Abstract
The evolution of canalized traits is a central question in evolutionary biology. Natural variation in highly conserved traits can provide clues about their evolutionary potential. Here we investigate natural variation in a conserved trait—even-skipped (eve) expression at the cellular blastoderm stage of embryonic development in Drosophila melanogaster. Expression of the pair-rule gene eve was quantitatively measured in three inbred lines derived from a natural population of D. melanogaster. One line showed marked differences in the spacing, amplitude and timing of formation of the characteristic seven-striped pattern over a fifty-minute period prior to the onset of gastrulation. Stripe 5 amplitude and the width of the interstripe between stripes 4 and 5 were both reduced in this line, while the interstripe distance between stripes 3 and 4 was increased. Engrailed expression in stage 10 embryos revealed a statistically significant increase in the length of parasegment 6 and a decrease in the length of parasegments 8 and 9. These changes are larger than those previously reported between D. melanogaster and D. pseudoobscura, two species that are thought to have diverged from a common ancestor over 25 million years ago. This line harbors a rare 448bp deletion in the first intron of knirps (kni). This finding suggested that reduced Kni levels caused the deviant eve expression, and indeed we observed lower levels of Kni protein at early cycle 14A in L2 compared to the other two lines. A second of the three lines displayed an approximately 20% greater level of expression for all seven eve stripes. The three lines are each viable and fertile, and none display a segmentation defect as adults, suggesting that early-acting variation in eve expression is ameliorated by developmental buffering mechanisms acting later in development. Canalization of the segmentation pathway may reduce the fitness consequences of genetic variation, thus allowing the persistence of mutations with unexpectedly strong gene expression phenotypes.
Keywords: Drosophila melanogaster, even-skipped, intraspecific variation, quantitative biology, deletion polymorphism, knirps
Introduction
In 1942, Conrad Waddington introduced the idea of canalization, which involves the conservation of phenotype in the presence of extensive genetic and environmental variation [Waddington, 1942]. The extent to which genetic variation can be buffered is currently unknown. Moreover, it is unclear if a canalized trait is maintained over evolution by phenotypically neutral mutations or a series of small compensatory phenotypic changes [Martinez et al., 2014, Bullaughey, 2011]. The conservation of gene expression driven by enhancers from highly diverged species [Hare et al., 2008, Fisher et al., 2006, Romano and Wray, 2003, Barrière et al., 2011, Ludwig et al., 1998] shows that functional conservation does not require sequence conservation, but these observations shed little light on the detailed process of evolutionary change which conserved the phenotype. Trans changes in the above process complicate matters further, and hence have received little attention. Natural variation acts on extant individuals, and therefore can reveal limits on developmental constraints and provide clues about the evolutionary potential of a conserved trait. Here we investigate natural variation in a conserved trait, the formation of the seven-striped pattern of even-skipped (eve) RNA expression at the blastoderm stage of embryonic development in Drosophila melanogaster.
eve is one of the most well-characterized genes in D. melanogaster. It is essential for the formation of segments [Nüsslein-Volhard and Wieschaus, 1980], and while classified as a pair-rule gene, it has the unique property that null mutations lead to a complete abolition of segments [Macdonald et al., 1986]. The segmentation function of eve is executed in the blastoderm stage of embryonic development. Transcripts can be reliably detected by cleavage cycle 12 and protein by cleavage cycle 13. After the 13th nuclear division, protein and RNA expression refine from a single broad domain to a characteristic pattern of seven transverse stripes [Surkova et al., 2008, Fowlkes et al., 2011]. These dynamic changes in expression are a consequence of the activation of eve expression by broadly distributed maternal factors and its repression by more localized domains of zygotic gap gene expression [Stanojevic et al., 1991, Reinitz and Sharp, 1995]. eve is necessary for the correct initiation of the expression of the segment polarity gene engrailed (en), which stably demarcates the future parasegmental borders [Gilbert, 2003].
The seven-stripe pattern of eve before gastrulation is conserved in the suborder Brachycera [Davis and Patel, 2002], albeit with different subcellular localizations in different species [Bullock et al., 2004]. Within the genus Drosophila, the dynamic pattern of eve expression in D. pseudoobscura is very similar to that of D. melanogaster despite the fact that these species diverged 25–55 million years ago [Fowlkes et al., 2011, Richards et al., 2005].
The individual enhancers of eve are very well characterized in terms of function [Harding et al., 1989, Goto et al., 1989, Small et al., 1992, 1993, 1996, Arnosti et al., 1996, Fujioka et al., 1999, Janssens et al., 2006, Kim et al., 2013]. A 15kb segment of DNA (−6.4kb to +8.6kb of eve) can provide a normal segmentation phenotype and rescue an eve null mutant to hatching [Fujioka et al., 1999, 2002]. If the stripe 2 enhancer is deleted from the construct, eve stripe 2 is greatly reduced in amplitude with a short parasegment 3 and vestigial En stripe 4, a lethal phenotype [Ludwig et al., 2005]. Other stripes have effects on survival that are marked but less severe. Transforming eve null flies with the eve locus bearing a deletion of the 4+6 enhancer results in viable flies missing certain abdominal segments [Fujioka et al., 2002]. The viability of these transformants may be a consequence of residual 4+6 expression driven from outside the classical enhancer, but it is equally possible that non-terminal abdominal segments are not absolutely required for viability.
Previous studies on the intraspecific variation of eve expression involved quantitative measurements of stripe placement but not amplitude in three lines of flies with differing egg size. eve expression among these lines scaled with egg size, demonstrating that intraspecific egg size variation can be compensated for by expression variation [Lott et al., 2007]. This point was reinforced by an experiment to artificially select for small or large embryos. In this case, the proportionality of eve stripe spacing to embryo length was not preserved, providing crucial evidence that eve stripe placement can be variable [Miles et al., 2011]. In both examples the phenotype under study was egg size, a complex genetic trait, with its consequences for eve expression a secondary effect. For this reason, the phenotypic alterations in these lines have not yet been fully mapped to sequence. In contrast, there exist two well characterized small deletions of the D. melanogaster eve cis-regulatory region in natural populations, but evidence linking them to phenotypic changes is ambiguous [Palsson et al., 2014].
In this work we characterize intraspecific variation in eve expression in terms of quantitative expression level, position, and timing. Our analysis of the three lines, while providing only a glimpse of the full range of eve phenotypic variation, demonstrates that significant quantitative variation exists, and can be provisionally assigned to specific changes in sequence.
Results
The dynamics of eve expression in three D. melanogaster lines
For reasons unrelated to the findings reported here, we examined eve expression in three lines from the Drosophila Genetic Reference Panel (DGRP) [Mackay et al., 2012]. These were RAL-437 (denoted as L1 in this work), RAL-502 (L2), and RAL-365 (L5). We unexpectedly found that eve expression from L2 differed from that of the other two lines much more strongly than the previously reported eve expression differences between D. melanogaster and D. pseudoobscura [Fowlkes et al., 2011], motivating the analysis presented here.
Image analysis of eve gene expression was carried out from 2D confocal scans of laterally-oriented embryos fluorescently stained for nuclei, Eve protein, and eve RNA. These scans were transformed into quantitative data at cellular resolution by image segmentation. The embryos were categorized into eight different time classes (T1–T8), each about 6.5 minutes long during cell cycle 14A [Surkova et al., 2008]. Background staining was removed, and 1D data from the central 10% of dorso-ventral values was used for the detection of quantitative features of the expression pattern [Janssens et al., 2005]. The analysis of eve expression in this paper was based on RNA expression. Protein expression resembles that of RNA with a lag of one time class (Supplementary Figure S1; S2).
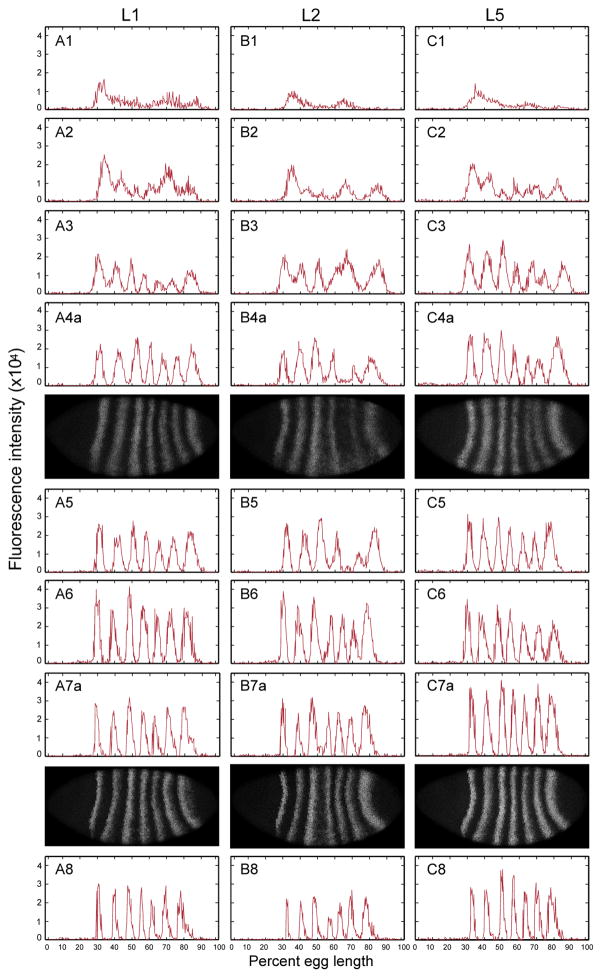
Figure 1 and Supplementary Figure S1 show the dynamics of eve RNA expression from typical individual embryos at each time class in the three lines during cell cycle 14A. The expression dynamics of eve in L2 are visibly different from the other two lines. For example, at T3 the presumptive stripes 4–6 are a single expression domain in L2 while L1 and L5 have already formed separate stripes. In T4 and T5, when L1 and L5 have formed 7 stripes, there is little or no stripe 5 eve expression in L2. By T6, stripe 5 eve expression in L2 rises to the levels seen in other stripes. Moreover, the 3/4 interstripe is wider in L2 than in the other two lines, with stripe 3 expressed at higher levels in L2.
Figure 1.
1D eve RNA expression from single embryo in cycle 14 from 3 lines. Each column shows eve RNA expression in one line (A: L1; B: L2; C: L5). The Arabic number following the letter represents the corresponding time class, T1 to T8. Each row shows the eve 1D RNA expression in its time class; for T4 and T7 embryos, both 1D expression (indicated by ‘a’) and the corresponding RNA fluorescent in situ embryo image (indicated by ‘b’) is shown. The expression is from 10% stripe from the middle of laterally-oriented embryo after segmentation and background removal.
We made a more precise analysis of differences in the eve expression patterns of the three lines by performing feature detection on the expression patterns. We performed the analysis on embryos from T4 to T8, because this is the period when reproducible features of the expression pattern can be characterized [Surkova et al., 2008]. We made pairwise comparisons on stripe morphology features using the Wilcoxon rank sum test. We performed inference as to whether gene expression differed with respect to each class of feature as follows. There are n = mk individual Wilcoxon tests, performed for each class of feature, where m is the number of such features (e.g. there are 6 interstripes) and k is the number of time classes they are measured in, which in this application is always 5 (T4–T8). After the tests, we did Bonferroni correction of n tests given the significance value of 0.05.
We considered three types of features: stripe height, interstripe width and stripe width (Supplementary Figure S3). Stripe height is measured from the peak of the stripe to the minimum of the adjacent interstripe [Ludwig et al., 2011, Manu et al., 2013]. Stripes 2 through 6 have two such height measurements, corresponding to the two adjacent interstripes; stripes 1 and 7 each has one. The A–P position at a point where expression was midway between stripe peak and interstripe minimum was taken to be location of the stripe border. We generated 6 interstripe distances and 7 stripe width from the 14 border positions.
We first calculated the absolute stripe height in the three lines (Supplementary Figure S4), and found that L5 had higher expression overall than the other two lines. The Wilcoxon rank sum test for pooled L5 stripe height was significantly different from L1 and L2 (in both cases, p < 2.2×10−16), while L1 and L2 show more modest differences (p = 0.045), which supports our observation (boxplot shown in Supplementary Figure S5). We also did pairwise Wilcoxon rank sum tests for the stripe height at each time class and each position (Supplementary Table S1). As expected, most differences arose from the higher overall expression in L5. Specifically, for stripe 2 (positions 2 and 3 in the table), L5 is expressed at a significantly higher level than L1 and L2. Notwithstanding this fact, the overall pattern of expression in L5 appeared very similar to L1 by visual inspection (Supplementary Figure S4). In order to compare the features for individual stripes more closely, we normalized stripe height in the three lines by dividing by the mean fluorescence intensity in that line in a given temporal class to detect changes in specific features. After normalizing stripe height, as expected, the most significant differences arose from L2 with respect to L1 and L5 in more specific features (Supplementary Figure S4, Supplementary Table S2).
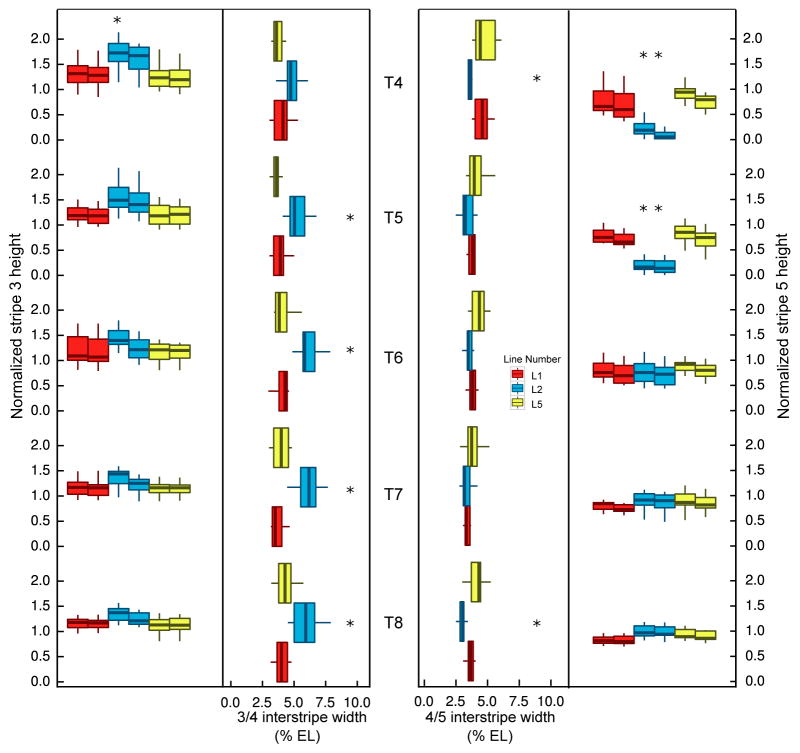
The major result from individual tests of quantitative features (Figure 2; Supplementary Tables S2–S4) was that L2 has a delay of maturation in stripe 5, with an increase of the 3/4 interstripe width and a decrease of the 4/5 interstripe width as a consequence. Specifically, for normalized stripe height, stripe 3 was significantly higher in L2 than L1 or L5 in T4, while stripe 5 was lower in both T4 and T5 but reached similar levels as the other two by T6. The interstripe distance between stripes 3 and 4 in L2 was significantly wider in T5–T8, and that between 4 and 5 is narrower at T4 and T8 compared to the other two lines (Supplementary Figure S4). Overall, the dynamic eve pattern in L1 and L5 resembled that seen in the lab stock Oregon R and Canton S [Surkova et al., 2008, Fowlkes et al., 2011], with that in L2 deviating from this pattern. These results indicate that eve expression differs among the individual lines.
Figure 2.
Quantification of normalized stripe height and interstripe width. Left to right: normalized stripe 3 height, 3/4 interstripe width, 4/5 interstripe width and normalized stripe 5 height from T4 to T8. If a feature is significantly different in two pairwise tests after Bonferroni correction, an asterisk is marked on top of the shared line for that feature. Two boxes represent each height, the one on the left shows the anterior height of the stripe, while the right shows the posterior height of the stripe. Outliers in boxplot are not shown.
Previous studies show that egg length varies within species [Houchmandzadeh et al., 2002, Gregor et al., 2007, Lott et al., 2007, Manu et al., 2009, Miles et al., 2011]. Therefore, we measured embryo size for the three lines from confocal-scanned images. Our results show that embryos from L2 have reduced AP axis length and increased DV axis length compared to the other two lines (Supplementary Figure S6). This change of embryo size in L2 could be related to the change of eve expression observed in L2, but we did not further address this possible relationship in this work.
Downstream effect of the eve pattern
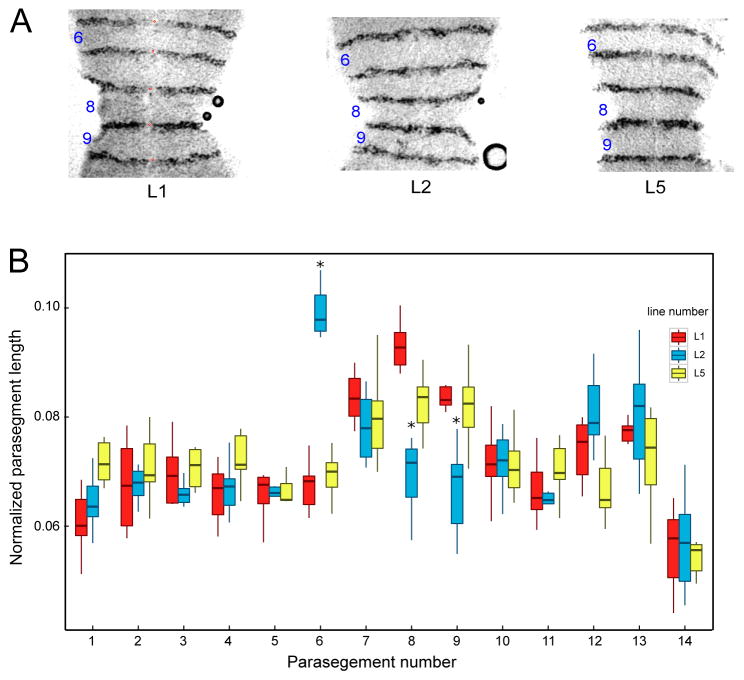
We sought to measure the functional effects of these expression differences by assaying expression of eve’s functional target, en, which is a marker of parasegment boundaries. Stage 10 embryos were stained for En protein (Figure 3A), and parasegment length was measured as the distance between the anterior margins of two successive En stripes, normalized to the sum of all such lengths. The results of these measurements (Figure 3B) showed that parasegment 6 is significantly longer while parasegments 8 and 9 are shorter in L2 compared to the two other lines. The alternation of eve stripe widths in L2 thus has specific functional consequences.
Figure 3.
Normalized parasegment lengths. A) Images of En expression in parasegments 6–9 of stage 10 dissected embryos from the three lines, as indicated. The red dots mark the anterior of the En stripes demarcating the parasegment, These fiducial marks were used to calculate the length of each parasegment. (B) shows the quantification of the normalized parasegment distance for each line. Significant differences of one line from the other two are marked with an asterisk on the boxplot. Outliers in boxplot are not shown.
Cause of altered expression in L2
We next explored the genetic basis of eve expression in L2. Given the marked alterations in stripe 5 expression, we first compared stripe 5 enhancer sequences [Gallo et al., 2011]. Sequence alignment (Supplementary Figure S7) revealed 5 SNPs in stripe 5 enhancer among the three lines, one of which is unique in L5. The remaining four SNPs are shared between L2 and L5 but differ with L1. Thus, there is no candidate SNP in the L2 stripe 5 enhancer to potentially account for the aberrant stripe 5 phenotype. For this reason, we consider it unlikely that the SNPs in the stripe 5 enhancer underlie the observed expression changes in L2.
In the absence of obvious differences in the stripe 5 enhancer, we checked the full eve sequence and the coding and flanking 2kb regions of plausible trans-regulators among the three lines (Supplementary Table S5) for large deletions (30 bp minimum length) that is unique in L2 [Karolchik et al., 2014]. By visualizing the alignment in clustalx [Larkin et al., 2007], we found large deletions or missing data in the DGRP sequences for eve, giant (gt), knirps (kni) and runt. Direct experimental checks by PCR revealed that of the four putative deletions, only the one in kni is real (Supplementary Table S7. We used BLAT [Kent, 2002] to map back the sequence and found that the deletion is 448bp in length and lies at the 5′ end of the first intron of kni (Supplementary Figure S8).
Several lines of evidence suggest that the deletion in the kni intron in L2 could be the cause of the altered eve expression. First, in kni mutants, only eve stripes 4–6 are abolished, while stripes 1–3 and 7 are present in T5–T8 at the protein level, although their amplitude is reduced [Surkova et al., 2013, Figure 1]. Second, in kni mutant embryos stripe 3 has a larger amplitude than other stripes. Finally, in kni heterozygotes, stripe 5 is completely absent until T5 but reaches a level close to wild type level by the onset of gastrulation [Frasch and Levine, 1987, Figure 5d], [Surkova et al., 2013, Figure S7]. L2 has altered expression in a subset of the stripe 4–6 region, increased amplitude of stripe 3 expression early, and a reduction in stripe 5 expression before T6, similar to that seen in kni/+. The deletion in L2 comprises 30% of the kni +1 enhancer, which drives expression in both the head and putative abdomen [Schroeder et al., 2004]. The L2 deletion also removes sequences in the kni proximal shadow enhancer [Perry et al., 2011]. These observations led us to predict that Kni expression would be reduced in L2.
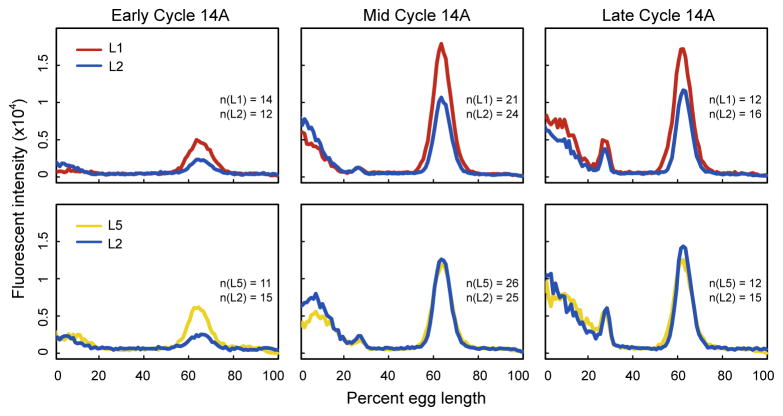
We tested our prediction by performing antibody staining of Kni in two independent experiments: L1 versus L2 and L2 versus L5. In each of these experiments, we compared embryos in early (T2–T3), middle (T4–T6), and late (T7–T8) cleavage cycle 14A. In early cleavage cycle 14A Kni expression is consistently lower in L2 than either L1 or L5 (Wilcoxon rank sum test of peak height between 45%–80% AP position gives p = 0.0001 for L1 versus L2, and p = 1.8×10−6 for L2 versus L5; see Figure 4). In mid to late cycle 14A, L2 and L5 show indistinguishable levels of Kni expression, but L1 gives slightly higher expression than the other two lines. There exist multiple lines of evidence that the time of formation of eve stripes is determined by rising levels of repressive gap gene products [Stanojevic et al., 1991, Small et al., 1992, Reinitz and Sharp, 1995, Fujioka et al., 1999, Janssens et al., 2006, Surkova et al., 2008], and hence it is extremely likely that the kni deletion in L2 is responsible for the eve phenotype we observe.
Figure 4.
Knirps expression in three lines. Average 1D Kni protein expression in three lines determined by two antibody staining experiments: L1 versus L2 and L2 versus L5 (upper and lower panel respectively). Expression from the middle 10% of D–V coordinates of laterally oriented embryos is shown. The early cycle 14A temporal class comprises T2 and T3, the mid cycle 14A temporal class comprises T4, T5 and T6, and the late cycle 14A temporal class comprises T7 and T8. Axes are as labeled; the numbers of embryos imaged in each time class are shown.
Only L2 bears this deletion among the DGRP collection of approximately 200 lines. In the core DPGP2 lines [Pool et al., 2012], which consist of lines originating from African and a few European populations, the full deletion is absent, although two small deletions (7–8 bp each) segregate in these populations. The 448bp deletion in L2 may, therefore, be a low frequency variant with limited geographic range.
Discussion
The three lines displayed three distinct eve stripe expression patterns. This variability was large enough to depart from the evolutionarily conserved reference pattern. The overall higher expression of eve in L5 could be caused by genetic variants controlling overall eve expression. In cis, these might involve variants in the eve autoregularoty elements or some unknown chromatin regulation mechanism at the whole-locus level. In trans, the cause could be the consequence of expression level changes of other genes in the network. In the absence of a clear genomic signiture, further experimentation will be required to elucidate the cause of increased expression. In addition to the higher overall expression of eve in L5, we observed in L2 the absence of eve stripe 5 formation at T4, the time at which stripe 5 typically becomes visible [Surkova et al., 2008]. Although the seven-stripe pattern is restored in the blastoderm embryo by T6, alternations in interstripe distances persist well into gastrulation. These differences are consistent from embryo to embryo, and are supported by relatively conservative statistical methods that do not assume normality and take into account multiple pairwise tests of significance. The unusual early eve expression pattern has measurable consequences later in the pattern formation cascade, as evidenced by a significant change in the spacing of En stripes. The altered pattern of eve expression in L2 strongly resembles that seen in a kni heterozygote [Surkova et al., 2013], and we found that this line bears a 448 bp deletion in its kni intron, which removes part of the kni +1 enhancer. This mutation appears to be rare in D. melanogaster, as it is not present in any other DGRP line, nor in any DPGP2 line, the majority of which are African lines representing the ancestral range of that species.
We predicted this deletion would lead to reduced kni expression, and verified this prediction by quantitative measurement of Kni expression. These observations strongly suggest the contribution of the trans background to eve stripe variation.
Odd numbered en stripes are expressed at the anterior margin of eve stripes, while the even numbered stripes are expressed at the anterior margins of ftz stripes [Hughes and Krause, 2001]. The expansion of the 3/4 eve interstripe leads to an increase of the distance between en stripes 6 and 7. The position of en stripe 6 relative to 5 and 7 depends on ftz expression [Hughes and Krause, 2001, Fujioka et al., 1995] and may indicate that ftz stripes are also altered in L2. Similarly, the reduction in the length of the eve 4/5 early interstripe causes a decrease in the total distance between the eve-dependent en stripes 7 and 9. The effect of the reduction of stripe 5 expression on parasegment 9 (the distance between en stripes 9 and 10) is reminiscent of that seen in enhancer deletions, but much milder [Fujioka et al., 2002]. This effect is also consistent with observations of ftz expression in kni heterozygotes [Carroll et al., 1988, Figure 3F]. In these embryos, ftz stripes 4 and 5 are markedly closer, consistent with reduction in distance between en stripes 8 and 10, while the distance between ftz stripes 3 and 4 increases, implying an increase of distance between en stripes 6 and 8.
In kni and Kr heterozygotes, altered gap and pair-rule expression are largely corrected by gastrulation [Surkova et al., 2013]. In L2 we did observe an En expression phenotype attributable to eve misexpression, but adults do not display any obvious phenotypic defect, and the line is viable and fertile. In bicoid copy number variants, the expansion of the head region in flies with extra bicoid copies is corrected by differential cell apoptosis [Busturia and Lawrence, 1994, Namba et al., 1997]. Multiple layers of canalization may buffer misexpression of early genes in the segmentation pathway, making variations more common than previously suspected based on the strong evolutionarily conservation of the pathway. Canalization theory predicts such variation in strongly buffered pathways [Meiklejohn and Hartl, 2002].
Evolutionary Implications
The most surprising result reported here is that phenotypic variation in eve expression within D. melanogaster is of larger magnitude than previously-reported inter-species variation between D. melanogaster and D. pseudoobsecura. We believe eve expression is under relatively strong stabilizing selection, and therefore it is generally conserved across relatively large evolutionary distance. However, the large variation within species indicates that conservation is not complete, and that there is a large potential for expression to change. There must be limits to natural variation, however, and the fact that the kni deletion is a rare variant suggests that it is deleterious in nature. The similarity of the L2 eve phenotype to that of kni heterozygote mutant embryos, which are viable, suggests that the permissive threshold may be around half of wild type expression. In natural populations, the kni deletion variant will be present almost entirely in heterozygotes because it is rare, and we expect that the phenotypic effect in heterozygotes will be subtler than the homozygous effects measured in L2.
Examples of cis and trans coevolution are pervasive [Barrière and Ruvinsky, 2014, Gordon and Ruvinsky, 2012, Ludwig et al., 2005], and binding sites turnover, rearrangement, and change of spacing within enhancers is rampant for phenotypically conserved traits. Both may be manifestations of compensatory evolution. There are also examples of developmental system drift [True and Haag, 2001], in which the underlying genetic network changes over evolutionary time while maintaining a specific phenotype. However, no detailed mechanism has been proposed for that phenomenon. Our work suggests that canalization of mutant phenotypes compresses the width of their phenotypic distribution. This reduces the consequences of genetic variance, allowing otherwise dramatic mutations, such as the partial loss of the kni +1 enhancer, to segregate in natural populations for a long enough time without being eliminated by selection to allow other compensatory mutations to occur. A computational model of enhancer evolution under stabilizing selection [Bullaughey, 2011] predicts rapid turnover of binding sites, and in many cases deleterious mutations persist before positive selection takes place. Buffering mechanisms will greatly reduce the fitness cost of a deleterious mutation, and therefore greatly increase the rate of compensatory evolution [Durrett and Schmidt, 2008]. Canalization, compensatory evolution, and developmental system drift may be multiple consequences of stabilizing selection acting to maintain the fidelity of developmental processes.
Variation in spatio-temporal patterns of gene expression in conserved developmental systems, as described in this work, is likely to be widespread as a consequence of buffering mechanisms that mitigate their developmental consequences on adult fitness. We note that strongly deleterious mutations, though rare in their individual frequency, are numerically abundant (though absent in the inbred DGRP lines). The kni deletion mutant may be an example of a non-lethal deleterious allele exhibiting a dramatic molecular phenotype. Deleterious mutations with molecular phenotype are expected to be much more common in natural populations than in the DGRP lines. We might also anticipate finding some of these variants to be common in populations if they co-occur with compensatory mutations. The continued development of models of transcription, such as [Kim et al., 2013], will be useful for predicting the effects of natural variation on gene expression.
Materials and Methods
Fly culture, embryo collection and fixation
Flies were grown and embryos were collected at 25°C. For eve in situ hybridization and antibody staining, embryos were collected after 1.5 hours, and aged for another 2 hours. Fixation was performed as described [Kosman et al., 2004] with a fixation time of 25 minutes. En antibody staining was performed on embryos aged for 8 hours and fixed for 10 minutes.
Fluorescent in situ hybridization and antibody staining
Fluorescent in situ hybridization followed the published protocol [Kosman et al., 2004]. FITC-labeled eve antisense RNA probe was generated from p48-X1.4 [Macdonald et al., 1986] using SP6 polymerase. Following hybridization, embryos were incubated with rabbit anti-FITC (1:1000) and Guinea Pig (1:1000) anti-Eve antibody [Azpiazu and Frasch, 1993]. After washing, embryos were incubated in Alexa Fluor 647 Goat Anti-Rabbit IgG (Life Technologies, 1:1000) and Alexa Fluor 555 Goat Anti-Guinea Pig IgG (Life Technologies, 1:1000). Nuclei were stained with DAPI (Life Technologies Cat. No. P36935). En protein is stained with mouse MAb 4D9 antibody (1:3) followed by Goat Anti-Mouse IgG-HRP (1:300) [Patel, 1994]. For Kni staining, embryos were incubated with anti-Kni Guinea Pig antibody (1:1000), followed by Alexa Fluor 555 Goat Anti-Guinea Pig IgG (1:1000) and anti-Eve Rabbit (1:2000) followed by Alexa Fluor 647 Goat Anti-Rabbit IgG (1:1000).
Imaging
Fluorescence data was acquired on a Leica SP5 confocal microscope. Gain was set to produce several saturated pixels after averaging, and offset was set so that approximately half of the background pixels outside the embryo displayed zero intensity and half non-zero intensity. This procedure was carried out with the 5 brightest embryos for each line, and the setting for the brightest line was used to standardize all data collection. Images were taken from the surface of lateral embryos, using a 20X apo objective (HC PL APO 20x/0.70NA lens [dry]). The images for eve are 12 bits per pixel and have 8 line averages. Note that 12 bit images are converted to 16 bit during processing, so that the fluorescent intensity is of 16 bit range. Five z-sections of 0.5 μm each were acquired for laterally-oriented embryos. From these 5 z-sections, the three that best traverse the layer of blastoderm nuclei that is close to cover slide were selected for further processing. A DIC image of the middle of the embryo was acquired to visualize membrane invagination to aid in time classification. Images for Kni staining were acquired under the same microscope, with 8 bit depth (converted to 16 bit showing in graph), two z-sections which are 1 μm apart.
Images of En staining were acquired using Zeiss Axioskop light microscope.
Time classification
Time classification of younger (T1–T3) embryos was performed by inspection of protein expression patterns as described [Surkova et al., 2008]. Older (T4–T8) embryos were categorized by the degree of cell membrane invagination. We noticed the membrane at the ventral side is more mature than the dorsal side, and we based on our membrane invagination on the ventral side membrane. The time classification by membrane invagination is quite robust, while the early patterns are not as reliable. For T1–T3 embryos, we noticed that characteristic stages of the RNA pattern occur about one time class earlier than the same stage of the protein pattern. This fact was useful in the classification of the altered patterns seen in some lines. T1 embryos could always be unambiguously classified by protein pattern. The RNA patterns of these embryos were indicative of the expected T2 protein pattern of a particular line. Continuation of this procedure provided consistent and reliable temporal classification for all lines.
Feature detection
For observations of eve and Kni expression, segmentation, background removal, and the extraction of data from the central 10% of dorso-ventral positional values were performed as described [Surkova et al., 2008]. Embryos stained for Kni were registered using Eve stripes as described [Surkova et al., 2008]. For investigations of eve expression, cubic splines were used to detect extrema and borders of the stripes for time class T4 to T8, when all seven eve stripes are detectable [Ludwig et al., 2011]. A border is defined to be the position with expression midway between that of a stripe peak and interstripe. Each stripe, except 1 and 7, has an anterior and posterior height defined by the difference between expression at the stripe peak and the adjacent interstripe. The width of a stripe was taken to be the distance between its borders in percent egg length (% EL), with 0% at the anterior pole. A small number of embryos that were detected to have fewer or more than the full set of 13 extrema were included in the analysis after manual addition or removal of extraneous features. The number of embryos collected for each time class is summarized in Supplementary Table S6.
Measurement of En parasegments
Points were marked at the midline of the anterior of each En stripe using imageJ [Schneider et al., 2012] (See the red dots in Figure 3A), and parasegment lengths were measured between these points. The normalized parasegment length was taken to be the length of each parasegment divided by the sum of length from all the parasegments in that embryo.
Measurement of embryo size
In the data processing step, we generate an outline of the entire embryo with the length in pixels of the major and minor axes. These are converted to microns using metadata from confocal imaging.
PCR primers
For validation of deletions for
eve: forward:GGTCGCTTGGAGAAGGAGTT reverse: CACACCCAGTCCGGTATAGC
gt: forward: TGGCACAAGAGCTCGATGTT reverse: TAAATGCAGGGGGTTCCGAC
kni: forward: CCTAAGTGTGAGCGAGCACA reverse: TGAGAAAACGTGCAGCAACG
runt: forward:ACATGACCTACGGCTATGCG reverse: TAATTTTTGCCCGCTTGCCG
Supplementary Material
Acknowledgments
We thank Chun Wai Kwan, Ah-Ram Kim, Hilde Janssens, Kenneth Barr, Vytas Bindokas, Stephen Small, Maira Arruda Cardoso, Ziyue Gao and Mengyu Xu for useful discussions and help. The work is supported by NIH grant NIH RO1 OD010936 (formerly RR07801), and the University of Chicago. Imaging was performed at the University of Chicago Integrated Light Microscopy Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. tinman and bagpipe: Two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes and Development. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Barrière A, Ruvinsky I. Pervasuve divergence of transcriptional gene regulation in Caenorhabditis Nematodes. PLoS Genetics. 2014;10:e1004435. doi: 10.1371/journal.pgen.1004435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A, Gordon KL, Ruvinsky I. Distinct functional constraints partition sequence conservation in a cis-regulatory element. PLoS Genetics. 2011;7:e1002095. doi: 10.1371/journal.pgen.1002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullaughey K. Changes in selective effects over time facilitate turnover of enhancer sequences. Genetics. 2011;187:567–582. doi: 10.1534/genetics.110.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Stauber M, Prell A, Hughes JR, Ish-Horowicz D, Schmidt-Ott U. Differential cytoplasmic mRNA localisation adjusts pair-rule transcription factor activity to cytoarchitecture in dipteran evolution. Development. 2004;131:4251–4261. doi: 10.1242/dev.01289. [DOI] [PubMed] [Google Scholar]
- Busturia A, Lawrence PA. Regulation of cell number in Drosophila. Nature. 1994;370:561–563. doi: 10.1038/370561a0. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Laughon A, Thalley BS. Expression, function and regulation of the hairy segmentation protein in the Drosophila embryo. Genes and Development. 1988;2:883–890. doi: 10.1101/gad.2.7.883. [DOI] [PubMed] [Google Scholar]
- Davis GK, Patel NH. Short, long, and beyond: Molecular and embryological approaches to insect segmentation. Annual Review of Entomology. 2002;47:669–699. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- Durrett R, Schmidt D. Waiting for two mutations: with applications to regulatory sequence evolution and the limits of Darwinian evolution. Genetics. 2008;180:1501–1509. doi: 10.1534/genetics.107.082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–9. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fowlkes CC, Eckenrode KB, Bragdon MD, Meyer M, Wunderlich Z, Simirenko L, et al. A conserved developmental patterning network produces quantitatively different output in multiple species of Drosophila. PLoS Genetics. 2011;7 doi: 10.1371/journal.pgen.1002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Levine M. Complementary patterns of even-skipped and fushi-tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes and Development. 1987;1:981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Yusibova GL, Patel NH, Brown SJ, Jaynes JB. The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries. Development. 2002;129:4411–21. doi: 10.1242/dev.129.19.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo SM, Gerrard DT, Miner D, Simich M, Des Soye B, Bergman CM, Halfon MS. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Research. 2011;39:D118–23. doi: 10.1093/nar/gkq999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. 7. Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- Gordon KL, Ruvinsky I. Tempo and Mode in evolution of transcriptional regulation. PLoS Genetics. 2012;8:e1002432. doi: 10.1371/journal.pgen.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, MacDonald P, Maniatis T. Early and late periodic patterns of even-skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K, Hoey T, Warrior R, Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. The EMBO Journal. 1989;8:1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosopila despite lack of sequence conservation. PLoS Genetics. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- Hughes SC, Krause HM. Establishment and maintenance of parasegmental compartments. Development. 2001;128:1109–18. doi: 10.1242/dev.128.7.1109. [DOI] [PubMed] [Google Scholar]
- Janssens H, Kosman D, Vanario-Alonso CE, Jaeger J, Samsonova M, Reinitz J. A high-throughput method for quantifying gene expression data from early Drosophila embryos. Development, Genes and Evolution. 2005;215:374–381. doi: 10.1007/s00427-005-0484-y. [DOI] [PubMed] [Google Scholar]
- Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nature Genetics. 2006;38:1159–1165. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Clawson H, Cline MS, Diekhans M, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Research. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. Blat–the blast-like alignment tool. Genome Research. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AR, Martinez C, Ionides J, Ramos AF, Ludwig MZ, Ogawa N, Sharp DH, Reinitz J. Rearrangements of 2.5 kilobases of noncoding DNA from the Drosophila even-skipped locus define predictive rules of genomic cis-regulatory logic. PLoS Genetics. 2013;9:e1003243. doi: 10.1371/journal.pgen.1003243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lott SE, Kreitman M, Palsson A, Alekseeva E, Ludwig MZ. Canalization of segmentation and its evolution in Drosophila. Proceedings of the National Academy of Sciences USA. 2007;104:10926–10931. doi: 10.1073/pnas.0701359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development. 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biology. 2005;3(4):e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Manu, Kittler R, White KP, Kreitman M. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genetics. 2011;7:e1002364. doi: 10.1371/journal.pgen.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky V, Janssens H, Kim A, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, Reinitz J. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biology. 2009;7:e1000049. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Ludwig Michael Z, Kreitman Martin. Sex-specific pattern formation during early Drosophila development. Genetics. 2013;194:163–173. doi: 10.1534/genetics.112.148205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Carlos, Kim Ah-Ram, Rest Joshua S, Ludwig Michael, Kreitman Martin, White Kevin, Reinitz John. Ancestral resurrection of the Drosophila S2E enhancer reveals accessible evolutionary paths through compensatory change. Molecular Biology and Evolution. 2014;31:903–916. doi: 10.1093/molbev/msu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL. A single mode of canalization. Trends in Ecology and Evolution. 2002;17(10):468–473. [Google Scholar]
- Miles CM, Lott SE, Luengo Hendriks CL, Ludwig MZ, Manu, Williams CL, Kreitman M. Artificial selection on egg size perturbs early pattern formation in Drosophila melanogaster. Evolution. 2011;65:33–42. doi: 10.1111/j.1558-5646.2010.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba R, Pazdera TM, Cerrone RL, Minden JS. Drosophila embryonic pattern repair: how embryos respond to bicoid dosage alternation. Development. 1997;124:1393–1403. doi: 10.1242/dev.124.7.1393. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Palsson A, Wesolowska N, Reynisdóttir S, Ludwig MZ, Kreitman M. Naturally occurring deletions of hunchback binding sites in the even-skipped stripe 3+7 enhancer. PLoS ONE. 2014;9:e91924. doi: 10.1371/journal.pone.0091924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods in Cell Biology. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Perry M, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, Duchen P, Emerson JJ, Saelao P, Begun DJ, Langley CH. Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genetics. 2012;8:e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinitz J, Sharp DH. Mechanism of eve stripe formation. Mechanisms of Development. 1995;49:133–158. doi: 10.1016/0925-4773(94)00310-j. [DOI] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Research. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano LA, Wray GA. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130:4187–4199. doi: 10.1242/dev.00611. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, Schindelin J, Arganda-Carreras I, et al. NIH image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan HQ, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biology. 2004;2:e271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. The EMBO Journal. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Arnosti DN, Levine M. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development. 1993;119:767–772. [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Developmental Biology. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Surkova S, Kosman D, Kozlov K, Manu, Myasnikova E, Samsonova A, Spirov A, Vanario-Alonso CE, Samsonova M, Reinitz J. Characterization of the Drosophila segment determination morphome. Developmental Biology. 2008;313(2):844–862. doi: 10.1016/j.ydbio.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkova S, Golubkova E, Manu, Panok L, Mamon L, Reinitz J, Samsonova M. Quantitative dynamics and increased variability of segmentation gene expression in the Drosophila Krüppel and knirps mutants. Developmental Biology. 2013;376:99–112. doi: 10.1016/j.ydbio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evolution & Development. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.