Abstract
The unexpected repurposing of nuclear transport proteins from their function in interphase to an equally vital and very different set of functions in mitosis was very surprising. The multi-talented cast when first revealed included the import receptors, importin alpha and beta, the small regulatory GTPase RanGTP, and a subset of nuclear pore proteins. In this review, we report that recent years have revealed new discoveries in each area of this expanding story in vertebrates: (a) The cast of nuclear transport receptors playing a role in mitotic spindle regulation has expanded: both transportin, a nuclear import receptor, and Crm1/Xpo1, an export receptor, are involved in different aspects of spindle assembly. Importin beta and transportin also regulate nuclear envelope and pore assembly. (b) The role of nucleoporins has grown to include recruiting the key microtubule nucleator the γ-TuRC complex and the exportin Crm1 to the mitotic kinetochores of humans. Together they nucleate microtubule formation from the kinetochores towards the centrosomes. (c) New research finds that the original importin beta/RanGTP team have been further co-opted by evolution to help regulate other cellular and organismal activities, ranging from the actual positioning of the spindle within the cell perimeter, to regulation of a newly discovered spindle microtubule branching activity, to regulation of the interaction of microtubule structures with specific actin structures. (d) Lastly, because of the multitudinous roles of karyopherins throughout the cell cycle, a recent large push toward testing their potential as chemotherapeutic targets has begun to yield burgeoning progress in the clinic.
Karyopherins and RanGTP in mitosis: An evolutionary tour-de-force of repurposing
Each dividing eukaryotic cell cycles with elegant choreography between interphase and mitosis. Mitosis in higher eukaryotes, the focus of this review, involves the set up and breakdown of multiple “scenes”, each with a different purpose or theme. First, there is assembly of a large mitotic spindle with duplicated chromosomes aligned via their kinetochores (metaphase). Then a poignant separation of the duplicated chromosomes occurs (anaphase), and finally a triumphant reassembly of nuclear envelopes and nuclear pores around the sets of separated chromosomes (telophase). At the molecular level, each structure involves the choreographed assembly of hundreds (kinetochores and NPCs) to thousands (spindles and nuclei) of individual proteins. Early on in cell cycle research, it was found that kinases and phosphatases determine the timing of the above events. This set of regulatory phosphorylations was further aided by key ubiquitination and proteolytic events in order to convey irreversibility, such as in the shift from metaphase to anaphase. But what spatial regulation directs each of the large mitotic structures to assemble in the correct place?
The answer was unexpected. If the first act of cell cycle proliferation is interphase, the second act is mitosis. Instead of changing the protagonists, in an ingenious tour-de-force, evolution kept the main actors of nuclear transport -- karyopherins and RanGTP -- and assigned them new roles for mitosis. Other cast members came on stage for the second act. These different assembly factors, regulated by karyopherins and RanGTP, proved to be proteins directly involved in forming the mitotic structures to be assembled: spindle assembly factors (SAFs) for the mitotic spindle, nucleoporins for nuclear pore assembly, and quite a few surprises.
Initial studies: In vitro spindle and nuclear reconstitution
Prior to 1999, little thought of a mitotic role for karyopherins was envisioned. Then, an impressive set of seminal studies by multiple labs broke upon the scene from 1999–2002. Using in vitro mitotic Xenopus egg extracts, long known to be capable of spindle assembly [1], multiple groups discovered that importin beta and RanGTP together determine where spindle assembly occurs [2–8]. Importin beta, often with the aid of its NLS-binding adaptor protein, importin alpha, binds to and masks Spindle Assembly Factors (SAFs) in cytoplasmic areas distant from the chromatin. By such binding, importin beta prevents spatially inappropriate spindle formation. In the vicinity of the mitotic chromosomes, however, importin β releases its bound SAFs, which sets spindle assembly in motion. Why only around chromosomes? Strikingly, a localized RanGTP “cloud” is produced around the mitotic chromosomes. This localized RanGTP cloud results from the fact that active RCC1, the RanGEF that stimulates the production of RanGTP, is a chromatin- and DNA-binding protein [9–11]. In addition, RanGAP is cytoplasmic and converts any RanGTP that diffuses away from the chromatin into RanGDP. Thus, importin beta (or α/β-) bound SAFs are released from inhibition by the high levels of RanGTP near the mitotic chromosomes and the mitotic spindle in all its complexity and beauty forms solely in that locale [9,10] (Figure 1B).
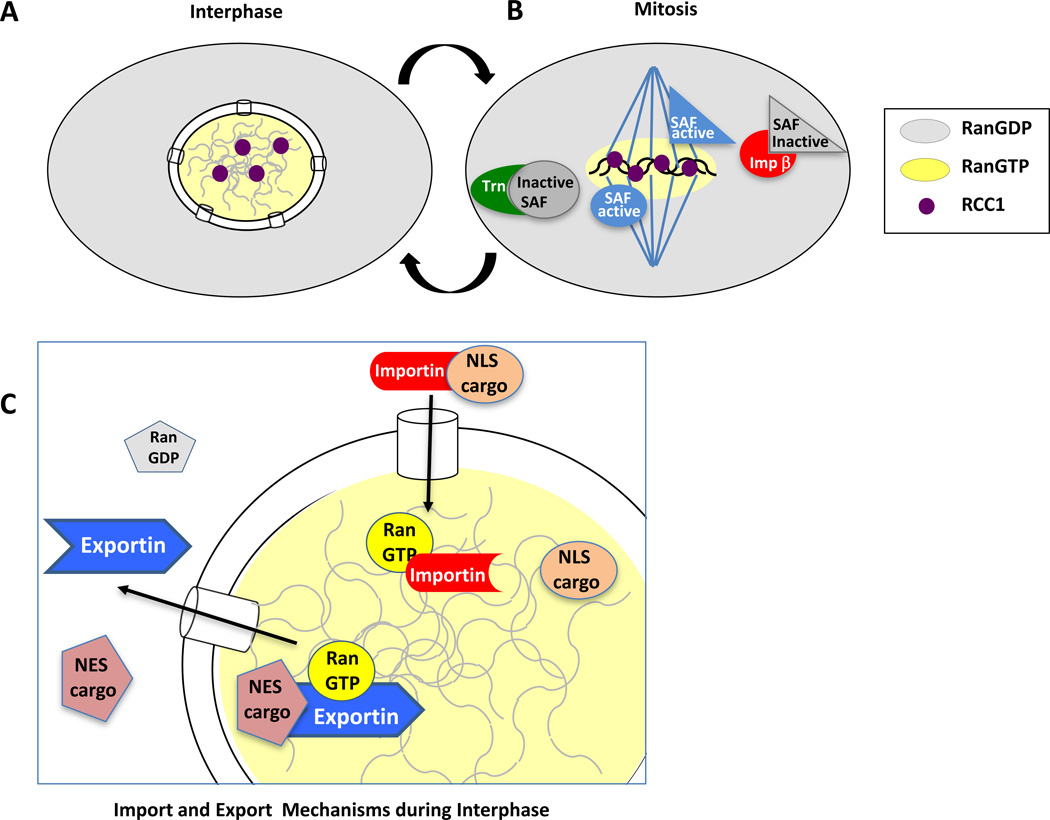
Figure 1. Karyopherins and RanGTP in interphase and mitosis.
(A) In interphase, RanGTP (yellow) is primarily found only in the nucleus, due to the localization of the RanGEF, RCC1 (purple), to chromatin. RanGDP (light grey) is found in the cytoplasm where the RanGAP and its accessory protein RanBP1 (both not shown) are also localized and induce RanGTP hydrolysis. (B) In mitosis, RCC1 continues to be bound to the chromatin of the mitotic chromosomes and produces a “cloud” of RanGTP, which dissociates any adjacent importin beta/SAF or transportin/SAF pairs. The freed spindle assembly factors or SAFs thus promote spindle assembly only around the mitotic chromosomes. At a distance from the chromosomes, the SAFs are held inactive by the binding of the transport receptors importin beta and transportin [9,49,52]. Thus, overall RanGTP appears to act as a spatial cue for assembly of the mitotic spindle and, later in mitosis, for assembly of the nuclear membranes and nuclear pores around chromatin. (C) For reference, the schematic shows the details of nuclear import of an NLS cargo protein by a generic importin receptor. The importin/NLS cargo complex is dissociated by nuclear RanGTP. Also shown is the export of an NES cargo protein by a generic exportin receptor. In this case, the export complex requires RanGTP as a co-factor in its formation. After export, the exportin/NES cargo/RanGTP complex is dissociated upon RanGTP hydrolysis by cytoplasmic RanGAP/RanBP1 (not shown).
The flurry of initial studies showed that RanGTP and importin beta act as dueling positive (RanGTP) and negative (importin beta) regulators. At heart, RanGTP acts as an all-powerful “GPS” or “genome-positioning signal” for mitotic assembly, counteracting the overall micromolar concentrations of karyopherins, albeit in a very localized area. In this manner, the production of RanGTP acts as a spatial cue that directs the major mitotic structures to assemble around the chromosomes -- and not elsewhere (Figure 1B) [9]. Depending on the phase of mitosis, the dueling karyopherin/RanGTP team regulates assembly of the mitotic spindle, the nuclear envelope, and the nuclear pores (see left panels, Figure 3 A–C). Indeed, it was the easy manipulability of Xenopus egg extracts that facilitated the in vitro study of all these assembly events [10,12–16].
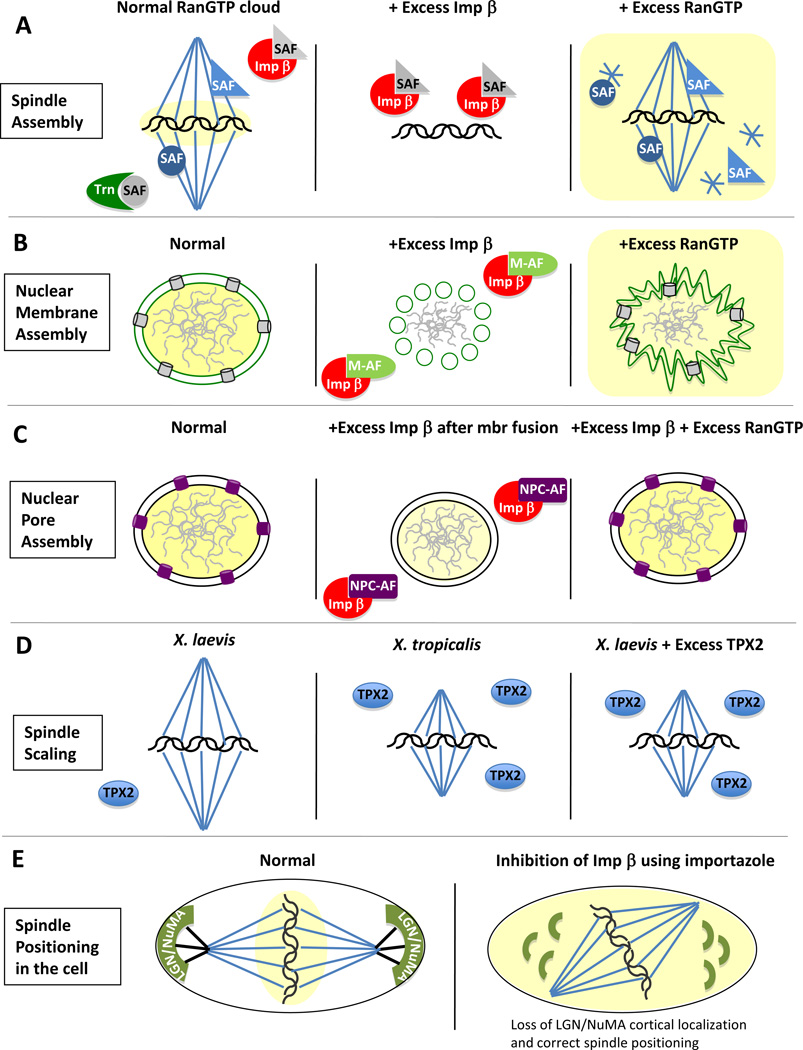
Figure 3. Multiple different arenas are regulated by karyopherins and RanGTP.
(A) Spindle assembly. During normal mitotic conditions (left panel), as also described in Legend 1B, importin beta (Imp β, red) and transportin (Trn, green) bind to and inhibit Spindle Assembly Factors (inactive SAFs; grey) in areas far from chromatin. This inhibition prevents mitotic microtubule assembly at a distance from the chromosomes. A RanGTP cloud (yellow area), produced by the RanGEF RCC1 (not shown), around the mitotic chromosomes causes localized release of the SAFs from adjacent importins; the released SAFs are now active and promote spindle assembly (active SAF; blue) in the correct location, i.e.,, around the chromosomes. In extracts, as in cells, the initial balance of RanGTP, karyopherins AND SAFs determines the influence of karyopherins and Ran on spindle assembly. If excess importin beta is added (middle panel), the importin beta (red) overwhelms the amount of RanGTP produced (not shown for clarity) and sequesters even the SAFs close to the chromosomes preventing formation of a mitotic spindle. In contrast, if excess RanGTP is added (right panel, yellow), the release of SAFs from importin beta and transportin (not shown) occurs throughout the extract, independently of their position with respect to chromatin. Thus, microtubules nucleate throughout the reaction, generating not only a mitotic spindle, but abundant microtubule asters (stars). (B) Nuclear membrane assembly. Normally the coordinated action of RanGTP and importin beta promote the formation of a double nuclear membrane surrounding the chromatin during telophase (green lines; left panel). If excess importin beta is added (middle panel), this is found to prevent the fusion of the ER membrane vesicles and tubules that normally form the double nuclear membrane, resulting in unfused vesicles (green circles). This occurs presumably by inhibiting one or more “membrane assembly factors” (M-AFs), whose nature is still unknown and could be either soluble or membrane-bound. If instead too much RanGTP is added (right panel), this causes excess nuclear membrane production, which appears as invaginated nuclear membranes replete with nuclear pores (light grey) around the chromatin. (C) Nuclear pore assembly. In normal conditions, cells possess a double nuclear membrane studded with nuclear pore complexes (purple cylinders; right panel). When excess importin beta is added to a pore-free nuclear assembly intermediate (containing fused nuclear membranes but no nuclear pores that has been induced by BAPTA [52]; middle panel), the excess importin beta binds to and inhibits nuclear pore complex assembly factors (NPC-AFs: purple color), and thus the nucleus remains devoid of nuclear pores. These NPC-AFs are disassembled nuclear pore proteins that act as NPC assembly factors at the end of mitosis. If, instead, excess RanGTP is included with the excess importin beta (left panel), the balance between the two is restored and nuclear pore assembly occurs as normal. Note: In (A), (B) and (C), an excess of transportin would cause the same effect that an excess of importin beta does (middle panels). (D) Spindle scaling. In nature, mitotic spindle size mirrors organismal size. For example, the spindle of the large frog, X laevis (left panel), is larger than the spindle of a smaller frog, X tropicalis (middle panel). This spindle scaling has been shown to be dependent on TPX2 levels. Indeed, adding an excess of TPX2 to X laevis spindle assembly reactions reduces the size of the X laevis spindle to that of X. tropicalis (right panel). Note: Karyopherins and RanGTP are present, but are dominated by higher TPX2 concentrations [65]. (E) Spindle positioning in the cell. The mitotic spindle orients lengthwise in a normal cell (left panel) due to the pulling forces that cortically-bound LGN and NuMA (green) generate on the astral microtubules (black lines). Inhibition of importin β, using the inhibitor importazole, causes misoriented spindles due to a loss of LGN and NuMA from the cortex (right panel).
Spindle assembly factors: Extensive regulation
The first molecular targets of karyopherin/Ran regulation were in the area of spindle assembly. The identification of a number of importin beta- (and alpha/beta-) inhibited spindle assembly factors burst upon the scene (Figure 1B). In our review, the definition of SAF is taken to mean any factor that promotes the assembly (or nucleation) of microtubules into a bipolar spindle at mitosis. Many of the SAFs identified (Table 1) were found, quite logically, to regulate microtubule nucleation, growth, stability and organization. Microtubule-associated proteins (MAPs) with specific spindle assembly functions include the MAPs TPX2, NuMA, Xnf7, HURP, Maskin, and NuSAP. Other SAFs were kinesins (XCTK2, Kid), which affect spindle bipolarity and chromosome orientation, and Cdk11, a cyclin-L-dependent Kinase, responsible for microtubule stabilization and microtubule-kinetochore interaction [9,17]. Still other relevant players were found to regulate RanGTP production and modulation. These include the RanGEF RCC1, the RanGAP, its activating partner RanBP1, and RanBP2 (Figure 2A) [18,19]. Interestingly, some proteins that are nuclear in interphase and SAFs in mitosis, such as lamin B and Rae1, are part of a mitotic spindle matrix, an entity that includes all proteins encompassed in the cytoplasmic region of the spindle [9,20–22].
Table 1.
| Spindle Assembly Factors (SAFs) | Examples | Refs. |
|---|---|---|
| MAPs | TPX2, NuMA, Xnf7, HURP, Maskin, NuSAP | [9] |
| Kinesins | (1)XCTK2(2)Kid | [9] |
| Spindle Matrix Proteins | Lamin B, Rae1 | [9] [20–22] |
| K-fiber stabilization | MCRS1* | [26] |
| Tumor Suppressor | Adenomatous Polyposis Coli (Apc)* | [23] |
| Chromatin Remodeling ATPases | CHD4*, ISWI* | [24] [25] |
| Multiple cellular functions | Nucleophosmin, Survivin, Cdk11 | [9] [17] |
| NPC Proteins | Nup107–160 complex, Nup98*, ELYS/Mel28* | [38] [39] [42] [44] |
| NPC Assembly Factors | Examples | Refs. |
| Importin Beta and/or Transportin Regulated Nups |
Nup107–160 complex, ELYS FG Nups (Nup358, Nup214, Nup153, Nup98, Nup62, Nup50) |
[39][40] [50–52] |
| Ran Modulation Factors | Examples | Refs. |
| RCC1, RanGAP, RanBP1*, RanBP2, Sumo | [9] [19] [63] | |
Spindle-Kinesin
Chromokinesin
The star (*) indicates recently characterized Ran- and/or Karyopherin- regulated SAFs
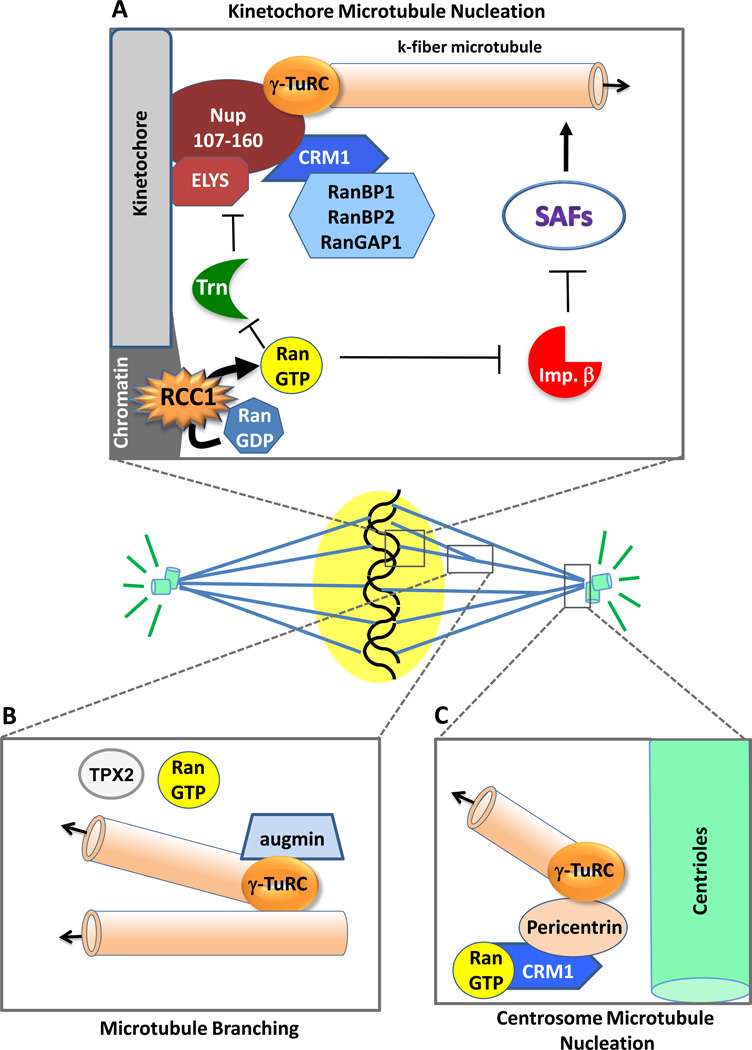
Figure 2. Microtubule nucleation occurs at multiple locales within the spindle and is regulated by RanGTP in each.
(A) In mitosis, microtubules have been shown to grow from γ-TuRC complexes recruited to the kinetochore by the Nup107–160 nucleoporin complex [42]. In humans, the export receptor Crm1 is also present at the kinetochore with its binding partners RanGap1, RanBP1, and RanBP2. The RanGEF RCC1 is present on mitotic chromosomes and, as described in Legend 1B, produces a gradient (“cloud”) of RanGTP around the chromosomes. RanGTP frees local SAFs from inhibition by importin beta (multiple SAFs) and transportin (the Nup107/160 complex and Elys SAFs) [9,10,49,52], such that k-fiber microtubules can grow from the kinetochores. (B) A newly discovered branching mechanism acting in spindle microtubule assembly is depicted. Here assembly can occur not only from centrosomes and kinetochores, as has long been known, but also from the sides of existing spindle microtubules. Augmin and γ-TuRC complexes are involved in the initiation of branching; RanGTP and TPX2 are also involved, but their exact mechanistic roles remain unknown [53–55]. (C) Spindle microtubules also initiate strongly from the centrosomes in many instances [35], nucleated by γ-TuRC complexes. Exportin Crm1/RanGTP aids in the recruitment of pericentrin to the centrosome region, which then recruits the γ-TuRC complexes [91].
Since 2008, more SAFs controlled by importin beta and Ran have been identified (Table 1). These include: (a) the tumor suppressor Adenomatous polyposis coli (Apc), which helps assemble and bundle microtubules [23], (b) two chromatin remodeling ATPases, CHD4 and ISWI, that act as Ran-dependent microtubule stabilizers [24,25], (c) a k-fiber stabilizer, MCRS1 [26], and (d) a set of nucleoporins (see below) [27,28]. Each of the above SAFs, both old and new, are hypothesized to be bound and inhibited by importin beta (or α/β) away from chromatin and released by RanGTP near chromatin.
Nuclear pore proteins act in spindle microtubule nucleation
A very unexpected group of spindle assembly factors were revealed to be nuclear pore proteins or nucleoporins (Nups). It turns out that a subset of nucleoporins, like importin beta and Ran, lead a double life during mitosis. In interphase, the nucleoporins in question reside largely at the nuclear pore (and to a lesser level at certain chromatin sites) [29–31]. In mitosis, however, this set of Nups transits broadly to the kinetochores, centrosomes, and/or fibers of the mitotic spindle to carry out mitotic functions [27,28].
In spindle assembly in animal cells, it is known that microtubules can grow from the centrosomes and from the kinetochores. Depending on the cell type or situation, one or the other source of microtubule growth can dominate [32–35]. It is now clear that Ran regulates both types of microtubule assembly [36].
An expanding list of nucleoporins has been found to regulate multiple aspects of mitosis [27–29]. A full description is not possible here, but among them the most intriguing is the 9-member nuclear pore subunit termed the Nup107–160 or Y complex and its closely associated partner, ELYS. These Nups clearly localize to the kinetochore at mitosis and have proven essential for functional kinetochores and for microtubule nucleation into spindles (Figure 2A) [37–44]. In fact, depletion of the Y complex was shown to allow initiation of centrosomal microtubules in in vitro spindle reconstitution extracts, but proper connection to the chromosomes was not possible, and the microtubules disassembled with time [38].
Surprisingly, it was later discovered that the Nup107–160 subcomplex acts to recruit γ-TuRCs (gamma-tubulin ring complexes) to the kinetochore at mitosis, which initiate “k-fiber” or kinetochore-initiated spindle microtubules [42]. This startling discovery made symmetrical sense from a nucleation point of view in that γ-TuRC ring complexes were first discovered at the poles of the spindle and act there as the microtubule-nucleating components of the centrosomes. RanGTP has also been found to be present at both kinetochores and centrosomes [34,36,42,45,46]. Importantly, the presence of RanGTP at the kinetochore is vital for Nup107–160/γ-TuRC-microtubule nucleation and counteracts the effects of importin beta (Figure 2A) [42,45,47]. In different studies, it was shown that importin β binds to the Nup107–160 protein complex and the nucleoporin ELYS/Mel-28 and prevents their interaction both with chromatin [40,43,48–51] and with kinetochores [52] in Xenopus in vitro extracts.
Microtubule branching occurs in spindles and is regulated by RanGTP
A newer discovery was in the works: it had long been thought that microtubule assembly occurs by the linear addition of tubulin α/β hetero-dimers to existing microtubules to produce entirely linear microtubules. A recent surprising study has shown that during in vitro spindle assembly in Xenopus egg extracts, a branching microtubule assembly mechanism occurs [53– 55]. Petry et al (2013), using total internal fluorescence (TIRF) microscopy, visualized microtubules initiating on the sides of existing microtubules: branch points could clearly be seen. This branching requires the microtubule nucleating protein augmin, γ-tubulin and TPX2, and is stimulated by RanGTP (Figure 2B), the latter of which was shown to act by freeing the spindle assembly factor TPX2, presumably from importin α/β [53,54,56]. The branching leads to an increased density of bundles of spindle microtubules and the authors suggest this may be for the purpose of amplifying the effect of the RanGTP gradient around the mitotic chromosomes. Nup98 also can induce a phenotype of excess microtubules in mitotic spindle assays, via excess Nup98 C-terminal fragment addition. The Nup98 fragment appears to inhibit MCAK (the microtubule-depolymerizing mitotic centromere-associated kinesin) [57]. It will be interesting to determine whether Nup98 or MCAK in some way influence the hitherto unsuspected branching microtubule mechanism that enhances spindle assembly.
RanGTP and its modifiers: New findings and variations on the theme
It is now well established that a gradient of RanGTP is crucial for spatially assembling the right mitotic structures at the right place: microtubule nucleation occurs where RanGTP is sufficiently high, which in most in vivo instances is around chromatin (Figure 3A, left panel). Indeed, addition of excess RanGTP to Xenopus mitotic extracts causes starbursts of microtubule nucleation (i.e., asters) to occur throughout the extract, as karyopherin inhibition is released everywhere (Figure 3A, right panel). Another study further showed that simply tethering RanGTP-generating machinery to the plasma membrane causes organized microtubule arrays to form at that locale [58]. The discovery that RanGTP is found at other sites in the cell, such as at the centrosomes and kinetochores where microtubules nucleate (see Crm1 section below) or inside of primary or motile cilia [59,60]), leads one to believe that we have not heard the last of Ran’s molecular talents.
Probing the regulatory mechanism for generating local RanGTP in mitosis is thus of great interest. Localization of RCC1 on chromatin has been seen to increase during mitosis [61,62], an increase tightly regulated by RanBP1 [19]. RanBP1, already known to increase the activity of the RanGAP, has been shown to bind to RCC1 and modulate RCC1's enzymatic activity, thus controlling the spatial organization and amplitude of the mitotic spindle [19]. Other experiments using beads coated with purified RCC1 in Xenopus egg extracts demonstrate that RCC1 is one of the minimal chromosome components able to generate a spindle [63].
This is perhaps a good place to bring forth the fact that a set of recent studies find that certain cell types and developmental stages appear to have become less reliant on spatial control by Ran/karyopherin gradients [35,64]. One example is in spindle size scaling [65–67]. Xenopus frog species differ in overall size, a difference mirrored in their mitotic spindles. Eggs of the small frog X. tropicalis contain 3-fold as much SAF TPX2 as eggs of the larger X. laevis. Interestingly, artificially increasing TPX2 concentration in X. laevis egg extracts (without increasing importin alpha/beta) produces smaller spindles [65] (Figure 3D). It may be that simply increasing SAF concentration in some cell types, via evolutionary change, could decrease normal control by karyopherin/RanGTP if the latter’s concentrations remained lower. However, an in-depth analysis of diverse adaptations such as this can be better obtained from more extensive reviews [35,64]
Nuclear membrane and nuclear pore assembly: Karyopherin/RanGTP regulation in in vitro reconstitution systems
Equally compelling were discoveries that the importin beta/Ran pair of dueling regulators also spatially controlled the major assembly events of late mitosis, i.e., nuclear membrane assembly and nuclear pore assembly [10,68–72]. Following recruitment of membrane vesicles to mitotic chromatin in interphase nuclear reconstitution extracts, importin beta/RanGTP were found to regulate the vesicle-vesicle fusion reaction required to form a double nuclear membrane. Excess importin beta inhibited fusion, while excess RanGTP promoted it. The balance of their activity was essential: excess importin beta resulted in unfused membrane vesicles, while excess RanGTP resulted in excessive, invaginated nuclear membranes around the chromatin [10,69] (Figure 3B). One recent study implicates the Lamin B receptor, LBR, as an importin beta-regulated target involved in nuclear membrane formation [73].
A similar regulatory scheme controls nuclear pore assembly in late mitosis: excess importin beta blocks nuclear pore assembly, while added RanGTP allows NPC assembly [10,69–72,74] (Figure 3C). For example, importin beta regulates the seeding of initiation sites for NPC assembly on chromatin via binding to and inhibiting ELYS and Nup107–160 complex, the initiators of telophase NPC assembly (aka “postmitotic” assembly), from anchoring to chromatin in vitro [40,48,50,75]. Importin beta regulation of NPC assembly is also observed to take place in vivo in interphase pore assembly [76]. Further description of these events and more complete references are reviewed in [10,77,78].
An expanding karyopherin network: Transportin
With 21 importin beta family members in humans [79], a key question is whether karyopherins other than importin beta regulate mitotic assembly events. It is widely thought that importin beta binds the SAFs it regulates in mitosis by binding to their nuclear localization sequences (NLSs). Transportin, the first identified relative of importin beta, recognizes a different class of NLS that, although varying in length and sequence, is often characterized by a Proline-Tyrosine (PY) dipeptide [80,81]. PG, PV or PL NLS motifs as well as very different Lys/Arg basic NLSs are also recognized by transportin [82,83]. An extensive review of transportin in normal and disease contexts is available [84].
Transportin has been found to negatively regulate spindle assembly, nuclear membrane assembly, and nuclear pore assembly [49,52], by directly binding and inhibiting targets, in a manner parallel to importin beta. Addition of a super-affinity Transportin NLS (M9M) causes aster assembly throughout the extract cytoplasm, indicating that simply freeing transportin’s cargo is enough to initiate spindle and aster assembly. It was found that transportin also regulates nuclear envelope and NPC assembly via a direct inhibition model [52]. What are transportin’s assembly factor targets? For spindle assembly, a major likely target is the Nup107/160 Y complex, given that mitotic extracts depleted of the Nup107/160 Y complex fail to form bipolar spindles, and transportin blocks the binding of the Y complex to kinetochores in vitro [52] [27,38,41,85]. Thus, the network of regulatory importins is expanded to include both transportin and importin beta.
The exportin Crm1 is a cell cycle regulator and chemotherapy target
Crm1, also known as Exportin-1 or Xpo-1, is the major nuclear export receptor for protein cargos in interphase. Crm1 recognizes leucine-rich nuclear export signals (NESs) on a multitude of gene regulatory and nucleocytoplasmic shuttling proteins. Crm1 can only export its cargos in the form of a ternary complex, Crm1/NEScargo/RanGTP. Upon reaching the cytoplasmic face of the pore (where in mammals RanGAP is bound), RanGAP together with RanBP1 and RanBP2 stimulate RanGTP hydrolysis to disassemble the export complex (Figure 1C) [86].
In the past decade, a growing body of evidence has revealed that Crm1 plays essential roles in mitosis. However, instead of releasing assembly factors in areas of high RanGTP as do importin β and transportin, Crm1 binds to both RanGTP and key mitotic proteins to target those proteins to specific areas of the spindle.
Crm1 and Kinetochores
One key area is the kinetochore of mitotic chromosomes. Surprisingly, Crm1/Xpo1, localizes to the mitotic kinetochores of both yeast and humans [18]. Such Crm1 presence was shown to be needed for functional kinetochore nucleation of microtubules in humans [18,34,87]. Specifically, Crm1 targets a complex of RanBP2, RanGAP1 and RanGTP to the kinetochores of human cells (Figure 2A) [18,87,88]. With its partners, Crm1 is proposed to stabilize the connection of microtubule kinetochore fibers (k-fibers) to the kinetochore and, by doing so, promote proper chromosome segregation. In addition, Crm1 in human cells has been implicated in tethering the Chromosomal Passenger Complex (CPC) to the centromeric region of chromosomes, via the CPC Survivin protein’s NES domain [89]. A recent study delineating antagonistic roles of importin beta and Crm1 at human kinetochores reveals that overlapping karyopherin regulatory webs exist [90]. Lastly, it should be noted that Xenopus kinetochores have been mentioned to lack Crm1 and/or RanGAP presence (mentioned, but not shown in ref 87), but it is not known whether this apparent lack is due to antigen inaccessibility, less stable k-fibers, or actual absence of Crm1 (M. Dasso, personal communication).
Crm1 and centrosomes
Interestingly, Crm1 is also observed to be present at the centrosomes throughout the cell cycle (Figure 2C) [91]. It is proposed that Crm1 binds to RanGTP present in the centrosome, then recruits the major centrosomal scaffold protein, pericentrin. Pericentrin in turn is known to recruit γ-TuRC complexes and together these act to nucleate the centrosome-initiated spindle microtubules. Either RNAi depletion of Crm1 or overexpression of the N-terminal RanGTP-binding domain of Crm1 causes reduction in both pericentrin and γ-TuRCs at centrosomes and disrupts the mitotic spindles in cultured cells [91]. Also, Prior to its targeting, Crm1 is mitotically phosphorylated by the mitotic kinase CDK1/cyclin-B (Ser391), which enhances its ability to target RanBP2/RanGAP1 to the mitotic spindle [92].
Crm1 has also been observed to be involved in the targeting of NES-bearing proteins to the centrosome. BRCA1 and BARD1, an E3 ubiquitin ligase when heterodimerized, are both targeted to the centrosome by Crm1 independently of one another [93,94]. Normally, the BRCA1/BARD1 protein complex plays roles in DNA damage response and centrosome duplication, thus ensuring proper centrosome duplication. Correct centrosomal targeting of these proteins is critical, since perturbations to centrosome duplication can lead to inherited genetic defects and aneuploidy. For example, inhibition of Crm1 in early metaphase results in excess, acentriolar spindle poles [95].
Crm1, cancer and chemotherapy
Crm1 is the major nuclear export receptor for many DNA damage monitors and tumor repressor proteins, including p53, Rb, and FOXO [96,97]. The observed overexpression of Crm1 in many cancers results in preferential localization of these tumor suppressors to the cytoplasm where it is thought they are unable to function to subvert DNA damage and inappropriate cell proliferation [97]. Newly developed Small Inhibitors of Nuclear Export or SINEs represent a promising treatment for many cancer types. Modeled on an older inhibitor of Crm1 (Leptomycin B/LMB), SINEs prevent NES-cargo binding to vertebrate Crm1 by covalently binding to Cysteine 528 in the NES-binding cleft. SINEs are less toxic than LMB, as they are more highly specific to Crm1 [98]. Currently it is thought that the fact that SINEs block the export of many tumor suppressor proteins and other functionally relevant cell cycle inhibitors from the nucleus explains how SINEs act as cancer inhibitors in cultured cell studies, mouse studies, and an increasing number of human clinical trials. However, it would appear from the above considerations equally possible that SINEs could interfere with the mitotic roles of Crm1 delineated above, and thereby affect their observed block to cell proliferation in cancer trials.
In vivo evidence
Strong corroborative evidence for the karyopherin/Ran control of spindle assembly comes from a number of in vivo studies, only a few of which can be mentioned here. The presence of a RanGTP cloud around mitotic chromatin has been demonstrated in vivo using the fluorescent biosensor Rango, which increases its FRET signal when released from importin beta by RanGTP [99–101]. Increased RanGTP has also been observed using RanGTP biosensors around spindles assembled in vitro in mitotic Xenopus egg extracts [102]. Similarly, a RanGTP gradient has been observed in vivo in living mouse oocytes using FRET [64,103].
Excellent reviews of a number of seminal in vivo studies on the karyopherin/Ran dueling regulators by the Lavia group and others include [64,104]. Recently, Hasegawa et al [105] found a steep RanGTP gradient exists around the mitotic chromosomes of rapidly growing cells, while reduced RanGTP gradients are observed around the chromosomes in primary cells (HFF-1 cells). Interestingly, overexpression of the RanGEF RCC1 in these primary cells causes induction of a steep RanGTP gradient. Further, cell-cell fusion studies lead the authors to propose that chromosome gain can also increase the RanGTP gradient [105], a gain that might also be seen in cancer cells.
Strong in vivo evidence for importin beta’s role in mitosis comes from microinjection of different importin beta protein fragments into cells at prophase or prometaphase: a fragment of importin beta (aa 71–876) lacking the Ran-GTP binding domain caused blockage of spindle assembly and/or proper chromosome segregation in a majority of cells [4,106]. Thus, the above in vivo findings reinforce the in vitro findings: RanGTP and karyopherins are dueling regulators and a correct balance helps coordinate correct mitotic assembly events.
New cellular arenas for karyopherin/RanGTP regulation
Karyopherin/Ran and the timing of anaphase
Rape and colleagues [107,108] have discovered that -- once anaphase begins and the Anaphase Promoting Complex (APC/C) is activated by successful chromosome attachment -- a higher level of regulation takes over. They found that, unlike cyclin B1 which is ubiquitinated and degraded at the very onset of anaphase, SAFs such as TPX2, HURP, and NuSAP, remain stable through anaphase. These SAFs, presumably originally freed from importin beta by RanGTP, are now protected from ubiquitination and degradation through anaphase by their physical association with the spindle microtubules. Later, when released from microtubules, the SAFs are quickly modified by the APC/C and degraded. This protective mechanism ensures that these SAFs are maintained as long as the spindle needs them, and their subsequent degradation prepares conditions for the next phase of the cell cycle [107–110].
Spindle positioning along the cell axis
Often in cell division, the spindle in mitotic cells orients along the long axis of the cell. In a study designed to reveal why this positioning occurs, a new role for karyopherins and RanGTP was discovered. Proteins that generate pulling forces on the astral microtubules, LGN and NuMA, the latter of which was an early SAF target of importin α/β [9], were also found to be key for spindle positioning [111]. When an inhibitor of the RanGTP/importin beta interaction, importazole, was added to human cells, a misoriented spindle resulted [111]. It turned out that LGN and NuMA had become mislocalized. The addition of CLASP1, a protein that stabilizes aster microtubules, restored correct spindle orientation. It appears that the karyopherin/Ran system works with LGN and NuMA to define proper spindle orientation along the long axis of the cell [111,112] (Figure 3E).
Actin cytoskeleton regulation
Up to this point the cytoskeletal elements influenced by the karyopherin/RanGTP team have been microtubule-related. Now Samwer et al [113] have found an actin connection. They identify a novel actin-bundling kinesin, NabKin (for Nuclear and meiotic actin-bundling Kinesin), that binds to and stabilizes nuclear actin bundles in interphase and also stabilizes the actin-based cortical ring structure that divides cells during cytokinesis. In vitro, they find that importin beta blocks NabKin kinase interaction with filamentous actin, while RanGTP reverses this inhibition. They conclude that NabKin directly links microtubules to F-actin and does so in a classical karyopherin/Ran-regulated manner [113].
These new arenas can be added to the previous most unexpected area for karyopherin regulation, that of synapse-to-nucleus communication, where importin beta mediates the retrotranslocation of damage signals from an injured nerve terminus to its neuronal cell nucleus [10,114,115].
Perspectives
The karyopherins, importin beta and transportin, and their RanGTP counterpart are in fact a unique way to impose a wide-reaching regulatory regime over disparate cellular events. While kinase/phosphatase and ubiquitinase/proteolysis pairs depend on enzymatic amplification, the karyopherin/Ran paradigm depends instead on: (a) the high and pervasive concentrations (micromolar) of these transport receptors throughout the cytoplasm during mitosis, (b) the ability of each karyopherin to bind to a very broad spectrum of motifs and molecules, and (c) importantly, the focused production of RanGTP in specified locales for targeted activation of assembly factors and pathways. Perhaps rivaled only by the hsp70 chaperone family for versatility of binding, the added element of localized RanGTP production renders the evolutionary power of the karyopherin/Ran regulation even greater. It now stands out as one to be watched by all.
Acknowledgments
The current studies concerning karyopherins and RanGTP number in the thousands with an increasing fraction concerning mitotic roles. The overall mitotic literature is even larger; we very much regret that only a fraction could be cited here. This work was supported by a National Institutes of Health grant, R01-GM033279, to D.J. F. The authors would like to thank Dennis Garland for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- 2. Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9.. • References [2–7] represent the flurry of studies that first revealed karyopherin/RanGTP regulation as a cellular strategy for mitotic regulation.
- 3.Zhang C, Hughes M, Clarke PR. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci. 1999;112(Pt 14):2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]
- 4.Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 5.Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- 6.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 7.Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quimby BB, Dasso M. The small GTPase Ran: interpreting the signs. Curr Opin Cell Biol. 2003;15:338–344. doi: 10.1016/s0955-0674(03)00046-2. [DOI] [PubMed] [Google Scholar]
- 9.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026.. • A comprehensive and easy to read review of the first 10 years of karyopherin/RanGTP regulation of spindle assembly.
- 11.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohka MJ, Maller JL. Induction of nuclear envelope breakdown, chromosome condensation, and spindle formation in cell-free extracts. J Cell Biol. 1985;101:518–523. doi: 10.1083/jcb.101.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- 14.Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987;48:219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- 15.Chan RC, Forbes DI. In vitro study of nuclear assembly and nuclear import using Xenopus egg extracts. Methods Mol Biol. 2006;322:289–300. doi: 10.1007/978-1-59745-000-3_20. [DOI] [PubMed] [Google Scholar]
- 16.Bernis C, Forbes DJ. Analysis of nuclear reconstitution, nuclear envelope assembly, and nuclear pore assembly using Xenopus in vitro assays. Methods Cell Biol. 2014;122:165–191. doi: 10.1016/B978-0-12-417160-2.00008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama H, Gruss OJ, Rybina S, Caudron Mw, Schelder M, Wilm M, Mattaj IW, Karsenti E. Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. The Journal of Cell Biology. 2008;180:867–875. doi: 10.1083/jcb.200706189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J, Dasso M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol. 2005;7:626–632. doi: 10.1038/ncb1263. [DOI] [PubMed] [Google Scholar]
- 19. Zhang MS, Arnaoutov A, Dasso M. RanBP1 governs spindle assembly by defining mitotic Ran-GTP production. Dev Cell. 2014;31:393–404. doi: 10.1016/j.devcel.2014.10.014.. • This paper shows a new role for RanBP1 during mitosis. Using Xenopus M phase egg extracts, the authors determine that RanBP1 (normally a co-factor of RanGAP in interphase) binds to the RanGEF RCC1 and regulates both RCC1’s enzymatic activity and the spatial distribution of RanGTP production during mitosis.
- 20.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- 22.Schweizer N, Weiss M, Maiato H. The dynamic spindle matrix. Curr Opin Cell Biol. 2014;28:1–7. doi: 10.1016/j.ceb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Dikovskaya D, Li Z, Newton IP, Davidson I, Hutchins JR, Kalab P, Clarke PR, Nathke IS. Microtubule assembly by the Apc protein is regulated by importin-beta—RanGTP. J Cell Sci. 2010;123:736–746. doi: 10.1242/jcs.060806. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama H, Nakos K, Santarella-Mellwig R, Rybina S, Krijgsveld J, Koffa MD, Mattaj IW. CHD4 is a RanGTP-dependent MAP that stabilizes microtubules and regulates bipolar spindle formation. Curr Biol. 2013;23:2443–2451. doi: 10.1016/j.cub.2013.09.062. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama H, Rybina S, Santarella-Mellwig R, Mattaj IW, Karsenti E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J Cell Biol. 2009;187:813–829. doi: 10.1083/jcb.200906020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meunier S, Vernos I. K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat Cell Biol. 2011;13:1406–1414. doi: 10.1038/ncb2372. [DOI] [PubMed] [Google Scholar]
- 27.Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–2230. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatel G, Fahrenkrog B. Nucleoporins: Leaving the nuclear pore complex for a successful mitosis. Cellular Signalling. 2011;23:1555–1562. doi: 10.1016/j.cellsig.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Imamoto N, Funakoshi T. Nuclear pore dynamics during the cell cycle. Curr Opin Cell Biol. 2012;24:453–459. doi: 10.1016/j.ceb.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Sood V, Brickner JH. Nuclear pore interactions with the genome. Curr Opin Genet Dev. 2014;25:43–49. doi: 10.1016/j.gde.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pascual-Garcia P, Capelson M. Nuclear pores as versatile platforms for gene regulation. Curr Opin Genet Dev. 2014;25:110–117. doi: 10.1016/j.gde.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Mitchison TJ. Mechanism and function of poleward flux in Xenopus extract meiotic spindles. Philos Trans R Soc Lond B Biol Sci. 2005;360:623–629. doi: 10.1098/rstb.2004.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:R749–R761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell. 2008;19:1873–1882. doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helmke KJ, Heald R, Wilbur JD. Interplay between spindle architecture and function. Int Rev Cell Mol Biol. 2013;306:83–125. doi: 10.1016/B978-0-12-407694-5.00003-1. [DOI] [PubMed] [Google Scholar]
- 36.Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci. 2007;64:1891–1914. doi: 10.1007/s00018-007-6568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proceedings of the National Academy of Sciences. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich Chromatin by ELYS Recruits POM121 and NDC1 to Initiate Nuclear Pore Assembly. Mol. Biol. Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107–160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12:164–169. doi: 10.1038/ncb2016.. •• This article described a novel mitotic role for the nucleoporin Nup107–160 complex. Specifically, the Nup107–160 complex recruits γ-TuRCs (gamma-tubulin ring complexes) to the kinetochores of HeLa cells, and thus the Nup107–160 complex promotes spindle assembly through microtubule nucleation via γ-TuRCs at kinetochores.
- 43.Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yokoyama H, Koch B, Walczak R, Ciray-Duygu F, Gonzalez-Sanchez JC, Devos DP, Mattaj IW, Gruss OJ. The nucleoporin MEL-28 promotes RanGTP-dependent gamma-tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat Commun. 2014;5:3270. doi: 10.1038/ncomms4270.. • The authors of this study show that the nucleoporin MEL-28/ELYS functions as a RanGTP-dependent MAP (microtubule-associated protein) during mitosis, independent of its role in nuclear pore complex assembly. In Xenopus egg extracts, MEL-28/ELYS recruits Nup107–160 and γ-TuRC to the spindle and is required to promote microtubule nucleation and spindle assembly. References [38–42] suggested this role.
- 45.Scrofani J, Sardon T, Meunier S, Vernos I. Microtubule Nucleation in Mitosis by a RanGTP-Dependent Protein Complex. Curr Biol. 2015;25:131–140. doi: 10.1016/j.cub.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 46.Di Fiore B, Ciciarello M, Lavia P. Mitotic functions of the Ran GTPase network: the importance of being in the right place at the right time. Cell Cycle. 2004;3:305–313. [PubMed] [Google Scholar]
- 47.Pinyol R, Scrofani J, Vernos I. The role of NEDD1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr Biol. 2013;23:143–149. doi: 10.1016/j.cub.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 48.Shaulov L, Gruber R, Cohen I, Harel A. A dominant-negative form of POM121 binds chromatin and disrupts the two separate modes of nuclear pore assembly. J Cell Sci. 2011;124:3822–3834. doi: 10.1242/jcs.086660. [DOI] [PubMed] [Google Scholar]
- 49.Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, Rasala BA, Forbes DJ. Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol Biol Cell. 2009;20:4043–4058. doi: 10.1091/mbc.E09-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell. 2009;20:4031–4042. doi: 10.1091/mbc.E09-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 52. Bernis C, Swift-Taylor B, Nord M, Carmona S, Chook YM, Forbes DJ. Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration. Mol Biol Cell. 2014;25:992–1009. doi: 10.1091/mbc.E13-08-0506.. •• This paper, following up the work of Ref. [49], addresses mechanism by which transportin acts in mitosis. It demonstrates that the nuclear import receptor, transportin, regulates the major mitotic events of spindle assembly, nuclear membrane assembly, and nuclear pore assembly using a direct inhibition mechanism, binding to and inhibiting assembly factors in a way parallel to that of importin beta.
- 53. Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044.. •• A new and surprising mechanism for microtubule nucleation was described using Xenopus mitotic egg extracts. With TIRF microscopy, this paper shows for the first time how new microtubules can be initiated by branching from existing spindle microtubules. This process is dependent on augmin, γ-tubulin, and TPX2, and is stimulated by RanGTP.
- 54.Zheng Y, Iglesias PA. Nucleating new branches from old. Cell. 2013;152:669–670. doi: 10.1016/j.cell.2013.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petry S, Pugieux C, Nedelec FJ, Vale RD. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc Natl Acad Sci U S A. 2011;108:14473–14478. doi: 10.1073/pnas.1110412108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petry S, Vale RD. A new cap for kinetochore fibre minus ends. Nat Cell Biol. 2011;13:1389–1391. doi: 10.1038/ncb2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cross MK, Powers MA. Nup98 regulates bipolar spindle assembly through association with microtubules and opposition of MCAK. Mol Biol Cell. 2011;22:661–672. doi: 10.1091/mbc.E10-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan KJ, McCaffery JM, Wente SR. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J Cell Biol. 2003;160:1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450.. See annotation to Ref. [60•].
- 60. Fan S, Whiteman EL, Hurd TW, McIntyre JC, Dishinger JF, Liu CJ, Martens JR, Verhey KJ, Sajjan U, Margolis B. Induction of Ran GTP drives ciliogenesis. Mol Biol Cell. 2011;22:4539–4548. doi: 10.1091/mbc.E11-03-0267.. •• These papers discuss a set of recent findings showing that cilia are compartments containing high RanGTP, that cargo enters the ciliary compartment using repurposed nuclear import receptors such as transportin, and that the “gate” they enter through is constructed in part of nuclear pore proteins. New roles for RanGTP are described not only in regulating ciliary protein transport, but also in stimulating cilia formation.
- 61.Arnaoutov A, Dasso M. The Ran GTPase Regulates Kinetochore Function. Developmental Cell. 2003;5:99–111. doi: 10.1016/s1534-5807(03)00194-1. [DOI] [PubMed] [Google Scholar]
- 62.Hutchins JR, Moore WJ, Hood FE, Wilson JS, Andrews PD, Swedlow JR, Clarke PR. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr Biol. 2004;14:1099–1104. doi: 10.1016/j.cub.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 63. Halpin D, Kalab P, Wang J, Weis K, Heald R. Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts. PLoS Biol. 2011;9:e1001225. doi: 10.1371/journal.pbio.1001225.. • Using purified RCC1-coated beads in Xenopus egg extracts, the authors show that RCC1 alone is sufficient to induce bipolar spindle formation in vitro.
- 64.Kalab P, Solc P, Motlik J. The role of RanGTP gradient in vertebrate oocyte maturation. Results Probl Cell Differ. 2011;53:235–267. doi: 10.1007/978-3-642-19065-0_12. [DOI] [PubMed] [Google Scholar]
- 65.Helmke KJ, Heald R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J Cell Biol. 2014;206:385–393. doi: 10.1083/jcb.201401014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440.. See annotation to Ref. [111•].
- 68.Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- 69.Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hachet V, Kocher T, Wilm M, Mattaj IW. Importin alpha associates with membranes and participates in nuclear envelope assembly in vitro. Embo J. 2004;23:1526–1535. doi: 10.1038/sj.emboj.7600154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 72.Hetzer M, Gruss OJ, Mattaj IW. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol. 2002;4:E177–E184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- 73.Lu X, Shi Y, Lu Q, Ma Y, Luo J, Wang Q, Ji J, Jiang Q, Zhang C. Requirement for lamin B receptor and its regulation by importin {beta} and phosphorylation in nuclear envelope assembly during mitotic exit. J Biol Chem. 2010;285:33281–33293. doi: 10.1074/jbc.M110.102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delmar VA, Chan RC, Forbes DJ. Xenopus importin beta validates human importin beta as a cell cycle negative regulator. BMC Cell Biol. 2008;9:14. doi: 10.1186/1471-2121-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doucet CM, Talamas JA, Hetzer MW. Cell Cycle-Dependent Differences in Nuclear Pore Complex Assembly in Metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell. 2011;22:1058–1069. doi: 10.1091/mbc.E10-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wandke C, Kutay U. Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell. 2013;152:1222–1225. doi: 10.1016/j.cell.2013.02.046.. • An excellent review of nuclear assembly.
- 78.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends in Cell Biology. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 80.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM. Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kimura M, Kose S, Okumura N, Imai K, Furuta M, Sakiyama N, Tomii K, Horton P, Takao T, Imamoto N. Identification of cargo proteins specific for the nucleocytoplasmic transport carrier transportin by combination of an in vitro transport system and stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics. Mol Cell Proteomics. 2013;12:145–157. doi: 10.1074/mcp.M112.019414.. • An innovative technique for identifying NLS cargos for the import receptor transportin. New NLS types are also identified.
- 84.Twyffels L, Gueydan C, Kruys V. Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett. 2014;588:1857–1868. doi: 10.1016/j.febslet.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 85.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, et al. The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. Embo J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bischoff FR, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 87.Arnaoutov A, Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 2005;4:1161–1165. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- 88.Clarke PR. The Crm de la creme of mitosis. Nat Cell Biol. 2005;7:551–552. doi: 10.1038/ncb0605-551. [DOI] [PubMed] [Google Scholar]
- 89.Knauer SK, Bier C, Habtemichael N, Stauber RH. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 2006;7:1259–1265. doi: 10.1038/sj.embor.7400824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Roscioli E, Di Francesco L, Bolognesi A, Giubettini M, Orlando S, Harel A, Schinina ME, Lavia P. Importin-beta negatively regulates multiple aspects of mitosis including RANGAP1 recruitment to kinetochores. J Cell Biol. 2012;196:435–450. doi: 10.1083/jcb.201109104.. • This article finds that importin beta inhibits RanGAP1 binding to the kinetochores in human cells, an inhibition that can be rescued by overexpression of Crm1. These results reveal a new antagonistic role between importin beta and Crm1 in the delivery of RanGAP1 to the kinetochores.
- 91.Liu Q, Jiang Q, Zhang C. A fraction of Crm1 locates at centrosomes by its CRIME domain and regulates the centrosomal localization of pericentrin. Biochem Biophys Res Commun. 2009;384:383–388. doi: 10.1016/j.bbrc.2009.04.154. [DOI] [PubMed] [Google Scholar]
- 92.Wu Z, Jiang Q, Clarke PR, Zhang C. Phosphorylation of Crm1 by CDK1-cyclin-B promotes Ran-dependent mitotic spindle assembly. J Cell Sci. 2013;126:3417–3428. doi: 10.1242/jcs.126854. [DOI] [PubMed] [Google Scholar]
- 93.Brodie KM, Henderson BR. Characterization of BRCA1 protein targeting, dynamics, and function at the centrosome: a role for the nuclear export signal, CRM1, and Aurora A kinase. J Biol Chem. 2012;287:7701–7716. doi: 10.1074/jbc.M111.327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brodie KM, Mok MT, Henderson BR. Characterization of BARD1 targeting and dynamics at the centrosome: the role of CRM1, BRCA1 and the Q564H mutation. Cell Signal. 2012;24:451–459. doi: 10.1016/j.cellsig.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 95.Rousselet A. Inhibiting Crm1 causes the formation of excess acentriolar spindle poles containing NuMA and B23, but does not affect centrosome numbers. Biol Cell. 2009;101:679–693. doi: 10.1042/BC20080218. [DOI] [PubMed] [Google Scholar]
- 96. Mor A, White MA, Fontoura BM. Nuclear trafficking in health and disease. Curr Opin Cell Biol. 2014;28:28–35. doi: 10.1016/j.ceb.2014.01.007.. See comment to Ref. [98•].
- 97. Senapedis WT, Baloglu E, Landesman Y. Clinical translation of nuclear export inhibitors in cancer. Semin Cancer Biol. 2014;27:74–86. doi: 10.1016/j.semcancer.2014.04.005.. See annotation to Ref. [98•].
- 98. Tan DS, Bedard PL, Kuruvilla J, Siu LL, Razak AR. Promising SINEs for embargoing nuclear-cytoplasmic export as an anticancer strategy. Cancer Discov. 2014;4:527–537. doi: 10.1158/2159-8290.CD-13-1005.. •• These articles describe the role of Crm1 in cancer and the latest advances made in targeting nuclear export as an anticancer strategy using small inhibitors of nuclear export (SINEs). Clinical advances are summarized.
- 99.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 100.Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee YP, Wong CH, Chan KS, Lai SK, Koh CG, Li HY. In vivo FRET imaging revealed a regulatory role of RanGTP in kinetochore-microtubule attachments via Aurora B kinase. PLoS One. 2012;7:e45836. doi: 10.1371/journal.pone.0045836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 103.Dumont J, Verlhac MH. Using FRET to study RanGTP gradients in live mouse oocytes. Methods Mol Biol. 2013;957:107–120. doi: 10.1007/978-1-62703-191-2_7. [DOI] [PubMed] [Google Scholar]
- 104.Roscioli E, Bolognesi A, Guarguaglini G, Lavia P. Ran control of mitosis in human cells: gradients and local signals. Biochem Soc Trans. 2010;38:1709–1714. doi: 10.1042/BST0381709. [DOI] [PubMed] [Google Scholar]
- 105. Hasegawa K, Ryu SJ, Kalab P. Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J Cell Biol. 2013;200:151–161. doi: 10.1083/jcb.201206142.. • By comparing highly proliferative HeLa cells with slow-growing primary HFF-1 fibroblasts, the authors find that the RanGTP gradient over the mitotic chromosomes of HeLa cells is a steep gradient, but a less steep gradient is found in primary cells. Overexpressing the RanGEF RCC1 or, alternatively, increasing the number of chromosomes/cell by means of cell-cell fusion was observed to now induce a steep mitotic RanGTP gradient in HFF-1 primary cells, one comparable to that in proliferative HeLa cells.
- 106.Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. Embo J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038.. See annotation to Ref. [108•].
- 108. Song L, Craney A, Rape M. Microtubule-dependent regulation of mitotic protein degradation. Mol Cell. 2014;53:179–192. doi: 10.1016/j.molcel.2013.12.022.. •• This work reveals that the binding to spindle microtubules protects different spindle assembly factors (SAFs), such as TPX2, HURP and NuSAP, from degradation at anaphase via the APC/C and proteosome. This protective mechanism allows for selective disposal of SAFs only after they have performed their mitotic functions.
- 109.Vaites LL, Harper JW. Spindle assembly factor protection. Mol Cell. 2014;53:165–166. doi: 10.1016/j.molcel.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 110.Wrighton KH. Mitosis: Microtubules protect spindle assembly factors. Nat Rev Mol Cell Biol. 2014;15:150–151. doi: 10.1038/nrm3759. [DOI] [PubMed] [Google Scholar]
- 111. Bird SL, Heald R, Weis K. RanGTP and CLASP1 cooperate to position the mitotic spindle. Mol Biol Cell. 2013;24:2506–2514. doi: 10.1091/mbc.E13-03-0150.. •• This Ref. [111] and Ref. [67] show that the positioning of the mitotic spindle in human cells is regulated by importin beta and RanGTP through their control of the cortical localization of two proteins that generate pull on the astral microtubules of the spindle, LGN and NuMA. CLASP1 also plays a role in this regulation by stabilizing astral microtubule interactions with the cortex.
- 112.McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Samwer M, Dehne HJ, Spira F, Kollmar M, Gerlich DW, Urlaub H, Gorlich D. The nuclear F-actin interactome of Xenopus oocytes reveals an actin-bundling kinesin that is essential for meiotic cytokinesis. Embo j. 2013;32:1886–1902. doi: 10.1038/emboj.2013.108.. •• The study reveals the role of importin beta and RanGTP in regulating NabKin (Nuclear and meiotic actin-bundling Kinesin), which helps mediate actin bundle interactions with microtubules during cytokinesis. Interference leads to cytokinesis defects. This novel instance of importin beta and RanGTP regulation of an actin cytoskeletal event is not expected to be the last.
- 114.Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 115. Panayotis N, Karpova A, Kreutz MR, Fainzilber M. Macromolecular transport in synapse to nucleus communication. Trends Neurosci. 2015;38:108–116. doi: 10.1016/j.tins.2014.12.001.. • The review highlights the recent findings involving the role of importins in the larger field of synapse-to-nucleus signaling.