Abstract
Peroxisomes are remarkably responsive organelles. Their composition, abundance and even their mechanism of biogenesis are influenced strongly by cell type and the environment. This plasticity underlies peroxisomal functions in metabolism and the detoxification of dangerous reactive oxygen species. However, peroxisomes are integrated into the cellular system as a whole such that they communicate intimately with other organelles, control signaling dynamics as in the case of innate immune responses to infectious disease, and contribute to processes as fundamental as longevity. The increasing evidence for peroxisomes having roles in various cellular and organismal functions, combined with their malleability, suggests complex mechanisms operate to control cellular dynamics and the specificity of cellular responses and functions extending well beyond the peroxisome itself. A deeper understanding of the functions of peroxisomes and the mechanisms that control their plasticity could offer opportunities for exploiting changes in peroxisome abundance to control cellular function.
Introduction
Peroxisomes are spherical compartments delimited by a single phospholipid bilayer and are found distributed throughout the cytoplasm of most eukaryotic cells. In most cell types investigated to date, peroxisomes exhibit remarkable plasticity, responding to various environmental stimuli to alter their size and number per cell and their metabolic functions [1]. Peroxisomes are formed by two separate, and possibly complementary, biogenesis pathways: de novo budding from the endoplasmic reticulum (ER), and growth and division of existing peroxisomes [1,2]. They possess a posttranslational protein translocation system, termed the peroxisomal importomer [3], which imports exclusively fully folded, and sometimes oligomeric, protein complexes composed of enzymes destined for the peroxisomal matrix together with their peroxisome-targeting chaperone [4–6]. Peroxisomes are metabolically plastic, which is due in part to the enzyme-mediated production of, and protection from, reactive oxygen species (ROS) and the broad specificity in substrates these oxidative reactions confer [7]. Beyond their metabolic functions, and in alignment with an increasing recognition of the complexity and interconnectedness of various components of the cell, peroxisomes are increasingly being revealed as hubs or platforms for signaling in their own right, with roles critical for innate immunity, development and differentiation [8]. Therefore, the mechanisms controlling the plasticity of peroxisomes and the formation of signaling complexes on peroxisomes offer exciting avenues for research. In this review, we highlight recent findings from yeast and mammalian cells that reveal the coordinated control that gives rise to both the dynamic formation of peroxisomes and the signaling events carried out at the organelle.
Peroxisomes - Control at the level of transcription
Factors involved in the biogenesis and proliferation of peroxisomes have been well conserved during evolution [9], and particularly since the divergence of metazoan and fungal lineages some 1.5–1.2 billion years ago. PEX genes encode proteins called peroxins that facilitate the varied aspects of the peroxisome life cycle, including membrane protein targeting, matrix protein targeting and translocation, peroxisome division, peroxisome movement, and selected peroxisome turnover, or pexophagy. This conservation in cellular pathways regulating peroxisomal biogenesis extends to the underlying transcriptional response to environmental and metabolic signals that initiate peroxisome proliferation. Ligand-mediated regulation of genes coding for peroxisomal proteins in the budding yeast Saccharomyces cerevisiae starts with the fatty-acid-mediated activation of the oleate-activated transcription factor 1 and peroxisome induction pathway 2 (Oaf1/Pip2) heterodimer [10,11]. Upon its binding to a fatty acid, Oaf1 complexes with Pip2 to form a heterodimer, which binds to DNA sequences known as oleate response elements located in the upstream promoter regions of many peroxisomal genes, including PIP2 itself. Similarly, transcriptional regulation of peroxisomal genes in mammals was first discovered in rodent models where peroxisome proliferators such as fatty acids, but also hypolipidemic drugs, activate the peroxisome proliferator-activated receptor (PPAR) and retinoic acid receptor (RAR) family of nuclear receptors, leading to the upregulation of expression of genes encoding peroxisomal proteins and the proliferation of peroxisomes [12,13].
Closer examination of the kinetics of regulation of the Oaf1/Pip2 and PPAR/RAR heterodimers revealed that they function as asymmetric positive feedback loops, so named because ligand-mediated heterodimerization upregulates the expression of only one member of the heterodimer pair [14]. Asymmetric positive feedback is a core network motif and a prominent feature of many biomolecular regulatory systems, including systems involved in adipocyte differentiation, cholesterol homeostasis, myogenesis and cellular antiviral response [14]. Mathematical and experimental tests comparing asymmetric positive feedback, termed ASSURE for ASymmetric Self-UpREgulation, to a symmetric positive feedback (SPF) system where both regulators are upregulated upon activation revealed the ASSURE motif to be more robust. For example, the response time of ASSURE was robust to changes in ligand Kd and provided the cell with the ability to adapt to rapid changes in environmental conditions [14].
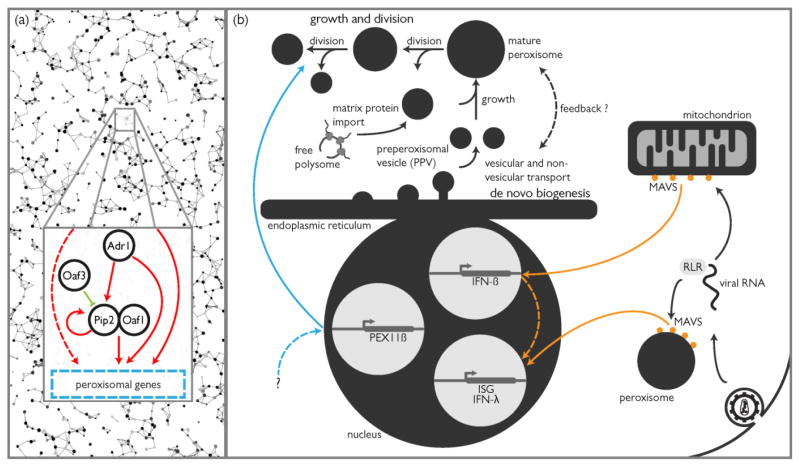
The Oaf1/Pip2 heterodimer does not regulate peroxisomal genes exclusively. Instead, this core regulatory motif functions within a larger regulatory network that coordinates peroxisome induction with many other activities (Figure 1a). For example, the network includes alcohol dehydrogenase regulator 1 (Adr1) [15,16], a global regulator of glucose-repressed genes [17,18], and Oaf3, a negative regulator of Oaf1/Pip2 that serves to dampen the cellular response to their autoactivation [19]. Yet, even these four regulators are insufficient to explain all of the transcriptional control of peroxisome biogenesis; whereas most of the metabolic machinery and fatty acid transporters required for the β-oxidation of oleate respond dramatically to oleate and are controlled by Oaf1/Pip2, of the 34 PEX genes in yeast, only PEX5, PEX6, PEX11 and PEX18 are similarly responsive [20].
Figure 1.
(a) Schematic representation of the transcriptional response of S. cerevisiae to oleic acid. The core Pip2-Oaf1 asymmetric network motif is nested within a larger network that includes the feedforward action of Adr1 and the dampening action of Oaf3. (b) Depending on organism, cell type, or even environmental condition, the proliferation of peroxisomes is achieved through two routes: growth and division of existing peroxisomes, and de novo biogenesis of preperoxisomal vesicles from the ER that mature via fusion into mature organelles. In both cases, peroxisomal matrix enzymes and membrane proteins are acquired by the peroxisome posttranslationally, either directly from the cytosol (some membrane proteins and all matrix proteins) or from the ER (some membrane proteins). Control over either pathway can occur through signaling and transcriptional responses to environmental stimuli or intracellular signaling and crosstalk from other organelles. In this example, an innate immune response is initiated by the detection of viral RNA by RLR that initiates a signal cascade by activating MAVS on mitochondria and peroxisomes. Mitochondrial MAVS activates a type I interferon response resulting in the upregulation of IFN-β expression and, via autocrine signaling, the upregulation of interferon-stimulated genes (ISG). Peroxisomal MAVS activates a type III interferon response that directly upregulates the expression of ISG and IFN-λ. Through an as yet unidentified mechanism, peroxisomal genes including PEX11β are also upregulated, resulting in the proliferation of peroxisomes.
A network model has been developed that predicts the transcriptional response of yeast to oleate exposure on a genome-wide level [20]. This network was generated by integrating data from a compendium of 1516 publicly available mRNA expression datasets, known network interactions, and common promoter regions for known transcription factor binding motifs. The topology of the network was then explored with a linear regression algorithm that made predictions for the ability of any given transcription factor to control the expression of a given bicluster, i.e. a collection of genes with coherent expression profiles, for a given environmental condition. Focusing on the transcriptional control of peroxisome biogenesis, predicted regulators were validated in more focused studies that included time course analysis of transcriptional responses, analysis of regulator deletion data, ChIP-chip data and transcription factor binding motif data. This analysis revealed complex transcriptional networks that coordinate transcriptional activities across the genome.
Given the complexity of transcriptional control of peroxisome proliferation in yeast, it is perhaps not surprising that mechanisms to control peroxisome numbers in mammalian cells are not well understood. For example, evidence for a role of the eponymously labeled PPARs in regulating the transcription of peroxisomal genes in humans is lacking [13]. The upstream promoter elements of human peroxisomal genes lack canonical PPAR-binding elements, and the evidence for the transcriptional regulation of peroxisomal genes by PPARs is indirect. For example, ChIP-chip analysis of PPARα chromatin binding in response to treatment with agonist found enrichment for the promoter region of the gene for the peroxisomal matrix enzyme acyl-CoA oxidase but not for PEX genes [21]. In a study of the molecular underpinnings of scarring alopecia, it was shown that loss of peroxisomes correlated with decreased expression of PPARγ and that treatment with PPARγ agonists induced the expression of genes for metabolic enzymes known to localize to peroxisomes [22].
The signaling networks controlling both biogenesis (perhaps directly) and transcription are also a means for cells to coordinate peroxisomes with various other cellular activities [23–25]. Components of these networks have been revealed in yeast, and similarly to transcriptional networks, they have proven highly complex, suggesting that cells invest considerable resources to control peroxisome number, while retaining the capacity to rapidly change peroxisome abundance. For example, a study that modeled organelle biogenesis mechanisms on organelle variance–the fluctuation in organelle number from cell to cell–concluded that peroxisomes in yeast switch from a de novo biogenesis mechanism to one primarily reliant on fission when yeast were transferred from a glucose-rich to a fatty-acid-rich environment [26]. This observation raises the intriguing possibility that cells respond appropriately to environmental signals through direct signaling and transcriptional mechanisms that act to control peroxisome production [1]. The temporal differences in responsiveness or molecular composition of differently produced peroxisomes could contribute to peroxisome heterogeneity and influence peroxisome controlled signaling dynamics.
Peroxisomes are not autonomous
Mitochondria and peroxisomes share proteins, some metabolic functions, and communicate through vesicular transport [27,28]. Indeed, it has been known for quite some time that cross-talk between mitochondria and peroxisomes manifests at the transcriptional level [29]. However, how the regulation of peroxisomes is integrated with other organelles, especially the ER, is poorly understood. Further evidence of the intimate association between peroxisomes, the ER, and mitochondria comes from a recent systems biology screen in yeast that uncovered an association between the ER and mitochondrion encounter structure (ERMES) and peroxisomes [30]. Peroxisomes themselves are intimately associated physically with mitochondria [31,32] and can be formed from the ER [33–35]. Furthermore, there are implied, but also direct, observations of cross-talk and feedback systems that function to coordinate peroxisome biogenesis and proliferation with the responses of other organelles. Here, we highlight two recent examples of coordination between peroxisomes and other organelles in innate immune responses and the metabolic requirements of different cell types.
Peroxisomes and the innate immune response
In response to intracellular pathogens, such as viruses and invading bacteria, RIG-I like receptors (RLRs) induce the expression of interferon-stimulated genes (ISGs) whose protein products carry out an innate immune response [36]. Type I interferons (IFN), such as IFNα or IFNβ, are secreted by cells for paracrine and autocrine signaling through the interferon α/β receptor (IFNAR), which activates the JAK/STAT pathway to amplify the innate immune response and also regulate a humoral response to the infectious disease [37]. In contrast to the Toll-like receptors, which detect the presence of pathogens extracellularly or from within endosomes, most RLRs are RNA helicases that detect viral or bacterial nucleic acids in the cytosol [38]. Upon detection, these RLRs recruit signaling components that activate transcription factors, such as NF-κB and interferon regulatory factors (IRFs). Different subsets of RLRs detect different types of nucleic acid, and this has been suggested to enable pathogen-specific immune responses. Adaptor proteins, such as mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, Cardif, or VISA), recruit activated RLRs and, through a process of aggregation, in turn activate transcription factors such as IRF3 [39].
In a pioneering study [40], Dixit and colleagues investigated the subcellular localization of the adaptor protein MAVS upon infection of cells with reovirus or vesicular stomatitis virus (VSV). They showed that MAVS was found on peroxisomes in addition to being localized to mitochondria (Figure 1b). To dissect the organelle-specific function of MAVS, organelle-specific targeting sequences were appended to MAVS, directing it exclusively to mitochondria, peroxisomes, or the cytosol. Peroxisome-targeted MAVS stimulated the expression of ISGs but not type I interferons. Analyzing the dynamics of signaling from peroxisomes versus signaling from mitochondria revealed that peroxisome-based signaling resulted in an immediate, but transient, response [40]. In contrast, mitochondrial signaling led to a sustained response in line with a type I interferon response that upregulates IFNβ and results in autocrine and paracrine signaling. As might be expected, signaling from both organelles was shown to be necessary for a maximum immune response, but the nature of the cross-talk and feedback mechanisms implied by this observation was not explored further. However, using mice infected with VSV, which is known to interfere with the IFN response initiated by mitochondria, the authors were able to show that peroxisome-MAVS-mediated upregulation of ISGs was achieved through IRF1 and IRF3 [40].
In a subsequent study, the role of peroxisomes in innate responses to intracellular infection was expanded to include Sendai virus, dengue virus and the bacterial pathogen, Listeria monocytogenes [41]. This study also showed that peroxisome-based RLR signaling activates the Jak-STAT pathway regulated by Jak2, leading to expression of IFN-λ, further suggesting that peroxisome-based innate responses activate type III interferons that are important for the innate immune response of mucosal surfaces. Surprisingly, epithelial cells of the intestine, liver and lung showed elevated numbers of peroxisomes, and the abundance of peroxisomes could be correlated with the efficiency of the innate response of these tissues [41]. Furthermore, Pex11β, one of three Pex11 isoforms in humans that regulate the proliferation of peroxisomes, was upregulated in response to pathogen infection (Figure 1b). Finally, cell lines from Zellweger patients with mutations in Pex19 or Pex16 and which lack morphologically observable peroxisomes, exhibited dysregulated expression of ISGs.
Peroxisomes and the regulation of mTORC
A central regulator of cellular growth is the mammalian target of rapamycin complex 1 (mTORC1) [42]. mTORC1 responds to diverse inputs such as the availability of amino acids, glucose, insulin or other mitogens to regulate the switch between cell growth and autophagy. In mammals, mTORC1 activation corresponds with its translocation to the lysosome and association with a Ras-GTPase homolog termed Rheb [43]. Rheb, in turn, is active when bound to GTP and farnesylated [43]. In a surprising study, components of the tuberous sclerosis complex (TSC), which is a GTPase-activating-protein (GAP) of Rheb and therefore a negative regulator of mTORC1, were found to localize to peroxisomes [44]. The TSC is heterotrimer composed of TSC1, TSC2 and TBC1D7. Curiously, TSC2 was recruited to peroxisomes by the peroxisomal matrix protein receptor Pex5, whereas TSC1 was recruited by the peroxisomal membrane protein receptor Pex19. From the peroxisome, TSC1 and TSC2 regulate mTORC1 in response to ROS, suggesting that the peroxisome is a cellular sensor for ROS and provides input into the switch between cell growth and autophagy by negatively regulating the action of mTORC1 [44].
Perspectives
Peroxisome plasticity, whether it be in composition, choice of proliferation mechanism, or transcriptional control, uniquely positions peroxisomes for many diverse, distinct and unexpected roles. Their recently discovered role as a scaffold for MAVS in innate signaling is perhaps not surprising, as there are many examples where molecular complex formation exploits cellular structures as spatial beacons. However, what is remarkable is that the shift in distribution of MAVS between mitochondria and peroxisomes confers signaling dynamics and specificity to the output, including transcriptional regulation. Similar spatiotemporal control and transcriptional regulation may apply to mTORC signaling. If one considers the plasticity of peroxisomes, it is not difficult to imagine additional control mechanisms that further impact signaling specificities and dynamics in different cell types or in the same cell exposed to different conditions.
Despite the evident functional connections between peroxisomes and mitochondria, it is rare for studies investigating the functions of mitochondria or other organelles to explore how peroxisomes contribute to the underlying phenotype of interest. The importance of this point was demonstrated recently in yeast where an increase in chronological lifespan that was initially attributed to an inhibition of mitochondrial division was actually the result of an inhibition of peroxisomal division [45]. Because the role of peroxisomes, whether it is in metabolism, innate immunity or longevity, is intimately intertwined with that of other organelles, further untangling the mechanisms of its regulation will require systems approaches [1].
In addition to the novel roles discussed here, peroxisomes have well established roles in protection of the nervous system and in detoxification in the liver and kidney [7]. Given this context-dependent plasticity, it is unlikely that the mechanisms of peroxisome-mediated regulation of cellular phenotypes will be consistent between all cell types. Rather, it is likely that the underlying network motifs that lead to these emergent properties will be similar and subsequently elaborated for each specific circumstance [46]. Discovery of new spatial and temporal network motifs, such as the recently described analog-to-digital conversion that occurs when epidermal growth factor receptor-mediated signal transduction is internalized [47], is ongoing, and looking for them within the context of peroxisome biology is an exciting avenue of pursuit.
Acknowledgments
F.D.M. is a Postdoctoral Fellow of the Canadian Institutes of Health Research (CIHR). Research in the Rachubinski laboratory is supported by grants 9208 and 53326 from the CIHR. Research in the Aitchison laboratory is supported by grants P50 GM076547, U54 GM103511, U01 GM098256, and R01 GM112108 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hettema EH, Erdmann R, van der Klei I, Veenhuis M. Evolving models for peroxisome biogenesis. Curr Opin Cell Biol. 2014;29:25–30. doi: 10.1016/j.ceb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meinecke M, Cizmowski C, Schliebs W, Kruger V, Beck S, Wagner R, Erdmann R. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12:273–277. doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- 4.Glover JR, Andrews DW, Rachubinski RA. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc Natl Acad Sci USA. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Titorenko VI, Nicaud JM, Wang H, Chan H, Rachubinski RA. Acyl-CoA oxidase is imported as a heteropentameric, cofactor-containing complex into peroxisomes of Yarrowia lipolytica. J Cell Biol. 2002;156:481–494. doi: 10.1083/jcb.200111075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mast FD, Fagarasanu A, Knoblach B, Rachubinski RA. Peroxisome biogenesis: something old, something new, something borrowed. Physiology. 2010;25:347–356. doi: 10.1152/physiol.00025.2010. [DOI] [PubMed] [Google Scholar]
- 8.Titorenko VI, Rachubinski RA. The peroxisome: orchestrating important developmental decisions from inside the cell. J Cell Biol. 2004;164:641–645. doi: 10.1083/jcb.200312081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schluter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol Biol Evol. 2006;23:838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- 10.Baumgartner U, Hamilton B, Piskacek M, Ruis H, Rottensteiner H. Functional analysis of the Zn2Cys6 transcription factors Oaf1p and Pip2p. Different roles in fatty acid induction of β-oxidation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:22208–22216. doi: 10.1074/jbc.274.32.22208. [DOI] [PubMed] [Google Scholar]
- 11.Phelps C, Gburcik V, Suslova E, Dudek P, Forafonov F, Bot N, MacLean M, Fagan RJ, Picard D. Fungi and animals may share a common ancestor to nuclear receptors. Proc Natl Acad Sci USA. 2006;103:7077–7081. doi: 10.1073/pnas.0510080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 13.Misra P, Reddy JK. Peroxisome proliferator-activated receptor-α activation and excess energy burning in hepatocarcinogenesis. Biochimie. 2014;98:63–74. doi: 10.1016/j.biochi.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 14*.Ratushny AV, Saleem RA, Sitko K, Ramsey SA, Aitchison JD. Asymmetric positive feedback loops reliably control biological responses. Mol Syst Biol. 2012;8:577. doi: 10.1038/msb.2012.10. In this study modeling-guided experimentation was used to examine the properties of prevalent transcriptional regulatory network motifs. Asymmetry in positive feedback systems confers a competitive advantage by increasing robustness, precision and responsiveness to environmental stimuli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rottensteiner H, Wabnegger L, Erdmann R, Hamilton B, Ruis H, Hartig A, Gurvitz A. Saccharomyces cerevisiae PIP2 mediating oleic acid induction and peroxisome proliferation is regulated by Adr1p and Pip2p-Oaf1p. J Biol Chem. 2003;278:27605–27611. doi: 10.1074/jbc.M304097200. [DOI] [PubMed] [Google Scholar]
- 16.Young ET, Kacherovsky N, Van Riper K. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J Biol Chem. 2002;277:38095–38103. doi: 10.1074/jbc.M206158200. [DOI] [PubMed] [Google Scholar]
- 17.Tachibana C, Yoo JY, Tagne JB, Kacherovsky N, Lee TI, Young ET. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25:2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JJ, Miller LR, Kreisberg R, Vazquez L, Wan Y, Aitchison JD. Environment-responsive transcription factors bind subtelomeric elements and regulate gene silencing. Mol Syst Biol. 2011;7:455. doi: 10.1038/msb.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratushny AV, Ramsey SA, Roda O, Wan Y, Smith JJ, Aitchison JD. Control of transcriptional variability by overlapping feed-forward regulatory motifs. Biophys J. 2008;95:3715–3723. doi: 10.1529/biophysj.108.134064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danziger SA, Ratushny AV, Smith JJ, Saleem RA, Wan Y, Arens CE, Armstrong AM, Sitko K, Chen WM, Chiang JH, et al. Molecular mechanisms of system responses to novel stimuli are predictable from public data. Nucleic Acids Res. 2014;42:1442–1460. doi: 10.1093/nar/gkt938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Meer DL, Degenhardt T, Vaisanen S, de Groot PJ, Heinaniemi M, de Vries SC, Muller M, Carlberg C, Kersten S. Profiling of promoter occupancy by PPARα in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 2010;38:2839–2850. doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karnik P, Tekeste Z, McCormick TS, Gilliam AC, Price VH, Cooper KD, Mirmirani P. Hair follicle stem cell-specific PPARγ deletion causes scarring alopecia. J Invest Dermatol. 2009;129:1243–1257. doi: 10.1038/jid.2008.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saleem RA, Rogers RS, Ratushny AV, Dilworth DJ, Shannon PT, Shteynberg D, Wan Y, Moritz RL, Nesvizhskii AI, Rachubinski RA, et al. Integrated phosphoproteomics analysis of a signaling network governing nutrient response and peroxisome induction. Mol Cell Proteomics. 2010;9:2076–2088. doi: 10.1074/mcp.M000116-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saleem RA, Long-O’Donnell R, Dilworth DJ, Armstrong AM, Jamakhandi AP, Wan Y, Knijnenburg TA, Niemisto A, Boyle J, Rachubinski RA, et al. Genome-wide analysis of effectors of peroxisome biogenesis. PLoS One. 2010;5:e11953. doi: 10.1371/journal.pone.0011953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saleem RA, Knoblach B, Mast FD, Smith JJ, Boyle J, Dobson CM, Long-O’Donnell R, Rachubinski RA, Aitchison JD. Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis. J Cell Biol. 2008;181:281–292. doi: 10.1083/jcb.200710009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Mukherji S, O’Shea EK. Mechanisms of organelle biogenesis govern stochastic fluctuations in organelle abundance. Elife. 2014;3:e02678. doi: 10.7554/eLife.02678. This study used modeling to explore the dynamics of organelle biogenesis and proposes that peroxisome biogenesis can be switched between de novo biogenesis from the ER and the fission of existing peroxisomes in response to environmental stimuli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-Navarro MA, McBride HM. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 28.Braschi E, Goyon V, Zunino R, Mohanty A, Xu L, McBride HM. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr Biol. 2010;20:1310–1315. doi: 10.1016/j.cub.2010.05.066. [DOI] [PubMed] [Google Scholar]
- 29.Epstein CB, Waddle JA, Hale Wt, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattiazzi Usaj M, Brloznik M, Kaferle P, Zitnik M, Wolinski H, Leitner F, Kohlwein SD, Zupan B, Petrovic U. Genome-wide localization study of yeast Pex11 identifies peroxisome-mitochondria interactions through the ERMES complex. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohanty A, McBride HM. Emerging roles of mitochondria in the evolution, biogenesis, and function of peroxisomes. Front Physiol. 2013;4:268. doi: 10.3389/fphys.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen Y, Klug YA, Dimitrov L, Erez Z, Chuartzman SG, Elinger D, Yofe I, Soliman K, Gartner J, Thoms S, et al. Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol Biosyst. 2014;10:1742–1748. doi: 10.1039/c4mb00001c. [DOI] [PubMed] [Google Scholar]
- 33.Agrawal G, Joshi S, Subramani S. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2011;108:9113–9118. doi: 10.1073/pnas.1018749108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam SK, Yoda N, Schekman R. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2010;107:21523–21528. doi: 10.1073/pnas.1013397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Zand A, Gent J, Braakman I, Tabak HF. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 2012;149:397–409. doi: 10.1016/j.cell.2012.01.054. [DOI] [PubMed] [Google Scholar]
- 36.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 40**.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. This paper and the following one [41**] establish peroxisomes as a site for MAVS signaling in innate immune responses to pathogenic infection by promoting a type III interferon response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. This study identifies the peroxisome as a signaling organelle involved in the regulation of mTORC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefevre SD, Kumar S, van der Klei IJ. Inhibition of peroxisome fission, but not mitochondrial fission, increases yeast chronological lifespan. Cell Cycle. 2015 doi: 10.1080/15384101.2015.1029685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mast FD, Ratushny AV, Aitchison JD. Systems cell biology. J Cell Biol. 2014;206:695–706. doi: 10.1083/jcb.201405027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villasenor R, Nonaka H, Del Conte-Zerial P, Kalaidzidis Y, Zerial M. Regulation of EGFR signal transduction by analogue-to-digital conversion in endosomes. eLife. 2015;4 doi: 10.7554/eLife.06156. [DOI] [PMC free article] [PubMed] [Google Scholar]