Abstract
We previously reported that the Wnt pathway is preferentially activated in basal-like breast cancer. However, the mechanisms by which the Wnt pathway regulates down-stream targets in basal-like breast cancer, and the biological significance of this regulation, are poorly understood. In this study, we found that c-Myc is highly expressed in the basal-like subtype by microarray analyses and immunohistochemical staining. After silencing β-catenin using siRNA, c-Myc expression was decreased in non-basal-like breast cancer cells. In contrast, c-Myc mRNA and protein expression was up-regulated in the basal-like breast cancer cell lines. Decreased c-Myc promoter activity was observed after inhibiting β-catenin by siRNA in non-basal-like breast cancer cells; however, inhibition of β-catenin or over-expression of dominant-negative LEF1 had no effect on c-Myc promoter activity in basal-like breast cancer cell lines. In addition, CDKN1A mRNA and p21 protein expression were significantly increased in all breast cancer cell lines upon β-catenin silencing. Interestingly, inhibiting β-catenin expression alone did not induce apoptosis in breast cancer cell lines despite c-Myc regulation, but we observed a modest increase of cells in the G1 phase of the cell cycle and decrease of cells in S phase upon β-catenin silencing. Our findings suggest that the regulation of c-Myc in breast cancer cells is dependent on the molecular subtype, and that β-catenin-mediated regulation of c-Myc and p21 may control the balance of cell death and proliferation in breast cancer.
Keywords: basal-like breast cancer, β-catenin, c-Myc, CDKN1A, Wnt
Introduction
Triple negative breast cancer (TNBC, estrogen receptor-negative, progesterone receptor-negative and Her2/Neu-negative) is well known for its poor prognosis and lack of therapeutic targets [1]. However, TNBC is quite heterogeneous. In a recent study with gene expression profiles from 21 breast cancer data sets and 587 TNBC cases, TNBC was classified into 6 subtypes, including two basal-like, an immunomodulatory, a mesenchymal, a mesenchymal stem-like, and a luminal androgen receptor subtype [2]. In immunohistochemical (IHC) staining, basal-like breast cancers usually are triple negative, cytokeratin 5/6 positive, and/or epidermal growth factor receptor positive [3]. Basal-like breast cancer constitutes one of the most challenging subtypes of breast cancers. Although it only accounts for about 10–15% of breast cancer cases, basal-like breast cancer is responsible for a disproportionate number of breast cancer deaths [4]. This subtype of tumors typically has an early age of onset [5, 6], a strong tendency to metastasize to other organs such as the brain and lung [7, 8] and a lack of therapeutic targets[9, 10]. The molecular pathways leading to the development of basal-like breast cancer are not well understood.
Wnt/β-catenin signaling is deregulated in most human cancers [11]. However, the role of Wnt/β-catenin in breast cancer is not as clear as it is in colon cancer and hepatocarcinoma. Mutations of Wnt pathway components, such as APC, CTNNB1 and Axin are rare in breast cancer [12–14]. In addition, results of IHC staining with β-catenin antibodies in breast tumors were inconsistent [15–18]. Moreover, no endogenous TCF reporter activity was detected in breast cancer cell lines [19, 20]. Much of the work has focused upstream of β-catenin, especially at the ligands level. Autocrine Wnt signaling was identified in breast cancer cell lines [21, 22]. In recent years, there is accumulation of subtype-based analysis of the Wnt pathway in breast cancer. We reported that nuclear and cytosolic accumulation of β-catenin was enriched in basal-like breast cancer and correlated with poor prognosis and metastasis, suggesting robust Wnt pathway activation in this specific subtype [23]. Reis-Filho’s group described that Wnt pathway activation in breast cancer is associated with the triple negative phenotype but not with CTNNB1 mutation [24]. Yang et al reported that Wnt component FZD7 over-expression is essential for tumorigenesis of TNBC [25]. Most recently, Dey et al. demonstrated that there is a subtype-specific up-regulation of the Wnt pathway in TNBC as compared to luminal (HR+) or HER2+ tumors. In contrast to CTNNB1 mRNA levels, β-catenin protein expression was significantly higher in TNBC cell lines compared with the other two subtypes [26, 27].
The proto-oncogene c-Myc is a potent activator of tumorigenesis and is deregulated in a variety of cancers [28]. The c-Myc gene is highly expressed in basal-like breast tumors based on gene expression analysis [29–31]. This suggests that c-Myc may play an important role in defining basal-like breast cancer. c-Myc is a downstream effector of β-catenin in colorectal cancer [32]. A study showed that c-Myc activates Wnt in breast cancer by suppressing the Wnt inhibitors DKK1 and SFRP1, which are strongly repressed in breast cancer cell lines [33]. However, exactly how the Wnt pathway regulates c-Myc and other down-stream targets in breast cancer and the biological significance are still unclear. In this study, we found that the regulation of c-Myc in breast cancer cells is dependent on the molecular subtype, and that β-catenin-mediated control of c-Myc and p21 may determine the balance of cell death and proliferation in breast cancer by TCF-independent mechanisms.
Materials and Methods
Patient materials
The research protocols were approved by the Institutional Review Board of the University of Chicago and University of North Carolina. 168 sporadic breast cancer cases were selected from the tumor bank based on tissue availability from cases diagnosed between 1992 and 2002. Tissue sections containing >50% tumor cells were selected after microscopic examination.
cDNA microarrays
RNAs were extracted from 168 fresh frozen invasive breast carcinomas. Microarray was performed in Dr Perou’s laboratory at University of North Carolina, Chapel Hill using Agilent oligo microarrays (Agilent Technologies, United States). All primary microarray data are in the Gene Expression Omnibus (GEO) under the accession number of GSE1992. Gene expression data were retrieved from the microarray database where the Lowess normalization procedure and data filtering was performed. In order to identify genes whose expression distinguishes basal-like tumors from all other tumor subtypes combined, we performed a two-class unpaired significance analysis of microarrays (SAM, http://www-stat.stanford.edu/~tibs/SAM/) with a false discovery rate (FDR) <5%.
IHC assays
IHC assays were performed as described [23], using a DAKO immunostainer (DAKO, Carpinteria, CA) with antibodies against c-Myc (N262, Santa Cruz Biotechnology, Santa Cruz, CA). 41 patients sample with gene expression data were used for the analysis. Archival formalin-fixed and paraffin-embedded tissues of breast cancer patients were obtained from the surgical pathology archive of the University of Chicago for tissue microarrays (TMA) construction. Cell lines HCC38 and UACC3199 were pelleted and processed by formalin-fixation and paraffin-embedding, then stained with c-Myc antibody and served as positive control. Immunostaining was scored semi-quantitatively by two observers (G. Khramtsova and A. Khramtsov). Scoring was based on intensity and percentage of positive stained cells (nuclei, cytoplasm and membrane respectively). All discrepancies were resolved by a second examination by two observers simultaneously using a multiheaded microscope. The results were scored using a scale from 0 to 3. Weak staining in <15% of tumor cell nuclei was considered as 0 (“negative”), whereas a score of 1 (“weak”) was assigned when >15% of the nuclei were weak positive. Scores 2 and 3 (strong immunoreactivity in ≥15% of the nuclei) were considered together as “strong” signal.
Cell lines
MCF-7, MDA-MB-231, HCC1937 and HCC38 cell lines were obtained from the American Type Culture Collection (Rockville, MD). UACC3199 was obtained from the University of Arizona Cancer Center (Tucson, AZ). MCF-7 was cultured in DMEM (Fisher Scientific, Hanover Park, IL). All other cell lines were grown in RPMI 1640 (Invitrogen, Grand Island, NY). The media were supplemented with 10% FBS and 1% penicillin/streptomycin.
siRNA transfection and cell growth assay
MDA-MB-231 or UACC3199 cells were plated at 1 × 105 per well in 12-well plate. The cells were transfected with si-β-catenin (Hs_CTNNB1_5_HP validated siRNA) or si-control using the HiPerfect transfection reagent (all from Qiagen, Valencia, CA), three days and six days after transfection, cell growth was measured by counting live cells after staining with trypan blue.
Cell cycle analysis
MDA-MB-231 or UACC-3199 cells were seeded at 5 × 105 cells/well in 6-well plates. Cells were transfected with si-β-catenin or si-control. 48 hours after transfection, the cells were harvested for cell cycle analysis in trypsin and washed twice with cold phosphate buffered saline (PBS). Cells were fixed in 100% ethanol for at least 24 hrs and stored at −20°C. Cells were centrifuged at 2000 rpm for 10mins at 4°C, washed twice with cold PBS, re-suspended in propidium iodide (PI) solution (0.1% triton X, 20 µg/ml PI and 0.4mg RNaseA), incubated at room temperature for 30mins. DNA content was measured using the LSRII by the Flow Cytometry Facility at the University of Chicago.
Apoptosis assay
Cells in 6-well plate were transfected with either si-β-catenin or si-control for 24 hrs. Cells were transferred to 96-well plates at the density of 10,000 cells/well. Apoptosis was measured 48 hrs later using the Caspase–glo 3/7 assay system (Promega, Madison, WI). 100 µL of caspase reagent was added directly to the treated cells, followed by one hour incubation, and luminescence activity was measure on a Bio-Tek plate reader.
RNA isolation and quantitative RT-PCR
RNA extraction and cDNA synthesis were carried out as described [34]. Real time PCR was performed on an ABI 7900 instrument with power SYBR Green (Applied Biosystems). The primer sequences for c-Myc: Forward 5’-CAG CTG CTT AGA CGC TGG ATT T-3’; Reverse 5’-ACC GAG TCG TAG TCG AGG TCA T-3’.The primer sequences for CTNNB1: Forward 5’-AAT ACC ATT CCA TTG TTT GTG CAG-3’; Reverse 5’-AGC TCA ACT GAA AGC CGT TT-3’. The primer sequences for CDKN1A: Forward 5’-GCG ATG GAA CTT CGA CTT TGT-3’; Reverse 5’-GGG CTT CCT CTT GGA GAA GAT-3’. The primer sequences for ACTB: Forward: 5′-CGG TCA GGT CAT CAC TAT CGG-3′; Reverse: 5′-CAC AGG ATT CCA TAC CCA GGA-3′. ACTB was used as a control for normalization.
Protein extraction and Western blot
Protein extraction and Western blot were carried out as previously described [34]. Western blot was repeated at least one time. Anti-β-catenin antibody was from BD Transduction (Cat. No. 610154, Franklin Lakes, NJ). Anti-c-Myc (N262) antibody was purchased from Santa Cruz Biotechnology (Cat. No. Sc-764, Santa Cruz, CA). Anti-p21 monoclonal antibody was from Millipore (Cat. No. 05–345, Temecula, CA). Anti-β-actin antibody was from Sigma-Aldrich (Cat. No. A2228, Santa Louis, MO)
Luciferase assay
Luciferase activity was measured with the Dual Luciferase Assay system (Promega, WI). A renilla luciferase reporter construct, pGL4.75 (Promega), was used as a transfection efficiency control. FuGENE HD (Roche Molecular Biochemicals) was used as the transfection reagent. The cell lysate was prepared using Passive Lysis Buffer (Promega). The Luc-c-Myc promoter was kindly provided by Dr. Kato of Tsukuba University, Japan [35]. Mutant β-cateninS37A was from Stephen Byers at Georgetown University.
Statistical analyses
Two-factor analysis of variance (ANOVA) was used for gene expression analysis with real-time PCR. Fisher's exact test was used to examine whether there is difference in IHC staining of c-Myc between basal-like breast cancer and non-basal-like breast cancer. The Student’s t-test was used for the cell growth and apoptosis assay. P<0.05 is considered statistically significant.
Results
c-Myc is highly expressed in basal-like breast cancer
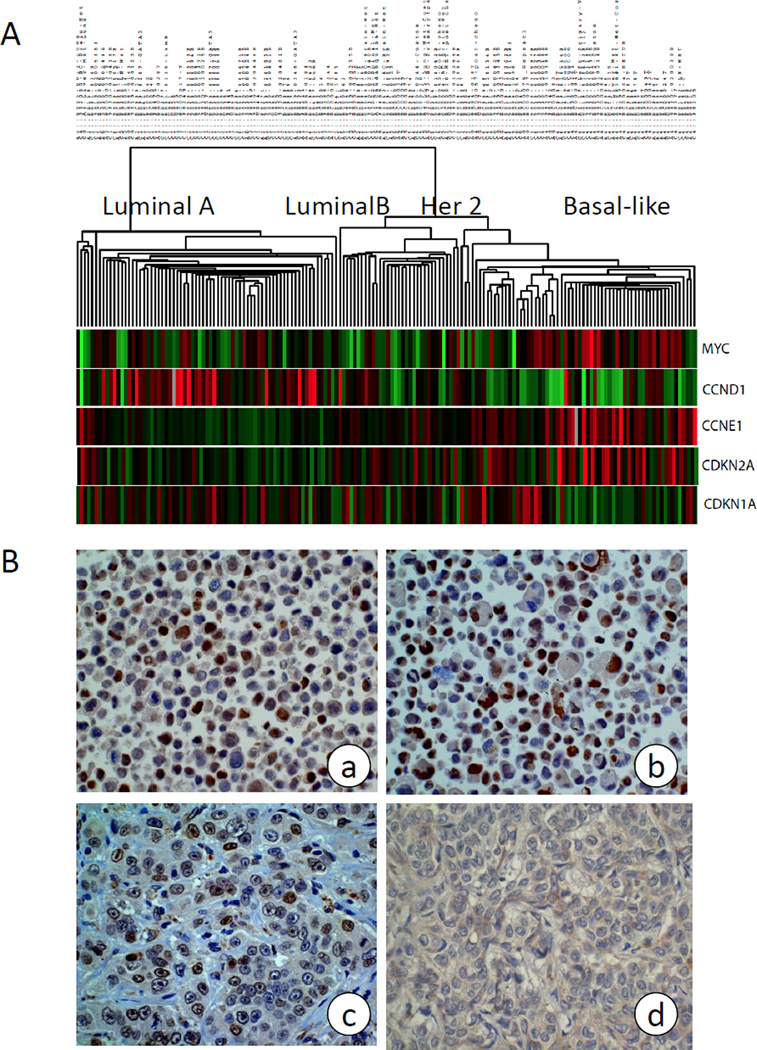
By analyzing the gene expression profiles of 168 breast tumors using DNA microarrays, we found that c-Myc was highly expressed in the basal-like subtype in comparison to the other four breast cancer subtypes [31] (Figure 1A). In addition, CCNE1 and CDKN2A were up-regulated in basal-like tumors as previously reported [36]. In contrast, CDKN1A was down-regulated. To validate this observation, we analyzed c-Myc protein expression by IHC in 41 primary breast tumors that were classified with DNA microarray analysis (Fig. 1B). Cell lines HCC38 and UACC3199 (Fig. 1B a and b) were used as positive control. As shown in Table 1, c-Myc was highly expressed in 79% of basal-like tumors; in contrast, only 26% of non-basal-like tumors showed high level c-Myc expression. This difference was statistically significant (p=0.005 by Fisher’s exact test). In addition, we confirmed the concordant expression of beta-catenin and c-Myc in 9 of 14 basal-like breast tumors although it did not reach statistical significance due to small sample size.
Figure 1. c-Myc is highly expressed in basal-like breast cancer.
(A) Microarray analysis of 168 primary tumors. Supervised clustering shows c-Myc, CCNE1 and CDKN2A are highly expressed, while expression of CCND1 and CDKN1A is low in basal-like breast cancer compared with other subtypes.
(B) c-Myc protein expression analyzed by IHC with basal-like breast cancer cell line UACC3199 (a) and HCC38 (b), and with Myc high (c) and Myc low (d) primary tumors. Cell lines HCC38 and UACC3199 were pelleted and processed by formalin-fixation and paraffin-embedding, then stained with c-Myc antibody and served as positive control.
Table 1.
c-Myc expression by IHC in 41 primary tumors with subtype status by microarray
| Non-basal-like | Basal-like | Total | |
|---|---|---|---|
| Myc-low | 20 [LumA: 9 ; LumB: 3 ; Her2: 8] (74%) | 3 (21%) | 23 (56%) |
| Myc-high | 7 [LumA: 4 ; LumB: 0 ; Her2: 3] (26%) | 11 (79%) | 18 (44%) |
| Total | 27 [LumA: 13 ; LumB: 3 ; Her2: 11] (66%) | 14 (34%) | 41 (100%) |
Basal-like vs. non-basal-like: P=0.005 by Fisher’s exact test.
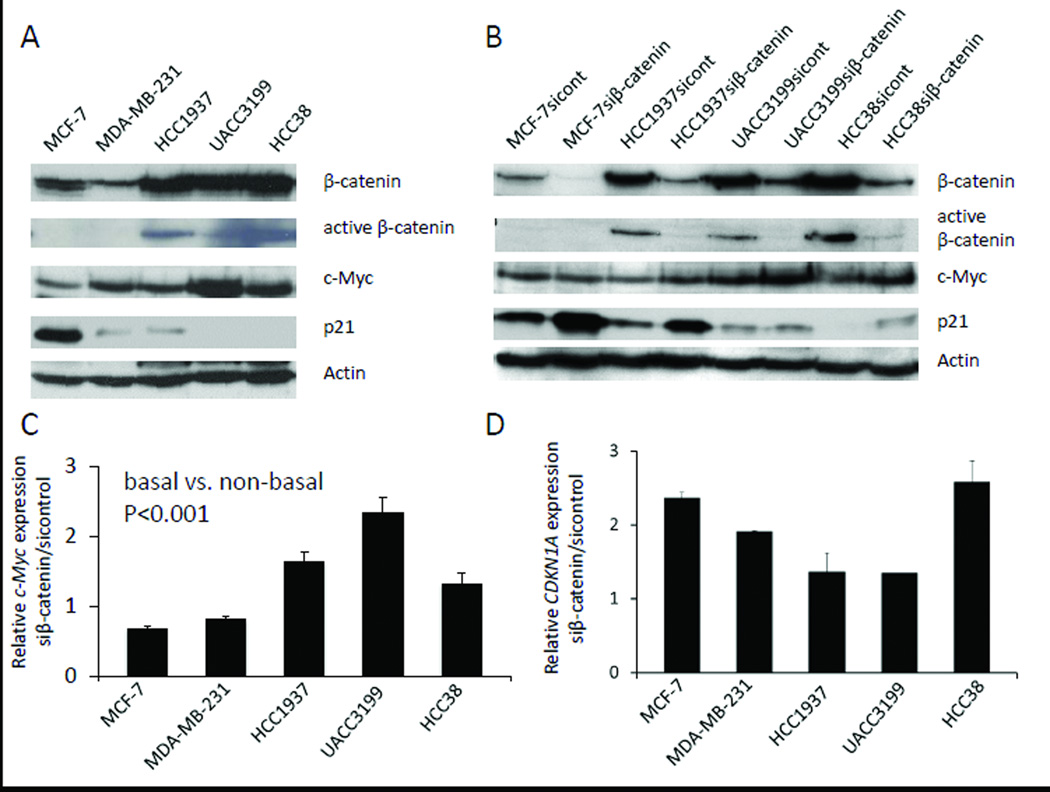
Protein expression of c-Myc was analyzed in five cell lines (MCF-7, MDA-MB-231, HCC1937, UACC3199 and HCC38) by Western blot. As shown in Figure 2A, c-Myc was highly expressed in the four TNBC cell lines (three basal-like cell lines HCC1937, UACC3199 and HCC38, and a claudin-low line MDA-MB-231) in comparison with the luminal cell line MCF-7. While, β-catenin proteins was highly expressed in the basal-like cell lines in comparison to non-basal-like cell lines MDA-MB-231and MCF-7, and the active form of β-catenin (unphosphorylated protein) was only detected in the basal-like breast cancer cells (Figure 2A and 2B). In contrast, no or very little p21 protein expression was detected in the three basal-like breast cancer cell lines and the claudin-low line MDA-MB-231compared to MCF-7 cells (Fig. 2A).
Figure 2. β-catenin regulates c-Myc and CDKN1A gene expression.
(A) Protein expression of β-catenin, active β-catenin, c-Myc and p21 in five breast cancer cell lines analyzed by Western blot. Actin was used as a loading control.
(B) Protein expression after knocking down β-catenin by siRNA in breast cancer cells. Scramble siRNA was used as a control.
(C) c-Myc mRNA expression by quantitative RT-PCR after 48 hr si-β-catenin treatment in breast cancer cells (basal vs. non-basal, P<0.001 by Two-factor ANOVA. For each cell lines, si-control vs. si-β-catenin P<0.05 by Two-factor ANOVA). Results are represented as the average of two independent experiments with triplicates. Bars represent mean ± SD.
(D) CDKN1A mRNA expression by quantitative RT-PCR after 48 hr si-β-catenin treatment in breast cancer cells. Results are represented as the average of two independent experiments with triplicates. Bars represent mean ± SD.
β-catenin regulates c-Myc and CDKN1A gene expression
To understand how β-catenin regulates c-Myc expression in breast cancer, we examined c-Myc mRNA and protein expression after knocking down β-catenin using siRNA transfection. c-Myc expression was decreased in MCF-7 and MDA-MB-231 cells by depletion of β-catenin. Surprisingly, c-Myc mRNA and protein expression were up-regulated in the three basal-like breast cancer cell lines after knocking down β-catenin (Figure 2C, 2B; Figure S1, S2). The difference between basal-like and non-basal-like was statistically significant (p<0.001). These data suggest that β-catenin functions as a repressor of c-Myc in basal-like breast cancer cells, but perhaps as an activator of c-Myc expression in non-basal-like breast cancer cells.
To examine how β-catenin regulates CDKN1A expression, we analyzed mRNA and protein expression of CDKN1A after depleting β-catenin in the same experimental setting. As shown in Fig. 2D and 2B, CDKN1A mRNA and protein expression levels were increased in all cell lines tested after the β-catenin knocking down, suggesting that β-catenin inhibits CDKN1A gene expression in breast cancer cells (Fig. 2B, Fig. S1 and S2). UACC3199 cells showed the least increase of CDKN1A mRNA and protein expression.
β-catenin regulates gene expression in a TCF-independent manner
To assess more completely the mechanism(s) by which β-catenin affects c-Myc transcription in more detail, luciferase assays were conducted with a c-Myc promoter construct in the five breast cancer cell lines (MCF-7, MDA-MB-231, HCC1937, UACC3199, and HCC38). We observed that inhibiting β-catenin expression caused reduction of c-Myc promoter activity in non-basal-like breast cancer cells but had no effect on c-Myc promoter activity in basal-like breast cancer cells (Fig. 3A). To understand whether β-catenin regulates c-Myc transcription through TCF/LEF activity, a dominant-negative form of LEF1 (dnLEF1) was co-transfected with the c-Myc promoter. Surprisingly, dnLEF1 had little effect on c-Myc promoter activity in all five cell lines that had been tested (Fig. 3B). This indicates that endogenous β-catenin regulates c-Myc transcription in a TCF-independent pathway in breast cancer. To further support this conclusion, luciferase assays were carried out in UACC3199 cells, with ectopic over-expression of S37A β-catenin, a stabilized mutant of β-catenin, we observed a four-fold increase of c-Myc promoter activity (Fig. 3C). Introduction of dnLEF1 suppressed S37A β-catenin induced-c-Myc promoter activity, but had no effect on the promoter activity if only endogenous β-catenin were present.
Figure 3. Luciferase promoter assay in breast cancer cell lines.
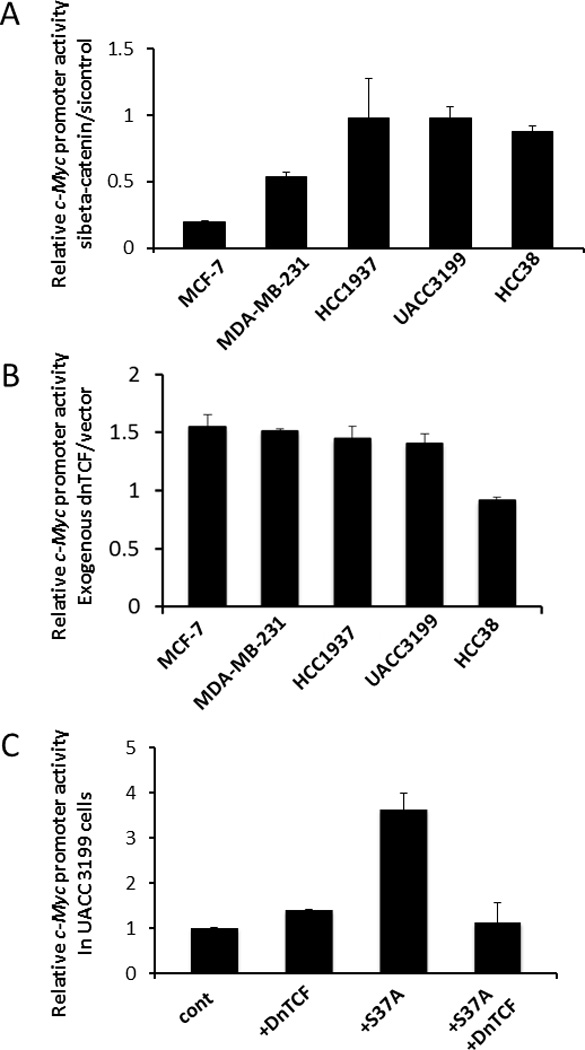
Luciferase activity was measured using a dual-luciferase reporter assay system. Results are represented as the average of two independent experiments with triplicates. Bars represent mean ± SD.
(A) c-Myc promoter assay after inhibiting β-catenin by siRNA for 48hr.
(B) Effect of dnLEF1 on c-Myc promoter activity.
(C) Effect of exogenous mutant β-catenin on c-Myc promoter activity in UACC3199 cells.
β-catenin inhibition does not induce c-Myc-dependent apoptosis in cultured breast cancer cells
Based on the observations that 1) β-catenin and c-Myc are highly expressed in basal-like breast cancer cells and 2) β-catenin functions as a repressor of c-Myc in basal-like breast cancer cells, we hypothesized that there was an antagonism between endogenous β-catenin and c-Myc in regulating apoptosis in basal-like breast cancer cells. We tested whether inhibiting β-catenin expression could sensitize the basal-like breast cancer cell lines to apoptosis. Apoptosis was measured with the caspase glo 3/7 assay after knocking down β-catenin by siRNA in the five breast cancer cell lines. As shown in Figure 4, suppressing β-catenin expression did not induce activation of caspase 3/7 in these breast cancer cell lines, suggesting that β-catenin inhibition alone was insufficient to induce c-Myc-dependent apoptosis in cultured breast cancer cells.
Figure 4. Suppression of β-catenin expression alone is insufficient to induce apoptosis in breast cancer cells.
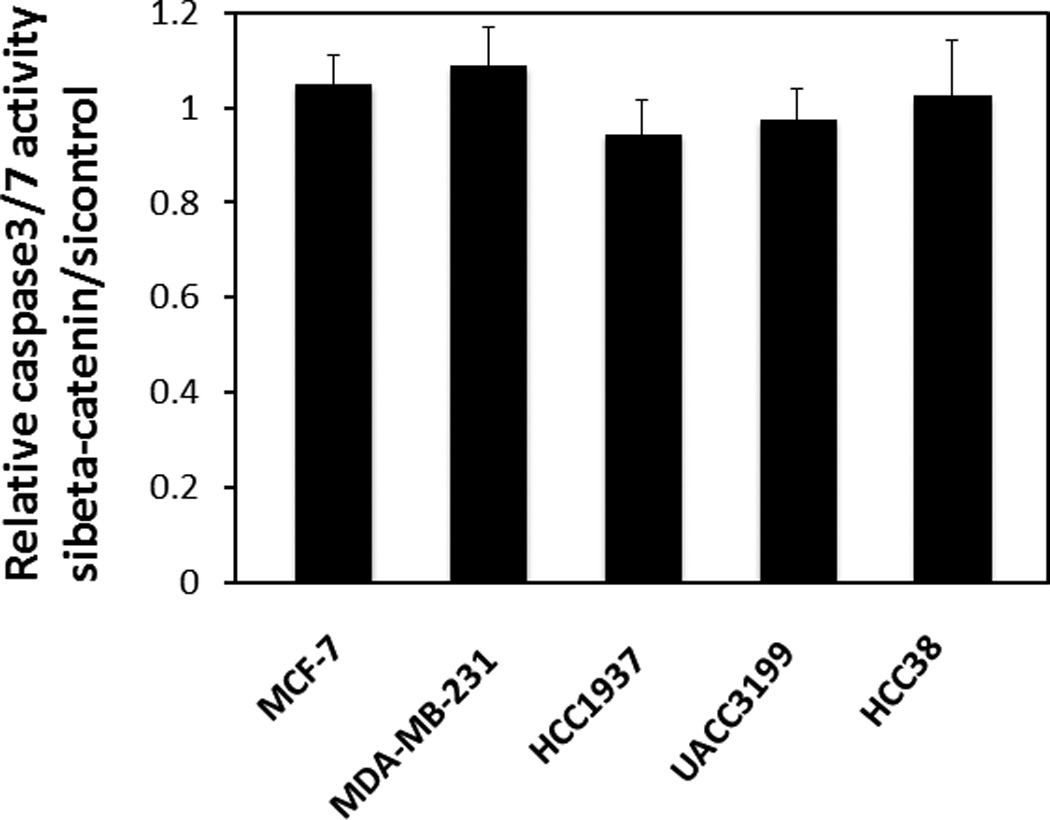
Apoptosis was analyzed by caspase-glo 3/7 assay. After siRNA treatment, the cells were starved in serum-free media for 48 hrs before apoptosis detection. The experiments were repeated three times. Bars represent mean ± SD.
β-catenin controls breast cancer cell cycle and growth
Because knocking down β-catenin induces up-regulation of CDKN1A gene expression, we tested the effect of β-catenin knockdown on cell cycle progression using MDA-MB-231 and UACC3199 cells. As shown in Table 2, in concordance with p21 up-regulation, we observed a subtle increase of cells in G1 and decrease of cells in S phase in both cell lines. This result indicates that inhibiting β-catenin partially blocks G1-S cell cycle progression.
Table 2.
Cell cycle analysis by FACS after 48 hr siRNA treatment
| MDA-MB-231 si-Control |
MDA-MB-231 si-β-catenin |
UACC3199 si-control |
UACC3199 si-β-catenin |
|
|---|---|---|---|---|
| G1% | 50.25±0.67 | 55±1.84 | 28.25±0.64 | 35.2±2.83 |
| S% | 28.25±3.46 | 22.3±4.1 | 42.8±1.56 | 35.4±2.4 |
| G2% | 15.3±0.57 | 17.7±1.84 | 21.15±0.49 | 20.7±2.4 |
Results are represented as the mean ± SD (Standard deviation) of two independent experiments.
Cell proliferation was analyzed by cell quantification three days and six days after siRNA transfection. There was no statistic difference of cell numbers with three days inhibition of β-catenin. We observed a 30% reduction of proliferation six days after β-catenin down-regulation (Fig. 5).
Figure 5. β-catenin knockdown by siRNA partially inhibits breast cancer cell proliferation.
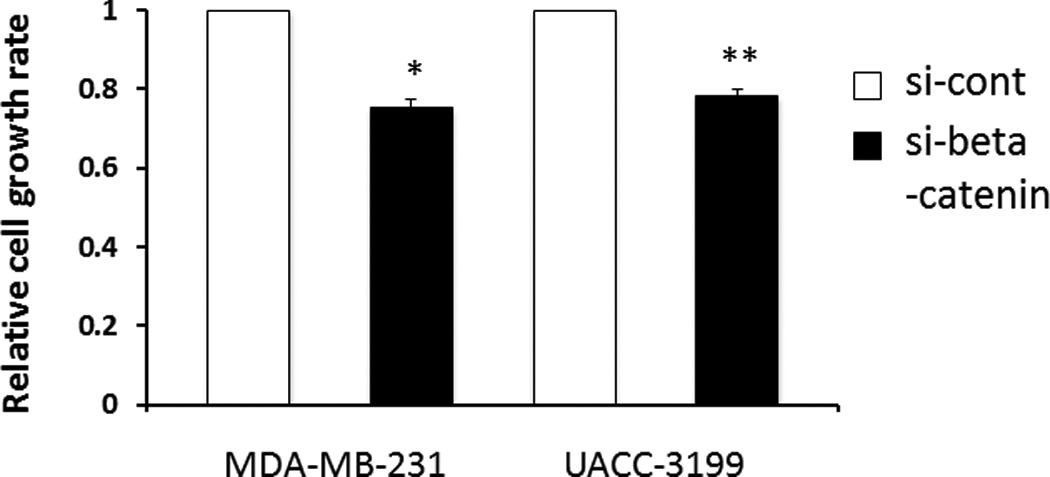
Cell proliferation was analyzed by cell counting at day 3 and day 6 after siRNA treatment. Relative growth at day 6 is shown (si-β-catenin vs. si-control, P<0.01 with the Student’s t-test). Results are represented as the average of two independent experiments performed in triplicate. Bars represent mean ± SD.
Discussion
In this study, using cell line models, we confirmed our previous observation that the Wnt pathway is preferentially activated in basal-like breast cancer [23]. We demonstrated that c-Myc is highly expressed in the basal-like subtype using microarray and IHC analyses. In contrast to the observation that β-catenin positively regulates c-Myc expression in colorectal cancer and hepatocarcinoma, β-catenin regulates c-Myc expression in a subtype dependent manner in breast cancer. After silencing β-catenin using siRNA, c-Myc expression was decreased in non-basal-like breast cancer cells. However, c-Myc mRNA and protein expression was up-regulated in the basal-like breast cancer cell lines. Interestingly, β-catenin regulates c-Myc in a TCF-independent manner. Furthermore, we found that β-catenin represses CDKN1A expression in a subtype-independent manner.
In general, c-Myc promotes cellular proliferation, which is necessary for tumorigenesis. However, over-expression of c-Myc causes apoptosis. Abrogation of c-Myc-induced apoptosis is crucial for cellular transformation and tumorigenesis. Basal-like tumor cell lines express high levels of both β-catenin and c-Myc. When β-catenin was knocked down with siRNA, we observed an unexpected increase of c-Myc expression. For that reason, we propose that β-catenin antagonizes c-Myc expression. While appearing contradictive in some regards, one could view this as a survival mechanism for tumor cells such that c-Myc level is tightly controlled so as not to be too high in basal-like tumor cells; otherwise, the cells would undergo apoptosis. Consistent with this interpretation, it has been reported that Wnt/β-catenin signaling suppressed apoptosis by inhibiting c-Myc-induced caspase activation when over-expressing both Wnt1 and c-Myc in Rat-1 and RIE cell lines [37]. On the other hand, we could not exclude the possibility that the antagonism only occurs when the expression of both β-catenin and c-Myc has reached certain levels. Since silencing β-catenin caused up-regulation of c-Myc expression, we hypothesized that inhibition of β-catenin in basal-like breast cancer may promote c-Myc-induced apoptosis. However, inhibition of endogenous β-catenin expression does not induce apoptosis in basal-like breast cancer cell lines as measured by caspase 3/7 activation. This suggests that alternative pathways exist to prevent c-Myc-induced apoptosis and inhibiting β-catenin alone is not enough to induce apoptosis. There are multiple mechanisms involved in c-Myc deregulation in breast cancer including gene amplification, transcriptional regulation, and mRNA and protein stabilization [31]. In addition, high level of c-Myc expression is associated with poor outcomes [31]. However, the mechanism by which c-Myc is highly expressed in basal-like breast tumors is still unknown. Furthermore, there are many genes expressed along with c-Myc, and comprehensive transcriptome analysis is needed in future to fully appreciate the upstream regulators and downstream effectors of c-Myc.
p21 has been shown to be a direct target of c-Myc, and c-Myc promotes cell proliferation through inhibition of p21 [38, 39]. In this study, we observed that β-catenin represses p21 transcription in addition to c-Myc. Inhibition of β-catenin caused up-regulation of p21 at mRNA and protein levels even though the c-Myc expression level was increased in basal-like breast cancer after treatment, resulting in a G1 cell cycle arrest and a reduction of cell proliferation. UACC3199 cells showed the most modest increase of CDKN1A mRNA and protein expression after β-catenin knockdown, probably due to repression of high levels of c-Myc in the cells. It has been reported that p21 was up-regulated in sFRP1-expressing tumors, suggesting that it is a downstream mediator of WNT signaling [40]. Our results demonstrate that β-catenin can directly regulate p21 expression. p21 has also been shown to have anti-apoptotic activity and probably plays a key role in cell survival following DNA damage by promoting the growth arrest that permits DNA repair [41]. This may partially explain why β-catenin knockdown did not influence apoptosis.
The central dogma of the canonical Wnt pathway is cytosolic β-catenin stabilization, nuclear translocation and transactivation of downstream gene targets after binding to TCF/LEF transcriptional factors. Surprisingly, our data suggest that β-catenin regulates c-Myc in a TCF-independent manner. Unlike most colon cancer cells, there was no TCF/LEF reporter activity in breast cancer cell lines [19, 20] irrespective of the presence of “active β-catenin” or not. Since silencing β-catenin in non-basal-like cells affects both endogenous c-Myc and c-Myc reporter activity, we suspect that a pool of β-catenin other than so-called “active β-catenin” is responsible for the regulation. These findings suggest that Wnt pathway regulation may be unique in breast cancer cells. Because the c-Myc reporter activity was extremely high (i.e., perhaps saturated) in basal-like cancer cells compared with non-basal-like, silencing β-catenin increased endogenous c-Myc expression in basal-like cells without increasing reporter activity. Exogenous S37A mutant activated the c-Myc reporter in basal-like UACC3199 cells in a TCF-dependent fashion, which could be antagonized by dnTCF1. These data collectively illustrate the complexity of β-catenin-mediated gene regulation and indicate that further study is warranted to understand the mechanism of TCF-independent regulation of gene expression via β-catenin, particularly if considering β-catenin as an anticancer therapeutic target.
In addition to recent reports concerning Wnt activation in basal-like and TNBC, Schade et al. reported that β-catenin signaling is a critical event in the basal category of ERBB2-mediated mammary tumor progression [42]. They proposed that targeting of β-catenin–dependent signaling may hold potential therapeutic value in the treatment of the basal category of ERBB2-positive breast cancer. Furthermore, MDA-MB-231 is the representative of the claudin-low breast cancer subtype, characterized by the enrichment of EMT and stem cell-like features, and significantly associated with disease recurrence [43]. Further underscoring the complexity regarding Wnt signalling in breast cancer and urgent need for personalized medicine for breast cancer treatment, we found that this cell line has a drastic difference in terms of Wnt pathway activation compared with basal-like breast cancer. And finally, while this work primarily focused on cell line models, it will be necessary and important to further validate the findings using primary tumors from patients, including patient-derived xenografts, in future work.
Supplementary Material
Protein expression of β-catenin, c-Myc and p21 in MDA-MB-231 cells analyzed by Western blot after β-catenin siRNA transfection. Actin was used as a loading control.
Quantitative analyses of protein expression by densitometry. c-Myc and p21 expression before or after β-catenin inhibition were quantified by image J and normalized to β-Actin.
Acknowledgments
We thank Dr. C. Perou of University of North Carolina, Chapel Hill for kindly providing microarray data. We thank Dr. M. Kato from Tsukuba University for the c-Myc reporter plasmid. We thank Dr. S. Byers at Georgetown University for mutant β-cateninS37A construct. We also thank Michelle Porcellino and Lise Sveen for critical reading of the manuscript.
Grant support: National Cancer Institute Cancer Center Breast Cancer SPORE at the University of Chicago, the Breast Cancer Research Foundation, the National Women’s Cancer Research Alliance, and the Falk Medical Research Trust (O.I. Olopade). Startup funding from Jianghan Univesity to J. Xu.
Definitions for abbreviations
- ANOVA
analysis of variance
- APC
Adenomatosis polyposis coli
- CCNE1
Cyclin E1
- CDKN1A
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
- CDKN2A
Cyclin-dependent kinase inhibitor 2A (p16)
- c-Myc
v-myc avian myelocytomatosis viral oncogene homolog
- CTNNB1
Catenin (cadherin-associated protein), beta 1
- FBS
fetal bovine serum
- FDR
false discovery rate
- FZD7
Frizzled family receptor 7
- G1 phase
gap phase 1
- G2 phase
gap phase 2
- GEO
Gene Expression Omnibus
- IHC
immunochemistry
- PBS
phosphate buffered saline
- PI
propidium iodide
- S phase
DNA synthesis phase
- SAM
significant analysis of microarrays
- TCF4/LEF1
T cell specific factor 4/ Lymphoid enhancer-binding factor 1
- TNBC
Triple negative breast cancer (Estrogen receptor-negative, Progesterone receptor-negative and Her2/Neu-negative breast cancer)
- Wnt
Wingless-type MMTV integration site family
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Turner NC, Reis-Filho JS. Tackling the diversity of triple-negative breast cancer. Clin Cancer Res. 2013;19(23):6380–6388. doi: 10.1158/1078-0432.CCR-13-0915. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 4.Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, Sledge GW, Carey LA. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14(24):8010–8018. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 5.Foulkes WD, Brunet JS, Stefansson IM, Straume O, Chappuis PO, Begin LR, Hamel N, Goffin JR, Wong N, Trudel M, et al. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res. 2004;64(3):830–835. doi: 10.1158/0008-5472.can-03-2970. [DOI] [PubMed] [Google Scholar]
- 6.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA : the journal of the American Medical Association. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 7.Fulford LG, Reis-Filho JS, Ryder K, Jones C, Gillett CE, Hanby A, Easton D, Lakhani SR. Basal-like grade III invasive ductal carcinoma of the breast: patterns of metastasis and long-term survival. Breast Cancer Res. 2007;9(1):R4. doi: 10.1186/bcr1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luck AA, Evans AJ, Green AR, Rakha EA, Paish C, Ellis IO. The influence of basal phenotype on the metastatic pattern of breast cancer. Clin Oncol (R Coll Radiol) 2008;20(1):40–45. doi: 10.1016/j.clon.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Voduc D, Nielsen TO. Basal and triple-negative breast cancers: impact on clinical decision-making and novel therapeutic options. Clinical breast cancer. 2008;8(Suppl 4):S171–S178. doi: 10.3816/CBC.2008.s.014. [DOI] [PubMed] [Google Scholar]
- 10.Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology. 2009;41(1):40–47. doi: 10.1080/00313020802563510. [DOI] [PubMed] [Google Scholar]
- 11.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Sorlie T, Bukholm I, Borresen-Dale AL. Truncating somatic mutation in exon 15 of the APC gene is a rare event in human breast carcinomas. Mutations in brief no. 179. Human mutation. 1998;12(3):215. Online. [PubMed] [Google Scholar]
- 13.Schlosshauer PW, Brown SA, Eisinger K, Yan Q, Guglielminetti ER, Parsons R, Ellenson LH, Kitajewski J. APC truncation and increased beta-catenin levels in a human breast cancer cell line. Carcinogenesis. 2000;21(7):1453–1456. [PubMed] [Google Scholar]
- 14.Webster MT, Rozycka M, Sara E, Davis E, Smalley M, Young N, Dale TC, Wooster R. Sequence variants of the axin gene in breast, colon, and other cancers: an analysis of mutations that interfere with GSK3 binding. Genes, chromosomes & cancer. 2000;28(4):443–453. [PubMed] [Google Scholar]
- 15.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97(8):4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong SC, Lo SF, Lee KC, Yam JW, Chan JK, Wendy Hsiao WL. Expression of frizzled-related protein and Wnt-signalling molecules in invasive human breast tumours. J Pathol. 2002;196(2):145–153. doi: 10.1002/path.1035. [DOI] [PubMed] [Google Scholar]
- 17.Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66(10):5487–5494. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Knowles E, Zardawi SJ, McNeil CM, Millar EK, Crea P, Musgrove EA, Sutherland RL, O'Toole SA. Cytoplasmic localization of beta-catenin is a marker of poor outcome in breast cancer patients. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19(1):301–309. doi: 10.1158/1055-9965.EPI-09-0741. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. British journal of cancer. 2008;98(6):1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng SS, Mahmoudi T, Danenberg E, Bejaoui I, de Lau W, Korswagen HC, Schutte M, Clevers H. Phosphatidylinositol 3-kinase signaling does not activate the wnt cascade. J Biol Chem. 2009;284(51):35308–35313. doi: 10.1074/jbc.M109.078261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6(5):497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 22.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Autocrine WNT signaling contributes to breast cancer cell proliferation via the canonical WNT pathway and EGFR transactivation. Breast Cancer Res. 2007;9(5):R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. The American journal of pathology. 2010;176(6):2911–2920. doi: 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, Natrajan R, Reis-Filho JS. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(2):209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, Deng X, Chen L, Kim CC, Lau S, et al. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene. 2011;30(43):4437–4446. doi: 10.1038/onc.2011.145. [DOI] [PubMed] [Google Scholar]
- 26.Dey N, Young B, Abramovitz M, Bouzyk M, Barwick B, De P, Leyland-Jones B. Differential activation of Wnt-beta-catenin pathway in triple negative breast cancer increases MMP7 in a PTEN dependent manner. PloS one. 2013;8(10):e77425. doi: 10.1371/journal.pone.0077425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey N, Barwick BG, Moreno CS, Ordanic-Kodani M, Chen Z, Oprea-Ilies G, Tang W, Catzavelos C, Kerstann KF, Sledge GW, Jr, et al. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer. 2013;13(1):537. doi: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandriani S, Frengen E, Cowling VH, Pendergrass SA, Perou CM, Whitfield ML, Cole MD. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PloS one. 2009;4(8):e6693. doi: 10.1371/journal.pone.0006693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes & cancer. 2010;1(6):629–640. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 33.Cowling VH, D'Cruz CM, Chodosh LA, Cole MD. c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol Cell Biol. 2007;27(14):5135–5146. doi: 10.1128/MCB.02282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Huo D, Chen Y, Nwachukwu C, Collins C, Rowell J, Slamon DJ, Olopade OI. CpG island methylation affects accessibility of the proximal BRCA1 promoter to transcription factors. Breast Cancer Res Treat. 2010;120(3):593–601. doi: 10.1007/s10549-009-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yagi K, Furuhashi M, Aoki H, Goto D, Kuwano H, Sugamura K, Miyazono K, Kato M. c-myc is a downstream target of the Smad pathway. J Biol Chem. 2002;277(1):854–861. doi: 10.1074/jbc.M104170200. [DOI] [PubMed] [Google Scholar]
- 36.Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12(5):479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You Z, Saims D, Chen S, Zhang Z, Guttridge DC, Guan KL, MacDougald OA, Brown AM, Evan G, Kitajewski J, et al. Wnt signaling promotes oncogenic transformation by inhibiting c-Myc-induced apoptosis. The Journal of cell biology. 2002;157(3):429–440. doi: 10.1083/jcb.200201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee S, Conrad SE. c-Myc suppresses p21WAF1/CIP1 expression during estrogen signaling and antiestrogen resistance in human breast cancer cells. J Biol Chem. 2005;280(18):17617–17625. doi: 10.1074/jbc.M502278200. [DOI] [PubMed] [Google Scholar]
- 39.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22(3):351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 40.Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res. 2009;11(3):R32. doi: 10.1186/bcr2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss RH. p21Waf1/Cip1 as a therapeutic target in breast and other cancers. Cancer Cell. 2003;4(6):425–429. doi: 10.1016/s1535-6108(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 42.Schade B, Lesurf R, Sanguin-Gendreau V, Bui T, Deblois G, O'Toole SA, Millar EK, Zardawi SJ, Lopez-Knowles E, Sutherland RL, et al. beta-Catenin signaling is a critical event in ErbB2-mediated mammary tumor progression. Cancer Res. 2013;73(14):4474–4487. doi: 10.1158/0008-5472.CAN-12-3925. [DOI] [PubMed] [Google Scholar]
- 43.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein expression of β-catenin, c-Myc and p21 in MDA-MB-231 cells analyzed by Western blot after β-catenin siRNA transfection. Actin was used as a loading control.
Quantitative analyses of protein expression by densitometry. c-Myc and p21 expression before or after β-catenin inhibition were quantified by image J and normalized to β-Actin.