Abstract
Castration resistant prostate cancer (CRPC) remains dependent on androgen receptor (AR) signaling. Alternative splicing of the AR to generate constitutively active, ligand-independent variants is one of the principal mechanisms that promote the development of resistance to next-generation anti-androgens such as enzalutamide. Here, we demonstrate that the splicing factor heterogeneous nuclear RNA-binding protein A1 (hnRNPA1) plays a pivotal role in the generation of AR splice variants such as AR-V7. HnRNPA1 is overexpressed in prostate tumors compared to benign prostates and its expression is regulated by NF-kappaB2/p52 and c-Myc. CRPC cells resistant to enzalutamide exhibit higher levels of NF-kappaB2/p52, c-Myc, hnRNPA1, and AR-V7. Levels of hnRNPA1 and of AR-V7 are positively correlated with each other in PCa. The regulatory circuit involving NF-kappaB2/p52, c-Myc and hnRNPA1 plays a central role in the generation of AR splice variants. Downregulation of hnRNPA1 and consequently of AR-V7 resensitizes enzalutamide-resistant cells to enzalutamide, indicating that enhanced expression of hnRNPA1 may confer resistance to AR-targeted therapies by promoting the generation of splice variants. These findings may provide a rationale for co-targeting these pathways to achieve better efficacy through AR blockade.
Keywords: hnRNPA1, AR-V7, c-Myc, Enzalutamide, Androgen Receptor
Introduction
Prostate cancer (PCa) remains the 2nd most lethal disease for males in western countries. The development of abiraterone and enzalutamide marked the continuing success of androgen deprivation therapy (ADT) practiced for over 70 years, reinforcing the concept that androgen receptor (AR) is the key factor for metastatic castration-resistant PCa (CRPC) progression and lethality. However, like earlier ADT, these new therapies have a short efficacy due to primary or acquired resistance. A major form of ADT-resistance in PCa is the generation of AR splicing variants that lack the ligand-binding-domain, thus evading binding of anti-androgens such as Bicalutamide and Enzalutamide. Several reports have documented the expression of alternatively spliced AR-Vs lacking the C-terminal ligand binding domain in PCa cells, which are constitutively nuclear and active even in the absence of androgens, thus indicating their potential role in the acquisition of the CRPC phenotype. Expression of these variants arises from the inclusion of cryptic exons located in intron 2 and 3 of the AR gene, which inserts premature stop codons and termination sites, yielding shorter AR proteins of 75–80 kDa lacking the androgen-binding domain (1, 2). Truncated AR-Vs such as AR-V7 (AR3) and ARv567es can function independently of full-length AR and their selective knockdown can suppress androgen-independent growth of CRPC cells. Alternatively, AR-Vs may play important roles in activating the full length AR in a ligand-independent manner (3). AR-Vs confer resistance to not only AR targeted therapies (4, 5) but to conventional chemotherapeutics such as taxanes used as first line therapies against CRPC (6). These splice variants are rapidly induced after androgen deprivation and are suppressed after restoration of androgen supply (7). The mechanisms mediating increased expression of aberrant AR-Vs in PCa are still largely unknown. One possible cause of defective splicing is the genomic rearrangement and/or intragenic deletions of the AR locus in CRPC (8). Alternatively, aberrant expression of specific splicing factors in PCa cells may also contribute to unbalanced splicing and aberrant recognition of cryptic exons in the AR gene. Understanding the molecular mechanism of AR-Vs production will facilitate the design of mechanism-based inhibitors, extending the efficacy of current ADT, and possibly treating progression of CRPC and prolonging patient survival.
The importance of alternative messenger RNA splicing in regulatory circuits is underscored by the fact that >90% of human genes encode transcripts that undergo at least one alternative splicing event with a frequency higher than 10% (9, 10). Alternative splicing plays important roles in development, physiology, and disease and is often disturbed in inflammatory disorders and cancers (11, 12). Alternative splicing modulates the generation of protein isoforms with distinct structural and functional properties or affects mRNA stability, by the insertion of premature stop codons, and translatability, by altering microRNA target sites (13). Two nuclear RNA-binding protein families, heterogeneous nuclear ribonucleoproteins (hnRNP) and serine/arginine-rich proteins (SR), play pivotal roles in regulation of alternative splicing. The hnRNP family consists of ~20 members which bind to splicing silencers located in exons or introns to promote exon exclusion and act as splicing repressors (13). The best characterized proteins of this group are hnRNPA1 and hnRNPA2, which share a high degree of sequence and functional homology (14). HnRNPA1 and hnRNPA2 are over-expressed in various kinds of tumors and serve as early tumor biomarkers (15–17). The SR family includes >20 members which bind to splicing enhancers and predominantly function to counterbalance the activity of hnRNP proteins (18). Splicing factor 2/alternative splicing factor (SF2/ASF), the best characterized member of the SR family, is up-regulated in multiple human cancers, including lung and cervical cancers, and plays important roles in the establishment and maintenance of cellular transformation (19). During tumor progression, stimuli from the tumor microenvironment may affect the expression and/or activity of splicing regulatory factors, thus perturbing the physiological splicing program of genes involved in cellular processes. An increasing body of evidence indicates that splicing variants of many cancer-related genes can directly contribute to the oncogenic phenotype and to the acquisition of resistance to therapeutic treatments (11, 12, 20). Hence, understanding the functional role(s) of cancer-associated alternative splicing variants and the mechanisms underlying their production offers the potential to develop novel diagnostic, prognostic and more specific anticancer therapies.
In the present study, we investigated the mechanisms involved in aberrant splicing of AR transcripts in a constitutively occurring setting as well as in response to chronic treatment with enzalutamide. Our results show that the splicing factor hnRNPA1 plays a major role in generation of AR-Vs. We also demonstrate that enhanced expression of hnRNPA1 may be mediated by c-Myc and NF-kappaB2/p52, thus paving the way for increase in transcript numbers of constitutively active splice variants and contributing to CRPC therapy resistance.
Materials and Methods
Cell lines and reagents
LNCaP, CWR22Rv1 and VCaP cells were obtained in 2001 from the American Type Culture Collection (ATCC, Manassas, VA) and were cultured in RPMI containing 10% complete FBS and penicillin/streptomycin. ATCC uses Short Tandem Repeat (STR) profiling for testing and authentication of cell lines. C4-2B cells were kindly provided and authenticated by Dr. Leland Chung, Cedars-Sinai Medical Center, Los Angeles, CA in 2006. All experiments with these cell lines were performed within 6 months of resuscitation after cryopreservation. LNCaP cells stably expressing NF-kappaB2/p52 were generated by stable transfection of LNCaP cells with plasmids expressing NF-kappaB2/p52 as described previously (21) and were not authenticated further. 22Rv1 and C4-2B cells resistant to enzalutamide (22Rv1-Enza-R and C4-2B-Enza-R) were generated by chronic culture of 22Rv1 and C4-2B cells in enzalutamide as described previously (22, 23) and were not authenticated further. Antibodies against NF-kappaB2/p52 (K-27), AR (441; mouse monoclonal), HA, Tubulin, U2AF65 and ASF/SF2 were from Santa Cruz Biotechnologies. Antibodies against splicing factors hnRNPA1 (9H10) and hnRNPA2B1 (DP3B3) were from Sigma-Aldrich and AbCam respectively. Sso Fast™ Eva Green qPCR Supermix was from Bio-Rad. All other reagents were of analytical grade and obtained from local suppliers.
Cell growth assays
Plasmid transfections were performed using Attractene transfection reagent (Qiagen). Oligonucleotide siRNA transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen). Viable cell numbers were determined using a Coulter cell counter (Beckman Coulter).
Western Blot Analysis
Cells were lysed in high salt buffer containing 50 mM Hepes pH 7.9, 250 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM PMSF, 1 mM Na Vanadate, 1 mM NaF and protease inhibitor cocktail (Roche). Total protein was estimated using the Coomassie Protein Assay Reagent (Pierce). Equal amounts of protein were loaded on 10% SDS–PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk in PBST (1x PBS+0.1% Tween-20) and probed with indicated primary antibodies in 1% BSA. The signal was detected by ECL (Millipore) after incubation with the appropriate HRP-conjugated secondary antibodies.
Real-Time quantitative RT-PCR
Total RNAs were extracted using TriZOL reagent (Invitrogen). 1 μg of total RNAs were subjected to digestion with RNase-free RQ1 DNase (Promega) and cDNAs were prepared using ImPromII Reverse Transcriptase (Promega) according to manufacturer’s instructions. The cDNAs were analyzed by real-time reverse transcription-PCR (RT-PCR) using Sso Fast™ Eva Green Supermix (Bio-Rad) as described previously (24). Each reaction was normalized by co-amplification of actin. Triplicates of samples were run on a Bio-Rad CFX-96 real-time cycler.
RNA Immunoprecipitation Assay (RIP)
RIP assays were performed as described earlier (25). RNA-protein complexes were crosslinked using 1% formaldehyde. Nuclear extracts were immunoprecipitated with antibodies against indicated RNA-binding splicing factors. Isotype-matched IgG was used as control. Bound RNAs were purified, reverse transcribed and the levels of indicated transcripts were analyzed by qPCR. Splice sites in the full length AR pre-mRNA were detected using ESRsearch and ESEFinder programs (Suppl. Fig. 1).
Human clinical specimens
Paired benign and tumor prostate tissue extracts were described previously (26). Total RNAs from human clinical specimens used for measurement of splicing factor transcript levels were described previously (27).
Gene Expression Omnibus Analysis
Two separate datasets from NCBI GEO were screened independently for expression levels of hnRNPA1, hnRNPA2, U2AF65 and SF2/ASF. GDS1439 (28) compared specimens of benign prostatic hyperplasia with clinically localized primary PCa and metastatic PCa. GDS2545 (29) compared normal prostate specimens without any pathology, normal prostate adjacent to tumor, primary prostate tumor, and metastatic PCa. Significant differences between groups were determined using Microsoft Excel Tools.
Oncomine Analysis
Data sets generated from four comparisons of normal prostate tissue with prostate carcinoma: Lapointe_prostate (30), Wallace_prostate (31), Singh_prostate (32) and Yu_prostate (33) were analyzed using the differential expression function of Oncomine.
Statistical Analyses
Data are shown as means ± SD. Multiple group comparison was performed by one-way ANOVA followed by the Scheffe procedure for comparison of means. P≤0.05 was considered significant.
Results
HnRNPA1 regulates the expression of AR variants
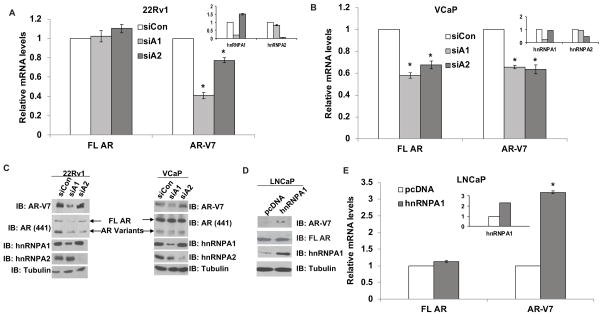
To test whether generation of AR variants by alternative splicing is dependent upon expression of hnRNPs, we analyzed expression levels of full length AR and of variants such as AR-V7, AR-V1, AR-V5, AR-1/2/2b and AR-1/2/3/2b using qRT-PCR in 22Rv1 and VCaP PCa cells transfected with siRNAs against hnRNPA1 and hnRNPA2. Downregulation of hnRNPA1 and hnRNPA2 decreased the expression levels of AR variants in 22Rv1 and VCaP cells (Fig. 1A, 1B & Suppl. Fig. 2A, 2B). Insets in Fig. 1A and B confirm the downregulation of hnRNPA1 and hnRNPA2 by specific siRNAs. The downregulation of hnRNPA1 and the resultant suppression of AR-V7 protein levels were confirmed by Western blot analysis (Fig. 1C). The protein levels of AR-V7 variant were decreased in both 22Rv1 and VCaP cells transfected with hnRNPA1 siRNA. These results indicate that hnRNPA1 may regulate the generation of AR-Vs in PCa cells.
Figure 1.
HnRNPA1 promotes generation of AR splice variants. qPCR to determine the expression levels of AR-V7 and FL AR in A) 22Rv1 and B) VCaP cells transfected with hnRNPA1 or hnRNPA2 siRNAs. C) Western analysis in 22Rv1 and VCaP cells transfected with hnRNPA1 or hnRNPA2 siRNAs. D) Western blotting for AR-V7 in LNCaP cells transfected with hnRNPA1 cDNA. E) qPCR analysis in LNCaP cells transfected with hnRNPA1 cDNA. Results are presented as means ± SD of 3 experiments performed in triplicate. * denotes p≤0.05.
Next, we tested whether overexpression of hnRNPA1 affects the expression levels of AR-Vs. LNCaP cells were transfected with full length hnRNPA1 cDNA and levels of AR-Vs were analyzed by Western blotting and qRT-PCR. Overexpression of hnRNPA1 enhanced AR-V7 protein levels in LNCaP cells which possess undetectable endogenous levels of AR-V7 protein (Fig. 1D). qRT-PCR confirmed that overexpression of hnRNPA1 significantly enhanced the mRNA levels of AR-V7, AR-V5, AR-1/2/2b and AR-1/2/3/2b variants in LNCaP cells (Fig. 1E & Suppl. Fig. 2C). Inset in Fig. 1E confirms the overexpression of hnRNPA1 after transfection in LNCaP cells. These results using downregulation as well as overexpression of hnRNPA1 suggest that hnRNPA1 plays an important role in the generation of AR splice variants.
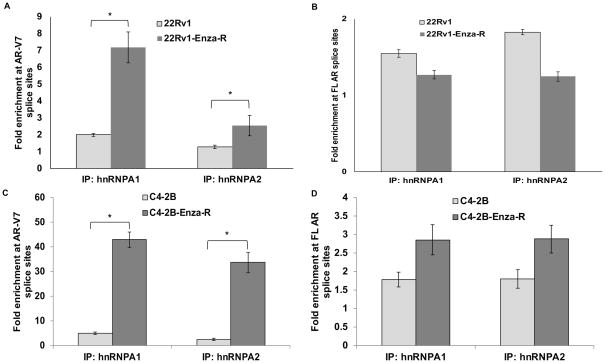
Recruitment of hnRNPA1 to splice sites in AR pre-mRNA is increased in enzalutamide-resistant cells
We found hnRNP binding sites (UAGGGA) in the full length AR mRNA using sequence analysis and ESRSearch program (Suppl. Fig. 1). To determine whether hnRNPA1 is recruited to splice sites in the AR pre-mRNA, we performed RNA Immunoprecipitation (RIP) assays using specific antibodies against hnRNPA1 and hnRNPA2 in 22Rv1 vs. 22Rv1-Enza-R and C4-2B vs. C4-2B-Enza-R cell lines. The 22Rv1-Enza-R and C4-2B-Enza-R cell lines were generated by chronic exposure to enzalutamide and display resistance to enzalutamide (22, 23). The degree of recruitment of hnRNPA1 to AR-V7 splice sites was significantly higher in 22Rv1-Enza-R cells compared to parental 22Rv1 cells (Fig. 2A), indicating that hnRNPA1 may promote generation of AR-V7 in PCa cells resistant to enzalutamide. Even though the recruitment of hnRNPA2 to AR-V7 splice sites was also enhanced in 22Rv1-Enza-R cells compared to 22Rv1 cells, the recruitment of hnRNPA1 was several fold higher than that of hnRNPA2 (Fig. 2A). No significant differences were observed in the recruitment of either hnRNPA1 or hnRNPA2 to FL AR splice sites between 22Rv1 parental and 22Rv1-Enza-R cells (Fig. 2B). We also analyzed recruitment of hnRNPA1 and hnRNPA2 to splice sites for other AR-Vs such as AR-V1, AR-V5, AR-1/2/2b and AR-1/2/3/2b (Suppl. Fig. 3A, B, C & D). Recruitment of hnRNPA1 to AR-1/2/2b splice sites was significantly higher in 22Rv1-Enza-R cells (Suppl. Fig. 3C), indicating that hnRNPA1 may play a selective role in generation of AR splice variants. In addition, recruitment of hnRNPA2 to AR-1/2/3/2b splice sites was significantly enhanced in 22Rv1-Enza-R cells (Suppl. Fig. 3D). These results imply that different splicing factors may function co-operatively to promote generation of AR-Vs in enzalutamide-resistant PCa cells. These results also confirm that hnRNP proteins are physically recruited to splice sites in the AR pre-mRNA with the degree of recruitment increasing in PCa cells resistant to enzalutamide, indicating that hnRNP proteins may drive the generation of AR splice variants leading to enzalutamide resistance.
Figure 2.
Recruitment of hnRNPA1 and hnRNPA2 to splice sites in AR pre-mRNA is enhanced in 22Rv1-Enza-R and C4-2B-Enza-R enzalutamide-resistant PCa cells compared to 22Rv1 and C4-2B parental cells respectively. RIP assays for splice sites of A) AR-V7, B) AR in 22Rv1-Enza-R compared to 22Rv1 and C) AR-V7, D) AR in C4-2B-Enza-R compared to C4-2B. Results are presented as means ± SD of 2 experiments performed in duplicate. * denotes p≤0.05.
To confirm the above results in another enzalutamide-resistant cell line, we analyzed the C4-2B vs. C4-2B-Enza-R cell line pair. Our results showed that recruitment of hnRNPA1 and hnRNPA2 to AR-V7 splice sites was significantly enhanced in C4-2B-Enza-R cells compared to parental C4-2B cells (Fig. 2C), while, similar to 22Rv1-Enza-R cells, recruitment of either hnRNPA1 or hnRNPA2 to FL AR splice sites was not altered significantly in C4-2B-Enza-R cells compared to parental C4-2B cells (Fig. 2D). In all cases, the fold enrichment of hnRNPA1 at splice sites on AR pre-mRNA was much higher compared to hnRNPA2, indicating that hnRNPA1 may play a more central role in promoting the expression of AR-Vs (Suppl. Fig. 3E, F, G & H). These results collectively demonstrate that different splicing factors may play context- and cell type-dependent roles in PCa cells in alternative splicing of the AR.
Expression levels of hnRNPA1 are elevated in PCa tissues
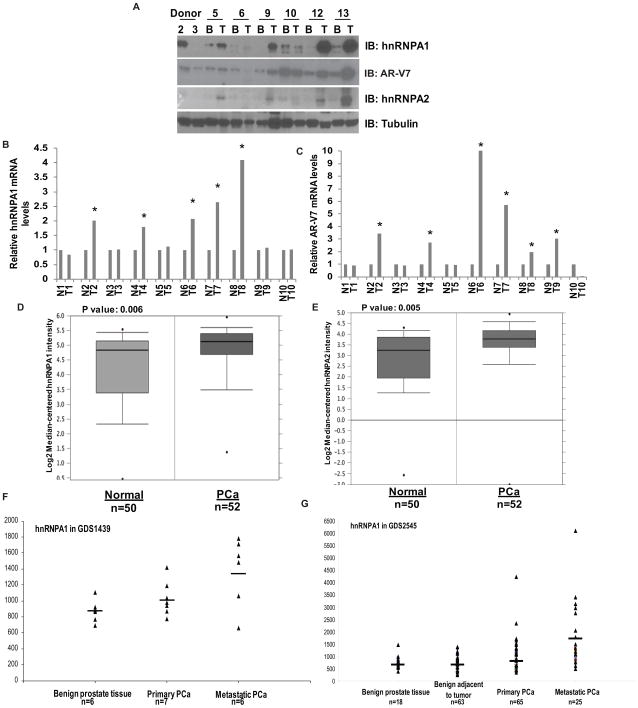
To determine whether increased expression of splicing factors and AR-Vs is associated with PCa, we examined the expression levels of hnRNPA1 and hnRNPA2 by immunoblotting in lysates from 27 archived paired benign and tumor PCa clinical samples. Levels of hnRNPA1 and hnRNPA2 were elevated in ~44% of tumor tissues compared to matched benign tissues (Fig. 3A and Suppl. Fig. 4A). These results were correlated positively with the protein expression levels of AR-V7, which were enhanced in ~ 48% of tumor tissues compared to their benign counterparts (Fig. 3A and Suppl. Fig. 4A). In addition, expression levels of hnRNPA1 and hnRNPA2 were low or undetectable in 9/12 and 6/12 of donor prostates respectively. These observations were also correlated with expression levels of AR-V7 which were low or undetectable in 8/12 donor tissues (Table 1).
Figure 3.
Expression levels of hnRNPA1 and AR-V7 are positively correlated with each other. A) Representative immunoblot for hnRNPA1, hnRNPA2 and AR-V7 in lysates from 27 paired benign and tumor patient samples. qRT-PCR of mRNA levels of (B) hnRNPA1 and (C) AR-V7 in 10 paired normal and tumor clinical prostate samples. Results are presented as means ± SD of 2 experiments performed in triplicate. * denotes p≤0.05. Relative expression levels of hnRNPA1 (D) and hnRNPA2 (E) in the representative Singh_prostate (n=102) dataset from Oncomine. Relative expression levels of hnRNPA1 in GDS1439 dataset (F) and in GDS2545 dataset (G) from GEO.
Table 1.
Summary of the Western analysis of levels of hnRNPA1, hnRNPA2 and AR-V7 in 27 paired benign and tumor prostate clinical samples.
| Gene | T>B | T<B | T=B | High in donor | Low in donor |
|---|---|---|---|---|---|
| hnRNPA1 | 12 (44%) | 8 (29%) | 7 (26%) | 3/12 | 9/12 |
| AR-V7 | 13 (48%) | 7 (25%) | 7 (26%) | 4/12 | 8/12 |
| hnRNPA2 | 12 (44%) | 8 (29%) | 7 (26%) | 6/12 | 6/12 |
We also analyzed the mRNA levels of hnRNPA1, hnRNPA2 and AR-V7 in archival total RNAs extracted from 10 pairs of matched benign and tumor clinical PCa specimens (27, 34). Transcript levels of hnRNPA1 were elevated in 5/10 of tumor tissues compared to matched benign tissues with no appreciable differences between tumor and benign being observed in the other 5/10 of samples (Fig. 3B). Transcript levels of AR-V7 were elevated in 6/10 tumor tissues compared to their matched benign counterparts (Fig. 3C), demonstrating that expression of hnRNPA1 and AR-V7 may be positively correlated with each other in human PCa. No significant differences were observed in mRNA levels of hnRNPA2 between matched tumor and benign prostate tissues (Suppl. Fig. 4B).
To further confirm our findings, we analyzed expression levels of hnRNPA1 and hnRNPA2 in clinical PCa tissues using publicly available datasets from Gene Expression Omnibus (GEO) and Oncomine. Results from an analysis of Oncomine datasets revealed that expression levels of hnRNPA1 and hnRNPA2 are significantly elevated in prostate tumor tissues compared to benign prostates in 17/21 and 15/21 datasets respectively (Suppl. Fig. 4C). Results from a representative dataset, Singh_prostate (n=102) from Oncomine are shown (Fig. 3D & E). An analysis of GEO revealed that expression levels of hnRNPA1 and hnRNPA2 were elevated in primary as well as metastatic PCa compared to benign prostates (Fig. 3F & G and Suppl. Fig. 4D). Data regarding expression levels of AR-Vs were not available in these datasets, but nonetheless, these results indicate that elevated levels of hnRNPA1 may contribute to PCa development and progression. Our findings correlate well with studies showing that expression levels of AR-V7 are elevated in ~40% of CRPC tissues (2, 35), indicating that elevated expression of hnRNPA1 in prostate tumors may contribute to generation of higher levels of AR-Vs.
Expression of hnRNPA1 is regulated by c-Myc
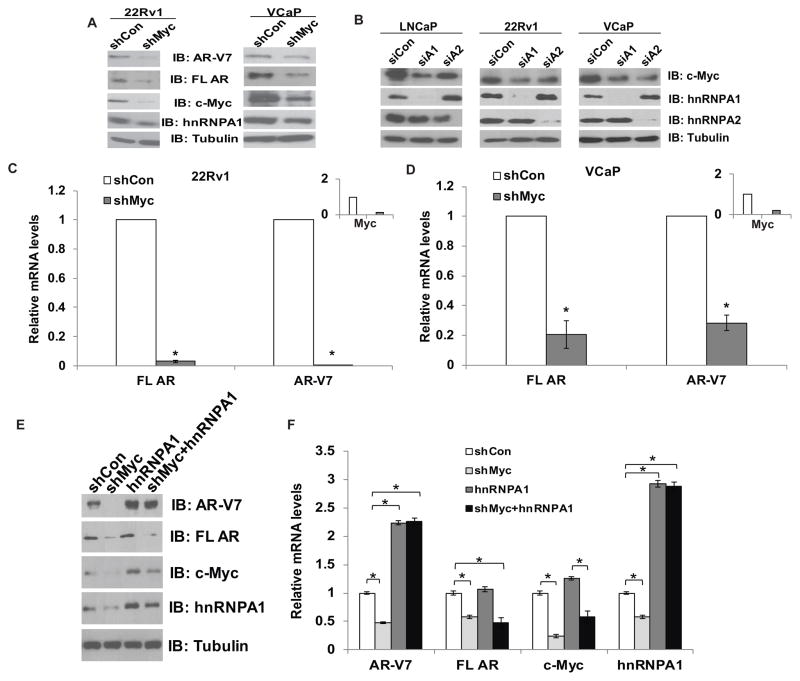
Previous studies indicated that hnRNPA1 and c-Myc exhibit positive reciprocal regulation (36). C-Myc enhances hnRNPA1 expression transcriptionally, while hnRNPA1 regulates c-Myc via alternative splicing. Previous studies also showed that c-Myc is one of the transcription factors which regulate transcription of the AR (37). Hence, we analyzed the status of c-Myc or hnRNPA1 when the expression of either was downregulated in PCa cells. Lysates from 22Rv1 and VCaP cells transfected with shRNA against c-Myc were analyzed using specific antibodies against hnRNPA1. Downregulation of c-Myc reduced protein levels of hnRNPA1 significantly (Fig. 4A). Similarly, lysates from LNCaP, 22Rv1 and VCaP cells transfected with siRNA against hnRNPA1 were examined by immunoblotting using specific antibodies against c-Myc. Downregulation of hnRNPA1 reduced protein levels of c-Myc (Fig. 4B). These results confirm that hnRNPA1 and c-Myc exhibit reciprocal regulation in PCa cells. We also analyzed whether reduction in hnRNPA1 levels by c-Myc shRNA affects expression of AR-Vs in 22Rv1 and VCaP cells. Western blotting and qRT-PCR analyses showed that levels of AR-Vs, including that of AR-V7, were abrogated due to depletion of hnRNPA1 caused by downregulation of c-Myc (Fig. 4A, C & D and Suppl. Fig. 5). These findings support an important role for c-Myc in the generation of AR splice variants.
Figure 4.
Reciprocal regulation between c-Myc and hnRNPA1 is responsible for the generation of AR splice variants. A) Immunoblotting for hnRNPA1 and AR-V7 in 22Rv1 and VCaP cells transfected with c-Myc shRNA. B) Immunoblotting for c-Myc in LNCaP, 22Rv1 and VCaP cells transfected with hnRNPA1 or hnRNPA2 siRNAs. qRT-PCR for mRNA levels of full length AR and AR-V7 in 22Rv1 (C) and VCaP (D) cells transfected with c-Myc shRNA. Insets show the expression of c-Myc mRNA in cells transfected with c-Myc shRNA. (E) Protein and (F) mRNA levels of AR-V7, FL AR, c-Myc and hnRNPA1 in 22Rv1 cells transfected with c-Myc shRNA with or without overexpression of hnRNPA1. Results are presented as means ± SD of 2 experiments performed in triplicate. * denotes p≤0.05.
In order to analyze whether overexpression of hnRNPA1 can overcome the effects of downregulation of Myc, we suppressed expression of c-Myc using shRNA in 22Rv1 cells followed by overexpression of hnRNPA1. The results showed that even though suppression of c-Myc reduced mRNA as well as protein levels of both FL AR and AR-V7, subsequent overexpression of hnRNPA1 restored the expression of AR-V7 fully while having minimal effect on FL AR (Fig. 4E & F). These results indicate that hnRNPA1 primarily regulates generation of alternative splice variants of the AR and not the generation of the FL AR.
NF-kappaB2/p52 regulates AR-V7 expression via hnRNPA1 and c-Myc
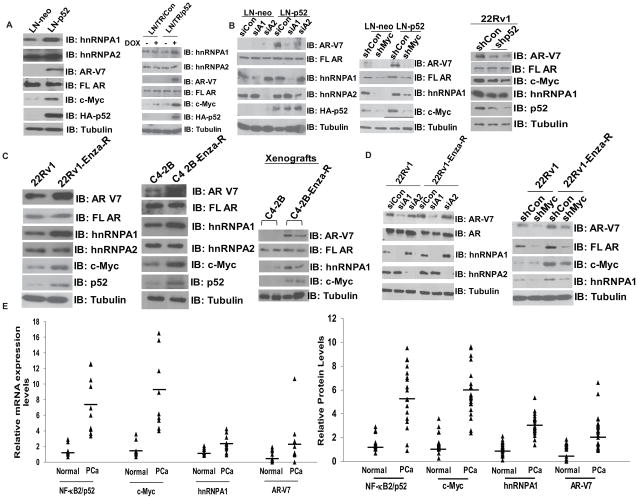
We reported earlier that activation of NF-kappaB2/p52 promotes progression to CRPC and enzalutamide resistance via the generation of AR variants, specifically AR-V7 (21, 23, 24). Our previous findings also indicated that NF-kappaB2/p52 may regulate c-Myc expression. Hence, we examined whether NF-kappaB2/p52 plays a role in the elevated expression of hnRNPA1 and c-Myc in PCa using lysates from LNCaP cells stably expressing p52 (LN-p52). Protein levels of AR-V7, hnRNPA1 and c-Myc were elevated in LN-p52 cells, while no appreciable differences were found in the expression of hnRNPA2 (Fig. 5A, left panel). These results were confirmed using LNCaP cells expressing p52 under the control of a Tet-inducible promoter (LN/TR/p52). Induction of p52 expression led to increases in expression levels of AR-V7, hnRNPA1 and c-Myc (Fig. 5A, right panel), indicating that upregulation of AR-V7 by p52 may be mediated by hnRNPA1 and c-Myc. To examine the relationship between hnRNPA1, c-Myc and AR-V7 in LN-p52 cells, we analyzed levels of AR-V7 by Western blotting in LN-p52 cells transfected with hnRNPA1 or hnRNPA2 siRNAs. Downregulation of hnRNPA1 abrogated the expression of AR-V7 in LN-p52 cells, while downregulation of hnRNPA2 did not have an appreciable effect on AR-V7 protein levels (Fig. 5B, left panel). Protein levels of c-Myc were also downregulated, keeping in line with earlier findings that hnRNPA1 and c-Myc regulate each other (36). In addition, expression of hnRNPA1 and AR-V7 were abolished as a result of downregulation of c-Myc expression in LN-p52 cells transfected with c-Myc shRNA (Fig. 5B, middle panel).
Figure 5.
NF-kappaB2/p52 regulates expression of c-Myc and hnRNPA1. A) Left panel, Immunoblotting for hnRNPA1, c-Myc and AR-V7 in LN-p52 cells. Right panel, Immunoblotting for hnRNPA1, c-Myc and AR-V7 in LN/TR/p52 cells. B) Left panel, Western blotting for AR-V7 in LN-p52 cells transfected with hnRNPA1 siRNA. Middle panel, Immunoblotting for AR-V7 and hnRNPA1 in LN-p52 cells transfected with c-Myc shRNA. Right panel, Immunoblotting for AR-V7, hnRNPA1 and c-Myc in 22Rv1 cells transfected with p52 shRNA. C) Left panel, 22Rv1 cells resistant to enzalutamide (22Rv1-Enza-R) express higher levels of AR-V7, hnRNPA1, c-Myc and NF-kappaB2/p52. Middle panel, C4-2B cells resistant to enzalutamide (C4-2B-Enza-R) express higher levels of hnRNPA1, AR-V7, c-Myc and NF-kappaB2/p52. Right panel, Xenografts from C4-2B-Enza-R cells exhibit higher levels of AR-V7, hnRNPA1 and c-Myc. D) Left panel, Western analysis of AR-V7 in 22Rv1-Enza-R cells transfected with hnRNPA1 siRNA. Right panel, Expression of AR-V7 and hnRNPA1 in 22Rv1-Enza-R cells transfected with c-Myc shRNA. All results are shown as representative images from 2 experiments performed in duplicate. E) Left panel, Chart depicting the positive correlation between mRNA levels of NF-kappaB2/p52, c-Myc, hnRNPA1 and AR-V7 in 10 paired benign and tumor prostate clinical samples. Right panel, Chart depicting the correlation between relative protein levels of NF-kappaB2/p52, c-Myc, hnRNPA1 and AR-V7 in 27 paired benign and tumor prostate clinical samples. Band intensities in immunoblots were quantified using ImageJ software and plotted as arbitrary units. The horizontal lines represent the median of each series.
To confirm these findings in a cell line with constitutive expression of both p52 and AR-V7, we analyzed levels of AR-V7, hnRNPA1 and c-Myc by immunoblotting in 22Rv1 cells transfected with p52 shRNA. Downregulation of p52 in 22Rv1 cells abrogated expression of AR-V7, hnRNPA1 and c-Myc (Fig. 5B, right panel). These results demonstrate that NF-kappaB2/p52 may modulate generation of AR-Vs by regulation of hnRNPA1 and c-Myc.
PCa cells resistant to enzalutamide exhibit higher levels of splicing factors
As our results demonstrate that expression of hnRNPA1 and AR variants may be positively correlated with each other in PCa cells, and AR-V7 expression has been shown to be involved in the acquisition of resistance to enzalutamide (3, 23), we tested the correlation between levels of AR variants and hnRNPA1 in PCa cells that have acquired resistance to enzalutamide. 22Rv1-Enza-R and C4-2B-Enza-R cell lines exhibited higher levels of AR-V7 and hnRNPA1 (Fig. 5C), indicating that expression of hnRNPA1 may be positively correlated with expression of AR-Vs. Furthermore, expression levels of c-Myc and NF-kappaB2/p52 were also elevated in enzalutamide-resistant cells, confirming the importance of the NF-kappaB2/p52:c-Myc:hnRNPA1:AR-V7 axis in enzalutamide resistance. No significant differences were observed in the expression of hnRNPA2 in 22Rv1-Enza-R and C4-2B-Enza-R cells compared to their parental cells (Fig. 5C right & middle panels). To confirm these results in vivo, we analyzed extracts from xenograft tumors derived from C4-2B and C4-2B-Enza-R cells using antibodies against AR-V7 and hnRNPA1. Higher levels of AR-V7 were observed in xenografts derived from C4-2B-Enza-R cells, which was correlated well with higher levels of hnRNPA1 and c-Myc (Fig. 5C right panel), confirming our observations that expression of AR-Vs is positively correlated with expression of hnRNPA1 in PCa cell lines resistant to enzalutamide.
Next, we tested whether downregulation of hnRNPA1 affects endogenous levels of AR-Vs in 22Rv1-Enza-R cells. Transfection of hnRNPA1 siRNA abrogated levels of AR-V7 in 22Rv1-Enza-R cells (Fig. 5D left panel). Similarly, downregulation of c-Myc by specific shRNA reduced expression levels of AR-V7 and hnRNPA1 in 22Rv1 and 22Rv1-Enza-R cells (Fig. 5D right panel), confirming the c-Myc: hnRNPA1: AR-V7 axis in PCa cells.
To confirm the importance of the link between NF-kappaB2/p52, c-Myc, hnRNPA1 and AR-V7 in PCa, we analyzed the correlation between their expression levels at mRNA and protein levels in paired benign and tumor prostate clinical samples from Fig. 3. Transcript (left panel) and protein (right panel) levels of NF-kappaB2/p52, c-Myc, hnRNPA1 and AR-V7 were positively correlated with each other (Fig. 5E), demonstrating that the NF-kappaB2/p52:c-Myc:hnRNPA1:AR-V7 axis plays a vital role in PCa and in the development of castration and therapy resistance.
Suppression of hnRNPA1 resensitizes enzalutamide-resistant PCa cells to enzalutamide
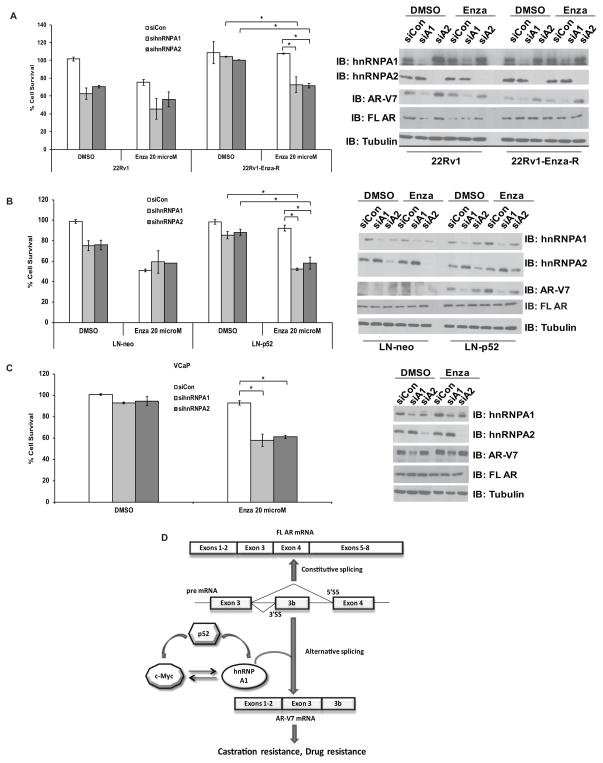
To examine the functional relevance of regulation of AR alternative splicing by hnRNPA1, we tested whether downregulation of hnRNPA1 resulting in decreased levels of AR-V7 resensitizes enzalutamide-resistant cells to enzalutamide. We examined cell survival in 22Rv1 and 22Rv1-Enza-R cells transfected with hnRNPA1 or hnRNPA2 siRNAs and treated with 0 and 20 μM enzalutamide for 24 h. Reduced expression of hnRNPA1 enhanced the sensitivity of enzalutamide-resistant 22Rv1-Enza-R cells to enzalutamide (Fig. 6A), indicating that upregulation of AR-V7 expression by hnRNPA1 may be required to sustain the acquired resistance of 22Rv1-Enza-R cells to enzalutamide. Downregulation of hnRNPA1 and hnRNPA2 also enhanced the sensitivity of LN-neo and LN-p52 cells transfected with hnRNPA1 or hnRNPA2 siRNAs and treated with 20 μM enzalutamide to enzalutamide (Fig. 6B). Suppression of hnRNPA1 expression reduced cell survival by ~40–50% when enzalutamide-resistant LN-p52 cells were treated with enzalutamide. Suppression of hnRNPA1 expression also reduced survival of VCaP cells when treated with enzalutamide (Fig. 6C), confirming the essential nature of AR variants in these cells.
Figure 6.
Suppression of hnRNPA1 restores enzalutamide sensitivity of enzalutamide-resistant PCa cells. A) Left panel, Cell survival in 22Rv1-Enza-R cells transfected with hnRNPA1 or hnRNPA2 siRNAs and treated with vehicle or 20 μM enzalutamide. Cell numbers were counted after 48 h. Right panel, immunoblots confirm the downregulation of hnRNPA1 or hnRNPA2 and of AR-V7. B) Left panel, Cell survival in LN-p52 cells transfected with hnRNPA1 or hnRNPA2 siRNAs and treated with vehicle or 20 μM enzalutamide. Cell numbers were counted after 48 h. Right panel, immunoblots confirm the downregulation of hnRNPA1 or hnRNPA2 and AR-V7. C) Left panel, Cell survival in VCaP cells transfected with hnRNPA1 or hnRNPA2 siRNAs and treated with vehicle or 20 μM enzalutamide. Cell numbers were counted after 48 h. Right panel, immunoblots confirm the downregulation of hnRNPA1 or hnRNPA2 and of AR-V7. Results are presented as means ± SD of 3 experiments performed in triplicate. * denotes p≤0.05. D) Schematic representation of the alternative splicing of AR mRNA regulated by the NF-kappaB2:c-Myc:hnRNPA1 axis.
Our findings collectively demonstrate that hnRNPA1 plays a major role in the generation of AR splice variants. Expression of hnRNPA1 is modulated by NF-kappaB2/p52 via c-Myc (Fig. 6D). Our results point to the enhanced expression of hnRNPA1 in prostate tumors being instrumental in inducing alternative splicing of the precursor AR mRNA.
Discussion
A large number of previous studies have shown that C-terminally truncated AR-Vs are expressed in PCa cells and even in normal prostate epithelial cells (35, 38) and promote CRPC progression under androgen deprivation (39, 40). In addition, enhanced expression of AR-V7 confers resistance to next generation therapeutics such as enzalutamide and abiraterone (4, 5). These studies attest to the importance of AR-Vs in CRPC and co-targeting the mechanisms which contribute to their generation may increase the efficacy of currently used AR-targeted therapies and prolong time to development of resistance. Our findings in this study demonstrate that the splicing factor hnRNPA1 plays a major role in the alternative splicing of AR mRNA.
HnRNPA1 is a multifunctional RNA-binding protein involved in the regulation of RNA biogenesis. HnRNPA1 is under the transcriptional control of the c-Myc proto-oncogene and modulates the splicing of PKM2, activating the metabolic switch to aerobic glycolysis that is a hallmark of cancer cells (11, 36). HnRNPA1 also regulates alternative splicing of genes involved in invasion and metastasis such as Rac1 and Ron (11). Increased expression of hnRNPA1 has been documented in proliferating and transformed cells (20) and in lung, breast, colon, renal cell carcinomas and gliomas (41–46). HnRNPs co-operate with other splicing factors to generate pro-oncogenic and pro-inflammatory molecules in cancers (41, 47). Silencing of hnRNPA1 and A2 promotes apoptosis in human and mouse cancer cell lines, while having no effect on normal epithelial and fibroblastic cell lines (48).
In this study, hnRNPA1 binding sites (UAGGGA) were identified in AR mRNA using sequence analysis and ESRSearch program. Downregulation of hnRNPA1 significantly reduced the expression of AR-Vs such as AR-V7, while not affecting full length AR. HnRNPA1 binding sites were not detected at full length AR splice sites in the AR pre-mRNA. Moreover, the slight decrease seen in either full length AR mRNA or protein levels in VCaP or 22Rv1 cells respectively (Fig. 1) was not observed consistently in all experiments. In consideration of these observations, we concluded that hnRNPA1 does not play a significant role in splicing of full length AR from AR pre-mRNA. RNA-binding assays revealed that hnRNPA1 is recruited to splice sites for AR splice variants. Enhanced expression of hnRNPA1 was observed in PCa tissues compared to their benign counterparts, which was correlated positively with expression of AR-V7. Increased expression of hnRNPA1 was also correlated with higher levels of AR-V7 in PCa cells with acquired resistance to enzalutamide. Exploration of the mechanisms revealed that c-Myc and NF-kappaB2/p52 contribute to the development of therapy resistance in PCa cells by inducing hnRNPA1 expression and thereby ligand-independent AR-Vs. Downregulation of hnRNPA1 resensitized enzalutamide-resistant cells to enzalutamide, indicating that suppression of hnRNPA1 resulting in suppression of AR-Vs reversed the acquired resistance to enzalutamide. These data led us to conclude that hnRNPA1 is the central player in a splicing regulatory circuit involving c-Myc, NF-kappaB2/p52 and AR.
Our study demonstrates that elevated levels of splicing factors such as hnRNPA1 promote expression of alternative splice forms of AR. Relative amounts of splicing factors have been proposed to determine alternative splicing (45). A recent study by Liu et al (49) showed that recruitment, and not expression, of splicing factors SF2/ASF and U2AF65 determines the generation of AR splice variants in enzalutamide resistance. We found higher levels of hnRNPA1 in PCa clinical samples compared to SF2/ASF (data not shown), indicating that higher expression of a splicing factor, and not simply its recruitment to splice sites under certain conditions, may determine the levels of alternative splice forms. These results are supported by previous studies which showed that the relative levels of hnRNPA1 expression increased to a greater extent than those of SF2/ASF in lung tumors (44). Of note, analysis of expression levels of U2AF65 and SF2/ASF in PCa tissues using GEO and Oncomine datasets revealed that expression levels of both splicing factors are elevated in PCa tissues compared to benign counterparts (Suppl. Fig. 6), lending credence to our observations that enhanced expression of splicing factors may be one of the principal mechanisms driving generation of AR-Vs. Elevated levels of hnRNPA1 may conceivably change the splicing milieu of a broad spectrum of proteins in addition to that of the AR, with splicing of CD44 being an example. But splice variants of AR have been demonstrated to play major roles in resistance to enzalutamide, underlining the importance of our results in splicing regulation of the AR. Furthermore, earlier studies indicated that the splice variant AR-1/2/3/2b is also generated by intragenic genomic rearrangement of the AR gene due to duplication of exon 3 (1). Our studies consistently found transcripts corresponding to this splice variant in LNCaP, C4-2B and VCaP cells, albeit at extremely low levels. These observations warrant further exploration of the mechanisms involved in generation of this splice variant and further validation.
In summary, we demonstrated that hnRNPA1, in concert with NF-kappaB2/p52 and c-Myc, regulates the generation of AR-Vs in PCa cells and that the NF-kappaB2/p52:c-Myc:hnRNPA1:AR-V7 axis (Fig. 6D) plays a pivotal role in the development and maintenance of resistance to androgen blockade. These findings may have important implications in targeting AR-Vs and the splicing factors responsible to overcome acquired enzalutamide resistance in PCa.
Supplementary Material
Acknowledgments
Financial Support
This work was supported by grants NIH/NCI CA140468, CA168601, CA179970 (A.C. Gao), DOD PC100502 (N. Nadiminty), and US Department of Veterans Affairs, ORD VA Merits I01 BX000526 (A.C. Gao), and by a Stand Up To Cancer—Prostate Cancer Foundation-Prostate Dream Team Translational Cancer Research Grant SU2C-AACR-PCF DT0812, made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer research.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
References
- 1.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a Novel Androgen Receptor Exon Generates a Constitutively Active Androgen Receptor that Mediates Prostate Cancer Therapy Resistance. Cancer Research. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-Independent Androgen Receptor Variants Derived from Splicing of Cryptic Exons Signify Hormone-Refractory Prostate Cancer. Cancer Research. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–56. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mostaghel EA, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM, et al. Resistance to CYP17A1 Inhibition with Abiraterone in Castration-Resistant Prostate Cancer: Induction of Steroidogenesis and Androgen Receptor Splice Variants. Clinical Cancer Research. 2011;17:5913–25. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KAT, Dehm SM. Androgen Receptor Splice Variants Mediate Enzalutamide Resistance in Castration-Resistant Prostate Cancer Cell Lines. Cancer Research. 2013;73:483–9. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, et al. Androgen Receptor Splice Variants Determine Taxane Sensitivity in Prostate Cancer. Cancer Research. 2014;74:2270–82. doi: 10.1158/0008-5472.CAN-13-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Z, Chen S, Sowalsky AG, Voznesensky OS, Mostaghel EA, Nelson PS, et al. Rapid Induction of Androgen Receptor Splice Variants by Androgen Deprivation in Prostate Cancer. Clinical Cancer Research. 2014;20:1590–600. doi: 10.1158/1078-0432.CCR-13-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–67. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, et al. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Research. 2013;41:8665–79. doi: 10.1093/nar/gkt579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 11.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes & Development. 2010;24:2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9:556–70. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, et al. Genome-wide Analysis of Alternative Pre-mRNA Splicing and RNA-Binding Specificities of the Drosophila hnRNP A/B Family Members. Molecular Cell. 2009;33:438–49. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golan-Gerstl R, Cohen M, Shilo A, Suh S-S, Bakàcs A, Coppola L, et al. Splicing Factor hnRNP A2/B1 Regulates Tumor Suppressor Gene Splicing and Is an Oncogenic Driver in Glioblastoma. Cancer Research. 2011;71:4464–72. doi: 10.1158/0008-5472.CAN-10-4410. [DOI] [PubMed] [Google Scholar]
- 16.Mayeda A, Helfman DM, Krainer AR. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Molecular and Cellular Biology. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL. hnRNP A2/B1 Modulates Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. Cancer Research. 2010;70:7137–47. doi: 10.1158/0008-5472.CAN-10-0860. [DOI] [PubMed] [Google Scholar]
- 18.Liu H-X, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR-proteins. Genes & Development. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biamonti G, Bonomi S, Gallo S, Ghigna C. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT) Cell Mol Life Sci. 2012;69:2515–26. doi: 10.1007/s00018-012-0931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nadiminty N, Chun JY, Lou W, Lin X, Gao AC. NF-κB2/p52 enhances androgen-independent growth of human LNCaP cells via protection from apoptotic cell death and cell cycle arrest induced by androgen-deprivation. The Prostate. 2008;68:1725–33. doi: 10.1002/pros.20839. [DOI] [PubMed] [Google Scholar]
- 22.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, et al. Niclosamide Inhibits Androgen Receptor Variants Expression and Overcomes Enzalutamide Resistance in Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2014;20:3198–210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadiminty N, Tummala R, Liu C, Yang J, Lou W, Evans CP, et al. NF-κB2/p52 Induces Resistance to Enzalutamide in Prostate Cancer: Role of Androgen Receptor and Its Variants. Molecular Cancer Therapeutics. 2013;12:1629–37. doi: 10.1158/1535-7163.MCT-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadiminty N, Lou W, Sun M, Chen J, Yue J, Kung H-J, et al. Aberrant Activation of the Androgen Receptor by NF-kB2/p52 in Prostate Cancer Cells. Cancer Research. 2010;70:3309–19. doi: 10.1158/0008-5472.CAN-09-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selth LA, Close P, Svejstrup JQ. Studying RNA-protein interactions In vivo by RNA immunoprecipitation. Methods in Molecular Biology. 2011;791:253–64. doi: 10.1007/978-1-61779-316-5_19. [DOI] [PubMed] [Google Scholar]
- 26.Dhir R, Ni Z, Lou W, DeMiguel F, Grandis JR, Gao AC. Stat3 activation in prostatic carcinomas. The Prostate. 2002;51:241–6. doi: 10.1002/pros.10079. [DOI] [PubMed] [Google Scholar]
- 27.Nadiminty N, Tummala R, Lou W, Zhu Y, Shi X-B, Zou JX, et al. MicroRNA let-7c Is Downregulated in Prostate Cancer and Suppresses Prostate Cancer Growth. PLoS ONE. 2012;7:e32832. doi: 10.1371/journal.pone.0032832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Chandran U, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, et al. Tumor Immunobiological Differences in Prostate Cancer between African-American and European-American Men. Cancer Research. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 32.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–9. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 33.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene Expression Alterations in Prostate Cancer Predicting Tumor Aggression and Preceding Development of Malignancy. Journal of Clinical Oncology. 2004;22:2790–9. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 34.Nadiminty N, Tummala R, Lou W, Zhu Y, Zhang J, Chen X, et al. MicroRNA let-7c Suppresses Androgen Receptor Expression and Activity via Regulation of Myc Expression in Prostate Cancer Cells. Journal of Biological Chemistry. 2012;287:1527–37. doi: 10.1074/jbc.M111.278705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A Novel Androgen Receptor Splice Variant Is Up-regulated during Prostate Cancer Progression and Promotes Androgen Depletion–Resistant Growth. Cancer Research. 2009;69:2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni M, Chen Y, Fei T, Li D, Lim E, Liu XS, et al. Amplitude modulation of androgen signaling by c-MYC. Genes & Development. 2013;27:734–48. doi: 10.1101/gad.209569.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun S, Sprenger CCT, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. The Journal of Clinical Investigation. 2010;120:2715–30. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mostaghel EA, Plymate SR, Montgomery B. Molecular Pathways: Targeting Resistance in the Androgen Receptor for Therapeutic Benefit. Clinical Cancer Research. 2014;20:791–8. doi: 10.1158/1078-0432.CCR-12-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, Sprenger C, Sun S, Epilepsia KS, Haugk K, Zhang X, et al. AR Variant ARv567es Induces Carcinogenesis in a Novel Transgenic Mouse Model of Prostate Cancer. Neoplasia. 2013;15:1009–17. doi: 10.1593/neo.13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo R, Li Y, Ning J, Sun D, Lin L, Liu X. HnRNP A1/A2 and SF2/ASF Regulate Alternative Splicing of Interferon Regulatory Factor-3 and Affect Immunomodulatory Functions in Human Non-Small Cell Lung Cancer Cells. PLoS ONE. 2013;8:e62729. doi: 10.1371/journal.pone.0062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeFave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. The EMBO Journal. 2011;30:4084–97. doi: 10.1038/emboj.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Allred DC, Avis I, Martinez A, Vos MD, Smith L, et al. Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer. Breast Cancer Res Treat. 2001;66:217–24. doi: 10.1023/a:1010631915831. [DOI] [PubMed] [Google Scholar]
- 44.Zerbe LK, Pino I, Pio R, Cosper PF, Dwyer-Nield LD, Meyer AM, et al. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: Implications for pre-mRNA processing. Molecular Carcinogenesis. 2004;41:187–96. doi: 10.1002/mc.20053. [DOI] [PubMed] [Google Scholar]
- 45.Piekielko-Witkowska A, Wiszomirska H, Wojcicka A, Poplawski P, Boguslawska J, Tanski Z, et al. Disturbed Expression of Splicing Factors in Renal Cancer Affects Alternative Splicing of Apoptosis Regulators, Oncogenes, and Tumor Suppressors. PLoS ONE. 2010;5:e13690. doi: 10.1371/journal.pone.0013690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boukakis G, Patrinou-Georgoula M, Lekarakou M, Valavanis C, Guialis A. Deregulated expression of hnRNP A/B proteins in human non-small cell lung cancer: parallel assessment of protein and mRNA levels in paired tumour/non-tumour tissues. BMC Cancer. 2010;10:434. doi: 10.1186/1471-2407-10-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mole S, McFarlane M, Chuen-Im T, Milligan SG, Millan D, Graham SV. RNA splicing factors regulated by HPV16 during cervical tumour progression. The Journal of Pathology. 2009;219:383–91. doi: 10.1002/path.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patry C, Bouchard L, Labrecque P, Gendron D, Lemieux B, Toutant J, et al. Small Interfering RNA-Mediated Reduction in Heterogeneous Nuclear Ribonucleoparticule A1/A2 Proteins Induces Apoptosis in Human Cancer Cells but not in Normal Mortal Cell Lines. Cancer Research. 2003;63:7679–88. [PubMed] [Google Scholar]
- 49.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2013;33:3140–50. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.