Abstract
Tropomyosin is an elongated α-helical coiled-coil that binds to seven consecutive actin subunits along the long-pitch helix of actin filaments. Once bound, tropomyosin polymerizes end-to-end and both stabilizes F-actin and regulates access of various actin binding proteins including myosin in muscle and non-muscle cells. Single tropomyosin molecules bind weakly to F-actin with millimolar Kd, whereas the end-to-end linked tropomyosin associates with about a one thousand-fold greater affinity. Despite years of study, the assembly mechanism of tropomyosin onto actin filaments remains unclear. In the current study, we used total internal reflection fluorescence (TIRF) microscopy to directly monitor the cooperative binding of fluorescently labeled tropomyosin molecules to phalloidin-stabilized actin filaments. We find that tropomyosin molecules assemble from multiple growth sites following random low affinity binding of single molecules to actin. As the length of the tropomyosin chain increases, the probability of detachment decreases, which leads to further chain growth. Tropomyosin chain extension is linearly dependent on tropomyosin concentration, occurring at approximately 100 monomers/(μM*s). The random tropomyosin binding to F-actin leads to discontinuous end-to-end association where gaps in the chain continuity smaller than the required seven sequential actin monomers are available. Direct observation of tropomyosin detachment revealed the number of gaps in actin-bound tropomyosin, the time course of gap annealing, and the eventual filament saturation process.
Indexing: TIRF Microscopy, Single-molecule detection, Actin-binding proteins, Muscle Thin Filament Regulation
Introduction
Tropomyosin is a double-stranded alpha-helical coiled coil that binds seven consecutive actin subunits along the actin long-pitch helix. Tropomyosin molecules interact end-to-end to form continuous chains that run the length of the actin filament. In skeletal and cardiac muscle, calcium binding to troponin causes a shift in tropomyosin’s position along actin, exposing myosin binding sites and thereby facilitating muscle contraction (Lehman, Craig et al. 1994). In smooth muscle cells, tropomyosin plays a more modulatory role on actin to regulate contraction (Wang and Coluccio 2010). Moreover, tropomyosin in all cells plays multiple roles on actin in such diverse processes as cellular motility, nuclear division, and embryogenesis (Gunning, Weinberger et al. 1997, Gunning, O’Neill et al. 2008). Not surprisingly, mutations in tropomyosin associated with ineffective actin-binding and faulty muscle thin filament regulation lead to a number of myopathies including congenital fiber type distortion, nemaline myopathy, distal arthrogryposis, and familial hypertrophic and dilated cardiomyopathies (Thierfelder, Watkins et al. 1994, Kee and Hardeman 2008, Lawlor, Dechene et al. 2010, Marston, Memo et al. 2013).
It is well-known that each tropomyosin coiled-coil monomer contains seven consensus pseudorepeats that interact with seven successive actin subunits along thin filaments (McLachlan and Stewart 1975, Singh and Hitchcock-DeGregori 2007). Therefore in order to form a continuous chain on actin filaments that is free of gaps, tropomyosin must bind end-to-end along the entire filament. In fact, unpolymerized single tropomyosin molecules bind extremely weakly to actin filaments with millimolar binding affinity (Ka ~ 3 × 103 M−1) (Wegner 1980, Weigt, Wegner et al. 1991, Tobacman 2008), while the polymerized tropomyosin chain is tightly bound with affinity in the micromolar range (Wegner 1980, Heald and Hitchcock-DeGregori 1988). A nucleation-polymerization binding model, proposed by Wegner and colleagues, attempted to explain how low affinity tropomyosin monomers assemble on actin to form a tightly bound chain (Wegner 1979, Wegner and Ruhnau 1988, Weigt, Wegner et al. 1991). Their model suggests that weak, transient monomer binding leads to the random formation and growth of multiple tropomyosin chains along the actin filament. At these binding sites, actin-bound tropomyosin monomers “link” together via end-to-end bonds and bind actin with a greater affinity for the filament. Additional monomer binding to these growing chains adds to and further extends the length of the tropomyosin chain (polymerization). However, the model failed to describe how gaps between randomly initiated chains were prevented or were annealed if the chains were separated from each other by less than the requisite seven actin subunits. Later, Monte Carlo simulations by Vilfan attempted to rationalize how long tropomyosin chains become dominant at the expense of shorter ones, resulting in the annealing of gaps on the actin filament (Vilfan 2001) in a stochastic mechanism later articulated by Holmes and Lehman (Holmes and Lehman 2008).
In the current study, we present the first direct observations of the early stages of skeletal and smooth muscle tropomyosin binding to actin filaments, and describe the underlying mechanism of assembly. Our data show that individual tropomyosin molecules bind randomly to actin. The probability of stable tropomyosin chain formation increases as the number of adjacently bound tropomyosin molecules increases. Since the binding of individual tropomyosin molecules appears to be random, most growing chains will be out of phase with respect to the underlying seven actin monomer template. Therefore, gaps will occur between growing chains, as is observed. We find that tropomyosin binding to actin filaments is a dynamic process of chain growth, reduction and occasional annihilation occurring primarily at the ends of chains viz. at gaps between chains. However, the decreasing number of gaps observed over time indicates a competitive sorting process that leads to near full filament coverage by tropomyosin as consistent with a stochastic sorting mechanism (Vilfan 2001, Holmes and Lehman 2008).
Results
Mechanism of Tropomyosin Binding
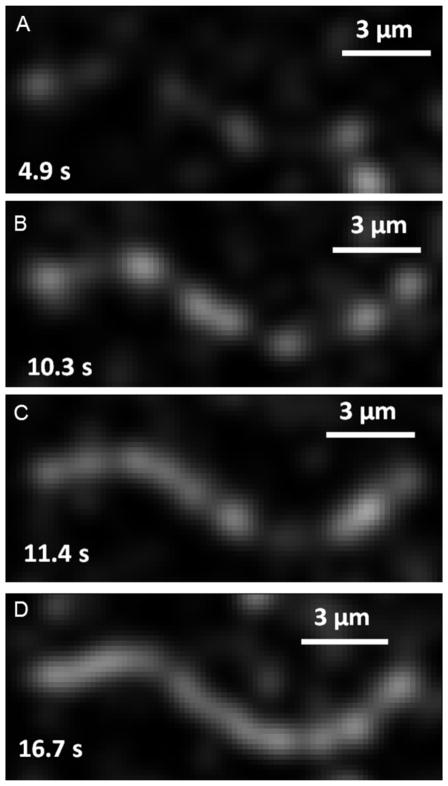
Using total internal reflection fluorescence microscopy, we followed the binding to and assembly of tropomyosin on surface immobilized actin filaments. Four representative images of the tropomyosin binding process are shown in Figure 1 along with the lapse between tropomyosin addition to the flow cell and image acquisition (also see Supplementary Video). The first image (Figure 1A) illustrates the initial binding of tropomyosin along the actin filament. Subsequent images (Figure 1B–1C) show increased fluorescence that extends from these initial sites of binding, while the final image (Figure 1D) shows an almost completely tropomyosin-decorated actin filament. Labeled tropomyosin patches were not observed to translate linearly along the actin filament, although small movements would be difficult to distinguish given the dynamic nature of tropomyosin chain ends and the pixel resolution of the camera (0.16 μm). The observations are consistent with a model where tropomyosin molecules initially bind with low affinity, while subsequent binding is bolstered by end-to-end tropomyosin binding and the corresponding increased affinity of oligomeric tropomyosin for actin.
Figure 1. Binding Mechanism of Tropomyosin.
Skeletal tropomyosin binding to F-actin at 75 mM [KCl]. (A) Tropomyosin binds with low affinity at random lattice sites along F-actin. (B–C) Adjacent monomer binding causes buildup of high affinity tropomyosin-bound sites followed by rapid chain elongation. (D) Chain elongation continues until near saturation of actin with tropomyosin.
Tropomyosin Chain Growth and Isolated Binding Rates
Because tropomyosin chain initiation shows no preferential distance from the filament ends, the binding of tropomyosin molecules appears to be a random process. Growth or shrinkage of the tropomyosin chain is expected to be stochastic and depend on the ratio of the binding and detachment rates of tropomyosin molecules. Therefore, the position of the ends of chains are in constant flux as monomers continually add to and detach from the chains’ ends. Although it is formally possible for tropomyosin molecules to bind to and detach from the middle of the growing chains, this is unlikely since assembly or disassembly of internal tropomyosins in the chain would involve the breaking of two instead of one end-to-end linkage from their defined binding sites on the surface of actin (Li, Tobacman et al. 2011).
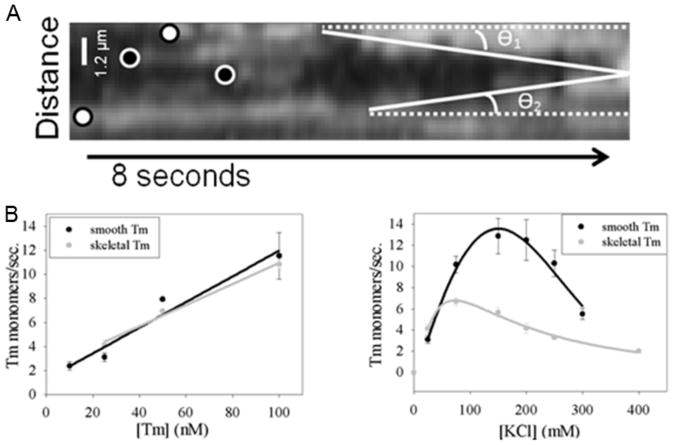
The rate of addition of tropomyosin monomers to surface-immobilized actin filaments was measured from kymographs, and the average growth rate (microns/second) was determined by tracing a straight line that measured the overall increase in chain length that occurred on time scales that ranged from tens of seconds to a few hundred seconds depending on binding conditions. A representative kymograph is shown in Figure 2A. Our TIRF microscopy assay allows tropomyosin binding rate (monomers/second) to be determined for a range of tropomyosin concentrations (Figure 2B, left) between 10 and 100 nM smooth (black) and skeletal (grey) muscle tropomyosin. Figure 2B illustrates the linear relationship between binding rate and tropomyosin concentration for both isoforms expected of a second-order binding reaction. The binding rates for skeletal and smooth muscle tropomyosin were 90 and 110 monomers/([μM]*s) respectively (p > 0.05).
Figure 2. Tropomyosin Elongation Rates and Binding Events.
(A) Representative kymograph illustrating how elongation rates and binding events were counted. Examples of binding events resulting in elongation are marked by white circles with black trim, while events that did not lead to elongation are marked with black circles and white trim. Two elongation rates are traced with solid white lines. (B) Rate of smooth (black lines) and skeletal (grey lines) muscle tropomyosin monomer addition versus tropomyosin concentration at 25 mM KCl buffer concentration (left). Rate of 25 nM smooth (black lines) and skeletal (grey lines) muscle tropomyosin monomer addition versus KCl concentration (right).
The binding rates observed here agree with previously computed rate constants modeled by Wegner (Wegner 1979) and are slightly higher than values (40 molecules/(μM*sec) estimated from fluorescence stopped flow and light scattering experiments (Wegner and Ruhnau 1988, Weigt, Wegner et al. 1991). The small differences between data sets are attributable, at least in part, to the ionic conditions, owing to the effects of both monovalent and divalent cations (Eaton, Kominz et al. 1975, Wegner 1979). It is important to note that binding rates represent an average rate at which monomers bind to, and extend, the length of the leading chain of tropomyosin, and experimental limitations prevent the determination of individual growth rates of chains on opposite sides of the filament. Therefore, we cannot verify if the two chains grow simultaneously or independently. However, due to tropomyosin’s high binding cooperativity (Hill, Mehegan et al. 1992, Lehrer, Golitsina et al. 1997, Tobacman 2008) and structural symmetry on actin filaments (Li, Tobacman et al. 2011, von der Ecken, Muller et al. 2014), it is reasonable to assume that we are observing the growth of a single chain and the growth rates of the two chains are nearly equivalent.
Initial binding of tropomyosin to actin was determined for both skeletal and smooth muscle tropomyosin by counting the number of binding events that occurred at previously unoccupied sites on actin. Fluorescent events caused by simple diffusion of labeled tropomyosin molecules into the detection area were discounted by including only molecules that remained bound for greater than one half second. The isolated binding rates of skeletal and smooth muscle tropomyosin were 0.52 ± 0.033 (n=22) and 0.57 ± 0.058 (n=20) molecules/(μM*s) respectively, and were not significantly different from each other. The near 200-fold decrease in isolated binding rates for skeletal and smooth muscle tropomyosin isoforms when compared to contiguous binding rates to an actin-bound tropomyosin chain agrees with previous equilibrium data, suggesting that single tropomyosin molecules bind weakly with low affinity (Wegner 1979, Wegner 1980, Weigt, Wegner et al. 1991).
Effect of Buffer Salt on Tropomyosin Binding
Binding rates of contiguously associated smooth and skeletal muscle tropomyosin were compared at varying KCl concentrations (Figure 2B, right). As previously reported, the relationship between binding rate and KCl concentration is complex (Eaton, Kominz et al. 1975). For skeletal muscle tropomyosin (Figure 2B, grey) the rate of binding to actin increases with [KCl] and reaches a maximum at 75 mM, after which it steadily decreases. These results are consistent with maximal binding of tropomyosin at ~100 mM KCl (Tanaka and Oosawa 1971, Eaton, Kominz et al. 1975) and with decreased binding of tropomyosin as monovalent ion concentration is lowered to 2 mM KCl (Kominz 1966, Eaton, Kominz et al. 1975). Presumably an optimal salt concentration is required to screen ionic interactions between actin and tropomyosin, whereas at high salt concentrations electrostatic contacts become compromised (Drabikowski and Nowak 1968, Hill, Mehegan et al. 1992, Li, Tobacman et al. 2011). The dependence of smooth muscle tropomyosin–actin binding rate on ionic strength (Figure 2B, black) is analogous; however the binding rate plateaus at 150 mM [KCl] instead of the 75 mM [KCl] optimum found for skeletal muscle tropomyosin. This difference is likely related to the well-known stronger end-to-end binding for smooth muscle tropomyosin compared to skeletal muscle tropomyosin (Sanders and Smillie 1984, Lehrer, Golitsina et al. 1997, Sousa, Cammarato et al. 2010). Thus, the direct visualization of tropomyosin binding to actin filaments quantifies the early stages of binding and reveals characteristics of the assembly process previously predicted from equilibrium binding data.
Probability of Sustained Chain Growth Increases with the Number of Tropomyosin Molecules Bound
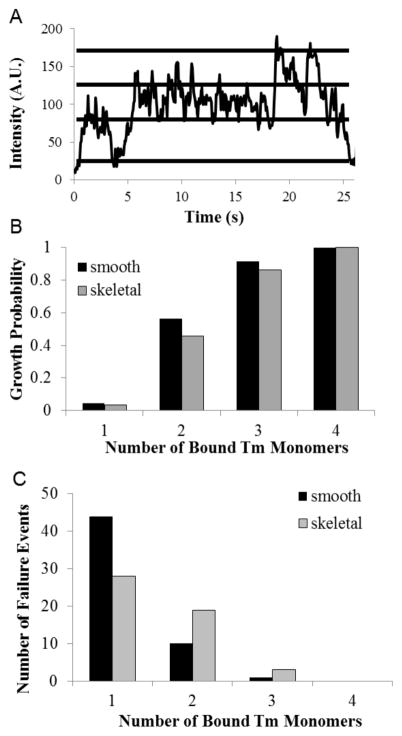
For each binding event determined from kymograph records (Figure 2) resulting in elongation of a nascent chain, a line was traced through the center of the binding event and the intensity plotted over time (Figure 3A). Given the fluorophore:tropomyosin labeling ratio (1.2:1), a single quantal intensity increase represented the binding of one tropomyosin molecule conjugated to a single fluorophore, while a double quantal intensity increase was judged to represent the binding of a single tropomyosin molecule conjugated to two fluorophores, one on each chain of tropomyosin. This interpretation was verified by measuring the percentage of double step events relative to total tropomyosin binding events, which was determined to be ~20 percent, as expected from our labeling ratio. Although we cannot completely rule out the possibility that two singly-labeled tropomyosin molecules bind actin within 50 milliseconds of each other (the frame rate of the camera), the statistical data combined with measured binding rates of tropomyosin on actin (1–10 molecules/second) suggest that this alternative is unlikely. Furthermore, the data suggest that within the spatial and temporal limits of the analysis defined by pixel size (160 nm) and the mean lifetime of the molecular fluorophore (< 3.5 s), we are observing the initial growth of a single chain. For example, only a maximum number of 4 successive, quantal intensity increases within a single pixel was observed. If tropomyosin binding occurred on both sides of the filament simultaneously, then 8 successive increases could theoretically occur. This suggests that in the initial stages of binding, one chain begins elongating prior to the second, perhaps due to a slightly more exposed actin binding site that accelerates formation of an isolated binding site followed by rapid, cooperative chain growth. Although it is possible that tropomyosin molecules simultaneously bind at isolated sites on both sides of the actin filament, we presume this scenario unlikely due to the high binding cooperativity of end-to-end linked tropomyosin molecules (Hill, Mehegan et al. 1992, Lehrer, Golitsina et al. 1997, Tobacman 2008) and significantly lower isolated binding rates relative to end-to-end tropomyosin binding (Supplemental Figure S1).
Figure 3. Detection of Single Molecule Binding Events.
(A) Representative plot of the intensity variation that occurs within a single binding event over time for a tropomyosin binding event. In this example the quantal variation is ~45 units. Stepwise intensity variations were used to determine the maximum number of tropomyosin monomers bound for each failure and elongation event. (B) The probability that tropomyosin chain elongation will occur following the binding of a given number of tropomyosin monomers. (C) Histograms of the number of failure events that occur when a given number of tropomyosin monomers bind successively to actin. Note the exponential decrease in failure events as more tropomyosin molecules bind.
Figure 3B shows the probability of chain elongation for skeletal and smooth muscle tropomyosin following the successive binding of one to four tropomyosin monomers. An elongation event occurred when binding at an isolated site was followed by continued polymerization of the tropomyosin chain, as indicated on the kymographs by a continuous line of fluorescence originating from the initial growth site and extending in the y-axis direction. The number of chain elongation events was divided by the sum of both the elongation events and binding events that failed to elongate, to parameterize the overall probability of elongation. The data indicate that when two monomers bind, an almost equal probability of chain elongation and failure (45%–55% growth probability) results, but when three or more bind, the probability of overall chain growth without detachment is enhanced (> 85% growth probability).
The number of tropomyosin failure events was also plotted as a function of bound monomers, and results for skeletal and smooth muscle tropomyosin compared (Figure 3C). The histogram shows a decrease in the number of failure events once single monomer binding is followed by a second binding event. The binding of three monomers shows a dramatically further decrease which rarely results in detachment of the tropomyosin chain (≤ 6% failure rate), while the binding of four invariably caused continued net chain growth. Comparison of failure rates for skeletal (41%) and smooth (18%) muscle isoforms (after 2 monomers bound) is consistent with previous indications of stronger end-to-end bond strength in smooth muscle tropomyosin (Lehrer, Golitsina et al. 1997, Coulton, Koka et al. 2008, Sousa, Cammarato et al. 2010).
Tropomyosin Gap Formation and Detachment from F-actin
As mentioned, a random initial association of tropomyosin with actin will likely result in the formation of tropomyosin-free regions on actin filaments. Our semi-quantitation predicts that, on average, 3 to 4 gaps would form per thin filament (Supplemental Figure S1). Whether or not gaps occur in situ in muscle is more difficult to evaluate, although they are not obvious (Matsumura and Lin 1982, Ohtsuki and Shiraishi 2002). In any case, the overall effect of a low gap number on the collective performance of muscle might be minimal given the low muscle myosin duty ratio on actin (Holmes and Geeves 2000, Rosenfeld, Xing et al. 2003). In contrast, gaps in tropomyosin strands may well occur and serve a functional role in non-muscle systems, where cooperative activation and relaxation of myosin ATPase may not be critical as suggested by others (Lees, Bach et al. 2011, Choi, Kim et al. 2012, Vindin and Gunning 2013). We therefore measured the number of gaps in non-muscle Tpm1.6cy (a.b.b.d) (Geeves, Hitchcock-DeGregori et al. 2014) (i.e. common name Tm2) tropomyosin chains, an isoform that has been shown to differentially regulate the access of other actin binding proteins (Dabrowska, Hinssen et al. 1996, Khaitlina, Fitz et al. 2013, Vindin and Gunning 2013).
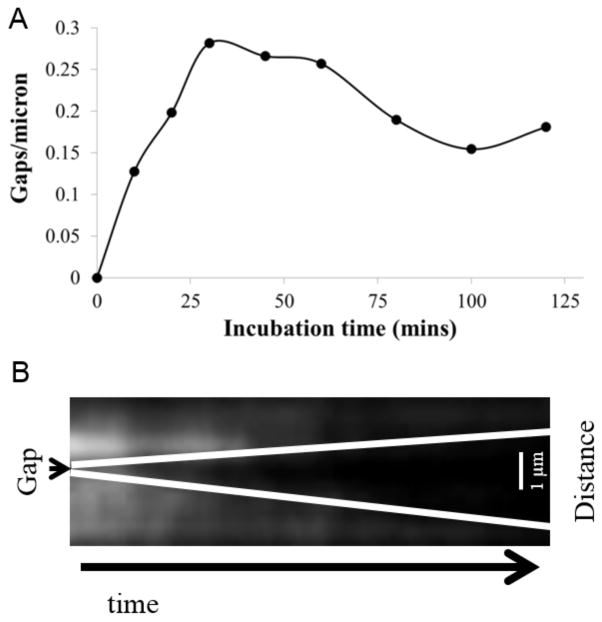
Because of resolution limitations imposed by the photon spread function of the fluorescence source, gaps present in a tropomyosin chain are not detectable following the initial phases of tropomyosin binding to actin filaments. However, since tropomyosin detachment is more likely from the ends than the middle of a chain, observation of what appears to be a new gap in a chain following rapid dilution will provide an estimate (Figure 4A) and location (Figure 4B) of preexisting but undetected tropomyosin gaps. We therefore exploited this phenomenon to measure the number of gaps in tropomyosin chains bound to F-actin by first mixing Cy3-labeled Tm2 with phalloidin-stabilized F-actin and monitoring tropomyosin detachment.
Figure 4. Time Dependence of Tropomyosin Gap Density and Observation of Detachment.
(A) Plot of gap density vs. tropomyosin-actin incubation time measured as explained in Materials and Methods. (B) Kymograph showing the detachment of tropomyosin from an actin filament. The total time elapsed is 57 seconds (x-axis) and length is 4 microns (y-axis). The average detachment rate of a tropomyosin molecule from an actin-bound chain was calculated to be approximately 0.33 molecules/second. Note the presence of a tropomyosin “gap” on the actin filament, and the directional detachment from that gap. This suggests that tropomyosin randomly binds along the actin filament, leaving gaps where there are not exactly seven actin monomers available to bind.
The kymograph in Figure 4B shows a tropomyosin gap in the middle of a Cy3-Tm2 labeled actin filament that grows directionally over time after rapid tropomyosin dilution, showing progressive tropomyosin detachment from the chains’ ends. To ensure that fluorescence loss is due to tropomyosin detachment and not photobleaching, we measured the Cy3 bleaching rate (Supplemental Figure S2) at the laser intensity used for the experiment, and fluorophore bleaching rate (τ ~ 69 sec.) was significantly longer than average tropomyosin detachment times observed in kymographs (~0.33 monomers/second) indicating that fluorescence loss was due to tropomyosin detachment and not Cy3 fluorophore bleaching. Gaps in the tropomyosin chain were counted for actin-tropomyosin incubation time points ranging between 10 and 120 minutes and normalized to total actin filament length (Figure 4A). The number of gaps increases with incubation time, followed by a plateau between 30 and 60 minutes, and then a slow decline until stabilizing at times over 80 minutes, consistent with a stochastic sorting process of tropomyosin chains on actin (Vilfan 2001, Holmes and Lehman 2008).
During the detachment experiments, it is likely that the detachment of tropomyosin occurs from a single chain on actin. Although allosteric cross-talk between actin strands induced by the detachment of a single tropomyosin may be possible, the distance between the binding location of tropomyosin on opposite sides of actin (Li, Tobacman et al. 2011, von der Ecken, Muller et al. 2014) suggests this possibility is unlikely. Given the ~30 second lapse between buffer washout and imaging, significant detachment of tropomyosin chains has already occurred, and therefore the observed tropomyosin detachment is likely to have been observed from multiple locations where only one chain is bound to actin.
Discussion
Tropomyosin binding to actin filaments is well-adapted to tropomyosin function. Tight global binding is essential for steric regulation of acto-myosin interaction; however, weak local binding is required to allow azimuthal movements to neutralize the steric hindrance in response to Ca2+-binding to troponin and myosin binding to actin during muscle activation. To accomplish this, individual tropomyosin molecules weakly bind to actin with extremely low affinity and thus can oscillate among regulatory states, while end-to-end bonds between neighboring tropomyosins result in a high collective affinity of tropomyosin on actin biased to one or another regulatory state by troponin or myosin interactions. The data presented here provide the first direct observation of the initial steps of tropomyosin binding to actin marked by the formation of multiple tropomyosin growth sites and chain elongation. Our data indicate that once two or three successively linked tropomyosin molecules bind to actin, the probability for tropomyosin chain growth greatly exceeds the probability of chain detachment. Because tropomyosin binds seven sequential actin monomers along the actin filament helix, a mechanism involving the random binding of tropomyosin along the length of the actin filament will produce gaps between polymerizing strands of tropomyosin that can be from 1 to 6 actin monomers in length. The observation that tropomyosin polymerization and depolymerization occur continuously suggests that tropomyosin–free gaps on actin must either anneal or continuously redistribute by a stochastic process.
Our data can be explained by a reaction scheme presented in Figure 5. Here, single tropomyosin molecules bind weakly to seven consecutive actin monomers at a rate, kAT+[Tm], and in the absence of end-to-end bonds detach at a rate of kAT−. Under such conditions, individual tropomyosin molecules transiently bind to the actin filament with low affinity. Tropomyosin end-to-end bonding rate to adjacent tropomyosin monomers on actin is given by kTT+. Whether a tropomyosin molecule detaches from the end of a growing chain or contributes to its elongation depends on the ratio of tropomyosin detachment, kTT−, and attachment, kTT+ of additional molecules at the ends of the growing tropomyosin chain. Our data show that once two or three tropomyosin molecules are bound, complete chain dissociation is improbable and elongation is favored (Figure 3). It is likely that detachment of tropomyosin molecules from a chain’s interior is low because detachment of centrally located tropomyosin molecules requires breakage of two end-to-end bonds instead of one. It follows that tropomyosin binding and assembly on an actin filament is predominantly observed at the ends of the tropomyosin polymer, with a constant flux that depends on actin-tropomyosin affinity, tropomyosin end-to-end bond strength, and free tropomyosin concentration.
Figure 5. Kinetic Binding Model of Tropomyosin Molecules to F-actin.
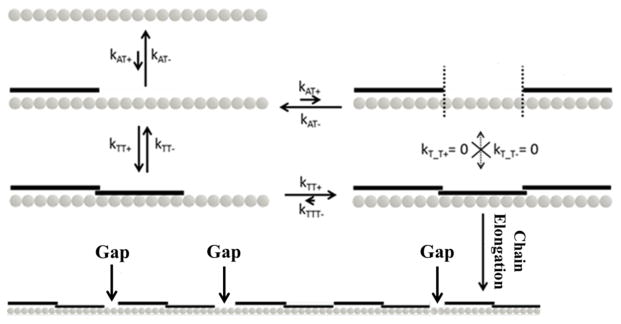
The relative quantities of various kinetic constants are represented by arrow lengths. A single tropomyosin molecule initially binds bare F-actin with low affinity. Once one binds (kAT+), a second can contiguously bind via end-to-end bond formation (kTT+), bind at a separate lattice site (kAT+), or fall off the filament (kAT−). Once two are contiguously bound, a monomer can detach (kTT−) or a third can contiguously add to the chain (kTT+). If three tropomyosin monomers contiguously bind, sustained chain growth is favored because it is unlikely that the rate of detachment from an end (kTTT−) or middle molecule (kT_T−) will exceed kTT+. The formation of multiple growth sites along the actin filament, and subsequent chain elongation from them, results in the buildup of gaps where there are not exactly seven actin monomers sandwiched between adjacent chains.
Since two separate strands of tropomyosin wrap continuously around the actin helix, the geometry of the binding surface must permit all actin binding sites equal accessibility to tropomyosin molecules. For visualization, it was necessary to use myosin as a surface linker in order to increase the binding rate of tropomyosin, presumably caused by an increase in binding affinity for myosin-bound F-actin (Eaton 1976), beyond that of the fluorophore bleaching rate. Although it is possible that this linker strategy may have an effect on the relative accessibility of actin binding sites based on spatial orientation, we do not anticipate any actin binding sites would be completely inaccessible to tropomyosin monomer binding based on structural data of the rigor myosin S1-actin complex and the length and flexibility of the S2 hinge region (Rayment, Rypniewski et al. 1993, Rayment and Holden 1994, Schmitz, Reedy et al. 1996, Koubassova and Tsaturyan 2002, Holmes, Schroder et al. 2004, Gundapaneni, Xu et al. 2005). The specific attachment of myosin to the cover slip surface is not known. However, in vitro motility is impaired when the proteolytic S1 fragment of myosin is directly attached to the surface (Hynes, Block et al. 1987) when compared to the attachment of whole or single headed myosin containing the coiled-coil rod domain (Tyska, Dupuis et al. 1999) suggesting that whole myosin attaches to the nitrocellulose surface via the rod or tail domain. Assuming the rod portion of the myosin molecule lays flats on the nitrocellulose surface and the average angle of rigor S1 to the actin filament is slightly less than 90° (Rayment, Rypniewski et al. 1993), then S2 need only be oriented at a 20–25 degree angle relative to the surface in order for the actin filament to be a minimum perpendicular distance of 40 nm away from the surface. Thus we do not anticipate that the experimental geometry would prevent the tropomyosin monomer access to actin-binding sites that directly face the surface. However, we cannot dismiss the possibility that tropomyosin dimers and polymers may experience steric hindrances to F-actin binding, which could contribute to the differences observed in binding rates between the skeletal and smooth muscle isoforms (Figure 2).
Opposing Effects of Tropomyosin End-to-End Bonds
The sequences of the skeletal and smooth muscle tropomyosin isoforms are conserved (~82% sequence homology (Girjes, Keriakous et al. 2002) with the largest variability occurring in pseudorepeat 7 (derived from exon 9) at the C-terminus of the molecule, a region responsible for end-to-end association with the N-terminus of a second tropomyosin molecule (Li, Mui et al. 2002, Greenfield, Huang et al. 2006, Frye, Klenchin et al. 2010, Rao, Rivera-Santiago et al. 2012), the ability to polymerize in solution, actin affinity, and troponin T1 binding (Coulton, Koka et al. 2008). Given the C-terminal sequence variation, it is not surprising that the end-to-end interaction will differ, which is supported by the differences in binding rates for skeletal and smooth muscle tropomyosin (Figure 2). Considerable experimental evidence based on multiple types of assessment suggest that end-to-end bond strength in smooth muscle tropomyosin is stronger than skeletal muscle tropomyosin, with smooth muscle tropomyosin having a greater propensity to form polymers in solution (Lehrer, Golitsina et al. 1997, Coulton, Koka et al. 2008, Sousa, Cammarato et al. 2010). The comparatively fast rate of smooth muscle tropomyosin binding to actin observed here at nearly all ionic strengths likely results from differences in tropomyosin end-to-end bond strength.
The increased end-to-end bond strength would tend to increase the association between the growing tropomyosin chain and actin. However, the presence of long polymeric chains in solution (Sousa, Cammarato et al. 2010), as a result of end-to-end binding of tropomyosin molecules, might oppose this effect due to the increased entropic cost of wrapping the elongated tropomyosin polymer around the actin filament. The interplay between these two competing effects likely defines the complex relationship between ionic strength and tropomyosin binding (Figure 2B, right), although we cannot dismiss the possibility that some actin binding sites are inaccessible to long tropomyosin molecules due to surface effects. Increasing the ionic strength of the buffer weakens tropomyosin end-to-end bonds and shifts the solution equilibrium for tropomyosin toward the monomer. This is consistent with an inverse relationship between solution salt concentration and tropomyosin polymerization in solutions of tropomyosin, where long polymers are incapable of binding to actin, while monomeric (or potentially dimeric) tropomyosin are binding-competent. The polymerization rate for smooth muscle tropomyosin maximizes at higher KCl concentration than skeletal muscle tropomyosin presumably because much of the smooth muscle tropomyosin is in a polymerized form in low buffer salt (Sousa, Cammarato et al. 2010).
Tropomyosin Gaps
The number of gaps formed during tropomyosin binding will depend on the number of growth sites formed during the initial binding of tropomyosin to actin. Large gaps greater than the seven requisite actin binding sites will rapidly fill between tropomyosin chains but smaller gaps require a relatively slow process of nascent tropomyosin chain detachment and reattachment. Here we quantified the number (Figure 4A) and locations (Figure 4B) of tropomyosin gaps by observation of tropomyosin detachment after rapid dilution and provided a measurement of gap number for several actin-tropomyosin incubation times (Figure 4A). Our data provide direct experimental support for models of stochastic binding and redistribution of tropomyosin chains (Vilfan 2001, Holmes and Lehman 2008).
Tropomyosin is thought of as a “gatekeeper” of actin filaments because it competitively regulates the binding of several actin-binding proteins (Holmes and Lehman 2008). Thus, depending on their binding characteristics, each of the over 40 non-muscle tropomyosins may differentially control intracellular function. For example, the non-muscle tropomyosin isoform, Tm2 is an important regulator for the binding of various ABP’s (Dabrowska, Hinssen et al. 1996, Khaitlina, Fitz et al. 2013, Vindin and Gunning 2013), and tropomyosin gap number might be one important determinant of ABP association.
Cells may exploit the presence of gaps to regulate the accessibility of actin-binding sites from other actin-binding proteins that remodel actin filament networks. For example, stress fibers are stable F-actin networks in the cytoskeleton that disassemble to sponsor motility. Following cellular locomotion, tropomyosin plays an essential role by binding actin to promote re-formation of F-actin stress fiber networks and preventing the binding of other actin depolymerizing proteins (Khaitlina, Fitz et al. 2013). Consistent with this notion, RNAi-induced silencing of tropomyosin expression in TGF-beta induced epithelial cells has been shown to cause uncontrolled cell motility and tumor formation (Bakin, Safina et al. 2004). The reduced tropomyosin expression in these cells may lead to an increase in the number of gaps on actin filaments making those sites available for other ABP’s to bind and remodel the actin filament.
Summary and Conclusions
In this work, we have provided the first direct visual evidence leading to the mechanism of tropomyosin binding to pre-formed actin filaments. This process allows tropomyosin chains to bind strongly to actin but also allows their azimuthal position to be easily perturbed in response to calcium activation and deactivation. The random site-polymerization process observed here inevitably leads to the formation of gaps on the actin filament lacking the seven actin monomers required for binding. The process of gap annealing is slow compared to the initial binding of tropomyosin to the actin filament, and it is intriguing to speculate that gaps may serve a vital regulatory role in cytoskeletal actin filament remodeling. We would expect gap number to be a function of tropomyosin concentration, actin affinity, and isolated and contiguous binding rates, all of which would be affected by end-to-end bond strength. Future studies will determine tropomyosin binding rates and gap formation for other non-muscle tropomyosin isoforms and correlate those data with in vivo regulatory data to further dissect the role that gaps could play in the regulation of binding by other actin-binding proteins.
Materials and Methods
Through-the-Objective TIRF Apparatus
A custom-built TIR (Total Internal Reflection) fluorescence microscope was used to measure single tropomyosin molecules binding to actin. To achieve TIR illumination, a frequency-doubled Yittrium Aluminum Garnet (YAG, 532 nm) laser (Crysta Laser, Reno, NV) is circularly polarized by a λ/4 wave plate and focused at the edge of the back of a 100x 1.45 NA objective (Nikon, Tokyo, Japan.) via a system of lenses and mirrors (Supplemental Figure S3).
Myosin and Actin Purification
Full-length chicken skeletal muscle myosin was purified (Margossian and Lowey 1982, Pollard 1982) and stored in a 50% glycerol. Chicken skeletal muscle actin was then extracted from acetone powder as described originally in Spudich and Watt, 1971 (Spudich and Watt 1971) and converted to F-actin by dialysis against high salt storage buffer (4 mM Imidazole, 100 mM KCl, 2 mM MgCl2, 1 mM NaN3, 0.5 mM ATP, 1 mM DTT). Dried stock (stored at −20 °C) FITC labeled phalloidin was dissolved in 10 μl of methanol (100 μM) and added to 1 ml unlabeled F-actin at 1 μM concentration. The FITC-phalloidin labeled actin was stored at 4°C overnight.
Tropomyosin Purification and Labeling
Rabbit skeletal and chicken gizzard smooth muscle tropomyosin was isolated and purified according to previously published procedures (Jancso and Graceffa 1991, Graceffa 1999, Graceffa 2000). Purified skeletal muscle tropomyosin consists of either two α-α chains (Tpm1.1st (a.b.b.a)) or one α and one β (Tpm2.2st (a.b.b.a)) chain, while smooth muscle tropomyosin contains mostly heterodimers with one α (Tpm1.4sm (a.a.b.d)) and one β (Tpm2.1sm/cy (a.b.a.d)) chain (Geeves, Hitchcock-DeGregori et al. 2014). Non-muscle tropomyosin isoform Tpm1.6cy (a.b.b.d) (Geeves, Hitchcock-DeGregori et al. 2014), common name Tm2, was generously provided by Dr. Alan Huang. All three isoforms were dissolved in Phosphate Buffered Saline and reduced by reacting with 5 mM DTT for 3 hours followed by dialysis against 3–4 changes of buffer. Tissue purified tropomyosin was labeled at Cys-190 with a small molecule fluorescent probe, Alexa-532, which has little or no effect on actin binding (Bacchiocchi and Lehrer 2002, Bacchiocchi, Graceffa et al. 2004). Labeling on cysteine-190 was achieved by incubation of tropomyosin with a 5-fold molar excess of the maleimide derivative of AlexaFluor-532 (Invitrogen) or Cy3 (Lumiprobe) overnight at 4°C. The labeled proteins were purified by gel filtration chromatography using Sephadex G25 (Supplemental Figure S4).
Labeling Ratio of Flurophore:Tropomyosin
The molar ratio of Alexa-532 conjugated to skeletal and smooth muscle tropomyosin at Cys-190 was determined by measuring the quantal bleaching of single tropomyosin molecules excited by TIRF. Labeled tropomyosin at 2 nM concentration was added to a flow cell containing either 20 nM actin filaments tethered to a nitrocellulose-coated surface by monomeric myosin molecules or a nitrocellulose-coated surface only at high salt (175–350 mM KCl). These conditions favored formation of single tropomyosin molecules in solution; and therefore, the number of stepwise intensity decreases corresponded to the number of bound fluorophores per molecule. The average number of fluorophores bound to a tropomyosin molecule was 1.23 ± 0.12 (n = 13) and 1.20 ± 0.11 (n= 15) for skeletal and smooth muscle tropomyosin respectively.
TIRF Tm Binding Assay
A 30mm × 22mm nitrocellulose-coated cover glass was affixed to a 22mm × 22mm cover glass, forming a 25 μl flow cell with 4mm clearance on either side. The flow cell was affixed using vacuum grease (Fisherbrand™ Cello-Seal™) to a custom-made aluminum microscope slide which contained a rectangular hole to allow contact between the TIRF objective and cover glass. The nitrocellulose-coated surface of the flow cell was incubated with 100 μg/ml myosin dissolved in high salt buffer (300 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, and 1 mM DTT) followed by incubation with 1 mg/ml BSA in high salt buffer to prevent non-specific binding to the surface. Unbound BSA was removed with two volumes of low salt buffer (25 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, and 1 mM DTT), followed by addition of 10 nM FITC-phalloidin labeled actin filaments. FITC-phalloidin labeled actin was incubated for five minutes. Unbound filaments were washed out with two volumes actin buffer. FITC-labeled actin filaments were excited with a mercury lamp using 480/40 excitation and 535/50 emission filters (Chroma Technology Corp., Bellows Falls, VT) and observed on a Nikon Eclipse TE-2000 microscope with an Andor iXon3 897 EMCCD camera. A region of the field containing two to four actin filaments was selected and light from the mercury lamp was shuttered. Excitation and emission filters were manually changed to 545/30 and 600/45 respectively and the 532 nm YAG TIRF laser was turned on. Alexa-532-labeled tropomyosin dissolved in actin buffer and oxygen scavengers (17 units/ml glucose oxidase, 125 units/ml catalase), with variable tropomyosin or KCl concentration was flowed into the flow cell and fluorescent tropomyosin binding was recorded. Optimal working tropomyosin concentrations ranged between 10 and 100 nM. Tropomyosin binding at concentrations below 10 nM and above 100 nM failed to occur at a rate that exceeded fluorophore bleaching or yielded too low a signal to noise ratio for accurate filament identification respectively.
Post-acquisition image processing
Videos were recorded as 16.7 frames per second audio video interleave (.avi) files and were imported into ImageJ for processing. Tropomyosin-bound actin filaments were cropped out of each video. The background was subtracted and the contrast enhanced in order to more clearly define the Tm-bound actin filament. A Gaussian blur was applied with a rolling ball radius of 2.00 to reduce speckled noise. After image processing, a line was drawn through the center, along the entire length of the Tm-bound filament. The “Reslice” command was performed to produce a kymograph, which plotted the individual intensities, in every movie frame, for each pixel that intersected the line.
Elongation Rates and Single Molecule Binding Analysis
For each tropomyosin binding condition (e.g. tropomyosin or salt concentration), the rate of addition of tropomyosin molecules to actin was determined from the average polymerization rate of labeled tropomyosin molecules on the actin filament using the kymograph. Chain length and elongation rate were determined from videos using pixel size (0.16 μm/pixel) and frame rate (16.7 frames/second). Each elongation rate expressed in μm/s was converted to monomers per second by dividing by .040 microns, the length of a tropomyosin monomer (Censullo and Cheung 1994). In each case, two to four elongation rates for one or two filaments each were averaged and standard deviations and errors calculated. Skeletal and smooth muscle tropomyosin elongation rates as a function of tropomyosin concentration were determined from the slopes of the linear regression fits of elongation rates plotted against tropomyosin concentration. The difference in the slopes was tested for significance by performing a two-tailed t-test.
The time between tropomyosin addition to the flow cell and initial random actin binding was determined for each tropomyosin binding event that occurred at an unbound area of the actin filament. Each binding rate (molecules/second) was divided by tropomyosin concentration (10nM–100nM) and normalized to the difference between the total number of non-fluorescent pixels just prior to the binding event and the number of pixels illuminated by the binding event (typically 4–5). Normalization accounted for the increased number of binding sites available to tropomyosin along the entire actin filament as opposed to end-to-end continuous binding at the end of an actin-bound tropomyosin chain. Events that occurred late in the binding process, where accurate determination of unbound areas was not possible, were discounted from the analysis along with events that occurred prior to video recording.
The number of bound tropomyosin monomers was determined by eye via stepwise increases in fluorescence intensity upon tropomyosin binding to F-actin. Due to variations in the intensity of the excitation field (Mattheyses, Simon et al. 2010), the quantal value for a single tropomyosin molecule varied depending on the location of the binding event on the actin filament. The quantal intensity value was determined by the stepwise intensity variations (both increases due to binding and decreases due to bleaching or detachment) over the entire length of the recorded video (an average duration of 2 minutes). Stepwise variations in intensity were then used to determine the number of tropomyosin molecules bound to the actin filament in various regions of the excitation field.
Since the labeling ratio of Alexa-532:tropomyosin was approximately 1.2:1 for skeletal and smooth muscle tropomyosin isoforms, 80% of the molecules contained one fluorophore while the remaining 20% contained two. Therefore, a “double jump” in intensity could be the binding of two singly-labeled molecules or one doubly-labeled molecule. To reconcile this ambiguity, the number of “double-jumps” was counted for each binding event of tropomyosin, and the ratio of “double jumps” to total quantal intensity increases was determined. The “double jump” ratio for skeletal tropomyosin was 0.22 (n = 174) and smooth muscle tropomyosin was 0.19 (n = 146). The observation that a “double jump” occurred approximately 20% of the time correlated with a 1.2:1 fluorophore:tropomyosin labeling ratio and suggested that it represented the binding of a single tropomyosin molecule containing two fluorophores
Gap Density and Tm2 Detachment Experiments
Mixtures of 500 nM Cy3-labeled Tm2 and phalloidin-stabilized actin filaments were prepared and incubated at room temperature in standard actin buffer (100 mM KCl). A flow cell was loaded with 5 μg/ml myosin, blocked with 1 mg/ml BSA as described above, and washed thrice with low salt buffer (100 mM KCl). At each incubation time point (10, 20, 30, 45, 60, 80, 100, 120 mins.), each mixture was quickly diluted 1:25, 20 nM final actin and tropomyosin concentration, in actin buffer (100 mM KCl) and immediately injected into the flow cell. After two minutes, unbound actin-Tm was removed by three flow cell volumes of low salt buffer (100 mM KCl) containing oxygen scavengers with 1 mM Trolox. Following buffer washout, the flow cell was immediately placed on the microscope and observed with 532 nm TIRF illumination. Cy3-Tm2 bound actin filaments were located and tropomyosin detachment was observed and recorded at 25 frames per second using an Andor iXon897 EMCCD camera. After video acquisition, .avi files were imported into ImageJ and kymographs were generated as described previously. Tropomyosin gaps on the actin filament were identified on the kymograph, counted, and normalized to the total length of filaments analyzed. Total filament lengths analyzed for each time point ranged between 10 and 40 microns.
Cy3 Flurophore Bleaching Rate Determination
To ensure that fluorescence intensity loss was due to tropomyosin detachment and not Cy3 flurophore bleaching, the bleaching rate of Cy3 was determined at the laser intensity employed to visualize tropomyosin detachment. Sixty-seven μg/ml Cy3-Tm2 (1 μM) was flowed into a lysine-coated flow cell and incubated for 10 minutes then washed out with scavenger buffer. The positively charged lysine surface interacts with the solvent-exposed negative charges of tropomyosin and sticks the Cy3-Tm2 to the surface. The flow cell was placed on the microscope, TIRF laser activated, and video of fluorophore bleaching was recorded using NIS Elements software. After importing raw video into ImageJ, two non-overlapping fields of view were selected of approximately 125 pixels2 (187 × 375 pixels total field of view) and the mean pixel intensity decays were monitored. The data were plotted and fit to the following exponential decay equation: f= y0 + a*e−b*x, and the mean lifetime τ was calculated by taking the reciprocal of b, the decay constant (Supplemental Figure S2).
Supplementary Material
Substituting [Tm] = 14 μM, the average tropomyosin concentration in muscle (Borina, Pellegrino et al. 2010), binding time = 1 second, the expected time for 14 μM tropomyosin to cover all available binding sites on actin based on our observations, and actin filament length = 1 μm, the average length of a muscle thin filament, the total number of binding sites was estimated to be ~4.5. This is an obvious overestimation as the free tropomyosin concentration in muscle is likely much lower than 14 μM. Therefore, the number of gaps predicted to form on a thin filament was estimated to be between 3 and 4.
Supplemental Figure 2. Cy3-Tm2 Bleaching Rate. Cy3 has been shown to be more photostable with reduced bleaching rates compared to Alexa-dyes (Aitken, Marshall et al. 2008), and we confirmed its increased photostability under TIRF illumination. The mean fluorophore lifetimes for the solid and dashed traces are 70.4 seconds and 61.3 seconds respectively. This is considerably larger than the calculated detachment rate for Cy3-Tm2 tropomyosin (~0.33 monomers/second), suggesting that fluorescence loss was due to tropomyosin detachment.
Supplemental Figure 3. TIRF laser optics. Schematic of the custom built TIRF apparatus viewed from above the optical table.
Supplemental Figure 4. Labeling of Tropomyosin at Cys-190. Purified tropomyosin was dissolved in Phosphate Buffered Saline (PBS) and reduced by reacting with 5 mM DTT for 3 hours followed by dialysis against several changes of PBS. It was then labeled on cysteine-190 with a 5-fold molar excess of the maleimide derivative of AlexaFluor-532 (Invitrogen) or Cy3 (Lumiprobe) overnight at 4°C. The labeled protein was purified by gel filtration chromatography on a Sephadex G25 column.
Acknowledgments
We would like to thank Dr. Paul Leavis for labeling smooth and skeletal muscle tropomyosin, and Dr. Alan Huang for labeling the Tm2 tropomyosin isoform. This work was supported by NIH grants R01HL077280 (to J.R.M.) and R37HL036153 (to W.L.).
References
- Aitken CE, Marshall RA, Puglisi JD. An Oxygen Scavenging System for Improvement of Dye Stability in Single-Molecule Fluorescence Experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchiocchi C, Graceffa P, Lehrer SS. Myosin-Induced Movement of Alphaalpha, Alphabeta, and Betabeta Smooth Muscle Tropomyosin on Actin Observed by Multisite Fret. Biophys J. 2004;86:2295–2307. doi: 10.1016/S0006-3495(04)74287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchiocchi C, Lehrer SS. Ca(2+)-Induced Movement of Tropomyosin in Skeletal Muscle Thin Filaments Observed by Multi-Site Fret. Biophys J. 2002;82:1524–1536. doi: 10.1016/S0006-3495(02)75505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin AV, Safina A, Rinehart C, Daroqui C, Darbary H, Helfman DM. A critical role of tropomyosins in TGF-beta regulation of the actin cytoskeleton and cell motility in epithelial cells. Mol Biol Cell. 2004;15(10):4682–4694. doi: 10.1091/mbc.E04-04-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borina E, Pellegrino MA, D’Antona G, Bottinelli R. Myosin and Actin Content of Human Skeletal Muscle Fibers Following 35 Days Bed Rest. Scand J Med Sci Sports. 2010;20:65–73. doi: 10.1111/j.1600-0838.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- Censullo R, Cheung HC. Tropomyosin length and two-stranded F-actin flexibility in the thin filament. J Mol Biol. 1994;243(3):520–529. doi: 10.1006/jmbi.1994.1677. [DOI] [PubMed] [Google Scholar]
- Choi C, Kim D, Kim S, Jeong S, Song E, Helfman DM. From Skeletal Muscle to Cancer: Insights Learned Elucidating the Function of Tropomyosin. J Struct Biol. 2012;177:63–69. doi: 10.1016/j.jsb.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Coulton AT, Koka K, Lehrer SS, Geeves MA. Role of the head-to-tail overlap region in smooth and skeletal muscle beta-tropomyosin. Biochemistry. 2008;47(1):388–397. doi: 10.1021/bi701144g. [DOI] [PubMed] [Google Scholar]
- Dabrowska R, Hinssen H, Galazkiewicz B, Nowak E. Modulation of Gelsolin-Induced Actin-Filament Severing by Caldesmon and Tropomyosin and the Effect of These Proteins on the Actin Activation of Myosin Mg(2+)-Atpase Activity. Biochem J. 1996;315:753–759. doi: 10.1042/bj3150753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski W, Nowak E. Studies on the Interaction of F-Actin with Tropomyosin. Eur J Biochem. 1968;5:376–384. doi: 10.1111/j.1432-1033.1968.tb00380.x. [DOI] [PubMed] [Google Scholar]
- Eaton BL. Tropomyosin binding to F-actin induced by myosin heads. Science. 1976;192(4246):1337–1339. doi: 10.1126/science.131972. [DOI] [PubMed] [Google Scholar]
- Eaton BL, Kominz DR, Eisenberg E. Correlation between the Inhibition of the Acto-Heavy Meromyosin Atpase and the Binding of Tropomyosin to F-Actin: Effects of Mg2+, Kcl, Troponin I, and Troponin C. Biochemistry. 1975;14:2718–2725. doi: 10.1021/bi00683a025. [DOI] [PubMed] [Google Scholar]
- Frye J, Klenchin VA, Rayment I. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition. Biochemistry. 2010;49(23):4908–4920. doi: 10.1021/bi100349a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil. 2014 doi: 10.1007/s10974-014-9389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girjes AA, Keriakous D, Cockerill GW, Hayward IP, Campbell GR, Campbell JH. Cloning of a Differentially Expressed Tropomyosin Isoform from Cultured Rabbit Aortic Smooth Muscle Cells. Int J Biochem Cell Biol. 2002;34:505–515. doi: 10.1016/s1357-2725(01)00148-0. [DOI] [PubMed] [Google Scholar]
- Graceffa P. Movement of Smooth Muscle Tropomyosin by Myosin Heads. Biochemistry. 1999;38:11984–11992. doi: 10.1021/bi9825495. [DOI] [PubMed] [Google Scholar]
- Graceffa P. Phosphorylation of Smooth Muscle Myosin Heads Regulates the Head-Induced Movement of Tropomyosin. J Biol Chem. 2000;275:17143–17148. doi: 10.1074/jbc.M001979200. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol. 2006;364(1):80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Gundapaneni D, Xu J, Root DD. High flexibility of the actomyosin crossbridge resides in skeletal muscle myosin subfragment-2 as demonstrated by a new single molecule assay. J Struct Biol. 2005;149(2):117–126. doi: 10.1016/j.jsb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Gunning P, O’Neill G, Hardeman E. Tropomyosin-Based Regulation of the Actin Cytoskeleton in Time and Space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Gunning P, Weinberger R, Jeffrey P. Actin and Tropomyosin Isoforms in Morphogenesis. Anat Embryol (Berl) 1997;195:311–315. doi: 10.1007/s004290050050. [DOI] [PubMed] [Google Scholar]
- Heald RW, Hitchcock-DeGregori SE. The Structure of the Amino Terminus of Tropomyosin Is Critical for Binding to Actin in the Absence and Presence of Troponin. J Biol Chem. 1988;263:5254–5259. [PubMed] [Google Scholar]
- Hill LE, Mehegan JP, Butters CA, Tobacman LS. Analysis of Troponin-Tropomyosin Binding to Actin. Troponin Does Not Promote Interactions between Tropomyosin Molecules. J Biol Chem. 1992;267:16106–16113. [PubMed] [Google Scholar]
- Holmes KC, Geeves MA. The structural basis of muscle contraction. Philos Trans R Soc Lond B Biol Sci. 2000;355(1396):419–431. doi: 10.1098/rstb.2000.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29(6–8):213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Schroder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359(1452):1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes TR, Block SM, White BT, Spudich JA. Movement of Myosin Fragments Invitro - Domains Involved in Force Production. Cell. 1987;48(6):953–963. doi: 10.1016/0092-8674(87)90704-5. [DOI] [PubMed] [Google Scholar]
- Jancso A, Graceffa P. Smooth muscle tropomyosin coiled-coil dimers. Subunit composition, assembly, and end-to-end interaction. J Biol Chem. 1991;266(9):5891–5897. [PubMed] [Google Scholar]
- Kee AJ, Hardeman EC. Tropomyosins in skeletal muscle diseases. Adv Exp Med Biol. 2008;644:143–157. doi: 10.1007/978-0-387-85766-4_12. [DOI] [PubMed] [Google Scholar]
- Khaitlina S, Fitz H, Hinssen H. The Interaction of Gelsolin with Tropomyosin Modulates Actin Dynamics. Febs J. 2013;280:4600–4611. doi: 10.1111/febs.12431. [DOI] [PubMed] [Google Scholar]
- Kominz DR. Interactions of calcium and native tropomyosin with myosin and heavy meromyosin. Arch Biochem Biophys. 1966;115(3):583–592. doi: 10.1016/0003-9861(66)90078-6. [DOI] [PubMed] [Google Scholar]
- Koubassova NA, Tsaturyan AK. Direct modeling of x-ray diffraction pattern from skeletal muscle in rigor. Biophys J. 2002;83(2):1082–1097. doi: 10.1016/S0006-3495(02)75232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MW, Dechene ET, Roumm E, Geggel AS, Moghadaszadeh B, Beggs AH. Mutations of tropomyosin 3 (TPM3) are common and associated with type 1 myofiber hypotrophy in congenital fiber type disproportion. Hum Mutat. 2010;31(2):176–183. doi: 10.1002/humu.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JG, Bach CT, O’Neill GM. Interior Decoration: Tropomyosin in Actin Dynamics and Cell Migration. Cell Adh Migr. 2011;5:181–186. doi: 10.4161/cam.5.2.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman W, Craig R, Vibert P. Ca(2+)-Induced Tropomyosin Movement in Limulus Thin Filaments Revealed by Three-Dimensional Reconstruction. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- Lehrer SS, Golitsina NL, Geeves MA. Actin-Tropomyosin Activation of Myosin Subfragment 1 Atpase and Thin Filament Cooperativity. The Role of Tropomyosin Flexibility and End-to-End Interactions. Biochemistry. 1997;36:13449–13454. doi: 10.1021/bi971568w. [DOI] [PubMed] [Google Scholar]
- Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin Position on F-Actin Revealed by Em Reconstruction and Computational Chemistry. Biophys J. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mui S, Brown JH, Strand J, Reshetnikova L, Tobacman LS, Cohen C. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc Natl Acad Sci U S A. 2002;99(11):7378–7383. doi: 10.1073/pnas.102179999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margossian SS, Lowey S. Preparation of Myosin and Its Subfragments from Rabbit Skeletal Muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- Marston S, Memo M, Messer A, Papadaki M, Nowak K, McNamara E, Ong R, El-Mezgueldi M, Li X, Lehman W. Mutations in Repeating Structural Motifs of Tropomyosin Cause Gain of Function in Skeletal Muscle Myopathy Patients. Hum Mol Genet. 2013;22:4978–4987. doi: 10.1093/hmg/ddt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura F, Lin JJ. Visualization of monoclonal antibody binding to tropomyosin on native smooth muscle thin filaments by electron microscopy. J Mol Biol. 1982;157(1):163–171. doi: 10.1016/0022-2836(82)90520-4. [DOI] [PubMed] [Google Scholar]
- Mattheyses AL, Simon SM, Rappoport JZ. Imaging with Total Internal Reflection Fluorescence Microscopy for the Cell Biologist. J Cell Sci. 2010;123:3621–3628. doi: 10.1242/jcs.056218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan AD, Stewart M. Tropomyosin Coiled-Coil Interactions: Evidence for an Unstaggered Structure. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- Ohtsuki I, Shiraishi F. Periodic binding of troponin C.I and troponin I to tropomyosin-actin filaments. J Biochem. 2002;131(5):739–743. doi: 10.1093/oxfordjournals.jbchem.a003159. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Myosin Purification and Characterization. Methods Cell Biol. 1982;24:333–371. doi: 10.1016/s0091-679x(08)60665-2. [DOI] [PubMed] [Google Scholar]
- Rao JN, Rivera-Santiago R, Li XE, Lehman W, Dominguez R. Structural analysis of smooth muscle tropomyosin alpha and beta isoforms. J Biol Chem. 2012;287(5):3165–3174. doi: 10.1074/jbc.M111.307330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Holden HM. The three-dimensional structure of a molecular motor. Trends Biochem Sci. 1994;19(3):129–134. doi: 10.1016/0968-0004(94)90206-2. [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-Dimensional Structure of Myosin Subfragment-1: A Molecular Motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Xing J, Chen LQ, Sweeney HL. Myosin IIb is unconventionally conventional. J Biol Chem. 2003;278(30):27449–27455. doi: 10.1074/jbc.M302555200. [DOI] [PubMed] [Google Scholar]
- Sanders C, Smillie LB. Chicken gizzard tropomyosin: head-to-tail assembly and interaction with F-actin and troponin. Can J Biochem Cell Biol. 1984;62(6):443–448. doi: 10.1139/o84-060. [DOI] [PubMed] [Google Scholar]
- Schmitz H, Reedy MC, Reedy MK, Tregear RT, Winkler H, Taylor KA. Electron tomography of insect flight muscle in rigor and AMPPNP at 23 degrees C. J Mol Biol. 1996;264(2):279–301. doi: 10.1006/jmbi.1996.0641. [DOI] [PubMed] [Google Scholar]
- Singh A, Hitchcock-DeGregori SE. Tropomyosin’s periods are quasi-equivalent for actin binding but have specific regulatory functions. Biochemistry. 2007;46(51):14917–14927. doi: 10.1021/bi701570b. [DOI] [PubMed] [Google Scholar]
- Sousa D, Cammarato A, Jang K, Graceffa P, Tobacman LS, Li XE, Lehman W. Electron Microscopy and Persistence Length Analysis of Semi-Rigid Smooth Muscle Tropomyosin Strands. Biophys J. 2010;99:862–868. doi: 10.1016/j.bpj.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The Regulation of Rabbit Skeletal Muscle Contraction. I. Biochemical Studies of the Interaction of the Tropomyosin-Troponin Complex with Actin and the Proteolytic Fragments of Myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Tanaka H, Oosawa F. The effect of temperature on the interaction between F-actin and tropomyosin. Biochim Biophys Acta. 1971;253(1):274–283. doi: 10.1016/0005-2728(71)90253-2. [DOI] [PubMed] [Google Scholar]
- Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, Seidman JG, Seidman CE. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77(5):701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Tobacman LS. Cooperative binding of tropomyosin to actin. Adv Exp Med Biol. 2008;644:85–94. doi: 10.1007/978-0-387-85766-4_7. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Dupuis DE, Guilford WH, Patlak JB, Waller GS, Trybus KM, Warshaw DM, Lowey S. Two heads of myosin are better than one for generating force and motion. Proc Natl Acad Sci U S A. 1999;96(8):4402–4407. doi: 10.1073/pnas.96.8.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilfan A. The Binding Dynamics of Tropomyosin on Actin. Biophys J. 2001;81:3146–3155. doi: 10.1016/S0006-3495(01)75951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindin H, Gunning P. Cytoskeletal Tropomyosins: Choreographers of Actin Filament Functional Diversity. J Muscle Res Cell Motil. 2013;34:261–274. doi: 10.1007/s10974-013-9355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin--tropomyosin complex. Nature. 2014 doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Coluccio LM. New Insights into the Regulation of the Actin Cytoskeleton by Tropomyosin. Int Rev Cell Mol Biol. 2010;281:91–128. doi: 10.1016/S1937-6448(10)81003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner A. Equilibrium of the Actin-Tropomyosin Interaction. J Mol Biol. 1979;131:839–853. doi: 10.1016/0022-2836(79)90204-3. [DOI] [PubMed] [Google Scholar]
- Wegner A. The Interaction of Alpha, Alpha- and Alpha, Beta-Tropomyosin with Actin Filaments. FEBS Lett. 1980;119:245–248. doi: 10.1016/0014-5793(80)80263-8. [DOI] [PubMed] [Google Scholar]
- Wegner A, Ruhnau K. Rate of binding of tropomyosin to actin filaments. Biochemistry. 1988;27(18):6994–7000. doi: 10.1021/bi00418a049. [DOI] [PubMed] [Google Scholar]
- Weigt C, Wegner A, Koch MH. Rate and mechanism of the assembly of tropomyosin with actin filaments. Biochemistry. 1991;30(44):10700–10707. doi: 10.1021/bi00108a014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Substituting [Tm] = 14 μM, the average tropomyosin concentration in muscle (Borina, Pellegrino et al. 2010), binding time = 1 second, the expected time for 14 μM tropomyosin to cover all available binding sites on actin based on our observations, and actin filament length = 1 μm, the average length of a muscle thin filament, the total number of binding sites was estimated to be ~4.5. This is an obvious overestimation as the free tropomyosin concentration in muscle is likely much lower than 14 μM. Therefore, the number of gaps predicted to form on a thin filament was estimated to be between 3 and 4.
Supplemental Figure 2. Cy3-Tm2 Bleaching Rate. Cy3 has been shown to be more photostable with reduced bleaching rates compared to Alexa-dyes (Aitken, Marshall et al. 2008), and we confirmed its increased photostability under TIRF illumination. The mean fluorophore lifetimes for the solid and dashed traces are 70.4 seconds and 61.3 seconds respectively. This is considerably larger than the calculated detachment rate for Cy3-Tm2 tropomyosin (~0.33 monomers/second), suggesting that fluorescence loss was due to tropomyosin detachment.
Supplemental Figure 3. TIRF laser optics. Schematic of the custom built TIRF apparatus viewed from above the optical table.
Supplemental Figure 4. Labeling of Tropomyosin at Cys-190. Purified tropomyosin was dissolved in Phosphate Buffered Saline (PBS) and reduced by reacting with 5 mM DTT for 3 hours followed by dialysis against several changes of PBS. It was then labeled on cysteine-190 with a 5-fold molar excess of the maleimide derivative of AlexaFluor-532 (Invitrogen) or Cy3 (Lumiprobe) overnight at 4°C. The labeled protein was purified by gel filtration chromatography on a Sephadex G25 column.