Abstract
Helicobacter pylon (H. pylori) infection is the strongest known risk factor for gastric non-cardia adenocarcinoma (GNCA). We used multiplex serology to determine whether seropositivity to 15 H. pylori proteins is associated with the subsequent development of non-cardia gastric cancer in Linxian, China.
We included 448 GNCA cases and 1242 controls from two time-points within the Linxian General Population Nutrition Intervention Trial, Linxian. H. pylori multiplex seropositivity was defined as positivity to ≥4 of the 15 included antigens. Odds ratios (OR) and 95% confidence intervals were adjusted for major GNCA risk factors. Additionally, we undertook a meta-analysis combining H. pylori multiplex serology data from both timepoints.
H. pylori multiplex seropositivity was associated with a significant increase in risk of GNCA at one time-point (1985; OR: 3.44, 95% CI: 1.91, 6.19) and this association remained significant following adjustment for H. pylori or CagA ELISA seropositivity (OR: 2.92, 95% CI: 1.56, 5.47). Combining data from both timepoints in a meta-analysis H. pylori multiplex seropositivity was associated with an increased risk of GNCA, as were 6 individual antigens: GroEL, HP0305, CagA, VacA, HcpC and Omp. CagM was inversely associated with risk of GNCA.
We identified 6 individual antigens which confer an increase in risk of GNCA within this population of high H. pylori seroprevalence, as well as a single antigen which may be inversely associated with GNCA risk. We further determined that the H. pylori multiplex assay provides additional information to the conventional ELISA methods on risk of GNCA.
Keywords: Helicobacter pylori, multiplex serology, gastric cancer, esophageal cancer
Introduction
Helicobacter pylori (H. pylori) infection is the strongest known risk factor for gastric non-cardia adenocarcinoma (GNCA 1). The discrepancy between the large number of people infected with H. pylori and the minority of individuals who develop this malignancy is thought to relate to a complex interplay of environmental risk factors, host susceptibility, and differing virulence among H. pylori strains. Identifying virulence factors is important both for understanding the mechanism of H. pylori induced carcinogenesis and for better risk-stratification of H. pylori infected individuals. Cytotoxin-associated gene A (CagA) and vacuolating toxin (VacA) have been associated with an increased risk of atrophic gastritis, intestinal metaplasia, ulcer and cancer2-4 and recent studies have suggested additional H. pylori antigens which may be associated with malignant potential5-7. Studies using a recently developed multiplex serologic assay that can detect host antibodies generated against CagA, VacA, and 13 other H. pylori protein antigens also suggest that other H. pylori proteins may be important virulence factors 8, 9.
We sought to determine whether seropositivity to the 15 H. pylori proteins included in the multiplex assay were associated with the development of non-cardia gastric cancer in Linxian, China, a region with a very high prevalence of H. pylori infection and high rates of esophageal squamous cell carcinoma and gastric cancer. Further, we sought to determine whether the multiplex assay conferred any information additional to that provided by conventional ELISA assays against whole cell H. pylori and CagA protein.
Methods
Subjects in this study were selected from the participants of the Linxian General Population Nutrition Intervention Trial (NIT). Details of the trial have been published elsewhere 10-12. Briefly, 29,584 healthy adult trial participants aged between 40-69 years were enrolled from four Linxian communes. In 1985, before the trial intervention began, each participant was interviewed, given a brief physical examination and had 10mL of blood drawn. Serum was separated and stored at −80°C until analyzed. Beginning in March 1986, participants were randomized into 8 groups and given daily vitamin/mineral combinations for 5.25 years, through May 1991. After the intervention, the participants were followed as a cohort13. Between 1999 and 2000, all living NIT participants were invited to a blood collection survey, and ~16,000 (~80% of all eligible participants) participated. Plasma was separated and stored at −80°C until analyzed.
Diagnoses of cancer were made at local commune and county hospitals and supplemented by a study team which provided clinical and diagnostic services, including endoscopy, for patients with symptoms suggestive of oesophageal or gastric cancer. A panel of U.S. and Chinese experts reviewed the diagnostic materials and confirmed the cancer cases from 1986 through 1996, and Chinese experts performed similar reviews of the cases from 1996-2007. Case ascertainment is considered complete and loss to follow-up minimal (n = 176 or 1%)10. Written informed consent was obtained from each participant prior to enrolment, and human subject protection procedures approved by the institutional review boards of the U.S. National Institutes of Health and the Chinese Academy of Medical Sciences were followed throughout the trial and subsequent follow-up.
Selection of study participants
Between baseline (1985) and 2001, 363 cases of GNCA were diagnosed; of these 330 had serum available for this analysis. In a previous analysis we selected a random sample of 1050 subjects from the entire baseline cohort14, for the current analysis we randomly selected 330 controls from this group, frequency matched to the sex of the 330 GNCA cases. At the time of original selection follow-up for cases and controls was available through 05/31/2001.
In order to replicate our results, we also evaluated all GNCA cases diagnosed between the 1999/2000 blood collection and the end of 2006 who had plasma available for analysis (n=118). An age- and sex-stratified random sample of 1000 subjects from the NIT cohort members who participated in the 1999/2000 blood collection survey had previously been selected to serve as a reference subcohort in another earlier analysis 15, and 912 of these subcohort members had plasma available for the current analysis. At the time of original selection follow-up for cases and controls was available through 12/31/2006. There was no overlap, either for cases or controls, between the 1999/2000 analytic set and the 1985 analytic set.
Laboratory Analysis
H. pylori multiplex serology assay
H. pylori multiplex serology was performed as previously described 16. Results are based on glutathione S-transferase (GST) capture immunosorbent assays combined with fluorescent bead technology 17 and simultaneously quantify antibodies to multiple protein antigens. Fifteen H. pylori proteins (UreA, Catalase, GroEL, Nap A, CagA, CagM, Cagδ, HP0231, VacA, HpaA, Cad, HyuA, Omp, HcpC, and HP0305) that were bacterially-expressed and affinity-purified recombinant glutathione-S transferase (GST) -H. pylori tag fusion proteins were used as antigens per previous studies 8, 9. Assay validation was described previously 9. H. pylori positivity was defined as those seropositive for ≥4 antigens 16, however as in previous analyses18, results in our study were similar whether a definition of ≥3, ≥4, or ≥5 antigens was used.
In addition to the assay controls, we inserted 4 blinded replicate QC (32 serum and 76 plasma) samples among the 90 test samples in each 96-well plate. Using the dichotomous measure of presence/absence for each antigen (calculated using the median reporter fluorescence intensity + 3 standard deviations, excluding positive outliers), we calculated Fleiss’ kappa to assess the reliability of agreement between repeated measures among the quality control samples. Among the serum samples the kappa value for CagM was 0.50 and for catalase was 0.43, the kappas for Omp, HcpC, GroEl, UreA, HP0231 ranged from 0.70 to 0.87. All other antigens had a kappa value of one across blinded replicates, indicating perfect agreement. Among the plasma samples the kappa value for GroEL was 0.17, for HpaA was 0.47 and for VacA was 0.53, for CagA was 1 and for catalase was 1. The kappa values for the remaining antigens ranged from 0.63 to 0.93.
H. pylori enzyme-linked immunosorbent assay
All serum samples from the 1985 baseline study had been previously evaluated for IgG antibodies to whole-cell and CagA H. pylori antigens using enzyme-linked immunosorbent assays as previously described14, 19. Experienced technicians who were unaware of participants’ case-control status performed the serologic assays in duplicate. When two assayed aliquots provided indeterminate results, additional aliquots were analysed and the average of all results was used to determine serologic status. One hundred and twelve external quality control serum samples, aliquoted from a single large pooled serum sample from Linxian, were equally distributed among 56 different batches. On the basis of these samples, the coefficients of variation were 15 and 20% for the H. pylori whole cell and CagA assays, respectively.
We had information from previously-conducted conventional H. pylori whole cell or CagA ELISA assays and the H. pylori multiplex assay for the individuals included in the 1985 baseline study. Of the 330 control sera, 276 were positive by H. pylori multiplex serology and 220 of these were positive for H. pylori or CagA by previous ELISA. Of the 54 sera negative by H. pylori multiplex, 41 were negative for both H. pylori whole cell and CagA ELISA, so that agreement between the two approaches was 79% (κ=0.42).
Statistical Analysis
The distributions of characteristics between GNCA cases and control subjects were compared using Student t tests for continuous variables and Pearson χ2 tests for categorical variables. Correlations between the 15 individual H. pylori antigens were investigated using a Spearman correlation matrix (Supplementary Table). Odds ratios (ORs) and 95% confidence intervals (95% CIs) for associations between each antigen and the risk of GNCA were calculated by unconditional logistic regression models. Multivariable models were adjusted for age at randomization, sex, ever smoking (answered yes to “have you ever smoked cigarettes for 6 months of longer?” or “Do you smoke cigarettes now?”), alcohol (yes/no for alcohol consumption within the past 12 months), and body mass index (kg/m2). Stratified analyses were undertaken by sex, median age and median follow-up time. Using data from the 1985 blood draw, we also performed analyses where the multivariable model was additionally adjusted for H. pylori whole cell or CagA seropositivity as determined by ELISA. We also investigated whether results were altered after excluding cases diagnosed within a year of blood draw.
We used principal components analysis as a possible data reduction method, because it is likely there is a high degree of correlation between antigens. Eigenvalues of the correlation matrix were inspected and the minimum number of principal components sufficient to explain >50% of the variance was selected. We performed a meta-analysis to combine the baseline and follow-up measurements for overall H. pylori positivity, and for each individual antigen. As fixed and random effects models produced similar estimates, the point estimates presented here are based on the fixed effects model. We undertook exploratory risk prediction analyses using the lasso method which incorporates an I1 penalty to produce a model with minimal prediction error and recover the “true” (sparse) model20. We assessed the risk prediction models using area under the curve (AUC) averaged from 10 simulations of 5-fold cross-validation.
All statistical analyses were performed using STATA version 11.2 (Stata Corp, College Station, TX), except for the risk prediction models and the Fleiss kappa analyses which were conducted using R. All P-values were two-sided. A P-value < 0.05 was considered statistically significant.
Results
Characteristics of GNCA cases and controls from both the 1985 (serum) and 1999/2000 (plasma; replication set) blood collections are shown in Table 1. GNCA cases from the 1985 blood draw were more likely to be older and less likely to have drunk alcohol in the previous 12 months than their controls. In the replication analyses, cases from the 1999/2000 blood draw were similar to the 1999/2000 sub-cohort members in all respects except that they had a slightly lower BMI.
Table 1.
Descriptive characteristics of non-cardia gastric cancer cases and controls from Linxian General Population Nutrition Intervention Trial, from the baseline (1985) and 1999/2000 collections.
| Characteristic | Serum: Baseline/1985 collection | Plasma: 1999/2000 collection | ||||
|---|---|---|---|---|---|---|
| GNCA cases (n=330) |
Controls (n=330) |
P value | GNCA cases (n=118) |
Sub-cohort (n=912) |
P value | |
| Male, N (%) | 222 (67) | 222 (67) | 1.001 | 65 (55) | 448 (49) | 0.221 |
| Age at blood draw, yrs, mean (SD) | 56 (8) | 52 (9) | <0.0012 | 65 (7) | 64 (8) | 0.272 |
| Ever smokers, N (%) | 158 (48) | 165 (50) | 0.561 | 41 (34) | 291 (32) | 0.561 |
| Drank alcohol in last 12 months, N (%) | 78 (24) | 103 (31) | 0.031 | 25 (21) | 242 (27) | 0.201 |
| Body mass index, mean kg/m2 (SD) | 21.52 (2.23) | 21.88 (2.27) | 0.042 | 21.30 (2.04) | 21.93 (2.38) | 0.0072 |
| Median time to diagnosis/death/end of follow-up, yrs, (IQR) |
6.99 (3.89-11.25) |
16.05 (13.54-16.09) |
<0.0012 | 3.60 (1.72, 5.18) |
6.74 (6.67, 7.10) |
<0.0012 |
P values were calculated by Pearson χ2 test
P values were calculated by Student t test
The relationships between H. pylori serostatus (as defined by positivity to ≥4 antigens) and baseline characteristics in controls/sub-cohort members are shown in Table 2. No significant differences were noted in baseline characteristics by H. pylori serostatus for either sample-set. Correlations between individual antigens in controls are shown in Supplementary Table 1.
Table 2.
Descriptive characteristics of controls from participants in the 1985 and 1999/2000 collections of the NIT study, according to multiplex H. pylori status1.
| Characteristic | 1985 | 1999/2000 | ||||
|---|---|---|---|---|---|---|
|
H. pylori − (n=54) |
H. pylori + (n=276) |
P |
H. pylori − (n=56) |
H. pylori + (n=856) |
P | |
| Age at blood draw, mean yrs, (SD) | 52 (10) | 52 (9) | 0.992 | 66 (8) | 64 (8) | 0.082 |
| Male, N (%) | 38 (70) | 184 (67) | 0.603 | 31 (55) | 417 (49) | 0.343 |
| Ever smokers, N (%) | 27 (51) | 138 (50) | 0.903 | 19 (35) | 272 (32) | 0.683 |
| Drank alcohol in last 12 months, N (%) | 20 (38) | 83 (30) | 0.273 | 18 (33) | 224 (26) | 0.293 |
| Body mass index, mean kg/m2 (SD) | 21.69 (1.81) | 21.91 (2.35) | 0.512 | 22.15 (2.08) | 21.91 (2.40) | 0.472 |
H. pylori seropositivity is defined as positivity to ≥ 4 H. pylori antigens.
P values were calculated by Student t test
P values were calculated by Pearson χ2 test
84% of controls from the 1985 blood draw were found to be H. pylori seropositive, compared to 95% of GNCA cases. In the 1999/2000 replication set (Table 3a), 94% of sub-cohort members were H. pylori seropositive, compared to 97% of GNCA cases (Table 3b). Multivariate adjusted odds ratios (adjusted for age, sex, alcohol, smoking and BMI) for GNCA were 3.44 (95%CI: 1.91, 6.19) for the 1985 blood draw and 1.82 (95% CI: 0.64, 5.14) for the 1999/2000 replication set.
Table 3a.
Odds ratios (OR) and 95% confidence intervals (CI) for seropositivity to H. pylori antigens and risk of GNCA in NIT in samples from the 1985 baseline collection.
| Serostatus | 1985: GNCA N (%) |
Control N (%) |
Odds Ratio (95% CI) (age adjusted) |
Odds Ratio (95% CI) (fully adjusted2) |
Odds Ratio (95% CI) (fully adjusted + H. pylori and/or CagA ELISA) |
|---|---|---|---|---|---|
| ELISA: H. pylori or CagA+ |
264 (80) | 233 (71) | 1.80 (1.24, 2.61)3 | 1.78 (1.22, 2.58)3 | 1.36 (0.91, 2.04) |
| H. pylori +1 | 313 (95) | 276 (84) | 3.47 (1.94, 6.23) 3 | 3.44 (1.91, 6.19)3 | 2.92 (1.56, 5.47) 3 |
| GroEL Chaperonin GroEL |
266 (81) | 241 (73) | 1.57 (1.08, 2.29) | 1.56 (1.07, 2.28) | 1.30 (0.86, 1.96) |
| UreA Urease alpha subunit |
142 (43) | 125 (38) | 1.21 (0.88, 1.68) | 1.21 (0.88, 1.67) | 1.18 (0.85, 1.63) |
| HP0231 Hypothetical protein |
95 (29) | 102 (31) | 0.88 (0.63, 1.24) | 0.88 (0.63, 1.25) | 0.76 (0.53, 1.09) |
| NapA Neutrophil activating protein |
182 (55) | 161 (49) | 1.21 (0.89, 1.67) | 1.24 (0.90, 1.71) | 1.08 (0.77, 1.51) |
| HP0305 Hypothetical protein |
228 (69) | 177 (54) | 2.11 (1.52, 2.95)3 | 2.16 (1.54, 3.02) 3 | 1.97 (1.35, 2.88) 3 |
| HpaA Neuraminyllactose- binding hemagglutinin homolog, surface |
99 (30) | 91 (28) | 1.13 (0.80, 1.60) | 1.11 (0.78, 1.58) | 1.03 (0.72, 1.47) |
| Cag delta Cag island protein 3 |
169 (51) | 145 (44) | 1.24 (0.91, 1.71) | 1.22 (0.88, 1.68) | 1.20 (0.87, 1.66) |
| CagM Cag island protein 16 |
49 (15) | 54 (16) | 0.87 (0.56, 1.35) | 0.83 (0.54, 1.29) | 0.83 (0.53, 1.28) |
| CagA Cytotoxin-associated gene A |
286 (87) | 249 (75) | 2.26 (1.49, 3.44) 3 | 2.28 (1.49, 3.48) 3 | 1.96 (1.21, 3.15) |
| HyuA Hydantoin utilization protein A, |
196 (59) | 190 (58) | 0.99 (0.72, 1.36) | 1.00 (0.73, 1.39) | 0.93 (0.67, 1.29) |
| Catalase Detoxification |
215 (65) | 191 (58) | 1.34 (0.97, 1.85) | 1.37 (0.99, 1.89) | 1.32 (0.95, 1.83) |
| VacA Vacuolating cytotoxin |
286 (87) | 255 (77) | 1.80 (1.18, 2.75) | 1.75 (1.14, 2.68) | 1.42 (0.89, 2.26) |
| HcpC Conserved hypothetical secreted protein |
156 (47) | 141 (43) | 1.26 (0.92, 1.73) | 1.26 (0.91, 1.73) | 1.05 (0.74, 1.49) |
| Cad Cinnamyl alcohol dehydrogenase ELI3-2 |
57 (17) | 54 (16) | 0.96 (0.63, 1.46) | 0.94 (0.61, 1.43) | 0.91 (0.59, 1.39) |
| Omp Outer membrane protein |
286 (87) | 249 (75) | 2.13 (1.40, 3.24) 3 | 2.09 (1.37, 3.18) 3 | 1.75 (1.10, 2.81) |
H. pylori seropositivity is defined here as positivity to ≥ 4 H. pylori antigens.
Full adjustment: age at blood draw, sex, ever smoking, alcohol, BMI
Associations passing Bonferroni correction (0.05/15: p<0.003)
Table 3b.
Odds ratios (OR) and 95% confidence intervals (CI) for seropositivity to H. pylori antigens and risk of GNCA in NIT in samples from the 1999/2000 collection.
| Serostatus | 1999/2000: GNCA N (%) |
Control N (%) |
Odds Ratio (95% CI) (age adjusted) |
Odds Ratio (95% CI) (fully adjusted2) |
|---|---|---|---|---|
| H. pylori +1 | 114 (97) | 856 (94) | 1.90 (0.67, 5.35) | 1.82 (0.64, 5.14) |
| GroEL Chaperonin GroEL |
98 (83) | 715 (78) | 1.38 (0.83, 2.30) | 1.45 (0.87, 2.42) |
| UreA Urease alpha subunit |
60 (51) | 469 (51) | 0.96 (0.66, 1.41) | 0.97 (0.66, 1.43) |
| HP0231 Hypothetical protein |
64 (54) | 441 (48) | 1.27 (0.86, 1.86) | 1.26 (0.86, 1.86) |
| NapA Neutrophil activating protein |
76 (64) | 594 (65) | 0.97 (0.65, 1.45) | 0.97 (0.65, 1.46) |
| HP0305 Hypothetical protein |
87 (74) | 518 (57) | 2.17 (1.41, 3.33) 3 | 2.16 (1.40, 3.33) 3 |
| HpaA Neuraminyllactose- binding hemagglutinin homolog, surface |
50 (42) | 425 (47) | 0.84 (0.57, 1.24) | 0.83 (0.56, 1.23) |
| Cag delta Cag island protein 3 |
58 (49) | 446 (49) | 1.02 (0.70, 1.49) | 0.99 (0.67, 1.45) |
| CagM Cag island protein 16 |
23 (20) | 272 (30) | 0.56 (0.35, 0.91) | 0.54 (0.34, 0.88) |
| CagA Cytotoxin-associated gene A |
102 (86) | 730 (80) | 1.66 (0.95, 2.89) | 1.70 (0.97, 2.98) |
| HyuA Hydantoin utilization protein A, |
83 (70) | 569 (62) | 1.43 (0.94, 2.16) | 1.43 (0.94, 2.17) |
| Catalase Detoxification |
91 (77) | 695 (76) | 1.05 (0.67, 1.66) | 1.03 (0.65, 1.64) |
| VacA Vacuolating cytotoxinA |
107 (91) | 734 (81) | 2.38 (1.25, 4.53) | 2.37 (1.24, 4.53) |
| HcpC Conserved hypothetical secreted protein |
71 (60) | 489 (54) | 1.30 (0.88, 1.92) | 1.31 (0.88, 1.94) |
| Cad Cinnamyl alcohol dehydrogenase ELI3-2 |
33 (28) | 203 (22) | 1.33 (0.87, 2.06) | 1.39 (0.89, 2.15) |
| Omp Outer membrane protein |
100 (85) | 643 (71) | 2.36 (1.40, 3.99) 3 | 2.30 (1.36, 3.88) 3 |
H. pylori seropositivity is defined here as positivity to ≥4 H. pylori antigens.
Full adjustment: age at blood draw, sex, ever smoking, alcohol, BMI
Associations passing Bonferroni correction (0.05/15: p<0.003)
In multivariate adjusted models, five individual antigens were significantly associated with increased risk of GNCA at the 1985 blood draw (Table 3a): GroEL (OR: 1.56, 95% CI: 1.07, 2.28), HP0305 (OR: 2.16, 95% CI: 1.54, 3.02), CagA (OR: 2.28, 95% CI: 1.49, 3.48), VacA (OR: 1.75, 95% CI: 1.14, 2.68) and Omp (OR: 2.09, 95% CI: 1.37, 3.18). The multivariate adjusted models of the data from the 1999/2000 blood draw show similar significant associations (Table 3b): HP0305 (OR: 2.16, 95% CI: 1.40, 3.33), VacA (OR: 2.37, 95% CI: 1.24, 4.53) and Omp (OR: 2.30, 95% CI: 1.36, 3.88), as well as a significant inverse association between CagM and GNCA (OR: 0.54, 95% CI: 0.34, 0.88). Principal components analysis did not provide anything additional by means of data reduction (Supplementary Table 2). Stratification by gender, age and length of follow-up did not significantly alter these associations (Supplementary Table 3).
H. pylori is believed to disappear as gastric disease and cancer progress so we then investigated the effect of excluding cases developing within one year of blood draw were from the analysis. In the 1985 blood draw group exclusion of cases developing within a year (n=7) had no effect on risk estimates. In the 1999/2000 blood draw estimates were similar after excluding cases developing within a year (n=19), and the positive associations between CagA and HyuA became statistically significant (OR: 1.95; 95% CI: 1.03, 3.67 and OR: 1.61; 95% CI: 1.01, 2.56, respectively).
Data from conventional H. pylori whole cell and CagA ELISA assays was available for all 660 individuals included in the 1985 blood draw. When the results for these ELIS A assays were combined and added to the multivariate adjusted model the association between H. pylori multiplex seropositivity and GNCA was slightly attenuated (OR: 2.92, 95% CI: 1.56, 5.47; Table 3a), but remained significant. The association between ELISA seropositivity (either H. pylori whole cell or CagA) and GNCA (OR: 1.78, 95%) CI: 1.22, 2.58) was also attenuated and no longer significant when it was included in the multivariate adjusted model for H. pylori multiplex seropositivity (OR: 1.36, 95% CI: 0.91, 2.04; Table 3a). When the results for the ELISA assays were added to the multivariate adjusted model for each individual antigen only 3 of the individual antigens remained significantly associated with risk of GNCA: HP0305 (OR: 1.97, 95% CI: 1.35, 2.88), CagA (OR: 1.96, 95% CI: 1.21, 3.15) and Omp (OR: 1.75, 95%) CI: 1.10, 2.81)( Table 3a) and in each of these models the association between ELISA H. pylori whole cell or CagA seropositivity and risk of GNCA was no longer significant (data not shown).
We undertook exploratory risk prediction analyses which used the area under the curve (AUC) averaged from 10 simulations of 5-fold cross-validation. Using the lasso method we explored gastric cancer risk prediction, based on positivity to any of the 15 measured antigens. Using the 1985 blood draw data, the AUC was calculated as 0.574 (SE= 0.0089). HP0305, CagA and Omp were selected in all of the highest scoring models for each of the 10 simulations (consistent with Table 3a). GroEL and VacA, which were significantly associated with gastric cancer in our logistic regression models were not selected in the highest scoring lasso models, however they are highly correlated with HP0305 (Pearson’s Chi-square p=4.9e−20& p=5.0 e−23, respectively). Using the 1999/2000 dataset the AUC was calculated as 0.617 (SE=0.0067) and HP0305, CagM, VacA and Omp were selected in the highest scoring models.
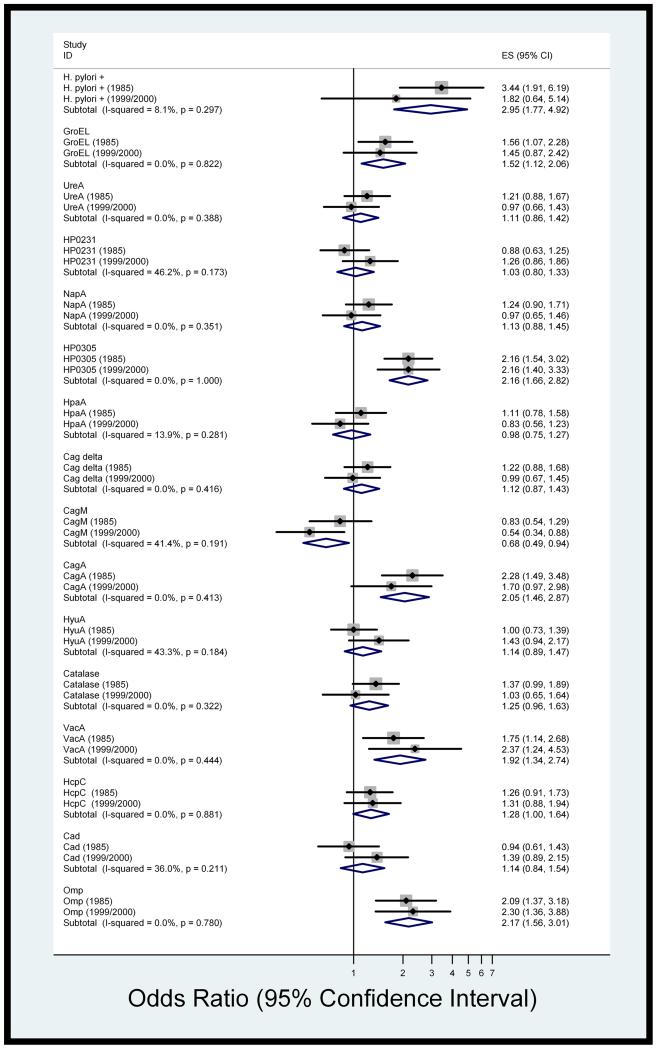
To combine the information taken from different individuals from the same population at two different time points, we undertook at meta-analysis of the 1985 baseline data and the 1999/2000 replication set (Figure 1). The combined risk estimate for H. pylori seropositivity and risk of GNCA was 2.95 (95% CI: 1.77, 4.92). Six individual antigens were significantly positively associated with GNCA: GroEL (OR: 1.52, 95% CI: 1.12, 2.06), HP0305 (OR: 2.16, 95% CI: 1.66, 2.82), CagA (OR: 2.05, 95% CI: 1.46, 2.87), VacA (OR: 1.92, 95% CI: 1.34, 2.74), HcpC (OR: 1.28, 95% CI: 1.00, 1.64) and OMP (OR: 2.17, 95%) CI: 1.56, 3.01). In addition, CagM was significantly inversely associated with risk of GNCA, with a combined estimate of 0.68 (95% CI: 0.49, 0.94).
Figure 1.
Forest plot (fixed-effects model) of risk estimates (fully-adjusted) for H. pylori seropositivity and individual antigen positivity at each timepoint. Rhomboids indicate pooled estimates.
Discussion
In this analysis of H. pylori seropositivity at two time points within the Linxian General Population Nutrition Intervention Trial Cohort we found that overall seropositivity was associated with a nearly threefold increase in risk of subsequently developing GNCA. In addition, we report that six specific antigens were significantly positively associated with GNCA development and one specific antigen was inversely associated with risk of GNCA. To date, this is the largest prospective analysis to use this multiplex assay to study the effects of individual H. pylori antigens on GNCA risk and the only analysis that has also included data from both conventional H. pylori (whole-cell) and/or CagA ELISA.
Our group previously used ELISA serologic assays to investigate the relationship between H. pylori (whole-cell) and/or CagA seropositivity and subsequent risk of GNCA in the NIT cohort14. This previous analysis, which compared cases identified from the 1985 blood draw to a subcohort, reported H. pylori seroprevalences of 73% in the subcohort and 80% in the GNCA cases (adjusted hazard ratio: 1.60, 95%) CI: 1.15, 2.21). In that study, the H. pylori (whole-cell) ELISA detected seropositivity against a cell lysate of the entire H. pylori bacterium, whereas in the current multiplex analysis, our definition of H. pylori positivity required seropositivity to four or more specific H. pylori antigens. We note that although we report a higher seroprevalence of infection in the current study, the magnitude of difference in seropositivity in GNCA cases relative to the subcohort is similar in both studies. It is also striking that the background prevalence of infection measured by the multiplex assay rose between the 1985 and 1999/2000 timepoints (84% vs. 94% in the controls/subcohort), given the falling rates of infection noted in developed countries during the same time period 21. It is likely that this discrepancy may be a function of antibody degradation and the likelihood of additional freeze-thaw cycles, over time, in the older samples.
Our analysis of multivariate models including both H. pylori multiplex and ELISA data suggests that the multiplex panel conveys additional information, beyond and independent of traditional ELISA methods. When the multivariate model was simultaneously adjusted for ELISA and multiplex H. pylori positivity, the positive association for ELISA was reduced and no longer significant while the multiplex association remained significant, albeit slightly attenuated. Also of interest, 3 specific antigens: HP0305, CagA and Omp, were found to confer a stronger signal, in these fully adjusted multiplex models, than ELISA positivity.
The very high prevalence of H. pylori infection we observed here was also noted previously when the same multiplex serology assay was used in a study of 226 GNCA cases and 451 matched controls from the Shanghai Men’s Health Study (SMHS) in China 22. In that study the seroprevalence of H. pylori infection was 94% among controls and 98% among GNCA cases. Similar to our findings, the authors also reported significant associations for GNCA with the individual antigens HP0305, CagA, VacA and Omp. While CagA and VacA are long established H. pylori virulence factors 23-25, little is known about HP0305 or Omp (also called HP1564, a putative outer membrane protein) and cancer. In contrast to the Shanghai study, we also found positive associations for GNCA with GroEL and HcpC. The molecular chaperone GroEL was also previously identified as a marker of gastric cancer risk in a German study 8 and is proposed to influence the binding of the bacterium to the gastrointestinal mucosa 26, 27. HcpC belongs to the family of Helicobacter cysteine-rich proteins that induce proinflammatory cytokines 28 and it has previously been associated with chronic atrophic gastritis 9. While our risk prediction analysis was purely exploratory, it is interesting to note that models using the multiplex data performed relatively well and were in general agreement with results of disease association models.
Interestingly, while CagA is generally associated with an increased risk of GNCA, we found that another of the Cag pathogenicity island proteins, CagM, was associated with a significant reduction in GNCA risk in our study (OR: 0.68; 95% CI: 0.49, 0.94). CagM is necessary for the translocation of the CagA protein into the host cell via the type IV secretion system 29. Further work is needed to replicate this finding, and to examine potential mechanisms.
Our study has a number of strengths, including the prospective study design, the large number of cancer cases, and virtually complete follow-up of all study participants. We applied a novel technology in a large nested case-control study and replicated our results. However, we also note several limitations to our study; though our study included two time-points there is no overlap between the individuals measured at baseline and those from 1999/2000 meaning that it is not possible to follow the serostatus of the same individuals over time, within this analysis. The population in Linxian, China, where these studies were undertaken, is known to be deficient in several essential nutrients, as well as having a high prevalence of H. pylori meaning that our results may not be generalizable to other, particularly Western, populations. Lastly, the population is remarkably homogenous in terms of socioeconomic status and lifestyle so that it is not possible to study how the range of these factors might influence H. pylori serostatus or disease in this population.
This is the largest prospective study to date to investigate the association between GNCA and individual antibodies to H. pylori using multiplex serology, and it is the only analysis to also include data from both conventional ELISA for H. pylori and CagA. Our analysis identified 6 individual antigens which confer increased risk of GNCA within this population of high H. pylori seroprevalence, and a single antigen which was inversely associated with GNCA risk. We further determined that the H. pylori multiplex assay provides additional information to traditional ELISA methods on risk of GNCA. Given the high prevalence of H. pylori infection in this region identifying particular strains, or antigens, which may influence GNCA risk may be valuable in understanding the disease etiology and tailoring public health strategies accordingly.
Supplementary Material
Novelty & Impact Statement.
We identified 6 individual antigens associated with increased risk of cancer within this population, and a single antigen that was inversely associated with cancer risk. The H. pylori multiplex assay provides additional information to traditional ELISA methods on risk of cancer.
Given the high prevalence of H. pylori infection in this region, identifying particular strains, or antigens, which may influence cancer risk may be valuable in understanding the disease etiology and tailoring public health strategies accordingly.
Acknowledgments
Funding: Intramural Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Department of Health and Human Services.
Abbreviations
- BMI
Body mass index
- CI
confidence interval
- H. pylori
Helicobacter pylori
- OR
odds ratio
Footnotes
Disclosure of Potential Conflicts of Interest: None to declare.
References
- 1.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori Infection and the Development of Gastric Cancer. N Engl J Med. 2001;345:784–89. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 2.Kuipers EJ, Perez-Perez Gl, Meuwissen SG, Blaser MJ. Helicobacter pylori and atrophic gastritis: importance of the cagA status. Journal of the National Cancer Institute. 1995;87:1777–80. doi: 10.1093/jnci/87.23.1777. [DOI] [PubMed] [Google Scholar]
- 3.Atherton JC, Cao P, Peek RM, Jr., Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–7. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 4.Kamangar F, Dawsey SM, Blaser MJ, Perez-Perez Gl, Pietinen P, Newschaffer G, Abnet CC, Albanes D, Virtamo J, Taylor PR. Opposing risks of gastric cardia and noncardia gastric adenocarcinomas associated with Helicobacter pylori seropositivity. J Natl Cancer Inst. 2006;98:1445–52. doi: 10.1093/jnci/djj393. [DOI] [PubMed] [Google Scholar]
- 5.Haas G, Karaali G, Ebermayer K, Metzger WG, Lamer S, Zimny-Arndt U, Diescher S, Goebel UB, Vogt K, Roznowski AB, Wiedenmann BJ, Meyer TF, et al. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics. 2002;2:313–24. doi: 10.1002/1615-9861(200203)2:3<313::aid-prot313>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 6.Krah A, Miehlke S, Pleissner KP, Zimny-Arndt U, Kirsch C, Lehn N, Meyer TF, Jungblut PR, Aebischer T. Identification of candidate antigens for serologic detection of Helicobacter pylori-infected patients with gastric carcinoma. International journal of cancer. 2004;108:456–63. doi: 10.1002/ijc.11557. [DOI] [PubMed] [Google Scholar]
- 7.Mini R, Bernardini G, Salzano AM, Renzone G, Scaloni A, Figura N, Santucci A. Comparative proteomics and immunoproteomics of Helicobacter pylori related to different gastric pathologies. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:63–79. doi: 10.1016/j.jchromb.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 8.Gao L, Michel A, Week MN, Arndt V, Pawlita M, Brenner H. Helicobacter pylori infection and gastric cancer risk: evaluation of 15 H. pylori proteins determined by novel multiplex serology. Cancer Res. 2009;69:6164–70. doi: 10.1158/0008-5472.CAN-09-0596. [DOI] [PubMed] [Google Scholar]
- 9.Gao L, Week MN, Michel A, Pawlita M, Brenner H. Association between chronic atrophic gastritis and serum antibodies to 15 Helicobacter pylori proteins measured by multiplex serology. Cancer Res. 2009;69:2973–80. doi: 10.1158/0008-5472.CAN-08-3477. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Taylor PR, Li JY, Dawsey SM, Wang W, Tangrea JA, Liu BQ, Ershow AG, Zheng SF, Fraumeni JF, Jr., et al. Linxian nutrition intervention trials. Design, methods, participant characteristics, and compliance. Ann Epidemiol. 1993;3:577–85. doi: 10.1016/1047-2797(93)90078-i. [DOI] [PubMed] [Google Scholar]
- 11.Blot WJ, Li JY, Taylor PR, Guo W, Dawsey S, Wang GQ, Yang CS, Zheng SF, Gail M, Li GY, Yu Y, Liu B, et al. Nutrition intervention trials in Linxian, China: supplementation with specific vitamin/mineral combinations, cancer incidence, and disease-specific mortality in the general population. Journal of the National Cancer Institute. 1993;85:1483–92. doi: 10.1093/jnci/85.18.1483. [DOI] [PubMed] [Google Scholar]
- 12.Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey SM, Dong ZW, Mark SD, Qiao YL, Taylor PR. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. International journal of cancer. 2005;113:456–63. doi: 10.1002/ijc.20616. [DOI] [PubMed] [Google Scholar]
- 13.Qiao YL, Dawsey SM, Kamangar F, Fan JH, Abnet CC, Sun XD, Johnson LL, Gail MH, Dong ZW, Yu B, Mark SD, Taylor PR. Total and cancer mortality after supplementation with vitamins and minerals: follow-up of the Linxian General Population Nutrition Intervention Trial. Journal of the National Cancer Institute. 2009;101:507–18. doi: 10.1093/jnci/djp037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamangar F, Qiao YL, Blaser MJ, Sun XD, Katki H, Fan JH, Perez-Perez Gl, Abnet CC, Zhao P, Mark SD, Taylor PR, Dawsey SM. Helicobacter pylori and oesophageal and gastric cancers in a prospective study in China. Br J Cancer. 2007;96:172–6. doi: 10.1038/sj.bjc.6603517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam TK, Freedman ND, Fan J-H, Qiao Y-L, Dawsey SM, Taylor PR, Abnet CC. Pre-diagnostic plasma vitamin C and risk of gastric adenocarcinoma and esophageal squamous cell carcinoma in the General Population Nutrition Intervention Trial. The American Journal of Clinical Nutrition. 2013 doi: 10.3945/ajcn.113.061267. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel A, Waterboer T, Kist M, Pawlita M. Helicobacter pylori multiplex serology. Helicobacter. 2009;14:525–35. doi: 10.1111/j.1523-5378.2009.00723.x. [DOI] [PubMed] [Google Scholar]
- 17.Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, Templin MF, Pawlita M. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin Chem. 2005;51:1845–53. doi: 10.1373/clinchem.2005.052381. [DOI] [PubMed] [Google Scholar]
- 18.Murphy G, Michel A, Taylor PR, Albanes D, Weinstein SJ, Virtamo J, Parisi D, Snyder K, Butt J, McGlynn KA, Koshiol J, Pawlita M, et al. Association of seropositivity to Helicobacter species and biliary tract cancer in the ATBC study. Hepatology. 2014 doi: 10.1002/hep.27193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limburg P, Qiao Y, Mark S, Wang G, Perez-Perez G, Blaser M, Wu Y, Zou X, Dong Z, Taylor P, Dawsey S. Helicobacter pylori seropositivity and subsite-specific gastric cancer risks in Linxian, China. Journal of the National Cancer Institute. 2001;93:226–33. doi: 10.1093/jnci/93.3.226. [DOI] [PubMed] [Google Scholar]
- 20.Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2011;73:273–82. [Google Scholar]
- 21.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nature reviews. Microbiology. 2009;7:887–94. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epplein M, Zheng W, Xiang YB, Peek RM, Jr., Li H, Correa P, Gao J, Michel A, Pawlita M, Cai Q, Shu XO. Prospective Study of Helicobacter pylori Biomarkers for Gastric Cancer Risk among Chinese Men. Cancer Epidemiol Biomarkers Prev. 2012;21:2185–92. doi: 10.1158/1055-9965.EPI-12-0792-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci U S A. 2000;97:1263–8. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–94. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 25.Nogueira C, Figueiredo C, Carneiro F, Gomes AT, Barreira R, Figueira P, Salgado C, Belo L, Peixoto A, Bravo JC, Bravo LE, Realpe JL, et al. Helicobacter pylori genotypes may determine gastric histopathology. Am J Pathol. 2001;158:647–54. doi: 10.1016/s0002-9440(10)64006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanet A, Labigne A. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect Immun. 1998;66:1023–7. doi: 10.1128/iai.66.3.1023-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, Corthesy-Theulaz IE. GroEL of Lactobacillus johnsonii Lai (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect Immun. 2006;74:425–34. doi: 10.1128/IAI.74.1.425-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demi L, Aigner M, Decker J, Eckhardt A, Schutz C, Mittl PR, Barabas S, Denk S, Knoll G, Lehn N, Schneider-Brachert W. Characterization of the Helicobacter pylori cysteine-rich protein A as a T-helper cell type 1 polarizing agent. Infect Immun. 2005;73:4732–42. doi: 10.1128/IAI.73.8.4732-4742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer W, Puis J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Molecular microbiology. 2001;42:1337–48. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.