Abstract
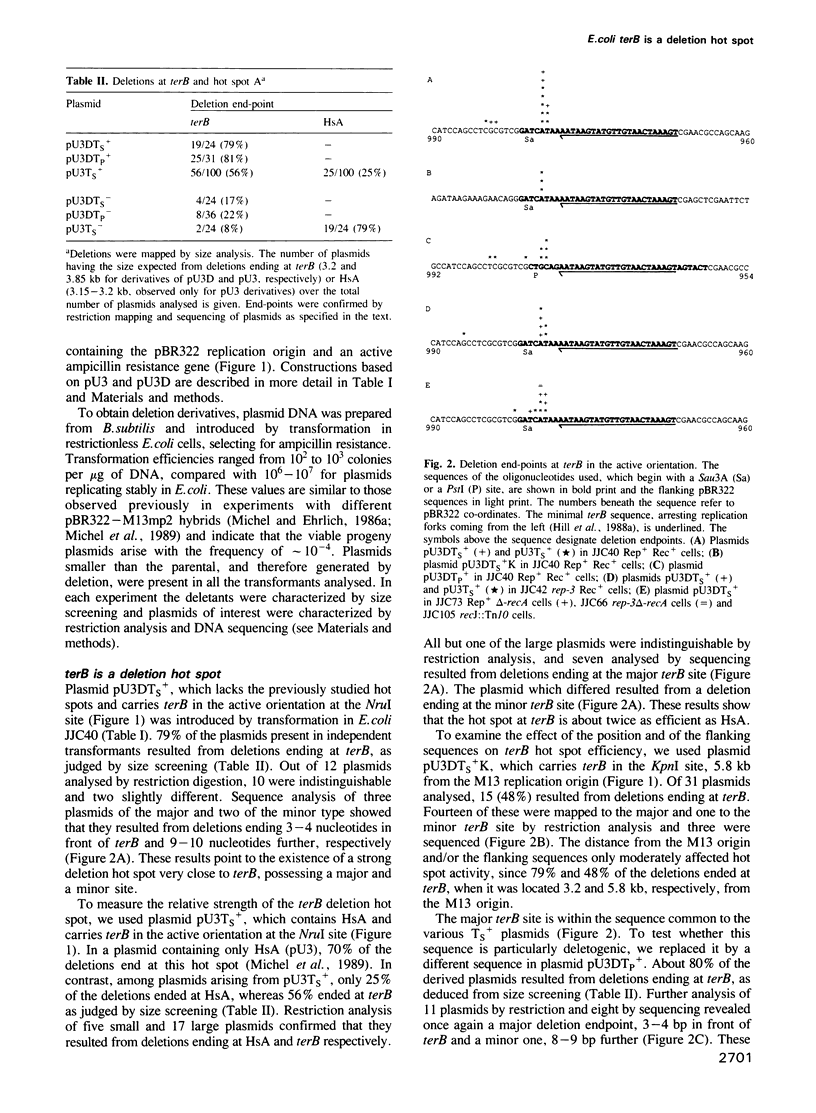
Hybrids composed of phage M13, plasmid pBR322 and the termination signal of Escherichia coli chromosome replication terB were used to show that arrest of DNA synthesis creates a very efficient deletion hot spot. Up to 80% of deletions occurring in these hybrids had one deletion end-point at terB provided that (i) terB was oriented to arrest M13 and pBR322 leading strand synthesis; and (ii) the host cells contained the Tus protein necessary for arresting DNA synthesis at terB. The position of terB and the flanking sequences had little effect on deletion hot spot activity. About 90% of the deletions at terB ended 5-6 nucleotides in front of the major replication arrest site. We propose two models to account for deletion formation and speculate that many genome rearrangements may be due to the pausing of DNA replication.
Full text
PDF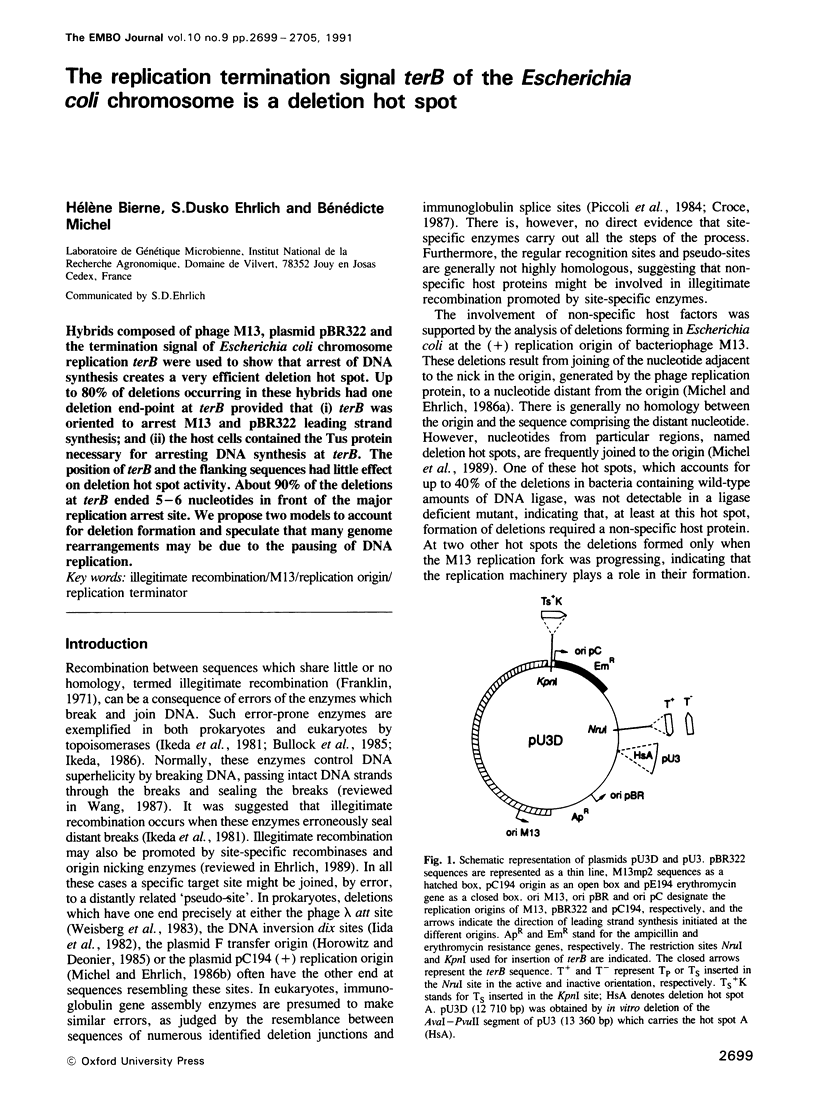





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Boyer H. W. Tetracycline resistance determined by pBR322 is mediated by one polypeptide. Gene. 1983 Dec;26(2-3):197–203. doi: 10.1016/0378-1119(83)90190-7. [DOI] [PubMed] [Google Scholar]
- Bastia D., Germino J., Crosa J. H., Ram J. The nucleotide sequence surrounding the replication terminus of R6K. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2095–2099. doi: 10.1073/pnas.78.4.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Carrigan C. M., Haarsma J. A., Smith M. T., Wake R. G. Sequence features of the replication terminus of the Bacillus subtilis chromosome. Nucleic Acids Res. 1987 Oct 26;15(20):8501–8509. doi: 10.1093/nar/15.20.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C. M. Role of chromosome translocations in human neoplasia. Cell. 1987 Apr 24;49(2):155–156. doi: 10.1016/0092-8674(87)90552-6. [DOI] [PubMed] [Google Scholar]
- Dagert M., Jones I., Goze A., Romac S., Niaudet B., Ehrlich S. D. Replication functions of pC194 are necessary for efficient plasmid transduction by M13 phage. EMBO J. 1984 Jan;3(1):81–86. doi: 10.1002/j.1460-2075.1984.tb01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S., Masukata H., Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987 Dec 24;51(6):1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. The functional origin of bacteriophage f1 DNA replication. Its signals and domains. J Mol Biol. 1984 Feb 5;172(4):507–521. doi: 10.1016/s0022-2836(84)80020-0. [DOI] [PubMed] [Google Scholar]
- Hill T. M., Kopp B. J., Kuempel P. L. Termination of DNA replication in Escherichia coli requires a trans-acting factor. J Bacteriol. 1988 Feb;170(2):662–668. doi: 10.1128/jb.170.2.662-668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. M., Marians K. J. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2481–2485. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. M., Pelletier A. J., Tecklenburg M. L., Kuempel P. L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988 Nov 4;55(3):459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Hidaka M. Core sequence of two separable terminus sites of the R6K plasmid that exhibit polar inhibition of replication is a 20 bp inverted repeat. Cell. 1988 Aug 12;54(4):515–523. doi: 10.1016/0092-8674(88)90073-6. [DOI] [PubMed] [Google Scholar]
- Ikeda H. Bacteriophage T4 DNA topoisomerase mediates illegitimate recombination in vitro. Proc Natl Acad Sci U S A. 1986 Feb;83(4):922–926. doi: 10.1073/pnas.83.4.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Moriya K., Matsumoto T. In vitro study of illegitimate recombination: involvement of DNA gyrase. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):399–408. doi: 10.1101/sqb.1981.045.01.054. [DOI] [PubMed] [Google Scholar]
- James A. A., Morrison P. T., Kolodner R. Genetic recombination of bacterial plasmid DNA. Analysis of the effect of recombination-deficient mutations on plasmid recombination. J Mol Biol. 1982 Sep 25;160(3):411–430. doi: 10.1016/0022-2836(82)90305-9. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Fishel R. A., Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985 Sep;163(3):1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaDuca R. J., Fay P. J., Chuang C., McHenry C. S., Bambara R. A. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry. 1983 Oct 25;22(22):5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- Lee E. H., Kornberg A., Hidaka M., Kobayashi T., Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., D'Alençon E., Ehrlich S. D. Deletion hot spots in chimeric Escherichia coli plasmids. J Bacteriol. 1989 Apr;171(4):1846–1853. doi: 10.1128/jb.171.4.1846-1853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Escherichia coli topoisomerase I can segregate replicating pBR322 daughter DNA molecules in vitro. J Biol Chem. 1986 Sep 5;261(25):11906–11917. [PubMed] [Google Scholar]
- Piccoli S. P., Caimi P. G., Cole M. D. A conserved sequence at c-myc oncogene chromosomal translocation breakpoints in plasmacytomas. 1984 Jul 26-Aug 1Nature. 310(5975):327–330. doi: 10.1038/310327a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J Mol Biol. 1985 Dec 5;186(3):515–525. doi: 10.1016/0022-2836(85)90126-3. [DOI] [PubMed] [Google Scholar]
- Weisberg R. A., Enquist L. W., Foeller C., Landy A. Role for DNA homology in site-specific recombination. The isolation and characterization of a site affinity mutant of coliphage lambda. J Mol Biol. 1983 Oct 25;170(2):319–342. doi: 10.1016/s0022-2836(83)80151-x. [DOI] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]



