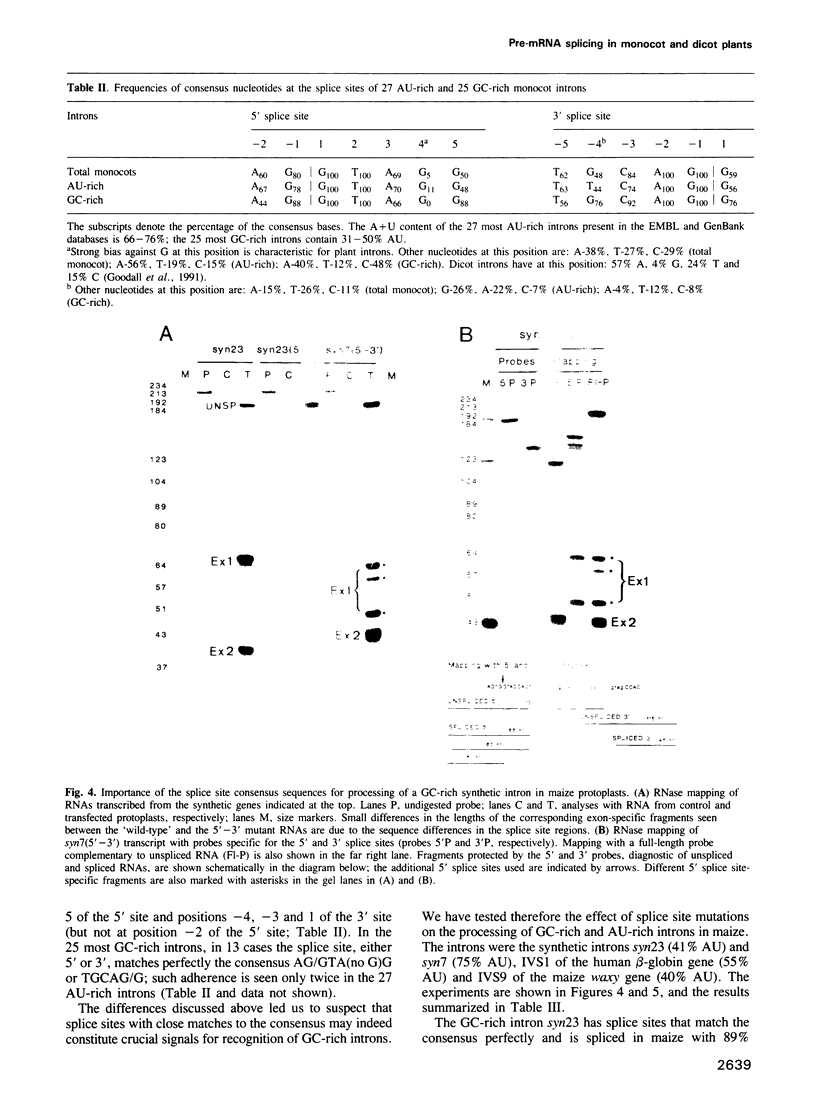
We have found previously that the sequences important for recognition of pre-mRNA introns in dicot plants differ from those in the introns of vertebrates and yeast. Neither a conserved branch point nor a polypyrimidine tract, found in yeast and vertebrate introns respectively, are required. Instead, AU-rich sequences, a characteristic feature of dicot plant introns, are essential. Here we show that splicing in protoplasts of maize, a monocot, differs significantly from splicing in a dicot, Nicotiana plumbaginifolia. As in the case of dicots, a conserved branch point and a polypyrimidine tract are not required for intron processing in maize. However, unlike in dicots, AU-rich sequences are not essential, although their presence facilitates splicing if the splice site sequences are not optimal. The lack of an absolute requirement for AU-rich stretches in monocot introns in reflected in the occurrence of GC-rich introns in monocots but not in dicots. We also show that maize protoplasts are able to process a mammalian intron and short introns containing stem--loops, neither of which are spliced in N.plumbaginifolia protoplasts. The ability of maize, but not of N.plumbaginifolia to process stem--loop-containing or GC-rich introns suggests that one of the functions of AU-rich sequences during splicing of dicot plant pre-mRNAs may be to minimize secondary structure within the intron.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Brisson N., Verma D. P. Soybean leghemoglobin gene family: normal, pseudo, and truncated genes. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4055–4059. doi: 10.1073/pnas.79.13.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W. A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res. 1986 Dec 22;14(24):9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Feix G., Frendewey D. Accurate in vitro splicing of two pre-mRNA plant introns in a HeLa cell nuclear extract. EMBO J. 1986 Nov;5(11):2749–2758. doi: 10.1002/j.1460-2075.1986.tb04563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C., Taylor F. M., Martindale D. W. Nuclear pre-mRNA introns: analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res. 1990 Sep 11;18(17):5133–5141. doi: 10.1093/nar/18.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol Biol. 1990 May;14(5):727–733. doi: 10.1007/BF00016505. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Wiebauer K., Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. In vitro processing of a plant pre-mRNA in a HeLa cell nuclear extract. Nucleic Acids Res. 1986 Oct 10;14(19):7513–7528. doi: 10.1093/nar/14.19.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988 May;8(5):2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Keith B., Chua N. H. Monocot and dicot pre-mRNAs are processed with different efficiencies in transgenic tobacco. EMBO J. 1986 Oct;5(10):2419–2425. doi: 10.1002/j.1460-2075.1986.tb04516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Walbot V. Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol Gen Genet. 1991 Jan;225(1):81–93. doi: 10.1007/BF00282645. [DOI] [PubMed] [Google Scholar]
- Masson P., Rutherford G., Banks J. A., Fedoroff N. Essential large transcripts of the maize Spm transposable element are generated by alternative splicing. Cell. 1989 Aug 25;58(4):755–765. doi: 10.1016/0092-8674(89)90109-8. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. Splicing stories and poly(A) tales: an update on RNA processing and transport. Curr Opin Cell Biol. 1990 Jun;2(3):528–538. doi: 10.1016/0955-0674(90)90138-5. [DOI] [PubMed] [Google Scholar]
- Montero L. M., Salinas J., Matassi G., Bernardi G. Gene distribution and isochore organization in the nuclear genome of plants. Nucleic Acids Res. 1990 Apr 11;18(7):1859–1867. doi: 10.1093/nar/18.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash J., Luehrsen K. R., Walbot V. Bronze-2 gene of maize: reconstruction of a wild-type allele and analysis of transcription and splicing. Plant Cell. 1990 Nov;2(11):1039–1049. doi: 10.1105/tpc.2.11.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Pautot V., Brzezinski R., Tepfer M. Expression of a mouse metallothionein gene in transgenic plant tissues. Gene. 1989 Apr 15;77(1):133–140. doi: 10.1016/0378-1119(89)90367-3. [DOI] [PubMed] [Google Scholar]
- Pereira A., Saedler H. Transpositional behavior of the maize En/Spm element in transgenic tobacco. EMBO J. 1989 May;8(5):1315–1321. doi: 10.1002/j.1460-2075.1989.tb03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Shah D. M., Hightower R. C., Meagher R. B. Genes encoding actin in higher plants: intron positions are highly conserved but the coding sequences are not. J Mol Appl Genet. 1983;2(1):111–126. [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Sun S. M., Hall T. C. Complete nucleotide sequence of a French bean storage protein gene: Phaseolin. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1897–1901. doi: 10.1073/pnas.80.7.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Waibel F., Filipowicz W. U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res. 1990 Jun 25;18(12):3451–3458. doi: 10.1093/nar/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. RNA splicing. Alive with DEAD proteins. Nature. 1991 Feb 7;349(6309):463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- Whittier R. F., Dean D. A., Rogers J. C. Nucleotide sequence analysis of alpha-amylase and thiol protease genes that are hormonally regulated in barley aleurone cells. Nucleic Acids Res. 1987 Mar 25;15(6):2515–2535. doi: 10.1093/nar/15.6.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K., Herrero J. J., Filipowicz W. Nuclear pre-mRNA processing in plants: distinct modes of 3'-splice-site selection in plants and animals. Mol Cell Biol. 1988 May;8(5):2042–2051. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu T., Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science. 1989 Jun 16;244(4910):1346–1348. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991 Jan;10(1):207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- van Santen V. L., Spritz R. A. Splicing of plant pre-mRNAs in animal systems and vice versa. Gene. 1987;56(2-3):253–265. doi: 10.1016/0378-1119(87)90142-9. [DOI] [PubMed] [Google Scholar]








